Abstract
Background
Chlamydia trachomatis is the most common sexually transmitted bacterial infection globally. Currently, there are no vaccines available despite the efforts made to develop a protective one. Polymorphic membrane protein D (PmpD) is an attractive immunogen candidate as it is conserved among strains and it is target of neutralizing antibodies. However, its high molecular weight and its complex structure make it difficult to handle by recombinant DNA techniques. Our aim is to predict B-cell and T-cell epitopes of PmpD.
Method
A sequence (Genbank AAK69391.2) having 99–100% identity with various serovars of C. trachomatis was used for predictions. NetMHC and NetMHCII were used for T-cell epitope linked to MHC I or MHC II alleles prediction, respectively. BepiPred predicted linear B-cell epitopes. For three dimensional epitopes, PmpD was homology-modeled by Raptor X. Surface epitopes were predicted on its globular structure using DiscoTope.
Results
NetMHC predicted 271 T-cell epitopes of 9-12aa with weak affinity, and 70 with strong affinity to MHC I molecules. NetMHCII predicted 2903 T-cell epitopes of 15aa with weak affinity, and 742 with strong affinity to MHC II molecules. Twenty four linear B-cell epitopes were predicted. Raptor X was able to model 91% of the three-dimensional structure whereas 57 residues of discontinuous epitopes were suggested by DiscoTope. Six regions containing B-cell and T-cell epitopes were identified by at least two predictors.
Conclusions
PmpD has potential B-cell and T-cell epitopes distributed throughout the sequence. Thus, several fragments were identified as valuable candidates for subunit vaccines against C. trachomatis.
Keywords: Chlamydia trachomatis, Vaccine, B-cell epitope, T-cell epitope, Molecular modeling, Epitope prediction
At a glance commentary
Scientific background on the subject
Chlamydia trachomatis infection has a high global prevalence and is associated with serious consequences on reproductive health. Antibiotic therapy is not successful at all, thus, vaccine development is strongly needed. One immunogen candidate is Polymorphic Membrane Protein D, a surface protein, highly conserved among serovars, and target of neutralizing antibodies.
What this study adds to the field
The identification of T- and B-cell epitopes on PmpD allows the selection of several regions that may be used to design subunits vaccines, potentially inducing both humoral and cellular immune responses. Bioinformatics presents powerful tools to the characterization of proteins favoring the rational design of vaccines.
Chlamydia trachomatis (Ct) is an intracellular bacterium that is an important cause of sexually transmitted infections (STI) with significant impact on public health. The World Health Organization (WHO) estimates that Ct is responsible of almost 106 million of the 500 million new cases of STI reported worldwide annually [1]. Ct includes three human biovars composed of different serovars [2] that can infect various cell types in humans. Serovars A-C are responsible for ocular infections that result in trachoma leading to blindness [3]. Serovars D-K causes sexually transmitted diseases such as cervicitis and pelvic inflammatory disease (PID), and globally are an important infectious cause of infertility, ectopic pregnancy [4] and chronic pelvic pain [5] in women. In men it is associated with urethritis, epididymitis and orchitis [6]. Moreover, serovars D-K cause urethritis and neonatal pneumonia [2]. The lymphogranuloma venereum (LGV) serovars L1–L3 not only cause sexually transmitted disease but can also infiltrate local lymph nodes, which ultimately results in systemic infection [7], [8]. Ct infections can be controlled by antibiotic therapy but the lack of compliance with treatment, the persistence of the infection even after a complete treatment, together with the high prevalence of asymptomatic cases [4] leading to severe reproductive complications strongly support the development of an effective Chlamydia vaccine. Currently, there are no vaccines available against Ct genital infection despite the many efforts that have been made throughout the years to develop a protective one. A failure of several vaccine designs may be attributed at least in part to the fact that protective immune response may result harmful for the host and the assumption that complete microorganisms could have components that induce both a protective and a immunopathogenic response. Safety concerns may be overcome by using subunit vaccines, but they require a thorough design in order to be efficient.
Among the antigen candidates that have been studied, members of the Polymorphic Membrane Protein family (Pmp A-I) have shown to be promising as vaccine components as they are dominant antigenic targets for cellular immune responses [9], [10], [11]. Pmps are a group of membrane bound surface exposed chlamydial proteins [12], [13], [14], [15], [16], [17], [18], [19] that have been characterized as autotransporter adhesins. These proteins are involved in the delivery of virulence factors involved in the initial phase of chlamydial infection [2], disease progression and immune evasion [18]. As typical type V autotransporters [20], all Pmps are characterized by containing conserved GGA (I, L, V) and FxxN tetrapeptide motifs, with an amino-terminal (N-terminal) dependent leader sequence, followed by a passenger domain and a carboxy-terminal (C-terminal) β-barrel [2], [21], [22]. The C-terminal region is incorporated into the outer membrane, forming a pore and allowing the translocation of the N-terminal passenger domain to the bacterial surface [22]. Pmp proteins may also undergo complex infection-dependent post-translational proteolytic processing [8], [17], [18], [23]. These proteins mediate in vitro chlamydial attachment to human epithelial and endothelial cells [8], [16].
PmpD is the second highest conserved Pmps demonstrating a 99.1% of amino acid identity among C. trachomatis serovars [24], and it is a target of broadly cross-reactive neutralizing antibodies [16]. The structural features of PmpD are relatively large size (1530 aa, 160.5 kDa), integrin-binding RGD motif (aa 698 to 670), and a putative nuclear localization signal (NLS; aa 783 to 798) [18] besides of N-terminal GGA(I/L/V) and FxxN tetrapeptide repeats as in all Pmps.
