Abstract
Inheritance of germline mutations in the tumor suppressor gene TP53 causes Li-Fraumeni syndrome (LFS), a cancer predisposition disorder. The arginine to histidine substitution at amino acid position 337 of p53 (R337H) is a founder mutation highly prevalent in southern and southeastern Brazil and is considered an LFS mutation. Although this mutation is of significant clinical interest, its role in tumorigenesis using animal models has not been described. Here we generate a knockin mouse model containing the homologous R337H mutation (mouse R334H). De novo tumorigenesis was not significantly increased in either heterozygous (p53334R/H) or homozygous (p53334H/H) p53 R334H knockin mice compared with wild-type mice. However, susceptibility to diethylnitrosamine (DEN)-induced liver carcinogenesis was increased in a mutant allele dose-dependent manner. In parallel, p53334H/H mice exposed to DEN exhibited increased DNA damage but decreased cell cycle regulation in the liver. Oligomerization of p53, which is required for transactivation of target genes, was reduced in R334H liver, consistent with its decreased nuclear activity compared to wild-type. By modeling a TP53 mutation in mice that has relatively weak cancer penetrance, this study provides in vivo evidence that the human R337H mutation can compromise p53 activity and promote tumorigenesis.
Keywords: Li-Fraumeni syndrome, p53, R337H, mouse model, carcinogenesis
Introduction
p53 transactivates many genes regulating cellular pathways important for maintaining genomic integrity and suppressing tumorigenesis (1,2). Somatic mutations of p53, encoded by the human gene TP53, are highly prevalent in human cancers and commonly alter specific “hotspot” amino acid residues located in its DNA-binding core domain (3). Because the tetramerization of p53 protein permits its cooperative and efficient binding to DNA, core domain mutants can demonstrate dominant negative activity by hetero-oligomerizing with wild-type protein and suppress the transactivation of its target genes (4,5). Clinically, germline transmission of these hotspot DNA-binding domain mutations causes the cancer predisposition disorder Li-Fraumeni syndrome (LFS) which has an estimated cancer risk of 90% by age 60 years, making it imperative to expand our understanding of the biology of LFS mutations (6–8).
Mutations in the oligomerization domain of p53, located within its carboxy-terminal domain (CTD), would also be predicted to diminish its transactivation activity. In contrast to the DNA-binding domain, LFS mutations in the CTD had not been frequently observed until the association of germline TP53 R337H mutation with adrenocortical carcinoma (9). The R337H mutation has turned out to be highly prevalent (~0.3%) in the general population of southern Brazil, affecting hundreds of thousands of individuals (10,11). Unlike the high penetrance of cancer observed in hotspot DNA-binding domain mutation carriers, the R337H mutation is considered an atypical variant of incomplete penetrance as only a small fraction of carriers develop cancer by age 30 although much higher than the general population (11–13). In vitro work overexpressing p53 R337H has not revealed significant differences in activity compared with wild-type protein despite overwhelming clinical evidence of increased cancer risk associated with this mutation, suggesting that these types of studies may not be sufficient and more physiologic approaches are needed (9,14). We reasoned that studying this mutation in vivo without human genetic and environmental confounders could provide new insights and serve as a vehicle for testing cancer prevention strategies. Here, we report generating a mouse model of the human TP53 R337H mutation to study its role in tumorigenesis which reveals low cancer penetrance in mice with the mutation but provides mechanistic insights into its in vivo biology.
Materials and Methods
Generation of p53 R334H knockin mouse
All animal studies were approved by the Institutional Animal Care and Use Committee of the NHBLI-NIH. The p53 R334H knockin mouse was made using the conventional embryonic stem (ES) cell-mediated strategy. Briefly, a gene-targeting construct containing the CGC (Arg) to CAC (His) missense mutation and neomycin-resistant gene flanked by loxP sites was electroporated into Prx mES cells derived from C57BL/6N strain (Jackson Laboratories). ES cells were cultured on inactivated MEF feeder cells in DMEM with 20% fetal bovine serum, non-essential amino acids, 2 mM glutamine, 0.2 mM beta-mercaptoethanol, 1000 units/ml leukemia inhibitory factor, and 200 µg/ml G418. G418-resistant colonies were genotyped by PCR, and two correctly targeted ES clones were injected into blastocysts collected from C57BL/6 mice. The resulting chimeras were then bred with wildtype C57BL/6 mice for germline transmission of the mutant allele and subsequently crossed with CMV-Cre mice to remove the neo-resistance gene. p53−/− and p53 hotspot mutant p53+/R172H (strain 01XL9) mice were obtained from the Jackson Laboratories and NCI Frederick Mouse Repository, respectively, and were of the C57BL/6 strain or backcrossed at least 5 generations into the C57BL/6 background.
Cancer survival studies
For cancer phenotype and survival studies, mice were euthanized if any external mass exceeded 2 cm in its largest dimension or when the mouse met moribund criteria upon assessment by investigators and veterinary technicians. Upon reaching these survival endpoints, qualified veterinary pathologists (Division of Veterinary Resources, NIH) performed all necropsies and made the histopathologic diagnoses.
Diethylnitrosamine-induced liver carcinogenesis and whole body p53 induction
Male mice (14 d old) were treated with DEN (25 mg/kg i.p.; cat. #N0258, Millipore Sigma) and examined ~42 wk later for liver tumor development as previously described (15,16). The livers were excised and weighed, and digital images with scale ruler were acquired. Tumors visible on the liver surface (≥ 1 mm in diameter) were counted using the analyze-scale/measure functions of ImageJ software (version 1.51). Formalin-fixed paraffin sections of the right medial lobe (4 to 5 sections at ~2–3 mm intervals per liver sample) were hematoxylin and eosin (H&E) stained for histopathologic diagnosis by a board certified veterinary pathologist. For mRNA and protein analyses, 6 wk old male mice were injected with DEN (100 mg/kg i.p.) 24 h before harvesting liver tissue. To induce p53, mice received doxorubicin (20 mg/kg i.p.) or 5 Gy total body γ-irradiation (TBI) from a Gammacell 40 (K2K 1 X 8; MDS Nordion, Ontario, Canada) (17) 6 h prior to harvesting tissues which were snap frozen in liquid nitrogen and stored at −80 °C.
