Abstract
Long noncoding RNAs (lncRNAs) are a class of noncoding RNA, involved in regulation of diverse physiological and pathological processes. Ovarian cancer is the leading cause of death among all gynecological malignancies in the world and its underlying mechanism is still unclear. lncRNAs exhibit multiple biological functions in various stages of ovarian cancer development. We will discuss and summarize the new and important lncRNAs and their involvement in disease, which might represent promising therapeutic targets. Therapeutic intervention based on silencing or functional inhibition of target lncRNAs will be beneficial for ovarian cancer patients.
Keywords: Long noncoding RNA, ovarian cancer, biomarker, therapeutics, drug resistance, prognosis
Introduction
Humans were supposed to have many more genes than less complex organisms. However, the number is not much different than the estimated number of genes (20 000) in roundworm Caenorhabditis elegans or mice, indicating complexity does not correlate with the number of protein-coding genes. However, the complexity definitely correlates with the percentage of non-protein-coding sequences. Ninety-seven percent of the human genome is non-protein-coding, consisting of introns, regulatory sequences and noncoding RNAs [1]. A type of noncoding RNA, long noncoding RNA (lncRNA) is longer than 200 nucleotide transcripts in length, has a variety of biological functions and is closely associated with tumor development. lncRNAs provide a novel way of regulating the gene expression and function at all levels of DNA, RNA or proteins. lncRNAs are at the epicenter of understanding how the vast sequence in the genome regulates different pathways including cancer. They hold an enormous potential to understand the participation of the noncoding genome in different processes and can help to bridge the vast gap that exists between cancer drug discovery and treatment. There has been an exponential rise in the number of publications during the past few years related to the role of lncRNAs in cancer biology (Figure 1). Not many lncRNAs have been functionally characterized, many of the lncRNAs have been identified by revisiting the array datasets on the publicly available resources [2,3], making it necessary to understand the mechanisms and biology of this new class of regulators, which is now at the center of various physiological and pathological processes. This information will be very useful in developing biomarker-driven cancer therapeutics.
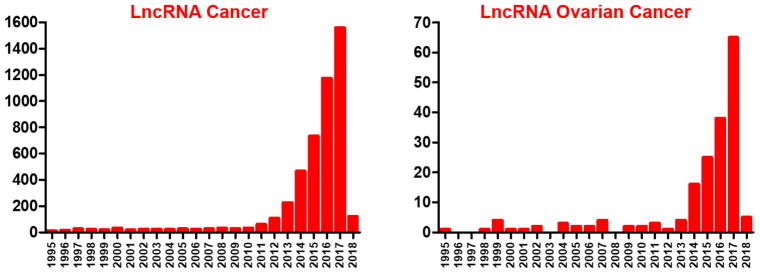
Figure 1.
Increasing number of articles about lncRNA and cancer, and lncRNA specific to ovarian cancer. PubMed was searched with keywords ‘lncRNA cancer’ or ‘lncRNA Ovarian Cancer’ (https://www.ncbi.nlm.nih.gov/pubmed).
Ovarian cancer
Ovarian cancer (OvCa) is the leading cause of death among gynecological malignancies in the world, and recurrent OvCa is almost always incurable [4]. Its underlying mechanism is still unclear. Several genetic and environmental factors have been shown to be implicated in the development of this type of cancer. The International Federation of Gynecology and Obstetrics (FIGO) Committee on Gynecologic Oncology has provided a staging system for OvCa [5]. The lack of specific signs and symptoms and the deficiency in screening programs have resulted in the late-stage diagnosis of OvCa, which in turn leads to the poor survival of these patients. The overall 5-year survival rate is low for the advanced stages OvCa owing to late diagnosis (because of its asymptomatic nature) and resistance to conventional carboplatin plus Taxol chemotherapy or relapse.
Better understanding of OvCa biology is needed to improve its management and early diagnosis. Such defects have necessitated the implementation of experimental approaches and clinical studies to discover and assess biomarkers associated with early-stage disease [6]. Compared with research on microRNAs, research on lncRNAs is still in its infancy. Studies in recent years have demonstrated that lncRNAs exhibit multiple biological functions in various stages of OvCa development. lncRNAs are closely involved in the pathogenesis of OvCa. The expression of lncRNAs indicates the early diagnosis, prognosis and response to chemotherapy of OvCa. Further research efforts are needed before fully identifying, characterizing and elucidating the actual functions of lncRNAs in OvCa at the molecular level and putting them into clinical practice [7]. Reports on lncRNAs associated with OvCa are very disjointed, which makes it necessary for an effort to study and develop a signature based on lncRNAs associated with OvCa recurrence to facilitate better OvCa therapy [4].
lncRNA-based therapeutics, biogenesis and mode of action
lncRNAs are important for the activation or repression of genes relevant to a variety of disorders. They work either as tumor suppressors or oncogenes or both. The lncRNAs are very difficult to identify from a suitable classification method. No unified mechanism of classification of lncRNAs exists. The most common relatively convenient way to classify lncRNAs depends on the genomic context, which is the position in the chromosome where the lncRNA is transcribed. The five major known classes are: natural antisense transcript, pseudogenes, large intergenic noncoding RNA, long intronic ncRNAs and other uncharacterized and divergent transcripts [8]. Figure 2 depicts the lncRNAs on the basis of their genomic context or biogenesis [9]. On the basis of targeting mechanisms, lncRNAs can be classified as: (i) Signal – show cell-type-specific expression and respond to diverse stimuli; (ii) Decoy – bind and titrate away a protein target, but do not exert any additional functions; (iii) Guide – bind proteins and then direct the localization of ribonucleoprotein complex to specific targets; (iv) Scaffold – serve as central platforms to bring together multiple proteins to form ribonucleoprotein complexes; and (v) Enhancer – loops chromosomes to bring the effective proteins together [8,10,11]. Figure 3 describes different modes of action of lncRNAs except signal mechanism. Based on different modes of action, strategies based on silencing (siRNAs, antisense oligos, ribozymes, CRISPR, ZNFs and TALENs) and functional inhibition (small molecules, nanobodies, aptamers and RNA decoys) can be utilized for lncRNA therapeutics [12].
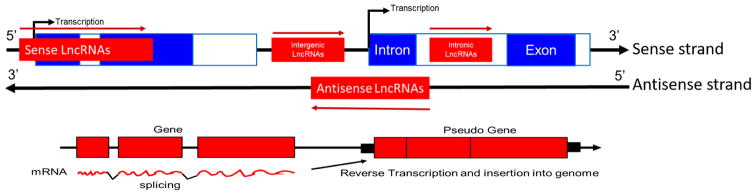
Figure 2.