Due to its conserved nature, surface localization, and immunological importance PmpD is an attractive vaccine candidate for the prevention of human infections [16]. However, it has a high molecular weight with a complex structure which makes it difficult to handle by recombinant DNA techniques. Thus, an in-depth study of the molecule is needed in order to make a rational choice of the immunogen taking into consideration the critical epitopes to induce the appropriate immunological reaction. Several authors suggest that immunity against C. trachomatis requires CD4+ T cells, (mainly Th1) with INF-γ production and, to a lesser degree CD8+ T cells. Besides, neutralizing antibodies are now the focus of immune protection and vaccine development [16]. One of the major difficulties in developing an effective chlamydial vaccine is identifying the B-cell epitopes and the MHC-bound chlamydial protein epitopes that are recognized by T-cells.
In this context, bioinformatic approaches can contribute to the design of epitope-based vaccines. Therefore, the aim of this study was to perform in silico prediction of B and T epitopes within the amino acid sequence of PmpD for the design of a subunit vaccine against C. trachomatis.
Materials and methods
Datasets
An amino acid sequence of C. trachomatis serovar L2 PmpD available from NCBI (Genbank AAK69391.2) [25] was used for computational prediction. PmpD genes are one the most conserved between C. trachomatis serovars (with 99.1% similarities at the amino acid sequence level) [24]. This was confirmed by Clustal Omega, an algorithm of multiple amino acid sequence alignment available at http://www.ebi.ac.uk/Tools/msa/clustalo/.
3D structure prediction
The online tool integrated at Raptor X (http://raptorx.uchicago.edu/) was used for homology modeling of the PmpD in order to predict B cell epitopes in 3D conformation. Raptor X is a protein structure prediction server for protein sequences without close homologs in the Protein Data Bank (PDB) (Källberg et al., 2012). The server uses PSI-BLAST to find homologue templates and model the 3D structure on the basis of solvent accessibility, secondary and tertiary structures, and disordered regions. The following confidence scores are used to indicate the quality of a predicted 3D model: P-value for the relative global quality and uGDT (un-normalized GDT) for the absolute global quality.
Moreover, programs for structural analysis were used to check the 3D model. ANOLEA was used for energy calculations at the atomic level in protein structure [26] and RAMPAGE to generate the Ramachandran plot, showing protein residues in the favored, allowed and outliers regions [27].
B-cell epitope prediction
Linear B-cell epitope prediction
BepiPred was used to determine B cell linear epitope of PmpD. It is a method made by combining the predictions of a hidden Markov model and the propensity scale by Parker et al. [28], [29]. BepiPred assigns a score value to each protein residue. The method's specificity and sensitivity depend on the score threshold. For this analysis the threshold of 0.35 was employed. BepiPred is available at www.cbs.dtu.dk/services/BepiPred.
Discontinous B-cell epitopes
DiscoTope was employed to predict the surface epitopes on a globular structure. The method is based on amino acid statistics, spatial information, and surface accessibility considering Parker's hydrophilicity value [29]. These analyses are carried out taking into account a compiled data set of discontinuous epitopes determined by X-ray crystallography of antibody/antigen protein complexes [30], [31]. The updated version of DiscoTope is available at www.cbs.dtu.dk/services/DiscoTope-2.0.
T-cell epitope prediction
NetMHC [32], [33], [34] version 3.4 and NetMHCII [35], [36] version 2.2 were used for T-cell epitope linked to mayor histocompatibility complex (MHC) I or MHC II alleles prediction, respectively. The predictions are based on artificial neural networks (ANN). The servers are free to use and available at: http://www.cbs.dtu.dk/services/NetMHC and http://www.cbs.dtu.dk/services/NetMHCII.
Results
3D structure prediction
PmpD modeling was performed using Raptor X, which makes alignments between a target sequence and one or multiple distantly related template proteins (especially those with sparse sequence profiles) and employs a nonlinear scoring function and a probabilistic-consistency algorithm [37].
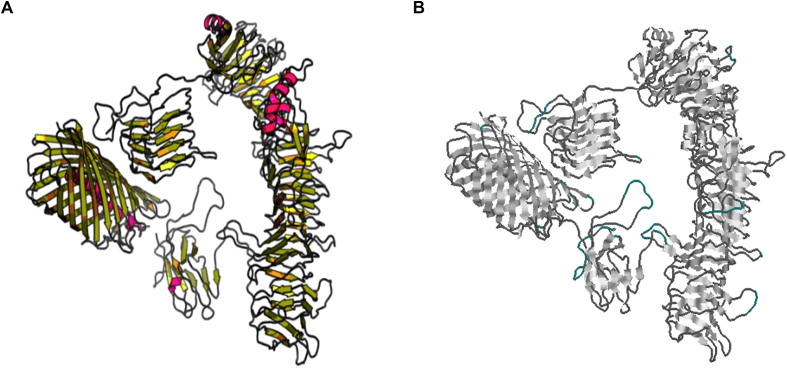
The Raptor X software modeled 1398 residues of 1530 (91%) of the PmpD sequence and identified four domains. To model domains 1 and 4, Raptor X used the passenger domain of the Escherichia coli autotransporter EspP (PDB entry: 3sze A) as a template, rhamnogalacturonase A from Aspergillus aculeatus (PDB entry: 1rmg A) as a template of domain 2, and autotransporter EstA from Pseudomonas aeruginosa (PDB entry: 3kvn A) as a template of domain 3. The software predicts that the protein has β sheets (31%), some α helices structure (9%) and several loops (58%) (Fig. 1A). According to the prediction, a 34% of PmpD is exposed to the surface while a 22% of PmpD is half exposed and a 43% is buried. The model seems to be good quality as suggested by confidence scores assigned by the software: P-value = 2.59 × 10−17, and uGDT = 409.