Antibodies
Antibodies were obtained from the following sources: p53 mouse mAb (1C12, #2524) for oligomerization assay, phospho-S15 p53 mouse mAb (#9284), BAX rabbit pAb (#2772) (Cell Signaling Technology); p53 rabbit pAb (FL393, #sc-6243) for chromatin immunoprecipitation (Santa Cruz Biotechnology); GAPDH mouse mAb (6C5, #AM4300) (Thermo Fisher Scientific); phospho-S139 histone H2AX mouse mAb (#05-636), p21 mouse mAb (#OP76) (Millipore Sigma).
Western blot analysis
Protein samples were solubilized in cold RIPA buffer with protease and phosphatase inhibitors, resolved using SDS/PAGE protein gel (Bio-Rad Laboratories), immunoblotted using Immobilon-P membrane (Millipore Sigma) and chemiluminescence developer (Thermo Fisher Scientific), and detected using ChemiDoc imaging system (Bio-Rad Laboratories). Due to the interference by endogenous mouse IgG, p53 immunoblotting was performed after depleting IgG using protein G-agarose (#sc-2002, Santa Cruz Biotechnology) in tissue lysates (18).
p53 oligomerization assay
The oligomerization state of p53 in tissue samples was determined using a glutaraldehyde-based cross-linking assay as previously described with some minor modifications (19). Frozen liver tissue samples (~50 mg) were homogenized in 1 ml cold lysis buffer (20 mM Tris-HCl (pH 7.4), 137 mM NaCl, 1% NP-40, 2 mM EDTA, 10% glycerol, protease/phosphatase inhibitor cocktail (#04-693-116-001 and #04-906-837-001, Roche)) with a IKA Ultra-Turrax homogenizer (#Z404519, Millipore Sigma) for 15 s and centrifuged at 12,000× g at 4 °C for 10 min. The supernatant was depleted of mouse IgG with protein G-agarose (#sc-2002, Santa Cruz Biotechnology). Cross-linking was performed by adding glutaraldehyde (0.1% final concentration; cat. #16120, Electron Microscopy Sciences) to the protein lysate (~2 µg protein/µl lysis buffer), incubating on ice for 15 min, and terminating the reaction with SDS/PAGE protein sample buffer. The protein samples were resolved on 4–20% gradient SDS/PAGE gels (Bio-Rad Laboratories) and immunoblotted with anti-p53 mouse monoclonal antibody (#2524, Cell Signaling Technology). Within each lane, the fraction of p53 immunoreactivity in the oligomer bands (monomer, dimer and tetramer) was determined using Image Lab software (Bio-Rad Laboratories).
Cell cycle analysis
Cell cycle analysis from frozen tissues were performed as previously described (20). Frozen liver tissue (~50 mg) was thawed in cold PBS containing 3.4 mM EDTA, dispersed through a 100 µm nylon mesh using a syringe plunger, filtered through a 30 µm cell strainer, and centrifuged at 300× g for 5 min at 4 °C. The cells were resuspended in PBS/EDTA and fixed with methanol at −20 °C overnight, spun down, and resuspended in room temperature PBS/EDTA. Nuclei were stained with FxCycle™ Propidium Iodide/RNase solution (#F10979, Thermo Fisher Scientific) and analyzed using FACSCanto II system (BD Biosciences).
TUNEL apoptosis assay
Terminal deoxynucleotidyl transferase-mediated dUTP-digoxigenin nick-end labelling (TUNEL) assay was performed per protocol using In Situ Cell Death Detection Kit, Fluorescein (#11 684 795 910, Roche). TUNEL positive nuclei were counted in 5 random non-overlapping fields per section using 20× objective lens magnification.
mRNA quantification
Total tissue RNA was purified using the RNeasy kit (Qiagen), bound to poly(dT) magnetic beads (Invitrogen), and reverse transcribed (RT) using Superscript III (Invitrogen). RT2 Profiler PCR arrays were used to screen for the expression of 84 genes related to p53-mediated signal transduction normalized to actin per manufacturer’s protocol (#330231 PAMM-027ZA, Qiagen). mRNA levels were quantified by real-time RT-PCR using SYBR green fluorescence on the LightCycler 96 Real-Time PCR system (Roche). The cycle threshold (Ct) values were normalized to a housekeeping gene eukaryotic translation initiation factor (TIF) (21). Color-coding of gene expression levels was done using Microsoft Excel. PCR primer sequences are listed in Supplementary Table 1.
Chromatin immunoprecipitation assay
Chromatin immunoprecipitation (ChIP) assay was performed using ChIP-IT High Sensitivity kit (#53040, Active Motif) according to the manufacturer’s protocol. Liver tissues from whole body irradiated mice were minced into small pieces and cross-linked with 1% formaldehyde for 10 min. Nuclear extracts were sonicated and immunoprecipitated with anti-p53 (FL393, #sc-6243, Santa Cruz) or control rabbit IgG (#sc-2027, Santa Cruz) overnight at 4 °C. Antibody-bound DNA fragments were purified and quantified by real-time PCR (primers listed in Supplementary Table 1). Ct values were normalized to input DNA Ct values and displayed relative to wild-type non-specific IgG samples.
Statistical analysis
Statistical analyses were carried out using Prism 7 software (GraphPad Software). Unless otherwise specified, all values are shown as mean ± SD and data were analyzed using the 2-tailed unpaired Student’s t test. One-way ANOVA with Tukey’s post-test analysis was performed for data containing more than two experimental groups. P value less that 0.05 was considered statistically significant.