Five classes of lncRNAs based on biogenesis. Sense lncRNAs overlap with coding genes on the same strand. Antisense lncRNAs overlap with protein-coding genes on the opposite strand. Intronic lncRNAs occur completely within an intron. Intergenic lncRNAs occur between two genes. Pseudogene lncRNAs are the product of reverse transcribed mRNA and inserted into genome. Adapted, with permission from [9].
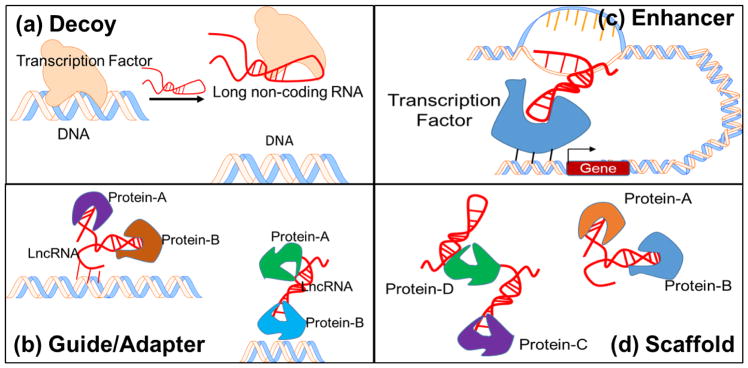
Figure 3.
Targeting mechanisms of lncRNA. (a) Decoy, titrate away DNA-binding proteins, such as transcription factors; (b) Guide/Adapter, chromatin modification enzymes, to DNA, RNA–DNA interactions, RNA interaction with a DNA-binding protein; (c) Enhancer, chromosome looping in an enhancer-like model, where looping defines the cis nature and spread of the lncRNA effect. (d) Scaffolds, bring two or more proteins into a complex or spatial proximity. Modified, with permission, from [11].
lncRNAs identified in ovarian cancer
lncRNAs are a new aspect to understanding OvCa – to diagnose and design new therapeutic approaches. Because the information on lncRNAs and OvCa is very fragmented, we made a comprehensive search for available information on lncRNAs related to OvCa on different databases such as NIH, ENCODE, Lnc2cancer, LncRNA disease database (cuilab), lncRNAdb and NONCODE. Transcription factor binding sites were analyzed using QIAGEN (GeneCards.org) and the ChIPseq database on the UCSC genome browser (ENCODE). In this review, we will summarize lncRNAs and their involvement in OvCa. The potential translation of this knowledge to diagnose and design therapeutic approaches for OvCa therapy will be very encouraging with the advent of cutting-edge translational research. Table 1 summarizes all the lncRNAs reported to date in relation to OvCa [4,13–57].
Table 1.
| lncRNA | Name | Alias | Chromosom e location: (strand) |
NCBI /ENSMBL |
Expression pattern |
Functional /clinical info |
|---|---|---|---|---|---|---|
| AB073614 [13] | NA | NA | Chr3:148891375-148893279(−) | NA | Up | Tumorigenesis, poor prognosis (OS) |
| AL132709.8 [4] | NA | NA | Chr14q32.31 | NA | Recurrence marker | |
| AC104699.1.1 [14] | NA | NA | NA | NA | Predictive survival | |
| ANRIL [15] | Antisense noncoding RNA in the INK4 locus (ANRIL) | CDKN2B-AS1; p15AS; CDKN2BAS; CDKN2B-AS; NCRNA00089 | Chr9:21994778-22121097(+) | NR_003529 | Up | Metastasis, poor prognosis-survival |
| ASAP1-IT1 [16] | ASAP1 intronic transcript 1 | ASAP1-IT; ASAP1IT; DDEF1IT1; HSPC054; NCRNA00050 | Chr8:130295355-130296533(−) | NR_002765 | Fav OS | |
| BACE1-AS [17] | β-site APP cleaving enzyme-1 antisense strand | BACE1-AS1; NCRNA00177 | Chr11:117291346-117292170(+) | NR_037803 | Down | Tumor suppressor |
| BCYRN1 [22] | Brain cytoplasmic RNA1 | BC200; BC200a; LINC00004; NCRNA00004 | ChrX:70430,035-70948962(−) | NR_001568; AF20057 | Up and down | Tumor suppressor |
| CCAT1 [20] | Colon cancer associated transcript 1 | CARLo-5 | Chr8:127207866-127219088(??) | NR_108049 | Down | Metastatic |
| CCAT2 [19] | Colon cancer associated transcript 2 | NCCP1; LINC00873 | Chr8:127400399-127402150(+) | NR_109834 | Up | Short OS and DFS, positive with tumor grade and distant metastasis |
| CRNDE [21] | NA | NA | Chr16:54845189-5492918912(−) | ENST00000613942 | High | Oncogenic, poor prognosis |
| FALEC [18] | Focally amplified long noncoding RNA in epithelial cancer | FAL1 | Chr1:150488233-150490508(+) | NR_051960 | Up | |
| FAM215A [16] | Family with sequence similarity 215 member A | APR-2; C17orf88; LINC00530 | Chr17:43917208-43917987(+) | NR_026770 | Up | Fav OS |
| GACAT3 [23] | Gastric cancer associated transcript 3 | LINC01458; lncRNA-AC130710 | Chr2:16050427-16085689(+) | NR_126559 | Nonequivalent outcome | |
| GAS5 [54,56] | Growth arrest specific 5 | GAS5; SNHG2; NCRNA00030 | Chr1:173863900-173868882(−) | NR_002578 | Down | |
| H19 [20,39,54,56] | H19, imprinted maternally expressed transcript | ASM; ASM1; BWS; D11S813E; LINC00008; NCRNA00008; WT2 | Chr11:1995163-2001470(−) | NR_131223 | Up | Tumorigenesis, recurrence marker, metastasis |
| HOST2 [29] | Human ovarian-cancer-specific transcript 2 | CERNA2 | Chr10:84167228-84172076(−) | NR_134505 | Up or down | Risk factor |
| HOTAIR [28,35,40,43,44] | HOX transcript antisense RNA | HOXAS; HOXC-AS4; HOXC11-AS1; NCRNA00072 | Chr12:53962308-53974956(−) | NR_003716 | Up/differential | Prognostic metastatic marker |
| HOXA11-AS [46] | HOXA11 antisense RNA | HOXA-AS51; HOXA11AS; HOXA11S; NCRNA00076; HOXA11-AS | Chr7:27185408-27189293(+) | NR_002795 | Down | Oncogenic |
| HOTAIRM1 [4] | HOXA transcript antisense RNA, myeloid-specific 1 | HOXA1-AS1; HOXA-AS1; NCRNA00179 | Chr7:27135713-27139877(−) | NC_000007.14 | Recurrence marker | |
| LINC00472 [16] | Long intergenic non-protein-coding RNA 472 | C6orf155 | Chr6:71407864-71420745(−) | NR_121612 | Fav OS | |
| LINC00515 [54] | NA | LINC00515; PRED21; C21orf71 | Chr21:25582770-25583326(−) | ENSG00000260583 | Down | |
