This tumor database study examines the prevalence of PDL1 amplification and its utility as a response biomarker to PD-1/PD-L1 blockade in solid tumors.
Key Points
Question
What is the prevalence and utility of programmed cell death ligand 1 (PDL1) gene amplification as a response biomarker to programmed cell death/programmed cell death ligand 1 blockade in solid tumors?
Findings
In this study of 118 187 tumor samples from a deidentified database, including a subset of 2039 samples from a clinically annotated database, the prevalence of PDL1 amplification was 0.7%. The objective response rate for patients with solid tumors that harbored PDL1 amplification was 66.7%, with a median progression-free survival of 15.2 months.
Meaning
The results of this study suggest that PDL1 amplification occurs in a small subset of malignant tumors; however, testing for this alteration may be warranted because of the frequent and durable responses to programmed cell death/programmed cell death ligand 1 blockade.
Abstract
Importance
Copy number alterations in programmed cell death ligand 1 (PDL1 or CD274), programmed cell death 1 ligand 2 (PDCD1LG2 or PDL2), and Janus kinase 2 (JAK2) genes (chromosome 9p24.1) characterize Hodgkin lymphoma, resulting in high response rates to programmed cell death 1 (PD-1)/programmed cell death ligand 1 (PD-L1) blockade. The prevalence and utility of PDL1 amplification as a response biomarker to PD-1/PD-L1 blockade are unknown in other tumors.
Objectives
To examine the prevalence of PDL1 amplification and its utility as a response biomarker to PD-1/PD-L1 blockade in solid tumors.
Design, Setting, and Participants
This retrospective study (October 1, 2012, to October 1, 2017) used a deidentified tumor database from a commercial company and annotated clinical records from a subset of patients treated at a university tertiary referral center. The study analyzed 118 187 tumors from the deidentified database, including a clinically annotated subgroup of 2039 malignant tumors.
Interventions
Comprehensive genomic profiling was performed on all samples to determine PDL1 amplification, microsatellite instability, and tumor mutational burden (TMB). A subset of patients was treated with PD-1/PD-L1 blockade.
Main Outcomes and Measures
The prevalence of PDL1 amplification was determined among 118 187 patient samples that underwent next-generation sequencing. Solid tumors treated with checkpoint blockade were evaluated for response and progression-free survival (PFS).
Results
Of the 118 187 deidentified tumor samples, PDL1 amplifications were identified in 843 (0.7%), including more than 100 types of solid tumors. Most PDL1-amplified tumors (84.8%) had a low to intermediate TMB. PDL1 amplification did not always correlate with high-positive PD-L1 expression by immunohistochemical analysis. Six of 9 patients (66.7%) from 1 center with PDL1-amplified solid tumors had objective responses after checkpoint blockade administration. The median PFS among all treated patients was 15.2 months. Responders included 1 patient with glioblastoma (PFS, ≥5.2 months), 2 patients with head and neck squamous cell cancer (PFS, ≥9 and 15.2 months), 2 patients with metastatic basal cell cancer (PFS, 3.8 and ≥24.1 months), and 1 patient with urothelial cancer (PFS, ≥17.8 months).
Conclusions and Relevance
The results of this study suggest that PDL1 amplification occurs in a small subset of malignant tumors. Additional large-scale, prospective studies of PDL1-amplified cancers are warranted to confirm the responses to checkpoint blockade described herein, even in the absence of microsatellite instability, high PD-L1 expression, and a high TMB.
Introduction
Checkpoint blockade with anti–programmed cell death 1 (PD-1)/programmed cell death ligand 1 (PD-L1) and anti–cytotoxic T-lymphocyte–associated protein 4 (CTLA4) antibodies has revolutionized the treatment of solid and hematologic malignant tumors. However, immune checkpoint inhibitors are only effective in a subset of patients. Biomarkers for determining response to PD-1/PD-L1 blockade include PD-L1 expression,1,2 microsatellite instability (MSI),3 and a high tumor mutational burden (TMB).4,5,6
Response rates of 65% to 87% have been reported in patients with refractory classic Hodgkin lymphoma treated with checkpoint inhibitors.7,8 In nodular sclerosing Hodgkin lymphoma, amplification of the chromosomal region 9p24.1, which contains the genes programmed cell death ligand 1 (PDL1 or CD274) (OMIM 605402), programmed cell death ligand 2 (PDCD1LG2 or PDL2) (OMIM 605723), and Janus kinase 2 (JAK2) (OMIM 147796), is directly correlated with increased expression of these proteins on Reed-Sternberg cells.9 Overall, 105 of 108 biopsy specimens (97.2%) from patients with newly diagnosed classic Hodgkin lymphoma10 have had increased PDL1 and PDCD1LG2 copy numbers. This increase is attributable to 9p24.1 amplifications, copy number alterations (CNAs), or polysomy of chromosome 9p. In addition, expression and activation of JAK2, which is also encoded by a gene residing on the 9p24.1 locus, are increased in Hodgkin lymphoma Reed-Sternberg cells, further augmenting transcription of the PDL1 gene.9
The CNAs of the 9p24.1 locus have also been detected in 63% of primary mediastinal large B-cell lymphomas and 50% of primary central nervous system large B-cell lymphomas and are associated with high PD-L1 and PD-L2 expression on immunohistochemical analysis.9,11,12 Recently, a study13 found that all 5 patients with relapsed or refractory primary central nervous system large B-cell lymphoma or testicular large B-cell lymphoma treated with PD-1 blockade experienced an objective response, and 60% remained progression free at 13 to 17 months. Taken together, in certain lymphomas, chromosome 9p24.1 alterations, which include PDL1, are relatively common and are associated with high susceptibility to PD-1 blockade.
