Abstract
Background
Clear cell sarcoma of soft tissue (CCSST) and clear cell sarcoma-like gastrointestinal tumor (CCSLGT) are malignant mesenchymal tumors that share some pathological features, but they also have several different characteristics. They are well known to express chimeric fusions of Ewing sarcoma breakpoint region 1 (EWSR1) and cAMP response element-binding protein (CREB) family members; namely, EWSR1-activating transcription factor 1 (ATF1) and EWSR1-CREB1. In addition, recent studies have suggested the presence of other fusions.
Methods
We used fluorescence in situ hybridization to detect specific rearrangements including EWSR1, ATF1, CREB1, and cAMP response element modulator (CREM) in 16 CCSST and 6 CCSLGT cases. We also used reverse transcription polymerase chain reaction (RT-PCR) to detect specific chimeric fusions of EWSR1-ATF1 and EWSR1-CREB1 using fresh tumor samples in available cases.
Results
A total of 15 of 16 CCSST cases (93.8%) had EWSR1 rearrangement, of which 11 (68.8%) also had ATF1 rearrangement, suggestive of the presence of EWSR1-ATF1 fusions. One CCSST case (6.3%) was found to have EWSR1 and CREM rearrangements, and 4 of 6 CCSLGT cases (66.7%) had EWSR1 rearrangement, of which 2 (33.3%) showed ATF1 rearrangement and the other 2 cases (33.3%) showed CREB1 rearrangement. These cases most likely had EWSR1-ATF1 and EWSR1-CREB1 fusions, respectively. RT-PCR was performed in 8 available cases, including 6 CCSSTs and 2 CCSLGTs. All CCSSTs showed EWSR1-ATF1 fusions. Among the 2 CCSLGT cases, one had EWSR1-ATF1 fusion and the other had EWSR1-CREB1 fusion.
Conclusions
Rearrangements of EWSR1 and ATF1 or EWSR1-ATF1 fusion were predominantly found in CCSST, whereas those of EWSR1 and CREB1 or EWSR1-CREB1 tended to be detected in CCSLGT. A novel CREM fusion was also detected in a few cases of CCSST and CCSLGT. The cases in which EWSR1 rearrangement was detected without definitive partner genes should be considered for the presence of CREM rearrangement.
Keywords: Clear cell sarcoma of soft tissue (CCSST), Clear cell sarcoma-like gastrointestinal tumor (CCSLGT), EWSR1-ATF1, EWSR1-CREB1, EWSR1-CREM
Background
Clear cell sarcoma of soft tissue (CCSST) is a malignant mesenchymal tumor that mostly affects young adults and tends to affect the lower extremities, close to the tendon and aponeuroses [1]. Histologically, CCSSTs have epithelioid tumor nests accompanied by some spindling areas, and wreath-like multinucleated giant cells. CCSSTs present with a melanocytic differentiation and often express melanocytic markers including S-100 protein, melanoma antigen (Melan-A), human melanoma black 45 (HMB45), microphthalmia-associated transcription factor (MITF), and SRY-Box 10 (SOX-10) on immunohistochemistry (IHC). Ultrastructurally, CCSST has premelanosomes in the cytoplasm of tumor cells and shares some characteristic features with malignant melanomas (MMs). MMs genetically have BRAF mutations, although CCSST lacks this mutation. Clear cell sarcoma-like gastrointestinal tumor (CCSLGT) is also a malignant mesenchymal tumor that shares some pathological features with CCSST and arises from the gastrointestinal tract, such as the small and large intestine, and stomach. CCSLGT was originally reported to be an “osteoclast-rich tumor of the gastrointestinal tract with features resembling clear cell sarcoma of the soft parts” [2] and the first case of CCSLGT was reported by Alpers et al. [3] as a “malignant neuroendocrine tumor of the jejunum with osteoclast-like giant cells” in 1985. Subsequently, the term CCSLGT was first used by Kosemehmetoglu et al. [4] in their review, which included 13 CCSLGT cases. However, some authors have proposed using the term “malignant gastrointestinal neuroectodermal tumor,” because CCSLGTs lack melanocytic differentiation on IHC and ultrastructural examination and appear to have poorer prognosis [5]. Although CCSLGT has a similar histology to CCSST in some respects, such as a clear cytoplasm and epithelioid cells, there are some differing characteristics. CCSLGT has a pseudo-papillary growth pattern and many osteoclast-type giant cells, and the tumor cells tend to be positive for S-100 protein but show less expression of melanocytic markers on IHC [6]. Genetically, CCSST and CCSLGT usually have characteristic chimeric fusions of Ewing sarcoma breakpoint region 1 (EWSR1) with cAMP response element-binding protein (CREB) gene family members, EWSR1-activating transcription factor 1 (ATF1) and EWSR1-CREB1, which were derived from each translocation of t(12;22)(q13;q12) and t(2;22)(q34;q12), respectively [7–10]. EWSR1-ATF1 fusion is much more frequent than EWSR1-CREB1 fusion, but EWSR1-CREB1 fusion of CCSLGT is comparatively often observed.
