Abstract
Background
XAGE-1b is shown to be overexpressed in lung adenocarcinoma and to be a strong immunogenic antigen among non-small cell lung cancer (NSCLC) patients. However, 3D structure of XAGE-1b is not available and its confirmation has not been solved yet.
Methods
Multiple sequence alignment was run to select the most reliable templates. Homology modeling technique was performed using computer-based tool to generate 3-dimensional structure models, eight models were generated and assessed on basis of local and global quality. Immune Epitope Database (IEDB) tools were then used to determine potential B-Cell epitopes while NetMHCpan algorithms were used to enhance the determination for potential epitopes of both Cytotoxic T-lymphocytes and T-helper cells.
Results
Computational prediction was performed for B-Cell epitopes, prediction results generated; 3 linear epitopes where XAGE-1b (13-21) possessed the best score of 0.67, 5 discontinuous epitopes where XAGE-1b (40-52) possessed the best score of 0.67 based on the predicted model of the finest quality. For a potential vaccine design, computational prediction yielded potential Human Leukocyte Antigen (HLA) class I epitopes including HLA-B*08:01-restricted XAGE-1b (3-11) epitope which was the best with 0.2 percentile rank. Regarding HLA Class II epitopes, HLA-DRB1*12:01-restricted XAGE-1b (25-33) was the most antigenic epitope with 5.91 IC50 value. IC50 values were compared with experimental values and population coverage percentages of epitopes were computed.
Conclusions
This study predicted a model of XAGE-1b tertiary structure which could explain its antigenic function and facilitate usage of predicted peptides for experimental validation towards designing immunotherapies against NSCLC.
Keywords: Epitope prediction, NSCLC immunotherapy, Bioinformatics, Peptide vaccines, Cancer/testis antigen, XAGE-1b
At a glance commentary
Scientific background on the subject
XAGE-1b has been identified as an overexpressed surface antigen in NSCLC cells and has been shown to be immunogenic. The quest for designing immunotherapies as peptide vaccines based on XAGE-1b has been challenged by the lack of detailed structural and sequence based information regarding its immunogenic properties.
What this study adds to the field
This study provides an extensive computational analysis of XAGE-1b that provides essential antigenic data that could aid in the development of immunotherapies against NSCLC. The paper also provides detailed structural as well as sequence based properties for each antigenic determinants on XAGE-1b.
Lung cancer is the primary cause of cancer death among both genders. An estimated number of 158,080 deaths are expected to occur in 2016 approximately quarter of the expected number of deaths to be caused by cancer. The most common type of lung cancers is Non-small cell lung cancer (NSCLC) as it accounts for about 84% of lung cancer cases [1]. About 70 cancer/testis (CT) antigens have been identified by immunological or genetic approaches [2], [3], [4] and have been shown to generate humoral and cell-mediated immune responses in cancer patients [5]. Because they are limitedly expressed in normal tissues and show great antigenicity, CT antigens are interestingly being attractive targets for cancer vaccines [6], [7], [8], [9], [10]. PAGE/GAGE-related genes were investigated using an expression sequence tag database [11] where XAGE-1 was found to have similar characteristics to CT antigens [12], [13], [14]. Four transcript variants of XAGE-1 were identified which are XAGE-1a, XAGE-1b, XAGE-1c, and XAGE-1d and were shown to be expressed in lung cancers as well as metastatic melanoma, breast cancer, Ewing sarcoma and prostate cancers [15], [16], [17], [18]. XAGE-1b protein has been shown to be located in the cellular nuclei, using Immuno-histochemical nuclear staining methods. XAGE-1b was observed to be expressed strongly in 53% of lung adenocarcinomas; XAGE-1b expression was also immunogenic where most mRNA-positive specimens have been shown to express XAGE-1b protein [19]. The relevance of the XAGE-1b antigen expression was observed in Caucasian NSCLC patients [20]. XAGE-1b overexpression in adenocarcinoma cases of NSCLC is considered one of the most immunogenic antigens and a promising target for lung adenocarcinoma immunotherapy which requires determination of antigenic epitopes and tertiary structure of that antigen that may be addressed by computational tools which could pave the way for experimental validation and designing an epitope-based vaccine for NSCLC [21], [22].
Materials and methods
Retrieval of 9kD cancer/testis-associated protein XAGE-1b protein sequence
The antigenic protein sequence of 9kD cancer/testis-associated protein XAGE-1b protein was retrieved by accessing the NCBI databases [23] with 81 amino acid sequence in order to study the antigenicity and solvent accessible regions which permits potential vaccine targets to recognize active sites against NSCLC and then submitted to IEDB tools and other bioinformatics methods in order to computationally predict 3-dimensional model, assess modeling quality, and perform computational epitope prediction.
Homology modelling
Synthesizing peptides does not depend on the secondary structure of the protein, instead of that, it completely depends on the primary structure. Subsequently it does not indicate that the tertiary structures of the Antigen. Synthesis of XAGE-1b Antigenic peptides have been reported in the literature [20] However, peptide synthesis does not indicate that the 3D structure of XAGE-1b conformational epitopes have been solved. Furthermore in order to be able to study the immune interaction between XAGE-1b epitopes and HLA molecules, we need to study the 3D structure of the Surface antigen and the conformations of the epitopes. The prevalence of XAGE-1b antigen in different populations including Caucasians [20] supports that screening capabilities of immunoinformatics approaches could be helpful to provide large scale immune interaction analysis of XAGE-1b interaction with HLA class I and II as well as the structural analysis that may be implemented for further immune interaction studies or more interestingly peptide vaccine design.
Homology modeling is a comparative method of building an atomic-resolution model of a target protein sequence using an experimental 3-dimensional structure of a homologous Template. It depends on the identification of template structures resembling the target structure of the input sequence, doing an alignment that compares the input sequence to the template sequence. The rationale behind homology modelling is that Protein structures are proved to be more conserved than protein sequences among homologues, but proteins of sequence identity less than 20% can have non-homologous structure. Evolutionarily related proteins show similar amino acids sequences while naturally occurring homologous proteins have correlated 3-dimensional structure, so 3-dimensional protein structure is more conserved than expected on the basis of sequence conservation alone. Multiple Sequence alignment and template structure could be both used to produce a structural model of the target.
Despite its importance in Cancer research, the 3-dimensional structure of this protein is not available on bioinformatics databases (NCBI) (PDB) and it is confirmation has not been solved experimentally yet. Similarity alignment was done to select different reliable modeling templates and Top models were generated and tested according to SWISS-Model [24] parameters such as QMEAN which is a combined scoring function for estimating global and local model quality. QMEAN4 scores are directly related to a set of high-resolution PDB structures (Z-score). GMQE (Global Model Quality Estimation) parameter was also used for quality estimation which combines properties from the target-template alignment [25], [26].
