Abstract
Cryptochromes (CRY) are flavoproteins that direct a diverse array of developmental processes in response to blue light in plants. Conformational changes in CRY are induced by the absorption of photons and result in the propagation of light signals to downstream components. In Arabidopsis, CRY1 and CRY2 serve both distinct and partially overlapping functions in regulating photomorphogenic responses and photoperiodic flowering. For example, both CRY1 and CRY2 regulate the abundance of transcription factors by directly reversing the activity of E3 ubiquitin ligase on CONSTITUTIVE PHOTOMORPHOGENIC 1 and SUPPRESSOR OF PHYA-105 1 complexes in a blue light-dependent manner. CRY2 also specifically governs a photoperiodic flowering mechanism by directly interacting with a transcription factor called CRYPTOCHROMEINTERACTING BASIC-HELIX-LOOP-HELIX. Recently, structure/function analysis of CRY1 revealed that the CONSTITUTIVE PHOTOMORPHOGENIC 1 independent pathway is also involved in CRY1-mediated inhibition of hypocotyl elongation. CRY1 and CRY2 thus not only share a common pathway but also relay light signals through distinct pathways, which may lead to altered developmental programs in plants.
Keywords: Cryptochrome, De-etiolation, Flowering, Photomorphogenesis, Transcription
Introduction
Because plants rely on light as a source of energy, they have evolved sophisticated mechanisms to maximize the use of available light by regulating their own growth and development in response to surrounding light environments. Plants possess a variety of photoreceptors that allow them to recognize changes in the direction, intensity, and quality of light, including UV RESISTANCE LOCUS 8, phototropins, CRYs, FLAVIN-BINDING KELCH REPEAT F-BOX 1, ZEITLUPE (ZTL), LOV KELCH PROTEIN 2 (LKP2) and phytochromes (Cashmore et al. 1999; Smith 2000; Briggs et al. 2001; Kami et al. 2010; Yu et al. 2010). Among these, CRYs are the only photoreceptors present in all major evolutionary lineages (Lin and Todo 2005). CRYs are flavoproteins that share sequence similarity to DNA photolyases, which repair UV-induced DNA damage by removing pyrimidine dimers from DNA under blue light (Lin and Shalitin 2003; Sancar 2003). CRYs together with DNA photolyases make up the CRY/photolyase superfamily, which consists of five major subgroups: cyclobutane pyrimidine photolyase, 6-4 photolyase, CRY-DASH, plant CRY, and animal CRY (Sancar 2003; Chaves et al. 2011). Animal CRYs are further divided into light-dependent type 1 and light-independent type 2 CRYs (Zhu et al. 2005; Yuan et al. 2007; Chaves et al. 2011). CRY-DASH may play a role in the repair of cyclobutane pyrimidine dimers in damaged single stranded DNA (Brudler et al. 2003; Selby and Sancar 2006) or in the loop structures of double-stranded DNA (Pokorny et al. 2008). Plant and animal CRYs have lost DNA-repair activity but evolved to have other biochemical functions (Lin et al. 1995; Malhotra et al. 1995). For example, they regulate photomorphogenic growth and development in plants, and the circadian clock in both plants and animals (Cashmore 2003; Lin and Shalitin 2003; Sancar 2003). CRYs have also been reported to serve as magnetoreceptors in animals (Ritz et al. 2000; Gegear et al. 2010; Ritz et al. 2010).
The functions of CRY
Plant CRYs regulate a variety of blue-light-induced responses (Wang et al. 2015b), including de-etiolation, photoperiodic flowering (Ahmad and Cashmore 1993; Guo et al. 1998; El-Assal et al. 2001), guard cell development, stomata opening (Mao et al. 2005; Kang et al. 2009; Wang et al. 2010), leaf senescence (Meng et al. 2013), pathogenic responses (Jeong et al. 2010; Wu and Yang 2010) and so on. Arabidopsis thaliana has been used to study plant CRYs most extensively. Three CRYs, CRY1, CRY2 and CRY3, are encoded in the Arabidopsis genome. CRY3 is a CRY-DASH type CRY that is detected in mitochondrion or chloroplasts and functions to repair UV-induced damage on single-stranded DNA (Kleine et al. 2003; Pokorny et al. 2008). CRY1 and CRY2 function as major blue light receptors regulating blue light induced de-etiolation and photoperiodic flowering (Ahmad and Cashmore 1993; Guo et al. 1998; El-Assal et al. 2001). This review focuses on CRY1 and CRY2 mediated mechanisms of blue light signal transduction in Arabidopsis.
Both CRY1 and CRY2 function as dimers in plant cells (Sang et al. 2005; Yu et al. 2007b; Rosenfeldt et al. 2008) with monomers that consist of two major domains: the N-terminal Photolyase Homologous Region (PHR) domain and the diverged CRY C-terminal Extension (CCE) domain. The PHR domain is a highly conserved, 500 amino acid long domain that can be further divided into the N-terminal α/β and C-terminal α subdomains. The primary CRY chromophore, flavin adenine dinucleotide (FAD), binds non-covalently to the α subdomain of the PHR domain, allowing for blue light absorption (Lin et al. 1995; Malhotra et al. 1995; Banerjee et al. 2007). The PHR domain has been successfully co-crystallized with a non-hydrolysable ATP analogue, adenylyl-imidodi-phosphate (AMP-PNP), from a CRY1 protein lacking the CCE domain (Brautigam et al. 2004). In the crystal structure, AMP-PNP was found in the vicinity of FAD at sites where photolyases dock to pyrimidine-dimer lesions in the FAD binding pocket. As is the case with photolyases, the PHR domain possibly possesses a second chromophore, 5,10-methenyltetra-hydrofolate, in the α/β subdomain, although it seems to be lost during purification of CRY (Lin et al. 1995; Malhotra et al. 1995; Banerjee et al. 2007; Hoang et al. 2008). In addition to perceiving blue light, the PHR domain is also responsible for the dimerization of CRY (Sang et al. 2005; Rosenfeldt et al. 2008). The CCE domains of Arabidopsis CRY1 and CRY2 are 180 and 110 amino acid residues in length, respectively. CCE domains vary considerably in their length and sequence among plant CRYs and they are poorly structured (Partch et al. 2005). However, it is conceivable that the CCE domains of CRY1 and CRY2 are critical for the function and regulation of CRYs (Yang et al. 2000; Wang et al. 2001; Yang et al. 2001; Yu et al. 2007b).
The blue light induced inhibition of hypocotyl elongation and photoperiodic flowering are often analyzed to clarify the signal transduction mechanism and function of CRYs in Arabidopsis. Genetic analyses with Arabidopsis cry1 and cry2 mutants reveal that CRY1 primarily mediates blue light induced inhibition of hypocotyl elongation while CRY2 predominantly regulates photoperiodic control of floral initiation (Ahmad and Cashmore 1993; Guo et al. 1998). Different from CRY1, which is stable, CRY2 undergoes ubiquitination and is degraded immediately by the 26S proteasome system under blue light conditions (Ahmad et al. 1998a, b; Lin et al. 1998; Shalitin et al. 2002; Yu et al. 2007a, 2009; Weidler et al. 2012). The protein stability of CRY2 is consistent with the fluence rate dependence of CRY2 in the regulation of blue light induced inhibition of hypocotyl elongation. Namely, CRY2 regulates the hypocotyl response preferentially under dim blue light (Lin et al. 1998). On the other hand, CRY1 may partially function in the regulation of flowering time. The cry1 single mutants flower late under white light in some experimental conditions but not in all reported so far (Goto et al. 1991; Mozley and Thomas 1995; Bagnall et al. 1996; Bruggemann et al. 1996; Blazquez et al. 2003; Mockler et al. 2003). The conditions resulting in varied phenotypes between differing experiments is unknown. The gain-of-function mutant alleles of CRY1 exhibited an early flowering phenotype in long day conditions (Exner et al. 2010; Gu et al. 2012). Additionally, the cry1cry2 double mutant flowered later compared to wild type plants in continuous blue light, whereas cry1 and cry2 single mutants did not vary from wild type, suggesting that CRY1 acts redundantly with CRY2 to promote flowering under these conditions (Mockler et al. 1999; Liu et al. 2008a). Therefore, CRY1 and CRY2 have distinct but partially overlapping functions in the regulation of hypocotyl elongation and photoperiodic floral initiation.
