Abstract
N 6-methyladenosine (m6A) and N6, 2′-O-Dimethyladenosine (m6Am) modifications (m6A/m) of messenger RNA mediate diverse cellular functions. Oncogenic Kaposi’s sarcoma-associated herpesvirus (KSHV) has latent and lytic replication phases that are essential for the development of KSHV-associated cancers. To date, the role of m6A/m in KSHV replication and tumorigenesis is unclear. Here, we provide mechanistic insights by examining the viral and cellular m6A/m epitranscriptomes during KSHV latent and lytic infection. KSHV transcripts contain abundant m6A/m modifications during latent and lytic replication, and these modifications are highly conserved among different cell types and infection systems. Knockdown of YTHDF2 enhanced lytic replication by impeding KSHV RNA degradation. YTHDF2 binds to viral transcripts and differentially mediates their stability. KSHV latent infection induces 5′UTR hypomethylation and 3′UTR hypermethylation of the cellular epitranscriptome, regulating oncogenic and epithelial-mesenchymal transition pathways. KSHV lytic replication induces dynamic reprograming of epitranscriptome, regulating pathways that control lytic replication. These results reveal a critical role of m6A/m modifications in KSHV lifecycle and provide rich resources for future investigations.
Kaposi’s sarcoma-associated herpesvirus (KSHV) is etiologically linked to several malignancies including Kaposi’s sarcoma (KS) and primary effusion lymphoma (PEL)1. KSHV lifecycle has latent and lytic replication phases1. KSHV establishes latent infection in host cells replicating as an episome in the nucleus and expressing only several latent genes. Latently infected cells can be reactivated into lytic replication expressing most viral genes and producing virions. In KS, most tumor cells are latently infected by KSHV indicating an essential role of latent infection in this malignancy1. However, a few KSHV-infected tumor cells undergo spontaneous lytic replication contributing to tumor progression. Defining mechanisms regulating KSHV latent and lytic infection is essential for understanding KSHV lifecycle and pathogenesis of KSHV-associated malignancies. Despite intensive studies, these molecular events remain ill-defined.
N6-methyladenosine (m6A), the most prevalent modification on mRNA, was identified in 1970s2. The advent of methylated RNA immunoprecipitation followed by sequencing (m6A-seq) technology has enabled transcriptome-wide mapping of m6A3,4. However, the recent development of another technique, m6A individual-nucleotide-resolution cross-linking and immunoprecipitation (miCLIP), which allows precise transcriptome-wide mapping of m6A and another modification, N6, 2′-O-dimethyladenosine (m6Am) found mostly on the first nucleotide of mRNA, has revealed that the original m6A-seq mapping technique, in fact, detects both m6A and m6Am (m6A/m)5.
The large methyltransferase complex, which consists of METTL3, METTL14, WTAP, KIAA1429, RBM15, and other undiscovered subunits, catalyzes m6A methylation (“writers”)6–11. The two demethylases (“erasers”) ALKBH5 and FTO were initially reported to remove m6A on mRNA12,13. However, a recent study has revealed that FTO is actually a m6Am eraser14. m6A/m is functionally linked to mRNA degradation, translation, splicing, microRNA (miRNA) processing, and nuclear export, which are mediated by the “reader” proteins15–18. Among the “readers”, proteins of the YTH domain-containing family are the most heavily studied. YTHDF1 enhances translation of mRNA19. YTHDF2 was the only reader initially found to mediate mRNA degradation20. However, YTHDF1 and YTHDF3 are now also known to induce mRNA degradation and deadenylation21,22. In addition, YTHDF3, cooperates with either YTHDF1 or YTHDF2 to promote translation or degradation of transcripts, respectively23,24. YTHDC1 has been implicated in splicing whereas YTHDC2 is implicated in both translation and degradation25,26.
Recently, m6A/m modifications in several RNA viruses including human immunodeficiency virus type 1 (HIV-1), Zika virus (ZIKA), and hepatitis C virus (HCV) have been profiled, and in some cases shown to mediate viral replication22,27–30. While this manuscript was in preparation, a study on the role of m6A/m in KSHV lytic replication was published, however, this study lacks transcriptome-wide profiling of m6A/m31. Here, we have examined both viral and cellular m6A/m epitranscriptomes in diverse cell types latently infected by KSHV and in KSHV-infected cells undergoing lytic replication. We have found wide-spread m6A/m modifications on KSHV transcripts during latent and lytic replication. Importantly, YTHDF2 negatively affects lytic replication by enhancing decay of KSHV transcripts. During latent infection, KSHV reprograms cellular m6A/m epitranscriptome preferentially at the 5′UTR and 3′UTR to deregulate pathways critical for latency and tumorigenesis. During lytic replication, a subset of cellular genes that regulate KSHV lytic replication is dynamically methylated. These results reveal an important role of m6A/m in KSHV lifecycle and provide rich resources for further understanding the molecular basis of KSHV infection and KSHV-induced oncogenesis.
RESULTS
Viral latent transcripts are marked with m6A/m
To determine the role of m6A/m modifications in KSHV lifecycle, we began by mapping the KSHV epitranscriptomes in five types of cells latently infected by KSHV using the m6A-seq technology3,4, which is referred to as m6A/m-seq in this study since it is now known to detect both m6A and m6Am (m6A/m)5. These cell types include a PEL line BCBL1-R stably expressing doxycycline-inducible replication and transcription activator (RTA); KTIME, a telomerase-immortalized human microvascular endothelial cell line (TIME) infected by recombinant KSHV BAC36; KMSC, which are primary human adipose tissue-derived mesenchymal stem cells (MSC) infected and transformed by BAC36; KMM, which are primary rat metanephric mesenchymal precursor cells (MM) infected and transformed by BAC36; and KiSLK, a renal carcinoma cell line stably expressing doxycycline-inducible RTA (iSLK) infected by recombinant KSHV BAC16.
KSHV infection did not significantly alter the expression levels of transcripts of m6A/m “writers”, “erasers”, and “readers” in 4 pairs of cell types examined (Supplementary Fig. 1a). Upregulation at protein level by KSHV was observed with METTL14, FTO, ALKBH5, and YTHDF2 in iSLK cells (KiSLK vs iSLK); ALKBH5 and YTHDF1 in TIME cells (KTIME vs TIME); and YTHDF1 in MSC cells (KMSC vs MSC) (Supplementary Fig. 1b). All m6A/m-related enzymes were also expressed in latent BCBL1-R cells (Supplementary Fig. 1c,d). These results indicated that m6A/m-related proteins were likely functional in these cells.
Only a few KSHV genes including ORF71 (vFLIP), ORF72 (vCyclin), and ORF73 (LANA) were expressed in KMM indicating tight viral latency (Fig. 1a). However, robust spontaneous lytic replication was observed in BCBL1-R cells but to a lesser extent in KiSLK cells followed by KTIME and KMSC cells, resulting in reads from lytic transcripts during latency. Spurious transcripts spanning the ORF17-ORF20 region in KTIME, KMSC, and KMM were likely due to the insertion of a CMV promoter driven bacterial artificial chromosome (BAC) cassette containing a GFP gene and a hygromycin resistance gene32.
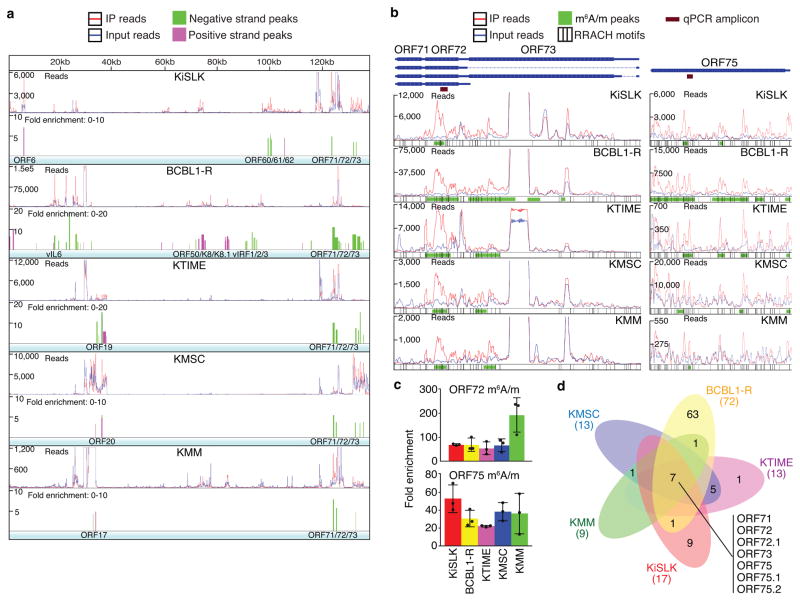
Figure 1. KSHV m6A/m epitranscriptome during viral latent infection.
a, Transcriptome-wide maps of KSHV m6A/m-IP reads, input reads, and m6A/m peaks in KiSLK, BCBL1-R, KTIME, KMSC, and KMM cells latently infected by KSHV. Selected genes containing m6A/m peaks are listed below each track. Reads were normalized to KiSLK for ease of comparison. b, Enlarged regions of ORF71, ORF72 and ORF73 (left), and ORF75 (right) from (a) containing the positions of qPCR amplicons and RRACH motifs. c, Validation of m6A/m peaks in ORF72 and ORF75 by MeRIP-qPCR. Fold enrichment was determined by calculating the fold change of IP to input Ct values. Experiments were independently repeated three times, and results are presented as mean +/− SD from the three experiments. d, Venn diagram showing the overlaps of methylated viral genes in all latently infected cells.
We subjected three biological replicates of poly-A purified RNA of each cell type to m6A/m-seq followed by peak calling using the exomePeak package with a stringent peak calling setting33. The results of three biological replicates were highly consistent (Supplementary Fig. 2). The most prominent m6A/m peaks conserved among all cell types were detected in transcripts of latent genes with the most enriched region centered in vCyclin coding region and extended into LANA C-terminus (Fig. 1b, left, Supplementary Table 1). Because these latent genes are essential for KSHV latency and cellular transformation, m6A/m modifications in this locus might regulate their expression and functions. The transcript of tegument protein ORF75, which is essential for lytic replication and silencing immune surveillance, is also methylated across all cell lines (Fig. 1b, right). It is highly expressed in KMSC, BCBL1-R and KiSLK cells but at lower levels in KTIME and KMM cells. Multiple m6A/m peaks are present on ORF75 transcript in all cell types except KMM cells. The m6A/m peaks on vCyclin and ORF75 transcripts were confirmed by methylated RNA immunoprecipitation quantitative real-time PCR (MeRIP-qPCR) (Fig. 1c). Furthermore, vFLIP, vCyclin, LANA, and ORF75 transcripts had m6A/m peaks conserved across all five cell types during KSHV latency (Fig. 1d).
