Abstract
The marine actinomycete genus Salinispora is a remarkably prolific source of structurally diverse and biologically active secondary metabolites. Herein, we select the model organism Salinispora tropica CNB-440 for development as a heterologous host for the expression of biosynthetic gene clusters (BGCs) to complement well-established Streptomyces host strains. In order to create an integratable host with a clean background of secondary metabolism, we replaced three genes (salA–C) essential for salinosporamide biosynthesis with a cassette containing the Streptomyces coelicolor ΦC31 phage attachment site attB to generate the mutant S. tropica CNB-4401 via double-crossover recombination. This mutagenesis not only knocks-in the attachment site attB in the genome of S. tropica CNB-440 but also abolishes production of the salinosporamides, thereby simplifying the strain’s chemical background. We validated this new heterologous host with the successful integration and expression of the thiolactomycin BGC that we recently identified in several S. pacifica strains. When compared to the extensively engineered superhost S. coelicolor M1152, the production of thiolactomycins from S. tropica CNB-4401 was approximately 3-fold higher.To the best of our knowledge, this is the first example of using a marine actinomycete as a heterologous host for natural product BGC expression. The established heterologous host may provide a useful platform to accelerate the discovery of novel natural products and engineer biosynthetic pathways.
Keywords: Heterolgous expression, Salinispora, Natural products, Genetic engineering
Introduction
Microbially produced natural products are of paramount importance in human medicine. Not only are the majority of antimicrobial and anticancer drugs derived directly or inspired by natural products, many other branches of medicine, such as immunology, neurology, and cardiology, have similarly benefited from them (Newman and Cragg 2016). Bacteria in the order Actinomycetales, commonly called actinomycetes, are the undisputed leading source for natural product discovery, accounting for approximately 75% of the microbial natural products used in human therapy (Berdy 2005). Among the actinomycetes, the genus Streptomyces has been the major source of bioactive compounds discovered to date and has thus become the model organism for the study of natural product biosynthesis, pathway engineering, and genome mining (Baltz 2016; Chater 2016; Horinouchi 2007). Recently, however, common soil-derived Streptomyces bacteria have fallen out of favor as a resource for drug discovery due in part to the common re-discovery of well-known compounds. This has led to a search for other Actinobacteria and the exploration of actinomycetes from poorly exploited environments such as the ocean (Jensen et al. 2005; Schorn et al. 2016).
The marine actinomycete genus Salinispora was initially isolated from marine sediments and subsequently described as the first obligate marine actinomycete genus (Jensen et al. 2015). Since then, Salinispora species have displayed the ability to produce a diverse range of secondary metabolites, including salinosporamide A, which is currently in clinical trials as an anticancer agent (Gulder and Moore 2010). To date, 25 classes of compounds have been discovered from this genus, with 16 representing new carbon skeletons, correlating to virtually all known biosynthetic classes (Jensen et al. 2015). Recent genome sequencing efforts have revealed that only a small fraction of the natural product biosynthetic potential of Salinispora species has been identified (Amos et al. 2017; Letzel et al. 2017; Ziemert et al. 2014). In a recent study, the genomes of 75 Salinispora strains were queried for nonribosomal peptide synthetase (NRPS) and polyketide synthase (PKS) biosynthetic gene clusters (BGCs), revealing 124 distinct biosynthetic pathways predicted to encode 124 distinct natural products (Ziemert et al. 2014). This provides an unprecedented opportunity to explore the biosynthetic potential of Salinispora to discover new compounds.
Heterologous expression of whole BGCs has become a routine approach not only for the discovery of metabolites from “orphan” or “cryptic” gene clusters (Ren et al. 2017; Rutledge and Challis 2015), but also to probe biosynthetic logic involved in natural product biosynthesis (Gomez-Escribano and Bibb 2014; Ongley et al. 2013). This approach relies on two factors: first, the ability to clone BGCs into a suitable expression vector, and second, the availability of a compatible heterologous host. Recently, several whole-pathway direct cloning approaches, such as full-length RecE-mediated recombination in E. coli (Fu et al. 2012), transformation-associated recombination (TAR) in Saccharomyces cerevisiae (Kim et al. 2010; Yamanaka et al. 2014), and Cas9-Assisted in vitro assembly method (Jiang et al. 2015), have been developed, and these have greatly accelerated the BGC isolation procedure. On the other hand, several Streptomyces strains have been established for heterologous expression of natural product biosynthesis genes (Gomez-Escribano and Bibb 2011; Komatsu et al. 2013; Komatsu et al. 2010; Zhang et al. 2017b). However, empirically, we have found that some Salinispora gene clusters, such as those encoding the salinosporamide (Eustaquio et al. 2009), lymphostin (Miyanaga et al. 2011), and lomaiviticin pathways (Kersten et al. 2013), are active in S. tropica strains but inactive when introduced to a Streptomyces heterologous host such as S. coelicolor or S. lividans. The expression of a BGC is a complicated cellular process that is highly dependent on the genetics and biochemistry of the producing organism (Baltz 2010). We believe that the ability to easily test the expression of BGCs across various host organisms is a valuable asset to maximize the likelihood of achieving successful heterologous production of natural product small molecules (Zhang et al. 2017a). Therefore, establishing a biosynthetically versatile organism such as Salinispora as a heterologous host may facilitate the expression of additional BGCs for future discovery and engineering efforts.
S. tropica CNB-440 was the first whole genome-sequenced marine actinomycete and has been used as a model organism to investigate secondary metabolites (Udwary et al. 2007). So far, eight classes of compounds have been discovered from this strain alone, including the beta-lactone proteasome inhibitor salinosporamide A (Eustaquio et al. 2009) and the polyketide cytotoxin lomaiviticin A (Kersten et al. 2013). Here, we report the engineering of S. tropica CNB-440 as a heterologous host by introducing the phage ΦC31 attachment site attB into the genome for integratable expression and simultaneously abolishing background salinisporamide production. The new Salinispora host is directly compatible with all previously established genetic toolkits leveraging the phage ΦC31 integrase for Streptomyces expression (Tang et al. 2015; Yamanaka et al. 2014). We validated this new heterologous platform with the successful expression of the thiolactomycin BGC recently identified in several S. pacifica strains but absent in S. tropica (Tang et al. 2015). To the best of our knowledge, this is the first showcase of the engineering and use of a marine actinomycete as a heterologous host for natural product pathway expression.