In contrast, data are limited regarding PDL1 amplifications in solid tumors. To date, such amplifications have only been detected in small studies of head and neck squamous cell carcinoma,14 cervical squamous cell carcinoma,15 triple-negative breast cancer,16,17,18 and non–small cell lung cancer.19 Consistent with the aforementioned data on lymphomas, recent case reports found responses to PD-1 blockade in patients with PDL1-amplified, microsatellite-stable colon cancer20 and metastatic basal cell carcinoma,21 suggesting the need for further interrogation of the potential utility of PDL1 amplifications as a biomarker for immune checkpoint blockade response. We describe, to our knowledge, the largest cohort of tumor samples (N = 118 187) evaluated for PDL1 CNAs and report the frequency of PDL1 amplification across a variety of solid tumors.
Methods
Patients and Samples
We analyzed 118 187 deidentified tumor samples from the Foundation Medicine (https://www.foundationmedicine.com/) database, including a subset of 2039 clinically annotated patient tumors from the University of California, San Diego (UCSD) Moores Center for Personalized Cancer Therapy from October 1, 2012, to October 1, 2017 (eFigure 1 in Supplement 1). This study was performed in accordance with UCSD Institutional Review Board guidelines for data analysis22,23 and for any investigational treatments for which patients gave written informed consent.
Profiling and Assessment of PDL1 Amplification, MSI, and TMB
Comprehensive Genomic Profiling and PDL1 (CD274) Assessment
Comprehensive genomic profiling was performed using the FoundationOne and FoundationOneHeme assay (Foundation Medicine), as previously described in detail.24,25 In brief, the pathologic diagnosis of each case was confirmed by review of hematoxylin-eosin–stained slides, and all samples that advanced to DNA extraction contained a minimum of 20% tumor cells. The fail rate was approximately 1%. Hybridization capture of exonic regions from 315, 327, or 405 cancer-related genes was applied to 50 ng or more of DNA extracted from formalin-fixed, paraffin-embedded cancer specimens. These libraries were sequenced to high, uniform median coverage (>500 times) and assessed for base substitutions, short insertions and deletions, CNAs, and gene fusions and rearrangements. Sequencing was performed from October 1, 2012, to October 1, 2017.24 PDL1 amplification was performed for 6 or more CNAs.
TMB Evaluation
For TMB (mutations per megabase), the number of somatic mutations detected on comprehensive genomic profiling (interrogating 1.2 Mb of the genome) were quantified, and that value was extrapolated to the whole exome using a validated algorithm.26 Alterations likely or known to be oncogenic drivers and germline polymorphisms were excluded. A TMB of 5 mutations per megabase or more was designated as low; 6 to 19, intermediate; and 20 or more, high.
MSI Assessment
The MSI status was calculated using 114 loci determined to be useful in detecting evidence of polymerase slippage and therefore MSI.27 The information from these loci were then used in principal component analysis to produce an MSI score. Ranges of MSI scores were assigned as high MSI (MSI-H), microsatellite stable, or intermediate or ambiguous MSI.
Database Analysis for PDL1 Amplification and TMB
To understand the large-scale prevalence of PDL1 amplification and its relevant associations, we analyzed 118 187 patient samples with cancer from the Foundation Medicine deidentified database. Only patients with chromosome 9p24.1 alterations in PDL1, PDL2, and/or JAK2 alterations were further reviewed (eFigure 1 in Supplement 1 and Supplement 2). We focused on patients with solid tumors.
Patient and Sample Selection
To retrieve data that would provide clinical correlations of PDL1 CNAs with checkpoint inhibitor response, we evaluated 2039 consecutive cancer samples from patients at the UCSD Moores Center for Personalized Cancer Therapy (October 1, 2012, to October 1, 2017). All patients had undergone comprehensive genomic profiling (Foundation Medicine; https://www.foundationmedicine.com/).
Pathology for TILs and Immunohistochemistry for PD-L1
Tumor samples, when available, were reviewed by a pathologist (H.-Y.W.) for enumeration of tumor-infiltrating lymphocytes (TILs) as described by Salgado et al.28,29 The mean percentage of TILs was quantified from evaluating 3 high-power fields (original magnification, ×400). Macrophages were excluded from the TIL count. Immunohistochemical analysis for PD-L1 expression was performed using commercially available assays (eTable 1 and eTable 2 in Supplement 1).
Outcomes
Responses were assessed based on Response Evaluation Criteria In Solid Tumors (RECIST) criteria.30 Progression-free survival (PFS) and overall survival (OS) were calculated using the Kaplan-Meier method (P values by log-rank test) (starting from the first day of immunotherapy). The PFS and OS are censored at the date that the patient was last seen provided that the patient’s cancer had not progressed (for PFS) and the patient had not died (for OS).
Statistical Analysis
The Fisher exact test was used to assess categorical variables. Bonferroni correction was applied as a multitesting correction. Statistical analyses were performed using GraphPad Prism, version 7.0. A 2-sided P ≤ .05 and Q ≤ .05 were considered to be statistically significant.