CRY2 localizes exclusively in the nucleus (Guo et al. 1999; Kleiner et al. 1999; Yu et al. 2007b), whereas CRY1 localizes both in the nucleus and cytoplasm (Cashmore et al. 1999; Wu and Spalding 2007). It is nuclear-targeted CRY1 and CRY2 that regulate most CRY mediated responses, with the exception of CRY1-mediated cotyledon expansion and root elongation (Wu and Spalding 2007), suggesting that CRY1 and CRY2 signal transduction occurs primarily in the nucleus. Furthermore, CRY1 and CRY2 regulate the transcription of 5–25 % of genes in response to blue light in Arabidopsis seedlings initially grown in the dark (Folta et al. 2003; Ma et al. 2001; Ohgishi et al. 2004; Sellaro et al. 2009). A number of CRY-dependent genes encode signaling proteins such as kinases, transcription factors and cell cycle regulators, phytohormone signaling factors and enzymes involved in the cell wall metabolism and phytohormone biosynthesis. Changes in the expression of these genes partially account for the morphological changes observed during the CRY-mediated de-etiolation process. Thus, it is generally accepted that blue light-induced structural rearrangements and modifications of CRY proteins occur dynamically in the nucleus whereby protein–protein interactions trigger changes in gene expression and physiological responses in plants. CRY2 differs from CRY1 in that it results in the formation of nuclear structures called photobodies in response to blue light (Más et al. 2000; Yu et al. 2009; Zuo et al. 2012). Photobodies have been a proposed site for CRY2 signal transduction and desensitization via degradation, because there is a correlation between photobody formation and CRY2 function and degradation (Yu et al. 2009; Zuo et al. 2012). However, the function and mechanism of CRY2 photobody formation are still not fully understood.
Light sensing of CRY
The redox status of FAD in the PHR domain of CRY is believed to play an important role in CRY photoexitation (Liu et al. 2010). Based on in vitro spectroscopic analysis of CRY1 and CRY2 purified from baculovirus-infected insect cells, both CRY1 and CRY2 possess the oxidized form of FAD, which primarily absorbs UV-A and blue light, in the resting state (Lin et al. 1995; Malhotra et al. 1995; Banerjee et al. 2007; Bouly et al. 2007; Zeugner et al. 2005). Upon blue light absorption, FAD in CRY undergoes rapid photoreduction (within microseconds) with incorporation of an electron followed by protonation, thereby turning into the semi-reduced neutral FAD radical (FADH˙), which absorbs a broader spectrum of light. FADH˙ is unstable and can be converted back to the oxidized FAD within minutes in the absence of light. Therefore, both CRY1 and CRY2 undergo a photocycle as FAD is reduced to form FADH˙ and then transformed into its oxidized state in vitro (Lin et al. 1995; Malhotra et al. 1995; Banerjee et al. 2007; Bouly et al. 2007; Zeugner et al. 2005).
Oxidized FAD exhibits light absorption peaks at the UV-A and blue light regions of the spectrum. The fact that CRY mediated responses are most efficiently induced by blue light is consistent with the observation that CRY1 and CRY2 bind with the oxidized, blue light absorbing form of FAD. Hence, it is most likely that inactive CRYs possess oxidized FAD that will be activated by blue light. Following CRY excitation involving the oxidized form of FAD, rapid photoreduction converts FAD into FADH˙ allowing for a broader spectrum of light absorption, including green light. It is then conceivable that the signaling state of activated CRY1 and CRY2, on the other hand, may possess the semireduced flavin, FADH˙ (Chaves et al. 2011). Interestingly, partial inhibitory effects of green light have been reported in blue light induced inhibition of hypocotyl elongation, anthocyanin accumulation, flowering and blue light induced CRY2 protein degradation (Zeugner et al. 2005; Banerjee et al. 2007; Bouly et al. 2007), presumably because the photoequilibrium between oxidized FAD and FADH˙ shifts toward decreased levels of FADH˙ as green light is absorbed, thereby diminishing the amount of active CRY. However, the action model that FADH˙ constitutes the signaling state of CRY in vivo is still under debate (Liu et al. 2010; Li et al. 2011).
It is proposed that the Trp-triad, which is composed of three highly conserved tryptophan residues in the α subdomain region of CRY, serves as an electron donor in the photoreduction process from oxidized FAD to FADH˙. In this model, the electron is relayed from W324 (W321 in CRY2), which is exposed to solvent on the CRY1 protein surface, to W377 (W374 in CRY2), which exists between W324 and W400, and on to W400 (W397 in CRY2), which is in close proximity to FAD in the FAD binding pocket where FAD is then reduced (Liu et al. 2010; Chaves et al. 2011). The importance of the Trp-triad in photoreduction of CRY in vitro has been repeatedly reported. The substitutions of Trp-triad amino acids with alanine or phenylalanine, which are redox-inert yet cause minimal structural disturbance, abolished the photoreduction of CRY1 and CRY2 purified from baculovirus-infected insect cells, suggesting that this triple tryptophan cascade is the electron donor needed for photoreduction from oxidized FAD to FADH˙, at least in vitro (Zeugner et al. 2005; Bouly et al. 2007; Li et al. 2011; Engelhard et al. 2014; Gao et al. 2015). It is also reported that the aspartic acid near FAD in the FAD binding pocket (D396 in CRY1) works as a proton donor (Kottke et al. 2006; Immeln et al. 2010; Cailliez et al. 2014; Hense et al. 2015). In addition, ATP binding to the FAD binding pocket of CRY was found to affect the protonation status of D396 and enhances photoreduction in vitro by reducing reoxidation (Immeln et al. 2007; Burney et al. 2009; Cailliez et al. 2014; Müller et al. 2014).
The physiological relevance of the Trp-triad in vivo has been tested in CRY1. Zeugner et al. (2005) reported that the overexpression of two mutant CRY1 proteins with mutations in tryptophan residues of the Trp-triad (at W324 and W400), which abolish the photoreduction in vitro, failed to complement the long hypocotyl and low anthocyanin accumulation phenotypes of cry1 mutant. This suggests that photoreduction is associated with physiological functions of CRY1 and that the Trp-triad serves as the electron donor in CRY1 photoreduction in vivo. However, this hypothesis is highly controversial. The amino acid substitutions of W377 and W394 in CRY2 resulted in constitutively active CRY2 in Arabidopsis (Li et al. 2011), although the amino acid substitution on W321, which is exposed to the protein surface of CRY2, resulted in the inactivation of CRY2 as was also reported in CRY1 by Zeugner et al. (2005). All CRY2 Trp-triad mutants retained blue light responsiveness during proteolytic degradation (Li et al. 2011), despite showing drastically decreased photoreduction in vitro. In particular, degradation of CRY2 protein in the W374A mutant revealed similar kinetic results to that of wild type CRY2 protein, although in vitro results did not show significant photoreduction. More recently, Gao et al. (2015) reported that all non-photoreducing CRY1 mutants, including W324F and W400F, could rescue cry1 mutant phenotypes such as blue light induced inhibition of hypocotyl elongation and anthocyanin accumulation, even though the mutants had reportedly reduced biological function (Zeugner et al. 2005). These data strongly argue that Trp-triad dependent photoreduction is not required for CRY function in vivo. It should be noted that Trp-triad dependent photoreduction is not required for the function of most CRY/photolyase superfamily proteins reported so far, including Drosophila CRY, light-dependent animal type 1 CRY (Song et al. 2007; Oztürk et al. 2008; Gegear et al. 2010; Ozturk et al. 2011; Fedele et al. 2014), and Escherichia coli photolyase (Li and Sancar 1990; Li et al. 1991; Kavakli and Sancar 2004). It was recently reported that the addition of ATP partially rescued defects in CRY2 photoreduction induced by mutations in Trp-triad residues (Engelhard et al. 2014). These reports suggest that there are alternative, Trp-triad independent electron pathways active in CRY2 photoreduction in vivo. In CRY1, alternatively, defective photoreduction in W324A and W400F mutants were not rescued by the addition of ATP in vitro (Gao et al. 2015), yet biological function was not disrupted in vivo. Strong evidence to elucidate the CRY photoexcitation mechanism is still missing, and further intensive efforts in this area are truly necessary to understand the physiological implications of CRY photoreduction and function.
Phosphorylation of CRY
CRYs are phosphoproteins that are phosphorylated at multiple sites along the CCE domain in a blue light dependent manner (Shalitin et al. 2002, 2003; Yu et al. 2007b). Blue light-dependent phosphorylation is well correlated with the physiological activities and functions of CRYs: (1) both CRY1 and CRY2 are phosphorylated in response to blue light and the level of phosphorylation increases relative to blue light fluence rate and irradiation periods (Shalitin et al.2002,2003; Yu et al. 2007a; Tan et al. 2013). (2) Amino acid substitutions that impair the physiological activities of CRY1 decrease blue light dependent phosphorylation (Shalitin et al. 2003). (3) Constitutively active CRY2 CCE domains dimerized by fusion with β-glucuronidase (GUS-CRY2 CCE) are also constitutively phosphorylated (Yang et al. 2000; Sang et al. 2005; Yu et al. 2007b). (4) Mutations on several serine residues of CRY2 that have been shown to be phosphorylated in a blue light dependent manner, including S588, S599 and S605, result in decreased phosphorylation of CRY2 along with decreased blue light mediated physiological activities and degradation of CRY2 (Wang et al. 2015a). These findings suggest that CRY phosphorylation plays an important role in CRY photoactivation and function.