Most KSHV transcripts are methylated during viral lytic replication
We further mapped the KSHV m6A/m epitranscriptome during lytic replication. Treatment with doxycycline efficiently triggers KSHV lytic replication in KiSLK and BCBL1-R cells34,35. Upon induction of lytic replication, m6A/m-related enzymes remained largely unchanged at both mRNA and protein levels in KiSLK cells; however, they declined at both mRNA and protein levels in BCBL1-R cells with ALKBH5, YTHDF1, and YTHDF2 proteins having the sharpest decreases 24h after induction (Supplementary Fig. 1c,d).
Purified mRNA from three biological replicates of lytically induced cells at 24h and 48h from KiSLK cells and at 48h from BCBL1-R cells were subjected to m6A/m mapping. Again, we observed highly consistent results among three biological replicates (Supplementary Fig. 2). During viral lytic replication, transcripts of most KSHV genes were expressed (Fig. 2a) with increase in reads by 76- and 119-fold at 24h and 48h, respectively, in KiSLK cells, and by 46-fold at 48h in BCBL1-R cells. We detected abundant m6A/m peaks on transcripts after lytic induction (Fig. 2a, Supplementary Table 2).
Figure 2. KSHV m6A/m epitranscriptome during viral lytic replication.
a, Transcriptome-wide maps of KSHV m6A/m-IP reads, input reads, and m6A/m peaks in KiSLK cells before (latent) and after induction for lytic replication for 24 h or 48 h, and in BCBL1-R cells before (latent) and after induction for lytic replication for 48 h. Selected genes containing m6A/m peaks are listed below each track. The latent datasets were reproduced from Fig. 1a for ease of comparison with the lytic datasets. Reads were normalized to KiSLK latent for ease of comparison. b, Enlarged regions of ORF71, ORF72, ORF73, RTA, ORF-K8, ORF-K8.1, ORF-K1, ORF4, ORF6, ORF-K3, ORF70, ORF8, ORF9, ORF10, ORF11, and ORF57 containing the positions of qPCR amplicons and RRACH motifs. c, Validation of m6A/m peaks by MeRIP-qPCR. Fold enrichment was determined by calculating the fold change of IP to input Ct values. Experiments were independently repeated three times, and results are presented as mean +/− SD from the three experiments. d, Venn diagrams comparing the number of methylated viral genes before (latent) and after induction for lytic replication in KiSLK (left) and BCBL1-R (right) cells. e, Comparison of methylated genes at 48 h after induction for lytic replication between KiSLK and BCBL1-R cells.
First, we observed gains of additional m6A/m peaks on vFLIP, vCyclin and LANA latent transcripts during lytic replication in both KiSLK and BCBL1-R cells, particularly new m6A/m peaks in LANA central and N-terminus regions, which were absent during latency (Fig. 2b). The latent locus contained many immunoprecipitated (IP) reads spanning its entire region, which might be due to multiple m6A/m bases that could not be sufficiently resolved by this approach. Second, we examined the RTA locus, which expresses multiple transcripts coding for RTA, an early protein K8, and a late protein K8.1 (Fig. 2b). Because of the RTA cassette, reads from the region contained those from the cassette and those endogenously transcribed. m6A/m peaks were detected on the N- and C-termini, and central region of RTA in lytic KiSLK cells but only on the central and C-terminal regions in lytic BCBL1-R cells. Interestingly, the expression of K8.1 was already robust in latent BCBL1-R cells compared to KiSLK cells and was significantly increased in both lytic BCBL1-R and KiSLK cells. An m6A/m peak was detected on the C-terminal region of K8.1 transcripts. Third, we observed other conserved m6A/m peaks between the two cell lines on transcripts of ORF-K1, ORF4, ORF-K3, ORF8, ORF11, and ORF57 (Fig. 2b). Numerous m6A/m peaks were cell type specific. For example, the m6A/m peak on ORF6 transcript was present in latent and lytic KiSLK cells but not in BCBL1-R cells. Fourth, all the selected m6A/m peaks were confirmed by MeRIP-qPCR (Fig. 2c).
Compared to latent KiSLK cells, there were significant gains in methylated viral genes in lytic KiSLK cells reflecting the increased expression levels of viral genes (Fig. 2d, left). In contrast, similar methylated viral genes were observed in latent and lytic BCBL1-R cells, reflecting high spontaneous lytic replication in these latent cells (Fig. 2d, right). Importantly, most of the methylated viral genes at 48h lytic replication were conserved between KiSLK and BCBL1-R cells (Fig. 2e). The abundance of m6A/m peaks on viral transcripts during lytic replication and their conserved nature between two cell types point to an important role of m6A/m modifications in KSHV lytic replication.
YTHDF2 inhibits KSHV lytic replication by promoting degradation of viral transcripts
Since the functions of m6A/m modifications are executed by the “reader” proteins, we investigated their roles in KSHV lytic replication. Knockdown of YTHDF1, YTHDC1 or YTHDC2 had no significant or consistent effect on viral lytic replication (Supplementary Fig. 3a–c). While knockdown of YTHDF3 reduced viral lytic replication by 40% and 70% with the two siRNAs, respectively, examination of viral transcripts and proteins did not yield consistent results (Supplementary Fig. 3d,e). In contrast, we observed consistent effect of YTHDF2 knockdown on KSHV lytic replication. Efficient knockdown of YTHDF2 was achieved two days after siRNA transfection (Fig. 3a,b), which consistently led to a 4-fold increase in viral production (Fig. 3c). Accordingly, we observed 2- to 6-fold increases in RTA, ORF57, ORF-K8, and ORF65 lytic transcripts (Fig. 3d), which also led to increases in their proteins (Fig. 3e). Overexpression of YTHDF2 led to decreased viral production and reduced levels of viral proteins (Fig. 3f–h). RNA-binding protein immunoprecipitation and quantitative real-time PCR (RIP-qPCR) showed that YTHDF2 strongly bound to RTA, vIL6, ORF-K8 and ORF57 lytic transcripts and, to lesser extents, LANA, PAN RNA and ORF65 transcripts (Fig. 3i).
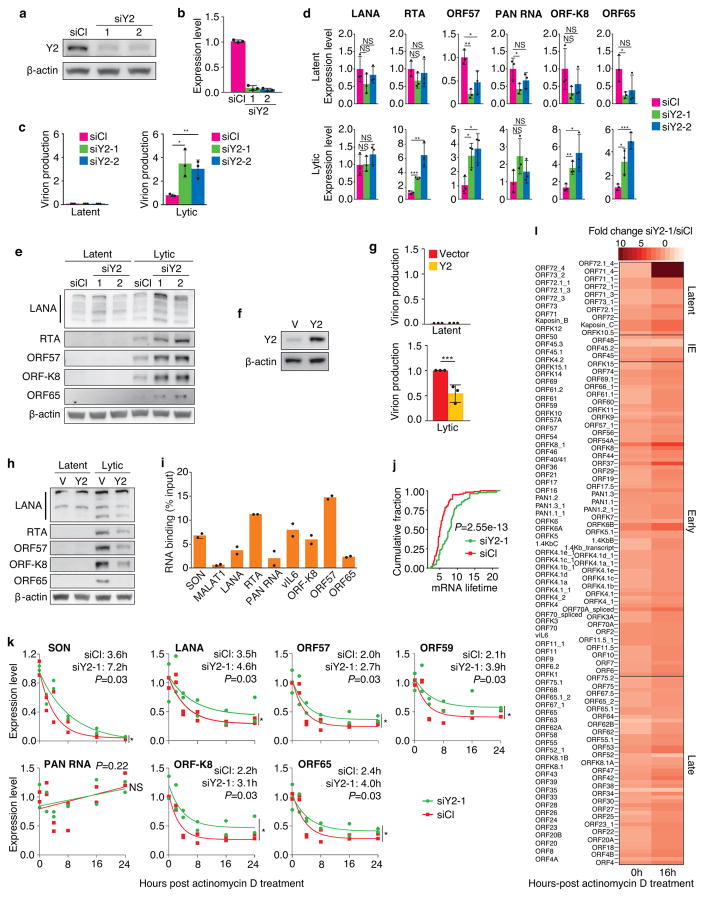
Figure 3. Silencing of YTHDF2 enhances KSHV lytic replication.
a,b, Knockdown of YTHDF2 shown at the protein (a) and mRNA (b) levels in KiSLK cells at day 2 post-transfection of siRNAs. c–e, Quantification of KSHV virions in culture supernatant by qPCR (c), and levels of viral transcripts (d) and proteins (e) were examined by RT-qPCR and Western-blotting, respectively, at day 3 after induction of lytic replication. Experiments were independently repeated three times, and results are presented as mean +/− SD from the three experiments (b–d) except (a,e) where representative results from one experiment are presented. NS = not significant, * p<0.05, ** p<0.01, *** p<0.001. f, YTHDF2 overexpression in KiSLK cells two days after lentiviral transduction. g,h, Quantification of KSHV virions in culture supernatant by qPCR (g), and levels of viral proteins (h) were examined by Western-blotting at day 3 after induction of lytic replication. Experiments were independently repeated three times, and results are presented as mean +/− SD from the three experiments (g) except (f,h) where representative results from one experiment are presented. i, KiSLK cells overexpressing Flag-YTHDF2 were induced for lytic replication and cell lysate was collected at 48 h to detect YTHDF2 binding of viral RNAs by RIP-qPCR. SON and MALAT1 are cellular positive and negative controls, respectively. Experiments were independently repeated twice, and results are presented as mean from the two experiments. j, Lifetimes of KSHV transcripts were measured in cells transfected with YTHDF2 siRNA (siY2-1) or a control siRNA (siCl). Results are from two independent experiments. k, Quantification of levels of viral and cellular transcripts following treatment with actinomycin D at day 3 after induction of lytic replication in KiSLK cells transfected with an YTHDF2 siRNA (siY2-1) or a control siRNA (siCl). The half-lives of the transcripts in hours were calculated. Experiments were independently repeated twice, and results are presented as mean +/− SD from the two experiments. l, Fold changes of KSHV transcripts in cells transfected with YTHDF2 siRNA (siY2-1) and control siRNA (siCl) at 0 h and 16 h post-actinomycin D treatment sorted by transcript class. Results are from two independent experiments.
