Abstract
Objectives: The authors discuss the usability of an automated tool that supports entry, by clinical experts, of the knowledge necessary for forming high-level concepts and patterns from raw time-oriented clinical data.
Design: Based on their previous work on the RESUMÉ system for forming high-level concepts from raw time-oriented clinical data, the authors designed a graphical knowledge acquisition (KA) tool that acquires the knowledge required by RÉSUMÉ. This tool was designed using Protégé, a general framework and set of tools for the construction of knowledge-based systems. The usability of the KA tool was evaluated by three expert physicians and three knowledge engineers in three domains—the monitoring of children's growth, the care of patients with diabetes, and protocol-based care in oncology and in experimental therapy for AIDS. The study evaluated the usability of the KA tool for the entry of previously elicited knowledge.
Measurements: The authors recorded the time required to understand the methodology and the KA tool and to enter the knowledge; they examined the subjects' qualitative comments; and they compared the output abstractions with benchmark abstractions computed from the same data and a version of the same knowledge entered manually by RÉSUMÉ experts.
Results: Understanding RÉSUMÉ required 6 to 20 hours (median, 15 to 20 hours); learning to use the KA tool required 2 to 6 hours (median, 3 to 4 hours). Entry times for physicians varied by domain—2 to 20 hours for growth monitoring (median, 3 hours), 6 and 12 hours for diabetes care, and 5 to 60 hours for protocol-based care (median, 10 hours). An increase in speed of up to 25 times (median, 3 times) was demonstrated for all participants when the KA process was repeated. On their first attempt at using the tool to enter the knowledge, the knowledge engineers recorded entry times similar to those of the expert physicians' second attempt at entering the same knowledge. In all cases RÉSUMÉ, using knowledge entered by means of the KA tool, generated abstractions that were almost identical to those generated using the same knowledge entered manually.
Conclusion: The authors demonstrate that the KA tool is usable and effective for expert physicians and knowledge engineers to enter clinical temporal-abstraction knowledge and that the resulting knowledge bases are as valid as those produced by manual entry.
Abstraction of Time-oriented Clinical Data
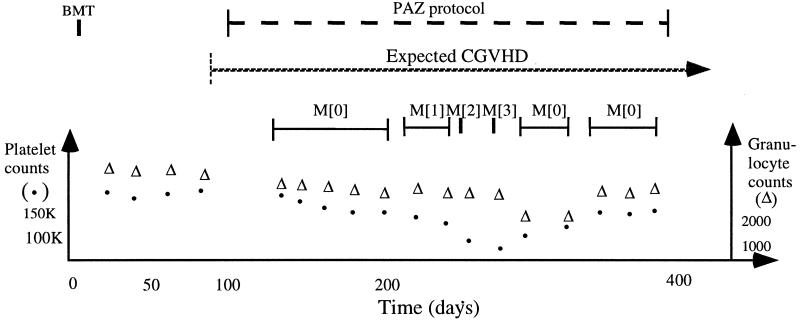
Caring for patients often requires integration and interpretation of a substantial amount of data over time and abstraction of those data into higher-level concepts and patterns that are meaningful for that domain. We refer to this common task as temporal abstraction.1 It is impossible to consider data trends, evolution of processes, or causality without an explicit representation of time, especially in clinical domains that involve care for patients who have chronic illness (e.g., cancer, AIDS, or diabetes), in particular when they require monitoring and therapy planning (▶).
Figure 1.
Temporal abstraction in a clinical domain. Raw patient data are plotted over time at the bottom. External events (interventions) and the abstractions computed from the data are plotted as intervals at the top. BMT indicates a bone-marrow-transplantation event (external intervention); PAZ, a prednizone/azathioprine therapy protocol for treating chronic graft-versus-host disease (CGVHD), a complication of BMT. Solid dots indicate platelet counts; triangles, granulocyte counts. The staggered interval bar (“PAZ protocol”) indicates an event interval; the hatched right-pointing arrow (“expected CGVHD”), a context interval; and the solid interval bars, abstraction intervals. M[n] indicates bone marrow toxicity (myelotoxicity) grande n.
As we show in the next section, performance of the temporal-abstraction task requires access to a considerable amount of highly specified, task-specific clinical knowledge. In this paper, we focus on the issue of acquiring this knowledge from expert physicians using an automated graphical tool. We discuss how we designed the tool and how we evaluated its usability for entry of the required knowledge by a group of expert physicians and knowledge engineers.
Summarization and interpretation of large volumes of raw time-stamped data into meaningful abstractions that hold over time periods is essential for effective clinical decision making. Examples include deciding, during therapy under a particular clinical guideline, whether a patient who had previous bone marrow transplantation has had more than two episodes of bone marrow toxicity of severity grade 1 or higher, each lasting at least one week. Notice that the value of a temporal abstraction, such as the severity classification of bone marrow toxicity, is often specific to the context of therapy under a particular clinical guideline (▶).
Thus, in ▶, the definitions of the bone marrow toxicity levels, such as grade 2, are specific to the particular context in which the hematologic data were acquired—namely, patients who have undergone bone marrow transplantation and who are on a prednizone/azathioprine (PAZ) protocol for prophylaxis or therapy for chronic graft-versus-host disease (GVHD). In this case, the level is a context-specific function of the platelet and granulocyte toxicity levels, which in turn are functions, respectively, of the platelet and granulocyte raw counts. Furthermore, as much as possible (using knowledge about the relevant hematology parameters), the abstractions are aggregated into intervals, such as periods of one or more weeks. Such a conceptual representation is at a level useful for residents and attending physicians who need succinct summaries of patient data for such purposes as support of a therapy decision. (An overview of our temporal-abstraction framework is presented in the next section.)
The ability to automatically create interval-based abstractions of large numbers of time-stamped clinical data in a context-sensitive manner has many useful applications, which include:
Automated summary of time-oriented electronic patient records
Support of recommendations by intelligent medical decision-support systems2
Monitoring the execution of therapy plans
Generation of context-specific abstractions and maintenance of several interpretations of the same clinical data within different meaningful contexts
Explanation of recommended actions by intelligent medical decision-support systems
Representing intentions of therapy plans, such as goals of clinical guidelines, as temporal patterns to achieve or avoid, thus supporting automated quality-assessment3
Support of intelligent visualization and exploration of time-oriented clinical data and their multiple-level abstractions, enabling interactive data mining in patient records4
Abstractions must be specific to the medical domain and context in which the data were acquired, because only then can they reduce considerably the information overload on the care provider. To reduce that overload further, succinct summaries must include not only data abstractions that hold over time points, such as “bone marrow toxicity of grade 2 at 11:00 A.M. on July 17, 1998,” but also conclusions that hold over time intervals, such as “six months of a pattern of quiescent GVHD, in the context of post-bone-marrow transplantation.”
Domain-specific abstraction of time-oriented clinical data requires acquisition and disciplined maintenance of complex types of knowledge about domain-specific time-oriented properties of the particular data. Examples of such domain-specific knowledge include functions that classify raw data or intermediate conclusions into more abstract concepts, specification of constraints that define meaningful temporal patterns, and knowledge of whether similar abstractions can be joined over time or should be considered separate episodes.
In this paper, we first summarize briefly our previous work on a computational method for forming high-level concepts from raw time-oriented clinical data (the knowledge-based temporal abstraction method) and on RÉSUMÉ, a software system that implements that method. Then, we present a graphical knowledge-acquisition (KA) tool that we have developed recently, which acquires the knowledge required by RÉSUMÉ. Finally, we describe an evaluation of the usability of the KA tool by expert physicians and knowledge engineers for entry of clinical temporal-abstraction knowledge in three clinical domains—the monitoring of children's growth, the care of patients with diabetes, and protocol-based care. In all three domains, we had benchmark knowledge bases (entered by a knowledge engineer using a text editor), data sets, and resultant abstraction from these data sets, based on our previous work. Those knowledge bases had been elicited and entered manually by knowledge engineers. We describe the quantitative and qualitative results of enabling expert physicians to enter the same knowledge by direct interaction with the new graphical KA tool, and analyze our results.
Background
Knowledge-based Temporal Abstraction
In this section, we describe briefly our previous work on the knowledge-based temporal-abstraction method, its implementation as a software system, and several applications using it. We emphasize the ontology (terms, properties, and relations) of the method and its knowledge requirements. These requirements influenced substantially the design of the KA tool that we describe in the next section. We present only a bare sketch of the computational framework, since it has been described elsewhere.1,5 We emphasize, however, the domain-specific knowledge that is required by our methodology and that needs to be acquired from expert physicians.
The temporal-abstraction task6 is an interpretation task. The input to that task is time-stamped clinical measurable data (e.g., hemoglobin values), external events (typically, clinical interventions such as administration of medications), and the user's goals (e.g., therapy for insulin-dependent diabetes, which can change the context for the abstraction process). The output of that task is a set of interval-based abstractions of the data (e.g., a two-week interval of level-2 bone marrow toxicity in the context of PAZ therapy). These abstractions interpret past and present states and trends that are relevant in the context of the care-provider's goals (see ▶).
