Abstract
Background
Lung cancer is the leading cause of cancer-related death worldwide and it is critical to discover specific biomarkers to provide better individualized treatment and subsequently better prognosis. The sirtuins (SIRT1–7) have been reported to be involved in cancers including non-small cell lung cancer (NCSLC), however, the results are not consistent and not all the seven sirtuins are explored and compared.
Methods
TCGA data was downloaded and used to investigate and compare the associations of sirtuins mRNA levels with clinicopathological parameters and prognosis in NSCLC.
Results
Our results suggested SIRT1, SIRT3, SIRT4, and SIRT7 were highly expressed in adeno-carcinoma (ADC) patients and female patients while SIRT5 were highly expressed in squamous cell carcinoma (SCC) patients and male patients. Associations of high SIRT7 with younger onset age, high SIRT1 with distant metastasis and low T stage, and high SIRT4 with high T stage and TNM stage were also found. Kaplan-Meier plot curves and univariate Cox proportional regression analyses indicated that high SIRT2, SIRT4, and SIRT6 expressions were associated with longer overall survival (OS) time. Multivariate analyses indicated that SIRT2 and SIRT6 were still associated with OS. For recurrence-free survival (RFS), high SIRT1 expression was significantly associated with shorter RFS time while high SIRT2-3 and SIRT5-7 expressions were associated with longer RFS time in univariate analyses. After adjusting the confounding factors, significant associations were still found in SIRT1-2 and SIRT5-7 but not in SIRT3. We also stratified the patients by combining SIRT1 and SIRT2 and revealed that the combination of SIRT1 and SIRT2 was a better prediction model for RFS in NSCLC. To preliminarily understand the potential mechanisms of sirtuins in NSCLC carcinogenesis, the genes co-expressed with sirtuins were analyzed and annotated.
Conclusion
sirtuins might be the potential therapy targets and prognostic biomarkers in NSCLC.
Keywords: sirtuins, NSCLC, clinical features, overall survival, recurrence-free survival
Introduction
Lung cancer is the leading cause of cancer-related death worldwide with a low 5-year survival rate of 18%.1 Small cell lung cancer (SCLC) and non–small cell lung cancer (NSCLC) are the two major subtypes of lung cancer.2 NSCLC accounts for ~85% of all lung cancers.2 Surgical resection is an effective approach to achieve long-term survival; however, the prognosis of NSCLC remains poor due to late diagnosis and postoperative relapse.3 Thus, it is critical to discover specific biomarkers to provide better individualized treatment and better prognosis.
In the past decades, a larger number of cellular pathways such as the EGFR signaling pathway have been identified in the carcinogenesis of a series of malignancies including lung cancer and are used as targets in drug development and predictive factors of prognosis.4,5 This encourages researchers to continue to identify novel therapy targets and prognostic biomarkers.
The sirtuins are a family of mammalian proteins that are homologous to the yeast silent information regulator 2 (Sir2).6,7 Seven different sirtuins (SIRT1–7) have been found to be widely expressed in mammals and exert nicotinamide adenine dinucleotide (NAD+)-dependent protein deacylase and/or ADP ribosyltransferase activities.8–16 In addition to their deacylase activities, recent studies have suggested SIRT5 can mediate protein desuccinylation, demalonylation, and deglutarylation,17–20 and SIRT7 can mediate histone desuccinylation.21 The sirtuins have been linked to the modulation of numerous critical biological processes including cell metabolism, genomic stability, cell cycle, cell division, differentiation, and transcriptional regulation and are also correlated with the pathogenesis of a variety of diseases such as metabolic diseases,22 neurode-generative diseases,23 aging,24 and cardiovascular diseases.25 Currently, accumulating evidence has shown that the aberrant epigenetic activation of sirtuin signaling pathways contributes to tumor carcinogenesis and may be potential therapeutic targets for future treatments and biomarkers in predicting prognosis in cancers.26,27 Furthermore, some previous studies have been conducted to explore the clinical significance of sirtuins in lung cancer. However, the results are not consistent, the sample sizes in the previous studies are limited, and not all of the seven sirtuins were explored and compared.28–42
In the present study, we utilized the mRNA profile dataset of 1017 NSCLC patients from The Cancer Genome Atlas (TCGA) to investigate and compare the associations of the seven sirtuins (SIRT1–7) with clinicopathological parameters and prognosis in NSCLC and preliminarily explore the molecular mechanisms of sirtuins in carcinogenesis.
Materials and methods
Analysis of TCGA data
To investigate the associations of sirtuin mRNA levels with clinicopathological parameters and clinical outcomes in NSCLC, we downloaded NSCLC mRNA profile data and corresponding clinical data from the TCGA database. The patient samples were divided into two groups by the optimal cutoff value of sirtuin mRNA levels that was obtained on the basis of prognosis. The correlations between sirtuin expression and clinicopathological parameters were analyzed by the chi-square or Fisher’s exact test. Kaplan–Meier plot curves and Cox proportional regression analysis were used to analyze the effects of sirtuins on overall survival (OS) and recurrence-free survival (OS) in NSCLC. Statistical analyses were conducted with the software GraphPad Prism 6 and SPSS 19.0. P-value <0.05 was considered to be statistically significant. To investigate the related genes during sirtuin alteration that might be involved in lung cancer carcinogenesis, the expression correlations of sirtuins with other genes were examined by Pearson’s correlation analyses. Then, the top 500 genes that correlated with sirtuins were subjected to gene ontology (GO) analysis in the GeneCoDis3 online database (http://genecodis.cnb.csic.es).
Literature review
A systematic literature search was conducted in three English databases (PubMed, EMBASE, and ISI Web of science) for related reports on the associations of sirtuin expression and OS in NSCLC up to January 2018. The following terms were used: “lung OR pulmonary”, “cancer OR tumor OR carcinoma”, and “Sirtuins OR SIRT1 OR SIRT2 OR SIRT3 OR SIRT4 OR SIRT5 OR SIRT6 OR SIRT7”. The retrieved documents were screened by two independent authors by reviewing the article titles, abstracts, and full texts. In addition, the review articles were screened to identify additional eligible studies. The sample size, sirtuin detection method, and the HRs with 95% CIs of each included study were extracted by two investigators independently.
Examination of the concordance of SIRT2 mRNA and protein expressions in NSCLC
To investigate the concordance of SIRT2 mRNA and protein expressions in NSCLC, the SIRT2 protein and mRNA expressions in NSCLC and normal lung tissues were initially compared. Results of SIRT2 immunohistochemical (IHC) staining of NSCLC and normal tissues were evaluated using the data from the Human Protein Atlas (HPA).43 SIRT2 mRNA expression in NSCLC and normal adjacent tissues was compared with the TCGA data by the Mann–Whitney test. In addition, we undertook a re-analysis of the results from Li et al’s study, which detected SIRT2 mRNA expressions by quantitative real-time polymerase chain reaction (qRT-PCR) and protein expressions by Western blotting in seven NSCLC tissues and corresponding normal tissues.44 The mRNA data were obtained by reading the histograms with Plot Digitizer software. In addition, Image J 1.8.0 software was used to scan the Western blotting bands to obtain expression levels of SIRT2 protein. Then, SIRT2 mRNA and protein expressions in NSCLC and corresponding normal tissues were compared with the paired t-test. Next, the correlations of SIRT2 mRNA with protein expressions in Li et al’ report were explored by Pearson’s correlation analysis. At the same time, studies reporting both proteomics and transcription profiles in the same lung cancer patients were screened by searching the PRIDE database and reading the corresponding publications. Two studies were identified.45,46 However, only one study reported SIRT2.46 Then, SIRT2 mRNA and protein expression data were extracted and subjected to Pearson’s correlation analysis. P<0.05 was considered as indicative of statistical significance.
Results
Correlations of sirtuins with clinicopathological parameters in NSCLC
In order to explore the associations of sirtuin mRNA expressions with the clinicopathological parameters of NSCLC patients, we downloaded the related datasets from the TCGA database. In total, 1017 NSCLC patients with basal clinical characteristics and prognosis information were identified. The sirtuin expressions of the patients were split into the high-expression and low-expression group according to the optimal cutoff value that was obtained on the basis of prognosis. The chi-square test was used to clarify the associations of sirtuins with the clinicopathological parameter of NSCLC patients. The results of analysis suggested SIRT1, SIRT3, SIRT4, and SIRT7 were highly expressed in adenocarcinoma (ADC) whereas SIRT2 and SIRT5 were highly expressed in squamous cell carcinoma (SCC; Table 1). SIRT1, SIRT3, SIRT4, and SIRT7 were highly expressed in female patients whereas SIRT5 was highly expressed in male patients. High SIRT7 expression was associated with younger age at onset (<70 years vs ≥70 years). High SIRT1 expression was associated with distant metastasis (M1 vs M0) and low T stage (T1 vs T2–4). High SIRT4 was associated with high T stage (T2–4 vs T1) and TNM stage (II–IV vs I). High SIRT1 and SIRT4 was associated with non-smoking whereas high SIRT5 was associated with smoking.
Table 1.
