Abstract
Nonmuscle myosin II is an actin-based motor that executes numerous mechanical tasks in cells including spatiotemporal organization of the actin cytoskeleton, adhesion, migration, cytokinesis, tissue remodeling, and membrane trafficking. Nonmuscle myosin II is ubiquitously expressed in mammalian cells as a tissue-specific combination of three paralogs. Recent studies reveal novel specific aspects of their kinetics, intracellular regulation and functions. On the other hand, the three paralogs also can copolymerize and cooperate in cells. Here we review the recent advances from the prospective of how distinct features of the three myosin II paralogs adapt them to perform specialized and joint tasks in the cell.
Introduction
Myosins constitute a family of molecular motors that use the energy of ATP hydrolysis to move along actin filaments. At present, over 30 distinct myosin classes are known in eukaryotes. The human genome contains 38 myosin genes from 12 of these classes. Class II myosins are unique in their ability to polymerize into bipolar filaments, which can contract an array of oppositely oriented actin filaments and exert large mechanical forces in cells via their ability to act as multimotor ensembles. Mammalian class II myosins include multiple sarcomeric paralogs, as well as one smooth muscle and three nonmuscle myosins, which are closely related to each other and more distinct from sarcomeric myosins. Nonmuscle myosin II (NMII) is present in virtually all animal cell types and involved in numerous cell functions, including migration, adhesion, cytokinesis, intracellular transport, organelle morphogenesis, as well as organization and remodeling of the actin cytoskeleton. Three mammalian NMII heavy chain genes (MYH9, MYH10, and MYH14) encode NMIIA, IIB, and IIC, respectively. Despite overall similarity, NMII paralogs exhibit significant differences in motor kinetics, structure and dynamics of bipolar filaments and cellular functions. NMII paralogs are expressed in cells as various cell type- and tissue-specific combinations [1, 2]. NMIIA and NMIIB are relatively broadly expressed, whereas NMIIC expression is limited to some differentiated tissues, but generally low in fetal tissues and stem cells. Comparative analyses of NMIIA and NMIIB in vitro and in cells represent an active focus of current research, whereas studies of NMIIC are still in early stages.
In this review, we focus on our current knowledge of how differences in properties and regulation of mammalian NMII paralogs translate into their specific intracellular functions, and how these paralogs cooperate in cell. Several recent reviews have discussed other aspects of NMII activity, such as kinetics [3], regulation [4], and roles in development and disease [2, 5].
Features of the NMII molecule
Structure
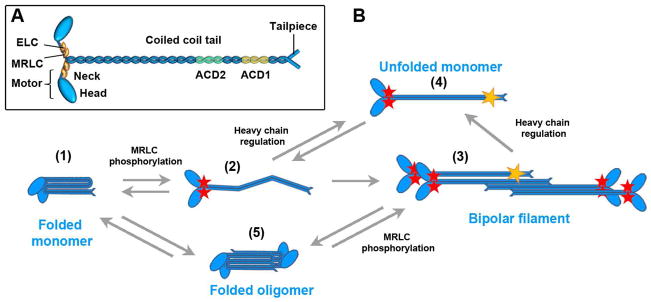
The NMII molecule is a hexamer (Fig. 1A). NMII heavy chains form parallel homodimers that associate with two pairs of light chains – essential and regulatory (myosin regulatory light chain, MRLC) – to form the NMII holoenzyme referred to as NMIIA, NMIIB, and NMIIC depending on the type of the heavy chain. As in all other myosins, the NMII heavy chain contains a conserved N-terminal motor domain, which consists of a globular head followed by an α-helical neck, which is stabilized by light chains and serves as a lever arm to amplify the ATP hydrolysis-dependent conformational change in the head to make a step along the actin filament. The NMII heavy chain C-terminus consists of a long α-helical rod domain, which is responsible for heavy chain dimerization through coiled-coil formation, and a short nonhelical tailpiece, which is most divergent among NMII paralogs. Two hinge regions in the coiled-coil rod allow the NMII molecule to acquire a folded autoinhibitory conformation.
Figure 1.
Structure and dynamics of NMII molecules
A. Structure of a hexameric NMII molecule. ELC, essential light chain; MRLC, myosin regulatory light chain; ACD, assembly competence domains.
B. The basic lifecycle of NMII.
(1) In the autoinhibited conformation, the NMII rod folds onto the heads and blocks motor activity.
(2) Phosphorylation on MRLC (red stars) disrupts the autoinhibition, releases the motors and allows for straightening of the rod.
(3) MRLC-phosphorylated NMII monomers are able to polymerize into bipolar filaments.
(4) Filament disassembly is promoted by heavy chain phosphorylation or protein-protein interaction (yellow star). Combinatorial MRLC phosphorylation and heavy chain regulation may lead to formation of pool of motor-active monomers in an extended conformation.
(5) Folded NMII molecules can associate into antiparallel dimers (or oligomers) that would unfold and join a bipolar filament upon RLC phosphorylation. Alternatively, MRLC dephosphorylation within bipolar filament may lead to formation of folded monomers or oligomers that could serve as storage/transported form of NMII.
Motor
Mechanoenzymatic properties of NMII motors vary among NMII paralogs and are tailored to specific intracellular functions [3]. In general, class II myosins have a relatively high ATPase rate and a low duty ratio (the proportion of the ATPase cycle that a motor spends strongly attached to its track), which makes them non-processive motors. However, since NMII in cells works in ensembles (bipolar filaments), NMII can stay associated with actin tracks over multiple ATP cycles as a processive motor [6, 7]. At the same time, the low duty ratio ensures that myosin heads in the filament do not interfere with each other during movement.
NMII paralogs are kinetically distinct. NMIIA is the fastest NMII paralog with highest ATPase rate. NMIIB moves slower with a relatively high duty ratio due to high binding affinity for ADP [8]. Additionally, force resisting the motor powerstroke can increase the duration of actin-myosin interaction, leading to catch-bond behavior [9]. The extent of mechanosensitivity varies among myosins. For example, NMIIB exhibits much stronger catch-bond behavior than NMIIA [10]. Kinetic diversity of NMII motors can be further increased due to alternative splicing of NMIIB and NMIIC heavy chains at two sites in the motor domains [2, 11]. Furthermore, intrinsic mechanokinetic properties of NMII paralogs can be modulated by external factors, such the actin isoform that forms the track [12], the presence of tropomyosins [13], and viscosity of the environment [7, 14].
Polymerization
Polymerization of NMII molecules into bipolar filaments occurs through staggered parallel and anti-parallel interactions between their rod domains. In assembled bipolar filaments, motor domains face opposite directions from the midzone. The filament nucleation depends on two conserved assembly competence domains (ACD1 and 2) at the end of the heavy chain rod. ACDs are thought to induce antiparallel dimerization of NMII molecules via electrostatic interactions between their complementary charges [15, 16]. Folded NMII monomers also can form antiparallel dimers in vitro, suggesting that NMII unfolding may occur after filament nucleation [17, 18]. Subsequent addition of NMII subunits to dimeric nuclei depends on interactions between periodically alternating positively and negatively charged segments of the rod [19]. These interactions may also promote unfolding of subunits added in the folded state [18]. The resulting NMII bipolar filament consists of up to 30 molecules for NMIIA and NMIIB and ~14 molecules for NMIIC [17].
Regulation of NMII turnover cycle
NMII undergoes constant polymerization-depolymerization cycles in cells. In the autoinhibitory (folded, 10S) conformation, the coiled coil rod folds at two hinge regions so that the second hinge binds MRLCs at the neck [20–22]. This interaction inhibits both NMII motor activity and polymerization [4]. The NMII dynamic cycle includes activation of autoinhibited molecules and their assembly into bipolar filaments followed by filament disassembly and subunit recycling (Fig. 1B). This cycle allows NMII to build and dismantle the contractile system as needed. Individual steps of the NMII cycle are controlled by phosphorylation and protein-protein interactions.
Activation of the motor
The ATPase activity of autoinhibited NMII molecules is restored by MRLC phosphorylation on Ser19, whereas additional phosphorylation of Thr18 further increases the actin-activated ATPase activity [23]. MRLC can be phosphorylated by multiple kinases, including ROCK, MLCK, MRCK, PAK, and citron kinase [4]. Individual kinases are thought to activate NMII at different subcellular locations and/or in response to different signals. MRLC can also be phosphorylated at Ser1/Ser2/Thr9 by protein kinase C (PKC). Phosphorylation of these residues decreases the rate of MRLC phosphorylation by MLCK in vitro, thereby indirectly inhibiting NMII activity [24]. This regulation was shown to promote PDGF-induced stress fiber disassembly [25] and cell chemorepulsion [26], but not affect NMII assembly in another study [27]. Since MRLC is shared by NMII paralogs, NMII regulation through MRLC phosphorylation is not expected to be paralog-specific, unless the enzymes can recognize paralog-specific sequences in the second hinge region of the heavy chain, which interacts with MRLC in the folded molecule [28, 29].
Regulation of NMII polymerization and depolymerization
Besides restoring NMII motor activity, MRLC phosphorylation releases the MRLC-rod interaction, thus permitting, although not imposing the rod unfolding [18]. Experimental abrogation of the MRLC-rod interaction by deleting the MRLC binding site [30] or eliminating the MRLC-interacting hinge region [31] caused over-assembly of these mutants in cells, thus confirming an inhibitory role of this interaction for NMII filament assembly.