The Knowledge-based Temporal-abstraction Problem-solving Method
We have developed a general problem-solving method7 for interpreting data in time-oriented domains, with clear semantics for both the problem-solving method and its domain-specific knowledge requirements—the knowledge-based temporal-abstraction method.1,6 This method comprises a knowledge-level8 representation of the temporal-abstraction task and the knowledge required to solve that task. The knowledge-based temporal-abstraction method has a formal model of input and output entities, their relations, and properties associated with these entities.1
The knowledge-based temporal-abstraction method decomposes the temporal-abstraction task into five parallel subtasks, which are solved respectively by five temporal-abstraction mechanisms (nondecomposable computational modules).
The temporal-abstraction mechanisms produce output abstractions of several abstraction types—state (e.g., MODERATE ANEMIA), gradient (e.g., INCREASING BLOOD PRESSURE), rate (e.g., FAST-CHANGING HEART RATE), and pattern (e.g., the linear pattern QUIESCENT-ONSET GRAFT-VERSUS-HOST DISEASE or the periodic pattern WEEKEND POST-LUNCH HYPERGLYCEMIA).
The temporal-abstraction mechanisms require four domain-specific knowledge types for any particular domain; these knowledge types need to be acquired from expert physicians in each domain—structural knowledge (e.g., ABSTRACTED-FROM [derived-from] relations); classification knowledge (e.g., definition of a parameter range as LOW, or a specification of a pattern); temporal-semantic knowledge (e.g., unlike two consecutive periods of anemia, two episodes of 9-month pregnancies cannot be summarized as an episode of an 18-month pregnancy, since they are not concatenable, a temporal-semantic property9); and temporal-dynamic knowledge (e.g., persistence of some characterization over time when data are unavailable, which can be represented by a function that specifies maximal gaps that can be bridged to connect two intervals into one larger one10). As we show when we discuss the KA tool that we designed for acquisition of temporal-abstraction knowledge, physicians' understanding of the role of each knowledge type is important for elicitation of that knowledge from these experts. The understanding can be general, however, and the experts do not need to know the computational details of the processes using the knowledge.
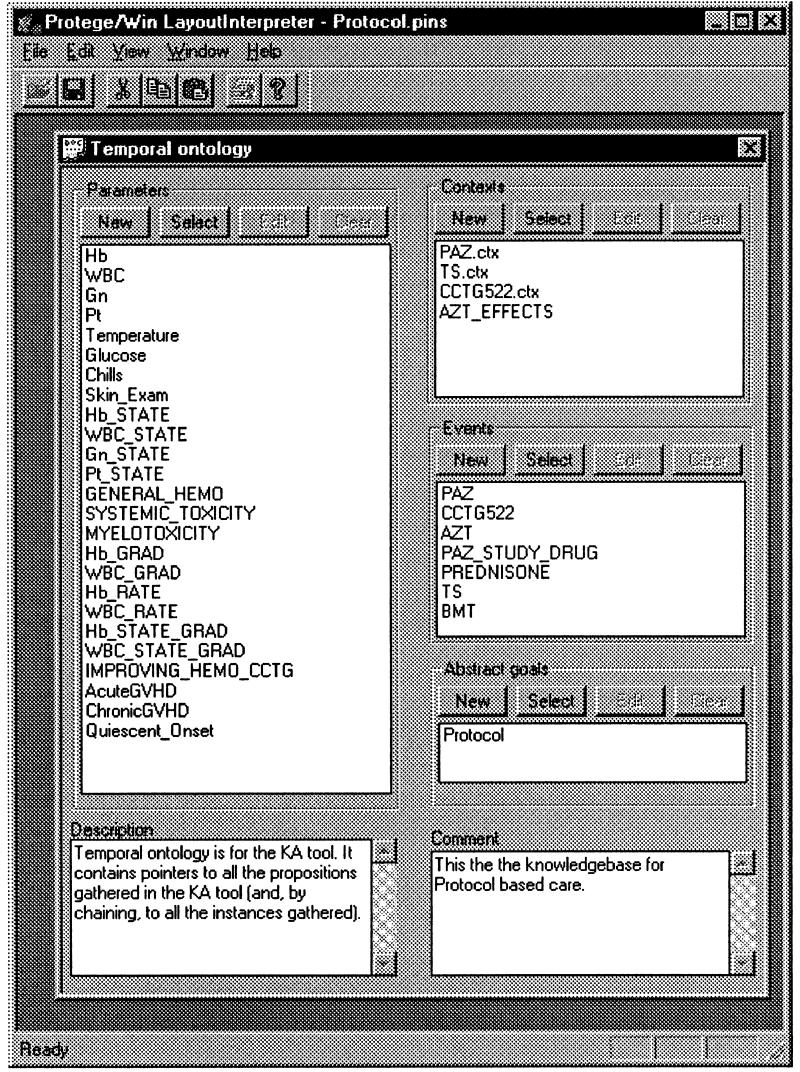
The knowledge needed to abstract time-oriented data in any particular clinical domain is represented as the temporal-abstraction ontology (a theory of terms and relations) of that domain. A temporal-abstraction ontology includes a parameter ontology, which is a theory of the relevant parameters and their temporal properties in the domain and the relations among these parameters (e.g., IS-A, ABSTRACTED-FROM); a pattern ontology, which defines all patterns and their relations to other patterns, parameters, and events (e.g., a linear pattern can have a COMPOSED-FROM relation to two abstractions; for practical purposes, it can be viewed as an ABSTRACTED-FROM relation); an event ontology, which includes external events (e.g., medications), their interrelations (e.g., PART-OF relations), and their properties; a context ontology, which includes interpretation contexts (e.g., the temporal context defined by the effect of a drug) and relations (e.g., SUBCONTEXT) among interpretation contexts11; an abstraction-goal ontology, which includes the user's potential goals for the abstraction process (these can induce contexts—e.g., monitoring of diabetes therapy) and their IS-A relations; and all relations between inducing propositions and induced contexts (e.g., INDUCED-CONTEXTS).
The RÉSUMÉ Knowledge-based Temporal-abstraction System
In our previous research, we implemented the knowledge-based temporal-abstraction method as the RÉSUMÉ system5,12 in the CLIPS language.13 RÉSUMÉ generates temporal abstractions, given time-stamped data and events and the domain's temporal-abstraction ontology.
The RÉSUMÉ system is not a rule-based system but rather is an implementation of a problem-solving method. The problem-solving method assumes during run time the existence of certain types of domain-specific clinical knowledge, which can be represented in various forms (e.g., classification rules, persistence functions, and temporal-semantic properties). Part of the implementation of the computational aspect of the original RÉSUMé system happened to use rules,12 but unlike those in rule-based expert systems, these rules were used simply as a high-level programming language construct, referred to the assumed domain-specific knowledge that parameterizes the rules, and were the same for all application domains. Indeed, other parts of the implementation incorporated object-oriented design and methods.
The internal organization of the knowledge assumed by the RÉSUMÉ system (i.e., the clinical domain's temporal-abstraction ontology) does not use rules either, but rather uses classes (e.g., events, parameters, laboratory parameters) and instances (e.g., properties of platelets in the context of PAZ therapy) of knowledge, represented in a frame-based language.5 Slots in the classes or instances typically correspond to various knowledge types and subtypes (e.g., temporal-semantic properties) and may include various types of representations, such as a classification table.
Applications of the RÉSUMÉ System
As part of our previous research, we tested the RÉSUMÉ system in several different clinical domains—protocol-based care (experimental therapy for patients who have AIDS and those who have chronic graft-versus-host disease, and prevention of AIDS-related complications),6,12 the monitoring of children's growth,6,14 therapy for patients who have insulin-dependent diabetes,5 support for application of clinical guidelines,2 and assessment of the quality of guideline-based care, when the intentions of the guideline designers are expressed as temporal abstractions to be achieved or avoided and the patient data are abstracted for comparison with the guideline's intentions.3
Methods
Design and Evaluation of the Knowledge-acquisition Tool
Our methodology in this study had three phases—design of a new KA tool to enter and maintain temporal-abstraction knowledge; evaluation of the usability of that tool for knowledge entry, which consisted of having a group of expert physicians and a group of knowledge engineers create, using the tool, knowledge bases in three clinical domains, when given a text-based declarative specification of the knowledge in an object-oriented language; and determination whether the abstractions created by the RÉSUMÉ system using the resulting knowledge bases and a benchmark set of data in each domain were valid, by comparison of these abstractions to the output of RÉSUMÉ for the same data set, using the original, text-based, manually entered knowledge.