In this study, we used fluorescence in situ hybridization (FISH) and reverse transcription polymerase chain reaction (RT-PCR) to perform genetic analyses of 22 cases of CCSSTs and CCSLGTs, and compared their different chimeric fusion types.
Methods
Case selection
The study protocol for the collection of tumor samples and clinical information were approved by the Institutional Review Board of Sapporo Medical University Hospital (Sapporo, Japan; No. 292–3012). We selected 22 cases of clear cell sarcoma (CCS) including 16 CCSST and 6 CCSLGT cases from the clinicopathological archive at the Department of Surgical Pathology, Sapporo Medical University Hospital. We reviewed all hematoxylin and eosin-stained sections and confirmed that each case fulfilled the histologic criteria of CCSST and CCSLGT.
Immunohistochemistry
We evaluated previously reported IHC findings of melanocytic markers, including S-100 protein, Melan-A, HMB45, and SOX-10, and assessed their positivity. We also performed additional IHC using representative sections from formalin-fixed and paraffin-embedded tissues in some cases. These tissues were sliced into 3-mm-thick sections and examined with an automated IHC system at Sapporo Medical University Hospital. All slides were loaded into a PT Link module (Agilent Technologies, Santa Clara, CA) and subjected to a heat-induced antigen-retrieval protocol with EnVision FLEX Target Retrieval Solution (Agilent Technologies) before being transferred to the Autostainer Link 48 instrument (Agilent Technologies) and Dako Omnis (Agilent Technologies). We used antibodies against the following antigens: S-100 protein (polyclonal; Agilent Technologies), Melan-A (A103; Agilent Technologies), HMB45 (HMB45; Agilent Technologies), and SOX-10 (N-20; Santa Cruz Biotechnology, Santa Cruz, CA).
Fluorescence in situ hybridization
We performed FISH using the specimens obtained from tumor materials and 4 μm slices on glass slides. First, we selected an area showing representative histology and marked a 5-mm-diameter circle with a marker on the glass slides for FISH analyses. We performed FISH using dual color break apart probe for EWSR1 (Abbott, Abbott Park, IL), ATF1 (Empire Genomics, Buffalo, NY), CREB1 (Empire Genomics), and CREM (Empire Genomics). FISH was conducted as previously described [11], with the following modifications: baking (1 h at 60 °C), deparaffinization, target gene activation (20 min with 0.2 M HCl followed by 30 min with pretreatment solution at 80 °C), enzyme treatment (60 min with protease solution at 37 °C), re-fixation (10 min in 10% formalin neutral buffer solution), denaturation (5 min with denaturation solution at 72 °C), washing and dehydration (1 min each in 70%, 85%, and 100% ethanol), hybridization with 10 mL DNA probe solution (5 min at 90 °C followed by 48 h at 37 °C), and washing with post-hybridization wash buffer (2 min at 72 °C). As a counterstain, 10 μL 4,6-diamidino-2-phenylindole was added. Slides were coverslipped for viewing under a fluorescence microscope.
To detect the presence of EWSR1, ATF1, CREB1, and CREM rearrangements, we counted 50 nuclei in tumor cells that showed a pair of fused and split signals, and calculated the percentage of split signals. The signals were considered split when the distance between the red and green signals was at least twice the estimated signal diameter. We did not evaluate any truncated and overlapping cells in FISH analysis. We considered the specimen to be “split positive” if split signals were observed in more than 10% of tumor cells [12].
Reverse transcription-polymerase chain reaction
We detected chimeric fusions by RT-PCR using fresh tumor samples in several available cases. RT-PCR analysis was performed for EWSR1-ATF1 and EWSR1-CREB1 fusions. For RT-PCR detection of the EWSR1-ATF1 and EWSR1-CREB1 fusions, we used the forward primer EWSex7-F1 with either the CREB1ex7-RevA primer (binds both CREB1 and ATF1; sequence: TCCATCAGTGGTCTGTGCATACTG) or the CREB1ex7-RevC primer (specific for CREB1; sequence: GTACCCCATCGGTACCATTGT) [1, 7, 13].
Results
Clinical findings
This study involved 8 male and 14 female patients with a mean age of 40 years (range, 8–78 years). Mean tumor size was 4.6 cm (range, 2–10). The anatomical locations were deep soft tissue of the upper (n = 6) and lower (n = 8) extremities, esophagus (n = 1), small intestine (n = 5), abdominal wall (n = 1), and skin (n = 1). The primary site in Case 13 was the upper extremity, but a specimen was not available, so we used lymph node specimens of metastatic lesions. Mean follow-up duration was 38 months (range, 3–249 months; Table 1).
Table 1.
