Abstract
Background
Probiotics, live cells with different beneficiary characteristics, have been extensively studied and explored commercially in many different products in the world. Their benefits to human and animal health have proven in hundreds of scientific studies. Based on rich bibliographic material, Curd is the potential source of probiotic Lactobacilli.
Method
The aim of the present study was to observe Lactobacilli with probiotic potential activities from different curd samples for isolation, identification and characterization of Lactobacillus species.
Results
Among the samples, thirty lactic acid bacterial strains were isolated, sixteen (16/30) best Lactobacillus isolates were selected by preliminary screening as potential probiotic for acid and bile tolerance, further confirmed using 16s rRNA identification. All the selected Lactobacillus isolates were then characterized in vitro for their probiotic characteristics and antimicrobial activities against pathogens and aggregation studies. The results indicated that selected potential probiotic isolates (T2, T4 and T16) were screened and confirmed as Lactobacillus. The isolates produced positive tolerance to excited pH, NaCl and bile salts, also revealed noticeable antimicrobial activities against pathogens. All the Lactobacillus isolates were susceptible to clinical antibiotics used. Besides, T2 isolate was constituted to retain stronger auto and co-aggregation and cell surface hydrophobicity capacity.
Conclusion
Based on the drawn results, T2, T4 and T16 Lactobacillus isolates were recognised as ideal, potential in vitro antimicrobial probiotic isolates against pathogens and studies are needed further in-vivo assessment and human health benefits in their real-life situations.
Keywords: Lactic acid bacteria, Lactobacillus species, Curd, Bacteriocin, Antimicrobial activity
At a glance commentary
Scientific background on the subject
Probiotic products consist of live bacteria that have potentially favourable health effects. A number of studies provide evidence that milk products with probiotics beneficial for digestive health and improve digestive problems. Present study investigate Lactobacillus species with potential activities isolated from different local made and commercially available at local market.
What this study adds to the field
These areas of research suggest that isolates from curd samples have promising properties that are important for potential probiotics. Hence, more research is needed to exploit other potential probiotic properties. Further, in-vivo trials are needed to determine whether they function as probiotics in real-life situations for human health benefits.
Probiotics have been with us for as long as people have eaten fermented milk, but their association with health benefits dates only from the turn of the last century, growing awareness of the health benefits of consuming microorganisms as probiotics has encouraged consumer's worldwide [1]. These probiotic bacteria are essential for beneficial effect on particular organism's health and host nutrition for healthy gastrointestinal function. Probiotics are defined as “live microorganisms when administered in adequate amounts; confer a health benefit on the host [2]. This definition acknowledges a possible role for probiotics in medical practice, but does not acknowledge that probiotics (albeit rarely) can cause disease. Over the last 10 years there has been increasing public and scientific interest in the administration of these live micro-organisms to prevent or treat disease. Twenty-three publications were retrieved from PubMed for the year 1995 using the search term ‘probiotic’ compared with about 200 in the year 2000 and more than 600 for the first half of 2012. Much of the focus of this research has been on the use of probiotics for the prevention or treatment of gastrointestinal conditions such as inflammatory bowel disease and inflammatory bowel syndrome [3], [4]. More recent studies have suggested that the influence of probiotics extends well beyond the gut and may even extend to modulation of emotional states and neurological functioning [5], [6].
The vast majority (>90%) of the total cells in the body are present as bacteria in the colon, reaching 1012 for every gram of large intestinal contents. Under natural conditions, a protective gut microflora develops and there is no need for a bacterial supplement. But the changing food habits and lifestyle force us to take processed and sterile food, which affects our access to, and colonisation, by certain type of bacteria.
The genus Lactobacillus belongs to the normal mucosal microbiota of humans and animals [7]. This group of bacteria is important for maintaining the stability of the gastrointestinal tract, preventing intestinal infections and generally supporting intestinal health [8]. Several species of lactobacilli have generally regarded as safe status and some can interact with intestinal epithelial cells. An important group is the Lactobacillus bacteria, predominantly isolated (43.48%) from locally made from households and commercially available at milk parlours. Lactobacilli, primarily facultative or strict anaerobes generally has fastidious growth requirement. They prefer an acidic environment by producing lactic and other acids. In general, Lactobacilli have not been associated with disease and have been regarded as non-pathogenic and isolates were able to tolerate the acidic condition of the environment, NaCl concentration and resistance to bile.
Lactic acid bacteria (LAB) strains are potentially promising because they generate bactericidal bioactive agents that are able to control the growth of the pathogens. Beneficial effects conferred by Lactobacilli, including inhibition of gram negative and positive pathogenic bacteria described by Maragkoudakis et al., 2006 [9] and Charlier et al., 2008 [10]. Sustaining the antimicrobial activities of probiotics will affirm their use in the development of functional foods for the betterment of the health of the consuming health [11]. The isolated Lactobacillus isolates in the present study, exhibited very remarkable and noticeable antimicrobial activity against pathogenic bacteria, several suggestions have been proposed for inhibition of pathogenic bacteria, these might be Lactobacillus isolates were responsible for production of antimicrobial compounds like; bacteriocins, hydrogen peroxide and organic acids [10], [12], [13].
Multiple mechanisms of action for the beneficial effect of probiotics have been proposed [14], [15], [16]. The ability to adhesion of probiotic microorganism to the intestinal mucosa is considered important for many of the observed probiotic health effects. The ability to adhere to epithelial cells and mucosal surfaces has been suggested as being an important property of many bacterial strains used as probiotics. Adherence is an important prerequisite for the colonization of probiotics in the intestinal cavity, providing a competitive advantage in this ecosystem [16]. Several workers have suggested that the ability of beneficial micro-organisms to aggregate and adhere aids in colonization of the gut and in the establishment of a barrier which prevents enteropathogens from establishing an infection. Factors which prevent infection include the physical presence of beneficial micro-organisms and modulation of the gut immune system by these organisms [15], [17], [18]. Among the Lactobacillus isolates T2 isolate was the most dominantly evaluated the qualitatively and quantitatively the aggregation and co-aggregation capacities of collected Lactobacillus isolates.
The objective of this study was to isolate and characterize effective probiotic Lactobacillus isolates isolated from different local made from households and commercially available at local market of milk parlours. In vitro probiotic properties such as pH, NaCl tolerance, bile tolerance, antibiotic susceptibility profile, antimicrobial activity, auto- and co-aggregation abilities, time-kill assay and cell-surface hydrophobicity capacity of the three effectively selected probiotic isolates were investigated with selected indicator strains (pathogens) and its role in controlling the pathogen growth.
Materials and methods
Isolation of probiotic bacteria from different dairy products
Collection of samples
This cross-sectional study was conducted at Department of Biotechnology, Gulbarga University, Kalaburagi, between September 2015 and May 2016. The different curd samples which are randomly collected from different local made from households and commercially available at local market of milk parlours. These samples were collected in clean, sterile, wide-mouthed containers, without disinfectant or detergent residue and tight-fitting leak-proof lids. Immediately after collection, the samples were transferred to the laboratory for microbiological analysis and stored aseptically in low temperature (−4 °C) refrigerator to protect from contamination and deterioration.
Media
The standard media accepted by the International Dairy Federation for lactic acid bacteria differential the fermented dairy products, is deMan Rogosa Sharp (MRS) IDF 1983. The bacteria Lactobacillus spp. was isolated from curd samples by using modified MRS broth and MRS agar media [19]. Additionally, 0.05% cysteine was added to MRS to improve the specificity of this medium for isolation of Lactobacillus [20]. The pH of the media adjusted to 6.5 ± 0.2.
Isolation of probiotic bacteria
In the present study, Bacteria were isolated from curd samples by using MRS medium. Ten gram of each collected samples were diluted with sterilized phosphate-buffered saline (PBS) and transferred to 100 ml of MRS broth at pH 6.5 MRS (deMan, Rogosa and Sharpe) medium was used for primary isolation of probiotic Lactobacillus bacteria by diluting the sample with normal saline solution. These solutions were added to the MRS broth and streaked on to the MRS agar plates after 6 h of incubation. The plates were aerobically incubated at 37 °C for 18–24 h. Cells were grown under a cool-white light. After incubation, white colonies that formed were selected for single-colony isolation and to isolate different strains of Lactobacillus species.
Identification of Lactobacillus spp.
The isolated colony formed on the MRS agar (Hi-media pvt ltd) plates was identified by phenotypically (gram stain and biochemical tests) and genotypically (16s rRNA sequencing). The identification was performed according to Bergey's manual of determinative of bacteriology. The culture was kept in MRS agar slant and stored at 4 °C. For long term storage, glycerol stocks were maintained and stored at −20 °C.
