Abstract
Small GTPases of the Ras superfamily are master regulators of intracellular trafficking and constitute essential signaling components in all eukaryotes. Aberrant small GTPase signaling is associated with a wide spectrum of human diseases including cancer. Here we developed a high-throughput, multiple-reaction monitoring-based workflow coupled with stable isotope labeling by amino acids in cell culture, for targeted quantification of approximately 100 small GTPases in cultured human cells. Using this method, we investigated the differential expression of small GTPases in three pairs of primary and metastatic melanoma cell lines. Bioinformatic analyses of The Cancer Genome Atlas data and other publicly available data as well as cell-based assays revealed previously unrecognized roles of RAB38 in promoting melanoma metastasis. Diminished promoter methylation and the subsequent augmented binding of transcription factor MITF contributed to elevated expression of RAB38 gene in metastatic versus primary melanoma cells. Moreover, RAB38 promoted invasion of cultured melanoma cells by modulating the expression and activities of matrix metalloproteinases-2 and -9. Together, these data establish a novel targeted proteomic method for interrogating the small GTPase proteome in human cells and identify epigenetic reactivation of RAB38 as a contributing factor to metastatic transformation in melanoma.
Keywords: Targeted Proteomics, Multiple Reaction Monitoring, Small GTPases of Ras Superfamily, Melanoma Metastasis
Introduction
Small GTPases of the Ras superfamily are highly conserved in eukaryotes, including more than 100 members that could be divided into six subfamilies, i.e. Ras, Rho, Rab, Sar1/Arf, Ran and others (1). They can exist in the GTP-bound active state or GDP-bound inactive state, which are modulated by guanine nucleotide exchange factors (GEFs), GTPase-activating proteins (GAPs) and guanine nucleotide dissociation inhibitors (GDIs) (2). Small GTPases serve as master regulators of cellular trafficking and are involved in numerous cell signaling cascades (1,3). In addition, emerging evidence has linked aberrant expression of small GTPases with cancer progression, including RHOC and RAB27A in melanoma (4,5).
Despite the importance of small GTPases in cell signaling and human diseases, very few studies have been conducted to assess quantitatively the small GTPases at the proteome-wide scale. In recent years, multiple-reaction monitoring (MRM)-based targeted proteomic method has emerged as a powerful approach for analyzing proteins and peptides of interest with high specificity and sensitivity (6,7). We reason that a targeted proteomic method for the measurement of small GTPases may enable mechanistic studies of small GTPase signaling and facilitate the discovery of novel roles of small GTPases in the etiology of human diseases.
In the present study, we developed a facile and effective MRM-based method for high-throughput profiling of small GTPases in cultured human cells and we also applied the method for assessing the roles of small GTPases in melanoma metastasis. We chose to examine the roles of small GTPases in melanoma metastasis because melanoma is one of the most aggressive and treatment-resistant types of human cancers. In this vein, an estimated 91,270 new cases of melanoma and 9320 deaths are expected in the United States in 2018 (8), and the high mortality rate of melanoma is attributed to its high probability to metastasize (9).
Materials and Methods
Cell Culture
HCT-116 human colorectal cancer cells, HEK293T human embryonic kidney cells, HL-60 human promyelocytic leukemia cells, Jurkat-T human T lymphocytic leukemia cells, MCF-7 human breast cancer cells, WM-115 and WM-266-4 human melanoma cells were purchased from American Type Culture Collection (ATCC; Manassas, VA). GM00637 human skin fibroblasts were kindly provided by Prof. Gerd P. Pfeifer (the City of Hope). IGR39 and IGR37 human melanoma cells were generous gifts from Prof. Peter H. Duesberg (University of California, Berkeley). WM793 and 1205Lu human melanoma cells were purchased from Wistar Institute. HCT-116, HEK293T, GM00637, MCF-7, WM-115 and WM-266-4 cells were cultured in Dulbecco’s Modified Eagle Medium (DMEM; Invitrogen-Gibco, Carlsbad, CA). HL-60, Jurkat-T, IGR39, IGR37, WM793 and 1205Lu cells were cultured in RPMI 1640 Medium (Invitrogen-Gibco). All culture media were supplemented with 10% fetal bovine serum (FBS; Invitrogen-Gibco) and penicillin/streptomycin (100 IU/mL). Cells were maintained at 37°C in a humidified atmosphere containing 5% CO2, and the culture medium was changed in every 2 to 3 days as necessary.
The initial passage numbers for melanoma cells used were: WM-115 (p9), WM-266-4 (p6), IGR39 (p4), IGR37 (p7), WM793 (p16), and 1205Lu (p70). All the relevant experiments were conducted within 20 passages from revival of the initial frozen seeds. LookOut Mycoplasma PCR Detection Kit (MP0035, Sigma-Aldrich, MO) for detection of 19 mycoplasma species was used following the manufacturer’s instructions. PCRs were performed using HotStart Taq Polymerase. Results were visualized on a 1.2% agarose gel, where mycoplasma-positive samples would show a band at 261 bp, and internal control DNA showed a band at 500 bp. WM-115, WM-266-4, IGR39, IGR37, WM793, and 1205Lu melanoma cell lines were tested by this method to be free of mycoplasma on May 9, 2018. In addition, these six melanoma cell lines were authenticated by ATCC on May 24, 2018 using Short Tandem Repeat (STR) analysis as described in 2012 in ANSI Standard (ASN-0002) Authentication of Human Cell Lines.
For SILAC experiments, [13C6,15N2]-L-lysine and [13C6]-L-arginine (Cambridge Isotopes Inc., MA), or the corresponding unlabeled lysine and arginine, were added to SILAC DMEM media depleted of L-Lysine and L-Arginine (Thermo Scientific Pierce, MA) until their final concentrations reached 0.398 and 0.798 mM, respectively, to yield ‘heavy’ and ‘light’ media. The SILAC RPMI-1640 media were prepared in a similar fashion except that the final concentrations of the added lysine and arginine were 0.274 mM and 1.15 mM, respectively. The SILAC media were again supplemented with 10% dialyzed FBS (Corning, NY). WM-115 and WM-266-4 cells were cultured in the heavy-DMEM medium, and IGR39, IGR37, WM793 and 1205Lu cells were cultured in the heavy-RPMI medium for at least six cell doublings to ensure complete heavy-isotope incorporation.
Gene Ontology (GO) Analysis and Data Source for Bioinformatic Analyses
Gene Ontology analyses were conducted using the web-based Database for Annotation, Visualization and Integrated Discovery (DAVID, version 6.7; https://david.ncifcrf.gov/) (10). Patient RNAseq data were obtained from The Cancer Genome Atlas (TCGA) via cBioPortal (11). We used data from 458 melanoma patients in the TCGA-SKCM project for bioinformatic analyses. The Cancer Cell Line Encyclopedia (CCLE) (http://www.broadinstitute.org/ccle/home) were employed for the comprehensive evaluation of mRNA expression for candidate genes among more than 1,000 cell lines representing 37 cancer types (12). Multi-tumor RAB38 mRNA expression box plot and scatter plot for melanoma cell lines were retrieved from the CCLE database using cBioPortal. Publicly available transcriptomic profiles with accession numbers GSE7553, GSE7929, GSE8401, GSE22153, GSE44662, GSE46522 and GSE70621 were downloaded from the National Center for Biotechnology Information (NCBI) Gene Expression Omnibus (GEO) database and analyzed using R (version 3.4.3).
Immunoblotting
Total protein was extracted from cell pellet using ice-cold CelLytic M cell lysis reagent (Sigma-Aldrich, MO) containing 1% (v/v) protease inhibitor cocktail (Sigma-Aldrich, MO). After cell lysis, the protein concentration was determined by the Quick Start™ Bradford Protein Assay (Bio-Rad, CA). Approximately 10–50 μg whole cell lysates, mixed with 4×Laemmli SDS loading buffer, were electrophoresed in 10% SDS-PAGE gels and transferred to nitrocellulose membranes. The membranes were incubated with primary antibodies against human RAB12 (Thermo Fisher; rabbit polyclonal, 1:2,000), RAB27A (Abcam; rabbit polyclonal, 1:5,000), RAB31 (4D12. Santa Cruz; rabbit polyclonal, 1:2,000), RAB32 (Thermo Fisher; rabbit polyclonal, 1:2,000), RAB38 (A-8. Santa Cruz; mouse polyclonal, 1:2,000), MITF (D-9, Santa Cruz; mouse polyclonal, 1:5,000), or β-actin (Thermo Fisher; rabbit polyclonal, 1:10,000), followed by incubation with peroxidase-labeled donkey anti-rabbit secondary antibody (Thermo Fisher; 1:10,000) or mouse m-IgGκ BP-HRP (Santa Cruz; 1:10,000). Amersham ECL Prime Western Blot Detecting Reagent (GE Healthcare, CA) was used to visualize the protein bands.
Migration and Invasion Assays
For transwell migration assay, cells (0.5−1 × 105) were placed in the upper chamber of transwell inserts (Corning, NY) with serum-free DMEM medium. DMEM medium containing 10% FBS was added to the lower chamber as chemoattractants and the cells were incubated at 37°C for 24 h. After removal of unmigrated cells, the cells attached to the reverse side of the membrane were stained with 0.5% crystal violet, and 5 randomly selected fields were counted under an inverted microscope in each experiment. The invasion assay was conducted under the same conditions except that the transwell membranes were pre-coated with Matrigel (Corning, NY).