Results
p53 R334H mutation does not significantly affect tumor development or lifespan in mice
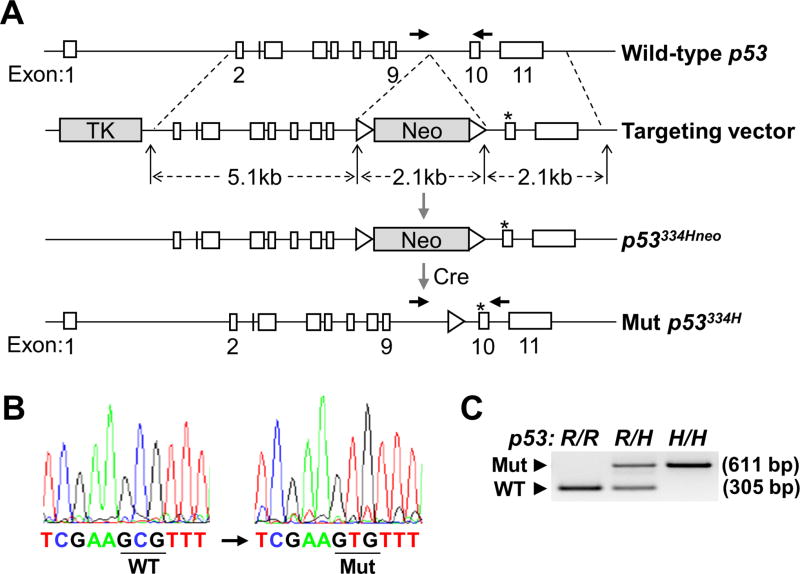
The p53 R334H amino acid mutation, homologous to the human R337H mutation, was knocked into the p53 gene locus of C57BL/6 mice using a homologous recombination strategy (Fig. 1A). The G>A (c.1001) transition mutation was confirmed by genomic DNA sequencing and PCR (Fig. 1B and C). Mice with the p53 R334H mutation were generally born at the expected Mendelian frequency although there was a trend of lower than expected number of female homozygous mutant mice (Supplementary Table 2). This was notable because the number of female p53 null mice born are known to be significantly less than expected (22). Nonetheless, compared with wild-type both the heterozygous and homozygous mutant mice developed normally, exhibiting similar body weight, body mass composition, and aerobic metabolic capacity as assessed by blood lactate levels and running endurance (Supplementary Figs. S1 and S2). We next compared the survival time of heterozygous (p53334R/H) and homozygous (p53334H/H) mutant mice with wild-type mice (p53334R/R) and performed necropsies at the end of their survival time for cancer diagnosis. Although the R334H mutant mice showed a trend of decreased median lifespan and increased cancer incidence, they were not statistically different for either gender (Fig. 2A to C). Cancer diagnosis across all 3 genotypes was relatively high but consistent with the reported incidence of histiocytic sarcomas in the C57BL strain (23). Given the previous reports of increased incidence of pediatric adrenocortical carcinoma in TP53 R337H mutation carriers, we specifically examined the adrenal glands of the knockin mice for abnormalities at the endpoint of the survival study. The prevalence of adrenal hyperplasia or hypertrophy was ~50–70% for all 3 genotypes with no statistically significant differences being observed (Supplementary Table 3).
Figure 1.
p53 R334H mutation knockin construct and screening strategy. A, conventional ES cell-mediated strategy was used to knockin the c.1001G>A missense mutation into the p53 gene (p53334H; mouse R334H corresponding to human R337H). Mice with germline transmission of the mutation were crossed with cre-expressing mice to remove the neomycin (Neo) resistance gene flanked by loxP sites (triangles). Asterisk indicates the mutated nucleotide position in exon 10 and horizontal arrowheads indicate forward and reverse primers for genotype screening. B, sequencing of the complementary strand confirmed correct integration of the mutation. The wild-type (WT) and mutant (Mut) codon positions are underlined. C, genomic DNA PCR was also performed to confirm the genotypes of mice. The patterns of WT (305 bp) and Mut (611 bp) PCR products indicate wild-type (R/R), heterozygous mutant (R/H), and homozygous mutant (H/H) genotypes.
Figure 2.
p53 R334H does not significantly affect lifespan or cancer incidence in mice. A, Kaplan-Meier survival plot of male and female p53334R/R, p53334R/H, and p53334H/H mice; the respective median survival times (male/female) for these 3 genotypes were 114/99, 103/103, and 103/101 wk. No significant difference in survival time was observed by log-rank test (n = 9–29). B, incidence of cancer by p53 R334 genotype at necropsy after reaching survival endpoint (n = 24–39). C, spectrum of cancer diagnosis by p53 R334 genotype (n = 24–39). p53 R334 genotypes: wild-type (R/R); heterozygous mutant (R/H); and homozygous mutant (H/H). Both incidence and spectrum of cancer were not significantly different compared with wild-type mice by Fisher’s exact test.
p53 R334H increases the susceptibility of mice to liver carcinogenesis
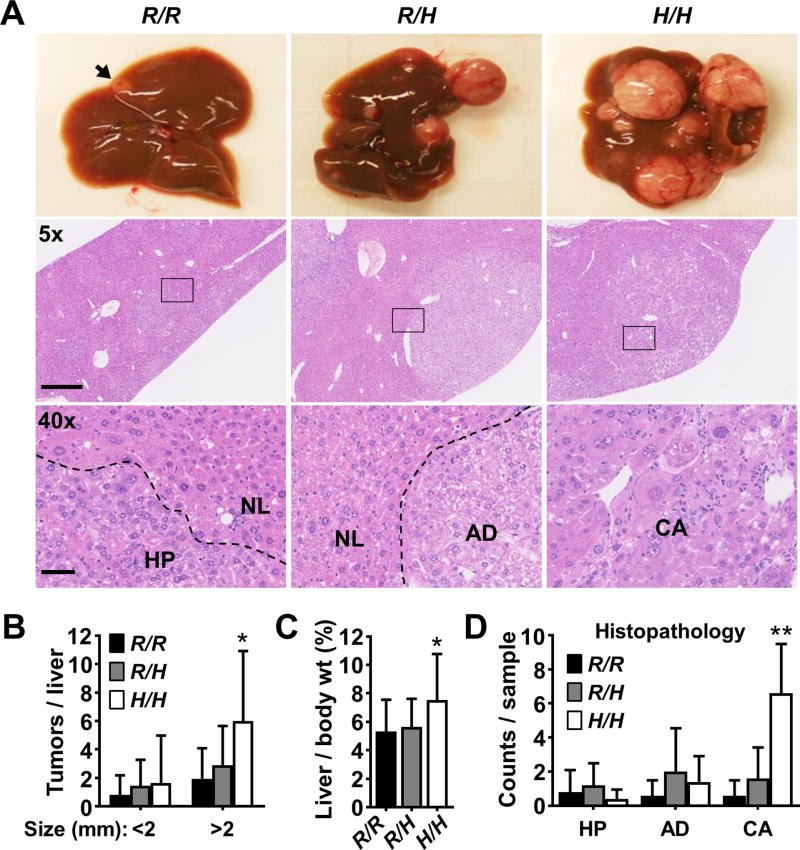
We next evaluated whether the p53 R334H mutation could confer increased susceptibility to tumorigenesis under conditions of genotoxic stress. In both mouse and rat models of hepatocarcinogenesis, p53 deficiency appears to promote tumorigenesis induced by diethylnitrosamine (DEN), a genotoxic chemical present in tobacco smoke and dietary meat (15,24,25). We reasoned that this carcinogenesis model may provide a sensitive readout of whether the R334H mutation compromises wild-type p53 activity in vivo and can promote de novo tumor formation. Liver carcinogenesis depends on the bioactivation of DEN by cytochrome P450 CYP2E1 which was expressed at similar levels independent of p53 genotype (Supplementary Fig. S3) (26). Wild-type and mutant mice (14 d old) were injected intraperitoneally with DEN and examined for tumor formation in the liver after 42 wk. Indeed, there was a mutant allele dose-dependent increase in the number and size of tumors studding the surface of the liver (total tumors per liver: 2.8 ± 2.2, 4.4 ± 3.9, and 7.6 ± 6.5 for the p53334R/R, p53334R/H, and p53334H/H genotypes, respectively) (Fig. 3A and B). Accordingly, the livers of homozygous R334H mutant mice were significantly heavier than that of wild-type mice (7.5 ± 3.2% versus 5.3 ± 2.2% of body weight, respectively) (Fig. 3C). Histopathologic analysis of the liver tumors also showed significantly increased incidence of carcinomas in homozygous mutant p53334H/H livers (Fig. 3A lower panel and D). On the other hand, genomic analysis of heterozygous p53334R/H liver tumors did not reveal loss of heterozygosity, consistent with their smaller size and low incidence of carcinoma (Supplementary Fig. S4). These in vivo observations indicated that the R334H mutation can compromise the tumor suppressive activity of p53 in a mutant allele dose-dependent manner.
