Abstract
Environmental antibiotic-resistant bacteria (ARB) can be transferred to humans through foods. Fresh produce in particular is an ideal vector due to frequent raw consumption. A major contamination source of fresh produce is irrigation water. We hypothesized that water quality significantly affects loads of ARB and their diversity on fresh produce despite various other contamination sources present under agricultural practice conditions. Chive irrigated from an open-top reservoir or sterile-filtered water (control) was examined. Heterotrophic plate counts (HPC) and ARB were determined for water and chive with emphasis on Escherichia coli and Enterococcus spp. High HPC of freshly planted chive decreased over time and were significantly lower on control- vs. reservoir-irrigated chive at harvest (1.3 log (CFU/g) lower). Ciprofloxacin- and ceftazidime-resistant bacteria were significantly lower on control-irrigated chive at harvest and end of shelf life (up to 1.8 log (CFU/g) lower). Escherichia coli and Enterococcus spp. repeatedly isolated from water and chive proved resistant to up to six or four antibiotic classes (80% or 49% multidrug-resistant, respectively). Microbial source tracking identified E. coli-ST1056 along the irrigation chain and on chive. Whole-genome sequencing revealed that E. coli-ST1056 from both environments were clonal and carried the same transmissible multidrug-resistance plasmid, proving water as source of chive contamination. These findings emphasize the urgent need for guidelines concerning ARB in irrigation water and development of affordable water disinfection technologies to diminish ARB on irrigated produce.
Keywords: irrigation water, fresh produce, antibiotic resistance, E. coli, Enterococcus spp, microbial source tracking
Irrigation water quality impacts on antibiotic-resistant bacteria of chive representing raw consumed fresh produce grown under agricultural practice conditions.
INTRODUCTION
Consumer demand for fresh produce has increased in the past decades, as it is being associated with a healthy lifestyle and offers the advantage of convenient and economic meals with minimal preparation (Hu et al. 2014; Li et al. 2014; Wang et al. 2014b). Fresh produce consumption is actively promoted by public institutions such as the European Commission (EC) through actions like the school fruit scheme encouraging consumption by children or free distribution of produce withdrawn as remains from the market to various public service bodies (EC 2017). Such increased consumption of mostly raw or minimally processed fresh produce including fresh herbs comes with an increase in related foodborne outbreaks (Callejón et al. 2015). Apart from pathogenic bacteria causing such outbreaks and the naturally present harmless microbiota, it is increasingly recognized that raw or minimally processed fresh produce can deliver several antibiotic-resistant bacteria (ARB) to the consumer (Falomir, Gozalbo and Rico 2010; Pesavento et al. 2014; Nüesch-Inderbinen et al. 2015; Araújo et al. 2017). Antibiotic resistances, even if harbored in non-pathogenic bacteria, can potentially be spread through horizontal gene transfer to other species including opportunistic pathogens present in the environment or—upon consumption of ARB-contaminated fresh produce—the gut microbiome (Devirgiliis, Barile and Perozzi 2011) thereby contributing to the pool of antibiotic-resistance genes (ARG) in the human gut (Hu et al. 2013; Thanner, Drissner and Walsh 2016).
Contamination of fresh produce can occur before or after harvest through a variety of sources such as soil, irrigation water, wild or domestic animals, and manure (pre-harvest), or harvesting and processing equipment, dust, rinse water and transport vehicles (post-harvest) (Olaimat and Holley 2012). Of all these probable contamination sources, irrigation water has the potential of spreading localized contamination by directly reaching the edible plant parts, especially when applied through sprinkler irrigation (Fonseca et al. 2011). This is of particular interest as the microbiological quality of irrigation water can vary considerably from potable water over groundwater to various surface waters (Allende and Monaghan 2015; Uyttendaele et al. 2015). Irrigation water quality deserves great attention since the use of low-quality waters such as reclaimed wastewaters potentially contaminated not only with pathogenic bacteria but also with antibiotics, ARB and ARG is unavoidable (Czekalski et al. 2012; Fahrenfeld et al. 2013; Christou et al. 2017). This is of particular concern in parts of the world with limited access to potable water (Gemmell and Schmidt 2012) or where surface water sources such as dedicated canals are used. The presence of ARB in surface waters such as lakes, rivers or irrigation ponds has been highlighted recently (Micallef et al. 2013; Zurfluh et al. 2014a; Blaustein et al. 2015), and in some instances the prevalence of multidrug-resistant (MDR) bacterial strains has been found to be higher in surface waters than in wastewaters (Farkas, Bocoş and Butiuc-Keul 2016). Moreover, ARB and ARG have been detected in drinking water treatment and distribution systems (Schwartz et al. 2003; Xi et al. 2009), and the drinking water treatment process has been pinpointed as potentially increasing bacterial antibiotic resistance (Czekalski et al. 2012; Bai et al. 2015).
A direct link between irrigation water containing ARB and ARG and contaminated fresh produce has been suggested in various studies. Many of these studies have focused on indicator bacteria. Escherichia coli and Enterococcus spp. are species commonly used as indicators of fecal contamination when assessing quality of water but also of foods (Anderson, Whitlock and Harwood 2005; Jay, Loessner and Golden 2005; Pappas et al. 2008), and both species are also used as indicators in antibiotic resistance monitoring of foods (EFSA 2008). Moreover, they are regularly detected on fresh produce (Giraffa 2002; Johnston and Jaykus 2004; Pesavento et al. 2014; Faour-Klingbeil et al. 2016; Gekenidis et al. 2017). Escherichia coli is acknowledged to be a central player in the spread of antibiotic resistance due to its genetic flexibility and adaptability (Szmolka and Nagy 2013), and Enterococcus spp. have long been notorious for harboring and spreading ARG (Franz, Holzapfel and Stiles 1999; Leisibach 2004; Palmer, Kos and Gilmore 2010). To link contaminated water to fresh produce contamination, investigations have been based on isolation and characterization of antibiotic-resistant E. coli (Holvoet et al. 2013). In a very recent study, Araújo and colleagues (2017) investigated the prevalence of antibiotic-resistant E. coli in irrigation water and on vegetables from 16 household farms, and by typing isolates obtained from both sources using repetitive elements PCR (rep-PCR), suggested a link between water and vegetables. Other studies have also combined strain typing with characterization of antibiotic resistance for E. coli source tracking, however, with these methods only a potential link can be determined (Du Plessis, Duvenage and Korsten 2015; Jongman and Korsten 2016a). Concerning antibiotic-resistant Enterococcus spp., some studies have investigated fresh produce (Johnston and Jaykus 2004; Gomes et al. 2008; Pesavento et al. 2014) or potential irrigation water (Goldstein et al. 2014), but studies investigating both sources are scarce and did not establish a link between water and the irrigated produce (Abriouel et al. 2008; Micallef et al. 2013).
From a legal perspective, guidelines including critical values for indicator bacteria such as E. coli and Enterococcus spp. in irrigation water have been established in many countries. In particular, for safe use of wastewater in agriculture, the WHO has described risk assessment and management approaches taking into consideration attributable risks (WHO 2006). However, until now no guidelines addressing ARB in irrigation water exist.
There is an urgent need for in-depth understanding of the role of irrigation water in contamination of fresh produce with ARB, in order to apply appropriate mitigation strategies comprising preventive measures and technological sanitation of irrigation water before usage in the field. Overall, there is a lack of controlled studies proving the link between ARB detected on the produce and irrigation water by tracing them back to their source. The presented greenhouse study pursued two aims: (1) to investigate the impact of irrigation water quality on ARB detected on fresh produce (exemplified by chive) under agricultural practice conditions by describing diversity and antibiotic resistance of total ARB and target ARB E. coli and Enterococcus spp. from water and plants, and (2) to investigate whether selected isolates of ARB from plants originate from the applied irrigation water, by tracing them back through the irrigation chain to the water source. Notably, the cultivation conditions under which chive was grown are commonly used for other leafy greens as well, e.g. rocket salad or lamb's lettuce. To investigate to what extent irrigation water quality influences the diversity of ARB detected on fresh produce, two types of water were used for overhead irrigation of the chive: (a) sterile-filtered water obtained from a three-stage filtration unit and (b) water sourced from an open-top reservoir collecting rain water, greenhouse rooftop run-off water and surface drainage water.
MATERIALS AND METHODS
Bacterial culture conditions
The following media were used for bacterial cultivation: R2A (Sigma-Aldrich, St. Louis, USA) for determination of total heterotrophic plate count (HPC); eosine methylene blue (EMB) agar (Becton Dickinson, Franklin Lakes, NJ, USA) for Enterobacteriaceae; CHROMagar E. coli (CHROMagar, Paris, France) and ready-to-use Brilliance ESBL plates for E. coli; and m-Enterococcus agar (mEA) (Sigma-Aldrich), Compact Dry ETC plates (HyServe, Uffing, Germany) for direct incubation of water filters, and ready-to-use Brilliance VRE plates (Oxoid Ltd., Hampshire, UK) for isolation of Enterococcus spp.
For isolation of ARB, all above-mentioned media apart from ESBL and VRE were supplemented with antibiotics. With the exception of high-concentrated ampicillin chosen to avoid overgrowth of non-target bacteria, antibiotic concentrations were based on epidemiological cutoffs (ECOFFs) defined by the European Committee on Antimicrobial Susceptibility Testing (EUCAST), as recommended by the European Food Safety Authority (EFSA 2012), while minimizing detection of false positives when using broad-spectrum antibiotics. For isolation of total ARB, R2A was supplemented with broad-spectrum antibiotics: combined trimethoprim and sulfamethoxazole (SXT, 4 and 76 mg L–1, respectively) and tetracycline (TE, 8 mg L–1) representing ancient antibiotics (Czekalski et al. 2012), ciprofloxacin (CIP, 1 mg L–1) or ceftazidime (CAZ, 8 mg L–1). Additionally, pimaricin was added (25 mg L–1, ASA Spezialenzyme GmbH, Wolfenbüttel, Germany) to each of the R2A/antibiotics combinations to suppress fungal growth. For isolation of target ARB, EMB agar and CHROMagar were supplemented with either ampicillin (AM, 100 mg L–1), kanamycin (K, 16 mg L–1), ciprofloxacin (CIP, 1 mg L–1) or ceftazidime (CAZ, 8 mg L–1), while mEA and ETC plates were supplemented with either erythromycin (ERY, 4 mg L–1) or ciprofloxacin (CIP, 1 mg L–1).
Total HPC and total ARB were determined by direct plating. For detection of target ARB, samples were enriched (24 h, 37°C) prior to cultivation: Buffered peptone water (BPW, 10.0 g of peptone, 5.0 g of NaCl, 3.5 g of anhydrous Na2HPO4 and 1.5 g of KH2PO4 (Sigma-Aldrich) per 1 liter of water, pH 7.0) was used for enrichment of Enterococcus spp. and EE broth Mossel (Becton Dickinson) for enrichment of Enterobacteriaceae and E. coli. Using a 10 μl loop, the enriched samples were then streaked onto the respective antibiotic-containing agar plates. R2A plates were incubated at room temperature for 72 h; ETC plates, EMB agar and CHROMagar at 37°C for 24 h; ESBL, mEA and VRE at 37°C for 48 h. All media were incubated under aerobic conditions.