Gelatin Zymography Assay
At 24 h following plasmid transfection or 72 h following siRNA transfection, the culture medium was removed, and the cells were washed twice with, and reconstituted in, serum-free DMEM medium. After a 24-h incubation, conditioned medium (CM) was collected by centrifugation to remove cell debris. The collected CM was further concentrated using Microcon centrifugal filter units with a molecular weight cutoff of 30 kDa (EMD Millipore, CA) and the Quick Start™ Bradford Protein Assay was used to determine the total protein concentration. Subsequently, 5–10 μg total CM proteins were separated using 7.5% SDS-PAGE gels containing 0.1% gelatin. After electrophoresis, the gels were incubated with zymography washing buffer (2.5% Triton X-100, 50 mM Tris-HCl, pH 7.5) at room temperature for 1 h to remove excess SDS and renature the matrix metalloproteinases (MMPs). The gels were then incubated at 37°C for 24 h in zymography developing buffer (1.0% Triton X-100, 50 mM Tris-HCl, pH 7.5) to induce gelatin digestion by the renatured enzymes. The gels were subsequently stained with 0.5% Coomassie blue G-250 and destained until clear bands were visible against the dark background, indicative of proteolytic activities of MMPs.
Chromatin immunoprecipitation (ChIP) and real-time quantitative PCR (RT-qPCR)
For ChIP, approximately 1 × 107 WM-115 and WM-266-4 cells were harvested and fixed in PBS with 1% formaldehyde at room temperature for 10 min. After cross-linking, the cell pellets were resuspended in 1 mL of lysis buffer (50 mM Tris-HCl, pH 8.0, 150 mM NaCl, 5 mM EDTA, 1% Triton X-100, and 0.1% sodium deoxycholate) and sonicated to obtain DNA fragments of 300–500 bp in length. Anti-MITF antibody (ab12039, Abcam) or normal IgG (2729S, Cell Signaling Technology) was used to precipitate the chromatin. The precipitated DNA was purified using the QIAquick PCR purification Kit (Qiagen, MD) and used for RT-qPCR analysis. The primers used in RT-qPCR coupled to ChIP were: RAB38 forward, GCCACAAACTTGTGAGGTGT; RAB38 reverse, CTTCAGACCTGTGGTCAACG; TBC1D16 forward, GGCCACATACAAAGGGATCG; TBC1D16 reverse, CTCGCGGAGGCAATCTGA.
Bisulfite Sequencing
Approximately 5 × 103 cells collected from six melanoma cell lines: WM-115, WM-266-4, IGR39, IGR37, WM793 and 1205Lu, respectively, were lysed and treated with bisulfite using the EZ DNA Methylation-Direct Kit (Zymo Research, CA). The resulting DNA was subsequently amplified using ZymoTaq DNA Polymerase (Zymo Research, CA). RAB38 primers were designed using the MethPrimer 2.0 online tool (http://www.urogene.org/methprimer2/) to amplify the 179 bp fragment of the promoter region of the bisulfite converted-RAB38 gene (Chr11: 87,908,686–87,908,864, UCSC Genome Browser Human Feb. 2009 Assembly, GRCh37/hg19). The outer PCR was set up using the following primers: RAB38 forward primer, 5´-GGTTAGGGTTATAGGTGAAAATAGT-3´, RAB38 reverse primer, 5´- AACTCCTCCCCTAAAAATTAATCC-3´. The reaction mixture was heated to 95°C for 10 min followed by 40 cycles, denaturing at 95°C for 30 sec, annealing at 55°C for 45 sec and elongating at 72°C for 1 min, followed by a final elongation step at 72°C for 7 min. The successful amplification and the right size of the amplicon were validated using 2% agarose gel. Amplicons were purified using the E.Z.N.A. Gel Extraction Kit (Omega Bio-tek, GA). The purified PCR products were cloned into the pGEM®-T vector (Promega Corporation, WI) and the ligation products were selected by blue/white colony screening. Ten white colonies selected for each cell line were grown in liquid Lysogeny broth (LB) media overnight and the plasmids were extracted using GeneJET Plasmid Miniprep kit (Thermo Fishier Scientific, MA). The plasmids were then subjected to Sanger sequencing and the methylation status of each individual CpG dinucleotides in the amplicon was subsequently determined. Sequencing results were analyzed by the BiQ Analyzer software (http://biq-analyzer.bioinf.mpi-inf.mpg.de/) to generate the lollipop-representation.
5-Aza-2´-deoxycytidine (5-Aza) Treatment
WM-115 and IGR39 cells were seeded at a density of 2 × 104 cells/mL and treated at 24 h later with 5 μM 5-Aza (Sigma-Aldrich, MO). Dimethyl sulfoxide (DMSO) was used as the vehicle control. The cells were replenished with freshly prepared DMSO/5-Aza in complete growth medium in every 24 h for up to 96 h of treatment (four pulses). Total RNAs were isolated for cDNA conversion and RT-qPCR analyses. The primers used for RT-qPCR analyses are listed in Table S1.
Results
Development of a High-throughput LC-MRM Assay for Targeted Quantitative Profiling of Small GTPases in Cultured Human Cancer Cells
We set out to develop a high-throughput, multiplexed MRM-based targeted proteomics method for interrogating small GTPases in the entire human proteome. In this context, Halvey et al. (13) developed an LC-MRM method for quantifying wild-type and mutant K-RAS proteins in cultured cancer cells and pancreatic cyst fluids after enrichment of low-molecular weight (20–25 kDa) proteins using sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). In addition, Zhang et al. (14) described the use of MRM in combination with SDS-PAGE-based enrichment of proteins in the molecular weight range of 15–25 kDa to measure simultaneously the activities of 12 small GTPases after affinity enrichment using the GTPase-binding domains of four effector proteins. Building upon these previous studies, we developed an SDS-PAGE fractionation coupled with LC-MRM workflow for targeted quantification of small GTPases at the entire proteome scale.
A high-throughput LC-MRM workflow for the proteome-wide interrogation of small GTPases requires the collection of tandem mass spectra and chromatographic retention time of unique (or signature) peptides derived from the targeted small GTPases. Because the expression of small GTPases differs among different cell lines, we established an MRM spectral library based on the data collected from shotgun proteomic analyses of tryptic digestion mixtures of low-molecular weight proteins (15–37 kDa) from the lysates of 9 human cell lines of different tissue origins. These included GM00637 (skin), HCT-116 (colon), HEK293T (kidney), HL-60 (peripheral blood), Jurkat T (peripheral blood), K562 (bone marrow), MCF-7 (breast), WM-115 (skin), and WM-266-4 (skin). To this end, we fractionated the whole-cell protein lysate using SDS-PAGE, excised the gel bands in the molecular weight region of 15–37 kDa, reduced the cysteine residues in proteins with dithiothreitol and alkylated them with iodoacetamide. The proteins were then digested in gel with trypsin and the resulting peptide mixtures subjected to LC-MS/MS analysis in the data-dependent acquisition (DDA) mode. The identified proteins (> 5000) were then filtered using the DAVID bioinformatic tool with the Gene Ontology (GO) term of ‘small GTPase’ (10). The tandem mass spectra of all peptides from small GTPases along with their retention time information were subsequently imported into Skyline (version 3.6) (15) to establish the MRM spectral library.
To achieve reliable MRM-based quantification, we selected an average of three peptides that are unique for each small GTPase, and when necessary, to its specific isoform(s). To maximize selectivity and sensitivity for the MRM measurements, we chose the transitions corresponding to the formation of the three most abundant y-ions based on the MS/MS acquired from shotgun proteomic analyses (Fig. 1A and 1B) (16). Table S2 shows the complete list of small GTPases of the Ras superfamily, which are organized according to the individual subfamilies (Ras, Rho, Rab, Sar1/Arf, Ran, and others). The current version of the MRM spectral library encompassed 432 distinct peptides representing 113 non-redundant small GTPases encoded by unique genes (Table S3A). To our knowledge, this is the first targeted proteomic method developed for profiling comprehensively the Ras superfamily of small GTPases.
Fig. 1. A targeted proteomic strategy for high-throughput quantitative profiling of small GTPases in paired primary/metastatic melanoma cells. (A).
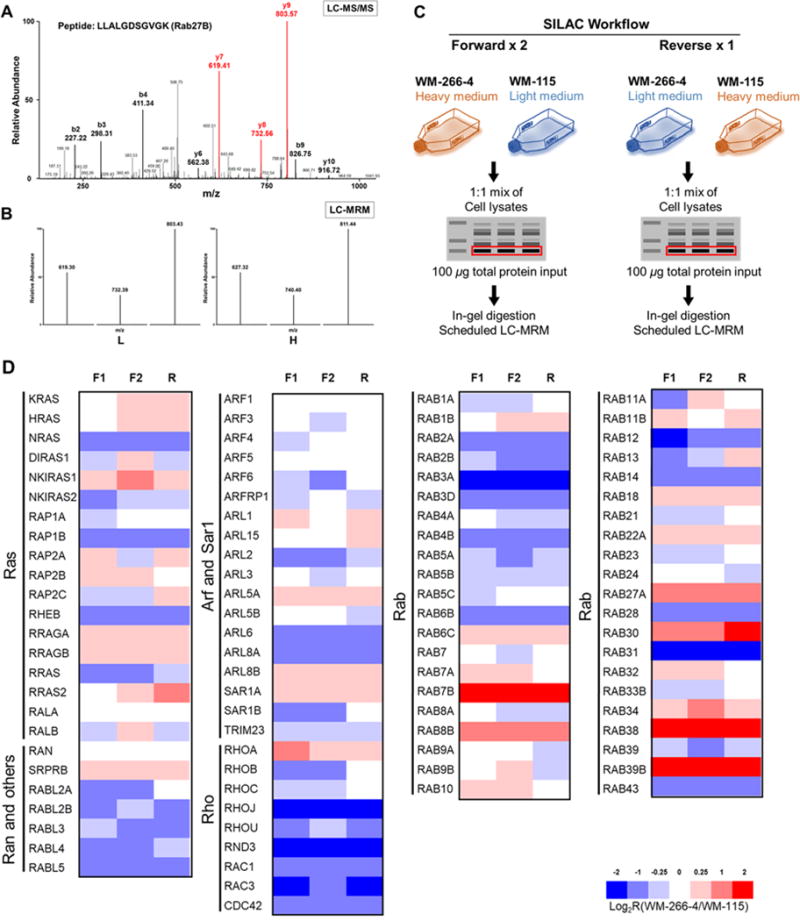
A representative MS/MS obtained from data-dependent acquisition supporting the reliable identification of the peptide LLALGDSGVGK from RAB27B; (B) LC-MRM spectra for the same peptide from targeted analysis with light- and heavy-labeled lysine on the C terminus, respectively. The distribution of the peak intensities was consistent with the theoretical distribution found in the MS/MS from the MRM spectral library; (C) A schematic diagram showing the targeted proteomic workflow, relying on metabolic labeling with SILAC, SDS-PAGE fractionation, and LC-MRM analysis, for quantifying the differential expressions of small GTPases in WM-115 (primary) and WM-266-4 (metastatic) melanoma cells; (D) A heatmap showing the differential expression of small GTPases in paired WM-115 and WM-266-4 melanoma cells. Shown are the Log2R(WM-266-4/WM-115) values obtained from scheduled LC-MRM analyses of samples from two forward and one reverse SILAC labeling experiments. The red and blue bars designate those small GTPases that are up-and down-regulated, respectively, in the WM-266-4 metastatic melanoma cells relative to the WM-115 primary melanoma cells, as indicated by the scale bar.