Microscopic observation and colony characterization
Microscopic observation
Gram's staining
Prepare a smear of isolated culture on a grease free cleaned slide then fix the smear by light warming. Then slowly cover the smear using a crystal violet stain. Keep it for 1 min. Then wash off the stain with clean tap water or distilled water. Then pour a drop of Gram's iodine on the smear and keep it for 1 min. Then wash with water and decolourising agent absolute alcohol and again immediately wash with water. At last dry the smear and observe the smear under microscope, first with 10× objective to check staining and then under oil immersion 100×. Record the result.
Colony characterization
Gram positive, rod shaped bacilli cells were observed. Aerobic and facultative anaerobes, optimum temperature for growth is 35–37 °C.
Biochemical tests
Suspected colonies on the primary or subculture plates resembling those of Lactobacillus spp. were selected for further identification by standard procedures. All the isolates were subjected to the biochemical test to identify the isolates. Identification was done on the basis of carbohydrate fermentation test, motility test, catalase and oxidase test. The performance and readings of the tests were quality controlled using the reference strains Lactobacillus fermentum NCDC 141, Lactobacillus casei NCDC297.
Characteristics of Lactobacillus spp.
Determination of optimal growth at different pH
Determination of optimal growth and pH of Lactobacillus spp., 1% (v/v) fresh over night culture (a single isolated colony was subcultured in MRS broth) of Lactobacillus were inoculated into MRS broth with varying pH ranging from 2 to 6.5. The pH was adjusted with concentrated acetic acid (99%) and 5 N NaOH. The inoculated broths were incubated in anaerobic condition 24 h at 37 °C. After 24 h of incubation growth of the bacteria were measured using a spectrophotometer, reading the optical density at 560 nm (OD560) against the uninoculated broth.
Bile salt tolerance test
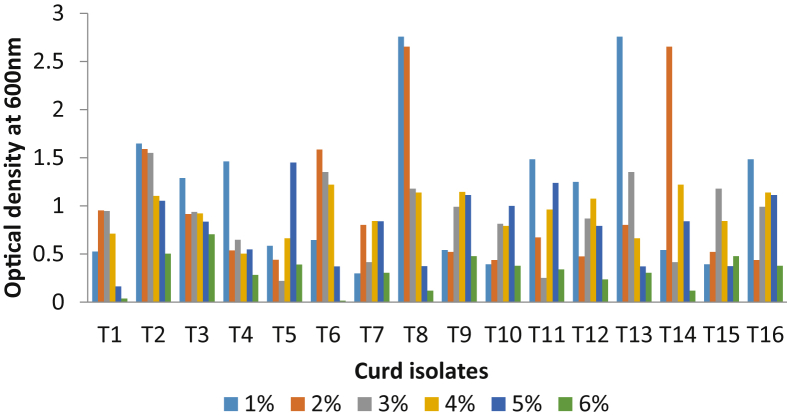
The ability of the strains to tolerate bile salts was determined according to the modified method described by Gilliland and colleagues [21]. This bile salt tolerance test was examined for optimum growth by inoculating the various isolates separately into MRS broth tubes containing 0.5%, 1%, 1.5%, 2% and 2.5% bile salts. Bacterial growth was monitored by measuring absorbance at 600 nm after incubation for 18–24 h at 37 °C. Bile salt-free MRS was used as control for this experiment.
NaCl tolerance test
For determination of NaCl tolerance, all the isolates were grown in MRS broth supplemented with different concentrations of NaCl (1–6%). The broth were inoculated with 10 μl overnight culture of the isolates and incubated anaerobically at 37 °C for 18–24 h of incubation, bacterial growth was monitored by measuring absorbance at 600 nm [22] and NaCl free MRS broth used as control.
Molecular identification by 16S rRNA
In the present study the determination of the 16S rRNA sequencing has been employed as a tool in arriving at identification and confirmation of bacterial strains by sequencing of 16S rRNA isolated from dairy samples. This method is fast and valid technique for molecular identification. An isolated Lactobacillus isolate sequence was commercially sequenced at “Chromous Biotech Pvt. Ltd, Bangalore.” Crude sequence attained by sequencing the amplified for the BLASTn (https://blast.ncbi.nlm.nih.gov/Blast.cgi) search at NCBI server and the homologous hits were studied. Such hits sequence across the species were recovered and issued for multiple alignments using ClustalW at EBI server. Based on the scores of multiple alignments the dendogram was produced and predicted using PHYLIP 3.6 version and studied for the evolutionary distance with other similar sequence retrieved from different species.
Antibiotic susceptibility test
Antibacterial susceptibility testing in the clinical laboratory is the most often performed test using the disk diffusion method [23]. The method was originally standardised according to the International Organization for Standardization (ISO) and quality assurance guidelines of World Health Organization (WHO). The activated cultures were swabbed on to the Muller Hinton agar plates. In this study, various antibiotics were supplied in the form of dodeca discs (Hi Media, India) which included Ampicillin, Amoxyclav, Amikacin, Azithromycin, Cefuroxime, Cephalothin, Clindamycin, Co-trimoxazole, Chloramphenicol, Ciprofloxacin, Ceftriaxone, Erythromycin, Gentamycin, Neomycin, Novobiocin, Oxacillin, Sulfamethizole, Tetracycline and Vancomycin. The zones of inhibition were measured after incubation at 37 °C for 24 h.
Antimicrobial activity
Antagonistic activity against pathogens
Seven strains that are pathogenic to humans were used as test pathogens to investigate the antagonistic activity of the Lactobacillus spp. They are Staphylococcus aureus (MTCC 96), Enterococcus faecalis (MTCC439), Klebsiella pneumonia (MTCC 432), Pseudomonas aeruginosa (MTCC 7925), E. coli (MTCC 443), Salmonella typhii (MTCC734), and Shigella spp. (MTCC 13313) were obtained from the culture collection of Prof. C.K. lab, Department of Biotechnology, Gulbarga University, Kalaburagi.
Antimicrobial activity of selected probiotic Lactobacillus isolates against the test pathogenic strains were assessed using the agar spot test described by Mami et al., 2012 [24], with modifications. An agar-well diffusion assay was used, aliquots of 60–80 μl of the sterile cell free supernatant were placed in 7 mm diameter wells on Muller–Hinton-agar plates previously seeded with the respective test pathogens. After 18 h of incubation at 37 °C, the diameters of the zones of growth inhibition were measured. Inhibition zones more than 20 mm, 10–20 mm and less than 10 mm were reviewed as strong, intermediate and low inhibition, respectively. The test was performed twice, each in triplicate.
Characterization of antimicrobial substances
The selected probiotic Lactobacillus spp. (T2, T4 & T16) were assayed for production of antimicrobial substances such as bacteriocins, hydrogen peroxide and organic acids using the agar well diffusion technique described by Toure et al., 2003 [25], with modifications. The bacterial strains were grown in 25 ml of MRS broth at 37 °C overnight, after which the cultures were centrifuged at 4000×g for 10 min at 4 °C. The supernatant of each strain was diverged into equal portion of different assays. For Bacteriocin assay, the supernatant (5 ml) treated with 1 mg/ml pronase or 1 mg/ml trypsin. For Organic acids assay, the supernatant (5 ml) was adjusted to pH 6.5 ± 0.1 using 1 N NaOH and for hydrogen peroxide assay, the supernatant (5 ml) was treated with 0.5 mg/ml catalase (Hi-media pvt ltd). Treated supernatant were filter sterilised through 0.22 μm pore-size filters (Axiva pvt ltd) and 100 μl was placed into 7 mm diameter wells, the plates were seeded with 1% (v/v) overnight culture of each indicator strain (test pathogen). The plates were kept at 4 °C for 30 min or better diffusion of the treated supernatant and then incubated for 24 h at 37 °C and diameter of inhibition zones (including 7 mm well diameter) were measured.
Determination of minimum inhibitory concentration
Minimum inhibitory concentration (MIC) was determined to evaluate the phenotypic antimicrobial resistance of a strain to a certain probiotic Lactobacillus spp. (Cell free culture supernatant). MIC was defined as lowest Lactobacillus spp. concentration that resulted in no visible growth. This MIC test was determined by broth-dilution technique by following the reference standard established by ISO 2010 [23]. Serial two fold dilutions (Higher and lower) of the CFCS Lactobacillus spp. were inoculated with an overnight culture at a final concentration of 107−8 colony forming unit (cfu/ml). MIC level was determined by measuring the test pathogen's absorbance at 600 nm and Lactobacillus free broth used as control.
Auto-aggregation of probiotic Lactobacillus spp.
Aggregation study was examined for the three selected effective probiotic Lactobacillus spp. from curd samples on the basis of their sedimentation characteristics. Overnight culture, 108 cfu/ml each Lactobacillus spp. from were harvested by centrifugation at 6000×g for 20 min, 4 °C washed three times with Phosphate Buffer Saline (pH 7.3) and eliminated in the same buffer. Then, the mixture was vortexed and incubated at 37 °C for 4 h without agitation. The auto-aggregation percentage was asserted of three different Lactobacillus spp. as
where, Atime and AInitial measured at 600 nm, represents the absorbance of the mixture at 0 h and 4 h.