Since YTHDF proteins enhance degradation and deadenylation of m6A/m-containing transcripts21,22, we examined the half-lives of cellular transcripts with and without YTHDF2 knockdown. As expected, the presence of m6A/m shortened the half-lives of transcripts (Supplementary Fig. 4a). Interestingly, YTHDF2 knockdown did not alter the negative effect of m6A/m on mRNA stability though all the transcripts became more stable (Supplementary Fig. 4b). Hence, YTHDF2 knockdown increased the half-lives of cellular transcripts (Supplementary Fig. 4c). These results could be due to other reader proteins such as YTHDF1, YTHDF3 and YTHDC2, which redundantly mediate m6A/m-dependent mRNA stability21,22,26. These reader proteins might compensate its function following YTHDF2 knockdown. Indeed, the YTHDF proteins have been reported to redundantly mediate the genome stability of several RNA viruses, including HCV, ZIKA and HIV-122,28–30. The fact that we only detected a consistent effect of single knockdown in YTHDF2 but not other reader proteins on KSHV lytic replication could be due to its relatively higher expression level in cells20.
We hypothesized that knockdown of YTHDF2 would prolong the half-lives of viral transcripts, leading to their accumulation. Indeed, silencing YTHDF2 caused an increase of the overall half-life of KSHV transcripts (Fig. 3j). Reverse transcription quantitative real-time PCR (RT-qPCR) confirmed that the half-lives of LANA, ORF57, ORF59, ORF-K8, and ORF65 transcripts were increased by an average of ~1.5 fold (Fig. 3k). The half-life of PAN RNA, a long non-coding RNA localized to the nucleus, was not significantly affected as it might escape YTHDF2-mediated degradation in the cytoplasm. Examination of transcript stability by functional class showed that latent transcripts coding for LANA, vCyclin, vFLIP, and early transcripts coding for ORF-K8, ORF37 and ORF-K6 had the largest increases in half-life following YTHDF2 knockdown, whereas immediate-early transcripts were least affected (Fig. 3l). Taken together, our results indicated that YTHDF2 might inhibit KSHV lytic replication by promoting the degradation of viral lytic transcripts. It is possible that YTHDF2 might act as a cellular defense mechanism to inhibit viral replication by restricting the expression of viral transcripts.
Cellular m6A/m epitranscriptome is reprogramed during KSHV latent infection
To determine whether KSHV modulates the cellular epitranscriptome, we examined the four pairs of uninfected and KSHV latently infected cells (Supplementary Fig. 1a). We included BCBL1-R because of its unique B-cell origin representing PEL. The three biological replicates showed excellent m6A/m-seq overlaps (Supplementary Table 3). First, we compared the uninfected cells and identified 3,233 methylated genes that were conserved among TIME, MSC, MM, and iSLK cells, accounting for about 40–60% of methylated genes in each cell type (Supplementary Fig. 5a). These results are expected as the m6A epitranscriptome is conserved among organisms. An m6A/m peak in Dicer1 and another in JunB as well as an unmethylated Dicer1 region conserved among all four cell types were selected and confirmed by MeRIP-qPCR (Supplementary Fig. 6a,b). Among the three human cell types, another 2,685 methylated genes (34–35%) were conserved, indicating that over 74–76% of the methylated genes were conserved among the human cells. MM cells had the highest number of uniquely methylated genes (1,156), indicating that the effect of species was stronger than cell type in specifying m6A/m methylation pattern. MM and MSC cells also shared many methylated genes (3,887), which could be attributed to both being primary precursor cells. Further clustering analysis confirmed the results, and identified conserved and specific m6A/m patterns among these cells (Supplementary Fig. 5b).
Next, we compared the methylated genes among the five latently infected cells. Like the uninfected cells, 3,042 methylated genes (33–51% of total genes) were conserved among all types of cells with more conservation observed among the human cells (5,227 genes or ~56–72% of total genes) (Fig. 4a). Of the human cells, BCBL1-R cells had 1,329 uniquely methylated genes compared to 245, 454, and 218 genes in KiSLK, KTIME, and KMSC cells, respectively. Clustering analysis confirmed the results, and identified conserved and specific m6A/m patterns among these cells (Supplementary Fig. 7a). The same m6A/m peaks in Dicer1 and JunB were also confirmed by MeRIP-qPCR (Supplementary Fig. 6a,b).
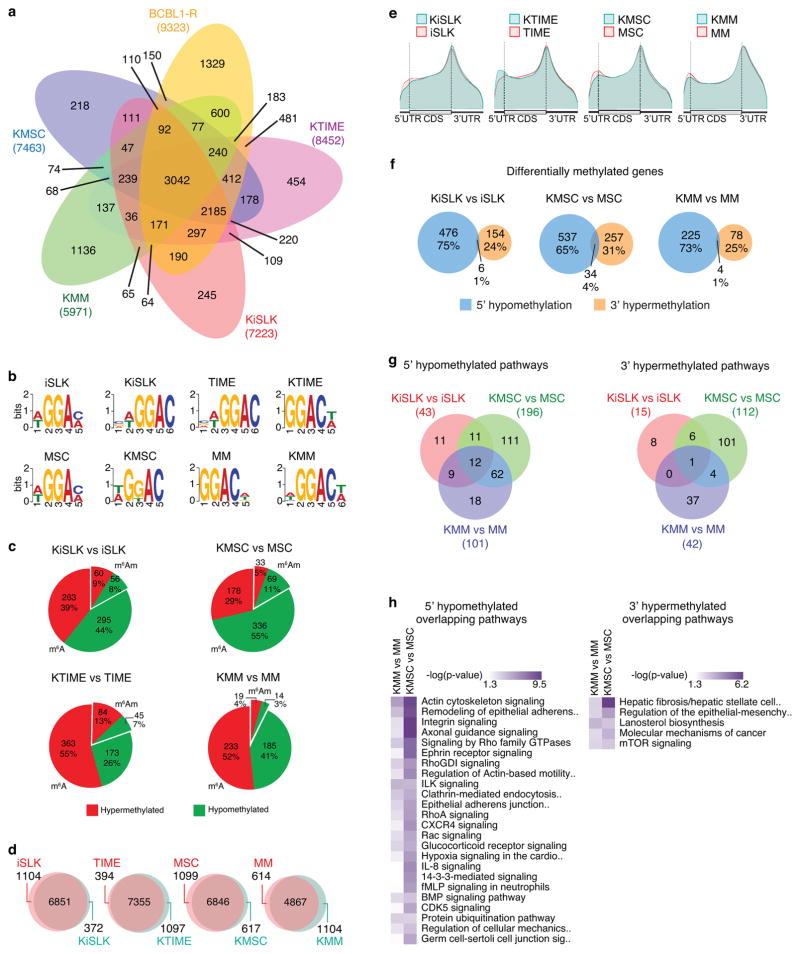
Figure 4. Reprograming of cellular m6A/m epitranscriptome during KSHV latency.
a, Venn diagram showing the overlaps of methylated cellular genes in all five types of cells latently infected by KSHV. b, Most significant motifs in cellular m6A/m peaks identified by MEME in uninfected cells and cells latently infected by KSHV. c, The predicted proportions of m6A and m6Am methylated transcripts, and the percentages of hypermethylated and hypomethylated genes. d, Comparison of cellular m6A/m genes in different pairs of uninfected cells and cells latently infected by KSHV. e, Distribution of cellular m6A/m peaks on transcripts in different pairs of uninfected cells and cells latently infected by KSHV as plotted by the Guitar software package. f, Comparisons between cellular 5′ hypomethylated genes and 3′ hypermethylated genes in different pairs of uninfected cells and cells latently infected by KSHV. g, Venn diagram showing overlaps of significantly enriched pathways of cellular 5′ hypomethylated genes (top) and 3′ hypermethylated genes (bottom) as a result of KSHV latent infection in different types of cells. h, Heat maps of significantly enriched pathways of 5′ hypomethylated genes (left) and 3′ hypermethylated genes (right) sorted by P-values as a result of KSHV latent infection in different types of cells. The results are from three biological replicates.
We detected the canonical “GGAC” m6A motif as the most common motif in both uninfected and infected cells, indicating that most m6A/m peaks were likely m6A peaks (Fig. 4b). Because this mapping approach cannot directly identify the m6Am peaks, we predicted the potential m6Am peaks as those “A”s at the beginning of a transcript, containing the m6Am conserved BCA motif5, and inside exomePeak predicted peaks. Only 7–20% of the peaks were predicted to be putative m6Am peaks (Fig. 4c).
Because we could not precisely map and distinguish m6A and m6Am motifs, we analyzed the overall m6A/m methylated genes without separating both types of peaks. Most methylated genes in uninfected cells remained methylated in KSHV-infected cells (Fig. 4d). Surprisingly, among the small number of differentially methylated genes, most occurred in 5′UTR and 3′UTR (Fig. 4e). We detected a loss of 5′UTR methylation and a slight gain of 3′UTR methylation following KSHV infection in KiSLK, KMSC, and KMM cells. The distributions of m6A/m in KTIME cells were different from other cells, possibly reflecting their untransformed state. Hence, we focused further analyses on KiSLK, KMSC, and KMM cells. Most 5′UTR hypomethylated and 3′UTR hypermethylated genes were unique (Fig. 4f, Supplementary Table 4), indicating that m6A/m might affect expression or translation of these two groups of genes by different mechanisms. Pathway analysis identified 12 conserved pathways that were 5′UTR hypomethylated and 1 conserved pathway that were 3′UTR hypermethylated across all three pairs of cell types (Fig. 4g). However, it was obvious that KMSC and KMM cells shared more common 5′UTR hypomethylated pathways compared to KiSLK cells. KMSC and KMM are primary cells transformed by KSHV, which are excellent models for studying KSHV-induced oncogenesis36,37. Indeed, the top 5′UTR hypomethylated and 3′UTR hypermethylated pathways in both KMM vs MM and KMSC vs MSC were broadly related to oncogenic/mitogenic signaling, cytoskeleton and extracellular signaling, endocytosis, loss of contact inhibition, remodeling of adherens junction, and cellular adhesion/invasion, all of which have been implicated in KSHV latency and cellular transformation (Fig. 4h). These findings indicated that m6A/m modifications might mediate cellular pathways implicated in KSHV-induced cellular transformation. We did not find any correlation of the extent and site of methylation (5′UTR vs 3′UTR) with gene expression, suggesting that m6A/m might mediate post-transcriptional gene regulation such as splicing, nuclear export, or translation.
