Abstract
The human ACKR1 gene encodes a glycoprotein expressing the Duffy blood group antigens (Fy). The Duffy protein acts as a receptor for distinct pro-inflammatory cytokines and malaria parasites. We determined the haplotypes of the ACKR1 gene in a population inhabiting a malaria-endemic area. We collected blood samples from 60 healthy volunteers in Ethiopia’s southwestern low-altitude tropical region. An assay was devised to amplify the ACKR1 gene as a single amplicon and determine its genomic sequence. All haplotypes were resolved at 5178 nucleotides each, covering the coding sequence (CDS) of the ACKR1 gene and including the 5′- and 3′-untranslated regions (UTR), intron 1, and the 5′- and 3′-flanking regions. When necessary, allele-specific PCR with nucleotide sequencing or length polymorphism analysis was applied. Among the 120 chromosomes analyzed, 18 ACKR1 alleles were confirmed without ambiguity. We found 18 single-nucleotide polymorphisms (SNPs); only one SNP was novel. The non-coding sequences harbored 14 SNPs. No SNP, other than c.-67T>C, indicative of a non-functional allele, was detected. We described haplotypes of the ACKR1 gene in an autochthonous East-African population and found 18 distinct ACKR1 alleles. These long-range alleles are useful as templates to phase and analyze next-generation sequencing data, thus enhancing the reliability of clinical diagnostics.
Malaria: Seeking resistant blood types in Ethiopia
Researchers have surveyed genetic diversity related to malaria resistance in a region of Ethiopia where malaria is endemic. Duffy antigens, a component of blood type, are encoded by the ACKR1 gene, and individuals with the Duffy-negative blood type are resistant to malaria. Although the genes encoding resistant types have been identified, they have not been fully surveyed in malaria-endemic regions, where novel types are most likely to be found. Willy Flegel at the National Institutes of Health, Bethesda, USA, and co-workers sequenced ACKR1 in 60 people from Gambela, Ethiopia, where malaria is endemic. They detected 18 variants, including one never before documented. Almost all (16 of 18) of the variants encoded the Duffy-negative blood type. The authors plan to compare the genetic diversity in this region with a nearby region where malaria is not endemic.
Introduction
The human atypical chemokine receptor 1 gene (ACKR1, MIM #613665)1,2 encodes a multi-pass trans-membrane glycoprotein. It is a receptor for pro-inflammatory cytokines, such as interleukin-6 and -83,4, and the malaria parasites Plasmodium vivax and Plasmodium knowlesi5–7. Other than its expression in erythroid cells, the ACKR1 glycoprotein (also known as Duffy) is expressed on Purkinje neurons8, venular endothelial cells in skin9, the epithelial cells of renal collecting ducts, and pulmonary alveoli10, and on the endothelial cells lining postcapillary venules throughout the body, except in the liver10,11.
The ACKR1 glycoprotein carries the five antigens of the Duffy blood group system (Fy)12. The two major antithetical antigens Fya and Fyb, encoded by the co-dominant alleles FY*A (FY*01) and FY*B (FY*02), are among the clinically most significant blood group antigens, involved in severe hemolytic transfusion reactions and hemolytic disease of the fetus and newborn13–17. FY*A and FY*B allele frequencies range from only 0 to 5% in East Africa18,19.
Fy(a-b-) is the most common phenotype in West Africans20,21, East Africans22,23, and African Americans24. The cause of this prevalent Duffy-null phenotype is a homozygous inheritance of a point mutation (−67T>C; rs2814778) in a regulatory element of the FY*B allele promoter5,24–28, found in the recessive allele FY*02N.01. This GATA box mutation disrupts a binding site for the GATA-1 erythroid transcription factor and abolishes the expression of the ACKR1 protein on red blood cells only. The ACKR1 protein remains expressed on cells of non-erythroid tissues13,29, which prevents patients from forming alloantibodies to Fyb and Fy327.
Despite the importance of ACKR1 in malaria infection, only one study has systematically analyzed the ACKR1 gene at the haplotype level30, and no long-range ACKR1 haplotype has been confirmed in any malaria-endemic area. The population in Gambela, a southwestern region of Ethiopia, is indigenous and has been exposed to malaria for many generations31. The existence of a strong selective pressure for malaria resistance implies that the Ethiopian population has a propensity to develop and maintain distinct malaria-resistant ACKR1 haplotypes. We identified long-range haplotypes and variations of the ACKR1 gene, including potential regulatory elements, without ambiguity, in an autochthonous population from a malaria-endemic area.
