Abstract
Several polymorphisms in the vitamin D receptor (VDR) are associated with the occurrence of chronic liver disease. Here, we investigated the association between BsmI, ApaI, TaqI and FokI VDR polymorphisms and the severity of liver cirrhosis in relation to serum cytokine and lipopolysaccharide binding protein (LBP) levels and their role on survival in cirrhotic patients. We found that patients harboring the BB genotype had higher MELD score, and they were mainly at CP stage C; patients harboring the AA genotype had increased LBP, IL-1β and IL-8 levels, and they were mostly at CP stage C; TT genotype carriers had higher MELD score and they were mainly at CP stage C and FF genotype carriers had lower IL-1β levels when compared to Bb/bb, Aa/aa, Tt/tt and Ff/ff genotypes respectively. In the multivariate analysis ApaI, BsmI and TaqI polymorphisms were independently associated with liver cirrhosis severity. In the survival analysis, the independent prognostic factors were CP score, MELD and the FF genotype. Our results indicate that the ApaI, TaqI and BsmI polymorphisms are associated with the severity of liver cirrhosis, through the immunoregulatory process. Survival is related to the FF genotype of FokI polymorphism, imparting a possible protective role in liver cirrhosis.
Introduction
Liver cirrhosis is defined as the histological development of regenerative nodules surrounded by fibrous bands in response to chronic liver injury, and is associated with the development of liver failure and portal hypertension1,2. Infection with Hepatitis B (HBV) or C (HCV), alcohol abuse and nonalcoholic fatty liver disease (NAFLD) are the main etiologic factors of liver cirrhosis worldwide1,2. However, certain genetic polymorphisms may influence the progression of liver fibrosis3.
The vitamin D receptor (VDR) is a DNA-binding transcription factor that is expressed on peripheral blood (PB) monocytes and activated T lymphocytes. VDR belongs to the nuclear receptor superfamily and is associated with many physiological processes4–6. The most common polymorphisms of the VDR gene are the BsmI, FokI, TaqI and ApaI. FokI, is located in exon 2 of the VDR gene and the presence of this polymorphism results in a shortened VDR protein due to an alteration in the start codon7. The ApaI and the BsmI polymorphisms are located in intron 8 at the 3′ end of the VDR gene. These polymorphisms do not change the amino acid sequence of the VDR protein. However, BsmI and ApaI may affect gene expression through the alteration of mRNA stability, the disruption of splice sites for mRNA transcription, or a change in intronic regulatory elements8,9. The TaqI polymorphism is located in exon 9 at the 3′ end of the human VDR gene and results in a synonymous change due to a nucleotide substitution. The presence of TaqI polymorphism does not modify the VDR protein but is involved in the regulation of the stability of VDR mRNA8,9. Recent studies have shown that there is a genetic association of VDR polymorphisms to autoimmune hepatitis (AIH), primary biliary cirrhosis (PBC), HBV infection and hepatocellular carcinoma (HCC)8,10–17. Moreover, the progression of liver fibrosis has been associated with the existence of VDR polymorphisms in patients with PBC10 and HCV18 and with reduced full-length VDR protein expression, but increased VDR protein fragments in patients with NAFLD10,18,19.
Cytokines are key mediators in the pathophysiology of liver disease as they play an essential role in hepatic regeneration and fibrosis20. The hepatic non parenchymal cells which are involved in liver fibrosis development, can rapidly produce profibrogenic cytokines which lead to hepatic inflammation and fibrosis21. In contrast, antifibrogenic cytokines downregulate the pro-inflammatory response promoting the hepatic regeneration20,21. VDR polymorphisms may influence the immune regulation by affecting cytokine levels and, thus, they might play a role in the progression of liver disease11,13.
In this study, we have investigated the potential associations between VDR gene polymorphisms and the severity of liver cirrhosis, in relation to the cytokine and bacterial profiles, vitamin D and vitamin D binding protein (VDBP) levels, and their role on patient survival.
Results
The main demographic and clinical characteristics of the examined patients are presented in Table 1 and the main characteristics of the examined VDR polymorphisms are presented in Table 2.
Table 1.
Patients’ demographic and main clinical baseline characteristics.
| Mean | Range | |
|---|---|---|
| Age (years) | 60.74 | 29–84 |
| N | Percentage (%) | |
| Sex (M/F) | 60/29 | 67.4/32.6 |
| Etiology of liver cirrhosis | N | Percentage (%) |
| Alcohol consumption | 32 | 36.4 |
| HBV ± HDV infection | 29 | 32.5 |
| HCV infection | 10 | 10.4 |
| Cryptogenic cirrhosis | 6 | 6.5 |
| Autoimmune hepatitis | 5 | 5.2 |
| Primary biliary cirrhosis | 3 | 3.9 |
| Nonalcoholic steatohepatitis | 2 | 2.6 |
| Primary sclerosing cholangitis | 1 | 1.3 |
| HBV infection + HCV infection | 1 | 1.3 |
| Median (IQR) | ||
| Vitamin D levels (ng/mL) | 21.1 (14.7, 31.6) | |
| Hb (g/dL) | 12.7 (11.2, 13.8) | |
| Plt (K/uL) | 117.5 (66, 160) | |
| INR | 1.3 (1.1, 1.6) | |
| Creatinine (mg/dL) | 0.9 (0.7, 1.1) | |
| SGPT (U/L) | 31.0 (21.0, 58.0) | |
| G-GT (U/L) | 54.0 (29.0, 109.0) | |
| ALP (U/L) | 101.5 (76.0, 142.0) | |
| Albumin (g/dL) | 3.6 (3.0, 4.0) | |
| Total Bilirubin (mg/dL) | 1.3 (0.7, 2.5) | |
| K (mmol/L) | 4.3 (3.9, 4.7) | |
| Na (mmol/L) | 138.4 (135.2, 141.0) | |
| Ca (mmol/L) | 9.0 (8.6, 9.4) | |
| Mg (mmol/L) | 1.9 (1.6, 2.1) | |
| Systolic pressure (mmHg) | 130.0 (115.0, 150.0) | |
| Diastolic pressure (mmHg) | 75.0 (70.0, 80.0) | |
| CP score | 6.0 (5.0, 9.0) | |
| MELD | 11.0 (8.0, 15.5) | |
| VDBP (µg/mL) | 160.8 (99.3, 257.9) | |
| IL-12 (pg/mL) | 7.5 (2.1, 8.7) | |
| TNF-a (pg/mL) | 4.7 (1.2, 5.7) | |
| IL-1β (pg/mL) | 8.0 (3.1, 11.6) | |
| IL-6 (pg/mL) | 7.8 (4.3, 22.0) | |
| IL-8 (pg/mL) | 35.7 (20.6, 90.7) | |
| IL-10 (pg/mL) | 3.9 (3.1, 4.7) | |
| LBP (μg/mL) | 10.8 (8.3, 11.4) | |
Abbreviations: M, male; F, female; HBV, hepatis B virus; HDV, hepatis D virus; HCV, hepatis C virus; IQR, Interquartile range; Hb, Hemoglobin; Plt, Platelets; INR, International normalized ratio; SGPT, Alanine aminotransferase; G-GT, Gamma-Glutamyl Transferase; ALP, Alkaline phosphatase; CP, Child Pugh; MELD, Model for end-stage liver disease; VDBP, Vitamin D binding protein; LBP, Lipopolysaccharide binding protein.
Table 2.
Characterization of VDR polymorphisms.
| SNP name | SNP ID | SNP location | Nucleotide change | Correspondence of nomenclature of SNP alleles |
|---|---|---|---|---|
| FokI | rs2228570 | Exon 2 | C > T | F > f |
| BsmI | rs1544410 | Intron 8 | A > G | B > b |
| TaqI | rs731236 | Exon 9 | C > T | T > t |
| ApaI | rs7975232 | Intron 8 | A > C | A > a |
Abbreviations: SNP, single nucleotide polymorphism.
Distribution of clinical variables and serum cytokine expression according to the VDR genotypes
As shown in Table 3 the presence of BsmI polymorphism, in particular the BB genotype, was associated with advanced Child-Pugh (CP) stage (p = 0.044) and higher model for the end-stage liver disease (MELD) score (p = 0.045). The AA genotype of the ApaI polymorphism was associated with advanced CP stage (p = 0.001) and increased LBP levels (p = 0.014). The presence of TaqI polymorphism (TT genotype) was associated with advanced CP stage (p = 0.027) and MELD score (p = 0.025). As regards to the FokI polymorphism, the FF genotype was associated with lower levels of the pro-inflammatory cytokine IL-1β (p = 0.045).
Table 3.
