Abstract
Background
Although miR-221/222 cluster plays an important role in many human malignancies, the correlation between miR-221/222 cluster overexpression and tumor prognosis remains controversial. Therefore, an updated meta-analysis was conducted to clarify its prognostic value in malignancy.
Methods
We conducted a search of literature in English electronic databases of PubMed, Embase, and Cochrane Library, and Chinese electronic databases of China Biology Medicine disc and China National Knowledge Infrastructure to obtain appropriate studies. Besides, we extracted hazard ratios (HRs) and 95% CIs to evaluate the strength of the correlations. In addition, the results of different subgroups analyses and publication bias test were also shown in this article.
Results
32 publications, including 15 tumor types and 2,693 patients were embraced in this meta-analysis. The results of univariate (HR =1.69, 95% CI: 1.18–2.44, P<0.01) and multivariate (HR =2.10, 95% CI: 1.63–2.69, P<0.01) analyses revealed that miR-221/222 cluster high expression in various tumors was significantly associated with adverse overall survival (OS). Correspondingly, we also found subgroups analyses consisted of country, miR-221/222 cluster component, sample size, and test method have similar results.
Conclusion
miR-221/222 cluster overexpression was closely related to adverse OS in human carcinoma, while overexpression of miRNA-221/222 cluster could be viewed as a protection factor in prostate cancer. Blood-derived miR-221/222 cluster was not proper to assess OS.
Keywords: MiRNA-221/222 cluster, cancer, prognosis, meta-analysis
Introduction
miRNAs are an abundant class of endogenous small non-coding RNAs. Since the discovery of miRNAs, emerging studies have demonstrated that miRNAs could serve as oncogenes or tumor suppressors participating in initiation and progression of various cancers.1 It is well documented that miRNAs can inhibit translation and/or induce degradation of target mRNAs, thereby negatively regulated gene expression. The misexpression of miRNAs has a great impact on extensive biological functions consisting of cell differentiation, apoptosis, tumorigenesis, and metabolism.2 Thus, aberrantly expressed miRNAs can be viewed as a new type of biomarkers for monitoring therapeutic efficacy and predicting prognosis.
Two highly homologous miRNAs (miR-221, miR-222), commonly acting as a gene cluster (miR-221/222 cluster), have been extensively studied in human malignancies.3 miR-221/222 cluster overexpression was observed in hepatocellular cancer, pancreatic cancer (PC), breast cancer (BC), etc4–6; while the lowly expressed miR-221/222 cluster was found in tongue squamous cell carcinoma and prostate cancer (PCa).7,8 Several studies have shown that the expression levels of miR-221/222 cluster have close connections with occurrence, progression, and prognosis in several tumors. For instance, a receiver operating characteristic curve analysis conducted by Eissa et al revealed that the higher miR-221 expression, the worse 5-year relapse-free survival (RFS) in patients with BC (P=0.0124)6; a recent study indicated that overall survival (OS) of higher miR-221 expression group was obviously lower than that of the lower miR-221 expression group (P<0.05).9 Whereas, according to the log-rank test, including 125 triple-negative BC (TNBC) patients, Deng et al found that patients with high miR-221 expression have a better 5-year disease-free survival (DFS).10 The aforementioned studies showed that miR-221/222 cluster could be potentially used in predicting the prognostic value of cancer. Meanwhile, the details of over-expressed miR-221/222 cluster’s prognostic value in various cancer types are still controversial.
Meta-analysis is capable to obtain relatively accurate estimation by integrating all available evidence to explore authentic and comprehensive results.11 The only meta-analysis regarding the prognostic value of miRNA-221/222 cluster in cancers was published in 2013 by Wang et al.12 Thus, we carried out an updated meta-analysis with larger sample size, more cancer types, more ethnicities, and different analysis models to verify the previous conclusions and especially uncover some novel findings, which may contribute to the exploration of new promising biomarkers for assessing the therapeutic efficacy and prognostic value of different types of cancers.
Methods
Search strategy
To search eligible studies, we explored literature resources consisting of PubMed, Cochrane Library, Embase, Chinese National Knowledge Infrastructure, and Chinese Biomedical Literature database, with the terms (“microRNA OR miRNA OR miR-221 OR miR-222 OR miR-221/222 cluster OR miR-221/222 family”), (“survival OR prognosis OR prognostic”) and (“cancer OR tumor OR tumor OR neoplasm OR neo-plasma OR neoplasia OR carcinoma OR Malignancy”). The deadline for current investigation was on January 30, 2018. Meanwhile, the language of publication was only limited to English and Chinese. To avoid omissions as much as possible, we checked through the references list in search of potentially relevant researches.
Inclusion and exclusion
Available studies must obey the following criteria: 1) Clinical study about the correlation of miR-221/222 family with human cancer prognosis; 2) Study with relevant data of hazard ratios (HRs) with corresponding 95% CIs. Besides, studies met one of the criteria should be excluded as follows: 1) Studies with deficient data; 2) Duplicate studies (we select the study with relatively complete data); 3) Articles with other types, such as reviews and abstracts; and 4) Cell lines or animal models as research objects.
Data extraction
At the criteria of inclusion and exclusion, the corresponding data of qualified studies were extracted. If we noticed disagreements, a discussion would be conducted by PZ and MZ, or further reviewed by RH. The data, including first author, publication year, research country, test method, cancer type, miRNAs category, sample source, survival outcome, HR (95% CI), sample size, and the cutoff value were extracted to assess the tumor prognosis. Moreover, patient sources came from Asia, Africa, Europe, and North America. In situ hybridization (ISH) and reverse transcription-polymerase chain reaction (RT-PCR) were included in test methods; sample sources were divided into tissue, formalin-fixed and paraffin-embedded (FFPE), and blood; sample sizes included ≥100 and <100 groups; we just divided cancer types into 3 groups of hepatocellular carcinoma (HCC), non-small cell lung cancer (NSCLC), and others due to insufficient data; analyses methods included univariate and multivariate analyses. Patients’ prognostic outcomes consisted of OS, DFS, RFS, and progression-free survival (PFS).
