Abstract
The physiological role of autophagy in the catabolic process of the body involves protein synthesis and degradation in homeostasis under normal and stressed conditions. In hepatocellular carcinoma (HCC), the role of tumor microenvironment (TME) has been concerned as the main issue in fighting against this deadly malignancy. During the last decade, the crosstalk between tumor cells and their TME in HCC extensively accumulated. However, a deeper knowledge for the actual function of autophagy in this interconnection which involved in supporting tumor development, progression and chemoresistance in HCC is needed but still largely unknown. Recent studies have shown that coagulants tissue factor (TF) and factor VII (FVII) has a pathological role in promoting tumor growth by activating protease-activated receptor 2 (PAR2). Autophagy-associated LC3A/B-II formation was selectively suppressed by FVII/PAR2 signaling which mediated by mTOR activation through Atg7 but not Atg5/Atg12 axis. The coagulant-derived autophagic suppression seemed potentiate a vicious circle of malignancy in producing more FVII and PAR2 which facilitate in vivo and in vitro tumor progression of HCC and the investigations are consistent with the clinical observations. In this review, we briefly summarize the current understanding of autophagy and discuss recent evidence for its role in HCC malignancy.
Keywords: Autophagy, Coagulation, Factor VII, Hepatitis B virus, Hepatocellular carcinoma, Microenvironment
Autophagy constitutes some of the most basic reactions in which cells sequesters part of their own cytosolic components and organelles into membrane-bound vesicles for degradation and, upon energy deprivation, convert the protein and lipid contents into life-preserving fuel while delivering to lysosomes. In addition, autophagy also participates in removal of malfolded protein aggregates and dysfunctional organelles under stressed conditions, whereas its cargo contents are ultimately broken down and recycled back for supporting metabolic processes [1], [2]. Autophagy process is evolutionally conserved which have been categorized into distinct steps with more than 30 autophagy-related proteins (ATG) involved [3], [4], [5]. This “self-eating” process is induced by a variety of intracellular and extracellular stimuli, such as oxidative stress, assorted pathogens cytokine stimulation, damaged organelles and accumulated protein aggregates especially in low-nutrient environments [6], [7].
The mammalian target of rapamycin (mTOR), a key molecule lies at the heart of nutrient-sensing cascades which has been identified as a main regulator of autophagy activity and plays a pivotal role in coordination of cell growth and progression in various cancers [8], [9]. Solid tumors generally experience both nutrient deprivation and hypoxia while lacking vascularization, and induce a proliferation response in cancer cells [10], [11], [12] in which mTOR acts as a restriction point regulator between proliferation and differentiation in response to their environmental change [13]. Thus, cancer cells are optimized for growth and survival through the ability to surpass metabolic hurdle, thereby alter cell metabolism toward anabolic conditions, anaerobic glycolysis and acidosis to meet their advantageous demands [14], [15].
However, hepatocellular carcinoma (HCC), the third leading cause of death from cancer worldwide is a highly vascularized solid tumor with a rapid annual growth rate to be diagnosed and poor prognosis. Although the incidence of HCC decreased to the third from the second leading cancer causes since 2012, HCC patients are still facing the high risk of recurrence and mortality in Taiwan [16]. In one center experience of Kaohsiung Chang Gung Memorial Hospital, most cases of HCC are due to liver disorders with chronic viral infection (hepatitis B, C and B + C, more than 90%). Furthermore, hospital-based analysis for identifying prognostic factors of HCC was earlier conducted. In a large retrospective cohort of 6381 HCC cases diagnosed from 1986 to 2002 were enrolled and the independent factors influencing survival were revealed by multivariate analysis. Besides those well-known prognostic factors, such as alpha-fetoprotein, HBV surface antigen positivity, degree of liver function impairment and tumor status, relative high platelet counts were identified as a poor prognostic factor [17]. Hence, the special etiologies identified in our clinical observations imply critical tumor microenvironments may be involved in the malignant progression of HCC in this HBV endemic area.
Autophagy and HBV
Chronic HBV infection has been epidemiologically associated to the development of HCC for almost half a century and HBV X protein (HBx) has been found to play critical roles in this hepatocellular carcinogenesis [18], [19], however the underlying mechanisms by which infection to drive HCC malignancy are still largely unclear. HBx subverts a variety of cellular activities such as transcription, autophagy and proliferation by interacting with transcription factors without direct DNA binding [20], [21]. Autophagy can be triggered by HBx directly through up-regulation of autophagic protein expression, or indirectly through activating class III phosphatidylinositol 3-kinase (PtdIns3K) and ER stress response [22], [23], [24]. The relevance of HBx-promoted autophagy has been suggested to promote viral replication, however the mechanisms to affect the cytoplasmic modulation of signal transduction pathways by autophagy are still controversial [22], [23], [25]. Interestingly, a different group has shown that autophagy-deficient mice with systemic mosaic deletion of Atg5 or liver-specific loss of Atg7 develop multiple tumors [26]. A recent study also demonstrated HBx inhibits autophagy degradation by interfering lysosomal maturation, although the number of autophagosomes was increased in tumor cells. These findings indicates an important suppressive role of autophagy in tumorigenesis of HCC, and that HBx restrained autophagic flux leading to the accumulation of autophagosomes which might contribute to further malignancy.