Correlations of sirtuins and clinicopathological parameters in patients with NSCLC
| Parameters | N | SIRT1
|
SIRT2
|
SIRT3
|
SIRT4
|
SIRT5
|
SIRT6
|
SIRT7
|
||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Low | High | P-value | Low | High | P-value | Low | High | P-value | Low | High | P-value | Low | High | P-value | Low | High | P-value | Low | High | P-value | ||
| Histology | ||||||||||||||||||||||
| ADC | 515 | 109 (21.17%) |
406 (78.83%) |
0.000 | 237 (46.02%) |
278 (53.98%) |
0.000 | 293 (56.89%) |
222 (43.11%) |
0.000 | 308 (59.81%) |
207 (40.19%) |
0.000 | 331 (64.27%) |
184 (35.73%) |
0.000 | 361 (70.1%) |
154 (29.9%) |
0.788 | 202 (39.22%) |
313 (60.78%) |
0.046 |
| SCC | 502 | 313 (62.35%) |
189 (37.65%) |
127 (25.30%) |
375 (74.70%) |
417 (83.07%) |
85 (16.93%) |
445 (88.65%) |
57 (11.35%) |
196 (39.04%) |
306 (60.96%) |
348 (69.32%) |
154 (30.68%) |
228 (45.42%) |
274 (54.58%) |
|||||||
| Gender | ||||||||||||||||||||||
| Female | 402 | 133 (33.08%) |
269 (66.92%) |
0.000 | 138 (34.33%) |
264 (65.67%) |
0.409 | 258 (64.18%) |
144 (35.82%) |
0.001 | 272 (67.66%) |
130 (32.34%) |
0.000 | 243 (60.45%) |
159 (39.55%) |
0.000 | 280 (69.65%) |
122 (30.35%) |
0.924 | 170 (42.29%) |
232 (57.71%) |
0.976 |
| Male | 602 | 280 (46.51%) |
322 (53.49%) |
222 (36.88%) |
380 (63.12%) |
445 (73.92%) |
157 (26.08%) |
472 (78.41%) |
130 (21.59%) |
280 (46.51%) |
322 (53.49%) |
421 (69.93%) |
181 (30.07%) |
254 (42.19%) |
348 (57.81%) |
|||||||
| Age | ||||||||||||||||||||||
| <70 years | 627 | 258 (41.15%) |
369 (58.85%) |
0.999 | 234 (37.32%) |
393 (62.68%) |
0.192 | 444 (70.81%) |
183 (29.19%) |
0.522 | 454 (72.41%) |
173 (27.59%) |
0.155 | 314 (50.08%) |
313 (49.92%) |
0.178 | 433 (69.06%) |
194 (30.94%) |
0.831 | 244 (38.92%) |
383 (61.08%) |
0.012 |
| ≥70 years | 350 | 144 (41.14%) |
206 (58.86%) |
116 (33.14%) |
234 (66.86%) |
241 (68.86%) |
109 (31.14%) |
268 (76.57%) |
82 (23.43%) |
191 (54.57%) |
159 (45.43%) |
244 (69.71%) |
106 (30.29%) |
165 (47.14%) |
185 (52.86%) |
|||||||
| M stage | ||||||||||||||||||||||
| M0 | 747 | 327 (43.78%) |
420 (56.22%) |
0.014 | 252 (33.73%) |
495 (66.27%) |
0.125 | 535 (71.62%) |
212 (28.38%) |
0.463 | 551 (73.76%) |
196 (26.24%) |
0.876 | 375 (50.20%) |
372 (49.80%) |
0.715 | 529 (70.82%) |
218 (29.18%) |
0.202 | 320 (42.84%) |
427 (57.16%) |
0.343 |
| M1 | 32 | 7 (21.88%) |
25 (78.13%) |
15 (46.88%) |
17 (53.13%) |
21 (65.63%) |
11 (34.38%) |
24 (75.00%) |
8 (25.00%) |
15 (46.88%) |
17 (53.13%) |
26 (81.25%) |
6 (18.75%) |
11 (34.38%) |
21 (65.63%) |
|||||||
| N stage | ||||||||||||||||||||||
| N0 | 642 | 258 (40.19%) |
384 (59.81%) |
0.299 | 225 (35.05%) |
417 (64.95%) |
0.444 | 452 (70.4%) |
190 (29.6%) |
0.910 | 469 (73.05%) |
173 (26.95%) |
0.142 | 326 (50.78%) |
316 (49.22%) |
0.246 | 455 (70.87%) |
187 (29.13%) |
0.353 | 273 (42.52%) |
369 (57.48%) |
0.521 |
| N1–3 | 344 | 150 (43.6%) |
194 (56.4%) |
129 (37.5%) |
215 (62.5%) |
241 (70.06%) |
103 (29.94%) |
266 (77.33%) |
78 (22.67%) |
188 (54.65%) |
156 (45.35%) |
234 (68.02%) |
110 (31.98%) |
139 (40.41%) |
205 (59.59%) |
|||||||
| T stage | ||||||||||||||||||||||
| T1 | 280 | 95 (33.93%) |
185 (66.07%) |
0.003 | 96 (34.29%) |
184 (65.71%) |
0.516 | 188 (67.14%) |
92 (32.86%) |
0.214 | 195 (69.64%) |
85 (30.36%) |
0.039 | 148 (52.86%) |
132 (47.14%) |
0.780 | 203 (72.50%) |
77 (27.50%) |
0.235 | 115 (41.07%) |
165 (58.93%) |
0.665 |
| T2–4 | 721 | 318 (44.11%) |
403 (55.89%) |
263 (36.48%) |
458 (63.52%) |
513 (71.15%) |
208 (28.85%) |
548 (76.01%) |
173 (23.99%) |
374 (51.87%) |
347 (48.13%) |
495 (68.65%) |
226 (31.35%) |
307 (42.58%) |
414 (57.42%) |
|||||||
| Stage | ||||||||||||||||||||||
| I–II | 518 | 207 (39.96%) |
311 (60.04%) |
0.448 | 174 (33.59%) |
344 (66.41%) |
0.131 | 352 (67.95%) |
166 (32.05%) |
0.143 | 368 (71.04%) |
150 (28.96%) |
0.022 | 259 (50.00%) |
259 (50.00%) |
0.131 | 373 (72.01%) |
145 (27.99%) |
0.132 | 228 (44.02%) |
290 (55.98%) |
0.228 |
| III–IV | 482 | 204 (42.32%) |
278 (57.68%) |
184 (38.17%) |
298 (61.83%) |
348 (72.2%) |
134 (27.8%) |
373 (77.39%) |
109 (22.61%) |
264 (54.77%) |
218 (45.23%) |
326 (67.63%) |
156 (32.37%) |
194 (40.25%) |
288 (59.75%) |
|||||||
| Smoking status | ||||||||||||||||||||||
| Yes | 890 | 381 (42.81%) |
509 (57.19%) |
0.001 | 314 (35.28%) |
576 (64.72%) |
0.388 | 631 (70.90%) |
259 (29.10%) |
0.054 | 672 (75.51%) |
218 (24.49%) |
0.006 | 447 (50.22%) |
443 (49.78%) |
0.009 | 624 (70.11%) |
266 (29.89%) |
0.698 | 369 (41.46%) |
521 (58.54%) |
0.059 |
| No | 93 | 23 (24.73%) |
70 (75.27%) |
37 (39.78%) |
56 (60.22%) |
57 (61.29%) |
36 (38.71%) |
58 (62.37%) |
35 (37.63%) |
60 (64.52%) |
33 (35.48%) |
67 (72.04%) |
26 (27.96%) |
48 (51.61%) |
45 (48.39%) |
|||||||
Note: Bold: P<0.05 demonstrated by chi-square test.
Abbreviations: NSCLC, non–small cell lung cancer; SIRT1, sirtuin 1; SIRT2, sirtuin 2; SIRT3, sirtuin 3; SIRT4, sirtuin 4; SIRT5, sirtuin 5; SIRT6, sirtuin 6; SIRT7, sirtuin 7; ADC, adenocarcinoma; SCC, squamous cell carcinoma.