Figure 3.
p53 R334H confers increased susceptibility to DEN-induced liver tumorigenesis in mice. A, representative images showing liver tumor formation 42 wk after DEN treatment (top panel). Small tumor indicated by arrowhead. Lower panels show representative H&E stained photomicrographs of liver tumor sections under low (5×) and high (40×, boxed areas in 5×) objective magnification. Broken lines delineate the following regions of histopathology: normal (NL), hyperplasia (HP), adenoma (AD), and carcinoma (CA). Scale bar = 500 µm and 50 µm. B, quantification of tumors visible on the surface of the liver by size (≥ 1 mm diameter) (n = 14–20). C, liver weight was normalized by body weight to provide an index of tumor burden (n = 14–20). D, quantification of liver tumor histopathologic diagnosis as abbreviated in (A). Counts indicate the number of specific diagnosis made in each liver sample (n = 5). p53 R334 genotypes: wild-type (R/R); heterozygous mutant (R/H); and homozygous mutant (H/H). Values are mean ± SD. Statistical differences were tested by 1-way ANOVA. *P < 0.05 and **P < 0.01
p53 R334H displays a partial reduction in transcriptional activity
To further characterize the effect of the R334H mutation on the in vivo activity of p53, liver samples were harvested from p53334R/R, p53334R/H, p53334H/H and p53−/− mice 24 h after DEN treatment and analyzed for p53 target gene expression using a RT-PCR screening array. Notably, only a small subset of p53-regulated genes were induced by DEN, and the pattern of mildly decreased transcriptional activity depended on the mutant allele copy number (Fig. 4A). Out of the 11 genes that were significantly induced in the liver of DEN-treated versus control wild-type mice (≥ 2-fold and P < 0.05), the expression levels of 10 genes were mildly reduced with statistical significance in homozygous p53334H/H mice indicating a partial decrease in p53 transcriptional activity (Fig. 4B). The decreased transcriptional activity of the p53 R334H mutant was further demonstrated in the DEN-treated liver samples by performing independent RT-PCR of 4 prototypical p53 target genes p21, BAX, MDM2 and PUMA (Fig. 4C). The pattern of decreased expression of the queried p53 target genes in a mutant allele dose-dependent manner in vivo revealed its compromised transcriptional activity and suggested that loss of heterozygosity may promote tumorigenesis.
Figure 4.
Partial loss of p53 transcriptional activity in liver tissue of homozygous mutant p53334H/H mice treated with DEN. A, color-coded relative expression of 84 p53-regulated genes in untreated (control) and DEN-treated (24 h) mouse livers of the indicated p53 genotype analyzed by RT-PCR screening array (n = 3). Red and green indicate induced and repressed mRNA levels, respectively. Expression levels were normalized to control R/R samples. B, the screening array was filtered for genes significantly induced (≥ 2-fold) in wild-type mice by DEN treatment. Of the 11 genes that were identified, 10 were significantly lower in the homozygous mutant compared with wild-type livers (n = 3). p21, DR5, WIG, NOXA, and PUMA are referred to as CDKN1A, TNFRSF10B, ZMAT3, PMAIP1, and BBC3, respectively, in (A). C, the mRNA expression levels of 4 prototypical p53-regulated genes were confirmed by independent real-time RT-PCR (n = 4). Expression levels were normalized relative to wild-type samples. p53 R334 genotypes: wild-type (R/R); heterozygous mutant (R/H); homozygous mutant (H/H); and p53 null (−/−). Values are mean ± SD. Statistical differences were tested by 1-way ANOVA. *P < 0.05.