Experimental setup and plant growth conditions
The experiment was setup in a greenhouse covering an area of 1000 m2 (10 × 100 m, 6 beds) and equipped with an overhead irrigation system. Water from an open-top reservoir as also applied standardly in practice was used for irrigation. To investigate the influence of irrigation water quality on ARB detected on plants, a second irrigation system (control irrigation) was installed by disconnecting the existing tubing and installing new tubing and sprinklers for control irrigation. To provide clean water for control irrigation, a closed water tank was filled with fresh tap water daily and the tank water was pumped through a three-stage filtration unit (pore sizes 1.2 μm, 0.65 μm and 0.2 μm; Sartorius AG, Goettingen, Germany) to generate sterile water before supplying the sprinkler system. Chive plants (Allium schoenoprasum L.) were grown from untreated seeds by placing about 25 seeds into 4 × 4 cm pots containing substrate compliant with Bio Suisse standards (Bio Suisse 2015). After a germination period of approximately 10 days at 20°C (relative humidity: 75%), the temperature was lowered to 12°C for 7 days (relative humidity: 60%). Thereafter, temperature was further lowered (5–10°C) for another 14 days to increase plant durability. Finally, the seedlings were transplanted to the greenhouse containing agricultural field soil. To minimize plant–soil contact, the beds were overlaid with an organic foil before planting the seedlings. After planting, the chive plants were irrigated overhead at different frequencies depending on their growth phase: three to four times per day during the first three weeks (each irrigation about 2 L m–2), once per day thereafter for two weeks (about 5 liters m–2) and twice per day during the last week before harvest (each about 1 L m–2). The total field output at harvest was 600 kg.
Field sampling and bacterial culture preparation
Irrigation water and plant material were sampled in summer 2016 (July–August). The greenhouse planted with chive was sampled every two weeks during a whole growth period, i.e. from freshly planted seedlings to harvest of marketable plants, resulting in a total of four samplings for analysis of total heterotrophic bacteria as well as total and target ARB. Of note, plant material was collected before running overhead irrigation to collect the water samples.
Irrigation water
At each sampling, irrigation water samples were collected in sterile water sampling bottles (VWR, Radnor, USA) along the complete irrigation chain. For the control-water chain, water was sampled from the municipal tap (tap), the water tank (tank), the three-stage filtration unit (inF) and the corresponding greenhouse sprinklers (spF). For the reservoir-water chain, water was sampled from the tube draining surface waters into the open-top reservoir (drain), the open-top reservoir itself (R) and the greenhouse inlet (inR) and sprinklers (spR) sourced from the open-top reservoir. Notably, reservoir water (R) was pumped through a particle filter (F-600 Gravel Filter; Netafim, Tel Aviv, Israel) before entering the greenhouse (inR). Water samples were transported at approximately 8°C and processed within 10 h. For determination of HPC, serial 10-fold dilutions were plated in duplicate on R2A. Further, depending on water sample clarity largely varying from sterile-filtered water to surface water, 5–500 mL were concentrated through nitrocellulose filters (0.22 μm pore size, EMD Millipore, Billerica, USA) for direct incubation on the surface of antibiotic-containing R2A or ETC plates. Additionally, 300 or 500 mL were filtered depending on water clarity for subsequent enrichment in 5 mL BPW and EE broth. For cultivation of target ARB, the enrichment broths were finally streaked onto the respective antibiotic-containing selective media.
Seeds and seedlings
Chive seeds as well as seedlings before entering the greenhouse were sampled and analyzed. Twenty grams of seeds or seedling leaves were weighed into a stomacher bag containing 100 mL of either BPW or EE broth and homogenized in a Smasher (Biomérieux, Marcy l'Etoile, France) for 3 min. From the resulting BPW supernatant, appropriate volumes and dilutions as determined in pre-experiments were plated on R2A plates with and without antibiotics. Both BPW and EE homogenates were then incubated for enrichment and streaked onto antibiotic-containing selective media as described above.
Plant
For sampling of plant material, the field area was divided into six plots, three plots per treatment (control- or reservoir-irrigation). A sample of at least 120 g plant material was collected per plot by randomly sampling parts of about 50 plants, resulting in three plant samples per treatment. The samples were transported at approximately 8°C and processed within 10 h: of each sample, 20 g were weighed into 100 mL of either BPW or EE broth and processed as described for seeds and seedlings. For analysis of chive at the end of its shelf life, plant material from the last field sample was stored for 6 days at 4°C.
MALDI biotyping
Representative colonies from all media containing antibiotics were identified by MALDI biotyping. Attention was paid to picking the different morphotypes from R2A (up to 10 colonies per sample and antibiotic) and each morphotype from the selective media at least once (minimum 3 colonies per sample and antibiotic). MALDI biotyping was performed by direct smearing as described previously (Gekenidis et al. 2014) using a microflex LT MALDI-TOF mass spectrometer (Bruker Daltonics, Bremen, Germany) and the associated MALDI biotyper RTC Software (Version 3.1).
Antibiotic susceptibility tests
Escherichia coli and Enterococcus spp. were screened for antibiotic resistance by disk diffusion assays against 32 and 11 clinically relevant antibiotics, respectively. Each bacterial strain was subcultured on Columbia agar with 5% sheep blood (BioMérieux, Marcy l'Etoile, France) at 37°C and 7.5% CO2. Disk diffusion assays were performed according to the European Committee of Antimicrobial Susceptibility Testing (EUCAST 2012): (1) bacterial suspensions with a turbidity corresponding to 0.5 McFarland were produced in saline (9 g L–1) and streaked on Mueller Hinton E (MHE) agar (Becton Dickinson), (2) antibiotic disks were applied (i2a, Montpellier, France) and (3) the plates were incubated at 35°C for 18 h ± 2 h or 24 h (E. coli or Enterococcus spp., respectively). Special attention was paid to meeting the 3 × 15 min rule, i.e. none of the three steps should exceed 15 min. Finally, the inhibition zones were measured with a Sirscan instrument (i2a) (Hombach, Zbinden and Böttger 2013) and manually corrected on-screen whenever needed. For determination of antibiotic susceptibility, EUCAST's epidemiological cutoff (ECOFF) values were used based on EFSA recommendations for epidemiological antibiotic resistance screening (EFSA 2012). For antibiotics with no defined ECOFF value (cefpodoxime and fosfomycin for E. coli), EUCAST's clinical breakpoints were used. Where EUCAST guidelines did not define any value, breakpoints from the Clinical and Laboratory Standards Institute (CLSI) were applied (i.e. for colistin, minocycline, kanamycin, sulfonamide, tetracycline, temocillin and cefalotin for E. coli; gentamicin high concentration, erythromycin, tetracycline and chloramphenicol for Enterococcus spp.) (CLSI 2016). Species with intrinsic resistances as defined by EUCAST expert rules were considered resistant (Enterococcus faecalis and Enterococcus faecium to erythromycin; Enterococcus gallinarum and Enterococcus casseliflavus to erythromycin and vancomycin) (Leclercq et al. 2013).
Phylogenetic groups (PG)
Escherichia coli phylogenetic groups (PG) were determined as described by Clermont and colleagues (2013) by quadruplex PCR amplification of three genes (arpA, chuA and yjaA) and a DNA fragment (TspE4.C2) using custom-synthesized primers (Microsynth, Balgach, Switzerland) and a DreamTaq hot start PCR master mix (Thermo Fisher Scientific, Waltham, USA). PCR conditions were as described by Clermont and colleagues (2013). Bands were visualized with GelRed (Biotium Inc., Fremont, USA) on a TBE gel (2% agarose, 35 min, 100 V), and strains with ambiguous band patterns were subjected to confirmatory C- or E-PCR.
Multilocus sequence typing (MLST)
Sequence types (ST) of MDR E. coli were determined by amplifying and sequencing seven housekeeping gene fragments (adk, fumC, gyrB, icd, mdh, purA and recA) as described by Wirth and colleagues (2006). Briefly, custom-synthesized primers (Microsynth) were used for amplification by Phusion high-fidelity DNA polymerase (NEB, Ipswich, USA). PCR products were verified on an agarose gel and purified with a PCR clean-up kit (Macherey-Nagel, Düren, Germany). The purified amplicons were Sanger sequenced (Microsynth), and alleles and ST were determined using the MLST tool of BioNumerics 7.5 (Applied Maths NV, Keistraat, Belgium) and the E. coli MLST database (Warwick Medical School 2017).
Whole-genome sequencing and bioinformatics
Genomic DNA was extracted from two MDR E. coli strains of same sequence type, one isolated from drain water and one from irrigated chive plants, using the commercial GenElute Bacterial Genomic DNA Kit (Sigma-Aldrich) according to the manufacturer's recommendations. Genomic DNA was sequenced on a Pacific Biosciences RSII instrument (20-kb insert library, P6/C4 chemistry, 360 min movie) at the Functional Genomics Center Zurich. The genomes were assembled de novo using the Hierarchical Genome Assembly Process (HGAP) (Chin et al. 2013) from SMRT Analysis run through the online platform SUSHI (Hatakeyama et al. 2016) with default settings. The resulting contigs (two per strain) were circularized and trimmed using AMOS and minimus2 to get the complete circular chromosome or plasmid (Schatz 2006; Sommer et al. 2007) and subsequently polished by remapping raw reads and creating consensus sequences using pbalign and Quiver (Huguet-Tapia et al. 2016). The generated fasta files, each containing two contigs, were submitted to Basic Local Alignment Search Tool (BLAST). The closest hits for each contig (one chromosome and one plasmid, respectively) were combined to generate a reference sequence (Ecoli_1943_pH2291) for subsequent usage in CSI Phylogeny 1.4, a freely available online tool for inferring phylogeny from single nucleotide polymorphisms (SNPs) developed by the Center for Genomic Epidemiology (CGE) of the Technical University of Denmark (DTU) (Kaas et al. 2014). Briefly, CSI Phylogeny is calling SNPs, then filtering them to remove low-quality SNPs, validating the sites, and finally inferring a phylogeny using the concatenated SNP alignment. Additionally to phylogenetic analysis of the two strains, the CGE online tools MLST 1.8, ResFinder 3.0, PlasmidFinder 1.3 and pMLST 1.4 were used to confirm sequence type and identify antibiotic resistance determinants and plasmid replicons (Larsen et al. 2012; Zankari et al. 2012; Carattoli et al. 2014). All CGE tools were used with default settings (CSI Phylogeny: SNP pruning minimum distance 10 bp, minimum SNP quality 30; ResFinder: minimum 90% ID, minimum 60% length; PlasmidFinder: minimum 95% ID, minimum 60% length). Finally, the online service for Rapid Annotation using Subsystem Technology (RAST) was used to elucidate the potential of the plasmid to be transmitted to other bacteria (Aziz et al. 2008). The PacBio raw reads (ERR2535305, ERR2535306) and assembled nucleotide sequences (ERZ535013, ERZ535014) were submitted to the European Nucleotide Archive under BioProject PRJEB26426.