It was shown that CRY1 proteins purified from baculovirus-infected insect cells could be phosphorylated by phytochrome A (phyA), one of the major red/far-red photoreceptor that has been shown to have kinase activity (Yeh and Lagarias 1998), in vitro (Ahmad et al. 1998a, b). However, endogenous CRY1 is phosphorylated in phytochrome deficient Arabidopsis mutants showing regular kinetic activity (Shalitin et al. 2003). Moreover, CRY1 purified from baculovirus-infected insect cells can be phosphorylated in response to blue light in the absence of phytochromes and kinases added in vitro (Bouly et al. 2003; Shalitin et al. Ozgur and Sancar 2006). Whereas CRY1 does not have any kinase related domain, CRY1 was found to bind with an ATP analogue, AMP-PNP, in close proximity to FAD in the FAD binding pocket, as demonstrated by the crystal structure of CRY1 PHR domain (Brautigam et al. 2004). It was also shown that plant CRY1 stoichiometrically binds with ATP in vitro (Bouly et al. 2003; Brautigam et al. 2004; Ozgur and Sancar 2006). These data have led to the proposal that CRY1 phosphorylates itself, at least in part, through its autophosphorylation activity coupled with photoexcitation. In addition to autophosphorylation, one or more kinases that have yet to be identified may also be involved in CRY1 phosphorylation, because multiple serine residues of CRY1 are phosphorylated in response to blue light and recombinant CRY1 protein, which is autophosphorylated in response to blue light, did not show a mobility up-shift in western blot analyses as is seen in the case of phospohorylated CRY1 protein extracted from plants (Shalitin et al. 2003). In contrast to CRY1, clear autophospohorylation activity has not been detected for CRY2 (Ozgur and Sancar 2006). Instead, it has been recently reported that Casein kinase 1, CK1.3 and CK1.4, phosphorylate CRY2 at S587 and T603 in vitro, affecting the physiological activities and functions of CRY2 (Tan et al. 2013). However, other unidentified kinases are expected to be involved in CRY2 phosphorylation because the role of CK1.3 and CK1.4 in vivo does not demonstrate a noticeable effect on CRY2 phosphorylation.
Signal transduction pathways of CRY
CRYs induce plant responses to blue light primarily through the transcriptional regulation of a great number of genes (Chaves et al. 2011; Liu et al. 2011a, b). Two mechanisms have been elucidated for CRY-mediated transcriptional regulation. One mechanism includes the CONSTITUTIVE PHOTOMORPHOGENIC 1 (COP1)-mediated proteolytic regulation of transcription factors. Other mechanism involves the direct binding of CRY2 to basic helix-loop-helix transcription factor proteins.
COP1 together with SUPPRESSOR OF PHYA-105 (SPA) 1 and SPA1 related proteins (SPA2-SPA4 in Arabidopsis) comprise an E3 ubiquitin ligase complex (Lau and Deng 2012). Cop1 mutant and spa quadruple mutants grown in the dark display phenotypes that resemble wild type plants grown in the light (Laubinger et al. 2004, 2006), indicating that COP1/SPA complexes repress photomorphogenesis. COP1/SPA complexes control the light-regulated abundance of LONG HYPOCOTYL 5 (HY5) and other transcription factors (Osterlund et al. 2000; Lau and Deng 2012). HY5 is a basic-leucine zipper transcription factor that binds directly to the promoters of numerous light-regulated genes to regulate transcription, promoting photomorphogenic development (Yi and Deng 2005). Under dark conditions, the COP1/SPA complex targets HY5 for ubiquitination, inducing proteasomal degradation. Light inactivates the COP1/SPA complex so that HY5 accumulates in the nucleus, triggering photomorphogenic responses. CRY1, as well as multiple other photoreceptors, mediate suppression of COP1-dependent degradation of HY5 (Osterlund et al. 2000; Jiao et al. 2007; Favory et al. 2009). Recently, it has been shown that CRY1 plays a role in inactivating COP1/SPA1 complexes by binding directly to SPA1 protein in a blue light dependent manner (Lian et al. 2011; Liu et al. 2011a, b). Similarly, the blue light dependent binding of CRY2 to SPA1 also inactivates COP1/SPA1 complexes (Zuo et al. 2011) and results in the accumulation of CONSTANS (CO), a key transcription factor regulating flowering time. CO binds to the promoter of the florigen coding gene, FLOWERING LOCUS T (FT), to induce the expression of FT under long day conditions, regulating photoperiodic flowering (Searle and Coupland 2004; Jang et al. 2008; Liu et al. 2008b).
Both CRY1 and CRY2 interact directly with SPA1 in a blue light dependent manner to inactivate COP1/SPA complexes. However, the mechanisms underlying the inactivation of COP1/SPA1 complexes differ between CRY1 and CRY2 (Lian et al. 2011; Liu et al. 2011a, b; Zuo et al. 2011). It was reported that SPA1 interacts with COP1 to positively regulate the E3 ubiquitin ligase activity of COP1, though light suppresses the interaction between COP1 and SPA1, consequently decreasing E3 ubiquitin ligase activity of COP1 (Saijo et al. 2003; Seo et al. 2003). In line with this, blue light dependent binding of CRY1 to SPA1 inhibits the interaction between COP1 and SPA1 (Lian et al. 2011; Liu et al. 2011a, b). This CRY1 mediated sequestering of SPA1 from COP1 via direct binding with SPA1 inactivates COP1/SPA1 complexes, thereby allowing accumulation of HY5 and initiation of photomorphogenesis (Fankhauser and Ulm 2011). On the other hand, the binding of CRY2 to SPA1 does not change the affinity between COP1 and SPA1. Instead, the binding of CRY2 to SPA1 stimulates interaction between CRY2 and COP1 (Zuo et al. 2011). Increased affinity between CRY2 and COP1 resulting from blue light dependent binding of CRY2 to SPA1 is thought to inactivate the E3 ubiquitin ligase activity of COP1. The exact mechanism of how the E3 ubiquitin ligase activity of COP1 is suppressed by strengthened interaction between CRY2 and COP1 remains unclear. CRY1 and CRY2 also bind with the other SPA proteins (SPA2-4), but binding affinities to other SPA proteins vary between CRY1 and CRY2 (Lian et al. 2011; Liu et al. 2011a, b; Zuo et al. 2011; Chen et al. 2015), although the results are inconsistent among the experiments. The difference in CRY1 and CRY2 binding affinity to SPA proteins may explain the functional specificity of CRY1 and CRY2. SPA proteins not only have overlapping function but also distinct functions in plant development (Laubinger et al. 2004). The structural requirements of CRY1 and CRY2 for blue light dependent binding to SPA are also different (see below).
In addition to regulating transcription factor protein accumulation through COP1/SPA complex binding, CRY2 can also directly bind with CRYPTOCHROME-INTERACTING BASIC-HELIX-LOOP-HELIX (CIB) proteins to regulate their function, promoting photoperiodic flowering (Liu et al. 2008a). CIB1 was initially isolated as blue-light dependent CRY2-interacting protein in the yeast-two-hybrid screening, and later four of six CIB1-related basic helix-loop-helix proteins (CIB2-CIB5) were also shown to bind with CRY2 in a blue light dependent manner (Liu et al. 2008a, 2013b). Among the five CRY2 binding CIBs, overexpression of CIB1, CIB2, CIB3 and CIB5, but not CIB4, can confer an early flowering phenotype under long day conditions (Liu et al., 2013b). The early flowering phenotype of CIB overexpression lines was CRY dependent. Conversely to early flowering time, cib1cib2cib5 as well as cib1cib5 knockout mutants flowered late, but not every individual mutant showed a late flowering phenotype, suggesting that these CIBs redundantly promote CRY-mediated photoperiodic flowering. CIB1 and other CIBs form heterodimers and bind directly to non-canonical E-box DNA sequences on the FT promoter, initiating FT expression (Liu et al. 2008a, 2013b). Whereas it is still unknown exactly how CRY2/CIB binding regulates CIB function, it was reported that CIB proteins are stabilized under blue light conditions by ZTL and LKP2, the other blue light receptors, which are required for CIB protein function in CRY2-mediated photoperiodic flowering (Liu et al. 2013a). Plants have thus evolved exquisite blue light dependent regulation of photoperiodic flowering whereby CRY2 governs two distinct pathways to control flowering time: (1) the COP1 dependent pathway shared with CRY1, and (2) the CIB dependent pathway that converges signals from two evolutionarily different blue light receptors.
Aside from COP1/SPA complexes and CIBs, phytochromes have also been reported to be CRY binding proteins (Ahmad et al. 1998a, b, Más et al. 2000). However, CRYs remain functional in Arabidopsis mutants that lack all phytochrome species (Strasser et al. 2010), suggesting that CRY function and signaling is only modified by phytochrome-CRY binding rather than being determined by it. Notably, it has also been recently reported that phytochromes inactivate COP1/SPA1 complexes by dissociating SPA1 from COP1 via red light dependent direct binding of phytochromes to SPA1 (Lu et al. 2015; Sheerin et al. 2015), indicating that the COP1/SPA1 complex is a common intermediate for CRY and phytochrome signaling. Functional interactions have been seen between CRYs and phytochromes in various light responses including inhibition of anthocyanin accumulation, photoperiodic flowering, root greening and so on (Mohr 1994; Neff and Chory 1998; Mockler et al. 1999, 2003; Casal 2000; Usami et al. 2004). Photoreceptor interactions and the convergence of photoreceptor signaling at the COP1/SPA complex provide strong clues for molecular mechanism. Additionally, it has been recently reported that phyA predominantly degrades SPA2 protein to inactivate the COP1/SPA2 complex not only in far-red light but also in blue light (Chen et al. 2015). Both phyA and phytochrome B, another major phytochrome species, are required for red light induced SPA2 degradation. Taken together with the fact that CRY1 and CRY2 have varying affinities to differing SPA proteins, it is apparent that we need to understand how CRYs and phytochromes selectively or collectively recognize SPA family proteins to modulate COP1/SPA complex activity in heterogeneous light environments in nature.