Co-aggregation of Lactobacillus spp. with different pathogenic cells
The co-aggregation analysis was performed according to slight modified method to Collado et al., 2008. The three (T2, T4 & T16) selected probiotic Lactobacillus spp. and seven different test pathogens were separately cultured at 37 °C for 24 h in MRS and TSB medium. Bacterial suspension (108 cfu/ml) were formulated as described in the auto-aggregation in above method, equal volume of cells of the different probiotic Lactobacillus spp. and pathogenic strains (1:1 v/v) were mixed and incubated at 37 °C without agitation. Absorbance, A600 of the mixture represent above, was supervised during incubation at 4 h, percentage of co-aggregation were directed as
where, Apathogen and Alactobacillus and Amix represent the A600 of individual pathogen, Lactobacillus spp. and their mixture after incubation for 4 h, respectfully.
Time-kill assay with cell free culture supernatant (CFCS) of Lactobacillus spp. on various pathogen
The time-kill assay was executed by co-culture of the each pathogenic cells and Cell free culture supernatant (CFCS) of Lactobacillus spp. 300 μl of pathogenic suspension, 108 cfu/ml were added into 15 ml CFCS, CFCS adjusted to be pH 6.5 and MRS broth (6.5), respectively and were incubated at 37 °C. At initial and predetermined intervals, aliquots were separated, serially diluting and plating on LB agar to determine the surviving cells of individual pathogens.
Cell surface hydrophobicity
Cell surface hydrophobicity was determined following to the capacity of the three different Lactobacillus spp. and the seven test pathogens to individually partition into xylene from PBS. The cells were washed twice with PBS and the optical density (A) at 540 nm adjusted to 0.5 ± 0.01–1.0 ml of bacterial suspension, 60 μl xylene was added and vortexed for 1 min and the optical density of the water phase was determined. Percentage hydrophobicity was calculated according to formula [26].
Quantification of organic acid and determination of pH value
One percent (v/v) 24 h active culture of Lactobacillus isolates (T2, T4 and T16) was utilised to inoculate 10% sterilized skim milk existed from Hi-media pvt ltd Bangalore and initial pH 6.76 was determined by digital electrode pH meter. The inoculated skim milk was incubated at 37 °C for 72 h and samples were assembled in every 12 h, 24 h, 48 h and 72 h and liquids of coagulated milk were disparate by filtration. pH of the separated liquid was recorded using a digital electrode pH meter and quantification organic acid was performed through titration with 0.1 N NaOH.
Scanning electron microscopic study
As it is difficult to notice small changes in cell morphologies of bacteria under the light microscope, SEM was used in the present investigation to review the changes or any rapture in cell morphology of the populations by the effect of Lactobacillus spp. [27]. The preferred pathogen grown in TSB media with increasing antibiotic concentrations and normal pathogen. The bacterial cells from each culture were recovered by centrifugation at 6000 rpm/min and the cells were washed twice with potassium phosphate buffer (50 mM, pH 7.0). Bacterial cells were then fixed by drown in 2.5% glutaraldehyde in potassium phosphate buffer (50 mM, pH 7.0) for overnight at 4 °C. Then the specimens were washed twice with buffer and dehydrated by ethanol series (v/v) ranging from 30%, 40%, 50%, 60%, 70%, 80% and 90%–100% and stored in 100% ethanol. For SEM, the specimens were dried to critical point, coated with gold and inspected with an S-200C scanning electron microscope. Results were correlated with standard pathogenic culture and control selected pathogens.
Results
Isolation and identification of Lactobacillus spp.
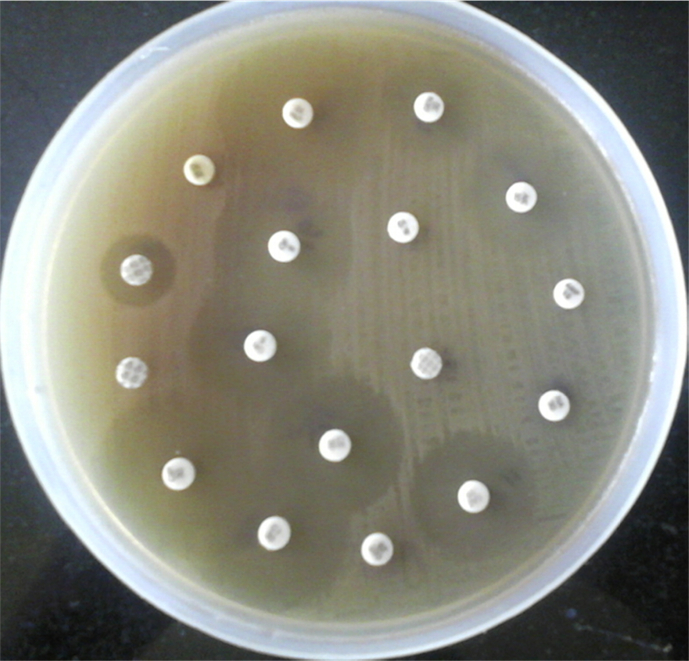
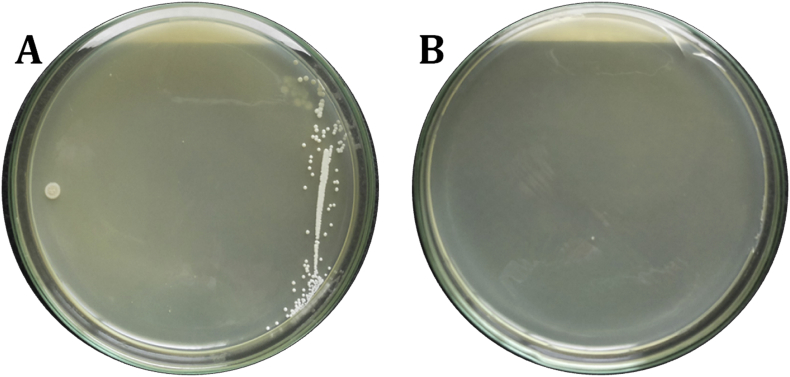
Over the period of 7 months, a total of 16 home made curd samples were collected from surrounding region to the campus and local Gulbarga city. Thirty strains were isolated, after culturing for 18–24 h, 16 (48.48%) strains were selected as forming wide and white colonies on the MRS agar plates; among the Lactic acid bacteria, the most common and predominant isolates were Lactobacillus spp. (Fig. 1), by noticing their colony morphology, physiological and as well as some biochemical characteristics (Table 1). Microscopically they were Gram-positive (Fig. 2), rod shaped, non-motile, catalase negative and absence of Endospore. These were subcultured on MRS with glycerol broth and stored at −20 °C. The isolated strains were named T1, T2, T3, T4, T5, T6, T7, T8, T9, T10, T11, T12, T13, T14, T15 and T16.
Fig. 1.
Typical characteristics of the isolates grown on MRS agar medium.
Table 1.
Morphological, cultural and biochemical characteristics of isolated Lactobacillus spp. from curd samples.
| Sl. No | Selected Lactobacillus spp. | Morphological and cultural characteristics | Gram's staining | Motility test | Catalase test | Carbohydrate fermentation test |
|||
|---|---|---|---|---|---|---|---|---|---|
| Glucose |
Sorbitol | Sucrose | |||||||
| Acid | Gas | ||||||||
| 1 | T1 | 1.0 mm white, rough, irregular and round | Gram +ve, bacilli | Non motile | Negative | +ve | −ve | −ve | −ve |
| 2 | T2 | Small, 0.1–0.5 mm, rough dull and round | Gram +ve, bacilli | Non motile | Negative | −ve | −ve | −ve | −ve |
| 3 | T3 | 1 mm, White, shiny smooth | Gram +ve, bacilli | Non motile | Negative | −ve | −ve | −ve | −ve |
| 4 | T4 | Small circular, white creamy | Gram +ve, bacilli | Non motile | Negative | −ve | −ve | −ve | −ve |
| 5 | T5 | 1.0 mm white, rough, irregular and round | Gram +ve, bacilli | Non motile | Negative | −ve | −ve | −ve | −ve |
| 6 | T6 | Small circular, white creamy | Gram +ve, bacilli | Non motile | Negative | +ve | −ve | −ve | −ve |
| 7 | T7 | Small, 0.1–0.5 mm, rough dull and round | Gram +ve, bacilli | Non motile | Negative | +ve | −ve | +ve | +ve |
| 8 | T8 | 1.0 mm white, rough, irregular and round | Gram +ve, bacilli | Non motile | Negative | +ve | −ve | +ve | +ve |
| 9 | T9 | Small circular, white creamy | Gram +ve, bacilli | Non motile | Negative | +ve | −ve | +ve | −ve |
| 10 | T10 | Small, 0.1–0.5 mm, rough dull and round | Gram +ve, bacilli | Non motile | Negative | +ve | −ve | +ve | +ve |
| 11 | T11 | Small circular, white creamy | Gram +ve, bacilli | Non motile | Negative | +ve | −ve | +ve | +ve |
| 12 | T12 | 1.0 mm white, rough, irregular and round | Gram +ve, bacilli | Non motile | Negative | +ve | −ve | +ve | +ve |
| 13 | T13 | Small, 0.1–0.5 mm, rough dull and round | Gram +ve, bacilli | Non motile | Negative | −ve | −ve | +ve | +ve |
| 14 | T14 | Small, irregular, smooth and circular | Gram +ve, bacilli | Non motile | Negative | +ve | −ve | −ve | +ve |
| 15 | T15 | Small circular, white creamy | Gram +ve, bacilli | Non motile | Negative | +ve | −ve | −ve | −ve |
| 16 | T16 | 1.0 mm white, rough, irregular and round | Gram +ve, bacilli | Non motile | Negative | −ve | −ve | +ve | −ve |
Fig. 2.