Disassembly of bipolar filaments is largely regulated through the NMII heavy chain, primarily, through the nonhelical tailpiece and adjacent regions of the coiled coil rod. These regions contain paralog-specific phosphorylation sites and can also bind regulatory proteins [4, 16]. Differences in the C-terminal regions of the NMII heavy chain are largely responsible for different filament dynamics in cells, as well as distinct intracellular localization of the paralogs.
In vitro studies showed that phosphorylation or phosphomimetic mutations of the nonhelical tailpiece inhibit polymerization of rod domains of mammalian NMIIA, NMIIB and NMIIC [32, 33]. Despite similar effects of phosphorylation, deletion of the tailpiece promotes assembly of the NMIIA and NMIIB rods, but weakens assembly of the NMIIC rod [33, 34]. In NMIIC, the dephosphorylated tailpiece flips onto and binds the coiled coil, which promotes rod polymerization, but this activity is lost upon tailpiece phosphorylation [34]. The scenario is likely opposite for NMIIA: the tailpiece may gain affinity for the coiled coil upon phosphorylation, which could inhibit filament assembly [16].
Phosphorylation sites in the NMIIA heavy chain include a putative PKC target site at Ser1916 (human numbering) just before the tailpiece, and a putative casein kinase II (CKII) site at Ser1943 in the tailpiece. Notably, CKII depletion from cells did not affect the level of S1943 phosphorylation suggesting involvement of other kinase(s) [35]. The main regulatory site for the NMIIB heavy chain is a stretch of five serine residues (1935–1941) that can be phosphorylated by PKCγ [36] and aPKCζ [37]. Among several C-terminal phosphorylation sites in NMIIC, only phosphomimetic mutations of PKC sites (T1957D/T1960D) in the tailpiece inhibited polymerization of NMIIC rods in vitro [33].
Cell-based assays, in general, support the insights from in vitro studies about regulation of NMII assembly. Expression of NMIIA heavy chains either lacking the tailpiece or containing the S1943A substitution resulted in over-assembly of NMII in cells [30]. In NMIIB, deletion or phosphomimetic mutations of the tailpiece serine cluster increased NMIIB dynamics in cells [38] and decreased the insoluble (polymerized) NMIIB fraction in cells [36]. Notably, heavy chain phosphorylation unlikely functions as an on/off switch, because NMII heavy chains with phosphomimetic mutations could be found in association with the cytoskeleton in cells [39]. Most likely, heavy chain phosphorylation shifts the balance toward NMII filament disassembly by weakening subunit interactions.
In addition to heavy chain phosphorylation, assembly of NMII filaments is regulated by interacting proteins. The best characterized regulator is S100A4/Mts1, which specifically regulates disassembly of NMIIA [16, 32]. The binding mechanism includes initial recognition of the nonhelical tailpiece and subsequent binding to and partial unwinding of the ACD1-proximal coiled coil, which could eventually lead to dissociation of the NMIIA subunit [40, 41]. Another protein from the same family, S100P, can dissociate NMIIA and NMIIC filaments by a similar mechanism [42]. The cancer suppressor Lgl1 can bind directly to the coiled coil of NMIIA between ACD1 and ACD2, potentially through electrostatic interactions, and block filament assembly in vitro [43]. This study also showed that ectopically expressed Lgl1 and NMIIA could be coimmunoprecipitated from cells. However, another study revealed an interaction of Lgl1 only with NMIIB, but not NMIIA, using coimmunoprecipitation of endogenous proteins [44]. This discrepancy remains to be resolved, especially because all charged residues within the proposed Lgl1-binding site of NMIIA are conserved in NMIIB. Recently, it was shown that motor-inactive myosin 18A can copolymerize with NMII and might regulate the degree of NMII assembly and/or the mechanical output by reducing the number of force generating NMII heads per bipolar filament [45]. Direct interaction with the NMIIA rod have been also reported for gelsolin [46], gelsolin-like protein flightless-1 [47], Arf GAP ASAP1 [48], and Rho GAP Dlc1 [49], with the latter two interactions affecting stress fiber assembly in cells. The direct interaction between rod domains of NMIIB and kinesin 12 regulated migration of astrocytes [50]. Other interaction partners of NMII, especially of NMIIA, have been reported, but it is not clear whether any of these interactions affect NMII polymerization.
Although the NMII heavy chain-dependent regulatory mechanisms are generally thought to promote disassembly of NMII filaments, in principle, they also can prevent filament assembly. Simultaneous phosphorylation of MRLC and the heavy chain has a potential to produce unfolded motor-active NMII molecules. Indeed, unfolded MRLC-phosphorylated NMIIA and NMIIB monomers were detected in cells and appeared to be functionally important [51–53].
General principles of assembly and remodeling of NMII-containing structures in cells
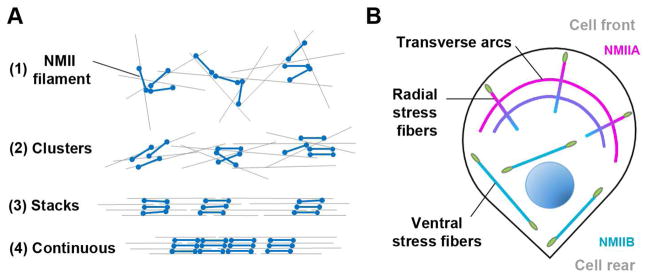
The major function of NMII filaments in cells is contraction, which is performed in cooperation with actin filaments. The main principles of assembly and dynamics of contractile structures are similar for different NMII paralogs, at least for the best studied ones, NMIIA and NMIIB. The actin-NMII contractile systems consist of non-aligned networks, aligned bundles and intermediate arrays of actin and NMII filaments. These systems gradually evolve in the processes of actin and NMII polymerization and actin-NMII interaction (Fig. 2A).
Figure 2.
Development of the actin-NMII contractile system
A. Stages of contractile system evolution.
(1) Newly assembled bipolar filaments form clusters within randomly oriented actin filaments producing an actin-NMII contractile network.
(2) NMII sliding along actin filaments results in coalignment of actin and NMI filaments producing incipient bundles.
(3) Progressive bundling together with gradual registration of NMII filaments into stacks leads to development of quazi-sarcomers in bundles.
(4) Longitudinal contraction of the aging bundle brings stacks of NMII filaments close together resulting in their continuous distribution.
B. Types of stress fibers formed by mesenchymal cells on flat substrate.
Transverse arcs form behind leading edge in the course of actin retrograde flow and NMII contraction. Radial stress fibers have a focal adhesion (green) at the distal end near the leading edge; their proximal ends are often incorporated into transvers arcs. Ventral stress fibers are localized at the basal cell surface and anchored to the substrate by focal adhesions at both ends. They typically develop from merging and straightening of two radial stress fibers and interconnecting arcs.
Assembly of new NMII filaments in cells often begins behind the protrusive cell edges. The NMII filaments then drift away from the cell edge with the actin retrograde flow simultaneously forming clusters, in which NMII filaments often interact at their ends [54, 55], which seems to be an intrinsic property of NMII filaments also observed in vitro [7, 17]. Clusters of NMII filaments embedded into disordered actin filament arrays constitute actin-NMII networks capable of large-scale contractile activities including cell body translocation in migrating keratocytes [55], apical constriction of epithelial cells [56], cytokinesis [57], and many others. In the course of contraction, actin-NMII networks can disassemble with subsequent recycling of NMII monomers [56, 58]. The local assembly-disassembly cycles producing pulsatile contraction in the cell lamella are characteristic for NMIIA, but not exhibited by NMIIB, and results from the differences in their motor domains [58]. Alternatively, actin-NMII networks in the course of contraction can reorganize into aligned actin-NMII bundles [55, 57, 59, 60]. NMIIA and NMIIB both participate in this type of actin-NMII remodeling, but NMIIB typically remains associated with actin bundles for longer time [61].
Stress fibers and circular bundles are two main types of aligned bundles of actin and NMII filaments, which can develop greater contractile forces than networks due to their superior organization. Stress fibers participate in cell migration and are often organized into a complex system attached to the substratum by focal adhesions. Circular actin-NMII bundles do not interact with focal adhesions, but apply force to plasma membrane through other protein complexes. They function in cytokinesis, cell-cell adhesion, wound closure and cell extrusion [62]. In actin-NMII bundles, NMII filaments often form registered stacks [54] that can be arranged in a discontinuous manner and alternate with α-actinin-rich zones, thus resembling sarcomeric organization [63]. In the course of contraction, the spacing between the NMII stacks in stress fibers decreases [64] up to a complete loss of gaps (Fig. 2A). Eventually, even long-lived actin-NMII bundles can be disassembled and recycled.
Dynamics and sorting of NMII paralogs
Intracellular segregation of NMIIA and NMIIB
In cells cultured on 2D substrate, NMIIA and NMIIB have overlapping but distinct distributions. At steady state, NMIIB typically acquires more central (in unpolarized cells) or posterior (in front-back polarized cells) localization relative to NMIIA [65, 66]. This phenomenon was initially interpreted as reflecting different sites and/or timing of NMIIA and NMIIB polymerization. However, when the contractile system was allowed to assemble de novo [53, 67], NMIIA and NMIIB initially exhibited indistinguishable distribution and segregated much later, suggesting essentially similar assembly pathways for both paralogs that are followed by their sorting during system maturation.