Prediction of B-Cell epitopes
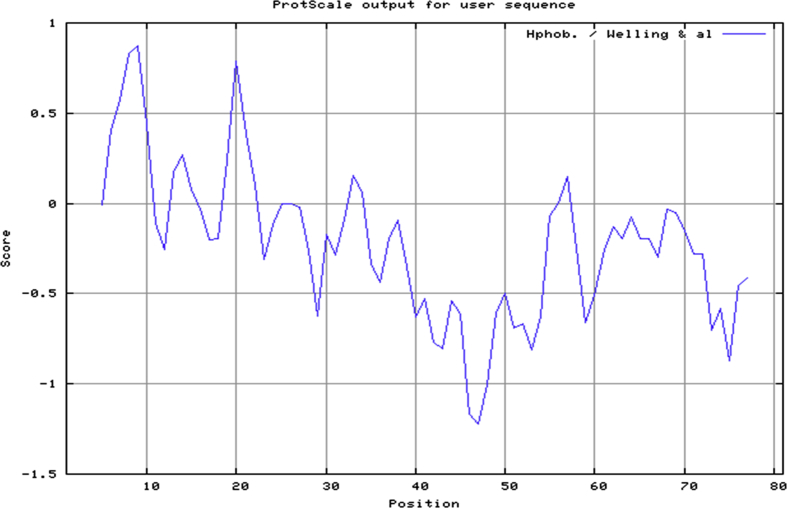
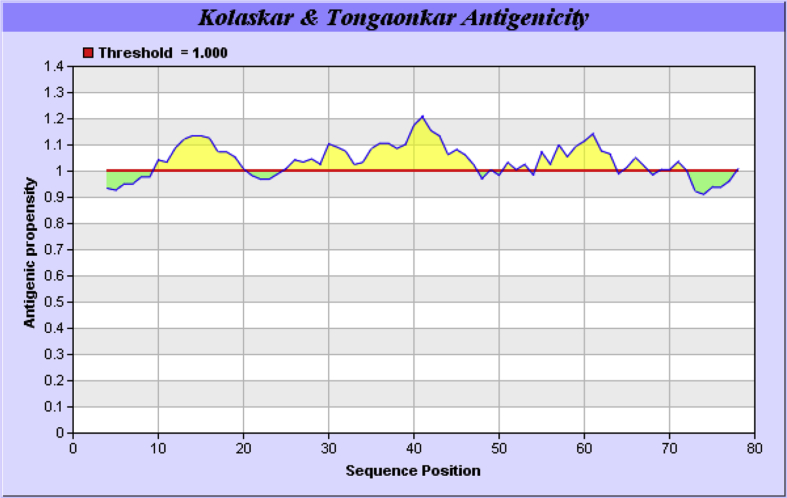
The amino acid sequence of XAGE-1b was submitted to IEDB tools to implement Kolaskar and Tongaonkar Method that can computationally determine linear B-Cell epitopes. Developers have applied this tool to a variety of proteins to predict B-Cell epitopes and results came out with 75% prediction accuracy [27]. Therefore, it is more favorable over other methods in the field. B-Cell epitopes properties including hydrophilicity, surface accessibility and flexibility have shown to be key players in epitope antigenicity; therefore, they were computationally determined by Emini surface accessibility (see Supplementary Fig. 2) prediction with 1.0 average propensity and flexibility prediction of Karplus and Schulz with 1.013 average propensity (see Supplementary Fig. 3). Conformational discontinuous B-Cell epitopes were computationally determined by ElliPro tool at IEDB with 0.7 as a minimum Protrusion Index and 6 Å as a maximum distance. The ElliPro tool allowed identification of B-Cell epitopes in a submitted protein 3-dimensional structure as a PDB file after homology modeling. ElliPro runs on a database of discontinuous determinants produced from 3-dimensional structures of antigen–antibody complexes have shown to have highest accuracy in comparison to six adjacent tools [28]. welling et al. [Fig. 3] hydrophobicity methods which compute the locations of antigenic epitopes in XAGE-1b were also plotted to detect Antigenic epitopes which are exposed on the protein surface so they would be distributed in hydrophilic regions, their values were computed from the free energies of side chain residues between ethanol and H2O molecules [29].
Fig. 3.
Welling et al. (1985) hydrophobicity plot of 9kD cancer/testis-associated protein XAGE-1b [29].
Prediction of human leukocyte antigen HLA class II [CD4 T-helper cells] epitopes
In order to predict T-helper cell, XAGE-1b amino acid sequence was submitted to NetMHCIIpan 3.0 server [30]. The 50 nM (IC50) Threshold for potential binding peptides was set to predict the binding affinities between helper T-Cell peptide epitopes and human leukocyte antigen class II alleles. It is interesting that NetMHCIIpan-3.0 tool was the first to cover all HLA class II molecules including HLA-DP, HLA-DR and HLA-DQ. The tool also predicts peptide binding to any HLA-II allele in a submitted sequence of amino acids. The method was evaluated and has given significant results over other allele-specific tools; therefore, the tool is considered to have the highest accuracy regarding MHC- II epitope prediction [31].
The epitopes that elicited the maximum Antigenicity of binding HLA Class II alleles were designated as recognized peptide epitopes in Table 4.
Table 4.
The most antigenic cytotoxic t-lymphocytes epitopes predicted by IEDB MHC-I recommended methods.
| Epitope | Length | From | To | Score/Percentile Rank | Restricted Allele |
|---|---|---|---|---|---|
| SPKKKNQQL | 9 | 3 | 11 | 0.2 | HLA-B*08:01 |
| GVKVKIIPK | 9 | 57 | 65 | 0.3 | HLA-A*30:01 |
| ILHLGSRQK | 9 | 15 | 23 | 0.35 | HLA-A*03:01 |
| RQKKIRIQL | 9 | 21 | 29 | 0.45 | HLA-A*31:01 |
| RSQCATWKV | 9 | 30 | 38 | 2.9 | HLA-B*57:01 |
| KIRIQLRSQ | 9 | 24 | 32 | 5.2 | HLA-B*07:02 |
| GSGVKVKII | 9 | 55 | 63 | 6.7 | HLA-B*58:01 |
| KSCISQTPG | 9 | 41 | 49 | 13 | HLA-B*15:01 |
| KKKNQQLKV | 9 | 5 | 13 | 30 | HLA-B*51:01 |
| VKVKIIPKE | 9 | 58 | 66 | 38 | HLA-B*53:01 |
Prediction of human leukocyte antigen HLA class I [CD8 cytotoxic T-Lymphocytes] epitopes
IEDB prediction server including NetMHCpan 2.8 [32] tool With 0.5 percentile rank threshold and Consensus tool which includes different methods such as; ANN [33] aka Comblib [34], SMM [35], and Consensus [36] to computationally determine The cytotoxic T-lymphocyte (CTL) epitopes. A computational high-throughput tool NetMHCpan is designed for In-silico prediction of peptides binding to HLA-I alleles, as the tool includes the majority of HLA class I alleles. Therefore, it is able to generate a powerful analysis for interaction of the epitope peptides and HLA-I alleles to establish potential vaccine design. The tool has an average accuracy of [75%–80%] for epitope peptides binding to HLA-I alleles. In this in-silico study, the most antigenic epitopes that elicited the highest binding affinity to HLA-A alleles were designated as potential epitope peptide candidates. Furthermore, it is well known that peptides with high antigenicity are more suspected to be Cytotoxic T-Lymphocyte epitopes than those of weak antigenicity. Therefore, the IEDB antigenicity prediction tool was accessed to predict the antigenicity of the proposed peptide epitopes. This tool computationally determines the antigenicity of (pHLA) complex depending on residues positions in a given sequence and their chemical and physical characteristics. Epitope conservancy was also checked using IEDB tools to calculate the conservancy level of an epitope in aligned amino acid sequences of XAGE-1b.
Prediction of population coverage
Peptides restricted to diverse HLA binding alleles indicate more coverage of different populations in specific geographical areas where the designed peptide vaccine is expected to be implemented. Considering this fact, we had used population coverage tool of IEDB to calculate the rate of coverage for each single epitope. Each epitope and its interacting HLA alleles were submitted (see Supplementary Tables 1 and 2), and analyzed in 2 ethnic groups; worldwide and North African population where we are further interested to implement our vaccine design [37], [38].