Microscopic observation of Gram's stained Lactobacillus spp.
Characteristics of Lactobacillus isolates
Growth at different pH
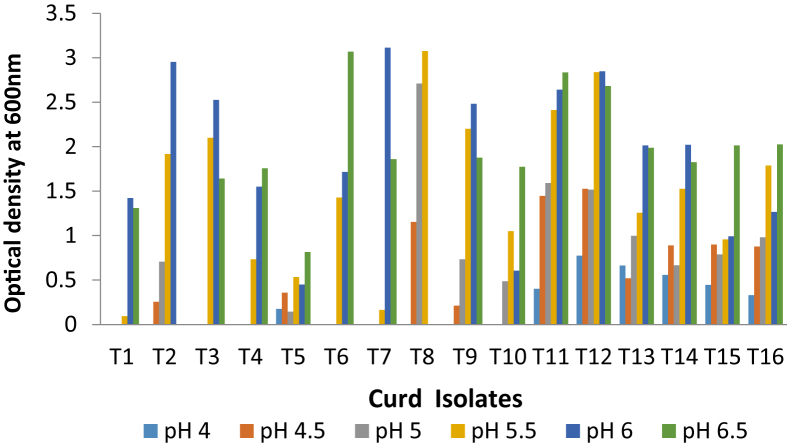
Lactobacillus isolates continuance in low pH is very important for tolerating initial stress in the stomach; Fig. 3 shows the results of the growth of Lactobacillus isolates at various pH values. The growth noted for various pH values in the range of 4.0–6.5. Representing that the bacteria adopted to grow in both acidic and neutral environment.
Fig. 3.
Optimal growth and pH of isolated Lactobacillus isolates from curd samples.
Tolerance to bile salt and NaCl
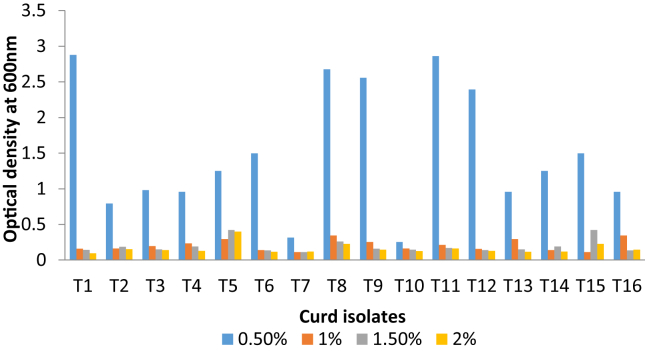
The isolated Lactobacillus isolates to be called as probiotics should be capable to resist inhibitory ingredient in the gastrointestinal tract like; bile salt [2]. For this intent, the effect of different concentrations of bile salts in MRS broth containing 0.5–2%. The attained results suggest that all isolates of Lactobacillus were resistance to bile salt during the incubation period (Fig. 4). Also, Lactobacilli from curd samples were able to tolerate 1–6% NaCl as shown in Fig. 5.
Fig. 4.
Bile acid tolerance of Lactobacillus isolates from curd samples.
Fig. 5.
NaCl tolerance of Lactobacillus isolates from curd samples.
Molecular identification of Lactobacillus spp.
The lactobacillus strain ckcs01 was identified by 16s rRNA sequencing, after the initial analysis at NCBI and RDP II (http//:rdp.cme.msu.edu) website, the relevant sequences were downloaded and Phylogenetic analysis was done and tree was constructed using ClustalW software and showed the Phylogenetic relationship of the isolates (Fig. 6). In analysis of the sequence showed 99% sequence similarity with the sequence reported on Lactobacillus spp. and consequently isolates of ckcs01 were Phylogenetic studies indicate that the L. fermentum. The 16s rRNA isolate sequence is deposited in the GenBank with the accession number KX242349.
Fig. 6.
Phylogenetic tree based on Neighbour-joining method of 16s rRNA gene sequencing.
Antibiotic susceptibility test
The antibiotic susceptibility test was carried out for all the 16 positive potentially probiotic Lactobacillus spp. against the 15 antibiotics consisted different groups (Fig. 7 and Table 2). Isolates were shown sensitive to amoxyclav, cephalothin, co-trimoxazole, erythromycin, tetracycline, sulfamethizole, amikacin, some isolates were displayed intermediate resistance to cefuroxime, clindamycin, gentamycin, vancomycin, chloramphenicol, ciprofloxacin and prominent observation was resistance towards ampicillin, oxacillin denoted by the collect Lactobacillus spp.
Fig. 7.
Antibiotic susceptibility pattern of Lactobacillus isolates.
Table 2.
Antibiotic susceptibility test for Lactobacillus isolates curd samples.
| Sl. No. | Antibiotics used | No. of resistance | No. of sensitive | No. of intermediate |
|---|---|---|---|---|
| 1 | Ampicillin | 10 (87.50%) | 3 (18.75%) | – |
| 2 | Amikacin | 5 (31.25%) | 11 (68.75%) | – |
| 3 | Amoxyclav | 5 (31.25%) | 11 (68.75%) | – |
| 4 | Cefuroxime | 6 (37.50%) | 9 (56.25%) | 1 (6.25%) |
| 5 | Cephalothin | 5 (31.25)% | 11 (68.75)% | – |
| 6 | Chloramphenicol | 8 (50%) | 8 (50%) | – |
| 7 | Ciprofloxacin | 7 (37.5%) | 10 (62.5%) | – |
| 8 | Clindamycin | 9 (56.25%) | 7 (43.75%) | – |
| 9 | Cotrimoxazole | 5 (31.25%) | 11 (68.75%) | – |
| 10 | Erythromycin | 5 (31.25%) | 11 (68.75%) | – |
| 11 | Gentamycin | 5 (31.25%) | 11 (68.75%) | – |
| 12 | Oxacillin | 16 (100%) | – | – |
| 13 | Sulfamethizole | 5 (31.25%) | 8 (50%) | 2 (12.5%) |
| 14 | Tetracycline | 7 (37.5%) | 10 (62.5%) | – |
| 15 | Vancomycin | 8 (50%) | 5 (18.75%) | 3 (31.25%) |
Antimicrobial activity
Antagonistic activity
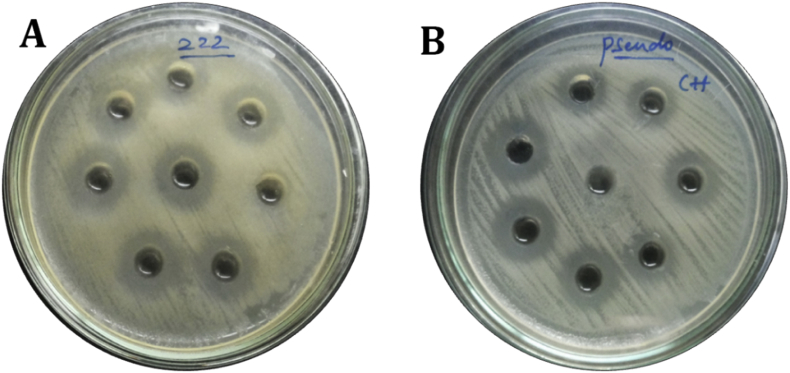
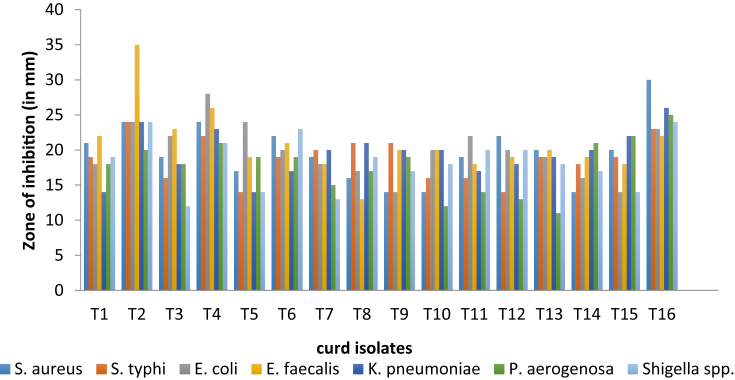
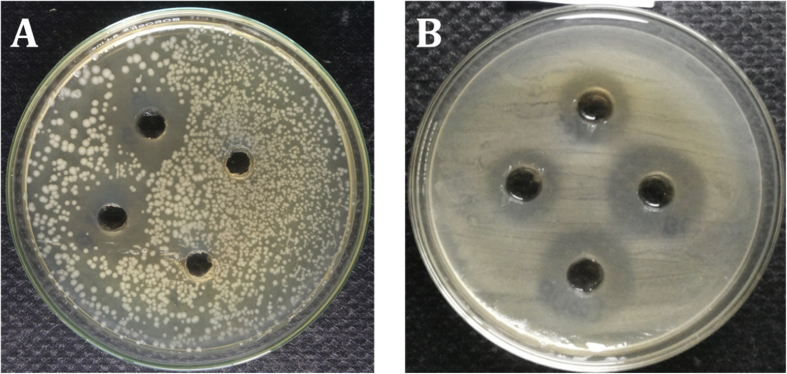
The selected Lactobacillus isolates were noticeable to consider their antimicrobial activity by modified agar-well diffusion method. For this point, the results of antagonistic effects of the Lactobacillus strains were subjected against the indicator microorganisms such as S. aureus, E. faecalis, K. pneumonia, P. aeruginosa, Escherichia coli, S. typhii and S. spp. All sixteen Lactobacillus strains and reference strain showed antagonistic effects against all indicator microorganisms tested, but degrees of antagonism varied among the Lactobacillus strains. The results revealed that all the isolated Lactobacillus strains, exhibited the average inhibition (15–25 mm) on the growth of test pathogens, but the Lactobacillus strain T2, T4 and T16 was the most effective noticeable strains in inhibiting the growth of the test pathogens (28–32 mm). Overall, many of the isolated Lactobacillus strains showed better antagonistic activities against the test pathogen than the reference strain L. fermentum (Fig. 8, Fig. 9 and Table 3).