| LINC00961 [54] | NA | NA | Chr9: 35909483-35937153(+) | ENSG00000235387 | Up | |
| linc-CARS2-2 [54] | Cysteinyl-tRNA synthetase 2, mitochondrial | FLJ12118 | Chr13:110641584-10644681(−) | ENST00000542774 | Up | |
| lin-RECK-3 [54] | reversion inducing cysteine rich protein with kazal motifs | ST15, hRECK | Chr9:36036924-36086181(+) | ENST00000475774 | Up | |
| linc-TNFRSF19-1 [54] | TNF receptor superfamily member 19 | TAJ-alpha, TROY, TAJ, TRADE | Chr13:23579359-23594411(+) | ENST00000464735 | Down | |
| LOC100128881 [20] | VPS9D1 antisense RNA1 | LOC100128881; VPS9D1-AS1 | Chr16:89711856-89718165(+) | NR_036480 | Down | Metastasis |
| LOC100190986 [4] | Uncharacterized | NA | Chr16:21432003-21434455(−) | NA | Recurrence marker | |
| LOC100292680 [20] | Long intergenic non-protein-coding RNA 942 | LOC100292680; LINC00942 | Chr12:1500525-1507318(−) | NR_028415 | Down | |
| LSINCT5 [49] | Long stress-induced noncoding transcript 5 | NA | Chr5:2712591-2715237(−) | NR_145480 | Up | |
| LUCAT1 [54] | Lung cancer associated transcript 1 | LUCAT1; SCAL1 | Chr5:91303029-91314402(−) | NR_103548 | Up | |
| MALAT1 [4,20,27,33,41,55,57] | Metastasis-associated lung adenocarcinoma transcript 1 | HCN; LINC00047; NCRNA00047; NEAT2; PRO2853 | Chr11:65497679-65504494(+) | NR_002819 | Up | Recurrence marker |
| MEG3 [48,51,53] | Maternally expressed 3 | FP504; GTL2; LINC00023; NCRNA00023; PRO0518; PRO2160; onco-lncRNA-83; prebp1 | Chr14:100826108-100861026(+) | NR_046472 | ||
| MIR22HG [36] | NA | C17orf91 | Chr17:1711504-1716272(−) | NR_028502 | Oncogene, prognostic marker | |
| MNX1-AS1 [20] | MNX1 antisense RNA 1 | CCAT5; MNX1-AS1; LOC645249 | Chr7:157010805-157016426 ?? | NR_038835 | Down | |
| NEAT1 [8,26] | Nuclear paraspeckle assembly transcript 1 | LINC00084; NCRNA00084; TncRNA; VINC | Chr11:65422798-65445540(+) | NR_131012 | Clinical biomarker | |
| NRCP [47] | NA | NA | NA | NA | ||
| OVAAL [24] | Ovarian adenocarcinoma-amplified long noncoding RNA | LINC01131; OVAL | Chr1:180558974-180566518(+) | NR_125716 | ||
| P15INK4B [15] | Cyclin-dependent kinase inhibitor 2B | P15; MTS2; TP15; CDK4I; INK4B; p15INK4b; CDKN2B | Chr9:22002903-22009363(−) | ENSG00000147883 | Down | |
| PTPRD-AS1 [23] | PTPRD antisense RNA1 | Chr9:8858018-8861727(+) | NR_121600 | Nonequivalent outcome | ||
| PVT1 [31,37,38] | Pvt1 oncogene | LINC00079; MYC; NCRNA00079; onco-lncRNA-10; PVT1; LINC00079; NCRNA00084 | Chr8:127794533-128101253(+) | NR_003367 | Up | Oncogene |
| RP1-223E5.4 [23] | NA | AL441883.1 | Chr6:13614111-13615155(−) | ENST00000566170NOTFOUND | Nonequivalent outcome | |
| RP4-799P18.3 [23] | NA | AL122008.4 | Chr1:234268583-234272500(−) | ENST00000446433 | Nonequivalent outcome | |
| RP11-254122.1 [23] | NA | MIR583HG | Chr5:96050115-96215519(+) | ENST00000507997 | Nonequivalent outcome | |
| RP11-284N8.3.1 [14] | NA | NA | NA | NA | Predictive survival | |
| RP11-307C12.11 [23] | NA | DCST1-AS1 | Chr1:155045191-155046118(−) | ENST00000452962 | Nonequivalent outcome | |
| RP11-57P19.1 [23] | NA | AC009432.1 | Chr15:94600014-94600821(+0 | ENST00000560391 | Nonequivalent outcome | |
| RP11-80H5.7 [23] | NA | AL157400.4 | Chr10:89694295-89697928(−) | ENST00000455699 | Nonequivalent outcome | |
| RUNX1-IT1 [4] | NA | Chr21q22.12 | NA | Recurrence marker | ||
| TC0100223 [45] | NA | NA | NA | NA | Down | |
| TC0101441 [42,45] | NA | NA | Chr1:202377159-202378011 | NA | Down | Prognostic factor OS |
| TC010686 [45] | NA | NA | Chr1:244341256-244343791 | NA | Up | |
| TC1500845 [45] | NA | NA | Chr15:38773376-38774597 | NA | Up | |
| TUG1 [34] | Taurine upregulated 1 | LINC00080; NCRNA00080; TI-227H | Chr22:30969211-30979395(+) | NR_002323 | ||
| UCA1 [20,50,52] | Urothelial cancer associated 1 | CUDR; LINC00178; NCRNA00178; UCAT1; onco-lncRNA-36 | Chr19:15828947-15836321(+) | NR_015379 | Down | |
| XIST [32] | X inactive specific transcript | DXS1089; DXS399E; LINC00001; NCRNA00001; SXI1; swd66 | ChrX:73820651-73852753(−) | NR_001564 | Down | |
| ZNF300P1 [30] | NA | NA | Chr5:150930645-150946289(−) | ENST00000356555 |
H19
H19 is a metastatic lncRNA that induces cell cycle arrest and apoptosis through certain cell-cycle-related and apoptosis-related proteins, it was identified by profiling 70 pairs of OvCa tissue [20,56]. H19 also showed relation to acquired drug resistance toward cisplatin chemotherapy; and high-grade serous ovarian cancer (HGSC) tissues showed strong correlation with cancer recurrence with H19 expression levels [54,58,59]. H19 RNA was detected in 90% of patients with ovarian cancer ascites fluid (OCAF) so, in an effort to develop targeted therapy for OvCa, the therapeutic potential of the toxin vector DTA-H19 was tested in ovarian carcinoma cell lines and in a heterotopic animal model for OvCa. DTA-H19 is a plasmid expressing diphtheria toxin under the control of H19 regulatory sequences [39].