Materials and methods
Bacterial strains, plasmids, and culture conditions.
Chemicals and microbiological and molecular biological agents were purchased from standard commercial sources. Strains and plasmids used in this study are summarized in Table 1. Salinispora tropica CNB-440 (ATCC BAA-916 and DSM 44818) and its respective derivatives were maintained and grown on A1 agar (10 g soluble starch, 4 g yeast extract, 2 g peptone, 16 g agar, 1 liter natural or artificial seawater) and/or A1 liquid medium (10 g soluble starch, 4 g yeast extract, 2 g peptone, 1 liter natural or artificial seawater). Escherichia coli strains were cultivated in LB medium (components purchased from BD Biosciences or Fisher Scientific) supplemented with the appropriate antibiotics. DNA isolation and manipulations were carried out according to standard methods for E. coli and Salinispora.
Table 1.
Plasmids and bacterial strains used in this work
| Plasmid/Strains | Description | References |
|---|---|---|
| Plasmids | ||
| BHXS1782 | pCC1FOS fosmid that carries a 44 kb genomic region containing the entire salinosporamide BGC from S. tropica CNB-440 |
Eustaquio et al. 2009 |
| pMXT13 | pCAP03 derivative that carries a 26 kb genomic region containing the entire thiolactomycin BGC from S. pacifica CNS- 863 |
Tang et al. 2015 |
| pIJ773 | A plasmid containing the cassette aac(3)IV (ApraR) + oriT | Gust et al. 2003 |
| pKD20 | λ-RED (gam, bet, exo), bla, araC, oriR101, rep101ts | Datsenko and Wanner 2000 |
| pUZ8002 | neo, RP4 | Paget et al. 1999 |
| pUB307 | Self-transmissible plasmid that mobilizes other plasmids in trans for DNA transfer into hosts: RP4, neo |
Flett et al. 1997 |
| pMXT01 | acc(3)IV, PermE, FRT, bla | This study |
| pMXT04 | BHXS1782 derivative in which salA-C has been replaced by the knockout cassette from pMXT01 |
This study |
| pMXT10 | pMXT04 derivative in which acc(3)IV (ApraR) has been removed by FLP-FRT recombination |
This study |
| pMXT11 | pMXT10 derivative in which chloramphenicol resistance gene cat has been replace by the cassette containing oriT and aac(3)IV (ApraR) from pIJ773 |
This study |
| Strains | ||
| S. tropica CNB-440 | Wild-type, ATCC BAA-916 / DSM 44818 | Udwary et al. 2007 |
| S. tropica CNB-4401 | S. tropica CNB-440 derivative, Δsal and ΦC31attB | This study |
|
S. tropica CNB-4401/tlm |
S. tropica CNB-4401 derivative, Δsal and ΦC31attB with tlm integration |
This study |
| E. coli Top10 | Host strain for routine cloning: F-, Δ(araA-leu)7697, (araD139)B/r, Δ(codBlacI)3, ϕ80dlacZ58(M15), galK0, mcrA0, galU−, recA1, endA1, nupG−, rpsL-(strR), Δ(mcrC-mrr)715 |
Invitrogen |
| E. coli BW25113 | Host strain for λ-RED recombination, K12 derivative: F-, Δ(araDaraB)567, ΔlacZ4787(::rrnB-3), λ-, rph-1, Δ(rhaD- rhaB)568, hsdR514 |
Datsenko and Wanner 2000 |
| E. coli BT340 | Host strain for FLP recombination: F-, Δ(argF-lac)169, ϕ80dlacZ58(M15), glnV44(AS), λ-, rfbC1, gyrA96(NalR), recA1, endA1, spoT1, thi-1, hsdR17, pCP20 |
Cherepanov and Wackernagel 1995 |
| E. coli ET12567 | DNA methylation deficient donor strain for conjugation: F2 dam13::Tn9, dcm, 6 hsdM, hsdR, recF143, zjj-202::Tn10, galK2, galT22 ara-14, laacY1, xyl-5, leuB6, thi-1, tonA31, rpsL 136, hisG4, tsx-78, mtl-1, glnV44 |
Flett et al. 1997 |
Construction of a gene disruption cassette containing the attB attachment site.
A 500 bp fragment containing the ΦC31 phage attachment site attB was amplified from the gDNA of Streptomyces coelicolor M1152 (Gomez-Escribano and Bibb 2011) with primers attBreg-F and attBreg-R (Table S1). The gene disruption cassette acc(3)IV (ApraR) and oriTRK2 were jointly amplified from pIJ773 (Gust et al. 2003) with the primers P1reg-F and P2reg-R (Table S1). These two PCR products were assembled by an overlap extension PCR reaction using the primers attBreg-F and P2reg-R (Table S1). The final disruption cassette was further cloned into pCR™2.1-TOPO® vector (Fisher Scientific, Hampton, NH, USA) to give pMXT01 and was verified by restriction analysis and sequencing.
Gene replacement on the fosmid BHXS1782 containing the salinosporamide gene cluster.
The disruption cassette containing the ΦC31 phage attachment site attB, acc(3)IV (ApraR), and oriTRK2 were jointly amplified from the 1,961-bp NotI/SpeI fragment of pMXT01 with the primer pair of salA-C_KO-F/salA-C_KO-R (Table S1). The PCR products were used to replace the genes salA-C on the fosmid BHXS1782 (Eustaquio et al. 2009) in E. coli BW25113/pKD20 (Datsenko and Wanner 2000) using the PCR targeting system. The resulting fosmid pMXT04 was confirmed by restriction analysis. Excision of the apramycin resistance cassette was performed in E. coli BT340 (Cherepanov and Wackernagel 1995), taking advantage of the FLP recognition sites surrounding the resistance cassette. E. coli clones containing the fosmid pMXT10 were screened for apramycin sensitivity and verified by restriction analysis and PCR using the primer pair SalA-C-test650-F/SalA-C-test650-R (Table S1). The chloramphenicol resistance marker cat on pMXT10 was replaced by the cassette containing oriTRK2 and aac(3)IV (ApraR), which was amplified from pIJ773 with primers Cat-knockout-F1 and Cat-knockout-R1 (Table S1), to give pMXT11.