p53 R334H mutation is associated with increased DNA damage marker and decreased p53-mediated responses
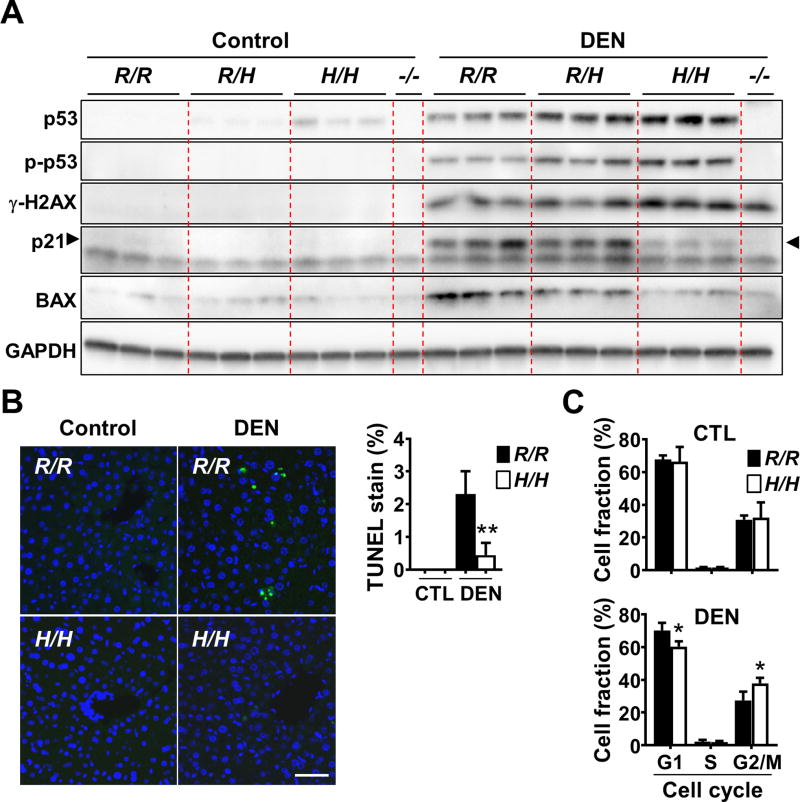
In addition to the transcriptional changes caused by the R334H mutation, we examined its effect on the cellular response to DEN which is known to alkylate DNA after metabolism in the liver (27). Both the DNA damage marker γ-H2AX and p53 were consistently higher in the homozygous mutant state, suggesting increased DNA damage as in the p53 null state although the relative contribution of this finding versus other mechanisms to DEN-induced tumorigenesis is unclear (Fig. 5A; Supplementary Fig. S5) (28,29). Furthermore, although the basal levels of p21 and BAX proteins in homozygous mutant liver samples were similar to that of wild-type, they were induced to a lower level after DEN treatment in parallel with attenuated cell cycle arrest and apoptosis in vivo (Fig. 5A to C; Supplementary Fig. S5). Impaired senescence response to oncogenic stress has been suggested to promote liver tumorigenesis, but we did not observe significant senescence changes associated with the p53 R334H mutation (Supplementary Fig. S6) (30). Nonetheless, these modest changes in cellular response to genotoxic stress were consistent with the mildly lower levels of BAX, PUMA and p21 mRNA induced in p53334H/H liver compared with wild-type after DEN treatment (Fig. 4C). Taken together, the deregulation of some tumor suppressor functions of p53 in the mutant R334H state could contribute to enhanced liver carcinogenesis after DEN exposure.
Figure 5.
Liver tissues of p53334H/H mice exposed to DEN show increased DNA damage response markers but diminished p53 activity. A, liver tissue samples from untreated (control) and DEN-treated (24 h) mice were immunoblotted with the indicated antibodies. Phospho-p53 (S15) and histone γ-H2AX (S139) serve as DNA damage response markers. Each lane represents an independent replicate mouse of untreated control and DEN-treated conditions. p53−/− mouse liver serves as negative control for p53 protein and activity. Arrowheads indicate p53-dependent p21 induced by DEN; note absence of this band in the p53−/− lane. B, apoptosis imaging and quantification in liver sections before or after DEN treatment. Fluorescent images of TUNEL stained sections of the indicated p53 genotype are shown. The fractions of apoptotic TUNEL-positive nuclei (green) versus total nuclei stained with Hoechst 33342 (blue) were determined (n = 3). Scale bar = 50 µm. C, FACS cell cycle analysis of untreated control and DEN-treated liver cells stained with propidium iodide. The relative fraction of cells in the different cell cycle stages (G1, S and G2/M) were quantified (n = 5). p53 R334 genotypes: wild-type (R/R); heterozygous mutant (R/H); and homozygous mutant (H/H). Values are mean ± SD. *P < 0.01 and **P < 0.001.
p53 R334H mutation disrupts its oligomerization in vivo
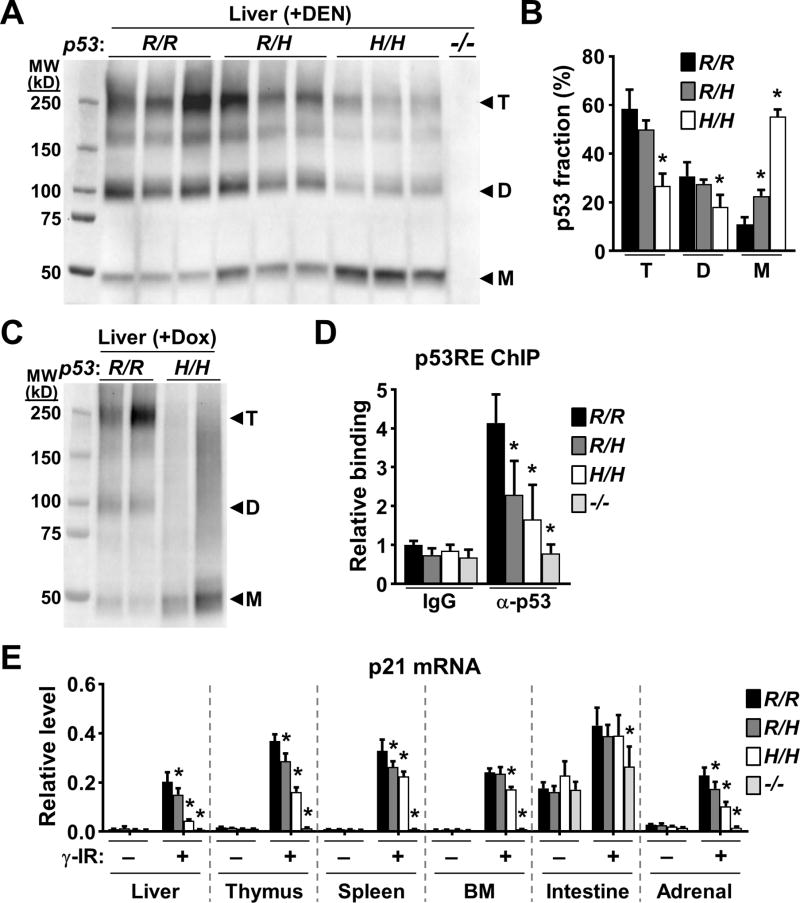
We further investigated the mechanism underlying the decreased transcriptional activity of the R334H mutation which did not appear to be due to decreased mutant p53 levels. The basal and DEN-induced levels of p53 were both higher in the heterozygous and homozygous mutant states (Fig. 5A; Supplementary Fig. S7). The higher basal levels of p53 protein associated with the R334H mutation, also seen in tumor, could be contributed by lower expression of MDM2 which has recently been shown to preferentially degrade oligomerized p53 (Fig. 4B and C; Supplementary Fig. S7) (31). Additionally, the subcellular fractionation of liver tissue after DEN treatment showed proportionally higher levels of p53 in the nucleus of homozygous mutant liver compared with wild-type, indicating that the R334H mutation did not affect its nuclear localization (Supplementary Fig. S8). Although in vitro studies have shown that the human R337H mutation can disrupt p53 oligomerization in a pH-dependent manner (32), the lack of a mouse model has precluded testing its properties in vivo. To assess the oligomerization state of mutant p53, liver samples were harvested from mice 24 h after DEN treatment and cross-linked with glutaraldehyde to capture in vivo protein-protein interactions. Compared with wild-type samples, immunoblotting of the homozygous mutant cross-linked lysates revealed a striking decrease in both the tetramer and dimer forms of p53 R334H with a concomitant increase in the monomer form (Fig. 6A). Consistent with the p53 target gene expression results, quantification of the oligomer bands showed a statistically significant reduction in p53 tetramers in the homozygous but not in heterozygous mutant liver samples (Fig. 6B). Treatment with a different DNA damaging agent doxorubicin to induce p53 in liver also showed reduced levels of tetramers in the homozygous R334H mutant state (Fig. 6C). To confirm these results, non-denaturing protein gel electrophoresis was used to show decreased high molecular weight p53 complex in the R334H mutant liver sample (Supplementary Fig. S9).