Results
Homology modelling
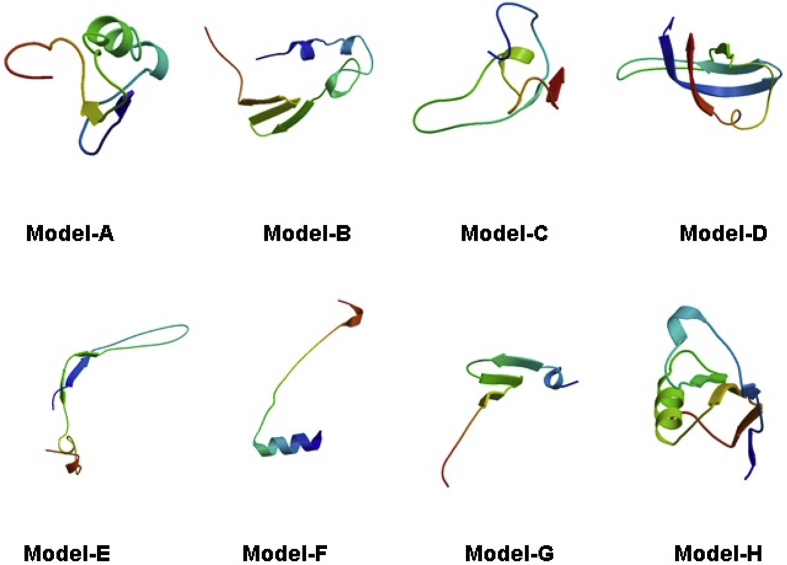
Using SWISS-Model web server the modelling process was initiated by template recognition process where templates were selected according to the maximum sequence similarity, 4ifd.1 was of highest sequence identity (Sequence identity: 42.00), and lowest sequence similarity found 4pbn with sequence identity of 25.00, Templates were selected for building XAGE-1b model. The software used PDB: 4IFD chain A as a template and homology modeled the structure. Criteria for reliable model selection included models in Fig. 1 range between 2–4Å RMSD from native Template structure. Whereas, identity percentage between sequence and template >30–40% indicated highly accurate model, Model A also showed high degree of compatibility between 3D model and primary amino acid sequence using verify3D. The SWISS-MODEL template library (SMTL) was mined with Blast [39] as well as HHBlits [40] in order to evolutionary relate structures matching the sequence of XAGE-1b. Models were built using ProMod3 [41] based on the target-template alignment, Coordinates which were conserved between XAGE-1b and suggested templates were copied from the template to the model. Fragment library was used to remodel Insertions and deletions. Side chains were then rebuilt. Finally, a force field was used to regularize the geometry of the resulting model. The QMEAN scoring function was used to assess the global and per-residue model quality has been assessed. For improved performance, weights of the individual QMEAN terms have been trained specifically for SWISS-MODEL. Models were selected based on their sequence identity as well as Swiss-MODEL quality assessment parameters; GMQE and QMEAN4 that assess both per residues and global quality of each model as well as Ramachandran plot calculations using structure quality evaluation methods including, PROCHECK [42], [43], Verifiy3D [44], [45] and RAMPAGE [46] structure validation tools for each model including number of residues in most favorable regions of Ramachandran plot which is an indicator of the level of torsion angles quality of model local structure. Model A was selected according to quality estimation parameters and was submitted to PMDB Protein Model Database [47] at PMDB ID PM0080777.
Fig. 1.
Generated models of Highest Accuracy of XAGE-1b using SWISS-MODEL tool.
Results of B-Cell epitope prediction
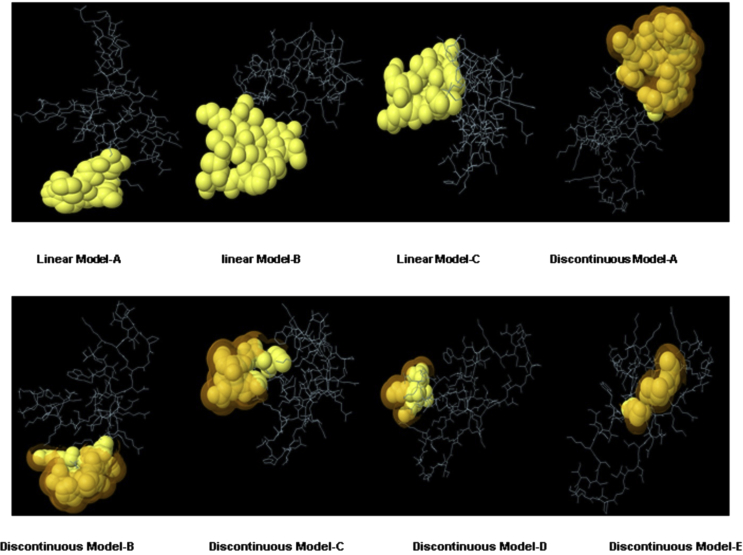
Using the IEDB database tools including Kolaskar and Tongaonkar's method [Fig. 2] (see Supplementary Fig. 1), ten linear B-Cell epitopes of XAGE-1b were computationally determined as Hexamer epitopes with 100% conservancy level within XAGE-1b sequences which had been calculated using IEDB conservancy tool as mentioned previously. Furthermore, the results declared an average propensity value of 1.040 between minimum value of 0.912 and a maximum of 1.208. Results also considered fragment flexibility as well as Surface accessibility to be crucial for predicting B-Cell epitopes, so both aspects were analyzed on basis of submitting the amino acid sequence of XAGE-1b to IEDB showing highest surface probability of 6.451 at residues from 1 to 10. The sequence of hexa-peptide is 5KKKNQQ10, such that the surface residue is K7. The smallest value of surface probability was 0.063 for peptide 39ICKSCI44, such that the surface residue is S42. Similarly, flexibility analysis of predicted epitopes declared that the highest flexibility value was 1.100 at residues ranging from 4 to 22, at 3SPKKKNQ9, where K6 is the flexible residue. Peptide 13RVGILHLG19 has the smallest flexibility value of 0.927, where the flexibility residue is L16. it is also very important to consider conformational discontinuous epitopes to be detected on basis of tertiary structure model, so 3 linear [Table 2] and 5 discontinuous [Table 3] B-Cell epitopes with more than 0.7 Protrusion Index were identified using ElliPro by analyzing XAGE-1b model A generated from homology modeling procedure [Fig. 4]. The highest score of a linear epitope was 0.67 and that of the best discontinuous epitope was 0.66 with a PI score of 0.965.
Fig. 2.
Kolaskar and Tongaonkar antigenicity plot showing antibody recognized antigenicity for the 9kD cancer/testis-associated protein XAGE-1b (see Supplementary Fig. 1) [27].
Table 2.
linear epitopes generated by ElliPro based on model A of XAGE-1b generated by homology modelling.
| No. | Chain | Start | End | Peptide | No. of residues | Score |
|---|---|---|---|---|---|---|
| 1 | A | 13 | 21 | VGILHLGSR | 9 | 0.67 |
| 2 | A | 40 | 52 | CKSCISQTPGINL | 13 | 0.66 |
| 3 | A | 59 | 69 | KSCISQTPGHC | 11 | 0.544 |
Table 3.
discontinuous epitopes generated by Ellipro based on model A of XAGE-1b generated by homology modelling.
| No. | Residues | No. of residues | Score |
|---|---|---|---|
| 1 | A:C40, A:K41, A:S42, A:I44, A:S45, A:Q46, A:T47, A:P48, A:G49 A:I50, A:N51, A:L52 | 13 | 0.66 |
| 2 | A:V13, A:G14, A:I15, A:L16, A:H17, A:L18, A:G19, A:S20, A:R21, A:K23 | 10 | 0.652 |
| 3 | A:I63, A:P64, A:K65, A:E66, A:E67, A:H68 | 6 | 0.644 |
| 4 | A:K59, A:V60, A:K61 | 3 | 0.6 |
| 5 | A:I25, A:I27, A:S31 | 3 | 0.589 |
Fig. 4.
Visualization using Jmol of generated epitopes using Ellipro, the first 3 are linear epitopes, while the other 5 models are discontinuous epitopes.