Maturation of individual stress fibers in cells expressing both NMIIA and NMIIB proceeds through stereotypic temporal changes in their NMII contents [61]. Stress fibers newly formed near the leading edge are enriched with NMIIA, but also contain NMIIB. Over time, while the stress fiber undergoes retrograde flow, it progressively loses NMIIA and becomes enriched with NMIIB. Eventually, NMIIB-rich stress fibers either disassemble or form long-lived ventral stress fibers – the most mature type of stress fibers [68] – at the cell center or rear. In contrast, NMIIA is enriched in the younger stress fiber types – transverse arcs and radial stress fibers – formed at earlier stages of network-to-bundle reorganization of actin-NMII assemblies [55, 64, 69] (Fig. 2B).
Dynamics of NMIIA and NMIIB
Gradual replacement of NMIIA by NMIIB in stress fibers can be explained by similar polymerization and distinct depolymerization mechanisms for two paralogs [61]. The NMII assembly mechanisms are very similar, because they involve phosphorylation of the shared MRLC and interaction between conserved ACDs, which exhibit ~80% identity and ~88% homology between respective NMIIA and NMIIB sequences. Accordingly, NMII paralogs copolymerize in cells [53, 67]. The similar assembly properties explain equivalent incorporation of NMIIA and NMIIB into nascent homo- and heterotypic NMII filaments, which likely occurs according to availability of respective polymerization-competent monomers.
Current ideas about the NMII disassembly mechanisms, on the other hand, implicate the most divergent sequences of the NMII heavy chain – the nonhelical tailpiece [16]. Consistent with this, NMII paralogs exhibit different turnover rates in cells. Specifically, NMIIB was found to exhibit slower rates of fluorescence recovery after photobleaching and larger immobile fractions than NMIIA [70–72]. Analysis of NMIIA/NMIIB chimeras revealed that these differences in NMII turnover depended on the C-terminus of the NMII heavy chain [70]. These findings support the idea that NMII turnover is controlled by the divergent NMII tails, which can be affected by distinct depolymerization mechanisms, such as phosphorylation or binding of interaction partners, to stimulate dissociation of specific NMII subunits from either homotypic or heterotypic bipolar NMII filaments.
Shifting the NMII dynamics toward disassembly often correlates with increased cell migration and invasion, likely due to an increased NMII turnover, which promotes cytoskeleton reorganization. Such behavior was observed upon phosphorylation of S1916 [73] or S1943 [39] on NMIIA. Similarly, several accelerators of NMIIA disassembly positively regulate cell migration and are often upregulated in cancer cells [16, 42, 43, 74]. Phosphomimetic mutations in the NMIIB tailpiece compromised stability of actomyosin bundles and induced protrusive activity at the cell rear [38].
Mechanistic model of NMII self-sorting
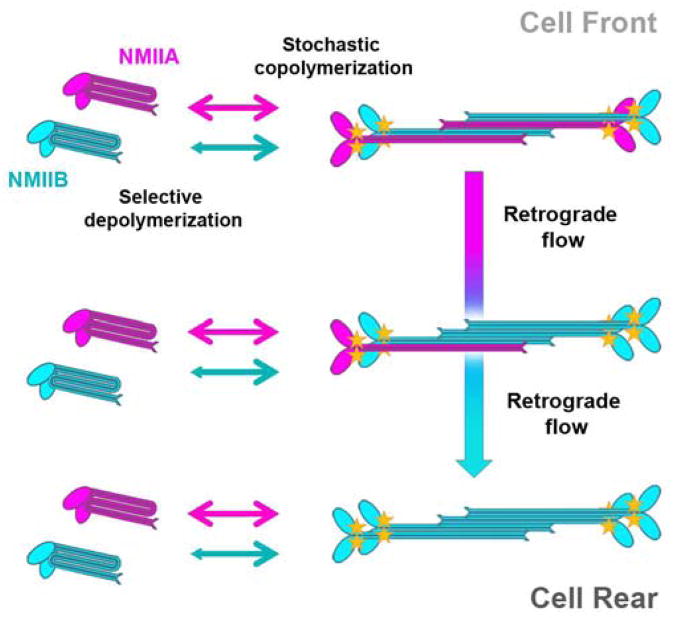
The higher rates of NMIIA turnover between monomeric and polymeric states, as compared with that of NMIIB, suggest that NMIIA subunits would dissociate from NMIIA/NMIIB heteropolymers more readily than NMIIB, because of the differences in their tail sequences targeted by the regulators of bipolar filament disassembly. However, new subunits should be added proportionally to the abundance of each paralog in the monomer pool due to their similar assembly properties. Repeating cycles of preferential dissociation of NMIIA subunits and unselective recruitment of new subunits will gradually increase the fraction of NMIIB in the older NMII filaments. Because NMII filaments, as a component of stress fibers, undergo retrograde flow over time, the older NMIIB-enriched filaments become concentrated farther away from the leading edge, as compared with the younger NMIIA-enriched filaments, thus generating the polarized anterior-posterior NMIIA-NMIIB distribution (Fig. 3). This mechanism also explains why NMII chimeras are sorted according to the identity of their C-terminal tails [70]. Since NMIIB is less dynamic, a substantial fraction of NMIIB becomes sequestered in the long-lived stress fibers at steady state, thus decreasing the pool of NMIIB monomers and exacerbating enrichment of NMIIA in nascent stress fibers.
Figure 3.
Self-sorting of NMIIA and NMIIB paralogs during front-back cell polarization, modified from [61]. Monomers of NMIIA (magenta) and NMIIB (blue) incorporate into bipolar filaments with equal efficiency (forward arrows), while the dissociation rates (reverse arrows) are greater for NMIIA than for NMIIB. Faster dissociation of NMIIA subunits together with equivalent addition of new NMIIA and NMIIB subunits leads to gradual enrichment of NMIIB in old filaments that accumulate at the cell rear due to retrograde flow.
Different motor properties of NMII paralogs can also contribute to paralog segregation. Fast motility and a short duty ratio of NMIIA can accelerate NMIIA dynamics by allowing the dissociated subunits to quickly diffuse away from the parent filament. Conversely, because of its high duty ratio, which further increases under resisting load [6, 8], NMIIB spends much time bound to actin [75], which can further reduce its turnover. Consistent with this idea, NMIIB polarizes toward the cell rear only on stiff, but not soft substrates [72], suggesting that tension generated by the cell augments paralog segregation.
Distinct properties of actin-NMIIA and actin-NMIIB arrays
Different dynamics and kinetics of NMII paralogs translate into different properties of actin-NMII structures formed by these paralogs [61]. For example, NMIIB favors formation of stable and long-lived ventral stress fibers, whereas NMIIA promotes formation of highly dynamic transverse arcs and radial stress fibers. When different paralogs are present simultaneously in cells, their copolymerization allows for the formation of bipolar filaments with a continuous range of dynamic properties between the extremes characteristic for homotypic NMII filaments. In cells expressing NMIIA, NMIIB exhibits faster dynamics and acquires more disperse distribution compared with cells lacking NMIIA [61]. On the other hand, by forming mixed filaments with NMIIA, NMIIB makes them more processive runners in vitro than the NMIIA-only filaments [7].
Functions in cells
The main function of NMII in cells is generation of contractile forces, which are used in most cell types for many purposes. NMII is best known to function in cell migration, where it contributes to regulation of leading edge protrusion, cell-substrate adhesion, cell body translocation and cell polarity. NMII is also important for cytokinesis, remodeling of the extracellular matrix (ECM), formation of cell-cell adhesions and cell shape determination [76]. Novel NMII roles have been recently revealed in endocytosis, exocytosis and vesicular transport, some of which appear to include functions of motor-active NMII monomers.
Cell-ECM adhesion
Cell adhesion to ECM provides traction to migrating cells. It is typically mediated by adhesion receptors of the integrin family. Integrin-mediated adhesion exhibits catch-bond behavior due to mechanosensitive properties of the adhesion complex [77]. The adhesion-strengthening force is largely generated by NMII, which pulls on actin filaments anchored to integrins through adaptor proteins. In some cell types, this mechanism can lead to the formation of large focal adhesions at the tips of stress fibers. The size and fate of a focal adhesion depends both on how strongly NMII pulls on the attached actin bundle and to what extent actin polymerization at the focal adhesion alleviates the tension. For example, fast actin polymerization at the ends of radial stress fibers delays maturation of the associated focal adhesions until actin elongation is inhibited through phosphorylation of VASP [68].
As a fast motor able to better cope with rapid actin polymerization, NMIIA is expected to have greater contribution to focal adhesion assembly near the leading edge, whereas NMIIB is better posed to stabilize focal adhesions in more central cellular regions, where NMIIB-dependent isometric tension could be sufficient [78]. Nonetheless, NMIIB or NMIIC are able to initiate adhesion formation in cells lacking NMIIA. Compared with NMIIB, NMIIA is more capable of traction force generation, so that NMIIA depletion dramatically decreases the forces that the cell exerts on the substratum [61, 79, 80].