Data analysis
STATA (Version 12.0) and Review Manager (RevMan 5) soft-wares were utilized in this study. HR and corresponding 95% CI were extracted to evaluate the potential prognostic value of miR-221/222 cluster overexpression in human malignancy. Moreover, chi-square-based Q and I2 tests have been applied in this study to assess the heterogeneity. (I2<25% means no heterogeneity, I2>50% means extreme heterogeneity).13 Generally, we used fixed-effects model in studies with no or moderate heterogeneity. When I2>50% or P<0.01 for Q test occurred in studies, we always used random-effects model to avoid obvious heterogeneity.14 Publications were individually deleted to assess the stability of the results and investigated the effect of each study on pooled HR. We evaluated the publication bias and attached the corresponding Begg’s funnel plot. P<0.05 indicated a bias of study.15 Correspondingly, we also carried out similar statistical analysis in different subgroups included country category, sample source, test method, sample size, miR-221/222 component, and cancer type.
Results
Summary of included studies
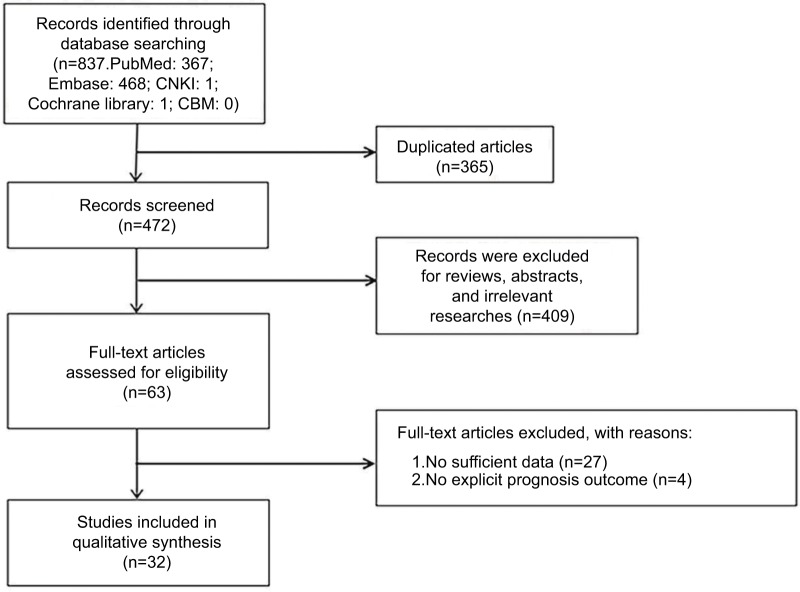
Thirty-two studies consisted of 2,693 samples satisfied the eligible studies4–6,8–10,16–41 (Figure 1). Table 1 listed the main features of those available studies. Among those studies, Amankwah et al’s27 study involved in tumor samples of 37 non-recurrent and 28 recurrent PCa patients after radical prostatectomy aimed to explore whether the expression levels of miR-221 and miR-222 were related to PCa recurrence. Gyöngyösi et al29 analyzed the association between the various miRNA of HCC and survival rate of patients treated with sorafenib after fine-needle aspiration biopsy. Mao et al33 compared 2 different cohorts of regional lymph node involvement and no regional lymph node involvement to explore the prognostic potential of miR-222 expression in NSCLC. Cai et al34 evaluated the prognostic value of miR-221 from 182 colon cancer (CC) patients after surgical resection both in the squamous cell carcinoma and adenocarcinoma cohorts. Finally, we integrated 32 studies independently into meta-analysis and established in Table 1.
Figure 1.
Flow diagram of the study selection process in the meta-analysis.
Abbreviations: CBM, Chinese Biomedical Literature Database; CNKI, Chinese National Knowledge Infrastructure.
Table 1.
The main characteristics and survival data of included 32 studies
| First author | Year | Country | Test method | Cancer type | miRNA | Sample source | Outcome | HR (95% CI) | Sample size | Cutoff value |
|---|---|---|---|---|---|---|---|---|---|---|
| Gramantieri L4 | 2009 | Italy | qRT-PCR | HCC | miR-221 | Tissue | OS/DFS | (U)1.12 (0.81–1.56)/(U)1.56 (1.20–2.03) | 51 | Median |
| Greither T5 | 2010 | Germany | qRT-PCR | PC | miR-222 | Tissue | OS | (M)2.05 (1.05–4.00) | 56 | Median |
| Schaefer A8 | 2010 | Germany | qRT-PCR | PCa | miR-221/miR-222 | Tissue | DFS | (U)0.93 (0.30–2.89)/(U)0.69 (0.22–2.17) | 76 | Median |
| Wong QW16 | 2010 | China | qRT-PCR | HCC | miR-222 | Tissue | OS/DFS | (U)2.73 (1.41–5.30)/(U)2.18 (1.20–3.98) | 76 | Median |
| Wang Y17 | 2010 | China | qRT-PCR | ALL | miR-221 | Tissue | OS | (U)0.54 (0.30–0.97) | 32 | Median |
| Spahn M18 | 2010 | Germany | qRT-PCR | PCa | miR-221 | FFPE | DFS | (M) 0.53 (0.29–0.95) | 92 | Median |
| Pu XX19 | 2010 | China | qRT-PCR | CRC | miR-221 | Blood | OS | (M) 3.48 (1.04–11.65) | 103 | Youden index |
| Guo HQ20 | 2010 | China | qRT-PCR | N/T L | miR-221 | Blood | OS | (U) 0.40 (0.17–0.95)/(M) 0.18 (0.06–0.56) | 79 | Youden index |
| Wang RM21 | 2011 | China | qRT-PCR | GC | miR-221 | Tissue | OS | (U) 5.85 (2.32–14.72) | 96 | Median |
| Li JP9 | 2011 | China | qRT-PCR | HCC | miR-221 | Blood | OS | (M) 1.90 (1.24–2.98) | 46 | Average fold change |
| Yoon SO22 | 2011 | Korea | qRT-PCR | HCC | miR-221 | FFPE | DFS | (M) 3.07 (1.56–6.07) | 115 | Fold change =1 |
| Karakatsanis A23 | 2011 | Greece | qRT-PCR | HCC | miR-221 | FFPE | OS | (M) 1.79 (1.24–2.58) | 60 | Average fold change |
| Liu K24 | 2012 | China | qRT-PCR | GC | miR-221 | Tissue | OS | (M) 2.32 (1.11–4.85) | 92 | Mean |
| Zhang C25 | 2012 | China | ISH | GM | miR-221/miR-222 | Tissue | OS | (U) 2.03 (1.18–3.50)/(U) 2.56 (1.16–5.65) | 50 | Final score =3 |