Fig. 1.
3D structure of PmpD. (A) Molecular model obtained with Raptor X. (B) Conformational B-cell epitopes predicted by DiscoTope (cyan regions).
Additional checking was performed with ANOLEA and RAMPAGE. ANOLEA analysis indicated that 45% of amino acids modeled by Raptor X have high energy. Ramachandran plot showed that 91.3% residues in the PmpD model were within the most favored regions, 5.8% residues in the allowed region and 2.9% residues in the outlier region. Homology modeling was also performed with Phyre [38] and Swiss-Model [39] softwares. However, unlike Raptor X, these only predicted the β barrel forming a single domain (data not shown).
B-cell epitope prediction
Linear B-cell epitope prediction
BepiPred predicted 48 B-cell linear epitopes (that have more than four residues) with a sensitivity and a specificity of 75% and 50%, respectively. As can be seen in Fig. 2, these epitopes are abundant and are homogeneously distributed throughout the protein sequence. Forty motifs characteristic of Pmps (21 GGA and 19 FxxN) are present in the amino acid sequence, ten of them are included in epitope regions (sequences are highlighted in bold (FxxN) and underlined (GGA) in Fig. 2). Larsen et al. found that BepiPred had the highest prediction accuracy on the test data set, and it was shown to perform significantly better than all other methods tested on the validation data set [28], [40].
Fig. 2.
PmpD amino acid sequence. BepiPred predicted 48 B-cell linear epitopes (highlighted in gray). FxxN motifs are indicated in bold, GGA sequences are underlined.
Discontinous B-cell epitopes
DiscoTope identified 57 residues, as part of epitope regions. A threshold of −3.7 for epitope identification was selected; therefore B-cell discontinuous epitopes were predicted with 0.75 specificity and 0.47 sensitivity. As the theory predicts, conformational epitopes were observed in loops, mainly in the regions not included in the β-barrel domain (C-terminal) (Fig. 1B).
Although confidence scores assigned by Raptor X indicated a good quality model, the reliability of the 3D model was further analyzed at the localization of the predicted conformational epitopes. The analysis performed by ANOLEA indicated that 85% of the 57 residues have high energy. The Ramachandran plot showed that 86% of the epitope residues are included in favorable regions, 12.3% in permitted regions and 1.7% in outlier regions.
T-cell epitope prediction
The ability of an epitope for binding to MHC molecules is expressed as IC50 value (nM). Strong binding peptides (SB) have an IC50 value below 50 nM, and weak binding peptides (WB) an IC50 value between 50 and 500 nM. The location of T-cell epitopes with strong and weak affinity for human MHC-I and MHC-II alleles was predicted using IEDB resource. As can be seen in Table 1, NetMHC predicted 271 T-cell epitopes of different lengths (9aa, 10aa, 11aa, 12aa) with weak affinity to molecules of MHC I and 70 T-cell epitopes of different lengths (9aa, 10aa, 11aa, 12aa) with strong affinity to molecules of MHC I (Table 1A). On the other hand, NetMHCII predicted 2903 T-cell epitopes of 15aa with weak affinity and 742 T-cell epitopes of 15aa with strong affinity to molecules of MHC class II for the most prevalent MHC alleles in Argentina according to Perusco et al. [40] (Table 1B). These epitopes are abundant and are homogeneously distributed throughout the protein sequence. Multiple alleles of MHC Class II are able to bind the same epitope. This was observed with several epitopes.
Table 1.
(A) Number of T-cell epitopes of different lengths (9aa, 10aa, 11aa, 12aa) predicted with Strong Binding (SB) and Weak Binding (WB) to MHC class I. (B) Number of T-cell epitopes of 15aa predicted with Strong Binding (SB) and Weak Binding (WB) to MHC class II.