Results
Patient Characteristics
Overall, 843 of 118 187 patient samples (0.7%) that had undergone comprehensive genomic profiling had 6 or more CNAs in PDL1 (Table). A total of 405 gene panels were performed on 15 982 tumor samples, 327 gene panels on 450 tumor samples, and 315 gene panels on 101 755 tumor samples. PDL1 amplification was identified in 88 samples from the 405 gene panels, 5 samples from the 327 gene panels, and 750 samples from the 315 gene panels (total of 843 samples). PDL1 CNAs were identified in more than 100 solid tumor histologic types (eTable 3 in Supplement 1). The tumor type with the highest percentage of PDL1 amplification was mixed hepatocellular cholangiocarcinoma (10.5% of samples). Solid tumors with a significantly increased percentage of PDL1 amplification included breast carcinoma (111 [1.9%]; P < .001), head and neck squamous cell carcinoma (39 [3.1%]; P < .001), lung squamous cell carcinoma (50 [1.7%]; P < .001), undifferentiated soft-tissue sarcoma (13 [3.9%]; P < .001), thyroid anaplastic carcinoma (9 [5.1%]; P < .001), unknown primary squamous cell carcinoma (16 [2.0%]; P = .01), nasopharyngeal carcinoma (5 [5.1%]; P = .03), and kidney sarcomatoid carcinoma (4 [6.1%]; P = .04) (Table). Neoplasms notable for having a lower frequency of PDL1 CNAs included colorectal, pancreatic, and prostate cancer and melanoma (Table).
Table. Frequency of PDL1 Amplificationsa .
| Diagnosis | Total Patients, No. (%) (N = 118 187) | Patients With PDL1 Amplification | |
|---|---|---|---|
| Q Valueb | OR (95% CI) | ||
| Tumors with the highest prevalence of PDL1 amplification | |||
| Breast carcinoma (NOS) | 111/5838 (1.9) | <0.0001 | 3.0 (2.4-3.6) |
| Head and neck squamous cell carcinoma | 39/1275 (3.1) | <0.0001 | 4.6 (3.3-6.3) |
| Lung squamous cell carcinoma | 50/2952 (1.7) | <0.0001 | 2.5 (1.9-3.3) |
| Undifferentiated soft-tissue sarcoma | 13/330 (3.9) | <0.0001 | 5.8 (3.3-10.1) |
| Thyroid anaplastic carcinoma | 9/177 (5.1) | 0.0004 | 7.5 (3.8-14.8) |
| Soft-tissue sarcoma (NOS) | 18/903 (2.0) | 0.0069 | 2.9 (1.8-4.6) |
| Unknown primary squamous cell carcinoma | 16/788 (2.0) | 0.0119 | 2.9 (1.8-4.8) |
| Cervix squamous cell carcinoma | 10/374 (2.7) | 0.0188 | 3.9 (2.1-7.3) |
| Nasopharyngeal carcinoma | 5/99 (5.1) | 0.0293 | 7.4 (3.0-18.3) |
| Renal sarcomatoid carcinoma | 4/66 (6.1) | 0.0449 | 9.0 (3.3-24.9) |
| Bladder squamous cell carcinoma | 3/40 (7.5) | 0.0905 | 11.3 (3.5-36.8) |
| Liver mixed hepatocellular cholangiocarcinoma | 2/19 (10.5) | 0.1807 | 16.41 (3.8-71.2) |
| Lung sarcomatoid carcinoma | 5/187 (2.7) | 0.2226 | 3.8 (1.6-9.4) |
| Tumors with the lowest prevalence of PDL1 amplification | |||
| Colorectal adenocarcinoma | 18/9851 (0.18) | <0.0001 | 0.2 (0.1-0.4) |
| Pancreatic cancer | 1/3294 (0.03) | <0.0001 | 0.04 (0.01-0.3) |
| Multiple myeloma | 2/2707 (0.07) | <0.0001 | 0.1 (0.03-0.4) |
| Acute myeloid leukemia | 0/1273 | 0.0133 | 0 (0-0.9) |
| Prostate cancer | 5/2461 (0.2) | 0.0337 | 0.3 (0.1-0.7) |
| Myelodysplastic syndromes | 0/861 | 0.0984 | 0 (0-1.3) |
| Cutaneous melanoma | 1/1090 (0.09) | 0.1426 | 0.1 (0.02-0.9) |
| PDL1 amplification in common tumor histologic types | |||
| Lung adenocarcinoma | 90/14 910 (0.6) | 1.0000 | 0.8 (0.4-1.7) |
| Glioblastoma | 11/3199 (0.3) | 0.2095 | 0.5 (0.3-0.9) |
| Gastroesophageal junction adenocarcinoma | 5/1956 (0.3) | 0.2095 | 0.4 (0.2-0.9) |
| Lung small cell carcinoma | 14/1071 (1.3) | 0.4373 | 1.9 (1.1-3.2) |
| Ovarian epithelial carcinoma | 14/1052 (1.3) | 0.4100 | 1.9 (1.1-3.2) |
| Renal cell carcinoma | 4/766 (0.5) | 1.0000 | 0.7 (0.3-2.0) |
| Stomach adenocarcinoma | 8/1325 (0.6) | 1.0000 | 0.8 (0.4-1.7) |
| Endometrial adenocarcinoma | 6/1223 (0.2) | 1.0000 | 0.2 (0.03-1.7) |
| Hepatocellular carcinoma | 3/691 (0.4) | 1.0000 | 0.6 (0.2-1.9) |
| Cholangiocarcinoma | 7/1867 (0.4) | 1.0000 | 0.5 (0.3-1.1) |
Abbreviations: NOS, not otherwise specified; OR, odds ratio; PDL1, programmed cell death ligand 1.
Data are provided for solid tumors with the highest percentile of PDL1 amplification. Nonsolid tumors, such as Hodgkin lymphoma, also had PDL1 amplification in 97% of patients.10 Tumors with a significant percentage of PDL1 amplification and all tumor types with a low-percentile PDL1 amplification are reported.
Calculated using Fisher’s exact test. Q ≤ 0.05 is considered to be significant when using Bonferroni correction.