| Rong M26 | 2013 | China | qRT-PCR | HCC | miR-221 | FFPE | DFS | (U) 1.40 (0.91–2.15) | 48 | Median |
| Amankwah EK27 | 2013 | USA | qRT-PCR | PCa | miR-221 | FFPE | DFS | (U) 1.79 (0.67–4.77) | 65 | Median |
| Kim BH28 | 2013 | Korea | qRT-PCR | GC | miR-221 | FFPE | OS | (M) 1.50 (0.70–2.90) | 91 | Fold change =3 |
| Gyöngyösi B29 | 2014 | Italy | qRT-PCR | HCC | miR-221 | FFPE | OS/PFS | (U) 1.92 (0.61–6.10)/(U) 1.32 (0.47–3.66) | 20 | Median |
| miR-222 | OS/PFS | (U) 2.04 (0.64–6.46)/(U) 1.43 (0.51–3.99) | ||||||||
| Tao K30 | 2014 | China | qRT-PCR | CRC | miR-221 | FFPE | OS | (U) 2.42 (1.31–4.45)/(M) 2.04 (1.10–3.81) | 90 | Median |
| Fu ZC31 | 2014 | China | qRT-PCR | GC | miR-222 | Blood | DFS | (U) 4.49 (2.68–7.49)/(M) 3.41 (1.84–6.16) | 114 | 2.23 |
| Li P32 | 2014 | China | qRT-PCR | CMM | miR-221 | Blood | OS/DFS | (M) 3.19 (1.78–6.78)/(M) 2.12 (1.96–8.55) | 72 | 2.95 |
| Mao KP33 | 2014 | China | qRT-PCR | NSCLC | miR-222 | Tissue | OS | (M) 3.31 (1.97–5.58) | 100 | Median |
| Cai K34 | 2015 | China | qRT-PCR | CC | miR-221 | Tissue | OS | (U) 2.19 (1.11–4.43)/(M) 2.39 (1.21–4.91) | 182 | Median |
| Zhang YH35 | 2015 | China | qRT-PCR | NSCLC | miR-221 | Tissue | OS | (U) 1.87 (1.27–2.76)/(M) 1.87 (1.27–2.77) | 104 | Mean |
| Yang Z36 | 2015 | China | qRT-PCR | OSM | miR-221 | Blood | OS/RFS | (M) 7.66 (1.83–15.92)/(M) 6.82 (1.33–13.69) | 108 | Median |
| Goto Y37 | 2015 | Japan | qRT-PCR | PCa | miR-222 | Tissue | PFS | (U) 0.35 (0.14–0.92)/(M) 0.21 (0.07–0.64) | 92 | NM |
| Eissa S6 | 2015 | Egypt | qRT-PCR | BC | miR-221 | Tissue | RFS | (M) 14.84 (1.80–9.50) | 76 | 1.03 |
| Liao L38 | 2016 | China | qRT-PCR | NSCLC | miR-221/miR-222 | Tissue | PFS | (U) 2.52 (1.07–5.92)/(U) 3.15 (1.30–7.65) | 55 | Median |
| Xie DF39 | 2017 | China | qRT-PCR | HCC | miR-221 | Tissue | OS | (M) 1.74 (1.00–3.77) | 70 | ≥1.795 |
| Liu Y40 | 2017 | China | qRT-PCR | NSCLC | miR-221 | Tissue | OS | (M) 2.58 (1.41–4.85) | 151 | Median |
| Deng L10 | 2017 | China | qRT-PCR | TNBC | miR-221 | Tissue | DFS | (U) 0.49 (0.27–0.88)/(M) 0.48 (0.26–0.88) | 125 | NM |
| Zhao H41 | 2017 | USA | qRT-PCR | GBM | miR-222 | Blood | DFS | (U) 1.71 (1.07–3.63) | 106 | Median |
Abbreviations: TNBC, triple-negative breast cancer; BC, breast cancer; NSCLC, non-small cell lung cancer; HCC, hepatocellular carcinoma; GC, gastric cancer; CRC, colorectal cancer; ALL, acute lymphoblastic leukemia; CMM, cutaneous malignant melanoma; CC, colon cancer; N/T L, NK/T-cell lymphoma; GBM, glioblastoma; GM, glioma; OSM, osteosarcoma; PC, pancreatic cancer; PCa, prostate cancer; NM, not mentioned; qRT-PCR, quantitative reverse transcription-polymerase chain reaction; OS, overall survival; DFS, disease-free survival; ISH, in situ hybridization; PFS, progression-free survival; RFS, relapse-free survival.
There are 31 studies published in English and 1 study written in Chinese in current study. The maximum sample size was 180 and the minimum was 20. Tumor types were as follows: 1 triple-negative BC, 1 osteosarcoma, 1 PC, 4 gastric cancer (GC), 1 NK/T-cell lymphoma, 1 glioma, 4 NSCLC, 1 cutaneous malignant melanoma, 1 acute lymphoblastic leukemia, 2 colorectal cancer, 1 CC, 1 BC, 8 HCC, 1 glioblastoma (GBM), and 4 PCa. Besides, 1 ISH and 31 RT-PCR were applied in this study. Meanwhile, there were 8 FFPE; 7 blood; and 17 tissue. As for the survival outcomes, we split 32 eligible studies into 50 datasets: 27 for OS, 13 for DFS, 6 for PFS, and 4 for RFS. However, the cutoff value of included studies was inconsistent partly (Table 1).
Meta-analysis in OS
Twelve studies were included in univariate analysis to evaluate the prognostic value of overexpression of miR-221/222 cluster in human malignancy. Highly expressed miR-221/222 cluster of tumors was connected with poor OS (HR =1.69, 95% CI: 1.18–2.44, P<0.01) (Figure 2A). In addition, sub-analyses results revealed that there were certain correlations (Table 2). 15 studies were involved in multivariate analysis to conduct an evaluation regarding the prognostic value of miR-221/222 cluster. Meanwhile, tumor-associated miR-221/222 cluster overexpression also connected with poor OS (HR =2.10, 95% CI: 1.63–2.69, P<0.01) (Figure 2B). Similarly, several subgroups have an analogous result (Table 2).
Figure 2.