Structure requirement of CRYs for their signal transduction
The two major domains of CRY1 and CRY2, the PHR domain and the CCE domain, play roles in the CRY signal transduction mechanism that has turned out to be more complicated than initially proposed. In the original model, the blue light signal perceived in the PHR domain is transduced through the CCE domain to a downstream component of CRY signal transduction (Yang et al. 2000; Li and Yang 2007). This simple model arose from the observation that the overexpression of truncated CRY1 CCE domain fused to β-glucuronidase (GUS-CRY1 CCE) as well as GUS-CRY2 CCE gave rise to the constitutive photomorphogenic phenotype when grown in the dark (Yang et al. 2000). Consistent with this observation, both the CRY1 CCE domain and the CRY2 CCE domain can constitutively bind with COP1 (Wang et al. 2001; Yang et al. 2001). In addition, numerous mutations in the CCE domain prevented inhibition of hypocotyl elongation and anthocyanin accumulation by eliminating CRY mediation (Ahmad et al. 1995; Shalitin et al. 2003; Ruckle et al. 2007), suggesting that the CCE domain is of functional importance in CRY signal transduction. In line with this, CRY1 interacts with SPA1 through the CCE domain (Lian et al. 2011; Liu et al. 2011a, b). In contrast, the binding interface between CRY2 and SPA1 does not occur at the CCE domain, rather at the PHR domain (Zuo et al. 2011), suggesting a different mode of action between CRY1 and CRY2 in the regulation of COP1/SPA1 complexes. Furthermore, the binding interface between CRY2 and CIBs is also mapped to the PHR domain (Liu et al. 2008a; Kennedy et al. 2010). These data suggest that the PHR domain is of functional importance in the signal transduction pathway of CRY2.
Recently, the CRY1 PHR domain was shown to mediate the inhibition of hypocotyl elongation in response to blue light when targeted to the nucleus by fusion with a nuclear localization signal (CRY1 PHR-NLS) (He et al. 2015). CRY1 PHR-NLS neither binds to COP1 nor allows HY5 protein to accumulate, suggesting that it mediates blue light induced inhibition of hypocotyl elongation independently of the COP1 pathway. Although the binding partners of the CRY1 PHR domain remain unknown, the transcriptome analysis revealed that CRY1 PHR-NLS regulates 34 % of CRY-regulated genes. Also in most cases, CRY1 PHR-NLS acts antagonistically with phytohormones such as auxin, brassinosteroid and gibberellin, to regulate gene expression, which may lead the inhibition of hypocotyl elongation. This implies that transcription factors that regulate such phytohormone signaling pathways are directly involved in the CRY1 PHR domain-mediated response. Importantly, the CRY1 PHR domain-mediated response is strikingly different from the CIBs-mediated response in that neither the overexpresser nor the loss-of-function mutants of CIBs showed any phenotype that deviates from wild type in deetiolation responses, but they did show changes in flowering phenotype (Liu et al. 2008a; 2013b; He et al. 2015). Therefore, both CRY1 and CRY2 apparently constitute higher order complexes to act as hubs that drive at least two distinct pathways: (1) the CCE-domain-dependent COP1 pathway, which is common between CRY1 and CRY2, and (2) the COP1-independent, PHR-domain-dependent pathways that differ between CRY1 and CRY2, and may confer the functional specificities to these two CRYs (Fig. 1).
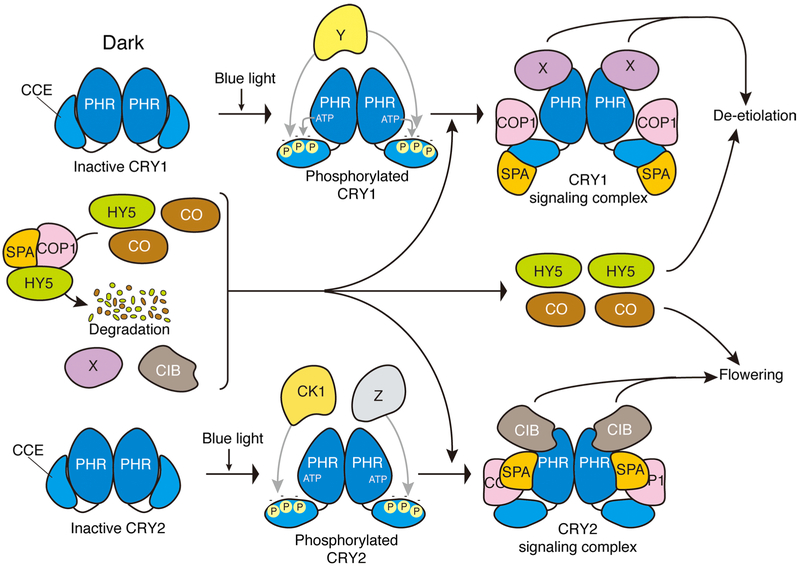
Fig. 1.
A model depicting cryptochrome signal transduction mechanisms in Arabidopsis. Cryptochromes (CRY) are composed of two domains, the PHR domain and the CCE domain, and form homodimers through the PHR domains. Upon blue light activation, CRYs undergo conformational changes in both the PHR and CCE domains to transduce signals to downstream components. Light-dependent phosphorylation at multiple sites of the CCE domain produce negative charges (−) at these sites via autophosphorylation and possibly by unknown kinases (Y) in the case of CRY1, and by CK1 and unknown kinases (Z) in the case of CRY2. The negative charges result in the electrostatic repulsion of CCE domain from PHR domain. In the dark, COP1 and SPA constitute an active complex to promote E3 ubiquitin ligase activity, leading the degradation of transcription factors such as HY5 and CO, positive regulaters for de-etiolation and flowering, respectively. In the light, photoactivated CRY1 constitutes the CRY1 signaling complex, in which CRY1 dissociates COP1 from SPA by direct binding via the CRY1 CCE domain to inactivate COP1/SPA complex. This results in the accumulation of HY5 and probably CO, and CRY1 binds with unknown factor (X) via the CRY1 PHR domain to activate the COP1 independent pathway, to promote primarily de-etiolation. Photoactivated CRY2 directly binds with SPA via the PHR domain to inactivate COP1/SPA complex by tightening the connection between COP1 and CRY2. CRY2 also binds with CIBs through the PHR domain to promote flowering. CK1 Casein kinase1, CCE cryptochrome C-terminal extension, CIB CRYinteracting basic-helix-loop-helix, CO CONSTANS, COP1 CONSTITUTIVE PHOTOMORPHOGENIC 1, FT FLOWERING LOCUS T, HY5 LONG HYPOCOTYL5, PHR photolyase-homologous region, SPA SUPPRESSOR OF PHYA-105
Photoactivation of CRYs
The way that information is transferred from photoexcited FAD to the effecter domains of CRY is largely unknown. The most important discovery in our understanding of the intramolecular signaling of CRY is the overexpression of GUS-CRY1 CCE and GUS-CRY2 CCE in the constitutively photomorphogenic phenotype of Arabidopsis (Yang et al. 2000). This observation has led to the closed/open model, in which the PHR domain binds to the CCE domain, repressing CCE domain activity under dark conditions, while under light conditions the PHR domain absorbs light, releasing the CCE domain and terminating repression to activate CRY. Consistent with this action model for CRY photoactivation, the CCE domain was shown to be sensitive to blue light dependent protease digestion (Partch et al. 2005). Transient grating spectroscopy revealed a large decrease in the diffusion coefficient of full-length CRY1 protein in response to blue light, which was not seen for the PHR domain of CRY1 (Kondoh et al. 2011), suggesting that dynamic conformational change takes place in the CCE domain during photoactivation. The mechanism, by which blue light de-represses the CCE domain from the PHR domain, is not fully understood, but it is thought to be associated with the electrostatic potential on the surface of the PHR domain. The crystal structure reveals that the CRY1 PHR domain is predominantly negatively charged (Brautigam et al. 2004). Given that the phosphate group is negatively charged, the blue light dependent phosphorylation at the multiple serine residues on the CCE domain could cause the electrostatic repulsion of the CCE domain from the PHR domain (Liu et al. 2011a, b). GUS-CRY2 CCE, in which CCE domain presumably has an opened form, does not require phosphorylation to show the constitutively photomorphogenic phenotype (Yu et al. 2007b), implying that CRY phosphorylation plays a role in the derepression of CCE domain from PHR domain. Blue light dependent autophosphorylation could trigger partial detachment of the CCE domain from the PHR domain, thereby making multiple sites accessible for phosphorylation by unidentified kinases (Bouly et al. 2003; Shalitin et al. 2003; Ozgur and Sancar 2006; Liu et al. 2011b). However, this initial de-repression mechanism prior to multiple phospohrylations cannot be applicable to CRY2 due to its apparent loss of autophosphorylation activity (Ozgur and Sancar 2006). In addition, the mechanism of how photoexcitation triggers autophosphorylation of CRY1 is still unknown.