Correlations of sirtuins with prognosis in NSCLC patients
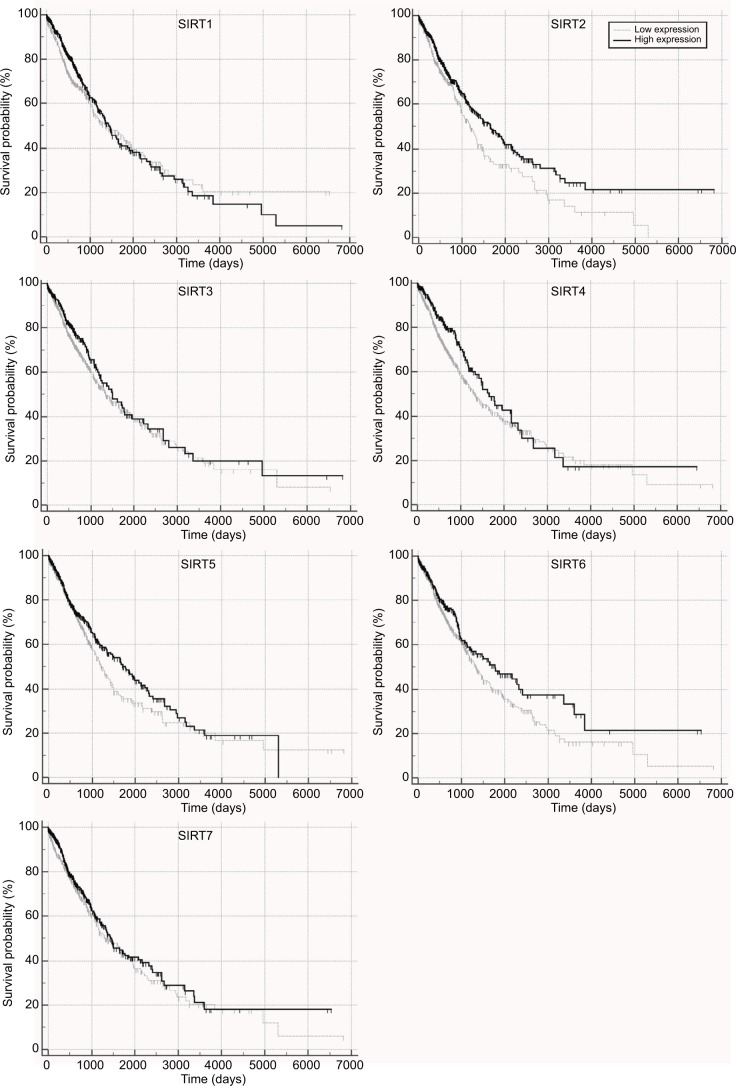
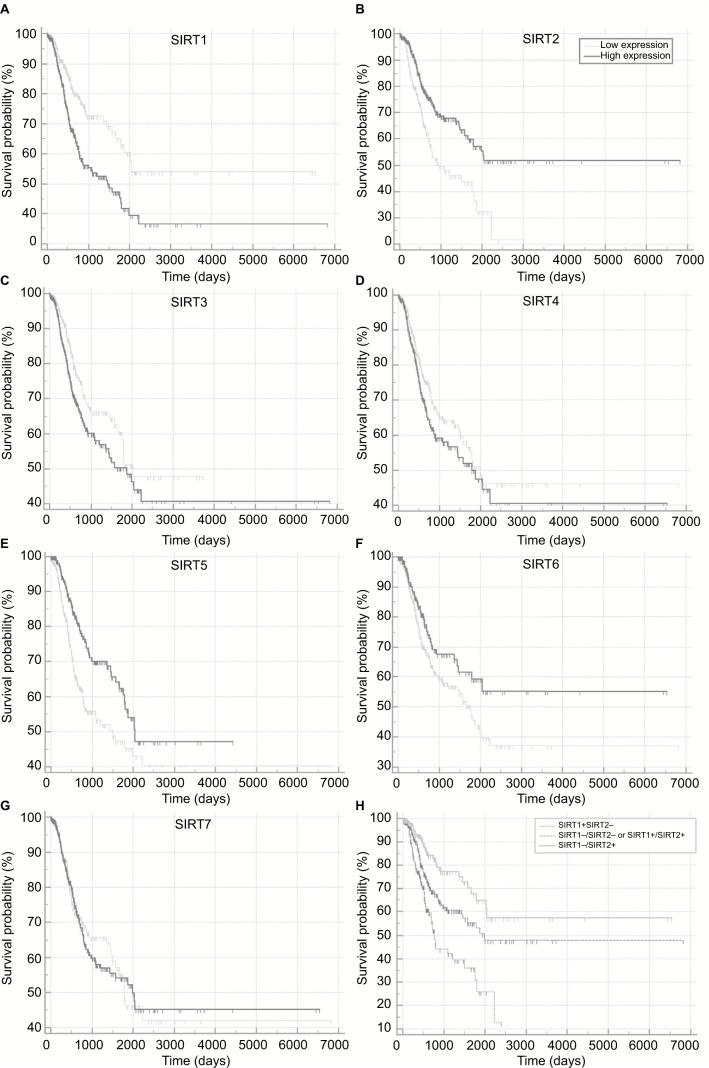
The correlations of sirtuins mRNA expressions with prognosis in NSCLC patients were investigated by Kaplan–Meier plot curves and Cox proportional regression analyses, wherein high SIRT2, SIRT4, and SIRT6 expressions were found to be associated with longer OS whereas no significant association of OS with other sirtuins was observed (Figure 1 and Table 2). Multivariate analyses indicated that SIRT2 and SIRT6 was still significantly associated with OS after adjusting for potential confounding factors including age, M, N, and T stages. For RFS, univariate analyses suggested high SIRT1 expression was significantly associated with shorter RFS time whereas high expressions of SIRT2, SIRT3, SIRT5, SIRT6, and SIRT7 were significantly associated with longer RFS time (Figure 2A–G and Table 3). After adjusting for the confounding factors, including histology, N, and T stages, significant associations were still found in SIRT1, SIRT2, SIRT5, SIRT6, and SIRT7 but not in SIRT3.
Figure 1.
Overall survival in TCGA NSCLC patients with high and low sirtuin expression (SIRT1–7) were compared with Kaplan–Meier plot curves.
Abbreviations: TCGA, The Cancer Genome Atlas; NSCLC, non–small cell lung cancer; SIRT1, sirtuin 1; SIRT2, sirtuin 2; SIRT3, sirtuin 3; SIRT4, sirtuin 4; SIRT5, sirtuin 5; SIRT6, sirtuin 6; SIRT7, sirtuin 7.
Table 2.
Cox proportional regression analysis to investigate the correlations of sirtuins and overall survival in patients with NSCLC
| SIRTs | Univariate analysis
|
Multivariate analysis*
|
||||||
|---|---|---|---|---|---|---|---|---|
| P-value | HR | 95% CI | P-value | HR | 95% CI | |||
| SIRT1 | 0.352 | 0.902 | 0.725 | 1.121 | 0.956 | 1.007 | 0.787 | 1.288 |
| SIRT2 | 0.007 | 0.736 | 0.588 | 0.920 | 0.005 | 0.697 | 0.543 | 0.894 |
| SIRT3 | 0.244 | 0.865 | 0.677 | 1.104 | 0.568 | 0.922 | 0.696 | 1.220 |
| SIRT4 | 0.043 | 0.761 | 0.584 | 0.992 | 0.255 | 0.842 | 0.626 | 1.132 |
| SIRT5 | 0.109 | 0.837 | 0.673 | 1.040 | 0.128 | 0.828 | 0.650 | 1.056 |
| SIRT6 | 0.044 | 0.780 | 0.612 | 0.994 | 0.009 | 0.684 | 0.514 | 0.911 |
| SIRT7 | 0.253 | 0.881 | 0.708 | 1.095 | 0.305 | 0.878 | 0.686 | 1.125 |
Note:
The potential confounding factors were age, M, N, and T stage. Bold font: P<0.05 demonstrated by Cox proportional regression analysis.
Abbreviation: TCGA, The Cancer Genome Atlas; NSCLC, non-small cell lung cancer; SIRT1, sirtuin 1; SIRT2, sirtuin 2; SIRT3, sirtuin 3; SIRT4, sirtuin 4; SIRT5, sirtuin 5; SIRT6, sirtuin 6; SIRT7, sirtuin 7; HR, hazard ratio; CI, confidence interval.
Figure 2.
Recurrence-free survival in TCGA NSCLC patients stratified by high and low sirtuin expression (SIRT1–7) or a combination of SIRT1 and SIRT2 were compared with Kaplan–Meier plot curves. (A) SIRT1, (B) SIRT2, (C) SIRT3, (D) SIRT4, (E) SIRT5, (F) SIRT6, (G) SIRT7, and (H) Combination of SIRT1 and SIRT2.
Abbreviations: TCGA, The Cancer Genome Atlas; NSCLC, non–small cell lung cancer; SIRT1, sirtuin 1; SIRT2, sirtuin 2; SIRT3, sirtuin 3; SIRT4, sirtuin 4; SIRT5, sirtuin 5; SIRT6, sirtuin 6; SIRT7, sirtuin 7; +, high expression; –, low expression.
Table 3.
Cox proportional regression analysis to investigate the correlations of sirtuins and recurrence-free survival in patients with NSCLC
| SIRTs | Univariate analysis
|
Multivariate analysis*
|
||||||
|---|---|---|---|---|---|---|---|---|
| P-value | HR | 95% CI | P-value | HR | 95% CI | |||
| SIRT1 | 0.000 | 1.899 | 1.366 | 2.639 | 0.001 | 1.852 | 1.294 | 2.650 |
| SIRT2 | 0.000 | 0.531 | 0.394 | 0.717 | 0.001 | 0.588 | 0.432 | 0.800 |
| SIRT3 | 0.034 | 1.410 | 1.026 | 1.938 | 0.107 | 1.316 | 0.943 | 1.837 |
| SIRT4 | 0.099 | 1.284 | 0.954 | 1.730 | 0.416 | 1.144 | 0.827 | 1.583 |
| SIRT5 | 0.000 | 0.565 | 0.415 | 0.769 | 0.007 | 0.636 | 0.459 | 0.883 |
| SIRT6 | 0.028 | 0.687 | 0.492 | 0.961 | 0.004 | 0.606 | 0.431 | 0.852 |
| SIRT7 | 0.013 | 0.625 | 0.432 | 0.906 | 0.005 | 0.585 | 0.403 | 0.848 |
Note:
The potential confounding factors were histology, N, and T stage. Bold font: P<0.05 demonstrated by Cox proportional regression analysis.
Abbreviation: TCGA, The Cancer Genome Atlas; NSCLC, non-small cell lung cancer; SIRT1, sirtuin 1; SIRT2, sirtuin 2; SIRT3, sirtuin 3; SIRT4, sirtuin 4; SIRT5, sirtuin 5; SIRT6, sirtuin 6; SIRT7, sirtuin 7; HR, hazard ratio; CI, confidence interval.
Prediction of RFS with the combination of SIRT1 and SIRT2
Considering that SIRT1 and SIRT2 were the most significant biomarkers that were associated with RFS among the seven sirtuins and SIRT1 was a poor prognostic factor while SIRT2 was a favorable factor, we stratified the patients by combining SIRT1 and SIRT2 and then compared their RFS in order to provide a better prediction model for RFS in NSCLC. Kaplan–Meier plot curves and Cox proportional analyses suggested that, compared to patients with the SIRT1−/SIRT2+ genotype, patients with SIRT1−/SIRT2− or SIRT1+/SIRT2+ genotypes had poor RFS and patients with the SIRT1+/SIRT2− genotype had poorer prognosis (Figure 2H and Table 4).
Table 4.
Cox proportional regression analysis to investigate the correlations of the combination of SIRT1 and SIRT2 with recurrence-free survival in patients with NSCLC
| SIRTs | Univariate analysis
|
Multivariate analysis* | ||||||
|---|---|---|---|---|---|---|---|---|
| P-value | HR | 95% CI | P-value | HR | 95% CI | |||
| SIRT1−/SIRT2+ | Reference | |||||||
| SIRT1−/SIRT2− or SIRT1+/SIRT2+ | 0.007 | 1.758 | 1.167 | 2.648 | 0.010 | 1.746 | 1.144 | 2.665 |
| SIRT1+/SIRT2− | 0.000 | 3.134 | 2.037 | 4.820 | 0.000 | 3.021 | 1.883 | 4.849 |
Note:
The potential confounding factors were histology, N, and T stage. Bold font: P<0.05 demonstrated by Cox proportional regression analysis.