We investigated the role of m6Am in the deregulation of cellular pathways during KSHV latency because of its presence at the start of transcripts, which might contribute to the 5′UTR hypomethylation. The ratios of the predicted m6Am hypermethylated to hypomethylated genes were similar to that of the overall m6A/m containing genes (Fig. 4c). Pathway analysis of m6Am containing genes did not yield any significantly enriched pathways, indicating a minimal contribution of m6Am to the altered oncogenic pathways of 5′UTR hypomethylation in KSHV latent cells.
KSHV lytic replication induces dynamic changes in cellular m6A/m epitranscriptome
We analyzed cellular m6A/m epitranscriptome during KSHV lytic replication (Supplementary Table 6). Over 83% and 41% of m6A/m genes remained methylated when KSHV was reactivated from latency in KiSLK and BCBL1-R cells, respectively (Fig. 5a). We confirmed the m6A/m peaks in Dicer1 and JunB by MeRIP-qPCR (Supplementary Fig. 8a,b). There were more de novo methylated genes in KiSLK cells than BCBL1-R cells (1,138 vs 51) at 48h after lytic induction (Fig. 5a). However, the m6A/m epitranscriptomes were clustered by cell type rather than replication status (Supplementary Fig. 7b). KiSLK and BCBL1-R cells shared most of the m6A/m genes during latent and lytic replication, respectively, albeit we observed small subsets of cell type specific methylated genes (Fig. 5b). Similar to latently infected cells, we detected “GGAC” as the most common motif in the lytic cells (Fig. 5c).
Figure 5. Reprograming of cellular m6A/m epitranscriptome during KSHV lytic replication.
a, Venn diagram comparing the number of methylated cellular genes before (latent) and after induction for lytic replication in KiSLK (left) and BCBL1-R (right) cells. b, Comparison of methylated cellular genes between KiSLK and BCBL1-R cells during latency (left) and lytic replication (right) at 48 h after induction. c, Most significant motifs in cellular m6A/m peaks identified by MEME in latent and lytic KiSLK and BCBL1-R cells. The latent KiSLK motifs are the same as in Fig. 4b and are replicated in this panel for ease of comparison. d, Distribution of cellular m6A/m peaks on transcripts in latent vs lytic cells in KiSLK cells (left) and BCBL1-R cells (right) as plotted by the Guitar software package. e, Comparison of significantly enriched pathways of cellular genes that are hypermethylated (left) or hypomethylated (right) as a result of reactivation from latency in KiSLK and BCBL1-R cells. f, Heat map of conserved hypermethylated pathways between KiSLK and BCBL1-R cells sorted by P-value. g, Heat map of conserved hypomethylated pathways between KiSLK and BCBL1-R cells sorted by P-value. h, The proportions of m6A and m6Am methylated transcripts, and the percentages of hypermethylated and hypomethylated genes. The results are from three biological replicates.
The most apparent difference between the two cell types was the overall change of m6A/m distribution on the transcripts when cells were reactivated from latency with KiSLK cells being 5′UTR hypermethylated and 3′UTR hypomethylated while BCBL1-R cells being 5′UTR hypomethylated and 3′UTR hypermethylated (Fig. 5d). Interestingly, of all significantly hypermethylated or hypomethylated genes in both cell types, over 50% of the enriched pathways in BCBL1-R cells overlapped with those in KiSLK cells (Fig. 5e, Supplementary Table 7). The common enriched pathways in both cell types included protein ubiquitination, ERK/MAPK signaling, integrin signaling, hypoxia signaling and sumoylation pathways (Fig. 5f.g), which have been implicated in KSHV lytic replication38–41.
Since YTHDF2 mediated the degradation of transcripts (Supplementary Fig. 4c), we further examined whether it might target genes according to m6A/m distribution. We did not find that different locations of m6A/m on the transcripts affected the half-lives of cellular transcripts. Nevertheless, it remains possible that YTHDF2 binding to different positions on the transcripts might differentially affect their stability.
Similarly, we investigated m6Am during KSHV lytic replication. Only 26% and 16% of peaks were predicted m6Am in KiSLK and BCBL1-R cells, respectively (Fig. 5h). Furthermore, few enriched pathways of the hypermethylated and hypomethylated m6Am genes overlap with those of m6A/m. Hence, m6Am had a minimal contribution to the overall m6A/m-altered pathways during KSHV lytic replication. While the presence of m6A/m shortened the half-lives of cellular transcripts, m6Am only had a marginal effect (Supplementary Fig. 4a), which was abolished following YTHDF2 knockdown (Supplementary Fig. 4b). Hence, YTHDF2 might be involved in m6Am regulation of mRNA stability in a small subset of transcripts. Among the viral transcripts, m6Am was only predicted to be present in ORF65, ORF66 and ORF67 transcripts in lytic KiSLK cells, and both latent and lytic BCBL1-R cells; and in ORF34, ORF35 and ORF36 transcripts in lytic BCBL1-R cells, indicating that m6Am was minimally involved in regulating the stability of viral transcripts.
DISCUSSION
Systematic profiling and characterization have revealed conservation across different kingdoms and diverse cellular functions of m6A/m modifications on mRNA42. m6A has been found on HSV-1 mRNA for decades but the transcript and position containing the modification as well as the underlying function remain unclear43. Numerous studies have recently profiled m6A/m modifications in genomes of RNA viruses including HIV-1, ZIKA, and HCV22,27–30. We have mapped m6A/m modifications on KSHV and cellular transcripts in diverse KSHV-infected cell types undergoing latent and lytic infection. This is the first effort to systematically profile viral and cellular m6A/m epitranscriptomes in cells infected by a large DNA virus.
Because of the limited resolution of the technique used in this study, we cannot distinguish the m6A and m6Am peaks and precisely map their positions. Hence, we have analyzed their combined m6A/m peaks. Nevertheless, based on the feature of m6Am, we have predicted that less than 20% and 26% of the methylation peaks are m6Am peaks on the cellular transcripts in latent and lytic KSHV infected cells, respectively.
We have identified a highly conserved m6A/m peak within the latent transcript, which contains as many as 10 potentially methylated RRACH motifs during latent infection (Fig. 1). During lytic replication, there are widespread m6A/m modifications on almost all viral transcripts (Fig. 2a), reflecting the overall expression of viral transcripts albeit there is no correlation between a m6A/m peak or peak level with the actual expression level of a transcript. m6A/m peaks on KSHV transcripts are conserved across KiSLK and BCBL1-R cells undergoing lytic replication (Fig. 2e), suggesting an essential role for m6A/m in KSHV lifecycle.
We show that YTHDF2 suppresses KSHV lytic replication (Fig. 3). Since YTHDF2 mediates the degradation and deadenylation of mRNA20,21, we propose that YTHDF2 might act as a cellular restriction factor of KSHV lytic replication by mediating degradation of lytic transcripts. YTHDF2 could transiently shuttle mRNA to P-bodies20; however, it might also directly shuttle viral transcripts to exonucleases, deadenylases or decapping enzymes such as XRN1, PARN, CCR4-NOT or DCP1/2 for degradation in a P-body-independent mechanism. Indeed, an example of P-body-independent mRNA degradation occurs during KSHV lytic replication via the viral SOX protein, an endonuclease that cleaves host mRNA at specific sites and recruits the 5′-3′ exonuclease XRN1 to complete the degradation of host mRNA44. Coincidently, we have observed that mRNA levels of m6A/m-related enzymes rapidly decline in BCBL1-R cells during KSHV lytic replication (Supplementary Fig. 1c,d), which could be attributed to the effect of SOX-mediated degradation44. Results of a microarray study also show that METTL3, WTAP, FTO, YTHDF1, and YTHDF2 are degraded during KSHV lytic replication, presumably by SOX45. However, we have observed more potent SOX activity in BCBL1-R than KiSLK cells (Supplementary Fig. 1c). It is currently unclear whether YTHDF2 and m6A/m modifications are involved in SOX-mediated RNA degradation.
We propose that the host cells might restrict KSHV lytic replication by degrading methylated viral transcripts via YTHDF2. Alternatively, KSHV might have hijacked this host function in favoring viral latency. Recent studies showed that YTHDF1, YTHDF2, and YTHDF3 suppressed ZIKA and HCV replication but enhanced HIV-1 replication22,28–30. While we did not detect any consistent roles of other reader proteins in KSHV lytic replication, more comprehensive works are required to exclude the possibility that they might regulate different stages of viral replication. As KSHV is a large DNA virus with a complex viral replication program involving multiple stages that are regulated by many viral and cellular factors, fine-tuning of each of these stages might be necessary to ensure optimal outcomes for its lifecycle.
In cells that are latently infected by KSHV, the most apparent phenotypes are consistent 5′UTR hypomethylation and 3′UTR hypermethylation in all pairs of cells except KTIME vs TIME cells (Fig. 4e). Of these genes, numerous pathways implicated in cellular transformation are enriched. In particular, the epithelial-mesenchymal transition (EMT) process is critical for embryogenesis and wound healing; however, KS and other cancer cells have hijacked this process to promote tumor growth, invasion, vascular extravasation, and metastasis36,37,46. EMT can be induced by various stimuli including growth factors and oncogenic signaling46,47. Indeed, the common enriched pathways among KMM and KMSC cells that are dynamically methylated include oncogenic pathways such as ephrin receptor signaling, ILK signaling, hypoxia signaling, BMP signaling, hepatic fibrosis, mTOR signaling, as well as pathways of remodeling of adherens junctions, Rho GTPase, Rac signaling, and regulation of actin-based motility by Rho directly involved in EMT in cancer46,47. Therefore, KSHV might induce cellular transformation by manipulating post-transcriptional mRNA modification. Since 5′UTR m6A/m methylation has been shown to mediate translation through an eIF4E-dependent pathway48 while the 3′UTR is preferentially targeted by miRNAs and is often enriched for m6A/m3,4, it would be interesting to investigate how m6A modifications at the 5′UTR or 3′UTR might mediate translation and miRNA targeting of the transcripts during KSHV latency. Indeed, we have previously shown that multiple KSHV miRNAs are essential for KSHV-induced cellular transformation and tumorigenesis49. Examination of KMM vs MM and KMSC vs MSC have not found any significant correlation of differential methylation in total, 3′UTR and 5′UTR with targets of KSHV miRNAs. It remains possible that another mechanism such as specific m6A/m location, secondary structure of the transcript and miRNA binding site, or recruitment of other cellular proteins might be involved, which has evaded our detection.