To increase the throughput of the assay, we employed scheduled LC-MRM with the use of normalized retention time (iRT) (17). The iRT is a dimensionless score for a peptide derived from its empirical retention time observed in shotgun proteomic analysis and the retention times for a set of standard peptides analyzed under the same LC conditions. In scheduled LC-MRM analysis, the mass spectrometer could be scheduled to collect subsets of transitions in predefined retention time windows according to the chromatographic setup, where the retention times for the targeted peptides were predicted from their iRT values in the library and from the actual retention times observed for the standard peptides. In doing so, we established a robust and high-throughput MRM-based targeted proteomic workflow for the Ras superfamily of small GTPases, where the 432 unique peptides from small GTPases could be monitored in a single LC-MRM run.
Scheduled LC-MRM Analysis Revealed Differential Expression of Small GTPases in Paired Primary/Metastatic Melanoma Cells
Paired cell lines derived from the same cancer patients are powerful resources for investigating the mechanisms of cancer progression. Here we employed three pairs of primary/metastatic melanoma cell lines for the targeted analyses of small GTPases: The ‘WM’ pair consists of WM-115 and WM-266-4, which were derived from the primary tumor site and the right thigh skin metastatic site of the same melanoma patient, respectively (18); the ‘IGR’ pair comprises IGR39 and IGR37, which were respectively derived from the primary tumor site and the groin metastatic site of another individual (19); in the ‘WMLu’ pair, WM793 was initiated from a superficial spreading melanoma, and 1205Lu was derived from a lung metastasis of WM793 cells after subcutaneous injection into the tail vein of an immune-deficient mouse (20).
We employed stable isotope-labeling by amino acid in cell culture (SILAC) (21), in conjunction with the above-described SDS-PAGE fractionation and LC-MRM analysis, for assessing the differential expression of small GTPases in the three pairs of matched primary/metastatic melanoma cells (Fig. 1C). To this end, we first modified the Skyline MRM library by incorporating the corresponding transitions for the ‘heavy’ forms of precursor and fragment ions. By using this approach, we were able to quantify approximately 100 small GTPases in each of the three paired melanoma cell lines (Table S3B). Among the 101 small GTPases quantified for the WM pair (Fig. 1D), 14 and 10 were substantially up- and down-regulated (by at least 1.5-fold), respectively, in the metastatic (WM-266-4) relative to the primary (WM-115) melanoma cells (Fig. S1A). In addition, among the 93 small GTPases quantified for the IGR pair, 20 and 12 were considerably up- and down-regulated, respectively, in the metastatic (IGR37) relative to the primary (IGR39) melanoma cells (Fig. S1B). Of the 93 small GTPases quantified for the third pair, 9 and 24 were up- and down-regulated by at least 1.5-fold in the metastatic (1205Lu) compared to the primary (WM793) melanoma cells, respectively (Fig. S1C).
We also explored the similarities and differences in the expression profiles of small GTPases by hierarchical clustering analysis of the quantitative proteomics data. It turned out that the features in differential expression of small GTPases, induced by metastatic transformation, were more similar for the WM and IGR pairs than between either of the two pairs and the WMLu pair (Fig. S1D). This might be attributed in part to the differences in transcriptional and/or epigenetic regulations in the three pairs of melanoma cell lines (vide infra) and the fact that the metastatic lines in the first two pairs were derived from melanoma patients, whereas that of the last pair was obtained from experimental metastasis in mouse, as noted above.
It is worth noting that, by utilizing the iRT algorithms, we were able to accurately predict the actual retention times for the targeted peptides with the use of a 6-min retention time window. The R2 values were 0.992 and 0.996 for LC-MRM measurements of peptide samples obtained from the WM-115/WM266-4 (Fig. S2A) and IGR39/IGR37 (Fig. S2B) paired cell lines, respectively. Moreover, this method also displayed excellent reproducibility between different replicates (Fig. S2C−S2F).
For comparison, we also analyzed the peptide samples from the WM-115/WM-266-4 cells using shotgun proteomic approach on an LTQ Orbitrap Velos mass spectrometer. The results showed that the LC-MRM method outperformed the shotgun proteomic method in reproducibility and sensitivity, the latter of which is reflected by the pronouncedly larger numbers of small GTPases quantified by the former method (101 vs. 59, Fig. S2G). The excellent reproducibility of the MRM-based quantification is manifested by the observation that 101 small GTPase proteins could be reproducibly quantified in all three sets of SILAC labeling experiments. In contrast, among the 59 small GTPases detected by the shotgun proteomic method, 9 and 4 were exclusively detected in the forward and reverse SILAC experiments, respectively (Fig. S2H). Taken together, the established targeted proteomic workflow provided excellent sensitivity and reproducibility, and it allowed for robust and high-throughput quantifications of small GTPases in melanoma cells.
Targeted Proteomics Revealed the Up-regulation of RAB27A and RAB38 in WM-266-4 and IGR37 Metastatic Melanoma Cells
One goal of the present study was to uncover small GTPases that drive and/or suppress melanoma metastasis. Hence, we expected to confirm the differential expression of some previously reported drivers and/or suppressors for melanoma metastasis. In this vein, RAB27A was shown to promote melanoma metastasis through the regulation of the MET network (22). Indeed our LC-MRM data revealed significant up-regulations of RAB27A in the WM-266-4 and IGR37 metastatic melanoma cell lines relative to the corresponding primary melanoma cells, though similar observation was not made for the WMLu pair (Fig. 2A−2C).
Fig. 2. LC-MRM and Western blots revealed consistently higher levels of expression of RAB27A and RAB38 proteins in metastatic melanoma cell lines (WM-266-4 and IGR37) than the paired primary melanoma cell lines (WM-115 and ICR39). (A).
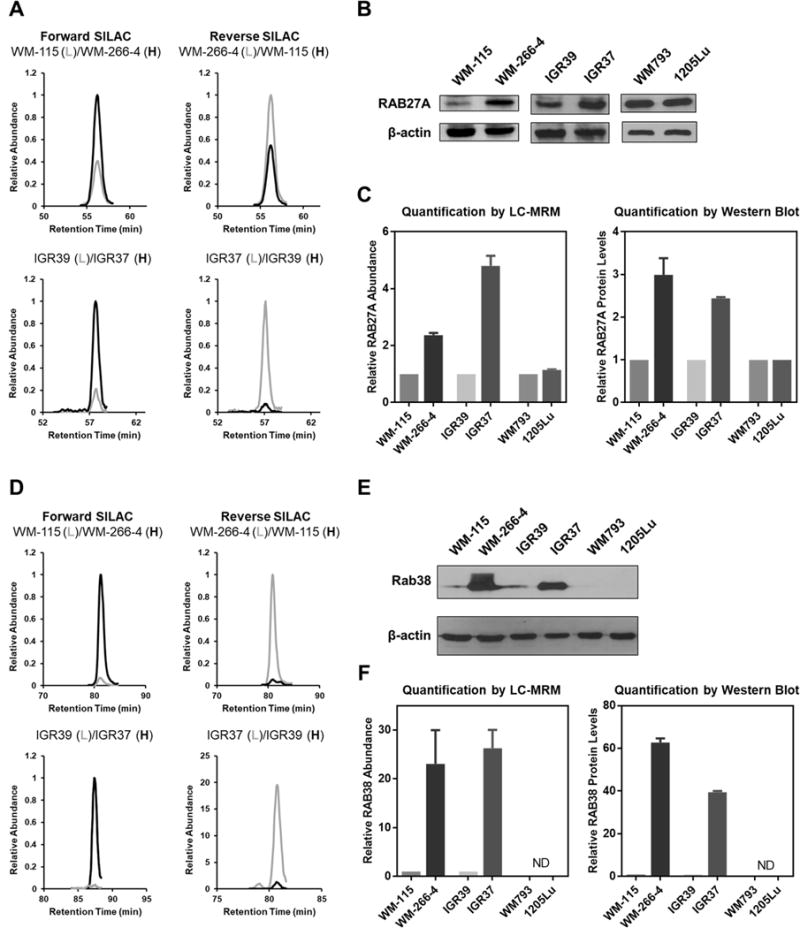
Extracted MRM traces for three transitions (y8, y7, and y6) monitored for a unique tryptic peptide from RAB27A, i.e. TSVLYQYTDGK, with light (grey) and heavy (black) labels in forward and reverse SILAC experiments for both WM-115/WM-266-4 and IGR39/IGR37 paired melanoma cells; (B) Western blot analysis confirmed the elevated expression of RAB27A in WM-266-4 and IGR37 cells; (C) Quantification results for RAB27B from LC-MRM and Western blot analyses; (D) Extracted MRM traces for three transitions (y9, y8, and y7) monitored for a unique tryptic peptide from RAB38, i.e. LLVIGDLGVGK, with light (grey) and heavy (black) labels in forward and reverse SILAC experiments for both WM-115/WM-266-4 and IGR39/IGR37 paired melanoma cells; (E) Western blot analysis confirmed the elevated expression of RAB38 in WM-266-4 and IGR37 cells; (F) Quantification results for RAB38 from LC-MRM and Western blot analyses. The values represent the mean and standard deviation of results obtained from three independent experiments.