Results of Human leukocyte antigen HLA class I [CD8 Cytotoxic T-Lymphocytes] Epitopes Prediction
Prediction of Cytotoxic T-Lymphocyte epitopes is crucial for epitope-based vaccine design as it helps to understand mechanisms of T-Cell stimulation. HLA-I restricted alleles play a vital immunity role in taking advantage of Carcinogenesis process. Herein, 46 HLA class I antigenic XAGE-1b peptides having nonamer sequences were computationally determined using the Interaction prediction of the recommended IEDB tool to be potential CTL epitopes as peptides computationally bind a wide variety of HLA-I alleles eliciting very strong binding affinities. HLA-B*08:01-restricted XAGE-1b (3–11) was the best of 0.2 percentile rank. Most antigenic epitopes should bind the maximum number of HLA-I alleles to be considered as Promising CTL epitope peptides. Epitope RSQCATWKV hold the biggest number of HLA-I alleles interaction (25 alleles) and KSCISQTPG with 24 alleles. Oppositely, epitopes SPKKKNQQL (14 alleles) and VKVKIIPKE (13 alleles) had the smallest number of interacting HLA-I alleles. Cytotoxic T-Lymphocyte epitope peptides which possess a high antigenicity score are more likely to induce strong immune response (see Supplementary Table 2). Furthermore, these Cytotoxic T-Lymphocyte epitope candidates owned 100% conservancy level between available XAGE-1b aligned sequences, the most antigenic epitopes were ranked according to their percentile ranks calculated regarding their affinities to HLA-I alleles in [Table 4].
Results of Human leukocyte antigen HLA class II [CD4 T-helper cells] Epitopes prediction
HLA-II restricted CD4+ T-Cells stimulation is vital for potential vaccine design in order to induce Cytotoxic T-Lymphocyte response and effective T-helper recruitment. In-silico prediction using the NetMHCIIpan 3.0 server resulted in; 85 XAGE-1b potential antigenic peptides with nonamer core sequences that were computationally determined to be HLA-II restricted. Based on their ability to computationally bind a wide variety of HLA-DR alleles having IC50 smaller than 50 nM, it was indicated that they elicited strong binding affinities to HLA-DR alleles. A potential T-helper cell epitope should binds HLA Class II alleles as possible. Consequently, the most antigenic epitopes with the maximum number of interacting HLA-DR alleles were designated as promising T-helper cell epitope peptides. HLA-DRB1*12:01-restricted XAGE-1b (25–33) was the most antigenic epitope with 5.91 IC50. Both Epitopes LRSQCATWK and INLDLGSGV were interacting with [27 HLA-DR alleles] holding the highest number of interacting HLA-DR alleles. Oppositely, RIQLRSQCA peptide was predicted to be interacting with 11 HLA-DR alleles. KKIRIQLRS was also predicted to be interacting with 6 HLA-DR alleles. Therefore, RIQLRSQCA and KKIRIQLRS were the epitopes holding the smallest number of interacting HLA-DR alleles. Furthermore, all designated Potential epitope peptides have 100% conversancy level among available XAGE-1b sequences. The most antigenic epitopes were ranked according to their calculated IC50 values regarding their affinities to HLA-II alleles in [Table 5].
Table 5.
Most antigenic T-helper epitopes predicted by NetMHCIIpan 3.0.
| Core Epitope | Length | From | To | IC50 | Restricted Allele | Peptide |
|---|---|---|---|---|---|---|
| IRIQLRSQC | 9 | 25 | 33 | 5.91 | HLA-DRB1*12:01 | RQKKIRIQLRSQCAT |
| ILHLGSRQK | 9 | 15 | 23 | 6.26 | HLA-DRB5*01:01 | KVGILHLGSRQKKIR |
| INLDLGSGV | 9 | 50 | 58 | 54.38 | HLA-DRB1*13:02 | GINLDLGSGVKVKII |
| LRSQCATWK | 9 | 29 | 37 | 84.35 | HLA-DRB1*11:01 | IRIQLRSQCATWKVI |
| LDLGSGVKV | 9 | 52 | 60 | 100.79 | HLA-DRB1*07:01 | GINLDLGSGVKVKII |
| RIQLRSQCA | 9 | 26 | 34 | 210.77 | H LA-DRB1*04:05 | KIRIQLRSQCATWKV |
| GILHLGSRQ | 9 | 14 | 22 | 329.88 | HLA-DRB1*09:01 | LKVGILHLGSRQKKI |
| QLKVGILHL | 9 | 10 | 18 | 421.45 | HLA-DRB1*15:01 | KNQQLKVGILHLGSR |
| KKIRIQLRS | 9 | 23 | 31 | 698.29 | HLA-DRB1*08:02 | CATWKVICKSCISQT |
| KIRIQLRSQ | 9 | 24 | 32 | 1547.88 | HLA-DRB1*03:01 | LGSRQKKIRIQLRSQ |
Allele restricted epitope prediction IC50 values comparison
Computationally predicted antigenic Allele restricted epitopes in this in-silico study have been compared to experimentally predicted XAGE-1b epitopes in terms of their binding affinities to HLA Alleles represented by their IC50 values. We found that Prediction algorithms have predicted various peptides to be having more affinity to the same alleles compared to experimentally identified peptides that may reflects the limitation of experimental approaches in terms of screening capabilities which is considered a major advantage of computational approaches [50]. In (see Supplementary File 1), Computationally predicted IC50 values of Allele restricted XAGE-1b peptide epitopes predicted several peptides that were characterized computationally by having better affinities - to the same alleles - than experimentally determined epitopes mentioned previously. XAGE-1b (25–29) was predicted to be DRB1*04:05-restricted as well as having higher affinity compared to experimentally determined peptides including XAGE-1b (18–31) which has been identified as a DRB1*04:05-restricted epitope [22]. XAGE-1b (49–63) was predicted to be HLA-DRB1*09:01-restricted as well as having higher affinity compared to experimentally determined HLA-DRB1*09:01-restricted peptides including XAGE-1b (33–49) epitope which has been identified to be restricted by DR *0901 [48]. XAGE-1b (24–38) was predicted to be HLA-DRB1*04:10-restricted as well as having higher affinity compared to experimentally determined HLA-DRB1*04:10-restricted peptides including XAGE-1b (37–48) [49]. XAGE-1b (47–61) was predicted to be HLA-DRB1*13:02-restricted as well as having higher affinity compared to experimentally determined peptides to be restricted by the HLA-DRB1*13 allele including XAGE-1b (9–24), XAGE-1b (13–28) and XAGE-1b (17–32) [19]. XAGE-1b (49–58) was predicted to be HLA-A*02:06-restricted as well as having higher affinity compared to experimentally determined HLA-A*02:06-restricted peptides including XAGE-1b (21–29) which has been identified as a CD8 T-Cell epitope [22]. Although these results seem interesting, experimental validation is crucial to investigate the probability of false-positive predictions of computational algorithms [50].
Population coverage and distribution of epitopes
Due to Allele coverage differences among ethnicities, HLA distribution coverage to a specific population was considered to design a vaccine that covers a wide range of populations and possess a significant coverage in the population of interest. Herein, all determined alleles in (see Supplementary Tables 1 and 2) were recognized as potential alleles to interact with predicted epitopes and subsequently used to calculate population coverage percentage for each epitope. All recognized t-helper cell and CTL epitopes possessed High population coverage percentage in world-wide geographic regions as well as in North African population. For t-helper cell, top ten epitopes obtained world-wide population coverage of (87.17%) whereas in North Africa they obtained (75.06%) coverage. For CTL epitopes world-wide coverage was (98.55%) while coverage in North African population was (96.03%) Generally, our results demonstrated that recognized HLA-I epitopes and HLA-II epitopes can bind with predominant HLA alleles, provoking required immunogenic response in population of interest.
Limitations of experimental determination of B-Cell and T-cell epitopes
Experimental procedures for determining B-Cell epitopes are facing some limitations. Monoclonal antibody (MAb) resistant variant studies [51], MAb antigen contact studies and peptide scanning [52] may be of limited feasibility related to peptide screening capabilities where immunoinformatics studies could offer to predict epitopes on a huge scale. These limitations might be due to limited availability of X-ray solved crystal structures of surface antigens which limits the reliability of monoclonal antibody antigen contact research studies [53], where Immunoinformatics presents different approaches to overcome that problem such as homology modelling techniques that have been used in this study. At this point computational immunoinformatics studies provide promising alternative on both aspects of time and cost effectiveness [54].