Distribution of clinical variables according to the VDR genotypes.
| BsmI A > G (rs1544410) | |||||
|---|---|---|---|---|---|
| bb | Bb | BB | Overall | p-value | |
| N (%) | |||||
| CP stage | 0.044 | ||||
| A | 12 (45.8) | 32 (60.4) | 2 (22.2) | 46 (51.3) | |
| B | 11 (41.7) | 13 (23.3) | 2 (22.2) | 26 (28.9) | |
| C | 3 (12.5) | 9 (16.3) | 5 (55.6) | 17 (19.8) | |
| Median (IQR) | |||||
| Vitamin D levels (ng/mL) | 20.6 (14.8, 29.6) | 21.5 (14.7, 31.6) | 29.8 (11.2, 46.7) | 21.1 (14.7, 31.6) | 0.828 |
| MELD | 12.0 (8.5, 17.0) | 10.0 (8.0, 14.0) | 16.0 (12.0, 19.0) | 11.0 (8.0, 15.5) | 0.045 |
| VDBP (μg/mL) | 177.6 (143.9, 259.0) | 136.4 (89.7, 256.7) | 199.3 (126.8, 209.6) | 160.8 (99.3, 257.9) | 0.307 |
| IL-12 (pg/mL) | 6.9 (0.0, 8.7) | 8.0 (5.6, 8.7) | 5.9 (0.0, 6.8) | 7.5 (2.1, 8.7) | 0.438 |
| TNF-a (pg/mL) | 4.9 (1.0, 5.9) | 4.6 (1.5, 5.3) | 5.2 (0.3, 6.0) | 4.7 (1.2, 5.7) | 0.764 |
| IL-1β (pg/mL) | 8.1 (6.5, 12.6) | 7.9 (6.2, 11.5) | 7.5 (0.0, 11.2) | 8.0 (3.1, 11.6) | 0.752 |
| IL-6 (pg/mL) | 5.8 (5.1, 12.8) | 7.8 (4.3, 27.2) | 13.0 (6.5, 49.4) | 7.8 (4.3, 22.0) | 0.520 |
| IL-8 (pg/mL) | 33.5 (16.0, 101.6) | 41.3 (23.0, 87.9) | 29.3 (20.9, 44.3) | 35.7 (20.6, 90.7) | 0.698 |
| IL-10 (pg/mL) | 3.7 (2.0, 4.2) | 4.0 (3.3, 5.2) | 4.0 (3.7, 6.3) | 3.9 (3.1, 4.7) | 0.257 |
| LBP (μg/mL) | 9.8 (4.9, 11.3) | 10.8 (8.4, 11.6) | 11.3 (11, 11.4) | 10.8 (8.3, 11.4) | 0.128 |
| FokI C > T (rs10735810) | |||||
| ff | Ff | FF | Overall | p- value | |
| N (%) | |||||
| CP stage | 0.846 | ||||
| A | 4 (42.8) | 21 (53) | 21 (51.4) | 46 (51.3) | |
| B | 4 (42.8) | 9 (23.5) | 13 (31.4) | 26 (28.9) | |
| C | 1 (14.2) | 9 (23.5) | 7 (17.2) | 17 (19.8) | |
| Median (IQR) | |||||
| Vitamin D levels (ng/mL) | 30.3 (21.3, 46.7) | 22.0 (14.7, 32.2) | 18.4 (13.9, 26.4) | 21.1 (14.7, 31.6) | 0.139 |
| MELD | 12.0 (9.0, 13.0) | 11.0 (8.0, 16.0) | 11.0 (8.0, 17.0) | 11.0 (8.0, 15.5) | 0.983 |
| VDBP (μg/mL) | 217.8 (189.0, 282.9) | 147.3 (112.2, 270.4) | 159.3 (89.7, 220.8) | 160.8 (99.3, 257.9) | 0.355 |
| IL-12 (pg/mL) | 5.3 (0.0, 8.2) | 7.5 (5.3, 8.9) | 8.0 (0.0, 8.5) | 7.5 (2.1, 8.7) | 0.382 |
| TNF-a (pg/mL) | 4.9 (1.6, 5.8) | 4.6 (1.5, 5.7) | 4.7 (0.4, 5.7) | 4.7 (1.2, 5.7) | 0.846 |
| IL-1β (pg/mL) | 9.3 (7.5, 11.8) | 9.4 (7.4, 12.3) | 7.2 (0.0, 10.1) | 8.0 (3.1, 11.6) | 0.045 |
| IL-6 (pg/mL) | 5.8 (5.1, 7.7) | 7.8 (4.2, 20.1) | 9.8 (4.9, 29.4) | 7.8 (4.3, 22.0) | 0.712 |
| IL-8 (pg/mL) | 33.7 (20.8, 87.9) | 44.7 (22.6, 110.4) | 33.0 (20.4, 85.8) | 35.7 (20.6, 90.7) | 0.812 |
| IL-10 (pg/mL) | 3.7 (1.8, 4.7) | 3.8 (3.2, 4.6) | 4.0 (3.2, 4.6) | 3.9 (3.1, 4.7) | 0.970 |
| LBP (μg/mL) | 10.8 (9.6, 11.3) | 10.5 (8.7, 11.3) | 10.9 (4.3, 11.4) | 10.8 (8.3, 11.4) | 0.995 |
| ApaI G > T (rs7975232) | |||||
| aa | Aa | AA | Overall | p- value | |
| N (%) | |||||
| CP stage | 0.001 | ||||
| A | 3 (30) | 29 (63.4) | 13 (40) | 45 (51.3) | |
| B | 7 (60) | 14 (29.3) | 5 (16) | 26 (28.9) | |
| C | 1 (10) | 3 (7.3) | 14 (44) | 18 (19.8) | |
| Median (IQR) | |||||
| Vitamin D levels (ng/mL) | 20.0 (9.5, 26.2) | 20.8 (14.9, 30.4) | 24.1 (14.1, 33.6) | 21.1 (14.7, 31.6) | 0.754 |
| Diastolic pressure (mmHg) | 60.0 (60.0, 70.0) | 77.5 (70.0, 90.0) | 77.5 (70.0, 80.0) | 75.0 (70.0, 80.0) | 0.011 |
| MELD | 9.5 (8.0, 14.0) | 11.0 (8.0, 14.0) | 13.0 (9.0, 17.0) | 11.0 (8.0, 15.5) | 0.377 |
| VDBP (μg/mL) | 244.9 (157.6, 288.6) | 170.8 (101.1, 256.7) | 130.2 (81.7, 215.3) | 160.8 (99.3, 257.9) | 0.140 |
| IL-12 (pg/mL) | 8.5 (5.1, 9.0) | 7.7 (0.0, 8.5) | 7.0 (4.7, 8.5) | 7.5 (2.1, 8.7) | 0.794 |
| TNF-a (pg/mL) | 5.6 (1.0, 5.9) | 4.4 (1.5, 5.2) | 5.0 (1.1, 6.1) | 4.7 (1.2, 5.7) | 0.394 |
| IL-1β (pg/mL) | 8.1 (0.0, 12.6) | 7.3 (0.0, 9.6) | 10.1 (7.5, 11.7) | 8.0 (3.1, 11.6) | 0.076 |
| IL-6 (pg/mL) | 7.7 (5.5, 12.3) | 5.8 (4.1, 17.6) | 12.9 (5.2, 29.8) | 7.8 (4.3, 22.0) | 0.177 |
| IL-8 (pg/mL) | 33.7 (19.9, 146.9) | 27.9 (19.5, 83.5) | 45.5 (24.4, 200.2) | 35.7 (20.6, 90.7) | 0.076 |
| IL-10 (pg/mL) | 3.7 (0.8, 4.0) | 3.9 (3.1, 4.6) | 4.1 (3.4, 5.4) | 3.9 (3.1, 4.7) | 0.297 |
| LBP (μg/mL) | 9.4 (3.8, 11.3) | 10.2 (4.6, 11.3) | 11.3 (10.5, 11.4) | 10.8 (8.3, 11.4) | 0.014 |
| TaqI C > T (rs731236) | |||||
| tt | Tt | TT | Overall | p- value | |
| N (%) | |||||
| CP stage | 0.027 | ||||
| A | 12 (42.3) | 33 (63.4) | 2 (22.2) | 47 (51.3) | |
| B | 12 (42.3) | 11 (22) | 2 (22.2) | 25 (28.9) | |
| C | 4 (15.4) | 8 (14.6) | 5 (55.6) | 17 (19.8) | |
| Median (IQR) | |||||
| Vitamin D levels (ng/mL) | 21.2 (15.5, 30.8) | 20.8 (14.7, 30.3) | 29.8 (11.2, 46.7) | 21.1 (14.7, 31.6) | 0.719 |
| MELD | 12.0 (9.0, 17.0) | 10.0 (8.0, 14.0) | 16.0 (12.0, 19.0) | 11.0 (8.0, 15.5) | 0.025 |
| VDBP (μg/mL) | 175.8 (123.8, 259.0) | 140.5 (90.9, 259.3) | 199.3 (126.8, 209.6) | 160.8 (99.3, 257.9) | 0.597 |
| IL-12 (pg/mL) | 8.0 (0.0, 9.0) | 7.8 (5.4, 8.6) | 5.9 (0.0, 6.8) | 7.5 (2.1, 8.7) | 0.475 |
| TNF-a (pg/mL) | 5.2 (1.0, 5.9) | 4.5 (1.5, 5.3) | 5.2 (0.3, 6.0) | 4.7 (1.2, 5.7) | 0.595 |
| IL-1β (pg/mL) | 8.1 (6.5, 12.6) | 7.9 (3.1, 11.6) | 7.5 (0.0, 11.2) | 8.0 (3.1, 11.6) | 0.786 |
| IL-6 (pg/mL) | 7.7 (5.1, 13.8) | 7.6 (4.3, 23.6) | 13.0 (6.5, 49.4) | 7.8 (4.3, 22.0) | 0.569 |
| IL-8 (pg/mL) | 33.7 (16.0, 101.6) | 41.3 (22.9, 90.7) | 29.3 (20.9, 44.3) | 35.7 (20.6, 90.7) | 0.759 |
| IL-10 (pg/mL) | 3.8 (2.0, 4.4) | 3.9 (3.3, 5.0) | 4.0 (3.7, 6.3) | 3.9 (3.1, 4.7) | 0.468 |
| LBP (μg/mL) | 10.2 (5.2, 11.3) | 10.7 (8.2, 11.4) | 11.3 (11.0, 11.4) | 10.8 (8.3, 11.4) | 0.200 |
Abbreviations: IQR, Interquartile range; CP, Child Pugh; MELD, Model for end-stage liver disease; VDBP, Vitamin D binding protein; LBP, Lipopolysaccharide binding protein.