Forest plot of the association between high expression of miR-221/222 family in various tumors and OS under different types of analysis.
Note: (A) univariate analysis; (B) multivariate analysis.
Abbreviations: HR, hazard ratio; OS, overall survival; SE, standard error.
Table 2.
Stratified analysis of the high expression of miR-221/222 family and overall survival
| Categories | Subgroups | Univariate analyses
|
multivariate analyses
|
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| No of dataset | HR (95% CI) | P-value | I2 | Ph | No of dataset | HR (95% CI) | P-value | I2 | Ph | ||
| All | 12 | 1.26 (1.07–1.47) | 0.004 | 75% | 0.000 | 15 | 2.10 (1.63–2.69) | 0.000 | 56% | 0.004 | |
| Country | China | 9 | 1.28 (1.05–1.56) | 0.02 | 79% | 0.000 | 12 | 2.19 (1.60–3.01) | 0.000 | 64% | 0.001 |
| Others | 3 | 1.09 (0.95–1.24) | 0.21 | 0% | 0.45 | 3 | 1.28 (1.12–1.48) | 0.000 | 0% | 0.82 | |
| Test method | qRT-PCR | 10 | 1.23 (1.02–1.48) | 0.03 | 78% | 0.000 | 15 | 2.10 (1.63–2.69) | 0.000 | 56% | 0.004 |
| ISH | 2 | 1.40 (1.16–1.71) | 0.000 | 0% | 0.64 | / | / | / | / | / | |
| Sample source | FFPE | 3 | 1.42 (1.15–1.76) | 0.001 | 0% | 0.93 | 3 | 1.29 (1.12–1.48) | 0.000 | 0% | 0.82 |
| Tissue | 8 | 1.30 (1.08–1.58) | 0.005 | 78% | 0.000 | 7 | 1.37 (1.24–1.52) | 0.000 | 0% | 0.94 | |
| Blood | 1 | 0.67 (0.46–0.98) | 0.04 | / | / | 5 | 1.35 (0.90–2.03) | 0.15 | 85% | 0.000 | |
| miR221/222 | miR221 | 9 | 1.20 (1.00–1.45) | 0.05 | 79% | 0.000 | 13 | 2.01 (1.52–2.65) | 0.000 | 58% | 0.004 |
| Component | miR222 | 3 | 1.50 (1.23–1.84) | 0.000 | 0% | 0.91 | 2 | 2.73 (1.72–4.33) | 0.000 | 19% | 0.27 |
| Sample size | ≥100 | 2 | 1.33 (1.15–1.55) | 0.000 | 0% | 0.70 | 6 | 2.72 (1.96–3.79) | 0.000 | 35% | 0.18 |
| <100 | 10 | 1.24 (1.02–1.51) | 0.03 | 78% | 0.000 | 9 | 1.27 (1.10–1.47) | 0.001 | 60% | 0.01 | |
| Cancer type | NSCLC | 1 | 1.87 (1.27–2.76) | 0.002 | / | / | 3 | 2.42 (1.69–3.45) | 0.000 | 35% | 0.22 |
| HCC | 4 | 1.71 (1.00–2.90) | 0.05 | 53% | 0.09 | 3 | 1.83 (1.38–2.42) | 0.000 | 0% | 0.97 | |
| others | 7 | 1.64 (0.87–3.10) | 0.13 | 83% | 0.000 | 9 | 2.05 (1.26–3.33) | 0.004 | 71% | 0.000 | |
Note: The bold font indicated P-value of the HR was <0.05.
Abbreviations: NSCLC, non-small cell lung cancer; HCC, hepatocellular carcinoma; qRT-PCR, quantitative reverse transcription-polymerase chain reaction; ISH, in situ hybridization; FFPE, formalin-fixed and paraffin-embedded; HR, hazard ratio; CI, confidence interval; Ph, P-value of heterogeneity test.
Meta-analysis in DFS/PFS/RFS
There were 9, 5, and zero studies about DFS/PFS/RFS involved in univariate analysis. Correspondingly, 5, 1, and 4 studies about DFS/PFS/RFS were collected in multivariate analysis, respectively. Ultimately, we detected no association between highly expressed miR-221/222 cluster with DFS (univariate: HR =1.45, 95% CI: 0.94–2.25, P=0.10; multivariate: HR =1.40, 95% CI: 0.59–3.34, P=0.45) (Figure 3), PFS (univariate: HR =1.41, 95% CI: 0.65–3.05, P=0.38; multivariate: HR =0.21, 95% CI: 0.07–0.63, P=0.006) (Figure S1), and RFS (multivariate: HR =2.18, 95% CI:0.34–13.89, P=0.41) (Figure S2).
Figure 3.
Forest plot of the association between high expression of miR-221/222 family in various tumors and DFS under different types of analysis.
Note: (A) univariate analysis; (B) multivariate analysis.
Abbreviations: DFS, disease-free survival; HR, hazard ratio; SE, standard error.
Sensitivity analysis
We removed each single study respectively to evaluate its specific effect on the mixed HRs, and sensitivity analysis indicated a relatively stable mixed result. Univariate and multivariate analyses of the pooled results (OS and DFS) were performed in Figure 4A, B and Figure S3A, B, but the result of PFS and RFS only in single-variate analysis was not shown.
Figure 4.
One-way sensitivity analysis of high expression of miR-221/222 family in various tumors with OS under different types of analysis.
Note: (A) univariate analysis; (B) multivariate analysis.
Abbreviation: OS, overall survival.
Publication bias
According to Begg’s funnel plot, there was no publication bias of OS (P=0.784>0.05) (Figure 5A), DFS (P=0.322>0.05), and PFS (P=0.624>0.05) among univariate analysis, and no publication bias of OS (P=0.324>0.05) (Figure 5B), DFS (P=0.624>0.05), and RFS (P=0.497>0.05) among multivariate analysis. Similar to that, we also performed evaluations of subgroup of OS. There was no evidence for publication bias in any subgroup. However, the publication bias of PFS or RFS was not evaluated due to the fewer or no datasets for meta-analysis.
Figure 5.
Funnel plot analysis of publication bias of high expression of miR-221/222 family in various tumors with OS under different types of analysis.
Notes: (A) univariate analysis; (B) multivariate analysis.
Abbreviations: HR, hazard ratio; OS, overall survival.
