Abstract
Background
Fusobacterium nucleatum is a Gram-negative anaerobic bacterium associated with periodontal disease. Some oral bacteria, like Porphyromonas gingivalis, evade the host immune response by inhibiting inflammation. On the other hand, F. nucleatum triggers inflammasome activation and release of danger-associated molecular patterns (DAMPs) in infected gingival epithelial cells.
Methods
In this study, we characterized the pro-inflammatory response to F. nucleatum oral infection in BALB/c mice. Western blots and ELISA were used to measure cytokine and DAMP (HMGB1) levels in the oral cavity after infection. Histology and flow cytometry were used to observe recruitment of immune cells to infected tissue and pathology.
Results
Our results show increased expression and production of pro-inflammatory cytokines during infection. Furthermore, we observe that F. nucleatum infection leads to recruitment of macrophages in different tissues of the oral cavity. Infection also contributes to osteoclast recruitment, which could be involved in the observed bone resorption.
Conclusions
Overall, our findings suggest that F. nucleatum infection rapidly induces inflammation, release of DAMPs, and macrophage infiltration in gingival tissues and suggest that osteoclasts may drive bone resorption at early stages of the inflammatory process.
Keywords: Immunology, Inflammation, Periodontal disease, Dental, Innate immunity
At a glance commentary
Scientific background on the subject
The effect of Fusobacterium nucleatum on the immune response remains poorly understood in mouse models of oral infection.
What this study adds to the field
This study showed that oral infection with F. nucleatum stimulates inflammation and infiltration of macrophages in gingival tissue, which could lead to bone loss in the oral cavity.
The oral cavity is colonized with hundreds of different species of bacteria which compose the oral microbiome [1], [2]. Some common bacteria found in individuals afflicted with periodontitis include Fusobacterium nucleatum, Porphyromonas gingivalis, and Aggregatibacter actinomycetemcomitans [3], [4]. Gingivitis is diagnosed when the gingiva, or gums, reveals signs of swelling, redness, or chronic bleeding [5], usually associated with gingival infection. However, chronic inflammation can lead to development of periodontitis with signs of deep periodontal pockets, alveolar bone resorption, and tooth loss [6], [7].
The tooth is surrounded by the gingival epithelium. This microenvironment is optimal for growth of anaerobic bacteria and provides an opportunity for pathogenic bacteria to attach and coaggregate into biofilms [8]. F. nucleatum is one of the predominant bacteria and contributors to biofilm formation [9], [10], [11]. The bacteria utilize adhesion mechanisms of lectin-like and non-lectin-like interactions and adhesion peptides, such as FadA (Fusobacterium adhesin A) for attachment [12], [13], [14], [15], [16]. These interactions facilitate coaggregation or infiltration into lymphocytes, polymorphonuclear neutrophils, erythrocytes, epithelial cells, and fibroblasts [11], [12], [14], [17], [18].
The oral epithelium defends against bacterial colonization by secretion of antimicrobial peptides called defensins [4], [19], [20], [21]. β-defensins target bacteria as the peptides are electrostatically attracted to their negative charged membranes and induce pore formation [4], [19], [22]. Antimicrobial peptides can also act as chemoattractants and recruit other immune cells, neutrophils or T cells [4], [7], [19]. Thus, β-defensins play an active role as part of innate and adaptive responses to oral infection.
Gingival epithelial cells (GECs) represent a major barrier to infection by invasive bacteria, and also contribute to immune recognition of the pathogens and the immune response [23], [24]. When pathogen-associated molecular patterns (PAMPs) of bacteria are recognized by host pathogen recognition receptors (PRRs) on GECs, they activate NF-κB and induce expression of cytokines and chemokines, and recruit neutrophils and macrophages [25], [26], [27]. Recognition of F. nucleatum and A. actinomycetemcomitans infection results in production of cytokines such as IL-1β [28], [29], [30]. TNF-α and IL-17 can also synergize with IL-1β to enhance expression and production of other cytokines (e.g., IL-6) and defensins, along with endothelial activation to enhance the immune response [31], [32], [33], [34], [35], [36].
Although the goal of inflammation is to resolve oral infection, it can also lead to bone resorption. Alveolar bone is one of the most dynamic bones in the body, as osteoclasts and osteoblasts continually induce bone remodeling to maintain homeostasis [37], [38], [39], [40]. Osteoclasts are resorptive cells that are activated and differentiated by macrophage-colony stimulating factor, receptor activator of nuclear factor kappa-B ligand (RANKL)-RANK signaling, interleukins, and TNF-α [38], [39], [41], [42], [43]. Once osteoclasts adhere to bone, a ruffled border is created between the activated osteoclast and bone [38], [40], and osteoclasts are able to degrade the mineral matrix [38], [40]. Degraded bone matrix is removed as it is transcytosed in vesicles through osteoclasts, and fuses with cytoplasmic vesicles containing tartrate-resistant acid phosphatase (TRAP) to be released in the extracellular matrix [38], [44]. Phagocytes remove the debris and osteoblasts are recruited for bone formation after osteoclasts detach from the bone [38].
F. nucleatum mechanisms for invasion and host response have been evaluated both in vitro and in vivo [3], [45], [46], [47], [48]. We have previously reported that F. nucleatum infection induces inflammasome activation and release of cytokines and danger signals in human GECs in vitro [29], [46]. In this study, we examined the immune response to F. nucleatum oral infection in BALB/c mice, which had not been previously characterized.
Materials and methods
Bacteria
F. nucleatum (ATCC 25586) was cultured at 37 °C under anaerobic conditions in brain-heart infusion broth supplemented with yeast extract (5 mg/mL), hemin (5 μg/mL), and menadione (1 μg/mL). Erythromycin (5 μg/ml) was used as a selective agent for F. nucleatum as previously described [49]. After 24 h of growth, bacteria were collected by centrifugation at 6000 × g for 10 min at 4 °C, washed twice and resuspended with phosphate-buffered saline (PBS). Quantification of bacteria was measured by optical density (OD) to obtain a concentration of 109 colony-forming units (CFU)/ml using a reference standard.
Mice and oral challenge with F.nucleatum
BALB/c mice were obtained from the animal facility of the Institute of Biophysics Carlos Chagas Filho at the Federal University of Rio de Janeiro. All protocols used in this study followed the guidelines and were approved by the Institutional Animal Care and Use Committee at the Federal University of Rio de Janeiro (CEUA-UFRJ 076/15).
Six-to eight-week-old male BALB/c mice were given ad libitum water containing 10 mL of Bactrim (Roche) comprised of sulfamethoxazole/trimethoprim for 10 days. Then antibiotic-free water was given to the mice for 3 days prior to infection. The protocol for oral infection was adapted from Baker et al. [50]. On days of infection, mice were anesthetized with 100 μl of ketamine-xylazine solution (100 mg/ml and 20 mg/ml) by intraperitoneal injection. Anesthetized mice were orally challenged with F. nucleatum at 109 bacteria in 100 μl of PBS with 2% carboxymethylcellulose (Sigma), or were sham-infected with the same solution containing no bacteria three times over 2-day intervals.
Collection of maxilla
Maxillas were surgically removed collected at days 1, 4, and 7 after the last infection day. Half maxilla was placed in TRIzol reagent (Life Technologies) for PCR analysis and the other half in cell lysis buffer (Sigma) containing protease inhibitor (Roche) for western blotting analysis. Then the samples were macerated and homogenized using TissueLyser LT (Qiagen) for 5 min at 50 Hz. After centrifugation at 1000 × g for 10 min supernatants were transferred to new tubes and used for experiments. Prior to protein assays, maxilla halves from each group of mice were pooled together and quantified using Pierce BCA Protein Assay Kit (Thermo Fisher Scientific) for equal loading.
