Abstract
Plant viruses are still one of the main contributors to economic losses in agriculture. It has been estimated that plant viruses can cause as much as 50 billion euros loss worldwide, per year. This situation may be worsened by recent climate change events and the associated changes in disease epidemiology. Reliable and early detection methods are still one of the main and most effective actions to develop control strategies for plant viral diseases. During the last years, considerable progress has been made to develop tools with high specificity and low detection limits for use in the detection of these plant pathogens. Time and cost reductions have been some of the main objectives pursued during the last few years as these increase their feasibility for routine use. Among other strategies, these objectives can be achieved by the simultaneous detection and (or) identification of several viruses in a single assay. Nucleic acid-based detection techniques are especially suitable for this purpose. Polyvalent detection has allowed the detection of multiple plant viruses at the genus level. Multiplexing RT polymerase chain reaction (PCR) has been optimized for the simultaneous detection of more than 10 plant viruses/viroids. In this short review, we provide an update on the progress made during the last decade on techniques such as multiplex PCR, polyvalent PCR, non-isotopic molecular hybridization techniques, real-time PCR, and array technologies to allow simultaneous detection of multiple plant viruses. Also, the potential and benefits of the powerful new technique of deep sequencing/next-generation sequencing are described.
Keywords: multiplex, polymerase chain reaction, molecular hybridization, microarrays, polyprobes, nextgeneration sequencing, plant viruses, viroids
Introduction
Plant viruses and viroids are still a major concern in modern agriculture. They cause substantial economic losses in many important crops, especially those for which no virus-resistant varieties are available. Thus, early detection of these pathogens is still one of the main ways to control the development of the disease. Serology-based methods for virus detection have contributed significantly to evaluation of the sanitary status of these crops during the last 30 years and are still the methods of choice for a large number of laboratories involved in certification schemes (López et al., 2003; Boonham et al., 2014). The advent of nucleic acid-based technologies has allowed improving sensitivity to limits below the potential pathogenic thresholds solving some of the challenges posed by the need for specific and sensitive detection of plant viruses and viroids (Lopez et al., 2009). One of the most important challenges in plant virus/viroid diagnosis during the last decade has been the implementation of polyvalent and/or multiplex detection methods as these contribute to cost reductions, increased efficiency, and routine use (James et al., 2006). Different approaches, based on different biochemical principles, can be used to detect simultaneously multiple plant viruses or viroids including the following: (i) multiplex or polyvalent polymerase chain reaction (PCR), (ii) molecular hybridization including array techniques, and (iii) next-generation sequencing (NGS) technologies. The latter is revolutionizing the way plant virus diagnosticians are addressing the identification and characterization of new viruses and viroids and is having a profound impact on plant pathology in general (see Barba et al., 2014; Massart et al., 2014; Wu et al., 2015 for comprehensive reviews of NGS). However, PCR-based and molecular hybridization methods are still used frequently in most diagnostic laboratories due to years of validation, knowledge of their specificity and sensitivity, ease of implementation, and relatively low cost. In this short review, we update the progress made during the last 12 years on the multiplex or broad-spectrum detection of plant viruses and viroids.
Molecular Hybridization
This technique is based on the complementarity of base pairs of nucleic acids that results in a stable hybrid formed by part (or the totality) of the nucleic acid sequence of the pathogen to be detected (target molecule), and the labeled complementary sequence (probe). Probes can be synthesized in the form of RNA (riboprobes) or DNA (DNA probes) molecules. Most plant viruses and all viroids have RNA as their genetic material. Since RNA–RNA hybrids are more stable than RNA–DNA or DNA–DNA hybrids and, consequently, more stringent conditions can be used with riboprobes detecting pathogenic RNAs, it is not surprising that riboprobes are the most frequent probes used in phytodiagnosis. In fact, molecular hybridization as a diagnostic tool in plant virology was first applied for the detection of viroids (Owens and Diener, 1981) for which no serological method could be used due to the lack of any protein component in their structural constituents. Subsequently, the technique was used for the detection of plant viruses (Garger et al., 1983; Maule et al., 1983). The stability of the resultant hybrid, and therefore the technical reliability, depend on both electrostatic and hydrophobic forces, which in turn depend on the reaction conditions such as temperature, salt concentration, and length of the probe, among other factors. Non-radioactive probes can detect RNA target molecules to the femtomole level when properly prepared and quantified. Detailed procedures for the synthesis of the labeled probe, sample preparation, hybridization, and detection have been described in previous reviews (e.g., Hull, 1993; Pallás et al., 1998, 2017; Mühlbach et al., 2003). Molecular hybridization has an intermediate sensitivity level between serological and PCR-based methods but maintains the user friendliness of the former and lacks the main disadvantages of the latter (higher possibility of false positives and contamination). It is not surprising that today more and more companies are increasingly offering molecular hybridization-based tests among their services to detect plant pathogens. As yet though there is no industry standard in terms of the format used. Although molecular hybridization is considered a very robust and reliable procedure, it is not exempt of some weaknesses. For instance, in some pathogen/host combinations, spurious hybridization signals can be observed due to host RNAs with sequence similarity with the plant virus/viroid to be detected (e.g., Cañizares et al., 1999). The potential false positives can be eliminated through incubation of the membranes with RNase at high ionic strength after hybridization. When molecular hybridization is applied in a tissue printing format, probes can bind to host proteins causing false positive signals as observed for the detection of peach latent mosaic viroid (PLMVd) in peach samples (WenXing et al., 2009). An extra step for removing proteins should be included to avoid inaccurate results (WenXing et al., 2009).
Remarkably, molecular hybridization can be easily adapted to simultaneously detect different viruses and/or viroids in a single hybridization assay (James et al., 2006). This can be addressed by mixing in the same probe solution different DNA or RNA probes or by synthesizing a unique probe that harbors in tandem the corresponding partial RNA (riboprobes) or DNA (DNA probes) complementary sequences to the plant viruses/viroids to be detected.
Probe Mix
The first molecular hybridization approach used to detect several viruses at the same time involved the mixture of different probes, each specific for a different target, in the same assay. Probe mixtures have been successfully applied to phytosanitary certification of tomato in Italy (Saldarelli et al., 1996), to the simultaneous detection of five viruses affecting carnation (Sánchez-Navarro et al., 1999), three ilarviruses affecting stone fruit trees (Saade et al., 2000), two viruses affecting geranium plants (Ivars et al., 2004), and of 10 artichoke viruses (Minutillo et al., 2012) (Table 1). In all these cases, probes (DNA or RNA) were mixed in the hybridization solution and reliably detected all target viruses, with high specificity and identical sensitivity to that obtained using individual probes. The main disadvantage of this approach is that mixtures of many riboprobes can result in undesirable background that can make the results indecipherable. Treating the membranes with RNase A after the washing steps can overcome, at least in part, this drawback (Sánchez-Navarro et al., 1999). In addition, this disadvantage can be totally overcome by the use of a unique probe, polyprobe, containing in tandem several partial sequences of different viruses and/or viroids (see next section). A mixture of two DNA probes has been recently used that allowed the simultaneous visualization of two Citrus tristeza virus genotypes in vascular and non-vascular tissues of citrus trees (Bergua et al., 2016). This approach can be of great interest to study the basis for the interactions between different components of virus populations and for getting a deeper knowledge of the superinfection exclusion phenomenon at the cellular level.
Table 1.