Biolog
Additionally to whole-genome sequencing, the two MDR E. coli strains were phenotypically characterized using Biolog Phenotype Microarrays in duplicate (Biolog Inc., Hayward, USA). Two microarrays for carbon sources (PM-1 and PM-2A), one for nitrogen sources supplemented with sodium succinate/ferric citrate as a carbon source (2 M/200 μM, PM-3B), and one for osmotic stress to simulate dry conditions of the phyllosphere as opposed to the water environment (PM-9) were selected. All microarray plates were prepared as described previously for E. coli (Mackie et al. 2014) and after incubation for 48 h at 37°C, the data were evaluated using the opm package in R (Vaas et al. 2013; R Core Team 2017).
Statistical analysis
For changes in bacterial numbers along irrigation chains or over time, ANOVA was performed on log-transformed data (Hirano et al. 1982) using GraphPad Prism 6 (GraphPad Software, La Jolla, USA) after replacing values below the limit of detection with the limit of detection of the measurement (Lorimer and Kiermeier 2007). Additionally, Tukey's multiple comparison test was applied. To compare the effect of the two irrigation regimes on bacterial numbers, Student's t test (unpaired, two-tailed, homoscedastic) was applied to log-transformed data. Significant differences are reported (P < 0.05).
RESULTS
Greenhouse climatic conditions
To detect whether growth conditions were comparable for all plants, climatic conditions in two distant parts of the greenhouse were recorded (one for control- and one for reservoir-irrigated plants) using data loggers measuring temperature and relative humidity (RH) at 15 min intervals. Overall, temperature and RH were virtually the same during the complete sampling period, as shown by overall maximal, minimal and average values (Fig. S1, Supporting Information). Development over time showed that daily average temperature and RH values were equal, although one sector displayed overall higher maximal temperatures and sometimes lower maximal RH values. Of note, samplings were conducted in the morning, long before the daily maximal temperature was reached (Fig. S1 A, Supporting Information).
Total HPC along the two irrigation water chains and on chive
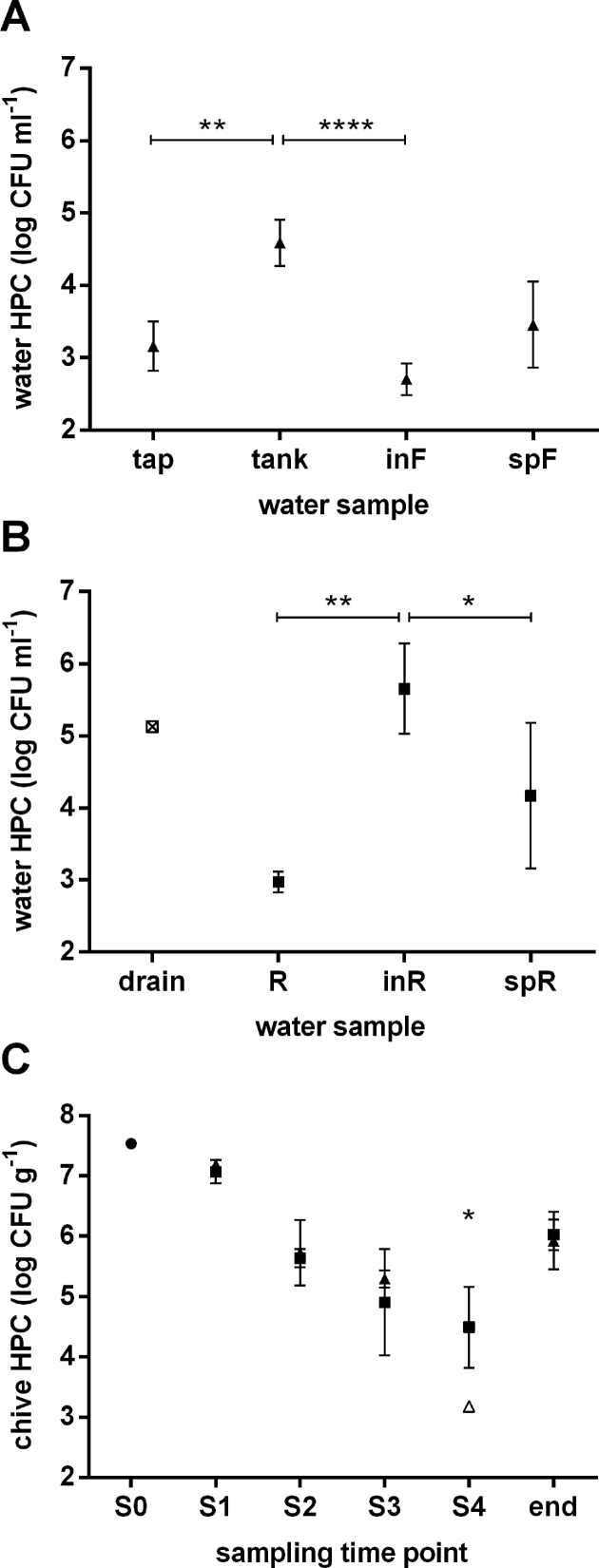
Total HPC was determined for water from both irrigation systems each sampled at four sites, from water source to greenhouse sprinklers. Mean HPC values of the four sampling time points are displayed in Fig. 1 for control-water and reservoir-water system (Fig. 1A and B, respectively). Of note, the first site of the reservoir-water system (drain, Fig. 1B) could be sampled only once (S1), since the tube draining water into the open-top reservoir was submerged in water at the other three sampling occasions. Overall, total HPC varied greatly along both irrigation chains with average values between 2.4 and 6.0 log CFU mL–1 (Fig. 1). At the beginning of the control-water chain (Fig. 1A), the sampled tap water displayed low average HPC (3.2 log CFU mL–1) with small deviations between sampling time points. Numbers in water from the tank were significantly increased (4.6 log CFU mL–1, P < 0.01) as compared to tap water and very constant over the four sampling time points. Filtration resulted in a significant reduction of HPC to 2.7 log CFU mL–1 in filtered water compared to tank water (P < 0.0001), reaching levels below average HPC of tap water. Upon entering the corresponding greenhouse sprinkler system, a tendency towards increased HPC as compared to filtered water was observed (3.5 log CFU mL–1, not significant (ns)). For the first sampling site of the reservoir-water chain (drain, Fig. 1B), HPC was around 5.1 log CFU mL–1. Water from the open-top reservoir displayed an average HPC of 3.0 log CFU mL–1 with very small variation throughout all samplings (Fig. 1B). Counts were significantly increased in the water arriving at the greenhouse inlet as compared to open-top reservoir water (5.7 log CFU mL–1, P = 0.001). Interestingly, HPC values from water recovered from the respective greenhouse sprinklers were reduced as compared to inlet water (4.2 log CFU mL–1, P < 0.05), displaying, however, a large variability between samplings.
Figure 1.
Total HPC for water samples from the control-water chain (triangles; A), the reservoir-water chain (squares; B), and from seedlings (dot) as well as greenhouse-grown control- and reservoir-irrigated chive plants (triangles and squares, respectively; C). Square with cross: value from a single sampling; open symbol: sample mean containing samples below the limit of detection. Tap, tap water; tank, tank water; inF, filter-sourced inlet water; spF, filter-sourced sprinkler water; drain, drain water; R, open-top reservoir water; inR, reservoir-sourced inlet water; spR, reservoir-sourced sprinkler water; S0, seedling sampling. S1–S4, sampling 1 to sampling 4; end, end of shelf life. Results are displayed as mean values (n = 4 and n = 3 for water and chive, respectively) and error bars show standard deviations. *P < 0.05; **P < 0.01; ****P < 0.0001.
For control- and reservoir-irrigated plants, total HPC was determined in triplicate per treatment and sampling time point (Fig. 1C). HPC for seedlings before entering the greenhouse was around 7.5 log CFU g–1 (S0, Fig. 1C). After planting, HPC values were comparable for chive plants from both treatments, from sampling 1 to sampling 3 (S1–S3, Fig. 1C). As the plants grew during this period, counts decreased significantly from approximately 7.1 log CFU g–1 (S1) to an average of 5.3 and 4.9 log CFU g–1 (S3) for control- and reservoir-irrigated plants, respectively (P < 0.001 and P < 0.01). Only in the last sampling (S4), in which plants were harvested to be packaged and shipped to retailers, did the HPC of control-irrigated plants drop significantly below that of reservoir-irrigated plants (3.2 and 4.5 log CFU g–1, respectively, P < 0.05; S4, Fig. 1C). Notably, HPC values of harvested chive at the end of shelf life (6 days) were significantly increased as compared to the last field sample (P < 0.0001 and P < 0.05 for control- and reservoir-irrigated plants, respectively), lying again around 6.0 log CFU g–1 for both treatments (end, Fig. 1C).
Counts of ARB on chive plants
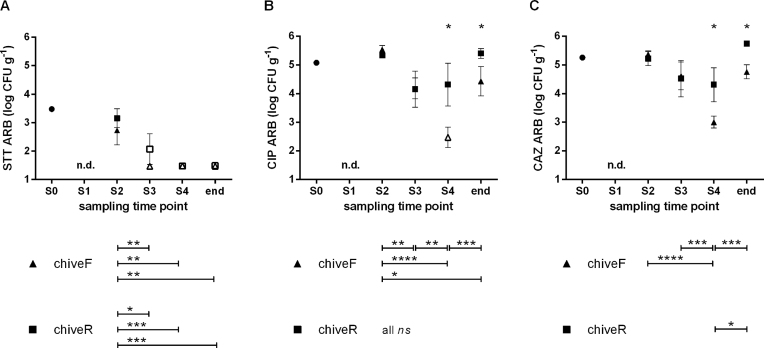
Bacteria resistant to antibiotic combination sulfamethoxazole-trimethoprim-tetracycline (STT) displayed the lowest counts (Fig. 2A). Starting at 3.5 log CFU g–1 on seedlings, counts were still around 3.0 log CFU g–1 in the second sampling (S2) and decreased to undetectable levels thereafter (P < 0.01 and P < 0.001 for control- and reservoir-irrigated chive, respectively). Control-chive had lower loads of STT-resistant bacteria as compared to reservoir-chive (ns). For CIP-resistant bacteria compared to STT-resistant ones, overall higher counts were detected. Starting at 5.1 log CFU g–1 on seedlings, counts were around 5.4 log CFU g–1 in sampling 2 and decreased to 4.2 log CFU g–1 in sampling 3 for both control- and reservoir-irrigated plants (S2 and S3, Fig. 2B). In the last sampling (S4), CIP-resistant bacteria had further decreased by 1.7 log units on control-chive, while counts on reservoir-chive had slightly increased (0.2 log units). CIP-resistant bacteria were thus significantly less abundant on control- than on reservoir-chive (2.5 vs. 4.4 log CFU g–1; P < 0.05). A similar trend was observed for CAZ-resistant bacteria, where numbers were around 5.3 log CFU g–1 on seedlings and were in the same range for plants from both treatments in samplings 2 and 3 (around 5.3 log CFU g–1 and 4.6 log CFU g–1, respectively; Fig. 2C). As observed for ciprofloxacin, CAZ-resistant bacteria from sampling 4 were on average significantly lower for control- than for reservoir-chive (3.0 vs. 4.3 log CFU g–1; P < 0.05). Overall, both CIP- and CAZ-resistant bacteria decreased significantly over the sampling period for control- but not for reservoir-chive (Fig. 2B and C). Finally, on chive at the end of shelf life (end, Fig. 2B and C), CIP- as well as CAZ-resistant bacteria were still significantly reduced on control- as compared to reservoir-chive (1.0 log unit lower; P < 0.05).