Fig. 8.
Antagonistic activity of Lactobacillus isolates against different test pathogens. (A) Gram-positive (B) Gram-negative.
Fig. 9.
Antagonistic activity of Lactobacillus isolates from Curd samples.
Table 3.
Antagonistic activity of Lactobacillus isolates against test pathogens from curd samples.
| Sl. No | Lactobacillus isolates | Zone of inhibition in mm (from outer edge of Lactobacillus colony to outer edge of clear zone) |
||||||
|---|---|---|---|---|---|---|---|---|
| S. aureus | S. typhi | E. coli | E. faecalis | K. pneumoniae | P. aeruginosa | Shigella dysentriae | ||
| 1 | T1 | 21 | 19 | 18 | 22 | 14 | 18 | 19 |
| 2 | T2 | 24 | 24 | 24 | 35 | 24 | 20 | 24 |
| 3 | T3 | 19 | 16 | 22 | 23 | 18 | 18 | 12 |
| 4 | T4 | 24 | 22 | 28 | 26 | 23 | 21 | 21 |
| 5 | T5 | 17 | 14 | 24 | 19 | 14 | 19 | 14 |
| 6 | T6 | 22 | 19 | 20 | 21 | 17 | 19 | 23 |
| 7 | T7 | 19 | 20 | 18 | 18 | 20 | 15 | 13 |
| 8 | T8 | 16 | 21 | 17 | 13 | 21 | 17 | 19 |
| 9 | T9 | 14 | 21 | 14 | 20 | 20 | 19 | 17 |
| 10 | T10 | 14 | 16 | 20 | 20 | 20 | 12 | 18 |
| 11 | T11 | 19 | 16 | 22 | 18 | 17 | 14 | 20 |
| 12 | T12 | 22 | 14 | 20 | 19 | 18 | 13 | 20 |
| 13 | T13 | 20 | 19 | 19 | 20 | 19 | 11 | 18 |
| 14 | T14 | 14 | 18 | 16 | 19 | 20 | 21 | 17 |
| 15 | T15 | 20 | 19 | 14 | 18 | 22 | 22 | 14 |
| 16 | T16 | 30 | 23 | 23 | 22 | 26 | 25 | 24 |
To know, whether the isolates are bacteriostatic or bacteriocidal, confirmation test was done by modified agar overlay method were conducted; in this case swabs were taken from each clear zone of the test organism and were streaked on to nutrient agar plates for growth. Depending on the growth, the bacteriostatic and bacteriocidal activities are tabulated in that Table 4. Presence of growth of the indicator organism was elucidated as an inhibitory activity, called bacteriostatic, while no growth was expounded as bacteriocidal.
Table 4.
Bacteriostatic and bactericidal activity of curd isolates (T2, T4 and T16).
| Name of the pathogens | T2 isolate | T4 isolate | T16 isolate |
|---|---|---|---|
| S. aureus | + | + | − |
| E. faecalis | + | + | − |
| E. coli | − | − | − |
| P. aeruginosa | − | − | − |
| K. pneumoniae | − | − | − |
| S. typhii | − | + | − |
| Shigella spp | − | + | + |
+: Bacteriostatic; −: Bacteriocidal.
Characterization of inhibitory substance
The effective Lactobacillus strains (T2, T4 and T16) were considered for the characterization of inhibitory substances like bacteriocin, organic acid and hydrogen peroxide. The antimicrobial substance provided by the three Lactobacillus strains was characterized by agar well diffusion assay against different test pathogens. The results presented that culture supernatants of all three isolated Lactobacillus strains and the reference strain treated with pronase (1 mg/ml) or trypsin (1 mg/ml) did not have any inhibitory activities effects of the Lactobacillus spp. This indicated that inhibitory effect of Lactobacillus strains were due to bacteriocin production. Culture supernatants treated with catalase also did not affect the inhibitory activities of the Lactobacillus strains against the test pathogens. This showed that inhibition by the Lactobacillus strains was not due to hydrogen peroxides production. However, neutralized supernatant (pH 6.5) of all three Lactobacillus strains did not have any inhibitory activity effects of the Lactobacillus strains were due to their organic acid production. Hence, this study concludes that among three Lactobacillus isolates T2 isolate (ckcs01) was bacteriocin and T4, T16 isolates were responsible for organic acid production respectfully (Fig. 10 and Table 5).
Fig. 10.
Antimicrobial activity of Lactobacillus (CFCS) Inhibitory substances against test pathogen. (A) Bacteriocin (B) Organic acid.
Table 5.
Characterization of antimicrobial substances of selected curd isolates.
| Sl. No. | Shigella selected strains | T2 (in mm) |
T4 (in mm) |
T16 (in mm) |
|||
|---|---|---|---|---|---|---|---|
| Bacteriocin assay | Organic acid assay | Bacteriocin assay | Organic acid assay | Bacteriocin assay | Organic acid assay | ||
| 1 | S. aureus | 21 | – | – | 14 | – | 13 |
| 2 | E. faecalis | – | 19 | – | 13 | – | 12 |
| 3 | E. coli | 22 | – | – | 18 | – | 19 |
| 4 | P. aeruginosa | – | 23 | – | 17 | – | 17 |
| 5 | K. pneumoniae | 19 | – | – | 12 | – | 16 |
| 6 | S. typhii | – | 21 | – | 18 | – | 21 |
| 7 | Shigella spp | – | 17 | – | 17 | – | 22 |
Minimal inhibitory concentration of cell free culture supernatant (cfcs) Lactobacillus spp.
All the selected CFCS of Lactobacillus isolates were used for MIC assay, the results indicated that MIC for T2 (cfcs01) isolate was 50 μl against S. aureus, K. pneumoniae, E. coli and P. aeruginosa, 100 μl against E. faecalis, S. typhii and Shigella spp.; T4 isolates showed MIC of 100 μl against E. faecalis, S. typhii, S. aureus, K. pneumoniae, E. coli and 120 μl against P. aeruginosa and S. spp. and T16 isolate showed MIC of 100 μl against P. aeruginosa, S. spp. E. faecalis, S. typhii, S. aureus, K. pneumoniae and 128 μl against E. coli.
Auto- and co-aggregation assays of Lactobacillus spp
The auto-aggregation assay was examined for three effective selected Lactobacillus strains and the seven test pathogens on the basis of their sublimate characteristics. The results of this study revealed that, auto-aggregation expanded as a concern of time and were highest at the 24 h of incubation time period (Table 6). The percentage of auto-aggregation for all indicator strains ranged between 5% and 23%, which is 2.5 fold below the range for the Lactobacillus, isolates (40% and 51%) after the 24 h in the similar form. Among the Lactobacillus strain assay, T2 was most predominantly exhibited the highest percentage of auto-aggregation after 24 h (51%).