Polyvalent molecular hybridization assays for the detection of plant viruses and viroids.
| Probe type | Crop applied or virus type | No. of targets | Nature of targets | Reference |
|---|---|---|---|---|
| Mixed riboprobes | Tomato | 6 | Viruses | Saldarelli et al., 1996 |
| Mixed riboprobes | Carnation | 5 | Viruses | Sánchez-Navarro et al., 1999 |
| Mixed riboprobes | Stone fruits | 3 | Viruses | Saade et al., 2000 |
| Mixed riboprobes | Geranium | 2 | Viruses | Ivars et al., 2004 |
| Mixed DNA probes | Artichoke | 10 | Viruses | Minutillo et al., 2012 |
| Polyprobe | Stone fruits | 6 | Viruses | Herranz et al., 2005 |
| Polyprobe | Citrus | 3 | Viroids | Cohen et al., 2006 |
| Polyprobe | Tomato | 6 | Viruses | Aparicio et al., 2009 |
| Polyprobe | Pome and stone fruits | 6 | Viroids | Lin et al., 2011 |
| Polyprobe | Stone fruits | 10 | 8 Viruses + 2 viroids | Peiró et al., 2012 |
| Polyprobe | Grapevine | 4 | Viroids | Zhang et al., 2012 |
| Polyprobe | Ornamentals and vegetables | 8 | Viroids | Torchetti et al., 2012 |
| Polyprobe | Tomato | 3 | 1 virus + 1 viroid + 1 bacteria | Zamora-Macorra et al., 2015 |
| Polyprobe | Coleus | 8 | Viroids | Jiang et al., 2013 |
| Polyprobe | Apple and pear | 4 | Viruses | Fajardo and Nickel, 2014 |
| Polyprobe | Grapevine | 18 | 13 Viruses + 5 viroids | Sánchez-Navarro et al., 2018b |
| Polyprobe | Potyvirus genus | 32 | Viruses | Sánchez-Navarro et al., 2018a |
| Arrays | Potato | 6 | Viruses | Boonham et al., 2003; Bystricka et al., 2003 |
| Arrays | Cucurbits | 4 | Viruses | Lee et al., 2003 |
| Arrays | Fruit trees | 7 | Viruses | Lenz et al., 2008 |
| Arrays | Potyviruses | 4 | Viruses | Wei et al., 2009 |
| Arrays | Tomato | 10 | Viruses | Tiberini et al., 2010 |
| Arrays | Tomato | 16 | Viruses + viroids | Tiberini and Barba, 2012 |
| Arrays | Artichoke | 14 | Viruses | Tiberini and Barba, 2013 |
| Arrays | Grapevine | 44 | Viruses | Engel et al., 2010 |
| Arrays | Grapevine | 15 | Viruses | Abdullahi et al., 2011 |
| Arrays | General | 52 | Viruses | Nicolaisen, 2011 |
| Arrays | General | 37 | Viroids | Zhang et al., 2013 |
Polyprobes
This approach, developed in 2005 by Herranz et al. (2005) uses a unique polyprobe for the polyvalent detection of different pathogens in a single assay. A polyprobe results from the cloning, in tandem, of complementary partial sequences, usually between 200 and 400 nucleotide residues, of different viruses/viroids after the promoter region of a RNA polymerase. After digestion with the appropriate restriction enzyme, a long transcript is synthesized in the presence of a non-radioactive precursor. Hybridization and immunological detection steps have been previously explained in detail (e.g., Pallás et al., 1998; Mühlbach et al., 2003) and will not be described here.
Polyprobes have been applied successfully for the detection of an array of combinations of plant viruses and/or viroids affecting different crops (Table 1). Polyprobes have been developed and used to detect four viroids affecting citrus trees (Cohen et al., 2006), the six main viruses infecting tomato (Aparicio et al., 2009), six viruses affecting pome and stone fruits (Lin et al., 2011), four viroids affecting grapevine (Zhang et al., 2012), eight viroids affecting ornamentals and vegetables (Torchetti et al., 2012), eight viroids infecting coleus plants (Jiang et al., 2013), and four viruses of apple and pear trees (Fajardo and Nickel, 2014). Remarkably, this technology has great potential for simultaneous multipathogen detection. Using polyprobes, Peiró et al. (2012) were able to detect eight viruses and two viroids infecting stone fruit trees. In addition, three pathogens with very different life cycle styles (bacteria, virus, and viroid) were simultaneously detected in a single assay in tomato plants (Zamora-Macorra et al., 2015). In general, the polyprobes permit the detection of several pathogens with comparable detection limit to the individual probes, although with long polyprobes (e.g., 10 probes in tandem or more) a reduction of the hybridization temperature is required (e.g., Peiró et al., 2012). In our hands, up to 18-mer polyprobes (detecting 13 viruses plus 5 viroids of grapevine; Sánchez-Navarro et al., 2018b) gave clear and reliable results without loss of sensitivity. Recently, a unique polyprobe with the capacity to detect all members of the Potyvirus genus was developed (Sánchez-Navarro et al., 2018a). The authors observed that sequences of the different potyvirus species showing a percentage similarity of 68% or higher, could be detected with the same probe by hybridizing at 50–55°C, with a detection limit of picograms of viral RNA comparable to the specific individual probes. The developed polyprobe contains seven different 500-nt fragments of a conserved region of the NIb gene and was able to detect all 32 potyviruses assayed with no signal in the healthy tissue. This assay is being considered as a genus probe for general potyvirus detection.
Arrays
Another broad-spectrum approach for parallel detection of multiple plant viruses relies on the DNA microarray technology. The technical details as well as the historical concept and development of DNA microarrays have been described exhaustively in previous reviews (Hadidi et al., 2004; Boonham et al., 2007; Zhu et al., 2017). Due to the high-throughput nature of the array technology, this approach was expected to have, in principle, a great potential for broad-spectrum diagnostics of plant viruses and viroids. However, the application of this technology has been limited so far. This is mainly due to the relative complexity of the different steps required to accomplish implementation of the test. This approach was first applied for the detection and identification of six potato viruses (Boonham et al., 2003; Bystricka et al., 2003, 2005) and four species of selected cucurbit-infecting tobamoviruses (Lee et al., 2003). An oligonucleotide microarray for the detection of some fruit tree viruses was designed and its theoretical detection limit was assessed (Lenz et al., 2008). The authors concluded that the sensitivity of detection is, among others, influenced by the proximity of the probe hybridization site to the unlabeled end of the targets. Wei et al. (2009) developed a 25-mer oligonucleotide microarray targeting four distinct potyviruses that included 85 probes designed from conserved and variable sequence regions of the nuclear inclusion b (NIb) gene, RNA-dependent RNA polymerase (RdRp) gene, coat protein (CP) gene, and the 3′ untranslated region (UTR), specific to the four targeted potyviruses at both species and strain levels. Using “Combimatrix” platform 40-mer oligonucleotide probes, Tiberini et al. (2010) designed a DNA microarray chip for screening 10 major economically important tomato viruses and later on this platform was optimized to include six pospiviroid species (Tiberini and Barba, 2012). This same platform was used to develop an oligonucleotide-based microarray for detection of multiple artichoke viruses (Tiberini and Barba, 2013). This diagnostic array demonstrated its applicability for routine diagnostic use in artichoke germplasm as it detected simultaneously 14 viruses in one single hybridization event. Two oligonucleotide microarrays have been developed for detecting grapevine viruses. Engel et al. (2010) used a 70-mer microarray containing 570 unique probes designed against highly conserved and species-specific regions of 44 grapevine viral genomes, whereas Abdullahi et al. (2011) used a range between 27 and 75 nucleotides in length for oligonucleotides and detected eight nepoviruses, two vitiviruses, and one each of closterovirus, foveavirus, ampelovirus, maculavirus, and sadwavirus. Nicolaisen (2011) developed a microarray with 150 probes potentially capable of detecting 52 viruses from a broad range of genera. Forty nine of the 52 species tested were identified correctly to species level. Finally, Zhang et al. (2013) designed a microarray with a minimal number of probes that can detect a wide spectrum of all 8 reported viroid genera including 37 known plant viroid species.
Microarrays can be used not only for diagnostics but for phylogenetic or taxonomic purposes. An oligonucleotide-based microarray was developed to detect and differentiate cucumber mosaic virus (CMV) serogroups and subgroups (Deyong et al., 2005). A long 70-mer oligonucleotide DNA microarray was developed that was capable of simultaneously detecting and genotyping plum pox virus strains (Pasquini et al., 2008).