We have observed alterations of m6A/m modifications in a subset of the cellular transcripts during KSHV reactivation from latency with more de novo methylated genes observed in KiSLK cells than BCBL1-R cells (Fig. 5a). This is not due to different sequencing depth among the samples (Supplementary Table 8). Furthermore, there was a striking cell type specific difference in the distribution of m6A/m on these cellular transcripts, with 5′UTR manifesting hypermethylation in KiSLK cells and hypomethylation in BCBL1-R cells (Fig. 5d). This differential effect could be due to the more robust SOX activity in BCBL1-R cells than KiSLK cells that might cause preferential degradation of 5′UTR methylated transcripts (Supplementary Fig. 1c,d). Importantly, the enriched pathways that are hypermethylated in KiSLK or hypomethylated in BCBL1-R cells are highly overlapped (Fig. 5e). First, the ubiquitin/proteasome pathway, which is hypermethylated during lytic replication, is hijacked by KSHV RTA, ORF-K3, ORF-K5, and ORF-K7, ubiquitin ligases that downregulate the host cell immune surveillance by inducing proteasomal degradation of key immune response molecules50. Our results suggest that hypermethylation of gene transcripts might play a role in promoting the activity of this pathway. Second, pathways including integrin, actin nucleation, PI3K/AKT, hypoxia, ERK/MAPK, and ATM signaling implicated in lytic replication38–41 are both hypermethylated or hypomethylated, indicating that KSHV might deregulate these pathways by regulating m6A/m modifications to favor viral lytic replication.
In conclusion, as an additional layer of gene regulation at post-transcriptional level, it is not surprising that m6A/m is utilized by KSHV to mediate different stages of its lifecycle. While we have established the landscape of viral and cellular m6A/m epitranscriptomes during KSHV latent and lytic replication, and identified novel roles of m6A/m in KSHV lifecycle (Supplementary Fig. 9), the functions of specific m6A/m modifications and m6A/m-related factors in different stages of KSHV lifecycle, as well as how KSHV manipulates the m6A/m epitranscriptome to facilitate its infection and replication leading to the induction of KSHV-associated malignancies remain to be further investigated.
MATERIALS AND METHODS
Cell culture
iSLK, a renal carcinoma cell line containing a stable doxycycline-inducible cassette of RTA (ORF50), a KSHV immediate-early gene, which is essential and sufficient for activating KSHV lytic replication, was grown in HyClone DMEM supplemented with 10% fetal bovine serum (FBS, Sigma-Aldrich, St. Louis, MO), 1 μg/ml puromycin, 250 μg/ml G418, and 1% penicillin-streptomycin. KiSLK cells derived from iSLK cells by infection with a recombinant KSHV BAC1634 were grown as iSLK cells except that the medium also contained 1.2 mg/ml hygromycin B. BCBL1-R, a KSHV-infected PEL cell line BCBL1 stably expressing doxycycline-inducible RTA, was grown in RPMI1640 supplemented with 10% FBS, 1% penicillin-streptomycin, and 20 μg/ml hygromycin B35. MSC cells, which are primary human adipose tissue-derived mesenchymal stem cells, were cultured in MSC medium (ScienCell Research Laboratories, Carlsbad, CA) containing 20% FBS as previously described37. KMSC cells derived from MSC cells by infection and transformation with BAC3632,37 were cultured in the same media with the addition of 100 μg/ml hygromycin B. TIME, a telomerase-immortalized human microvascular endothelial cell line, and KTIME cells derived from TIME cells by infection with BAC36, were cultured as previously described51. MM cells, which are primary rat metanephric mesenchymal precursor cells, and KMM cells derived from MM cells by infection and transformation with BAC36, were cultured as previously described36.
No further authentication of the cell lines was performed for this study. iSLK, KiSLK, and BCBL1-R cells were obtained from Dr. Jae Jung. TIME cells were obtained from Dr. Don Ganem. KMM, MM, KMSC, MSC, and KTIME cells were generated in our laboratory. iSLK cells were isolated from a KS lesion of an AIDS patient. However, they were later found to be of renal cell carcinoma origin. Since iSLK cells support efficient and robust KSHV lytic replication, they have been used for studying KSHV lytic replication. In this study, we employed iSLK cells to study KSHV lytic replication. iSLK, KiSLK, BCBL1-R, KMM, MM, KMSC, MSC, TIME, and KTIME cells were tested for mycoplasma and showed to be negative.
KSHV lytic replication was induced in KiSLK and BCBL1-R cells by adding 1 μg/ml doxycycline to the culture media as previously described34,35. Infectious virions, viral transcripts and proteins were examined at day 3 after induction of lytic replication unless noted otherwise.
Antibodies
Antibodies used in this study included a rabbit anti-m6A/m antibody (202-003, Synaptic Systems, Goettingen, Germany); a rabbit anti-human METTL3 antibody (A301-567A, Bethyl Laboratories, Inc., Montgomery, TX); a rabbit anti-human METTL14 antibody (HPA038002, Sigma, St. Louis, MO); a rabbit anti-human WTAP antibody (NBP1-83040, Novus Biologicals, LLC, Littleton, CO); a mouse anti-FTO antibody (ab92821, Abcam, Cambridge, MA); a rabbit anti-ALKBH5 antibody (HPA007196, Sigma, St. Louis, MO); a rabbit anti-human YTHDF1 antibody (ab99080, Abcam, Cambridge, MA); a rabbit anti-human YTHDF3 antibody (ab103328, Abcam, Cambridge, MA); a goat anti-mouse YTHDF3 antibody (sc-87503, Santa Cruz Inc., Dallas, TX); a rat anti-LANA antibody (ab4103, Abcam, Cambridge, MA); a rabbit anti-human YTHDF2 antibody (24744-1-AP, Proteintech Group, Rosemont, IL); a mouse anti-ORFK8 antibody (sc-57889, Santa Cruz Inc., Dallas, TX); a rabbit anti-human YTHDC1 antibody (ab133836, Abcam, Cambridge, MA); a rabbit anti-human YTHDC2 antibody (ab176846, Abcam, Cambridge, MA); a mouse anti-human β-actin antibody (sc-47778, Santa Cruz Inc., Dallas, TX). An anti-ORF57 antibody was generated by Sigma-Aldrich by immunizing a rabbit with the peptide IDGESPRFDDSIIP. The rabbit antibody to RTA and the mouse antibody to ORF65 were previously described52,53.
Isolation of m6A/m RNA fragments
Isolation of m6A/m-containing fragments was performed as previously described with minor modifications54. Briefly, total RNA was extracted from cells using TRI Reagent® (Sigma-Aldrich) followed by one round of polyA purification with Dynabeads® mRNA DIRECT™ Kit according to manufacturers’ instructions (ThermoFisher Scientific, Waltham, MA). The mRNA was fragmented in a buffer containing 100 mM Tris-HCl at pH 7.0 and 100 mM ZnCl2 followed by incubation at 94˚C for 3 min. Successful fragmentation of mRNA with sizes close to 100 nucleotides was validated using a BioRad Experion™ Automated Electrophoresis Station (Bio-Rad, Hercules, CA). Prior to immunoprecipitation, 10 μg of anti-m6A/m antibody was incubated with 30 μl slurry of Pierce™ Protein A Agarose beads (ThermoFisher Scientific) by rocking in 250 μl PBS at 4˚C for 3 h. The beads were washed 3 times in cold PBS followed by one wash in an immunoprecipitation (IP) buffer containing 200 mM Tris-HCl at pH 7.4, 1 M NaCl, and 1% Igepal CA-630 (Sigma-Aldrich). To isolate the m6A/m-containing fragments, 120 μg of fragmented mRNA was added to the antibody bound beads in 250 μl IP buffer supplemented with RNasin Plus RNase inhibitor (Promega, Madison, WI), and the mixture was mixed at 4˚C for 2 h. The beads were washed 7 times with 1 ml IP buffer prior to elution with 100 μl IP buffer supplemented with 20 mM of m6A salt (M2780, Sigma-Aldrich). The mixture was incubated for 1 h at 4˚C and the eluate collected. A second elution was carried out and the eluates were pooled together before purification with TRI Reagent®.
Preparation of m6A/m-seq cDNA library
Purified eluate and input samples were used for preparation of libraries and sequencing at the Genome Sequencing Facility at the Greehey Children’s Cancer Research Institute at the University of Texas Health Science Center at San Antonio. Approximately 10–25 ng of mRNA was used for RNA-seq library preparation using the TruSeq stranded mRNA kit (Illumina, San Diego, CA) according to manufacturer’s protocol with two modifications. First, the elute-frag-prime stage was done at 80°C for 2 min to allow annealing without causing fragmentation. RNA was reverse transcribed into first strand cDNA using reverse transcriptase and random primers. This was followed by the second strand cDNA synthesis using DNA Polymerase I and RNase H. The cDNA fragments then went through an end repair process with the addition of a single ‘A’ base followed by ligation of adapters. The products were then purified and enriched by PCR amplification for 10 cycles to generate the final RNA-Seq library. The second modification was done to adjust the bead-DNA ratio to preserve smaller fragments during the adapter ligation double beads clean up step. A beads:DNA ratio of 1:3 instead of 1:1 was used. cDNA libraries were quantified and pooled for cBot amplification and subsequent sequencing on an Illumina HiSeq 2000 platform 50 bp single read sequencing module. After the sequencing run, demultiplexing with CASAVA was employed to generate a fastq file for each sample.