Table 6.
Percentage of auto-aggregation of Probiotic selected isolates with pathogenic strains.
| Lactobacillus Strains | Auto-aggregation (%) |
||
|---|---|---|---|
| 4 h | 18 h | 24 h | |
| T2 isolate | 23 ± 0.7 | 34 ± 0.2 | 51 ± 0.4 |
| T4 isolate | 17 ± 0.9 | 26 ± 0.8 | 40 ± 0.2 |
| T16 isolate | 13 ± 1.1 | 28 ± 1.2 | 48 ± 0.7 |
| Pathogenic strains | |||
| Staphylococcus aureus | 3.2 ± 1.0 | 4.2 ± 0.1 | 6.0 ± 0.9 |
| Enterococcus faecalis | 2.9 ± 1.4 | 3.6 ± 0.4 | 4.3 ± 0.8 |
| E. coli | 7.8 ± 1.2 | 14.1 ± 0.8 | 18 ± 1.2 |
| Pseudomonas aeruginosa | 4.5 ± 0.8 | 12.1 ± 1.1 | 21 ± 1.5 |
| Klebsiella pneumoniae | 6.1 ± 1.1 | 13 ± 1.3 | 19 ± 1.1 |
| Salmonella typhi | 3.1 ± 0.8 | 11 ± 0.9 | 23 ± 0.1 |
| Shigella spp. | 2.9 ± 0.4 | 10.8 ± 0.1 | 22 ± 1.0 |
The co-aggregation results of the three Lactobacillus stains tested with seven diverge test pathogens as shown in Table 7. Interrogation of all selected strains manifest their aggregation abilities with the test pathogens tested, but the percentage of the co-aggregation was strain-specific. Among the isolated strains, T2, T4 and T16 strains, revealed the notable co-aggregation with S. spp. as 27.3, 23.4 and 21.4% respectively, at the same state, T4 isolate exhibited the less co-aggregation abilities with P. aerogenosa, as well as with rest indicator strains used.
Table 7.
Percentage of cell-surface hydrophobicity of bacterial strains.
| Selected Lactobacillus strains | % of co-aggregation with indicator strains |
||||||
|---|---|---|---|---|---|---|---|
| S. aureus | E. faecalis | E. coli | P. aeruginosa | K. pneumoniae | S. typhii | Shigella spp. | |
| T2 isolate | 25.6 ± 2.2 | 12.8 ± 0.8 | 19.6 ± 0.8 | 16 ± 0.8 | 18 ± 1.8 | 23 ± 0.6 | 27.3 ± 6.2 |
| T4 isolate | 5.4 ± 4.2 | 11.4 ± 0.6 | 7.1 ± 1.2 | 3.6 ± 0.9 | 12 ± 0.8 | 10 ± 0.4 | 21.4 ± 5.9 |
| T16 isolate | 11.2 ± 1.8 | 18.57 ± 0.7 | 16.8 ± 0.6 | 10.2 ± 1.2 | 14 ± 1.2 | 14 ± 0.3 | 23.4 ± 1.8 |
Cell surface hydrophobicity
Cell-surface hydrophobicity was resoluted in order to study attainable association between this physic-chemical property and the potential to adhere to the intestinal mucus. Hydrophobic cell surface was denoted by high adherence to xylene, an apolar solvent. The hydrophobicity percentage of effective probiotic and indicator strains to xylene as shown in Table 8. The cell-surface hydrophobicity disparate with the strains in the case of selected probiotic strains, T2 isolate (73%) was most notable hydrophobic nature. The results indicated that all the other selected strains were lesser or no hydrophobic nature towards xylene from the control taken as 0%. Among the indicator strains, S. spp. and E. coli (33 & 28%) showed high hydrophobicity percentage, P. aerogenosa and S. typhii (20 & 19%), but S. aureus and E. faecalis (9 & 4%) formed less hydrophobicity percentages. However, no or less attachment was observed between the cell-surface hydrophobicity and the potential to adhere to the intestinal mucus.
Table 8.
Percentage of cell-surface hydrophobicity of bacterial strains.
| Selected strains | Cell-surface hydrophobicity (%) |
|---|---|
| T2 | 73.0 ± 0.6 |
| T4 | 37 ± 0.7 |
| T16 | 51 ± 1.2 |
| Indicator strains | |
| Staphylococcus aureus | 9.1 ± 0.2 |
| Enterococcus faecalis | 4 ± 0.1 |
| E. coli | 28.4 ± 0.6 |
| Pseudomonas aeruginosa | 20.6 ± 1.2 |
| Klebsiella pneumoniae | 10.1 ± 0.3 |
| Salmonella typhi | 19.4 ± 0.8 |
| Shigella spp. | 33.1 ± 0.2 |
Time-kill assay with cell free culture supernatant (CFCS) of Lactobacillus spp. on various pathogen
Time-kill assay studies, exhibits the reduction in the cell counts of the different pathogens observed in the presence of concentrated CFCS of effective probiotic T2, T4 and T16 isolates ranged of 2–3 aliquots for different incubation periods (6 h, 12 h, 18 h & 24 h). The killing effect was more evident in the case of T2 isolate. In contrast, a significant decrease in the viability of indicator strains (log3-7 dilutions) was seen when CFCS of different probiotic cultures of T4 and T16 and T2 isolate were effective in killing most of pathogens as increase in the time period (Fig. 11). The outcome of the results suggests that an antibacterial substance; organic acid and bacteriocin present in the CFCS of these probiotic isolates are the responsible.
Fig. 11.
Time kill assay of Lactobacillus (CFCS) isolate against test pathogen. (A) Reduction in pathogen colonies (B) Complete killing of pathogen colonies.
Quantification of organic acid and determination of pH value
The identified Lactobacillus species from curd samples (T2, T4 & T16) coagulated the skim milk and produced organic acids in the sterilized skim milks which were detected by titrimetric method. The results are presented in Table 9.
Table 9.
Quantification of organic acid and determination of pH value of selected Lactobacillus spp.
| Sources of bacteria | Name of the bacteria | Incubation time (Hour) | Incubation temp. (°C) | Organic acid (%) | pH |
|---|---|---|---|---|---|
| Curd | T2 (Lactobacillus fermentum) | 12 | 37 | 0.2 | 5.47 |
| 24 | 37 | 0.28 | 5.11 | ||
| 48 | 37 | 0.31 | 4.91 | ||
| 72 | 37 | 0.33 | 4.62 | ||
| Curd | T4 | 12 | 37 | 0.12 | 5.36 |
| 24 | 37 | 0.22 | 5.23 | ||
| 48 | 37 | 0.21 | 4.72 | ||
| 72 | 37 | 0.30 | 4.51 | ||
| Curd | T16 | 12 | 37 | 0.21 | 5.8 8 |
| 24 | 37 | 0.23 | 5.34 | ||
| 48 | 37 | 0.29 | 4.92 | ||
| 72 | 37 | 0.31 | 4.45 |
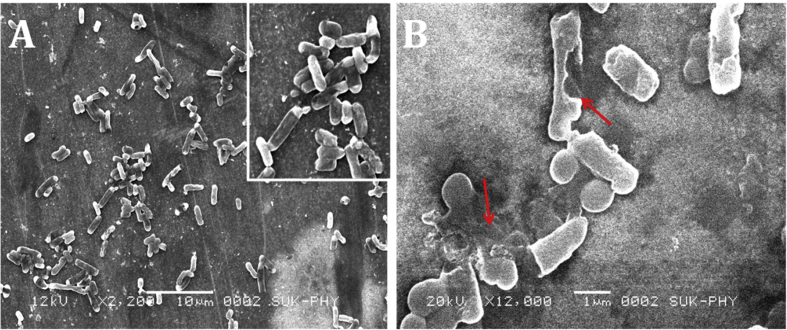
Scanning electron microscopic study
The upshots of cell morphology of test pathogen treated with cell free supernatant of Lactobacillus species were inspected by SEM, alteration or any rupture in the cell morphology was observed and shown in Fig. 12.
Fig. 12.
Scanning Electron Microscopy images. (A) Control. (B) Treated with Lactobacillus isolate (CFCS).
Discussion
The goal of the research work was to isolate, identify and characterize the potential probiotic Lactobacillus spp. from curd samples, locally made from households and commercially available at local market of Gulbarga city. Present study describes that, T2, T4 & T16 Lactobacillus isolates, considered as effective and novel probiotic bacteria to assess their anti-bacterial activity against some common human pathogens. Based on the cultural and morphological characteristics [28] of each three Lactobacillus spp individually isolated from curd samples and these isolates were genotypically (16s rRNA) identified as L. fermentum and isolate were named as ckcs01. After gram staining the isolated bacteria were rod shaped, convex, rough, smooth, shiny, irregular, circular, gram positive, facultative anaerobic, non spore forming which indicate them to be the member of Lactobacillus spp [29] (Table 1).