Multiplex PCR
Multiplex PCR (DNA targets) and multiplex RT-PCR (mRT-PCR) (RNA targets) are quick, reliable, and cost-effective methods that have been used successfully for detecting a variety of pathogens simultaneously in a single assay. Whereas uniplex RT-PCR is potentially expensive and resource intensive (requiring time and resources to test for each virus or viroid separately), mRT-PCR incorporates different sets of specific primers for two to more targets in one reaction tube and enables simultaneous amplification of different target nucleic acids in a single test. This reduces material costs, labor, and time. It is indeed possible also to amplify simultaneously several regions of a target virus thereby improving the reliability of detection. The main approach for this purpose is the mRT-PCR which uses specific or degenerative primers that amplify and allow identification of the size and species-specific amplicons by agarose gel analysis. This approach represents 71.8% of all multiplex detection reports since 2005 (Figure 1), allowing the simultaneous detection of as many as six (Park et al., 2005; Cating et al., 2015), seven (Roy et al., 2005; Tao et al., 2012; Kwon et al., 2014; Zhao et al., 2015), eight (Sánchez-Navarro et al., 2005; Deb and Anderson, 2008; Kwak et al., 2014), and up to nine (Gambino and Gribaudo, 2006; Gambino, 2015) pathogens. However, the sensitivity of this technique is influenced by the number of targets to be detected (Sánchez-Navarro et al., 2005; Tao et al., 2012; Kwon et al., 2014; Nam et al., 2015), mainly due to the number of different primer pairs instead of the total amount of primer present in the cocktail. Sánchez-Navarro et al. (2005) showed that the use of a cocktail of five primer pairs do not affect the detection limit of mRT-PCR, while seven pairs does affect the detection limit. In agreement with this observation, 91.4% of the multiplex reactions reported since 2005 describe the detection of two to five pathogens (Figure 1). Some of these limitations have been overcome by improving the quality of the nucleic acid extraction procedures, the products obtained and/or different modalities of the PCR technology. Magnetic nanobeads (Deng et al., 2014), dual priming oligonucleotide (DPO) primers (Kwon et al., 2014), or a nested reaction (Foissac et al., 2005; Maliogka et al., 2007; Papayiannis et al., 2011) have all contributed to increased levels of specificity and sensitivity.
FIGURE 1.
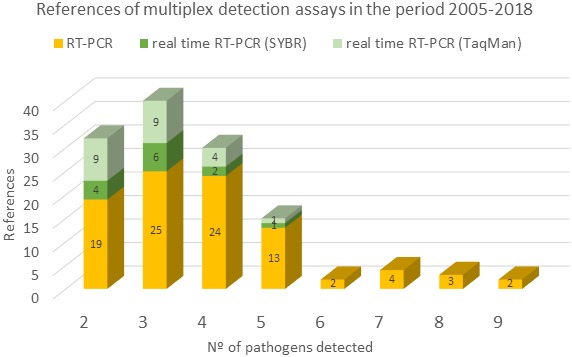
References of multiplex detection assays in the period 2005–2018. Number of references that use RT-PCR (yellow), real-time RT-PCR SYBR (green), and real-time RT-PCR TaqMan (light green) are represented against the number of pathogens detected by these techniques. Source used was the Web of Science (all database) and the parameters used for the searching were: (multiplex or simultaneous or polyvalent) and detection and (plant virus or viroid).
Other limitations of the mRT-PCR reaction are the use of the agarose gel-based detection for discriminating the size-specific amplicons with its putative optical error and the confirmation that the amplified DNA fragments correspond to the target sequences. Some alternative approaches have eliminated the use of agarose gels such as the use of species-specific biotinylated probes in streptavidin-coated microtiter wells (Charoenvilaisiri et al., 2014) or, more recently, the use of a platform for multiplexed nucleic acid detection such as the microsphere-based flow cytometric system developed by Luminex (Austin, TX, United States). The Luminex xMAP system incorporates 5.6 μm polystyrene microspheres that are internally dyed with two spectrally distinct fluorochromes. Using precise amounts of each of these fluorochromes, an array is created consisting of 100 different microsphere sets with specific spectral addresses. Each microsphere set can possess a different reactant on its surface (e.g., a unique anti-MagPlex-TAG oligonucleotide sequence). Because microsphere sets can be distinguished by their spectral addresses, they can be combined, allowing up to 100 different targets to be measured simultaneously in a single reaction vessel. A third fluorochrome coupled to a reporter molecule quantifies the biomolecular interaction that has occurred at the microsphere surface. Microspheres are detected individually in a rapidly flowing fluid stream as they pass by two separate lasers in the Luminex® 100TM analyzer. Thousands of microspheres per second could be detected, resulting in an analysis system capable of analyzing and reporting up to 100 different reactions in a single reaction vessel in just a few seconds per sample. The LUMINEX bead-based array has been applied for the multiple detection of begomovirus (van Brunschot et al., 2014a), pospiviroids (van Brunschot et al., 2014b), and lily viruses (Lim et al., 2016). This luminex approach implies the amplification of the target sequences by mRT-PCR reaction, which could be a bottleneck if many primers should be used, as commented above. Although this technology has been successfully used to detect related pathogens by using universal primers in the RT-PCR reaction (e.g., begomovirus, pospiviroids), it now remains to know if this technology could be adjusted to detect many unrelated pathogens.
Another alternative to the analysis of the amplicons by agarose gel-based detection, is the multiplex real-time PCR in which, the amplified fragments are detected directly during the reaction by using non-probe based fluorescent dyes such as SYBRGreen or EvaGreen (Zipper et al., 2004; Mao et al., 2007) or specific fluorescent probes such as TaqMan® probes (Livak et al., 1995), molecular beacons (Tyagi and Kramer, 1996), or the minor groove binding (MGB) probes (de Kok et al., 2002). Real-time PCR allows quantification of the pathogen and reduced significantly the detection limit to as little as a few molecules (Torres et al., 2005; Mortimer-Jones et al., 2009; Tuo et al., 2014; Huo et al., 2015). However, despite the clear advantages of the real-time procedure, the references of multiplex detection assays based on this technique, either using non-probe fluorescent days or fluorescent probes, represent only 28.1% of such test since 2005 (Figure 1). The tests have been adapted to allow the simultaneous detection of two to five pathogens. The problems derived from the necessity of discriminating between different labeled amplicons by melting curve analysis or the availability of fluorescent dyes with overlapping excitation/emission spectra, have hampered the number of pathogens detected by multiplex real-time PCR. The few multiplex real-time PCR assays that use melting curve analysis based on the SYBR Green I, detect three targets at most, due to the shortcomings of the dye (Guion et al., 2008). This problem has been overcome by using the EvaGreen or SYTO dyes (Eischeid, 2011), which are less inhibitory towards PCR and provide better peak resolution, allowing the detection of up to five viruses (Bester et al., 2012; Cheng et al., 2013; Aloisio et al., 2018).
Multiplex detection of three or more targets by fluorescent probes is influenced by the availability of dyes with compatible excitation/emission spectra. Simultaneous detection of four pathogens has been reported for four retroviruses (Vet et al., 1999) or four potato (Agindotan et al., 2007; Mortimer-Jones et al., 2009) or cassava (Otti et al., 2016) viruses. In addition, this technology was adapted for the detection of five grapevine viruses, although it was necessary to perform a compensation color assay, which aimed to minimize the emission interference among the five fluorescent dyes (Lopez-Fabuel et al., 2013).
Multiplex RT-PCR tests have also been developed to detect all members of a specific genus by designing degenerative or specific primers that target conserved regions (Table 2). Obviously, a previous step for using this approach is the identification of conserved genomic regions that can be targeted by the amplification primers. Thus, this procedure has allowed the detection of virus species in the genera Allexvirus (Kumar et al., 2010; Majumder and Baranwal, 2014), Ilarvirus (Maliogka et al., 2007), Bromovirus and Cucumovirus (Seo et al., 2014), Begomovirus (van Brunschot et al., 2014a), Potyvirus (Zheng et al., 2010), and members of the viroid genus Pospiviroids (Botermans et al., 2013; Luigi et al., 2014; Olivier et al., 2014; van Brunschot et al., 2014b) or Trichovirus, Capillovirus and Foveavirus (Foissac et al., 2005), or viroids and phytoplasmas (Malandraki et al., 2015). In the majority of cases, the species-specific identification was performed by restriction fragment length polymorphism (RFLP) analysis or by sequencing the corresponding amplicons. However, the introduction of the Luminex technology opens an interesting tool to allow the direct identification of the pathogen without any further analysis (van Brunschot et al., 2014a,b).
Table 2.