Isolation, processing and analysis of murine gingival cells
The isolation, processing and analysis of murine maxilla gingival cells collected from uninfected or F. nucleatum-infected mice was as described by Mizraji et al., 2013 [51]. Briefly, after the protocol for oral infection, mice were euthanized and both upper and lower mandibles were collected. The mandibles were cut into hemi-maxillae and the palatal tissue was trimmed until reaching the alveolar bone. Gingival tissues were peeled using forceps (without teeth), placed in 1 ml PBS + 2% FCS, 2 mg/mL of collagenase type II and 1 mg/mL of DNAse type 1, well minced and incubated in a shaker incubator for 20 min at 37 °C, 200 rpm, plus an additional 10 min after adding 20 μl of EDTA 0.5 M. Samples were washed with 12 ml of PBS + 2% FCS and centrifuged at 4 °C, 400 × g for 8 min. Cells were resuspended with 2 ml PBS + 2% FCS and filtered on a 70 μm cell-strainer. The collected samples were centrifuged at 4 °C, 320 × g for 5 min and resuspended with 300 μL PBS + 2% FCS for cellular quantification and antibody staining.
Flow cytometry analysis
Freshly isolated murine gingival cells were immunostained for flow cytometry analysis. 5 × 105 cells were stained with 0.1 μg/mL Fixable Viability Stain 510 (FVS510) – BD Horizon™ for 15 min at room temperature to distinguish live and dead cells. After washing with PBS, cells were blocked for nonspecific binding with 5 μg/mL with CD16/32 mAb for 20 min on ice and stained with the following monoclonal antibodies (eBioscience) for 30 min at 4 °C: 1 μg/mL anti-CD90 (Thy-1.2) APC (Clone 53–2.1), 1 μg/mL anti- F4/80 Antigen eFluor® 450 (Clone BM8), 1 μg/mL anti-Ly-6G PE (clone 1A8-Ly6g), 2.5 μg/mL anti-CD11b Alexa Fluor® 488 (Clone M1/70). Cells were washed with PBS, fixed with 4% paraformaldehyde for 20 min at room temperature and kept at PBS until their acquisition by flow cytometry. Fluorescence was evaluated by acquiring 50,000 events/sample using FACSCanto II (BD Biosciences, San Jose, California, USA). Results were analyzed using the FACSDiva (BD Biosciences, San Jose, California, USA) and presented as percentage of positive events.
RNA extraction and quantitative PCR
Following the manufacturer's instructions, total RNA was extracted using Trizol Reagent (Life Technologies). Total RNA was quantified by using NanoDrop (Thermo Fisher Scientific). RNA was converted to cDNA using High-Capacity cDNA Reverse Transcription Kit (Life Technologies). Quantitative PCR was performed using SYBR-green fluorescence quantification system (SYBR Select Master Mix (Life Technologies)). Real-time PCR cycling parameters were as follows: 95 °C for 10 min and then 40 cycles of 95 °C for 30 s, 60 °C for 1 min, and 72 °C for 1 min. The following primers were used as previously described: IL-1β forward, 5′-TTCAGGCAGGCAGTATCACTC-3′; IL-1β reverse, 5′-CCACGGGAAAGACACAGGTAG-3′; TNFα forward, 5′-TTCTATGGCCCAGACCCTCA-3′; TNFα reverse, 5′-GTGGTTTGCTACGACGTGGG-3′; IL-6 forward, 5′-TCCTCTCTGCAAGAGACTTCC-3′; IL-6 reverse, 5′-TTGTGAAGTAGGGAAGGCCG-3′; IL-17 forward, 5′-TCAGCGTGTCCAAACACTGAG-3′, IL-17 reverse, 5′-GACTTTGAGGTTGACCTTCACAT-3′; GAPDH forward, 5′-GGTCATCCCAGAGCTGAACG-3′; GAPDH reverse, 5′- TTGCTGTTGAAGTCGCAGGA-3′ [52]. Relative expression levels were calculated against GAPDH as the reference gene using the comparative cycle threshold method. Quantification of infected mice results were normalized against control mice.
PCR
The PCR was performed following the protocol from Liu et al. [53]. Maxilla RNA targeted a 360-bp region using F. nucleatum forward, 5′-AGAGTTTGATCCTGGCTCAG-3′ and reverse, 5′-GTCATCGTGCACACAGAATTGCTG-3′ primer sequences [53]. The samples were amplified using GoTaq Green Master Mix (Promega) under the same conditions of 5 min at 94 °C and 30 cycles, with each cycle consisting of denaturation at 94 °C for 30 s, annealing at 58 °C for 30 s, extension at 72 °C for 1 min, and final extension for 10 min. PCR products were loaded onto a 2% gel in Tris-acetate buffer with EDTA. The gel was stained with GelRed nucleic acid gel stain (Biotium) and visualized under UV light.
ELISA
Homogenized maxilla were used for ELISA experiments. IL-1β, IFN-γ, and TNF-α cytokine levels were measured using Mouse IL-1β, IFN-γ, and TNF-α ELISA kits (R&D Systems). ELISAs were performed following manufacturer's instructions.
Western blot
Protein samples were dissolved in 6X Laemmli buffer and boiled. Then they were run on SDS-PAGE gels and transferred to PVDF membranes. Membranes were blocked with 5% BSA and incubated with anti-HMGB1 (Abcam) overnight. After primary incubation, the membranes were washed and incubated with HRP conjugated anti-goat IgG antibody (Millipore). Finally, the membranes were developed with Luminata Forte (Millipore) substrate. Images were acquired using ImageQuant LAS 4000 system and analyzed using NIH-ImageJ.
Histopathology and immunohistochemistry
For morphological studies mandibles were collected and were immediately immersed in zinc-formaldehyde for fixation for 72 h. Next, tissues were decalcified in Morse's solution for 7 days. The complete decalcification of bones was manually assessed. Tissues were then washed in water and dehydrated crescent solutions of ethanol, clarified in xylene and embedded in paraffin. Five-micrometer sections were cut and stained with hematoxylin-eosin (H&E). For immunohistochemistry, paraffin sections were collected onto charged histological slides. A rat monoclonal antibody F4/80 (Abd Serotec) was used to detect macrophages. Briefly, after dewaxing and rehydrating, sections were submitted to endogenous peroxidase inhibition (15 min with 3% H2O2 in methanol), followed by an enzymatic antigen retrieval, with a 0.1% trypsin solution containing 0.01% calcium chloride in Tris-buffer pH 7.4 (Sigma-Aldrich) for 5 min. After blocking nonspecific binding of immunoglobulins, primary antibody was incubated for 14–18 h, at 4 °C, in a humid chamber. The sections were then washed in 0.25% Tween-phosphate saline buffer (PBS) solution for 5 min and then the secondary antibody conjugated to peroxidase were incubated for 1 h at room temperature (Nichirei). The chromogen substrate was diaminobenzidine (Dako). Negative control slides were incubated with rat nonimmune serum or with the antibody diluent solution.
Histomorphometry
Histomorphometry was performed using a computer-assisted image analysis system comprising a Nikon Eclipse E-800 microscope connected via a digital camera (Evolution, Media Cybernetics Inc., Bethesda, MD) to a computer. The graphical interface software Q-Capture 2.95.0, version 2.0.5 (Silicon Graphic Inc., Milpitas, CA) was used. Ten high quality photomicrographs (high-quality images, 2048 × 1536 pixel buffer) were randomly captured from tissues of each animal using the 40× objective lens.
Quantification of the number of F4/80 macrophages
The number of macrophages was quantified in F4/80-stained sections. Results were expressed as the number of macrophages/histological field.
AFM force spectroscopy
For force spectroscopy studies, uninfected and 1, 4 and 7 days infected mandibles were collected and immediately immersed in PBS. After, tissues were cleaned, and the region of alveolar bone was exposed to start the measurement. Bone region was examined in a Dimension Icon Scanning Probe Microscopy (Bruker, Santa Barbara – CA). RTESPA-300 AFM probe (Bruker, Camarillo – CA) was used and its cantilever elastic constant was obtained by thermal noise method. Force curves were acquired in air using contact mode and young modulus elasticity was extracted by those curves using NanoScope Analysis 1.5 software (Bruker, Santa Barbara – CA).