Discussion
Currently, exploring the clinically available cancer signatures is still the hotspot of researches due to the complexity of tumor. Dysregulations of distinct miRNA fingerprints often occur in specific types of human malignant tumors and obviously associated with cancer diagnosis, therapy, and even prognosis.42,43 Numerous studies have demonstrated that miR-221/222 cluster plays a significantly regulatory role in the prognosis of several types of tumors. For instance, miR-222 overexpression promoted HCC cell migratory through activating AKT phosphorylation assisted by the regulation of protein phosphatase 2A subunit B, contributing to both the development and poor prognosis of HCC.16 Besides, it is reported that miR-221 negatively regulates poly ADP-Ribose polymerase 1 expression levels by 3′-untranslated region binding, thereby affecting TNBC patients’ prognoses.10 Meanwhile, overexpression of miRNA-221 enhanced cell proliferation via downregulating the target gene apoptotic protease activating factor-1 (APAF-1), while APAF-1 expression was associated with a poor prognosis.44 It is reported that the upregulation of miR-221/222 cluster in GBM may be a key factor in the decrease of p27Kip1 expression levels and as such, will correlate with adverse prognosis.45 Similarly, miRNA-222 could target HIPK2 to promote GC cell proliferation, invasion, and inhibited apoptosis, revealing a poor survival outcome for GC patients.46 Moreover, the expression of miR-221 exhibited modulatory effect on BIM-Bax/Bak axis, upregulated target gene BIM, induced by miR-221 knockdown, can promote cisplatin-eliciting apoptosis through inducing mitochondrial dysfunction, revealing a poor survival outcome for BC patients.47 Hence, we proposed that activation or inhibition of multiple pathways might play synergistic roles in the impact of miRNA-221/222 cluster expression on prognosis. However, the clinically prognostic value of dysregulated miR-221/222 expression often affected by small sample-size studies with insufficient data and thus inconsistent and unconvincing.
Meta-analysis is a useful tool, which can get a relatively precise result and provide convincing evidence through integrating and assessing inconsistent outcomes from different studies. We have explored the potential associations in overall population and the corresponding subgroups via combining univariate and multivariate analyses. Likewise, several subgroups have similar statistical results. However, no association of miR-221/222 family was detected with DFS/PFS/RFS.
Generally, we only explored the potential associations in corresponding subgroups of OS due to the sufficient data. When stratified by sample source, given the result in multivariate analysis, blood miR-221/222 expression was unlikely as a promising marker on tumor prognosis. Meanwhile, the result was also not convincing in univariate analysis based on only one study. However, the miR-221/222 expression in FFPE or tissue might be associated with poor OS of carcinoma. When stratified by the sample size, we discovered that high expression of miR-221/222 cluster connected with unfavorable OS significantly in both groups with different sample sizes. When stratified by miRNA, the result indicated that the over expression of miR-221 and miR-222 both can predict poor OS apparently in multivariate analysis. However, we proposed overexpression miRNA-221/222 cluster could be viewed as a protection factor in PCa after integrating 4 studies on PCa48 (Figure S4). It is supposed that overexpression miRNA-221/222 cluster could repress PCa cell invasion37 and decrease prostate-specific antigen expression mediated by androgen receptor,49 which may participate in the protection process. Besides, lack of sufficient data indeed has a certain impact on the intriguing result. Future studies from multicenter comprising larger cohort size are needed to verify the current findings.
Actually, there were some limitations in our study that we should notice in the current meta-analysis. First, as for OS, part of the outcome failed to be analyzed separately due to the small sample size and no publication bias could be assessed in the current meta-analysis. In consideration of the limitations of qualified studies quantities, we did not process subgroup analysis for DFS/PFS/RFS. But the relative DFS/PFS/RFS data presented in this study may improve our understanding of cancer progression and relapse. Moreover, the design of studies, cutoff values, and measure methods being distinct in different studies, we could not consolidate those inconsistent objective elements, which were influential factors to the results. In addition, we did not compare the difference of ethnicities because of the limited number of different countries or races.
Conclusion
A significant correlation was explored in overall population and corresponding subgroups. Concretely, it presented that miR-221/222 family overexpression was significantly linked with poor OS, while no relationship was found between higher miR-221/222 family expression and DFS/PFS/RFS. Besides, sample sizes had no effect on our results, majority of subgroup analysis result was consistent with overall conclusion. Besides, miRNA-221/222 cluster from tissue or FFPE was more convincing in prediction of OS of tumor patients compared with blood-derived miRNA-221/222 cluster. Moreover, miRNA-221/222 cluster overexpression could be viewed as a protection factor in PCa to some extent. The elucidation of deregulated miR-221/222 cluster is expected to improve the understanding on tumor and promote the further development of biomarkers in cancer prognosis.
Supplementary materials
Forest plot of the association between high expression of miR-221/222 family in various tumors and PFS under different types of analysis. (A: univariate analysis; B: multivariate analysis).
Abbreviations: HR, hazard ratio; PFS, progression-free survival; SE, standard error.
Forest plot of the association between high expression of miR-221/222 family in various tumors and RFS under multivariate analysis.
Abbreviations: HR, hazard ratio; RFS, relapse free survival; SE, standard error.
One-way sensitivity analysis of high expression of miR-221/222 family in various tumors with DFS under different types of analysis.
Note: (A) univariate analysis; (B) multivariate analysis.
Abbreviation: DFS, disease-free survival.
Forest plot of the association between high expression of miR-221/222 family in prostate cancer and DFS/RFS.
Abbreviations: HR, hazard ratio; DFS, disease-free survival; RFS, relapse free survival; SE, standard error.