Polyvalent PCR assays for the detection of plant viruses and/or viroids at the genus level.
| Genus | Primers | Region | Amplicon (bp) | Species identification | Reference |
|---|---|---|---|---|---|
| Ilarvirus | Degenerated | RNA2/RdRp gene | 381 | Amplicon sequencing/RFLP | Maliogka et al., 2007 |
| Pospiviroid | Degenerated | 270 | Luminex | van Brunschot et al., 2014b | |
| Pospiviroid | Specific/several primers | Terminal conserved region (TCR) and terminal right domain (TR) | 170–180 | TaqMan probe only for genus | Botermans et al., 2013 |
| Bromovirus/cucumovirus | Degenerated with adaptor sequences | RNA1 | 337 | Amplicon sequencing | Seo et al., 2014 |
| Pospiviroid | Degenerated | 200 | Amplicon sequencing | Olivier et al., 2014 | |
| Pospiviroid | Degenerated | The terminal left and the pathogenesis domains | 300 | Amplicon sequencing/RFLP | Luigi et al., 2014 |
| Begomovirus | Degenerated | C3 ORF | 290 | Luminex | van Brunschot et al., 2014a |
| Potyvirus | Degenerated | NIb | 350 | Amplicon sequencing | Zheng et al., 2010 |
| Trichoviruses, capilloviruses, foveaviruses | Degenerated | RdRp | 362 | Amplicon sequencing | Foissac et al., 2005 |
| Phytoplasmas | Specific | 16S rDNA | 200 | TaqMan only for phytoplasmas | Christensen et al., 2013; Malandraki et al., 2015 |
| Allexivirus | Degenerated | 3′-end of ORF6 | 183–192 | Amplicon sequencing | Kumar et al., 2010; Majumder and Baranwal, 2014 |
Multiplexing Potential of Next-Generation Sequencing
Next-generation sequencing, known also as massively parallel sequencing or deep sequencing, is a powerful technology that allows the generation of massive amounts of sequence data. There are various approaches or NGS platforms, each with different characteristics and with the potential in some cases to generate as many as three billion reads per run with read lengths that vary from approximately 35 to 800 nucleotides depending on the platform (Barzon et al., 2011; Buermans and den Dunnen, 2014; Wu et al., 2015; Jones et al., 2017; Rott et al., 2017). The technology has been proposed as a valuable tool for diagnostic virology (Adams et al., 2009; Barzon et al., 2011). NGS is highly sensitive and has the potential to detect the full spectrum of viruses infecting a given host, including known and even unknown viruses (Barzon et al., 2011; Villamor et al., 2016; Rott et al., 2017; Jo et al., 2018). The broad-spectrum and unbiased nature of the technology makes it a valuable tool for plant-based metagenomics, allowing the simultaneous screening and detection of populations of graft transmissible agents that include viruses (RNA and DNA), viroids, and phytoplasma in a sample (Adams et al., 2009; Kreuze et al., 2009). Jones et al. (2017) indicated that the multiplexing potential of NGS (RNA-seq) might allow a researcher to answer the question of how many different viruses are present in a crop plant. NGS even has the power to detect plant viruses that were not detected using current and standard tools that were based on biological, serological, and molecular tests (Villamor et al., 2016). See Jones et al. (2017) for an excellent review describing the use of RNA-seq for the detection of multiple viruses in each of various host plants.
Kreuze et al. (2009) used NGS to simultaneously detect/identify the viruses sweet potato feathery mottle virus (SPFMV, family Potyviridae), and sweet potato chlorotic stunt virus (family Closteroviridae). They targeted small RNAs (sRNA, 20–24 nt) in total RNA extracts from co-infected plants. Using this approach, they were able to assemble the complete genome of SPFMV and detected unexpectedly also two new and distinct badnaviruses (family Caulimoviridae) and a new mastrevirus (family Geminiviridae). This approach allowed the simultaneous detection of viruses with RNA and DNA genomes and viruses from distinctly different families. Verdin et al. (2017) used also a similar strategy targeting small RNAs from a range of ornamental plants, with pooling of samples, to detect (+) and (–) ssRNA viruses, dsRNA viruses, dsDNA viruses, an ssDNA virus, and a viroid. NGS allows simultaneous detection and identification of viruses belonging to different families and genera, but also multiple isolates or variants of the same virus co-infecting a single host. James et al. (2017) detected, by NGS analysis of total RNA extracted from a single apple plant, an infection complex that included apple chlorotic leaf spot virus (ACLSV, genus Trichovirus) and apple stem pitting virus (ASPV, genus Foveavirus). Five isolates/variants of ACLSV as well as 14 definite (but perhaps as many as 29) isolates/variants of ASPV were identified, all in a single sample. Rott et al. (2017) described the detection of 12 distinct genotypes of ASPV in sample #103 of their analyses. This shows the incredible ability of NGS to allow simultaneous detection and differentiation even of similar and closely related genomic sequences, which is not easily achieved with other diagnostic tools. Multiplex detection that allows accurate identification of variants or isolates of a single virus present in a sample may be desirable in some circumstances as this may have biological significance, influencing disease symptoms (James et al., 2017).
The nucleic acid template used for NGS analysis influences the reliability of simultaneous and broad-spectrum detection of plant viruses. Total RNA and small RNAs are effective targets for broad spectrum and even simultaneous detection of DNA viruses, RNA viruses and viroids (Kreuze et al., 2009; Wylie et al., 2012; Massart et al., 2014; Zhang et al., 2014; Wu et al., 2015; Verdin et al., 2017). Wu et al. (2015) suggest that only NGS analysis of total small RNAs is suitable for simultaneous detection of DNA viruses, RNA viruses, and viroids. For broad-spectrum virus detection Wylie et al. (2012) used a strategy of pooling total RNA from several plants, targeting polyadenylated RNA by using oligo-d(T) primers and was able to simultaneously detect by NGS 16 virus species belonging to various genera. Double-stranded (ds) RNA, the replicative form of RNA viruses (Dodds et al., 1984), as a template for NGS may be limited in that it may allow only reliable detection of RNA viruses and viroids (Massart et al., 2014). However, in comparing total nucleic acid extracts (TNA) to dsRNA for the analysis of infected grapevine, Al Rwahnih et al. (2009) obtained 54,605 viral hits from the dsRNA template versus 1,275 viral hits for the TNA template. If the targets for detection are known RNA viruses, perhaps the use of dsRNA for analysis will improve sensitivity. Jo et al. (2018) indicated that mRNA targets enriched by oligo dT were suitable for the detection and identification of different types of viral genomes including DNA viruses, ds RNA viruses, and viroids. The authors suggested that (U)-rich regions in viruses without a poly-A tail can be amplified by oligo dT.
As with any technology, there are concerns and/or limitations associated with the use of NGS as a diagnostic tool for plant viruses. There is a need for suitable bioinformatics tools and expertise to extract the required information from the enormous amounts of data generated (Wu et al., 2015; Visser et al., 2016; Roossinck, 2017). Small or low-resourced research groups may not always have access to bioinformaticians or access to a bioinformatics facility and this may influence their ability to utilize effectively the technology (Jones et al., 2017). The high sensitivity of NGS makes it susceptible to cross contamination including contamination with samples containing mycoviruses and insect viruses (Rott et al., 2017). To minimize the occurrence of false positives by NGS analysis, it is proposed that at least two different approaches be used for virus detection and identification (Rott et al., 2017; Jo et al., 2018). On the other end of the sensitivity spectrum, Jo et al. (2018) reported that NGS did not detect viruses or viroids in low titer that could be detected by RT-PCR. There is the possibility that virus sequences detected by NGS may be the remnants of sequences incorporated into host genomes (Martin et al., 2016); also, the biological significance of novel viruses or in some cases partial sequences detected need to be determined. There is a need for validation data, for more information on the sensitivity of NGS-based detection, compared to established diagnostic techniques such as real-time RT-PCR, and the definition of thresholds for a positive detection are needed (Massart et al., 2014).
In their review of various publications describing NGS analysis for plant virus detection, Jones et al. (2017) identified a number of issues, at least two of which have special significance for reliable diagnosis. These include the detection of different viruses or levels of viruses associated with different parts of the plants and the fact that different analytical tools can give different results for viruses being detected. Consistent and appropriate sampling and the choice of appropriate analytical tools used are crucial therefore for obtaining consistent and perhaps reliable results by NGS analysis.
In attempts to simplify the analysis of the enormous amount of NGS data generated, an e-probe based approach was utilized by Stobbe et al. (2014) and Visser et al. (2016). E-probes are pathogen-specific sequences that are rigorously assessed for their specificity and fitness for purpose (Stobbe et al., 2014). When used to screen 18 NGS data sets generated from dsRNA extracted from grapevines, e-probe detection using the program Truffle for data analysis (with e-probes developed for 55 known viruses) was as sensitive as a de novo assembly-based NGS data analysis pipeline. In some cases, e-probe detection seemed to be more reliable (Visser et al., 2016).