Coagulant microenvironment in HCC
Tumor-associated inflammation especially procoagulant-driven inflammation is now widely recognized as one of key determinants of various type of cancer [27], [28], [29], [30], including HCC [31], [32], [33]. The transmembrane tissue factor (TF) triggers downstream signaling upon binding with blood coagulation factor VII (FVII) and transmitting signals through protease-activated receptors (PARs) activation, especially PAR2 [34], [35], [36]. In general, a hypoxic microenvironment involves in tumor progression which includes local invasion, distant metastasis and therapeutic resistance [37]. Both TF and FVII are found to be induced in response to hypoxia, and known to initiate key pathogenesis in cancer [38], however their regulatory mechanisms are distinct in ovarian cancer cells [30], [39], [40], [41], [42]. In addition to ovarian cancer, it has also been demonstrated by immunohistochemical analyses that TF is high expressed in breast cancer and pancreatic cancer tissues [43], [44], [45]. Although numerous studies have suggested that TF-fVIIa complex formation on the cell surface contributes to the malignant phenotypes of cancer, TF expression varies among different types of malignancies; some may be more pro-thrombotic than others. Inhibition using specific antibodies or peptide inhibitors concludes that blockade of the FVII/TF/PAR2 signaling independent of the coagulation response can suppress cancer progression [25], [29]. The detailed regulatory mechanisms are not clear; however transcriptional activation appears to be a major mechanism of TF over-expression [30]. Therefore, therapeutic strategies targeting TF has been considered to be advantageous to cancer progression, although the possible impairment of the homeostasis of coagulant physiology should be considered.
Mechanisms of coagulant initiation in HCC are not completed revealed. Interestingly, extremely low level of TF mRNA is expressed in liver compared to other tissues, however the physiological reason is also not clear [46], [47]. Angiogenesis has well known to be an important factor in the development, progression and recurrence of HCC and its targeting has been vigorously studied for potential therapeutic strategies [48], [49], [50]. Poon et al. have evaluated the correlation between TF expression with tumor angiogenesis and invasiveness in HCC which was the first suggesting a significant association of TF levels with microvascular density, venous invasion, microsatellite nodules, tumor staging, and survival [51]. A recent study also demonstrates that circular and local regional TF is overexpressed in HCC patients, and it is closely related to the invasive and metastasis indexes [52]. Our recent studies have demonstrated that activation of the TF/FVII/PAR2 axis is associated with increased migration and invasiveness in hepatoma cells in vitro [32], [33], [52]. This finding could be consistently observed in tumor tissues from HCC patients, where we found that increased FVII and PAR2 levels were significantly associated with clinical stage, increased invasion, and poor disease-free survival, however none of any significant association with TF expression. It is noteworthy that the signals driving FVII/PAR2 stimulation of cell migration are mainly via ERK-TSC, independent of other coagulation effectors such as thrombin/PAR1. Furthermore, only FVII, but not soluble TF, activates ERK1/2 via PAR2 signaling in our cellular model which indicates that FVII/PAR2 signaling could be regulated by other mechanisms distinct from TF activation. Additionally, both activated FVII and the PAR2 agonist drastically induce FVII and PAR2 expression whereas no significant effects on TF expression were observed. Hu et al. have shown that MAPK/ERK signals inhibit PI3K/Akt-mediated TF expression in breast cancer and ovarian cancer cells [53]. Their findings indicate opposite regulation by PI3K/Akt and MAPK/ERK pathways of tissue factor expression existing in tumor-bearing coagulant microenvironment. Therefore, the amount of activated FVII would determine the ratio that is engaged in activating PAR2 for tumor progression. Although TF expression is closely related to the aggressiveness of several cancers, our results are more consistent with the findings from Rullier et al., in which no association between TF levels and clinicopathological characteristics of HCC was observed [54]. Some studies have also shown that TF contributes minimally to tumor growth [55], [56], [57]. Therefore, the role of TF in cancer progression may be essential in some but not all cancers, and TF may not be a reliable prognostic marker at least for HCC progression in some local geographic variations of viral etiology. In addition, animal model using mouse xenografts can well reflect our clinical findings, where we find that FVIIa administration only positively affects vascular density, not the size and number of inoculated tumors. The results are consistent with the expression of FVII in liver tissue of patients with HCC, which is related to vascular invasion and envelope capsulations, but not to the number and size of tumors.