Recently, ATP binding of the PHR domain was shown to promote the protonation of D396, a potential proton donor to FAD, which results in the increase of yield and lifetime of FADH˙ (Immeln et al. 2007; Burney et al. 2009; Cailliez et al. 2014; Müller et al. 2014). A strong negative charge then occurs in D396 as it donates a proton to FAD, resulting in the expulsion of negatively charged ATP from the FAD binding pocket (Müller et al. 2014; Müller and Bouly 2015). Because CCE domain covers the FAD binding pocket, the expulsion of ATP from the FAD binding pocket would trigger the detachment of the CCE domain and thereby facilitate phosphorylation by CK1.3, CK1.4 and other unidentified kinases. It should be noted, however, that the protonation of D396 is accompanied by the Trp-triad dependent photoreduction of FAD (Müller et al. 2014), which is not necessary for in vivo activity and function of CRYs (Li et al. 2011; Gao et al. 2015).
On the other hand, the COP1 independent signaling pathway of CRY1 and the blue-light dependent binding of CRY2 to SPA1 and CIBs would not rely on the electrostatic repulsion based closed/open model, because the PHR domain can trigger these responses without any aid by the CCE domain in a blue light dependent manner (Liu et al. 2008a; Kennedy et al. 2010; He et al. 2015). Blue light induced changes in the PHR domain were observed via Fourier Transform Infrared Spectroscopy and by the time-resolved step-scan Fourier Transform Infrared Spectroscopy complemented by UV–vis spectroscopy (Kottke et al. 2006; Thöing et al. 2015). The loss of β-sheets in the α/β subdomain of plant CRY from green algae in response to blue light is of great interest, because it has been reported that one mutation in the α/β subdomain of mouse CRY 1, a light-independent animal type 2 CRY, resulted in the inability of CRY to bind to BMAL1 and CLOCK, both of which are E-box binding basic helix-loop-helix transcription factors (Nangle et al. 2014). In particular, the interaction of this subdomain with BMAL1 and CLOCK is reminiscent of the blue light dependent binding of CIBs, the E-box binding basic helix-loop-helix transcription factors in the CRY2 PHR domain (Liu et al. 2008a; Liu et al. 2013b), although CRYs evolved independently in different evolutionary lineages (Lin and Todo 2005). In addition, two mutations that impair CRY1 mediated inhibition of hypocotyl elongation were found in this subdomain (Shalitin et al. 2003; Ruckle et al. 2007). The blue light induced change in the α/β subdomain might therefore contribute to the PHR dependent protein–protein interaction with the downstream components in the PHR dependent pathways. However, it remains speculative and conformational changes in the PHR domain induced by photoexcitation may possibly also be involved in the unfolding of PHR/CCE domains.
Concluding remarks and future outlook
Since CRY1 was first discovered in 1993, significant progress has been made in characterizing the action mode of plant CRYs. Blue light dependent phosphorylation of CRYs has turned out to play a critical role in the regulatory mechanism of this photoreceptor. Light dependent inactivation of the COP1/SPA complex has been identified as a regulatory element of CRY-mediated responses. Additionally, the COP1 independent pathway has also emerged as the primary mechanism of CRY signal transduction. Still, many questions with respect to CRY photoexcitation, signal transduction and photoactivation mechanisms remain unanswered. The biggest enigma awaiting elucidation is the photoexcitation mechanism of this photoreceptor. Because both the Trp-triad dependent photoreduction and the ATP dependent enhancement of photoreduction models have been shown to be irrelevant to CRY physiological function in vivo, an alternative mechanism underlying photoexcitation must be identified. With regard to CRY signal transduction and function, the protein kinases (and potentially protein phosphatases) involved in CRY1 and CRY2 phosphorylation also need to be identified. Furthermore, the binding partner of the CRY1 PHR domain and CRY2 binding factors other than CIBs and COP1/SPA complex also need to be identified to fully understand the CRY signal transduction mechanism. On the other hand, the expansion of crystallographic resolution of the CRY1 PHR domain, full-length CRY protein, and photoexcited CRY can be expected to reveal CRY structure and to uncover mechanisms of CRY conformational changes in response to blue light. Continued discoveries to answer these unresolved questions about CRY will provide invaluable insight into the action mechanism of CRY and the general understanding of plant photomorphogenesis.
Acknowledgments
This work was supported by the Program for New Century Excellent Talents in Fujian Province University and the School Special Development program of Fujian Agriculture and Forestry University (6112C035001).
References
- Ahmad M, Cashmore AR (1993) HY4 gene of A. thaliana encodes a protein with characteristics of a blue-light photoreceptor. Nature 366:162–166 [DOI] [PubMed] [Google Scholar]
- Ahmad M, Lin C, Cashmore AR (1995) Mutations throughout an Arabidopsis blue-light photoreceptor impair blue-light-responsive anthocyanin accumulation and inhibition of hypocotyl elongation. Plant J 8:653–658 [DOI] [PubMed] [Google Scholar]
- Ahmad M, Jarillo JA, Cashmore AR (1998a) Chimeric proteins between cry1 and cry2 Arabidopsis blue light photoreceptors indicate overlapping functions and varying protein stability. Plant Cell 10:197–208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmad M, Jarillo JA, Smirnova O, Cashmore AR (1998b) The CRY1 blue light photoreceptor of Arabidopsis interacts with phytochrome A in vitro. Mol Cell 1:939–948 [DOI] [PubMed] [Google Scholar]
- Bagnall DJ, King RW, Hangarter RP (1996) Blue-light promotion of flowering is absent in hy4 mutants of Arabidopsis. Planta 200:278–280 [DOI] [PubMed] [Google Scholar]
- Banerjee R, Schleicher E, Meier S, Viana RM, Pokorny R, Ahmad M, Bittl R, Batschauer A (2007) The signaling state of Arabidopsis cryptochrome 2 contains flavin semiquinone. J Biol Chem 282:14916–14922 [DOI] [PubMed] [Google Scholar]
- Blazquez MA, Ahn JH, Weigel D (2003) A thermosensory pathway controlling flowering time in Arabidopsis thaliana. Nat Genet 33:168–171 [DOI] [PubMed] [Google Scholar]
- Bouly JP, Giovani B, Djamei A, Mueller M, Zeugner A, Dudkin EA, Batschauer A, Ahmad M (2003) Novel ATP-binding and autophosphorylation activity associated with Arabidopsis and human cryptochrome-1. Eur J Biochem 270:2921–2928 [DOI] [PubMed] [Google Scholar]
- Bouly JP, Schleicher E, Dionisio-Sese M, Vandenbussche F, Van Der Straeten D, Bakrim N, Meier S, Batschauer A, Galland P, Bittl R, Ahmad M (2007) Cryptochrome blue light photoreceptors are activated through interconversion of flavin redox states. J Biol Chem 282:9383–9391 [DOI] [PubMed] [Google Scholar]
- Brautigam CA, Smith BS, Ma Z, Palnitkar M, Tomchick DR, Machius M, Deisenhofer J (2004) Structure of the photolyaselike domain of cryptochrome 1 from Arabidopsis thaliana. Proc Natl Acad Sci USA 101:12142–12147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briggs WR, Christie JM, Salomon M (2001) Phototropins: a new family of flavin-binding blue light receptors in plants. Antioxid Redox Signal 3:775–788 [DOI] [PubMed] [Google Scholar]
- Brudler R, Hitomi K, Daiyasu H, Toh H, Kucho K, Ishiura M, Kanehisa M, Roberts VA, Todo T, Tainer JA, Getzoff ED (2003) Identification of a new cryptochrome class. Structure, function, and evolution. Mol Cell 11:59–67 [DOI] [PubMed] [Google Scholar]
- Bruggemann E, Handwerger K, Essex C, Storz G (1996) Analysis of fast neutron-generated mutants at the Arabidopsis thaliana HY4 locus. Plant J 10:755–760 [DOI] [PubMed] [Google Scholar]
- Burney S, Hoang N, Caruso M, Dudkin EA, Ahmad M, Bouly JP (2009) Conformational change induced by ATP binding correlates with enhanced biological function of Arabidopsis cryptochrome. FEBS Lett 583:1427–1433 [DOI] [PubMed] [Google Scholar]
- Cailliez F, Müller P, Gallois M, de la Lande A (2014) ATP binding and aspartate protonation enhance photoinduced electron transfer in plant cryptochrome. J Am Chem Soc 136:12974–17986 [DOI] [PubMed] [Google Scholar]