Cost and complexity are major impediments to the implementation of molecular techniques, but simultaneous detection of various pathogens contributes to cost reduction (Martin et al., 2000; James et al., 2006). The potential to pool samples from different plants, even different plant species, for NGS analysis (Wylie et al., 2012; Villamor et al., 2016; Rott et al., 2017; Verdin et al., 2017) can contribute also to reductions in the time and costs of diagnostics. Also, NGS has the potential to identify novel plant pathogens that may be the causal agents of diseases of unknown etiology that otherwise could not be determined (Kreuze et al., 2009; Barba and Hadidi, 2015; James and Phelan, 2017). Any successful implementation of a new technology for routine use for reliable pest diagnosis requires an understanding of the limitations of the technique. Issues such as the uneven distribution of viruses in plants (Jones et al., 2017) and the fact that RT-PCR was reported to be more reliable than NGS in some cases where viruses or viroids were in low titer (Jo et al., 2018) might indicate the need for caution in implementation and interpretation of NGS results.
Author Contributions
All authors listed have made a substantial, direct, and intellectual contribution to the work and approved it for publication.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We want to acknowledge the excellent technical assistance of Lorena Corachan in the different research lines that made possible this review. We apologize to those whose work could not be cited due to space limitations.
Footnotes
Funding. This work was funded by grant BIO2017-88321-R from the Spanish Direccion General de Investigacion Cientifica y Tecnica (DGICYT) and the Prometeo Program GV2014/010 from the Generalitat Valenciana.
References
- Abdullahi I., Gryshan Y., Rott M. (2011). Amplification-free detection of grapevine viruses using an oligonucleotide microarray. J. Virol. Methods 178 1–15. 10.1016/j.jviromet.2011.07.009 [DOI] [PubMed] [Google Scholar]
- Adams I. P., Glover R. H., Monger W. A., Mumford R., Jackeviciene E., Navalinskiene M., et al. (2009). Next-generation sequencing and metagenomic analysis: a universal diagnostic tool in plant virology. Mol. Plant Pathol. 10 537–545. 10.1111/j.1364-3703.2009.00545.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agindotan B. O., Shiel P. J., Berger P. H. (2007). Simultaneous detection of potato viruses, PLRV, PVA, PVX and PVY from dormant potato tubers by TaqMan((R)) real-time RT-PCR. J. Virol. Methods 142 1–9. 10.1016/j.jviromet.2006.12.012 [DOI] [PubMed] [Google Scholar]
- Al Rwahnih M., Daubert S., Golino D., Rowhani A. (2009). Deep sequencing analysis of RNAs from a grapevine showing Syrah decline symptoms reveals a multiple virus infection that includes a novel virus. Virology 387 395–401. 10.1016/j.virol.2009.02.028 [DOI] [PubMed] [Google Scholar]
- Aloisio M., Morelli M., Elicio V., Saldarelli P., Ura B., Bortot B., et al. (2018). Detection of four regulated grapevine viruses in a qualitative, single tube real-time PCR with melting curve analysis. J. Virol. Methods 257 42–47. 10.1016/j.jviromet.2018.04.008 [DOI] [PubMed] [Google Scholar]
- Aparicio F., Soler S., Aramburu J., Galipienso L., Nuez F., Pallás V., et al. (2009). Simultaneous detection of six RNA plant viruses affecting tomato crops using a single digoxigenin-labelled polyprobe. Eur. J. Plant Pathol. 123 117–123. 10.1007/s10658-008-9347-5 [DOI] [Google Scholar]
- Barba M., Czosnek H., Hadidi A. (2014). Historical perspective, development and applications of next-generation sequencing in plant virology. Viruses 6 106–136. 10.3390/v6010106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barba M., Hadidi A. (2015). An overview of plant pathology and application of next-generation sequencing technologies. CAB Rev. 10 1–21. 10.1079/PAVSNNR201510005 [DOI] [Google Scholar]
- Barzon L., Lavezzo E., Militello V., Toppo S., Palù G. (2011). Applications of next-generation sequencing technologies to diagnostic virology. Int. J. Mol. Sci. 12 7861–7884. 10.3390/ijms12117861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergua M., Phelan D. M., Bak A., Bloom D. C., Folimonova S. Y. (2016). Simultaneous visualization of two Citrus tristeza virus genotypes provides new insights into the structure of multi-component virus populations in a host. Virology 491 10–19. 10.1016/j.virol.2016.01.017 [DOI] [PubMed] [Google Scholar]
- Bester R., Jooste A. E. C., Maree H. J., Burger J. T. (2012). Real-time RT-PCR high-resolution melting curve analysis and multiplex RT-PCR to detect and differentiate Grapevine leafroll-associated virus 3 variant groups I, II, III and VI. Virology J. 9:219. 10.1186/1743-422X-9-219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boonham N., Kreuze J., Winter S., van der Vlugt R., Bergervoet J., Tomlinson J., et al. (2014). Methods in virus diagnostics: from ELISA to next generation sequencing. Virus Res. 186 20–31. 10.1016/j.virusres.2013.12.007 [DOI] [PubMed] [Google Scholar]
- Boonham N., Tomlinson J., Mumford R. (2007). Microarrays for rapid identification of plant viruses. Annu. Rev. Phytopeth. 45 307–328. 10.1146/annurev.phyto.45.062806.094349 [DOI] [PubMed] [Google Scholar]
- Boonham N., Walsh K., Smith P., Madagan K., Graham I., Barker I. (2003). Detection of potato viruses using microarray technology: towards a generic method for plant viral disease diagnosis. J. Virol. Methods 108 181–187. 10.1016/S0166-0934(02)00284-7 [DOI] [PubMed] [Google Scholar]
- Botermans M., van de Vossenberg B. T., Verhoeven J. T., Roenhorst J. W., Hooftman M., Dekter R., et al. (2013). Development and validation of a real-time RT-PCR assay for generic detection of pospiviroids. J. Virol. Methods 187 43–50. 10.1016/j.jviromet.2012.09.004 [DOI] [PubMed] [Google Scholar]
- Buermans H. P. J., den Dunnen J. T. (2014). Next generation sequencing technology: advances and applications. Biochim. Biophys. Acta 1842 1932–1941. 10.1016/j.bbadis.2014.06.015 [DOI] [PubMed] [Google Scholar]
- Bystricka D., Lenz O., Mraz I., Dedic P., Sip M. (2003). DNA microarray: parallel detection of potato viruses. Acta Virol. 47 41–44. [PubMed] [Google Scholar]
- Bystricka D., Lenz O., Mraz I., Piherova L., Kmoch S., Sip M. (2005). Oligonucleotide-based microarray: a new improvement in microarray detection of plant viruses. J. Virol. Meth. 128 176–182. 10.1016/j.jviromet.2005.04.009 [DOI] [PubMed] [Google Scholar]
- Cañizares M. C., Marcos J. F., Pállas V. (1999). Molecular characterization of an almond isolate of hop stunt viroid (HSVd) and conditions for eliminating spurious hybridization in its diagnosis in almond samples. Eur. J. Plant Pathol. 105 553–558. 10.1023/A:1008794531725 [DOI] [Google Scholar]
- Cating R. A., Funke C. N., Kaur N., Hamm P. B., Frost K. E. (2015). A multiplex reverse transcription (RT) high-fidelity PCR protocol for the detection of six viruses that cause potato tuber necrosis. Am. J. Pot. Res. 92 536–540. 10.1007/s12230-015-9457-5 [DOI] [Google Scholar]