Figure 2.
ARB determined for seedlings (dot) and for greenhouse-grown control- and reservoir-irrigated chive (triangles and squares, respectively). ARB isolated from R2A containing (A) sulfamethoxazole-trimethoprim-tetracycline (STT), (B) ciprofloxacin (CIP) or (C) ceftazidime (CAZ). S0, seedling sampling. S1–S4, sampling 1 to sampling 4; end, end of shelf life; chiveF, control-irrigated chive; chiveR, reservoir-irrigated chive; n.d., not determined; ns, not significant; *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001. Results are displayed as mean values (n = 3) and error bars show standard deviations. Open symbols mark sample mean containing samples below the limit of detection.
Assignment of ARB to species in the two systems
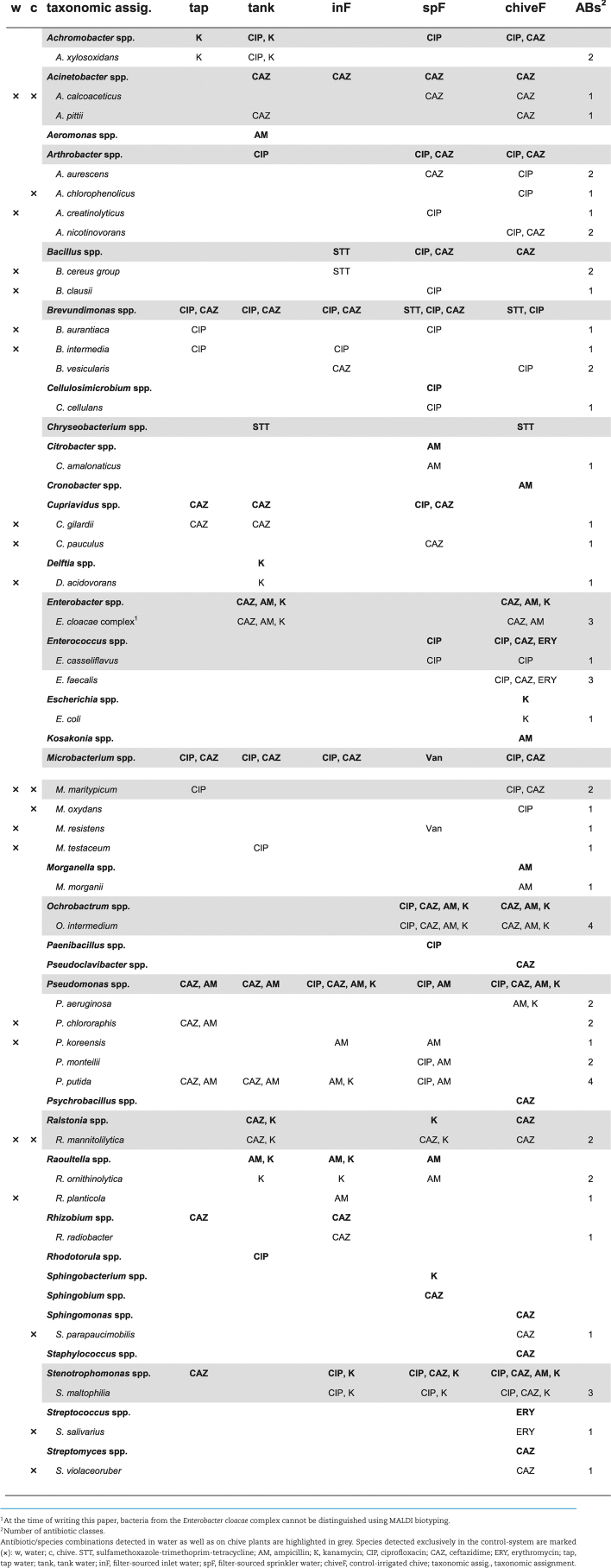
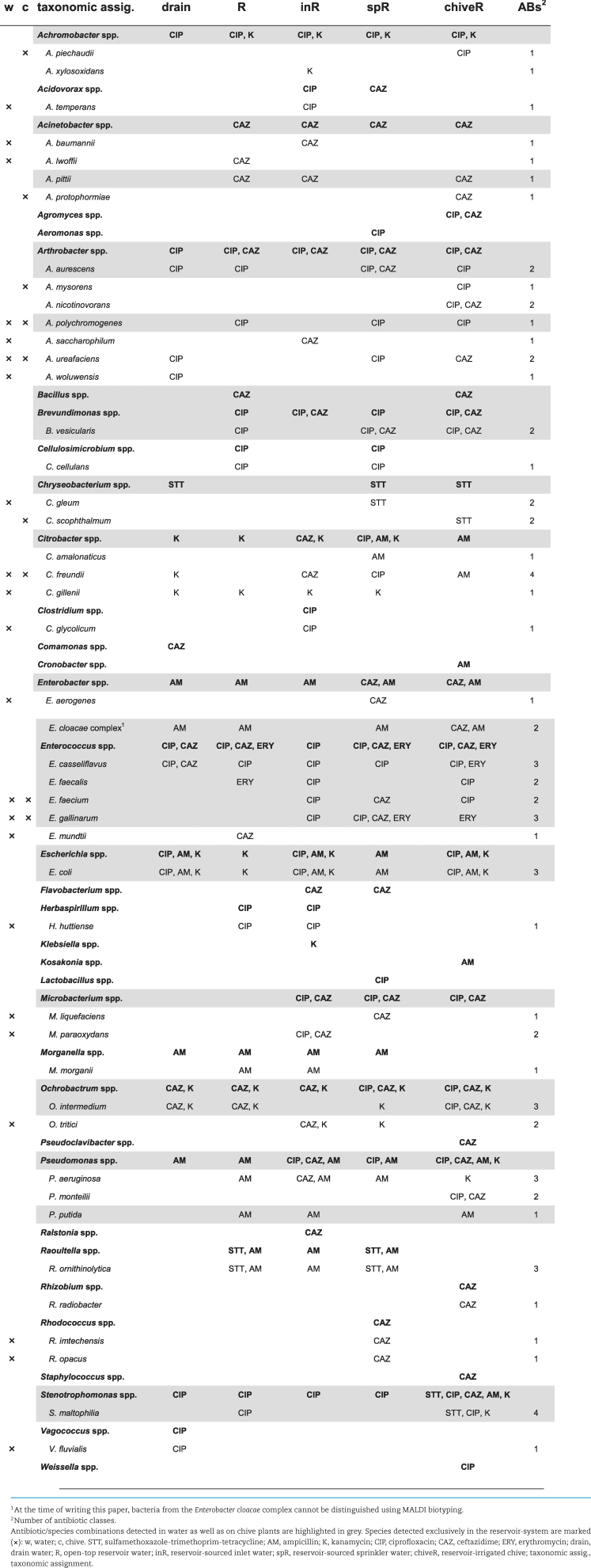
For qualitative comparison of ARB along the two irrigation chains as well as on the respective chive plants, total ARB and target ARB (Enterobacteriaceae, E. coli and Enterococcus spp.) were identified (Tables 1 and 2 for control- and reservoir-irrigation, respectively).
Table 1.
Total and target ARB from control-water chain and irrigated chive plants.
Table 2.
Total and target ARB from reservoir-water chain and irrigated chive plants.
Overall, 40 and 45 different species of ARB were identified by MALDI biotyping in the control- and reservoir-system, respectively. From the 40 control-system species, half (20 species) were detected in water exclusively, 10 species on chive plants exclusively, and 10 species both in water and on chive plants (Table 1). From the 45 reservoir-system species, again half (22 species) originated exclusively from water, 7 species from chive plants exclusively and 16 species were detected in both water and on chive plants (Table 2). Among the species recovered from both water and chive—representing interesting candidates for microbial source tracking—were various Enterococcus spp. (one or four species from control- or reservoir-system, respectively) and E. coli (reservoir-system only). However, while antibiotic-resistant Enterococcus spp. were already isolated from chive seeds and seedlings, E. coli were not detected before the first irrigation event (data not shown). Regarding species unique to one of the two systems, 13 or 17 of the detected species from water and 5 or 4 of the detected species from chive plants were unique to the control- or the reservoir-system, respectively. Of the species detected both in water and on chive plants, 3 or 5 species were unique to the control- or the reservoir-system, respectively, with E. faecium and E. gallinarum among the five reservoir-species. In terms of antibiotic resistance, 5 or 8 species from the control- or reservoir-system, respectively, were isolated on at least three different antibiotics along the chain (two counts for STT; Tables 1 and 2). These included species from our target ARB, namely E. faecalis for the control-system and E. casseliflavus, E. gallinarum and E. coli for the reservoir-system. Interestingly, for the ones from the reservoir-system the same antibiotic/species combination was detected both in water and on chive (grey in Table 2), while E. faecalis from the control-system was isolated solely from chive. Notably, no ESBL E. coli or VRE Enterococcus spp. were detected at any time.
Antibiotic resistance profiling of target ARB
For selected antibiotic-resistant E. coli and Enterococcus spp., resistance to clinically relevant antibiotics was determined in disk diffusion assays. Of note, for Enterococcus spp. the focus was on clinically relevant species (E. faecalis, E. faecium, E. casseliflavus and E. gallinarum) (Leclercq et al. 2013).