Coagulants and autophagy in HCC
In our clinical investigation for HCC, we were the first to demonstrate that both FVII and PAR2 expressions were inversely correlated with the amount of autophagic proteins LC3A/B-II [31]. We also showed that TF, FVII and PAR2 agonist decreased expression of LC3A/B-II proteins in cultured Hep3B cells suggesting a crucial impact of the TF/FVII/PAR2 coagulation pathway on autophagic suppression. The dependence of mTOR activation on thrombosis has been seen in various pathological conditions, in which the risk of thrombotic events while patients were receiving organ transplants [58], [59], [60], [61], [62], [63]and coronary stents [64]was associated with TF expression. In our study, TF, FVII as well as the PAR2 peptide agonist, rather than thrombin and PAR1 agonist, induced mTOR activation and expression whereas silencing of TF, FVII or PAR2 by siRNAs repressed its phosphorylation and expression in hepatoma cell lines. Furthermore, suppressed LC3A/B-II expression levels by activation of TF, FVII and PAR2 were fully rescued by mTOR knockdown or treatment of mTOR inhibitors in vitro. Our results illustrated the mTOR dependence incoagulant-mediated repression of autophagy. However, mTOR targeting may result in differential outcomes. Administration of metformin, which negatively regulates mTOR by activating adenosine monophosphate kinases (AMPK)has been found to elicit a contradictory effect, suggesting that mTOR blockade does not always lead to autophagy [65].
The miRNA-mediated autophagic repression in HCC
Numerous studies have shown that aberrant miRNA expression can be used as a potential diagnostic or prognostic tool for malignant tumors including HCC because tumor-derived miRNAs circulate in relatively stable form [66], [67]. For instance, miR-122, miR-223 and miR-199a/b-3p are those of the most highly expressed miRNAs in the liver, were found to be down-regulated in HCC [68], [69]. In addition, miRNAs act as oncogenes including miR-221, miR-130b and miR-494 have been shown to regulate HCC growth and invasion [70], [71], [72] and emerging evidences suggest that miRNAs could mediate the survival, progression, invasiveness and chemoresistance of hepatic cancer stem cells [73], [74], [75]. Thus, recent studies have focused upon investigating critical mechanisms regulated by miRNAs expression underlying the “loss of stemness” to characterize epithelial mesenchymal transition (EMT) and serving as potent biomarkers or therapeutic tools to control neoplastic behaviors of HCC [76].
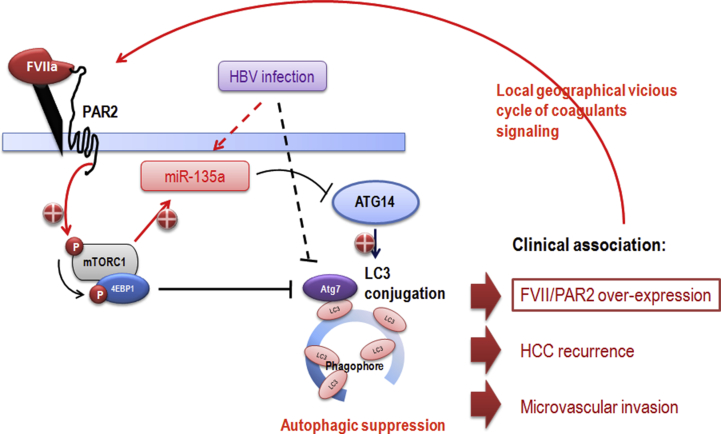
For the regulation of miRNA homeostasis, it has been discovered that the miRNA processing components DICER1 and EIF2C are targeted by selective autophagy which establishes a “checkpoint” for maintenance of miRNA abundance and their proper activity [77], [78], [79]. This finding suggests pathological inter-dependence of dysregulated autophagy with defects in miRNA expression. Nonetheless, miRNAs that modulate autophagy and their involvement in HCC progression have not been much addressed. A recent study has identified that miR-7 is down-regulated and plays an onco-suppressor role by direct targeting mTOR in HCC [80]. In this study, autophagic suppression enhances the growth inhibition effect of miR-7 in HepG2 cells suggesting that modulation of autophagy could be practical approach to promoting onco-suppressive activity of miR-7. Our current study identified miR-135a as a highly expressed miRNA of tumor tissues in association with higher FVII/PAR2 expression and in response to FVII/PAR2 activation [81]. Interestingly, increased miR-135a levels in HCC are significantly associated with viral etiology, hepatitis B surface antigen (HBsAg), tumor staging, recurrence, microvascular invasion as well as decreased disease-free survival. We also identified a key autophagic component ATG14 which orchestrates the autophagy process by regulating autophagosome formation is direct targeted by miR-135a. Over-expression of miR-135a resulted in an apparent decrease in Atg14 protein, and its downstream LC3A/B was also decreased in Hep3B cells. We also demonstrate a strong correlation between FVII, miR-135a and Atg14 levels in 103 clinical cases of HCC. This study is also the first to reveal the association among coagulant-mediated autophagic suppression, viral etiology, microvascular invasion and tumor malignancies in a translational approach of HCC. It implies that the suppressive role in autophagy by miR-135a could be crucial for tumor progression within a local geographical microenvironment affected by both viral infection and circulating factors such as FVII in the liver sinusoids. The regional autophagic suppression might induce a vicious cycle in producing more FVII/PAR2/miR-135a molecules and might be an earlier molecular event for tumor invasion of HCC [Fig. 1]. Detailed investigation for the vast of other differentially expressed miRNAs affected by the local cellular context of coagulation is required to be carefully examined for potential future diagnostic and therapeutic purposes.