Summary of clinical, immunohistochemical, and genetic findings of CCSST and CCSLGT cases
| No. | Age (years) /Sex | Location | Tumor size (cm) | Outcome (months) | Immunohistochemistry | Fluorescence in situ hybridization (%) | Expected fusion genes by FISH | RT-PCR findings | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| S-100 protein | Melan-A and/or HMB45 | SOX-10 | EWSR1 | ATF1 | CREB1 | CREM | |||||||
| 1 | 29/F | Leg | 5 | DOD (73) | + | + | NP | 72 | 48 | 2 | NP | EWSR1-ATF1 | NP |
| 2 | 41/M | Leg | 4 | DOD (62) | + | + | NP | 68 | ND | ND | NP | EWSR1- (unknown partner) | NP |
| 3 | 25/M | Leg | 8 | DOD (29) | + | + | NP | 50 | 32 | 4 | NP | EWSR1-ATF1 | EWSR1 exon 8-ATF1 exon 4 |
| 4 | 62/F | Leg | 2 | NED (6) | + | + | NP | 8 | 6 | 0 | NP | Unknown | EWSR1 exon 8-ATF1 exon 4 |
| 5 | 33/M | Leg | 2.5 | DOD (8) | + | + | NP | 16 | 0 | 0 | NP | EWSR1- (unknown partner) | EWSR1 exon 10-ATF1 exon 5 |
| 6 | 34/F | Leg | NA | AWD (16) | + | + | NP | 62 | 32 | 0 | NP | EWSR1-ATF1 | NP |
| 7 | 34/M | Leg | 6 | AWD (9) | + | + | NP | 76 | 66 | 6 | NP | EWSR1-ATF1 | NP |
| 8 | 12/F | Leg | 2 | NED (96) | + | – | – | 36 | 0 | 2 | 2 | EWSR1-(unknown partner) | NP |
| 9 | 39/F | Arm | 3 | DOD (10) | + | + | NP | 74 | 54 | ND | NP | EWSR1-ATF1 | NP |
| 10 | 69/F | Arm | 2 | DOD (26) | + | + | NP | 50 | 58 | 0 | NP | EWSR1-ATF1 | EWSR1 exon 8-ATF1 exon 4 |
| 11 | 41/M | Arm | 2.5 | AWD (11) | + | + | NP | 58 | 58 | 2 | NP | EWSR1-ATF1 | EWSR1 exon 10-ATF1 exon 5 |
| 12 | 40/M | Arm | 5.5 | AWD (23) | + | + | NP | 70 | 36 | 8 | NP | EWSR1-ATF1 | EWSR1 exon 8-ATF1 exon 5 |
| 13 | 49/F | Arm | 4 | AWD (24) | + | + | NP | 70 | 8 | 4 | 36 | EWSR1-CREM | * |
| 14 | 56/F | Arm | 3.9 | AWD (3) | + | + | NP | 80 | 86 | 2 | 0 | EWSR1-ATF1 | NP |
| 15 | 8/F | Abdominal wall | 9.5 | DOD (249) | + | + | NP | 50 | 36 | 0 | NP | EWSR1-ATF1 | NP |
| 16 | 43/M | Skin | 3 | NED (29) | + | – | NP | 84 | 76 | 0 | NP | EWSR1-ATF1 | NP |
| 17 | 41/F | Ileum | 4 | NED (28) | + | – | NP | 72 | 58 | 2 | NP | EWSR1-ATF1 | NP |
| 18 | 78/F | Ileum | 9 | NED (48) | + | – | + | 2 | 2 | 0 | 12 | (unknown partner)-CREM | NP |
| 19 | 38/F | Small intestine | 10 | DOD (17) | + | – | NP | 56 | 6 | 42 | NP | EWSR1-CREB1 | NP |
| 20 | 20/F | Ileum | 4 | NED (7) | + | – | + | 80 | 94 | 4 | NP | EWSR1-ATF1 | EWSR1 exon 8-ATF1 exon 4 |
| 21 | 47/F | Ileum | 4.5 | AWD (34) | + | – | + | 74 | 0 | 62 | NP | EWSR1-CREB1 | EWSR1 exon 7-CREB1 exon 7 |