| A | ||||||||
|---|---|---|---|---|---|---|---|---|
| MHC class I Prediction | 9aa |
10aa |
11aa |
12aa |
||||
| SB | WB | SB | WB | SB | WB | SB | WB | |
| HLA-A*02:01 | 14 | 25 | 4 | 20 | 1 | 15 | 7 | 21 |
| HLA-B*35:01 | 9 | 33 | 1 | 17 | 10 | 26 | 10 | 22 |
| HLA-B*35:03 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 1 |
| HLA-C*07:01 | 2 | 15 | 3 | 11 | 3 | 12 | 4 | 6 |
| HLA-C*07:02 | 0 | 10 | 0 | 10 | 1 | 12 | 0 | 14 |
| B | ||
|---|---|---|
| MHC class II Prediction | 15aa |
|
| SB | WB | |
| HLA-DRβ1*01:01 | 375 | 619 |
| HLA-DRβ1*04:01 | 39 | 479 |
| HLA-DRβ1*04:04 | 40 | 442 |
| HLA-DRβ1*04:05 | 47 | 365 |
| HLA-DRβ1*07:01 | 99 | 402 |
| HLA-DRβ1*11:01 | 20 | 264 |
| HLA-DRβ1*13:02 | 122 | 332 |
Vaccine candidates
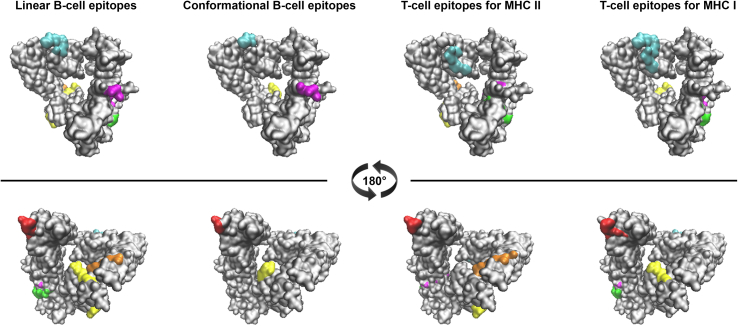
Giving that potential B- and T-cell epitopes were found to be distributed throughout the whole sequence, numerous fragments could be selected as candidates for the design of a vaccine. Nevertheless, it is necessary to define both B- and T-cell epitopes as candidates, to thereby generate a vaccine inducing both humoral and cellular response. Therefore, a more exhaustive analysis was performed to identify regions comprising both types of epitopes. The analysis consisted of simultaneously comparing the results obtained by the predictors. As shown in Table 2, residues of several epitopes have been identified simultaneously by four predictors: BepiPred (linear B-cell), DiscoTope (conformational B-cell), NetMHC (T-cell for MHCI) and NetMHCII (T-cell for MHC II). The data indicate that there are six regions with epitopes predicted by at least two predictors (Table 2, Fig. 3). An additional analysis of sequence conservancy showed that these regions are conserved among serovars, with the exception of aa 782 (V in serovars A-C, and A in serovars D-L). Regarding other proteins from the PmpD family, there is a variable degree of similarity among the selected fragments and PmpA, -B, -C, -E, -F, and -H (0–55%), and with Pmp21 from Clamydia pneumoniae (15–55%). No significant similarity was observed with human proteins.
Table 2.
B- and T-cell epitopes that include residues predicted simultaneously by at least two predictors among BepiPred (B-cell lineal), DiscoTope (B-cell conformational), NetMHCI (T-cell for MHCI) and NetMHCII (T-cell for MHCI) are highlighted in bold. Core amino acids of MHC II peptides are underlined.
| Regions | Linear B-cell epitopes | Conformational B-cell epitopes | T-cell epitopes for MHC II (SB) | T-cell epitopes for MHC I (WB) |
|---|---|---|---|---|
| 1 |
PSSFQEKDADTLPGKVE (82–98) |
FQEKD (85–89) |
HLA-DRβ1*13:02 GKVEQSTLFSVTNPV (95–109) |
HLA-B*35:01 LIVGDPSSF (77–85), HLA-B*35:01 LPGKVEQSTLF (93–103) HLA-A*02:01 KVEQSTLFSV (96–105) HLA-C*07:02 SFQEKDADTL (84–93) |
| 2 |
NNQAGISFEGGKASFG (412–427) |
GIS (416–418) E (420) |
HLA-DRβ1*01:01 AGISFEGGKASFGGG (415–429) |
HLA-B*35:01 VSIQNNQAGISF (408–419) |
| 3 |
FASEDGDLSPESSISSEELA (763–782) |
SEELAL (778–783) |
HLA-DRβ1*01:01 FITAANQALFASEDG (754–768) |
HLA-B*35:01 FASEDGDLSPES (763–774) |
| 4 | LREEDKLDGQ (825–834) |
HLA-DRβ1*13:02 GQIPEVLISGNAGDV (833–848) |
HLA-A*02:01 KLDGQIPEVLI (830–840) |
|
| 5 |
PEIEEDTYGHMGDWSEAKIQ (1171–1190) |
IHVATPEI (1166–1173) |
HLA-DRβ1*01:01 WSEAKIQDGTLVINW (1184–1198) |
HLA-B*35:01 TPEIEEDTY (1170–1178) |
| 6 |
LGESSASWT (1353–1361) |
HLA.DRβ1*01:01 RYGVLGESSASWTSR (1349–1363) |
Fig. 3.
Regions containing epitopes predicted simultaneously by more than one predictor. B- and T-cell epitopes. Regions are highlighted in different colors: 1: cyan, 2: red, 3: magenta, 4: green, 5: yellow, and 6: orange.
Discussion
C. trachomatis is the leading cause of bacterial sexually transmitted disease worldwide. Despite the fact that antibiotic treatment is effective in eliminating the pathogen, up to a 70% of all infections are asymptomatic. Untreated Chlamydia infections can lead to severe reproductive problems. Several C. trachomatis vaccine designs have been tested, but unfortunately there is no vaccine for Chlamydia despite the many efforts that have been made throughout the years. In part, this is due to an incomplete understanding of the immune response to Chlamydia urogenital infection [41] and of the critical immune factors required for resolving infections and protecting against reinfection [42], [43]. Many researchers suggest that both humoral and cell mediated immunity for a successful Chlamydia vaccine are required. Olsen et al. [44] describe that the primary role of neutralizing antibodies will be to reduce initial infectious load once intra-cellular, remaining bacteria can be targeted by a bactericidal cell mediated immune (CMI) response. For this reason, a rationally designed Chlamydia vaccine requires identification of optimal cell T- and B-cell antigens. Considering that the immunoinformatics is a discipline which can predict a priori whether an antigen is immunogenic through epitope prediction, our aim was to perform in silico prediction of B- and T-cell epitopes within the amino acid sequence of PmpD for the design of a subunit vaccine against C. trachomatis.