Construction of the Knowledge-acquisition Tool
We developed a graphical KA tool to automate the process of entry of temporal-abstraction knowledge by expert physicians in multiple clinical domains. The goal of the tool is to minimize intervention by knowledge engineers, a well-known limiting factor in the development of knowledge-based systems, and to facilitate the maintenance of the knowledge base, once acquired, by the same or other domain experts (or, in some cases, by knowledge engineers). We constructed the KA tool using software tools from the Protégé project.15,16,17,18 As a software artifact, the KA tool is technically an automated tool. More accurately, it is a semi-automated tool in two senses—in its mode of generation and in its mode of use. First, it is a software tool that is generated automatically by the Protégé system and provides multiple automated support services for KA; however, custom tailoring of the final interface is performed manually by the developer of the KA tool, using one of the Protégé tools. Second, the resultant KA tool is an automated graphical tool, but it requires (unlike, for example, machine-learning techniques that learn only from data) interaction with a human domain expert (i.e., an expert physician) who enters the domain-specific knowledge using the dialog implicit in the structure of the KA tool.
The Protégé System
The Protégé framework15,16,17,18 is a workbench for constructing and using ontologies, for creating domain-specific knowledge-acquisition tools derived from ontologies, and for entering of domain knowledge into these knowledge-acquisition tools. The current system (Protégé/Win) runs in a Windows 95 or Windows NT environment and is in routine use at more than a dozen academic and commercial sites worldwide.
The Protégé approach assumes that domain ontologies (e.g., the ontology of the protocol-based-care domain) and the knowledge bases derived from them will serve as input to a problem-solving method. Problem-solving methods are reusable, because they encode stereotypic problem-solving strategies, such as classification, constraint satisfaction, or skeletal planning. The knowledge-based temporal-abstraction method is an example of a problem-solving method. We have applied the Protégé methodology to multiple domains, from automated support of guideline-based therapy to the determination of ribosomal structure.19
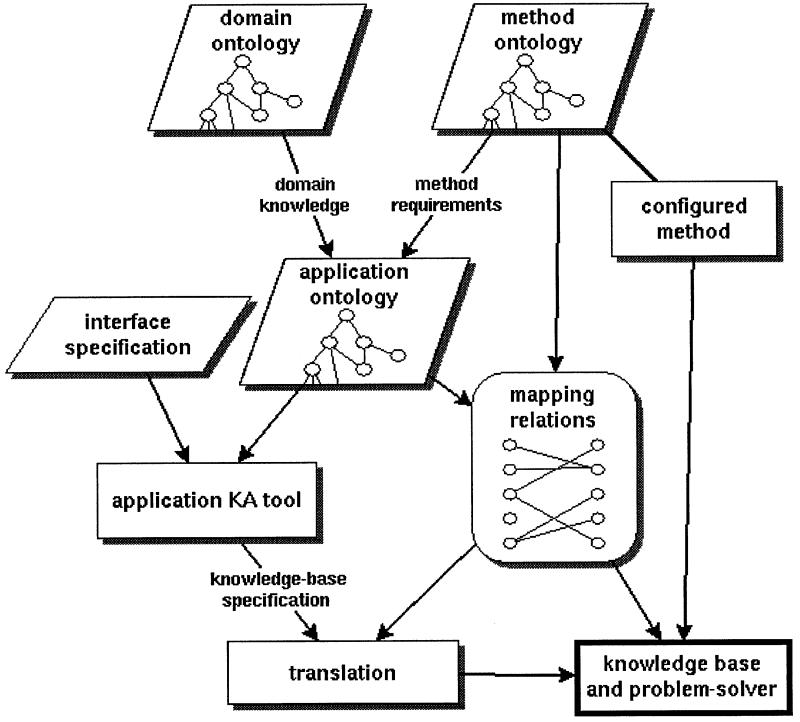
Thus, a major advantage of the Protégé approach is the production, given the relevant problem-solving method (e.g., the knowledge-based temporal-abstraction method) and domain ontology (e.g., protocol-based care), of automated knowledge-acquisition tools, tailored to the selected problem-solving method and domain. The tools allow knowledge-base authors to define instances of the classes in the ontology by various methods, such as by filling out graphical forms or by drawing pictures on the computer screen. ▶ summarizes the complete development process of an application within the Protégé framework.
Figure 2.
Application development in the Protégé framework. The developer selects a problem-solving method for the task. On the basis of the knowledge requirements of the method (the method's ontology) he or she transforms, or custom tailors, a domain ontology into an application ontology. The domain-specific terms and relations in the application ontology are mapped to those in the domain-independent method ontology. From the application ontology, Protégé generates a domain- and task-specific graphical knowledge-acquisition tool for entry of domain knowledge, which is translated into objects in the implementation system.
Design of the Knowledge-acquisition Tool with the Protégé System
The first phase in our experiment was the creation of the temporal-abstraction KA tool in the Protégé framework. In preparation for the knowledge-entry evaluation phase and the output evaluation phase, this phase was performed by the designers of the KA tool (Y.S. and H.C.) and involved two steps:
Creation and editing of the KA tool target (method) ontology, which is similar in this case to that of the knowledge-based temporal-abstraction method underlying the RÉSUMÉ system, but is different in several practical aspects, and
Custom tailoring of the layout of the default graphical KA tool, which is generated automatically by Protégé.
Once a layout specification exists, the layout-interpreter module of the Protégé system can display it and enable users to start creating knowledge bases using the KA tool.
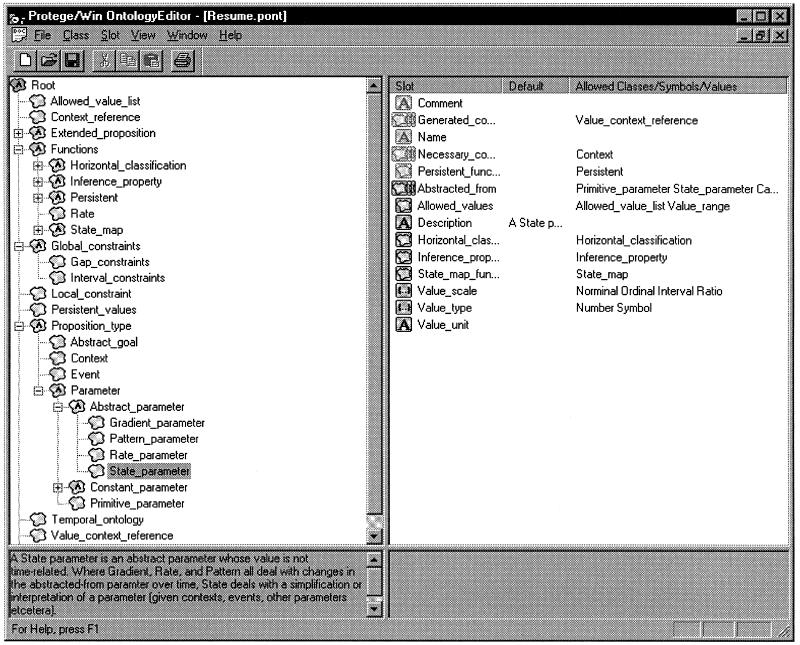
▶ shows the KA tool ontology, displayed in the Protégé ontology editor.
Figure 3.
The ontology of the temporal-abstraction knowledge-acquisition tool. This ontology was used by the Protégé system to generate a default graphical layout for that tool; it drives the knowledge-acquisition process.
In general, there are four major classes of concepts in the KA tool ontology—abstraction goals, contexts, events (interventions), and measured or derived parameters. (The class of patterns was represented in the KA tool as a subclass of the Parameter class). The RÉSUMÉ system uses the knowledge entered as instances of these classes to interpret patient data. Besides these four major classes, the KA ontology includes also multiple auxiliary classes that are useful for the construction of the major classes. The design of the KA tool ontology, which directly affects the interface generated by the Protégé system, reflects a delicate tradeoff between avoiding a too-complex structure and avoiding an overcrowded interface.
After we designed the KA tool ontology, the Protégé system generated a default graphical interface for acquisition of instances of that ontology. Typically, different classes in the ontology correspond to separate windows in the graphical interface, whereas slots of classes correspond to panels in each window. Using the Protégé layout-editing tools, we could easily custom tailor the interface by modifying sizes and rear-ranging relationships of windows and panels, changing certain labels, and so on.
The top level of the resultant KA tool, after we defined the ontology and customized the graphical layout within Protégé layout editor, is shown in ▶. Every knowledge-acquisition or knowledge-maintenance session starts at that top level. There are six panels in this frame. Four of them are the four major ontology classes. In addition, there are two help panels, which exist in almost all the frames in the KA tool. The Description panel of each frame provides domain-independent help for the expert who enters the knowledge. It typically explains the meaning of each frame or of specific slots in that frame and may contain instructions on how to fill out the frame. The Description panel is a slot maintained at the level of the KA tool ontology, and it therefore shows up in every frame generated from that ontology. The Comment panel is used by the expert to document the particular domain-specific knowledge that the expert enters.
Figure 4.