| 22 | 57/M | Esophagus | 3 | NED (18) | + | – | + | 2 | 2 | 0 | 8 | Unknown | NP |
+ Positive, − Negative, AWD Alive with disease, NED No evidence of disease, DOD Died of disease, NA Not available, ND Not detected, NP Not performed, FISH Fluorescence in situ hybridization, RT-PCR Reverse transcription PCR; *, fusion gene was not detected by RT-PCR in formalin-fixed, paraffin-embedded materials
Histological and IHC findings
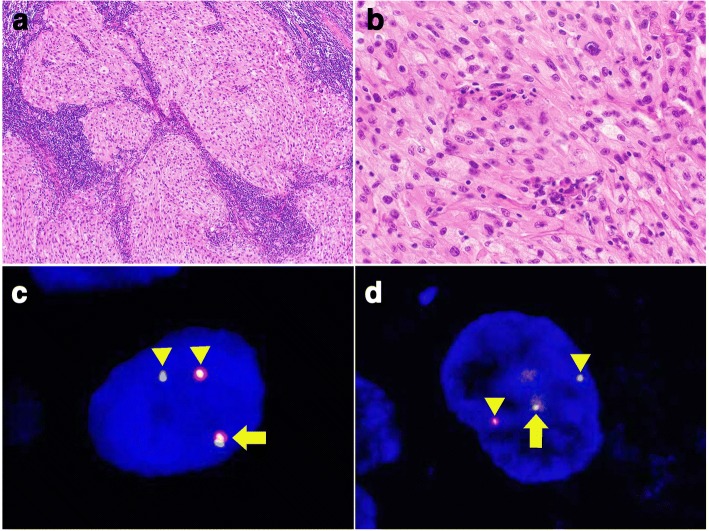
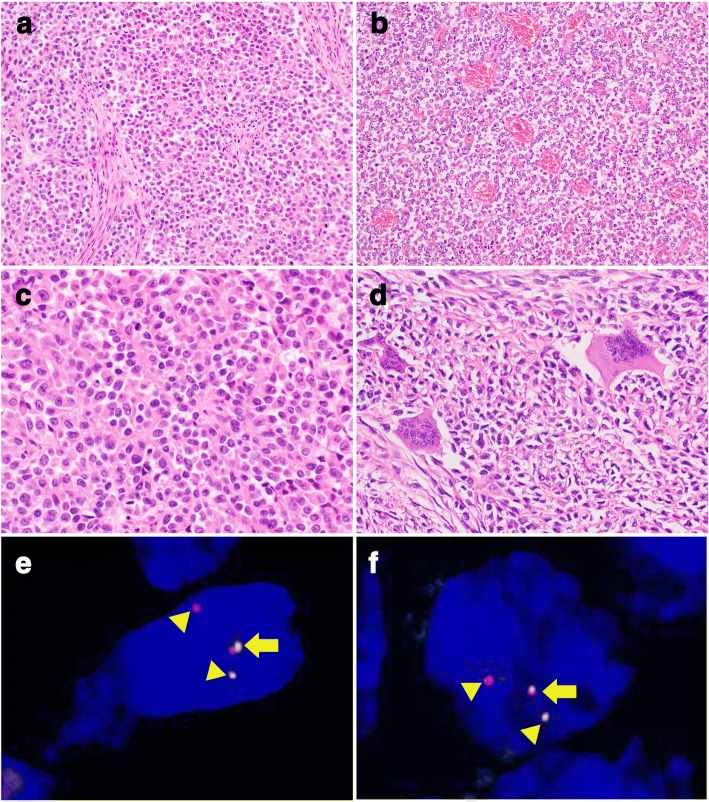
Histologically, the majority of CCSSTs showed sheet-like and nested growth patterns of epithelioid and/or spindle tumor cells that had round to mildly irregularly shaped nuclei with conspicuous nucleoli and clear to pale eosinophilic or amphophilic cytoplasm (Fig. 1a, and b). Among the 16 CCSSTs, 13 cases showed predominant epithelioid cytology and 3 cases exhibited predominant spindle cytology. Multinucleated giant cells were sparsely found in 14 cases. Visible melanocytic differentiation (melanin pigmentation) was observed in 4 cases. The predominant architecture was sheet-like and nested in 12 cases, fascicular in 3 cases, and pseudo-papillary in 1 case. Nucleolar prominence was found in 14 cases. Case 13, which had both EWSR1 and CREM rearrangements (metastatic lesions in lymph nodes), showed sheet-like and fascicular proliferation of epithelioid and spindle tumor cells that had round nuclei with conspicuous nucleoli and abundant pale eosinophilic cytoplasm (Fig. 2a, and b); no myxoid change was found. On the other hand, CCSLGTs often exhibited pseudo-papillary patterns of epithelioid tumor cells, with round to irregular-shaped nuclei showing a coarse chromatin pattern and having a slightly eosinophilic to less clear cytoplasm (Fig. 3a, and b). All 6 CCSLGT cases showed predominant epithelioid cytology and did not exhibit visible melanocytic differentiation. The predominant architecture was pseudo-papillary in 3 cases and sheet-like in 3 cases. Nucleoli were less conspicuous than those from CCSSTs (Fig. 3c), and nucleolar prominence was observed in 2 cases. Osteoclast-type giant cells were scattered in 5 cases (Fig. 3d). Case 18, which had CREM rearrangement, showed pseudo-papillary proliferation of epithelioid cells that had round nuclei without conspicuous nucleoli and pale eosinophilic cytoplasm. There were no histologic patterns indicating any correlation between ATF1 and CREB1 or indicating cases with CREM rearrangement.