In the present study, different tools were used to analyze antigenic epitopes for PmpD of C. trachomatis. The complete amino acid sequence of PmpD was acquired from the data base of NCBI (http://www.ncbi.nlm.nih.gov/) corresponding to serovar L2. PmpD is one of the most conserved protein among C. trachomatis serovars (with 99.1% similarity at the amino acid sequence level) [24]. We confirmed that serovars A to L share 99–100% identity of amino acid sequence by Clustal Omega (Multiple Alignment).
Considering that PmpD plays a critical role in chlamydial biology and pathogenesis, particularly at early host–cell interaction [45], it is very important to determine the presence and localization of B-cell epitopes in the molecule. In this study, B-cell epitopes with a sensitivity and a specificity of 75% and 50%, respectively were predicted employing two different algorithms, BepiPred (linear epitopes) and DiscoTope (conformational epitopes). BepiPred predicted 48 B-cell linear epitopes and DiscoTope identified 57 residues which could account for 12 B-cell conformational epitopes. Regarding linear epitopes, ten of the GGA and FxxN motifs are included in epitope regions. These motifs can modulate the protein conformation implicated in adhesion or can modulate adhesin interaction with the eukaryotic receptors [2].
Considering that the conformational epitopes are predicted using a 3D model, the reliability of the model must be taken into account, particularly in the predicted regions as conformational B-cell epitopes. Quality parameters given by Raptor X suggest an overall good quality model and RAMPAGE indicated that a high proportion (86%) of the residues predicted to be part of B-cell epitopes are in favored regions. On the other hand, ANOLEA analysis indicated that 45% of amino acids modeled by RaptorX and 85% of the residues in conformational epitopes have high energy. This is not surprising giving that most of the conformational epitopes were predicted in loops, which are a big challenge in protein modeling because they often represent insertions or deletions in homologue proteins, they are less confined by alignment derived restraints [26], and they are usually structures with positive energy as predicted by ANOLEA or other servers [46], [47].
Simultaneously, BepiPred and DiscoTope predicted 9 Epitopes. The analysis performed using a bioinformatic approach contributed to identify numerous B-cell epitopes, and this correlates well with several previous studies where PmpD was able to elicit antibody production. Human serum studies have found anti-Pmps antibodies in C. trachomatis-infected patients [2]. Moreover, Tan et al. indicate that some Pmps, including PmpD, may be more abundantly expressed or specifically exposed at the chlamydial surface to elicit a relatively stronger antibody response in adolescents with and without pelvic inflammatory disease (PID) [11]. Several researchers have also shown the neutralizing ability of anti-PmpD antibodies [15], [16], [18]. In vitro neutralization assays showed that serovars Ba, D, E, and L2 were more efficiently neutralized than serovars A, C, F, G and K by antibodies against recombinant PmpD [16]. Although the high percentage of similarity among sequences, these differences could be related to little changes in specific amino acid. Then, defining epitope containing regions which are simultaneously conserved may allow choosing an immunogen able to induce neutralizing antibodies against all serovars. In this work, we identified six regions containing B- and T-cell epitopes, which additionally are conserved among serovars A-L. Besides being useful for vaccine design, predicting B-cell epitopes can contribute to the design of various diagnostic methods.
An adequate humoral response depends upon appropriate CD4+ T-cell response. Moreover, vaccines that induce cellular immune responses are essential for infections caused by intracellular pathogens. One of the major impediments in developing such vaccines includes difficulty in identifying relevant T-cell antigens that elicit protective cellular immunity [48]. CD4+ T-cell-mediated immunity is a major component of host defense against C. trachomatis infection [49] and to a lesser degree CD8+ T cells that recognize specific chlamydial antigens on MHC molecules [2]. The identification of epitopes presented by MHC class I and II molecules should enable the development of an effective T-cell response against C. trachomatis. In this study, NetMHC and NetMHCII were employed for T-cell epitope prediction. Regarding the prediction of T-cell epitopes in the PmpD sequence performed, NetMHC predicted 271 T-cell epitopes of different lengths (9aa, 10aa, 11aa, 12aa) with weak affinity to MHC I molecules and 70 T-cell epitopes of different lengths (9aa, 10aa, 11aa, 12aa) with strong affinity to MHC I molecules (Table 1A). Moreover, NetMHCII predicted 742 (Table 1B) T-cell epitopes of 15aa with strong affinity and 2903 T-cell epitopes of 15aa with weak affinity to MHC class II molecules for the most prevalent MHC alleles in Argentina according to Perusco et al. [40], as well as in North America and Europe (http://www.allelefrequencies.net/default.asp).These epitopes are abundant and homogeneously distributed throughout the protein sequence. Multiple alleles of MHC Class II are able to bind the same epitope. This was observed with several epitopes.
CD4+ and CD8+ T-cell immune responses in humans and mice are induced by Chlamydia infection, but the role of CD8+ T-cells in protective immunity is not clear. In the murine model, CD8+ T-cells contribute significantly to upper genial tract pathology [50], [51] and infertility [52] but CD8+ T-cell are not necessary for clearing genital tract infection or protecting against reinfection [53], [54]. Therefore, the results obtained in this paper are promising due to the fact that we found more epitopes able to link with strong affinity to MHC class II with respect to epitopes predicted to bind to MHC class I, which are less and with weak affinity. The results allow choosing a PmpD sequence containing strong affinity to molecules of MHC class II, which would favor a desired response of CD4+ type against C. trachomatis.
Comparative analyses performed by simultaneous predictors identifies six regions that have lineal and conformational B-cell epitopes and affinity to MHC I (WB) and MHC II (SB) molecules. This allows selecting sequences that may activate T-cells and induce cytokine and antibody production which contribute in the resolution of a chlamydial infection. Nevertheless, experimental assessment of the immunogenic ability of such regions both in vitro and in vivo should be done to verify this prediction or to select the best fragment.