Abbreviations: TCGA, The Cancer Genome Atlas; NSCLC, non–small cell lung cancer; SIRT1, sirtuin 1; SIRT2, sirtuin 2; HR, hazard ratio; CI, confidence interval; +, high expression; –, low expression.
Integrated analysis of TCGA dataset and other publicly available datasets
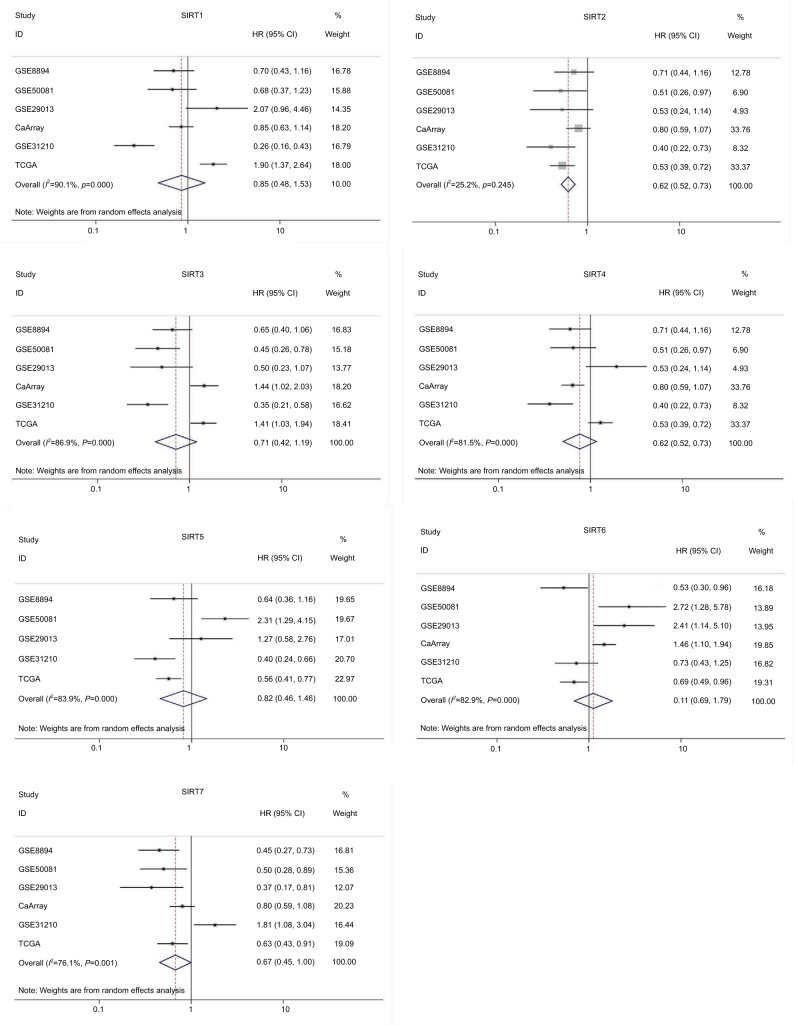
To further validate our results, we conducted an integrated analysis by pooling the TCGA NSCLC dataset and other public datasets. The Kaplan–Meier plotter (KM plotter) is an online tool to analyze correlations of individual genes with prognosis of patients in previous microarray datasets that comprised a total of 2,437 lung cancer cases. HRs with 95% CIs of the associations between sirtuins and RFS from each separate dataset in the database were obtained. After pooling these results by a meta-analysis method, high SIRT2 and SIRT7 expressions were found to be associated with favorable RFS in lung cancer (Figure 3).
Figure 3.
Forest plot for the correlations of sirtuins (SIRT1–7) and recurrence-free survival in patients with NSCLC by pooling the results of the datasets from TCGA and Kaplan–Meier plotter databases.
Abbreviations: TCGA, The Cancer Genome Atlas; NSCLC, non–small cell lung cancer; SIRT1, sirtuin 1; SIRT2, sirtuin 2; SIRT3, sirtuin 3; SIRT4, sirtuin 4; SIRT5, sirtuin 5; SIRT6, sirtuin 6; SIRT7, sirtuin 7.
Prognostic value of sirtuins in NSCLC reported in the published literature
Some articles have reported the associations of sirtuin expressions with prognosis in NSCLC patients. Here, we conducted a systematic literature search in three English databases (PubMed, EMBASE, and ISI Web of science) up to January 2018. In total, 14 articles published from 2013 to 2017 were identified after screening (Table 5), of which seven, two, two, one, and three articles reported the associations of SIRT1,28–34 SIRT2,30,36 SIRT3,37,38 SIRT5,39 and SIRT6,40–42 respectively, with OS in NSCLC. Moreover, no article has reported SIRT4 and SIRT7 associations. High SIRT1 expression was found to be a poor prognostic factor in four reports, and no association was found in the remaining three articles. High SIRT2 was found to be associated with longer OS time in one article and high expressions of SIRT3, SIRT5, and SIRT6 were associated with poor OS. However, these reports were rare and their sample size was limited (SIRT3: N=2, n=133; SIRT5, N=1, n=39; SIRT6, N=3, n=394). Notably, the results for SIRT6 in the previous reports and our study were inconsistent, which needs to be clarified with a large sample size in the future.
Table 5.
Associations of high sirtuins expressions with overall survival in NSCLC in previous reports
| Study | Sirtuins | HR | P value | Sample size | Detection method |
|---|---|---|---|---|---|
| Noh et al28 | SIRT1 | NS | 144 | IHC | |
| Zhang et al29 | SIRT1 | HR>1 | 0.003 | 295 | IHC |
| Grbesa et al30 | SIRT1 | NS | 105 | IHC | |
| Li et al31 | SIRT1 | HR>1 | 0.009 | 75 | IHC |
| Gharabaghi32 | SIRT1 | HR>1 | 0.001 | 40 | IHC |
| Lin and Peng33 | SIRT1 | NS | 260 | IHC | |
| Wang and Wang34 | SIRT1 | HR>1 | <0.01 | 60 | IHC |
| Grbesa et al30 | SIRT2 | NS | 105 | IHC | |
| Li et al36 | SIRT2 | HR<1 | <0.01 | 28 | PCR |
| Yang et al37 | SIRT3 | HR>1 | 0.001 | 131 | IHC |
| Xiong et al38 | SIRT3 | HR>1 | 0.028 | 70 | IHC |
| Lu et al39 | SIRT5 | HR>1 | 0.03 | 39 | PCR |
| Azuma et al40 | SIRT6 | HR>1 | 0.022 | 98 | IHC |
| Bai et al41 | SIRT6 | HR>1 | 0.034 | 174 | IHC |
| Chen et al42 | SIRT6 | HR>1 | <0.01 | 122 | IHC |
Abbreviations: NSCLC, non–small cell lung cancer; NS, not significant; SIRT1, sirtuin 1; SIRT2, sirtuin 2; SIRT3, sirtuin 3; SIRT5, sirtuin 5; SIRT6, sirtuin 6; HR, hazard ratio; IHC, immunohistochemistry; PCR, polymerase chain reaction.
