Abstract
Background
The prognostic value of EGFR and KRAS mutations in resected non-small cell lung cancer (NSCLC) has been reported. However, conflicting results were reported in these studies. The effect of mutations in these two genes in resected NSCLC remains controversial.
Methods
We searched Internet databases for studies reporting disease-free survival (DFS) and overall survival (OS) in resected NSCLC patients with EGFR or KRAS mutations. A meta-analysis calculating the pooled hazard ratio (HR) for DFS and OS was used to measure the association of EGFR or KRAS mutations with the prognosis of patients after surgery.
Results
A total of 9,635 patients from 32 studies were included in this analysis. The combined HR for EGFR mutations on DFS was 0.77 (95% CI 0.66–0.90, p=0.001) and on OS was 0.72 (95% CI 0.66–0.80, p<0.00001). In addition, the combined HR for KRAS mutations on DFS was 1.5 (95% CI 1.15–1.96, p=0.002) and on OS was 1.49 (95% CI 1.28–1.73, p<0.00001). Sensitivity analysis, subgroup analysis, and bias analysis proved the stability of the results.
Conclusion
The analysis showed that EGFR mutations were significantly associated with DFS and OS. These findings indicated that surgically treated NSCLC patients with EGFR mutations were inclined to exhibit a prolonged DFS and OS. In addition, the results indicated that KRAS mutations predicted worse DFS and OS in patients with resected NSCLC.
Keywords: EGFR mutations, KRAS mutations, meta-analysis, non-small cell lung cancer, prognosis, resected
Introduction
Lung cancer is one of the most common malignancies in the world and the main cause of cancer-related death.1 Lung cancer is generally classified into small cell lung cancer and non-small cell lung cancer (NSCLC) according to its pathology and treatment, and NSCLC accounts for more than 80% of all lung cancer cases.2 Although the treatment for NSCLC has made great strides, the 5-year survival rate is only approximately 15%.1 The principal treatments for NSCLC are surgery, chemotherapy, radiotherapy, and targeted drug therapy. Among these treatments, surgery is recognized as the most efficient treatment, but relapse after surgical treatment occurs in 20–50% of all cases, and the prognosis remains elusive.3–6
Numerous studies have reported prognostic factors that could predict survival and recurrence of NSCLC. Lee et al7 discovered that gene mutations can sensibly predict postoperative recurrence. Many mutant genes in NSCLC have been identified, including KRAS, EGFR, HER2, and FGFR1.8 Among them, the most well-studied mutant genes are EGFR and KRAS.
EGFR is a stimulatory factor and driving gene in NSCLC. EGFR mutations lead to abnormal activation of receptors and downstream molecules in the absence of ligands. EGFR mutations promote tumorigenesis by increasing cell proliferation and reducing cell apoptosis, angiogenesis, and metastasis.9 The discovery of EGFR has led to a completely new phase of systemic treatment of NSCLC. Identification of mutations in NSCLC molecular pathways and the continuous improvement in genetic testing methods in clinical research have prompted the individualized treatment trend in NSCLC. EGFR mutations are the predicting factor for EGFR-TKI.10 However, the predictive value of EGFR mutations on postoperative survival and recurrence of resected NSCLC remains unclear. The results of studies about the prognostic impact of EGFR mutations in resected NSCLC are inconsistent. Kim et al11 suggested that EGFR is not a prognostic factor for resected NSCLC, whereas Ma et al12 suggested that EGFR mutations seem to be more likely a predictive marker for EGFR-TKI treatment than a prognostic marker for overall survival (OS). However, the study by Izar et al13 demonstrated that EGFR mutations are positive prognostic markers in completely resected stage I NSCLC.
KRAS is involved in several solid tumors, including colorectal cancer and NSCLC. KRAS is a signal transducer downstream of tyrosine kinase receptors including EGFR, which is a complex signaling cascade involved in the development of cancer. Mutated KRAS can activate this pathway automatically and initiate transduction of downstream signals in the absence of EGFR signaling to allow NSCLC to further develop. Mutated KRAS also renders the EGFR-targeted drug in upstream of tyrosine kinase receptors ineffective.14 It is generally believed that KRAS mutations are contraindications to the use of anti-EGFR antibody therapy in colorectal cancer,15 but the effect of these mutations in NSCLC is unclear. The prognostic value of KRAS mutations in NSCLC in each study is inconsistent, and the considerable heterogeneity is noted among studies. Kadota et al16 studied the effect of KRAS mutations on the prognosis of 129 NSCLC patients undergoing surgical resection. The results showed that the 5-year OS of KRAS-mutated NSCLC patients was significantly reduced compared with that of wild-type KRAS patients, and the relapse rate of patients with KRAS mutations increased. However, in a retrospective study17 assessing KRAS mutations in postoperative NSCLC, the results revealed no significant difference between recurrence-free survival (RFS) and OS in patients with KRAS mutations and wild-type KRAS. Some studies suggest that KRAS mutations are prognostic factors of NSCLC, whereas other studies demonstrate no relationship between KRAS mutations and NSCLC patient survival. In addition, a meta-analysis assessing KRAS mutations in the surgical treatment of NSCLC has not been reported to date. Although EGFR and KRAS are hotspot studies on NSCLC, their real prognostic value in resected NSCLC remains unknown. To elucidate the prognostic significance, we performed meta-analysis to explain the prognostic value of EGFR and KRAS mutations in resected NSCLC patients.
Methods
Search strategy and selection criteria
We searched PubMed, the Cochrane Library, and Web of Science as well as the references of included studies. The literature search was completed in July 2017. The articles must meet the following criteria for inclusion in our study: 1) all patients were pathologically confirmed to have NSCLC; 2) all patients underwent complete excision operations; 3) all patients harbored EFGR or KRAS mutations; and 4) the hazard ratio (HR) of disease-free survival (DFS) and OS is reported in the article or can be calculated from the relevant parameters. If the same researcher reported the results of the same patient population, we used most recent study or the study for which the data were most complete.
Quality assessment of articles
We used the European Lung Cancer Working Group (ELCWP) Quality Scale used by Steels et al18 to ensure the quality of the included studies. There were scientific design, laboratory methods, reproducibility, and result analysis in the list, and also there were some specific items in each category. Maximum of 2 points awarded in each item. One point was given for an incomplete or unclear description, and an item that was not defined was given 0 point. Then, we employed SPSS (www.spss.com) analysis to ensure the accuracy of the score.