The entry screen (top level) of the temporal-abstraction knowledge-acquisition tool. Shown are the four main ontological classes and two Help panels.
In the current version of our KA tool, using the Protégé/Win system, users create and edit only knowledge instances, whose type (e.g., Event or Parameter) must be one of those determined in the KA tool ontology. Domain-specific classes, such as Laboratory Parameters, are typically created by the knowledge engineer designing the KA tool. The knowledge engineer creates these classes because the addition of such domain-specific classes to the KA tool ontology requires the use of the generic ontology editing tools, which is more suitable for the knowledge engineers than for the domain experts. Once a class is defined in the ontology, a “tailorable” interface is generated for it automatically, and instances of the new class can be acquired.
Once the user of the KA tool decides to edit a knowledge instance that belongs to one of the major classes (e.g., a particular parameter) or create a new one, the KA tool will create an instance of that class. Using the conventions of automatically generated interfaces in Protégé, users can use the “New” button (▶) to define new instances or “Select” to select an instance that has already been defined. They can use the “Edit” button to modify a previously defined instance. The “Clear” button deletes a previously defined instance from the list.
Figure 5.
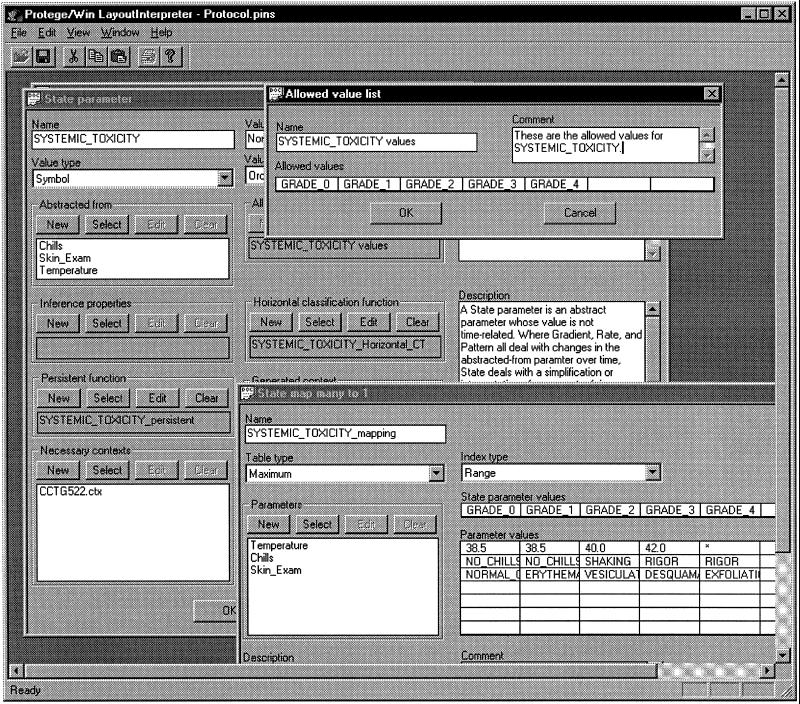
Acquisition of a state-abstraction parameter with the temporal-abstraction knowledge-acquisition tool. The Systemic toxicity parameter is abstracted, in the context of therapy by the CCTG522 AIDS therapy protocol, from values of the Temperature, Chills, and Skin_Exam parameters. The classification scheme defined by the expert is shown in the table displayed in the bottom-right subwindow. In this case, the expert has chosen to define the classification function using a maximum OR table; that is, after the toxicity value is determined individually for each input parameter (i.e., in each row), the KA tool selects the maximum value (i.e., the column whose output toxicity grade is maximum). The resultant output values of the Systemic toxicity parameter (Grade_0, Grade_1, etc.) are constrained by the Allowed value list, shown in the top-right subwindow. The values in this list (like the allowed values for each of the input parameters) have been defined by the user as part of the essential knowledge about each clinical parameter.
▶ shows the process of acquisition of knowledge regarding a state parameter—Systemic toxicity in the protocol-based care domain, in the context of therapy according to the CCTG522 experimental AIDS-therapy protocol. We can see that this parameter is abstracted from the parameters Temperature, Chills, and Skin_Exam, all of which have been defined previously by the expert, or exist in the knowledge base, as primitive (raw-data) parameters. The state abstraction, like all abstract parameters, has a Persistence function property (represented as a slot in the knowledge instance) (see legend for ▶). The Necessary contexts slot, which has the value CCTG522.ctx, defines the context or combination of contexts in which the abstraction is relevant. The two open subwindows show the values of an Allowed value list slot and the State mapping function slot. In the Allowed value list window we can see that the list of values legitimate for the parameter whose properties are being acquired is being defined. In the State map many to 1 window, the expert defines the values of the systemic toxicity parameter as a function of the values of the primitive parameters in the Abstracted from slot.
Figure 6.
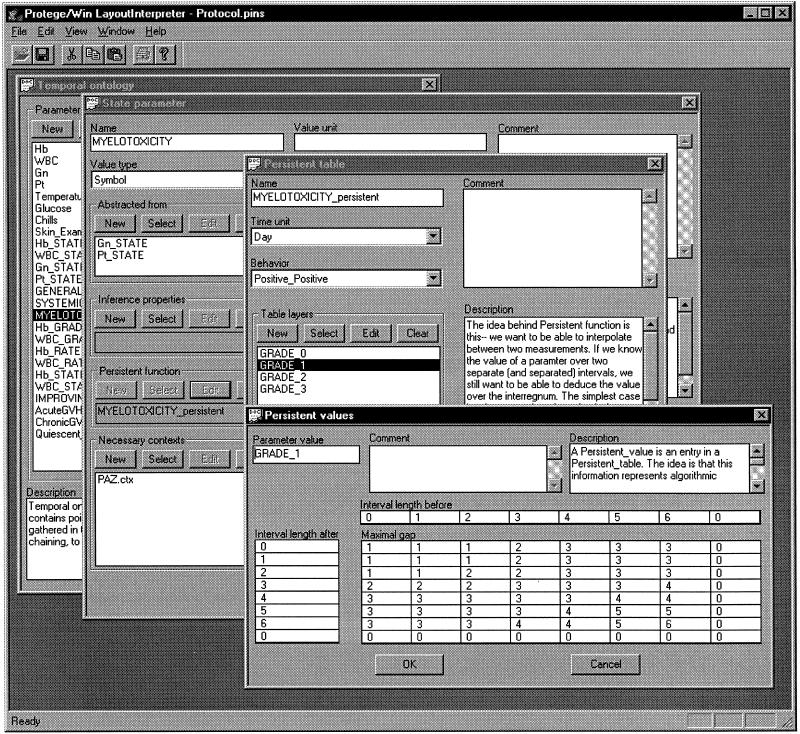
Specification of the persistence properties of the grade 1 value of the Myelotoxicity (bone marrow toxicity) parameter in the context of the PAZ protocol, as used in the temporal-abstraction knowledge-acquisition tool. The expert has selected the Grade_1 layer of the (conceptually) three-dimensional persistence table, and specifies, in the bottom-right subwindow, a two-dimensional table that defines the maximal gap that allows concatenation of two intervals of grade 1 bone marrow toxicity, as a function of the duration (Interval length) of the interval before the gap and the duration of the interval after the gap. The lengths of the two intervals are the indices of the two-dimensional table; the value of the parameter (in this case, Grade_1) is the layer index for the three-dimensional table.
▶ shows the acquisition of persistence knowledge from a domain expert—in this case, for the bone-marrow-toxicity (myelotoxicity) parameter. Persistence knowledge enables interpolation between points or intervals of clinical data and their abstractions and thus supports the creation of intervals that summarize the data as succinctly as possible, without concatenation of intervals that are too far apart.
For each given parameter (here, myelotoxicity) and context (in this case, therapy by the PAZ protocol), a persistence table can be specified by the expert. The table is, conceptually, a three-dimensional function; for ease of elicitation, however, it is displayed in the KA tool as a series of two-dimensional tables. For each layer (one value from the allowed values of the parameter) of the three-dimensional conceptual table, the expert specifies one two-dimensional table. This table lists the maximal temporal gap that can be bridged to connect two intervals (over both of which the clinical parameter has that value) to create a longer interval with the given value. The input arguments in each two-dimensional table are the duration of each interval before and after the gap; the output is the maximal duration of the gap that still allows aggregation of the intervals (see ▶). This methodology, which conforms to the global-persistence-function model,10 has certain theoretic advantages (e.g., given certain conditions, the resultant intervals do not depend on the order of arrival of data or performance of computations) as well as simplicity in specification.
Like many other properties, persistence tables are often inherited from higher-level classes (e.g., properties of the hemoglobin parameter or even of the hematologic class of parameters in general) but can be modified for each context (e.g., therapy by PAZ).