Statistical analysis
Results are shown as mean ± standard deviation (SD). Statistical significance was calculated by two-tailed Student's t-test and differences were considered significant at P < 0.05.
SPM results were evaluated by one-way Anova followed by Dunnett's test and differences were considered significant at p < 0.05.
Results
F. nucleatum infection induced pro-inflammatory cytokine expression in maxilla
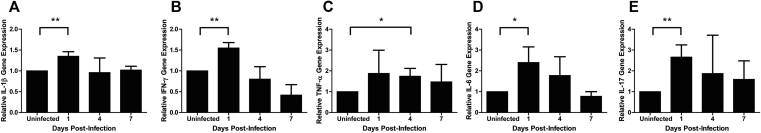
To assess cytokine expression in F. nucleatum-infected mice, we isolated RNA from the maxilla at various time points post-infection. Since F. nucleatum infection in GECs led to a time-dependent increase of IL-1β gene expression in vitro [46], we determined whether oral infection of F. nucleatum in mice would similarly upregulate IL-1β gene expression over a course of 7 d.p.i. (days post infection) [Fig. 1A]. Indeed, we observed a significant induction of IL-1β 1 d.p.i. compared to uninfected mice followed by a decrease in the transcriptional response thereafter. Because it was already known that F. nucleatum-infected BMDMs trigger cytokine production (such as IL-1β, TNF-α, and IL-6) through the Toll-like receptor (TLR) signaling pathway [54], we analyzed mRNA expression of other pro-inflammatory cytokines in our in vivo model of F. nucleatum infection. IFN-γ and IL-6 mRNA expression followed the same time-dependent response described for IL-1β, with an increase of gene expression at 1 d.p.i., decreasing thereafter [Fig. 1B and D]. On the other hand, TNF-α enhancement was delayed (at 4 d.p.i) but also declined thereafter [Fig. 1C]. Since IL-17 is implicated in bone resorption [52], [53], [54], [55], we examine its transcriptional response and found that it was upregulated early during infection, at 1 d.p.i., as observed for IL-1β, IFN-γ and IL-6 [Fig. 1E]. The results indicated that multiple pro-inflammatory cytokines were upregulated during the course of F. nucleatum infection.
Fig. 1.
Gene expression is upregulated in the maxilla during F. nucleatum infection. (A–E) Relative IL-1β, IFN-γ, TNF-α, IL-6, and IL-17 mRNA gene expression compared with control was evaluated by real-time PCR (qPCR) from the maxilla of F. nucleatum infected BALB/c mice. Days 1, 4, and 7 post-infection were tested. Results represent an average of three independent experiments with at least 4 mice in each group per experiment. Error bars represent the mean ± SD. (*< 0.05, **<0.01, Student's t-test).
Cytokine production is enhanced in the maxilla during F. nucleatum infection
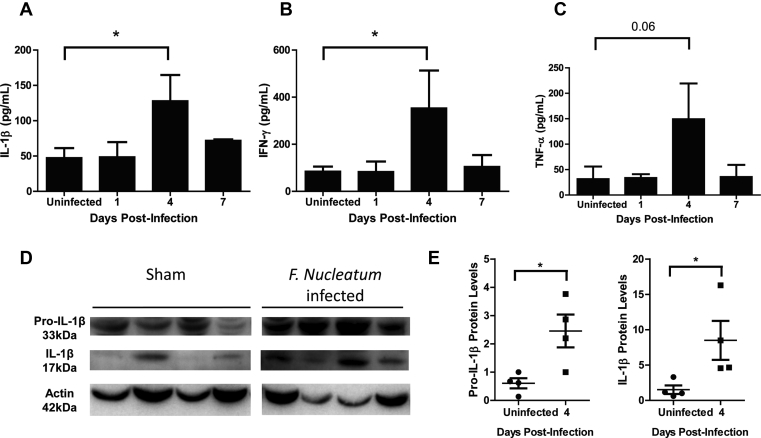
To directly evaluate cytokine production, we performed ELISA analysis using maxilla samples of infected mice and found that IL-1β and IFN-γ levels increased at 4 d.p.i.; i.e., 3 d after the transcriptional activation of these genes [Fig. 2A and B]. Additionally, a slight (non-statistically significant) increase of TNF-α was noted at 4 d pi [Fig. 2C].
Fig. 2.
F.nucleatum infection induces increased cytokine production in the maxilla. (A–C) The cytokines IL-1β, IFN-γ, and TNF-α were measured by ELISA. Maxilla from days 1, 4, and 7 post-infection with F. nucleatum were evaluated. (D) Western blotting to evaluate immature pro-IL-1β (33 kDa) and mature IL-1β (17 kDa) protein expression from the maxilla of F. nucleatum-infected BALB/c mice at day 4. Actin (42 kDa) was used as a loading control. (E) Relative protein was measured by quantification of densitometry using NIH-ImageJ. (A–C) represent an average of three independent experiments with at least 4 mice in each group per experiment. (D and E) represent average of 4 mice in each group. Error bars represent the mean ± SD. (*< 0.05, **<0.01, Student's t-test).
To further analyze the cytokine production, we determined by Western blotting the presence of the immature and mature forms of IL-1β in the maxilla of mice at the peak of cytokine production (4 d.p.i.). There was an enhancement of pro-IL-1β and IL-1β protein expression in the maxilla of F. nucleatum-infected mice, compared with sham-infected mice [Fig. 2D]. Confirmed by densitometry analysis [Fig. 2E], these data suggest that the early (1 d.p.i.) transcriptional response induced by F. nucleatum translated into sustained production of the pro-IL-1β precursor, which underwent cleavage at this time-point (4 d.p.i.). These in vivo results are consistent with our previous findings showing that IL-1β production by F. nucleatum-infected GECs is driven by the NRLP3 inflammasome-caspase 1 pathway [29].
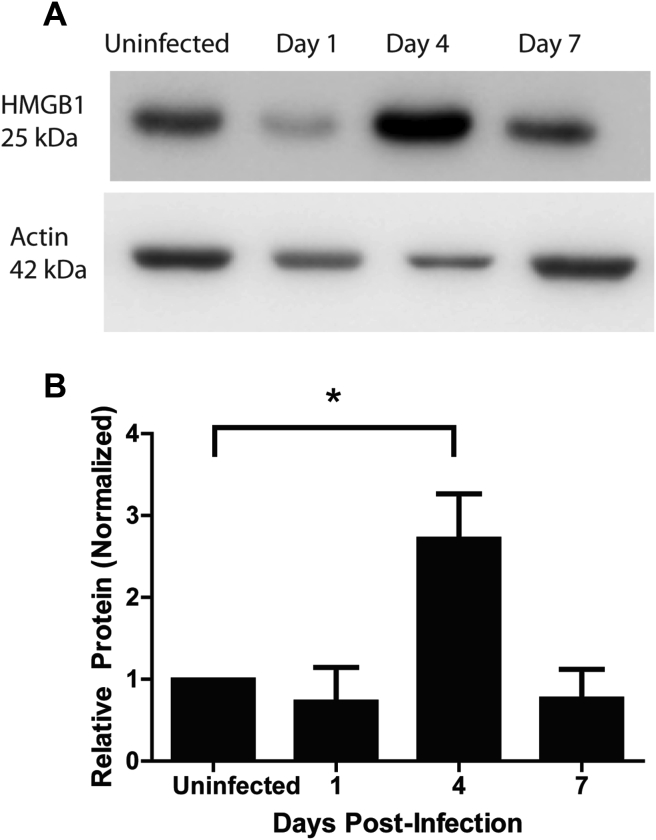
Next, we looked at high mobility group box 1 (HMGB1) expression because this damage-associated motif pattern (DAMP) acts as a transcription regulator, pro-inflammatory cytokine, and macrophage activator [55], [56], [57]. IFN-γ, TNF-α, and TGF-β have been shown to augment expression of HMGB1 mRNA in THP-1 macrophages and human peripheral-blood monocytes [56], [58]. As we observed an increase in two of these cytokines during F. nucleatum oral infection, it seemed plausible that these pro-inflammatory responses were linked to HMGB1 production. Using Western blot analysis, we found a significant increase of HMGB1 expression at 4 d.p.i. as compared with the sham-infected mice [Fig. 3A and B].