Fig. 1.
Schematic representation shows that autophagic suppression may be mediated by mTOR-dependent coagulation signaling via direct ATG7 inhibition, miR-135a-targeting of ATG14 under specific viral etiology, which results in geographical vicious cycle of coagulant accumulation and facilitate malignancies of HCC.
Conflicts of interest
The authors have no conflicts of interest to declare regarding this manuscript.
Acknowledgements
This work was supported by the grants from Chang Gung Memorial Hospital [CMRPG8D0561, CMRPG8D0751, CMRPG8D1022 and CMRPG8D1023 to KDC; CMRPG8A1203 and CMRPG8E1651 to CCL; CMRPG8F0661 and CMRPG8F0662 to MCT; CMRPG8C1151, CMRPG8D1032 and CMRPG8D1033 to KTH; CMRPG8D1001 and CMRPG8F0621 to Chiu KW].
Footnotes
Peer review under responsibility of Chang Gung University.
Supplementary data related to this article can be found at https://doi.org/10.1016/j.bj.2018.03.002.
Appendix A. Supplementary data
The following is the supplementary data related to this article:
References
- 1.Baehrecke E.H. Autophagy: dual roles in life and death? Nat Rev Mol Cell Biol. 2005;6:505–510. doi: 10.1038/nrm1666. [DOI] [PubMed] [Google Scholar]
- 2.Komatsu M., Waguri S., Ueno T., Iwata J., Murata S., Tanida I. Impairment of starvation-induced and constitutive autophagy in Atg7-deficient mice. J Cell Biol. 2005;169:425–434. doi: 10.1083/jcb.200412022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nakatogawa H., Suzuki K., Kamada Y., Ohsumi Y. Dynamics and diversity in autophagy mechanisms: lessons from yeast. Nat Rev Mol Cell Biol. 2009;10:458–467. doi: 10.1038/nrm2708. [DOI] [PubMed] [Google Scholar]
- 4.Feng Y., He D., Yao Z., Klionsky D.J. The machinery of macroautophagy. Cell Res. 2014;24:24–41. doi: 10.1038/cr.2013.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Klionsky D.J., Emr S.D. Autophagy as a regulated pathway of cellular degradation. Science. 2000;290:1717–1721. doi: 10.1126/science.290.5497.1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rautou P.E., Mansouri A., Lebrec D., Durand F., Valla D., Moreau R. Autophagy in liver diseases. J Hepatol. 2010;53:1123–1134. doi: 10.1016/j.jhep.2010.07.006. [DOI] [PubMed] [Google Scholar]
- 7.Czaja M.J. Functions of autophagy in hepatic and pancreatic physiology and disease. Gastroenterology. 2011;140:1895–1908. doi: 10.1053/j.gastro.2011.04.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Beauchamp E.M., Platanias L.C. The evolution of the TOR pathway and its role in cancer. Oncogene. 2013;32:3923–3932. doi: 10.1038/onc.2012.567. [DOI] [PubMed] [Google Scholar]
- 9.Dunlop E.A., Tee A.R. mTOR and autophagy: a dynamic relationship governed by nutrients and energy. Sem Cell Dev Biol. 2014;36:121–129. doi: 10.1016/j.semcdb.2014.08.006. [DOI] [PubMed] [Google Scholar]
- 10.Howell J.J., Manning B.D. mTOR couples cellular nutrient sensing to organismal metabolic homeostasis. Trends Endocrinol Metab. 2011;22:94–102. doi: 10.1016/j.tem.2010.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dang C.V. Links between metabolism and cancer. Genes Dev. 2012;26:877–890. doi: 10.1101/gad.189365.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vander Heiden M.G. Targeting cancer metabolism: a therapeutic window opens. Nat Rev Drug Discov. 2011;10:671–684. doi: 10.1038/nrd3504. [DOI] [PubMed] [Google Scholar]
- 13.Love N.K., Keshavan N., Lewis R., Harris W.A., Agathocleous M. A nutrient-sensitive restriction point is active during retinal progenitor cell differentiation. Development. 2014;141:697–706. doi: 10.1242/dev.103978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vander Heiden M.G., Cantley L.C., Thompson C.B. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. 2009;324:1029–1033. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Agathocleous M., Love N.K., Randlett O., Harris J.J., Liu J., Murray A.J. Metabolic differentiation in the embryonic retina. Nat Cell Biol. 2012;14:859–864. doi: 10.1038/ncb2531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lin C.C., Chen C.L. Living donor liver transplantation for hepatocellular carcinoma achieves better outcomes. Hepatobiliary Surg Nutr. 2016;5:415–421. doi: 10.21037/hbsn.2016.08.02. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Changchien C.S., Chen C.L., Yen Y.H., Wang J.H., Hu T.H., Lee C.M. Analysis of 6381 hepatocellular carcinoma patients in southern Taiwan: prognostic features, treatment outcome, and survival. J Gastroenterol. 2008;43:159–170. doi: 10.1007/s00535-007-2134-9. [DOI] [PubMed] [Google Scholar]