- Charoenvilaisiri S., Seepiban C., Bhunchoth A., Warin N., Luxananil P., Gajanandana O. (2014). Development of a multiplex RT-PCR-ELISA to identify four distinct species of tospovirus. J. Virol. Methods 202 54–63. 10.1016/j.jviromet.2014.03.003 [DOI] [PubMed] [Google Scholar]
- Cheng J., Jiang Y., Rao P., Wu H., Dong Q., Wu Z., et al. (2013). Development of a single-tube multiplex real-time PCR for detection and identification of five pathogenic targets by using melting-curve analysis with EvaGreen. Arch. Virol. 158 379–386. 10.1007/s00705-012-1493-6 [DOI] [PubMed] [Google Scholar]
- Christensen N. M., Nyskjold H., Nicolaisen M. (2013). Real-time PCR for universal phytoplasma detection and quantification. Methods Mol. Biol. 938 245–252. 10.1007/978-1-62703-089-2_21 [DOI] [PubMed] [Google Scholar]
- Cohen O., Batuman O., Stanbekova G., Sano T., Mawassi M., Bar-Joseph M. (2006). Construction of a multiprobe for the simultaneous detection of viroids infecting citrus trees. Virus Genes 33 287–292. [DOI] [PubMed] [Google Scholar]
- de Kok J. B., Wiegerinck E. T., Giesendorf B. A., Swinkels D. W. (2002). Rapid genotyping of single nucleotide polymorphisms using novel minor groove binding DNA oligonucleotides (MGB probes). Hum. Mutat. 19 554–559. 10.1002/humu.10076 [DOI] [PubMed] [Google Scholar]
- Deb M., Anderson J. M. (2008). Development of a multiplexed PCR detection method for Barley and Cereal yellow dwarf viruses, Wheat spindle streak virus, Wheat streak mosaic virus and Soil-borne wheat mosaic virus. J. Virol. Methods 148 17–24. 10.1016/j.jviromet.2007.10.015 [DOI] [PubMed] [Google Scholar]
- Deng X.-G., Zhu F., Chen Y.-J., Liu J., Zhu T., Li J.-Y., et al. (2014). A more sensitive and rapid multiplex RT-PCR assay combining with magnetic nanobeads for simultaneous detection of viruses in sweet potato. Eur. J. Plant Pathol. 140 111–117. 10.1007/s10658-014-0447-0 [DOI] [Google Scholar]
- Deyong Z., Willingmann P., Heinze C., Adam G., Pfunder M., Frey B., et al. (2005). Differentiation of Cucumber mosaic virus isolates by hybridization to oligonucleotides in a microarray format. J. Virol. Methods 123 101–108. 10.1016/j.jviromet.2004.09.021 [DOI] [PubMed] [Google Scholar]
- Dodds J. A., Morris T. J., Jordan R. L. (1984). Plant virus double-stranded RNA. Ann. Rev. Phytopathol. 22 151–168. 10.1146/annurev.py.22.090184.001055 [DOI] [Google Scholar]
- Eischeid A. C. (2011). SYTO dyes and EvaGreen outperform SYBR Green in real-time PCR. BMC Res. Notes 4:263. 10.1186/1756-0500-4-263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engel E. A., Escobar P. F., Rojas L. A., Rivera P. A., Fiore N., Valenzuela P. D. (2010). A diagnostic oligonucleotide microarray for simultaneous detection of grapevine viruses. J. Virol. Methods 163 445–451. 10.1016/j.jviromet.2009.11.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fajardo T. V. M., Nickel O. (2014). Simultaneous detection of four viruses affecting apple and pear by molecular hybridization using a polyprobe. Cienc. Rural 44 1711–1714. 10.1590/0103-8478cr20131629 [DOI] [Google Scholar]
- Foissac X., Svanella-Dumas L., Gentit P., Dulucq M. J., Marais A., Candresse T. (2005). Polyvalent degenerate oligonucleotides reverse transcription-polymerase chain reaction: a polyvalent detection and characterization tool for trichoviruses, capilloviruses, and foveaviruses. Phytopathology 95 617–625. 10.1094/PHYTO-95-0617 [DOI] [PubMed] [Google Scholar]
- Gambino G. (2015). “Multiplex RT-PCR method for the simultaneous detection of nine grapevine viruses,” in Plant Virology Protocols: New Approaches to Detect Viruses and Host Responses, 3rd Edn, eds Uyeda I., Masuta C. (New York, NY: Humana Press; ), 39–47. [DOI] [PubMed] [Google Scholar]
- Gambino G., Gribaudo I. (2006). Simultaneous detection of nine grapevine viruses by multiplex reverse transcription-polymerase chain reaction with coamplification of a plant RNA as internal control. Phytopathology 96 1223–1229. 10.1094/PHYTO-96-1223 [DOI] [PubMed] [Google Scholar]
- Garger S. J., Turpen T., Carrington J. C., Morris T. J., Dodds J. A., Jordan R. L., et al. (1983). Rapid detection of plant RNA viruses by dot blot hybridization. Plant Mol. Biol. Rep. 1 21–25. 10.1007/BF02680258 [DOI] [Google Scholar]
- Guion C. E., Ochoa T. J., Walker C. M., Barletta F., Cleary T. G. (2008). Detection of diarrheagenic Escherichia coli by use of melting-curve analysis and real-time multiplex PCR. J. Clin. Microb. 46 1752–1757. 10.1128/JCM.02341-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadidi A., Czosnek H., Barba M. (2004). DNA microarray and their potential applications for the detection of plant viruses, viroids and phytoplasmas. J. Plant Pathol. 86 97–104. [Google Scholar]
- Herranz M. C., Sánchez-Navarro J. A., Aparicio F., Pallás V. (2005). Simultaneous detection of six stone fruit viruses by non-isotopic molecular hybridization using a unique riboprobe or ‘polyprobe’. J. Virol. Methods 124 49–55. 10.1016/j.jviromet.2004.11.003 [DOI] [PubMed] [Google Scholar]
- Hull R. (1993). “Nucleic acid hybridization procedures,” in Diagnosis of Plant Virus Diseases, ed. Matthews R. E. F. (Boca Raton, FL: CRC Press, Inc.). [Google Scholar]
- Huo P., Shen W. T., Yan P., Tuo D. C., Li X. Y., Zhou P. (2015). Simultaneous detection of Papaya ringspot virus, Papaya leaf distortion mosaic virus, and Papaya mosaic virus by multiplex real-time reverse transcription PCR. Acta Virol. 59 380–388. 10.4149/av_2015_04_380 [DOI] [PubMed] [Google Scholar]
- Ivars P., Alonso M., Borja M., Hernandez C. (2004). Development of a non-radioactive dot-blot hybridisation assay for the detection of Pelargonium flower break virus and Pelargonium line pattern virus. Eur. J. Plant Pathol. 110 275–283. 10.1023/B:EJPP.0000019798.87567.22 [DOI] [Google Scholar]
- James D., Phelan D. (2017). Complete genome sequence and analysis of blackcurrant leaf chlorosis associated virus; a new member of the genus Idaeovirus. Arch. Virol. 162 1705–1709. 10.1007/s00705-017-3257-9 [DOI] [PubMed] [Google Scholar]
- James D., Phelan J., Jesperson G. (2017). “The Geneva complex – an interesting disease perspective based on the results of NGS analysis,” in Proceedings of the 24th International Conference on Virus and Other Graft Transmissible Diseases of Fruit Crops (ICVF), Thessaloniki, 43. [Google Scholar]
- James D., Varga A., Pállas V., Candresse T. (2006). Strategies for simultaneous detection of multiple plant viruses. Can. J. Plant Pathol. 28 16–29. 28687754 [Google Scholar]
- Jiang D., Hou W., Sano T., Kang N., Qin L., Wu Z., et al. (2013). Rapid detection and identification of viroids in the genus Coleviroid using a universal probe. J. Virol. Methods 187 321–326. 10.1016/j.jviromet.2012.10.006 [DOI] [PubMed] [Google Scholar]
- Jo Y., Lian S., Chu H., Cho J. K., Yoo A.-H., Choi H., et al. (2018). Peach RNA viromes in six different peach cultivars. Sci. Rep. 8:1844. 10.1038/s41598-018-20256-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones S., Baizan-Edge A., Macfarlane S., Torrance L. (2017). Viral diagnostics in plants using next generation sequencing: computational analysis in practice. Front. Plant Sci. 8:1770. 10.3389/fpls.2017.01770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreuze J. F., Perez A., Untiveros M., Quispe D., Fuentes S., Barker I., et al. (2009). Complete viral genome sequence and discovery of novel viruses by deep sequencing of small RNAs: a generic method for diagnosis, discovery and sequencing of viruses. Virology 388 1–7. 10.1016/j.virol.2009.03.024 [DOI] [PubMed] [Google Scholar]