References
- 1.Goto Y, Kojima S, Nishikawa R, et al. microRNA expression signature of castration-resistant prostate cancer: the microRNA-221/222 cluster functions as a tumour suppressor and disease progression marker. Br J Cancer. 2015;113(7):1055–1065. doi: 10.1038/bjc.2015.300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gyöngyösi B, Végh É, Járay B, et al. Pretreatment microRNA Level and Outcome in Sorafenib-treated Hepatocellular Carcinoma. J Histochem Cytochem. 2014;62(8):547–555. doi: 10.1369/0022155414537277. [DOI] [PubMed] [Google Scholar]
- 3.Liao L, Wang J, Feng S. The expression of MIR-221 and MIR-222 in non-small cell lung cancer and their significances. Cancer Res Clinic. 2016;28:590–594. [Google Scholar]
- 4.Amankwah EK, Anegbe E, Park H, Pow-Sang J, Hakam A, Park JY. miR-21, miR-221 and miR-222 expression and prostate cancer recurrence among obese and non-obese cases. Asian J Androl. 2013;15(2):226–230. doi: 10.1038/aja.2012.160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Eissa S, Matboli M, Sharawy A, El-Sharkawi F. Prognostic and biological significance of microRNA-221 in breast cancer. Gene. 2015;574(1):163–167. doi: 10.1016/j.gene.2015.08.004. [DOI] [PubMed] [Google Scholar]
- 6.Yang Z, Zhang Y, Zhang X, et al. Serum microRNA-221 functions as a potential diagnostic and prognostic marker for patients with osteosarcoma. Biomed Pharmacother. 2015;75:153–158. doi: 10.1016/j.biopha.2015.07.018. [DOI] [PubMed] [Google Scholar]
- 7.Deng L, Lei Q, Wang Y, et al. Downregulation of miR-221-3p and upregulation of its target gene PARP1 are prognostic biomarkers for triple negative breast cancer patients and associated with poor prognosis. Oncotarget. 2017;8(65):108712–108725. doi: 10.18632/oncotarget.21561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fu Z, Qian F, Yang X, Jiang H, Chen Y, Liu S. Circulating miR-222 in plasma and its potential diagnostic and prognostic value in gastric cancer. Med Oncol. 2014;31(9):164. doi: 10.1007/s12032-014-0164-8. [DOI] [PubMed] [Google Scholar]
- 9.Gramantieri L, Fornari F, Ferracin M, et al. microRNA-221 targets Bmf in hepatocellular carcinoma and correlates with tumor multifocality. Clin Cancer Res. 2009;15(16):5073–5081. doi: 10.1158/1078-0432.CCR-09-0092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rong M, Chen G, Dang Y. Increased miR-221 expression in hepatocellular carcinoma tissues and its role in enhancing cell growth and inhibiting apoptosis in vitro. BMC Cancer. 2013;13:21. doi: 10.1186/1471-2407-13-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schaefer A, Jung M, Mollenkopf HJ, et al. Diagnostic and prognostic implications of microRNA profiling in prostate carcinoma. Int J Cancer. 2010;126(5):1166–1176. doi: 10.1002/ijc.24827. [DOI] [PubMed] [Google Scholar]
- 12.Wong QW, Ching AK, Chan AW, et al. miR-222 overexpression confers cell migratory advantages in hepatocellular carcinoma through enhancing AKT signaling. Clin Cancer Res. 2010;16(3):867–875. doi: 10.1158/1078-0432.CCR-09-1840. [DOI] [PubMed] [Google Scholar]
- 13.Zhao H, Shen J, Hodges TR, Song R, Fuller GN, Heimberger AB. Serum microRNA profiling in patients with glioblastoma: a survival analysis. Mol Cancer. 2017;16(1):59. doi: 10.1186/s12943-017-0628-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li P, He QY, Luo CQ, Qian LY. Circulating miR-221 expression level and prognosis of cutaneous malignant melanoma. Med Sci Monit. 2014;20:2472–2477. doi: 10.12659/MSM.891327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Spahn M, Kneitz S, Scholz CJ, et al. Expression of microRNA-221 is progressively reduced in aggressive prostate cancer and metastasis and predicts clinical recurrence. Int J Cancer. 2010;127(2):394–403. doi: 10.1002/ijc.24715. [DOI] [PubMed] [Google Scholar]
- 16.Yoon SO, Chun SM, Han EH, et al. Deregulated expression of microRNA-221 with the potential for prognostic biomarkers in surgically resected hepatocellular carcinoma. Hum Pathol. 2011;42(10):1391–1400. doi: 10.1016/j.humpath.2010.12.010. [DOI] [PubMed] [Google Scholar]
Acknowledgments
This work was supported by the National Natural Science Foundation of China (Grant nos. 81401518, 81630019), Anhui Provincial Institutes for Translational Medicine (Grant no. 2017ZHYX02), and Cultivation Project of Young Top-Notch Talent Support from Anhui Medical University (AHMU). Received Funding for Distinguished Young Scientists of the First Affiliated Hospital of AHMU.
Footnotes
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Esquela-Kerscher A, Slack FJ. Oncomirs – microRNAs with a role in cancer. Nat Rev Cancer. 2006;6(4):259–269. doi: 10.1038/nrc1840. [DOI] [PubMed] [Google Scholar]
- 2.Kong YW, Ferland-McCollough D, Jackson TJ, Bushell M. microRNAs in cancer management. Lancet Oncol. 2012;13(6):e249–258. doi: 10.1016/S1470-2045(12)70073-6. [DOI] [PubMed] [Google Scholar]
- 3.Sun T, Yang M, Kantoff P, Lee GS. Role of microRNA-221/-222 in cancer development and progression. Cell Cycle. 2009;8(15):2315–2316. doi: 10.4161/cc.8.15.9221. [DOI] [PubMed] [Google Scholar]
- 4.Gramantieri L, Fornari F, Ferracin M, et al. microRNA-221 targets Bmf in hepatocellular carcinoma and correlates with tumor multifocality. Clin Cancer Res. 2009;15(16):5073–5081. doi: 10.1158/1078-0432.CCR-09-0092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Greither T, Grochola LF, Udelnow A, Lautenschläger C, Würl P, Taubert H. Elevated expression of microRNAs 155, 203, 210 and 222 in pancreatic tumors is associated with poorer survival. Int J Cancer. 2010;126(1):73–80. doi: 10.1002/ijc.24687. [DOI] [PubMed] [Google Scholar]