Data extraction and summary effect analysis
The main data we extracted from the literature included the following: first author, year of publication, source of patients, number of patients, stage, EGFR mutation rate, KRAS mutation rate, KRAS mutation state, EGFR mutation state, detection method, and HR. We set DFS as the first end point and OS served as the second end point. A p<0.05 indicated that the result was statistically significant. The analysis utilized Review Manager 5.3 (http://community.cochrane.org/help/tools-and-software/revman-5) and stata12 (https://www.stata.com/). The results were combined with p-values for HR. The fixed-effect model (I2<50%) and the random-effect model (I2≥50%) were chosen based on heterogeneity. We used sensitivity analysis to identify studies that caused heterogeneity. Subgroup analysis was used to further explore the source of heterogeneity. The grouping was based on statistical methods of the study, NSCLC stages of patients, the detection methods of gene mutations, and the population origin of samples. We used Begg’s test and Egger’s test to explore publication bias among the items that included more than 10 studies.
Results
Selection of studies
A total of 2,501 potential studies were defined, and 2,463 studies were excluded after screening. Moreover, the full texts of 38 articles were intensively scrutinized and five studies were excluded due to incomplete data. Finally, 33 studies11,13,16,17,19–47 fulfilling all of the inclusion criteria were eligible for meta-analysis. Figure 1 shows the flowchart of the search results.
Figure 1.
Flowchart of publication search and selection.
Abbreviation: NSCLC, non-small cell lung cancer.
Study description and quality assessment
The total number of NSCLC patients was 10,869, including 3,651 harboring EGFR mutations and 1,687 harboring KRAS mutations. The EGFR mutation rate was 9.6–82.2%, and the KRAS mutation rate was 3.5–75.2%. We concluded that the average mutation frequency of EGFR in Asian populations (43.5%) was higher than that in other races (37.9%), whereas the average frequency of KRAS mutations in Asian populations (12.7%) was much lower than that in other races (46.1%). The main mutation site of EGFR involves exons 18–21, and the main mutation site of KRAS is exon 2. Among these studies, three studies28,36,45 mentioned other KRAS mutation sites (exons 3 and 6). Table 1 presents the primary characteristics of these included studies.
Table 1.
Characteristics of the included studies
| Reference | Year | Source | Patients (N) | Stage | Mutation number (%) | Mutation type (locus/exon) | Gene testing method | Statistical methods |
|---|---|---|---|---|---|---|---|---|
| Na et al19 | 2007 | Korea | 133 | I–III | EGFR 32 (24) | EGFR (18–21) | SEQ | Univariate |
| Suehisa et al20 | 2007 | Japan | 187 | I–IIIA | EGFR 79 (43) | EGFR (19, 21) | PCR, SEQ | Multivariate |
| Marks et al21 | 2008 | USA | 296 | I–III | EGFR 40 (13.6), KRAS 50 (17) | EGFR (18–21), KRAS (2) | PCR, SEQ | Multivariate |
| Kobayashi et al22 | 2008 | Japan | 127 | I | EGFR 64 (50.4) | EGFR (19, 21) | Mutant-enriched PCR | Multivariate |
| Woo et al24 | 2009 | Japan | 190 | I | KRAS 24 (12.6) | KRAS (2) | PCR | Multivariate |
| Hosokawa et al23 | 2009 | Japan | 93 | I–III | EGFR 37 (40) | EGFR (18–21) | PCR, SEQ | Univariate |
| Lee et al25 | 2009 | Korea | 117 | I–IIIA | EGFR 48 (41.8) | EGFR (18–21) | Nested PCR | Univariate |
| Galleges Ruiz et al26 | 2009 | The Netherlands | 178 | I–III | KRAS 25 (14) | No data | Nested PCR | Multivariate |
| Kosaka et al27 | 2009 | Japan | 397 | I–IV | EGFR 196 (49), KRAS 142 (38) | EGFR (18–21), KRAS (2) | PCR, SEQ | Multivariate |
| Liu et al28 | 2010 | Taiwan (China) | 164 | I–IIIA | EGFR 52 (31.7), KRAS 7 (4.3) | EGFR (18–21), KRAS (2, 3) | SEQ | Multivariate |
| Tsao et al29 | 2011 | Canada | 436 | IB–II | EGFR 27 (12.2) | EGFR (19, 21) | ARMS | Multivariate |
| D’Angelo et al30 | 2012 | USA | 1,118 | I–III | EGFR 896 (80.1), KRAS 841 (75.2) | EGFR (19, 21), KRAS (2) | PCR, SEQ | Multivariate |
| Kim et al31 | 2012 | Korea | 229 | I–IV | EGFR 110 (48), KRAS 8 (3.5) | EGFR (18–21), KRAS (2) | Nested PCR | Multivariate |
| Scoccianti et al32 | 2012 | Europe (multinational) | 152 | I–IV | EGFR 18 (11.8) | EGFR (18–21) | PCR, SEQ | Multivariate |
| 249 | I–IV | KRAS 46 (18.5) | KRAS (2) | Mutant-enriched PCR, SEQ | Multivariate | |||
| Sonobe et al33 | 2012 | Japan | 180 | I–III | EGFR 148 (82.2) | EGFR (18–21) | RT-PCR | Multivariate |
| Izar et al13 | 2013 | USA | 307 | I | EGFR 62 (19.9), KRAS 127 (40.7) | EGFR (18–21), KRAS (2) | SEQ, multiplex PCR | Multivariate |
| Sun et al35 | 2013 | China | 150 | IIIA | EGFR 43 (28.7) | EGFR (19, 21) | RT-PCR | Multivariate |
| Maki et al34 | 2013 | Japan | 105 | IA | EGFR 51 (49) | No data | PCR, SEQ | Univariate |
| Kim et al11 | 2013 | Korea | 863 | IB–IIA | EGFR 354 (41) | EGFR (18–21) | PCR, SEQ | Univariate |
| Ragusa et al36 | 2014 | Italy | 230 | I–III | EGFR 22 (9.6), KRAS 39 (16.9) | EGFR (18–21), KRAS (2, 3) | Nested PCR | Univariate |
| Ohba et al37 | 2014 | Japan | 354 | I | EGFR 122 (34.4) | EGFR (19, 21) | PCR, SEQ (EGFR)/RFLP (KRAS) | Univariate |
| Liu et al38 | 2014 | China | 131 | I–IIIA | EGFR 58 (44.3) | EGFR (18–21) | Nested PCR | Multivariate |
| Ayyoub et al39 | 2014 | Spain | 216 | I–III | EGFR 21 (9.7), KRAS 29 (3.4) | EGFR (18–21), KRAS (2) | RT-PCR | Univariate |
| Kudo et al40 | 2015 | Japan | 198 | I–III | EGFR 57 (28.7) | EGFR (18, 19, 21) | PCR, SEQ | Univariate |
| Nadal et al41 | 2015 | USA | 179 | I–IV | KRAS 85 (47.5) | KRAS (2) | PCR, SEQ | Multivariate |
| Nishii et al42 | 2017 | Japan | 388 | I | EGFR 185 (47.7) | EGFR (19–21) | PCR | Multivariate |
| Kadota et al16 | 2016 | USA | 378 | I | EGFR 85 (22.5) | EGFR (18–21) | Sequenom | Univariate |
| 482 | I–II | KRAS 129 (27) | KRAS (2) | PCR | Multivariate | |||
| Isaka et al43 | 2016 | Japan | 202 | II–IIIA | EGFR 100 (49.5) | EGFR (19, 21) PCR | Multivariate | |
| Zheng et al17 | 2016 | China | 1,368 | I–IV | KRAS 118 (8.26) | KRAS (2) | RT-PCR, SEQ | Multivariate |
| Kaseda et al44 | 2017 | Japan | 162 | I | EGFR 81 (50), KRAS 17 (10.5) | EGFR (19, 21), KRAS (2) | PCR | Univariate |
| Sullivan et al45 | 2017 | USA | 131 | IA–IIB | No data | KRAS (6) | RT-PCR | Multivariate |
| Takamochi et al47 | 2017 | Japan | 939 | I–IV | EGFR 418 (44.5) | EGFR (18, 19, 21) | PCR | Multivariate |
| Yotsukura et al46 | 2017 | Japan | 369 | I–II | EGFR 160 (46.9) | EGFR (19, 21) | PCR | Multivariate |
Abbreviations: ARMS, amplification refractory mutation system; PCR, polymerase chain reaction; RFLP, restriction fragment length polymorphism; RT, reverse transcription; SEQ, sequencing.