Genome annotation
For KSHV BAC36 and BAC16 genomes, we collected annotation information from previous studies of RNA-seq, 5′ and 3′ RACE, and transcript isoform mapping to generate a comprehensive annotation of viral transcripts55–60. UCSC hg19 and rn5 reference genomes of human and rat, and annotation files were downloaded from Illumina iGenome.
m6A/m-seq data analysis
Reads of the IP/Input samples were aligned to the corresponding genomes using Tophat2 Aligner v2.0.6, which implicitly calls Bowtie2 with default options61,62. Following this, peak calling and differential m6A/m methylation analyses were performed using the exomePeak R/Bioconductor package, a software specifically designed for m6A-seq data analysis33,63. The peak calling is based on the Przyborowski and Wilenski’s method for comparing the means of two Poisson distributions (C-test)64, which computes the methylation enrichment of normalized IP reads over normalized input reads (or mRNA abundance) as the test statistics. Only the loci that show significant methylation enrichment are determined as peak regions. For differential m6A/m analysis, the fold change of methylation enrichments between two conditions are calculated and a rescaled hypergeometric test is applied to determine the significance of differential fold enrichment. The output of exomePeak includes the loci of the (differential) m6A/m peaks, the gene symbol of the transcripts to which peaks localize, the detection p-values, false discovery rates (FDR), and methylation enrichment (for peak calling) or enrichment fold change (for differential m6A/m analysis). For both m6A/m peak calling and differential peak discovery, an FDR threshold of 0.05 is used. m6A/m peaks were annotated with identifiers such as Gene Symbol and RefSeq ID as well as regional overlapping status (5′UTR/CDS/3′UTR). Regional distribution of m6A/m was plotted using the Guitar package65. Motif analysis of m6A/m peaks was performed using the MEME package66.
The m6A/m-seq technology cannot measure the stoichiometry of m6A/m methylation and the predicted peaks could have low stoichiometry. However, exomPeak predicts m6A/m peaks based on their enrichment of IP reads over input reads. While this enrichment cannot directly estimate the stoichiometry, it preserves, to some extent, the relationship of the stoichiometry between different sites. That is, the higher the enrichment, the larger the stoichiometry the predicted site is likely to have. As a result, the significant peaks predicted by exomePeak are likely to have relatively higher stoichiometry. Nevertheless, because of the experimental and modeling noise, there is always a chance that the predicted peaks might have low stoichiometry.
m6Am analysis
Using the m6A/m-IP datasets, we predicted m6Am sites as “A”s that locate at the beginning of RefSeq-annotated transcripts, contain the BCA motif5, and are inside the exomePeak predicted peaks. If a peak was mapped to a RefSeq transcript with multiple transcript start sites, only the “A” that is closest to the peak center was predicted as an m6Am sites. Because of the limitation of m6A/m-seq, these predicted m6Am sites are only putative m6Am sites in the transcriptome and this method would miss the true m6Am sites without the BCA motifs. Furthermore, the predicted results could change if a different transcript annotation system is used.
Clustering analysis
To investigate the methylation behavior of genes across samples, the highest fold enrichment among all peaks within a gene was considered as the methylation fold enrichment of the gene. Hierarchical clustering with Euclidean distance was used to group similarly methylated genes according to sample.
Gene expression analysis
The expression of transcripts and isoforms, and differential expression levels were calculated using cufflinks and cuffdiff, respectively, based on the input samples67,68. To avoid underestimating the expression of viral genes during lytic replication, we used reads mapped to both cellular and viral genomes to calculate the FPKM. All the other bioinformatics analyses were performed on Matlab, R or Perl.
siRNA knockdown
siRNA silencing was performed by transfecting 2.5 pmol of each siRNA per well in a 12-well plate into the KiSLK cells using Lipofectamine® RNAi Max according to manufacturer’s instructions (ThermoFisher Scientific). Two days after transfection, the cells were monitored for knockdown efficiency of the target gene by RT-qPCR and Western-blotting, and induced for lytic replication with 1 μg/ml of doxycycline. siRNAs were purchased from Sigma-Aldrich. Their product numbers and sequences are as follows. YTHDF1 si1: SASI_Hs01_00233686 (5′-CAGAGCUCCGCGUAUGGGA-3′); YTHDF1 si2: SASI_Hs01_00233688 (5′-GUCAAUGGGAGUGGGCAUU-3′); YTHDF1 si3: SASI_Hs01_00233689 (5′-GAAACGCGGCGUUUGGGCA-3′); YTHDF2 si1: SASI_Hs01_00133215 (5′-GUUCCAUUAAGUAUAAUAU-3′); YTHDF2 si2: SASI_Hs01_00133214 (5′-CUGCUUAUCGUUCCAUGAA-3′); YTHDF3 si1: SASI_Hs01_00202277 (5′-CAAACCUCAACCGAAACUU-3′); YTHDF3 si2: SASI_Hs01_00202278 (5′-CAAUUCAAGGGACACUCAA-3′); YTHDC1 si1 SASI_Hs01_00115890 (5′-CUUGUAUCAGGUCAUUCAU-3′); YTHDC1 si2 SASI_Hs01_00363062 (5′-GUGAUGGACAGGAAAUUGA-3′); YTHDC2 si1 SASI_Hs01_00161045 (5′-CAACUUAGAGCAUCAGGUU-3′); YTHDC2 si2 SASI_Hs01_00161046 (5′-GCCUAUUUAUAACCACUGA-3′); and siControl (siCl): Sigma siRNA Universal Negative Control #1 (SIC001-10NMOL).
RT-qPCR for gene expression and MeRIP-qPCR for m6A/m-seq validation
Total RNA was isolated with TRI Reagent® according to manufacturer’s protocol. Reverse transcription was performed with 1 μg of total RNA using Maxima H Minus First Strand cDNA Synthesis Kit (Cat.# K1652, Thermo Fisher Scientific). qPCR was done using SsoAdvanced™ Universal SYBR® Green Supermix (BioRad) and analyzed with a CFX Connect™ Real-Time PCR Detection System (BioRad). Relative gene expression levels were obtained by normalizing the CT values to those of 18S to yield 2−ΔΔCt values. For validation of m6A/m-seq, 5 ng of eluate or input mRNA was subjected to RT-qPCR. Fold enrichment was calculated by calculating the 2−ΔCt of eluate in relative to the input sample. The primers used for gene expression are: ATCAACTTTCGATGGTAGTCG (forward) and TCCTTGGATGTGGTAGCCG (reverse) for human 18S; CATTCGAACGTCTGCCCTAT (forward) and GTTTCTCAGGCTCCCTCTCC (reverse) for rat 18S; CGACAGCGCTGGAATCCTAT (forward) and GCCATCAAGGGATCCACTCC (reverse) for human SON; GCAAGAAGGTAGGGGTCATGT (forward) and AGGGGTAAATCCCTGTCAAACA (reverse) for human METTL3; GATAGCCGCTTGCAGGAGAT (forward) and ACTTTCAGCTCCCAACTGCT (reverse) for human METTL14; ATCAGGCAAGTGAAAGCCTCA (forward) and GTCCAGGATGACCTTATGGGTT (reverse) for human WTAP; GTGGAGACTTCTCTTGGCCC (forward) and GGGCACCATTTCCTAGCTGT (reverse) for human FTO; TCAAGCCTATTCGGGTGTCG (forward) and ATCCACTGAGCACAGTCACG (reverse) for human ALKBH5; CTCAATGAGCGACCCCTACC (forward) and AGTAGACCACGGAGCCTCAT (reverse) for human YTHDF1; TGTTGGAGAAGCTTCGGTCC (forward) and ACCCGGCCATGTTTCAGATT (reverse) for human YTHDF2; ATTGTGGACCCGAGAAGCAG (forward) and GAAGAGGCCCGTCTTTTCCG (reverse) for human YTHDF3; ACGTCCATCCCGTCGAGAAC (forward) and ACACATCTCGGCGAACTCCT (reverse) for human YTHDC1; AAGGGGCTGAAGGACATTCG (forward) and CCATTTCTCTCTGGTCCCCG (reverse) for human YTHDC2; AAGCAGCAGTTCGTGGTGAA (forward) and TCGTTAGCGCTCCTTCCTTC (reverse) for human MALAT1; ATGTGCAGCCCAACTGGATT (forward) and CTGTGCTTAAACCGGGCAAC (reverse) for rat METTL3; GGGGAAGGATTGGACCTTGG (forward) and GCAGTGCTCCTTTGTCCTCT (reverse) for rat METTL14; AGCAGCAACAGCAGGAATCT (forward) and GGTGCACTCTTGCATCTCCT (reverse) for rat WTAP; AGAAGGCCAATGAAGACGCT (forward) and CTTCATCATCGCAGGACGGT (reverse) for rat FTO; ACGGCCTCAGGACATCAAAG (forward) and AAGCATAGCTGGGTGGCAAT (reverse) for rat ALKBH5; TCAGGACAAGTGGAAGGGGA (forward) and TCTGATGTGCCGCAGTTGAT (reverse) for rat YTHDF1; GGACACTCAGGAAGTGCCTC (forward) and TGGCTTCCTCCTCCTCTTGA (reverse) for rat YTHDF2; ACTTTCAAGCACACCACCTCA (forward) and TGGCTTCCTCCTCCTCTTGA (reverse) for rat YTHDF3; CCAGGAAGTCCCACAGTGTT (forward) and AGACACAGGATGGGATGGAG (reverse) for LANA; AGGTCCCCCTCACCAGTAAA (forward) and GAGGACGTGTGTTTTGACCG (reverse) for ORF57; CATGCTGATGCGAATGTGC (forward) and AGCTTCAACATGGTGGGAGTG (reverse) for ORF-K8; TTTAGCACTGGGACTGCCC (forward) and CAAGAAGGCAAGCAGCGAG (reverse) for PAN RNA; AATGTCAGCGTCCACTCCTG (forward) and GAAGAGGGGGCACAGGTAAC (reverse) for RTA; AAGGTGAGAGACCCCGTGAT (forward) and AGGGTATTCATGCGAGCCAC (reverse) for ORF65; and TTCTTAACCCCAGAACGCCAG (forward) and CAAGTGCACGGATCGGCTT (reverse) for ORF59; TAAAAAGCTCGCCGATGGCT (forward) and ACTGATTTTCCAAACTCCGTCG (reverse) for vIL6. The LANA primers were also used for quantification of virion. The primers used for validation of m6A/m peaks are: GCTGCACATCAAGGTGCTAA (forward) and GCAACGTTCTGCAGTTCACA (reverse) for the human Dicer1 m6A/m peak; AAACGAAGGCAGTGCTACCC (forward) and GGGCTGATCAGGTCTGGGATA (reverse) for the human Dicer1 without m6A/m peak; GAACGCCTGATTGTCCCCAA (forward) and AAAAGTACTGTCCCGGGGGT (reverse) for the human JUNB m6A/m peak; AGGTGCTACTAGACCCTCCTT (forward) and GAGCTTAGCAGGTGACTCGG (reverse) for rat the Dicer1 m6A/m peak; AAACGAAGGCAGTGCTACCC (forward) and CAGGGTTGATCGGGTTTGGG (reverse) for the rat Dicer1 without m6A/m peak; AACTGGAGCGCTTGATCGTC (forward) and TAAAAGTACTGTCCCGGAGGC (reverse) for the rat JUNB m6A/m peak; GACTCCTTTTCCCGCCAAGA (forward) and AAGTGACGTCCGTCGCTAAG (reverse) for ORF72 m6A/m peak; TCTGCAAAACCGTGACGTTG (forward) and TAGGGACTACCGCTGCGTG (reverse) for ORF75 m6A/m peak; GCGGTCAAATTTGGGTGGAC (forward) and TGGAGCTTCTGACGAAGACC (reverse) for vIL6 m6A/m peak; TCCATGGTAGACCTCAGCGA (forward) and ATGTTGGGATGGGGTTTGCT (reverse) for RTA m6A/m peak; AGAGGAAGAGACGCGCACTA (forward) and GCAATAAACCCACAGCCCAT (reverse) for ORF-K8.1 m6A/m peak; AGTTGGACCACATTCCATTGC (forward) and CCTGCGAGTTCACAGGTTGG (reverse) for ORF4 m6A/m peak; CCGGCAGGTCTGTAACCATT (forward) and CACCAGTGGCACGGTAATGA (reverse) for ORF6 m6A/m peak; AAGCGGGAGAACCAACACAT (forward) and AGCGCCCAAGTTGTTACAGT (reverse) for ORF-K3 m6A/m peak; GAACTTCCTGGCGGGGTAAA (forward) and GGGAACCGGACACCTAACTG (reverse) for ORF11 m6A/m peak; and CACCATGGCGCATGTTTCAA (forward) and CCCTGTCCGTAAACACCTCC (reverse) for ORF57 m6A/m peak.