Analyses of the genes involved in NSCLC during sirtuin alteration
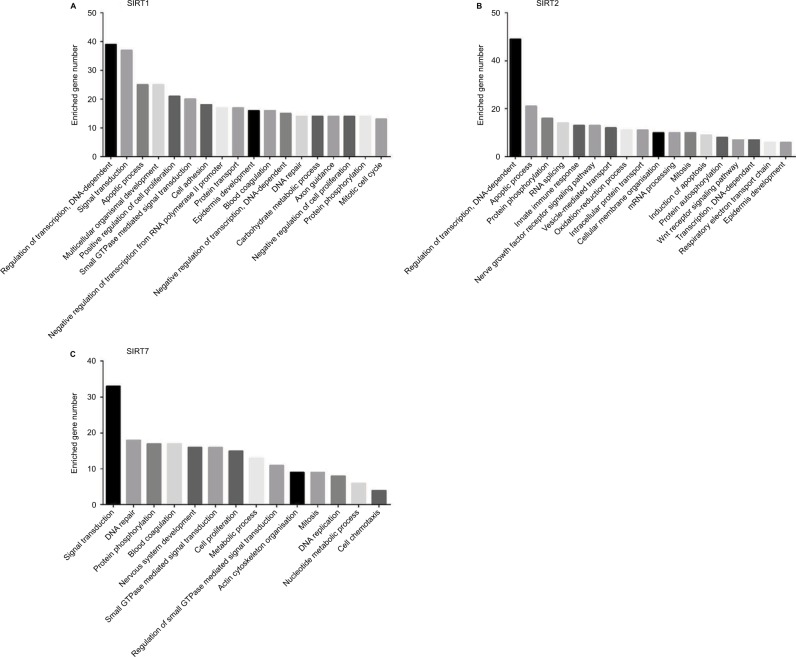
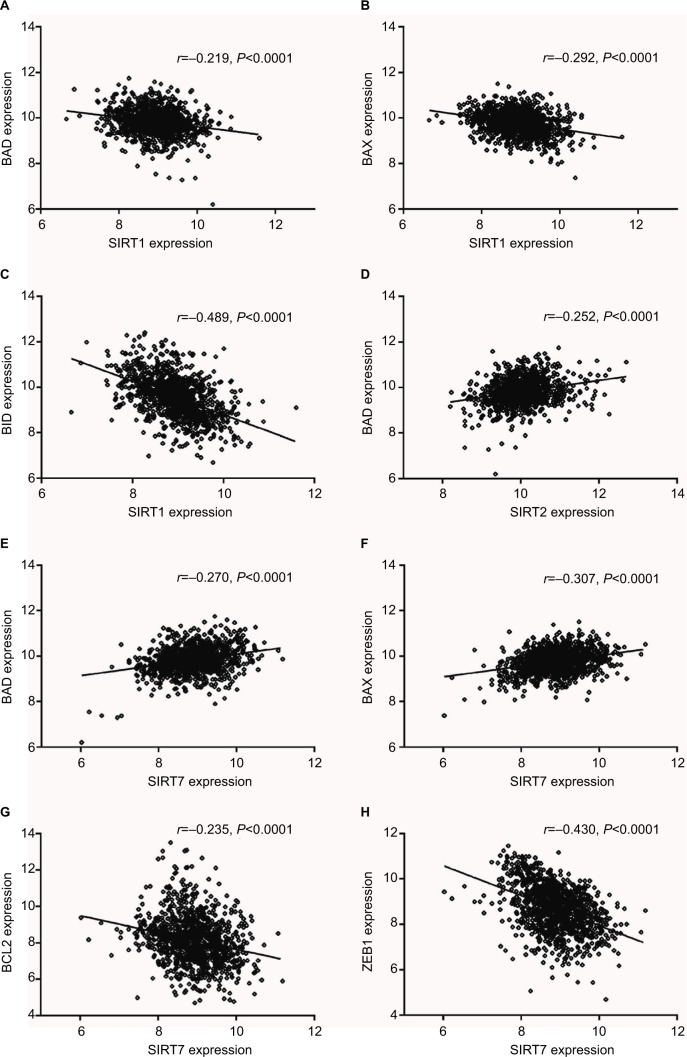
According to our screening of the literature, we found 18 articles reported the underlying mechanisms of sirtuins in lung cancer (Table S1). There were five, five, four, one, and four articles exploring the molecular mechanisms of SIRT1, SIRT2, SIRT3, SIRT5, and SIRT6 in lung cancer, respectively, whereas SIRT4 and SIRT7 were not investigated. To further understand the potential mechanisms of sirtuins in NSCLC carcinogenesis, genes that were positively or negatively associated with sirtuin expressions were analyzed and annotated. Pearson’s correlation analyses were conducted to obtain genes that were co-expressed with sirtuins in the 1017 TCGA NSCLC patients. The top 500 significant genes were selected and subjected to GO analysis with the GeneCoDis3 online tool (http://genecodis.cnb.csic.es). SIRT1, SIRT2, and SIRT7 were used as examples. Our results suggest that the top enriched biological processes were: regulation of transcription, signal transduction, apoptotic process, multicellular organismal development, positive regulation of cell proliferation, small GTPase-mediated signal transduction, and cell adhesion for SIRT1 (Figure 4A); regulation of transcription, apoptotic process, protein phosphorylation, RNA splicing, innate immune response, nerve growth factor receptor signaling, and vesicle-mediated transport for SIRT2 (Figure 4B); and signal transduction, DNA repair, protein phosphorylation, blood coagulation, nervous system development, small GTPase-mediated signal transduction, and cell proliferation for SIRT7 (Figure 4C). Cell apoptotic or proliferation processes are overlapping processes and have been reported to be involved in the mechanisms of SIRT1 and SIRT2 in NSCLC. Thereafter, the associations of SIRT1, SIRT2, and SIRT7 with the key genes in the apoptotic process including BAD, BAX, BCL2, and BID were investigated. The results suggested that SIRT1 was negatively associated with proapoptotic factors BAD, BAX, and BID whereas SIRT2 was positively associated with the proapoptotic factor, BAD (Figure 5A–D). Furthermore, SIRT7 was positively associated with proapoptotic factors BAD and BAX, and negatively associated with the antiapoptotic factor BCL2 (Figure 5E–G). In addition, SIRT7 was negatively asso ciated with ZEB1, a pro-proliferative factor (Figure 5H). To some extent, this explained the different functions of SIRT1, SIRT2, and SIRT7 in NSCLC carcinogenesis.
Figure 4.
Biological processes regulated by SIRT1, SIRT2, and SIRT7 in TCGA NSCLC patients: (A) SIRT1, (B) SIRT2, and (C). SIRT7.
Abbreviations: TCGA, The Cancer Genome Atlas; NSCLC, non–small cell lung cancer; SIRT1, sirtuin 1; SIRT2, sirtuin 2; SIRT7, sirtuin 7.
Figure 5.
Expression correlations of SIRT1, SIRT2, and SIRT7 with BAD, BAX, BID, BCL2, and ZEB1 in TCGA NSCLC patients. (A–C). Correlations of SIRT1 with BAD (A), BAX (B), and BID (C). (D) Correlation of SIRT2 with BAD. (E–H) Correlations of SIRT7 with BAD (E), BAX (F), BCL2 (G), and ZEB1 (H).
Abbreviations: SIRT1, sirtuin 1; SIRT2, sirtuin 2; SIRT7, sirtuin 7; BAX, Bcl-2 associated X; BAD, Bcl-2 associated agonist of cell death; BID, BH3 interacting domain death agonist; ZEB1, zinc finger E-box binding homeobox 1; TCGA, The Cancer Genome Atlas; NSCLC, non-small cell lung cancer.
SIRT2 protein expression was in concordance with its mRNA expression in NSCLC
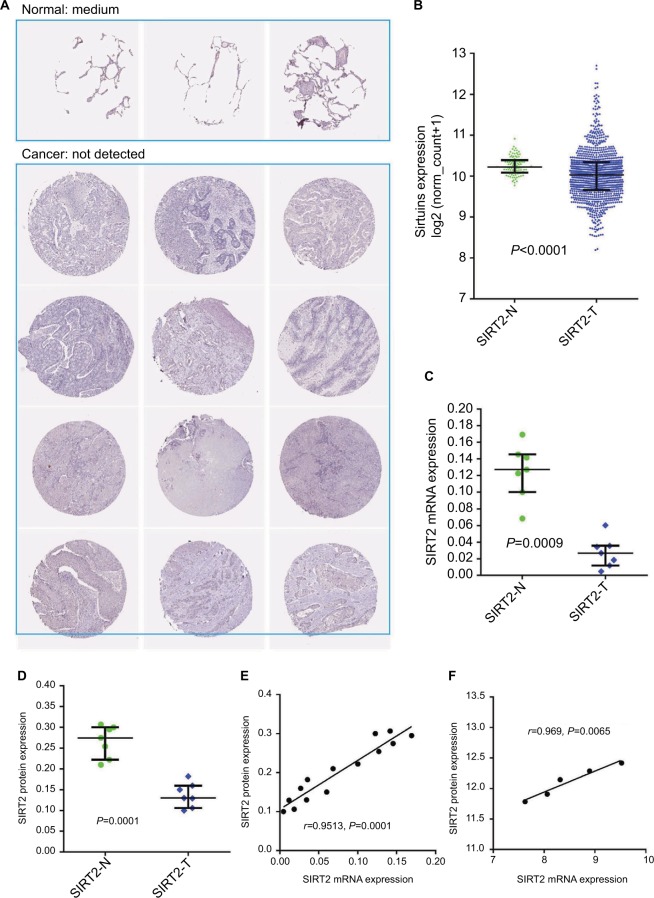
Genes generally function at the protein level; therefore, we investigated the concordance between sirtuin mRNA and protein expressions in NSCLC, with SIRT2 as an example. SIRT2 protein and mRNA expressions were initially compared between NSCLC tissues and corresponding controls. IHC staining data from the HPA suggested that SIRT2 protein expression was downregulated in NSCLC tissues compared with normal tissues (Figure 6A). The TCGA NSCLC data revealed that the SIRT2 mRNA expression was downregulated in NSCLC tissues (Figure 6B). In addition, we re-analyzed the results from Li et al’s study,44 which detected SIRT2 mRNA expression by qRT-PCR, and protein expression by Western blotting in the same NSCLC patients; moreover, we found SIRT2 mRNA and protein expressions were downregulated in lung cancers compared with paired normal tissues (Figure 6C and D). Then, the correlation of SIRT2 mRNA and protein expressions was examined and a high positive correlation was found (r=0.9513, P<0.0001; Figure 6E). Furthermore, we identified one study reporting both NSCLC proteomics and transcription profiles from the PRIDE database46 and also found a high concordance between SIRT2 mRNA and protein expressions (r=0.9689, P=0.0065; Figure 6F). The above results revealed that SIRT2 protein and mRNA expressions shared high concordance in NSCLC.
Figure 6.
Concordance of SIRT2 mRNA and protein expressions in NSCLC. (A). SIRT2 immunohistochemical staining results in normal lung tissues (n=3) and lung cancer tissues (n=12). The images were obtained from the Human Protein Atlas.43 (B). SIRT2 mRNA expression in normal lung tissues (n=110) and lung cancer tissues (n=1017) in the TCGA NSCLC cohort. (C–E). SIRT2 mRNA expression detected by qRT-PCR (C) and protein expression detected by Western blotting (D) in seven lung cancers and paired adjacent normal tissues (n=7); consistency of SIRT2 mRNA and protein expression were explored by Pearson’s correlation analysis (E). These were re-analyses results of Li et al’s report.44 (F). Associations of SIRT2 mRNA with protein expressions in lung cancers. SIRT2 protein expression and mRNA expression data were extracted from proteomics and gene microarray profiles, respectively, which was reported by Stewart et al.46
Abbreviations: SIRT2, sirtuin 2; N, normal; T, tumor; qRT-PCR, quantitative real-time polymerase chain reaction.
