Abstract
The complete genome sequence data of S. aureus SJTUF_J27 isolated from seaweed in China is reported here. The size of the genome is 2.8 Mbp with 32.9% G + C content, consisting of 2614 coding sequences and 77 RNAs. A number of virulence factors, including antimicrobial resistance genes (fluoroquinolone, beta-lactams, fosfomycin, mupirocin, trimethoprim, and aminocoumarin) and the egc enterotoxin cluster, were found in the genome. In addition, the genes encoding metal-binding proteins and associated heavy metal resistance were identified. Phylogenetic data analysis, based upon genome-wide single nucleotide polymorphisms (SNPs), and comparative genomic evaluation with BLAST Ring Image Generator (BRIG) were performed for SJTUF_J27 and four S. aureus strains isolated from food. The completed genome data was deposited in NCBI׳s GenBank under the accession number CP019117, https://www.ncbi.nlm.nih.gov/nuccore/CP019117.
Keywords: Staphylococcus aureus, Genome assembly, Whole genome sequencing (WGS), Virulence factor
Specifications Table
| Subject area | Biology |
| More specific subject area | Microbial genomics |
| Type of data | Completed genome sequence in FASTA format, figures |
| How data was acquired | Illumina Miseq sequencing platform |
| Data format | Analyzed |
| Experimental factors | Staphylococcus aureus SJTUF_J27 isolated from seaweed |
| Experimental features | Whole genome sequencing, de novo assembly, and annotation |
| Data source location | Shanghai, China (Latitude 31.23 N and Longitude 121.47 E) |
| Data accessibility | Data is with this article and available online at https://www.ncbi.nlm.nih.gov/nuccore/CP019117 |
Value of the data
-
•
The complete genome sequence of S. aureus SJTUF_J27, which was isolated from Chinese seaweed, provides a genetic basis for understanding the epidemiology of food-associated staphylococci.
-
•
The sequence data will be useful for comparative genomic study of S. aureus.
-
•
Analyses of virulence and antibiotic resistance genes can be used to predict the probability of the organism being a multidrug resistance pathogen.
-
•
The genome-wide SNP analysis generated a high-resolution phylogenetic tree of S. aureus food isolates, which is a useful tool for accurately discriminating closely related species.
1. Data
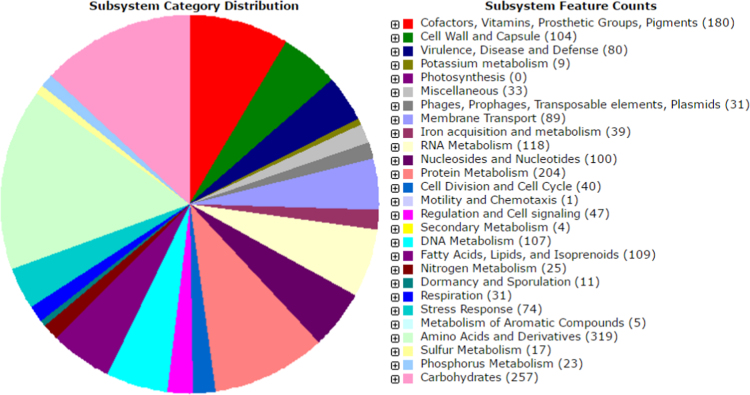
With an average of 331-fold sequencing coverage, a genome size of 2,804,759 bp constituting 32.9% of G + C content was generated. RAST annotation of the genome revealed a total of 399 subsystems, 2614 coding sequences (80 of them related to virulence, disease and defense), and 77 RNAs (Fig. 1). PathogenFinder showed the probability of this strain being a human pathogen was 98%.
Fig. 1.
Subsystem categories and distribution of the S. aureus SJTUF_J27 genome annotated by RAST.