Fig. 3.
HMGB1 levels augmented at day 4 post-infection. (A) HMGB1 (25 kDa) was detected by Western blot from the maxilla of BALB/c mice infected with F. nucleatum over a time-course of day 1, 4, and 7 post-infection. Actin (42 kDa) was used as a loading control. (B) Relative protein was measured by quantification of densitometry using NIH-ImageJ. Results represent an average of three independent experiments with at least 4 mice in each group per experiment. Error bars represent the mean ± SD. (*< 0.05 Student's t-test).
F. nucleatum infection results in recruitment of immune cells to the maxilla
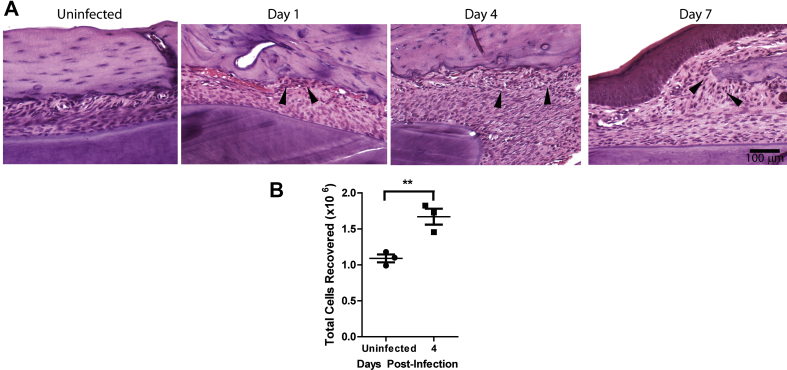
Given that inflammatory cytokine production is increased over time in F. nucleatum infected mice, we next characterized the dynamics of leukocyte infiltration in tissue sections obtained from the mandible. Analysis by H&E showed increased numbers of infiltrating leukocytes near the alveolar bone at 1, 4 and 7 d.p.i., when compared with uninfected mice [Fig. 4A]. To confirm this result, we quantified total inflammatory cells isolated from murine gingival tissues surrounding the teeth and found increased infiltration at 4 d.p.i. as compared with the uninfected group [Fig. 4B].
Fig. 4.
Immune cells are recruited near the alveolar bone during F. nucleatum infection. (A) Sections from mandible of infected BALB/c mice were stained with H&E. Arrows indicate area of immune cells localized near the bone in dark stain. Bar represents 100 μm. (B) Total immune cells recruited to the murine gingival tissue of F. nucleatum-infected BALB/c mice at day 4. Graph shows mean ± SD of immune cell numbers found in the murine gingival tissue of infected mice. Results show an average of 6 mice in each group (**<0.01, Student's t-test).
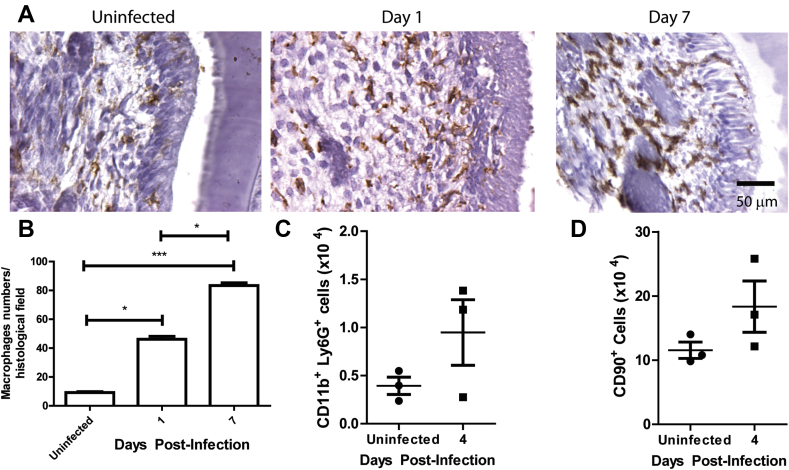
To further characterize the infiltrated cells after F. nucleatum infection in vivo, we used immunohistochemistry images to observe macrophage infiltration of the infected gingival tissues. F4/80+ staining revealed that macrophage recruitment is increased in the infected gingiva (1 and 7 d.p.i.) when compared with uninfected mice [Fig. 5A and B]. We next quantified CD11b+ Ly6G+ neutrophils [Fig. 5C] and CD90 + lymphocytes [Fig. 5D] and found a modest increase (not statistically significant) in both subsets in the infected mice group compared to controls [59]. Activated macrophages were detected in adipose tissues, skeletal muscle fibers, and periosteal localization [59]. Noteworthy, however, macrophages were more concentrated in the dental pulp, around the periodontal ligaments, and also between odontoblasts.
Fig. 5.
F. nucleatum infection increases immune cell recruitment in the dental pulp and in gingival tissue. (A) Immunohistochemistry for F4/80+ macrophages in the dental pulp of infected BALB/c mice. Bar represents 50 μm. (B) Quantification of macrophages from 10 histological fields. Total (C) neutrophils (CD11b+ Ly6G+) and (D) lymphocytes (CD90+) recruited to the murine gingival tissue of F. nucleatum-infected BALB/c mice at day 4. Graphs show the mean ± SD of immune cells numbers found in the murine gingival tissue of infected or uninfected mice. (C and D) each point represents 2 mice in each group. Error bars represent the mean ± SD. (*<0.05, ***<0.001, Student's t-test).
Bone resorption during F. nucleatum infection
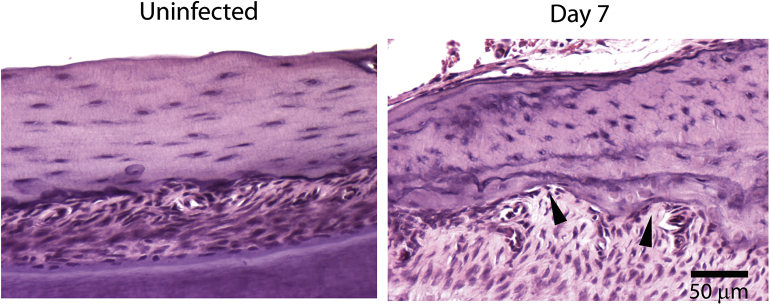
Osteoclast activation leads to their attachment to the bone, resorption of the bone through its secretory factors, and detachment from the resorption site [60], [61]. Previous studies have shown that IL-1β, and TNF-α enhance osteoclast development [41], [62], [63]. Since the production of these cytokines was elevated during F. nucleatum infection, we determined whether these inflammatory responses were coupled to bone resorption by osteoclasts. Indeed, we detected bone resorption pits in the alveolar bone (7 d.p.i.) compared to the uninfected group [Fig. 6], which may have resulted from osteoclast involvement.
Fig. 6.
F. nucleatum infection induces bone resorption. Mandibles of infected BALB/c mice were stained with H&E. Arrows indicate areas of bone resorption. Bar represents 50 μm.
F. nucleatum infection is associated with softened alveolar bone
To analyze the impact of the infection on bone structure, we measured the mechanical properties of the alveolar bone by Force Spectroscopy using an atomic force microscope. Uninfected mice prepared for these experiments expressed an elastic constant average of 7.95 ± 10.73 GPa [Fig. 7]. Infected mice had a significantly lower elasticity when compared with uninfected mice. These differences were more obvious between uninfected and infected mice 1 d.p.i, when elasticity decreased to 2.76 ± 5.56 GPa. Elasticity values at days 4 and 7 were 5.61 ± 7.88 GPa and 3.44 ± 3.42 GPa, respectively [Fig. 7].