TMB and MSI
The mean TMB for PDL1-amplified tumors was 13.3 mutations per megabase, and the median was 6.3 mutations per megabase. For unamplified tumors, the mean was 7.4 mutations per megabase and the median was 3.6 mutations per megabase (eTable 3 in Supplement 1). Overall, 128 PDL1-amplified tumors (15.2%) were classified as having high TMB compared with 7510 unamplified tumors (6.4%). Most PDL1-amplified tumors had a low to intermediate TMB (84.8%). For some tumors (ie, kidney sarcomatoid carcinoma [n = 4], pancreas ductal carcinoma [n = 1], and prostate cancer [n = 5]), 100% of PDL1-amplified tumors had a low TMB (≤5 mutations per megabase). The MSI-H and PDL1 amplification were not mutually exclusive. Five of 741 patients (0.7%) with PDL1 amplification (2 gastrointestinal tumors and 3 carcinomas of unknown primary) who were tested for microsatellite status were MSI-H; 1435 of 103 373 patients (1.4%) who did not have PDL1 amplification and were tested for microsatellite status were MSI-H. In the UCSD cohort (n = 13), the median TMB for PDL1-amplified tumors was 9 mutations per megabase vs 4 mutations per megabase for non–PDL1-amplified tumors (P = .007). Nine of the 13 patients (69.2%) had an intermediate to high TMB, whereas 4 patients (30.8%) had a low TMB (1-5 mutations per megabase). Finally, 11 of 13 tumors (84.6%) tested for MSI were stable.
Clinical Characteristics of the Cohort With PDL1 CNAs
Thirteen patients were identified with PDL1 CNAs from the 2039 patients who had undergone comprehensive genomic profiling (eTable 4 and eFigure 1 in Supplement 1). All 13 patients had coamplification of PDCD1LG2 (PDL2), and all but 1 (92.3%) had coamplification of JAK2. All 13 (100%) had locally advanced (3 [23.1%]) or metastatic (10 [76.9%]) disease (2 with hematologic malignant tumors and 11 with solid tumors). The median time alive with locally advanced or metastatic disease was 26.5 months (range, 7.9-65.5 months). Nine different malignant tumors were identified that harbored PDL1 CNAs, including head and neck tumors (3 patients) and a glioblastoma (eTable 4 in Supplement 1). Nine patients (69.2%) (all with solid tumors) received therapy with a PD-1/PD-L1 inhibitor.
Genomics, PD-L1 Expression, and TILs in the Cohort With PDL1 CNAs
A total of 70 genes with 143 alterations were identified among the 13 patients with PDL1 CNAs (Figure 1 and eTable 5 in Supplement 1). Among the 13 patients, only 5 samples from 5 patients were available for pathologic evaluation for TILs (4 of them were stromal TILs and 1 [B-cell lymphoma] was intratumoral TILs). The TILs ranged from a mean of 10% to 60% per high-power field (original magnification, ×400). Four of 6 tumors (66.7%) tested expressed PD-L1 (immunohistochemical analysis) (eTable 1 in Supplement 1). Of note, one patient with glioblastoma and another patient with metastatic basal carcinoma had undetectable PD-L1 expression by immunohistochemical analysis, but both responded to checkpoint blockade.
Figure 1. Genomic Alterations.
Total genomic alterations in 13 patients with cancer with alterations that involve programmed cell death ligand 1 gene (PDL1).
Additional Alterations in PDL1 and PDCD1L2G (PDL2)
Eleven (0.001%) of the 118 187 samples harbored PDL1 exon 7 truncations. Two individuals in the UCSD cohort had other alterations that involved PDL1 and PDCD1LG2 (eTable 6 in Supplement 1). One patient had metastatic head and neck squamous cell carcinoma that harbored a PDL1 exon 7 truncation. This alteration disrupts the 3′ untranslated region of PD-L1.31 The patient achieved a partial response to treatment with durvalumab, a PD-L1 inhibitor. The other patient had metastatic cholangiocarcinoma with a PKD1P1-PDCD1LG2 rearrangement (but was not treated with checkpoint blockade). This alteration was not identified in any of the other 118 186 samples.
Response to Checkpoint Blockade
Nine of the 13 patients (69.2%) with PDL1 amplification were treated with checkpoint blockade (all solid tumors) (eTable 5 in Supplement 1). The median number of prior therapies in these 9 patients was 4 (range, 1-7). Five patients were treated with PD-1/PD-L1 inhibitor monotherapy, 3 with a PD-1/PD-L1 inhibitor plus an investigational agent, and 1 with anti–PD-1 and anti-CTLA4 combination therapy. The response rate was 66.7%. The median PFS among the 9 patients was 15.2 months (range, 1.6 to ≥24.1 months); median OS was not reached from the start of checkpoint blockade (range, 1.6 to ≥24.1 months) (Figure 2). Responders included 1 patient with glioblastoma (PFS, ≥5.2 months), 2 patients with head and neck squamous cell cancer (PFS, ≥9 and 15.2 months), 2 patients with metastatic basal cell cancer (PFS, 3.8 and ≥24.1 months), and 1 patient with urothelial cancer (PFS, ≥17.8 months). In addition, a patient with primary mediastinal lymphoma that was refractory to chemotherapy, including high-dose chemotherapy followed by autologous stem cell rescue, had an ongoing complete response to allogeneic stem cell transplantation at 24.1 months.
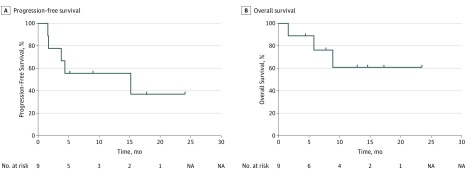
Figure 2. Patient Survival.
Progression-free survival among 9 patients with PDL1 amplification treated with checkpoint blockade (median, 15.2 months; range, 1.2 to ≥24.1 months). Median overall survival among patients with PDL1 amplification was not reached from start of checkpoint blockade (range, 1.6 to ≥24.1 months). NA indicates not applicable.