- 18.Tang H., Oishi N., Kaneko S., Murakami S. Molecular functions and biological roles of hepatitis B virus x protein. Cancer Sci. 2006;97:977–983. doi: 10.1111/j.1349-7006.2006.00299.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang T., Zhang J., You X., Liu Q., Du Y., Gao Y. Hepatitis B virus X protein modulates oncogene yes-associated protein by CREB to promote growth of hepatoma cells. Hepatology. 2012;56:2051–2059. doi: 10.1002/hep.25899. [DOI] [PubMed] [Google Scholar]
- 20.Ng S.A., Lee C. Hepatitis B virus X gene and hepatocarcinogenesis. J Gastroenterol. 2011;46:974–990. doi: 10.1007/s00535-011-0415-9. [DOI] [PubMed] [Google Scholar]
- 21.Zhang X., Zhang H., Ye L. Effects of hepatitis B virus X protein on the development of liver cancer. J Lab Clin Med. 2006;147:58–66. doi: 10.1016/j.lab.2005.10.003. [DOI] [PubMed] [Google Scholar]
- 22.Li J., Liu Y., Wang Z., Liu K., Wang Y., Liu J. Subversion of cellular autophagy machinery by hepatitis B virus for viral envelopment. J Virol. 2011;85:6319–6333. doi: 10.1128/JVI.02627-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sir D., Tian Y., Chen W.L., Ann D.K., Yen T.S., Ou J.H. The early autophagic pathway is activated by hepatitis B virus and required for viral DNA replication. Proc Natl Acad Sci U S A. 2010;107:4383–4388. doi: 10.1073/pnas.0911373107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tang H., Da L., Mao Y., Li Y., Li D., Xu Z. Hepatitis B virus X protein sensitizes cells to starvation-induced autophagy via up-regulation of beclin 1 expression. Hepatology. 2009;49:60–71. doi: 10.1002/hep.22581. [DOI] [PubMed] [Google Scholar]
- 25.Tang S.W., Ducroux A., Jeang K.T., Neuveut C. Impact of cellular autophagy on viruses: insights from hepatitis B virus and human retroviruses. J Biomedical Sci. 2012;19:92. doi: 10.1186/1423-0127-19-92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Takamura A., Komatsu M., Hara T., Sakamoto A., Kishi C., Waguri S. Autophagy-deficient mice develop multiple liver tumors. Genes Dev. 2011;25:795–800. doi: 10.1101/gad.2016211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Elinav E., Nowarski R., Thaiss C.A., Hu B., Jin C., Flavell R.A. Inflammation-induced cancer: crosstalk between tumours, immune cells and microorganisms. Nat Rev Cancer. 2013;13:759–771. doi: 10.1038/nrc3611. [DOI] [PubMed] [Google Scholar]
- 28.Crusz S.M., Balkwill F.R. Inflammation and cancer: advances and new agents. Nat Rev Clin Oncol. 2015;12:584–596. doi: 10.1038/nrclinonc.2015.105. [DOI] [PubMed] [Google Scholar]
- 29.Balkwill F.R., Mantovani A. Cancer-related inflammation: common themes and therapeutic opportunities. Semin Cancer Biol. 2012;22:33–40. doi: 10.1016/j.semcancer.2011.12.005. [DOI] [PubMed] [Google Scholar]
- 30.Koizume S., Miyagi Y. Potential coagulation factor-driven pro-inflammatory responses in ovarian cancer tissues associated with insufficient O(2) and plasma supply. Int J Mol Sci. 2017;18 doi: 10.3390/ijms18040809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen K.D., Wang C.C., Tsai M.C., Wu C.H., Yang H.J., Chen L.Y. Interconnections between autophagy and the coagulation cascade in hepatocellular carcinoma. Cell Death Dis. 2014;5:e1244. doi: 10.1038/cddis.2014.212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen K.D., Huang K.T., Tsai M.C., Wu C.H., Kuo I.Y., Chen L.Y. Coagulation factor VII and malignant progression of hepatocellular carcinoma. Cell Death Dis. 2016;7:e2110. doi: 10.1038/cddis.2015.395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tsai M.C., Chen K.D., Wang C.C., Huang K.T., Wu C.H., Kuo I.Y. Factor VII promotes hepatocellular carcinoma progression through ERK-TSC signaling. Cell Death Discov. 2015;1:15051. doi: 10.1038/cddiscovery.2015.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Camerer E., Huang W., Coughlin S.R. Tissue factor- and factor X-dependent activation of protease-activated receptor 2 by factor VIIa. Proc Natl Acad Sci U S A. 2000;97:5255–5260. doi: 10.1073/pnas.97.10.5255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mackman N. Role of tissue factor in hemostasis, thrombosis, and vascular development. Arterioscler Thromb Vasc Biol. 2004;24:1015–1022. doi: 10.1161/01.ATV.0000130465.23430.74. [DOI] [PubMed] [Google Scholar]