Fig. 7.
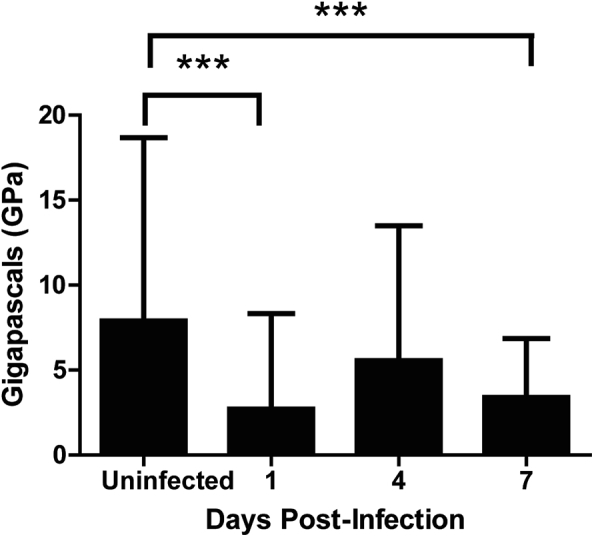
Alveolar bone is more elastic in F. nucleatum-infected mice. Young modulus of alveolar bone was measured with an atomic force microscope. Results represent an average of at least 160 measurements from 4 different mice per group. Error bars represent the mean ± SD. Multiple comparisons were corrected with Dunnett's method.
Discussion
Previous studies have examined the mechanisms involved in the adhesion and metabolic growth interactions between F. nucleatum and other periodontal bacteria, such as P. gingivalis and A. actinomycetemcomitans [64], [65], [66], [67], but monomicrobial oral infection with F. nucleatum in BALB/c mice had never been reported.
Our studies demonstrated that F. nucleatum can rapidly trigger inflammatory responses, manifested as increased mRNA for pro-inflammatory cytokines such as IL-1β, IFN-γ, IL-6 and IL-17 at 1 d.p.i., and secreted high levels of IL-1β and IFN-γ and the DAMP, HMGB1, in the maxilla at 4 d.p.i. These effects were followed by immune cell infiltration in gingival tissue observed from 1 d.p.i. on, culminating in bone resorption detected as early as 7 d.p.i. Alveolar bone softness was detected soon after, at 1 d.p.i.
Different oral bacteria can negatively or positively regulate the production of pro-inflammatory cytokines in GECs. While P. gingivalis subverts innate immunity by dampening IL-1β and HMGB1 release during infection in GECs, A. actinomycetemcomitans can stimulate IL-6 and TNF-α production in BMDMs [54], [68]. Likewise, infection with F. nucleatum stimulates cytokine production through the activation of Toll-like receptor (TLR)-2 and TLR-4 [54], [69], [70], or alternatively, through TLR-independent pathways [70], [71]. Our results show that F. nucleatum-infected mice upregulate IL-1β, IL-6, TNF-α, and HMGB1, consistent with previous in vitro studies on GECs and BMDMs infected with F. nucleatum [46], [54].
Given the heightened inflammatory response induced by F. nucleatum infection, it was not surprising to see a recruitment of immune cells to the alveolar bone during infection. Macrophages are known to reside in dental pulp [72]. Their activation relies on exposure to IFN-γ, TNF-α, and LPS [73], [74], [75]. IFN-γ is important for antibacterial activity and is augmented by stimulation with LPS [76], [77], [78]. We found that F. nucleatum infection induced significant levels of IFN-γ. In addition, immunohistochemistry revealed the recruitment of macrophages throughout the oral cavity.
It remains to be determined whether IFN-γ promotes or inhibits osteoclast activation [39], [62], [79], [80]. However, in our study, IFN-γ may have contributed to activation of osteoclasts as they were observed localized near the bone after F. nucleatum infection. Moreover, TNF-α and IL-17 synergize with IL-1β, and can facilitate osteoclast activation, further supporting a role for these cytokines in bone loss in the mandible [81], [82] during infection with F. nucleatum.
Conflicts of interest
The authors declare that they have no conflicts of interest.
Acknowledgements
This study was supported by intramural funds from the University of the Pacific, and the Fundação de Amparo à Pesquisa do Estado do Rio de Janeiro (FAPERJ). Cássio Luiz Coutinho Almeida-da-Silva received a PhD fellowship from CNPq (141715/2014-6) and FAPERJ (200417/2016; 200092/2016). Gustavo Miranda Rocha received a Postdoctoral fellowship from FAPERJ (217689/2015-2). We are grateful to Dr. Ana Morandini (University of the Pacific) for helpful discussions during preparation of this manuscript.
Footnotes
Peer review under responsibility of Chang Gung University.
Supplementary data related to this article can be found at https://doi.org/10.1016/j.bj.2018.05.001.
Appendix ASupplementary data
The following are the supplementary data related to this article:
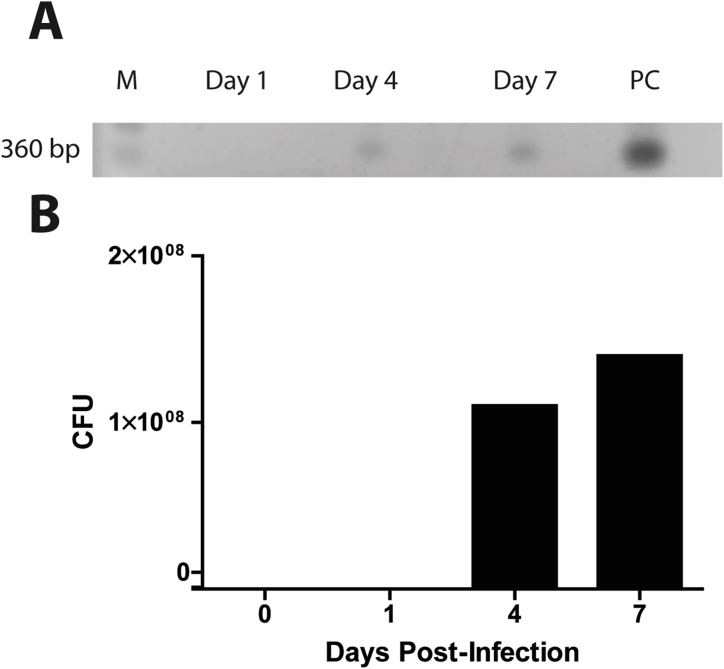
Supplementary Fig. 1.
Detection of F. nucleatum. (A) Gel electrophoresis of RT-PCR products from the maxilla of F. nucleatum infected BALB/c mice, measuring the F. nucleatum (band at 360 bp). 109 CFU were loaded for the positive control. (B) Band intensity was measured using NIH-ImageJ. CFUs for each day were calculated using relative band intensity, compared with the positive control. M: Marker, PC: Positive Control.
Supplementary Fig. 2.
Positive control for F4/80+staining of macrophages. BALB/c bone marrow was stained for F4/80+ macrophages. Negative control slide was prepared similarly to the immunostained ones except that, instead of specific antibody, the negative control slide was stained with the normal mice serum or the antibody dilution solution. Positive control slide: maxilla histological sections showing the positive bone marrow monocyte/macrophages for F4/80 antigen. Bar represents 50 μm.