The results of our quality assessment are presented in Table S1. We removed some items that were not suitable for our study. Studies with 20 or more points out of 38 points qualified for inclusion. The overall score of 31 studies was between 21 and 30, and the median score was 27 points. No significant difference (p=0.605>0.05) was noted between Asian and non-Asian studies, which is revealed in Table 2. The scores of studies that exclusively focused on EGFR or KRAS did not differ significantly from studies that researched both EGFR and KRAS (p=0.78>0.05). The included the studies because the scores indicated that the quality of those studies met our standards.
Table 2.
Statistical characteristics of quality assessment score
| Number of studies | Median score | Average score | Difference test (p-value) | |
|---|---|---|---|---|
| All studies | 33 | 26 | 26.1 | – |
| Asian | 22 | 26 | 25.95 | 0.605 |
| Non-Asian | 11 | 27 | 26.5 | |
| Onlya | 22 | 26.5 | 26.3 | 0.78 |
| Bothb | 11 | 26 | 26.4 |
Notes:
Studies of EGFR or KRAS.
Studies of both EGFR and KRAS.
Predictive value of EGFR mutations
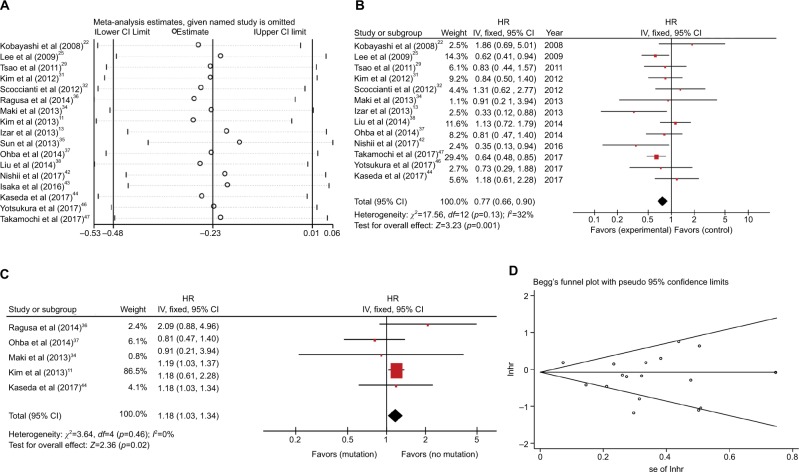
DFS Seventeen studies with 5,261 patients assessed the relationship between EGFR mutations and DFS,11,13,22,25,29,31,32,34–38,42–44,46,47 and six studies demonstrated that EGFR mutations positively influenced the DFS of resected NSCLC patients.13,25,35,42,43,47 Significant heterogeneity was observed between these studies (I2=72%, p<0.00001; Figure S1). We used sensitivity analysis to explore the sources of heterogeneity (Figure 2A). We identified four studies that may lead to heterogeneity.11,35,36,43 No obvious heterogeneity was noted among the studies after excluding four studies (I2=32%, p=0.13). The remaining 13 studies were subject to meta-analysis using fixed-effect model, and the combined HR was 0.77 (95% CI 0.66–0.90, p=0.001; Figure 2B). The results suggest that the effect of EGFR mutations on DFS is statistically significant and that EGFR mutations are prognostic factors for relapse in resected NSCLC patients.
Figure 2.
(A) Sensitivity analysis for combined HR of EGFR on DFS. (B) Fixed-effect model forest plot of DFS of EGFR mutations after removing the studies that caused the heterogeneity. (C) Fixed-effect model forest plot of DFS in univariate analysis subgroup according to EGFR mutation. (D) Begg’s funnel plot of enrolled studies for DFS of EGFR.
Abbreviations: DFS, disease-free survival; HR, hazard ratio; lnhr, logarithm of HR; IV, inverse variance; se, standard error.
Subgroup analysis was used to further explore heterogeneity. We considered the heterogeneity of stage, statistical methods, and source of study based on the four studies previ ously identified (Figure S2A–C). In the subgroup analysis, heterogeneity remained relatively large, and the value of I2 ranged from 53% to 75%. Among the subgroups, we found that the univariate analysis subgroup which included five studies that exhibited no significant heterogeneity revealed negative influence of EGFR mutations on DFS (HR 1.18, 95% CI 1.03–1.34, p=0.03; Figure 2C). In contrast, the multivariate analysis subgroup revealed an opposite result (HR 0.68, 95% CI 0.52–0.88, p=0.004; Figure S1B). Moreover, many studies11,25,30,31,37 have shown that the clinical impact of EGFR-TKIs cannot be ignored in EGFR-mutant patients. The data from the studies were divided into the EGFR-TKI subgroup and the no EGFR-TKI subgroup to verify the effects. The results revealed that significant heterogeneity remained in the EGFR-TKI group (EGFR-TKI: I2=72%, p=0.01; no EGFR-TKI: I2=65%, p=0.0007; Figure S2D). Moreover, we conducted bias analysis using funnel plot (Figure 2D), Begg’s test (p=0.537), and Egger’s test (p=0.116; Figure S3). No significant publication bias was observed in the studies.