One patient had progressive glioblastoma after tumor resection followed by adjuvant radiation therapy with concurrent temozolamide.32 Comprehensive genomic profiling identified 12 characterized alterations, including PDL1, PDCD1LG2, and JAK2 amplifications. MET protooncogene (MET) (OMIM 164860) and mouse double minute homolog 2 (MDM2) (OMIM 164785) amplifications were also identified. The case was presented at the molecular tumor board, and treatment with checkpoint inhibition was debated because of the presence of MDM2 amplification, which has been associated with hyperprogression.33 However, because of the grave prognosis of glioblastoma, the patient was prescribed combination therapy with nivolumab and the MET inhibitor cabozantinib (after signing consent for an institutional review board–approved protocol [Study of Molecular Profile-Related Evidence to Determine Individualized Therapy for Advanced or Poor Prognosis Cancers (I-PREDICT)23]). Brain magnetic resonance imaging (eFigure 2 in Supplement 1) performed 4 weeks after therapy initiation demonstrated a partial response with decreased enhancement within the primary mass and decreased mass effect. Cabozantinib (weeks 14-22) and nivolumab (weeks 11-23) were given secondary to the development of a transaminitis. Subsequent brain magnetic resonance imaging demonstrated an improving and ongoing response at 5.2 months.
Discussion
In our study, the prevalence of PDL1 CNAs in a large cohort of diverse tumors was 0.7%. These alterations were identified in a small subset of multiple solid tumor types, including rare neoplasms, such as bladder squamous cell carcinoma, undifferentiated soft-tissue sarcomas, and sarcomatoid renal cell carcinoma. Furthermore, we found that PDL1 CNAs can be associated with responses to checkpoint blockade across a diverse spectrum of tumors (eTable 4 in Supplement 1). Six of 9 patients (66.7%) with PDL1 amplification responded to immunotherapy vs 45 of 151 patients (29.8%) in the overall UCSD-treated cohort (P = .03).5
Although rare outside certain lymphomas, identification of amplifications in PDL1 is important because this subset of tumors appears to have a high likelihood of responding to checkpoint blockade. This situation is analogous to that in patients with lung cancer that harbors anaplastic lymphoma kinase (ALK) (OMIM 105590) and V-ROS avian UR2 sarcoma virus oncogene homolog 1 ROS1 (OMIM 165020) alterations, which both confer sensitivity to ALK inhibitors.34,35 Regarding histologic agnostic responsiveness, MSI-H confers response to checkpoint inhibitors across cancers and neurotrophic tyrosine receptor kinase fusions respond to neurotrophic tyrosine receptor kinase targeting in a tissue-agnostic fashion.3,36
Infection has been implicated in certain types of neoplasms identified to have a higher prevalence of PDL1 amplifications. These neoplasms include bladder squamous cell carcinoma associated with Schistosoma hematobium, nasopharyngeal carcinoma, Epstein-Barr virus, head and neck squamous cell carcinoma, human papillomavirus, and mixed cellularity variant of Hodgkin lymphoma, which is associated with Epstein-Barr infection.37 Viral-associated malignant neoplasms may be susceptible to tumor immune responses, perhaps through upregulation of APOBEC (apolipoprotein B messenger RNA editing enzyme, catalytic polypeptide-like), a family of cytidine deaminases that help protect from viral infections. APOBEC upregulation in turn correlates with high levels of PD-L1.2,38 It is plausible that these tumors are using PDL1 amplification as a mechanism of immune escape from an endogenous immune response.
Sarcomatoid renal cell carcinoma (6.5% of which had PDL1 amplification—one of the highest rates for solid tumors) (Table) is a rare subtype of renal cell carcinoma. Although only accounting for approximately 5% of renal cell carcinomas, the aggressive nature of this variant results in many patients having metastatic disease at diagnosis.39 In addition, these tumors are responsive to checkpoint blockade at least in a small series, with 2 of 6 patients achieving objective response to atezolizumab (PD-L1 inhibitor).40 PD-L1 expression in sarcomatoid renal cell carcinoma appears to be higher compared with standard renal cell carcinoma without sarcomatoid differentiation.41
Glioblastoma is a lethal tumor with limited effective treatment options. Outside MSI-H glioblastoma, checkpoint blockade has not been effective.42 In this report, we demonstrate, for the first time to our knowledge, a response to nivolumab in a PDL1-amplified glioblastoma.
Expression of PD-L1 was identified by immunohistochemical analysis in 4 of 6 patients who were tested. Of interest, 2 of the patients who lacked PD-L1 protein expression (1 with glioblastoma and 1 with metastatic basal cell carcinoma) (eTable 5 in Supplement 1) responded to PD-1 blockade. A recent report14 in head and neck squamous cell carcinoma also found that PDL1 CNAs were concordant with PD-L1 expression by immunohistochemical analysis only 73% of the time. Presence of gene amplification with no or low-level PD-L1 protein expression should make immune checkpoint blockade inhibitors less effective. Posttranscriptional splicing and methylation could be mechanisms that limit expression. However, insufficient sampling of tumor and other technical problems with immunohistochemical analysis, in part related to tumor heterogeneity and the presence of stroma or attributable to differences in affinity of distinct anti–PD-L1 antibodies, may limit the accuracy of the protein expression methods and may explain responses in patients who lacked PD-L1 expression on immunohistochemical analysis. Other mechanisms, such as expression of PD-L2 rather than PD-L1, may also be operative when patients respond to anti-PD1 agents in the absence of PD-L1 expression (because PD-L1 and PD-L2 interact with PD-1).43 All these issues merit in-depth exploration in larger cohorts of treated patients to better understand the association among PD-L1 expression, PDL1 amplification, and response to checkpoint blockade.