The roles of NMIIC are less understood, but could involve either positive [81] or negative [33, 82] regulation of cell adhesions. The C2 splice variant of NMIIC was found to interact and colocalize with β1 integrin and positively regulate adhesion in neuroblastoma cells [81]. NMIIA also interacts with some leukocyte-specific integrins, among which NMIIA interaction with α4-integrin was detected only at high salt conditions, when NMIIA filaments dissociate, suggesting a possible role of NMIIA monomers in adhesion regulation [83, 84].
Leading edge protrusion
Leading edge protrusion is driven by polymerization of actin filaments that push against the plasma membrane. The membrane resistance results in actin retrograde flow, which is also facilitated by NMII, especially in the cell lamella behind protrusions. How far the leading edge advances depends on a difference between rates of actin polymerization and retrograde flow. Accordingly, NMII activity negatively regulates leading edge protrusion [85–87]. On the other hand, efficient protrusion requires traction that is enabled by adhesion, which is a mechanosensitive process. Nascent adhesions are formed underneath lamellipodia and their formation is mechanically stimulated by retrograde flow [88, 89]. Although NMII appears dispensable for adhesion initiation in some cases [90], NMII motor activity contributes to adhesion initiation and productive leading edge advance in other situations [52]. In the latter case, the underlying mechanism was proposed to extend beyond the classic idea of bipolar filament-mediated contraction and involve motor-active individual NMII molecules.
The negative and positive roles of NMII in protrusion appear to be preferentially played by specific NMII paralogs. For example, NMIIA enables neurite retraction in cultured neuronal cells [91]. In non-neuronal cells, increased expression of NMIIA correlated with reduced cell spreading, probably, due to increased contraction [78, 79], while NMIIA-dependent periodic contractions in the lamella correlated with pauses in lamellipodium advance [92]. Conversely, NMIIB supported axon elongation in neurons [93], as well as cell spreading and lamellipodial protrusion in other cell types [78]. NMIIC also stimulated neurite outgrowth in neuroblastoma cells [82] and lamellipodial protrusion in epithelial cells [75].
Contractile forces
During cell migration, NMII-mediated contraction helps to detach obsolete adhesions, retract the cell rear, and translocate forward the cell body [55, 94, 95]. In neurons, NMII-mediated contraction enables consolidation of the axonal shaft behind the advancing growth cone [96], axon retraction in response to repulsive signals [97], and growth cone turning through its asymmetric retraction [98]. Similar contractile forces applied to compliant ECM contribute to ECM remodeling [99]. This function is particularly characteristic for fibroblasts – mesenchymal cells that organize the ECM in tissues. NMIIA is mainly responsible for generating large contractile forces for cell rear retraction [100] and ECM remodeling [101]. Contribution of NMIIB to these processes is minimal in 2D cultures, but becomes significant in 3D environment, where it promotes translocation of the nucleus through tight spaces [80, 102]. In neurons, however, growth cone retraction in response to a chemorepellent relied primarily on NMIIB [97].
Cell polarity
In migrating cells, actin-NMII bundles and networks undergo retrograde flow and accumulate at the cell rear, where they inhibit protrusive activity of lateral and posterior cell edges, thus supporting front-back cell polarity. This function is thought to largely depend on NMIIB, as it can maintain stable stress fibers [71, 85]. Preferential accumulation of NMIIB at the cell rear in the course of self-segregation of NMII paralogs facilitates the establishment of this polarity. However, excessive expression of NMIIB results in over-stabilized stress fibers and focal adhesions, which retard cell migration [61]. In cases of amoeboid type of cell migration, which is characterized by weak cell-substrate adhesions and an absence of stress fibers (for example, in neutrophils, which do not express NMIIB), NMIIA is responsible for the formation of a stable rear end (called uropod in neutrophils) [103].
Cytokinesis
The constriction of cleavage furrow during cytokinesis is another important NMII-dependent function [104]. Similar to interphase cells, NMII in mature cytokinetic contractile rings is organized into bipolar filaments arranged into stacks and aligned with the contraction axis. However, at earlier stages of cytokinesis, the assembly of the contractile ring follows the network contraction mechanism [57, 60]. Interestingly, motor-impaired mutants of NMIIA and NMIIB that are still able to bind actin filaments in an ATP-dependent manner were able to rescue cytokinesis defects in NMIIB-depleted COS-7 cells, which do not endogenously express NMIIA, although it is not clear how NMII-mediated crosslinking drives constriction of the cleavage furrow [105].
In general, each of NMII paralogs can execute cytokinesis [2]. The cellular preference in employing specific NMII paralogs for cytokinesis depends on their relative abundance in individual cell types and/or efficiency of their recruitment to the cleavage furrow. For example, in immature dividing megakaryocytes, which express both NMIIA and NMIIB, only NMIIB was recruited to the cleavage furrow, because NMIIB could respond to lower levels of RhoA activation than NMIIA [106]. The recruitment to the furrow of NMIIB in this system [107] or NMIIA in COS-7 cells [108] depended on the C-terminal heavy chain regions and did not require the motor domain suggesting an actin-independent targeting mechanism for NMII at the cleavage furrow.
Cell shape
Actin–NMII arrays define cell shape and mechanical properties of the cell surface. Here, NMII can function both as a cross-linker to generate isometric tension and as a motor to maintain dynamic actin-NMII networks. For example, in epithelial monolayers, circumferential actin-NMII bundles associated with apical adherens junctions generate tension to preserve junction integrity [109, 110] and stabilize the constricted shape of the apical domain during epithelium invagination. The apical constriction itself is driven by pulsed contractions of actin-NMII networks in the plane of the apical domain [56, 111], although other contractile mechanisms also contribute [112, 113]. The contractile forces at cell-cell junctions are counterbalanced by pushing forces generated by Arp2/3 complex-dependent polymerization of branched actin networks, which are required to maintain or expand the junction [110, 113].
The spherical shape and high cortical tension of mitotic cells are maintained by submembrane actin networks jointly assembled by NMII [114] and actin nucleators [115], suggesting their dynamic nature. An actin-NMII cytoskeleton at the dorsal surface of cultured cells contains both bundles and networks and undergoes constant remodeling between these states accompanied by corresponding changes in the mechanical properties of the surface [116].
Participation of stable and dynamic actin-NMII arrays in cell shape determination suggests contribution of NMIIB and NMIIA, respectively. In some epithelial cells, the relatively stable circumferential actin-NMII bundles at apical cell-cell junctions indeed required NMIIB functions [117–119], although NMIIA is also important [120], especially for the initial assembly of adherens junctions [117]. In individual cells, NMIIB and NMIIC promote stability of the cell cortex, which helps to reduce formation of surface blebs. In contrast, NMIIA has greater contribution to the cortex stiffness, contractility and bleb formation [75].
Membrane trafficking
Recent data increasingly point to roles of NMII in membrane organelle morphogenesis, such as exocytosis [121], endocytosis [122], post-Golgi and Golgi-to-ER trafficking [123, 124], and mitochondrion fission [125].
Roles of NMII in exocytosis are especially conspicuous during secretion of viscous cargos, such as salivary mucus [126, 127], lung surfactant [128], and endothelial von Willebrand factor [129, 130]. In these cases, NMII is thought to squeeze the cargo from the secretory vesicle. In salivary glands, both NMIIA and NMIIB are important for different aspects of this function. NMIIB prevents counterproductive expansion of the secretory granule immediately after its fusion, whereas NMIIA stimulates subsequent cargo expulsion [127].
In some membrane trafficking events, NMII might function in a monomeric form. For example, monomers of NMIIA have been found in association with lytic granules in the natural killer cells, where they promoted granule secretion [51], and with Golgi membranes isolated from the intestinal epithelium [123]. The association of NMIIA with the Golgi complex was mediated by its coiled coil rod [131]. It remains unclear whether this interaction is compatible with the rod-mediated NMII filament assembly.
Conclusions
Proper accomplishment of virtually every NMII mission requires fine tuning of the balance between the active contraction and tension maintenance. This task can be achieved through combinatorial engagement of NMII paralogs with distinct dynamic properties. The available data suggest that dynamic features of NMII paralogs often can be correlated with their functions in cells. In general, NMIIA is responsible for fast and powerful force generation in response to changing conditions, whereas NMIIB is more suitable to maintain long-lasting stresses and ensure cytoskeleton stability. Too little is known so far about NMIIC functions to propose what might be special about this paralog. Future research will bring new insights into paralog-specific regulation of NMII expression, intracellular dynamics, interactions with other proteins, and functions.
Highlights.