OS
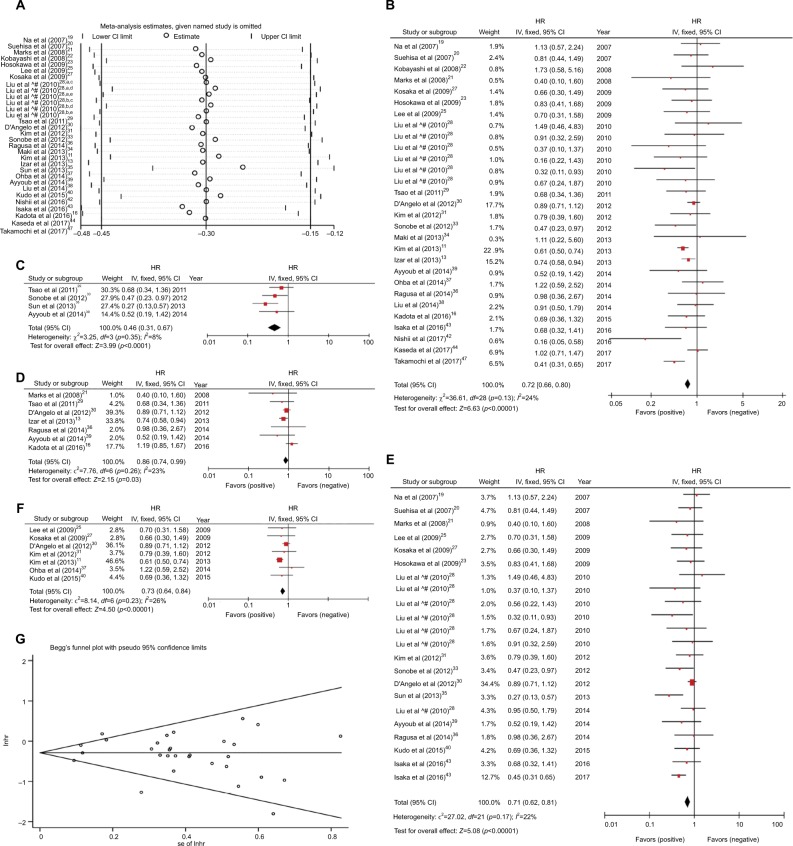
The relationship between EGFR mutations and OS was evaluated based on 26 studies,11,13,16,19–23,25,27–31,33–40,42–44,47 with 8,100 patients, and seven studies11,13,28,33,35,42,47 indicated that EGFR mutations were a favorable prognostic factor for OS in resected NSCLC patients. Some heterogeneity was noted between the studies (I2=42%, p=0.008; Figure S4A). We conducted a sensitivity analysis to identify the source of heterogeneity (Figure 3A). We identified that two studies16,35 that may cause heterogeneity; however, no significant heterogeneity was noted among the studies after excluding these two studies (I2=24%, p=0.13, HR 0.72, 95% CI 0.66–0.80, p<0.00001; Figure 3B). The pooled analysis indicated a better OS for NSCLC patients with EGFR mutations.
Figure 3.
(A) Sensitivity analysis for combined HR of EGFR on OS. aAll patients; bfive patients with no data at the EGFR mutation variable, and 18 patients who had received TKI treatment for tumor recurrence were not included; cmutation site: L858R; dmutation site: 19 Del; emutation site: others. (B) Fixed-effect model forest plot of OS of EGFR mutations after removing two studies that caused the heterogeneity. (C) Fixed-effect model forest plot of OS of EGFR mutations in RT-PCR subgroup (detection methods group). (D) Fixed-effect model forest plot of OS of EGFR mutations in other subgroups (research sources group). (E) Fixed-effect model forest plot of OS of EGFR mutations in other subgroups (stage group). (F) Fixed-effect model forest plot of OS of EGFR mutations in the EGFR-TKIs subgroup. (G) Begg’s funnel plot of enrolled studies for OS of EGFR.
Abbreviations: HR, hazard ratio; IV, inverse variance; lnhr, logarithm of HR; OS, overall survival; PCR, polymerase chain reaction; RT, reverse transcription; se, standard error.
We used subgroup analysis to continue to explore heterogeneity. We divided the study into different subgroups based on detection method, statistical analysis method, research source, pathological stage, and EGFR-TKIs (Figure S4B–F). Among them, the real-time polymerase chain reaction subgroup of the detection method group (I2=8%, p=0.35, HR 0.46, 95% CI 0.31–0.67, p<0.0001), the other subgroup of the research source group (I2=23, p=0.26, HR 0.86, 95% CI 0.74–0.99, p=0.03), the other subgroup of the stage group (I2=22, p=0.17, HR 0.71, 95% CI 0.62–0.81, p<0.00001), and the EGFR-TKI subgroup (I2=26%, p=0.23, HR 0.73, 95% CI 0.64–0.84, p<0.00001) exhibited no significant heterogeneity (Figure 3C–F). The results of these groups indicated that the EGFR mutation is a benign prognostic factor for OS. Sig nificant heterogeneity was noted in the early subgroup of the pathological stage group (I2=66%, p=0.02). Minimal heterogeneity existed in the sequencing subgroup of the detection method group (I2=38%, p=0.02), the analysis method group (multivariate: I2=38%, p=0.04, univariate: I2=49%, p=0.04), and the Asian subgroup of the source group (I2=38%, p=0.03). The multivariate analysis subgroup demonstrated that EGFR mutations had a positive effect on the OS, and the univariate analysis subgroup revealed no significant association between EGFR mutations and OS (Figure S4C). The Asian subgroup results indicated that EGFR mutations were benign factors of OS, and the other subgroup revealed that EGFR mutations did not significantly influence OS (Figure S4D). The results of the EGFR-TKI group revealed no significant heterogeneity in the EGFR-TKI subgroup (I2=26%, p=0.23), but heterogeneity was noted in the EGFR-TKI subgroup (I2=47%, p=0.006; Figure S4F). When the prognostic value of EGFR mutations is estimated, different analysis methods and different ethnic groups may influence the outcome of the study. No significant publication bias was observed in the funnel plot (Figure 3G), Begg’s test (p=0.175), and Egger’s test (p=0.595; Figure S5).