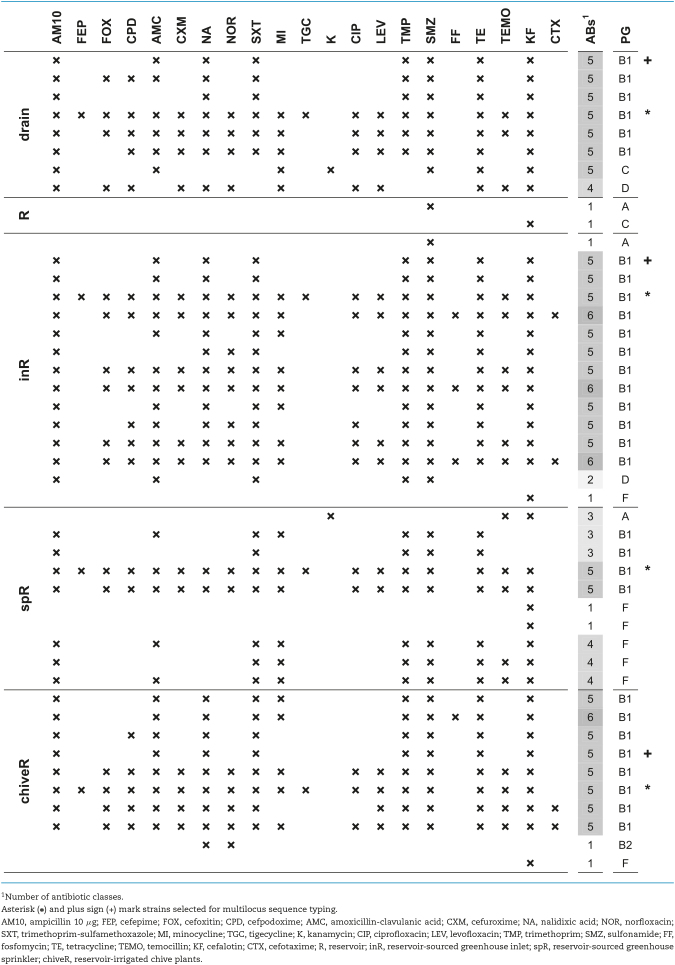
In the control-system, E. coli was isolated once from chive (Table 1) and displayed resistance to kanamycin only (data not shown). In contrast, E. coli was isolated repeatedly from water of the reservoir-system along the whole irrigation chain as well as on chive (Table 2) and displayed resistances to up to six of the seven screened antibiotic classes (Table 3). Of the 45 investigated isolates, 36 (80%) were MDR, that is, resistant to 3 or more different antibiotic classes. Most resistances were attributed to the β-lactam antibiotics ampicillin and cefalotin (36 and 39 strains or 80 and 87%, respectively); the sulfonamide antibiotic sulfamethoxazole (37 strains, 82%) and trimethoprim (34 strains, 76%) or their combination (34 strains); and tetracycline (35 strains, 78%). Resistance to the quinolone antibiotic nalidixic acid was detected in two third of the isolates (30 strains, 67%) while resistance to fluoroquinolones was less frequent (norfloxacin: 19 strains, 42%; ciprofloxacin and levofloxacin: 16 strains each, 36%). No resistance was detected to piperacillin-tazobactam, ceftazidime, ceftriaxone, meropenem, ertapenem, imipenem, tobramycin, gentamicin, amikacin, colistin and nitrofurantoin.
Table 3.
Antibiotic resistance of ARB E. coli determined in disk diffusion assays.
In contrast to E. coli detected only after planting and irrigation, Enterococcus spp. were already detected on chive seeds and seedlings before entering the greenhouse (E. faecium and E. mundtii on seeds; Enterococcus hirae and Enterococcus moraviensis on seedlings; data not shown) and displayed resistance to one or two antibiotics (seeds and seedlings, Table 4). After planting, E. casseliflavus and E. faecalis were isolated from the control-system (Table 1): while E. casseliflavus were all resistant to norfloxacin, vancomycin and erythromycin, E. faecalis were only erythromycin-resistant (chiveF, Table 4). A higher species diversity was observed in the reservoir-system including additionally E. faecium, E. gallinarum and Enterococcus mundtii (Table 2). Enterococcus casseliflavus from drain water, reservoir-water and reservoir-irrigated chive were resistant to norfloxacin, vancomycin and erythromycin (Table 4). Enterococcus faecalis from reservoir water were resistant to erythromycin, streptomycin, tetracycline and chloramphenicol, whereas E. faecalis from reservoir-irrigated chive showed resistance to erythromycin only. Enterococcus faecium were resistant to up to three antibiotics including one ampicillin-resistant strain, and E. gallinarum were resistant to vancomycin, erythromycin, streptomycin and tetracycline (Table 4). Of the 39 investigated strains, 19 (49%) were MDR. Resistance to erythromycin was the most frequent (all 39 tested isolates), followed by norfloxacin (22 strains, 56%) and vancomycin (16 strains, 41%). Of note, all vancomycin-resistant strains from this study are known to carry intrinsic resistance and are therefore not considered VRE strains (Leclercq et al. 2013). Streptomycin and tetracycline resistances were observed in 4 strains (10%), resistances to chloramphenicol and ampicillin were rare (2 and 1 strains or 5 and 3%, respectively), and no resistance was observed to gentamicin, linezolid or tigecycline.
Table 4.
Antibiotic resistance of ARB Enterococcus spp. determined in disk diffusion assays.
Tracing back MDR E. coli
Phylogenetic groups and sequence type
Escherichia coli was selected to investigate whether irrigation water constituted a source of MDR strains detected on the irrigated chive plants. To identify potential candidates for microbial source tracking, PG of 185 E. coli strains obtained from water or chive were determined. Strains belonging to groups A, B1, B2, C, D, E and F were detected (Clermont et al. 2013). The most frequent group was B1 (109 strains, 59%) followed by C (27 strains, 15%) and F (21 strains, 11%), while the remaining groups A, B2, D and E were represented by 3 to 5% of the isolates. PG of isolates from the antibiotic resistance profiling are shown in Table 3.
After determining PG, two sets of E. coli strains could be identified, each containing strains of same PG (group B1) and antibiotic resistance profiles (each set marked with asterisk or plus sign in Table 3, respectively). Within each set, strains originated from different water samples along the reservoir-irrigation chain as well as the respective chive plants. Multi-locus sequence typing (MLST) revealed that all strains—from drain water over greenhouse inlet and sprinkler water to chive plant on the field—belonged to the same sequence type (ST1056).
Whole-genome sequencing
To prove strain identity indicated by MLST, the full genomes of the first and the last E. coli-ST1056 strain of the chain (drain water and chive plant) were sequenced. The number of obtained reads was 95′649 and 109′630 for the strain from water and chive, respectively, with a mean read length of 15′863 and 17′175 bp. Genome assembly using HGAP resulted in two contigs for each strain. A BLAST search revealed that for both strains the respective larger contig's (4.725 Mbp) closest match was the chromosome of E. coli strain 1943 (GenBank number CP023359.1), and the respective smaller contig's (116.5 kbp) closest match was E. coli plasmid pH2291-144 (GenBank number KJ484628.1). These two sequences combined (reference sequence Ecoli_1943_pH2291) were used for SNP-based phylogenetic analysis.
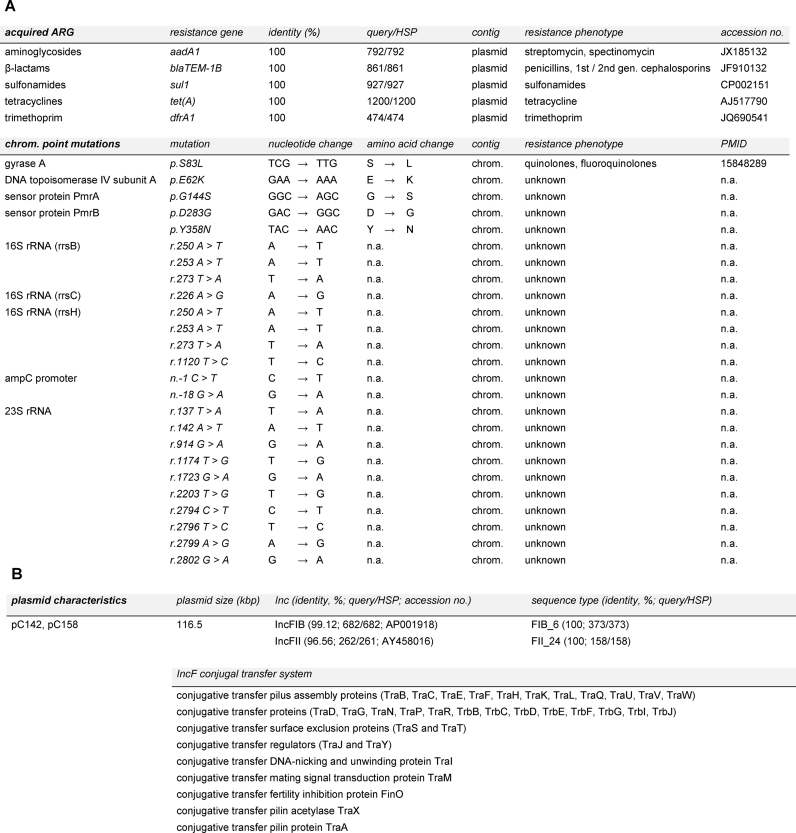
The sequence type of both strains was confirmed as ST1056 using the online tool MLST 1.8 (Larsen et al. 2012). The assembled sequences were then further analyzed using ResFinder 3.0 (known and unknown chromosomal point mutations and acquired ARG) as well as PlasmidFinder 1.3 and pMLST 1.4 (Zankari et al. 2012; Carattoli et al. 2014). Results generated by all tools were identical for the two strains. ResFinder results are shown in Fig. 3A. Acquired ARG were detected for aminoglycosides, β-lactams, sulfonamides, tetracyclines and trimethoprim on the contig matching the known plasmid pH2291-144 (Fig. 3A). Notably, all five genes were identical to the query sequence and covered its complete length (100% identity, query-to-HSP ratio of 1). A known chromosomal point mutation predicted to confer quinolone resistance was detected in both strains. Additionally, all detected unknown chromosomal point mutations were identical for both strains (Fig. 3A). Using PlasmidFinder, IncFIB and IncFII were detected in both strains (99.12% identity and 96.56%, query-to-HSP ratio 682/682 and 262/261, respectively; accession number AP001918 and AY458016; Fig. 3B). The replicon sequence type was identified as F24:A–:B6 using the pMLST tool (100% identity, query-to-HSP ratio of 1; Fig. 3B). Most relevantly, a complete and functional IncF plasmid conjugal transfer system consisting of 33 elements was identified using RAST on the plasmids of both strains (Fig. 3B).
Figure 3.
(A) Acquired antibiotic-resistance genes and chromosomal point mutation identified in drain water and chive E. coli ST1056 using CSI Phylogeny. Note that all results were identical for both strains. Identity: % identical bases between query sequence and E. coli ST1056 sequence; query/HSP: query sequence length compared to alignment length; contig: location of detected resistance gene or chromosomal point mutation; PMID: PubMed ID; n.a., not applicable; chrom., chromosome. (B) Plasmid characteristics determined by PlasmidFinder, pMLST and RAST. pC142 and pC158 are the plasmids from drain and chive isolate, respectively.
Finally, using Ecoli_1943_pH2291 as a reference and the two assembled genomes, a phylogenetic tree and a SNP pairwise comparison table was generated. The SNP distance between drain water and chive isolate was eight, whereas the SNP distance to the reference was 7173 and 7171, respectively. Notably, the reference used was the closest one available based on BLAST results.
Phenotypic comparison
Escherichia coli-ST1056 from drain water and chive were tested on Biolog phenotype microarrays for utilization of carbon sources, nitrogen sources and osmotic stress. In total, 190 carbon sources, 95 nitrogen sources and 96 osmotic stress conditions were tested. Evaluation of end-points after incubation for 48 h revealed that the two strains were phenotypically identical. The only exception was D-allose, a sugar which could be utilized by E. coli-ST1056 from drain water, but not by the strain from chive (data not shown). Genome analysis revealed a frameshift mutation between the two strains in the gene encoding the periplasmic sugar binding protein of a ribose ABC-type transport system. Since D-allose can bind to ribose-binding proteins (Kim, Song and Park 1997), such a frameshift might cause the observed difference in D-allose utilization.