Comparisons of clinical parameters and serum cytokine expression between VDR polymorphisms
As shown in Table 4, BsmI patients harboring the BB genotype had higher MELD score (p = 0.026) and were mainly at CP stage C (p = 0.020) compared to Bb/bb genotypes. ApaI patients harboring the AA genotype had increased levels of LBP (p = 0.004), IL-1β (p = 0.036) and IL-8 (p = 0.03) and were mostly at CP stage C (p = 0.001) compared to patients with the Aa/aa genotypes. The TT genotype carriers of the TaqI polymorphism had higher MELD score (p = 0.026) and were mainly at CP stage C (p = 0.02). Finally, FokI patients who had the FF genotype showed lower levels of IL-1β (p = 0.013) compared to patients with the Ff/ff genotypes. In the multivariate analysis, in the presence of other significant covariates, as well as cirrhosis’ etiology, AA genotype of ApaI polymorphism [OR: 5.5; 95% CI (1.3, 22.9), p = 0.019], BB genotype of BsmI polymorphism [OR: 9.6; 95% CI (1.3, 72.20), p = 0.027] and TT genotype of TaqI polymorphism [OR: 9.6; 95% CI (1.3, 72.20), p = 0.027] were found to be associated with increased odds of more advanced CP stage when compared to Aa/aa, Bb/bb and Tt/tt genotypes respectively (Table 5). FF genotype of FokI polymorphism was not found to be a significant predictor of disease severity.
Table 4.
Comparisons of clinical parameters and serum cytokine expression between VDR polymorphisms.
| BsmI A > G (rs1544410) | ||||
|---|---|---|---|---|
| Bb + bb | BB | Overall | p-value | |
| N (%) | ||||
| CP stage | 0.020 | |||
| A | 44 (55.3) | 2 (22.2) | 46 (51.3) | |
| B | 24 (29.8) | 2 (22.2) | 26 (28.9) | |
| C | 12 (14.9) | 5 (55.6) | 17 (19.8) | |
| Median (IQR) | ||||
| Vitamin D levels (ng/mL) | 21.0 (14.8, 30.3) | 29.8 (11.2, 46.7) | 21.1 (14.7, 31.6) | 0.540 |
| VDBP (μg/mL) | 157.6 (97.4, 259.0) | 199.3 (126.8, 209.6) | 160.8 (99.3, 257.9) | 0.890 |
| MELD | 11.0 (8.0, 14.0) | 16.0 (12.0, 19.0) | 11.0 (8.0, 15.5) | 0.026 |
| IL-12 (pg/mL) | 7.9 (5.1, 8.7) | 5.9 (0.0, 6.8) | 7.5 (2.1, 8.7) | 0.225 |
| TNF-a (pg/mL) | 4.6 (1.3, 5.7) | 5.2 (0.3, 6.0) | 4.7 (1.2, 5.7) | 0.804 |
| IL-1β (pg/mL) | 8.1 (6.2, 11.8) | 7.5 (0.0, 11.2) | 8.0 (3.1, 11.6) | 0.593 |
| IL-6 (pg/mL) | 7.7 (4.3, 18.1) | 13.0 (6.5, 49.4) | 7.8 (4.3, 22.0) | 0.298 |
| IL-8 (pg/mL) | 37.7 (20.4, 93.4) | 29.3 (20.9, 44.3) | 35.7 (20.6, 90.7) | 0.778 |
| IL-10 (pg/mL) | 3.8 (3.1, 4.6) | 4.0 (3.7, 6.3) | 3.9 (3.1, 4.7) | 0.449 |
| LBP (μg/mL) | 10.5 (7.0, 11.4) | 11.3 (11.0, 11.4) | 10.8 (8.3, 11.4) | 0.121 |
| FokI C > T (rs10735810) | ||||
| Ff + ff | FF | Overall | p- value | |
| N (%) | ||||
| CP stage | 0.829 | |||
| A | 25 (51.2) | 21 (51.5) | 46 (51.3) | |
| B | 13 (26.8) | 13 (31.4) | 26 (28.9) | |
| C | 10 (22) | 7 (17.1) | 17 (19.8) | |
| Median (IQR) | ||||
| Vitamin D levels (ng/mL) | 22.8 (15.9, 33.3) | 18.4 (13.9, 26.4) | 21.1 (14.7, 31.6) | 0.206 |
| VDBP (μg/mL) | 168.9 (113.7, 270.6) | 159.3 (89.7, 220.8) | 160.8 (99.3, 257.9) | 0.309 |
| MELD | 11.0 (8.0, 15.0) | 11.0 (8.0, 17.0) | 11.0 (8.0, 15.5) | 0.867 |
| IL-12 (pg/mL) | 7.0 (4.2, 8.9) | 8.0 (0.0, 8.5) | 7.5 (2.1, 8.7) | 0.840 |
| TNF-a (pg/mL) | 4.6 (1.5, 5.7) | 4.7 (0.4, 5.7) | 4.7 (1.2, 5.7) | 0.580 |
| IL-1β (pg/mL) | 9.3 (7.4, 12.3) | 7.2 (0.0, 10.1) | 8.0 (3.1, 11.6) | 0.013 |
| IL-6 (pg/mL) | 7.1 (4.2, 17.6) | 9.8 (4.9, 29.4) | 7.8 (4.3, 22.0) | 0.459 |
| IL-8 (pg/mL) | 38.1 (20.8, 110.4) | 33.0 (20.4, 85.8) | 35.7 (20.6, 90.7) | 0.555 |
| IL-10 (pg/mL) | 3.8 (3.0, 4.7) | 4.0 (3.1, 4.6) | 3.9 (3.1, 4.7) | 0.907 |
| LBP (μg/mL) | 10.5 (9.0, 11.3) | 10.9 (4.3, 11.4) | 10.8 (8.3, 11.4) | 0.983 |
| ApaI A > C (rs7975232) | ||||
| Aa + aa | AA | Overall | p- value | |
| N (%) | ||||
| CP stage | 0.001 | |||
| A | 32 (56.9) | 13 (40) | 45 (51.3) | |
| B | 20 (35.3) | 5 (16) | 25 (28.9) | |
| C | 5 (7.8) | 14 (44) | 19 (19.8) | |
| Median (IQR) | ||||
| Vitamin D levels (ng/mL) | 20.6 (14.8, 30.3) | 24.1 (14.1, 33.6) | 21.1 (14.7, 31.6) | 0.527 |
| VDBP (μg/mL) | 178.9 (123.4, 260.4) | 130.2 (81.7, 215.3) | 160.8 (99.3, 257.9) | 0.161 |
| MELD | 11.0 (8.0, 14.0) | 13.0 (9.0, 17.0) | 11.0 (8.0, 15.5) | 0.167 |
| IL-12 (pg/mL) | 7.8 (0.0, 8.7) | 7.0 (4.7, 8.5) | 7.5 (2.1, 8.7) | 0.704 |
| TNF-a (pg/mL) | 4.5 (1.3, 5.6) | 5.0 (1.1, 6.1) | 4.7 (1.2, 5.7) | 0.258 |
| IL-1β (pg/mL) | 7.5 (0.0, 11.4) | 10.1 (7.5, 11.7) | 8.0 (3.1, 11.6) | 0.036 |
| IL-6 (pg/mL) | 6.5 (4.1, 13.3) | 12.9 (5.2, 29.8) | 7.8 (4.3, 22.0) | 0.063 |
| IL-8 (pg/mL) | 29.8 (19.6, 84.4) | 45.5 (24.4, 200.2) | 35.7 (20.6, 90.7) | 0.030 |
| IL-10 (pg/mL) | 3.8 (3.0, 4.5) | 4.1 (3.5, 5.5) | 3.9 (3.1, 4.7) | 0.221 |
| LBP (μg/mL) | 9.9 (4.8, 11.3) | 11.3 (10.5, 11.4) | 10.8 (8.3, 11.4) | 0.004 |
| TaqI C > T (rs731236) | ||||
| Tt + tt | TT | Overall | p- value | |
| N (%) | ||||
| CP stage | 0.020 | |||
| A | 44 (55.3) | 2 (22.2) | 46 (51.3) | |
| B | 24 (29.8) | 2 (22.2) | 26 (28.9) | |
| C | 12 (14.9) | 5 (55.6) | 17 (19.8) | |