Allelic exchange in Salinispora tropica CNB-440.
The modified fosmid pMXT11 were introduced into E. coli ET12567/pUZ8002 (Paget et al. 1999) by electroporation and then transferred to S. tropica CNB-440 by conjugation. Apramycin resistant (ApraR) exconjugants were identified, indicating a single-crossover allelic exchange in S. tropica CNB-440. Single colonies were picked and streaked on A1 agar plates without any antibiotics. Apramycin sensitive colonies were further screened by PCR to obtain the double-crossover allelic exchange mutant S. tropica CNB-4401.
Heterologous expression of the thiolactomycin gene cluster.
The plasmid pMXT13 (Tang et al. 2015), containing the thiolactomycin BGC (tlm), was transferred into E. coli ET12567 and introduced into S. tropica CNB-4401 by triparental intergeneric conjugation with the help of E. coli ET12567/pUB307 (Flett et al. 1997). 200 μl overnight culture of E. coli was inoculated into 10 mL fresh LB medium plus appropriate antibiotics and grown for 3–4 h at 37°C until an OD600 of 0.4 was reached. The cells were washed twice with 10 mL of LB to remove antibiotics and resuspended in 0.5 mL of LB. Simultaneously, 10 μl (108) S. tropica spores were added to 200 μl A1 broth and heat shocked at 50°C for 10 min. The spore suspension was mixed with 100 μl E. coli ET12567/pMXT13 and 100 μl E. coli ET12567/pUB307. The mixture was plated on an A1 ager plate and incubated at 30°C for 16–20 hours. Each plate was then overlaid with 1 ml water containing 4 mg nalidixic acid (to selectively kill E. coli) and 2 mg kanamycin (for selection of mutants). Following incubation at 30°C for 7–9 days, kanamycin resistant clones were selected, confirmed by PCR and designated as S. tropica CNB-4401/tlm.
Production, extraction, and detection of salinosporamide A and thiolactomycins.
Five milliliters of A1 medium were inoculated with 10 μl (108) spore suspension of S. pacifica CNS-863, S. tropica CNB-440, S. tropica CNB-4401 or a derivative thereof. The cultures were incubated for 3–4 d at 30 °C at 220 rpm. One milliliter of preculture was inoculated into 50 mL A1 medium using wide bore pipet tips. The cultures were incubated for 9 d at 30 °C with 220 rpm shaking. S. coelicolor M1152/tlm was cultured as described previously (Tang et al. 2015). The culture supernatant was adjusted to pH 4 with acetic acid and subsequently extracted with an equal volume of ethyl acetate. The organic phase was evaporated, and extracts were dissolved in 0.5 mL methanol (MeOH) and filtered through Acrodisc MS PTFE Syringe filters (Pall Inc., Ann Arbor, MI, USA) prior to HPLC and LC-MS analysis. For HPLC analysis, 10 µl of the MeOH extract was injected into an Agilent 1200 series analytical HPLC system monitored by UV lamp using a Luna 100A-C18 column (5 μm, 250 × 4.6 mm; Phenomenex, US) as follows: 0–23 min, 10–60% B; 23–28 min, 60–100% B; 29–34 min, 100% B; 34–35 min, 100–10% B; 36–40 min, 10% B (solvent A: water/trifluoroacetic acid (999:1); solvent B: acetonitrile/trifluoroacetic acid (999:1)). For LC-MS analysis, 5 μL of each MeOH extract was injected onto a Phenomenex Luna C18 reverse-phase HPLC column (5 μm, 250 mm × 4.6 mm; Phenomenex, US.) and analyzed with an Agilent 6530 Accurate-Mass LC-MS coupled to an Agilent 1260 LC system by gradient elution (A: acetonitrile with 0.1% formic acid (FA); B: H2O with 0.1% FA: 35–70% A over 23 min, 70–100% A from 23 to 28 min, and 100% A from 28 to 33 min; 0.7 mL/min). Q-TOF MS settings during the LC gradient were as follows: Acquisition—mass range m/z 100–1700, MS scan rate 1 s−1, MS/MS scan rate 2 s−1, fixed collision energy 20 eV; Source—gas temperature 300 °C, gas flow 11 L/min;Nebulizer 45 psig, ion polarity positive; Scan source parameters—VCap 3000, Fragmentor 100, Skimmer 65, OctopoleRFPeak 750. The MS was autotuned using Agilent tuning solution in positive mode before each measurement. LC(DAD) data were analyzed with ChemStation software (Agilent), and MS data were analyzed with MassHunter software (Agilent).
Results
Adaptation of the ϕC31 int/attP-attB integration system in S. tropica CNB-440.
ϕC31 integrase, a site-specific bacteriophage serine recombinase, catalyzes the insertion of a DNA fragment containing an attP site into a chromosomal attB site via recombination (Smith et al. 2010). As attB sites are widely distributed in a broad range of Streptomyces species (Combes et al. 2002; Smith et al. 2004), ϕC31 int/attP has been heavily employed in constructing vectors such as pSET152 (Flett et al. 1997), pCAP01 (Yamanaka et al. 2014), and pCAP03 (Tang et al. 2015) that can be integrated into the chromosome of various Streptomyces species for stable maintenance. In contrast to terrestrial Streptomyces, marine Salinispora species lack the attB attachment site in their genomes (Lechner et al. 2011). Previously, we identified three pseudo-attB sites in the genome of S. tropica CNB-440 that were used to integrate a ϕC31 phage-based vector carrying a single gene (Lechner et al. 2011). However, despite multiple attempts, we could not obtain exconjugants upon transfer of ϕC31 phage-based vectors carrying modest or large size BGCs (>30kb) into S. tropica CNB-440. This observation is consistent with an earlier study, which reported that insertion into the attB site is 300 times more efficient than into a pseudo-attB site (Combes et al. 2002).