DISCUSSION
Irrigation water has been described as contamination source in agricultural systems (Olaimat and Holley 2012; Blaustein et al. 2015), and the microbiological quality of water used for fresh produce irrigation can vary considerably from tap water to various types of surface water such as rivers or lakes (Steele and Odumeru 2004). In addition to collecting various waters, rivers and lakes are prone to contamination through human activities or domestic and wild animals, and these pollutants can then be transferred to irrigated plants (Fonseca et al. 2011). Nevertheless, studies conducted under practice conditions are very scarce and mostly focus on human pathogens while testing artificially contaminated water (Erickson et al. 2010; Fonseca et al. 2011; Allende and Monaghan 2015). A very recent study on antibiotic-resistant E. coli suggested cross-contamination between water and vegetables but did not conclusively prove the link (Araújo et al. 2017). The present greenhouse study therefore described the population of ARB naturally occurring in irrigation water and chive under practice conditions and determined adequate ARB to demonstrate the potential of irrigation water as contamination source of fresh produce with ARB. Notably, while chive will not be consumed in such large quantities as other fresh produce such as lettuce it is a common ingredient in ready-made salads which can lead to a widespread distribution of the ARB.
From generic to ARB
Total HPC on chive continuously decreased over the growth period, as described previously in greenhouse-grown herbs (Gekenidis et al. 2017). A significant difference in total HPC, however, between control- and reservoir-chive was observed only at harvest (S4, Fig. 1C). Reasons for not detecting a significant difference earlier might be (a) greater soil proximity and thereby soil contamination of younger plants irrespective of irrigation regime, and (b) lower irrigation frequency of younger plants as compared to intensive irrigation one week before harvest—to reduce damage caused by Thrips tabaci (Poulsen 1989; Schuch, Redak and Bethke 1998)—both factors facilitating detection of an irrigation effect on older, that is, harvestable plants. It is worth noting that no significant difference was detected in total organic carbon, dissolved organic carbon and total nitrogen in sprinkler water of the two systems (data not shown). At the end of shelf life, total HPC on control-chive had risen to levels comparable to those of reservoir-chive. In contrast, numbers of bacteria resistant to ciprofloxacin and ceftazidime were significantly lower on control- compared to reservoir-chive at both harvest and end of shelf life (Fig. 2B and C). Thus, no conclusions could be drawn from total HPC on numbers of ARB, since ARB did not increase proportionally to total HPC on control-chive during storage.
Species diversity of ARB and resistance profiles
Comparing control- and reservoir-system in terms of species richness of ARB, the reservoir-system overall displayed only a slightly higher diversity (45 vs. 40 species), although the irrigation waters used were expected to differ greatly. The difference in species diversity of ARB was just as little pronounced on the irrigated plants: Merely three more species of ARB were detected on reservoir-chive (23 vs. 20 species). This is most probably due to numerous other contamination sources to which field-grown fresh produce is exposed beside irrigation water such as soil (Olaimat and Holley 2012) and which were common to both control- and reservoir-chive. Such might be for instance typical soil bacteria like Achromobacter spp., Arthrobacter spp., or Streptomyces spp. (Ma et al. 2011). Since no analogous studies describing the diversity of ARB along a complete irrigation chain from water source to irrigated plants seem to exist, direct comparisons to existing data cannot be drawn. In terms of ARB described to occur in different kinds of water and the environment, many of the species detected in our study have been described before: Achromobacter spp., Acinetobacter spp., Citrobacter freundii, Enterobacter cloacae, E. coli, Flavobacterium spp. and Pseudomonas spp. in water (McKeon, Calabrese and Bissonnette 1995; Messi, Guerrieri and Bondi 2005), or Acinetobacter spp., Enterobacteriaceae, Enterococcus spp., E. coli and Streptomyces spp. in different environments, including soil (Kümmerer 2004).
Potential candidates for bacterial tracing from plant to water source (10 and 16 in control- and reservoir-system, respectively) included both E. coli and Enterococcus spp. However, in the control-system only E. casseliflavus was isolated both from water and chive on ciprofloxacin (Table 1), whereas in the reservoir-system E. casseliflavus, E. faecalis, E. faecium, E. gallinarum, and E. coli were isolated both from water and chive and on all antibiotics tested (exception: no E. coli detected on ceftazidime; Table 2).
Enterococcus spp.
In disk diffusion assays of Enterococcus spp., norfloxacin and erythromycin were the most frequent resistances (incl. known intrinsic resistance to erythromycin (Leclercq et al. 2013)). Among chive isolates these two resistances were the only ones detected, apart from intrinsic vancomycin-resistance and one ampicillin-resistance (Table 4)—in good agreement with previous studies on Enterococcus spp. from fresh produce, detecting frequent resistance to erythromycin and fluoroquinolones (Johnston and Jaykus 2004; Abriouel et al. 2008). Among water isolates, a few displayed additional resistances to high-level streptomycin, tetracycline and/or chloramphenicol, which have equally been described previously for waterborne Enterococcus spp. (Abriouel et al. 2008). In terms of antibiotic/species combinations, norfloxacin- and/or erythromycin-resistant E. faecium were detected on reservoir-chive and on seeds. An ampicillin-resistant E. faecium was isolated from reservoir-chive (chiveR; Table 4), however, this combination was not detected in reservoir-water (drain, R, and spR; Table 4) to suggest water as contamination source. Enterococcus faecalis on the other hand was isolated in the control-system but only from chive, and E. faecalis from reservoir-chive strongly differed in antibiotic resistance profiles compared to E. faecalis from reservoir-water. Only E. casseliflavus resistant to vancomycin, norfloxacin and erythromycin might have originated from irrigation water (drain and reservoir water, Table 4).
E. coli
In E. coli antibiotic resistance was observed towards up to six of seven antibiotic classes with a high proportion of MDR strains (80%, Table 3). Such high proportions of MDR in environmental E. coli have been described previously (Marinescu et al. 2015). The resistances frequently detected in our study towards β-lactams (especially ampicillin and cefalotin), sulfamethoxazole, trimethoprim and tetracycline have been described in previous studies for E. coli from different waters directly or indirectly related to wastewater treatment plant effluents (Hu et al. 2008; Rizzo et al. 2013; Marinescu et al. 2015) as well as from fresh produce (Holvoet et al. 2013; Lima et al. 2017). Finally, as opposed to other studies describing occurrence of ESBL-producing E. coli in different waters and fresh produce (Zurfluh et al. 2013; Nüesch-Inderbinen et al. 2015), no ESBL-producing strains were detected on the investigated farm.
The two whole-genome sequenced MDR E. coli-ST1056 harbored plasmid-borne ARG as well as a chromosomal point mutation known to confer quinolone resistance (Fig. 3A), covering most resistances observed in disk diffusion assays. Only exception was resistance to the third and fourth generation cephalosporins (cefpodoxime and cefepime, respectively) and amoxicillin-clavulanic acid, which cannot be explained by the presence of blaTEM-1B alone. Notably, resistance to cefpodoxime and cefepime was marginal, i.e. one to three mm below the resistance cutoff. Additionally, plasmid-borne aadA1 encoding resistance to the aminoglycosides spectinomycin and streptomycin was identified, which were not tested in disk diffusion assays.
Tracing back E. coli
Escherichia coli was chosen for source tracking for various reasons. First, a variety of antibiotic-resistant Enterococcus spp. were already detected on chive seeds and seedlings before coming into contact with irrigation water. Second, E. coli displayed much more diverse and clinically relevant phenotypic resistance profiles, with resistance to up to six antibiotic classes (Table 3). Finally, antibiotic-resistant E. coli were not present at any time in control-water whereas they were detected at all stages of the reservoir-irrigation system (Tables 1 and 2). Sequence typing of E. coli revealed that selected strains from water and chive belonged to ST1056, described previously as the main sequence type in poultry (Zurfluh et al. 2014b; Maamar et al. 2016). Its presence in wild birds can therefore be assumed to be likely, from where it could easily have entered our investigated water system, e.g. from the greenhouse rooftops draining rain water into the reservoir. Roof-harvested rain water has been described recently to be contaminated often with E. coli (Jongman and Korsten 2016b). Notably, evidence for water being the source of chive contamination had been found previously in a pilot study (summer 2015), where MDR E. coli-ST1432 had been detected on the same farm in drain water as well as on the irrigated chive plants (unpublished).
MLST has been recognized as a reliable method reflecting microevolution of the E. coli core genome and has therefore been used to determine phylogenetic relationships (Guenther, Ewers and Wieler 2011). However, albeit being widely used for microbial source tracking (Foley, Lynne and Nayak 2009), it might fail to distinguish very closely related but non-clonal strains. We therefore fully sequenced two MDR E. coli-ST1056 from beginning and end of the chain (marked with asterisk in Table 3). One of the two contigs assembled for each strain matched a known transmissible plasmid (Wang et al. 2014a), which was also isolated in Switzerland and described to harbor the same ARG detected in this study. SNP pairwise comparison yielded a SNP distance of only eight between the two isolates, while SNP distance to the reference was 7173 and 7171, respectively. As described by the developers of CSI Phylogeny using Salmonella enterica outbreak strains, it is difficult to define a general cutoff for pairwise SNP comparison to determine clonality (Leekitcharoenphon et al. 2014). However, by comparing outbreak strains and closely related background strains, they could show that SNP distances within outbreak strains were smaller than between outbreak and background strains. Furthermore, analysis of an E. coli test set provided by the CGE (five E. coli strains, including three clonal outbreak strains) (Cavaco and Leekitcharoenphon 2017) yielded a maximum SNP distance within clonal strains of 73, whereas the minimum SNP distance between outbreak and non-outbreak strains was 461. The SNP distance of 8 between MDR E. coli-ST1056 from drain water and chive, combined with the fact of identical sequence type, ARG profile including unknown chromosomal point mutations, and presence of a completely assembled plasmid leads to the conclusion that the two strains are clonal. As for the plasmid it must be pointed out that the IncFII plasmid family can replicate in many species of the Enterobacteriaceae family and is important in the dissemination of plasmid mediated antimicrobial resistance, e.g. carbapenemase blaKPC in Klebsiella pneumoniae (Chen et al. 2014).
In conclusion, our findings show that while edible plants are exposed to several potential contamination sources, irrigation water quality significantly influences the frequency of ARB on fresh produce under normal agricultural practice conditions. This finding is vital in the control of ARB and ARG transmission from the environment to humans via the food chain. We could prove the transmission of a MDR E. coli-ST1056 carrying a completely assembled transmissible IncFII-IncFIB resistance plasmid from drain water to the irrigated chive plants through the complete irrigation chain, including open-top reservoir, particle filter, greenhouse inlet and overhead sprinklers. This underlines the urgent need to establish guidelines for agricultural practice as well as monitoring recommendations regarding ARB in irrigation water. Transmission of antibiotic resistances in non-pathogenic bacteria via food to humans may be an unfavorable event as they can—upon establishment in the intestine—spread ARG or contribute to inactivation of antibiotics, whereas transmission of antibiotic-resistant human pathogens is a risk. Currently, regulations are established to ensure the minimization of the risk from pathogenic bacteria, however, the long-term risk to human health from non-pathogenic ARB is currently unknown. Further, for regions where good-quality water is scarce, affordable sanitation technologies must be developed to eliminate ARB and ARG from the water before applying it to the plants. Such technologies might include UV-based disinfection, but should ideally be combined with other methods to improve their efficiency (McKinney and Pruden 2012). Wastewater treatment plants may also profit from such technological advances to mitigate the amount of ARB and ARG released by them into surface waters, subsequently used for irrigation. Finally, responsible use of antibiotics must be propagated and controlled better, to at least slow down if not stop the constant increase in environmental ARB of utmost clinical relevance.