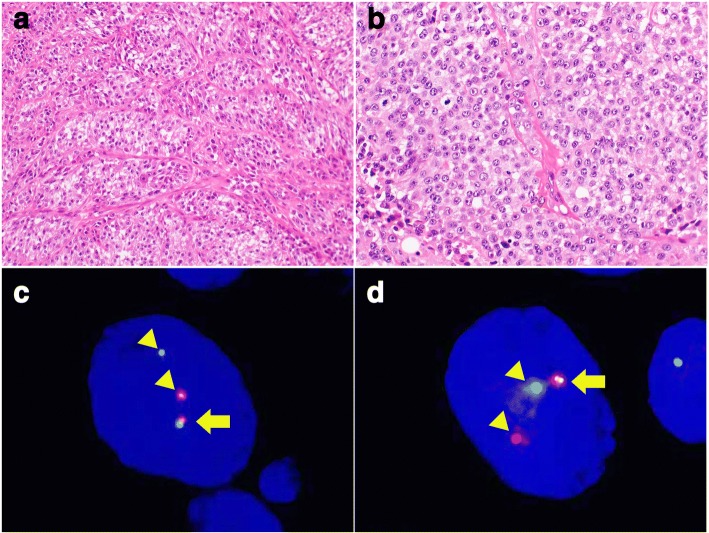
Fig. 1.
Pathological findings of CCSST with EWSR1 and ATF1 rearrangements. a CCSSTs showed sheet-like and nested growth patterns of polyhedral tumor cells (200×). b Tumor cells had round nuclei with conspicuous nucleoli and clear to pale eosinophilic cytoplasm (400×). c FISH of EWSR1 split signals. Tumor cells showed EWSR1 split signal with a pair of fused (arrow) and split (arrow head) patterns (1000×). d FISH of ATF1 split signals. Tumor cells showed EWSR1 split signal with a pair of fused (arrow) and split (arrow head) patterns (1000×)
Fig. 2.
Pathological findings of CCSST with EWSR1 and CREM rearrangements. a Metastatic CCSST of the lymph nodes (100×). b Tumor cells were polyhedral to spindle-shaped and had oval to round nuclei with pale to clear and eosinophilic cytoplasm (400×). c FISH of EWSR1 split signals. Tumor cells showed EWSR1 split signal with a pair of fused (arrow) and split (arrow head) patterns (1000×). d FISH of CREM split signals. Tumor cells showed CREM split signal with a pair of fused (arrow) and split (arrow head) patterns (1000×)
Fig. 3.
Pathological findings of CCSLGT with EWSR1 and CREB1 rearrangements. a CCSLGTs often exhibited sheet-like proliferation of polyhedral and epithelioid tumor cells (200×). b Areas of pseudo-papillary growth pattern around the vasculatures were also observed (200×). c Tumor cells had mildly irregular-shaped round nuclei showing a coarse chromatin pattern and lightly eosinophilic to less frequently clear cytoplasm. Nucleoli were inconspicuous (400×). d Osteoclast-type giant cells were scattered in the tumor (400×). e. FISH of EWSR1 split signals. Tumor cells showed EWSR1 split signal with a pair of fused (arrow) and split (arrow head) patterns (1000×). f. FISH of CREB1 split signals. Tumor cells showed CREB1 split signal with a pair of fused (arrow) and split (arrow head) patterns (1000×)
We performed IHC of melanocytic markers, including S-100 protein, Melan-A, HMB45, and SOX-10, and assessed their positivity (Table 1). In Cases 8 and 16, CCS of the deep soft tissue and skin showed no reactivity with Melan-A and HMB45 and Case 13 with both EWSR1 and CREM rearrangements was negative for Melan-A, but almost all of the CCSST cases were positive for all melanocytic markers. In contrast, CCSLGTs showed no reactivity with any melanocytic markers and were positive for only S-100 and SOX-10. All of the IHC results were compatible with the pathological diagnosis of CCSST and CCSLGT.
Fluorescence in situ hybridization
As shown in Table 1, 15 of the 16 CCSST cases (93.8%) had EWSR1 rearrangement, of which 11 (68.8%) also showed ATF1 rearrangement (Fig. 1c, and d), suggestive of the presence of EWSR1-ATF1 fusion. One CCSST (Case 13) (6.3%) exhibited EWSR1 and CREM rearrangements (Fig. 2c, and d), indicating EWSR1-CREM fusion although no fusion gene was proven by RT-PCR using formalin-fixed, paraffin-embedded materials (data not shown). Two CCSSTs with EWSR1 rearrangement had no partner genes: one (Case 8) showed no rearrangement of ATF1, CREB1, or CREM, and one (Case 2) did not exhibit any rearrangement. Four of 6 CCSLGT cases (66.7%) exhibited EWSR1 rearrangement: 2 (33.3%) showed ATF1 rearrangement and the other 2 (33.3%) showed CREB1 rearrangement (Fig. 3e, and f), suggestive of the presence of EWSR1-ATF1 and EWSR1-CREB1 fusions, respectively. One case (Case 18) was positive for split CREM signals, although no partner genes were detected. One case (Case 22) showed no rearrangement of EWSR1, ATF1, CREB1, or CREM.