We thus decided to introduce an authentic attB site into the chromosome of S. tropica CNB-440 by RP4-mediated intergeneric conjugation. We first amplified a 500 bp DNA fragment containing the attB site from the gDNA of Streptomyces coelicolor M1152. The PCR product was assembled with a cassette amplified from plasmid pIJ773 (Gust et al. 2003) to obtain an attB-containing disruptive cassette with an attB site, aac(3)IV (ApraR), oriTRK2, and two terminal FLP recognition target (FRT) sequences (Figure S1). This cassette was then cloned into pCR™2.1-TOPO® vector to form pMXT01 (Figure S1). pMXT01 was digested with NotI/SpeI, and the 1,961 bp fragment was purified by agarose gel electrophoresis to eliminate intact plasmid and used as a template for cassette amplification.
Deletion of endogenous BGCs has been used as a general strategy to 1) increase the metabolic flux of precursor availability and productivity of heterologously expressed BGCs, and 2) simplify the identification and characterization of heterologous products by activity screening and metabolite profiling (Gomez-Escribano and Bibb 2011; Komatsu et al. 2010). Thus, the fosmid BHXS1782 (Eustaquio et al. 2009) with a 44 kb insert containing the salinosporamide BGC (sal) was chosen to carry the attB-containing disruptive cassette into S. tropica CNB-440 to generate a double crossover mutant. We first replaced assembly line biosynthesis genes salA to salC on the fosmid BHXS1782 with the attB-containing disruptive cassette by Red/ET-mediated recombination, generating the fosmid pMXT04 (Figure 1). Subsequently, the region of aac(3)IV (ApraR) plus oriTRK2 was eliminated in vivo using FLP-mediated recombination to form pMXT10 (Figure 1). Additionally, the cassette containing aac(3)IV (ApraR) plus oriTRK2 sequence was used to replace the chloramphenicol resistance gene cat, as aac(3)IV is a better selection marker for generating single crossover mutants in Salinispora and oriTRK2 is essential for E. coli-mediated conjugation. The resulting fosmid, pMXT11, was introduced into S. tropica CNB-440 by conjugation (Figure S2). Single crossover mutants were selected for their apramycin resistance. Intergeneric conjugation between E. coli ET12567/pUZ8002/pMXT11 and S. tropica CNB-440 produced 10–20 colonies with apramycin resistance per conjugation using 10 μl (108) S. tropica spores. Although the efficiency is much lower than for conjugation with Streptomyces spores (104–105 using 108 spores), this might be due to the high concentration of sea salt in the A1 agar plate, which is essential for the growth of S. tropica.
Figure 1.
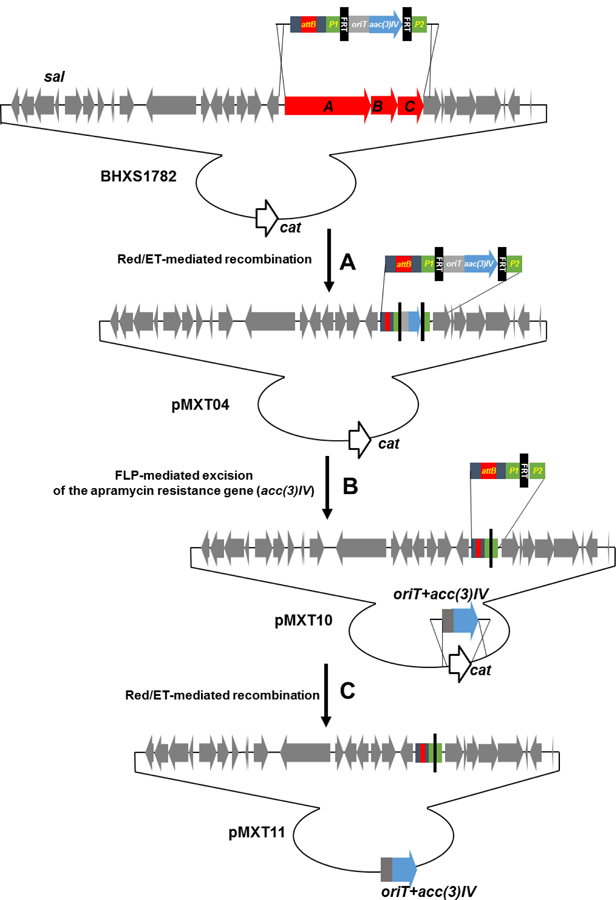
Schematic diagram depicting (A) the replacement of salA-C by Red/ET-mediated recombination with the attB-containing cassette; (B) subsequent removal of the resistance marker by FLP-mediated excision; and (C) replacement of the fosmid backbone chloramphenicol resistant gene cat with acc(3)IV (ApraR) and oriT of RK2 to generate fosmid pMXT11 to simultaneously knock-in the attB attachment site and knock-out salA-C in S. tropica CNB-440.
In order to obtain a double crossover mutant, single crossover mutants were streaked for single colonies on A1 agar plates without antibiotics. Fifty colonies were tested for apramycin sensitivity, and strains no longer resistant to apramycin were obtained for further verification. Total DNA was isolated from selected colonies, and a pair of primers flanking the outside of salA-C was designed for PCR confirmation. As designed, amplification of a double crossover deletion mutant gave a 626 bp PCR product (Figure 2). Furthermore, sequencing of the PCR product confirmed the presence of the attB site in the mutant strain, which was designated S. tropica CNB-4401. In the case of wild-type S. tropica CNB-440, no PCR products were obtained, as the targeted region was too large (9830 bp) (Figure 2).
Figure 2.