Western-blotting analysis
Protein samples were lysed in Laemmli buffer, separated by SDS-PAGE and transferred to a nitrocellulose membrane. The membrane was blocked with 5% milk and then incubated with the appropriate primary antibody overnight at 4˚C. The membrane was washed with TBS-Tween and probed with a secondary antibody conjugated to horseradish peroxidase (HRP). After further washing with TBS-T, the blot was visualized with Luminata Crescendo Western HRP substrate (Millipore, Billerica, MA) and imaged on a UVP BioSpectrum® Imaging System (UVP, LLC, Upland, CA). Quantification of band intensity was performed using the UVP BioSpectrum® Imaging System (UVP).
Measurement of transcript half-life
KiSLK cells induced for lytic replication were treated with 2 μg/ml actinomycin D. RNA was collected from at 0, 2, 4, 8, 16, and 24h after the treatment, and examined by RT-qPCR. RNA from 0, 4, and 16 h were poly(A) selected, converted to cDNA libraries and sequenced on an HiSeq 2000 platform using the 50 bp single read sequencing module. Reads were converted to FPKM by cuffdiff and normalized to GAPDH. mRNA lifetime profiling was calculated using methods previously described20. Fold changes of the half-lives of KSHV transcripts in the heat map were calculated by dividing siY2-1 FPKM by siCl FPKM based on values from 0 h and 16 h.
RNA-binding protein immunoprecipitation and qPCR (RIP-qPCR)
KiSLK cells were transduced with lentivirus carrying an empty vector or Flag-YTHDF2. One day after transduction, cells were split to a 10 cm tissue culture dish. Once 80% confluent, lytic replication was induced with 1 μg/ml doxycycline for 48h. Cells were lysed with lysis buffer containing 25 mM Tris-HCl pH 7.4, 150 mM NaCl, 1 mM, and 1% Igepal 630 for 20 min at 4˚C. A fraction of the lysate (10%) was saved as input. The cell lysate was incubated with mouse anti-flag M2 beads (Sigma-Aldrich) overnight. The next day, the beads were washed 5x with lysis buffer and RNA was collected by adding Trizol to the beads. Input lysate (1%) and all the immunoprecipitated RNA was used for RT-qPCR.
Pathway analysis
Significantly differentially methylated genes were identified by applying a false discovery rate (log10 FDR) filter of < −1.3 and enriched pathways were determined using the default settings of QIAGEN’s Ingenuity® Pathway Analysis (IPA®, QIAGEN Redwood City, www.qiagen.com/ingenuity). Comparison analysis was done by sorting the P-values of the commonly enriched pathways by score.
Statistical analyses
All the experiments were independently performed at least three times unless stated otherwise, and the results were analyzed and presented. Student’s t-test was used for RT-qPCR, MeRIP-qPCR, RIP-PCR, and Western-blotting band intensity analyses in Microsoft Excel. Transcript half-life was calculated using one phase decay nonlinear regression and the significance between the decay curves was determined using the Wilcoxon-signed rank test in GraphPad Prism 5 (GraphPad Software Inc., La Jolla, CA). The Mann-Whitney test was used to calculate the significance of curves of half-lives of transcripts with and without YTHDF2 knockdown. In all results, “NS” denotes “not significant”, * p<0.05, ** p<0.01, *** p<0.001.
Data availability statement
Sequencing data that support the findings of this study have been deposited in GEO with the accession code GSE93676.
Supplementary Material
Acknowledgments
We thank Dr. S-J Gao’s laboratory for technical assistance and helpful discussions. This work was supported by grants from NIH (CA096512, CA124332, CA132637, CA213275, CA177377, DE025465 and CA197153) to S-J Gao, and (GM113245) to Y. Huang.
Footnotes
AUTHOR CONTRIBUTIONS
Conceptualization, B.T., H.L., Y.H., S.-J.G.; Investigation, B.T., S.R.S., O.S., S.W.Z. H.F.; Methodology, B.T., H.L., S.Y.Z., Y.H., S.-J.G; Formal Analysis, H.L., S.Y.Z., L.Z., J.M., X.C.; Writing – Original Draft, B.T., H.L., S.-J.G.; Writing – Review & Editing, B.T., H.L., S.Y.Z., S.R.S., L.Z., J.M., X.C., O.S., H.F., S.W.Z., Y.H., S.-J.G.; Supervision and management, Y.H., S.-J.G.; Funding Acquisition, Y.H., S.-J.G.
Competing interests
The authors declare no competing financial interests.
Supplementary information is available for this paper.
References
- 1.Ye F, Lei X, Gao SJ. Mechanisms of Kaposi’s Sarcoma-Associated Herpesvirus Latency and Reactivation. Adv Virol. 2011;2011 doi: 10.1155/2011/193860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Desrosiers R, Friderici K, Rottman F. Identification of methylated nucleosides in messenger RNA from Novikoff hepatoma cells. Proc Natl Acad Sci USA. 1974;71:3971–3975. doi: 10.1073/pnas.71.10.3971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dominissini D, et al. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature. 2012;485:201–206. doi: 10.1038/nature11112. [DOI] [PubMed] [Google Scholar]
- 4.Meyer KD, et al. Comprehensive analysis of mRNA methylation reveals enrichment in 3′ UTRs and near stop codons. Cell. 2012;149:1635–1646. doi: 10.1016/j.cell.2012.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Linder B, et al. Single-nucleotide-resolution mapping of m6A and m6Am throughout the transcriptome. Nat Meth. 2015;12:767–772. doi: 10.1038/nmeth.3453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bokar JA, Rath-Shambaugh ME, Ludwiczak R, Narayan P, Rottman F. Characterization and partial purification of mRNA N6-adenosine methyltransferase from HeLa cell nuclei. Internal mRNA methylation requires a multisubunit complex. J Biol Chem. 1994;269:17697–17704. [PubMed] [Google Scholar]
- 7.Bokar JA, Shambaugh ME, Polayes D, Matera AG, Rottman FM. Purification and cDNA cloning of the AdoMet-binding subunit of the human mRNA (N6-adenosine)-methyltransferase. RNA. 1997;3:1233–1247. [PMC free article] [PubMed] [Google Scholar]
- 8.Schwartz S, et al. Perturbation of m6A writers reveals two distinct classes of mRNA methylation at internal and 5′ sites. Cell Rep. 2014;8:284–296. doi: 10.1016/j.celrep.2014.05.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ping XL, et al. Mammalian WTAP is a regulatory subunit of the RNA N6-methyladenosine methyltransferase. Cell Res. 2014;24:177–189. doi: 10.1038/cr.2014.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu J, et al. A METTL3-METTL14 complex mediates mammalian nuclear RNA N6-adenosine methylation. Nat Chem Biol. 2014;10:93–95. doi: 10.1038/nchembio.1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Patil DP, et al. m(6)A RNA methylation promotes XIST-mediated transcriptional repression. Nature. 2016;537:369–373. doi: 10.1038/nature19342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jia G, et al. N6-methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat Chem Biol. 2011;7:885–887. doi: 10.1038/nchembio.687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zheng G, et al. ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Mol Cell. 2013a;49:18–29. doi: 10.1016/j.molcel.2012.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mauer J, et al. Reversible methylation of m6Am in the 5′ cap controls mRNA stability. Nature. 2017;541:371–375. doi: 10.1038/nature21022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu N, et al. N(6)-methyladenosine-dependent RNA structural switches regulate RNA-protein interactions. Nature. 2015;518:560–564. doi: 10.1038/nature14234. nature14234 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhou KI, et al. N(6)-methyladenosine modification in a long noncoding RNA hairpin predisposes Its conformation to protein binding. J Mol Biol. 2016;428:822–833. doi: 10.1016/j.jmb.2015.08.021. S0022–2836(15)00486–6 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Konig J, et al. iCLIP reveals the function of hnRNP particles in splicing at individual nucleotide resolution. Nat Struct Mol Biol. 2010;17:909–915. doi: 10.1038/nsmb.1838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Alarcon CR, et al. HNRNPA2B1 is a mediator of m(6)A-dependent nuclear RNA processing events. Cell. 2015;162:1299–1308. doi: 10.1016/j.cell.2015.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang X, et al. N(6)-methyladenosine modulates messenger RNA translation efficiency. Cell. 2015;161:1388–1399. doi: 10.1016/j.cell.2015.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang X, et al. N6-methyladenosine-dependent regulation of messenger RNA stability. Nature. 2014;505:117–120. doi: 10.1038/nature12730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Du H, et al. YTHDF2 destabilizes m(6)A-containing RNA through direct recruitment of the CCR4-NOT deadenylase complex. Nat Commun. 2016;7:12626. doi: 10.1038/ncomms12626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kennedy EM, et al. Posttranscriptional m(6)A Editing of HIV-1 mRNAs Enhances Viral Gene Expression. Cell Host Microbe. 2016;19:675–685. doi: 10.1016/j.chom.2016.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shi H, et al. YTHDF3 facilitates translation and decay of N6-methyladenosine-modified RNA. Cell Res. 2017;27:315–328. doi: 10.1038/cr.2017.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li A, et al. Cytoplasmic m6A reader YTHDF3 promotes mRNA translation. Cell Res. 2017;27:444–447. doi: 10.1038/cr.2017.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xiao W, et al. Nuclear m(6)A reader YTHDC1 regulates mRNA splicing. Mol Cell. 2016;61:507–519. doi: 10.1016/j.molcel.2016.01.012. [DOI] [PubMed] [Google Scholar]