| Median (IQR) | ||||
| Vitamin D levels (ng/mL) | 21.0 (14.8, 30.3) | 29.8 (11.2, 46.7) | 21.1 (14.7, 31.6) | 0.540 |
| VDBP (μg/mL) | 157.6 (97.4, 259.0) | 199.3 (126.8, 209.6) | 160.8 (99.3, 257.9) | 0.890 |
| MELD | 11.0 (8.0, 14.0) | 16.0 (12.0, 19.0) | 11.0 (8.0, 15.5) | 0.026 |
| IL-12 (pg/mL) | 7.9 (5.1, 8.7) | 5.9 (0.0, 6.8) | 7.5 (2.1, 8.7) | 0.225 |
| TNF-a (pg/mL) | 4.6 (1.3, 5.7) | 5.2 (0.3, 6.0) | 4.7 (1.2, 5.7) | 0.804 |
| IL-1β (pg/mL) | 8.1 (6.2, 11.8) | 7.5 (0.0, 11.2) | 8.0 (3.1, 11.6) | 0.593 |
| IL-6 (pg/mL) | 7.7 (4.3, 18.1) | 13.0 (6.5, 49.4) | 7.8 (4.3, 22.0) | 0.298 |
| IL-8 (pg/mL) | 37.7 (20.4, 93.4) | 29.3 (20.9, 44.3) | 35.7 (20.6, 90.7) | 0.778 |
| IL-10 (pg/mL) | 3.8 (3.1, 4.6) | 4.0 (3.7, 6.3) | 3.9 (3.1, 4.7) | 0.449 |
| LBP (μg/mL) | 10.5 (7.0, 11.4) | 11.3 (11.0, 11.43) | 10.8 (8.3, 11.4) | 0.121 |
Abbreviations: IQR, Interquartile range; CP, Child Pugh; MELD, Model for end-stage liver disease; VDBP, Vitamin D binding protein; LBP, Lipopolysaccharide binding protein.
Table 5.
Multivariate analysis for the association of VDR polymorphisms with cirrhosis severity by means of CP stage.
| Factor | OR | 95% C.I. | p-value |
|---|---|---|---|
| SNP_ApaI | |||
| Aa + aa* | 1 | ||
| AA | 5.51 | (1.33, 22.89) | 0.019 |
| Etiology | |||
| alcohol* | 1 | ||
| viral | 5.01 | (1.11, 22.71) | 0.036 |
| other | 11.74 | (1.34, 102.94) | 0.026 |
| Sex | |||
| male* | 1 | ||
| female | 0.10 | (0.02, 0.53) | 0.007 |
| VDBP | |||
| per unit | 0.98 | (0.97, 0.99) | <0.001 |
| IL-8 | |||
| per unit | 1.01 | (1.00, 1.02) | 0.005 |
| LBP | |||
| per unit | 1.02 | (1.00, 1.05) | 0.031 |
| SNP_BsmI | |||
| Bb + bb* | 1 | ||
| BB | 9.64 | (1.29, 72.20) | 0.027 |
| Etiology | |||
| alcohol* | 1 | ||
| viral | 3.66 | (0.89, 15.05) | 0.072 |
| other | 7.78 | (0.94, 64.15) | 0.057 |
| Sex | |||
| male* | 1 | ||
| female | 0.12 | (0.02, 0.62) | 0.012 |
| VDBP | |||
| per unit | 0.98 | (0.97, 0.99) | <0.001 |
| IL-8 | |||
| per unit | 1.01 | (1.00, 1.02) | 0.003 |
| LBP | |||
| per unit | 1.02 | (1.00, 1.05) | 0.031 |
| SNP_TaqI | |||
| Tt + tt* | 1 | ||
| TT | 9.64 | (1.29, 72.20) | 0.027 |
| Etiology | |||
| alcohol* | 1 | ||
| viral | 3.66 | (0.89, 15.05) | 0.072 |
| other | 7.78 | (0.94, 64.15) | 0.057 |
| Sex | |||
| male* | 1 | ||
| female | 0.12 | (0.02, 0.62) | 0.012 |
| VDBP | |||
| per unit | 0.98 | (0.97, 0.99) | <0.001 |
| IL-8 | |||
| per unit | 1.01 | (1.00, 1.02) | 0.003 |
| LBP | |||
| per unit | 1.02 | (1.00, 1.05) | 0.031 |
| SNP_FokI | |||
| Ff + ff* | 1 | ||
| FF | 0.45 | (0.14, 1.49) | 0.191 |
| Etiology | |||
| alcohol* | 1 | ||
| viral | 3.15 | (0.77, 12.86) | 0.111 |
| other | 5.22 | (0.81, 33.69) | 0.082 |
| Sex | |||
| male* | 1 | ||
| female | 0.12 | (0.03, 0.58) | 0.008 |
| VDBP | |||
| per unit | 0.98 | (0.97, 0.99) | <0.001 |
| IL-8 | |||
| per unit | 1.01 | (1.00, 1.02) | 0.004 |
| LBP | |||
| per unit | 1.03 | (1.01, 1.06) | 0.011 |
*Reference category.
Abbreviations: OR, odds ratio; C.I., confidence interval; SNP, single nucleotide polymorphism; VDBP, vitamin D binding protein; LBP, lipopolysaccharide binding protein.
VDR polymorphisms and the etiology of liver cirrhosis
The grouping of cirrhotic population according to disease etiology was performed as follows: patients with cirrhosis of viral origin (n = 40, 44.2%), alcoholic origin (n = 32, 36.4%) and other etiologies (n = 17, 19.4%). None of the VDR polymorphisms interacted significantly with the etiology of the disease, indicating that the effect of the polymorphisms is similar across all groups regarding cirrhosis’ etiology.
Association between vitamin D and VDBP levels with VDR polymorphisms
We found no statistically significant differences between serum 25(OH) vitamin D levels and VDBP levels in relation to VDR polymorphisms.
Linkage disequilibrium of VDR polymorphisms in cirrhotic patients
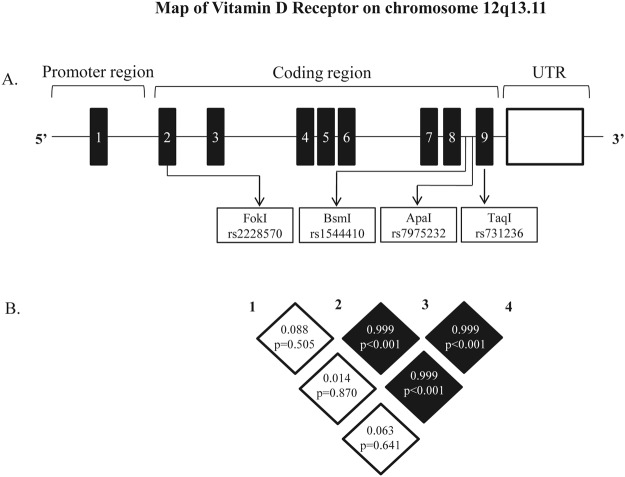
Linkage disequilibrium analysis revealed very strong LD between BsmI and TaqI (D’ = 0.999), BsmI and ApaI (D’ = 0.999) and TaqI and ApaI (D’ = 0.999) polymorphisms. In contrast, very weak LD was detected between FokI and BsmI (D’ = 0.088), FokI and TaqI (D’ = 0.063), FokI and ApaI (D’ = 0.014) polymorphisms (Fig. 1).
Figure 1.
(A) Structure of genomic region of the VDR gene on chromosome 12q13.11. (A) The black boxes demonstrate the exons of the VDR gene. The approximate locations of the examined polymorphisms are indicated by arrows. (B) Schematic representation of pairwise linkage disequilibrium (LD) pattern. (A) LD pattern of the VDR gene polymorphisms [BsmI A > G (B > b), ApaI A > C (A > a), TaqI C > T (T > t) and FokI C > T (F > f)] in the studied population (n = 89). Each square represents the D’ values and the p values between the pairs of polymorphisms. The intensity of the dark color of the blocks is proportional to the D’ value, indicating the strength LD between polymorphisms. Black boxes, high LD; white boxes, low LD.