Of interest, in addition to the 13 patients in the UCSD cohort who had PDL1 amplification, 2 patients harbored alterations that involved PDL1 and PDCD1LG2 (eTable 4 in Supplement 1) that were not CNAs. The first alteration, a PDL1 exon 7 truncation, is predicted to disrupt the 3′ untranslated region of PDL1. Similar alterations have been observed in many tumor types and correlate with increased PD-L1 expression, presumably via loss of inhibitory microRNA binding sites.31,44,45 This patient achieved a partial response that lasted 9 months with a durvalumab (anti–PD-L1)–based regimen. The other alteration, with a PKD1P1-PDCD1LG2 rearrangement, has not been previously reported or characterized. However, translocations that involve PDCD1LG2 and numerous partners have been highly characterized in primary mediastinal large B-cell lymphoma and result in increased PD-L2 expression.2
Limitations
The small number of patients precludes definitive conclusions regarding response rates, PFS, or OS except to suggest that further additional prospective clinical trials of checkpoint blockade in PDL1-amplified cancers are warranted. In addition, the current assay was validated for 6 or more copy numbers of PDL1, and future studies should determine the frequency of CNAs that are less than 6. This study also did not assess features of the tumor microenvironment, such as the presence of transforming growth factor β, which can have profound influences on the response to checkpoint blockade.46,47 Thus, application of checkpoint blockade and comparison to standard-of-care chemotherapy require properly designed randomized clinical trials with both PFS and OS end points.
Conclusions
Our data suggest that PDL1 CNAs are found in a small subgroup of diverse solid tumors and may correlate with responses to checkpoint blockade. Additional prospective studies are needed to validate this finding and to determine whether routine testing for this alteration is warranted.
eFigure. CONSORT Diagram
eTable 1. Biomarkers for Response to Immunotherapy in 13 Patients With PD-L1 Amplification Seen at University of California, San Diego Moores Cancer Center
eTable 2. Definitions for PD-L1 IHC Positivity
eTable 3. PDL1 Amplification by Solid Tumor Type (PDL1 Amplification Was Seen in 0.7% of 118 187 Samples With Malignancies)
eTable 4. Clinical Characteristics of 13 Patients With PDL1 and/or PDCD1LG2 (PDL2) Amplifications Seen at UCSD Center for Personalized Cancer Therapy
eTable 5. Patient Profiles and treatment Response
eTable 6. PDL1/L2 Alterations Other Than Amplifications
eFigure 2. Patient #4 With Glioblastoma
Complete Data Set
References
- 1.Patel SP, Kurzrock R. PD-L1 expression as a predictive biomarker in cancer immunotherapy. Mol Cancer Ther. 2015;14(4):847-856. doi: 10.1158/1535-7163.MCT-14-0983 [DOI] [PubMed] [Google Scholar]
- 2.Goodman A, Patel SP, Kurzrock R. PD-1-PD-L1 immune-checkpoint blockade in B-cell lymphomas. Nat Rev Clin Oncol. 2017;14(4):203-220. doi: 10.1038/nrclinonc.2016.168 [DOI] [PubMed] [Google Scholar]
- 3.Le DT, Uram JN, Wang H, et al. PD-1 blockade in tumors with mismatch-repair deficiency. N Engl J Med. 2015;372(26):2509-2520. doi: 10.1056/NEJMoa1500596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rizvi NA, Hellmann MD, Snyder A, et al. Cancer immunology: mutational landscape determines sensitivity to PD-1 blockade in non-small cell lung cancer. Science. 2015;348(6230):124-128. doi: 10.1126/science.aaa1348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Goodman AM, Kato S, Bazhenova L, et al. Tumor mutational burden as an independent predictor of response to immunotherapy in diverse cancers. Mol Cancer Ther. 2017;16(11):2598-2608. doi: 10.1158/1535-7163.MCT-17-0386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Khagi Y, Goodman AM, Daniels GA, et al. Hypermutated circulating tumor DNA: correlation with response to checkpoint inhibitor-based immunotherapy. Clin Cancer Res. 2017;23(19):5729-5736. doi: 10.1158/1078-0432.CCR-17-1439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ansell SM, Lesokhin AM, Borrello I, et al. PD-1 blockade with nivolumab in relapsed or refractory Hodgkin’s lymphoma. N Engl J Med. 2015;372(4):311-319. doi: 10.1056/NEJMoa1411087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Armand P, Shipp MA, Ribrag V, et al. Programmed death-1 blockade with pembrolizumab in patients with classical Hodgkin lymphoma after brentuximab vedotin failure. J Clin Oncol. 2016;34(31):3733-3739. doi: 10.1200/JCO.2016.67.3467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Green MR, Monti S, Rodig SJ, et al. Integrative analysis reveals selective 9p24.1 amplification, increased PD-1 ligand expression, and further induction via JAK2 in nodular sclerosing Hodgkin lymphoma and primary mediastinal large B-cell lymphoma. Blood. 2010;116(17):3268-3277. doi: 10.1182/blood-2010-05-282780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Roemer MGM, Advani RH, Ligon AH, et al. PD-L1 and PD-L2 genetic alterations define classical Hodgkin lymphoma and predict outcome. J Clin Oncol. 