Nonmuscle myosin II executes numerous mechanical tasks in cells including organization of the actin cytoskeleton, cell adhesion and migration
Three mammalian nonmuscle myosin II paralogs have distinct kinetic and dynamic properties in vitro and in vivo
Mammalian nonmuscle myosin II paralogs mix and match their abilities to perform both specialized and joint tasks in the cell
Acknowledgments
This work was supported by National Institutes of Health grant GM095977 to TMS. The authors declare no competing financial interests.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Golomb E, Ma X, Jana SS, Preston YA, Kawamoto S, Shoham NG, Goldin E, Conti MA, Sellers JR, Adelstein RS. Identification and characterization of nonmuscle myosin II-C, a new member of the myosin II family. J Biol Chem. 2004;279:2800–2808. doi: 10.1074/jbc.M309981200. [DOI] [PubMed] [Google Scholar]
- 2.Ma X, Adelstein RS. The role of vertebrate nonmuscle Myosin II in development and human disease. Bioarchitecture. 2014;4:88–102. doi: 10.4161/bioa.29766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Heissler SM, Sellers JR. Kinetic adaptations of myosins for their diverse cellular functions. Traffic. 2016;17:839–859. doi: 10.1111/tra.12388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Heissler SM, Sellers JR. Various themes of myosin regulation. J Mol Biol. 2016;428:1927–1946. doi: 10.1016/j.jmb.2016.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Peckham M. How myosin organization of the actin cytoskeleton contributes to the cancer phenotype. Biochem Soc Trans. 2016;44:1026–1034. doi: 10.1042/BST20160034. [DOI] [PubMed] [Google Scholar]
- 6.Nagy A, Takagi Y, Billington N, Sun SA, Hong DK, Homsher E, Wang A, Sellers JR. Kinetic characterization of nonmuscle Myosin IIB at the single molecule level. J Biol Chem. 2013;288:709–722. doi: 10.1074/jbc.M112.424671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Melli L, Billington N, Sun SA, Bird JE, Nagy A, Friedman TB, Takagi Y, Sellers JR. Bipolar filaments of human nonmuscle myosin 2-A and 2-B have distinct motile and mechanical properties. eLife. 2018;7:e32871. doi: 10.7554/eLife.32871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rosenfeld SS, Xing J, Chen LQ, Sweeney HL. Myosin IIb is unconventionally conventional. J Biol Chem. 2003;278:27449–27455. doi: 10.1074/jbc.M302555200. [DOI] [PubMed] [Google Scholar]
- 9.Greenberg MJ, Arpag G, Tuzel E, Ostap EM. A perspective on the role of myosins as mechanosensors. Biophys J. 2016;110:2568–2576. doi: 10.1016/j.bpj.2016.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kovacs M, Thirumurugan K, Knight PJ, Sellers JR. Load-dependent mechanism of nonmuscle myosin 2. Proc Natl Acad Sci U S A. 2007;104:9994–9999. doi: 10.1073/pnas.0701181104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Heissler SM, Manstein DJ. Comparative kinetic and functional characterization of the motor domains of human nonmuscle myosin-2C isoforms. J Biol Chem. 2011;286:21191–21202. doi: 10.1074/jbc.M110.212290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Muller M, Diensthuber RP, Chizhov I, Claus P, Heissler SM, Preller M, Taft MH, Manstein DJ. Distinct functional interactions between actin isoforms and nonsarcomeric myosins. PLoS One. 2013;8:e70636. doi: 10.1371/journal.pone.0070636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gateva G, Kremneva E, Reindl T, Kotila T, Kogan K, Gressin L, Gunning PW, Manstein DJ, Michelot A, Lappalainen P. Tropomyosin isoforms specify functionally distinct actin filament populations in vitro. Curr Biol. 2017;27:705–713. doi: 10.1016/j.cub.2017.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ge J, Bouriyaphone SD, Serebrennikova TA, Astashkin AV, Nesmelov YE. Macromolecular crowding modulates actomyosin kinetics. Biophys J. 2016;111:178–184. doi: 10.1016/j.bpj.2016.05.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nakasawa T, Takahashi M, Matsuzawa F, Aikawa S, Togashi Y, Saitoh T, Yamagishi A, Yazawa M. Critical regions for assembly of vertebrate nonmuscle myosin II. Biochemistry. 2005;44:174–183. doi: 10.1021/bi048807h. [DOI] [PubMed] [Google Scholar]
- 16.Dulyaninova NG, Bresnick AR. The heavy chain has its day: regulation of myosin-II assembly. Bioarchitecture. 2013;3:77–85. doi: 10.4161/bioa.26133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Billington N, Wang A, Mao J, Adelstein RS, Sellers JR. Characterization of three full-length human nonmuscle myosin II paralogs. J Biol Chem. 2013;288:33398–33410. doi: 10.1074/jbc.M113.499848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu X, Billington N, Shu S, Yu SH, Piszczek G, Sellers JR, Korn ED. Effect of ATP and regulatory light-chain phosphorylation on the polymerization of mammalian nonmuscle myosin II. Proc Natl Acad Sci U S A. 2017;114:E6516–E6525. doi: 10.1073/pnas.1702375114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rosenberg M, Straussman R, Ben-Ya’acov A, Ronen D, Ravid S. MHC-IIB filament assembly and cellular localization are governed by the rod net charge. PLoS One. 2008;3:e1496. doi: 10.1371/journal.pone.0001496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Salzameda B, Facemyer KC, Beck BW, Cremo CR. The N-terminal lobes of both regulatory light chains interact with the tail domain in the 10 S-inhibited conformation of smooth muscle myosin. J Biol Chem. 2006;281:38801–38811. doi: 10.1074/jbc.M606555200. [DOI] [PubMed] [Google Scholar]
- 21.Burgess SA, Yu S, Walker ML, Hawkins RJ, Chalovich JM, Knight PJ. Structures of smooth muscle myosin and heavy meromyosin in the folded, shutdown state. J Mol Biol. 2007;372:1165–1178. doi: 10.1016/j.jmb.2007.07.014. [DOI] [PubMed] [Google Scholar]
- 22.Jung HS, Billington N, Thirumurugan K, Salzameda B, Cremo CR, Chalovich JM, Chantler PD, Knight PJ. Role of the tail in the regulated state of Myosin 2. J Mol Biol. 2011;408:863–878. doi: 10.1016/j.jmb.2011.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Heissler SM, Sellers JR. Myosin light chains: Teaching old dogs new tricks. Bioarchitecture. 2014;4:169–188. doi: 10.1080/19490992.2015.1054092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nishikawa M, Sellers JR, Adelstein RS, Hidaka H. Protein kinase C modulates in vitro phosphorylation of the smooth muscle heavy meromyosin by myosin light chain kinase. J Biol Chem. 1984;259:8808–8814. [PubMed] [Google Scholar]
- 25.Komatsu S, Ikebe M. The phosphorylation of myosin II at the Ser1 and Ser2 is critical for normal platelet-derived growth factor induced reorganization of myosin filaments. Mol Biol Cell. 2007;18:5081–5090. doi: 10.1091/mbc.E06-12-1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Asokan SB, Johnson HE, Rahman A, King SJ, Rotty JD, Lebedeva IP, Haugh JM, Bear JE. Mesenchymal chemotaxis requires selective inactivation of myosin II at the leading edge via a noncanonical PLCgamma/PKCalpha pathway. Dev Cell. 2014;31:747–760. doi: 10.1016/j.devcel.2014.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Beach JR, Licate LS, Crish JF, Egelhoff TT. Analysis of the role of Ser1/Ser2/Thr9 phosphorylation on myosin II assembly and function in live cells. BMC Cell Biol. 2011;12:52. doi: 10.1186/1471-2121-12-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Beach JR, Hammer JA., 3rd Myosin II isoform co-assembly and differential regulation in mammalian systems. Exp Cell Res. 2015;334:2–9. doi: 10.1016/j.yexcr.2015.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chang CW, Kumar S. Differential contributions of nonmuscle myosin II isoforms and functional domains to stress fiber mechanics. Sci Rep. 2015;5:13736. doi: 10.1038/srep13736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Breckenridge MT, Dulyaninova NG, Egelhoff TT. Multiple regulatory steps control mammalian nonmuscle myosin II assembly in live cells. Mol Biol Cell. 2009;20:338–347. doi: 10.1091/mbc.E08-04-0372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kiboku T, Katoh T, Nakamura A, Kitamura A, Kinjo M, Murakami Y, Takahashi M. Nonmuscle myosin II folds into a 10S form via two portions of tail for dynamic subcellular localization. Genes Cells. 2013;18:90–109. doi: 10.1111/gtc.12021. [DOI] [PubMed] [Google Scholar]
- 32.Murakami N, Kotula L, Hwang YW. Two distinct mechanisms for regulation of nonmuscle myosin assembly via the heavy chain: phosphorylation for MIIB and mts 1 binding for MIIA. Biochemistry. 2000;39:11441–11451. doi: 10.1021/bi000347e. [DOI] [PubMed] [Google Scholar]