Materials and methods
Human research subjects
Healthy volunteers, age 18 and older, participated with informed consent in the NIH protocol NCT01282021. The blood samples from 57 individuals were collected at the Gambella Blood Bank, Gambela region, Ethiopia, and then transported to NIH. For comparison, three additional Ethiopian samples were drawn in Addis Ababa. The DNA was extracted (EZ1 DNA blood kit on a BioRobot EZ1 Workstation; Qiagen, Valencia, CA) from ethylenediaminetetraacetic acid (EDTA)-anticoagulated whole blood.
ACKR1 gene amplification
We devised a sequencing approach capturing the whole 2204-bp NM_002036.3 mRNA transcript, intron 1, and the 5′- and 3′-flanking regions harboring the promoter and other regulatory elements. A 12,125-nucleotide stretch of the ACKR1 gene was amplified as a single primary amplicon from 50 ng of genomic DNA using a long-range Taq polymerase (LongAmp Taq DNA Polymerase; New England Biolabs, Ipswich, MA, USA) and the first-round primers 5′-GCATTGCTTCCAGTTCTAAGCTC-3′ and 5′-CGTCTCAATCGGTCCCTAAATCC-3′ (Eurofins MWG Operon; Huntsville, AL). The thermocycling conditions were as follows: initial denaturation at 94 °C for 2 min; 30 cycles at 94 °C for 30 s, 55 °C for 1 min, and 65 °C for 13 min; and a final extension at 65 °C for 10 min (DNA Engine Tetrad 2 Peltier Thermal Cycler; Bio-Rad, Hercules, CA).
The first-round reaction product (1 µl) was inoculated into the second-round polymerase chain reaction (PCR) using the nested primers 5′-CAACCACTCCTCCCATGGCATT-3′ and 5′-GATGAGGAGGGGTTTCTGTCC-3′ (Eurofins MWG Operon) to generate an amplicon of 5782 nucleotides. The thermocycling conditions were as follows: initial denaturation at 94 °C for 2 min; 30 cycles at 94 °C for 30 s, 62 °C for 1 min, and 65 °C for 6 min; and a final extension at 65 °C for 10 min.
Nucleotide sequencing
The primers for sequencing were designed using Primer3 (Table S1). The nested amplicon was purified and sequenced as previously described30, with extensive confirmatory resequencing. The nucleotide sequences were aligned (CodonCode Aligner, CodonCode, Centerville, MA) to NCBI RefSeq NG_011626.3, and the nucleotide positions were defined using the first nucleotide of the coding sequence (CDS) of NM_002036.3. Our sequencing covered 5178 nucleotides of the ACKR1 gene (NM_002036.3), including 1011 nucleotides of CDS, 480 nucleotides of intron, 1947 nucleotides of the 5′-UTR, 50 nucleotides of the 3′-UTR, 2101 nucleotides of the 5′-flanking region, and 589 nucleotides of the 3′-flanking region. This sequencing strategy captured all 642 variable positions listed for the 3736 nucleotides of the ACKR1 gene in the dbSNP database32. It also covered 171 variable positions listed for another 1442 nucleotides of the 5′- and 3′-flanking regions of the ACKR1 gene with potential regulatory elements.
Fragment analysis
The nucleotide sequencing did not allow for the resolution of a short tandem repeat (STR) site in the 5′-flanking region of the ACKR1 gene, comprising either eight or nine copies of a TG repeat. To differentiate an STR with nine TG copies from an STR with the c.-2872_-2871 TG deletion, indicative of eight TG copies, a PCR-based fragment analysis assay (Genewiz, Frederick, MD) was applied to all 60 samples (Table S1). The fragment lengths were scored using GeneMapper Software v4.1 (Applied Biosystems, Foster City, CA). A sample was considered to have nine TG repeats when the peak was observed at a fragment size of 145 bp, or eight TG repeats when the peak was observed at 143 bp.
Physical confirmation of haplotypes (alleles)
Heterozygosity at a single site or complete homozygosity allowed for the unambiguous assignment of a haplotype as described previously33. Allele-specific PCR and subsequent sequencing of the PCR products were used to construct the haplotype structure in samples with more than one heterozygous site. Briefly, 34 allele-specific PCR primers were designed for the first and last heterozygous site found in the amplicons of 20 individuals (Table S1). Long-range allele-specific PCRs, nested in the secondary 5782-bp amplicon, were carried out, and all variant positions between the first and last heterozygous sites were sequenced.
Computational phasing (predicted haplotypes)
The unphased genotype data from the 60 Ethiopian individuals and from the 2504 individuals from the 1000 Genomes Project were used as input data in the Markov chain-based haplotyper MaCH 1.034 software. Due to the inherent uncertainty of computational phasing, we ran our Ethiopian genotype dataset using various MaCH program settings, evaluating several combinations of rounds and states. A stable number of predicted haplotypes were observed with 1000 and 2000 rounds and 500 to 12,000 states. Hence, the analysis in the Ethiopian and 1000 Genome datasets was performed with MaCH program settings of 2000 rounds and 500 states.