Classical approaches for determination of T-cell epitopes may include T-cloning that is followed by stimulation in an in vitro environment as well as epitope reconstitution assays. These classical approaches still have some inherent problems including the challenging process of peptide mixture separation that required development of alternative approaches like ex vivo functional assays including HLA-Transgenic mice for detecting and validating epitopes, at this level bioinformatics epitope prediction become a valuable tool that guides the selection process of epitopes for experimental verification, so experimental methods for T-Cell epitope discovery still have some pitfalls that are in need of further development [55].
These alternative experimental approaches will be valuable until the computational capabilities of epitope prediction as HLA ligands become successful enough at their prediction accuracy. There is an increasing interest in the mass spectrometry based approaches for T-Cell epitope experimental discovery where it can be also coupled with in vitro HLA binding assays including peptide trimming and proteasome cleavage. Despite all these advanced alternative approaches, we still do not have a completely reliable method to characterize HLA restricted epitopes [56].
Limitations of computational prediction of B-Cell and T-cell epitopes
B-Cell epitope prediction algorithms still face limitations that demand more improvement. Usually (receiver operating characteristic) ROC curve is used to evaluate the performance of these machine learning predictors [57], [58], [59], which is directly proportional to the predictive capability of the algorithm where those of best performance usually of an average AUC of 0.7 [59], [60], trying to overcome the limited predictive capability problem of individual tools, a consensus method should be used to overcome the limited capability of these predictive tools [61], so we tried to implement consensus algorithms provided by IEDB tools in our prediction pipeline. HLA peptide binding prediction algorithms have strongly succeeded in epitope discovery especially when followed by HLA binding assays in a stepwise manner that may be enriched by implementing biological assays including enzyme-linked immunosorbent spot-forming assays (ELISpot) or HLA-transgenic mice studies.
Computational approaches for prediction of T-cell epitopes have shown potential abilities as an entry for epitope discovery and vaccine design. Available databases for epitope prediction already cover a very wide range of peptides presented by MHC or HLA molecules [62], furthermore, it has been reported that top predicted epitopes suggested by sophisticated computational tools were more likely to induce experimental success.
With the wide range of epitopes being predicted by these algorithms and limited experimental resources to verify, ranking systems have been developed to select potent epitopes rather than IC50 values [50]. Despite being promising, T-Cell epitope prediction algorithms may be limited by high number of false-positives, where predicted potential epitopes fail to elicit experimental immune response. Furthermore, experimentally successful peptides may not be predicted by T-Cell prediction algorithm which increases the number of false-negatives. It is questionable that efficacy of recently developed as well as early developed tools have been shown to have high false-negatives that may be up to 25% of predicted epitopes which provides an intense need to develop novel approaches for refinement of prediction algorithms.
Interestingly, recent studies have shown that antigenic epitopes with TcR (T-cell receptor) contact residues that recruit wide diversity of TcR could make them able to survive negative predictions and increase their likelihood of experimental success of computer-aided epitope determination [63]. Integrated approaches combining computational, in vitro and ex vivo methods may be the optimal pipeline for epitope discovery. Despite the recent advances of computational prediction methods, they cannot completely replace the experimental methods even though they have been reported to decrease in vitro testing 20-fold at least [64].
Discussion
It is well established that promising vaccine should hold both categories of antigenic epitopes including HLA-II epitope, B-Cell epitope or a Cytotoxic T-Lymphocyte epitope [65]. Combination of these epitopes would generate specific immune response against predicted antigens. Therefore, On the basis of 3-dimensional structural Homology modeling and quality assessment, the predicted 3-dimensional structure of 9kD cancer/testis-associated protein XAGE-1b in model A possessed a 40.2 sequence identity as shown in [Table 1]. The Model also exhibited the highest Qmean score and the best Z-Score according to Qmean server quality estimation results among all predicted models visualized in [Fig. 1] with a high percentage of residues in most favorable regions according to Ramachandran plots structural validation. This model could significantly facilitate the structural design of XAGE-1b vaccine. Welling et al. and Kolaskar & Tongaonkar antigenicity plots were generated to predict the antigenicity of epitopes which demonstrate the possibility of their recognition by antibodies, which would be satisfactory to produce desired immune response against the locations of antigenic epitopes of the 9kD cancer/testis-associated protein XAGE-1b. Ellipro-predicted epitopes seem to be highly-competent binders as bigger fraction of their atoms were shown to be instantly sharing in antigenic interaction. XAGE-1b model A shows helices regions, which showed higher antigenic response than other regions of this Antigen besides showing high antigenicity. Regions of maximal hydrophilicity are more likely to be antigenic sites of prominent hydrophobic character. C- terminal of 9kD cancer/testis-associated protein XAGE-1b protein was shown to be unstructured as well as being solvent accessible; because of that, antibodies against these regions are most likely to identify the native structure. This In-silico data provides a comprehensible indication that B-Cells would target the C-terminal regions, whereas T-Cells would target the N-terminal regions of XAGE-1b.
Table 1.
Generated Models of XAGE-1b using SWISS-MODEL tool with specific template for each assessed according to the parameters of SWISS-Model (sequence similarity, sequence Identity, GMQE, QMEAN4).
| Property | Model A | Model B | Model C | Model D | Model E | Model F | Model G | Model H |
|---|---|---|---|---|---|---|---|---|
| Template PDB Acc. No. | 4IFD | 3U1K | 2AE8 | 4JTU | 1E3P | EJ43 | 2ZKQ | 4PBN |
| Sequence Identity | 42 | 38.3 | 32.39 | 32.2 | 30 | 28.57 | 28.17 | 25 |
| Sequence similarity | 0.4 | 0.38 | 0.36 | 0.38 | 0.37 | 0.34 | 0.35 | 0.36 |
| GMQE | 0.39 | 0.31 | 0.58 | 0.48 | 0.1 | 0.26 | 0.58 | 0.44 |
| QMEAN4 | −1.18 | −6.25 | −3.89 | −5.21 | −4.62 | −2.22 | −3.85 | −3.44 |
| Procheck G-factor e (phi/psi only | −0.76 | −0.75 | −1.45 | −1.11 | −1.00 | −0.47 | −1.03 | −0.66 |
| Procheck G-factor e (all dihedral angles) | −0.37 | −0.28 | −0.74 | −0.62 | −0.40 | −0.01 | −0.45 | −0.23 |
| Verify3D Score | 0.02 | 0.07 | 0.12 | 0.05 | −0.08 | −0.09 | −0.03 | 0.02 |
| Most favored regions | 78.4% | 88.9% | 63.3% | 71.2% | 79.6% | 77.8% | 80.0% | 91.8% |
| Additionally allowed regions | 21.6% | 9.3% | 30.6% | 25.8% | 20.4% | 18.5% | 17.1% | 6.6% |
| Generously allowed regions | 0.0% | 0.0% | 2.0% | 3.0% | 0.0% | 3.7% | 2.9% | 0.0% |
| Disallowed region | 0.0% | 1.9% | 4.1% | 0.0% | 0.0% | 0.0% | 0.0% | 1.6% |
| Most favored regions | 89.7% | 93.5% | 73.2% | 85.5% | 88.5% | 90% | 87.5% | 91.5% |
| Allowed regions | 6.9% | 3.2% | 19.6% | 10.5% | 9.8% | 10% | 10% | 7% |
| Disallowed regions | 3.4% | 3.2% | 7.1% | 3.9% | 1.6% | 0% | 2.5% | 1.4% |
| RAMPAGE server Ramachandran plot | 87.9% | 91.9% | 73.2% | 84.2% | 88.5% | 90.0% | 90.0% | 88.7% |
Previous Studies have determined specific epitopes for XAGE-1b for immune monitoring including DRB1*04:05-restricted XAGE-1b 18–31 peptide [ LGSRQKKIRIQLRS ] as a T-helper epitope of 3275.35 IC50 value which is considered of low affinity that is below 5000 nm, while in [Table 5], we can notice that the top ten epitopes possessed an IC50 lower than 70 nm especially the epitope XAGE-1b 26–34 [ RIQLRSQCA ] that was restricted to the same allele and possessed an 210.77 IC50 value which is considered of intermediate affinity. Studies also mentioned HLA-A*02:06-restricted XAGE-1b 21–29 peptide [ RQKKIRIQL ] as a cytotoxic T-lymphocytes epitope where we found that it scored 13.0 percentile rank, so in (Table 4) we found 3 epitopes that possessed better percentile ranks. It was also remarkable that epitope XAGE-1b 49–58 [GINLDLGSGV] restricted to the same allele (HLA-A*02:06) possessed a 2.6 percentile rank [22]. These results were also investigated In (see Supplementary File 1), where predicted IC50 values of Allele restricted XAGE-1b peptide epitopes were characterized computationally by having better affinities - to the same alleles - than experimentally determined epitopes. Herein, we found that there are more potential epitopes having higher scores than already mentioned epitopes as mentioned in [Table 4, Table 5], which would be promising for further immune assays to experimentally validate these predictions and avoid false-positives that may be a deceptive issue regarding epitope determination.