Haplotype analysis of VDR polymorphisms in relation to disease severity
Haplotype association with cirrhosis severity was evaluated by the distribution of VDR haplotypes in the different CP stages. Estimated VDR haplotype frequencies of FokI, BsmI, ApaI and TaqI polymorphisms are reported in Table 6. The results showed that in patients with CP stage C, BAT haplotype was more frequent suggesting a potential increased risk for advanced cirrhosis, whereas the complementary haplotype bat was more common in patients with CP stage A; however, this difference was no significant (LR test p = 0. 581).
Table 6.
Haplotype frequencies and association with cirrhosis severity.
| Haplotypes | CP stage | |||||
|---|---|---|---|---|---|---|
| A | B | C | ||||
| n | Frequency (%) | n | Frequency (%) | n | Frequency (%) | |
| BAT | 35 | 19.7 | 15 | 8.5 | 20 | 11.2 |
| bat | 37 | 20.8 | 24 | 13.5 | 7 | 3.9 |
| bAt | 18 | 10.1 | 10 | 5.6 | 10 | 5.6 |
| BAt | 0 | 0.0 | 1 | 0.01 | 1 | 0.01 |
| baT | 0 | 0.0 | 0 | 0.0 | 0 | 0.0 |
| bAT | 0 | 0.0 | 0 | 0.0 | 0 | 0.0 |
| Bat | 0 | 0.0 | 0 | 0.0 | 0 | 0.0 |
| BaT | 0 | 0.0 | 0 | 0.0 | 0 | 0.0 |
Abbreviations: CP stage, Child-Pugh stage.
Survival analysis
The cumulative mortality rate was 31.46% (28 out of 89 patients), after a median follow-up of 16 months (IQR: 3–40 months). The main causes of mortality were liver failure (n = 19, 67.9%), HCC (n = 4, 14.3%), renal failure (n = 3, 10.7%), bleeding (n = 1, 3.6%) and other causes (n = 1, 3.6%). In the univariable Cox regression analysis, the following factors were found to be significantly associated with mortality: CP score (p < 0.001), MELD (p < 0.001), VDBP levels (p = 0.003), IL-6 (p = 0.016), IL-8 (p = 0.001), LBP levels (p = 0.020) and the CP stage III (p < 0.001). In the multivariate analysis, CP score [HR: 1.26, 95% CI (1.02–1.56) p = 0.035], MELD [HR: 1.15, 95% CI (1.03–1.28) p = 0.012] and the presence of FF genotype [ff genotype vs FF: HR = 0.22 95% CI (0.06–0.77), p = 0.018)] were significant independent prognostic factors for patient survival (Table 7).
Table 7.
Univariate and multivariate cox regression analyses for cirrhotic patients’ survival.
| Factor | HR | 95% C.I. | p-value |
|---|---|---|---|
| Univariate analysis | |||
| CP score | |||
| per unit | 1.47 | (1.26, 1.71) | <0.001 |
| MELD | |||
| per unit | 1.20 | (1.11, 1.29) | <0.001 |
| VDBP | |||
| per unit | 0.99 | (0.99, 1.00) | 0.003 |
| IL-12 | |||
| per unit | 1.02 | (0.98, 1.06) | 0.287 |
| TNF-a | |||
| per unit | 1.04 | (0.94, 1.17) | 0.440 |
| IL-1β | |||
| per unit | 1.00 | (0.97, 1.04) | 0.827 |
| IL-6 | |||
| per unit | 1.00 | (1.00, 1.00) | 0.016 |
| IL-8 | |||
| per unit | 1.00 | (1.00, 1.00) | 0.001 |
| IL-10 | |||
| per unit | 1.01 | (0.98, 1.04) | 0.455 |
| LBP | |||
| per unit | 1.02 | (1.00, 1.04) | 0.020 |
| Age | |||
| per unit | 1.01 | (0.98, 1.05) | 0.438 |
| Sex | |||
| Male* | 1 | ||
| Female | 1.16 | (0.50, 2.71) | 0.735 |
| CP stage | |||
| A* | 1 | ||
| B | 2.87 | (0.99, 8.33) | 0.052 |
| C | 9.06 | (3.19, 25.76) | <0.001 |
| SNP_BsmI | |||
| bb* | 1 | ||
| Bb | 0.55 | (0.24, 1.30) | 0.175 |
| BB | 1.81 | (0.67, 4.86) | 0.242 |
| SNP_TaqI | |||
| tt* | 1 | ||
| Tt | 0.60 | (0.26, 1.42) | 0.249 |
| TT | 1.92 | (0.71, 5.18) | 0.195 |
| SNP_FokI | |||
| ff* | 1 | ||
| Ff | 0.72 | (0.26, 2.03) | 0.538 |
| FF | 0.51 | (0.18, 1.46) | 0.210 |
| SNP_ApaI | |||
| aa* | 1 | ||
| Aa | 1.21 | (0.27, 5.41) | 0.803 |
| AA | 2.11 | (0.48, 9.21) | 0.323 |
| Factor | HR | 95% C.I. | p - value |
| Multivariate analysis | |||
| CP score | |||
| per unit | 1.26 | (1.02, 1.56) | 0.035 |
| MELD | |||
| per unit | 1.15 | (1.03, 1.28) | 0.012 |
| SNP_FokI | |||
| ff* | 1 | ||
| Ff | 0.37 | (0.12, 1.13) | 0.080 |
| FF | 0.22 | (0.06, 0.77) | 0.018 |
*Reference category.
Abbreviations: HR, hazard ratio; C.I., confidence interval; CP, child pugh; MELD, model for end-stage liver disease; VDBP, vitamin D binding protein; LBP, lipopolysaccharide binding protein; SNP, single nucleotide polymorphism.
Discussion
This is the first report of an association between polymorphisms of the VDR gene and cytokine levels, severity of liver disease and survival in patients with liver cirrhosis. In particular, an independent association between BsmI, ApaI, and TaqI VDR polymorphisms and the severity of liver cirrhosis is clearly shown. Moreover, the presence of FF genotype of FokI polymorphism is associated with a better prognosis regarding survival in this cohort. These features appear to be independent of the etiology of liver cirrhosis, as they observed in patients of any cause.
Vitamin D promotes the stimulation of innate immunity, the differentiation of monocytes, the inhibition of lymphocyte proliferation and cytokine secretion by T and B cells22,23. VDR acts as a ligand-stimulated transcription factor and activates 1,25(OH)2D3 at the transcriptional level. The activation of VDR contributes to the regulation of immune response by inhibiting T helper 1 (Th1) cell proliferation and pro-inflammatory cytokine production and inducing Th2 cell proliferation and anti-inflammatory cytokine production7,22–26. The presence of VDR polymorphisms possibly leads to a dysfunctional receptor, affecting VDR activity and the subsequent vitamin D-mediated effects26.
The association between VDR polymorphisms and the occurrence of chronic liver disease from different etiologies such as autoimmune hepatitis, PBC, HCC or HBV infection has been investigated with conflicting results8,10–16,27. Previous reports have identified gene polymorphisms which affect the progression of liver fibrosis28–31. The relationship between liver fibrosis progression and the presence of VDR polymorphisms (ApaI, TaqI and BsmI) has been investigated, indicating that in PBC patients, BsmI and TaqI were associated with progressive cirrhosis10 and in NAFLD patients, VDR mRNA expression and profibrogenic genes were significantly affected by BsmI polymorphism18. The effect of bAt haplotype in fibrosis progression has been investigated in HCV patients as well, giving conflicting results18,19,32. Our results indicate that the presence of ApaI polymorphism (AA genotype) is related to significant higher levels of IL-1β and IL-8. The increased levels of these pro-inflammatory cytokines suggest that the ApaI VDR polymorphism leads to a less active VDR protein which may contribute to a disturbance of Th1/Th2 balance, a transition to Th1 polarization and a decreased activity of vitamin D-related signaling pathways.
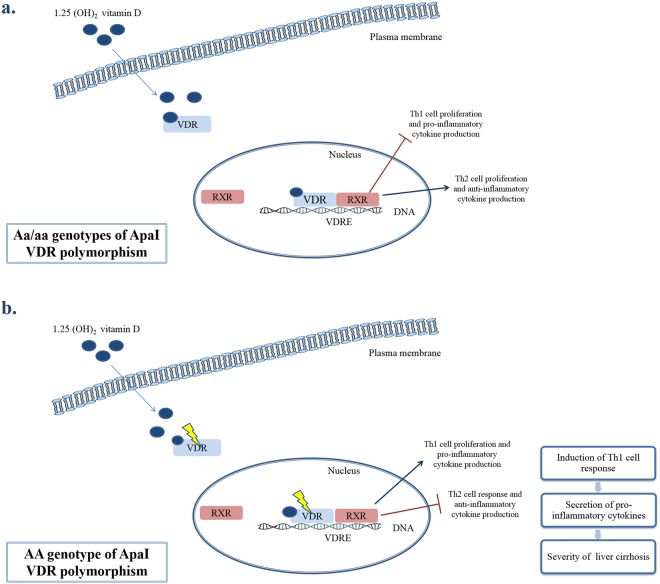
Several studies have demonstrated a positive correlation between higher pro-inflammatory cytokine levels and the severity of liver disease33–37. In this study, we have shown that the AA genotype of the ApaI polymorphism is related to decreased levels of platelets and increased levels of LBP, which are consistent with the progression of cirrhosis and portal hypertension development38,39. The presence of ApaI, TaqI and BsmI VDR polymorphisms could impede the interaction between vitamin D and VDR, resulting in ineffective vitamin D-VDR complex, impaired VDR-mediated transcription, decreased activity of vitamin D related signaling pathways, transition to a Th1 polarization, and consequently, to a more progressive form of liver cirrhosis (Fig. 2).