▶ shows the acquisition of a pattern parameter—Improving hemoglobin CCTG in the protocol-based AIDS-therapy domain. This pattern is linear; that is, it is composed of a single sequence of components (as opposed to a periodic pattern, which is typically composed of a repeating abstraction or linear pattern). To facilitate acquisition and maintenance, we define linear patterns by listing a set of one or more components that are instances of any of the four major classes, and defining local constraints on each component and global constraints between pairs of components. Each component (e.g., an abstract parameter) holds over a corresponding time interval. Local constraints include fuzzy intervals for the start time, end time, and duration, as well as a range of values for the parameter (or attribute of the event, such as dose of a drug). Global constraints include qualitative and quantitative temporal relations as well as value relations. Currently, the KA tool interface assumes that patterns are composed of up to four components, whose names, such as Input interval 1 (see ▶), are predefined for ease of acquisition. However, the number of components could be increased easily in the KA tool ontology, thus automatically modifying the resultant KA tool interface. The user can define more than one set of components that can fit the same top-level pattern (i.e., a disjunction of either of several sets of local and global constraints), but each set of constraints defines a conjunction of predicates, all of which should hold.
Figure 7.
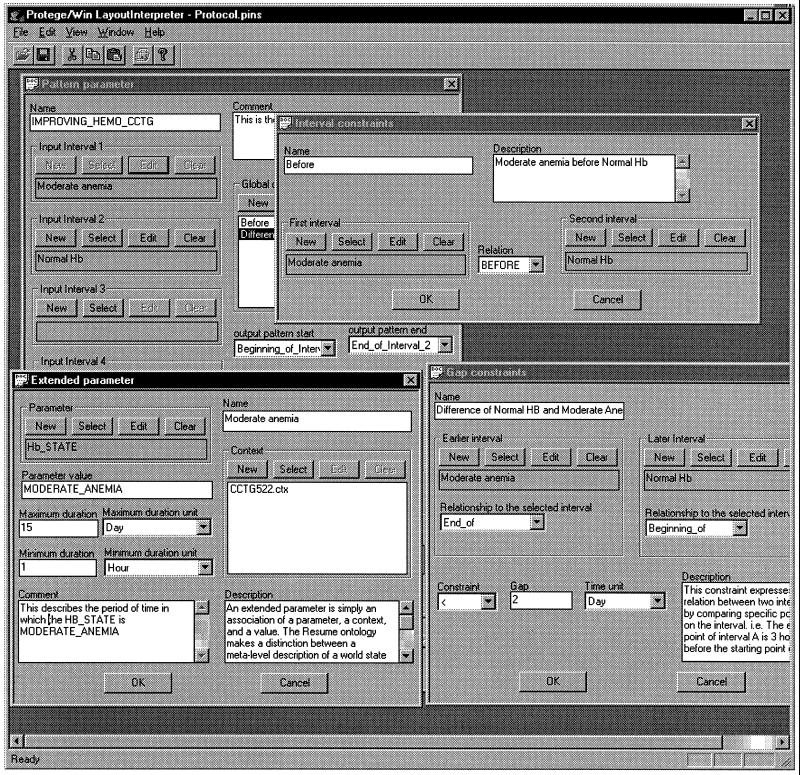
Specification of a temporal-pattern abstraction, as used in the temporal-abstraction knowledge-acquisition tool. The pattern of improving hematologic state in the context of therapy by the CCTG522 AIDS therapy protocol is composed of two components (input interval 1 and input interval 2), an interval of moderate anemia followed within two days by an interval of normal hemoglobin values. Both intervals are state abstractions of the Hemoglobin-value raw parameter. In the bottom-left subwindow, the expert defines the local constraints on interval 1. In the top-right sub-window, the expert defines a global qualitative temporal constraint over both intervals. In the bottom-right subwindow, the expert specifies a quantitative temporal constraint over the two intervals.
Thus, in ▶ we can see that the first component is called Moderate anemia. It is defined as a hemoglobin state (Hb_state) abstraction (a state abstraction of the hemoglobin-value raw parameter) whose local constraints include the moderate-anemia value as well as a maximum duration of 15 days and a minimum duration of 1 hour. The second interval, Normal Hb, is a hemoglobin-state abstraction whose local constraints include a normal-hemoglobin value. There are two global constraints:
Moderate anemia is before Normal Hb. This constraint is represented as an interval constraint: It defines the qualitative temporal relationship between two intervals.
Less than two days passed between the end of the Moderate anemia period and the beginning of the Normal Hb period. This is an example of a gap constraint: It defines the temporal gap between two intervals.
The Knowledge-acquisition Tool Filter
As we mentioned in the above section, the ontology of the KA tool is not a precise image of the RÉSUMÉ system's ontology. For instance, the Protégé/Win system cannot represent all arbitrary data types. Thus, a mapping between the ontologies is needed. For this and other significant reasons (such as knowledge validation and optimization of the representation), we developed a postprocessor module, the KA tool filter, that processes the output of the KA tool. Following the automated filter processing, the resultant knowledge base can be read directly into the RÉSUMÉ system, which can use that knowledge to interpret domain-specific data.
The KA tool filter is completely domain independent, since it refers to the data structures of the domain-independent KA tool and RÉSUMÉ system. The main task of the filter is to map the objects created by the KA tool to RÉSUMÉ objects. For example, the filter creates a three-dimensional table required by RÉSUMÉ from a set of lists, each representing one layer (or a two-dimensional table) created by the KA tool. The filter translates the KA tool output to a format that the RÉSUMÉ system can understand, performs integrity checking, and adds data structures (e.g., back pointers) that facilitate run-time computations by the RÉSUMÉ system.
The Knowledge-acquisition Tool's Online Manual
In addition to the two help slots described in the design section, we have created an extensive World Wide Web-based set of hypertext (HTML) files as an online manual. The manual help expert physicians understand the KA tool and the underlying knowledge-based temporal-abstraction methodology and its ontology. The hypertext files were available to the experts during the evaluation; all experts said that the files were useful.
Evaluation of the Knowledge-acquisition Tool
We conducted an experiment using as subjects three domain experts and three knowledge engineers. Our goal was to evaluate the usability of the KA tool for the specific purpose of entering existing (previously elicited) temporal-abstraction knowledge. Thus, we intentionally isolated the processes of knowledge entry and editing from the process of eliciting that knowledge, although the KA tool is probably helpful in the latter process, too. The reason for that separation is that the process of eliciting domain-specific knowledge often requires initial interviews of the domain experts by a knowledge engineer, to structure the knowledge before entry. Evaluation of the complete cycle of acquisition of temporal-abstraction knowledge is part of another, ongoing study.
The Knowledge Bases
We had previously acquired (through a series of interviews) and had entered manually temporal-abstraction knowledge for three clinical domains—the monitoring of children's growth,6,14 the care of patients with diabetes,5 and protocol-based care.6,12 We also had a small set of clinical data in each of these domains and the results of the RÉSUMÉ abstraction of those data (which were validated by domain experts), using the manually entered knowledge. In the current study, we used these three knowledge bases, the three representative data sets, and the respective output abstractions computed from them by RÉSUMÉ as the gold standards for verification and validation of the acquired knowledge bases.
In the domain of monitoring children's growth, two knowledge engineers (Y.S. and M. Kuilboer, MD, a collaborator in that domain) had previously conducted three interviews (about four hours overall) with a pediatric endocrinologist (D.M.W.), mainly to elicit and to structure manually the parameter ontology for that domain and to limit the scope of the resultant temporal-abstraction system.14 Then, two more hours of the expert's time were used for manual filling in the various tables (e.g., classification, interpolation). Finally, the knowledge was entered manually into the RÉSUMÉ knowledge base, which is in the CLIPS language, by the two knowledge engineers, a task that required ten more person-hours. The resultant knowledge base was tested on a data set representing three clinical cases, and the output abstractions correlated well with the abstractions created by the expert for the same data. We used these cases for the gold-standard data set.
In the domain of protocol-based care, we tested in previous experiments the knowledge representation capabilities of RÉSUMÉ using the temporal-abstraction aspects (as opposed to the therapy-planning aspects) of two protocols—the California Collaborative Treatment Group CCTG-522 experimental protocol for AIDS therapy [CCTG, personal communication], and the PAZ protocol for treating patients who have chronic GVHD (a complication of bone-marrow transplantation), which was developed by investigators at the Fred Hutchinson Cancer Center at the University of Washington in Seattle.20 In the domain of protocolbased care, reading the protocols and performing interviews with domain experts to elicit, structure, and acquire the temporal-abstraction knowledge portion of the protocols manually required ten hours (five hours for each protocol). About 16 additional hours (8 per protocol) were needed for one author (Y.S.) to enter the knowledge manually into CLIPS using a text editor. The abstractions created the knowledge and a data set that included a simulated patient whose record was constructed from two clinical cases, one in each protocol; they were judged by participating clinicians as correct according to the protocols, although no formal evaluation was done. We used the simulated case as the gold-standard data set for validation purposes.