Predictive value of KRAS mutations
DFS
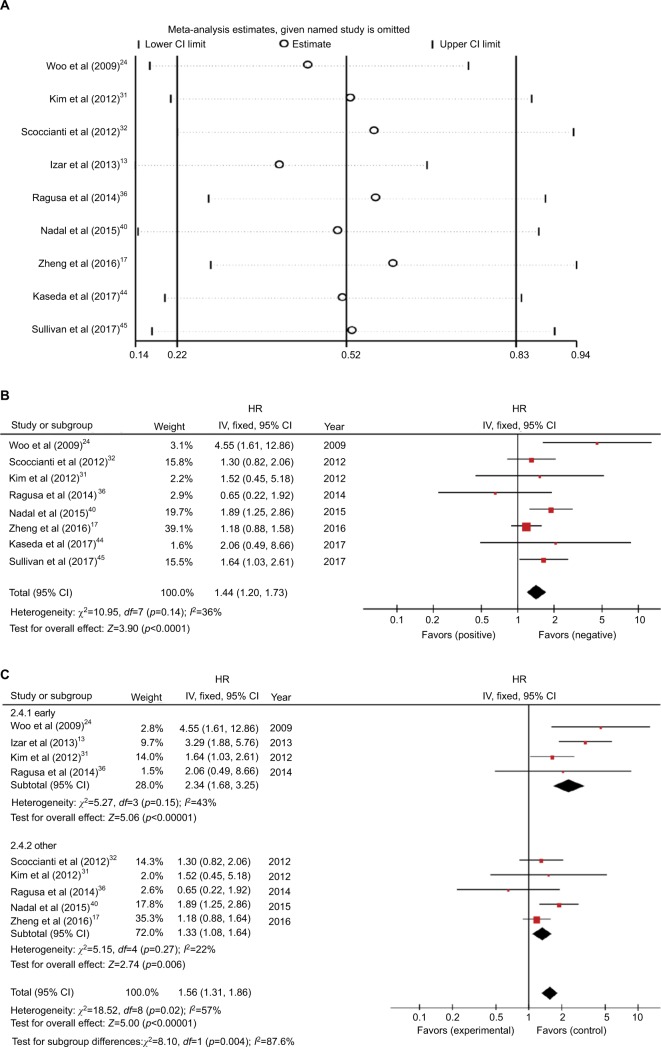
Nine studies with 3,045 patients were used to explain the relationship between EGFR mutations and DFS.13,17,24,31,32,36,41,44,45 Four studies demonstrated that KRAS mutations were not beneficial for recurrence of resected NSCLC patients.13,24,41,45 Significant heterogeneity was noted between the studies (I2=57%, p=0.02; Figure S6A). We found that one article13 was a source of heterogeneity based on sensitivity analysis (Figure 4A). Heterogeneity was significantly reduced after removing this article (I2=36%, p=0.14; Figure 4B). The merged HR was 1.5 (95% CI 1.15–1.96, p=0.002) based on fixed-effect model. The result indicated that KRAS mutations were a negative factor for DFS.
Figure 4.
(A) Sensitivity analysis for combined HR of KRAS on DFS. (B) Fixed-effect model forest plot of DFS of KRAS mutations after removing the study that caused the heterogeneity. (C) Random-effect model forest plot of DFS of KRAS mutations in stage subgroup analysis according to the patient’s pathological staging.
Abbreviations: DFS, disease-free survival; HR, hazard ratio; IV, inverse variance.
We grouped the studies based on pathological stage, research sources, and statistical methods to further explore the sources of heterogeneity (Figures 4C and S6B and C). No significant heterogeneity was noted in the early subgroup and other subgroups of the stage group in the subgroup analysis (Figure 4C). This finding indicated that data from patients with different pathological stages may generate heterogeneity.
OS
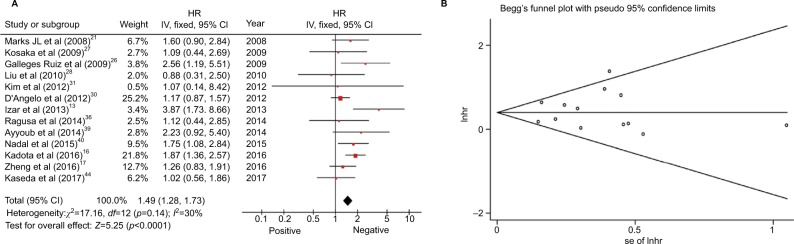
Thirteen studies with 5,326 patients were based on the connection between KRAS mutations and OS of resected NSCLC.13,16,17,21,26–28,30,31,36,39,41,44 Four of these studies indicated that KRAS mutations represented a risk factor for resected NSCLC.13,16,26,41 Significant heterogeneity was not noted in the studies (I2=30%, p=0.14). The overall HR was 1.49 (95% CI 1.28–1.73, p<0.00001; Figure 5A). The outcome indicated that patients with KRAS mutations exhibited shorter OS. No significant publication bias was observed in the funnel plot (Figure 5B), Begg’s test (p=1), and Egger’s test (p=0.74; Figure S7).
Figure 5.
(A) Random-effect model forest plot of OS of KRAS mutations. (B) Begg’s funnel plot of enrolled studies for OS of KRAS.
Abbreviations: HR, hazard ratio; IV, inverse variance; lnhr, logarithm of HR; OS, overall survival; se, standard error.
Discussion
Surgery is an effective method to treat patients with NSCLC. Both EGFR and KRAS are driver genes of NSCLC.8 Most studies suggest that EGFR and KRAS mutations are often mutually exclusive.48–51 Some clinical studies have reported that KRAS mutations can appear in patients with EGFR mutations, but the incidence of double mutations is <1%.52 Therefore, the simultaneous detection of EGFR and KRAS mutations is significant in guiding the individualized treatment of NSCLC patients. We assessed the prognostic signifi-cance of EGFR and KRAS mutations in postoperative NSCLC patients using meta-analysis that collect large amounts of data. Our meta-analysis reviewed thoroughly and released the latest data. Low heterogeneity was noted in this study, and no publication bias was found.