- 36.Mann K.G. Biochemistry and physiology of blood coagulation. Thromb Haemost. 1999;82:165–174. [PubMed] [Google Scholar]
- 37.Chang Q., Jurisica I., Do T., Hedley D.W. Hypoxia predicts aggressive growth and spontaneous metastasis formation from orthotopically grown primary xenografts of human pancreatic cancer. Cancer Res. 2011;71:3110–3120. doi: 10.1158/0008-5472.CAN-10-4049. [DOI] [PubMed] [Google Scholar]
- 38.Schaffner F., Ruf W. Tissue factor and PAR2 signaling in the tumor microenvironment. Arterioscler Thromb Vasc Biol. 2009;29:1999–2004. doi: 10.1161/ATVBAHA.108.177428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yokota N., Koizume S., Miyagi E., Hirahara F., Nakamura Y., Kikuchi K. Self-production of tissue factor-coagulation factor VII complex by ovarian cancer cells. Br J Cancer. 2009;101:2023–2029. doi: 10.1038/sj.bjc.6605406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Koizume S., Ito S., Yoshioka Y., Kanayama T., Nakamura Y., Yoshihara M. High-level secretion of tissue factor-rich extracellular vesicles from ovarian cancer cells mediated by filamin-A and protease-activated receptors. Thromb Haemost. 2016;115:299–310. doi: 10.1160/TH15-03-0213. [DOI] [PubMed] [Google Scholar]
- 41.Koizume S., Jin M.S., Miyagi E., Hirahara F., Nakamura Y., Piao J.H. Activation of cancer cell migration and invasion by ectopic synthesis of coagulation factor VII. Cancer Res. 2006;66:9453–9460. doi: 10.1158/0008-5472.CAN-06-1803. [DOI] [PubMed] [Google Scholar]
- 42.Koizume S., Ito S., Miyagi E., Hirahara F., Nakamura Y., Sakuma Y. HIF2alpha-Sp1 interaction mediates a deacetylation-dependent FVII-gene activation under hypoxic conditions in ovarian cancer cells. Nucleic Acids Res. 2012;40:5389–5401. doi: 10.1093/nar/gks201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Iodice S., Gandini S., Lohr M., Lowenfels A.B., Maisonneuve P. Venous thromboembolic events and organ-specific occult cancers: a review and meta-analysis. J Thromb Haemost. 2008;6:781–788. doi: 10.1111/j.1538-7836.2008.02928.x. [DOI] [PubMed] [Google Scholar]
- 44.Ueno T., Toi M., Koike M., Nakamura S., Tominaga T. Tissue factor expression in breast cancer tissues: its correlation with prognosis and plasma concentration. Br J Cancer. 2000;83:164–170. doi: 10.1054/bjoc.2000.1272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Contrino J., Hair G., Kreutzer D.L., Rickles F.R. In situ detection of tissue factor in vascular endothelial cells: correlation with the malignant phenotype of human breast disease. Nat Med. 1996;2:209–215. doi: 10.1038/nm0296-209. [DOI] [PubMed] [Google Scholar]
- 46.Mackman N., Sawdey M.S., Keeton M.R., Loskutoff D.J. Murine tissue factor gene expression in vivo. Tissue and cell specificity and regulation by lipopolysaccharide. Am J Pathol. 1993;143:76–84. [PMC free article] [PubMed] [Google Scholar]
- 47.Parry G.C., Erlich J.H., Carmeliet P., Luther T., Mackman N. Low levels of tissue factor are compatible with development and hemostasis in mice. J Clin Invest. 1998;101:560–569. doi: 10.1172/JCI814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Coulon S., Heindryckx F., Geerts A., Van Steenkiste C., Colle I., Van Vlierberghe H. Angiogenesis in chronic liver disease and its complications. Liver Int. 2011;31:146–162. doi: 10.1111/j.1478-3231.2010.02369.x. [DOI] [PubMed] [Google Scholar]
- 49.Sugimachi K., Tanaka S., Terashi T., Taguchi K., Rikimaru T. The mechanisms of angiogenesis in hepatocellular carcinoma: angiogenic switch during tumor progression. Surgery. 2002;131:S135–S141. doi: 10.1067/msy.2002.119365. [DOI] [PubMed] [Google Scholar]