Analysis of the genomic data showed the organism contains several antimicrobial resistance genes, including fluoroquinolone resistance-determining region of gyrA, gyrB, parC and parE, teicoplanin-associated operon of tcaR-tcaA-tcaB, beta-lactamase genes, and fosfomycin resistance gene fosB. Comprehensive Antibiotic Resistance Database (CARD) identified mupirocin resistance mediated by ileS, trimethoprim resistance mediated by dfrC, and aminocoumarin resistance mediated by alaS. The strain harbors heavy metal resistance genes and the enterotoxin gene cluster (egc) but lacks staphylococcal pathogenicity islands (SaPI).
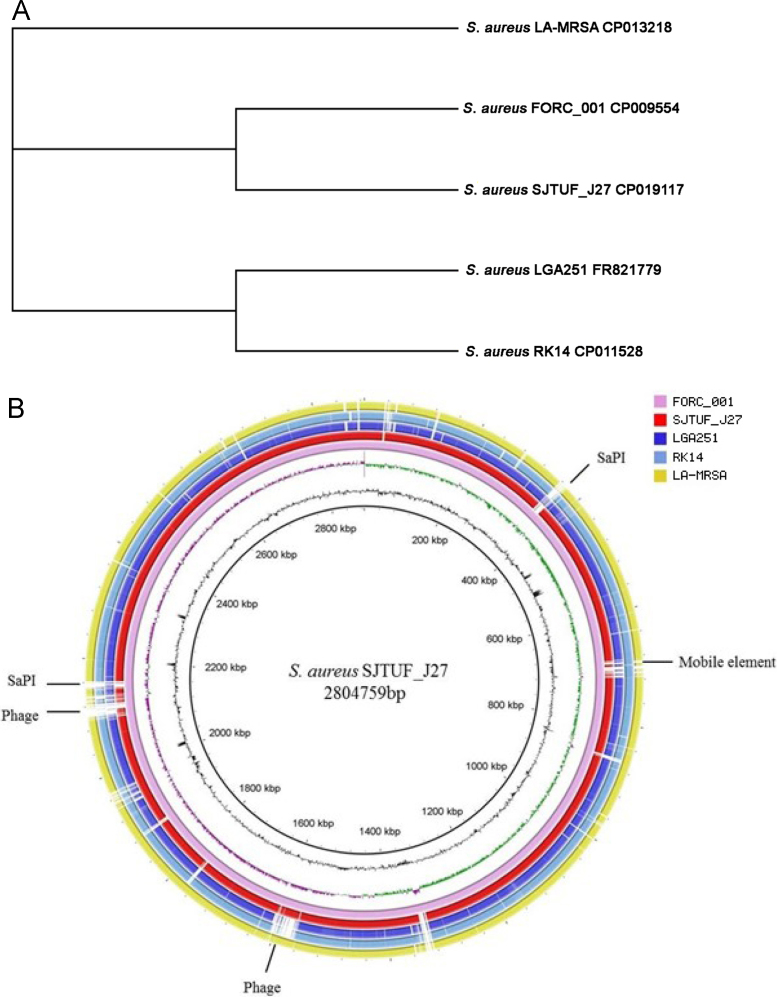
Genome-wide single nucleotide polymorphism analysis revealed the phylogenetic relationships of the strain to four food isolates (S. aureus FORC_001, LGA251, RK14 and LA-MRSA with accession numbers CP009554, FR821779, CP011528 and CP013218, respectively) (Fig. 2A). BRIG analysis showed the differences between these strains mainly in the mobile genetic elements of phage and SaPI (Fig. 2B). Furthermore, MLST (http://www.mlst.net) showed that SJTUF_J27 belongs to sequence type (ST)433, and all these food isolates belong to different STs (FORC_001 to ST30, LGA251 to ST425, RKI4 to ST8, and LA-MRSA to ST398).
Fig. 2.
A. Phylogenetic tree of S. aureus strains based on whole genome single-nucleotide polymorphisms. B. BRIG ring comparisons of the S. aureus strains SJTUF_J27, FORC_001, LGA251, RK14, and LA-MRSA. The main divergent regions are labeled with SaPI, phage and mobile element. The innermost rings represent the GC content (black) and GC skew (purple/green) of FORC_001.