Our LC-MRM quantification data showed that 14, 20 and 12 small GTPases were differentially expressed by at least two-fold in the metastatic (i.e. WM-266-4, IGR-37, and 1205Lu) compared to the corresponding paired primary (i.e. WM-115, IGR-39, and WM793) melanoma cells (Table S3B). Among other differentially expressed small GTPases, RAB38 was expressed at much lower levels in two out of the three primary melanoma cell lines (WM-115 and IGR39) than the corresponding metastatic lines (WM-266-4 and IGR37), as determined from LC-MRM analyses and confirmed by Western blot analyses (Fig. 2D−2F). RAB38 was, however, not detectable in the WM793 or 1205Lu cells by LC-MRM or Western blot analyses (Fig. 2E, 2F). We also validated, by using Western blot analyses, the LC-MRM quantification results for several other small GTPases, including RAB12, RAB31, and RAB32 (Fig. S3A−S3H, Fig. S4A−S4H). The highly consistent results obtained from LC-MRM and Western blot analyses underscored the robustness of the LC-MRM method in assessing quantitatively the differential expression of small GTPase proteins.
Potential Roles of RAB38 in Melanoma Progression
We next asked whether RAB38 expression level modulates prognosis in melanoma patients. We performed Kaplan-Meier survival analysis in melanoma patient cohort in the Cancer Genome Atlas (TCGA) database, and the results showed that poorer patient survival was significantly correlated with higher levels of mRNA expression of RAB38 gene (hazard ratio, HR = 1.323; 95% confidence interval, 95% CI = 1.009−1.736; Logrank p = 0.0402) (Fig. 3A). Furthermore, pan-cancer analysis of TCGA data using cBio Cancer Genomics Portal (cBioPortal: http://www.cbioportal.org/) revealed that the mRNA expression levels of RAB38 gene were highly up-regulated in two types of melanoma (skin cutaneous melanoma, SKCM; uveal melanoma, UVM) compared to other types of cancers (Fig. 3B) (23).
Fig. 3. Bioinformatic analyses revealed RAB38 as a potential driver for melanoma progression. (A).
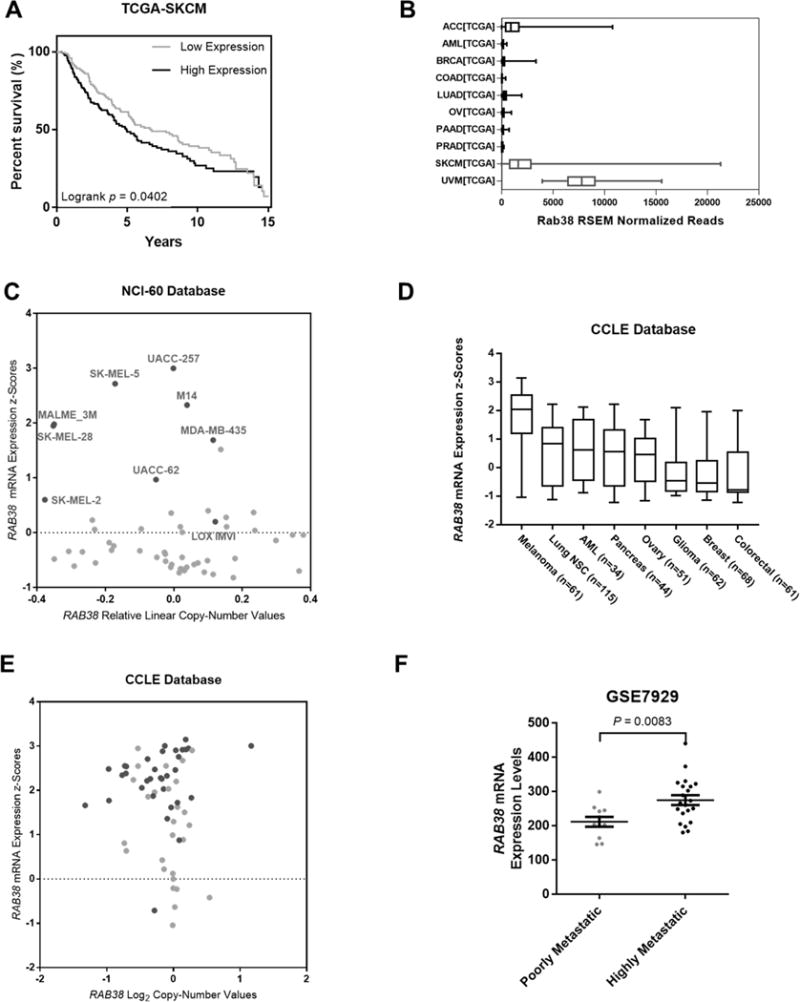
Kaplan-Meier plot of overall patient survival stratified by median RAB38 mRNA expression in the skin cutaneous melanoma (SKCM) cohort in The Cancer Genome Atlas (TCGA) database. Log-rank test p-value is displayed; (B) Box plot showing enriched RAB38 mRNA expressions in SKCM and uveal melanoma (UVM) in TCGA database; (C) Scatter plot showing up-regulated RAB38 mRNA expression in various metastatic melanoma cell lines (highlighted as black dots) in the NCI-60 Human Tumor Cell Lines Database; (D) Scatter plot showing up-regulated RAB38 mRNA expressions in melanoma cell lines in the Cancer Cell Line Encyclopedia (CCLE) Database; (E) Scattered plot showing up-regulated RAB38 mRNA expressions in various metastatic melanoma cell lines (highlighted as dark grey dots) in the CCLE Database; (F) RAB38 mRNA levels were significantly up-regulated in the highly metastatic derivatives of A375 cells cell lines compared to the poorly metastatic A375 parental cells (GEO data series: GSE7929). The p values were calculated by using an unpaired two-tailed Student’s t test.
We also queried public databases for the expression levels of RAB38 gene in other melanoma cell lines. First, analysis of the NCI-60 Human Tumor Cell Lines Database (https://dtp.cancer.gov/discovery_development/nci-60/) revealed the marked up-regulation of RAB38 gene in various metastatic melanoma cell lines (Fig. 3C). Likewise, the mRNA expression levels of RAB38 gene were up-regulated in the majority of 61 metastatic melanoma cell lines in the Cancer Cell Line Encyclopedia (CCLE) Database (http://www.broadinstitute.org/ccle/home) (Fig. 3D, 3E). Moreover, we utilized publicly accessible transcriptomic profiles in the Gene Expression Omnibus (GEO) database to analyze the mRNA expression of small GTPases in melanoma cells. In particular, we assessed previously published data about the differential gene expression between the highly metastatic human melanoma cell lines derived from an animal metastasis model and the poorly metastatic parental lines (accession number: GSE7929) (24). We found that RAB38 mRNA levels were again significantly up-regulated in the highly metastatic melanoma cells relative to the poorly metastatic counterparts (Fig. 3F). Taken together, the above results suggested RAB38 as a potential driver for melanoma metastasis.
RAB38 Promotes Invasion of Melanoma Cells through Up-regulation of Matrix Metalloproteinases (MMPs)
We next investigated, by employing transwell migration and invasion assay, whether the invasive phenotypes of melanoma cells could be modulated by the expression levels of RAB38 gene. Our results showed that ectopic overexpression of RAB38 protein in the WM-115 primary melanoma cells to a similar level as that in the metastatic WM-266-4 cells (Fig. S5A, S5B) resulted in a significant increase in the number of invaded cells (Fig. 4A, 4B). Reciprocal experiment with the metastatic WM-266-4 cells showed that the siRNA-mediated knockdown of RAB38 led to a significant decline in cell invasion, which is accompanied with a slight diminution of cell migration (Fig. 4C, 4D). In this vein, the knockdown efficiency of RAB38 gene by siRNA was confirmed by both real-time quantitative PCR (RT-qPCR) and immunoblot analysis (Fig. S5C, S5D). Collectively, we demonstrated that RAB38 promotes melanoma invasion in vitro.
Fig. 4. RAB38 enhanced melanoma metastasis in vitro. (A).
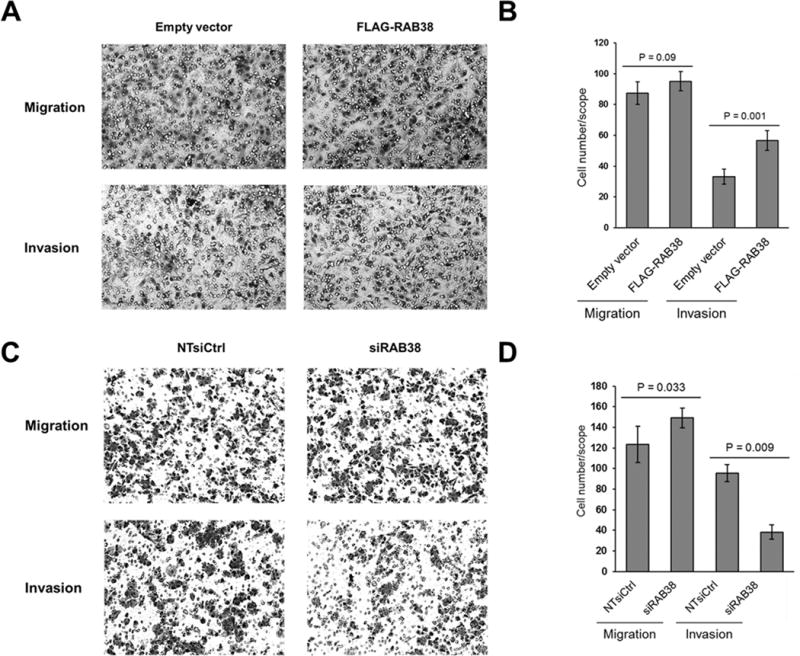
In vitro migration and invasion assays showed similar migration rates but significantly increased invasion rates in FLAG-RAB38-expressing WM-115 cells as compared to empty vector control. Migration and invasion capabilities were measured by using transwell migration and Matrigel-based invasion assays, respectively; (B) Quantification results for in vitro migration and invasion assay shown in panel (A). (C) In vitro migration and invasion assays showed similar migration rates but significantly decreased invasion rates for WM-266-4 cells with siRAB38 knockdown as compared to non-targeting siRNA control; (D) Quantification results for in vitro migration and invasion assay shown in panel (C). Error bars represent mean ± standard deviation (n = 3). The p values were calculated by using an unpaired, two-tailed Student’s t test.