- 33.Ronen D, Ravid S. Myosin II tailpiece determines its paracrystal structure, filament assembly properties, and cellular localization. J Biol Chem. 2009;284:24948–24957. doi: 10.1074/jbc.M109.023754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rosenberg MM, Ronen D, Lahav N, Nazirov E, Ravid S, Friedler A. High resolution characterization of myosin IIC protein tailpiece and its effect on filament assembly. J Biol Chem. 2013;288:9779–9789. doi: 10.1074/jbc.M112.430173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Betapudi V, Gokulrangan G, Chance MR, Egelhoff TT. A proteomic study of myosin II motor proteins during tumor cell migration. J Mol Biol. 2011;407:673–686. doi: 10.1016/j.jmb.2011.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rosenberg M, Ravid S. Protein kinase Cgamma regulates myosin IIB phosphorylation, cellular localization, and filament assembly. Mol Biol Cell. 2006;17:1364–1374. doi: 10.1091/mbc.E05-07-0597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Even-Faitelson L, Ravid S. PAK1 and aPKCzeta regulate myosin II-B phosphorylation: a novel signaling pathway regulating filament assembly. Mol Biol Cell. 2006;17:2869–2881. doi: 10.1091/mbc.E05-11-1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Juanes-Garcia A, Chapman JR, Aguilar-Cuenca R, Delgado-Arevalo C, Hodges J, Whitmore LA, Shabanowitz J, Hunt DF, Horwitz AR, Vicente-Manzanares M. A regulatory motif in nonmuscle myosin II-B regulates its role in migratory front-back polarity. J Cell Biol. 2015;209:23–32. doi: 10.1083/jcb.201407059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dulyaninova NG, House RP, Betapudi V, Bresnick AR. Myosin-IIA heavy-chain phosphorylation regulates the motility of MDA-MB-231 carcinoma cells. Mol Biol Cell. 2007;18:3144–3155. doi: 10.1091/mbc.E06-11-1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ramagopal UA, Dulyaninova NG, Varney KM, Wilder PT, Nallamsetty S, Brenowitz M, Weber DJ, Almo SC, Bresnick AR. Structure of the S100A4/myosin-IIA complex. BMC Struct Biol. 2013;13:31. doi: 10.1186/1472-6807-13-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kiss B, Kalmar L, Nyitray L, Pal G. Structural determinants governing S100A4-induced isoform-selective disassembly of nonmuscle myosin II filaments. FEBS J. 2016;283:2164–2180. doi: 10.1111/febs.13728. [DOI] [PubMed] [Google Scholar]
- 42.Du M, Wang G, Ismail TM, Gross S, Fernig DG, Barraclough R, Rudland PS. S100P dissociates myosin IIA filaments and focal adhesion sites to reduce cell adhesion and enhance cell migration. J Biol Chem. 2012;287:15330–15344. doi: 10.1074/jbc.M112.349787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dahan I, Yearim A, Touboul Y, Ravid S. The tumor suppressor Lgl1 regulates NMII-A cellular distribution and focal adhesion morphology to optimize cell migration. Mol Biol Cell. 2012;23:591–601. doi: 10.1091/mbc.E11-01-0015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Solinet S, Akpovi CD, Garcia CJ, Barry A, Vitale ML. Myosin IIB deficiency in embryonic fibroblasts affects regulators and core members of the par polarity complex. Histochem Cell Biol. 2011;136:245–266. doi: 10.1007/s00418-011-0840-0. [DOI] [PubMed] [Google Scholar]
- 45.Billington N, Beach JR, Heissler SM, Remmert K, Guzik-Lendrum S, Nagy A, Takagi Y, Shao L, Li D, Yang Y, Zhang Y, Barzik M, Betzig E, Hammer JA, 3rd, Sellers JR. Myosin 18A coassembles with nonmuscle myosin 2 to form mixed bipolar filaments. Curr Biol. 2015;25:942–948. doi: 10.1016/j.cub.2015.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Arora PD, Wang Y, Janmey PA, Bresnick A, Yin HL, McCulloch CA. Gelsolin and non-muscle myosin IIA interact to mediate calcium-regulated collagen phagocytosis. J Biol Chem. 2011;286:34184–34198. doi: 10.1074/jbc.M111.247783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Arora PD, Wang Y, Bresnick A, Janmey PA, McCulloch CA. Flightless I interacts with NMMIIA to promote cell extension formation, which enables collagen remodeling. Mol Biol Cell. 2015;26:2279–2297. doi: 10.1091/mbc.E14-11-1536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chen PW, Jian X, Heissler SM, Le K, Luo R, Jenkins LM, Nagy A, Moss J, Sellers JR, Randazzo PA. The Arf GTPase-activating protein, ASAP1, binds nonmuscle myosin 2A to control remodeling of the actomyosin network. J Biol Chem. 2016;291:7517–7526. doi: 10.1074/jbc.M115.701292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sabbir MG, Dillon R, Mowat MR. Dlc1 interaction with non-muscle myosin heavy chain II-A (Myh9) and Rac1 activation. Biol Open. 2016;5:452–460. doi: 10.1242/bio.015859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Feng J, Hu Z, Chen H, Hua J, Wu R, Dong Z, Qiang L, Liu Y, Baas PW, Liu M. Depletion of kinesin-12, a myosin-IIB-interacting protein, promotes migration of cortical astrocytes. J Cell Sci. 2016;129:2438–2447. doi: 10.1242/jcs.181867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sanborn KB, Mace EM, Rak GD, Difeo A, Martignetti JA, Pecci A, Bussel JB, Favier R, Orange JS. Phosphorylation of the myosin IIA tailpiece regulates single myosin IIA molecule association with lytic granules to promote NK-cell cytotoxicity. Blood. 2011;118:5862–5871. doi: 10.1182/blood-2011-03-344846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shutova M, Yang C, Vasiliev JM, Svitkina T. Functions of nonmuscle myosin II in assembly of the cellular contractile system. PLoS One. 2012;7:e40814. doi: 10.1371/journal.pone.0040814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shutova MS, Spessott WA, Giraudo CG, Svitkina T. Endogenous species of mammalian nonmuscle myosin IIA and IIB include activated monomers and heteropolymers. Curr Biol. 2014;24:1958–1968. doi: 10.1016/j.cub.2014.07.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Verkhovsky AB, Svitkina TM, Borisy GG. Myosin II filament assemblies in the active lamella of fibroblasts: their morphogenesis and role in the formation of actin filament bundles. J Cell Biol. 1995;131:989–1002. doi: 10.1083/jcb.131.4.989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Svitkina TM, Verkhovsky AB, McQuade KM, Borisy GG. Analysis of the actin-myosin II system in fish epidermal keratocytes: mechanism of cell body translocation. J Cell Biol. 1997;139:397–415. doi: 10.1083/jcb.139.2.397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Martin AC. Pulsation and stabilization: contractile forces that underlie morphogenesis. Dev Biol. 2010;341:114–125. doi: 10.1016/j.ydbio.2009.10.031. [DOI] [PubMed] [Google Scholar]
- 57.Spira F, Cuylen-Haering S, Mehta S, Samwer M, Reversat A, Verma A, Oldenbourg R, Sixt M, Gerlich DW. Cytokinesis in vertebrate cells initiates by contraction of an equatorial actomyosin network composed of randomly oriented filaments. Elife. 2017;6:e30867. doi: 10.7554/eLife.30867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Baird MA, Billington N, Wang A, Adelstein RS, Sellers JR, Fischer RS, Waterman CM. Local pulsatile contractions are an intrinsic property of the myosin 2A motor in the cortical cytoskeleton of adherent cells. Mol Biol Cell. 2017;28:240–251. doi: 10.1091/mbc.E16-05-0335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Burnette DT, Manley S, Sengupta P, Sougrat R, Davidson MW, Kachar B, Lippincott-Schwartz J. A role for actin arcs in the leading-edge advance of migrating cells. Nat Cell Biol. 2011;13:371–382. doi: 10.1038/ncb2205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Henson JH, Ditzler CE, Germain A, Irwin PM, Vogt ET, Yang S, Wu X, Shuster CB. The ultrastructural organization of actin and myosin II filaments in the contractile ring: new support for an old model of cytokinesis. Mol Biol Cell. 2017;28:613–623. doi: 10.1091/mbc.E16-06-0466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Shutova MS, Asokan SB, Talwar S, Assoian RK, Bear JE, Svitkina TM. Self-sorting of nonmuscle myosins IIA and IIB polarizes the cytoskeleton and modulates cell motility. J Cell Biol. 2017;216:2877–2889. doi: 10.1083/jcb.201705167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Schwayer C, Sikora M, Slovakova J, Kardos R, Heisenberg CP. Actin rings of power. Dev Cell. 2016;37:493–506. doi: 10.1016/j.devcel.2016.05.024. [DOI] [PubMed] [Google Scholar]
- 63.Svitkina T. The actin cytoskeleton and actin-based motility. Cold Spring Harb Perspect Biol. 2018;10:a018267. doi: 10.1101/cshperspect.a018267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tee YH, Shemesh T, Thiagarajan V, Hariadi RF, Anderson KL, Page C, Volkmann N, Hanein D, Sivaramakrishnan S, Kozlov MM, Bershadsky AD. Cellular chirality arising from the self-organization of the actin cytoskeleton. Nat Cell Biol. 2015;17:445–457. doi: 10.1038/ncb3137. [DOI] [PubMed] [Google Scholar]
- 65.Maupin P, Phillips CL, Adelstein RS, Pollard TD. Differential localization of myosin-II isozymes in human cultured cells and blood cells. J Cell Sci. 1994;107(Pt 11):3077–3090. doi: 10.1242/jcs.107.11.3077. [DOI] [PubMed] [Google Scholar]