Reverse transcription-polymerase chain reaction
RT-PCR was performed in 8 available cases including 6 CCSSTs and 2 CCSLGTs (Table 1). All 6 CCSST cases showed EWSR1-ATF1 fusion between EWSR1 exon 8 and ATF1 exon 4, EWSR1 exon 10 and ATF1 exon 5, or EWSR1 exon 8 and ATF1 exon 5. Although EWSR1-ATF1 fusion was confirmed by RT-PCR in 2 cases (Cases 4 and 5), adequate EWSR1 or ATF1 split signals were not detected by FISH. Among the 2 CCSLGT cases, one had EWSR1-ATF1 fusion between EWSR1 exon 8 and ATF1 exon 4, and the other had EWSR1-CREB1 fusion between EWSR1 exon 7 and CREB1 exon 7.
Discussion
Although CCSST and CCSLGT share similar pathological findings, there are apparent morphological and immunohistochemical differences between the two tumor types. We confirmed the differences in histology and IHC results in our cohort cases. Histologically, the cytological findings of tumor cells and architectural proliferation pattern differed. The tumor cells of CCSST were polyhedral to epithelioid, and spindle-shaped with round to mildly irregular-shaped nuclei and conspicuous nucleoli. In contrast, the tumor cells of CCSLGT had epithelioid tumor cells with irregular-shaped nuclei showing a coarse chromatin pattern and more eosinophilic cytoplasm. Nucleoli were not remarkable in CCSLGT compared to CCSST. CCSST exhibited sheet-like, solid, and nested tumor cell proliferation. In contrast, CCSLGT additionally showed a pseudo-papillary growth pattern. The existence of scattered osteoclast-type giant cells was also characteristic of CCSLGT. CCSSTs were positive for Melan-A and/or HMB45 melanocytic markers in addition to S-100 protein, as determined by IHC. On the other hand, CCSLGTs were not reactive for any melanocytic markers, with the exception of S-100 protein and SOX-10. As in previously reported studies, EWSR1-CREB1 fusion tended to be detected in CCSLGTs. This genetic tendency might reflect the morphological and immunohistochemical differences between the two tumor types.
The novel finding of the study was that we discovered CREM rearrangement in a few CCSs. Kao et al. [14–16] stated that an EWSR1-CREM fusion was previously detected by RNA sequencing in 2 melanoma cell lines (CHL-1 and COLO 699) and proposed that these cell lines may have originated from CCS because of the histological and immunohistochemical overlap between malignant melanoma and CCS. On the other hand, EWSR1-CREM fusion was found in a unique myxoid mesenchymal tumor that was recently described as a new entity [14, 17]. This myxoid tumor is thought to have an intracranial location, and 8 cases have previously been reported, of which 7 occurred in intracranial lesions like meninges, brain tumors, and ventricles, and one case arose in the pelvic/perirectal region. A genetic study revealed EWSR1 fusions with CREB family genes in all of these tumors. Among the 8 tumors, 3 had EWSR1-CREM fusion, 4 had EWSR1-CREB1 fusion, and one tumor showed EWSR1-ATF1 fusion. However, histological and immunohistochemical findings completely differed between this particular myxoid mesenchymal tumor and CCSST/CCSLGT, and interestingly, these genetic results corresponded to those of CCSST and CCSLGT.
CCSLGT was originally reported as an “osteoclast-rich tumor of the gastrointestinal tract with features resembling clear cell sarcoma of the soft parts” [2]. However, some authors prefer to refer to CCSLGT as a “malignant gastrointestinal neuroectodermal tumor” (GNET), because these tumors lack evidence of melanocytic differentiation [5]. A recent review discussed the relationship between clear cell sarcoma of the gastrointestinal tract (CCS-GIT) with GNET. There were differences in morphology and IHC findings between CCS-GIT and GNET. GNET tended to show a wider spectrum of growth patterns, including a pseudo-papillary growth pattern, and exhibited no evidence of melanocytic differentiation. Clinically, CCS-GIT affected males more often than females, and GNET occurred in younger patients although no significant differences existed in their biological behaviors [18]. It has been reported that GNET has poorer prognosis than CCS-GIT [5], but additional studies are needed to clarify the relationship between the two entities.
While a previous study revealed that EWSR1-ATF1 fusion was identified by RT-PCR in 91% of CCSST cases [1], we detected EWSR1-ATF1 fusion in 13 of 16 CCSST (81.3%) both by FISH and RT-PCR. Moreover, after excluding 1 case of EWSR1-CREM fusion, 13 of 15 CCSST cases (86.7%) had EWSR1-ATF1 fusion. The percentage of positive cases in the present study nearly reached that of the previous study. CCSLGT has been identified as having EWSR1-ATF1 or EWSR1-CREB1 fusion, with CCSLGT more frequently having EWSR1-CREB1 fusion. These fusions have also been detected in angiomatoid fibrous histiocytoma and primary pulmonary myxoid sarcoma despite the morphological and immunohistochemical differences between CCSST/CCSLGT and these tumors. To detect specific fusions, FISH is an effective and useful tool using formalin-fixed, paraffin-embedded sections in routine pathological work. In this study, we successfully detected EWSR1, ATF1, CREB1, and CREM rearrangements by FISH in the majority of cases. Some cases showed only one specific rearrangement and did not reveal any rearrangement of partner genes. In such cases, it is expected that certain unknown partner genes can form some novel chimeric genes, and that powerful analytic tools such as next-generation sequencing will be useful for detecting these novel fusions.