We also examined the roles of matrix metalloproteinases (MMPs) in RAB38-mediated alterations in invasiveness of melanoma cells. Degradation of extracellular matrix (ECM) proteins by MMPs, a family of zinc- and calcium-dependent proteolytic enzymes, constitutes a crucial initiating step in tumor invasion. Among the 23 members of the human MMP family, MMP2 (gelatinase A) and MMP9 (gelatinase B) are responsible for remodeling the ECM environment and facilitating cancer metastasis (25). Hence, we explored how the expression levels of RAB38 alter the mRNA expression and enzymatic activities of MMP2 and MMP9.
Consistent with the RT-qPCR results showing the diminished mRNA expression of MMP2 and MMP9 genes (Fig. 5A), gelatin zymography assay showed that the RNAi knockdown of RAB38 led to markedly diminished activities of both the pro-enzyme and active forms of MMP2 and MMP9 in the metastatic WM-266-4 cells (Fig. 5B, 5C). This result supports the role of RAB38 in modulating the mRNA expression and activities of MMP2 and MMP9, thereby altering the invasive potential of melanoma cells. In a reciprocal experiment, overexpression of RAB38 induced slight, yet significant increases in the enzymatic activities of MMP2 and MMP9 in WM-115 primary melanoma cells, which were in accordance with the heightened mRNA expression of these two genes as revealed by RT-qPCR analysis (Fig. 5D−5F). Together, these results demonstrated that RAB38 regulates the expression levels and activities of MMP2 and MMP9 in melanoma cells.
Fig. 5. RAB38 regulates melanoma metastasis by mediating the expression levels and activities of MMP2 and MMP9. (A).
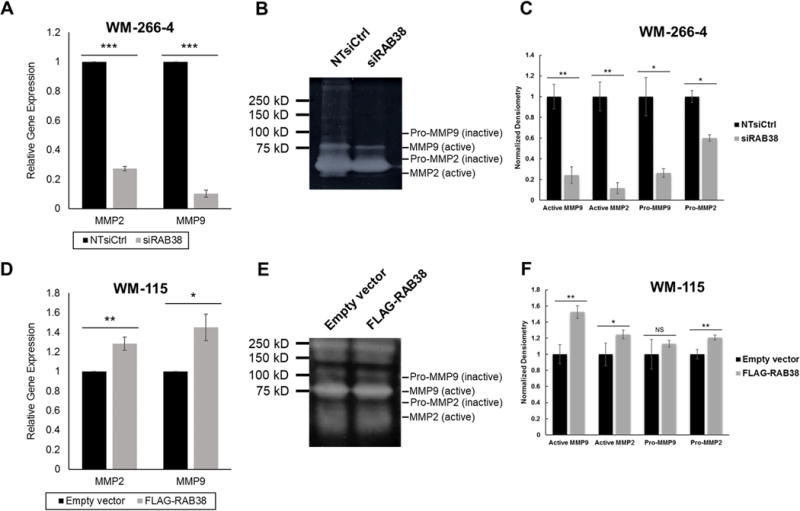
RT-qPCR assays showed decreased expression levels of MMP2 and MMP9 in WM-266-4 cells with siRAB38 knockdown as compared to non-targeting siRNA control. Error bars represent mean ± standard error of the mean (SEM) (n = 3); (B) Gelatin zymography assays revealed diminished enzymatic activities of MMP2 and MMP9 in WM-266-4 cells with siRAB38 knockdown as compared to non-targeting siRNA control; (C) Quantification results for gelatin zymography assays shown in (B). Error bars represent mean ± standard deviation (n = 3). (D) Real-time quantitative PCR (RT-qPCR) assays showed increased expression levels of MMP2 and MMP9 in FLAG-RAB38-expressing WM-115 cells as compared to empty vector control. Error bars represent mean ± SEM (n = 3); (E) Gelatin zymography assays revealed elevated enzymatic activities of MMP2 and MMP9 in FLAG-RAB38-expressing WM-115 cells as compared to empty vector control; (F) Quantification results for gelatin zymography assays shown in (E). Error bars represent mean ± standard deviation (n = 3). The p values for all figures are as follows: “*”, 0.01 < p < 0.05; “**”, 0.001 < p < 0.01; “***”, p < 0.001. The p values were calculated by using a paired, two-tailed Student’s t test.
Having revealed the regulatory roles of RAB38 in the secretion of MMPs in WM-115 and WM-266-4 melanoma cell lines, we extended our studies to IGR37 and M14 metastatic melanoma cells. In this context, M14 cells were also chosen for the study because these cells displayed pronounced expression of RAB38 (Fig. S6A). Consistent with our hypothesis, RT-qPCR experiments revealed down-regulations of MMP2 and MMP9 after RNAi knockdown of RAB38 in M14 cells (Fig. S6B). In addition, in IGR37 cells, we only observed diminished mRNA levels of MMP9, but not MMP2, after RNAi knockdown of RAB38 (Fig. S6C). Similar as what we observed for WM-266-4 cells, gelatin zymography assay results showed that RAB38 knockdown led to significantly decreased activities of MMP2 and MMP9 in M14 and IGR37 cells (Fig. S6D, S6E), lending further evidence to support that RAB38 regulates MMP2 and MMP9 activities in a range of metastatic melanoma cell lines. The observation of a decreased level of secreted MMP2 protein from IGR37 cells, but not the mRNA expression of the MMP2 gene in these cells, upon genetic depletion of RAB38 suggests that RAB38 modulates the level of secreted MMP2 through a post-transcriptional mechanism.
Epigenetic Reactivation of RAB38 in Metastatic Melanoma Cells
We next examined the mechanisms through which RAB38 gene was overexpressed in metastatic over primary melanoma cells. We first asked whether elevated RAB38 expression is accompanied with previously reported genetic alterations in melanoma, including mutations in BRAF, NRAS and TP53 genes. It turned out that, in the TCGA SKCM patient cohort, the expression levels of RAB38 gene did not exhibit any significant correlations with frequently observed mutations in BRAF, NRAS, or TP53 gene (Fig. S7A−S7E).
Numerous studies have underscored the significant roles of epigenetic and transcriptional regulations of oncogenes during cancer progression (26); hence, we next assessed whether these mechanisms contribute to elevated expression of RAB38 gene in metastatic melanoma cells. Microphthalmia-associated transcription factor (MITF) is the master regulator of melanocyte development, function, and survival through modulating many genes involved in differentiation and cell cycle progression (27). In this vein, earlier ChIP-Seq experiments conducted in 501Mel human melanoma cells identified MITF loci immediately upstream of the promoters of several Rab GTPase genes, including, among others, RAB27A and RAB38 (28). Therefore, we next asked whether elevated expression of RAB38 in the metastatic WM-266-4 and IGR37 cells are due to heightened transcriptional regulation mediated by MITF. We indeed observed the up-regulation of MITF at both the mRNA and protein levels in the two metastatic lines of melanoma cells (i.e. WM-266-4 and IGR37) relative to the corresponding primary melanoma cells (i.e. WM-115 and IGR39. Fig. S8A, S8B), though the mRNA expression of MITF was not detectable in 1205Lu cells.
To explore further the possible functional linkage between RAB38 expression and MITF regulation, we analyzed publicly available data for mouse models of experimental melanoma metastasis and cell lines. In this vein, both MITF and RAB38 were highly up-regulated at the mRNA levels in the highly metastatic derivatives of A375 human melanoma cells in comparison with the poorly metastatic parental lines (Fig. 3F and Fig. S8C). In addition, interrogation of the gene expression data of 120 melanoma cell lines (120Mel) and the Cancer Cell Line Encyclopedia (CCLE) Database (http://www.broadinstitute.org/ccle/home) revealed that the mRNA expression levels of RAB38 and MITF were positively correlated (Fig. S9A−S9D).
We also extended the bioinformatic analyses by examining RAB38/MITF expressions in different patient cohorts including the TCGA-SCKM cohort and two other patient cohorts (GSE7553 and GSE8401) retrieved from the GEO database. We again observed a clear positive correlation between MITF and RAB38 in both primary and metastatic melanoma tissues (Fig. 6A–6D, S9E−S9H). Notably, RAB38 mRNA expression was significantly up-regulated in the metastatic melanoma tissues relative to primary melanoma tissues from patients displaying MITF-high signature, namely for the patient population stratified with higher levels of MITF expression (Fig. 6E, 6F); however, an opposite trend was observed for patients exhibiting MITF-low signature (Fig. 6G, 6H), suggesting that the upregulation of RAB38 in metastatic melanoma is likely driven by MITF. Moreover, pathway analysis of the RAB38 gene co-expression signature in the TCGA-SKCM data showed that MITF is highly enriched and functionally involved with RAB38 (Fig. S10A).
Fig. 6. RAB38 expressions in large melanoma cell line or patient cohorts were highly correlated with the melanoma lineage-specific transcription factor MITF.