- 66.Kolega J. Asymmetric distribution of myosin IIB in migrating endothelial cells is regulated by a rho-dependent kinase and contributes to tail retraction. Mol Biol Cell. 2003;14:4745–4757. doi: 10.1091/mbc.E03-04-0205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Beach JR, Shao L, Remmert K, Li D, Betzig E, Hammer JA., 3rd Nonmuscle myosin II isoforms coassemble in living cells. Curr Biol. 2014;24:1160–1166. doi: 10.1016/j.cub.2014.03.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Tojkander S, Gateva G, Husain A, Krishnan R, Lappalainen P. Generation of contractile actomyosin bundles depends on mechanosensitive actin filament assembly and disassembly. Elife. 2015;4:e06126. doi: 10.7554/eLife.06126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hotulainen P, Lappalainen P. Stress fibers are generated by two distinct actin assembly mechanisms in motile cells. J Cell Biol. 2006;173:383–394. doi: 10.1083/jcb.200511093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sandquist JC, Means AR. The C-terminal tail region of nonmuscle myosin II directs isoform-specific distribution in migrating cells. Mol Biol Cell. 2008;19:5156–5167. doi: 10.1091/mbc.E08-05-0533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Vicente-Manzanares M, Koach MA, Whitmore L, Lamers ML, Horwitz AF. Segregation and activation of myosin IIB creates a rear in migrating cells. J Cell Biol. 2008;183:543–554. doi: 10.1083/jcb.200806030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Raab M, Swift J, PDPC, Shah P, Shin JW, Discher DE. Crawling from soft to stiff matrix polarizes the cytoskeleton and phosphoregulates myosin-II heavy chain. J Cell Biol. 2012;199:669–683. doi: 10.1083/jcb.201205056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Pasapera AM, Plotnikov SV, Fischer RS, Case LB, Egelhoff TT, Waterman CM. Rac1-dependent phosphorylation and focal adhesion recruitment of myosin IIA regulates migration and mechanosensing. Curr Biol. 2015;25:175–186. doi: 10.1016/j.cub.2014.11.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Chen Y, Yu Y, Sun S, Wang Z, Liu P, Liu S, Jiang J. Bradykinin promotes migration and invasion of hepatocellular carcinoma cells through TRPM7 and MMP2. Exp Cell Res. 2016;349:68–76. doi: 10.1016/j.yexcr.2016.09.022. [DOI] [PubMed] [Google Scholar]
- 75.Dey SK, Singh RK, Chattoraj S, Saha S, Das A, Bhattacharyya K, Sengupta K, Sen S, Jana SS. Differential role of nonmuscle myosin II isoforms during blebbing of MCF-7 cells. Mol Biol Cell. 2017;28:1034–1042. doi: 10.1091/mbc.E16-07-0524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Heissler SM, Manstein DJ. Nonmuscle myosin-2: mix and match. Cell Mol Life Sci. 2013;70:1–21. doi: 10.1007/s00018-012-1002-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Sun Z, Guo SS, Fassler R. Integrin-mediated mechanotransduction. J Cell Biol. 2016;215:445–456. doi: 10.1083/jcb.201609037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Betapudi V. Myosin II motor proteins with different functions determine the fate of lamellipodia extension during cell spreading. PLoS One. 2010;5:e8560. doi: 10.1371/journal.pone.0008560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Cai Y, Biais N, Giannone G, Tanase M, Jiang G, Hofman JM, Wiggins CH, Silberzan P, Buguin A, Ladoux B, Sheetz MP. Nonmuscle myosin IIA-dependent force inhibits cell spreading and drives F-actin flow. Biophys J. 2006;91:3907–3920. doi: 10.1529/biophysj.106.084806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Thomas DG, Yenepalli A, Denais CM, Rape A, Beach JR, Wang YL, Schiemann WP, Baskaran H, Lammerding J, Egelhoff TT. Non-muscle myosin IIB is critical for nuclear translocation during 3D invasion. J Cell Biol. 2015;210:583–594. doi: 10.1083/jcb.201502039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Saha S, Dey SK, Biswas A, Das P, Das MR, Jana SS. The effect of including the C2 insert of nonmuscle myosin II-C on neuritogenesis. J Biol Chem. 2013;288:7815–7828. doi: 10.1074/jbc.M112.417196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wylie SR, Chantler PD. Myosin IIC: a third molecular motor driving neuronal dynamics. Mol Biol Cell. 2008;19:3956–3968. doi: 10.1091/mbc.E07-08-0744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Morin NA, Oakes PW, Hyun YM, Lee D, Chin YE, King MR, Springer TA, Shimaoka M, Tang JX, Reichner JS, Kim M. Nonmuscle myosin heavy chain IIA mediates integrin LFA-1 de-adhesion during T lymphocyte migration. J Exp Med. 2008;205:195–205. doi: 10.1084/jem.20071543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Rosado LA, Horn TA, McGrath SC, Cotter RJ, Yang JT. Association between {alpha}4 integrin cytoplasmic tail and non-muscle myosin IIA regulates cell migration. J Cell Sci. 2011;124:483–492. doi: 10.1242/jcs.074211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Lo CM, Buxton DB, Chua GC, Dembo M, Adelstein RS, Wang YL. Nonmuscle myosin IIb is involved in the guidance of fibroblast migration. Mol Biol Cell. 2004;15:982–989. doi: 10.1091/mbc.E03-06-0359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Betapudi V, Licate LS, Egelhoff TT. Distinct roles of nonmuscle myosin II isoforms in the regulation of MDA-MB-231 breast cancer cell spreading and migration. Cancer Res. 2006;66:4725–4733. doi: 10.1158/0008-5472.CAN-05-4236. [DOI] [PubMed] [Google Scholar]
- 87.Shutova MS, Alexandrova AY, Vasiliev JM. Regulation of polarity in cells devoid of actin bundle system after treatment with inhibitors of myosin II activity. Cell Motil Cytoskeleton. 2008;65:734–746. doi: 10.1002/cm.20295. [DOI] [PubMed] [Google Scholar]
- 88.Alexandrova AY, Arnold K, Schaub S, Vasiliev JM, Meister JJ, Bershadsky AD, Verkhovsky AB. Comparative dynamics of retrograde actin flow and focal adhesions: formation of nascent adhesions triggers transition from fast to slow flow. PLoS One. 2008;3:e3234. doi: 10.1371/journal.pone.0003234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Gardel ML, Sabass B, Ji L, Danuser G, Schwarz US, Waterman CM. Traction stress in focal adhesions correlates biphasically with actin retrograde flow speed. J Cell Biol. 2008;183:999–1005. doi: 10.1083/jcb.200810060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Choi CK, Vicente-Manzanares M, Zareno J, Whitmore LA, Mogilner A, Horwitz AR. Actin and alpha-actinin orchestrate the assembly and maturation of nascent adhesions in a myosin II motor-independent manner. Nat Cell Biol. 2008;10:1039–1050. doi: 10.1038/ncb1763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Wylie SR, Chantler PD. Myosin IIA drives neurite retraction. Mol Biol Cell. 2003;14:4654–4666. doi: 10.1091/mbc.E03-03-0187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Giannone G, Dubin-Thaler BJ, Dobereiner HG, Kieffer N, Bresnick AR, Sheetz MP. Periodic lamellipodial contractions correlate with rearward actin waves. Cell. 2004;116:431–443. doi: 10.1016/s0092-8674(04)00058-3. [DOI] [PubMed] [Google Scholar]
- 93.Bridgman PC, Dave S, Asnes CF, Tullio AN, Adelstein RS. Myosin IIB is required for growth cone motility. J Neurosci. 2001;21:6159–6169. doi: 10.1523/JNEUROSCI.21-16-06159.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Chabaud M, Heuze ML, Bretou M, Vargas P, Maiuri P, Solanes P, Maurin M, Terriac E, Le Berre M, Lankar D, Piolot T, Adelstein RS, Zhang Y, Sixt M, Jacobelli J, Benichou O, Voituriez R, Piel M, Lennon-Dumenil AM. Cell migration and antigen capture are antagonistic processes coupled by myosin II in dendritic cells. Nat Commun. 2015;6:7526. doi: 10.1038/ncomms8526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Liu YJ, Le Berre M, Lautenschlaeger F, Maiuri P, Callan-Jones A, Heuze M, Takaki T, Voituriez R, Piel M. Confinement and low adhesion induce fast amoeboid migration of slow mesenchymal cells. Cell. 2015;160:659–672. doi: 10.1016/j.cell.2015.01.007. [DOI] [PubMed] [Google Scholar]
- 96.Loudon RP, Silver LD, Yee HF, Jr, Gallo G. RhoA-kinase and myosin II are required for the maintenance of growth cone polarity and guidance by nerve growth factor. J Neurobiol. 2006;66:847–867. doi: 10.1002/neu.20258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Brown JA, Wysolmerski RB, Bridgman PC. Dorsal root ganglion neurons react to Semaphorin 3A application through a biphasic response that requires multiple myosin II isoforms. Mol Biol Cell. 2009;20:1167–1179. doi: 10.1091/mbc.E08-01-0065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Turney SG, Bridgman PC. Laminin stimulates and guides axonal outgrowth via growth cone myosin II activity. Nat Neurosci. 2005;8:717–719. doi: 10.1038/nn1466. [DOI] [PubMed] [Google Scholar]