HOTAIR
Abnormal expression of HOX antisense intergenic RNA (HOTAIR) and common variants of HOTAIR are associated with risk of epithelial ovarian cancer (EOC). HOTAIR was significantly high in 44 OvCa tissues as compared with 14 normal ovary tissues [28]. Elevated HOTAIR expression leads to chemoresistance by activating the Wnt/β-catenin pathway in human OvCa [35], also overexpression of HOTAIR predicts poor patient prognosis and promotes tumor metastasis in epithelial OvCa. HOTAIR levels were highly positively correlated with the FIGO stage, the histological grade of the tumor, lymph node metastasis and reduced overall survival (OS) and disease-free survival (DFS). The pro-metastatic effects of HOTAIR were mediated by the regulation of the expression of a number of genes involved in cell metastasis and EMT, including matrix metalloproteinase (MMP)3, MMP9, E-cadherin, vimentin and Snail [43,58,59]. Upregulation of HOTAIR induced platinum resistance in OvCa, and increased HOTAIR levels were observed in recurrent platinum-resistant ovarian tumors vs primary ovarian tumors. The nuclear factor (NF)-κB-HOTAIR axis links DNA damage response, chemoresistance and cellular senescence in OvCa [40]. In a clinical study of serous ovarian cancer (SOC), HOTAIR overexpression was correlated with an advanced FIGO stage and a high histological grade [44].
MALAT1
Metastasis-specific lung adenocarcinoma transcript (MALAT)1 is a potential biomarker for tumor growth and metastasis, as well as a promising therapeutic target in OvCa. Differential RNA analysis of SKOV3 (parental) and SKOV3.ip1 (metastatic) cells identified it as a metastatic specific lncRNA, and overexpressed MALAT1 in SKOV3 cells promoted cell proliferation, migration and invasion [20,55]. MALAT1 expression was associated with the FIGO stage. Knockdown of MALAT1 expression in OVCAR3 cells inhibited cell proliferation, migration and invasion, leading to G0/G1 cell cycle arrest and apoptosis. Transforming growth factor (TGF)β1 has been shown to induce MALAT1 expression and subsequent phosphorylation of MEK1, ERK1, p38 and JNK1 suggested MALAT1 promoted OvCa cell proliferation, migration and invasion, and that mitogen-activated protein kinase (MAPK) pathways might be one of the regulatory mechanisms of MALAT1 [57]. MALAT1 also promotes proliferation and metastasis in EOC via the PI3/AKT pathway [33,59]. Plasma MALAT1 could act as a valuable biomarker for the diagnosis of metastasis. Plasma MALAT1 was significantly increased in the EOC/distant metastasis group compared with the EOC/NDM. Multivariate analysis indicated that overexpression of MALAT1, differentiation (poor), tumor-node-metastasis stage IV, lymph node metastasis (N3), peritoneal invasion (present) and higher serum carbohydrate antigen 125 levels were independent predictors of survival in patients with EOC. Survival analysis revealed that patients with increased MALAT1 expression had a poorer disease-free survival time [27]. MALAT1 also functions as a sponge for miR-200c and regulates miR-506 by targeting iASPP [60]. MALAT1 accumulates to high levels in the nucleus, where it has crucial roles in cancer progression and the formation of nuclear paraspeckles [61].
PVT1
PVT1 and Myc contribute independently to ovarian and breast pathogenesis when overexpressed because of genomic abnormalities. PVT1-mediated inhibition of apoptosis might explain why amplification of 8q24 is associated with reduced survival duration in patients treated with agents that act through apoptotic mechanisms. Amplification of PVT1 contributes to the pathophysiology of ovarian and breast cancer [31,58,59]. Overexpression of lncRNA PVT1 in OvCa cells promotes cisplatin resistance by regulating apoptotic pathways. The level of PVT1 was significantly higher in OvCa tissues of cisplatin-resistant patients and cisplatin-resistant cells. The mRNA levels and protein expression of TGFβ1, p-Smad4 and caspase-3 were much higher in cisplatin-resistant cells transfected with siPVT1 [38]. Carboplatin and docetaxel have been developed as the first-line drug treatment for ovarian carcinoma. lncRNA PVT1 could be a central downstream target of carboplatin plus docetaxel because expression of PVT1 positively correlates with anticancer action of carboplatin plus docetaxel. p53 and tissue inhibitor of matrix metalloproteinase (TIMP)1 were mediated by lncRNA PVT1, which could explain partially the anticancer activity of lncRNA PVT1 [37].
UCA1
Urothelial carcinoma associated (UCA)1 functions as an oncogene, which promotes cancer cell proliferation, invasion and metastasis, and is responsible for drug resistance. UCA1 overexpression predicts clinical outcome of patients with OvCa receiving adjuvant chemotherapy. High UCA1 expression was an independent prognostic marker of poor outcome. UCA1 could serve as an indicator of response to chemotherapy and prognosis of OvCa and plays an important part in the progression of OvCa. Differential RNA analysis of SKOV3 (parental) and SKOV3.ip1 (metastatic) cells also identified UCA1 as a metastatic specific lncRNA [20]. UCA1 is related to serine/threonine protein kinase (SRPK)1 in cisplatin resistance in human OvCa cells. UCA1 can improve cell migration, invasion and induce cisplatin resistance. SRPK1 and the apoptosis pathway proteins could be involved in the effect of UCA1 [50,59]. UCA1 expression has been correlated to metastasis in EOC. UCA1 was not only aberrantly upregulated in EOC tissues and cells but also correlated with status of lymph node metastasis and the FIGO stage. UCA1 was a prognostic factor for overall survival in EOC patients. UCA1 could also function as an endogenous sponge by directly binding to miR-485-5p. Depletion of UCA1 was involved in the downregulation of MMP14 expression, a target gene of miR-485-5p. UCA1 can be utilized as a prognostic biomarker and is connected to miR-485-5p and MMP14 in EOC metastasis [52].