2016;34(23):2690-2697. doi: 10.1200/JCO.2016.66.4482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shi M, Roemer MGM, Chapuy B, et al. Expression of programmed cell death 1 ligand 2 (PD-L2) is a distinguishing feature of primary mediastinal (thymic) large B-cell lymphoma and associated with PDCD1LG2 copy gain. Am J Surg Pathol. 2014;38(12):1715-1723. doi: 10.1097/PAS.0000000000000297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chapuy B, Roemer MGM, Stewart C, et al. Targetable genetic features of primary testicular and primary central nervous system lymphomas. Blood. 2016;127(7):869-881. doi: 10.1182/blood-2015-10-673236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nayak L, Iwamoto FM, LaCasce A, et al. PD-1 blockade with nivolumab in relapsed/refractory primary central nervous system and testicular lymphoma. Blood. 2017;129(23):3071-3073. doi: 10.1182/blood-2017-01-764209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Straub M, Drecoll E, Pfarr N, et al. CD274/PD-L1 gene amplification and PD-L1 protein expression are common events in squamous cell carcinoma of the oral cavity. Oncotarget. 2016;7(11):12024-12034. doi: 10.18632/oncotarget.7593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Howitt BE, Sun HH, Roemer MGM, et al. Genetic basis for PD-L1 expression in squamous cell carcinomas of the cervix and vulva. JAMA Oncol. 2016;2(4):518-522. doi: 10.1001/jamaoncol.2015.6326 [DOI] [PubMed] [Google Scholar]
- 16.Barrett MT, Anderson KS, Lenkiewicz E, et al. Genomic amplification of 9p24.1 targeting JAK2, PD-L1, and PD-L2 is enriched in high-risk triple negative breast cancer. Oncotarget. 2015;6(28):26483-26493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guo L, Li W, Zhu X, et al. PD-L1 expression and CD274 gene alteration in triple-negative breast cancer: implication for prognostic biomarker. Springerplus. 2016;5(1):805. doi: 10.1186/s40064-016-2513-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Balko JM, Schwarz LJ, Luo N, et al. Triple-negative breast cancers with amplification of JAK2 at the 9p24 locus demonstrate JAK2-specific dependence. Sci Transl Med. 2016;8(334):334ra53. doi: 10.1126/scitranslmed.aad3001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ikeda S, Okamoto T, Okano S, et al. PD-L1 Is upregulated by simultaneous amplification of the PD-L1 and JAK2 genes in non-small cell lung cancer. J Thorac Oncol. 2016;11(1):62-71. doi: 10.1016/j.jtho.2015.09.010 [DOI] [PubMed] [Google Scholar]
- 20.Sorscher S, Resnick J, Goodman M. First case report of a dramatic radiographic response to a checkpoint inhibitor in a patient with proficient mismatch repair gene expressing metastatic colorectal cancer. JCO Precis Oncol. 2017;1:1-4. doi: 10.1200/PO.16.00005 [DOI] [PubMed] [Google Scholar]
- 21.Ikeda S, Goodman AM, Cohen PR, et al. Metastatic basal cell carcinoma with amplification of PD-L1: exceptional response to anti-PD1 therapy. NPJ Genom Med. 2016;1:16037. doi: 10.1038/npjgenmed.2016.37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.ClinicalTrials.gov Study of Personalized Cancer Therapy to Determine Response and Toxicity (UCSD_PREDICT). NCT02478931. https://clinicaltrials.gov/ct2/show/NCT02478931. Accessed April 17, 2018.
- 23.ClinicalTrials.gov Study of Molecular Profile-Related Evidence to Determine Individualized Therapy for Advanced or Poor Prognosis Cancers (I-PREDICT). NCT02534675. https://clinicaltrials.gov/ct2/show/NCT02534675. Accessed April 17, 2018.
- 24.Frampton GM, Fichtenholtz A, Otto GA, et al. Development and validation of a clinical cancer genomic profiling test based on massively parallel DNA sequencing. Nat Biotechnol. 2013;31(11):1023-1031. doi: 10.1038/nbt.2696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.He J, Abdel-Wahab O, Nahas MK, et al. Integrated genomic DNA/RNA profiling of hematologic malignancies in the clinical setting. Blood. 2016;127(24):3004-3014. doi: 10.1182/blood-2015-08-664649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chalmers ZR, Connelly CF, Fabrizio D, et al. Analysis of 100,000 human cancer genomes reveals the landscape of tumor mutational burden. Genome Med. 2017;9(1):34. doi: 10.1186/s13073-017-0424-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hall MJ, Gowen K, Sanford EM, et al. Evaluation of microsatellite instability (MSI) status in 11,573 diverse solid tumors using comprehensive genomic profiling (CGP). J Clin Oncol. 2016;34(15):1523. [Google Scholar]
- 28.Salgado R, Denkert C, Campbell C, et al. Tumor-infiltrating lymphocytes and associations with pathological complete response and event-free survival in HER2-positive early-stage breast cancer treated with lapatinib and trastuzumab: a secondary analysis of the NeoALTTO trial. JAMA Oncol. 2015;1(4):448-454. doi: 10.1001/jamaoncol.2015.0830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Salgado R, Denkert C, Demaria S, et al. ; International TILs Working Group 2014 . The evaluation of tumor-infiltrating lymphocytes (TILs) in breast cancer: recommendations by an International TILs Working Group 2014. Ann Oncol. 2015;26(2):259-271. doi: 10.1093/annonc/mdu450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chalian H, Töre HG, Horowitz JM, Salem R, Miller FH, Yaghmai V. Radiologic assessment of response to therapy: comparison of RECIST versions 1.1 and 1.0. Radiographics. 2011;31(7):2093-2105. doi: 10.1148/rg.317115050 [DOI] [PubMed] [Google Scholar]