- 6.Eissa S, Matboli M, Sharawy A, El-Sharkawi F. Prognostic and biological significance of microRNA-221 in breast cancer. Gene. 2015;574(1):163–167. doi: 10.1016/j.gene.2015.08.004. [DOI] [PubMed] [Google Scholar]
- 7.Liu X, Yu J, Jiang L. microRNA-222 regulates cell invasion by targeting matrix metalloproteinase 1 (MMP1) and manganese superoxide dismutase 2 (SOD2) in tongue squamous cell carcinoma cell lines. Cancer Genom Proteom. 2009;6:131–139. [PMC free article] [PubMed] [Google Scholar]
- 8.Schaefer A, Jung M, Mollenkopf HJ, et al. Diagnostic and prognostic implications of microRNA profiling in prostate carcinoma. Int J Cancer. 2010;126(5):1166–1176. doi: 10.1002/ijc.24827. [DOI] [PubMed] [Google Scholar]
- 9.Li J, Wang Y, Yu W, Chen J, Luo J. Expression of serum miR-221 in human hepatocellular carcinoma and its prognostic significance. Biochem Biophys Res Commun. 2011;406(1):70–73. doi: 10.1016/j.bbrc.2011.01.111. [DOI] [PubMed] [Google Scholar]
- 10.Deng L, Lei Q, Wang Y, et al. Downregulation of miR-221-3p and upregulation of its target gene PARP1 are prognostic biomarkers for triple negative breast cancer patients and associated with poor prognosis. Oncotarget. 2017;8(65):108712–108725. doi: 10.18632/oncotarget.21561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Munafò MR, Flint J. Meta-analysis of genetic association studies. Trends Genet. 2004;20(9):439–444. doi: 10.1016/j.tig.2004.06.014. [DOI] [PubMed] [Google Scholar]
- 12.Wang J, Liu S, Sun GP, et al. Prognostic significance of microRNA-221/222 expression in cancers: evidence from 1,204 subjects. Int J Biol Markers. 2014;29(2):129–141. doi: 10.5301/jbm.5000058. [DOI] [PubMed] [Google Scholar]
- 13.Higgins JP, Thompson SG. Quantifying heterogeneity in a meta-analysis. Stat Med. 2002;21(11):1539–1558. doi: 10.1002/sim.1186. [DOI] [PubMed] [Google Scholar]
- 14.Dersimonian R, Laird N. Meta-analysis in clinical trials. Control Clin Trials. 1986;7(3):177–188. doi: 10.1016/0197-2456(86)90046-2. [DOI] [PubMed] [Google Scholar]
- 15.Begg CB, Berlin JA. Publication bias and dissemination of clinical research. J Natl Cancer Inst. 1989;81(2):107–115. doi: 10.1093/jnci/81.2.107. [DOI] [PubMed] [Google Scholar]
- 16.Wong QW, Ching AK, Chan AW, et al. miR-222 overexpression confers cell migratory advantages in hepatocellular carcinoma through enhancing AKT signaling. Clin Cancer Res. 2010;16(3):867–875. doi: 10.1158/1078-0432.CCR-09-1840. [DOI] [PubMed] [Google Scholar]
- 17.Wang Y, Li Z, He C, et al. microRNAs expression signatures are associated with lineage and survival in acute leukemias. Blood Cells Mol Dis. 2010;44(3):191–197. doi: 10.1016/j.bcmd.2009.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Spahn M, Kneitz S, Scholz CJ, et al. Expression of microRNA-221 is progressively reduced in aggressive prostate cancer and metastasis and predicts clinical recurrence. Int J Cancer. 2010;127(2):394–403. doi: 10.1002/ijc.24715. [DOI] [PubMed] [Google Scholar]
- 19.Pu XX, Huang GL, Guo HQ, et al. Circulating miR-221 directly amplified from plasma is a potential diagnostic and prognostic marker of colorectal cancer and is correlated with p53 expression. J Gastroenterol Hepatol. 2010;25(10):1674–1680. doi: 10.1111/j.1440-1746.2010.06417.x. [DOI] [PubMed] [Google Scholar]
- 20.Guo HQ, Huang GL, Guo CC, Pu XX, Lin TY. Diagnostic and prognostic value of circulating miR-221 for extranodal natural killer/T-cell lymphoma. Dis Markers. 2010;29(5):251–258. doi: 10.3233/DMA-2010-0755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang RM, Wang S, Zhang WB. Expression of microRNA-221 in gastric cancer and its clinical significance. Prac J Med. 2011;27:4424–4426. [Google Scholar]
- 22.Yoon SO, Chun SM, Han EH, et al. Deregulated expression of microRNA-221 with the potential for prognostic biomarkers in surgically resected hepatocellular carcinoma. Hum Pathol. 2011;42(10):1391–1400. doi: 10.1016/j.humpath.2010.12.010. [DOI] [PubMed] [Google Scholar]
- 23.Karakatsanis A, Papaconstantinou I, Gazouli M, Lyberopoulou A, Polyme-neas G, Voros D. Expression of microRNAs, miR-21, miR-31, miR-122, miR-145, miR-146a, miR-200c, miR-221, miR-222, and miR-223 in patients with hepatocellular carcinoma or intrahepatic cholangiocarcinoma and its prognostic significance. Mol Carcinog. 2013;52(4):297–303. doi: 10.1002/mc.21864. [DOI] [PubMed] [Google Scholar]
- 24.Liu K, Li G, Fan C, Diao Y, Wu B, Li J. Increased Expression of microRNA-221 in gastric cancer and its clinical significance. J Int Med Res. 2012;40(2):467–474. doi: 10.1177/147323001204000208. [DOI] [PubMed] [Google Scholar]
- 25.Zhang C, Zhang J, Hao J, et al. High level of miR-221/222 confers increased cell invasion and poor prognosis in glioma. J Transl Med. 2012;10:119. doi: 10.1186/1479-5876-10-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rong M, Chen G, Dang Y. Increased miR-221 expression in hepato-cellular carcinoma tissues and its role in enhancing cell growth and inhibiting apoptosis in vitro. BMC Cancer. 2013;13:21. doi: 10.1186/1471-2407-13-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Amankwah EK, Anegbe E, Park H, Pow-Sang J, Hakam A, Park JY. miR-21, miR-221 and miR-222 expression and prostate cancer recurrence among obese and non-obese cases. Asian J Androl. 2013;15(2):226–230. doi: 10.1038/aja.2012.160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim BH, Hong SW, Kim A, Choi SH, Yoon SO. Prognostic implications for high expression of oncogenic microRNAs in advanced gastric carcinoma. J Surg Oncol. 2013;107(5):505–510. doi: 10.1002/jso.23271. [DOI] [PubMed] [Google Scholar]
- 29.Gyöngyösi B, Végh É, Járay B, et al. Pretreatment microRNA Level and Outcome in Sorafenib-treated Hepatocellular Carcinoma. J Histochem Cytochem. 2014;62(8):547–555. doi: 10.1369/0022155414537277. [DOI] [PubMed] [Google Scholar]