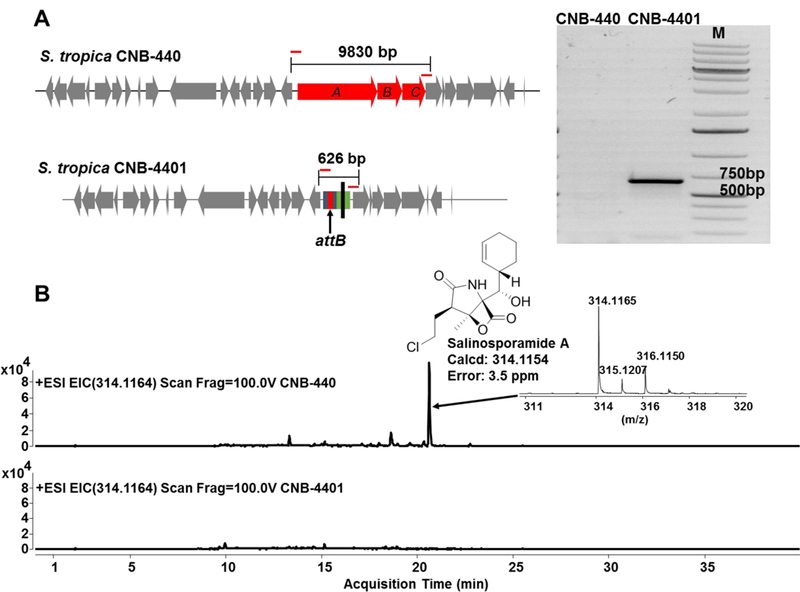
Verification of S. tropica CNB-4401 by (A) PCR amplification and (B) loss of production of salinosporamide A observed by LC-MS.
We also compared the production of salinosporamide A, the major product of the sal BGC from S. tropica CNB-440 and S. tropica CNB-4401. Because salA-C code for the salinosporamide enzymatic assembly line, S. tropica CNB-4401 should lose all ability to produce salinosporamide A. After cultivation, extracts were subjected to high resolution LC-ESI-MS analysis. In contrast to the wild-type strain S. tropica CNB-440, production of salinosporamide A (Figure 2) and all other analogs was completely abolished in the double crossover deletion mutant S. tropica CNB-4401. This result further confirmed the successful replacement of salA-C by the attB-containing disruptive cassette in the strain S. tropica CNB-4401.Additionally, the HPLC chromatogram of the ethyl acetate extract of the culture supernatant from S. tropica CNB-4401 contains a small number of peaks and a low baseline under various UV detection wavelengths (Figure S3). This would simplify downstream processes in detection and purification of the heterologously produced metabolites.
Heterologous expression of an exogenous biosynthetic gene cluster in S. tropica CNB-4401.
In order to assess the performance of S. tropica CNB-4401 as a heterologous host in expressing large BGCs, we selected the recently characterized BGC for the fatty acid synthase inhibitor thiolactomycin (tlm) that is only present in strains of S. pacifica. Other high-profile BGCs from Salinispora, such as the salinosporamide, lymphostin, and lomaiviticin BGCs, are endogenous to S. tropica CNB-440 and therefore were not selected for host validation. The tlm BGC encodes an unconventional polyketide synthase (PKS) and nonribosomal peptide synthetase (NRPS) hybrid assembly line, which represents an important class of natural products (Tang et al. 2015). Furthermore, the tlm genes were previously cloned into the integration vector pCAP03 to generate pMXT13 and successfully expressed in the Streptomyces coelicolor M1152 “superhost,” albeit at lower levels than the wild type producer (Gomez-Escribano and Bibb 2011). Thus, the tlm BGC was directly introduced to the S. tropica CNB-4401 host via integration into the newly introduced genomic ϕC31 attB site. Kanamycin resistant clones were selected and designated S. tropica CNB-4401/tlm. Intergeneric conjugation between E. coli ET12567/pUB307 (helper), E. coli ET12567/pMXT13, and S. tropica CNB-4401 yielded around 5 to 10 KanR exconjugants (experiment in sextuplicate) per conjugation using 10 μL (108) S. tropica CNB-4401 spores (Figure S4).
Although the integration efficiency is again much lower than for Streptomyces spores, our data are comparable with the allelic exchange rate for making single crossover mutants of S. tropica CNB-440 (10–20 mutants by biparental conjugation). This data thus supports the hypothesis that conjugation efficiency may be hampered by high salt concentrations necessary for the growth of some marine bacteria. No exconjugants were obtained from identical attempts to integrate the tlm BGC into the genome of S. tropica CNB-440 (Figure S4, experiment in sextuplicate). Therefore, our results strongly indicate that S. tropica CNB-4401 is capable of being readily integrated with large exogenous DNA fragments using the ϕC31 int/attP-attB integration system, while S. tropica CNB-440 is not. This strategy may be widely applicable in engineering other bacteria as integratable heterologous hosts that are directly compatible with established ϕC31-containing vectors (Martinez et al. 2004).
With the successful integration of the tlm biosynthetic pathway in S. tropica CNB-4401, we cultured, extracted, and analyzed the supernatant extracts of the S. tropica CNB-4401/tlm heterologous host by HPLC and LC-MS. The production of thiolactomycin (1) and three analogues (10-methyl thiolactomycin (2), 11-methyl thiolactomycin (3), and thiotetromycin (4)) were identified from the extracts of S. tropica CNB-4401/tlm by comparison with thiolactomycins produced by the native producer S. pacifica CNS-863 as well as the heterologous producer S. coelicolor M1152/tlm. All four thiolactomycin analogues were previously fully characterized by 1D and 2D NMR experiments (Figure 3). Corresponding parent mass peaks for (1) with m/z 211.0789 [M+H]+ (calcd for C11H15O2S, 211.0793), (2) with m/z 225.0944 [M+H]+ (calcd for C12H17O2S, 225.0949), (3) with m/z 225.0942 [M+H]+ (calcd for C13H19O2S, 225.0949), and (4) with m/z 239.1097 [M+H]+ (calcd for C12H17O2S, 239.1106) were detected by high resolution LC-MS (Figure S5). Thus, this is the first report of using the marine actinomycete Salinispora as an integratable host for natural product BGC heterologous expression.