Figure 2.
Schematic representation illustrating our proposed mechanism of how ApaI VDR polymorphisms potentially affect the progression of liver cirrhosis. (a) Presence of Aa/aa genotypes of ApaI VDR polymorphism. VDR is an intracellular ligand-activated transcription factor that specifically binds 1,25(OH)2D3 and regulates the expression of several target genes. Upon the activation of vitamin D, the ligated VDR heterodimerizes with retinoid X receptor (RXR) which is necessary for DNA binding, translocates to the nucleus, binds to vitamin D response elements (VDRE) and recruits other nuclear proteins to the transcriptional pre-initiation complex. This process results in the transcriptional activation or suppression of the target genes through the interaction with nuclear receptor co-activators or co-repressors. The binding of VDR with vitamin D may modulate cytokine responses by T cells, inhibiting Th1 cell proliferation and pro-inflammatory cytokine secretion and activating Th2 cell proliferation and anti-inflammatory cytokine secretion. (b) Presence of AA genotype of ApaI VDR polymorphism. The presence of polymorphisms may impair the activity of the VDR resulting in a dysfunctional receptor. The dimerization of the 1,25(OH)2D3-VDR with RXR may be hindered by the existence of genetic variations thus affecting VDR activity and subsequent downstream vitamin D-mediated effects. This impaired process may lead to disturbance of the Th1/Th2 balance, resulting in a transition to Th1 cell response and pro-inflammatory cytokine secretion that is closely related the progression of liver cirrhosis.
A second novel finding of this study, is the inverse association between the FokI polymorphism, particularly the FF genotype, with mortality in liver cirrhotic patients, imparting a protective role of this genotype in cirrhosis. The FokI polymorphism is located in the coding region of the VDR gene and results in a VDR protein with a different structure, creating a new start codon and consequently a VDR protein shortened by three amino acids38,40. This protein is more functional and has higher transcriptional activity compared to the long-length VDR protein38,40,41. FokI is the only polymorphism that was not associated with severity of liver cirrhosis in our study. The length of the VDR protein influences the regulation of gene transcription through occupation of recognition sites of other transcription factors and interference with their signaling pathways7. Therefore, a longer VDR protein may lead to a decreased transcriptional activity and an increased risk of susceptibility to disease40. These observations are in line with our study as we have shown that the presence of FokI polymorphism (FF genotype) is associated with significantly lower levels of IL-1β. Patients with the FF genotype produce a shorter form of VDR, leading to higher transcriptional activity, formation of more active complexes of VDR-vitamin D, inhibition of the Th1 response and induction of the Th2 cell response. Hence, patients with FokI FF genotype may have a better response to vitamin D resulting in a lower progression rate of cirrhosis. However, due to the fact that this hypothesis is of high interest, we suggest that it should be further explored in larger and more specific cohorts with more patients harboring the FokI polymorphism in order to be confirmed.
We have also shown the existence of strong linkage disequilibrium between the BsmI, ApaI and TaqI polymorphic sites in our cirrhotic population. These results are in agreement with previous reports suggesting an extensive LD between these genetic markers10,14. As these polymorphisms are in strong LD, it can be assumed that these single nucleotide polymorphisms (SNPs) contribute to the severity of cirrhosis in a dependent manner. Nevertheless, as these polymorphisms do not cause a functional change in the VDR gene, it is possible that BsmI, TaqI and ApaI are possibly genetic markers of other functional variations of the VDR gene or in other closely linked genes that are in linkage disequilibrium with the identified polymorphisms.
Some limitations of the current study should be acknowledged. The first limitation concerns the relatively small sample size, however our results are consistent with the reports on the association between VDR polymorphisms and the susceptibility to liver fibrosis10,18,19. Secondly, our study was performed on Caucasians patients and it would be interesting to perform the same analysis in different ethnic groups. Lastly, the single measurement of 25(OH)D at baseline may not be representative of the respective concentrations over time. However, there are reports supporting that although 25(OH)D levels present seasonal fluctuation, its levels remain stable over time42,43.
In conclusion, our results indicate that VDR polymorphisms are independently associated with the severity of liver cirrhosis and the survival of patients with liver disease, regardless of disease etiology, suggesting a potential influence of them in disease progression. Based on these results future studies will delineate causation between specific VDR polymorphisms and outcome/severity of liver cirrhosis, and the importance of VDR polymorphism analysis in clinical practice to identify patients at greater risk of disease progression and to modify patients’ surveillance and treatment accordingly.
Methods
Study design and participants
This study was a prospective cohort study, on 89 consecutive Caucasian patients with liver cirrhosis. During the recruitment, all cirrhotic patients were in stable clinical condition, without any severe complication of liver disease including gastro-intestinal bleeding, hepatorenal syndrome, moderate to severe hepatic encephalopathy, spontaneous bacterial peritonitis, malignancy, or organ failure. Patients with indications or history of bacterial infection at last 4 weeks prior to recruitment in the study, human immunodeficiency virus (HIV) infection and severe cardiopulmonary disease or renal failure were excluded. Severity of cirrhosis was assessed by the CP stage and the MELD score44. Diagnosis of cirrhosis was based on histological or compatible clinical, laboratory and imaging data45–47. After baseline examination, patients were followed in the hepatology clinic at regular intervals according to current guidelines48 until death, liver transplantation or completion of the study. The recruitment of the patients was performed at the University Hospital of Patras (Patras, Greece) between September 2009 and April 2013. Blood samples from all patients were collected throughout the year. Seasonal variability was defined as winter/spring from December to April and summer/autumn from May to October49. Sampling occurred mostly in winter/spring (70%) compared to summer/autumn (30%). All study participants, or their legal guardian, provided informed written consent prior to study enrollment. The study protocol was approved by Patras University Hospital Scientific Review Board and Ethics Committee. The Hospital abides by the 1975 Helsinki declaration on ethical principles for medical research involving human subjects. Further all the experiments were performed in accordance with relevant guidelines and regulations of the concerned ethical committee.
Vitamin D assay
Serum samples were collected from the patients and stored at −80 °C until analysis. Serum 25(OH)D levels were determined using a 25(OH)D vitamin D ELISA kit for serum and plasma (Enzo Life Sciences, NY, USA), according to the manufacturer’s instructions. Currently accepted standards for the definition of Vitamin D status are: optimal vitamin D levels ≥30 ng/mL, vitamin D deficiency ≤20 ng/mL and vitamin D insufficiency between 20 and 30 ng/mL50,51.
VDBP and LBP assays
Serum VDBP levels were determined using a human Vitamin D BP Quantikine ELISA Kit (R&D Systems, Minneapolis, MN, USA), and serum LBP levels using a human LBP ELISA kit (SunRed Biological Technology, Shanghai). Data analysis was performed using the Curve Expert 1.4 software.
Cytokine Assays
Serum interleukin-12 (IL-12), IL-1β, IL-6, IL-8, IL-10 and tumor necrosis factor alpha (TNF-a) levels were determined using a Cytometric Bead Array (CBA) assay (Human Inflammatory Cytokines Kit, BD Biosciences, San Diego, CA, USA) run on a BD FACS Array Bioanalyzer. Data were analyzed using the FlowJo V7.5 software (Tree Star Inc., Ashland, OR, USA).
DNA extraction
Genomic DNA was extracted using the NucleoSpin® Blood QuickPure kit (Macherey-Nagel, Germany). The DNA concentration of the samples was determined using a Nanodrop spectrophotometer (UV spectrophotometer Q3000, Quawell Technology, Inc., USA).
VDR Genotyping
Genotyping was carried out using TaqMan SNP Genotyping Assays (Applied Biosystems; Foster City, USA). The PCR reactions were carried out in MicroAmp® Fast Optical 96-Well Reaction Plates (Applied Biosystems) on the Step One Plus real-time PCR system (Applied Biosystems, CA, USA). The rs731236 (TaqI), rs1544410 (BsmI), rs7975232 (ApaI) and rs2228570 (FokI) probes were designed using TaqMan pre-designed SNP genotyping assays (Applied Biosystems). Two non-template-control wells were included on each plate. DNA amplification was performed as follows: 95 °C for 10 min, followed by 40 cycles of 92 °C for 15 sec and 60 °C for 1 min.