- 50.Wu X.Z. New strategy of antiangiogenic therapy for hepatocellular carcinoma. Neoplasma. 2008;55:472–481. [PubMed] [Google Scholar]
- 51.Poon R.T., Lau C.P., Ho J.W., Yu W.C., Fan S.T., Wong J. Tissue factor expression correlates with tumor angiogenesis and invasiveness in human hepatocellular carcinoma. Clin Cancer Res. 2003;9:5339–5345. [PubMed] [Google Scholar]
- 52.Zhou Q., Huang T., Wang Y.F., Zhou X.B., Liang L.J., Peng B.G. Role of tissue factor in hepatocellular carcinoma genesis, invasion and metastasis. Chin Med J. 2011;124:3746–3751. [PubMed] [Google Scholar]
- 53.Hu C., Huang L., Gest C., Xi X., Janin A., Soria C. Opposite regulation by PI3K/Akt and MAPK/ERK pathways of tissue factor expression, cell-associated procoagulant activity and invasiveness in MDA-MB-231 cells. J Hematol Oncol. 2012;5:16. doi: 10.1186/1756-8722-5-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rullier A., Senant N., Kisiel W., Bioulac-Sage P., Balabaud C., Le Bail B. Expression of protease-activated receptors and tissue factor in human liver. Virchows Arch. 2006;448:46–51. doi: 10.1007/s00428-005-0078-0. [DOI] [PubMed] [Google Scholar]
- 55.Bromberg M.E., Sundaram R., Homer R.J., Garen A., Konigsberg W.H. Role of tissue factor in metastasis: functions of the cytoplasmic and extracellular domains of the molecule. Thromb Haemost. 1999;82:88–92. [PubMed] [Google Scholar]
- 56.Palumbo J.S., Talmage K.E., Massari J.V., La Jeunesse C.M., Flick M.J., Kombrinck K.W. Tumor cell-associated tissue factor and circulating hemostatic factors cooperate to increase metastatic potential through natural killer cell-dependent and-independent mechanisms. Blood. 2007;110:133–141. doi: 10.1182/blood-2007-01-065995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Toomey J.R., Kratzer K.E., Lasky N.M., Broze G.J., Jr. Effect of tissue factor deficiency on mouse and tumor development. Proc Natl Acad Sci U S A. 1997;94:6922–6926. doi: 10.1073/pnas.94.13.6922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Guba M., Yezhelyev M., Eichhorn M.E., Schmid G., Ischenko I., Papyan A. Rapamycin induces tumor-specific thrombosis via tissue factor in the presence of VEGF. Blood. 2005;105:4463–4469. doi: 10.1182/blood-2004-09-3540. [DOI] [PubMed] [Google Scholar]
- 59.Trotter J.F. Sirolimus in liver transplantation. Transplant Proc. 2003;35:193S–200S. doi: 10.1016/s0041-1345(03)00234-3. [DOI] [PubMed] [Google Scholar]
- 60.Fortin M.C., Raymond M.A., Madore F., Fugere J.A., Paquet M., St-Louis G. Increased risk of thrombotic microangiopathy in patients receiving a cyclosporin-sirolimus combination. Am J Transplant. 2004;4:946–952. doi: 10.1111/j.1600-6143.2004.00428.x. [DOI] [PubMed] [Google Scholar]
- 61.Paramesh A.S., Grosskreutz C., Florman S.S., Gondolesi G.E., Sharma S., Kaufman S.S. Thrombotic microangiopathy associated with combined sirolimus and tacrolimus immunosuppression after intestinal transplantation. Transplantation. 2004;77:129–131. doi: 10.1097/01.TP.0000092522.36410.D0. [DOI] [PubMed] [Google Scholar]
- 62.Barone G.W., Gurley B.J., Abul-Ezz S.R., Gokden N. Sirolimus-induced thrombotic microangiopathy in a renal transplant recipient. J Kidney Dis. 2003;42:202–206. doi: 10.1016/s0272-6386(03)00424-4. [DOI] [PubMed] [Google Scholar]
- 63.Robson M., Cote I., Abbs I., Koffman G., Goldsmith D. Thrombotic micro-angiopathy with sirolimus-based immunosuppression: potentiation of calcineurin-inhibitor-induced endothelial damage? Am J Transplant. 2003;3:324–327. doi: 10.1034/j.1600-6143.2003.00051.x. [DOI] [PubMed] [Google Scholar]
- 64.Choi S.B. CYPHER coronary stents and risk of thrombosis. CMAJ. 2003;169:218. [PMC free article] [PubMed] [Google Scholar]
- 65.Ben Sahra I., Laurent K., Giuliano S., Larbret F., Ponzio G., Gounon P. Targeting cancer cell metabolism: the combination of metformin and 2-deoxyglucose induces p53-dependent apoptosis in prostate cancer cells. Cancer Res. 2010;70:2465–2475. doi: 10.1158/0008-5472.CAN-09-2782. [DOI] [PubMed] [Google Scholar]