Figure 3.
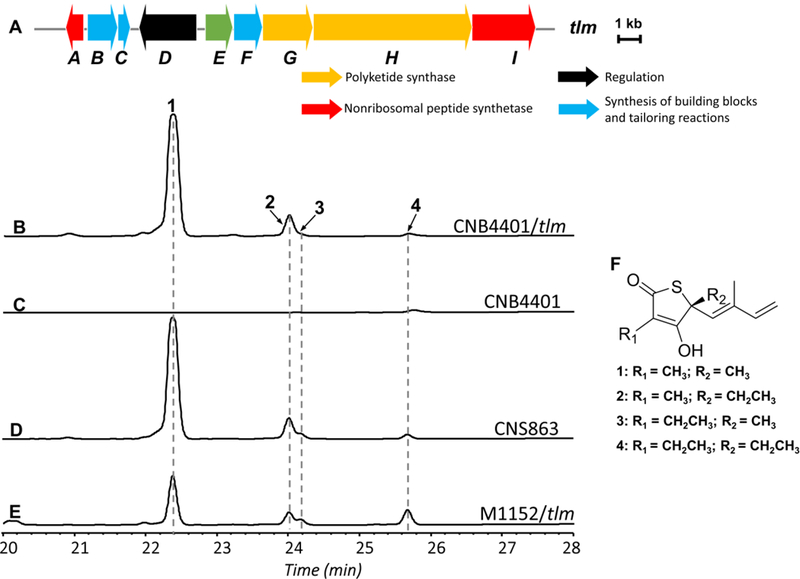
Detection of thiolactomycin and its analogues in various producing strains by HPLC. (A) Schematic illustrating the thiolactomycin (tlm) biosynthetic gene cluster. HPLC chromatograms of ethyl acetate extracts from (B) S. tropica CNB-4401/tlm, (C) S. tropica CNB-4401, (D) S. pacifica CNS-863, and (E) S. coelicolor M1152/tlm. Detection at 239 nm. (F) Structures of thiolactomycin (1), 10-methyl thiolactomycin (2), 11-methyl thiolactomycin (3), and thiotetromycin (4).
In contrast to S. coelicolor M1152/tlm, the production of thiolactomycin (1) from S. tropica CNB-4401/tlm and S. pacifica CNS-863 was approximately 3-fold higher (Figure 3), while production of 3 and 4 were only slightly lower. Overall, the production pattern of the four thiolactomycin analogues in S. tropica CNB-4401/tlm was very similar to that of the native producer, S. pacifica CNS-863, and subtly different from S. coelicolor M1152 (Figure 3). This difference in production levels as well as PKS extender unit incorporation suggests that heterologous host context with respect to genetics, biochemistry, and metabolism, influences heterologous thiolactomycin production and may also influence heterologous production of products associated with other BGCs.
Discussion
In this study, we selected S. tropica CNB-440 as the starting point for developing the first marine actinomycete heterologous host. In order to establish an integratable expression host with a cleaner background for convenient metabolic profiling, we introduced a disruptive cassette containing the well-established ΦC31 phage attachment site attB to replace three essential salinisporamide biosynthetic genes salA–C in S. tropica CNB-440. This newly established mutant strain S. tropica CNB-4401, therefore, possesses an authentic attB attachment site to integrate large exogenous BGCs through ϕC31 integrase activity for heterologous expression. We validated this new heterologous host with the successful integration and expression of the thiolactomycin BGC from several S. pacifica strains. To the best of our knowledge, this is the first example of using a marine actinomycete as a heterologous host for BGC expression.
The overall yield of all thiolactomycins produced heterologously by S. tropica CNB-4401 is very similar to the yield from the native producer S. pacifica CNS-863 and approximately 3 times higher than the yield from S. coelicolor M1152 (Figure 3). This is despite the fact that the S. coelicolor M1152 “superhost” has been subjected to about a dozen rounds of genetic manipluation to delete endogenous pathways and introduce a point mutation to rpoB that has been empirically shown to enhance antibiotic production in Streptomyces (Gomez-Escribano and Bibb 2011).
The different ratios of compounds 1–4 observed between S. tropica CNB-4401/tlm (or S. pacifica CNS-863) and S. coelicolor M1152/tlm may be due to differences in methylmalonyl-CoA and ethylmalonyl-CoA precursor supply in these strains. However, previous gene deletion and heterologous expression experiments in S. coelicolor M1152 suggest that the pool of ethylmalonyl-CoA precursors used by the TlmH PKS to generate compounds 2-4 is primarily supplied by the pathway specific crotonyl-CoA carboxylase (TlmB) and 3-hydroxybutyryl-CoA dehydrogenase (TlmC) (Tang et al. 2017). Thus, there may be additional mechanisms beyond precursor supply that influence TlmH precursor utilization, which results in a different profile of thiolactomycin production levels in S. coelicolor M1152 compared to S. tropica CNB-4401 and S. pacifica CNS-863. This supports our previous finding that host context can play an important role in heterologous coumpound production and provide biosynthetic insights (Zhang et al. 2017a).
S. tropica expresses the unusual crotonyl-CoA carboxylase SalG that not only catalyzes the construction of ethylmalonyl-CoA, but also the unique PKS precursor chloroethylmalonyl-CoA (Eustaquio et al. 2009) and the rare propylmalonyl-CoA (Liu et al. 2009). We employed LC-MS to interrogate thiolactomycin analogues that may have incorporated these alternate substrates, but no new thiolactomycin analogues were identified, perhaps due to substrate selectivity of the tlm PKS assembly line. Alternatively, insertion of a large DNA fragment into the engineered attachment site, which is upstream of the crotonyl-CoA carboxylase SalG, may cause polar effects that disrupt expression of salG. Heterologous expression of large BGCs remains challenging, particularly for pathways that require rare precursors and/or utilize uncertain regulatory, biosynthetic, or resistance factors. Our previous work to develop improved direct cloning and heterologous expression vectors demonstrated that having access to genetically diverse heterologous host organisms will enhance the likelihood of achieving successful heterologous expression for biosynthetic studies and identification of new compounds (Zhang et al. 2017a). As S. tropica CNB −4401 is a phylogenetically distinct actinomycete host that can now be easily leveraged using existing Streptomyces expression vectors for heterologous expression experiments, we believe that our study provides the natural product community an alternative and facile platform for discovering and engineering BGCs.