Other lncRNAs related to ovarian cancer etiology
ZNF-300P1 could play a part in promoting metastasis in OvCa cells [30]. TC0100223, TC0101686 and TC0101441 are aberrantly expressed in estrogen receptor (ER)α-positive EOC tissues, showing correlations with advanced FIGO stage and/or high histological grade and lymph node metastasis. Multivariate analysis indicated that TC0101441 was also an independent prognostic factor for overall survival. Knowledge of these E2-regulated lncRNAs could aid in the future understanding of the estrogenic effect on EOC progression and could assist in the clinical design of new target therapies based on a perspective of lncRNA [45]. CCAT1, LOC645249, LOC100128881 and LOC100292680 were identified as metastasis-specific lncRNAs, in a cluster of seven lncRNAs, in a model comparing the parental SKOV3 and metastatic variant SKOV3.ip1 cells by microarray [20].
AB073614 promotes development of OvCa via targeting ERK1/2 and the AKT-mediated signaling pathway and is a poor prognostic marker for overall survival [13]. Major transcription factors binding on the AB073614 promoter are RUNX3, MTA3, EBF1, NF1C and ATF2 (as per ENCODE). MR22HG is a potential prognostic marker and functions as an oncogene in OvCa, its repression impaired migration, invasion, viability and downregulated the pro-metastatic gene, MYC, at the mRNA and protein level [36]. NEAT1 is upregulated in OvCa patients and cell lines, and its expression is associated with the FIGO stage and lymph node metastasis. NEAT1 is suppressed by miR-124-3p and could serve as a potential target for antineoplastic therapies [26]. NEAT1 expression level was an independent factor in predicting the overall survival of OvCa patients. NEAT1 also contributes to paclitaxel resistance of OvCa cells by regulating ZEB1 expression via miR-194 [8]. lncRNA AC104699.1.1 and RP11-284N8.3.1 are immune-associated, and were identified after co-expression network construction using molecular profiles of 399 OvCa patients. These were differentially expressed, were independently predictive of the survival of patients with different stages and have their predictive roles in immune system activation and other antitumor processes in the OvCa microenvironment [14]. Antisense noncoding RNA in the INK4 locus (ANRIL) overexpression correlated with advanced FIGO stage and high histological grade, indicative of independent prognostic factor for overall survival, lymph node metastasis and poor prognosis. The proliferative effect (cell cycle progression, inhibition of apoptosis and senescence) of ANRIL was linked to downregulation of P15INK4B and upregulation of Bcl-2 [15]. Colon-cancer-associated transcript (CCAT)2 is involved in several cancers, including OvCa. High CCAT2 expression levels are associated with a shorter overall survival and disease-free survival in OvCa patients. CCAT2 expression positively correlates with the FIGO stage, tumor grade and distant metastasis [19]. Elevated levels of two different CRNDE transcripts were a negative prognostic factor; they increased the risk of death and recurrence in the group of patients treated with taxanes but not cyclophosphamide (DNA-damaging agents only) [21].
AL132709.8, HOTAIRM1, LOC100190986 and RUNX1-IT1 were identified as the OvCa recurrence signature to improve patient quality with personalized OvCa therapy [4]. Major transcription factors binding on the LOC100190986 promoter are Nkx3-1, HNF-1A, HNF-1, aMEF-2 and MEF-2A; and major transcription factors binding on the HOTAIRM1 promoter are PKNOX1, ARNT, SIN3A, FEZF1 and ZNF2 (as per ENCODE). Analyzed in 266 fresh frozen tumor samples of EOC, ASAP1-IT1, FAM215A and LINC00472 are associated with patient survival. They were more frequently highly expressed in low-grade tumors and early-stage disease compared with high-grade tumors and late-stage disease [16].
XIST lncRNA correlates with levels and disease-free periods of OvCa patients with Taxol in the therapeutic regimens. XIST expression could be a potential marker for chemotherapeutic responses in OvCa [32]. Taurine upregulated gene (TUG)1 is upregulated in OvCa and positively correlates with tumor grade and the FIGO stage [34]. OVAAL lncRNA was identified by genomic analysis of GENCODE lncRNAs in high-grade serous ovarian adenocarcinoma. It is a potent regulator of cell physiology and tumor development [24]. NRCP is highly upregulated in ovarian tumors, and has a role in cancer metabolism, promotes cancer growth by altering glycolysis and elucidates functional effects leading to increased tumor progression as a binding partner of signal transducer and activator of transcription (STAT)1 [47]. Maternally expressed gene (MEG)3 lncRNA is epigenetically silenced in EOC owing to promoter hypermethylation, which could contribute to the development of EOC, it also activates p53 [48]. MEG3 regulates ATG3 and induces autophagy [51]. As a therapeutic approach, curcumin suppresses cisplatin resistance development partly via modulating extracellular vesicle-mediated transfer of MEG3 and miR-214 in OvCa [53]. lncRNA-HOST2 (human ovarian cancer-specific transcript 2) promotes tumor cell migration, invasion and proliferation in EOC through a mechanism involving miR-let-7b. HOST2 harbors a let-7b binding site and modulates let-7b availability by acting as a molecular sponge [29]. HGSC shows primary and acquired drug resistance toward cisplatin chemotherapy. Lin-RECK-3, H19, LUCAT1, LINC00961 and linc-CARS2-2 showed significantly increased expression levels in cisplatin-resistant A2780-DR cells, whereas Linc-TNFRSF19-1 and LINC00515 showed significantly decreased expression levels [54]. LSINCT5 is a 2.6 Kb polyadenylated, long stress-induced noncoding transcript, localized in the nucleus and potentially transcribed by RNA polymerase III. Overexpressed in OvCa, it regulates cellular proliferation. lncRNA NEAT-1 and a protein-coding gene PSPC1 were significantly affected by knockdown of LSINCT5 [49]. β-site APP-cleaving enzyme 1 antisense strand (BACE1-AS) identified as a novel target for anisomycin is responsible for OvCa stem cell (OCSC) proliferation and invasion [17]. Focally amplified lncRNA on chromosome 1 (FAL1) promotes its oncogenic activity (cancer cell growth) via repression of p21, and regulates the transcription of CDKN1A via stabilization of epigenetic repressor BMI1. It was identified in a genome-wide survey on somatic copy-number alterations (SCNAs) of lncRNA in 2394 tumor specimens from 12 cancer types. FAL1 is an oncogene, whose copy number and expression are correlated with outcomes in OvCa [18].