Previously, we applied the knowledge-based temporal-abstraction methodology to the task of monitoring patients who have insulin-dependent diabetes mellitus.5 Two endocrinologists (L.V.B. and F. Kraemer, MD), who are experts in therapy for insulin-dependent diabetes, were the domain experts for that experiment. Creating the ontologies on paper (through elicitation interviews) required four two-hour meetings with the experts. In addition, 12 more hours were needed for a knowledge engineer (Y.S.) to enter the knowledge manually (with the assistance of a text editor).
A formal evaluation of the knowledge base was performed. The data used for the evaluation study were taken from a set of electronic clinical records of patients who have insulin-dependent diabetes and who were followed for several months. The data included mostly glucose and insulin codes. Special events (e.g., physical exercise and larger-than-usual meals) were sometimes reported, too, as were symptoms of hypoglycemia. The resultant RÉSUMÉ abstractions included 80.4 percent (132 of 164) of the temporal abstractions that were agreed on by two endocrinologists for the set of patients used in that study.5 When one of the experts, who eventually collaborated with us in the KA phase of the current study, had originally examined the detailed output for the first three cases, he agreed with 97 percent of the abstractions created by RÉSUMÉ.5 The high specificity was not surprising, since the diabetes domain's temporal-abstraction ontology reflected, at least partially, that expert's knowledge. We used these cases for the gold-standard data set.
▶ summarizes the characteristics of the knowledge elicitation and manual entry processes for the three knowledge bases used for the study.
Table 1.
Hours Required for Manual Acquisition and Entry of Temporal-abstraction Knowledge in Three Domains
| Monitoring Growth | Providing Diabetes Care | Providing Protocol-based Care | |
|---|---|---|---|
| Structuring interview of knowledge engineers with domain expert | 6 | 8 | 10 |
| Manual entry by knowledge engineer using text editor | 10 | 12 | 16 |
Evaluation of the Usability of the Knowledge-acquisition Tool for Knowledge Entry
To evaluate the feasibility of entering temporal-abstraction knowledge through the graphical KA tool, we provided three of the authors who are expert physicians (L.V.B., an endocrinologist; H.K., an oncologist; and D.P.S., an immunologist) and three knowledge engineers (all graduate students in computer science or medical informatics) with the manually entered, text-based versions of the three knowledge bases described in the previous section. The knowledge was represented declaratively as a unordered set of frames and slots that summarized the class definitions in the object-oriented CLIPS language13 version of the knowledge bases (which is the format in which the RÉSUMÉ system accesses these knowledge bases). The frames were highly declarative and self-explanatory, and corresponded to the class hierarchy that was typically elicited from the domain expert from which the knowledge was originally acquired.
Since the domain experts were familiar to some extent with the concept of class hierarchies, and since we provided them with syntactic assistance when necessary, we preferred using the CLIPS-like, frame-based representation to trying to create a natural-language representation of the temporal-abstraction knowledge bases, which were most naturally expressed as frames and slots, even when acquired manually from the original domain experts. The knowledge engineers were comfortable with the format (all were familiar with knowledge-based systems). Thus, the input forms given to participants simulated the stage in which the knowledge has been structured (either in the expert's mind or through assistance by a knowledge engineer) and needs to be entered into the semi-automated KA tool. Notice that, although the knowledge was given to the participants as a set of encapsulated instances, they had to solve the realistic problem of how best to use the automated KA tool to enter the instances. In particular, they had to determine where in the KA tool dialog (e.g., as a gradient abstraction) and how (e.g., as the value of the temporal-persistence slot) each instance should be entered. The relations among instances in the given text did not necessarily reflect links that might be useful in the automated KA tool, since the knowledge represented in the text was originally elicited by interviews and was entered manually (using the RÉSUMÉ system's internal format) by knowledge engineers who did not have the KA tool available at that time.
All participants were given copies of the text-based knowledge bases, instructions on downloading the Protégé/Win system from the Web, diskettes with the temporal-abstraction KA tool, one or more papers on the RÉSUMÉ system (in particular, a paper about its clinical evaluation5), and a pointer to the Web-based KA tool manual. They were then asked to enter at their convenience two or three knowledge bases using the graphical KA tool without outside assistance. In every case, we asked the participants to enter the growth knowledge base first, because we judged that task to be the least demanding. None of the physicians was an expert in that domain. Each physician then entered knowledge in a second domain (protocol-based care in oncology and AIDS, or diabetes care), the domain in which he is an expert. One author (H.C.), who was also one of the designers of the KA tool, provided online and live assistance in understanding the KA tool's functionality during the initial learning phase.
We asked all participants to record the time it took them to understand the essence of the knowledge-based temporal-abstraction methodology and, in particular, the ontology of the RÉSUMÉ system (excluding any computational details); to familiarize themselves with the KA tool; and to enter the knowledge in each knowledge base into the tool. In 10 of 14 knowledge-entry cases, we asked the participants, several weeks or months later, to enter the knowledge bases again, simulating a scenario likely to occur if the tool were used on a regular basis, so the experts would then have familiarity with both the domain and the KA tool. In several cases, the subjects performed the first entry using a slightly older version of the KA tool that did not have cut-and-paste editing facilities for strings and whole instances. In all these cases, a second entry was performed, and the two designers of the KA tool (Y.S. and H.C.) tested the entered knowledge bases using the gold-standard set of data and compared the abstractions created with these knowledge bases to the gold-standard set of abstractions created with the manually entered knowledge.
Although we did not provide domain-specific assistance beyond explaining the syntax and semantics of the text knowledge bases, we did provide in our meetings with the experts and in the online manual several guidelines to effective use of the KA tool. In theory, a user can define any instance of a major class in the KA tool ontology on the fly in each of the subframes by using the “New” button (see ▶). However, to avoid confusion and re-creation of instances that already exist (possibly with a slightly different name), we encouraged both expert physicians and knowledge engineers to adhere to the following approach when creating or maintaining a knowledge base:
Analyze the specific domain knowledge and categorize it into the framework provided by the KA tool (contexts, events, parameters). In this study, most of the analysis had been done, of course, when the original knowledge base was elicited; nevertheless, becoming familiar with the various knowledge classes in the given text knowledge base was important for facilitating efficient entry.
Define context instances first, so that they can be selected in the future from other frames. Examples include the regular_insulin_effect interpretation context.
Define the set of event instances. Examples include regular_insulin_administration. Events might be able to generate (induce) predefined contexts at run time, such as the regular_insulin_effect interpretation context. The precise temporal relationship can be specified when the event is defined.
Define the set of abstraction goals. These goals exist only for the induction of interpretation contexts at run time (e.g., the goal of care for insulin-dependent diabetes induces the top-level diabetes-care interpretation context, in which glucose and insulin data are interpreted accordingly).
Define parameter knowledge instances. This task is typically the main and most complex KA process, and it contributes the most to the knowledge base. Parameters have multiple properties, usually within a predefined interpretation context, which can be selected at acquisition time. Thus, the meaning of “grade 2 systemic toxicity” depends on the context. Furthermore, certain values of certain abstractions (e.g., “tachycardia” for the rate abstraction of the pulse) can be specified as generating another interpretation context, which would be useful for abstraction of other data types. That context also would be best selected from an existing context list.
These suggestions represented insights that we gained while designing the KA tool and can be seen as part of the methodology for best use of a KA tool generated by a Protégé system. The suggestions were not relevant for entering the knowledge using a text editor.
Results
The usability of the KA tool can be described by quantitative and qualitative measures.
Quantitative Measures
▶ summarizes the results of the experiment with respect to how long it took to learn the conceptual methodology and use of the KA tool (both a one-time cost) and to make first-pass and second-pass entries of all three knowledge bases for all participants.
Table 2.
Hours Required to Learn the KA Tool and Underlying Methodology and Use It for Semi-automated Entry of Temporal-abstraction Knowledge in Three Clinical Domains
| Expert Physicians |
Knowledge Engineers |
|||||
|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 1 | 2 | 3 | |
| Understanding the ontology | 6 | 20 | 6 | 20 | 20 | 15 |
| Understanding the KA tool | 6 | 4 | 2 | 2 | 5 | 3 |
| Monitoring growth | ||||||
| First entry | 6 | 20* | 2 | 1 | 3 | 25* |
| Second entry | 2 | 3 | - | - | 1.5 | 1 |
| Providing diabetes care | ||||||
| First entry | 12 | - | - | 2.5 | - | 2 |
| Second entry | 6 | - | - | - | - | 2 |
| Providing protocol-based care | ||||||
| First entry | - | 60* | 10 | 5 | 6 | 5 |
| Second entry |
- |
10 |
5 |
- |
2 |
1 |
| Note: Where two entries are given, the first is for first entry to the knowledge base and the second is for second entry to the same knowledge base. | ||||||
First entry to the knowledge base using an older version of the KA tool (e.g., no cut-and-paste editing functionality).