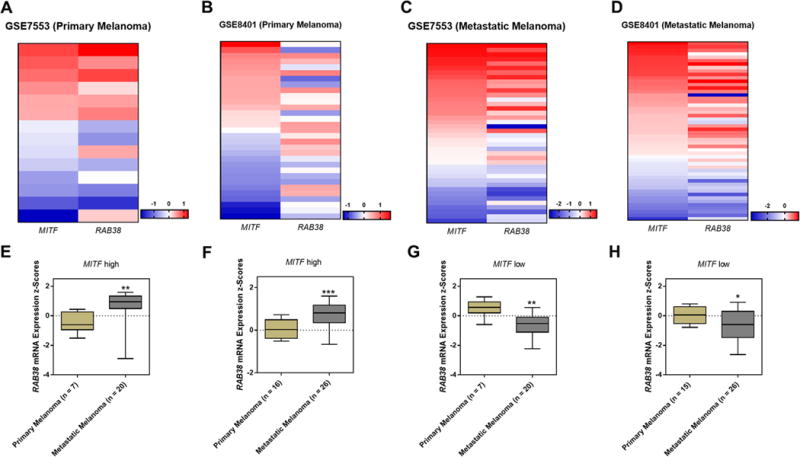
(A−D) Heatmaps showing the correlations between RAB38 and MITF mRNA expressions in the: (A) Primary melanoma tissues in the GSE7553 cohort; (B) Metastatic melanoma tissues in the GSE7553 cohort; (C) Primary melanoma tissues in the GSE8401 cohort; (D) Metastatic melanoma tissues in the GSE8401 cohort;
(E−H) Box plots showing the RAB38 mRNA expressions in the: (E) Metastatic patient tissues carrying MITF-high signature in the GSE7553 cohort; (F) Metastatic patient tissues carrying MITF-low signature in the GSE7553 cohort; (G) Metastatic patient tissues carrying MITF-high signature in the GSE8401 cohort; (H) Metastatic patient tissues carrying MITF-high signature in the GSE8401 cohort.
We also extended the analyses of the TCGA data to two known driver genes for melanoma metastasis, i.e. RAB27A and TBC1D16, both of which are enriched in the RAB38 gene co-expression signature and regulated by MITF (Fig. S10A). Similar to RAB38, in two additional patient cohorts (GSE7553 and GSE8401), we observed significantly higher levels of expression of RAB27A and TBC1D16 in the metastatic melanoma tissue samples carrying MITF-high signature (i.e. with high levels of MITF expression), but not in those with MITF-low signature (Fig. S10B−S10E).
To further substantiate the direct regulation of RAB38 by MITF, we performed chromatin immunoprecipitation (ChIP) followed by quantitative PCR (ChIP-qPCR) analysis to assess the occupancy of MITF protein in the promoter regions of RAB38 and TBC1D16 genes. In this respect, the 47-kDa isoform of TBC1D16 was observed to be regulated by MITF through binding to its remote promoter region (29,30). Indeed, our ChIP-qPCR results revealed higher levels of enrichment of MITF to the promoter elements of both RAB38 and TBC1D16 genes in WM-266-4 cells relative to WM-115 cells, suggesting that RAB38 is directly regulated by MITF in the metastatic WM-266-4 melanoma cells (Fig. S10F).
Having assessed the MITF-mediated transcriptional regulation of RAB38, we next asked whether the expression of RAB38 in these melanoma cells are epigenetically modulated. Analyses of the previously published methylation microarray data for IGR39/IGR37 (accession number: GSE46522) and WM-115/WM-266-4 (accession number: GSE70621) cells showed diminished levels of cytosine methylation at several CpG sites in the promoter region of RAB38 gene in the metastatic over primary melanoma cells (Fig. S11A, S11B). These results suggest that promoter hypomethylation and the ensuing epigenetic reactivation may elicit increased levels of RAB38 expression in the two metastatic lines (i.e. IGR37 and WM-266-4). To further substantiate this finding, we assessed the methylation status at 10 CpG sites in the promoter region of RAB38 gene in the three pairs of primary/metastatic melanoma cell lines by employing bisulfite sequencing. Strikingly, our results revealed that these 10 CpG sites were entirely unmethylated (0.0% methylation) in WM-266-4 and IGR37 cells, whereas the overall methylation levels at these sites were 98.0% and 41.0% in WM-115 and IGR39 cells, respectively (Fig. 7A). In contrast, these CpG sites are hypermethylated in both the WM793 (99.0%) and its matched metastatic melanoma line (i.e. 1205Lu, 93.0%) (Fig. 7A). These results support that epigenetic reactivation contributes to elevated expression of RAB38 in the metastatic lines of the WM and IGR pairs of melanoma cells, whereas epigenetic silencing led to lack of detectable levels of RAB38 protein in the primary or metastatic melanoma lines of the WMLu pair. To further validate that RAB38 expression is regulated by CpG methylation, we treated WM-115 and IGR39 cells with a DNA demethylating reagent, 5-aza-2´-deoxycytidine (5-Aza), for 96 h, and assessed the mRNA levels of RAB38 by RT-qPCR. Indeed our results showed that expression level of RAB38 was significantly increased upon 5-Aza treatment (Fig. 7B, 7C).
Fig. 7. Investigation of RAB38 methylation status in melanoma cell lines.
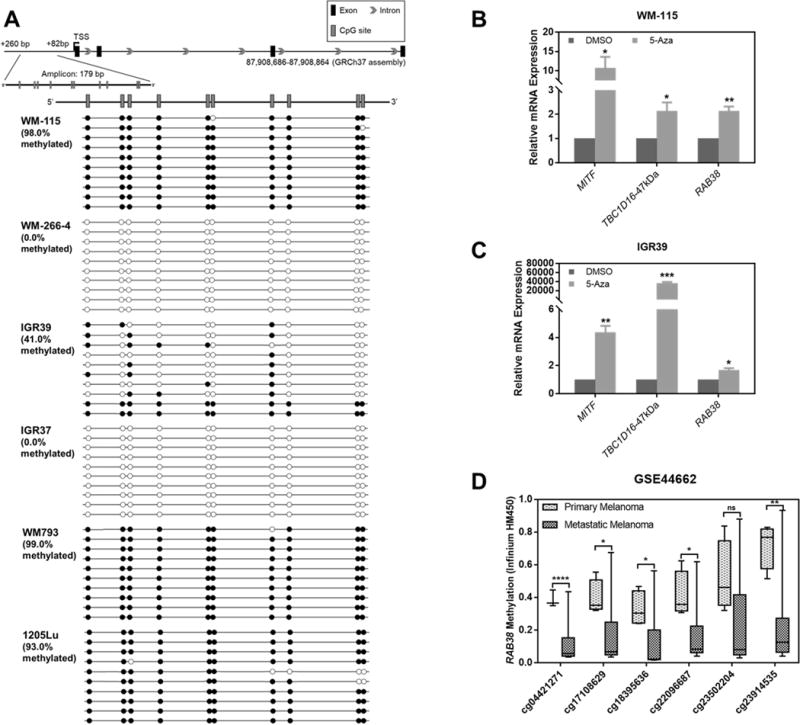
(A) Bisulfite sequencing demonstrated the methylation status of CpG sites in the promoter region of RAB38 gene in the three paired primary/metastatic cell lines, where high levels of methylation were observed for the WM-115, IGR39, WM793 and 1205Lu, but not for the WM-266-4 and IGR37 melanoma cell lines. CpG sites in the promoter region of RAB38 gene are indicated by short vertical bars, and exons are designated with black rectangles on the top. The arrow indicates the transcription start site (TSS). Each horizontal line represents one separate clone that was sequenced, and open and filled circles represent unmethylated and methylated CpG sites, respectively; (B) Increased mRNA expression levels of MITF, TBC1D16-47kDa and RAB38 after 5-aza-2′-deoxycytidine (5-Aza) treatment (96 h) in WM-115 cells; (C) Increased mRNA expression levels of MITF, TBC1D16-47kDa and RAB38 after 5-Aza treatment (96 h) in IGR39 cells; (D) Box plots representing DNA methylation in 4 primary melanoma and 33 metastatic melanoma samples (accession number: GSE44662). Metastatic melanomas contained lower RAB38 promoter methylation. The error bars in panels (B) and (C) represent mean ± SEM. The p values were calculated by using an unpaired two-tailed Student’s t test: “ns”, not significant; “*”, 0.01 < p < 0.05; “**”, 0.001 < p < 0.01; “***”, 0.0001 < p < 0.001; “****”, p < 0.0001.
We also assessed whether RAB38 hypomethylation occurs in metastatic melanoma patients of a previously reported melanoma cohort (accession number: GSE44662). It turned out that the promoter methylation of RAB38 gene was significantly lower in metastatic than primary melanoma tissues (Fig. 7D). In addition, analysis of the TCGA-SKCM cohort revealed that the mRNA expressions of MITF and RAB38 genes were inversely correlated with their promoter methylation levels, and promoter hypomethylation of the RAB38 gene is correlated with poor patient survival (HR, 0.6569; 95% Cl, 0.5017−0.8601; Logrank p = 0.0023) (Fig. S11C−S11E).
Together, our above results furnished evidence to support a model where loss of cytosine methylation in the promoter region of RAB38 gene leads to its epigenetic reactivation, which involves augmented binding of MITF transcription factor to the promoter region.
Discussion
Small GTPases of the Ras superfamily are master regulators of cellular trafficking. Here, we developed a novel targeted quantitative proteomic method for human small GTPase proteome with an unprecedented level of coverage. Our MRM-based targeted proteomic method enabled a powerful and high-throughput discovery of small GTPases that become aberrantly expressed during metastatic transformation of melanoma.
Our quantitative proteomic data, along with the results obtained from cell-based assays and from bioinformatic analyses of publicly available data, support the role of RAB38 in promoting melanoma metastasis. Thus, RAB38 joins other members of the small GTPase family that regulate melanoma metastasis, including RAB27A (5), RND3 (31), and ARF6 (32).