Conclusion
In this in-silico study, screening for XAGE-1b associated B-Cell, HLA-I, and HLA-II epitopes were implemented generating promising linear and discontinuous B-Cell epitopes. The majority of conformational discontinuous epitopes were identified among regions of XAGE-1b Model A showing high accessibility for inducing immune response. Remarkably, it is not enough just to neutralize antibodies in order to target any specific antigen, cytotoxic T-Lymphocytes are also of great importance due to their vital role in provoking desired immune response against the target antigen. Prediction results were given in percentile ranks and IC50 values, where there is an inverse relation between IC50 numbers and binding affinity, so that epitopes having less than 50 nM IC50 values are of high affinity, those of less than 500 nM are considered of intermediate affinity and less than 5000 nM IC50 values are considered of low affinity. The most antigenic HLA Class I epitope was HLA-B*08:01-restricted XAGE-1b (3–11) epitope of 0.2 percentile rank. Regarding HLA Class II epitopes, HLA-DRB1*12:01-restricted XAGE-1b (25–33) was the most antigenic epitope with 5.91 IC50 value. We might notice that there is no significant association between the immunogenic score of epitope and its coverage that may be due to neglecting the coverage percentage of the peptide on the prediction algorithm and mainly relying on its ranking score regarding specific restricted allele. In order to widen the population coverage, epitope peptides should interact with more HLA alleles so the most antigenic t-helper cell and cytotoxic T-lymphocytes epitopes that interact with the highest number of HLA alleles were recognized as potential vaccine epitopes. Population coverage analysis showed that all potential T-helper cell and cytotoxic T-lymphocyte epitopes possessed high population coverage percentages and could provide wider immune protection for world-wide regions as well as geographical areas of interest such as the North African population. Furthermore, the epitopes predicted in this in-silico study were strongly conserved between available aligned XAGE-1b isoforms sequences. This In-silico approach will be applicable for further experimental validation empowering integrative approaches that will facilitate epitope determination process and help overcoming experimental and computational limitations for future Vaccine design. Interdisciplinary approaches could help conducting clinical trials for XAGE-1b based immunotherapies with the promise that 9kD cancer/testis-associated protein XAGE-1b protein sequence contains several antigenic determinants to direct and enforce the immune system to generate strong response against NSCLC.
Conflicts of interest
The authors declare that there is no competing interest.
Footnotes
Peer review under responsibility of Chang Gung University.
Supplementary data related to this article can be found at https://doi.org/10.1016/j.bj.2018.04.002.
Appendix A. Supplementary data
The following are the supplementary data related to this article:
Tables representing computationally predicted IC50 values of Allele restricted XAGE-1b peptide epitopes that were computationally predicted in this study compared to previously identified in experimental studies.
Kolaskar and Tongaonkar's plot of XAGE-1b. Showing the immunogenic propensity of XAGE-1b residues with Average immunogenic score of 0.912. Area displayed above the threshold is considered as promising B-Cell epitopes against XAGE-1b Antigen.
Emini surface accessibility plot of XAGE-1b. Showing accessible propensity with Average antigenic score of 1. Area displayed above the threshold cutoff is reflecting promising B-Cell accessible segments.
Karplus and Schulz flexibility plot of XAGE-1b. Showing flexible propensity of XAGE-1b residues with Average antigenic score of 1.013. Area displayed above the threshold cutoff is reflecting promising flexible segments.
Nonamer peptides core sequences of XAGE-1b which are HLA-I predicted epitopes, ranked according to alleles' coverage.
Nonamer peptides core sequences in XAGE-1b which are HLA-II predicted epitopes with NetMHCIIpan 3.0 server, ranked according to alleles' coverage.
References
- 1.Cancer . 2016. American cancer society cancer facts and figures 2016.https://www.cancer.org/research/cancer-facts-statistics/all-cancer-facts-figures/cancer-facts-figures-2016/ [Google Scholar]
- 2.Old L.J., Chen Y.T. New paths in human cancer serology. J Exp Med. 1998;187:1163–1167. doi: 10.1084/jem.187.8.1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Simpson A.J.G., Caballero O.L., Jungbluth A., Chen Y.T., Old L.J. Cancer/testis antigens, gametogenesis and cancer. Nat Rev Cancer. 2005;5:615–625. doi: 10.1038/nrc1669. [DOI] [PubMed] [Google Scholar]
- 4.Hofmann O., Caballero O.L., Stevenson B.J., Chen Y.T., Cohen T., Chua R. Genome-wide analysis of cancer/testis gene expression. Proc Natl Acad Sci U S A. 2008;105:20422–20427. doi: 10.1073/pnas.0810777105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jäger E., Chen Y.T., Drijfhout J.W., Karbach J., Ringhoffer M., Jäger D. Simultaneous humoral and cellular immune response against cancer–testis antigen NY-ESO-1: definition of human histocompatibility leukocyte antigen (HLA)-A2–binding peptide epitopes. J Exp Med. 1998;187:265–270. doi: 10.1084/jem.187.2.265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gnjatic S., Wheeler C., Ebner M., Ritter E., Murray A., Altorki N.K. Seromic analysis of antibody responses in non-small cell lung cancer patients and healthy donors using conformational protein arrays. J Immunol Methods. 2009;341:50–58. doi: 10.1016/j.jim.2008.10.016. [DOI] [PubMed] [Google Scholar]
- 7.Rosenberg S.A. A new era for cancer immunotherapy based on the genes that encode cancer antigens. Immunity. 1999;10:281–287. doi: 10.1016/s1074-7613(00)80028-x. [DOI] [PubMed] [Google Scholar]
- 8.Scanlan M.J., Gure A.O., Jungbluth A.A., Old L.J., Chen Y.T. Cancer/testis antigens: an expanding family of targets for cancer immunotherapy. Immunol Rev. 2002;188:22–32. doi: 10.1034/j.1600-065x.2002.18803.x. [DOI] [PubMed] [Google Scholar]
- 9.Old L.J. Cancer vaccines: an overview. Cancer Immun. 2008;8:1. [PubMed] [Google Scholar]
- 10.Caballero O.L., Chen Y.T. Cancer/testis (CT) antigens: potential targets for immunotherapy. Cancer Sci. 2009;100:2014–2021. doi: 10.1111/j.1349-7006.2009.01303.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brinkmann U., Vasmatzis G., Lee B., Pastan I. Novel genes in the PAGE and GAGE family of tumor antigens found by homology walking in the dbEST database. Cancer Res. 1999;59:1445–1448. [PubMed] [Google Scholar]