Figure 6.
p53 oligomerization is decreased in p53334H/H mouse tissues. A, liver tissue lysates were cross-linked with glutaraldehyde, resolved in SDS-PAGE gel, and immunoblotted. Note p53 antibody specificity demonstrated by lack of immunoreactivity in the p53−/− sample. Protein standards are in kD. Triplicate lanes of each genotype represent liver samples from 3 separate mice. B, fraction of p53 oligomers (T, tetramer; D, dimer; M, monomer) within each lane of immunoblot (A) quantified by densitometry and compared with the respective wild-type oligomer (n = 3). C, p53 immunoblot of cross-linked liver lysates obtained from mice 6 h after p53 induction by doxorubicin treatment. Duplicate lanes of each genotype represent liver samples from 2 separate mice. D, p53 binding to the p53 response element (p53RE) of p21 in γ-irradiated (γ-IR) mouse liver. ChIP was performed using nonspecific IgG or anti-p53 antibody. p53RE binding is shown relative to wild-type non-specific IgG samples (n = 3). E, induction of p21 mRNA in the indicated tissues by γ-IR quantified by RT-PCR (n = 3). Levels were normalized relative to a housekeeping gene TIF. Bone marrow (BM); small intestine. p53 R334 genotypes: wild-type (R/R); heterozygous mutant (R/H); homozygous mutant (H/H); and p53 null (−/−). Values are mean ± SD. Compared to wild-type samples within each group, statistical differences were tested by 1-way ANOVA. *P < 0.05.
To further investigate the functional properties of p53 R334H protein in vivo, we employed whole body γ-irradiation, another widely used stimulus for stabilizing p53 protein, to examine whether this mutation affects its ability to interact with the prototypical p53 target gene p21. Chromatin immunoprecipitation of irradiated liver tissue showed significantly reduced interaction of the mutant protein with the p53 response element (p53RE) of p21 gene in a mutant allele dose-dependent manner (Fig. 6D). A tissue survey of p21 expression after whole body γ-irradiation also revealed a pattern of mutant allele dose-dependent decrease in p21 mRNA levels with some degree of tissue variability (Fig. 6E). These in vivo data obtained from freshly isolated tissues exposed to a different modality of DNA damage further supported the tumorigenic potential of the mutation and was consistent with the observation of higher frequency of diverse cancers in TP53 R337H carriers (33).
DISCUSSION
There are limited data on the human TP53 R337H mutation in de novo tumorigenesis or its biological properties in vivo. Here, we have demonstrated the pro-tumorigenic nature of the mouse homolog by creating a mouse model with knockin of the p53 R334H mutation that corresponds to the human TP53 R337H mutation. Using an established liver carcinogenesis model, the knockin mice showed increased susceptibility to tumor development, including higher grade carcinomas, in a mutant allele dose-dependent manner. Mechanistically, the increase in liver tumorigenesis was associated with decreased oligomerization, interaction with p53RE and transcriptional activity of the mutant p53 protein after DEN-induced DNA damage. These results provide new demonstration of the in vivo properties of a mouse homolog of the human TP53 R337H, a prevalent germline mutation in southern and southeastern Brazil that is estimated to affect hundreds of thousands of individuals.
TP53 R337H carriers have higher frequency of diverse cancers, and current data show a clear association with increased risk of childhood adrenocortical carcinoma and many typical LFS-associated cancers occurring in adulthood, albeit at later ages than in classic LFS (11,33). The variable cancer penetrance associated with this mutation is an area of active interest, thus, we reasoned that studying its biology in the laboratory may provide further insights into these clinical observations. Because p53 R334H mice displayed no overt phenotype, we explored whether this unique mutation would promote cancer under conditions of genotoxic stress. After exposure to the DNA alkylating agent DEN, there was evidence of increased DNA damage and mildly decreased apoptosis and cell cycle arrest in the liver tissue of p53334H/H mice compared with wild-type mice. This mild decrease in the tumor suppressive activity could contribute to tumorigenesis as small changes in cell birth and death rates have been proposed to be sufficient to confer growth advantages (34). Furthermore, the incidence of liver carcinomas was more significantly increased in the homozygous mutant state, paralleling observations from a large meta-analysis in which patients homozygous for the TP53 R337H allele had more cancer recurrences and shorter survival times (35).
Because individuals carrying the TP53 R337H mutation have a markedly higher incidence of pediatric adrenocortical carcinoma (9,36), we examined the adrenal glands of p53334H/H mice but did not observe significant neoplastic differences compared with that of wild-type mice. Structural studies of p53 have revealed that the Arg 337 and Asp 352 residues form an inter-molecular salt-bridge and that the R337H mutation can destabilize this interaction by deprotonating His 337 under physiologic pH (32,37). In this regard, the liver has one of the lowest concentrations of ascorbic acid amongst the various organs of the body which may facilitate the deprotonation and destabilization of mutant p53 tetramers and contribute to increased carcinogenesis as observed in our study (38). It is also noteworthy that, unlike mice, humans cannot synthesize vitamin C so that deficiency states are prevalent even in socioeconomically developed countries and that the plasma levels of ascorbic acid have been reported to be significantly reduced in TP53 R337H mutation carriers compared to non-carriers (39,40). Our p53 R334H mouse model reported here should facilitate future investigations designed to examine the biological and tumorigenic characteristics of this mutation under in vivo conditions similar to that observed in humans.