References
- 1.Dewhirst F.E., Chen T., Izard J., Paster B.J., Tanner A.C., Yu W.H. The human oral microbiome. J Bacteriol. 2010;192:5002–5017. doi: 10.1128/JB.00542-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bik E.M., Long C.D., Armitage G.C., Loomer P., Emerson J., Mongodin E.F. Bacterial diversity in the oral cavity of 10 healthy individuals. ISME J. 2010;4:962–974. doi: 10.1038/ismej.2010.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Han Y.W., Shi W., Huang G.T., Kinder Haake S., Park N.H., Kuramitsu H. Interactions between periodontal bacteria and human oral epithelial cells: Fusobacterium nucleatum adheres to and invades epithelial cells. Infect Immun. 2000;68:3140–3146. doi: 10.1128/iai.68.6.3140-3146.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Signat B., Roques C., Poulet P., Duffaut D. Fusobacterium nucleatum in periodontal health and disease. Curr Issues Mol Biol. 2011;13:25–36. [PubMed] [Google Scholar]
- 5.Armitage G.C. Periodontal diagnoses and classification of periodontal diseases. Periodontology. 2004;34:9–21. doi: 10.1046/j.0906-6713.2002.003421.x. [DOI] [PubMed] [Google Scholar]
- 6.Silva N., Abusleme L., Bravo D., Dutzan N., Garcia-Sesnich J., Vernal R. Host response mechanisms in periodontal diseases. J Appl Oral Sci: Revista FOB. 2015;23:329–355. doi: 10.1590/1678-775720140259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Arigbede A.O., Babatope B.O., Bamidele M.K. Periodontitis and systemic diseases: a literature review. J Indian Soc Periodontol. 2012;16:487–491. doi: 10.4103/0972-124X.106878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hasan A., Palmer R.M. A clinical guide to periodontology: pathology of periodontal disease. Br Dent J. 2014;216:457–461. doi: 10.1038/sj.bdj.2014.299. [DOI] [PubMed] [Google Scholar]
- 9.Karpathy S.E., Qin X., Gioia J., Jiang H., Liu Y., Petrosino J.F. Genome sequence of Fusobacterium nucleatum subspecies polymorphum – a genetically tractable fusobacterium. PLoS One. 2007;2:e659. doi: 10.1371/journal.pone.0000659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Al-Ahmad A., Wunder A., Auschill T.M., Follo M., Braun G., Hellwig E. The in vivo dynamics of Streptococcus spp., Actinomyces naeslundii, Fusobacterium nucleatum and Veillonella spp. in dental plaque biofilm as analysed by five-colour multiplex fluorescence in situ hybridization. J Med Microbiol. 2007;56:681–687. doi: 10.1099/jmm.0.47094-0. [DOI] [PubMed] [Google Scholar]
- 11.Zilm P.S., Rogers A.H. Co-adhesion and biofilm formation by Fusobacterium nucleatum in response to growth pH. Anaerobe. 2007;13:146–152. doi: 10.1016/j.anaerobe.2007.04.005. [DOI] [PubMed] [Google Scholar]
- 12.Han Y.W., Ikegami A., Rajanna C., Kawsar H.I., Zhou Y., Li M. Identification and characterization of a novel adhesin unique to oral fusobacteria. J Bacteriol. 2005;187:5330–5340. doi: 10.1128/JB.187.15.5330-5340.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tuttle R.S., Strubel N.A., Mourad J., Mangan D.F. A non-lectin-like mechanism by which Fusobacterium nucleatum 10953 adheres to and activates human lymphocytes. Oral Microbiol Immunol. 1992;7:78–83. doi: 10.1111/j.1399-302x.1992.tb00513.x. [DOI] [PubMed] [Google Scholar]
- 14.Mangan D.F., Novak M.J., Vora S.A., Mourad J., Kriger P.S. Lectinlike interactions of Fusobacterium nucleatum with human neutrophils. Infect Immun. 1989;57:3601–3611. doi: 10.1128/iai.57.11.3601-3611.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Temoin S., Wu K.L., Wu V., Shoham M., Han Y.W. Signal peptide of FadA adhesin from Fusobacterium nucleatum plays a novel structural role by modulating the filament's length and width. FEBS Lett. 2012;586:1–6. doi: 10.1016/j.febslet.2011.10.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xu M., Yamada M., Li M., Liu H., Chen S.G., Han Y.W. FadA from Fusobacterium nucleatum utilizes both secreted and nonsecreted forms for functional oligomerization for attachment and invasion of host cells. J Biol Chem. 2007;282:25000–25009. doi: 10.1074/jbc.M611567200. [DOI] [PubMed] [Google Scholar]
- 17.Martinon F., Burns K., Tschopp J. The inflammasome: a molecular platform triggering activation of inflammatory caspases and processing of proIL-beta. Mol Cell. 2002;10:417–426. doi: 10.1016/s1097-2765(02)00599-3. [DOI] [PubMed] [Google Scholar]
- 18.Ozaki M., Miyake Y., Shirakawa M., Takemoto T., Okamoto H., Suginaka H. Binding specificity of Fusobacterium nucleatum to human erythrocytes, polymorphonuclear leukocytes, fibroblasts, and HeLa cells. J Periodontal Res. 1990;25:129–134. doi: 10.1111/j.1600-0765.1990.tb01034.x. [DOI] [PubMed] [Google Scholar]
- 19.Diamond G., Ryan L. Beta-defensins: what are they really doing in the oral cavity? Oral Dis. 2011;17:628–635. doi: 10.1111/j.1601-0825.2011.01799.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mathews M., Jia H.P., Guthmiller J.M., Losh G., Graham S., Johnson G.K. Production of beta-defensin antimicrobial peptides by the oral mucosa and salivary glands. Infect Immun. 1999;67:2740–2745. doi: 10.1128/iai.67.6.2740-2745.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dale B.A., Krisanaprakornkit S. Defensin antimicrobial peptides in the oral cavity. J Oral Pathol Med : Off Publ Int Assoc Oral Pathol Am Acad Oral Pathol. 2001;30:321–327. doi: 10.1034/j.1600-0714.2001.300601.x. [DOI] [PubMed] [Google Scholar]
- 22.Gomes Pde S., Fernandes M.H. Defensins in the oral cavity: distribution and biological role. J Oral Pathol Med: Off Publ Int Assoc Oral Pathol Am Acad Oral Pathol. 2010;39:1–9. doi: 10.1111/j.1600-0714.2009.00832.x. [DOI] [PubMed] [Google Scholar]
- 23.Rao M.R., Preethi P.L., Sathish M., Madhusudhan P. Gingival epithelial cell responses to microbial infections – an overview. J Res Adv Dent. 2014;3:53–56. [Google Scholar]
- 24.Dale B.A. Periodontal epithelium: a newly recognized role in health and disease. Periodontology. 2002;30:70–78. doi: 10.1034/j.1600-0757.2002.03007.x. [DOI] [PubMed] [Google Scholar]
- 25.Laube D.M., Dongari-Bagtzoglou A., Kashleva H., Eskdale J., Gallagher G., Diamond G. Differential regulation of innate immune response genes in gingival epithelial cells stimulated with Aggregatibacter actinomycetemcomitans. J Periodontal Res. 2008;43:116–123. doi: 10.1111/j.1600-0765.2007.00998.x. [DOI] [PubMed] [Google Scholar]
- 26.Medzhitov R. Toll-like receptors and innate immunity. Nat Rev Immunol. 2001;1:135–145. doi: 10.1038/35100529. [DOI] [PubMed] [Google Scholar]
- 27.Kawai T., Akira S. Signaling to NF-kappaB by Toll-like receptors. Trends Mol Med. 2007;13:460–469. doi: 10.1016/j.molmed.2007.09.002. [DOI] [PubMed] [Google Scholar]
- 28.Kelk P., Claesson R., Chen C., Sjostedt A., Johansson A. IL-1beta secretion induced by Aggregatibacter (Actinobacillus) actinomycetemcomitans is mainly caused by the leukotoxin. Int J Medical Microbiol: IJMM. 2008;298:529–541. doi: 10.1016/j.ijmm.2007.06.005. [DOI] [PubMed] [Google Scholar]