Supplementary Material
ACKNOWLEDGEMENTS
We are grateful to Jürgen Krauss for assisting in field trial planning and setup, Pascal Gisler and Hannah Bruderer for assistance in field and laboratory work. We are indebted to Prof. Dr. med. Reinhard Zbinden (Institute of Medical Microbiology, University of Zurich) for the opportunity to conduct high-throughput disk diffusion assays and valuable discussions as well as Martina Marchesi for laboratory assistance. We are thankful for support in PacBio sequencing and bioinformatics analysis to Andrea Patrignani and Weihong Qi (Functional Genomics Center Zurich, ETH Zurich and University of Zurich). We acknowledge the Agroscope Research Programme ‘Reduction and Dynamics of Antibiotic-resistant and Persistent Microorganisms along Food Chains (REDYMO)’ and the National Research Programme ‘Antimicrobial Resistance’ (grant number 407240_167068) of the Swiss National Science Foundation for financial support.
Conflicts of interest. None declared.
REFERENCES
- Abriouel H, Ben Omar N, Molinos AC et al. Comparative analysis of genetic diversity and incidence of virulence factors and antibiotic resistance among enterococcal populations from raw fruit and vegetable foods, water and soil, and clinical samples. Int J Food Microbiol. 2008;123:38–49. [DOI] [PubMed] [Google Scholar]
- Allende A, Monaghan J. Irrigation water quality for leafy crops: a perspective of risks and potential solutions. Int J Environ Res Public Health. 2015;12:7457–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson KL, Whitlock JE, Harwood VJ. Persistence and differential survival of fecal indicator bacteria in subtropical waters and sediments. Appl Environ Microbiol. 2005;71:3041–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araújo S, Silva IAT, Tacão M et al. Characterization of antibiotic resistant and pathogenic Escherichia coli in irrigation water and vegetables in household farms. Int J Food Microbiol. 2017;257:192–200. [DOI] [PubMed] [Google Scholar]
- Aziz RK, Bartels D, Best AA et al. The RAST server: rapid annotations using subsystems technology. BMC Genomics. 2008;9:75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai X, Ma X, Xu F et al. The drinking water treatment process as a potential source of affecting the bacterial antibiotic resistance. Sci Total Environ. 2015;533:24–31. [DOI] [PubMed] [Google Scholar]
- Bio Suisse. Standards for the production, processing and marketing of ‘bud’ products: Association of Swiss Organic Agriculture Organisations, 2015.
- Blaustein R, Shelton D, Van Kessel J et al. Irrigation waters and pipe-based biofilms as sources for antibiotic-resistant bacteria. Environ Monit Assess. 2015;188:1–12. [DOI] [PubMed] [Google Scholar]
- Callejón RM, Rodríguez-Naranjo MI, Ubeda C et al. Reported foodborne outbreaks due to fresh produce in the United States and European Union: trends and causes. Foodborne Pathog Dis. 2015;12:32–8. [DOI] [PubMed] [Google Scholar]
- Carattoli A, Zankari E, Garcìa-Fernandez A et al. PlasmidFinder and pMLST: in silico detection and typing of plasmids. Antimicrob Agents Chemother. 2014;58:3895–903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavaco L, , Leekitcharoenphon P. Whole genome sequencing of bacterial genomes - tools and applications https://www.coursera.org/learn/wgs-bacteria Accessed October, 2017.
- Chen L, Mathema B, Chavda KD et al. Carbapenemase-producing Klebsiella pneumoniae: molecular and genetic decoding. Trends Microbiol. 2014;22:686–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin C-S, Alexander DH, Marks P et al. Nonhybrid, finished microbial genome assemblies from long-read SMRT sequencing data. Nat Methods. 2013;10:563–9. [DOI] [PubMed] [Google Scholar]
- Christou A, Agüera A, Bayona JM et al. The potential implications of reclaimed wastewater reuse for irrigation on the agricultural environment: The knowns and unknowns of the fate of antibiotics and antibiotic resistant bacteria and resistance genes - A review. Water Res. 2017;123:448–67. [DOI] [PubMed] [Google Scholar]
- Clermont O, Christenson JK, Denamur E et al. The Clermont Escherichia coli phylo-typing method revisited: improvement of specificity and detection of new phylo-groups. Environ Microbiol Rep. 2013;5:58–65. [DOI] [PubMed] [Google Scholar]
- Clinical and Laboratory Standards Institute (CLSI). Performance Standards for Antimicrobial Susceptibility Testing, M100S. 26th edn Wayne, PA, USA: Clinical and Laboratory Standards Institute, 2016. [Google Scholar]
- Czekalski N, Berthold T, Caucci S et al. Increased levels of multiresistant bacteria and resistance genes after wastewater treatment and their dissemination into lake Geneva, Switzerland. Front Microbiol. 2012;3:106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devirgiliis C, Barile S, Perozzi G. Antibiotic resistance determinants in the interplay between food and gut microbiota. Genes Nutr. 2011;6:275–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Plessis EM, Duvenage F, Korsten L. Determining the potential link between irrigation water quality and the microbiological quality of onions by phenotypic and genotypic characterization of Escherichia coli isolates. J Food Prot. 2015;78:643–51. [DOI] [PubMed] [Google Scholar]
- Erickson MC, Webb CC, Diaz-Perez JC et al. Surface and internalized Escherichia coli O157:H7 on field-grown spinach and lettuce treated with spray-contaminated irrigation water. J Food Prot. 2010;73:1023–9. [DOI] [PubMed] [Google Scholar]
- European Commission (EC). Agriculture and rural development. Fruit and vegetable regime https://ec.europa.eu/agriculture/fruit-and-vegetables_en Accessed December, 2017.
- European Committee on Antimicrobial Susceptibility Testing (EUCAST). Antimicrobial susceptibility testing EUCAST disk diffusion method. Version 2.0 2012.
- European Food Safety Authority (EFSA). Report from the Task Force on Zoonoses Data Collection including guidance for harmonized monitoring and reporting of antimicrobial resistance in commensal Escherichia coli and Enterococcus spp. from food animals. EFSA J. 2008;141:1–44. [Google Scholar]
- European Food Safety Authority (EFSA). Technical specifications for the analysis and reporting of data on antimicrobial resistance (AMR) in the European Union Summary Report. EFSA J. 2012;10:2587. [Google Scholar]
- Fahrenfeld N, Ma Y, O'Brien M et al. Reclaimed water as a reservoir of antibiotic resistance genes: distribution system and irrigation implications. Front Microbiol. 2013;4:130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falomir MP, Gozalbo D, Rico H. Coliform bacteria in fresh vegetables: from cultivated land to consumers. In: Méndez-Vilas A. (ed.) Current Research, Technology and Education Topics in Applied Microbiology and Microbial Biotechnology., Badajoz, Spain: FORMATEX, 2010, 1175–81. [Google Scholar]
- Faour-Klingbeil D, Kuri V, Fadlallah S et al. Prevalence of antimicrobial-resistant Escherichia coli from raw vegetables in Lebanon. J Infect. [DOI] [PubMed] [Google Scholar]
- Farkas A, Bocoş B, Butiuc-Keul A. Antibiotic resistance and intI1 carriage in waterborne Enterobacteriaceae. Water Air Soil Pollut. 2016;227:1–11. [Google Scholar]
- Foley SL, Lynne AM, Nayak R. Molecular typing methodologies for microbial source tracking and epidemiological investigations of Gram-negative bacterial foodborne pathogens?. Infect Genet Evol. 2009;9:430–40. [DOI] [PubMed] [Google Scholar]
- Fonseca JM, Fallon SD, Sanchez CA et al. Escherichia coli survival in lettuce fields following its introduction through different irrigation systems. J Appl Microbiol. 2011;110:893–902. [DOI] [PubMed] [Google Scholar]
- Franz CMAP, Holzapfel WH, Stiles ME. Enterococci at the crossroads of food safety?. Int J Food Microbiol. 1999;47:1–24. [DOI] [PubMed] [Google Scholar]
- Gekenidis MT, Gossin D, Schmelcher M et al. Dynamics of culturable mesophilic bacterial communities of three fresh herbs and their production environment. J Appl Microbiol. 2017;123:916–32. [DOI] [PubMed] [Google Scholar]
- Gekenidis MT, Studer P, Wüthrich S et al. Beyond the matrix-assisted laser desorption ionization (MALDI) biotyping workflow: in search of microorganism-specific tryptic peptides enabling discrimination of subspecies. Appl Environ Microbiol. 2014;80:4234–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gemmell ME, Schmidt S. Microbiological assessment of river water used for the irrigation of fresh produce in a sub-urban community in Sobantu, South Africa. Food Res Int. 2012;47:300–5. [Google Scholar]
- Giraffa G. Enterococci from foods. FEMS Microbiol Rev. 2002;26:163–71. [DOI] [PubMed] [Google Scholar]
- Goldstein RER, Micallef SA, Gibbs SG et al. Detection of vancomycin-resistant enterococci (VRE) at four U.S. wastewater treatment plants that provide effluent for reuse. Sci Total Environ. 2014;466:404–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomes BC, Esteves CT, Palazzo ICV et al. Prevalence and characterization of Enterococcus spp. isolated from Brazilian foods. Food Microbiol. 2008;25:668–75. [DOI] [PubMed] [Google Scholar]
- Guenther S, Ewers C, Wieler LH. Extended-spectrum beta-lactamases producing E. coli in wildlife, yet another form of environmental pollution?. Front Microbiol. 2011;2:246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatakeyama M, Opitz L, Russo G et al. SUSHI: an exquisite recipe for fully documented, reproducible and reusable NGS data analysis. BMC Bioinformatics. 2016;17:228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano SS, Nordheim EV, Arny DC et al. Lognormal distribution of epiphytic bacterial populations on leaf surfaces. Appl Environ Microbiol. 1982;44:695–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holvoet K, Sampers I, Callens B et al. Moderate prevalence of antimicrobial resistance in Escherichia coli isolates from lettuce, irrigation water, and soil. Appl Environ Microbiol. 2013;79:6677–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hombach M, Zbinden R, Böttger EC. Standardisation of disk diffusion results for antibiotic susceptibility testing using the sirscan automated zone reader. BMC Microbiol. 2013;13:225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu D, Huang J, Wang Y et al. Fruits and vegetables consumption and risk of stroke. A meta-analysis of prospective cohort studies. Stroke. 2014;45:1613–9. [DOI] [PubMed] [Google Scholar]
- Hu J, Shi J, Chang H et al. Phenotyping and genotyping of antibiotic-resistant Escherichia coliisolated from a natural river basin. Environ Sci Technol. 2008;42:3415–20. [DOI] [PubMed] [Google Scholar]
- Hu Y, Yang X, Qin J et al. Metagenome-wide analysis of antibiotic resistance genes in a large cohort of human gut microbiota. Nat Commun. 2013;4:2151. [DOI] [PubMed] [Google Scholar]
- Huguet-Tapia J, Peng Z, Yang B et al. Complete genome sequence of the African strain AXO1947 of Xanthomonas oryzae pv. oryzae. Genome Announc. 2016;4:e1730–15.. DOI: 10.1128/genomeA.01730-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jay JM, Loessner MJ, Golden DA. Indicators of Food Microbial Quality and SafetyModern Food Microbiology. 7th edn New York, NY, USA: Springer Science and Business Media, 2005, 473–95. [Google Scholar]
- Johnston LM, Jaykus LA. Antimicrobial resistance of Enterococcus species isolated from produce. Appl Environ Microbiol. 2004;70:3133–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jongman M, Korsten L. Genetic diversity and antibiotic resistance of Escherichia coli isolates from different leafy green production systems. J Food Prot. 2016a;79:1846–53. [DOI] [PubMed] [Google Scholar]
- Jongman M, Korsten L. Microbial quality and suitability of roof-harvested rainwater in rural villages for crop irrigation and domestic use. J Water Health. 2016b;6:961–71. [DOI] [PubMed] [Google Scholar]
- Kaas RS, Leekitcharoenphon P, Aarestrup FM et al. Solving the problem of comparing whole bacterial genomes across different sequencing platforms. PLoS One. 2014;9:e104984 DOI: 10.1371/journal.pone.0104984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim C, Song S, Park C. The D-allose operon of Escherichia coli K-12. J Bacteriol. 1997;179:7631–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kümmerer K. Resistance in the environment. J Antimicrob Chemother. 2004;54:311–20. [DOI] [PubMed] [Google Scholar]
- Larsen MV, Cosentino S, Rasmussen S et al. Multilocus sequence typing of total genome sequenced bacteria. J Clin Microbiol. 2012;50:1355–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leclercq R, Cantón R, Brown DF et al. EUCAST expert rules in antimicrobial susceptibility testing. Clin Microbiol Infect. 2013;19:141–60. [DOI] [PubMed] [Google Scholar]
- Leekitcharoenphon P, Nielsen EM, Kaas RS et al. Evaluation of whole genome sequencing for outbreak detection of Salmonella enterica. PLoS One. 2014;9:e87991 DOI: 10.1371/journal.pone.0087991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leisibach S. Antibiotic resistance genes in food: molecular identification and transfer between microorganisms with emphasis on enterococci [dissertation]: Swiss Federal Institute of Technology Zurich (ETHZ), 2004.