- Casal JJ (2000) Phytochromes, cryptochromes, phototropin: photoreceptor interactions in plants. Photochem Photobiol 71:1–11 [DOI] [PubMed] [Google Scholar]
- Cashmore AR (2003) Cryptochromes: enabling plants and animals to determine circadian time. Cell 114:537–543 [PubMed] [Google Scholar]
- Cashmore AR, Jarillo JA, Wu YJ, Liu D (1999) Cryptochromes: blue light receptors for plants and animals. Science 284:760–765 [DOI] [PubMed] [Google Scholar]
- Chaves I, Pokorny R, Byrdin M, Hoang N, Ritz T, Brettel K, Essen LO, van der Horst GT, Batschauer A, Ahmad M (2011) The cryptochromes: blue light photoreceptors in plants and animals. Annu Rev Plant Biol 62:335–364 [DOI] [PubMed] [Google Scholar]
- Chen S, Lory N, Stauber J, Hoecker U (2015) Photoreceptor specificity in the light-induced and COP1-mediated rapid degradation of the repressor of photomorphogenesis SPA2 in Arabidopsis. PLoS Genet 11(9):e1005516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Assal SE-D, Alonso-Blanco C, Peeters AJ, Raz V, Koornneef M (2001) A QTL for flowering time in Arabidopsis reveals a novel allele of CRY2. Nat Genet 29:435–440 [DOI] [PubMed] [Google Scholar]
- Engelhard C, Wang X, Robles D, Moldt J, Essen LO, Batschauer A, Bittl R, Ahmad M (2014) Cellular metabolites enhance the light sensitivity of Arabidopsis cryptochrome through alternate electron transfer pathways. Plant Cell 26:4519–4531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Exner V, Alexandre C, Rosenfeldt G, Alfarano P, Nater M, Caflisch A, Gruissem W, Batschauer A, Hennig L (2010) A gain-of-function mutation of Arabidopsis cryptochrome1 promotes flowering. Plant Physiol 154:1633–1645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fankhauser C, Ulm R (2011) Light-regulated interactions with SPA proteins underlie cryptochrome-mediated gene expression. Genes Dev 25:1004–1009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Favory JJ, Stec A, Gruber H, Rizzini L, Oravecz A, Funk M, Albert A, Cloix C, Jenkins GI, Oakeley EJ, Seidlitz HK, Nagy F, Ulm R (2009) Interaction of COP1 and UVR8 regulates UV-B-induced photomorphogenesis and stress acclimation in Arabidopsis. EMBO J 28:591–601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fedele G, Edwards MD, Bhutani S, Hares JM, Murbach M, Green EW, Dissel S, Hastings MH, Rosato E, Kyriacou CP (2014) Genetic analysis of circadian responses to low frequency electromagnetic fields in Drosophila melanogaster. PLoS Genet 10:e1004804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folta KM, Pontin MA, Karlin-Neumann G, Bottini R, Spalding EP (2003) Genomic and physiological studies of early cryptochrome 1 action demonstrate roles for auxin and gibberellin in the control of hypocotyl growth by blue light. Plant J 36:203–214 [DOI] [PubMed] [Google Scholar]
- Gao J, Wang X, Zhang M, Bian M, Deng W, Zuo Z, Yang Z, Zhong D, Lin C (2015) Trp triad-dependent rapid photoreduction is not required for the function of Arabidopsis CRY1. Proc Natl Acad Sci USA 112:9135–9140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gegear RJ, Foley LE, Casselman A, Reppert SM (2010) Animal cryptochromes mediate magnetoreception by an unconventional photochemical mechanism. Nature 463:804–807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goto N, Kumagai T, Koornneef M (1991) Flowering responses to light-breaks in photomorphogenic mutants of Arabidopsis thaliana, a long-day plant. Physiol Plant 83:209–215 [Google Scholar]
- Gu NN, Zhang YC, Yang HQ (2012) Substitution of a conserved glycine in the PHR domain of Arabidopsis cryptochrome 1 confers a constitutive light response. Mol Plant 5:85–97 [DOI] [PubMed] [Google Scholar]
- Guo H, Yang H, Mockler TC, Lin C (1998) Regulation of flowering time by Arabidopsis photoreceptors. Science 279:1360–1363 [DOI] [PubMed] [Google Scholar]
- Guo H, Duong H, Ma N, Lin C (1999) The Arabidopsis blue light receptor cryptochrome 2 is a nuclear protein regulated by a blue light-dependent post-transcriptional mechanism. Plant J 19:279–287 [DOI] [PubMed] [Google Scholar]
- He SB, Wang WX, Zhang JY, Xu F, Lian HL, Li L, Yang HQ (2015) The CNT1 domain of Arabidopsis CRY1 alone is sufficient to mediate blue light inhibition of hypocotyl elongation. Mol Plant 8:822–825 [DOI] [PubMed] [Google Scholar]
- Hense A, Herman E, Oldemeyer S, Kottke T (2015) Proton transfer to flavin stabilizes the signaling state of the blue light receptor plant cryptochrome. J Biol Chem 290:1743–1751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoang N, Bouly JP, Ahmad M (2008) Evidence of a light-sensing role for folate in Arabidopsis cryptochrome blue-light receptors. Mol Plant 1:68–74 [DOI] [PubMed] [Google Scholar]
- Immeln D, Schlesinger R, Heberle J, Kottke T (2007) Blue light induces radical formation and autophosphorylation in the light-sensitive domain of Chlamydomonas cryptochrome. J Biol Chem 282:21720–21728 [DOI] [PubMed] [Google Scholar]
- Immeln D, Pokorny R, Herman E, Moldt J, Batschauer A, Kottke T (2010) Photoreaction of plant and DASH cryptochromes probed by infrared spectroscopy: the neutral radical state of flavoproteins. J Phys Chem B 114:17155–17161 [DOI] [PubMed] [Google Scholar]
- Jang S, Marchal V, Panigrahi KC, Wenkel S, Soppe W, Deng XW, Valverde F, Coupland G (2008) Arabidopsis COP1 shapes the temporal pattern of CO accumulation conferring a photoperiodic flowering response. EMBO J 27:1277–1288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong RD, Chandra-Shekara AC, Barman SR, Navarre D, Klessig DF, Kachroo A, Kachroo P (2010) Cryptochrome 2 and phototropin 2 regulate resistance protein-mediated viral defense by negatively regulating an E3 ubiquitin ligase. Proc Natl Acad Sci USA 107:13538–13543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao Y, Lau OS, Deng XW (2007) Light-regulated transcriptional networks in higher plants. Nat Rev Genet 8:217–230 [DOI] [PubMed] [Google Scholar]
- Kami C, Lorrain S, Hornitschek P, Fankhauser C (2010) Light-regulated plant growth and development. Curr Top Dev Biol 91:29–66 [DOI] [PubMed] [Google Scholar]
- Kang CY, Lian HL, Wang FF, Huang JR, Yang HQ (2009) Cryptochromes, phytochromes, and COP1 regulate light-controlled stomatal development in Arabidopsis. Plant Cell 21:2624–2641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kavakli IH, Sancar A (2004) Analysis of the role of intraprotein electron transfer in photoreactivation by DNA photolyase in vivo. Biochemistry 43:15103–15110 [DOI] [PubMed] [Google Scholar]
- Kennedy MJ, Hughes RM, Peteya LA, Schwartz JW, Ehlers MD, Tucker CL (2010) Rapid blue-light-mediated induction of protein interactions in living cells. Nat Methods 7:973–975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleine T, Lockhart P, Batschauer A (2003) An Arabidopsis protein closely related to Synechocystis cryptochrome is targeted to organelles. Plant J 35:93–103 [DOI] [PubMed] [Google Scholar]
- Kleiner O, Kircher S, Harter K, Batschauer A (1999) Nuclear localization of the Arabidopsis blue light receptor cryptochrome 2. Plant J 19:289–296 [DOI] [PubMed] [Google Scholar]
- Kondoh M, Shiraishi C, Müller P, Ahmad M, Hitomi K, Getzoff ED, Terazima M (2011) Light-induced conformational changes in full-length Arabidopsis thaliana cryptochrome. J Mol Biol 413:128–137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kottke T, Batschauer A, Ahmad M, Heberle J (2006) Blue-light-induced changes in Arabidopsis cryptochrome 1 probed by FTIR difference spectroscopy. Biochemistry 45:2472–2479 [DOI] [PubMed] [Google Scholar]
- Lau OS, Deng XW (2012) The photomorphogenic repressors COP1 and DET1: 20 years later. Trends Plant Sci 17:584–593 [DOI] [PubMed] [Google Scholar]
- Laubinger S, Fittinghoff K, Hoecker U (2004) The SPA quartet: a family of WD-repeat proteins with a central role in suppression of photomorphogenesis in Arabidopsis. Plant Cell 16:2293–2306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laubinger S, Marchal V, Le Gourrierec J, Wenkel S, Adrian J, Jang S, Kulajta C, Braun H, Coupland G, Hoecker U (2006) Arabidopsis SPA proteins regulate photoperiodic flowering and interact with the floral inducer CONSTANS to regulate its stability. Development 133:3213–3222 [DOI] [PubMed] [Google Scholar]
- Li YF, Sancar A (1990) Active site of Escherichia coli DNA photolyase: mutations at Trp277 alter the selectivity of the enzyme without affecting the quantum yield of photorepair. Biochemistry 29:5698–5706 [DOI] [PubMed] [Google Scholar]