- 31.Kataoka K, Shiraishi Y, Takeda Y, et al. Aberrant PD-L1 expression through 3′-UTR disruption in multiple cancers. Nature. 2016;534(7607):402-406. doi: 10.1038/nature18294 [DOI] [PubMed] [Google Scholar]
- 32.Stupp R, Mason WP, van den Bent MJ, et al. ; European Organisation for Research and Treatment of Cancer Brain Tumor and Radiotherapy Groups; National Cancer Institute of Canada Clinical Trials Group . Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N Engl J Med. 2005;352(10):987-996. doi: 10.1056/NEJMoa043330 [DOI] [PubMed] [Google Scholar]
- 33.Kato S, Goodman A, Walavalkar V, Barkauskas DA, Sharabi A, Kurzrock R. Hyper-progressors after immunotherapy: analysis of genomic alterations associated with accelerated growth rate. Clin Cancer Res. 2017;23(15):4242-4250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shaw AT, Kim D-W, Mehra R, et al. Ceritinib in ALK-rearranged non–small-cell lung cancer. N Engl J Med. 2014;370(13):1189-1197. doi: 10.1056/NEJMoa1311107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shaw AT, Ou S-HI, Bang Y-J, et al. Crizotinib in ROS1-rearranged non–small-cell lung cancer. N Engl J Med. 2014;371(21):1963-1971. doi: 10.1056/NEJMoa1406766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hyman D, Laetsch T, Kummar S, et al. The efficacy of larotrectinib (LOXO-101), a selective tropomyosin receptor kinase (TRK) inhibitor, in adult and pediatric TRK fusion cancers [abstract]. J Clin Oncol. 2017;35(18):LBA2501. doi: 10.1200/JCO.2017.35.18_suppl.LBA2501 [DOI] [Google Scholar]
- 37.Green MR, Rodig S, Juszczynski P, et al. Constitutive AP-1 activity and EBV infection induce PD-L1 in Hodgkin lymphomas and posttransplant lymphoproliferative disorders: implications for targeted therapy. Clin Cancer Res. 2012;18(6):1611-1618. doi: 10.1158/1078-0432.CCR-11-1942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Boichard A, Tsigelny IF, Kurzrock R. High expression of PD-1 ligands is associated with kataegis mutational signature and APOBEC3 alterations. Oncoimmunology. 2017;6(3):e1284719. doi: 10.1080/2162402X.2017.1284719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shuch B, Bratslavsky G, Linehan WM, Srinivasan R. Sarcomatoid renal cell carcinoma: a comprehensive review of the biology and current treatment strategies. Oncologist. 2012;17(1):46-54. doi: 10.1634/theoncologist.2011-0227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.McDermott DF, Sosman JA, Sznol M, et al. Atezolizumab, an anti-programmed death-ligand 1 antibody, in metastatic renal cell carcinoma: long-term safety, clinical activity, and immune correlates from a phase Ia study. J Clin Oncol. 2016;34(8):833-842. doi: 10.1200/JCO.2015.63.7421 [DOI] [PubMed] [Google Scholar]
- 41.Joseph RW, Millis SZ, Carballido EM, et al. PD-1 and PD-L1 expression in renal cell carcinoma with sarcomatoid differentiation. Cancer Immunol Res. 2015;3(12):1303-1307. doi: 10.1158/2326-6066.CIR-15-0150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bouffet E, Larouche V, Campbell BB, et al. Immune checkpoint inhibition for hypermutant glioblastoma multiforme resulting from germline biallelic mismatch repair deficiency. J Clin Oncol. 2016;34(19):2206-2211. doi: 10.1200/JCO.2016.66.6552 [DOI] [PubMed] [Google Scholar]
- 43.Bhatti S, Heldstab J, Lehn C, et al. Clinical activity of pembrolizumab in a patient with metastatic triple-negative breast cancer without tumor programmed death-ligand 1 expression: a case report and correlative biomarker analysis. JCO Precis Oncol. 2017;(1):1-6. doi: 10.1200/PO.17.00032 [DOI] [PubMed] [Google Scholar]
- 44.Chong LC, Twa DDW, Mottok A, et al. Comprehensive characterization of programmed death ligand structural rearrangements in B-cell non-Hodgkin lymphomas. Blood. 2016;128(9):1206-1213. doi: 10.1182/blood-2015-11-683003 [DOI] [PubMed] [Google Scholar]
- 45.Twa DDW, Mottok A, Chan FC, et al. Recurrent genomic rearrangements in primary testicular lymphoma. J Pathol. 2015;236(2):136-141. doi: 10.1002/path.4522 [DOI] [PubMed] [Google Scholar]
- 46.Tauriello DVF, Palomo-Ponce S, Stork D, et al. TGFβ drives immune evasion in genetically reconstituted colon cancer metastasis. Nature. 2018;554(7693):538-543. doi: 10.1038/nature25492 [DOI] [PubMed] [Google Scholar]
- 47.Mariathasan S, Turley SJ, Nickles D, et al. TGFβ attenuates tumour response to PD-L1 blockade by contributing to exclusion of T cells. Nature. 2018;554(7693):544-548. doi: 10.1038/nature25501 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
eFigure. CONSORT Diagram
eTable 1. Biomarkers for Response to Immunotherapy in 13 Patients With PD-L1 Amplification Seen at University of California, San Diego Moores Cancer Center
eTable 2. Definitions for PD-L1 IHC Positivity
eTable 3. PDL1 Amplification by Solid Tumor Type (PDL1 Amplification Was Seen in 0.7% of 118 187 Samples With Malignancies)
eTable 4. Clinical Characteristics of 13 Patients With PDL1 and/or PDCD1LG2 (PDL2) Amplifications Seen at UCSD Center for Personalized Cancer Therapy
eTable 5. Patient Profiles and treatment Response
eTable 6. PDL1/L2 Alterations Other Than Amplifications
eFigure 2. Patient #4 With Glioblastoma
Complete Data Set