- 26.Hsu PJ, et al. Ythdc2 is an N6-methyladenosine binding protein that regulates mammalian spermatogenesis. Cell Res. 2017;27:1115–1127. doi: 10.1038/cr.2017.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lichinchi G, et al. Dynamics of the human and viral m(6)A RNA methylomes during HIV-1 infection of T cells. Nat Microbiol. 2016;1:16011. doi: 10.1038/nmicrobiol.2016.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gokhale NS, et al. N6-Methyladenosine in Flaviviridae Viral RNA Genomes Regulates Infection. Cell Host Microbe. 2016;20:654–665. doi: 10.1016/j.chom.2016.09.015. S1931–3128(16)30393–6 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lichinchi G, et al. Dynamics of human and viral RNA methylation during Zika virus infection. Cell Host Microbe. 2016;20:666–673. doi: 10.1016/j.chom.2016.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tirumuru N, et al. N(6)-methyladenosine of HIV-1 RNA regulates viral infection and HIV-1 Gag protein expression. Elife. 2016;5 doi: 10.7554/eLife.15528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ye F, Chen ER, Nilsen TW. Kaposi’s sarcoma-associated herpesvirus utilizes and manipulates RNA N6-adenosine methylation to promote lytic replication. J Virol. 2017 doi: 10.1128/JVI.00466-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhou FC, et al. Efficient infection by a recombinant Kaposi’s sarcoma-associated herpesvirus cloned in a bacterial artificial chromosome: application for genetic analysis. J Virol. 2002;76:6185–6196. doi: 10.1128/JVI.76.12.6185-6196.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Meng J, Cui X, Rao MK, Chen Y, Huang Y. Exome-based analysis for RNA epigenome sequencing data. Bioinformatics. 2013;29:1565–1567. doi: 10.1093/bioinformatics/btt171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Brulois KF, et al. Construction and manipulation of a new Kaposi’s sarcoma-associated herpesvirus bacterial artificial chromosome clone. J Virol. 2012;86:9708–9720. doi: 10.1128/JVI.01019-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nakamura H, et al. Global changes in Kaposi’s sarcoma-associated virus gene expression patterns following expression of a tetracycline-inducible Rta transactivator. J Virol. 2003;77:4205–4220. doi: 10.1128/JVI.77.7.4205-4220.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jones T, et al. Direct and efficient cellular transformation of primary rat mesenchymal precursor cells by KSHV. J Clin Invest. 2012;122:1076–1081. doi: 10.1172/JCI58530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lee MS, et al. Human mesenchymal stem cells of diverse origins support persistent infection with Kaposi’s sarcoma-associated herpesvirus and manifest distinct angiogenic, invasive, and transforming phenotypes. MBio. 2016;7:e02109–02115. doi: 10.1128/mBio.02109-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xie J, Ajibade AO, Ye F, Kuhne K, Gao SJ. Reactivation of Kaposi’s sarcoma-associated herpesvirus from latency requires MEK/ERK, JNK and p38 multiple mitogen-activated protein kinase pathways. Virology. 2008;371:139–154. doi: 10.1016/j.virol.2007.09.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Greene W, Gao SJ. Actin dynamics regulate multiple endosomal steps during Kaposi’s sarcoma-associated herpesvirus entry and trafficking in endothelial cells. PLoS Pathog. 2009;5:e1000512. doi: 10.1371/journal.ppat.1000512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Davis DA, et al. Hypoxia induces lytic replication of Kaposi’s sarcoma-associated herpesvirus. Blood. 2001;97:3244–3250. doi: 10.1182/blood.v97.10.3244. [DOI] [PubMed] [Google Scholar]
- 41.Hollingworth R, et al. Activation of DNA damage response pathways during lytic replication of KSHV. Viruses. 2015;7:2908–2927. doi: 10.3390/v7062752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Meyer KD, Jaffrey SR. Rethinking m6A readers, writers, and erasers. Annu Rev Cell Dev Biol. 2017 doi: 10.1146/annurev-cellbio-100616-060758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Moss B, Gershowitz A, Stringer JR, Holland LE, Wagner EK. 5′-Terminal and internal methylated nucleosides in herpes simplex virus type 1 mRNA. J Virol. 1977;23:234–239. doi: 10.1128/jvi.23.2.234-239.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Covarrubias S, et al. Coordinated destruction of cellular messages in translation complexes by the gammaherpesvirus host shutoff factor and the mammalian exonuclease Xrn1. PLoS Pathog. 2011;7:e1002339. doi: 10.1371/journal.ppat.1002339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chandriani S, Ganem D. Host transcript accumulation during lytic KSHV infection reveals several classes of host responses. PLoS One. 2007;2:e811. doi: 10.1371/journal.pone.0000811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chen T, You Y, Jiang H, Wang ZZ. Epithelial-mesenchymal transition (EMT): A biological process in the development, stem cell differentiation, and tumorigenesis. J Cell Physiol. 2017 doi: 10.1002/jcp.25797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Moustakas A, Heldin CH. Signaling networks guiding epithelial-mesenchymal transitions during embryogenesis and cancer progression. Cancer Sci. 2007;98:1512–1520. doi: 10.1111/j.1349-7006.2007.00550.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Meyer KD, et al. 5′ UTR m(6)A promotes cap-independent translation. Cell. 2015;163:999–1010. doi: 10.1016/j.cell.2015.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Moody R, et al. KSHV microRNAs mediate cellular transformation and tumorigenesis by redundantly targeting cell growth and survival pathways. PLoS Pathog. 2013;9:e1003857. doi: 10.1371/journal.ppat.1003857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Brulois K, Jung JU. Interplay between Kaposi’s sarcoma-associated herpesvirus and the innate immune system. Cytokine Growth Factor Rev. 2014;25:597–609. doi: 10.1016/j.cytogfr.2014.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lee MS, et al. Exploitation of the complement system by oncogenic Kaposi’s sarcoma-associated herpesvirus for cell survival and persistent infection. PLoS Pathog. 2014;10:e1004412. doi: 10.1371/journal.ppat.1004412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cheng F, et al. Screening of the human kinome identifies MSK1/2-CREB1 as an essential pathway mediating Kaposi’s sarcoma-associated herpesvirus lytic replication during primary infection. J Virol. 2015;89:9262–9280. doi: 10.1128/JVI.01098-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gao SJ, Deng JH, Zhou FC. Productive lytic replication of a recombinant Kaposi’s sarcoma-associated herpesvirus in efficient primary infection of primary human endothelial cells. J Virol. 2003;77:9738–9749. doi: 10.1128/JVI.77.18.9738-9749.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dominissini D, Moshitch-Moshkovitz S, Salmon-Divon M, Amariglio N, Rechavi G. Transcriptome-wide mapping of N(6)-methyladenosine by m(6)A-seq based on immunocapturing and massively parallel sequencing. Nat Protoc. 8:176–189. doi: 10.1038/nprot.2012.148. nprot.2012.148 [pii] [DOI] [PubMed] [Google Scholar]
- 55.Arias C, et al. KSHV 2.0: a comprehensive annotation of the Kaposi’s sarcoma-associated herpesvirus genome using next-generation sequencing reveals novel genomic and functional features. PLoS Pathog. 2014;10:e1003847. doi: 10.1371/journal.ppat.1003847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bai Z, et al. Genomewide mapping and screening of Kaposi’s sarcoma-associated herpesvirus (KSHV) 3′ untranslated regions identify bicistronic and polycistronic viral transcripts as frequent targets of KSHV microRNAs. J Virol. 2014;88:377–392. doi: 10.1128/JVI.02689-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Majerciak V, et al. A viral genome landscape of RNA polyadenylation from KSHV latent to lytic infection. PLoS Pathog. 2013;9:e1003749. doi: 10.1371/journal.ppat.1003749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pearce M, Matsumura S, Wilson AC. Transcripts encoding K12, v-FLIP, v-cyclin, and the microRNA cluster of Kaposi’s sarcoma-associated herpesvirus originate from a common promoter. J Virol. 2005;79:14457–14464. doi: 10.1128/JVI.79.22.14457-14464.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tang S, Zheng ZM. Kaposi’s sarcoma-associated herpesvirus K8 exon 3 contains three 5′-splice sites and harbors a K8.1 transcription start site. J Biol Chem. 2002;277:14547–14556. doi: 10.1074/jbc.M111308200. [DOI] [PubMed] [Google Scholar]
- 60.Yamanegi K, Tang S, Zheng ZM. Kaposi’s sarcoma-associated herpesvirus K8beta is derived from a spliced intermediate of K8 pre-mRNA and antagonizes K8alpha (K-bZIP) to induce p21 and p53 and blocks K8alpha-CDK2 interaction. J Virol. 2005;79:14207–14221. doi: 10.1128/JVI.79.22.14207-14221.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kim D, et al. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013;14:R36. doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Meth. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Meng J, et al. A protocol for RNA methylation differential analysis with MeRIP-Seq data and exomePeak R/Bioconductor package. Methods. 2014;69:274–281. doi: 10.1016/j.ymeth.2014.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Krishnamoorthy K, Thomson J. A more powerful test for comparing two Poisson means. J Stat Plan Inf. 2004;119:23–35. [Google Scholar]
- 65.Cui X, et al. Guitar: An R/Bioconductor package for gene annotation guided transcriptomic analysis of RNA-related genomic features. Biomed Res Int. 2016;2016:8367534. doi: 10.1155/2016/8367534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bailey TL, et al. MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 2009;37:W202–208. doi: 10.1093/nar/gkp335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Trapnell C, et al. Differential analysis of gene regulation at transcript resolution with RNA-seq. Nat Biotechnol. 2013;31:46–53. doi: 10.1038/nbt.2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Trapnell C, et al. Transcript assembly and quantification by RNA-seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol. 2010;28:511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Sequencing data that support the findings of this study have been deposited in GEO with the accession code GSE93676.