- Li QH, Yang HQ (2007) Cryptochrome signaling in plants. Photochem Photobiol 83:94–101 [DOI] [PubMed] [Google Scholar]
- Li YF, Heelis PF, Sancar A (1991) Active site of DNA photolyase: tryptophan-306 is the intrinsic hydrogen atom donor essential for flavin radical photoreduction and DNA repair in vitro. Biochemistry 30:6322–6329 [DOI] [PubMed] [Google Scholar]
- Li X, Wang Q, Yu X, Liu H, Yang H, Zhao C, Liu X, Tan C, Klejnot J, Zhong D, Lin C (2011) Arabidopsis cryptochrome 2 (CRY2) functions by the photoactivation mechanism distinct from the tryptophan (trp) triad-dependent photoreduction. Proc Natl Acad Sci USA 108:20844–20849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lian HL, He SB, Zhang YC, Zhu DM, Zhang JY, Jia KP, Sun SX, Li L, Yang HQ (2011) Blue-light-dependent interaction of cryptochrome 1 with SPA1 defines a dynamic signaling mechanism. Genes Dev 25:1023–1028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin C, Shalitin D (2003) Cryptochrome structure and signal transduction. Annu Rev Plant Biol 54:469–496 [DOI] [PubMed] [Google Scholar]
- Lin C, Todo T (2005) The cryptochromes. Genome Biol 6:220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin C, Robertson DE, Ahmad M, Raibekas AA, Jorns MS, Dutton PL, Cashmore AR (1995) Association of flavin adenine dinucleotide with the Arabidopsis blue light receptor CRY1. Science 269:968–970 [DOI] [PubMed] [Google Scholar]
- Lin C, Yang H, Guo H, Mockler T, Chen J, Cashmore AR (1998) Enhancement of blue-light sensitivity of Arabidopsis seedlings by a blue light receptor cryptochrome 2. Proc Natl Acad Sci USA 95:2686–2690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Yu X, Li K, Klejnot J, Yang H, Lisiero D, Lin C (2008a) Photoexcited CRY2 interacts with CIB1 to regulate transcription and floral initiation in Arabidopsis. Science 322:1535–1539 [DOI] [PubMed] [Google Scholar]
- Liu LJ, Zhang YC, Li QH, Sang Y, Mao J, Lian HL, Wang L, Yang HQ (2008b) COP1-mediated ubiquitination of CONSTANS is implicated in cryptochrome regulation of flowering in Arabidopsis. Plant Cell 20:292–306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B, Liu H, Zhong D, Lin C (2010) Searching for a photocycle of the cryptochrome photoreceptors. Curr Opin Plant Biol 13:578–586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B, Zuo Z, Liu H, Liu X, Lin C (2011a) Arabidopsis cryptochrome 1 interacts with SPA1 to suppress COP1 activity in response to blue light. Genes Dev 25:1029–1034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Liu B, Zhao C, Pepper M, Lin C (2011b) The action mechanisms of plant cryptochromes. Trends Plant Sci 16:684–691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Wang Q, Liu Y, Zhao X, Imaizumi T, Somers DE, Tobin EM, Lin C (2013a) Arabidopsis CRY2 and ZTL mediate blue-light regulation of the transcription factor CIB1 by distinct mechanisms. Proc Natl Acad Sci USA 110:17582–17587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Li X, Li K, Liu H, Lin C (2013b) Multiple bHLH proteins form heterodimers to mediate CRY2-dependent regulation of flowering-time in Arabidopsis. PLoS Genet 9:e1003861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu XD, Zhou CM, Xu PB, Luo Q, Lian HL, Yang HQ (2015) Red-light-dependent interaction of phyB with SPA1 promotes COP1-SPA1 dissociation and photomorphogenic development in Arabidopsis. Mol Plant 8(3):467–478 [DOI] [PubMed] [Google Scholar]
- Ma L, Li J, Qu L, Hager J, Chen Z, Zhao H, Deng XW (2001) Light control of Arabidopsis development entails coordinated regulation of genome expression and cellular pathways. Plant Cell 13:2589–2607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malhotra K, Kim ST, Batschauer A, Dawut L, Sancar A (1995) Putative blue-light photoreceptors from Arabidopsis thaliana and Sinapis alba with a high degree of sequence homology to DNA photolyase contain the two photolyase cofactors but lack DNA repair activity. Biochemistry 34:6892–6899 [DOI] [PubMed] [Google Scholar]
- Mao J, Zhang YC, Sang Y, Li QH, Yang HQ (2005) From the cover: a role for Arabidopsis cryptochromes and COP1 in the regulation of stomatal opening. Proc Natl Acad Sci USA 102:12270–12275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Más P, Devlin PF, Panda S, Kay SA (2000) Functional interaction of phytochrome B and cryptochrome 2. Nature 408:207–211 [DOI] [PubMed] [Google Scholar]
- Meng Y, Li H, Wang Q, Liu B, Lin C (2013) Blue light-dependent interaction between cryptochrome2 and CIB1 regulates transcription and leaf senescence in soybean. Plant Cell 25:4405–4420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mockler TC, Guo H, Yang H, Duong H, Lin C (1999) Antagonistic actions of Arabidopsis cryptochromes and phytochrome B in the regulation of floral induction. Development 126(10):2073–2082 [DOI] [PubMed] [Google Scholar]
- Mockler T, Yang H, Yu X, Parikh D, Cheng YC, Dolan S, Lin C (2003) Regulation of photoperiodic flowering by Arabidopsis photoreceptors. Proc Natl Acad Sci USA 100:2140–2145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohr H (1994) Coaction between pigment systems In: Kendrick RE, Kronenberg GHM (eds) Photomorphogenesis in plants, 2nd edn. Kluwer Academic Publishers, Dordrecht, pp 353–373 [Google Scholar]
- Mozley D, Thomas B (1995) Developmental and photobiological factors affecting photoperiodic induction in Arabidopsis thaliana Heynh. Landsberg erecta. J Exp Bot 46:173–179 [Google Scholar]
- Müller P, Bouly JP (2015) Searching for the mechanism of signalling by plant photoreceptor cryptochrome. FEBS Lett 589:189–192 [DOI] [PubMed] [Google Scholar]
- Müller P, Bouly JP, Hitomi K, Balland V, Getzoff ED, Ritz T, Brettel K (2014) ATP binding turns plant cryptochrome into an efficient natural photoswitch. Sci Rep 5:5175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nangle SN, Rosensweig C, Koike N, Tei H, Takahashi JS, Green CB, Zheng N (2014) Molecular assembly of the period-cryptochrome circadian transcriptional repressor complex. Elife 15:e03674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neff MM, Chory J (1998) Genetic interactions between phytochrome A, phytochrome B, and cryptochrome 1 during Arabidopsis development. Plant Physiol 118(1):27–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohgishi M, Saji K, Okada K, Sakai T (2004) Functional analysis of each blue light receptor, cry1, cry2, phot1, and phot2, by using combinatorial multiple mutants in Arabidopsis. Proc Natl Acad Sci USA 101:2223–2228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osterlund MT, Hardtke CS, Wei N, Deng XW (2000) Targeted destabilization of HY5 during light-regulated development of Arabidopsis. Nature 405:462–466 [DOI] [PubMed] [Google Scholar]
- Ozgur S, Sancar A (2006) Analysis of autophosphorylating kinase activities of Arabidopsis and human cryptochromes. Biochemistry 45:13369–13374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozturk N, Selby CP, Annayev Y, Zhong D, Sancar A (2011) Reaction mechanism of Drosophila cryptochrome. Proc Natl Acad Sci USA 108:516–521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oztürk N, Song SH, Selby CP, Sancar A (2008) Animal type 1 cryptochromes. Analysis of the redox state of the flavin cofactor by site-directed mutagenesis. J Biol Chem 283:3256–3263 [DOI] [PubMed] [Google Scholar]
- Partch CL, Clarkson MW, Ozgur S, Lee AL, Sancar A (2005) Role of structural plasticity in signal transduction by the cryptochrome blue-light photoreceptor. Biochemistry 44:3795–3805 [DOI] [PubMed] [Google Scholar]
- Pokorny R, Klar T, Hennecke U, Carell T, Batschauer A, Essen LO (2008) Recognition and repair of UV lesions in loop structures of duplex DNA by DASH-type cryptochrome. Proc Natl Acad Sci USA 105:21023–21027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritz T, Adem S, Schulten K (2000) A model for photoreceptor-based magnetoreception in birds. Biophys J 78:707–718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritz T, Yoshii T, Helfrich-Foerster C, Ahmad M (2010) Cryptochrome: a photoreceptor with the properties of a magnetoreceptor? Commun Integr Biol 3:24–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenfeldt G, Viana RM, Mootz HD, von Arnim AG, Batschauer A (2008) Chemically induced and light-independent cryptochrome photoreceptor activation. Mol Plant 1:4–14 [DOI] [PubMed] [Google Scholar]