Supplementary Material
Acknowledgments
We kindly thank W. Fenical and P. R. Jensen (Scripps Institution of Oceanography, UCSD) for providing the S. tropica strain, and P. R. Jensen and N. Ziemert (Scripps Institution of Oceanography, UCSD) for valuable discussion. We declare that we have no competing interests.
Funding This study was supported by a graduate fellowship from the National Science Foundation to J.J.Z. and National Institutes of Health grants F31-AI129299 to J.J.Z. and R01-GM085770 and R01-AI117712 to B.S.M.
Footnotes
Compliance with ethical standards
Conflict of interest The authors declare no competing financial interest.
Ethical statement This article does not contain any studies with human participants or animals performed by any of the authors.
References
- Amos GCA, Awakawa T, Tuttle RN, Letzel AC, Kim MC, Kudo Y, Fenical W, Moore BS, Jensen PR (2017) Comparative transcriptomics as a guide to natural product discovery and biosynthetic gene cluster functionality. Proc Natl Acad Sci U S A 114(52):E11121–E11130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baltz RH (2010) Streptomyces and Saccharopolyspora hosts for heterologous expression of secondary metabolite gene clusters. J Ind Microbiol Biotechnol 37(8):759–72 [DOI] [PubMed] [Google Scholar]
- Baltz RH (2016) Genetic manipulation of secondary metabolite biosynthesis for improved production in Streptomyces and other actinomycetes. J Ind Microbiol Biotechnol 43(2–3):343–70 [DOI] [PubMed] [Google Scholar]
- Berdy J (2005) Bioactive microbial metabolites. J Antibiot (Tokyo) 58(1):1–26 [DOI] [PubMed] [Google Scholar]
- Chater KF (2016) Recent advances in understanding Streptomyces. F1000Res 5:2795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherepanov PP, Wackernagel W (1995) Gene disruption in Escherichia coli: TcR and KmR cassettes with the option of Flp-catalyzed excision of the antibiotic-resistance determinant. Gene 158(1):9–14 [DOI] [PubMed] [Google Scholar]
- Combes P, Till R, Bee S, Smith MC (2002) The Streptomyces genome contains multiple pseudo-attB sites for the (phi)C31-encoded site-specific recombination system. J Bacteriol 184(20):5746–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datsenko KA, Wanner BL (2000) One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci U S A 97(12):6640–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eustaquio AS, McGlinchey RP, Liu Y, Hazzard C, Beer LL, Florova G, Alhamadsheh MM, Lechner A, Kale AJ, Kobayashi Y, Reynolds KA, Moore BS (2009) Biosynthesis of the salinosporamide A polyketide synthase substrate chloroethylmalonyl-coenzyme A from S-adenosyl-L-methionine. Proc Natl Acad Sci U S A 106(30):12295–300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flett F, Mersinias V, Smith CP (1997) High efficiency intergeneric conjugal transfer of plasmid DNA from Escherichia coli to methyl DNA-restricting streptomycetes. FEMS Microbiol Lett 155(2):223–9 [DOI] [PubMed] [Google Scholar]
- Fu J, Bian X, Hu S, Wang H, Huang F, Seibert PM, Plaza A, Xia L, Muller R, Stewart AF, Zhang Y (2012) Full-length RecE enhances linear-linear homologous recombination and facilitates direct cloning for bioprospecting. Nat Biotechnol 30(5):440–6 [DOI] [PubMed] [Google Scholar]
- Gomez-Escribano JP, Bibb MJ (2011) Engineering Streptomyces coelicolor for heterologous expression of secondary metabolite gene clusters. Microb Biotechnol 4(2):207–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez-Escribano JP, Bibb MJ (2014) Heterologous expression of natural product biosynthetic gene clusters in Streptomyces coelicolor: from genome mining to manipulation of biosynthetic pathways. J Ind Microbiol Biotechnol 41(2):425–31 [DOI] [PubMed] [Google Scholar]
- Gulder TA, Moore BS (2010) Salinosporamide natural products: Potent 20S proteasome inhibitors as promising cancer chemotherapeutics. Angew Chem Int Ed Engl 49(49):9346–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gust B, Challis GL, Fowler K, Kieser T, Chater KF (2003) PCR-targeted Streptomyces gene replacement identifies a protein domain needed for biosynthesis of the sesquiterpene soil odor geosmin. Proc Natl Acad Sci U S A 100(4):1541–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horinouchi S (2007) Mining and polishing of the treasure trove in the bacterial genus Streptomyces. Biosci Biotechnol Biochem 71(2):283–99 [DOI] [PubMed] [Google Scholar]
- Jensen PR, Gontang E, Mafnas C, Mincer TJ, Fenical W (2005) Culturable marine actinomycete diversity from tropical Pacific Ocean sediments. Environ Microbiol 7(7):1039–48 [DOI] [PubMed] [Google Scholar]
- Jensen PR, Moore BS, Fenical W (2015) The marine actinomycete genus Salinispora: a model organism for secondary metabolite discovery. Nat Prod Rep 32(5):738–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang W, Zhao X, Gabrieli T, Lou C, Ebenstein Y, Zhu TF (2015) Cas9-Assisted Targeting of CHromosome segments CATCH enables one-step targeted cloning of large gene clusters. Nat Commun 6:8101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kersten RD, Lane AL, Nett M, Richter TK, Duggan BM, Dorrestein PC, Moore BS (2013) Bioactivity-guided genome mining reveals the lomaiviticin biosynthetic gene cluster in Salinispora tropica. Chembiochem 14(8):955–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JH, Feng Z, Bauer JD, Kallifidas D, Calle PY, Brady SF (2010) Cloning large natural product gene clusters from the environment: piecing environmental DNA gene clusters back together with TAR. Biopolymers 93(9):833–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komatsu M, Komatsu K, Koiwai H, Yamada Y, Kozone I, Izumikawa M, Hashimoto J, Takagi