Discussion
Currently, a large number of cell signaling pathways have been discovered as being involved in tumor carcinogenesis and may serve as potential therapeutic targets for cancer treatment. Sirtuins are a family of mammalian proteins homologous to yeast Sir2.6,7 Seven different sirtuins (SIRT1–7) have been found and widely expressed in mammals. Previous reports have revealed the involvement of sirtuins in numerous critical biological processes including cell metabolism, genomic stability, cell cycle, cell division, differentiation, and transcriptional regulation and their functions in the pathogenesis of a variety of diseases such as metabolic diseases,22 neurodegenerative diseases,23 aging,24 cardiovascular diseases,25 and cancers.26,27 The clinical significance and potential molecular mechanisms of some sirtuins have been explored in lung cancer. However, the results are not consistent, the sample sizes in the previous studies are limited, and not all of the seven sirtuins have been explored and compared.28–42
In the present study, we utilized the mRNA profile dataset of 1017 NSCLC patients from the TCGA database to investigate and compare associations of the mRNA levels of the seven sirtuins (SIRT1–7) with clinicopathological parameters and prognosis in NSCLC. SIRT1, SIRT3, SIRT4, and SIRT7 were highly expressed in ADC and female patients whereas SIRT5 was highly expressed in SCC and male patients. Associations of high SIRT7 with younger age at onset, high SIRT1 with distant metastasis and low T stage, and high SIRT4 with high T stage and TNM stage were also found. Furthermore, the prognostic value of sirtuins in NSCLC was investigated, and the results indicated that high mRNA levels of SIRT2 and SIRT6 were associated with longer OS duration. Moreover, high SIRT1 expression was significantly associated with shorter RFS time whereas high SIRT2, SIRT5, SIRT6, and SIRT7 expressions were associated with longer RFS time.
To compare and validate our results with that from previous reports, we undertook a systematic literature search to identify the full-text of original articles on the associations of sirtuins with OS in NSCLC (Table 5). SIRT1 was the most studied sirtuin for evaluating the associations of sirtuins with OS in lung cancer. A significant association of SIRT1 with OS was found in four of the seven reports.28–34 A recent meta-analysis has pooled the results from the above seven reports and found that high SIRT1 expression was a poor factor for OS in NSCLC,35 whereas no significant association of SIRT1 with OS was found in our present study. This inconsistency might be attributed to the detection methods of SIRT1 and selection of cutoff value. Moreover, a discrepancy was found for the association of SIRT6 with OS. Three reports, with a total of 394 cases, suggested high SIRT6 expression was associated with poor OS, whereas our results suggested that high SIRT6 was associated with favorable OS.40–42 Until now, associations of SIRT4 or SIRT7 with prognosis of NSCLC patients have not been reported. More studies with large sample size should be conducted to clarify our results.
To date, some molecular mechanisms of sirtuins have been revealed in NSCLC (Table S1). SIRT1 can promote lung tumor growth through downregulating DLL4/Notch signaling and deacetylating the Notch1 intracellular domain;47 its inactivation exerts antitumor activities through the tumor suppressor p27 in NSCLC;48 and it is also involved in lung cancer development via the TNF-α/β-catenin axis under Benzo[a]pyrene (B[a]P) exposure – a carcinogen in cigarette smoke.49,50 SIRT2 can inhibit cell proliferation or induce cell apoptosis by increasing reactive oxygen species (ROS) production and p27 levels,44 downregulating JMJD2A,51 or deacetylating SKP2 followed by decreasing ubiquitination of p27.36 SIRT3 can promote tumor proliferation and inhibit apoptosis via interaction with NMNAT2, and promote NSCLC malignancy via activation Akt at the post-transcriptional level.38,52 SIRT5 can promote cell proliferation and chemotherapy resistance by inducing Nrf2 expression.39 An article reported that SIRT6 can increase tumor migration and invasion via enhancing ERK1/2 phosphorylation and MMP9 expression,41 whereas another report suggested that SIRT6 can suppress cell proliferation through inhibition of Twist1 expression.53 There is no report on the mechanisms of SIRT4 and SIRT7 in NSCLC. Modulation of cell proliferation and/ or apoptosis might be the common mechanisms of sirtuin action in NSCLC carcinogenesis. To further understand the potential mechanisms of sirtuins in NSCLC carcinogenesis, we investigated the cellular biological processes regulated by sirtuin alterations through annotating the sirtuin co-expressed genes by GO analysis. We took SIRT1, SIRT2, and SIRT7 as examples, because SIRT1 and SIRT2 were the mostly studied sirtuins and SIRT7 has not been investigated in NSCLC, and found they might affect tumor activities via modulating cell proliferation and apoptosis. The potential mechanisms explained the different prognostic value of SIRT1, SIRT2, and SIRT7. More in vitro and animal experiments should be undertaken to validate our findings.
Conclusion
Sirtuin expressions were significantly associated with clinicopathological parameters, especially tumor histology and gender, and clinical outcomes in NSCLC patients. For OS, high expressions of SIRT2 and SIRT6 were favorable prognostic factors. For RFS, high SIRT1 expression was a poor prognostic factor, whereas high SIRT2 and SIRT5–7 expressions were favorable prognostic factors. Combining SIRT1 and SIRT2 could provide a better prognostic model for RFS.
Supplementary material
Table S1.
Underlying mechanism of sirtuins in NSCLC in the previous reports
| Study | Sirtuins | Mechanisms |
|---|---|---|
| Xie et al47 | SIRT1 | SIRT1 promote lung tumor growth through downregulation of DLL4/Notch signaling and deacetylation of N1IC |
| Zhang et al29 | SIRT1 | High SIRT1 was associated with chemotherapy resistance |
| Han et al54 | SIRT1 | SIRT1 promotes cell migration |
| Zhu et al48 | SIRT1 | SIRT1 inactivation evokes antitumor activities in NSCLC through the tumor suppressor p27 |
| Grbesa et al30 | SIRT1 | Downregulation of SIRT1 decreased cell proliferation |
| Cheng et al55 | SIRT1 | K-Ras promotes the survival of non–small cell lung cancer cells by cooperating with sirtuin 1 and p27 under ROS stimulation |
| Lu et al49 | SIRT1 | SIRT1 is involved in Benzo[a]pyrene B[a]P-induced transformation associated with activation of the TNF-α/β-catenin axis |
| Li et al44 | SIRT2 | SIRT2 inhibits cell proliferation, induces apoptosis and cell-cycle arrest by increasing the ROS production and p27 levels, and is associated with increased chemotherapy sensitivity |
| Grbesa et al30 | SIRT2 | Downregulation of SIRT2 decreased cell proliferation |
| Xu et al51 | SIRT2 | SIRT2 inhibits cell proliferation and tumor growth via downregulating JMJD2A |
| Li et al36 | SIRT2 | Downregulation of SIRT2 increases the level of SKP2 acetylation, then increases p27 ubiquitination, and finally enhances tumor cell proliferation |
| Luo et al56 | SIRT2 | SPOP promotes SIRT2 degradation and suppresses non–small cell lung cancer cell growth |
| Xiao et al57 | SIRT3 | SIRT3 increased the bax/bcl-2 and bad/bcl-x/L ratios, and promoted AIF translocation to the nucleus. SIRT3 overexpression upregulated p53 and p21 protein levels, and decreased intracellular ROS levels |
| Li et al52 | SIRT3 | Low expression of SIRT3 significantly inhibited mitotic entry, growth, and proliferation of NSCLC cell lines and promoted apoptosis, which was related to energy metabolism involved in the interaction between SIRT3 and NMNAT2 |
| Xiong et al38 | SIRT3 | SIRT3 increases Akt activation by post-transcriptional modification and, then, promotes NSCLC malignancy |
| Xiong et al58 | SIRT3 | SIRT3 deacetylates and promotes degradation of P53 in PTENdefective non–small cell lung cancer |
| Lu et al39 | SIRT5 | SIRT5 promotes cell proliferation and chemotherapy resistance by induction of Nrf2 expression |
| Azuma et al40 | SIRT6 | SIRT6 increases NFkappaB and Beclin1, and is associated with chemotherapy resistance |
| Han et al53 | SIRT6 | SIRT6 suppresses cell proliferation through inhibition of Twist1 expression |
| Kim and Juhnn59 | SIRT6 | Cyclic AMP signaling reduces SIRT6 expression in non–small cell lung cancer cells by promoting ubiquitin-proteasomal degradation via inhibition of the Raf–MEK–ERK pathway |
| Bai et al41 | SIRT6 | SIRT6 increases tumor migration and invasion via increasing ERK1/2 phosphorylation and MMP9 expression |
Abbreviations: SIRT1, sirtuin 1; SIRT2, sirtuin 2; SIRT3, sirtuin 3; SIRT4, sirtuin 4; SIRT5, sirtuin 5; SIRT6, sirtuin 6; SIRT7, sirtuin 7; NSCLC, non-small cell lung cancer; DLL4, Delta-like ligand 4; N1IC, Notch1 intracellular domain; JMJD2A, lysine demethylase 4A; Skp2, S-phase kinase associated protein 2; SPOP, speckle type BTB/POZ protein; BAX, Bcl-2 associated X; BAD, Bcl-2 associated agonist of cell death; Bcl-x/L, Bcl-2 like 1; AIF, apoptosis inducing factor; NMNAT2, nicotinamide nucleotide adenylyltransferase 2; PTEN, phosphatase and tensin homolog; Nrf2, nuclear factor erythroid 2 like 2; Twist1, twist family bHLH transcription factor 1; MMP9, matrix metallopeptidase 9.
Acknowledgments
This study was supported by funding from the National Science Foundation for Young Scientists of China (grant no. 81601849) and Wenzhou Science and Technology Bureau (grant no. Y20160075).