- Kumar S., Baranwal V. K., Joshi S., Arya M., Majumder S. (2010). Simultaneous detection of mixed infection of Onion yellow dwarf virus and an Allexivirus in RT-PCR for ensuring virus free onion bulbs. Ind. J. Virol. 21 64–68. 10.1007/s13337-010-0008-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwak H.-R., Kim M.-K., Shin J.-C., Lee Y.-J., Seo J.-K., Lee H.-U., et al. (2014). The current incidence of viral disease in Korean sweet potatoes and development of multiplex RT-PCR assays for simultaneous detection of eight sweet potato viruses. Plant Pathol. J. 30 416–424. 10.5423/PPJ.OA.04.2014.0029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon J. Y., Hong J. S., Kim M. J., Choi S. H., Min B. E., Song E. G., et al. (2014). Simultaneous multiplex PCR detection of seven cucurbit-infecting viruses. J. Virol. Methods 206 133–139. 10.1016/j.jviromet.2014.06.009 [DOI] [PubMed] [Google Scholar]
- Lee G. P., Min B. E., Kim C. S., Choi S. H., Harn C. H., Kim S. U., et al. (2003). Plant virus cDNA chip hybridization for detection and differentiation of four cucurbit-infecting tobamoviruses. J. Virol. Methods 110 19–24. 10.1016/S0166-0934(03)00082-X [DOI] [PubMed] [Google Scholar]
- Lenz O., Petrzik K., Spak J. (2008). Investigating the sensitivity of a fluorescence-based microarray for the detection of fruit-tree viruses. J. Virol. Methods 148 96–105. 10.1016/j.jviromet.2007.10.018 [DOI] [PubMed] [Google Scholar]
- Lim M. S., Kim S. M., Choi S. H. (2016). Simultaneous detection of three lily-infecting viruses using a multiplex Luminex bead array. J. Virol. Methods 231 34–37. 10.1016/j.jviromet.2016.02.007 [DOI] [PubMed] [Google Scholar]
- Lin L., Li R., Mock R., Kinard G. (2011). Development of a polyprobe to detect six viroids of pome and stone fruit trees. J. Virol. Methods 171 91–97. 10.1016/j.jviromet.2010.10.006 [DOI] [PubMed] [Google Scholar]
- Livak K. J., Flood S. J., Marmaro J., Giusti W., Deetz K. (1995). Oligonucleotides with fluorescent dyes at opposite ends provide a quenched probe system useful for detecting PCR product and nucleic acid hybridization. PCR Meth. Appl. 4 357–362. 10.1101/gr.4.6.357 [DOI] [PubMed] [Google Scholar]
- López M. M., Bertolini E., Olmos A., Caruso P., Gorris M. T., Llop P., et al. (2003). Innovative tools for detection of plant pathogenic viruses and bacteria. Int. Microbiol. 6 233–243. 10.1007/s10123-003-0143-y [DOI] [PubMed] [Google Scholar]
- Lopez M. M., Llop P., Olmos E., Marco-Noales E., Cambra M., Bertolini E. (2009). Are molecular tools solving the challenges posed by detection of plant pathogenic bacteria and viruses? Curr. Issues Mol. Biol 11 13–46. [PubMed] [Google Scholar]
- Lopez-Fabuel I., Wetzel T., Bertolini E., Bassler A., Vidal E., Torres L. B., et al. (2013). Real-time multiplex RT-PCR for the simultaneous detection of the five main grapevine viruses. J. Virol. Methods 188 21–24. 10.1016/j.jviromet.2012.11.034 [DOI] [PubMed] [Google Scholar]
- Luigi M., Costantini E., Luison D., Mangiaracina P., Tomassoli L., Faggioli F. (2014). A diagnostic method for the simultaneous detection and identification of pospiviroids. J. Plant Pathol. 96 151–158. [Google Scholar]
- Majumder S., Baranwal V. K. (2014). Simultaneous detection of four garlic viruses by multiplex reverse transcription PCR and their distribution in Indian garlic accessions. J. Virol. Methods 202 34–38. 10.1016/j.jviromet.2014.02.019 [DOI] [PubMed] [Google Scholar]
- Malandraki I., Varveri C., Olmos A., Vassilakos N. (2015). One-step multiplex quantitative RT-PCR for the simultaneous detection of viroids and phytoplasmas of pome fruit trees. J. Virol. Methods 213 12–17. 10.1016/j.jviromet.2014.11.010 [DOI] [PubMed] [Google Scholar]
- Maliogka V. I., Dovas C. I., Katis N. I. (2007). Demarcation of ilarviruses based on the phylogeny of RNA2-encoded RdRp and a generic ramped annealing RT-PCR. Arch. Virol. 152 1687–1698. 10.1007/s00705-007-0995-0 [DOI] [PubMed] [Google Scholar]
- Mao F., Leung W. Y., Xin X. (2007). Characterization of EvaGreen and the implication of its physicochemical properties for qPCR applications. BMC Biotechnol. 7:76. 10.1186/1472-6750-7-76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin R. R., Constable F., Tzanetakis I. E. (2016). Quarantine regulations and the impact of modern detection methods. Annu. Rev. Phytopathol. 38 207–239. 10.1146/annurev-phyto-080615-100105 [DOI] [PubMed] [Google Scholar]
- Martin R. R., James D., Levesque C. A. (2000). Impacts of molecular diagnostic technologies on plant disease management. Annu. Rev. Phytopathol. 38 207–239. 10.1146/annurev.phyto.38.1.207 [DOI] [PubMed] [Google Scholar]
- Massart S., Olmos A., Jijakli H., Candresse T. (2014). Current impact and future directions of high throughput sequencing in plant virus diagnostics. Virus Res. 188 90–96. 10.1016/j.virusres.2014.03.029 [DOI] [PubMed] [Google Scholar]
- Maule A. J., Hull R., Donson J. (1983). The application of spot hybridization to the detection of DNA and RNA viruses in plant tissues. J. Virol. Methods 6 215–224. 10.1016/0166-0934(83)90048-4 [DOI] [PubMed] [Google Scholar]
- Minutillo S. A., Mascia T., Gallitelli D. (2012). A DNA probe mix for the multiplex detection of ten artichoke viruses. Eur. J. Plant Pathol. 134 459–465. 10.1007/s10658-012-0032-3 [DOI] [Google Scholar]
- Mortimer-Jones S. M., Jones M. G. K., Jones R. A. C., Thomson G., Dwyer G. I. (2009). A single tube, quantitative real-time RT-PCR assay that detects four potato viruses simultaneously. J. Virol. Methods 161 289–296. 10.1016/j.jviromet.2009.06.027 [DOI] [PubMed] [Google Scholar]
- Mühlbach H. P., Weber U., Gomez G., Pállas V., Duran-Vila N., Hadidi A. (2003). “Molecular hybridization,” in Viroids, eds Hadidi A., Flores R., Randles J. W., Semancik J. (Collingwood: CSIRO; ), 103–114. [Google Scholar]
- Nam M., Lee Y.-H., Park C. Y., Lee M.-A., Bae Y.-S., Lim S., et al. (2015). Development of multiplex RT-PCR for simultaneous detection of garlic viruses and the incidence of garlic viral disease in garlic genetic resources. Plant Pathol. J. 31 90–96. 10.5423/PPJ.NT.10.2014.0114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicolaisen M. (2011). An oligonucleotide-based microarray for detection of plant RNA viruses. J. Virol. Methods 173 137–143. 10.1016/j.jviromet.2011.01.022 [DOI] [PubMed] [Google Scholar]
- Olivier T., Demonty E., Fauche F., Steyer S. (2014). Generic detection and identification of pospiviroids. Arch. Virol. 159 2097–2102. 10.1007/s00705-014-1978-6 [DOI] [PubMed] [Google Scholar]
- Otti G., Bouvaine S., Kimata B., Mkamillo G., Kumar P. L., Tomlins K., et al. (2016). High-throughput multiplex real-time PCR assay for the simultaneous quantification of DNA and RNA viruses infecting cassava plants. J. Appl. Microb. 120 1346–1356. 10.1111/jam.13043 [DOI] [PubMed] [Google Scholar]