- Li M, , Fan Y, Zhang X et al. Fruit and vegetable intake and risk of type 2 diabetes mellitus: meta-analysis of prospective cohort studies. BMJ Open. 2014;4:e5497 DOI: 10.1136/bmjopen-2014-005497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lima CM, Souza I, dos Santos Alves T et al. Antimicrobial resistance in diarrheagenic Escherichia coli from ready-to-eat foods. J Food Sci Technol. 2017;54:3612–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorimer MF, Kiermeier A. Analysing microbiological data: Tobit or not Tobit?. Int J Food Microbiol. 2007;116:313–8. [DOI] [PubMed] [Google Scholar]
- Ma Y, Prasad M, Rajkumar M et al. Plant growth promoting rhizobacteria and endophytes accelerate phytoremediation of metalliferous soils. Biotechnol Adv. 2011;29:248–58. [DOI] [PubMed] [Google Scholar]
- Maamar E, Hammami S, Alonso CA et al. High prevalence of extended-spectrum and plasmidic AmpC beta-lactamase-producing Escherichia coli from poultry in Tunisia. Int J Food Microbiol. 2016;231:69–75. [DOI] [PubMed] [Google Scholar]
- Mackie AM, Hassan KA, Paulsen IT et al. Biolog phenotype MicroArrays for phenotypic characterization of microbial cells. In: Paulsen IT, Holmes AJ. (eds.) Environmental Microbiology: Methods and Protocols. Totowa, NJ, USA: Humana Press, 2014, 123–30. [DOI] [PubMed] [Google Scholar]
- Marinescu F, Marutescu L, Savin I et al. Antibiotic resistance markers among Gram-negative isolates from wastewater and receiving rivers in South Romania. Rom Biotechnol Lett. 2015;20:10055–69. [Google Scholar]
- McKeon DM, Calabrese JP, Bissonnette GK. Antibiotic resistant Gram-negative bacteria in rural groundwater supplies. Water Res. 1995;29:1902–8. [Google Scholar]
- McKinney CW, Pruden A. Ultraviolet disinfection of antibiotic resistant bacteria and their antibiotic resistance genes in water and wastewater. Environ Sci Technol. 2012;46:13393–400. [DOI] [PubMed] [Google Scholar]
- Messi P, Guerrieri E, Bondi M. Antibiotic resistance and antibacterial activity in heterotrophic bacteria of mineral water origin. Sci Total Environ. 2005;346:213–9. [DOI] [PubMed] [Google Scholar]
- Micallef SA, Goldstein RE, George A et al. Diversity, distribution and antibiotic resistance of Enterococcus spp. recovered from tomatoes, leaves, water and soil on U.S. Mid-Atlantic farms. Food Microbiol. 2013;36:465–74. [DOI] [PubMed] [Google Scholar]
- Nüesch-Inderbinen M, Zurfluh K, Peterhans S et al. Assessment of the prevalence of extended-spectrum beta-lactamase-producing Enterobacteriaceae in ready-to-eat salads, fresh-cut fruit, and sprouts from the Swiss market. J Food Prot. 2015;78:1178–81. [DOI] [PubMed] [Google Scholar]
- Olaimat AN, Holley RA. Factors influencing the microbial safety of fresh produce: A review. Food Microbiol. 2012;32:1–19. [DOI] [PubMed] [Google Scholar]
- Palmer KL, Kos VN, Gilmore MS. Horizontal gene transfer and the genomics of enterococcal antibiotic resistance. Curr Opin Microbiol. 2010;13:632–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pappas EA, Kanwar RS, Baker JL et al. Fecal indicator bacteria in subsurface drain water following swine manure application. Trans ASABE. 2008;51:1567–73. [Google Scholar]
- Pesavento G, Calonico C, Ducci B et al. Prevalence and antibiotic resistance of Enterococcus spp. isolated from retail cheese, ready-to-eat salads, ham, and raw meat. Food Microbiol. 2014;41:1–7. [DOI] [PubMed] [Google Scholar]
- Poulsen N. CHIVES Allium schoenoprasum L. In: Brewster JL, Rabinowitch HD. (eds.) Onions and Allied Crops: Biochemistry Food Science Minor Crops volume 3. Boca Raton, FL, USA: CRC Press, 1989, 231–47. [Google Scholar]
- R Core Team. R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria2017.
- Rizzo L, Manaia C, Merlin C et al. Urban wastewater treatment plants as hotspots for antibiotic resistant bacteria and genes spread into the environment: a review. Sci Total Environ. 2013;447:345–60. [DOI] [PubMed] [Google Scholar]
- Schatz M. AMOS: A Modular Open Source Assembler 2006, http://schatzlab.cshl.edu/teaching/AssemblyClass/. Accessed January 2018.
- Schuch UK, Redak RA, Bethke JA. Cultivar, fertilizer, and irrigation affect vegetative growth and susceptibility of chrysanthemum to western flower thrips. J Am Soc Hortic Sci. 1998;123:727–33. [Google Scholar]
- Schwartz T, Kohnen W, Jansen B et al. Detection of antibiotic-resistant bacteria and their resistance genes in wastewater, surface water, and drinking water biofilms. FEMS Microbiol Ecol. 2003;43:325–35. [DOI] [PubMed] [Google Scholar]
- Sommer DD, Delcher AL, Salzberg SL et al. Minimus: a fast, lightweight genome assembler. BMC Bioinformatics. 2007;8:64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steele M, Odumeru J. Irrigation water as source of foodborne pathogens on fruit and vegetables. J Food Prot. 2004;67:2839–49. [DOI] [PubMed] [Google Scholar]
- Szmolka A, Nagy B. Multidrug resistant commensal Escherichia coli in animals and its impact for public health. Front Microbiol. 2013;4:258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thanner S, Drissner D, Walsh F. Antimicrobial resistance in agriculture. MBio. 2016;7:e2227–15, DOI: 10.1128/mBio.02227-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uyttendaele M, Jaykus LA, Amoah P et al. Microbial hazards in irrigation water: Standards, norms, and testing to manage use of water in fresh produce primary production. Compr Rev Food Sci Food Saf. 2015;14:336–56. [Google Scholar]
- Vaas LA, Sikorski J, Hofner B et al. opm: an R package for analysing OmniLog(R) phenotype microarray data. Bioinformatics. 2013;29:1823–4. [DOI] [PubMed] [Google Scholar]
- Wang J, Stephan R, Power K et al. Nucleotide sequences of 16 transmissible plasmids identified in nine multidrug-resistant Escherichia coli isolates expressing an ESBL phenotype isolated from food-producing animals and healthy humans. J Antimicrob Chemother. 2014a;69:2658–68. [DOI] [PubMed] [Google Scholar]
- Wang X, Ouyang Y, Liu J et al. Fruit and vegetable consumption and mortality from all causes, cardiovascular disease, and cancer: systematic review and dose-response meta-analysis of prospective cohort studies. BMJ. 2014b;349:g4490, DOI: 10.1136/bmj.g4490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warwick Medical School. Escherichia coli MSLT Database. http://mlst.warwick.ac.uk/mlst/dbs/Ecoli. Accessed July, 2017.
- Wirth T, Falush D, Lan R et al. Sex and virulence in Escherichia coli: an evolutionary perspective. Mol Microbiol. 2006;60:1136–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Health Organization (WHO). Guidelines for the Safe Use of Wastewater, Excreta and Greywater. Volume II: Wastewater Use in Agriculture: Geneva: World Health Organization, 2006. [Google Scholar]
- Xi C, Zhang Y, Marrs CF et al. Prevalence of antibiotic resistance in drinking water treatment and distribution systems. Appl Environ Microbiol. 2009;75:5714–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zankari E, Hasman H, Cosentino S et al. Identification of acquired antimicrobial resistance genes. J Antimicrob Chemother. 2012;67:2640–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zurfluh K, Abgottspon H, Hächler H et al. Quinolone resistance mechanisms among extended-spectrum beta-lactamase (ESBL) producing Escherichia coli isolated from rivers and lakes in Switzerland. PLoS One. 2014a;9:e95864, DOI: 10.1371/journal.pone.0095864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zurfluh K, Hächler H, Nüesch-Inderbinen M et al. Characteristics of extended-spectrum beta-lactamase- and carbapenemase-producing Enterobacteriaceae isolates from rivers and lakes in Switzerland. Appl Environ Microbiol. 2013;79:3021–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zurfluh K, Wang J, Klumpp J et al. Vertical transmission of highly similar blaCTX-M-1-harboring IncI1 plasmids in Escherichia coli with different MLST types in the poultry production pyramid. Front Microbiol. 2014b;5:519. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.