Computational modeling of amino acid substitutions
PredictSNP was applied to predict the functional impact of non-synonymous nucleotide substitutions35.
Statistical analysis
Ninety-five percent confidence intervals (CI) for allele frequencies were calculated using the Poisson distribution36. The observed genotype frequencies were examined for deviation from the Hardy–Weinberg equilibrium (HWE) using a goodness-of-fit χ2-test with one degree of freedom.
Results
A random survey in 60 healthy volunteers was performed to describe the genetic variability of the ACKR1 gene for a large number of long-range haplotypes (alleles). We determined the ACKR1 genotype for 5178 nucleotides in each individual and resolved all alleles without ambiguity (Fig. 1).
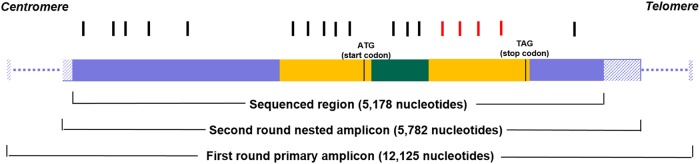
Fig. 1. Structure of the ACKR1 gene and SNPs found.
The sequenced region included the two exons (yellow) and intron 1 (green) along with the 5′- and 3′-flanking regions (blue), covering most of the nested amplicon. The primary amplicon was much larger (dotted line, not to scale). The 18 SNPs (bars; Table S2) were located in the coding (red bar) and non-coding sequence regions (black bar)
Nucleotide variations, genotype patterns, and alleles
We observed a total of 18 nucleotide positions where single-nucleotide polymorphisms (SNPs) were present (Table 1). Except for the GATA box mutation, no other SNP encoding a non-sense mutation or a frame-shift mutation was found, and only one SNP in the 5′-UTR was novel and had not been previously documented in the dbSNP database. Out of the 60 individuals analyzed and a total of 310,680 nucleotides sequenced (Table S2), 20 distinct genotype patterns were observed (Table S3). Physical evidence by allele-specific amplification and sequencing allowed us to discern 18 ACKR1 alleles (confirmed haplotypes) in the 120 chromosomes analyzed (Table 2).
Table 1.
Genetic variations detected in the ACKR1 gene
| Location | Nucleotide changea | dbSNP reference no. | Protein residue changeb | Observations (n = 60) | Global VAFc | HWE (p) | |||
|---|---|---|---|---|---|---|---|---|---|
| Homozygote reference | Heterozygote | Homozygote variant | VAF | ||||||
| 5′ Flanking region | −2872_−2871TG>del | rs5778112 | NA | 17 | 24 | 19 | 0.517 | 0.141 | 0.123 |
| −2456T>G | rs35432289 | NA | 56 | 4 | 0 | 0.033 | 0.005 | 0.789 | |
| −2212C>G | rs149599957 | NA | 59 | 1 | 0 | 0.008 | 0.003 | 0.948 | |
| −1982C>T | rs34190692 | NA | 48 | 11 | 1 | 0.108 | 0.006 | 0.693 | |
| −1310T>C | rs867811805 | NA | 59 | 1 | 0 | 0.008 | <0.01 | 0.948 | |
| 5′ UTR | −847C>T | rs114349581 | NA | 59 | 1 | 0 | 0.008 | 0.007 | 0.948 |
| −655A>G | rs3027011 | NA | 45 | 15 | 0 | 0.125 | 0.027 | 0.268 | |
| −436C>T | Novel | NA | 59 | 1 | 0 | 0.008 | NA | 0.948 | |
| −399_−398CT>del | rs71782098 | NA | 59 | 1 | 0 | 0.008 | 0.027 | 0.948 | |
| −67T>C | rs2814778 | NA | 0 | 2 | 58 | 0.983 | 0.266 | 0.896 | |
| Intron 1 | +115T>C | rs7550207 | NA | 43 | 14 | 3 | 0.208 | 0.118 | 0.215 |
| +150C>T | rs863002 | NA | 59 | 1 | 0 | 0.008 | 0.201 | 0.948 | |
| −243T>del | rs17838198 | NA | 0 | 0 | 60 | 1 | 0.222 | NA | |
| Exon 2 | 125G>A | rs12075 | Gly42Asp | 0 | 0 | 60 | 1 | 0.460 | NA |
| 265C>T | rs34599082 | Arg89Cys | 59 | 1 | 0 | 0.008 | 0.005 | 0.948 | |
| 298G>A | rs13962 | Ala100Thr | 59 | 1 | 0 | 0.008 | 0.069 | 0.948 | |
| 602C>T | rs758176489 | Thr201Met | 59 | 1 | 0 | 0.008 | 0.000 | 0.948 | |
| 3′ Flanking region | +268A>G | rs863003 | NA | 59 | 1 | 0 | 0.008 | 0.126 | 0.948 |
VAF variant allele frequency, HWE Hardy–Weinberg equilibrium, NA not applicable
aNucleotide substitutions are shown relative to the reference sequence (NG_011626.3). Nucleotide positions are defined using the first nucleotide of the coding sequence (CDS) of NM_002036.3 isoform as nucleotide position 1
bRelative to NCBI Reference Sequence NP_002027.2
cGlobal VAF from 1000Genome, TOPMed (nhlbiwgs.org), and gnomAD (http://gnomad.broadinstitute.org/) databases
Table 2.