Statistical analysis
Continuous variables were summarized as medians and interquartile ranges (IQRs) while counts and corresponding percentages were calculated for categorical variables. All comparisons were performed using non-parametric tests: Fisher’s exact tests in case of frequencies’ comparisons, Mann-Whitney and Kruskal-Wallis tests for the comparison of median values between two groups and more than two groups, respectively. Correlations between vitamin D and VDBP levels with VDR polymorphisms were assessed by the Spearman’s coefficient. Multivariable ordinal logistic regression models were fitted, to test the hypothesis that the VDR polymorphisms are associated with the CP stage. Further analysis was conducted to explore whether these polymorphisms’ effect interacts with the etiology of cirrhosis, i.e. whether the effect of the polymorphisms is different in the subgroups of viral, alcoholic or other etiology’s cirrhosis. The VDR gene polymorphisms’ Hardy-Weinberg equilibrium was examined by means of chi square test goodness of fit test, i.e by comparing observed and expected count in each of the polymorphisms groups (wt/wt, mt/wt, mt/mt). Pairwise linkage disequilibrium (LD) analysis between the VDR gene polymorphisms was performed using the genetics package of R software. Allelic frequencies were estimated by the hapipf stata command, based on the expectation-maximization (EM) algorithm. The hypothesis of allelic association with the CP stage was tested using the likelihood-ratio (LR) test. Time to death was analyzed using the Cox survival model. Before fitting the models, the proportional hazards assumption was assessed for all variables based on Schoenfeld residuals. Individuals’ baseline clinical and laboratory variables, including the VDR polymorphisms, were considered as potential risk factors. For all models selection, the Collett’s approach was followed52. More specifically, all variables with a p-value < 0.200 were initially included and then eliminated using backwards selection. When a model that included only significant covariates was reached, variables initially excluded entered the final model one by one and tested for significance in the presence of already included significant variables. Analysis was performed using Stata 13.1 (StataCorp LP, College Station, Texas, USA). Level of significance α was set at 0.05.
Acknowledgements
This work has been supported by “Hellenic Association for the Study of the Liver”.
Author Contributions
Triantos C.: study concept and design, drafting of the manuscript, critical revision of the manuscript for important intellectual content, final approval of the version to be published; Aggeletopoulou I.: acquisition of data; analysis and interpretation of data, drafting of the manuscript; Kalafateli M.: acquisition of data, analysis and interpretation of data; Spantidea P.: acquisition of data, analysis and interpretation of data; Vourli G.: analysis and interpretation of data, statistical analysis; Diamantopoulou G.: acquisition of data; Tapratzi D.: acquisition of data; Michalaki M.: analysis and interpretation of data, critical revision of the manuscript for important intellectual content; Manolakopoulos S.: analysis and interpretation of data, critical revision of the manuscript for important intellectual content; Gogos C.: analysis and interpretation of data, critical revision of the manuscript for important intellectual content; Kyriazopoulou V.: analysis and interpretation of data, critical revision of the manuscript for important intellectual content; Mouzaki A.: analysis and interpretation of data, critical revision of the manuscript for important intellectual content; Thomopoulos K.: analysis and interpretation of data, critical revision of the manuscript for important intellectual content.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Schuppan D, Afdhal NH. Liver cirrhosis. Lancet. 2008;371:838–851. doi: 10.1016/S0140-6736(08)60383-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sivanathan V, et al. Etiology and complications of liver cirrhosis: data from a German centre. Dtsch Med Wochenschr. 2014;139:1758–1762. doi: 10.1055/s-0034-1387240. [DOI] [PubMed] [Google Scholar]
- 3.Bataller R, North KE, Brenner DA. Genetic polymorphisms and the progression of liver fibrosis: a critical appraisal. Hepatology. 2003;37:493–503. doi: 10.1053/jhep.2003.50127. [DOI] [PubMed] [Google Scholar]
- 4.Bhalla AK, Amento EP, Clemens TL, Holick MF, Krane SM. Specific high-affinity receptors for 1,25-dihydroxyvitamin D3 in human peripheral blood mononuclear cells: presence in monocytes and induction in T lymphocytes following activation. J Clin Endocrinol Metab. 1983;57:1308–1310. doi: 10.1210/jcem-57-6-1308. [DOI] [PubMed] [Google Scholar]
- 5.Kato S. The function of vitamin D receptor in vitamin D action. J Biochem. 2000;127:717–722. doi: 10.1093/oxfordjournals.jbchem.a022662. [DOI] [PubMed] [Google Scholar]
- 6.Carlberg C, Campbell MJ. Vitamin D receptor signaling mechanisms: integrated actions of a well-defined transcription factor. Steroids. 2013;78:127–136. doi: 10.1016/j.steroids.2012.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.van Etten E, et al. The vitamin D receptor gene FokI polymorphism: functional impact on the immune system. Eur J Immunol. 2007;37:395–405. doi: 10.1002/eji.200636043. [DOI] [PubMed] [Google Scholar]
- 8.Vogel A, Strassburg CP, Manns MP. Genetic association of vitamin D receptor polymorphisms with primary biliary cirrhosis and autoimmune hepatitis. Hepatology. 2002;35:126–131. doi: 10.1053/jhep.2002.30084. [DOI] [PubMed] [Google Scholar]
- 9.Wang X, Cheng W, Ma Y, Zhu J. Vitamin D receptor gene FokI but not TaqI, ApaI, BsmI polymorphism is associated with Hashimoto’s thyroiditis: a meta-analysis. Sci Rep. 2017;7:41540. doi: 10.1038/srep41540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kempinska-Podhorecka A, et al. Vitamin d receptor polymorphisms predispose to primary biliary cirrhosis and severity of the disease in polish population. Gastroenterol Res Pract. 2012;408723:29. doi: 10.1155/2012/408723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Halmos B, et al. Association of primary biliary cirrhosis with vitamin D receptor BsmI genotype polymorphism in a Hungarian population. Dig Dis Sci. 2000;45:1091–1095. doi: 10.1023/A:1005581414918. [DOI] [PubMed] [Google Scholar]
- 12.Lakatos LP, et al. Vitamin D receptor, oestrogen receptor-alpha gene and interleukin-1 receptor antagonist gene polymorphisms in Hungarian patients with primary biliary cirrhosis. Eur J Gastroenterol Hepatol. 2002;14:733–740. doi: 10.1097/00042737-200207000-00004. [DOI] [PubMed] [Google Scholar]
- 13.Tanaka A, et al. Vitamin D receptor polymorphisms are associated with increased susceptibility to primary biliary cirrhosis in Japanese and Italian populations. J Hepatol. 2009;50:1202–1209. doi: 10.1016/j.jhep.2009.01.015. [DOI] [PubMed] [Google Scholar]
- 14.Falleti E, et al. Vitamin D receptor gene polymorphisms and hepatocellular carcinoma in alcoholic cirrhosis. World J Gastroenterol. 2010;16:3016–3024. doi: 10.3748/wjg.v16.i24.3016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yao X, et al. The associated ion between the VDR gene polymorphisms and susceptibility to hepatocellular carcinoma and the clinicopathological features in subjects infected with HBV. Biomed Res Int. 2013;953974:23. doi: 10.1155/2013/953974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Suneetha PV, et al. Association between vitamin D receptor, CCR5, TNF-alpha and TNF-beta gene polymorphisms and HBV infection and severity of liver disease. J Hepatol. 2006;44:856–863. doi: 10.1016/j.jhep.2006.01.028. [DOI] [PubMed] [Google Scholar]
- 17.Fan L, et al. Genetic association of vitamin D receptor polymorphisms with autoimmune hepatitis and primary biliary cirrhosis in the Chinese. J Gastroenterol Hepatol. 2005;20:249–255. doi: 10.1111/j.1440-1746.2005.03532.x. [DOI] [PubMed] [Google Scholar]
- 18.Baur K, et al. Combined effect of 25-OH vitamin D plasma levels and genetic vitamin D receptor (NR 1I1) variants on fibrosis progression rate in HCV patients. Liver Int. 2012;32:635–643. doi: 10.1111/j.1478-3231.2011.02674.x. [DOI] [PubMed] [Google Scholar]