- 66.Chen X., Ba Y., Ma L., Cai X., Yin Y., Wang K. Characterization of microRNAs in serum: a novel class of biomarkers for diagnosis of cancer and other diseases. Cell Res. 2008;18:997–1006. doi: 10.1038/cr.2008.282. [DOI] [PubMed] [Google Scholar]
- 67.Mitchell P.S., Parkin R.K., Kroh E.M., Fritz B.R., Wyman S.K., Pogosova-Agadjanyan E.L. Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci U S A. 2008;105:10513–10518. doi: 10.1073/pnas.0804549105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hou J., Lin L., Zhou W., Wang Z., Ding G., Dong Q. Identification of miRNomes in human liver and hepatocellular carcinoma reveals miR-199a/b-3p as therapeutic target for hepatocellular carcinoma. Cancer Cell. 2011;19:232–243. doi: 10.1016/j.ccr.2011.01.001. [DOI] [PubMed] [Google Scholar]
- 69.Qi P., Cheng S.Q., Wang H., Li N., Chen Y.F., Gao C.F. Serum microRNAs as biomarkers for hepatocellular carcinoma in Chinese patients with chronic hepatitis B virus infection. PLoS One. 2011;6:e28486. doi: 10.1371/journal.pone.0028486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Chuang K.H., Whitney-Miller C.L., Chu C.Y., Zhou Z., Dokus M.K., Schmit S. MicroRNA-494 is a master epigenetic regulator of multiple invasion-suppressor microRNAs by targeting ten eleven translocation 1 in invasive human hepatocellular carcinoma tumors. Hepatology. 2015;62:466–480. doi: 10.1002/hep.27816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ma S., Tang K.H., Chan Y.P., Lee T.K., Kwan P.S., Castilho A. miR-130b Promotes CD133(+) liver tumor-initiating cell growth and self-renewal via tumor protein 53-induced nuclear protein 1. Cell Stem Cell. 2010;7:694–707. doi: 10.1016/j.stem.2010.11.010. [DOI] [PubMed] [Google Scholar]
- 72.Yuan Q., Loya K., Rani B., Mobus S., Balakrishnan A., Lamle J. MicroRNA-221 overexpression accelerates hepatocyte proliferation during liver regeneration. Hepatology. 2013;57:299–310. doi: 10.1002/hep.25984. [DOI] [PubMed] [Google Scholar]
- 73.Kitisin K., Pishvaian M.J., Johnson L.B., Mishra L. Liver stem cells and molecular signaling pathways in hepatocellular carcinoma. Gastrointest Cancer Res. 2007;1:S13–S21. [PMC free article] [PubMed] [Google Scholar]
- 74.Ma S., Lee T.K., Zheng B.J., Chan K.W., Guan X.Y. CD133+ HCC cancer stem cells confer chemoresistance by preferential expression of the Akt/PKB survival pathway. Oncogene. 2008;27:1749–1758. doi: 10.1038/sj.onc.1210811. [DOI] [PubMed] [Google Scholar]
- 75.Bimonte S., Leongito M., Barbieri A., Del Vecchio V., Falco M., Giudice A. The therapeutic targets of miRNA in hepatic cancer stem cells. Stem Cells Int. 2016;2016:1065230. doi: 10.1155/2016/1065230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Chiba T., Iwama A., Yokosuka O. Cancer stem cells in hepatocellular carcinoma: therapeutic implications based on stem cell biology. Hepatol Res. 2016;46:50–57. doi: 10.1111/hepr.12548. [DOI] [PubMed] [Google Scholar]
- 77.Gibbings D., Mostowy S., Voinnet O. Autophagy selectively regulates miRNA homeostasis. Autophagy. 2013;9:781–783. doi: 10.4161/auto.23694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Gibbings D., Leblanc P., Jay F., Pontier D., Michel F., Schwab Y. Human prion protein binds Argonaute and promotes accumulation of microRNA effector complexes. Nat Struct Mol Biol. 2012;19:517–524. doi: 10.1038/nsmb.2273. S1. [DOI] [PubMed] [Google Scholar]
- 79.Gibbings D., Mostowy S., Jay F., Schwab Y., Cossart P., Voinnet O. Selective autophagy degrades DICER and AGO2 and regulates miRNA activity. Nat Cell Biol. 2012;14:1314–1321. doi: 10.1038/ncb2611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wang Y., Wang Q., Song J. Inhibition of autophagy potentiates the proliferation inhibition activity of microRNA-7 in human hepatocellular carcinoma cells. Oncol Lett. 2017;14:3566–3572. doi: 10.3892/ol.2017.6573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Huang K.T., Kuo I.Y., Tsai M.C., Wu C.H., Hsu L.W., Chen L.Y. Factor VII-induced microRNA-135a inhibits autophagy and is associated with poor prognosis in hepatocellular carcinoma. Mol Ther Nucleic Acids. 2017;9:274–283. doi: 10.1016/j.omtn.2017.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.