ACKR1 allele distribution in southwest Ethiopian individuals
| GenBank number | Allele (confirmed haplotype)a | Observations (n) | Allele frequency (%) | |
|---|---|---|---|---|
| Meanb | 95% CIc | |||
| NG_011626.3 | tgtcctcacctttctGCGCa | NA | NA | NA |
| MG932622 | tgtcctcacctctc-ACGCa | 36 | 30.0 | 21.2–40.6 |
| MG932623 | --tcctcacctctc-ACGCa | 41 | 34.3 | 24.1–45.8 |
| MG932624 | --tcttcacctctc-ACGCa | 9 | 7.5 | 3.7–14.0 |
| MG932625 | --gcctcacctctc-ACGCa | 3 | 2.5 | 0.7–6.8 |
| MG932626 | --tcctcacctccc-ACGCa | 2 | 1.7 | 0.3–5.6 |
| MG932627 | --tcttcacctccc-ACGCa | 2 | 1.7 | 0.3–5.6 |
| MG932628 | tgtcctcacctccc-ACGCa | 5 | 4.3 | 1.6–9.3 |
| MG932629 | tgtcctcgcctccc-ACGCa | 12 | 10.0 | 5.6–16.9 |
| MG932630 | --gcctcatctctc-ACGCa | 1 | 0.8 | 0.1–4.4 |
| MG932631 | --tgcttacctctc-ACGCa | 1 | 0.8 | 0.1–4.4 |
| MG932632 | tgtccccacctctc-ACGCa | 1 | 0.8 | 0.1–4.4 |
| MG932633 | tgtcctcacctttt-ATACa | 1 | 0.8 | 0.1–4.4 |
| MG932634 | tgtcctcgcctccc-ACGCg | 1 | 0.8 | 0.1–4.4 |
| MG932635 | tgtcctcacctttc-ACGCa | 1 | 0.8 | 0.1–4.4 |
| MG932636 | --tcttcac--ccc-ACGCa | 1 | 0.8 | 0.1–4.4 |
| MG932637 | --tcctcgcctccc-ACGCa | 1 | 0.8 | 0.1–4.4 |
| MG932638 | tgtcctcgcctctc-ACGCa | 1 | 0.8 | 0.1–4.4 |
| MG932639 | --tcttcacctctc-ACGTa | 1 | 0.8 | 0.1–4.4 |
| Total | 120 | 100 | NA | |
NA not applicable
aThe nucleotides at the 16 SNP and two dinucleotide repeat (rs5778112 and rs71782098) positions are shown in 5′- to 3′-orientation (see Table S2). The remaining 5158 nucleotide positions that we determined had no variation relative to the reference sequence NG_011626.3. The upper case nucleotides are located in the coding sequence while the lowercase nucleotides are located in the non-coding sequence of the ACKR1 gene
bNumber of observed alleles × 100/Total number of alleles
c95% confidence interval (CI), Poisson distribution, two sided36
Predicted blood group phenotype
All of the 18 alleles detected carried the variant (c.125A; p.Asp42) specific for the common Fy(b+) phenotype. The clinically relevant FY*02N.01 allele in Africans37, as defined only by the two SNPs at positions c.-67T>C (GATA box mutation) and c.125G>A (Fya/Fyb) in the promoter and coding sequence, respectively25, was consistent with 16 of the 18 alleles. The other two alleles represented the reference FY*02 allele of the Fy(b+) phenotype and an FY*02W.01 allele of the Fy(b+w) phenotype, respectively.
Predicted effect on protein structure
Only four non-synonymous SNPs were found (Table 3), and computational modeling by PredictSNP indicated that all four changes were neutral.
Table 3.