The results indicated that EGFR mutations not only extend the DFS of resected NSCLC but also contribute to the OS of patients. Zhang et al53 reported opposite conclusion demonstrating that the EGFR mutations were unrelated to the OS (HR 0.84, 95% CI 0.34–2.06, p=0.12) and DFS (HR 0.96, 95% CI 0.79–1.16, p=0.65). The cause involves deviations from the included studies or the differences in HRs from the survival curves. In this study, the meta results of EGFR DFS became statistically significant after excluding the four studies that caused heterogeneity. On the one hand, the great heterogeneity among studies may cause the results to be inaccurate. On the other hand, the excluded data also caused the results to change. Significant heterogeneity was discovered in the DFS studies. We identified four studies as the sources, but the heterogeneity was not resolved after subgroup analysis based on these studies. It is likely that we cannot accurately group based on some elements, such as gender and smoking. Moreover, the outcome of the multivariate subgroup was in complete opposition to the univariate subgroup. This finding suggests that different statistical approaches may affect the judgment of EGFR mutations in DFS. Therefore, we should carefully consider this point in subsequent research. The heterogeneity between OS study groups has not been accurately resolved. Subgroup analysis of OS revealed the difference between Asian and non-Asian studies. Heterogeneous results may be attributed to the fact that EGFR mutations play a different role in different races, so we need to consider this difference in the development of comprehensive treatment strategies. In addition, the statistical method group revealed that different statistical methods may affect the influence of EGFR mutations on OS. The results of the two subgroups of the stage group are also different, but this difference is likely caused by the great heterogeneity of the early subgroup. Moreover, we cannot consider the EGFR-TKIs as a source of heterogeneity of both DFS and OS according to the subgroup analysis of EGFR-TKIs. The reason may be that our existing data are not sufficiently comprehensive; we cannot completely separate patients who receive EGFR-TKI therapy from all patients. We are unable to conduct more rigorous analysis.
KRAS mutations are a negative factor of DFS and OS in patients with postoperative NSCLC. The meta-analysis revealed that the resected NSCLC patients with KRAS mutations exhibited reduced DFS and OS. This finding indicates that KRAS is an important indicator of the prognosis of patients with NSCLC. The DFS subgroup analysis expresses a difference of DFS between patients from different sources. This difference is likely because non-Asian patients are more affected by KRAS mutations than Asian patients. In addition, in the subgroup analysis of DFS, both the research sources group and the statistical methods group exhibited significant heterogeneity. On the one hand, this finding may indicate that neither of these two factors represent the source of DFS heterogeneity. On the other hand, given the relatively limited number of DFS studies, data from one study will lead to significant fluctuation of heterogeneity.
Despite all our efforts to provide accurate and comprehensive analysis, the meta-analysis still has some limitations. First, we did not conduct subgroup analyses due to insufficient data on age, gender, and smoking status to provide additional results. Second, we did not distinguish between patients who only underwent surgery or were subject to other treatments after surgical resection, which could result in bias. Moreover, additional and more complex studies based on different EGFR and KRAS mutation sites were not included. In future studies, these studies can be included in the analysis to provide more data available.
Despite these limitations, the meta-analysis revealed that EGFR mutations were associated with better OS and DFS in resected NSCLC patients. Patients with EGFR mutations who undergo surgical treatment exhibit an improved long-term prognosis for DFS and OS. KRAS mutations in NSCLC patients who undergo surgery predict worse DFS and OS.
Conclusion
Our meta-analysis found that EGFR mutations were associated with better DFS and OS, and EGFR mutations were a benign prognostic factor for DFS and OS of resected NSCLC. In addition, KRAS mutations indicate worse DFS and OS in resected NSCLC. The KRAS mutation is a poor prognostic factor for DFS and OS in patients with NSCLC after surgery.
Acknowledgments
This project was supported by the National Natural Science Foundation of China (grant number 61372151), the National Science Foundation for Young Scientists of China (grant number 81602483), the Fundamental Research Funds for the Central Universities (grant number xjj2017121), and the China Postdoctoral Science Foundation (grant numbers 2016M602798 and 2017T100737).
Footnotes
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2017. CA Cancer J Clin. 2017;67(1):7–30. doi: 10.3322/caac.21387. [DOI] [PubMed] [Google Scholar]
- 2.Lauro S, Onesti CE, Righini R, Marchetti P. The use of bevacizumab in non-small cell lung cancer: an update. Anticancer Res. 2014;34(4):1537–1545. [PubMed] [Google Scholar]
- 3.He B, Lu C, Zheng G, et al. Combination therapeutics in complex diseases. J Cell Mol Med. 2016;20(12):2231–2240. doi: 10.1111/jcmm.12930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Manser R, Wright G, Hart D, Byrnes G, Campbell DA. Surgery for early stage non-small cell lung cancer. Cochrane Database Syst Rev. 2005;1:CD004699. doi: 10.1002/14651858.CD004699.pub2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Saisho S, Yasuda K, Maeda A, et al. Post-recurrence survival of patients with non-small-cell lung cancer after curative resection with or without induction/adjuvant chemotherapy. Interact Cardiovasc Thorac Surg. 2013;16(2):166–172. doi: 10.1093/icvts/ivs450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Takenaka T, Takenoyama M, Yamaguchi M, et al. Impact of the epidermal growth factor receptor mutation status on the post-recurrence survival of patients with surgically resected non-small-cell lung cancer. Eur J Cardiothorac Surg. 2015;47(3):550–555. doi: 10.1093/ejcts/ezu227. [DOI] [PubMed] [Google Scholar]