Understanding the RÉSUMÉ ontology sufficiently for use of the KA tool, as judged by the participants, required from 6 to 20 hours (median, 15 to 20 hours). Notice that the knowledge engineers often took more time than the domain experts, since they wanted to understand the computational details in addition to the terms. Learning to use the KA tool, as judged by the participants, required from 2 to 6 hours (median, 3 to 4 hours); there did not seem to be significant differences between domain experts and knowledge engineers. Acquisition times for the three physicians varied by domain—from 2 to 20 hours for growth monitoring (median, 3 hours), 6 and 12 hours for diabetes care (only one expert physician entered knowledge into this domain), and from 5 to 60 hours for protocol-based care (median, 10 hours).
An increase in speed by a factor of up to 25 (median, a factor of 3) was shown for all participants when the KA process was repeated. The knowledge engineers recorded first-pass entry times similar to those recorded by the expert physicians on their second pass.
To assess the validity of the resulting knowledge bases, we ran the RÉSUMÉ system on the representative data sets in each domain, using each knowledge base. In all cases RÉSUMÉ, using the entered knowledge, generated an output that was judged by the two knowledge engineers (Y.S. and H.C.) to be almost identical to the output that it generated using the manually entered knowledge. In two cases, one abstraction was missing because the domain expert omitted a classification function or one of its values; otherwise, the output was identical. This result is not as surprising as it might seem, since all participants were given well-structured knowledge bases and the gold-standard cases included a relatively small number (typically 10 to 20) of raw time-stamped clinical parameters and events. Nevertheless, the results demonstrated that the knowledge bases acquired by use of the KA tool were as valid as the manually entered ones. We also verified the contents of the acquired knowledge by manual examination of the CLIPS instances generated by the KA tool and by comparison of these instances to the gold-standard knowledge bases, with similar results.
Qualitative Measures
The domain experts often needed significant support (provided by Y.S. and H.C. as well as by the papers and the online manual) during the initial phase of understanding the RÉSUMÉ methodology and ontology. The interpolation (persistence) functions, in particular, needed several explanations. The KA tool interface, however, was considered intuitive by all experts and knowledge engineers. All experts reported that, once they became familiar with the KA tool, they could enter any of our sample knowledge bases in several hours. A common comment was that it would have been useful if the knowledge, once entered, could have been used immediately to create abstractions on a sample data set, with the abstractions being visualized in some way. In this way, the knowledge could have been verified easily during entry time without painstaking checks. The online manual was cited by all expert physicians as highly useful.
Discussion
The temporal-abstraction task is common, in various direct or indirect forms, in many clinical applications, including patient monitoring, therapy planning, quality assessment, and visualization and exploration of time-oriented clinical data. Thus, automating this task as much as possible is highly desirable. Several researchers have previously examined the temporal-abstraction task or facets of it,21,22,23 usually without emphasizing the aspect of supporting acquisition and maintenance of the required knowledge. The knowledge-based temporal-abstraction method, with its well-defined ontology, can provide support for both performance of the temporal-abstraction task and creation of a formal framework that facilitates acquisition and maintenance of temporal-abstraction knowledge. However, it requires significant amounts of domain-specific knowledge to be effective. Thus, it is important to facilitate the process of acquisition of temporal-abstraction knowledge. In the past two decades, a common solution proposed by researchers was the use of semi-automated KA tools, although not many were evaluated for usability or were considered usable when evaluated.24
We created, prior to this study, a highly preliminary prototype of the current KA tool, using the Protégé-II system, a previous Protégé version which ran on a NeXT machine.25 No formal evaluation was performed at that time, although the experiment was encouraging. We have now demonstrated that the current temporal-abstraction KA tool, which we developed using the Protégé/Win system, it usable and effective, at least for both expert physicians and knowledge engineers to enter clinical temporal-abstraction knowledge in three clinical domains for which we had benchmark knowledge and data. Furthermore, the resulting knowledge bases were verified by comparison with the knowledge previously entered manually and were judged as valid as these knowledge bases are, in that they computationally produced almost the same abstractions when used by the RÉSUMÉ system to interpret a small set of data in each domain.
The evaluation described here did not include the crucial step of elicitation of the knowledge from the original domain experts who provided it and the structuring of that knowledge. As was shown in ▶, that step required a significant amount of time, spread over several meetings of a knowledge engineer with a domain expert. We consider that step, however, to be both unavoidable and justifiable in the case of complex knowledge-based systems aiming at provision of nontrivial conclusions based on large amounts of domain-specific knowledge. Similarly, we consider the one-time cost of understanding the RÉSUMÉ ontology and the KA tool interface reasonable for recurring, high-value tasks such as monitoring patients who have chronic diseases. In such domains, the resultant knowledge base is highly reusable, both for abstraction and for other purposes, such as visualization and exploration of the resultant abstractions.4 There was a partial overlap between the domain experts who participated in the elicitation phase and the knowledge-entry phase, which somewhat strengthened the results by presenting an almost complete, realistic life cycle of development of the knowledge base by the same expert. This overlap occurred for only one domain (diabetes) and one expert (L.V.B.).
In addition to eliminating the interviewing aspect of the elicitation phase, provision of the knowledge (to the experts using the KA tool) as text-based CLIPS classes eliminated much of the structuring aspect of that elicitation and provided a well-defined common ground. The object-oriented CLIPS format attempted to mimic the typical results of an elicitation process (i.e., an informal set of paper forms representing a set of domain-specific classes and objects with certain relations among them). All experts were familiar, in this particular case, with the ideas (if not details) of classes, objects, and slots (additional knowledge was available in the KA tool manual). However, the input format potentially could have positive (facilitating) and negative (detrimental) effects on the ease of the knowledge-entry process. On the positive side, the organization into classes and slots might have assisted the experts in determining how to structure the knowledge in the KA tool. On the negative side, CLIPS is a computer language with which medical experts are unfamiliar; furthermore, the format of the knowledge represented in the text given to the experts was similar to that found in actual RÉSUMÉ files (i.e., filtered knowledge, postprocessed from the KA tool), rather than to the format and general “look and feel” of the interface they eventually used in the KA tool. (It was that way because, to start the KA process when no knowledge had been acquired yet through the KA tool, we used the knowledge originally entered manually into RÉSUMÉ.) The experts might have done even better using, for instance, diagrams of their own knowledge as the input to be entered into the KA tool.
We represented the knowledge given to the participants using a uniform formal input format. The knowledge-entry methodology supported by the KA tool is also structured. Thus, the resulting knowledge instances were in an almost canonic form, leading to little variability in the representation once users understood the correct way to enter the knowledge. An experiment including the elicitation of the same type of knowledge de novo from different experts might have led to significant variability in the resulting knowledge bases. For example, in a previous experiment in the domain of caring for patients who have insulin-dependent diabetes, we noted significant differences between two experts with respect to their recommended management of the same patients.5 However, the two experts had been in agreement with high correlation with respect to the temporal abstractions that could be extracted from the patients' records, thus leaving open the possibility that temporal-abstraction knowledge is more likely to be a common ground than management knowledge, which often depends on subjective factors such as physician and patient preferences.5
Because of the different goal and design of this study, we did not aim to measure interauthor variability among the participating physicians or knowledge engineers. That goal was used, for instance, in the study of Giuse et al.26 within the framework of the QMR diagnostic system. That team focused on the creation, by seven internists, of a disease profile for a single disease (acute perinephric abscess), a profile comprising mainly sensitivity and positive predictive values for a series of findings. Unlike that study, ours used subjects who started with the same knowledge base in three different clinical domains. We also did not measure the rate of change in the knowledge base during maintenance, an assessment undertaken in another study within the QMR framework.27 Rather, we assessed the usability of the KA tool for entry and maintenance of highly complex types of knowledge implied by a particular problem-solving method.
The significant increase in speed (up to an order of magnitude) in knowledge entry times from the first pass to the second, especially in the case of the expert physicians, was interesting and suggests that any amount of training with the KA tool might have a significant positive effect on usability. There are several possible explanations for the increase in speed. First, the physicians might have become familiar with the domain's details (thus simulating a domain expert). Second, they might have become proficient in the use of the KA tool. Third, in several cases the first-pass knowledge entry used a slightly older version of the KA tool, which did not make it possible to cut and paste strings or whole knowledge instances, thus probably explaining the most marked improvements in entry time. (We estimate that cut-and-paste editing functionality would have been useful for entry of about 50 percent of the knowledge instances). Fourth, at least in the specific instances acquired, understanding the conceptual and mechanical underpinnings of the KA tool might have posed a major learning task, whereas entering temporal-abstraction knowledge might have been simpler. Thus, the second entry would be much faster. Such a conclusion would be highly encouraging with respect to the probability of success in entry of future knowledge or editing of existing knowledge by domain experts. The conclusion seems confirmed by our informal ongoing observations of the experts who wished to modify a knowledge base and who reported that the task was easy.