- Ruckle ME, DeMarco SM, Larkin RM (2007) Plastid signals remodel light signaling networks and are essential for efficient chloroplast biogenesis in Arabidopsis. Plant Cell 19:3944–3960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saijo Y, Sullivan JA, Wang H, Yang J, Shen Y, Rubio V, Ma L, Hoecker U, Deng XW (2003) The COP1-SPA1 interaction defines a critical step in phytochrome A-mediated regulation of HY5 activity. Genes Dev 17:2642–2647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sancar A (2003) Structure and function of DNA photolyase and cryptochrome blue-light photoreceptors. Chem Rev 103:2203–2237 [DOI] [PubMed] [Google Scholar]
- Sang Y, Li QH, Rubio V, Zhang YC, Mao J, Deng XW, Yang HQ (2005) N-terminal domain-mediated homodimerization is required for photoreceptor activity of Arabidopsis CRYPTOCHROME 1. Plant Cell 17:1569–1584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Searle I, Coupland G (2004) Induction of flowering by seasonal changes in photoperiod. EMBO J 23:1217–1222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selby CP, Sancar A (2006) A cryptochrome/photolyase class of enzymes with single-stranded DNA-specific photolyase activity. Proc Natl Acad Sci USA 103:17696–17700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sellaro R, Hoecker U, Yanovsky M, Chory J, Casal JJ (2009) Synergism of red and blue light in the control of Arabidopsis gene expression and development. Curr Biol 19:1216–1220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo HS, Yang JY, Ishikawa M, Bolle C, Ballesteros ML, Chua NH (2003) LAF1 ubiquitination by COP1 controls photomorphogenesis and is stimulated by SPA1. Nature 423:995–999 [DOI] [PubMed] [Google Scholar]
- Shalitin D, Yang H, Mockler TC, Maymon M, Guo H, Whitelam GC, Lin C (2002) Regulation of Arabidopsis cryptochrome 2 by blue-light-dependent phosphorylation. Nature 417:763–767 [DOI] [PubMed] [Google Scholar]
- Shalitin D, Yu X, Maymon M, Mockler T, Lin C (2003) Blue light-dependent in vivo and in vitro phosphorylation of Arabidopsis cryptochrome 1. Plant Cell 15:2421–2429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheerin DJ, Menon C, zur Oven-Krockhaus S, Enderle B, Zhu L, Johnen P, Schleifenbaum F, Stierhof YD, Huq E, Hiltbrunner A (2015) Light-activated phytochrome A and B interact with members of the SPA family to promote photomorphogenesis in Arabidopsis by reorganizing the COP1/SPA complex. Plant Cell 27(1):189–201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith H (2000) Phytochromes and light signal perception by plants—an emerging synthesis. Nature 407:585–591 [DOI] [PubMed] [Google Scholar]
- Song SH, Oztürk N, Denaro TR, Arat NO, Kao YT, Zhu H, Zhong D, Reppert SM, Sancar A (2007) Formation and function of flavin anion radical in cryptochrome 1 blue-light photoreceptor of monarch butterfly. J Biol Chem 282:17608–17612 [DOI] [PubMed] [Google Scholar]
- Strasser B, Sanchez-Lamas M, Yanovsky MJ, Casal JJ, Cerdan PD (2010) Arabidopsis thaliana life without phytochromes. Proc Natl Acad Sci USA 107:4776–4781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan ST, Dai C, Liu HT, Xue HW (2013) Arabidopsis casein kinase1 proteins CK1.3 and CK1.4 phosphorylate cryptochrome2 to regulate blue light signaling. Plant Cell 25:2618–2632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thöing C, Oldemeyer S, Kottke T (2015) Microsecond deprotonation of Aspartic Acid and response of the α/β subdomain precede C-terminal signaling in the blue light sensor plant cryptochrome. J Am Chem Soc 137:5990–5999 [DOI] [PubMed] [Google Scholar]
- Usami T, Mochizuki N, Kondo M, Nishimura M, Nagatani A (2004) Cryptochromes and phytochromes synergistically regulate Arabidopsis root greening under blue light. Plant Cell Physiol 45(12):1798–1808 [DOI] [PubMed] [Google Scholar]
- Wang H, Ma LG, Li JM, Zhao HY, Deng XW (2001) Direct interaction of Arabidopsis cryptochromes with COP1 in light control development. Science 294:154–158 [DOI] [PubMed] [Google Scholar]
- Wang FF, Lian HL, Kang CY, Yang HQ (2010) Phytochrome B is involved in mediating red light-induced stomatal opening in Arabidopsis thaliana. Mol Plant 3:246–259 [DOI] [PubMed] [Google Scholar]
- Wang Q, Barshop WD, Bian M, Vashisht AA, He R, Yu X, Liu B, Nguyen P, Liu X, Zhao X, Wohlschlegel JA, Lin C (2015a) The blue light-dependent phosphorylation of the CCE domain determines the photosensitivity of Arabidopsis CRY2. Mol Plant 8:631–643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Wang Q, Nguyen P, Lin C (2015b) Cryptochrome-mediated light responses in plants. Enzymes 35:167–189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weidler G, Zur Oven-Krockhaus S, Heunemann M, Orth C, Schleifenbaum F, Harter K, Hoecker U, Batschauer A (2012) Degradation of Arabidopsis CRY2 is regulated by SPA proteins and phytochrome A. Plant Cell 24:2610–2623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu G, Spalding EP (2007) Separate functions for nuclear and cytoplasmic cryptochrome 1 during photomorphogenesis of Arabidopsis seedlings. Proc Natl Acad Sci USA 104:18813–18818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L, Yang HQ (2010) CRYPTOCHROME 1 is implicated in promoting R protein-mediated plant resistance to Pseudomonas syringae in Arabidopsis. Mol Plant 3:539–548 [DOI] [PubMed] [Google Scholar]
- Yang HQ, Wu YJ, Tang RH, Liu D, Liu Y, Cashmore AR (2000) The C termini of Arabidopsis cryptochromes mediate a constitutive light response. Cell 103:815–827 [DOI] [PubMed] [Google Scholar]
- Yang HQ, Tang RH, Cashmore AR (2001) The signaling mechanism of Arabidopsis CRY1 involves direct interaction with COP1. Plant Cell 13:2573–2587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh KC, Lagarias JC (1998) Eukaryotic phytochromes: light-regulated serine/threonine protein kinases with histidine kinase ancestry. Proc Natl Acad Sci USA 95:13976–13981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi C, Deng XW (2005) COP1-from plant photomorphogenesis to mammalian tumorigenesis. Trends Cell Biol 15:618–625 [DOI] [PubMed] [Google Scholar]
- Yu X, Klejnot J, Zhao X, Shalitin D, Maymon M, Yang H, Lee J, Liu X, Lopez J, Lin C (2007a) Arabidopsis cryptochrome 2 completes its posttranslational life cycle in the nucleus. Plant Cell 19:3146–3156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X, Shalitin D, Liu X, Maymon M, Klejnot J, Yang H, Lopez J, Zhao X, Bendehakkalu KT, Lin C (2007b) Derepression of the NC80 motif is critical for the photoactivation of Arabidopsis CRY2. Proc Natl Acad Sci USA 104:7289–7294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X, Sayegh R, Maymon M, Warpeha K, Klejnot J, Yang H, Huang J, Lee J, Kaufman L, Lin C (2009) Formation of nuclear bodies of Arabidopsis CRY2 in response to blue light is associated with its blue light-dependent degradation. Plant Cell 21:118–130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X, Liu H, Klejnot J, Lin C (2010) The cryptochrome blue light receptors. Arabidopsis Book 23:e0135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan Q, Metterville D, Briscoe AD, Reppert SM (2007) Insect cryptochromes: gene duplication and loss define diverse ways to construct insect circadian clocks. Mol Biol Evol 24:948–955 [DOI] [PubMed] [Google Scholar]
- Zeugner A, Byrdin M, Bouly JP, Bakrim N, Giovani B, Brettel K, Ahmad M (2005) Light-induced electron transfer in Arabidopsis cryptochrome-1 correlates with in vivo function. J Biol Chem 280:19437–19440 [DOI] [PubMed] [Google Scholar]
- Zhu H, Yuan Q, Briscoe AD, Froy O, Casselman A, Reppert SM (2005) The two CRYs of the butterfly. Curr Biol 15:953–954 [DOI] [PubMed] [Google Scholar]
- Zuo Z, Liu H, Liu B, Liu X, Lin C (2011) Blue light-dependent interaction of CRY2 with SPA1 regulates COP1 activity and floral initiation in Arabidopsis. Curr Biol 21:841–847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo ZC, Meng YY, Yu XH, Zhang ZL, Feng DS, Sun SF, Liu B, Lin CT (2012) A study of the blue-light-dependent phosphorylation, degradation, and photobody formation of Arabidopsis CRY2. Mol Plant 5:726–733 [DOI] [PMC free article] [PubMed] [Google Scholar]