- 12.Brinkmann U., Vasmatzis G., Lee B., Yerushalmi N., Essand M., Pastan I. PAGE-1, an X chromosome-linked GAGE-like gene that is expressed in normal and neoplastic prostate, testis, and uterus. Proc Natl Acad Sci USA. 1998;95:10757–10762. doi: 10.1073/pnas.95.18.10757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu X.F., Helman L.J., Yeung C., Bera T.K., Lee B., Pastan I. XAGE-1, a new gene that is frequently expressed in Ewing's sarcoma. Cancer Res. 2000;60:4752–4755. [PubMed] [Google Scholar]
- 14.Nakagawa K., Noguchi Y., Uenaka A., Sato S., Okumura H., Tanaka M. XAGE-1 expression in non-small cell lung cancer and antibody response in patients. Clin Cancer Res. 2005;11:5496–5503. doi: 10.1158/1078-0432.CCR-05-0216. [DOI] [PubMed] [Google Scholar]
- 15.Zhou Q., Guo A.L., Xu C.R., An S.J., Wang Z., Yang S.Q. A dendritic cell-based tumour vaccine for lung cancer: full-length XAGE-1b protein-pulsed dendritic cells induce specific cytotoxic T lymphocytesin vitro. Clin Exp Immunol. 2008;153:392–400. doi: 10.1111/j.1365-2249.2008.03724.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Egland K.A., Kumar V., Duray P. PastanI. Characterization of overlapping XAGE-1 transcripts encoding a cancer testis antigen expressed in lung, breast, and other types of cancers. Mol Cancer Ther. 2002;1:441–450. [PubMed] [Google Scholar]
- 17.Zendman A.J., van Kraats A.A., den Hollander A.I., Weidle U.H., Ruiter D.J. Van MuijenGN. Characterization of XAGE-1b, a short major transcript of cancer/testis-associated gene XAGE-1, induced in melanoma metastasis. Int J Cancer. 2002;97:195–204. doi: 10.1002/ijc.1584. [DOI] [PubMed] [Google Scholar]
- 18.Zendman A.J., Van Kraats A.A., Weidle U.H., Ruiter D.J., Van Muijen G.N. The XAGE family of cancer/testis-associated genes: alignment and expression profile in normal tissues, melanoma lesions and Ewing's sarcoma. Int J Cancer. 2002;99:361–369. doi: 10.1002/ijc.10371. [DOI] [PubMed] [Google Scholar]
- 19.Sardaro A., Saito K., Nakayama E., Valmori D. Correction: immune responses to the cancer testis antigen XAGE-1b in non small cell lung cancer Caucasian patients. PLoS One. 2016;11 doi: 10.1371/journal.pone.0150623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sato S., Noguchi Y., Ohara N., Uenaka A., Shimono M., Nakagawa K. Identification of XAGE-1 isoforms: predominant expression of XAGE-1b in testis and tumors. Cancer Immun. 2007;7:5. [PMC free article] [PubMed] [Google Scholar]
- 21.Kikuchi E., Yamazaki K., Nakayama E., Sato S., Uenaka A., Yamada N. Prolonged survival of patients with lung adenocarcinoma expressing XAGE-1b and HLA class I antigens. Cancer Immun. 2008;8:13. [PMC free article] [PubMed] [Google Scholar]
- 22.Ohue Y., Eikawa S., Okazaki N., Mizote Y., Isobe M., Uenaka A. Spontaneous antibody, and CD4 and CD8 T-cell responses against XAGE-1b (GAGED2a) in non-small cell lung cancer patients. Int J Cancer. 2012;131:E649–E658. doi: 10.1002/ijc.27359. [DOI] [PubMed] [Google Scholar]
- 23.NCBI . 2016. The national center for biotechnology information.https://www.ncbi.com.gov/ [Google Scholar]
- 24.Arnold K., Bordoli L., Kopp J., Schwede T. The SWISS-MODEL workspace: a web-based environment for protein structure homology modelling. Bioinformatics. 2005;22:195–201. doi: 10.1093/bioinformatics/bti770. [DOI] [PubMed] [Google Scholar]
- 25.Benkert P., Künzli M., Schwede T. QMEAN server for protein model quality estimation. Nucleic Acids Res. 2009;37:W510–W514. doi: 10.1093/nar/gkp322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Benkert P., Biasini M., Schwede T. Toward the estimation of the absolute quality of individual protein structure models. Bioinformatics. 2010;27:343–350. doi: 10.1093/bioinformatics/btq662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kolaskar A., Tongaonkar P.C. A semi-empirical method for prediction of antigenic determinants on protein antigens. FEBS Lett. 1990;276:172–174. doi: 10.1016/0014-5793(90)80535-q. [DOI] [PubMed] [Google Scholar]
- 28.Ponomarenko J., Bui H.H., Li W., Fusseder N., Bourne P.E., Sette A. ElliPro: a new structure-based tool for the prediction of antibody epitopes. BMC Bioinf. 2008;9:514. doi: 10.1186/1471-2105-9-514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Welling G.W., Weijer W.J., van der Zee R., Welling-Wester S. Prediction of sequential antigenic regions in proteins. FEBS Lett. 1985;188:215–218. doi: 10.1016/0014-5793(85)80374-4. [DOI] [PubMed] [Google Scholar]
- 30.Nielsen M., Andreatta M. NetMHCpan-3.0; improved prediction of binding to MHC class I molecules integrating information from multiple receptor and peptide length datasets. Genome Med. 2016;8:33. doi: 10.1186/s13073-016-0288-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Karosiene E., Rasmussen M., Blicher T., Lund O., Buus S., Nielsen M. NetMHCIIpan-3.0, a common pan-specific MHC class II prediction method including all three human MHC class II isotypes, HLA-DR, HLA-DP and HLA-DQ. Immunogenetics. 2013;65:711–724. doi: 10.1007/s00251-013-0720-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hoof I., Peters B., Sidney J., Pedersen L.E., Sette A., Lund O. NetMHCpan, a method for MHC class I binding prediction beyond humans. Immunogenetics. 2009;61:1–13. doi: 10.1007/s00251-008-0341-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Andreatta M., Nielsen M. Gapped sequence alignment using artificial neural networks: application to the MHC class I system. Bioinformatics. 2015;32:511–517. doi: 10.1093/bioinformatics/btv639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sidney J., Assarsson E., Moore C., Ngo S., Pinilla C., Sette A. Quantitative peptide binding motifs for 19 human and mouse MHC class I molecules derived using positional scanning combinatorial peptide libraries. Immunome Res. 2008;4:2. doi: 10.1186/1745-7580-4-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Peters B., Sette A. Generating quantitative models describing the sequence specificity of biological processes with the stabilized matrix method. BMC Bioinf. 2005;6:132. doi: 10.1186/1471-2105-6-132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Moutaftsi M., Peters B., Pasquetto V., Tscharke D.C., Sidney J., Bui H.H. A consensus epitope prediction approach identifies the breadth of murine TCD8 -cell responses to vaccinia virus. Nat Biotechnol. 2006;24:817–819. doi: 10.1038/nbt1215. [DOI] [PubMed] [Google Scholar]
- 37.Bui H.H., Sidney J., Dinh K., Southwood S., Newman M.J., Sette A. Predicting population coverage of T-cell epitope-based diagnostics and vaccines. BMC Bioinf. 2006;7:153. doi: 10.1186/1471-2105-7-153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Doolan D.L., Southwood S., Chesnut R., Appella E., Gomez E., Richards A. HLA-dr-promiscuous T cell epitopes from plasmodium falciparum pre-erythrocytic-stage antigens restricted by multiple HLA class II alleles. J Immunol. 2000;165:1123–1137. doi: 10.4049/jimmunol.165.2.1123. [DOI] [PubMed] [Google Scholar]
- 39.Altschul S. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Remmert M., Biegert A., Hauser A., Söding J. HHblits: lightning-fast iterative protein sequence searching by HMM-HMM alignment. Nat Methods. 2011;9:173–175. doi: 10.1038/nmeth.1818. [DOI] [PubMed] [Google Scholar]