The knowledge-entry times of all participants in the study—even those of the expert physicians (in their second attempt, when they were more familiar with the tool)—were shorter than those required for the original manual entry for all knowledge bases by RÉSUMÉ experts (Y.S. and Dr. Kuilboer). Admittedly, the participants were starting from a more explicit description of the knowledge. Nevertheless, from the conceptual point of view, the participants' point of departure was similar to that of the knowledge engineers who entered the knowledge manually—that is, the knowledge had already been elicited and organized in a textual form and had to be entered in a way that was usable by the RÉSUMÉ system. Thus, the KA tool provided a significant advantage over manual revisions of program code, even though the latter were performed by the original developer of the RÉSUMÉ system and a knowledge engineer (Dr. Kuilboer) who knew that system.
The graduate-student knowledge engineers required shorter entry times than the expert physicians, in either their first or second pass, even though none of them had a medical background, probably because of their familiarity with knowledge-based systems and software tools. That domain experts are often unfamiliar with such tools has been noted previously as a major obstacle to the achievement of a KA process that does not require a knowledge engineer in the loop.24 We consider the involvement of knowledge engineers in the development of large knowledge bases to be unavoidable, but we seek to minimize that involvement. In this study, the expert physicians downloaded the Protégé tools and the KA tool manual from the Web and worked at their convenience on their own personal computers, thus demonstrating that distributed entry of clinical temporal-abstraction knowledge by domain experts is certainly feasible.
Future Work
Determination of the actual time and other costs involved in a complete development of a knowledge base de novo, including in particular the knowledge elicitation and structuring steps, is of considerable interest. Thus, we are now conducting a further study that includes the knowledge structuring process.
In addition, we are improving the expressiveness of the KA tool and the underlying computational framework. In particular, we are enhancing the expressiveness of the language for specification of linear patterns. We are also adding a highly expressive language for fuzzy periodic (and, in general, episodic) patterns.28 Such patterns are common in domains such as diabetes, where diurnal and weekly patterns are the norm; they are also useful, in many clinical domains, for interpretation and summarization purposes and for automated discovery in large time-oriented clinical databases.
Acknowledgments
The authors thank Dr. Fredric Kraemer for his time during the manual knowledge-acquisition process in the diabetes-care domain, Dr. Manon Kuilboer for her assistance in the acquisition of the knowledge in the growth-monitoring domain, Mr. Samson Tu for comments about the user interface of the original knowledge-acquisition tool, Mr. Arthur Menaker for maintaining the RÉSUMÉ system and participating in the knowledge-acquisition experiment, Ms. Xiaole Liu for assistance in creating the Help file and the questionnaire for the experts and for participating in the knowledge-acquisition experiment, Mr. Ray Fergerson for software development, and Ms. Lyn Dupré and Ms. Kathleen Jones for their assistance in editing the paper.
This work was supported by grants LM05708 and LM06245 from the National Library of Medicine and grant IRI-9528444 from the National Science Foundation.
References
- 1.Shahar Y. A framework for knowledge-based temporal abstraction. Artif Intell. 1997;90(1-2):79-133. [Google Scholar]
- 2.Musen MA, Tu SW, Das AK, Shahar Y. EON: a component-based approach to automation of protocol-directed therapy. J Am Med Inform Assoc. 1996;3:367-88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shahar Y, Miksch S, Johnson PD. A task-specific ontology for the application and critiquing of time-oriented clinical guidelines. Artif Intell Med. 1998;14:29-51. [DOI] [PubMed] [Google Scholar]
- 4.Shahar Y, Cheng C. Knowledge-based visualization of time-oriented clinical data. Proc AMIA Annu Fall Symp. 1998:155-9. [PMC free article] [PubMed]
- 5.Shahar Y, Musen MA. Knowledge-based temporal abstraction in clinical domains. Artif Intell Med. 1996;8(3):267-98. [DOI] [PubMed] [Google Scholar]
- 6.Shahar Y. A Knowledge-based Method for Temporal Abstraction of Clinical Data [PhD dissertation]. Stanford, Calif: Stanford University, 1994. Program in Medical Information Sciences, Knowledge Systems Laboratory report KSL-94-64, Department of Computer Science report STAN-CS-TR-94-1529.
- 7.Musen MA. Modern architectures for intelligent systems: reusable ontologies and problem-solving methods. Proc AMIA Annu Symp. 1998:46-52. [PMC free article] [PubMed]
- 8.Newell A. The knowledge level. Artif Intell. 1982;18(1):87-127. [Google Scholar]
- 9.Shoham Y. Temporal logics in AI: semantical and ontological considerations. Artif Intell. 1987;33(1):89-104. [Google Scholar]
- 10.Shahar Y. Knowledge-based temporal interpolation. J Exp Theor Artif Intell. 1999;11:123-44. [Google Scholar]
- 11.Shahar Y. Dynamic temporal interpretation contexts for temporal abstraction. Ann Math Artif Intell. 1998;22(1-2):159-92. [Google Scholar]
- 12.Shahar Y, Musen MA. RÉSUMÉ: a temporal-abstraction system for patient monitoring. Comput Biomed Res. 1993;26:255-73. Reprinted in: van Bemmel JH, McRay AT (eds). Yearbook of Medical Informatics 1994. Stuttgart, Germany: F.K. Schattauer and the International Medical Informatics Association, 1994:443-61. [DOI] [PubMed] [Google Scholar]
- 13.Giarratano J, Riley G. Expert systems: principles and programming. Boston, Mass: PWS Publishing, 1994.
- 14.Kuilboer MM, Shahar Y, Wilson DM, Musen MA. Knowledge reuse: temporal-abstraction mechanisms for the assessment of children's growth. Proc 17th Annu Symp Comput Appl Med Care. 1993:449-53. [PMC free article] [PubMed]
- 15.Musen MA. Dimensions of knowledge sharing and reuse. Comput Biomed Res. 1992;25(5):435-67. [DOI] [PubMed] [Google Scholar]
- 16.Musen MA, Gennari JH, Eriksson H, Tu SW, Puerta AR. Protégé-II: computer support for development of intelligent systems from libraries of components. Medinfo. 1995:766-70. [PubMed]
- 17.Tu SW, Eriksson H, Gennari J, Shahar Y, Musen MA. Ontology-based configuration of problem-solving methods and generation of knowledge-acquisition tools: application of protégé-II to protocol-based decision support. Artif Intell Med. 1995;7(3):257-89. [DOI] [PubMed] [Google Scholar]
- 18.Musen MA. Domain ontologies in software engineering: use of PROTÉGÉ with the EON architecture. Methods Inf Med. 1998;37:540-50. [PubMed] [Google Scholar]
- 19.Gennari JH, Cheng H, Altman RB, Musen MA. Reuse, CORBA, and knowledge-based systems. Int J Human-Comput Stud. 1998;49(4):523-46. [Google Scholar]
- 20.Sullivan KM, Witherspoon RP, Storb R, et al. Prednisone and azathioprine compared with prednisone and placebo for treatment of chronic graft-versus-host disease: prognostic influence of prolonged thrombocytopenia after allogeneic marrow transplantation. Blood. 1988;72:546-54. [PubMed] [Google Scholar]
- 21.Blum RL. Discovery and representation of causal relationships from a large time-oriented clinical database: the RX project. In: Lindberg DA, Reichartz PL (eds). Lecture Notes in Medical Informatics, vol. 19. New York: Springer-Verlag, 1982.
- 22.De Zegher-Geets IM, Freeman AG, Walker MG, Blum RL, Wiederhold G. Summarization and display of on-line medical records. MD Comput. 1988;5:38-46. [PubMed] [Google Scholar]
- 23.Russ TA. Use of data abstraction methods to simplify monitoring. Artif Intell Med. 1995;7(6):497-514. [DOI] [PubMed] [Google Scholar]
- 24.Kitto CM. Progress in automated knowledge acquisition tools: how close are we to replacing the knowledge engineer? Proceedings of the 3rd Knowledge Acquisition for Knowledge-based Systems Workshop; Banff, Alberta, Canada. 1988:14-1-13. [Google Scholar]
- 25.Stein A, Musen MA, Shahar Y. Knowledge acquisition for temporal abstraction. Proc AMIA Ann Fall Symp. 1996:204-8. [PMC free article] [PubMed]
- 26.Giuse NB, Giuse DA, Miller RA, et al. Evaluating consensus among physicians in medical knowledge base construction. Methods Inf Med. 1993;32(2):137-45. [PubMed] [Google Scholar]
- 27.Giuse DA, Giuse NB, Miller RA. Evaluation of long-term maintenance of a large medical knowledge base. J Am Med Inform Assoc. 1995;2(5):297-306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chakravarty S, Shahar Y. A constraint-based specification of periodic patterns in time-oriented data. Proceedings of the 6th International Workshop on Temporal Representation and Reasoning; Orlando, Florida; 1999:29-40.