2. Experimental design, materials and methods
S. aureus SJTUF_J27 was isolated from seaweed in China. Identification of the strain was carried out using the API Staph-Ident system (bioMerieux, Shanghai, China). The result was confirmed by 16s rRNA sequencing [1]. For genome sequencing, DNA was extracted using DNeasy Blood & Tissue Kit (Qiagen, Valencia, CA), quantified by a Qubit 2.0 fluorometer (Thermo Fisher Scientific, Waltham, MA), and then subjected to library construction using the Nextera XT sample preparation kit (Illumina, San Diego, CA). Next-generation sequencing was performed in Illumina Miseq platform with 2 × 300 paired-end sequencing chemistry. A total of 1,547,292 raw sequence reads were automatically generated, trimmed for quality, and then de novo assembled using the CLC genomics workbench v 9.5 and SPAdes 3.9. The assembled genome was validated by Sanger sequencing and mapping reads back to the assembly. The complete genome sequence of S. aureus SJTUF_J27 was deposited to NCBI under the accession number CP019117. Annotation of the genome was performed using the Rapid Annotation Subsystem Technology (RAST) sever (http://rast.nmpdr.org/) [2]. Pathogenicity and antibiotic resistance were predicted using PathogenFinder (https://cge.cbs.dtu.dk/services/PathogenFinder/) and Comprehensive Antibiotic Resistance Database (CARD) (https://card.mcmaster.ca/) [3], respectively. Single nucleotide polymorphism (https://cge.cbs.dtu.dk/services/CSIPhylogeny/) [4], BLAST Ring Image Generator (BRIG) (https://sourceforge.net/projects/brig/) [5], and MLST (http://www.mlst.net) were used for comparative analyses of the S. aureus food isolates.
Acknowledgement
This research was supported by the US Department of Agriculture, 8072-42000-071-00D Agricultural Research Service (USDA-ARS). We thank Dr. Xianming Shi at Shanghai Jiaotong University, Shanghai, China for providing the strain. SG is thankful to, the Department of Biotechnology (DBT), Ministry of Science & Technology, Government of India for Overseas Associateship Grant for the North Eastern Region (2015–16) and to the Indian Council of Agricultural Research for necessary support.
Footnotes
Transparency data associated with this article can be found in the online version at 10.1016/j.dib.2018.08.084.
Transparency document. Supplementary material
Supplementary material
References
- 1.Lane D.J. 16S/23S rRNA sequencing. In: Stackebrandt E., Goodfellow M., editors. Nucleic Acid Techniques in Bacterial Systematics. John Wiley & Sons; Chichester, UK: 1991. pp. 115–175. [Google Scholar]
- 2.Overbeek R., Olson R., Pusch G.D., Olsen G.J., Davis J.J., Disz T., Edwards R.A., Gerdes S., Parrello B., Shukla M. The SEED and the rapid annotation of microbial genomes using subsystems technology (RAST) Nucleic Acids Res. 2014;42:D206–D214. doi: 10.1093/nar/gkt1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.McArthur A.G., Waglechner N., Nizam F., Yan A., Azad M.A., Baylay A.J., Bhullar K., Canova M.J., de Pascale G., Ejim L. The comprehensive antibiotic resistance database. Antimicrob. Agents Chemother. 2013;57:3348–3357. doi: 10.1128/AAC.00419-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kaas R.S., Leekitcharoenphon P., Aarestrup F.M., Lund O. Solving the problem of comparing whole bacterial genomes across different sequencing platforms. PLoS One. 2014;9:e104984. doi: 10.1371/journal.pone.0104984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Alikhan N., Petty N.K., Zakour N.L.B., Beatson S.A. BLAST ring image generator (BRIG): simple prokaryote genome comparisons. BMC Genom. 2011;12:402–412. doi: 10.1186/1471-2164-12-402. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material