Conclusion
PmpD has potential B-cell and T-cell epitopes distributed throughout the whole sequence. Numerous fragments could be selected as candidates for the design of a vaccine against C. trachomatis. To this end, in this study six regions containing both B-cell and T-cell epitopes were identified that may be used to generate both humoral and cellular response. These antigenic peptides can be valuable candidates for diagnostic as well as for therapeutic and preventive vaccines against C. trachomatis. Moreover, for vaccine design it would be important to select those regions that have the structural features of PmpD such as N-terminal GGA(I/L/V) and FxxN tetrapeptide repeats, an integrin-binding RGD motif, and a putative nuclear localization signal besides both B-cell and T-cell epitopes.
Selected fragments of PmpD that contains B- and T-cell epitopes in conjunction with effective adjuvants and/or delivery systems may contribute to the development of an effective chlamydial vaccine that elicits a Th1-mediated and humoral protective immune response that does not induce adverse immunopathologies. The data obtained will be utilized to delineate a series of immunization strategies. Among them: the synthesis of single epitopes that could be administered as unique or in combination, the design of a multi-epitope chimeric molecule or the selection of fragments containing several immunogenic regions.
The results of our study provide computational data for the identification and screening of epitopes in PmpD, and may be used for the development of epitope vaccines that have an enhanced safety and efficacy.
Conflicts of interest
No conflicts of interests.
Acknowledgements
This work was supported by Universidad Nacional del Litoral (CAI+D Orientado 2014 to C.V.)
Footnotes
Peer review under responsibility of Chang Gung University.
References
- 1.WHO . World Health Organization; Geneva, Switzerland: 2012. Global incidence and prevalence of selected curable sexually transmitted infections-2008. [Google Scholar]
- 2.Vasilevsky S., Stojanov M., Greub G., Baud D. Chlamydial polymorphic membrane proteins: regulation, function and potential vaccine candidates. Virulence. 2016;7:11–22. doi: 10.1080/21505594.2015.1111509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vogel G. Infectious diseases. Tackling neglected diseases could offer more bang for the buck. Science. 2006;311:592–593. doi: 10.1126/science.311.5761.592a. [DOI] [PubMed] [Google Scholar]
- 4.Peipert J.F. Clinical practice. Genital chlamydial infections. N Engl J Med. 2003;349:2424–2430. doi: 10.1056/NEJMcp030542. [DOI] [PubMed] [Google Scholar]
- 5.Westrom L., Joesoef R., Reynolds G., Hagdu A., Thompson S.E. Pelvic inflammatory disease and fertility. A cohort study of 1,844 women with laparoscopically verified disease and 657 control women with normal laparoscopic results. Sex Transm Dis. 1992;19:185–192. [PubMed] [Google Scholar]
- 6.Redgrove K.A., McLaughlin E.A. The role of the immune response in Chlamydia trachomatis infection of the male genital tract: a double-edged sword. Front Immunol. 2014;5:534. doi: 10.3389/fimmu.2014.00534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schachter J. Chlamydial infections (third of three parts) N Engl J Med. 1978;298:540–549. doi: 10.1056/NEJM197803092981005. [DOI] [PubMed] [Google Scholar]
- 8.Becker E., Hegemann J.H. All subtypes of the Pmp adhesin family are implicated in chlamydial virulence and show species-specific function. Microbiologyopen. 2014;3:544–556. doi: 10.1002/mbo3.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Karunakaran K.P., Yu H., Jiang X., Chan Q., Moon K.M., Foster L.J. Outer membrane proteins preferentially load MHC class II peptides: implications for a Chlamydia trachomatis T cell vaccine. Vaccine. 2015;33:2159–2166. doi: 10.1016/j.vaccine.2015.02.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Johnson R.M., Yu H., Kerr M.S., Slaven J.E., Karunakaran K.P., Brunham R.C. PmpG303-311, a protective vaccine epitope that elicits persistent cellular immune responses in Chlamydia muridarum-immune mice. Infect Immun. 2012;80:2204–2211. doi: 10.1128/IAI.06339-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tan C., Hsia R.C., Shou H., Haggerty C.L., Ness R.B., Gaydos C.A. Chlamydia trachomatis-infected patients display variable antibody profiles against the nine-member polymorphic membrane protein family. Infect Immun. 2009;77:3218–3226. doi: 10.1128/IAI.01566-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Molleken K., Schmidt E., Hegemann J.H. Members of the Pmp protein family of Chlamydia pneumoniae mediate adhesion to human cells via short repetitive peptide motifs. Mol Microbiol. 2010;78:1004–1017. doi: 10.1111/j.1365-2958.2010.07386.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Montigiani S., Falugi F., Scarselli M., Finco O., Petracca R., Galli G. Genomic approach for analysis of surface proteins in Chlamydia pneumoniae. Infect Immun. 2002;70:368–379. doi: 10.1128/IAI.70.1.368-379.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vandahl B.B., Pedersen A.S., Gevaert K., Holm A., Vandekerckhove J., Christiansen G. The expression, processing and localization of polymorphic membrane proteins in Chlamydia pneumoniae strain CWL029. BMC Microbiol. 2002;2:36. doi: 10.1186/1471-2180-2-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wehrl W., Brinkmann V., Jungblut P.R., Meyer T.F., Szczepek A.J. From the inside out–processing of the Chlamydial autotransporter PmpD and its role in bacterial adhesion and activation of human host cells. Mol Microbiol. 2004;51:319–334. doi: 10.1046/j.1365-2958.2003.03838.x. [DOI] [PubMed] [Google Scholar]