RAB38 displays a unique tissue-specific expression pattern, with the highest levels being observed in the lung and skin (33). Together with RAB27A and RAB32, RAB38 has a well-established function in regulating the melanosome biogenesis and maturation (34). It was also found to be important for pigmentation in chocolate mice by regulating the trafficking of tyrosinase-related protein 1 (TYRP1) (35). Furthermore, previous studies unveiled the role of RAB38 in mesenchymal subtypes and malignant progression of glioma, where elevated expression of RAB38 confers poor prognosis in glioma patients (36). However, no reports have yet elucidated the mechanistic relationship between RAB38 and the invasive properties of any type of tumor. Here, we unveiled a previously unrecognized role of RAB38 in regulating melanoma metastasis. Furthermore, our results support that RAB38 promotes melanoma progression by regulating the secretion and activities of MMP2 and MMP9, which are essential for metastatic transformation of tumor cells.
To the best our knowledge, this is the first report to link RAB38 with matrix metalloproteinase pathways. Several small GTPases were previously reported to be involved in the regulation of the MMP pathways through their roles in trafficking. For instance, RAB37 was previously identified as a metastasis suppressor in lung adenocarcinoma by influencing the metallopeptidase inhibitor 1 (TIMP1)-MMP9 pathway (37). In particular, RAB37 was found to suppress metastasis through regulating the exocytotic trafficking of TIMP1, thereby inactivating MMP9 signaling and suppressing invasion. Moreover, RAB2A and RAB27B were shown to promote breast cancer invasion by stimulating endocytic trafficking of membrane type 1 (MT1)-MMP and MMP2, respectively (38,39). Thus, we reason that RAB38 may play a novel role in the endocytic or exocytotic trafficking of MMP enzymes and/or their regulators, and future studies are warranted for illustrating the exact mechanisms through which RAB38 regulates MMPs.
We also explored the potential upstream mechanisms of RAB38 regulation. In this connection, our results revealed a strong correlation between the mRNA expressions of RAB38 and MITF. Furthermore, we observed a complete loss of cytosine methylation, which is accompanied with elevated enrichment of MITF transcription factor, in the promoter of RAB38 gene in WM-266-4 cells relative to WM-115 cells, supporting that epigenetic reactivation contributes to the elevated expression of RAB38 gene in metastatic melanoma cells. The complete loss of promoter methylation was also observed for the metastatic IGR37 cells, but not for the metastatic 1205Lu cells or the paired WM793 primary melanoma cells. These findings are consistent with the relative levels of RAB38 proteins in the three paired melanoma cell lines. In this context, it is worth noting that Mueller et al. (40) observed, from Western blot analysis, higher levels of RAB38 in 2 human melanocyte samples than 3 primary and 3 metastatic melanoma tissues, though these cells and tissues were not derived from the same patients. These observations are in line with the notion that melanoma is a highly heterogeneous type of cancer (41). Further studies are therefore needed to reveal the mechanisms underlying the metastatic transformation for the WM793/1205Lu paired cell lines. Nevertheless, the interrogation of TCGA and other patient cohort data uncovered a significant correlation between the elevated mRNA expression level of RAB38 gene, or its promoter hypomethylation, and poor prognosis in melanoma patients. Moreover, a strong correlation between the expression levels of MITF and RAB38 genes was observed in a large number of melanoma cell lines and tumor tissues. Thus, this epigenetic and transcriptional mechanism might be at play for a substantial subset of melanoma patients.
Apart from RAB38, our targeted proteomic approach led to the discovery of other small GTPases that may function in melanoma metastasis. For instance, the consistent down-regulation of RAB12 in the WM-266-4 and 1205Lu metastatic melanoma cells relative to their primary melanoma counterparts suggests that this protein may serve as a suppressor for melanoma metastasis. RAB12 was found to regulate the constitutive degradation of transferrin receptor (42), and elevated levels of transferrin receptors were previously observed in melanoma cells metastasized to brain (43). Thus, RAB12 may suppress melanoma metastasis through elevated accumulation of transferrin receptors. Furthermore, we found that RAB31 was consistently down-regulated in all three metastatic melanoma lines relative to the corresponding primary lines. Grismayer et al. (44) demonstrated that the increased levels of RAB31 led to a switch of invasive to proliferative phenotype in breast cancer cells. It will be important to explore, in the future, the role of RAB31 in the metastatic transformation of other types of cancer, including melanoma.
Small GTPases, like other types of GTP-binding proteins, can shuffle between the GTP-bound active states and the GDP-bound inactive states, which are regulated by GEFs, GAPs and GDIs (2). The conformational alterations of small GTPases between these two states can modulate their binding towards different downstream effector proteins (2). In this vein, a limitation of our targeted proteomic approach is its inability in profiling the activities of small GTPases. This limitation can be overcome by further multiplexing the assay with affinity-based techniques. As discussed above, by combining affinity enrichment with gel-based fractionation, Zhang et al. (14) developed an MRM-based assay to profile the activities of 12 small GTPases; however, the throughput of this assay was relatively low. In addition, proteome-wide enrichment of active small GTPases using binding domains of their effector proteins is very challenging due to the tremendous structural diversity of effectors and the lack of knowledge about the effectors for some small GTPases. On the other hand, enrichment of small GTPases and other GTP-binding proteins with the use of acyl nucleotide affinity probes (45), together with LC-MRM analysis, may constitute an alternative approach for high-throughput profiling of activities of small GTPases. Such an approach is currently being explored in our laboratory.
In conclusion, we developed successfully a novel MRM-based targeted quantitative proteomic method for the comprehensive profiling of small GTPases. By using this method, we assessed the differential expression of small GTPases in paired primary/metastatic melanoma cell lines. The method, when combined with bioinformatic analysis of publicly available data and cell-based assays, constitutes an integrated and effective approach to discover small GTPase that serve as drivers or suppressors for melanoma metastasis. We found that RAB38 promotes melanoma metastasis in vitro through the regulation of matrix metalloproteinases, and the increased expression of RAB38 in metastatic melanoma cells arises from diminished promoter methylation and heightened binding of the MITF transcription factor. It can be envisaged that the targeted proteomic method can also be employed for studying small GTPase signaling (e.g. for discovering small GTPase substrates for GEFs and GAPs) and for investigating the implications of small GTPases in other aspects of cancer biology or cancer therapy (e.g. in therapeutic resistance).
Our finding that epigenetic reactivation of RAB38 gene stimulates melanoma metastasis suggests that the expression level of RAB38, in conjunction with the expression level of MITF, may serve as a biomarker for the prognosis of melanoma patients. In addition, targeting epigenetic modulation of RAB38 and/or its interactions with other proteins may serve as the basis for the therapeutic interventions of metastatic melanoma. In the latter respect, small-molecule inhibitors were previously reported for suppressing the interactions between small GTPases and their effector or GEF proteins (46,47).
Supplementary Material
Significance Statement.
A novel quantitative proteomic method leads to the discovery of RAB38 as a new driver of metastasis in melanoma.
Acknowledgments
We thank Drs. Gerd P. Pfeifer (Van Andel Research Institute), Mathew R. Bloomfield and Peter H. Duesberg (University of California, Berkeley) for providing cell lines.
Financial Support: This work was supported by the National Institutes of Health (R01 CA210072 to Y. Wang), and M. Huang was supported in part by an NRSA Institutional Training Grant (T32 ES018827).
Footnotes
Conflict of Interest Statement: The authors declared that there was no conflict of interest associated with this study.
Data Availability. PDF files containing the processed data are provided in Supplementary Materials. The Skyline MRM library (including the transition list for quantification and the iRT file) for human small GTPase proteome and the raw files obtained from the LC-MRM analyses of small GTPases for the three paired primary/metastatic melanoma cell lines were deposited into PeptideAtlas with the identifier number of PASS01194 (http://www.peptideatlas.org/PASS/PASS01194).