Functional significance of non-synonymous SNPs predicted by PredictSNP
| dbSNP reference number | Variation | Computational analysis results | ||
|---|---|---|---|---|
| Nucleotide changea | Amino acid substitutionb | Classification | Expected accuracy (%)c | |
| rs12075 | c.125G>A | p.Gly42Asp | Neutral | 83 |
| rs34599082 | c.265C>T | p.Arg89Cys | Neutral | 74 |
| rs13962 | c.298G>A | p.Ala100Thr | Neutral | 68 |
| rs758176489 | c.602C>T | p.Thr201Met | Neutral | 83 |
aRelative to NCBI Reference Sequence NM_002036.3
bRelative to NCBI Reference Sequence NP_002027.2
cNormalized confidence as calculated by the software (PredictSNP)
Predicted haplotypes by computational phasing
For comparison, we applied a common computational approach for haplotype prediction. The individual ACKR1 haplotypes were reconstructed by running 2000 iterations (rounds) and considering 500 haplotypes (states) as the MaCH program settings (Table S4). Using our genotype information (Table S2) as input data, the MaCH software predicted 17 ACKR1 haplotypes. Out of our 18 physically confirmed alleles, only 13 alleles (76.5%) were correctly predicted while five alleles (MG932630, MG932633 to MG932635, and MG932637) were missed (Table S5). Another four haplotypes (MaCH-01 to MaCH-04; 23.5%), which were not actually present in the 60 individuals, were predicted by MaCH as single occurrences (Table S5).
ACKR1 haplotypes in the 1000 Genomes project
Using the same MaCH program settings, we analyzed the ACKR1 unphased genotype data from the 1000 Genomes Project (1000GP, phase 3)38. Out of the 18 SNPs detected in Ethiopia (Table 1), only 13 SNPs were found in the 1000GP database (Table S6). The MaCH software predicted 20 haplotypes using the 13 SNPs. Among our 60 Ethiopian individuals and the 2504 individuals of the 1000GP, we observed only two shared alleles (MG932622 and MG932629), which were among the most prevalent alleles in both cohorts (Table S6).
Discussion
Previous population-based molecular studies on the malaria resistance-associated ACKR1 gene have been centered in sub-Saharan African22,39 and North African Arab populations40. We collected blood samples from 60 individuals from Gambela, a tropical malaria-endemic region of southwestern Ethiopia, and sequenced a 5178-nucleotide region of chromosome 1 encompassing the ACKR1 gene. This is the first study in an autochthonous African population to systematically categorize SNPs found at the ACKR1 gene locus into long-range alleles.
The dbSNP database32 lists 813 nucleotide variations in the 5178-nucleotide region that we analyzed. In the present study, we observed 17 known variations and one novel variation (Table 1). Many of the variants described in the dbSNP database but not observed in our study may not be polymorphic in our population or are so rare that our screening panel lacked adequate power to detect them. No variant associated with a non-functional ACKR1 protein was detected, besides the GATA box variant (rs2814778). A large fraction of the ACKR1 alleles (>70%) occurred with low prevalence (Table 2), correlating with the known diverse genetic background of African populations41. The ACKR1-null allele FY*02N.01 was prevalent with >95%, possibly explained by the endemic P. vivax malaria in the region18,31. Non-synonymous SNPs, which introduce amino acid changes, could affect protein structure and function;42 however, the four such variants identified in our study had no effect on the protein structure, as calculated by PredictSNP (Table 3). Therefore, these computer predictions should be interpreted with caution because the three-dimensional structure of the complete ACKR1 protein remains unknown.
The MaCH algorithm did not identify five of the actual alleles and incorrectly predicted four haplotypes that were not present (Table S5). All of the prediction errors concerned alleles with only one observation (0.8% each). Relying on only computerized allele calling would result in 6.6% incorrect allele calls, potentially affecting one out of 15 patients (Table 4). In one individual (no. 44 in Table S5), the infrequent MG932628 allele was missed and substituted by the more frequent, albeit incorrect, allele combination MG932626 + MG932629. Thus, although computerized allele prediction may replace physical sequencing approaches for determining common alleles, our observation of incorrect predictions warns that computational algorithms can falter when rare alleles are encountered, even in this era of effortlessly obtained big data.
Table 4.
MaCH prediction of the ACKR1 haplotypes
| Two haplotypes per individuala | MaCH prediction of | Rate (%) | |
|---|---|---|---|
| Haplotypes (n) | Individuals (n) | ||
| Both correct | 112 | 56 | 93.4 |
| Both incorrect | 8 | 4 | 6.6 |
| Total | 120 | 60 | 100 |
aAs compared to the 18 physically confirmed alleles (MG932622 to MG932639)
The most frequent haplotype in the 1000GP samples (22.2%; Table S6) carried a T nucleotide in intron 1 (rs17838198; Table 1) that all alleles in our Ethiopian samples lacked (Table 2 and S6). This is consistent with a low prevalence of this variant in the dbSNP database for African populations (1%) compared to non-African populations (23–37%) represented in the 1000GP samples. Each of the four SNPs without frequency information in the 1000GP (Table S6, footnote) are rare and were found only once in our Ethiopian samples. The dinucleotide repeat variation rs5778112, found in 48% of our alleles, was not found in the 1000GP because variations in microsatellite regions were difficult to accurately capture43,44.