- 16.Crane D.D., Carlson J.H., Fischer E.R., Bavoil P., Hsia R.C., Tan C. Chlamydia trachomatis polymorphic membrane protein D is a species-common pan-neutralizing antigen. Proc Natl Acad Sci U S A. 2006;103:1894–1899. doi: 10.1073/pnas.0508983103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kiselev A.O., Skinner M.C., Lampe M.F. Analysis of pmpD expression and PmpD post-translational processing during the life cycle of Chlamydia trachomatis serovars A, D, and L2. PLoS One. 2009;4:e5191. doi: 10.1371/journal.pone.0005191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Swanson K.A., Taylor L.D., Frank S.D., Sturdevant G.L., Fischer E.R., Carlson J.H. Chlamydia trachomatis polymorphic membrane protein D is an oligomeric autotransporter with a higher-order structure. Infect Immun. 2009;77:508–516. doi: 10.1128/IAI.01173-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tan C., Hsia R.C., Shou H., Carrasco J.A., Rank R.G., Bavoil P.M. Variable expression of surface-exposed polymorphic membrane proteins in in vitro-grown Chlamydia trachomatis. Cell Microbiol. 2010;12:174–187. doi: 10.1111/j.1462-5822.2009.01389.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Henderson I.R., Navarro-Garcia F., Desvaux M., Fernandez R.C., Ala'Aldeen D. Type V protein secretion pathway: the autotransporter story. Microbiol Mol Biol Rev. 2004;68:692–744. doi: 10.1128/MMBR.68.4.692-744.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Grimwood J., Stephens R.S. Computational analysis of the polymorphic membrane protein superfamily of Chlamydia trachomatis and Chlamydia pneumoniae. Microb Comp Genomics. 1999;4:187–201. doi: 10.1089/omi.1.1999.4.187. [DOI] [PubMed] [Google Scholar]
- 22.Henderson I.R., Lam A.C. Polymorphic proteins of Chlamydia spp.–autotransporters beyond the Proteobacteria. Trends Microbiol. 2001;9:573–578. doi: 10.1016/s0966-842x(01)02234-x. [DOI] [PubMed] [Google Scholar]
- 23.Saka H.A., Thompson J.W., Chen Y.S., Kumar Y., Dubois L.G., Moseley M.A. Quantitative proteomics reveals metabolic and pathogenic properties of Chlamydia trachomatis developmental forms. Mol Microbiol. 2011;82:1185–1203. doi: 10.1111/j.1365-2958.2011.07877.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Carrasco J.A., Tan C., Rank R.G., Hsia R.C., Bavoil P.M. Altered developmental expression of polymorphic membrane proteins in penicillin-stressed Chlamydia trachomatis. Cell Microbiol. 2011;13:1014–1025. doi: 10.1111/j.1462-5822.2011.01598.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kiselev A.O., Stamm W.E., Yates J.R., Lampe M.F. Expression, processing, and localization of PmpD of Chlamydia trachomatis Serovar L2 during the chlamydial developmental cycle. PLoS One. 2007;2:e568. doi: 10.1371/journal.pone.0000568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Melo F., Devos D., Depiereux E., Feytmans E. ANOLEA: a www server to assess protein structures. Proc Int Conf Intell Syst Mol Biol. 1997;5:187–190. [PubMed] [Google Scholar]
- 27.Lovell S.C., Davis I.W., Arendall W.B., 3rd, de Bakker P.I., Word J.M., Prisant M.G. Structure validation by Calpha geometry: phi, psi and Cbeta deviation. Proteins. 2003;50:437–450. doi: 10.1002/prot.10286. [DOI] [PubMed] [Google Scholar]
- 28.Larsen J.E., Lund O., Nielsen M. Improved method for predicting linear B-cell epitopes. Immunome Res. 2006;2:2. doi: 10.1186/1745-7580-2-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Parker J.M., Guo D., Hodges R.S. New hydrophilicity scale derived from high-performance liquid chromatography peptide retention data: correlation of predicted surface residues with antigenicity and X-ray-derived accessible sites. Biochemistry. 1986;25:5425–5432. doi: 10.1021/bi00367a013. [DOI] [PubMed] [Google Scholar]
- 30.Ponomarenko J.V., Van Regenmortel M.H. B-cell epitope prediction. In: Gu J., Bourne P.E., editors. Structural bioinformatics. John Wiley and Sons, Inc.; 2009. pp. 849–879. [Google Scholar]
- 31.Haste Andersen P., Nielsen M., Lund O. Prediction of residues in discontinuous B-cell epitopes using protein 3D structures. Protein Sci. 2006;15:2558–2567. doi: 10.1110/ps.062405906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nielsen M., Lundegaard C., Worning P., Lauemoller S.L., Lamberth K., Buus S. Reliable prediction of T-cell epitopes using neural networks with novel sequence representations. Protein Sci. 2003;12:1007–1017. doi: 10.1110/ps.0239403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lundegaard C., Lund O., Nielsen M. Accurate approximation method for prediction of class I MHC affinities for peptides of length 8, 10 and 11 using prediction tools trained on 9mers. Bioinformatics. 2008;24:1397–1398. doi: 10.1093/bioinformatics/btn128. [DOI] [PubMed] [Google Scholar]