Footnotes
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Torre LA, Siegel RL, Jemal A. Lung cancer statistics. Adv Exp Med Biol. 2016;893:1–19. doi: 10.1007/978-3-319-24223-1_1. [DOI] [PubMed] [Google Scholar]
- 2.Travis WD. Pathology of lung cancer. Clin Chest Med. 2011;32(4):669–692. doi: 10.1016/j.ccm.2011.08.005. [DOI] [PubMed] [Google Scholar]
- 3.Chen W, Zheng R, Zhang S, Zhao P, Zeng H, Zou X. Report of cancer incidence and mortality in China, 2010. Ann Transl Med. 2014;2(7):61. doi: 10.3978/j.issn.2305-5839.2014.04.05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pao W, Girard N. New driver mutations in non-small-cell lung cancer. Lancet Oncol. 2011;12(2):175–180. doi: 10.1016/S1470-2045(10)70087-5. [DOI] [PubMed] [Google Scholar]
- 5.Park SJ, More S, Murtuza A, Woodward BD, Husain H. New targets in non-small cell lung cancer. Hematol Oncol Clin North Am. 2017;31(1):113–129. doi: 10.1016/j.hoc.2016.08.010. [DOI] [PubMed] [Google Scholar]
- 6.Donmez G, Outeiro TF. SIRT1 and SIRT2: emerging targets in neurodegeneration. EMBO Mol Med. 2013;5(3):344–352. doi: 10.1002/emmm.201302451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Inoue T, Hiratsuka M, Osaki M, Oshimura M. The molecular biology of mammalian SIRT proteins: SIRT2 in cell cycle regulation. Cell Cycle. 2007;6(9):1011–1018. doi: 10.4161/cc.6.9.4219. [DOI] [PubMed] [Google Scholar]
- 8.Imai S, Armstrong CM, Kaeberlein M, Guarente L. Transcriptional silencing and longevity protein Sir2 is an NAD-dependent histone deacetylase. Nature. 2000;403(6771):795–800. doi: 10.1038/35001622. [DOI] [PubMed] [Google Scholar]
- 9.Landry J, Sutton A, Tafrov ST, et al. The silencing protein SIR2 and its homologs are NAD-dependent protein deacetylases. Proc Natl Acad Sci U S A. 2000;97(11):5807–5811. doi: 10.1073/pnas.110148297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.North BJ, Marshall BL, Borra MT, Denu JM, Verdin E. The human Sir2 ortholog, SIRT2, is an NAD+-dependent tubulin deacetylase. Mol Cell. 2003;11(2):437–444. doi: 10.1016/s1097-2765(03)00038-8. [DOI] [PubMed] [Google Scholar]
- 11.Tanner KG, Landry J, Sternglanz R, Denu JM. Silent information regulator 2 family of NAD- dependent histone/protein deacetylases generates a unique product, 1-O-acetyl-ADP-ribose. Proc Natl Acad Sci U S A. 2000;97(26):14178–14182. doi: 10.1073/pnas.250422697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Haigis MC, Mostoslavsky R, Haigis KM, et al. SIRT4 inhibits glutamate dehydrogenase and opposes the effects of calorie restriction in pancreatic beta cells. Cell. 2006;126(5):941–954. doi: 10.1016/j.cell.2006.06.057. [DOI] [PubMed] [Google Scholar]
- 13.Liszt G, Ford E, Kurtev M, Guarente L. Mouse Sir2 homolog SIRT6 is a nuclear ADP-ribosyltransferase. J Biol Chem. 2005;280(22):21313–21320. doi: 10.1074/jbc.M413296200. [DOI] [PubMed] [Google Scholar]
- 14.Barber MF, Michishita-Kioi E, Xi Y, et al. SIRT7 links H3K18 deacetylation to maintenance of oncogenic transformation. Nature. 2012;487(7405):114–118. doi: 10.1038/nature11043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Michishita E, McCord RA, Boxer LD, et al. Cell cycle-dependent deacetylation of telomeric histone H3 lysine K56 by human SIRT6. Cell Cycle. 2009;8(16):2664–2666. doi: 10.4161/cc.8.16.9367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kumar S, Lombard DB. Functions of the sirtuin deacylase SIRT5 in normal physiology and pathobiology. Crit Rev Biochem Mol Biol. 2018;53(3):311–334. doi: 10.1080/10409238.2018.1458071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Du J, Zhou Y, Su X, et al. Sirt5 is a NAD-dependent protein lysine demalonylase and desuccinylase. Science. 2011;334(6057):806–809. doi: 10.1126/science.1207861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Park J, Chen Y, Tishkoff DX, et al. SIRT5-mediated lysine desuccinylation impacts diverse metabolic pathways. Mol Cell. 2013;50(6):919–930. doi: 10.1016/j.molcel.2013.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nishida Y, Rardin MJ, Carrico C, et al. SIRT5 regulates both cytosolic and mitochondrial protein malonylation with glycolysis as a major target. Mol Cell. 2015;59(2):321–332. doi: 10.1016/j.molcel.2015.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tan M, Peng C, Anderson KA, et al. Lysine glutarylation is a protein posttranslational modification regulated by SIRT5. Cell Metab. 2014;19(4):605–617. doi: 10.1016/j.cmet.2014.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li L, Shi L, Yang S, et al. SIRT7 is a histone desuccinylase that functionally links to chromatin compaction and genome stability. Nat Commun. 2016;7:12235. doi: 10.1038/ncomms12235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Houtkooper RH, Pirinen E, Auwerx J. Sirtuins as regulators of metabolism and healthspan. Nat Rev Mol Cell Biol. 2012;13(4):225–238. doi: 10.1038/nrm3293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bonda DJ, Lee HG, Camins A, et al. The sirtuin pathway in ageing and Alzheimer disease: mechanistic and therapeutic considerations. Lancet Neurol. 2011;10(3):275–279. doi: 10.1016/S1474-4422(11)70013-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hall JA, Dominy JE, Lee Y, Puigserver P. The sirtuin family’s role in aging and age-associated pathologies. J Clin Invest. 2013;123(3):973–979. doi: 10.1172/JCI64094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Borradaile NM, Pickering JG. NAD(+), sirtuins, and cardiovascular disease. Curr Pharm Des. 2009;15(1):110–117. doi: 10.2174/138161209787185742. [DOI] [PubMed] [Google Scholar]
- 26.Bosch-Presegué L, Vaquero A. The dual role of sirtuins in cancer. Genes Cancer. 2011;2(6):648–662. doi: 10.1177/1947601911417862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yuan H, Su L, Chen WY. The emerging and diverse roles of sirtuins in cancer: a clinical perspective. Onco Targets Ther. 2013;6:1399–1416. doi: 10.2147/OTT.S37750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Noh SJ, Baek HA, Park HS, et al. Expression of SIRT1 and cortactin is associated with progression of non-small cell lung cancer. Pathol Res Pract. 2013;209(6):365–370. doi: 10.1016/j.prp.2013.03.011. [DOI] [PubMed] [Google Scholar]
- 29.Zhang T, Rong N, Chen J, et al. SIRT1 expression is associated with the chemotherapy response and prognosis of patients with advanced NSCLC. PLoS One. 2013;8(11):e79162. doi: 10.1371/journal.pone.0079162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Grbesa I, Pajares MJ, Martínez-Terroba E, et al. Expression of sirtuin 1 and 2 is associated with poor prognosis in non-small cell lung cancer patients. PLoS One. 2015;10(4):e0124670. doi: 10.1371/journal.pone.0124670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li C, Wang L, Zheng L, et al. SIRT1 expression is associated with poor prognosis of lung adenocarcinoma. Onco Targets Ther. 2015;8:977–984. doi: 10.2147/OTT.S82378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gharabaghi MA. Diagnostic investigation of BIRC6 and SIRT1 protein expression level as potential prognostic biomarkers in patients with non-small cell lung cancer. Clin Respir J. 2018;12(2):633–638. doi: 10.1111/crj.12572. [DOI] [PubMed] [Google Scholar]
- 33.Lin SY, Peng F. Association of SIRT1 and HMGA1 expression in non-small cell lung cancer. Oncol Lett. 2016;11(1):782–788. doi: 10.3892/ol.2015.3914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang J, Wang C. Prognostic and predictive role of sirtuin1 expression in lung adenocarcinoma. Clin Lab. 2016;62(10):1989–1994. doi: 10.7754/Clin.Lab.2016.160317. [DOI] [PubMed] [Google Scholar]
- 35.Chen Y, Wang T, Wang W, et al. Prognostic and clinicopathological significance of SIRT1 expression in NSCLC: a meta-analysis. Oncotarget. 2017;8(37):62537–62544. doi: 10.18632/oncotarget.19244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li Z, Huang J, Yuan H, Chen Z, Luo Q, Lu S. SIRT2 inhibits non-small cell lung cancer cell growth through impairing Skp2-mediated p27 degradation. Oncotarget. 2016;7(14):18927–18939. doi: 10.18632/oncotarget.7816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yang GC, Fu BC, Zhang DY, et al. The expression and related clinical significance of SIRT3 in non-small-cell lung cancer. Dis Markers. 2017;1953;2017:824. doi: 10.1155/2017/8241953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xiong Y, Wang M, Zhao J, et al. SIRT3 is correlated with the malignancy of non-small cell lung cancer. Int J Oncol. 2017;50(3):903–910. doi: 10.3892/ijo.2017.3868. [DOI] [PubMed] [Google Scholar]
- 39.Lu W, Zuo Y, Feng Y, Zhang M. SIRT5 facilitates cancer cell growth and drug resistance in non-small cell lung cancer. Tumour Biol. 2014;35(11):10699–10705. doi: 10.1007/s13277-014-2372-4. [DOI] [PubMed] [Google Scholar]