The significant growth of the isolates at pH 6.5 on MRS–agar plates in anaerobic conditions also confirmed their identification as Lactobacillus spp [30]. Oxidase and catalase test of the selected isolates were exhibited same results as Lactobacillus spp. All the isolates were Indole, MR, VP, Citrate, Oxidase and Catalase negative, the results are similar with the decree of Elizete and Carlos [31]. Among the carbohydrates used in this study, all the Lactobacillus isolates were able to ferment glucose, sucrose and Sorbitol. It indicates that they are able to grow in variety of habitats utilizing different type of carbohydrates. In order to be effective, probiotic bacteria must be able to survive in gastric acidic environments. In the present work, we found that incited gastric juice caused no relevant decreases in viability of Lactobacillus isolates. We therefore recommend these strains would likely survive in acidic environment of the stomach.
pH is an important factor which can consequentially affect bacterial growth. To be used as probiotic, organisms have to tolerate low pH of human gut. The isolated Lactobacillus spp. can tolerate a wide range of pH (2–8) and grow well at acidic pH (2–5) as shown in Fig. 3. NaCl is an inhibitory substance which may inhibit growth of certain types of bacteria and probiotic organisms have to withstand high salt concentration in human gut [21]. The current results revealed that Lactobacillus spp. isolated from curd was able to tolerate 1–6% of NaCl and excellent growth was perceived at 1–5% NaCl (Fig. 5). In this present study, 0.5–2% bile salt were supplemented in the growth media, as it is signify that found in the human intestinal tract and 0.5% is the maximum concentration that is present in healthy men [22]. Therefore, before selection of probiotic bacteria for human consumption it must be endurable to 0.5% bile concentration [32]. Lactobacillus spp. isolated in this study was resistant to 0.5% bile salt. All of the isolates are able to survive and grow in 0.5% bile salt concentration (Fig. 4).
The present evaluations reveal that organic acid production was increased with the incubation time and the pH of the media decreased with the increasing acid production. From the results table, highest acidity (0.3 ± 0.2) and lowest pH (4.45 ± 0.2) was observed after 72 h incubation at 37 °C for Lactobacillus sp. from all the collected dairy samples. This investigation noticed that, there is a slight variation in organic acid production by Lactobacilli due to their regional variation (Table 9).
All bacterial products anticipated for use as feed additives must be examined to establish the susceptibility of the component strain(s) to appropriate range of antimicrobials of human or veterinary importance [33]. In order to ensure the absence of transferable antibiotic resistance genes in any of the candidate probiotic strains, we assessed the antibiotic resistance profile. According to breakpoints levels established by EFSA, we found that all strains were susceptible to all antibiotics tested (except ampicillin and oxacillin), validating their safety as probiotic strains. The bacterial species used in the present study have already been recognized as well resistant to oxacillin [33] (Fig. 7 and Table 2). Susceptibility against antibiotics is look at to be the most essential probiotic characteristic. None of the strains of Lactobacilli tested in this study demonstrated antimicrobial resistance when tested according to EFSA guidelines.
Antimicrobial activity is one of the most crucial selection precedents for effective and novel probiotics. Antimicrobial effects of all Lactobacillus isolates are sustained by producing some substances such as organic acids (lactic, acetic, propionic acids, succinic acid etc), hydrogen peroxide, low molecular weight antimicrobial substances and bacteriocins [34]. Probiotics including Lactobacillus, Bifidobacterium and Streptococcus spp. are known to be inhibitory to the growth of a wide range of intestinal pathogens in human. In addition to the favourable effects against disease caused by an imbalance of the gut microflora, several experimental observations have revealed a potential protective effect of probiotic bacteria against the development of colon tumours [35].
In the study of Osuntoki et al., 2008 [28] Lactobacillus spp. isolated from fermented dairy products showed antibacterial activity against some clinically important pathogens such as Enterotoxigenic E. coli (4.2 mm), Salmonella typhimurium (4.3 mm) and Listeria monocytogenes (5.0 mm). Isolates of the present study have better antimicrobial capability than this Lactobacillus spp. isolates. Our isolates showed nearly similar antagonistic activity against E. coli and S. typhimurium as compared to Lactobacillus plantarum and Lactobacillus salivarius isolated by Murray et al., 2004, from a botanical probiotic. In the study by Gharaei-Fathabad and Eslamifar [36], [37] a strain of Lactobacillus paraplantarum isolated from tea leaves showed strong inhibitory activity against S. typhii (65 mm), E. coli (30 mm), S. aureus (56 mm), E. faecalis (55 mm) and Citrobacter spp. (60 mm). Isolates of the present study have almost similar antimicrobial capability. In our study, antagonistic activity of all selected Lactobacillus isolates against seven different test pathogens showed noticeable activity (as shown in Fig. 8, Fig. 9 and Table 3) and achieved that the activity of these Lactobacillus isolates due to organic acid and low molecular weight antimicrobial substances produced from the isolates was the responsible (Fig. 10).
Aggregation between microorganisms of the same strain (auto-aggregation) or between genetically different strains (co-aggregation) is of extensive importance in several ecological niches. Aggregating bacteria may achieve an adequate mass to form biofilms or adhere to the mucosal surfaces of the host and thus utilize their functions [38]. In the present study, the auto-aggregation assay was examined for three effective selected Lactobacillus strains and Shigella spp. on the basis of their sublimate characteristics. The results of this study revealed that, auto-aggregation expanded as a concern of time and were highest at the 4 h of incubation time period (Table 6, Table 7).
In conclusion of the work, Lactobacillus strains isolated in this study from the different dairy samples having in vitro properties that make them potential candidates for probiotic applications. Among the strains, Lactobacillus isolates from curd samples predominantly exhibited interesting probiotic properties such as excellent pH and bile tolerance, aggregations, suppression of pathogen growth under in vitro conditions. Moreover, all tested strains were susceptible to a number of clinically effective antibiotics. These results collectively suggest that isolates from curd samples have promising properties that are important for potential probiotics. Hence, more research is needed to exploit other potential probiotic properties of these strains. Further, in-vivo trials are needed to determine whether they function as probiotics in real-life situations for human health benefits.
Lactobacillus isolates also confirmed some probiotic properties which suggest their possible use in the medical field and most of the food industry. Indeed, a process for the incorporation of these isolates under some investigation by our research group. However, more studies are needed to complete the isolation and characterization of novel strains of Lactobacillus spp. and other probiotic bacteria that could be beneficial for the human health.
Conflicts of interest
We declare that no conflict of interest.
Ethical approval
This Medical Biotechnology and Phage Therapy Laboratory, Department of Biotechnology approved ethical clearance by Institutional Clearance Certificate (IECC) for in vitro and in-vivo studies.
Acknowledgments
The authors are (Kelmani Chandrakanth Revanasiddappa and Prabhurajeshwar C) profusely thankful to the Department of Biotechnology (grant from DBT, Govt. of India, BT/PR1812/SPD/24/577/2011) for funding the project and Department of Biotechnology, Gulbarga University, Gulbarga for providing facilities for pursuing the research work at the Department.
Footnotes
Peer review under responsibility of Chang Gung University.