- 7.Lee ES, Son DS, Kim SH, et al. Prediction of recurrence-free survival in postoperative non-small cell lung cancer patients by using an integrated model of clinical information and gene expression. Clin Cancer Res. 2008;14(22):7397–7404. doi: 10.1158/1078-0432.CCR-07-4937. [DOI] [PubMed] [Google Scholar]
- 8.Zhu QG, Zhang SM, Ding XX, He B, Zhang HQ. Driver genes in non-small cell lung cancer: characteristics, detection methods, and targeted therapies. Oncotarget. 2017;8(34):57680–57692. doi: 10.18632/oncotarget.17016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ciardiello F, Tortora G. EGFR antagonists in cancer treatment. N Engl J Med. 2008;358(11):1160–1174. doi: 10.1056/NEJMra0707704. [DOI] [PubMed] [Google Scholar]
- 10.Han SW, Kim TY, Hwang PG, et al. Predictive and prognostic impact of epidermal growth factor receptor mutation in non-small-cell lung cancer patients treated with gefitinib. J Clin Oncol. 2005;23(11):2493–2501. doi: 10.1200/JCO.2005.01.388. [DOI] [PubMed] [Google Scholar]
- 11.Kim YT, Seong YW, Jung YJ, et al. The presence of mutations in epidermal growth factor receptor gene is not a prognostic factor for long-term outcome after surgical resection of non-small-cell lung cancer. J Thorac Oncol. 2013;8(2):171–178. doi: 10.1097/JTO.0b013e318277a3bb. [DOI] [PubMed] [Google Scholar]
- 12.Ma L, Qiu B, Zhang J, et al. Survival and prognostic factors of non-small cell lung cancer patients with postoperative locoregional recurrence treated with radical radiotherapy. Chin J Cancer. 2017;36(1):93. doi: 10.1186/s40880-017-0261-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Izar B, Sequist L, Lee M, et al. The impact of EGFR mutation status on outcomes in patients with resected stage I non-small cell lung cancers. Ann Thorac Surg. 2013;96(3):962–968. doi: 10.1016/j.athoracsur.2013.05.091. [DOI] [PubMed] [Google Scholar]
- 14.Soulieres D, Greer W, Magliocco AM, et al. KRAS mutation testing in the treatment of metastatic colorectal cancer with anti-EGFR therapies. Curr Oncol. 2010;17(suppl 1):S31–S40. doi: 10.3747/co.v17is1.614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Allegra CJ, Jessup JM, Somerfield MR, et al. American Society of Clinical Oncology provisional clinical opinion: testing for KRAS gene mutations in patients with metastatic colorectal carcinoma to predict response to anti-epidermal growth factor receptor monoclonal antibody therapy. J Clin Oncol. 2009;27(12):2091–2096. doi: 10.1200/JCO.2009.21.9170. [DOI] [PubMed] [Google Scholar]
- 16.Kadota K, Sima CS, Arcila ME, et al. KRAS mutation is a significant prognostic factor in early-stage lung adenocarcinoma. Am J Surg Pathol. 2016;40(12):1579–1590. doi: 10.1097/PAS.0000000000000744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zheng D, Wang R, Zhang Y, et al. The prevalence and prognostic significance of KRAS mutation subtypes in lung adenocarcinomas from Chinese populations. Onco Targets Ther. 2016;9:833–843. doi: 10.2147/OTT.S96834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Steels E, Paesmans M, Berghmans T, et al. Role of p53 as a prognostic factor for survival in lung cancer: a systematic review of the literature with a meta-analysis. Eur Respir J. 2001;18(4):705–719. doi: 10.1183/09031936.01.00062201. [DOI] [PubMed] [Google Scholar]
- 19.Na II, Rho JK, Choi YJ, et al. Clinical features reflect exon sites of EGFR mutations in patients with resected non-small-cell lung cancer. J Korean Med Sci. 2007;22(3):393–399. doi: 10.3346/jkms.2007.22.3.393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Suehisa H, Toyooka S, Hotta K, et al. Epidermal growth factor receptor mutation status and adjuvant chemotherapy with uracil-tegafur for adenocarcinoma of the lung. J Clin Oncol. 2007;25(25):3952–3957. doi: 10.1200/JCO.2007.11.8646. [DOI] [PubMed] [Google Scholar]
- 21.Marks JL, Broderick S, Zhou Q, et al. Prognostic and therapeutic implications of EGFR and KRAS mutations in resected lung adenocarcinoma. J Thorac Oncol. 2008;3(2):111–116. doi: 10.1097/JTO.0b013e318160c607. [DOI] [PubMed] [Google Scholar]
- 22.Kobayashi N, Toyooka S, Ichimura K, et al. Non-BAC component but not epidermal growth factor receptor gene mutation is associated with poor outcomes in small adenocarcinoma of the lung. J Thorac Oncol. 2008;3(7):704–710. doi: 10.1097/JTO.0b013e31817c6080. [DOI] [PubMed] [Google Scholar]
- 23.Hosokawa S, Toyooka S, Fujiwara Y, et al. Comprehensive analysis of EGFR signaling pathways in Japanese patients with non-small cell lung cancer. Lung Cancer. 2009;66(1):107–113. doi: 10.1016/j.lungcan.2009.01.005. [DOI] [PubMed] [Google Scholar]
- 24.Woo T, Okudela K, Yazawa T, et al. Prognostic value of KRAS mutations and Ki-67 expression in stage I lung adenocarcinomas. Lung Cancer. 2009;65(3):355–362. doi: 10.1016/j.lungcan.2008.11.020. [DOI] [PubMed] [Google Scholar]
- 25.Lee YJ, Park IK, Park MS, et al. Activating mutations within the EGFR kinase domain: a molecular predictor of disease-free survival in resected pulmonary adenocarcinoma. J Cancer Res Clin Oncol. 2009;135(12):1647–1654. doi: 10.1007/s00432-009-0611-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Galleges Ruiz MI, Floor K, Steinberg SM, et al. Combined assessment of EGFR pathway-related molecular markers and prognosis of NSCLC patients. Br J Cancer. 2009;100(1):145–152. doi: 10.1038/sj.bjc.6604781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kosaka T, Yatabe Y, Onozato R, Kuwano H, Mitsudomi T. Prognostic implication of EGFR, KRAS, and TP53 gene mutations in a large cohort of Japanese patients with surgically treated lung adenocarcinoma. J Thorac Oncol. 2009;4(1):22–29. doi: 10.1097/JTO.0b013e3181914111. [DOI] [PubMed] [Google Scholar]
- 28.Liu HP, Isaac Wu HD, Chang JW, et al. Prognostic implications of epidermal growth factor receptor and KRAS gene mutations and epidermal growth factor receptor gene copy numbers in patients with surgically resectable non-small cell lung cancer in Taiwan. J Thorac Oncol. 2010;5(8):1175–1184. doi: 10.1097/JTO.0b013e3181e2f4d6. [DOI] [PubMed] [Google Scholar]
- 29.Tsao MS, Sakurada A, Ding K, et al. Prognostic and predictive value of epidermal growth factor receptor tyrosine kinase domain mutation status and gene copy number for adjuvant chemotherapy in non-small cell lung cancer. J Thorac Oncol. 2011;6(1):139–147. doi: 10.1097/JTO.0b013e3181fd83a4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.D’Angelo SP, Janjigian YY, Ahye N, et al. Distinct clinical course of EGFR-mutant resected lung cancers: results of testing of 1118 surgical specimens and effects of adjuvant gefitinib and erlotinib. J Thorac Oncol. 2012;7(12):1815–1822. doi: 10.1097/JTO.0b013e31826bb7b2. [DOI] [PubMed] [Google Scholar]