M, Omura S, Shinya K, Cane DE, Ikeda H (2013) Engineered Streptomyces avermitilis host for heterologous expression of biosynthetic gene cluster for secondary metabolites. ACS Synth Biol 2(7):384–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komatsu M, Uchiyama T, Omura S, Cane DE, Ikeda H (2010) Genome-minimized Streptomyces host for the heterologous expression of secondary metabolism. Proc Natl Acad Sci U S A 107(6):2646–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lechner A, Eustaquio AS, Gulder TA, Hafner M, Moore BS (2011) Selective overproduction of the proteasome inhibitor salinosporamide A via precursor pathway regulation. Chem Biol 18(12):1527–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letzel AC, Li J, Amos GCA, Millan-Aguinaga N, Ginigini J, Abdelmohsen UR, Gaudencio SP, Ziemert N, Moore BS, Jensen PR (2017) Genomic insights into specialized metabolism in the marine actinomycete Salinispora. Environ Microbiol 19(9):3660–3673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Hazzard C, Eustaquio AS, Reynolds KA, Moore BS (2009) Biosynthesis of salinosporamides from alpha, beta-unsaturated fatty acids: implications for extending polyketide synthase diversity. J Am Chem Soc 131(30):10376–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez A, Kolvek SJ, Yip CL, Hopke J, Brown KA, MacNeil IA, Osburne MS (2004) Genetically modified bacterial strains and novel bacterial artificial chromosome shuttle vectors for constructing environmental libraries and detecting heterologous natural products in multiple expression hosts. Appl Environ Microbiol 70(4):2452–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyanaga A, Janso JE, McDonald L, He M, Liu H, Barbieri L, Eustaquio AS, Fielding EN, Carter GT, Jensen PR, Feng X, Leighton M, Koehn FE, Moore BS (2011) Discovery and assembly-line biosynthesis of the lymphostin pyrroloquinoline alkaloid family of mTOR inhibitors in Salinispora bacteria. J Am Chem Soc 133(34):13311–3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman DJ, Cragg GM (2016) Natural products as sources of new drugs from 1981 to 2014. J Nat Prod 79(3):629–61 [DOI] [PubMed] [Google Scholar]
- Ongley SE, Bian X, Neilan BA, Muller R (2013) Recent advances in the heterologous expression of microbial natural product biosynthetic pathways. Nat Prod Rep 30(8):1121–38 [DOI] [PubMed] [Google Scholar]
- Paget MS, Chamberlin L, Atrih A, Foster SJ, Buttner MJ (1999) Evidence that the extracytoplasmic function sigma factor sigmaE is required for normal cell wall structure in Streptomyces coelicolor A3(2). J Bacteriol 181(1):204–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren H, Wang B, Zhao H (2017) Breaking the silence: new strategies for discovering novel natural products. Curr Opin Biotechnol 48:21–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutledge PJ, Challis GL (2015) Discovery of microbial natural products by activation of silent biosynthetic gene clusters. Nat Rev Microbiol 13(8):509–23 [DOI] [PubMed] [Google Scholar]
- Schorn MA, Alanjary MM, Aguinaldo K, Korobeynikov A, Podell S, Patin N, Lincecum T, Jensen PR, Ziemert N, Moore BS (2016) Sequencing rare marine actinomycete genomes reveals high density of unique natural product biosynthetic gene clusters. Microbiology 162(12):2075–2086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith MC, Brown WR, McEwan AR, Rowley PA (2010) Site-specific recombination by phiC31 integrase and other large serine recombinases. Biochem Soc Trans 38(2):388–94 [DOI] [PubMed] [Google Scholar]
- Smith MC, Till R, Brady K, Soultanas P, Thorpe H, Smith MC (2004) Synapsis and DNA cleavage in phiC31 integrase-mediated site-specific recombination. Nucleic Acids Res 32(8):2607–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang X, Li J, Millan-Aguinaga N, Zhang JJ, O’Neill EC, Ugalde JA, Jensen PR, Mantovani SM, Moore BS (2015) Identification of thiotetronic acid antibiotic biosynthetic pathways by target-directed genome mining. ACS Chem Biol 10(12):2841–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang X, Li J, Moore BS (2017) Minimization of the thiolactomycin biosynthetic pathway reveals that the cytochrome P450 enzyme TlmF is required for five-membered thiolactone ring formation. Chembiochem 18(12):1072–1076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Udwary DW, Zeigler L, Asolkar RN, Singan V, Lapidus A, Fenical W, Jensen PR, Moore BS (2007) Genome sequencing reveals complex secondary metabolome in the marine actinomycete Salinispora tropica. Proc Natl Acad Sci U S A 104(25):10376–81 4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamanaka K, Reynolds KA, Kersten RD, Ryan KS, Gonzalez DJ, Nizet V, Dorrestein PC, Moore BS (2014) Direct cloning and refactoring of a silent lipopeptide biosynthetic gene cluster yields the antibiotic taromycin A. Proc Natl Acad Sci U S A 111(5):1957–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang JJ, Tang X, Zhang M, Nguyen D, Moore BS (2017a) Broad-host-range expression reveals native and host regulatory elements that influence heterologous antibiotic production in gram-negative bacteria. MBio 8(5) e01291–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Lu C, Bai L (2017b) Conversion of the high-yield salinomycin producer Streptomyces albus BK3–25 into a surrogate host for polyketide production. Sci China Life Sci 60(9):1000–1009 [DOI] [PubMed] [Google Scholar]
- Ziemert N, Lechner A, Wietz M, Millan-Aguinaga N, Chavarria KL, Jensen PR (2014) Diversity and evolution of secondary metabolism in the marine actinomycete genus Salinispora. Proc Natl Acad Sci U S A 111(12):E1130–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