- 41.Guex N., Peitsch M.C. SWISS-MODEL and the Swiss-Pdb Viewer: an environment for comparative protein modeling. Electrophoresis. 1997;18:2714–2723. doi: 10.1002/elps.1150181505. [DOI] [PubMed] [Google Scholar]
- 42.Laskowski R., Rullmann J., Macarthur M., Kaptein R., Thornton J. AQUA and PROCHECK-NMR: programs for checking the quality of protein structures solved by NMR. J Biomolec NMR. 1996;8:477–486. doi: 10.1007/BF00228148. [DOI] [PubMed] [Google Scholar]
- 43.Laskowski R.A., Macarthur M.W., Moss D.S., Thornton J.M. PROCHECK: a program to check the stereochemical quality of protein structures. J Appl Cryst. 1993;26:283–291. [Google Scholar]
- 44.Lüthy R., Bowie J.U., Eisenberg D. Assessment of protein models with three-dimensional profiles. Nature. 1992;356:83–85. doi: 10.1038/356083a0. [DOI] [PubMed] [Google Scholar]
- 45.Bowie J., Luthy R., Eisenberg D. A method to identify protein sequences that fold into a known three-dimensional structure. Science. 1991;253:164–170. doi: 10.1126/science.1853201. [DOI] [PubMed] [Google Scholar]
- 46.Lovell S.C., Davis I.W., Arendall W.B., 3rd, de Bakker P.I., Word J.M., Prisant M.G. Structure validation by Cα geometry: ϕ,ψ and Cβ deviation. Proteins. 2003;50:437–450. doi: 10.1002/prot.10286. [DOI] [PubMed] [Google Scholar]
- 47.Castrignanò T., De Meo P.D., Cozzetto D., Talamo I.G., Tramontano A. The PMDB protein model database. Nucleic Acids Res. 2006;34:D306–D309. doi: 10.1093/nar/gkj105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shimono M., Uenaka A., Noguchi Y., Sato S., Okumura H., Nakagawa K. Identification of DR9-restricted XAGE antigen on lung adenocarcinoma recognized by autologous CD4 T-cells. Int J Oncol. 2007;30:835–840. [PubMed] [Google Scholar]
- 49.Morishita Y., Uenaka A., Kaya S., Sato S., Aji T., Nakayama E. HLA-DRB1*0410-Restricted recognition of XAGE-1b37-48 peptide by CD4 T cells. Microbiol Immunol. 2007;51:755–762. doi: 10.1111/j.1348-0421.2007.tb03965.x. [DOI] [PubMed] [Google Scholar]
- 50.Chaves F.A., Lee A.H., Nayak J.L., Richards K.A., Sant A.J. The utility and limitations of Current web-available algorithms to predict peptides recognized by CD4 T cells in response to pathogen infection. J Immunol. 2012;188:4235–4248. doi: 10.4049/jimmunol.1103640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Aktas S., Samuel A. Identification of anigenic epitopes on the foot and mouth disease virus isolate O1/Manisa/Turkey/69 using monoclonal antibodies. Rev Sci Tech. 2000;19:744–753. doi: 10.20506/rst.19.3.1244. [DOI] [PubMed] [Google Scholar]
- 52.Bittle J.L., Houghten R.A., Alexander H., Shinnick T.M., Sutcliffe J.G., Lerner R.A. Protection against foot-and-mouth disease by immunization with a chemically synthesized peptide predicted from the viral nucleotide sequence. Nature. 1982;298:30–33. doi: 10.1038/298030a0. [DOI] [PubMed] [Google Scholar]
- 53.Kuroda D., Shirai H., Jacobson M.P., Nakamura H. Computer-aided antibody design. Protein Eng Des Sel. 2012;25:507–522. doi: 10.1093/protein/gzs024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hunter P. Into the fold. EMBO Rep. 2006;7:249–252. doi: 10.1038/sj.embor.7400655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Scharnagl N.C., Klade C.S. Experimental discovery of T-cell epitopes: combining the best of classical and contemporary approaches. Exp Review Vaccines. 2007;6:605–615. doi: 10.1586/14760584.6.4.605. [DOI] [PubMed] [Google Scholar]
- 56.Jawa V., Cousens L.P., Awwad M., Wakshull E., Kropshofer H., De Groot A.S. T-cell dependent immunogenicity of protein therapeutics: preclinical assessment and mitigation. Clin Immunol. 2013;149:534–555. doi: 10.1016/j.clim.2013.09.006. [DOI] [PubMed] [Google Scholar]
- 57.Swets J. Measuring the accuracy of diagnostic systems. Science. 1988;240:1285–1293. doi: 10.1126/science.3287615. [DOI] [PubMed] [Google Scholar]
- 58.Spackman K.A. Proc. Sixth Int. Conf. on Machine Learning. 1989. Signal detection theory: valuable tools for evaluating inductive learning; pp. 160–163. [Google Scholar]
- 59.Fawcett T. An introduction to ROC analysis. Pattern Recognit Lett. 2006;27:861–874. [Google Scholar]
- 60.Liang S., Zheng D., Standley D.M., Yao B., Zacharias M., Zhang C. EPSVR and EPMeta: prediction of antigenic epitopes using support vector regression and multiple server results. BMC Bioinf. 2010;11:381. doi: 10.1186/1471-2105-11-381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Borley D.W., Mahapatra M., Paton D.J., Esnouf R.M., Stuart D.I., Fry E.E. Evaluation and use of in-silico structure-based epitope prediction with foot-and-mouth disease virus. PLoS One. 2013;8 doi: 10.1371/journal.pone.0061122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Deres K., Schumacher T.N., Wiesmuller K.H., Stevanovic S., Greiner G., Jung G. Preferred size of peptides that bind to H-2 Kb is sequence dependent. Eur J Immunol. 1992;22:1603–1608. doi: 10.1002/eji.1830220638. [DOI] [PubMed] [Google Scholar]
- 63.Kosmrlj A., Jha A.K., Huseby E.S., Kardar M., Chakraborty A.K. How the thymus designs antigen-specific and self-tolerant T cell receptor sequences. Proc Natl Acad Sci U S A. 2008;105:16671–16676. doi: 10.1073/pnas.0808081105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lundegaard C., Lund O., Nielsen M. Predictions versus high-throughput experiments in T-cell epitope discovery: competition or synergy? Exp Review Vaccines. 2012;11:43–54. doi: 10.1586/erv.11.160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Stevanovic S. Identification of tumour-associated t-cell epitopes for vaccine development. Nat Rev Cancer. 2002;2 doi: 10.1038/nrc841. 514–514. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Tables representing computationally predicted IC50 values of Allele restricted XAGE-1b peptide epitopes that were computationally predicted in this study compared to previously identified in experimental studies.
Kolaskar and Tongaonkar's plot of XAGE-1b. Showing the immunogenic propensity of XAGE-1b residues with Average immunogenic score of 0.912. Area displayed above the threshold is considered as promising B-Cell epitopes against XAGE-1b Antigen.
Emini surface accessibility plot of XAGE-1b. Showing accessible propensity with Average antigenic score of 1. Area displayed above the threshold cutoff is reflecting promising B-Cell accessible segments.
Karplus and Schulz flexibility plot of XAGE-1b. Showing flexible propensity of XAGE-1b residues with Average antigenic score of 1.013. Area displayed above the threshold cutoff is reflecting promising flexible segments.
Nonamer peptides core sequences of XAGE-1b which are HLA-I predicted epitopes, ranked according to alleles' coverage.
Nonamer peptides core sequences in XAGE-1b which are HLA-II predicted epitopes with NetMHCIIpan 3.0 server, ranked according to alleles' coverage.