Conclusions
Rearrangements of EWSR1 and ATF1 or EWSR1-ATF1 fusion were predominantly found in CCSST, whereas those of EWSR1 and CREB1 or EWSR1-CREB1 tended to be detected in CCSLGT. We detected a novel CREM rearrangement in surgical CCSST specimens by FISH. Although this novel rearrangement occurred in a minority of CCSST cases, further studies are needed to elucidate its pathological significance.
Acknowledgements
The authors thank the following pathologists for kindly contributing case materials and clinical follow-up information: Akiko Tonooka, Department of Pathology, Tokyo Metropolitan Cancer and Infectious Diseases Center Komagome Hospital, Tokyo, Japan; Misa Ishihara and Kimio Hashimoto, Department of Pathology, Nishi-Kobe Medical Center, Hyogo, Japan; Shigeo Hara, Department of Diagnostic Pathology, Kobe City Medical Center General Hospital, Hyogo, Japan; Koki Moriyoshi, Division of Clinical Pathology, National Hospital Organization Kyoto Medical Center, Kyoto, Japan; Shin Ichihara, Department of Surgical Pathology, Sapporo Kosei General Hospital, Hokkaido, Japan; Yukio Morishita, Department of Pathology, Tokyo Medical University Ibaraki Medical Center, Ibaraki, Japan; and Atsushi Uchida, Department of Pathology, Tsukuba Medical Center Hospital, Ibaraki, Japan.
Availability of data and material
All data were presented in this paper, and there are no additional supporting files.
Abbreviations
- CCS
Clear cell sarcoma
- CCSLGT
Clear cell sarcoma-like gastrointestinal tumor
- CCSST
Clear cell sarcoma of soft tissue
- FISH
Fluorescence in situ hybridization
- IHC
Immunohistochemistry
Authors’ contributions
KS participated in the design of the study, performed the pathological analysis, and drafted the manuscript. SS, TH, TS, YI, MT, and HF helped with the pathological analysis. TA and HA conducted the fluorescence in situ hybridization. TK performed genetic analysis using available surgical materials. ME examined the clinical data of cases. SS conceived the study, participated in its design and coordination, and helped draft the manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to participate
All patients, including legal parent or guardian of minor patients, gave general consent for the use of their tissue/data in research as authorized by the Institutional Review Board of Sapporo Medical University Hospital (No. 292-3012).
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Keiko Segawa, Email: k.segawa-1021@sapmed.ac.jp.
Shintaro Sugita, Email: ssugita@sapmed.ac.jp.
Tomoyuki Aoyama, Email: aoyamat@sapmed.ac.jp.
Terufumi Kubo, Email: kuboteru@sapmed.ac.jp.
Hiroko Asanuma, Email: asanuma@sapmed.ac.jp.
Taro Sugawara, Email: t.sugawara@sapmed.ac.jp.
Yumika Ito, Email: ito_yumika@sapmed.ac.jp.
Mitsuhiro Tsujiwaki, Email: tsujiwaki@sapmed.ac.jp.
Hiromi Fujita, Email: hsfujita@sapmed.ac.jp.
Makoto Emori, Email: memori@sapmed.ac.jp.
Tadashi Hasegawa, Email: hasetada@sapmed.ac.jp.
References
- 1.Antonescu CR, Tschernyavsky SJ, Woodruff JM, Jungbluth AA, Brennan MF, Ladanyi M. Molecular diagnosis of clear cell sarcoma: detection of EWS-ATF1 and MITF-M transcripts and histopathological and ultrastructural analysis of 12 cases. J Mol Diagn. 2002;4:44–52. doi: 10.1016/S1525-1578(10)60679-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zambrano E, Reyes-Mugica M, Franchi A, Rosai J. An osteoclast-rich tumor of the gastrointestinal tract with features resembling clear cell sarcoma of soft parts: reports of 6 cases of a GIST simulator. Int J Surg Pathol. 2003;11:75–81. doi: 10.1177/106689690301100202. [DOI] [PubMed] [Google Scholar]
- 3.Alpers CE, Beckstead JH. Malignant neuroendocrine tumor of the jejunum with osteoclast-like giant cells. Enzyme histochemistry distinguishes tumor cells from giant cells. Am J Surg Pathol. 1985;9:57–64. doi: 10.1097/00000478-198501000-00010. [DOI] [PubMed] [Google Scholar]
- 4.Kosemehmetoglu K, Folpe AL. Clear cell sarcoma of tendons and aponeuroses, and osteoclast-rich tumour of the gastrointestinal tract with features resembling clear cell sarcoma of soft parts: a review and update. J Clin Pathol. 2010;63:416–423. doi: 10.1136/jcp.2008.057471. [DOI] [PubMed] [Google Scholar]