- 99.Wolf K, Te Lindert M, Krause M, Alexander S, Te Riet J, Willis AL, Hoffman RM, Figdor CG, Weiss SJ, Friedl P. Physical limits of cell migration: control by ECM space and nuclear deformation and tuning by proteolysis and traction force. J Cell Biol. 2013;201:1069–1084. doi: 10.1083/jcb.201210152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Even-Ram S, Doyle AD, Conti MA, Matsumoto K, Adelstein RS, Yamada KM. Myosin IIA regulates cell motility and actomyosin-microtubule crosstalk. Nat Cell Biol. 2007;9:299–309. doi: 10.1038/ncb1540. [DOI] [PubMed] [Google Scholar]
- 101.Liu Z, Ho CH, Grinnell F. The different roles of myosin IIA and myosin IIB in contraction of 3D collagen matrices by human fibroblasts. Exp Cell Res. 2014;326:295–306. doi: 10.1016/j.yexcr.2014.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Beach JR, Hussey GS, Miller TE, Chaudhury A, Patel P, Monslow J, Zheng Q, Keri RA, Reizes O, Bresnick AR, Howe PH, Egelhoff TT. Myosin II isoform switching mediates invasiveness after TGF-beta-induced epithelial-mesenchymal transition. Proc Natl Acad Sci U S A. 2011;108:17991–17996. doi: 10.1073/pnas.1106499108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Hind LE, Vincent WJ, Huttenlocher A. Leading from the back: The role of the uropod in neutrophil polarization and migration. Dev Cell. 2016;38:161–169. doi: 10.1016/j.devcel.2016.06.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Mabuchi I, Okuno M. The effect of myosin antibody on the division of starfish blastomeres. J Cell Biol. 1977;74:251–263. doi: 10.1083/jcb.74.1.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Ma X, Kovacs M, Conti MA, Wang A, Zhang Y, Sellers JR, Adelstein RS. Nonmuscle myosin II exerts tension but does not translocate actin in vertebrate cytokinesis. Proc Natl Acad Sci U S A. 2012;109:4509–4514. doi: 10.1073/pnas.1116268109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Roy A, Lordier L, Mazzi S, Chang Y, Lapierre V, Larghero J, Debili N, Raslova H, Vainchenker W. Activity of nonmuscle myosin II isoforms determines localization at the cleavage furrow of megakaryocytes. Blood. 2016;128:3137–3145. doi: 10.1182/blood-2016-04-711630. [DOI] [PubMed] [Google Scholar]
- 107.Badirou I, Pan J, Legrand C, Wang A, Lordier L, Boukour S, Roy A, Vainchenker W, Chang Y. Carboxyl-terminal-dependent recruitment of nonmuscle myosin II to megakaryocyte contractile ring during polyploidization. Blood. 2014;124:2564–2568. doi: 10.1182/blood-2014-06-584995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Beach JR, Egelhoff TT. Myosin II recruitment during cytokinesis independent of centralspindlin-mediated phosphorylation. J Biol Chem. 2009;284:27377–27383. doi: 10.1074/jbc.M109.028316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Mui KL, Chen CS, Assoian RK. The mechanical regulation of integrin-cadherin crosstalk organizes cells, signaling and forces. J Cell Sci. 2016;129:1093–1100. doi: 10.1242/jcs.183699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Efimova N, Svitkina TM. Branched actin networks push against each other at adherens junctions to maintain cell–cell adhesion. J Cell Biol. 2018 doi: 10.1083/jcb.201708103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Vasquez CG, Heissler SM, Billington N, Sellers JR, Martin AC. Drosophila non-muscle myosin II motor activity determines the rate of tissue folding. eLife. 2016;5:e20828. doi: 10.7554/eLife.20828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Chung S, Kim S, Andrew DJ. Uncoupling apical constriction from tissue invagination. Elife. 2017;6:e22235. doi: 10.7554/eLife.22235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Del Signore SJ, Cilla R, Hatini V. The WAVE regulatory complex and branched F-Actin counterbalance contractile force to control cell shape and packing in the Drosophila eye. Dev Cell. 2018;44:471–483 e474. doi: 10.1016/j.devcel.2017.12.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Rosa A, Vlassaks E, Pichaud F, Baum B. Ect2/Pbl acts via Rho and polarity proteins to direct the assembly of an isotropic actomyosin cortex upon mitotic entry. Dev Cell. 2015;32:604–616. doi: 10.1016/j.devcel.2015.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Bovellan M, Romeo Y, Biro M, Boden A, Chugh P, Yonis A, Vaghela M, Fritzsche M, Moulding D, Thorogate R, Jegou A, Thrasher AJ, Romet-Lemonne G, Roux PP, Paluch EK, Charras G. Cellular control of cortical actin nucleation. Curr Biol. 2014;24:1628–1635. doi: 10.1016/j.cub.2014.05.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Eghiaian F, Rigato A, Scheuring S. Structural, mechanical, and dynamical variability of the actin cortex in living cells. Biophys J. 2015;108:1330–1340. doi: 10.1016/j.bpj.2015.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Smutny M, Cox HL, Leerberg JM, Kovacs EM, Conti MA, Ferguson C, Hamilton NA, Parton RG, Adelstein RS, Yap AS. Myosin II isoforms identify distinct functional modules that support integrity of the epithelial zonula adherens. Nat Cell Biol. 2010;12:696–702. doi: 10.1038/ncb2072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Gomez GA, McLachlan RW, Wu SK, Caldwell BJ, Moussa E, Verma S, Bastiani M, Priya R, Parton RG, Gaus K, Sap J, Yap AS. An RPTPalpha/Src family kinase/Rap1 signaling module recruits myosin IIB to support contractile tension at apical E-cadherin junctions. Mol Biol Cell. 2015;26:1249–1262. doi: 10.1091/mbc.E14-07-1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Ma X, Sung DC, Yang Y, Wakabayashi Y, Adelstein RS. Nonmuscle myosin IIB regulates epicardial integrity and epicardium-derived mesenchymal cell maturation. J Cell Sci. 2017;130:2696–2706. doi: 10.1242/jcs.202564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Ivanov AI, Naydenov NG. Dynamics and regulation of epithelial adherens junctions: recent discoveries and controversies. Int Rev Cell Mol Biol. 2013;303:27–99. doi: 10.1016/B978-0-12-407697-6.00002-7. [DOI] [PubMed] [Google Scholar]
- 121.Porat-Shliom N, Milberg O, Masedunskas A, Weigert R. Multiple roles for the actin cytoskeleton during regulated exocytosis. Cell Mol Life Sci. 2013;70:2099–2121. doi: 10.1007/s00018-012-1156-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Chandrasekar I, Goeckeler ZM, Turney SG, Wang P, Wysolmerski RB, Adelstein RS, Bridgman PC. Nonmuscle myosin II is a critical regulator of clathrin-mediated endocytosis. Traffic. 2014;15:418–432. doi: 10.1111/tra.12152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Fath KR. Characterization of myosin-II binding to Golgi stacks in vitro. Cell Motil Cytoskeleton. 2005;60:222–235. doi: 10.1002/cm.20060. [DOI] [PubMed] [Google Scholar]
- 124.Petrosyan A, Ali MF, Verma SK, Cheng H, Cheng PW. Non-muscle myosin IIA transports a Golgi glycosyltransferase to the endoplasmic reticulum by binding to its cytoplasmic tail. Int J Biochem Cell Biol. 2012;44:1153–1165. doi: 10.1016/j.biocel.2012.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Korobova F, Gauvin TJ, Higgs HN. A role for myosin II in mammalian mitochondrial fission. Curr Biol. 2014;24:409–414. doi: 10.1016/j.cub.2013.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Rousso T, Schejter ED, Shilo BZ. Orchestrated content release from Drosophila glue-protein vesicles by a contractile actomyosin network. Nat Cell Biol. 2016;18:181–190. doi: 10.1038/ncb3288. [DOI] [PubMed] [Google Scholar]
- 127.Milberg O, Shitara A, Ebrahim S, Masedunskas A, Tora M, Tran DT, Chen Y, Conti MA, Adelstein RS, Ten Hagen KG, Weigert R. Concerted actions of distinct nonmuscle myosin II isoforms drive intracellular membrane remodeling in live animals. J Cell Biol. 2017;216:1925–1936. doi: 10.1083/jcb.201612126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Miklavc P, Hecht E, Hobi N, Wittekindt OH, Dietl P, Kranz C, Frick M. Actin coating and compression of fused secretory vesicles are essential for surfactant secretion - a role for Rho, formins and myosin II. J Cell Sci. 2012;125:2765–2774. doi: 10.1242/jcs.105262. [DOI] [PubMed] [Google Scholar]
- 129.Nightingale TD, White IJ, Doyle EL, Turmaine M, Harrison-Lavoie KJ, Webb KF, Cramer LP, Cutler DF. Actomyosin II contractility expels von Willebrand factor from Weibel-Palade bodies during exocytosis. J Cell Biol. 2011;194:613–629. doi: 10.1083/jcb.201011119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Han X, Li P, Yang Z, Huang X, Wei G, Sun Y, Kang X, Hu X, Deng Q, Chen L, He A, Huo Y, Li D, Betzig E, Luo J. Zyxin regulates endothelial von Willebrand factor secretion by reorganizing actin filaments around exocytic granules. Nat Commun. 2017;8:14639. doi: 10.1038/ncomms14639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Miserey-Lenkei S, Bousquet H, Pylypenko O, Bardin S, Dimitrov A, Bressanelli G, Bonifay R, Fraisier V, Guillou C, Bougeret C, Houdusse A, Echard A, Goud B. Coupling fission and exit of RAB6 vesicles at Golgi hotspots through kinesin-myosin interactions. Nat Commun. 2017;8:1254. doi: 10.1038/s41467-017-01266-0. [DOI] [PMC free article] [PubMed] [Google Scholar]