We experimentally verified long-range haplotypes of the ACKR1 gene among an autochthonous Ethiopian population. As an adjunct to the human reference genome assembly (GRCh38), which is the gold standard reference45, our comprehensive, population-specific data and alleles are useful as template sequences for allele calling in high-throughput, next-generation sequencing and precision medicine approaches46,47. The design of our protocol will eventually allow us to compare populations from the Ethiopian highland and desert regions, which are not endemic for malaria, and to analyze more blood group system genes with high-throughput methods.
Web Resources
dbSNP database, Build ID: 151 (http://www.ncbi.nlm.nih.gov/SNP/)
Genome Aggregation Database (http://gnomad.broadinstitute.org/)
Hardy-Weinberg equilibrium calculator (http://www.tufts.edu/
~mcourt01/Documents/Court%20lab%20-%20HW%20calculator.xls)
MaCH, version 1.0 (http://www.sph.umich.edu/csg/abecasis/MACH/index.html)
PredictSNP, version 1.0 (http://loschmidt.chemi.muni.cz/predictsnp/)
Primer3 software, version 0.4.0 (http://bioinfo.ut.ee/primer3-0.4.0/)
TOPMed database (https://www.nhlbiwgs.org/)
Genome Aggregation Database (gnomAD; http://gnomad.broadinstitute.org/)
Ensembl genome browser (https://useast.ensembl.org/index.html)
Names for FY (ISBT 008) Blood Group Alleles (http://www.isbtweb.org/fileadmin/user_upload/files-2015/red cells/blood group allele terminology/allele tables/008 FY Alleles v3.0 140328.pdf).
Disclaimer
The views expressed do not necessarily represent the view of the National Institutes of Health, the U.S. Food and Drug Administration, the Department of Health and Human Services, or the U.S. Federal Government.
Electronic supplementary material
Acknowledgements
We thank Harvey G. Klein for review of the manuscript; the staff of HLA and TSL laboratories in our section for DNA extraction, sample freezing, and inventory management; and Elizabeth J. Furlong for English editing. This research of the NIH protocol 11-CC-N086 was supported by the Intramural Research Program of the NIH Clinical Center (ZIA CL002123-03).
Authors’ contributions
W.A.F. conceived the study. A.T.M. collected the samples and discussed the results. A.G. contributed to the sample collection. Q.Y. and W.A.F. designed the experiments. Q.Y. completed the experimental part of this study. Q.Y., K.S., and W.A.F. analyzed the data and wrote the manuscript.
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Supplementary information is available for this paper at 10.1038/s41439-018-0024-8.
References
- 1.Nibbs RJB, Graham GJ. Immune regulation by atypical chemokine receptors. Nat. Rev. Immunol. 2013;13:815–829. doi: 10.1038/nri3544. [DOI] [PubMed] [Google Scholar]
- 2.Bachelerie F, et al. International Union of Basic and Clinical Pharmacology. [corrected]. LXXXIX. Update on the extended family of chemokine receptors and introducing a new nomenclature for atypical chemokine receptors. Pharmacol. Rev. 2014;66:1–79. doi: 10.1124/pr.113.007724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Horuk R, et al. A receptor for the malarial parasite Plasmodium vivax: the erythrocyte chemokine receptor. Science. 1993;261:1182–1184. doi: 10.1126/science.7689250. [DOI] [PubMed] [Google Scholar]
- 4.Horuk R. The Duffy antigen receptor for chemokines DARC/ACKR1. Front. Immunol. 2015;6:279. doi: 10.3389/fimmu.2015.00279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hadley TJ, Peiper SC. From malaria to chemokine receptor: the emerging physiologic role of the Duffy blood group antigen. Blood. 1997;89:3077–3091. [PubMed] [Google Scholar]
- 6.Miller LH, Mason SJ, Dvorak JA, McGinniss MH, Rothman IK. Erythrocyte receptors for (Plasmodium knowlesi) malaria: Duffy blood group determinants. Science. 1975;189:561–563. doi: 10.1126/science.1145213. [DOI] [PubMed] [Google Scholar]
- 7.Mason SJ, Miller LH, Shiroishi T, Dvorak JA, McGinniss MH. The Duffy blood group determinants: their role in the susceptibility of human and animal erythrocytes to Plasmodium knowlesi malaria. Br. J. Haematol. 1977;36:327–335. doi: 10.1111/j.1365-2141.1977.tb00656.x. [DOI] [PubMed] [Google Scholar]
- 8.Horuk R, et al. Expression of chemokine receptors by subsets of neurons in the central nervous system. J. Immunol. 1997;158:2882–2890. [PubMed] [Google Scholar]