- 5.Stockman DL, Miettinen M, Suster S, Spagnolo D, Dominguez-Malagon H, Hornick JL, et al. Malignant gastrointestinal neuroectodermal tumor: clinicopathologic, immunohistochemical, ultrastructural, and molecular analysis of 16 cases with a reappraisal of clear cell sarcoma-like tumors of the gastrointestinal tract. Am J Surg Pathol. 2012;36:857–868. doi: 10.1097/PAS.0b013e31824644ac. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang J, Thway K. Clear cell sarcoma-like tumor of the gastrointestinal tract: an evolving entity. Arch Pathol Lab Med. 2015;139:407–412. doi: 10.5858/arpa.2013-0547-RS. [DOI] [PubMed] [Google Scholar]
- 7.Antonescu CR, Nafa K, Segal NH, Dal Cin P, Ladanyi M. EWS-CREB1: a recurrent variant fusion in clear cell sarcoma-association with gastrointestinal location and absence of melanocytic differentiation. Clin Cancer Res. 2006;12:5356–5362. doi: 10.1158/1078-0432.CCR-05-2811. [DOI] [PubMed] [Google Scholar]
- 8.Wang WL, Mayordomo E, Zhang W, Hernandez VS, Tuvin D, Garcia L, et al. Detection and characterization of EWSR1/ATF1 and EWSR1/CREB1 chimeric transcripts in clear cell sarcoma (melanoma of soft parts) Mod Pathol. 2009;22:1201–1209. doi: 10.1038/modpathol.2009.85. [DOI] [PubMed] [Google Scholar]
- 9.Hantschke M, Mentzel T, Rütten A, Palmedo G, Calonje E, Lazar AJ, et al. Cutaneous clear cell sarcoma: a clinicopathologic, immunohistochemical, and molecular analysis of 12 cases emphasizing its distinction from dermal melanoma. Am J Surg Pathol. 2010;34:216–222. doi: 10.1097/PAS.0b013e3181c7d8b2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Washimi K, Takagi M, Hisaoka M, Kawachi K, Takeyama M, Hiruma T, et al. Clear cell sarcoma-like tumor of the gastrointestinal tract: a clinicopathological review. Pathol Int. 2017;67:534–536. doi: 10.1111/pin.12573. [DOI] [PubMed] [Google Scholar]
- 11.Miura Y, Keira Y, Ogino J, Nakanishi K, Noguchi H, Inoue T, et al. Detection of specific genetic abnormalities by fluorescence in situ hybridization in soft tissue tumors. Pathol Int. 2012;62:16–27. doi: 10.1111/j.1440-1827.2011.02739.x. [DOI] [PubMed] [Google Scholar]
- 12.Sugita S, Asanuma H, Hasegawa T. Diagnostic use of fluorescence in situ hybridization in expert review in a phase 2 study of trabectedin monotherapy in patients with advanced, translocation-related sarcoma. Diagn Pathol. 2016;11:37. doi: 10.1186/s13000-016-0486-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kato T, Ichihara S, Gotoda H, Muraoka S, Kubo T, Sugita S, et al. Imprint cytology of clear cell sarcoma-like tumor of the gastrointestinal tract in the small intestine: a case report. Diagn Cytopathol. 2017;45:1137–1141. doi: 10.1002/dc.23786. [DOI] [PubMed] [Google Scholar]
- 14.Kao YC, Sung YS, Zhang L, Chen CL, Vaiyapuri S, Rosenblum MK, et al. EWSR1 fusions with CREB family transcription factors define a novel Myxoid mesenchymal tumor with predilection for intracranial location. Am J Surg Pathol. 2017;41:482–490. doi: 10.1097/PAS.0000000000000788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Klijn C, Durinck S, Stawiski EW, Haverty PM, Jiang Z, Liu H, et al. A comprehensive transcriptional portrait of human cancer cell lines. Nat Biotechnol. 2015;33:306–312. doi: 10.1038/nbt.3080. [DOI] [PubMed] [Google Scholar]
- 16.Giacomini CP, Sun S, Varma S, Shain AH, Giacomini MM, Balagtas J, et al. Breakpoint analysis of transcriptional and genomic profiles uncovers novel gene fusions spanning multiple human cancer types. PLoS Genet. 2013;9(4):e1003464. doi: 10.1371/journal.pgen.1003464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bale TA, Oviedo A, Kozakewich H, Giannini C, Davineni PK, Ligon K, et al. Intracranial myxoid mesenchymal tumors with EWSR1-CREB family gene fusions: myxoid variant of angiomatoid fibrous histiocytoma or novel entity? Brain Pathol. 2018;28:183–191. doi: 10.1111/bpa.12504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Green C, Spagnolo DV, Robbins PD, Fermoyle S, Wong DD. Clear cell sarcoma of the gastrointestinal tract and malignant gastrointestinal neuroectodermal tumour: distinct or related entities? A review. Pathology. 2018;50:490–498. doi: 10.1016/j.pathol.2018.05.001. [DOI] [PubMed] [Google Scholar]