Growth-arrest-specific (GAS)5 lncRNA acts as a tumor suppressor in OvCa. When GAS5 has decreased expression, this indicates poor prognosis in OvCa [62]. Lower GAS5 expression in 60 EOC patients was closely related to lymph node metastasis and tumor node metastasis. GAS5 also disrupts mitochondrial membrane potential, signifying its role in cell apoptosis through the mitochondria-mediated apoptosis pathway. It also promotes BAX, BAK, cleaved-caspase 3 and cleaved-caspase 9 expression. Overall, GAS5 can serve as a novel therapeutic target in patients with EOC [63]. The homeobox A (HOXA) region of protein-coding genes impacts the female reproductive system embryogenesis and ovarian carcinogenesis. The 5′ end of HOXA includes three lncRNAs: HOXA10-AS, HOXA11-AS and HOTTIP. GWAS data from 1201 serous EOC cases and 2009 controls identified HOXA11-AS, rs17427875 (A>T), which was marginally associated with reduced serous EOC risk. A functional variant of HOXA11-AS: HOXA11-AS minor allele-T, inhibits the oncogenic phenotype of EOC, compared with common allele-A expression in EOC. HOXA11-AS expression levels were significantly lower in human EOC tumors than normal ovarian tissues, suggesting a tumor suppressor function enhanced by the T allele [46]. BCYRNA or BC200 RNA downregulation leads to cancer cell proliferation and chemoresistance to carboplatin in SKOV3 and A2780 cells, it appears to have a role in the mediation of carboplatin-induced OvCa cell death [22].
Concluding remarks and future directions
OvCa is the leading cause of death among all gynecological malignancies. lncRNA has given a new facet to OvCa, to date not many lncRNAs reported have been functionally characterized in the OvCa disease process. Many of the lncRNAs have been identified by revisiting the array datasets on the publicly available resources, making it necessary to understand the mechanisms and biology of this new class of regulators, which is now at the center of various physiological and pathological processes. The information regarding OvCa-specific lncRNAs either alone or in combination with other types of markers (miRNAs, mRNAs, proteins) could prove useful to predict outcome or treatment follow-up to improve the therapeutic care of ovarian carcinoma patients. At present, more research is needed to elucidate the biological mechanisms [64] and clinical implications in tumor characterization as well as disease prognosis and treatment at the molecular level, putting them into clinical practice.
The field of precision medicine is getting traction and it would need analysis and identification of novel factors responsible for complex diseases like OvCa. The information about the current state of new lncRNAs reported in OvCa will help to identify new approaches to develop new therapies based on lncRNA status. lncRNAs are an attractive biomarker considering their expression pattern, tissue specificity and detectability using a small amount of sample. This comprehensive review summarizes the current state of new lncRNAs reported in OvCa along with the accessible information on their genomic locus and regulating transcription factors. This crucial information can be used to design therapeutics based on ‘silencing’ using siRNAs, antisense oligos, ribozymes, CRISPR, ZNFs and TALENs or ‘functional inhibition’ using small molecules, nanobodies, aptamers and RNA decoys for lncRNA therapeutics.
Highlights.
New roles of lncRNAs in ovarian cancer biology
lncRNA as new prognostic biomarkers for ovarian cancer patients
Role of lncRNA in metastasis, recurrence and drug-resistance pathways
Novel lncRNA as targets for personalized therapy in ovarian cancer
Acknowledgments
This work was supported by the National Institutes of Health Research Project Grant Program (R01CA204552, R01CA210192 and R01CA206069) to S.C.C. and UTHSC-CORNET award (M.K.T.). This work was also partially supported by UTHSC-College of Pharmacy-Dean’s Seed Grant and UTHSC New Grant Mechanism Award to S.C.C., M.M.Y. and M.J..
Footnotes
Conflicts of interest
The authors have no conflicts of interest to declare.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.ENCODE ProjectConsortium. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhan X, et al. Panel of seven long noncoding RNA as a candidate prognostic biomarker for ovarian cancer. Onco Targets Ther. 2017;10:2805–2813. doi: 10.2147/OTT.S128797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Liu R, et al. Long noncoding RNA expression signature to predict platinum-based chemotherapeutic sensitivity of ovarian cancer patients. Sci Rep. 2017;7:18. doi: 10.1038/s41598-017-00050-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yang K, et al. Identification of a six-lncRNA signature associated with recurrence of ovarian cancer. Sci Rep. 2017;7:752. doi: 10.1038/s41598-017-00763-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Prat J. FIGO’s staging classification for cancer of the ovary, fallopian tube, and peritoneum: abridged republication. J Gynecol Oncol. 2015;26:87–89. doi: 10.3802/jgo.2015.26.2.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Coleman RL, et al. Latest research and treatment of advanced-stage epithelial ovarian cancer. Nat Rev Clin Oncol. 2013;10:211–224. doi: 10.1038/nrclinonc.2013.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fu LL, et al. Role of lncRNAs as novel biomarkers and therapeutic targets in ovarian cancer. Crit Rev Eukaryot Gene Expr. 2017;27:183–195. doi: 10.1615/CritRevEukaryotGeneExpr.2017019244. [DOI] [PubMed] [Google Scholar]
- 8.Wang Q, et al. Long noncoding RNAs (lncRNAs) in triple negative breast cancer. J Cell Physiol. 2017;232:3226–3233. doi: 10.1002/jcp.25830. [DOI] [PubMed] [Google Scholar]
- 9.Quan M, et al. Exploring the secrets of long noncoding RNAs. Int J Mol Sci. 2015;16:5467–5496. doi: 10.3390/ijms16035467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ma L, et al. On the classification of long non-coding RNAs. RNA Biol. 2013;10:925–933. doi: 10.4161/rna.24604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rinn JL, Chang HY. Genome regulation by long noncoding RNAs. Annu Rev Biochem. 2012;81:145–166. doi: 10.1146/annurev-biochem-051410-092902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gutschner T, et al. From biomarkers to therapeutic targets-the promises and perils of long non-coding RNAs in cancer. Cancer Metastasis Rev. 2018;37:83–105. doi: 10.1007/s10555-017-9718-5. [DOI] [PubMed] [Google Scholar]