References
- 1.Takai Y, Sasaki T, Matozaki T. Small GTP-binding proteins. Physiol Rev. 2001;81:153–208. doi: 10.1152/physrev.2001.81.1.153. [DOI] [PubMed] [Google Scholar]
- 2.Bos JL, Rehmann H, Wittinghofer A. GEFs and GAPs: critical elements in the control of small G proteins. Cell. 2007;129:865–77. doi: 10.1016/j.cell.2007.05.018. [DOI] [PubMed] [Google Scholar]
- 3.Stenmark H. Rab GTPases as coordinators of vesicle traffic. Nat Rev Mol Cell Bio. 2009;10:513–25. doi: 10.1038/nrm2728. [DOI] [PubMed] [Google Scholar]
- 4.Clark EA, Golub TR, Lander ES, Hynes RO. Genomic analysis of metastasis reveals an essential role for RhoC. Nature. 2000;406:532–5. doi: 10.1038/35020106. [DOI] [PubMed] [Google Scholar]
- 5.Peinado H, Aleckovic M, Lavotshkin S, Matei I, Costa-Silva B, Moreno-Bueno G, et al. Melanoma exosomes educate bone marrow progenitor cells toward a pro-metastatic phenotype through MET. Nat Med. 2012;18:883–91. doi: 10.1038/nm.2753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lange V, Picotti P, Domon B, Aebersold R. Selected reaction monitoring for quantitative proteomics: a tutorial. Mol Syst Biol. 2008;4 doi: 10.1038/msb.2008.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Picotti P, Aebersold R. Selected reaction monitoring-based proteomics: workflows, potential, pitfalls and future directions. Nat Methods. 2012;9:555–66. doi: 10.1038/nmeth.2015. [DOI] [PubMed] [Google Scholar]
- 8.Society AC. Cancer Facts and Figures-2018. Atlanta, GA: American Cancer Society; 2018. pp. 1–52. [Google Scholar]
- 9.Lo JA, Fisher DE. The melanoma revolution: from UV carcinogenesis to a new era in therapeutics. Science. 2014;346:945–9. doi: 10.1126/science.1253735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 11.Cerami E, Gao JJ, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, et al. The cBio Cancer Genomics Portal: An Open Platform for Exploring Multidimensional Cancer Genomics Data. Cancer Discovery. 2012;2:401–4. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Barretina J, Caponigro G, Stransky N, Venkatesan K, Margolin AA, Kim S, et al. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature. 2012;483:603–7. doi: 10.1038/nature11003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Halvey PJ, Ferrone CR, Liebler DC. GeLC-MRM quantitation of mutant KRAS oncoprotein in complex biological samples. J Proteome Res. 2012;11:3908–13. doi: 10.1021/pr300161j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang CC, Li R, Jiang H, Lin S, Rogalski JC, Liu K, et al. Development and application of a quantitative multiplexed small GTPase activity assay using targeted proteomics. J Proteome Res. 2015;14:967–76. doi: 10.1021/pr501010v. [DOI] [PubMed] [Google Scholar]
- 15.MacLean B, Tomazela DM, Shulman N, Chambers M, Finney GL, Frewen B, et al. Skyline: an open source document editor for creating and analyzing targeted proteomics experiments. Bioinformatics. 2010;26:966–8. doi: 10.1093/bioinformatics/btq054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liebler DC, Zimmerman LJ. Targeted Quantitation of Proteins by Mass Spectrometry. Biochemistry. 2013;52:3797–806. doi: 10.1021/bi400110b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Escher C, Reiter L, MacLean B, Ossola R, Herzog F, Chilton J, et al. Using iRT, a normalized retention time for more targeted measurement of peptides. Proteomics. 2012;12:1111–21. doi: 10.1002/pmic.201100463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Balaban G, Herlyn M, Guerry Dt, Bartolo R, Koprowski H, Clark WH, et al. Cytogenetics of human malignant melanoma and premalignant lesions. Cancer Genet Cytogenet. 1984;11:429–39. doi: 10.1016/0165-4608(84)90024-4. [DOI] [PubMed] [Google Scholar]
- 19.Aubert C, Rouge F, Galindo JR. Tumorigenicity of Human-Malignant Melanocytes in Nude-Mice in Relation to Their Differentiation Invitro. J Natl Cancer Inst. 1980;64:1029–40. [PubMed] [Google Scholar]
- 20.Herlyn D, Iliopoulos D, Jensen PJ, Parmiter A, Baird J, Hotta H, et al. In vitro properties of human melanoma cells metastatic in nude mice. Cancer Res. 1990;50:2296–302. [PubMed] [Google Scholar]
- 21.Ong SE, Blagoev B, Kratchmarova I, Kristensen DB, Steen H, Pandey A, et al. Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol Cell Proteomics. 2002;1:376–86. doi: 10.1074/mcp.m200025-mcp200. [DOI] [PubMed] [Google Scholar]
- 22.Peinado H, Aleckovic M, Lavotshkin S, Matei I, Costa-Silva B, Moreno-Bueno G, et al. Melanoma exosomes educate bone marrow progenitor cells toward a pro-metastatic phenotype through MET. Nature Medicine. 2012;18:883. doi: 10.1038/nm.2753. + [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cancer Genome Atlas Research N. Weinstein JN, Collisson EA, Mills GB, Shaw KR, Ozenberger BA, et al. The Cancer Genome Atlas Pan-Cancer analysis project. Nat Genet. 2013;45:1113–20. doi: 10.1038/ng.2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xu L, Shen SS, Hoshida Y, Subramanian A, Ross K, Brunet JP, et al. Gene expression changes in an animal melanoma model correlate with aggressiveness of human melanoma metastases. Mol Cancer Res. 2008;6:760–9. doi: 10.1158/1541-7786.MCR-07-0344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Roomi MW, Monterrey JC, Kalinovsky T, Rath M, Niedzwiecki A. Patterns of MMP-2 and MMP-9 expression in human cancer cell lines. Oncol Rep. 2009;21:1323–33. doi: 10.3892/or_00000358. [DOI] [PubMed] [Google Scholar]
- 26.Wouters J, Vizoso M, Martinez-Cardus A, Carmona FJ, Govaere O, Laguna T, et al. Comprehensive DNA methylation study identifies novel progression-related and prognostic markers for cutaneous melanoma. BMC Med. 2017;15:101. doi: 10.1186/s12916-017-0851-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Levy C, Khaled M, Fisher DE. MITF: master regulator of melanocyte development and melanoma oncogene. Trends Mol Med. 2006;12:406–14. doi: 10.1016/j.molmed.2006.07.008. [DOI] [PubMed] [Google Scholar]
- 28.Strub T, Giuliano S, Ye T, Bonet C, Keime C, Kobi D, et al. Essential role of microphthalmia transcription factor for DNA replication, mitosis and genomic stability in melanoma. Oncogene. 2011;30:2319–32. doi: 10.1038/onc.2010.612. [DOI] [PubMed] [Google Scholar]
- 29.Gade P, Kalvakolanu DV. Chromatin Immunoprecipitation Assay as a Tool for Analyzing Transcription Factor Activity. Methods Mol Biol. 2012;809:85–104. doi: 10.1007/978-1-61779-376-9_6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vizoso M, Ferreira HJ, Lopez-Serra P, Carmona FJ, Martinez-Cardus A, Girotti MR, et al. Epigenetic activation of a cryptic TBC1D16 transcript enhances melanoma progression by targeting EGFR. Nat Med. 2015;21:741–50. doi: 10.1038/nm.3863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Klein RM, Higgins PJ. A switch in RND3-RHOA signaling is critical for melanoma cell invasion following mutant-BRAF inhibition. Mol Cancer. 2011;10 doi: 10.1186/1476-4598-10-114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Grossmann AH, Yoo JH, Clancy J, Sorensen LK, Sedgwick A, Tong Z, et al. The small GTPase ARF6 stimulates beta-catenin transcriptional activity during WNT5A-mediated melanoma invasion and metastasis. Sci Signal. 2013;6:ra14. doi: 10.1126/scisignal.2003398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Osanai K, Oikawa R, Higuchi J, Kobayashi M, Tsuchihara K, Iguchi M, et al. A Mutation in Rab38 Small GTPase Causes Abnormal Lung Surfactant Homeostasis and Aberrant Alveolar Structure in Mice. Am J Pathol. 2008;173:1265–74. doi: 10.2353/ajpath.2008.080056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hume AN, Collinson LM, Rapak A, Gomes AQ, Hopkins CR, Seabra MC. Rab27a regulates the peripheral distribution of melanosomes in melanocytes. J Cell Biol. 2001;152:795–808. doi: 10.1083/jcb.152.4.795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Loftus SK, Larson DM, Baxter LL, Antonellis A, Chen Y, Wu X, et al. Mutation of melanosome protein RAB38 in chocolate mice. Proc Natl Acad Sci U S A. 2002;99:4471–6. doi: 10.1073/pnas.072087599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang HJ, Jiang CL. RAB38 confers a poor prognosis, associated with malignant progression and subtype preference in glioma. Oncol Rep. 2013;30:2350–6. doi: 10.3892/or.2013.2730. [DOI] [PubMed] [Google Scholar]
- 37.Tsai CH, Cheng HC, Wang YS, Lin P, Jen J, Kuo IY, et al. Small GTPase Rab37 targets tissue inhibitor of metalloproteinase 1 for exocytosis and thus suppresses tumour metastasis. Nat Commun. 2014;5:4804. doi: 10.1038/ncomms5804. [DOI] [PubMed] [Google Scholar]
- 38.Kajiho H, Kajiho Y, Frittoli E, Confalonieri S, Bertalot G, Viale G, et al. RAB2A controls MT1-MMP endocytic and E-cadherin polarized Golgi trafficking to promote invasive breast cancer programs. Embo Reports. 2016;17:1061–80. doi: 10.15252/embr.201642032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hendrix A, Maynard D, Pauwels P, Braems G, Denys H, Van den Broecke R, et al. Effect of the secretory small GTPase Rab27B on breast cancer growth, invasion, and metastasis. J Natl Cancer Inst. 2010;102:866–80. doi: 10.1093/jnci/djq153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mueller DW, Rehli M, Bosserhoff AK. miRNA expression profiling in melanocytes and melanoma cell lines reveals miRNAs associated with formation and progression of malignant melanoma. J Invest Dermatol. 2009;129:1740–51. doi: 10.1038/jid.2008.452. [DOI] [PubMed] [Google Scholar]
- 41.Merlino G, Herlyn M, Fisher DE, Bastian BC, Flaherty KT, Davies MA, et al. The state of melanoma: challenges and opportunities. Pigment Cell Melanoma Res. 2016;29:404–16. doi: 10.1111/pcmr.12475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Matsui T, Itoh T, Fukuda M. Small GTPase Rab12 regulates constitutive degradation of transferrin receptor. Traffic. 2011;12:1432–43. doi: 10.1111/j.1600-0854.2011.01240.x. [DOI] [PubMed] [Google Scholar]
- 43.Nicolson GL, Nakajima M, Herrmann JL, Menter DG, Cavanaugh PG, Park JS, et al. Malignant-Melanoma Metastasis to Brain - Role of Degradative Enzymes and Responses to Paracrine Growth-Factors. J Neurooncol. 1994;18:139–49. doi: 10.1007/BF01050420. [DOI] [PubMed] [Google Scholar]
- 44.Grismayer B, Solch S, Seubert B, Kirchner T, Schafer S, Baretton G, et al. Rab31 expression levels modulate tumor-relevant characteristics of breast cancer cells. Mol Cancer. 2012;11 doi: 10.1186/1476-4598-11-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Xiao Y, Ji D, Guo L, Wang Y. Comprehensive characterization of (S)GTP-binding proteins by orthogonal quantitative (S)GTP-affinity profiling and (S)GTP/GTP competition assays. Anal Chem. 2014;86:4550–8. doi: 10.1021/ac500588q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gao Y, Dickerson JB, Guo F, Zheng J, Zheng Y. Rational design and characterization of a Rac GTPase-specific small molecule inhibitor. Proc Natl Acad Sci USA. 2004;101:7618–23. doi: 10.1073/pnas.0307512101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ostrem JM, Peters U, Sos ML, Wells JA, Shokat KM. K-Ras(G12C) inhibitors allosterically control GTP affinity and effector interactions. Nature. 2013;503:548–51. doi: 10.1038/nature12796. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