- 40.Azuma Y, Yokobori T, Mogi A, et al. SIRT6 expression is associated with poor prognosis and chemosensitivity in patients with non-small cell lung cancer. J Surg Oncol. 2015;112(2):231–237. doi: 10.1002/jso.23975. [DOI] [PubMed] [Google Scholar]
- 41.Bai L, Lin G, Sun L, et al. Upregulation of SIRT6 predicts poor prognosis and promotes metastasis of non-small cell lung cancer via the ERK1/2/MMP9 pathway. Oncotarget. 2016;7(26):40377–40386. doi: 10.18632/oncotarget.9750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chen T, Sun Z, Liu F, Wang Q. RASSF1A and SIRT6 in non-small cell lung cancer: relationship with clinical outcome. Oncol Lett. 2017;14(5):5759–5764. doi: 10.3892/ol.2017.6172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Uhlen M, Oksvold P, Fagerberg L, et al. Towards a knowledge-based Human Protein Atlas. Nat Biotechnol. 2010;28(12):1248–1250. doi: 10.1038/nbt1210-1248. [DOI] [PubMed] [Google Scholar]
- 44.Li Z, Xie QR, Chen Z, Lu S, Xia W. Regulation of SIRT2 levels for human non-small cell lung cancer therapy. Lung Cancer. 2013;82(1):9–15. doi: 10.1016/j.lungcan.2013.05.013. [DOI] [PubMed] [Google Scholar]
- 45.Li L, Wei Y, To C, et al. Integrated omic analysis of lung cancer reveals metabolism proteome signatures with prognostic impact. Nat Commun. 2014;5:5469. doi: 10.1038/ncomms6469. [DOI] [PubMed] [Google Scholar]
- 46.Stewart PA, Parapatics K, Welsh EA, et al. A pilot proteogenomic study with data integration identifies MCT1 and GLUT1 as prognostic markers in lung adenocarcinoma. PLoS One. 2015;10(11):e0142162. doi: 10.1371/journal.pone.0142162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Xie M, Liu M, He CS. SIRT1 regulates endothelial Notch signaling in lung cancer. PLoS One. 2012;7(9):e45331. doi: 10.1371/journal.pone.0045331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhu L, Chiao CY, Enzer KG, Stankiewicz AJ, Faller DV, Dai Y. SIRT1 inactivation evokes antitumor activities in NSCLC through the tumor suppressor p27. Mol Cancer Res. 2015;13(1):41–49. doi: 10.1158/1541-7786.MCR-14-0239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lu J, Zhang M, Huang Z, et al. SIRT1 in B[a]P-induced lung tumorigenesis. Oncotarget. 2015;6(29):27113–27129. doi: 10.18632/oncotarget.4729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Huang H, Pan X, Jin H, et al. PHLPP2 downregulation contributes to lung carcinogenesis following B[a]P/B[a]PDE exposure. Clin Cancer Res. 2015;21(16):3783–3793. doi: 10.1158/1078-0432.CCR-14-2829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Xu W, Jiang K, Shen M, Qian Y, Peng Y. SIRT2 suppresses non-small cell lung cancer growth by targeting JMJD2A. Biol Chem. 2015;396(8):929–936. doi: 10.1515/hsz-2014-0284. [DOI] [PubMed] [Google Scholar]
- 52.Li H, Feng Z, Wu W, Li J, Zhang J, Xia T. SIRT3 regulates cell proliferation and apoptosis related to energy metabolism in non-small cell lung cancer cells through deacetylation of NMNAT2. Int J Oncol. 2013;43(5):1420–1430. doi: 10.3892/ijo.2013.2103. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 53.Han Z, Liu L, Liu Y, Li S. Sirtuin SIRT6 suppresses cell proliferation through inhibition of Twist1 expression in non-small cell lung cancer. Int J Clin Exp Pathol. 2014;7(8):4774–4781. eCollection 2014. [PMC free article] [PubMed] [Google Scholar]
- 54.Han L, Liang XH, Chen LX, Bao SM, Yan ZQ. SIRT1 is highly expressed in brain metastasis tissues of non-small cell lung cancer (NSCLC) and in positive regulation of NSCLC cell migration. Int J Clin Exp Pathol. 2013;6(11):2357–2365. eCollection 2013. [PMC free article] [PubMed] [Google Scholar]
- 55.Cheng D, Zhao L, Xu Y, et al. K-Ras promotes the non-small lung cancer cells survival by cooperating with sirtuin 1 and p27 under ROS stimulation. Tumour Biol. 2015;36(9):7221–7232. doi: 10.1007/s13277-015-3429-8. [DOI] [PubMed] [Google Scholar]
- 56.Luo J, Bao YC, Ji XX, Chen B, Deng QF, Zhou SW. SPOP promotes SIRT2 degradation and suppresses non-small cell lung cancer cell growth. Biochem Biophys Res Commun. 2017;483(2):880–884. doi: 10.1016/j.bbrc.2017.01.027. [DOI] [PubMed] [Google Scholar]
- 57.Xiao K, Jiang J, Wang W, et al. Sirt3 is a tumor suppressor in lung adenocarcinoma cells. Oncol Rep. 2013;30(3):1323–1328. doi: 10.3892/or.2013.2604. [DOI] [PubMed] [Google Scholar]
- 58.Xiong Y, Wang L, Wang S, et al. SIRT3 deacetylates and promotes degradation of P53 in PTEN-defective non-small cell lung cancer. J Cancer Res Clin Oncol. 2018;144(2):189–198. doi: 10.1007/s00432-017-2537-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kim EJ, Juhnn YS. Cyclic AMP signaling reduces sirtuin 6 expression in non-small cell lung cancer cells by promoting ubiquitin-proteasomal degradation via inhibition of the Raf-MEK-ERK (Raf/mitogen-activated extracellular signal-regulated kinase/extracellular signal-regulated kinase) pathway. J Biol Chem. 2015;290(15):9604–9613. doi: 10.1074/jbc.M114.633198. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1.
Underlying mechanism of sirtuins in NSCLC in the previous reports
| Study | Sirtuins | Mechanisms |
|---|---|---|
| Xie et al47 | SIRT1 | SIRT1 promote lung tumor growth through downregulation of DLL4/Notch signaling and deacetylation of N1IC |
| Zhang et al29 | SIRT1 | High SIRT1 was associated with chemotherapy resistance |
| Han et al54 | SIRT1 | SIRT1 promotes cell migration |
| Zhu et al48 | SIRT1 | SIRT1 inactivation evokes antitumor activities in NSCLC through the tumor suppressor p27 |
| Grbesa et al30 | SIRT1 | Downregulation of SIRT1 decreased cell proliferation |
| Cheng et al55 | SIRT1 | K-Ras promotes the survival of non–small cell lung cancer cells by cooperating with sirtuin 1 and p27 under ROS stimulation |
| Lu et al49 | SIRT1 | SIRT1 is involved in Benzo[a]pyrene B[a]P-induced transformation associated with activation of the TNF-α/β-catenin axis |
| Li et al44 | SIRT2 | SIRT2 inhibits cell proliferation, induces apoptosis and cell-cycle arrest by increasing the ROS production and p27 levels, and is associated with increased chemotherapy sensitivity |
| Grbesa et al30 | SIRT2 | Downregulation of SIRT2 decreased cell proliferation |
| Xu et al51 | SIRT2 | SIRT2 inhibits cell proliferation and tumor growth via downregulating JMJD2A |
| Li et al36 | SIRT2 | Downregulation of SIRT2 increases the level of SKP2 acetylation, then increases p27 ubiquitination, and finally enhances tumor cell proliferation |
| Luo et al56 | SIRT2 | SPOP promotes SIRT2 degradation and suppresses non–small cell lung cancer cell growth |
| Xiao et al57 | SIRT3 | SIRT3 increased the bax/bcl-2 and bad/bcl-x/L ratios, and promoted AIF translocation to the nucleus. SIRT3 overexpression upregulated p53 and p21 protein levels, and decreased intracellular ROS levels |
| Li et al52 | SIRT3 | Low expression of SIRT3 significantly inhibited mitotic entry, growth, and proliferation of NSCLC cell lines and promoted apoptosis, which was related to energy metabolism involved in the interaction between SIRT3 and NMNAT2 |
| Xiong et al38 | SIRT3 | SIRT3 increases Akt activation by post-transcriptional modification and, then, promotes NSCLC malignancy |
| Xiong et al58 | SIRT3 | SIRT3 deacetylates and promotes degradation of P53 in PTENdefective non–small cell lung cancer |
| Lu et al39 | SIRT5 | SIRT5 promotes cell proliferation and chemotherapy resistance by induction of Nrf2 expression |
| Azuma et al40 | SIRT6 | SIRT6 increases NFkappaB and Beclin1, and is associated with chemotherapy resistance |
| Han et al53 | SIRT6 | SIRT6 suppresses cell proliferation through inhibition of Twist1 expression |
| Kim and Juhnn59 | SIRT6 | Cyclic AMP signaling reduces SIRT6 expression in non–small cell lung cancer cells by promoting ubiquitin-proteasomal degradation via inhibition of the Raf–MEK–ERK pathway |
| Bai et al41 | SIRT6 | SIRT6 increases tumor migration and invasion via increasing ERK1/2 phosphorylation and MMP9 expression |
Abbreviations: SIRT1, sirtuin 1; SIRT2, sirtuin 2; SIRT3, sirtuin 3; SIRT4, sirtuin 4; SIRT5, sirtuin 5; SIRT6, sirtuin 6; SIRT7, sirtuin 7; NSCLC, non-small cell lung cancer; DLL4, Delta-like ligand 4; N1IC, Notch1 intracellular domain; JMJD2A, lysine demethylase 4A; Skp2, S-phase kinase associated protein 2; SPOP, speckle type BTB/POZ protein; BAX, Bcl-2 associated X; BAD, Bcl-2 associated agonist of cell death; Bcl-x/L, Bcl-2 like 1; AIF, apoptosis inducing factor; NMNAT2, nicotinamide nucleotide adenylyltransferase 2; PTEN, phosphatase and tensin homolog; Nrf2, nuclear factor erythroid 2 like 2; Twist1, twist family bHLH transcription factor 1; MMP9, matrix metallopeptidase 9.