- 9.Pruenster M, et al. The Duffy antigen receptor for chemokines transports chemokines and supports their promigratory activity. Nat. Immunol. 2009;10:101–108. doi: 10.1038/ni.1675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chaudhuri A, et al. Detection of Duffy antigen in the plasma membranes and caveolae of vascular endothelial and epithelial cells of nonerythroid organs. Blood. 1997;89:701–712. [PubMed] [Google Scholar]
- 11.Hadley TJ, et al. Postcapillary venule endothelial cells in kidney express a multispecific chemokine receptor that is structurally and functionally identical to the erythroid isoform, which is the Duffy blood group antigen. J. Clin. Invest. 1994;94:985–991. doi: 10.1172/JCI117465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Meny GM. The Duffy blood group system: a review. Immunohematology. 2010;26:51–56. [PubMed] [Google Scholar]
- 13.Chaudhuri A, Polyakova J, Zbrzezna V, Pogo A. The coding sequence of Duffy blood group gene in humans and simians: restriction fragment length polymorphism, antibody and malarial parasite specificities, and expression in nonerythroid tissues in Duffy- negative individuals. Blood. 1995;85:615–621. [PubMed] [Google Scholar]
- 14.Iwamoto S, Omi T, Kajii E, Ikemoto S. Genomic organization of the glycoprotein D gene: Duffy blood group Fya/Fyb alloantigen system is associated with a polymorphism at the 44- amino acid residue. Blood. 1995;85:622–626. [PubMed] [Google Scholar]
- 15.Mallinson G, Soo KS, Schall TJ, Pisacka M, Anstee DJ. Mutations in the erythrocyte chemokine receptor (Duffy) gene: the molecular basis of the Fya/Fyb antigens and identification of a deletion in the Duffy gene of an apparently healthy individual with the Fy(a-b-) phenotype. Br. J. Haematol. 1995;90:823–829. doi: 10.1111/j.1365-2141.1995.tb05202.x. [DOI] [PubMed] [Google Scholar]
- 16.Ikin EW, Mourant AE, Pettenkofer HJ, Blumenthal G. Discovery of the expected haemagglutinin, anti-Fyb. Nature. 1951;168:1077–1078. doi: 10.1038/1681077b0. [DOI] [PubMed] [Google Scholar]
- 17.Cutbush M, Mollison PL, Parkin DM. A new human blood group. Nature. 1950;165:189. doi: 10.1038/165188b0. [DOI] [Google Scholar]
- 18.Howes RE, et al. The global distribution of the Duffy blood group. Nat. Commun. 2011;2:266. doi: 10.1038/ncomms1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mathews HM, Armstrong JC. Duffy blood types and vivax malaria in Ethiopia. Am. J. Trop. Med. Hyg. 1981;30:299–303. doi: 10.4269/ajtmh.1981.30.299. [DOI] [PubMed] [Google Scholar]
- 20.Cavalli-Sforza, L. L., Menozzi, P. & Piazza, A. The History And Geography Of Human Genes xi, Vol. 541, 18pp (Princeton University Press, Princeton, NJ, 1994).
- 21.Miller LH, Mason SJ, Clyde DF, McGinniss MH. The resistance factor to Plasmodium vivax in blacks. The Duffy-blood-group genotype, FyFy. N. Engl. J. Med. 1976;295:302–304. doi: 10.1056/NEJM197608052950602. [DOI] [PubMed] [Google Scholar]
- 22.Hamblin MT, Thompson EE, Di Rienzo A. Complex signatures of natural selection at the Duffy blood group locus. Am. J. Hum. Genet. 2002;70:369–383. doi: 10.1086/338628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hamblin MT, Di Rienzo A. Detection of the signature of natural selection in humans: evidence from the Duffy blood group locus. Am. J. Hum. Genet. 2000;66:1669–1679. doi: 10.1086/302879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sanger R, Race RR, Jack J. The Duffy blood groups of New York Negroes: the phenotype Fy (a–b–) Br. J. Haematol. 1955;1:370–374. doi: 10.1111/j.1365-2141.1955.tb05523.x. [DOI] [PubMed] [Google Scholar]
- 25.Tournamille C, Colin Y, Cartron JP, Le Van Kim C. Disruption of a GATA motif in the Duffy gene promoter abolishes erythroid gene expression in Duffy-negative individuals. Nat. Genet. 1995;10:224–228. doi: 10.1038/ng0695-224. [DOI] [PubMed] [Google Scholar]
- 26.Iwamoto S, Li J, Omi T, Ikemoto S, Kajii E. Identification of a novel exon and spliced form of Duffy mRNA that is the predominant transcript in both erythroid and postcapillary venule endothelium. Blood. 1996;87:378–385. [PubMed] [Google Scholar]
- 27.Castilho L. The value of DNA analysis for antigens in the Duffy blood group system. Transfusion. 2007;47:28S–31S. doi: 10.1111/j.1537-2995.2007.01307.x. [DOI] [PubMed] [Google Scholar]