References
- 1.Patel A.K., Ahire J.J., Pawar S.P., Chaudhari B.L., Chincholkar S.B. Comparative accounts of probiotic characteristics of Bacillus spp. isolated from food wastes. Food Res Int. 2009;42:505–510. [Google Scholar]
- 2.Food and Agriculture Organization of the United Nations and World Health Organization expert consultation report Rome. 2001. FAO/WHO evaluation of health and nutritional properties of powder milk and live lactic acid bacteria. [Google Scholar]
- 3.Miele E., Pascarella F., Giannetti E., Quaglietta L., Baldassano R.N., Staiano A. Effect of a probiotic preparation (VSL#3) on induction and maintenance of remission in children with ulcerative colitis. Am J Gastroenterol. 2009;104:437–443. doi: 10.1038/ajg.2008.118. [DOI] [PubMed] [Google Scholar]
- 4.Niedzielin K., Kordecki H., Birkenfeld B. A controlled double-blind randomized study on the efficacy of Lactobacillus plantarum 299V in patients with irritable bowel syndrome. Eur J Gastroenterol Hepatol. 2001;13:1143–1147. doi: 10.1097/00042737-200110000-00004. [DOI] [PubMed] [Google Scholar]
- 5.Gareau M.G., Wine E., Rodrigues D.M., Cho J.H., Whary M.T., Philpott D.J. Bacterial infection causes stress-induced memory dysfunction in mice. Gut. 2011;60:307–317. doi: 10.1136/gut.2009.202515. [DOI] [PubMed] [Google Scholar]
- 6.Bravo J.A., Forsythe P., Chew M.V., Escaravage E., Savignac H.M., Dinan T.G. Ingestion of Lactobacillus strain regulates emotional behaviour and central GABA receptor expression in a mouse via the vagus nerve. Proc Natl Acad Sci U S A. 2011;108:16050–16055. doi: 10.1073/pnas.1102999108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lahteinen T., Malinen E., Koort J.M., Mertaniemi-Hannus U., Hankimo T., Karikoski N. Probiotic properties of Lactobacillus isolates originating from porcine intestine and feces. Anaerobe. 2010;16:293–300. doi: 10.1016/j.anaerobe.2009.08.002. [DOI] [PubMed] [Google Scholar]
- 8.Gu R.X., Yang Z.Q., Li Z.H., Chen S.L., Luo Z.L. Probiotic properties of lactic acid bacteria isolated from stool samples of longevous people in regions of Hotan, Xinjiang and Bama, Guangxi, China. Anaerobe. 2008;14:313–317. doi: 10.1016/j.anaerobe.2008.06.001. [DOI] [PubMed] [Google Scholar]
- 9.Maragkoudakis P.A., Zoumpopoulou G., Miaris C., Kalantzopoulos G., Pot B., Tsakalidou E. Probiotic potential of Lactobacillus strains isolated from dairy products. Int Dairy J. 2006;16:189–199. [Google Scholar]
- 10.Charlier C., Even S., Gautier M., le Loir Y. Acidification is not involved in the early inhibition of Staphylococcus aureus growth by Lactococcus lactis in milk. Int Dairy J. 2008;18:197–203. [Google Scholar]
- 11.Eduardo L., Chuayana J.R., Carmina V.P., Rosanna M.A., Rivera B., Cabrera E.C. Antimicrobial activity of probiotics from milk products Phil. J Microbiol Infect Dis. 2003;32:71–74. [Google Scholar]
- 12.Lin W.H., Hwang L.W., Chen H.Y. Tsen variable counts characteristic evaluation for commercial lactic acid bacteria products. Food Microbiol. 2006;23:74–78. doi: 10.1016/j.fm.2005.01.013. [DOI] [PubMed] [Google Scholar]
- 13.Hernandez D., Cardell Zarate V. Antimicrobial activity of lactic acid bacteria isolated from Tenerife cheese: initial characterization of plantaricin TF711, a bacteriocin-like substance produced by Lactobacillus plantarum TF711. J Appl Microbiol. 2005;99:77–84. doi: 10.1111/j.1365-2672.2005.02576.x. [DOI] [PubMed] [Google Scholar]
- 14.Ng S.C., Hart A.L., Kamm M.A., Stagg A.J., Knight S.C. Mechanisms of action of probiotics: recent advances. Inflamm Bowel Dis. 2009;15:300–310. doi: 10.1002/ibd.20602. [DOI] [PubMed] [Google Scholar]
- 15.Sherman P.M., Ossa J.C., Johnson-Henry K. Unraveling mechanisms of action of probiotics. Nutr Clin Pract. 2009;24:10–14. doi: 10.1177/0884533608329231. [DOI] [PubMed] [Google Scholar]
- 16.Lebeer S., Vanderleyden J., De Keersmaecker S.C.J. Genes and molecules of lactobacilli supporting probiotic action. Microbiol Mol Biol Rev. 2008;72:728–764. doi: 10.1128/MMBR.00017-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cesena C., Morelli L., Alander M., Siljander T., Tuomola E., Salminen S. Lactobacillus crispatus and its no aggregating mutant in human colonization trials. J Dairy Sci. 2001;84:1001–1010. doi: 10.3168/jds.S0022-0302(01)74559-6. [DOI] [PubMed] [Google Scholar]
- 18.Collado M.C., Meriluoto J., Salminen S. Adhesion and aggregation properties of probiotic and pathogen strains. Eur Food Res Technol. 2008;226:1065–1073. [Google Scholar]
- 19.Hartemink R., Domenech V.R., Rombouts F.M. LAMVAB – a new selective medium for the isolation of lactobacilli from faeces. J Microbiol Methods. 1997;29:77–84. [Google Scholar]
- 20.Bauer A.W., Kirby W.M.N., Sherries J.L. Antibiotics susceptibility testing a standard disc method. Am J Clin Pathol. 1996;45:493–496. [PubMed] [Google Scholar]
- 21.Graciela F.V.D., Maria P.T. Spencer Humana Press Inc; Totowa: 2001. Food microbiology protocols probiotic properties of Lactobacilli. [Google Scholar]
- 22.Quwehand A.C., Vesterlund S. Antimicrobial components from lactic acid bacteria. In: Salminen Seppo, von Wright Atte, Ouwehand Arthur., editors. Lactic acid bacteria, microbiological and functional aspects. 2004. pp. 375–396. [Google Scholar]
- 23.ISO (International Organization for Standardization) ISO 10932/IDF 233 standard; 2010. Milk and milk products-determination of the minimal inhibitory concentration (MIC) of antibiotics applicable to bifidobacteria and non-enterococcal lactic acid bacteria (LAB) [Google Scholar]
- 24.Mami A.Z., Boumehira A.R., Hamedi J.E., Henni M. Screening of autochthonous Lactobacillus species from Algerian raw goats' milk for the production of bacteriocin-like compounds against Staphylococcus aureus. Afr J Microbiol Res. 2012;6:2888–2898. [Google Scholar]
- 25.Toure R., Kheadr E., Lacroix C., Moroni O., Fliss I. Production of antibacterial substances by bifidobacterial isolates from infant stool active against Listeria monocytogenes. J Appl Microbiol. 2003;95:1058–1069. doi: 10.1046/j.1365-2672.2003.02085.x. [DOI] [PubMed] [Google Scholar]
- 26.Savage D.C. Growth phase cellular hydrophobicity and adhesion in vitro of lactobacilli colonizing the keratinizing gastric epithelium in the mouse. Appl Environ Microbiol. 1992;58:1992–1995. doi: 10.1128/aem.58.6.1992-1995.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Raouf T., Marion B., Marielle G., Jean-Paul V. In vitro characterization of aggregation and adhesion properties of viable and heat-killed forms of two probiotic Lactobacillus strains and interaction with foodborne zoonotic bacteria, especially Campylobacter jejuni. J Med Microbiol. 2013;62:637–649. doi: 10.1099/jmm.0.049965-0. [DOI] [PubMed] [Google Scholar]
- 28.Osuntoki A.A., Ejide O.R., Omonigbehin E.A. Antagonistic effects on enteropathogenic and plasmid analysis of Lactobacilli isolated from fermented dairy products. Biotechnol. 2008;7:311–316. [Google Scholar]
- 29.Dhanasekaran D., Saha S., Thajuddin N., Rajalakshmi M., Panneerselvam A. Probiotic effect of Lactobacillus isolates against bacterial pathogen in fresh water fish. J Coast Dev. 2009;13:103–112. [Google Scholar]
- 30.Elizete D.F.R.P., Carlos R.S. Biochemical characterization and identification of probiotic Lactobacillus for swine. B CEPPA Curitiba. 2005;23:299–310. [Google Scholar]
- 31.Giilliland S.E., Staley T.E., Bush L.J. Importance of bile tolerance of Lactobacillus acidophilus used as a dietary adjunct. J Dairy Sci. 1984;67:3045–3051. doi: 10.3168/jds.S0022-0302(84)81670-7. [DOI] [PubMed] [Google Scholar]
- 32.EFSA Update of the criteria used in the assessment of bacterial resistance to antibiotics of human or veterinary importance. EFSA J. 2008;32:1–15. doi: 10.2903/j.efsa.2008.732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vankerckhoven V., Huys G., Vancanneyt M., Vael C., Klare I., Romond M.B. Biosafety assessment of probiotics used for human consumption: recommendations from the EU-PROSAFE project. Trends Food Sci Technol. 2008;19:102–114. [Google Scholar]
- 34.Savadogo A., Ouattara C.A.T., Bassole I.H.N., Traore A.S. Antimicrobial activities of lactic acid bacteria strains isolated from Burkina Faso fermented milk. Pak J Nutr. 2004;3:174–179. [Google Scholar]
- 35.Murry A.C., Hinton A., Morrison H. Inhibition of growth of Escherichia coli, Salmonella typhimurium and Clostridium perfringens on chicken feed media by Lactobacillus salivarius and Lactobacillus plantarum. Int J Poult Sci. 2004;3:603–607. [Google Scholar]
- 36.Gharaei-Fathabad E., Eslamifar M. Isolation and applications of one strain of Lactobacillus paraplantarum from tea leaves (Camellia sinensis) Am J Food Technol. 2011;6:429–434. [Google Scholar]
- 37.Grześkowiak L., Collado M.C., Salminen S. Evaluation of aggregation abilities between commensal fish bacteria and pathogens. Aquaculture. 2012;356:412–414. [Google Scholar]
- 38.De Man J.C., Rogosa M., Sharpe M.E. A medium for the cultivation of Lactobacilli. J Appl J Dairy Bacteriol. 1960;23:130–135. [Google Scholar]