- 19.Beilfuss A, et al. Vitamin D counteracts fibrogenic TGF-beta signalling in human hepatic stellate cells both receptor-dependently and independently. Gut. 2015;64:791–799. doi: 10.1136/gutjnl-2014-307024. [DOI] [PubMed] [Google Scholar]
- 20.Tilg H. Cytokines and liver diseases. Can J Gastroenterol. 2001;15:661–668. doi: 10.1155/2001/746736. [DOI] [PubMed] [Google Scholar]
- 21.Zhou WC, Zhang QB, Qiao L. Pathogenesis of liver cirrhosis. World J Gastroenterol. 2014;20:7312–7324. doi: 10.3748/wjg.v20.i23.7312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lemire JM, Archer DC, Beck L, Spiegelberg HL. Immunosuppressive actions of 1,25-dihydroxyvitaminD3: preferential inhibition of Th1 functions. J Nutr. 1995;125:1704S–1708S. doi: 10.1093/jn/125.suppl_6.1704S. [DOI] [PubMed] [Google Scholar]
- 23.Willheim M, et al. Regulatory effects of 1alpha,25-dihydroxyvitamin D3 on the cytokine production of human peripheral blood lymphocytes. J Clin Endocrinol Metab. 1999;84:3739–3744. doi: 10.1210/jcem.84.10.6054. [DOI] [PubMed] [Google Scholar]
- 24.Penna G, Adorini L. 1 Alpha,25-dihydroxyvitamin D3 inhibits differentiation, maturation, activation, and survival of dendritic cells leading to impaired alloreactive T cell activation. J Immunol. 2000;164:2405–2411. doi: 10.4049/jimmunol.164.5.2405. [DOI] [PubMed] [Google Scholar]
- 25.Valdivielso JM, Fernandez E. Vitamin D receptor polymorphisms and diseases. Clin Chim Acta. 2006;371:1–12. doi: 10.1016/j.cca.2006.02.016. [DOI] [PubMed] [Google Scholar]
- 26.Cao Y, Wang X, Cao Z, Cheng X. Association of Vitamin D receptor gene TaqI polymorphisms with tuberculosis susceptibility: a meta-analysis. Int J Clin Exp Med. 2015;8:10187–10203. [PMC free article] [PubMed] [Google Scholar]
- 27.Fan LY, et al. Genetic association of cytokines polymorphisms with autoimmune hepatitis and primary biliary cirrhosis in the Chinese. World J Gastroenterol. 2005;11:2768–2772. doi: 10.3748/wjg.v11.i18.2768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Uygun A, et al. The association of nonalcoholic fatty liver disease with genetic polymorphisms: a multicenter study. Eur J Gastroenterol Hepatol. 2017;29:441–447. doi: 10.1097/MEG.0000000000000813. [DOI] [PubMed] [Google Scholar]
- 29.Joshita S, Umemura T, Tanaka E, Ota M. Genetic Contribution to the Pathogenesis of Primary Biliary Cholangitis. J Immunol Res. 2017;2017:3073504. doi: 10.1155/2017/3073504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Motawi T, Shaker OG, Hussein RM, Houssen M. Polymorphisms of alpha1-antitrypsin and Interleukin-6 genes and the progression of hepatic cirrhosis in patients with a hepatitis C virus infection. Balkan J Med Genet. 2016;19:35–44. doi: 10.1515/bjmg-2016-0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Medrano LM, Jimenez-Sousa MA, Fernandez-Rodriguez A, Resino S. Genetic Polymorphisms Associated with Liver Disease Progression in HIV/HCV-Coinfected Patients. AIDS Rev. 2017;19:3–15. [PubMed] [Google Scholar]
- 32.de Azevedo LA, Matte U, da Silveira TR, Alvares-da-Silva MR. Genetic variants underlying vitamin D metabolism and VDR-TGFbeta-1-SMAD3 interaction may impact on HCV progression: a study based on dbGaP data from the HALT-C study. J Hum Genet. 2017;62:969–977. doi: 10.1038/jhg.2017.75. [DOI] [PubMed] [Google Scholar]
- 33.An L, Wang X, Cederbaum AI. Cytokines in alcoholic liver disease. Arch Toxicol. 2012;86:1337–1348. doi: 10.1007/s00204-012-0814-6. [DOI] [PubMed] [Google Scholar]
- 34.Giron-Gonzalez JA, et al. Implication of inflammation-related cytokines in the natural history of liver cirrhosis. Liver Int.l. 2004;24:437–445. doi: 10.1111/j.1478-3231.2004.0951.x. [DOI] [PubMed] [Google Scholar]
- 35.Goral V, Atayan Y, Kaplan A. The relation between pathogenesis of liver cirrhosis, hepatic encephalopathy and serum cytokine levels: what is the role of tumor necrosis factor alpha? Hepatogastroenterology. 2011;58:943–948. [PubMed] [Google Scholar]
- 36.Kawaratani H, et al. The effect of inflammatory cytokines in alcoholic liver disease. Mediators Inflamm. 2013;2013:495156. doi: 10.1155/2013/495156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li CP, et al. Plasma interleukin-8 levels in patients with post-hepatitic cirrhosis: relationship to severity of liver disease, portal hypertension and hyperdynamic circulation. J Gastroenterol Hepatol. 1996;11:635–640. doi: 10.1111/j.1440-1746.1996.tb00306.x. [DOI] [PubMed] [Google Scholar]
- 38.Alimirah F, Peng X, Murillo G, Mehta RG. Functional significance of vitamin D receptor FokI polymorphism in human breast cancer cells. PloS one. 2011;6:e16024. doi: 10.1371/journal.pone.0016024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Reiberger T, et al. Non-selective betablocker therapy decreases intestinal permeability and serum levels of LBP and IL-6 in patients with cirrhosis. J Hepatol. 2013;58:911–921. doi: 10.1016/j.jhep.2012.12.011. [DOI] [PubMed] [Google Scholar]
- 40.Whitfield GK, et al. Functionally relevant polymorphisms in the human nuclear vitamin D receptor gene. Mol Cell Endocrinol. 2001;177:145–159. doi: 10.1016/S0303-7207(01)00406-3. [DOI] [PubMed] [Google Scholar]
- 41.Elhoseiny SM, Morgan DS, Rabie AM, Bishay ST. Vitamin D Receptor (VDR) Gene Polymorphisms (FokI, BsmI) and their Relation to Vitamin D Status in Pediatrics betaeta Thalassemia Major. Indian J Hematol Blood Transfus. 2016;32:228–238. doi: 10.1007/s12288-015-0552-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jorde R, et al. Tracking of serum 25-hydroxyvitamin D levels during 14 years in a population-based study and during 12 months in an intervention study. Am J Epidemiol. 2010;171:903–908. doi: 10.1093/aje/kwq005. [DOI] [PubMed] [Google Scholar]
- 43.Sonderman JS, Munro HM, Blot WJ, Signorello LB. Reproducibility of serum 25-hydroxyvitamin d and vitamin D-binding protein levels over time in a prospective cohort study of black and white adults. Am J Epidemiol. 2012;176:615–621. doi: 10.1093/aje/kws141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chen K, Cao X, Zheng Y, Xu M, Peng J. Comparative study of the MELD-Na and Child-Turcotte-Pugh scores as short-term prognostic indicators of acute-on-chronic hepatitis B liver failure. Zhonghua Gan Zang Bing Za Zhi. 2014;22:801–805. doi: 10.3760/cma.j.issn.1007-3418.2014.11.001. [DOI] [PubMed] [Google Scholar]
- 45.Guan R, Lui HF. Treatment of hepatitis B in decompensated liver cirrhosis. Int J Hepatol. 2011;2011:918017. doi: 10.4061/2011/918017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Strauss E, Dias Teixeira MC. Quality of life in hepatitis C. Liver Int. 2006;26:755–765. doi: 10.1111/j.1478-3231.2006.01331.x. [DOI] [PubMed] [Google Scholar]
- 47.Gutteling JJ, de Man RA, Busschbach JJ, Darlington AS. Overview of research on health-related quality of life in patients with chronic liver disease. Neth J Med. 2007;65:227–234. [PubMed] [Google Scholar]
- 48.Garcia-Tsao G, Lim JK. Management and treatment of patients with cirrhosis and portal hypertension: recommendations from the Department of Veterans Affairs Hepatitis C Resource Center Program and the National Hepatitis C Program. Am J Gastroenterol. 2009;104:1802–1829. doi: 10.1038/ajg.2009.191. [DOI] [PubMed] [Google Scholar]
- 49.Webb AR, Kline L, Holick MF. Influence of season and latitude on the cutaneous synthesis of vitamin D3: exposure to winter sunlight in Boston and Edmonton will not promote vitamin D3 synthesis in human skin. J Clin Endocrinol Metab. 1988;67:373–378. doi: 10.1210/jcem-67-2-373. [DOI] [PubMed] [Google Scholar]
- 50.Wittnich C, Belanger MP, Askin N, Boscarino C, Wallen WJ. Lower liver transplant success in females: gender differences in metabolic response to global ischemia. Transplant Proc. 2004;36:1485–1488. doi: 10.1016/j.transproceed.2004.05.055. [DOI] [PubMed] [Google Scholar]
- 51.Konstantakis C, Tselekouni P, Kalafateli M, Triantos C. Vitamin D deficiency in patients with liver cirrhosis. Ann Gastroenterol. 2016;29:297–306. doi: 10.20524/aog.2016.0037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Collett, D. Modelling survival data in medical research. (CRC press, 2015).