Supplementary Material
Acknowledgments
We thank Zu-Xi Yu of the NHLBI Pathology Core and Seung-Kwon Ha and Ki-Sun Park for technical assistance and advice.
Grant Support
This work was supported by the National Heart, Lung, and Blood Institute, Division of Intramural Research HL006051-08 (to P.M. Hwang), and by a Bench-to-Bedside Award (to S.A. Savage, M.I. Achatz, and P.M. Hwang) of the National Institutes of Health. The work of S.A. Savage was supported by the intramural research program of the Division of Cancer Epidemiology and Genetics, National Cancer Institute. J.H. Park received postdoctoral fellowship support from the National Research Foundation of Korea (NRF) and the Korean Biomedical Scientist (KRIBB) program.
Footnotes
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- 1.Vousden KH, Prives C. Blinded by the Light: The Growing Complexity of p53. Cell. 2009;137:413–31. doi: 10.1016/j.cell.2009.04.037. [DOI] [PubMed] [Google Scholar]
- 2.Kastenhuber ER, Lowe SW. Putting p53 in Context. Cell. 2017;170:1062–78. doi: 10.1016/j.cell.2017.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Olivier M, Hollstein M, Hainaut P. TP53 mutations in human cancers: origins, consequences, and clinical use. Cold Spring Harb Perspect Biol. 2010;2:a001008. doi: 10.1101/cshperspect.a001008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chene P. The role of tetramerization in p53 function. Oncogene. 2001;20:2611–7. doi: 10.1038/sj.onc.1204373. [DOI] [PubMed] [Google Scholar]
- 5.Chillemi G, Kehrloesser S, Bernassola F, Desideri A, Dotsch V, Levine AJ, et al. Structural Evolution and Dynamics of the p53 Proteins. Cold Spring Harbor perspectives in medicine. 2017;7 doi: 10.1101/cshperspect.a028308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Malkin D, Li FP, Strong LC, Fraumeni JF, Jr, Nelson CE, Kim DH, et al. Germ line p53 mutations in a familial syndrome of breast cancer, sarcomas, and other neoplasms. Science. 1990;250:1233–8. doi: 10.1126/science.1978757. [DOI] [PubMed] [Google Scholar]
- 7.Schneider K, Garber J. Li-Fraumeni Syndrome. In: Pagon RA, et al., editors. GeneReviews [Internet] Seattle (WA): University of Washington; 2010. [PubMed] [Google Scholar]
- 8.Mai PL, Best AF, Peters JA, DeCastro RM, Khincha PP, Loud JT, et al. Risks of First and Subsequent Cancers Among TP53 Mutation Carriers in the National Cancer Institute Li-Fraumeni Syndrome Cohort. Cancer. 2016;122:3673–81. doi: 10.1002/cncr.30248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ribeiro RC, Sandrini F, Figueiredo B, Zambetti GP, Michalkiewicz E, Lafferty AR, et al. An inherited p53 mutation that contributes in a tissue-specific manner to pediatric adrenal cortical carcinoma. Proc Natl Acad Sci U S A. 2001;98:9330–5. doi: 10.1073/pnas.161479898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Achatz MI, Olivier M, Le Calvez F, Martel-Planche G, Lopes A, Rossi BM, et al. The TP53 mutation, R337H, is associated with Li-Fraumeni and Li-Fraumeni-like syndromes in Brazilian families. Cancer letters. 2007;245:96–102. doi: 10.1016/j.canlet.2005.12.039. [DOI] [PubMed] [Google Scholar]
- 11.Achatz MI, Zambetti GP. The Inherited p53 Mutation in the Brazilian Population. Cold Spring Harbor perspectives in medicine. 2016;6 doi: 10.1101/cshperspect.a026195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ribeiro RC, Rodriguez-Galindo C, Figueiredo BC, Mastellaro MJ, West AN, Kriwacki R, et al. Germline TP53 R337H mutation is not sufficient to establish Li-Fraumeni or Li-Fraumeni-like syndrome. Cancer letters. 2007;247:353–5. doi: 10.1016/j.canlet.2006.04.008. author reply 6–8. [DOI] [PubMed] [Google Scholar]
- 13.Amadou A, Waddington Achatz MI, Hainaut P. Revisiting tumor patterns and penetrance in germline TP53 mutation carriers: temporal phases of Li-Fraumeni syndrome. Curr Opin Oncol. 2018;30:23–9. doi: 10.1097/CCO.0000000000000423. [DOI] [PubMed] [Google Scholar]
- 14.Zambetti GP. Diving into in vivo p53 tumor suppressor studies using a new platform mouse model. Cell Cycle. 2011;10:2619. doi: 10.4161/cc.10.16.16546. [DOI] [PubMed] [Google Scholar]
- 15.Kemp CJ. Hepatocarcinogenesis in p53-deficient mice. Mol Carcinog. 1995;12:132–6. doi: 10.1002/mc.2940120304. [DOI] [PubMed] [Google Scholar]
- 16.Schneider C, Teufel A, Yevsa T, Staib F, Hohmeyer A, Walenda G, et al. Adaptive immunity suppresses formation and progression of diethylnitrosamine-induced liver cancer. Gut. 2012;61:1733–43. doi: 10.1136/gutjnl-2011-301116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen J, Lipovsky K, Ellison FM, Calado RT, Young NS. Bystander destruction of hematopoietic progenitor and stem cells in a mouse model of infusion-induced bone marrow failure. Blood. 2004;104:1671–8. doi: 10.1182/blood-2004-03-1115. [DOI] [PubMed] [Google Scholar]
- 18.Petry FR, Pelletier J, Bretteville A, Morin F, Calon F, Hebert SS, et al. Specificity of anti-tau antibodies when analyzing mice models of Alzheimer's disease: problems and solutions. PLoS One. 2014;9:e94251. doi: 10.1371/journal.pone.0094251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gu J, Nie L, Wiederschain D, Yuan ZM. Identification of p53 sequence elements that are required for MDM2-mediated nuclear export. Mol Cell Biol. 2001;21:8533–46. doi: 10.1128/MCB.21.24.8533-8546.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Heinlein C, Speidel D. High-resolution cell cycle and DNA ploidy analysis in tissue samples. Current protocols in cytometry. 2011;Chapter 7(Unit 7):39. doi: 10.1002/0471142956.cy0739s56. [DOI] [PubMed] [Google Scholar]