- 34.Lundegaard C., Lamberth K., Harndahl M., Buus S., Lund O., Nielsen M. NetMHC-3.0: accurate web accessible predictions of human, mouse and monkey MHC class I affinities for peptides of length 8-11. Nucleic Acids Res. 2008;1:509–512. doi: 10.1093/nar/gkn202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nielsen M., Lund O. NN-align. An artificial neural network-based alignment algorithm for MHC class II peptide binding prediction. BMC Bioinf. 2009;10:296. doi: 10.1186/1471-2105-10-296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nielsen M., Lundegaard C., Lund O. Prediction of MHC class II binding affinity using SMM-align, a novel stabilization matrix alignment method. BMC Bioinf. 2007;8:238. doi: 10.1186/1471-2105-8-238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kallberg M., Wang H., Wang S., Peng J., Wang Z., Lu H. Template-based protein structure modeling using the RaptorX web server. Nat Protoc. 2012;7:1511–1522. doi: 10.1038/nprot.2012.085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kelley L.A., Sternberg M.J. Protein structure prediction on the Web: a case study using the Phyre server. Nat Protoc. 2009;4:363–371. doi: 10.1038/nprot.2009.2. [DOI] [PubMed] [Google Scholar]
- 39.Biasini M., Bienert S., Waterhouse A., Arnold K., Studer G., Schmidt T. SWISS-MODEL: modelling protein tertiary and quaternary structure using evolutionary information. Nucleic Acids Res. 2014;42(Web Server issue):W252–W258. doi: 10.1093/nar/gku340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Perusco A., Gamba C., Galarza P., Rojas F., Kalapodis A., Onofri A. HLA antigenic and haplotype frequencies estimated in hematopoietic progenitor cell donors from Argentina. Transplant Proc. 2014;46:3064–3067. doi: 10.1016/j.transproceed.2014.07.029. [DOI] [PubMed] [Google Scholar]
- 41.Li L.X., McSorley S.J. A re-evaluation of the role of B cells in protective immunity to Chlamydia infection. Immunol Lett. 2015;164:88–93. doi: 10.1016/j.imlet.2015.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Morrison S.G., Morrison R.P. A predominant role for antibody in acquired immunity to chlamydial genital tract reinfection. J Immunol. 2005;175:7536–7542. doi: 10.4049/jimmunol.175.11.7536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Johansson M., Schon K., Ward M., Lycke N. Genital tract infection with Chlamydia trachomatis fails to induce protective immunity in gamma interferon receptor-deficient mice despite a strong local immunoglobulin A response. Infect Immun. 1997;65:1032–1044. doi: 10.1128/iai.65.3.1032-1044.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Olsen A.W., Follmann F., Erneholm K., Rosenkrands I., Andersen P. Protection against Chlamydia trachomatis infection and upper genital tract pathological changes by vaccine-promoted neutralizing antibodies directed to the VD4 of the major outer membrane protein. J Infect Dis. 2015;212:978–989. doi: 10.1093/infdis/jiv137. [DOI] [PubMed] [Google Scholar]
- 45.Kari L., Southern T.R., Downey C.J., Watkins H.S., Randall L.B., Taylor L.D. Chlamydia trachomatis polymorphic membrane protein D is a virulence factor involved in early host-cell interactions. Infect Immun. 2014;82:2756–2762. doi: 10.1128/IAI.01686-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Figueroa M., Oliveira N., Lejeune A., Kaufmann K.W., Dorr B.M., Matagne A. Octarellin VI: using rosetta to design a putative artificial (beta/alpha)8 protein. PLoS One. 2013;8:e71858. doi: 10.1371/journal.pone.0071858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chodanowski P., Grosdidier A., Feytmans E., Michielin O. Local alignment refinement using structural assessment. PLoS One. 2008;3:e2645. doi: 10.1371/journal.pone.0002645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Karunakaran K.P., Yu H., Foster L.J., Brunham R.C. Development of a Chlamydia trachomatis T cell vaccine. Hum Vaccine. 2010;6:676–680. doi: 10.4161/hv.6.8.12299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Brunham R.C., Rey-Ladino J. Immunology of Chlamydia infection: implications for a Chlamydia trachomatis vaccine. Nat Rev Immunol. 2005;5:149–161. doi: 10.1038/nri1551. [DOI] [PubMed] [Google Scholar]
- 50.Vlcek K.R., Li W., Manam S., Zanotti B., Nicholson B.J., Ramsey K.H. The contribution of Chlamydia-specific CD8(+) T cells to upper genital tract pathology. Immunol Cell Biol. 2015;94:208–212. doi: 10.1038/icb.2015.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Murthy A.K., Li W., Chaganty B.K., Kamalakaran S., Guentzel M.N., Seshu J. Tumor necrosis factor alpha production from CD8+ T cells mediates oviduct pathological sequelae following primary genital Chlamydia muridarum infection. Infect Immun. 2011;79:2928–2935. doi: 10.1128/IAI.05022-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Igietseme J.U., He Q., Joseph K., Eko F.O., Lyn D., Ananaba G. Role of T lymphocytes in the pathogenesis of Chlamydia disease. J Infect Dis. 2009;200:926–934. doi: 10.1086/605411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Morrison S.G., Morrison R.P. Resolution of secondary Chlamydia trachomatis genital tract infection in immune mice with depletion of both CD4+ and CD8+ T cells. Infect Immun. 2001;69:2643–2649. doi: 10.1128/IAI.69.4.2643-2649.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Morrison R.P., Feilzer K., Tumas D.B. Gene knockout mice establish a primary protective role for major histocompatibility complex class II-restricted responses in Chlamydia trachomatis genital tract infection. Infect Immun. 1995;63:4661–4668. doi: 10.1128/iai.63.12.4661-4668.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]