- 29.Hung S.C., Huang P.R., Almeida-da-Silva C.L.C., Atanasova K.R., Yilmaz O., Ojcius D.M. NLRX1 modulates differentially NLRP3 inflammasome activation and NF-kappaB signaling during Fusobacterium nucleatum infection. Microbes Infect. 2017:1–11. doi: 10.1016/j.micinf.2017.09.014. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bui F.Q., Johnson L., Roberts J., Hung S.C., Lee J., Atanasova K.R. Fusobacterium nucleatum infection of gingival epithelial cells leads to NLRP3 inflammasome-dependent secretion of IL-1β and the danger signals ASC and HMGB1. Cell Microbiol. 2016;18:970–981. doi: 10.1111/cmi.12560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Griffin G.K., Newton G., Tarrio M.L., Bu D.X., Maganto-Garcia E., Azcutia V. IL-17 and TNF-alpha sustain neutrophil recruitment during inflammation through synergistic effects on endothelial activation. J Immunol. 2012;188:6287–6299. doi: 10.4049/jimmunol.1200385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McGee D.W., Bamberg T., Vitkus S.J., McGhee J.R. A synergistic relationship between TNF-alpha, IL-1 beta, and TGF-beta 1 on IL-6 secretion by the IEC-6 intestinal epithelial cell line. Immunology. 1995;86:6–11. [PMC free article] [PubMed] [Google Scholar]
- 33.Guilloteau K., Paris I., Pedretti N., Boniface K., Juchaux F., Huguier V. Skin inflammation induced by the synergistic action of IL-17A, IL-22, Oncostatin M, IL-1{alpha}, and TNF-{alpha} recapitulates some features of psoriasis. J Immunol. 2010;184:5263–5270. doi: 10.4049/jimmunol.0902464. [DOI] [PubMed] [Google Scholar]
- 34.Awane M., Andres P.G., Li D.J., Reinecker H.C. NF-kappa B-inducing kinase is a common mediator of IL-17-, TNF-alpha-, and IL-1 beta-induced chemokine promoter activation in intestinal epithelial cells. J Immunol. 1999;162:5337–5344. [PubMed] [Google Scholar]
- 35.Palmqvist P., Lundberg P., Lundgren I., Hanstrom L., Lerner U.H. IL-1beta and TNF-alpha regulate IL-6-type cytokines in gingival fibroblasts. J Dental Res. 2008;87:558–563. doi: 10.1177/154405910808700614. [DOI] [PubMed] [Google Scholar]
- 36.Saperstein S., Chen L., Oakes D., Pryhuber G., Finkelstein J. IL-1beta augments TNF-alpha-mediated inflammatory responses from lung epithelial cells. J Interferon Cytokine Res: Off J Int Soc Interferon Cytokine Res. 2009;29:273–284. doi: 10.1089/jir.2008.0076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vignery A., Baron R. Dynamic histomorphometry of alveolar bone remodeling in the adult rat. Anat Rec. 1980;196:191–200. doi: 10.1002/ar.1091960210. [DOI] [PubMed] [Google Scholar]
- 38.Hienz S.A., Paliwal S., Ivanovski S. Mechanisms of bone resorption in periodontitis. J Immunol Research. 2015;2015:615486. doi: 10.1155/2015/615486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Soysa N.S., Alles N., Aoki K., Ohya K. Osteoclast formation and differentiation: an overview. J Med Dent Sci. 2012;59:65–74. [PubMed] [Google Scholar]
- 40.Goltzman D. Discoveries, drugs and skeletal disorders. Nat Rev. 2002;1:784–796. doi: 10.1038/nrd916. [DOI] [PubMed] [Google Scholar]
- 41.Boyce B.F., Li P., Yao Z., Zhang Q., Badell I.R., Schwarz E.M. TNF-alpha and pathologic bone resorption. Keio J Med. 2005;54:127–131. doi: 10.2302/kjm.54.127. [DOI] [PubMed] [Google Scholar]
- 42.Hwang S.Y., Putney J.W., Jr. Calcium signaling in osteoclasts. Biochim Biophys Acta. 2011;1813:979–983. doi: 10.1016/j.bbamcr.2010.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Blair H.C., Robinson L.J., Zaidi M. Osteoclast signalling pathways. Biochem Biophys Res Commun. 2005;328:728–738. doi: 10.1016/j.bbrc.2004.11.077. [DOI] [PubMed] [Google Scholar]
- 44.Hayman A.R. Tartrate-resistant acid phosphatase (TRAP) and the osteoclast/immune cell dichotomy. Autoimmunity. 2008;41:218–223. doi: 10.1080/08916930701694667. [DOI] [PubMed] [Google Scholar]
- 45.Lee H.R., Rhyu I.C., Kim H.D., Jun H.K., Min B.M., Lee S.H. In-vivo-induced antigenic determinants of Fusobacterium nucleatum subsp. nucleatum. Mol Oral Microbiol. 2011;26:164–172. doi: 10.1111/j.2041-1014.2010.00594.x. [DOI] [PubMed] [Google Scholar]
- 46.Bui F.Q., Johnson L., Roberts J., Hung S.C., Lee J., Atanasova K.R. Fusobacterium nucleatum infection of gingival epithelial cells leads to NLRP3 inflammasome-dependent secretion of IL-1beta and the danger signals ASC and HMGB1. Cell Microbiol. 2016;18:970–981. doi: 10.1111/cmi.12560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kostic A.D., Chun E., Robertson L., Glickman J.N., Gallini C.A., Michaud M. Fusobacterium nucleatum potentiates intestinal tumorigenesis and modulates the tumor-immune microenvironment. Cell Host Microbe. 2013;14:207–215. doi: 10.1016/j.chom.2013.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dharmani P., Strauss J., Ambrose C., Allen-Vercoe E., Chadee K. Fusobacterium nucleatum infection of colonic cells stimulates MUC2 mucin and tumor necrosis factor alpha. Infect Immun. 2011;79:2597–2607. doi: 10.1128/IAI.05118-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zaborina O., Li X., Cheng G., Kapatral V., Chakrabarty A.M. Secretion of ATP-utilizing enzymes, nucleoside diphosphate kinase and ATPase, by Mycobacterium bovis BCG: sequestration of ATP from macrophage P2Z receptors? Mol Microbiol. 1999;31:1333–1343. doi: 10.1046/j.1365-2958.1999.01240.x. [DOI] [PubMed] [Google Scholar]
- 50.Baker P.J., Dixon M., Evans R.T., Roopenian D.C. Heterogeneity of Porphyromonas gingivalis strains in the induction of alveolar bone loss in mice. Oral Microbiol Immunol. 2000;15:27–32. doi: 10.1034/j.1399-302x.2000.150105.x. [DOI] [PubMed] [Google Scholar]
- 51.Mizraji G., Segev H., Wilensky A., Hovav A.H. Isolation, processing and analysis of murine gingival cells. JoVE. 2013;77 doi: 10.3791/50388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ramos-Junior E.S., Morandini A.C., Almeida-da-Silva C.L., Franco E.J., Potempa J., Nguyen K.A. A dual role for P2X7 receptor during Porphyromonas gingivalis infection. J Dental Res. 2015;94:1233–1242. doi: 10.1177/0022034515593465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Liu P., Liu Y., Wang J., Guo Y., Zhang Y., Xiao S. Detection of fusobacterium nucleatum and fadA adhesin gene in patients with orthodontic gingivitis and non-orthodontic periodontal inflammation. PLoS One. 2014;9 doi: 10.1371/journal.pone.0085280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Park S.R., Kim D.J., Han S.H., Kang M.J., Lee J.Y., Jeong Y.J. Diverse Toll-like receptors mediate cytokine production by Fusobacterium nucleatum and Aggregatibacter actinomycetemcomitans in macrophages. Infect Immun. 2014;82:1914–1920. doi: 10.1128/IAI.01226-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Andersson U., Erlandsson-Harris H., Yang H., Tracey K.J. HMGB1 as a DNA-binding cytokine. J Leukoc Biol. 2002;72:1084–1091. [PubMed] [Google Scholar]