- 13.Cheng Z, et al. A long noncoding RNA AB073614 promotes tumorigenesis and predicts poor prognosis in ovarian cancer. Oncotarget. 2015;6:25381–25389. doi: 10.18632/oncotarget.4541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guo Q, et al. Comprehensive analysis of lncRNA-mRNA co-expression patterns identifies immune-associated lncRNA biomarkers in ovarian cancer malignant progression. Sci Rep. 2015;5:17683. doi: 10.1038/srep17683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Qiu JJ, et al. The long non-coding RNA ANRIL promotes proliferation and cell cycle progression and inhibits apoptosis and senescence in epithelial ovarian cancer. Oncotarget. 2016;7:32478–32492. doi: 10.18632/oncotarget.8744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fu Y, et al. Long non-coding RNAs, ASAP1-IT1, FAM215A, and LINC00472, in epithelial ovarian cancer. Gynecol Oncol. 2016;143:642–649. doi: 10.1016/j.ygyno.2016.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen Q, et al. Long non-coding RNA BACE1-AS is a novel target for anisomycin-mediated suppression of ovarian cancer stem cell proliferation and invasion. Oncol Rep. 2016;35:1916–1924. doi: 10.3892/or.2016.4571. [DOI] [PubMed] [Google Scholar]
- 18.Hu X, et al. A functional genomic approach identifies FAL1 as an oncogenic long noncoding RNA that associates with BMI1 and represses p21 expression in cancer. Cancer Cell. 2014;26:344–357. doi: 10.1016/j.ccr.2014.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huang S, et al. The long non-coding RNA CCAT2 is up-regulated in ovarian cancer and associated with poor prognosis. Diagn Pathol. 2016;11:49. doi: 10.1186/s13000-016-0499-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu SP, et al. Identification of differentially expressed long non-coding RNAs in human ovarian cancer cells with different metastatic potentials. Cancer Biol Med. 2013;10:138–141. doi: 10.7497/j.issn.2095-3941.2013.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Szafron LM, et al. The putative oncogene, CRNDE, is a negative prognostic factor in ovarian cancer patients. Oncotarget. 2015;6:43897–43910. doi: 10.18632/oncotarget.6016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wu DI, et al. Downregulation of BC200 in ovarian cancer contributes to cancer cell proliferation and chemoresistance to carboplatin. Oncol Lett. 2016;11:1189–1194. doi: 10.3892/ol.2015.3983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhou M, et al. Comprehensive analysis of lncRNA expression profiles reveals a novel lncRNA signature to discriminate nonequivalent outcomes in patients with ovarian cancer. Oncotarget. 2016;7:32433–32448. doi: 10.18632/oncotarget.8653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Akrami R, et al. Comprehensive analysis of long non-coding RNAs in ovarian cancer reveals global patterns and targeted DNA amplification. PLoS One. 2013;8:e80306. doi: 10.1371/journal.pone.0080306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.An J, et al. LncRNA NEAT1 contributes to paclitaxel resistance of ovarian cancer cells by regulating ZEB1 expression via miR-194. Onco Targets Ther. 2017;10:5377–5390. doi: 10.2147/OTT.S147586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chai Y, et al. HuR-regulated lncRNA NEAT1 stability in tumorigenesis and progression of ovarian cancer. Cancer Med. 2016;5:1588–1598. doi: 10.1002/cam4.710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen Q, et al. Plasma long non-coding RNA MALAT1 is associated with distant metastasis in patients with epithelial ovarian cancer. Oncol Lett. 2016;12:1361–1366. doi: 10.3892/ol.2016.4800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cui L, et al. Expression of long non-coding RNA HOTAIR mRNA in ovarian cancer. Sichuan Da Xue Xue Bao Yi Xue Ban. 2013;44:57–59. [PubMed] [Google Scholar]
- 29.Gao Y, et al. LncRNA-HOST2 regulates cell biological behaviors in epithelial ovarian cancer through a mechanism involving microRNA let-7b. Hum Mol Genet. 2015;24:841–852. doi: 10.1093/hmg/ddu502. [DOI] [PubMed] [Google Scholar]
- 30.Gloss B, et al. ZNF300P1 encodes a lincRNA that regulates cell polarity and is epigenetically silenced in type II epithelial ovarian cancer. Mol Cancer. 2014;13:3. doi: 10.1186/1476-4598-13-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Guan Y, et al. Amplification of PVT1 contributes to the pathophysiology of ovarian and breast cancer. Clin Cancer Res. 2007;13:5745–5755. doi: 10.1158/1078-0432.CCR-06-2882. [DOI] [PubMed] [Google Scholar]
- 32.Huang KC, et al. Relationship of XIST expression and responses of ovarian cancer to chemotherapy. Mol Cancer Ther. 2002;1:769–776. [PubMed] [Google Scholar]
- 33.Jin Y, et al. LncRNA MALAT1 promotes proliferation and metastasis in epithelial ovarian cancer via the PI3K-AKT pathway. Eur Rev Med Pharmacol Sci. 2017;21:3176–3184. [PubMed] [Google Scholar]
- 34.Kuang D, et al. Long non-coding RNA TUG1 regulates ovarian cancer proliferation and metastasis via affecting epithelial-mesenchymal transition. Exp Mol Pathol. 2016;101:267–273. doi: 10.1016/j.yexmp.2016.09.008. [DOI] [PubMed] [Google Scholar]
- 35.Li J, et al. Overexpression of long non-coding RNA HOTAIR leads to chemoresistance by activating the Wnt/beta-catenin pathway in human ovarian cancer. Tumour Biol. 2016;37:2057–2065. doi: 10.1007/s13277-015-3998-6. [DOI] [PubMed] [Google Scholar]
- 36.Li J, et al. Long noncoding RNA C17orf91 is a potential prognostic marker and functions as an oncogene in ovarian cancer. J Ovarian Res. 2016;9:49. doi: 10.1186/s13048-016-0258-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu E, et al. Carboplatin-docetaxel-induced activity against ovarian cancer is dependent on up-regulated lncRNA PVT1. Int J Clin Exp Pathol. 2015;8:3803–3810. [PMC free article] [PubMed] [Google Scholar]