- 30.Tao K, Yang J, Guo Z, et al. Prognostic value of miR-221-3p, miR-342-3p and miR-491-5p expression in colon cancer. Am J Transl Res. 2014;6(4):391. [PMC free article] [PubMed] [Google Scholar]
- 31.Fu Z, Qian F, Yang X, Jiang H, Chen Y, Liu S. Circulating miR-222 in plasma and its potential diagnostic and prognostic value in gastric cancer. Med Oncol. 2014;31(9):164. doi: 10.1007/s12032-014-0164-8. [DOI] [PubMed] [Google Scholar]
- 32.Li P, He QY, Luo CQ, Qian LY. Circulating miR-221 expression level and prognosis of cutaneous malignant melanoma. Med Sci Monit. 2014;20:2472–2477. doi: 10.12659/MSM.891327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mao KP, Zhang WN, Liang XM, Ma YR. microRNA-222 expression and its prognostic potential in non-small cell lung cancer. Scientific-WorldJournal. 2014;2014:908326. doi: 10.1155/2014/908326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cai K, Shen F, Cui JH, Yu Y, Pan HQ. Expression of miR-221 in colon cancer correlates with prognosis. Int J Clin Exp Med. 2015;8(2):2794–2798. [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang Y, Zhao Y, Sun S, Liu Z, Zhang Y, Jiao S. Overexpression of microRNA-221 is associated with poor prognosis in non-small cell lung cancer patients. Tumour Biol. 2016;37(8):10155–10160. doi: 10.1007/s13277-015-4662-x. [DOI] [PubMed] [Google Scholar]
- 36.Yang Z, Zhang Y, Zhang X, et al. Serum microRNA-221 functions as a potential diagnostic and prognostic marker for patients with osteosarcoma. Biomed Pharmacother. 2015;75:153–158. doi: 10.1016/j.biopha.2015.07.018. [DOI] [PubMed] [Google Scholar]
- 37.Goto Y, Kojima S, Nishikawa R, et al. microRNA expression signature of castration-resistant prostate cancer: the microRNA-221/222 cluster functions as a tumour suppressor and disease progression marker. Br J Cancer. 2015;113(7):1055–1065. doi: 10.1038/bjc.2015.300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liao L, Wang J, Feng S. The expression of MIR-221 and MIR-222 in non-small cell lung cancer and their significances. Cancer Res. Clinic. 2016;28:590–594. [Google Scholar]
- 39.Xie D, Yuan P, Wang D, Jin H, Chen H. Expression and prognostic significance of miR-375 and miR-221 in liver cancer. Oncol Lett. 2017;14(2):2305–2309. doi: 10.3892/ol.2017.6423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu Y, Wang J. High miRNA-221 expression is associated with poor overall survival in patients with non-small cell lung cancer. Biomed Res. 2017;28:2975–2980. [Google Scholar]
- 41.Zhao H, Shen J, Hodges TR, Song R, Fuller GN, Heimberger AB. Serum microRNA profiling in patients with glioblastoma: a survival analysis. Mol Cancer. 2017;16(1):59. doi: 10.1186/s12943-017-0628-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tomasetti M, Lee W, Santarelli L, Neuzil J. Exosome-derived microR-NAs in cancer metabolism: possible implications in cancer diagnostics and therapy. Exp Mol Med. 2017;49(1):e285. doi: 10.1038/emm.2016.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dong Y, Yu J, Ng SS. microRNA dysregulation as a prognostic biomarker in colorectal cancer. Cancer Manag Res. 2014;6:405–422. doi: 10.2147/CMAR.S35164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li J, Li Q, Huang H, et al. Overexpression of miRNA-221 promotes cell proliferation by targeting the apoptotic protease activating factor-1 and indicates a poor prognosis in ovarian cancer. Int J Oncol. 2017;50:1087–1096. doi: 10.3892/ijo.2017.3898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Le Sage C, Nagel R, Egan DA, et al. Regulation of the p27(Kip1) tumor suppressor by miR-221 and miR-222 promotes cancer cell proliferation. Embo J. 2007;26(15):3699–3708. doi: 10.1038/sj.emboj.7601790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tan X, Tang H, Bi J. microRNA-222-3p associated with Helicobacter pylori targets HIPK2 to promote cell proliferation, invasion and inhibits apoptosis in gastric cancer. J Cell Biochem. 2017 doi: 10.1002/jcb.26542. [DOI] [PubMed] [Google Scholar]
- 47.Ye Z, Hao R, Cai Y, Wang X, Huang G. Knockdown of miR-221 promotes the cisplatin-inducing apoptosis by targeting the BIM-Bax/Bak axis in breast cancer. Tumour Biol. 2016;37(4):4509–4515. doi: 10.1007/s13277-015-4267-4. [DOI] [PubMed] [Google Scholar]
- 48.Liu K, Zhao K, Wang L, Sun E. Prognostic value of microRNA-155 in human carcinomas: an updated meta-analysis. Clin Chim Acta. 2018;479:171–180. doi: 10.1016/j.cca.2018.01.036. [DOI] [PubMed] [Google Scholar]
- 49.Sun T, Wang Q, Balk S, Brown M, Lee GS, Kantoff P. The role of microRNA-221 and microRNA-222 in androgen-independent prostate cancer cell lines. Cancer Res. 2009;69(8):3356–3363. doi: 10.1158/0008-5472.CAN-08-4112. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Forest plot of the association between high expression of miR-221/222 family in various tumors and PFS under different types of analysis. (A: univariate analysis; B: multivariate analysis).
Abbreviations: HR, hazard ratio; PFS, progression-free survival; SE, standard error.
Forest plot of the association between high expression of miR-221/222 family in various tumors and RFS under multivariate analysis.
Abbreviations: HR, hazard ratio; RFS, relapse free survival; SE, standard error.
One-way sensitivity analysis of high expression of miR-221/222 family in various tumors with DFS under different types of analysis.
Note: (A) univariate analysis; (B) multivariate analysis.
Abbreviation: DFS, disease-free survival.
Forest plot of the association between high expression of miR-221/222 family in prostate cancer and DFS/RFS.
Abbreviations: HR, hazard ratio; DFS, disease-free survival; RFS, relapse free survival; SE, standard error.