- 21.Patino WD, Mian OY, Kang JG, Matoba S, Bartlett LD, Holbrook B, et al. Circulating transcriptome reveals markers of atherosclerosis. Proc Natl Acad Sci U S A. 2005;102:3423–8. doi: 10.1073/pnas.0408032102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sah VP, Attardi LD, Mulligan GJ, Williams BO, Bronson RT, Jacks T. A subset of p53-deficient embryos exhibit exencephaly. Nat Genet. 1995;10:175–80. doi: 10.1038/ng0695-175. [DOI] [PubMed] [Google Scholar]
- 23.Fox JG. The mouse in biomedical research. Amsterdam; Boston: Elsevier, AP; 2007. [Google Scholar]
- 24.Yan HX, Wu HP, Ashton C, Tong C, Ying QL. Rats deficient for p53 are susceptible to spontaneous and carcinogen-induced tumorigenesis. Carcinogenesis. 2012;33:2001–5. doi: 10.1093/carcin/bgs238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dhar D, Antonucci L, Nakagawa H, Kim JY, Glitzner E, Caruso S, et al. Liver Cancer Initiation Requires p53 Inhibition by CD44-Enhanced Growth Factor Signaling. Cancer Cell. 2018;33:1061–77. e6. doi: 10.1016/j.ccell.2018.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kang JS, Wanibuchi H, Morimura K, Gonzalez FJ, Fukushima S. Role of CYP2E1 in diethylnitrosamine-induced hepatocarcinogenesis in vivo. Cancer Res. 2007;67:11141–6. doi: 10.1158/0008-5472.CAN-07-1369. [DOI] [PubMed] [Google Scholar]
- 27.Verna L, Whysner J, Williams GM. N-nitrosodiethylamine mechanistic data and risk assessment: bioactivation, DNA-adduct formation, mutagenicity, and tumor initiation. Pharmacology & therapeutics. 1996;71:57–81. doi: 10.1016/0163-7258(96)00062-9. [DOI] [PubMed] [Google Scholar]
- 28.Buchmann A, Bauer-Hofmann R, Mahr J, Drinkwater NR, Luz A, Schwarz M. Mutational activation of the c-Ha-ras gene in liver tumors of different rodent strains: correlation with susceptibility to hepatocarcinogenesis. Proc Natl Acad Sci U S A. 1991;88:911–5. doi: 10.1073/pnas.88.3.911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jaworski M, Buchmann A, Bauer P, Riess O, Schwarz M. B-raf and Ha-ras mutations in chemically induced mouse liver tumors. Oncogene. 2005;24:1290–5. doi: 10.1038/sj.onc.1208265. [DOI] [PubMed] [Google Scholar]
- 30.Kang TW, Yevsa T, Woller N, Hoenicke L, Wuestefeld T, Dauch D, et al. Senescence surveillance of pre-malignant hepatocytes limits liver cancer development. Nature. 2011;479:547–51. doi: 10.1038/nature10599. [DOI] [PubMed] [Google Scholar]
- 31.Katz C, Low-Calle AM, Choe JH, Laptenko O, Tong D, Joseph-Chowdhury JN, et al. Wild-type and cancer-related p53 proteins are preferentially degraded by MDM2 as dimers rather than tetramers. Genes Dev. 2018;32:430–47. doi: 10.1101/gad.304071.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.DiGiammarino EL, Lee AS, Cadwell C, Zhang W, Bothner B, Ribeiro RC, et al. A novel mechanism of tumorigenesis involving pH-dependent destabilization of a mutant p53 tetramer. Nature structural biology. 2002;9:12–6. doi: 10.1038/nsb730. [DOI] [PubMed] [Google Scholar]
- 33.Mastellaro MJ, Seidinger AL, Kang G, Abrahao R, Miranda ECM, Pounds SB, et al. Contribution of the TP53 R337H mutation to the cancer burden in southern Brazil: Insights from the study of 55 families of children with adrenocortical tumors. Cancer. 2017;123:3150–8. doi: 10.1002/cncr.30703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vogelstein B, Papadopoulos N, Velculescu VE, Zhou S, Diaz LA, Jr, Kinzler KW. Cancer genome landscapes. Science. 2013;339:1546–58. doi: 10.1126/science.1235122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Borges LM, Ayres FM. R337H mutation of the TP53 gene as a clinical marker in cancer patients: a systematic review of literature. Genetics and molecular research : GMR. 2015;14:17034–43. doi: 10.4238/2015.December.16.4. [DOI] [PubMed] [Google Scholar]
- 36.Figueiredo BC, Sandrini R, Zambetti GP, Pereira RM, Cheng C, Liu W, et al. Penetrance of adrenocortical tumours associated with the germline TP53 R337H mutation. J Med Genet. 2006;43:91–6. doi: 10.1136/jmg.2004.030551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lwin TZ, Durant JJ, Bashford D. A fluid salt-bridging cluster and the stabilization of p53. J Mol Biol. 2007;373:1334–47. doi: 10.1016/j.jmb.2007.07.080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Corpe CP, Tu H, Eck P, Wang J, Faulhaber-Walter R, Schnermann J, et al. Vitamin C transporter Slc23a1 links renal reabsorption, vitamin C tissue accumulation, and perinatal survival in mice. J Clin Invest. 2010;120:1069–83. doi: 10.1172/JCI39191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schleicher RL, Carroll MD, Ford ES, Lacher DA. Serum vitamin C and the prevalence of vitamin C deficiency in the United States, 2003–2004 National Health and Nutrition Examination Survey (NHANES) Am J Clin Nutr. 2009;90:1252–63. doi: 10.3945/ajcn.2008.27016. [DOI] [PubMed] [Google Scholar]
- 40.Macedo GS, Lisboa da Motta L, Giacomazzi J, Netto CB, Manfredini V, C SV, et al. Increased Oxidative Damage in Carriers of the Germline TP53 p.R337H Mutation. PLoS ONE. 2012;7:e47010. doi: 10.1371/journal.pone.0047010. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.