- 38.Liu E, et al. Overexpression of long non-coding RNA PVT1 in ovarian cancer cells promotes cisplatin resistance by regulating apoptotic pathways. Int J Clin Exp Med. 2015;8:20565–20572. [PMC free article] [PubMed] [Google Scholar]
- 39.Mizrahi A, et al. Development of targeted therapy for ovarian cancer mediated by a plasmid expressing diphtheria toxin under the control of H19 regulatory sequences. J Transl Med. 2009;7:69. doi: 10.1186/1479-5876-7-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ozes AR, et al. NF-kappaB-HOTAIR axis links DNA damage response, chemoresistance and cellular senescence in ovarian cancer. Oncogene. 2016;35:5350–5361. doi: 10.1038/onc.2016.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pa M, et al. Long noncoding RNA MALAT1 functions as a sponge of MiR-200c in ovarian cancer. Oncol Res. 2017 doi: 10.3727/096504017X15049198963076. [DOI] [PubMed] [Google Scholar]
- 42.Qiu J, et al. Effects of oestrogen on long noncoding RNA expression in oestrogen receptor alpha-positive ovarian cancer cells. J Steroid Biochem Mol Biol. 2014;141:60–70. doi: 10.1016/j.jsbmb.2013.12.017. [DOI] [PubMed] [Google Scholar]
- 43.Qiu JJ, et al. Overexpression of long non-coding RNA HOTAIR predicts poor patient prognosis and promotes tumor metastasis in epithelial ovarian cancer. Gynecol Oncol. 2014;134:121–128. doi: 10.1016/j.ygyno.2014.03.556. [DOI] [PubMed] [Google Scholar]
- 44.Qiu JJ, et al. The long non-coding RNA HOTAIR promotes the proliferation of serous ovarian cancer cells through the regulation of cell cycle arrest and apoptosis. Exp Cell Res. 2015;333:238–248. doi: 10.1016/j.yexcr.2015.03.005. [DOI] [PubMed] [Google Scholar]
- 45.Qiu JJ, et al. Expression and clinical significance of estrogen-regulated long non-coding RNAs in estrogen receptor alpha-positive ovarian cancer progression. Oncol Rep. 2014;31:1613–1622. doi: 10.3892/or.2014.3000. [DOI] [PubMed] [Google Scholar]
- 46.Richards EJ, et al. A functional variant in HOXA11-AS, a novel long non-coding RNA, inhibits the oncogenic phenotype of epithelial ovarian cancer. Oncotarget. 2015;6:34745–34757. doi: 10.18632/oncotarget.5784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rupaimoole R, et al. Long noncoding RNA ceruloplasmin promotes cancer growth by altering glycolysis. Cell Rep. 2015;13:2395–2402. doi: 10.1016/j.celrep.2015.11.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sheng X, et al. Promoter hypermethylation influences the suppressive role of maternally expressed 3, a long non-coding RNA, in the development of epithelial ovarian cancer. Oncol Rep. 2014;32:277–285. doi: 10.3892/or.2014.3208. [DOI] [PubMed] [Google Scholar]
- 49.Silva JM, et al. LSINCT5 is over expressed in breast and ovarian cancer and affects cellular proliferation. RNA Biol. 2011;8:496–505. doi: 10.4161/rna.8.3.14800. [DOI] [PubMed] [Google Scholar]
- 50.Wang F, et al. Involvement of SRPK1 in cisplatin resistance related to long non-coding RNA UCA1 in human ovarian cancer cells. Neoplasma. 2015;62:432–438. [PubMed] [Google Scholar]
- 51.Xiu YL, et al. Upregulation of the lncRNA Meg3 induces autophagy to inhibit tumorigenesis and progression of epithelial ovarian carcinoma by regulating activity of ATG3. Oncotarget. 2017;8:31714–31725. doi: 10.18632/oncotarget.15955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yang Y, et al. UCA1 functions as a competing endogenous RNA to suppress epithelial ovarian cancer metastasis. Tumour Biol. 2016;37:10633–10641. doi: 10.1007/s13277-016-4917-1. [DOI] [PubMed] [Google Scholar]
- 53.Zhang J, et al. Curcumin suppresses cisplatin resistance development partly via modulating extracellular vesicle-mediated transfer of MEG3 and miR-214 in ovarian cancer. Cancer Chemother Pharmacol. 2017;79:479–487. doi: 10.1007/s00280-017-3238-4. [DOI] [PubMed] [Google Scholar]
- 54.Zheng ZG, et al. The essential role of H19 contributing to cisplatin resistance by regulating glutathione metabolism in high-grade serous ovarian cancer. Sci Rep. 2016;6:26093. doi: 10.1038/srep26093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhou Y, et al. The long noncoding RNA MALAT-1 is highly expressed in ovarian cancer and induces cell growth and migration. PLoS One. 2016;11:e0155250. doi: 10.1371/journal.pone.0155250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhu Z, et al. Ectopic expressed long non-coding RNA H19 contributes to malignant cell behavior of ovarian cancer. Int J Clin Exp Pathol. 2015;8:10082–10091. [PMC free article] [PubMed] [Google Scholar]
- 57.Zou A, et al. Long non-coding RNA MALAT1 is up-regulated in ovarian cancer tissue and promotes SK-OV-3 cell proliferation and invasion. Neoplasma. 2016;63:865–872. doi: 10.4149/neo_2016_605. [DOI] [PubMed] [Google Scholar]
- 58.Meryet-Figuiere M, et al. An overview of long non-coding RNAs in ovarian cancers. Oncotarget. 2016;7:44719–44734. doi: 10.18632/oncotarget.8089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Nikpayam E, et al. The role of long non-coding RNAs in ovarian cancer. Iran Biomed J. 2017;21:3–15. doi: 10.6091/.21.1.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lei R, et al. Long noncoding RNA MALAT1-regulated microRNA 506 modulates ovarian cancer growth by targeting iASPP. Onco Targets Ther. 2017;10:35–46. doi: 10.2147/OTT.S112686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wilusz JE. Long noncoding RNAs: re-writing dogmas of RNA processing and stability. Biochim Biophys Acta. 2016;1859:128–138. doi: 10.1016/j.bbagrm.2015.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Li J, et al. Decreased expression of long non-coding RNA GAS5 promotes cell proliferation, migration and invasion, and indicates a poor prognosis in ovarian cancer. Oncol Rep. 2016;36:3241–3250. doi: 10.3892/or.2016.5200. [DOI] [PubMed] [Google Scholar]
- 63.Gao J, et al. Long non-coding RNA growth arrest-specific transcript 5 is involved in ovarian cancer cell apoptosis through the mitochondria-mediated apoptosis pathway. Oncol Rep. 2015;34:3212–3221. doi: 10.3892/or.2015.4318. [DOI] [PubMed] [Google Scholar]
- 64.Kung JT, et al. Long noncoding RNAs: past, present, and future. Genetics. 2013;193:651–669. doi: 10.1534/genetics.112.146704. [DOI] [PMC free article] [PubMed] [Google Scholar]