- 28.Parasol N, et al. a novel mutation in the coding sequence of the FY*B allele of the Duffy chemokine receptor gene is associated with an altered erythrocyte phenotype. Blood. 1998;92:2237–2243. [PubMed] [Google Scholar]
- 29.Peiper SC, et al. The Duffy antigen/receptor for chemokines (DARC) is expressed in endothelial cells of Duffy negative individuals who lack the erythrocyte receptor. J. Exp. Med. 1995;181:1311–1317. doi: 10.1084/jem.181.4.1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schmid P, Ravenell KR, Sheldon SL, Flegel WA. DARC alleles and Duffy phenotypes in African Americans. Transfusion. 2012;52:1260–1267. doi: 10.1111/j.1537-2995.2011.03431.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Armstrong JC. Susceptibility to vivax malaria in Ethiopia. Trans. R. Soc. Trop. Med. Hyg. 1978;72:342–344. doi: 10.1016/0035-9203(78)90123-2. [DOI] [PubMed] [Google Scholar]
- 32.Sherry ST, et al. dbSNP: the NCBI database of genetic variation. Nucleic Acids Res. 2001;29:308–311. doi: 10.1093/nar/29.1.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Srivastava K, Lee E, Owens E, Rujirojindakul P, Flegel WA. Full-length nucleotide sequence of ERMAP alleles encoding Scianna (SC) antigens. Transfusion. 2016;56:3047–3054. doi: 10.1111/trf.13801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li, Y., Willer, C. J., Ding, J., Scheet, P. & Abecasis, G. R. MaCH: using sequence and genotype data to estimate haplotypes and unobserved genotypes. Genet. Epidemiol.34, 816–834 (2010). [DOI] [PMC free article] [PubMed]
- 35.Bendl J, et al. PredictSNP: robust and accurate consensus classifier for prediction of disease-related mutations. PLoS Comput. Biol. 2014;10:e1003440. doi: 10.1371/journal.pcbi.1003440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sachs, L. Angewandte Statistik - Anwendung Statistischer Methoden 7th edn, 446–447 (Springer-Verlag, Berlin, 1992).
- 37.Storry, J. R. et al. International Society of Blood Transfusion Working Party on red cell immunogenetics and blood group terminology: Cancun report (2012). Vox Sang.107, 90–96 (2014). [DOI] [PMC free article] [PubMed]
- 38.Auton A, et al. A global reference for human genetic variation. Nature. 2015;526:68–74. doi: 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Robinson HB. Academic freedom and the issue of expanded-duty auxiliaries. Dent. Surv. 1976;52:12–16. [PubMed] [Google Scholar]
- 40.Fernandez-Santander A, et al. Genetic relationships between southeastern Spain and Morocco: new data on ABO, RH, MNSs, and DUFFY polymorphisms. Am. J. Hum. Biol. 1999;11:745–752. doi: 10.1002/(SICI)1520-6300(199911/12)11:6<745::AID-AJHB4>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- 41.Campbell MC, Tishkoff SA. African genetic diversity: implications for human demographic history, modern human origins, and complex disease mapping. Annu. Rev. Genomics Hum. Genet. 2008;9:403–433. doi: 10.1146/annurev.genom.9.081307.164258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.de Coulgeans CD, et al. Synonymous nucleotide polymorphisms influence Dombrock blood group protein expression in K562 cells. Br. J. Haematol. 2014;164:131–141. doi: 10.1111/bjh.12597. [DOI] [PubMed] [Google Scholar]
- 43.McIver LJ, Fondon JW, 3rd, Skinner MA, Garner HR. Evaluation of microsatellite variation in the 1000 Genomes Project pilot studies is indicative of the quality and utility of the raw data and alignments. Genomics. 2011;97:193–199. doi: 10.1016/j.ygeno.2011.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Abecasis GR, et al. A map of human genome variation from population-scale sequencing. Nature. 2010;467:1061–1073. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schneider VA, et al. Evaluation of GRCh38 and de novo haploid genome assemblies demonstrates the enduring quality of the reference assembly. Genome Res. 2017;27:849–864. doi: 10.1101/gr.213611.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.McCarthy S, et al. A reference panel of 64,976 haplotypes for genotype imputation. Nat. Genet. 2016;48:1279–1283. doi: 10.1038/ng.3643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ansel, A. et al. Genetic variants related to disease susceptibility and immunotolerance in the Duffy antigen receptor for chemokines (DARC, Fy) gene in the black lion tamarin (Leontopithecus chrysopygus, primates). Am. J. Primatol.79, e22690:1–11 (2017). [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.