- 56.Lotze M.T., Tracey K.J. High-mobility group box 1 protein (HMGB1): nuclear weapon in the immune arsenal. Nat Rev Immunol. 2005;5:331–342. doi: 10.1038/nri1594. [DOI] [PubMed] [Google Scholar]
- 57.Erlandsson Harris H., Andersson U. Mini-review: the nuclear protein HMGB1 as a proinflammatory mediator. Eur J Immunol. 2004;34:1503–1512. doi: 10.1002/eji.200424916. [DOI] [PubMed] [Google Scholar]
- 58.Kalinina N., Agrotis A., Antropova Y., DiVitto G., Kanellakis P., Kostolias G. Increased expression of the DNA-binding cytokine HMGB1 in human atherosclerotic lesions: role of activated macrophages and cytokines. Arterioscler Thromb Vasc Biol. 2004;24:2320–2325. doi: 10.1161/01.ATV.0000145573.36113.8a. [DOI] [PubMed] [Google Scholar]
- 59.Lam R.S., O'Brien-Simpson N.M., Lenzo J.C., Holden J.A., Brammar G.C., Walsh K.A. Macrophage depletion abates Porphyromonas gingivalis-induced alveolar bone resorption in mice. J Immunol. 2014;193:2349–2362. doi: 10.4049/jimmunol.1400853. [DOI] [PubMed] [Google Scholar]
- 60.Vaananen K. Mechanism of osteoclast mediated bone resorption–rationale for the design of new therapeutics. Adv Drug Deliv Rev. 2005;57:959–971. doi: 10.1016/j.addr.2004.12.018. [DOI] [PubMed] [Google Scholar]
- 61.Teitelbaum S.L. Bone resorption by osteoclasts. Science. 2000;289:1504–1508. doi: 10.1126/science.289.5484.1504. [DOI] [PubMed] [Google Scholar]
- 62.Gao Y., Grassi F., Ryan M.R., Terauchi M., Page K., Yang X. IFN-gamma stimulates osteoclast formation and bone loss in vivo via antigen-driven T cell activation. J Clin Investig. 2007;117:122–132. doi: 10.1172/JCI30074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zupan J., Jeras M., Marc J. Osteoimmunology and the influence of pro-inflammatory cytokines on osteoclasts. Biochem Med. 2013;23:43–63. doi: 10.11613/BM.2013.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Diaz P.I., Zilm P.S., Rogers A.H. Fusobacterium nucleatum supports the growth of Porphyromonas gingivalis in oxygenated and carbon-dioxide-depleted environments. Microbiology. 2002;148:467–472. doi: 10.1099/00221287-148-2-467. [DOI] [PubMed] [Google Scholar]
- 65.Saito Y., Fujii R., Nakagawa K.I., Kuramitsu H.K., Okuda K., Ishihara K. Stimulation of Fusobacterium nucleatum biofilm formation by Porphyromonas gingivalis. Oral Microbiol Immunol. 2008;23:1–6. doi: 10.1111/j.1399-302X.2007.00380.x. [DOI] [PubMed] [Google Scholar]
- 66.Periasamy S., Kolenbrander P.E. Aggregatibacter actinomycetemcomitans builds mutualistic biofilm communities with Fusobacterium nucleatum and Veillonella species in saliva. Infect Immun. 2009;77:3542–3551. doi: 10.1128/IAI.00345-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Metzger Z., Lin Y.Y., Dimeo F., Ambrose W.W., Trope M., Arnold R.R. Synergistic pathogenicity of Porphyromonas gingivalis and Fusobacterium nucleatum in the mouse subcutaneous chamber model. J Endod. 2009;35:86–94. doi: 10.1016/j.joen.2008.10.015. [DOI] [PubMed] [Google Scholar]
- 68.Johnson L., Atanasova K.R., Bui P.Q., Lee J., Hung S.C., Yilmaz O. Porphyromonas gingivalis attenuates ATP-mediated inflammasome activation and HMGB1 release through expression of a nucleoside-diphosphate kinase. Microbes Infect. 2015;17:369–377. doi: 10.1016/j.micinf.2015.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Toussi D.N., Liu X., Massari P. The FomA porin from Fusobacterium nucleatum is a Toll-like receptor 2 agonist with immune adjuvant activity. Clin Vaccine Immunol. 2012;19:1093–1101. doi: 10.1128/CVI.00236-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Liu H., Redline R.W., Han Y.W. Fusobacterium nucleatum induces fetal death in mice via stimulation of TLR4-mediated placental inflammatory response. J Immunol. 2007;179:2501–2508. doi: 10.4049/jimmunol.179.4.2501. [DOI] [PubMed] [Google Scholar]
- 71.Quah S.Y., Bergenholtz G., Tan K.S. Fusobacterium nucleatum induces cytokine production through Toll-like-receptor-independent mechanism. Int Endod J. 2014;47:550–559. doi: 10.1111/iej.12185. [DOI] [PubMed] [Google Scholar]
- 72.Iwasaki Y., Otsuka H., Yanagisawa N., Hisamitsu H., Manabe A., Nonaka N. In situ proliferation and differentiation of macrophages in dental pulp. Cell Tissue Res. 2011;346:99–109. doi: 10.1007/s00441-011-1231-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Mosser D.M., Zhang X. Activation of murine macrophages. In: Coligan John E., editor. Current protocols in immunology. 2008. [Chapter 14:Unit 14 2] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Mosser D.M., Edwards J.P. Exploring the full spectrum of macrophage activation. Nat Rev Immunol. 2008;8:958–969. doi: 10.1038/nri2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Mosser D.M. The many faces of macrophage activation. J Leukoc Biol. 2003;73:209–212. doi: 10.1189/jlb.0602325. [DOI] [PubMed] [Google Scholar]
- 76.Schroder K., Hertzog P.J., Ravasi T., Hume D.A. Interferon-gamma: an overview of signals, mechanisms and functions. J Leukoc Biol. 2004;75:163–189. doi: 10.1189/jlb.0603252. [DOI] [PubMed] [Google Scholar]
- 77.Kamijo R., Le J., Shapiro D., Havell E.A., Huang S., Aguet M. Mice that lack the interferon-gamma receptor have profoundly altered responses to infection with Bacillus Calmette-Guerin and subsequent challenge with lipopolysaccharide. J Exp Med. 1993;178:1435–1440. doi: 10.1084/jem.178.4.1435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Shtrichman R., Samuel C.E. The role of gamma interferon in antimicrobial immunity. Curr Opin Microbiol. 2001;4:251–259. doi: 10.1016/s1369-5274(00)00199-5. [DOI] [PubMed] [Google Scholar]
- 79.Takayanagi H., Ogasawara K., Hida S., Chiba T., Murata S., Sato K. T-cell-mediated regulation of osteoclastogenesis by signalling cross-talk between RANKL and IFN-gamma. Nature. 2000;408:600–605. doi: 10.1038/35046102. [DOI] [PubMed] [Google Scholar]
- 80.Boyle W.J., Simonet W.S., Lacey D.L. Osteoclast differentiation and activation. Nature. 2003;423:337–342. doi: 10.1038/nature01658. [DOI] [PubMed] [Google Scholar]
- 81.Yago T., Nanke Y., Ichikawa N., Kobashigawa T., Mogi M., Kamatani N. IL-17 induces osteoclastogenesis from human monocytes alone in the absence of osteoblasts, which is potently inhibited by anti-TNF-alpha antibody: a novel mechanism of osteoclastogenesis by IL-17. J Cell Biochem. 2009;108:947–955. doi: 10.1002/jcb.22326. [DOI] [PubMed] [Google Scholar]
- 82.Kotake S., Udagawa N., Takahashi N., Matsuzaki K., Itoh K., Ishiyama S. IL-17 in synovial fluids from patients with rheumatoid arthritis is a potent stimulator of osteoclastogenesis. J Clin Investig. 1999;103:1345–1352. doi: 10.1172/JCI5703. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.