- 31.Kim HR, Shim HS, Chung JH, et al. Distinct clinical features and outcomes in never-smokers with nonsmall cell lung cancer who harbor EGFR or KRAS mutations or ALK rearrangement. Cancer. 2012;118(3):729–739. doi: 10.1002/cncr.26311. [DOI] [PubMed] [Google Scholar]
- 32.Scoccianti C, Vesin A, Martel G, et al. Prognostic value of TP53, KRAS and EGFR mutations in nonsmall cell lung cancer: the EUELC cohort. Eur Respir J. 2012;40(1):177–184. doi: 10.1183/09031936.00097311. [DOI] [PubMed] [Google Scholar]
- 33.Sonobe M, Kobayashi M, Ishikawa M, et al. Impact of KRAS and EGFR gene mutations on recurrence and survival in patients with surgically resected lung adenocarcinomas. Ann Surg Oncol. 2012;19(suppl 3):S347–S354. doi: 10.1245/s10434-011-1799-8. [DOI] [PubMed] [Google Scholar]
- 34.Maki Y, Soh J, Ichimura K, et al. Impact of GLUT1 and Ki-67 expression on earlystage lung adenocarcinoma diagnosed according to a new international multidisciplinary classification. Oncol Rep. 2013;29(1):133–140. doi: 10.3892/or.2012.2087. [DOI] [PubMed] [Google Scholar]
- 35.Sun HB, Ou W, Li Y, et al. Epidermal growth factor receptor mutation status and adjuvant chemotherapy in resected advanced non-small-cell lung cancer. Clin Lung Cancer. 2013;14(4):376–382. doi: 10.1016/j.cllc.2012.10.008. [DOI] [PubMed] [Google Scholar]
- 36.Ragusa M, Vannucci J, Ludovini V, et al. Impact of epidermal growth factor receptor and KRAS mutations on clinical outcome in resected non-small cell lung cancer patients. Am J Clin Oncol. 2014;37(4):343–349. doi: 10.1097/COC.0b013e31827a7e7a. [DOI] [PubMed] [Google Scholar]
- 37.Ohba T, Toyokawa G, Kometani T, et al. Mutations of the EGFR and K-ras genes in resected stage I lung adenocarcinoma and their clinical significance. Surg Today. 2014;44(3):478–486. doi: 10.1007/s00595-013-0589-2. [DOI] [PubMed] [Google Scholar]
- 38.Liu WS, Zhao LJ, Pang QS, Yuan ZY, Li B, Wang P. Prognostic value of epidermal growth factor receptor mutations in resected lung adenocarcinomas. Med Oncol. 2014;31(1):771. doi: 10.1007/s12032-013-0771-9. [DOI] [PubMed] [Google Scholar]
- 39.Ayyoub M, Memeo L, Alvarez-Fernandez E, et al. Assessment of MAGE-A expression in resected non-small cell lung cancer in relation to clinicopathologic features and mutational status of EGFR and KRAS. Cancer Immunol Res. 2014;2(10):943–948. doi: 10.1158/2326-6066.CIR-13-0211. [DOI] [PubMed] [Google Scholar]
- 40.Kudo Y, Shimada Y, Saji H, et al. Prognostic factors for survival after recurrence in patients with completely resected lung adenocarcinoma: important roles of epidermal growth factor receptor mutation status and the current staging system. Clin Lung Cancer. 2015;16(6):e213–e221. doi: 10.1016/j.cllc.2015.04.005. [DOI] [PubMed] [Google Scholar]
- 41.Nadal E, Beer DG, Ramnath N. KRAS-G12C mutation is associated with poor outcome in surgically resected lung adenocarcinoma. J Thorac Oncol. 2015;10(2):e9–e10. doi: 10.1097/JTO.0000000000000438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nishii T, Yokose T, Miyagi Y, et al. Prognostic value of EGFR mutations in surgically resected pathological stage I lung adenocarcinoma. Asia Pac J Clin Oncol. 2017:e204–e211. doi: 10.1111/ajco.12512. [DOI] [PubMed] [Google Scholar]
- 43.Isaka T, Nakayama H, Yokose T, et al. Epidermal growth factor receptor mutations and prognosis in pathologic N1-N2 pulmonary adenocarcinoma. Ann Thorac Surg. 2016;102(6):1821–1828. doi: 10.1016/j.athoracsur.2016.06.001. [DOI] [PubMed] [Google Scholar]
- 44.Kaseda K, Asakura K, Kazama A, Ozawa Y. Clinicopathological and prognostic features of surgically resected pathological stage I lung adenocarcinoma harboring epidermal growth factor receptor and K-ras mutation. Thorac Cancer. 2017;8(3):229–237. doi: 10.1111/1759-7714.12428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sullivan I, Salazar J, Arqueros C, et al. KRAS genetic variant as a prognostic factor for recurrence in resectable non-small cell lung cancer. Clin Transl Oncol. 2017;19(7):884–890. doi: 10.1007/s12094-017-1620-7. [DOI] [PubMed] [Google Scholar]
- 46.Yotsukura M, Yasuda H, Shigenobu T, et al. Clinical and pathological characteristics of EGFR mutation in operable early-stage lung adenocarcinoma. Lung Cancer. 2017;109:45–51. doi: 10.1016/j.lungcan.2017.04.014. [DOI] [PubMed] [Google Scholar]
- 47.Takamochi K, Oh S, Matsunaga T, Suzuki K. Prognostic impacts of EGFR mutation status and subtype in patients with surgically resected lung adenocarcinoma. J Thorac Cardiovasc Surg. 2017;154(5):1768–1774.e1. doi: 10.1016/j.jtcvs.2017.06.062. [DOI] [PubMed] [Google Scholar]
- 48.Do H, Krypuy M, Mitchell PL, Fox SB, Dobrovic A. High resolution melting analysis for rapid and sensitive EGFR and KRAS mutation detection in formalin fixed paraffin embedded biopsies. BMC Cancer. 2008;8:142. doi: 10.1186/1471-2407-8-142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Marchetti A, Martella C, Felicioni L, et al. EGFR mutations in non-small-cell lung cancer: analysis of a large series of cases and development of a rapid and sensitive method for diagnostic screening with potential implications on pharmacologic treatment. J Clin Oncol. 2005;23(4):857–865. doi: 10.1200/JCO.2005.08.043. [DOI] [PubMed] [Google Scholar]
- 50.He B, Qiu X, Li P, Wang L, Lv Q, Shi T. HCCNet: an integrated network database of hepatocellular carcinoma. Cell Res. 2010;20(6):732–734. doi: 10.1038/cr.2010.67. [DOI] [PubMed] [Google Scholar]
- 51.He B, Zhang H, Shi T. A comprehensive analysis of the dynamic biological networks in HCV induced hepatocarcinogenesis. PLoS One. 2011;6(4):e18516. doi: 10.1371/journal.pone.0018516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Marchetti A, Milella M, Felicioni L, et al. Clinical implications of KRAS mutations in lung cancer patients treated with tyrosine kinase inhibitors: an important role for mutations in minor clones. Neoplasia. 2009;11(10):1084–1092. doi: 10.1593/neo.09814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhang Z, Wang T, Zhang J, et al. Prognostic value of epidermal growth factor receptor mutations in resected non-small cell lung cancer: a systematic review with meta-analysis. PLoS One. 2014;9(8):e106053. doi: 10.1371/journal.pone.0106053. [DOI] [PMC free article] [PubMed] [Google Scholar]