- Owens R. A., Diener T. O. (1981). Sensitive and rapid diagnosis of potato spindle tuber viroid disease by nucleic acid hybridization. Science 213 670–672. 10.1126/science.213.4508.670 [DOI] [PubMed] [Google Scholar]
- Pallás V., Faggioli F., Aparico F., Sánchez-Navarro J. A. (2011). “Molecular hybridization techniques for detecting and studying fruit tree viruses and viroids,” in Virus and Virus-Like Diseases of Pome and Stone Fruits, eds Hadidi A., Barba M., Candresse T., Jelkmann W. (Saint Paul, MN: American Phytopathological Society; ), 335–342. [Google Scholar]
- Pallás V., Más P., Sánchez-Navarro J. A. (1998). “Detection of plant RNA viruses by non-isotopic dot-blot hybridization,” in Plant Virus Protocols: from Virus Isolation to Transgenic Resistance, eds Foster G., Taylor S. (Totowa, NJ: Humana Press; ), 461–468. [Google Scholar]
- Pallás V., Sánchez-Navarro J. A., Kinard G. K., Di Serio F. (2017). “Molecular hybridization techniques for detecting and studying viroids,” in Viroids and Satellites, eds Hadidi A., Flores R., Randles J. W., Palukaitis P. (London: Elsevier Inc.), 369–379. [Google Scholar]
- Papayiannis L. C., Harkou I. S., Markou Y. M., Demetriou C. N., Katis N. I. (2011). Rapid discrimination of Tomato chlorosis virus, Tomato infectious chlorosis virus and co-amplification of plant internal control using real-time RT-PCR. J. Virol. Methods 176 53–59. 10.1016/j.jviromet.2011.05.036 [DOI] [PubMed] [Google Scholar]
- Park K. S., Bae Y. J., Jung E. J., Kang S. J. (2005). RT-PCR-based detection of six garlic viruses and their phylogenetic relationships. J. Microbiol. Biotechnol. 15 1110–1114. [Google Scholar]
- Pasquini G., Barba M., Hadidi A., Faggioli F., Negri R., Sobol I., et al. (2008). Oligonucleotide microarray-based detection and genotyping of Plum pox virus. J. Virol. Methods 147 118–126. 10.1016/j.jviromet.2007.08.019 [DOI] [PubMed] [Google Scholar]
- Peiró A., Pallás V., Sánchez-Navarro J. A. (2012). Simultaneous detection of eight viruses and two viroids affecting stone fruit trees by using a unique polyprobe. Eur. J. Plant Pathol. 132 469–475. 10.1007/s10658-011-9893-0 [DOI] [Google Scholar]
- Roossinck M. J. (2017). Deep sequencing for discovery and evolutionary analysis of plant viruses. Virus Res. 239 82–86. 10.1016/j.virusres.2016.11.019 [DOI] [PubMed] [Google Scholar]
- Rott M., Xiang Y., Boyes I., Belton M., Saeed H., Kesanakurti P., et al. (2017). Application of next generation sequencing for diagnostic testing of tree fruit viruses and viroids. Plant Dis. 101 1489–1499. 10.1094/PDIS-03-17-0306-RE [DOI] [PubMed] [Google Scholar]
- Roy A., Fayad A., Barthe G., Brlansky R. H. (2005). A multiplex polymerase chain reaction method for reliable, sensitive and simultaneous detection of multiple viruses in citrus trees. J. Virol. Methods 129 47–55. 10.1016/j.jviromet.2005.05.008 [DOI] [PubMed] [Google Scholar]
- Saade M., Aparicio F., Sánchez-Navarro J. A., Herranz M. C., Myrta A., Di Terlizzi B., et al. (2000). Simultaneous detection of three ilarviruses affecting stone fruits by non-isotopic molecular hybridization and multiplex RT-PCR. Phytopathology 96 1330–1336. 10.1094/PHYTO.2000.90.12.1330 [DOI] [PubMed] [Google Scholar]
- Saldarelli P., Barbarossa L., Grieco F., Gallitelli D. (1996). Digoxigenin-labelled riboprobes applied to phytosanitary certification of tomato in Italy. Plant Dis. 80 1343–1346. 10.1094/PD-80-1343 [DOI] [Google Scholar]
- Sánchez-Navarro J. A., Aparicio F., Herranz M. C., Minafra A., Myrta A., Pállas V. (2005). Simultaneous detection and identification of eight stone fruit viruses by one-step RT-PCR. Eur. J. Plant Pathol. 111 77–84. 10.1007/s10658-004-1422-y [DOI] [Google Scholar]
- Sánchez-Navarro J. A., Cañizares M. C., Cano E. A., Pallás V. (1999). Simultaneous detection of five carnation viruses by non-isotopic molecular hybridization. J. Virol. Methods 82 167–175. 10.1016/S0166-0934(99)00097-X [DOI] [PubMed] [Google Scholar]
- Sánchez-Navarro J. A., Cooper C. N., Pallás V. (2018a). Polyvalent detection of members of the Potyvirus genus by molecular hybridization using a ‘Genus-probe’. Phytopathology 10.1094/PHYTO-04-18-0146-R [Epub ahead of print]. [DOI] [PubMed] [Google Scholar]
- Sánchez-Navarro J. A., Fiore N., Fajardo T. V. M., Pallás V. (2018b). “Simultaneous detection of the 13 viruses and 5 viroids affecting grapevine by molecular hybridization using a unique probe or ‘polyprobe’,” in Proceedings of the 19th Conference of the International Council for the Study of Virus and Virus-like Diseases of the Grapevine, Santiago. [Google Scholar]
- Seo J.-K., Lee Y.-J., Kim M.-K., Lee S.-H., Kim K.-H., Choi H.-S. (2014). A novel set of polyvalent primers that detect members of the genera Bromovirus and Cucumovirus. J. Virol. Methods 203 112–115. 10.1016/j.jviromet.2014.03.026 [DOI] [PubMed] [Google Scholar]
- Stobbe A. H., Schneider W. L., Hoyt P. R., Melcher U. (2014). Screening metagenomic data for viruses using the e-probe diagnostic nucleic acid assay. Phytopathology 104 1125–1129. 10.1094/PHYTO-11-13-0310-R [DOI] [PubMed] [Google Scholar]
- Tao Y., Man J., Wu Y. (2012). Development of a multiplex polymerase chain reaction for simultaneous detection of wheat viruses and a Phytoplasma in China. Arch. Virol. 157 1261–1267. 10.1007/s00705-012-1294-y [DOI] [PubMed] [Google Scholar]
- Tiberini A., Barba M. (2012). Optimization and improvement of oligonucleotide microarray-based detection of tomato viruses and pospiviroids. J. Virol. Methods 185 43–51. 10.1016/j.jviromet.2012.05.028 [DOI] [PubMed] [Google Scholar]
- Tiberini A., Barba M. (2013). Development of a oligonucleotide-based microarray for detection of multiple artichoke viruses. J. Plant Pathol. 95 145–154. [Google Scholar]
- Tiberini A., Tomassolia L., Barbaa M., Hadidic A. (2010). Oligonucleotide microarray-based detection and identification of 10 major tomato viruses. J. Virol. Methods 168 133–140. 10.1016/j.jviromet.2010.05.003 [DOI] [PubMed] [Google Scholar]
- Torchetti E., Navarro B., Di Serio F. (2012). A single polyprobe for detecting simultaneously eight pospiviroids infecting ornamentals and vegetables. J. Virol. Methods 186 141–146. 10.1016/j.jviromet.2012.08.005 [DOI] [PubMed] [Google Scholar]
- Torres E., Bertolini E., Cambra M., Monton C., Martin M. P. (2005). Real-time PCR for simultaneous and quantitative detection of quarantine phytoplasmas from apple proliferation (16SrX) group. Mol. Cell. Probes 19 334–340. 10.1016/j.mcp.2005.06.002 [DOI] [PubMed] [Google Scholar]
- Tuo D., Shen W., Yang Y., Yan P., Li X., Zhou P. (2014). Development and validation of a multiplex reverse transcription PCR assay for simultaneous detection of three papaya viruses. Viruses 6 3893–3906. 10.3390/v6103893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyagi S., Kramer F. R. (1996). Molecular beacons: probes that fluoresce upon hybridization. Nat. Biotechnol. 14 303–308. 10.1038/nbt0396-303 [DOI] [PubMed] [Google Scholar]
- van Brunschot S. L., Bergervoet J. H. W., Pagendam D. E., de Weerdt M., Geering A. D. W., Drenth A., et al. (2014a). A bead-based suspension array for the multiplexed detection of begomoviruses and their whitefly vectors. J. Virol. Methods 198 86–94. 10.1016/j.jviromet.2013.12.014 [DOI] [PubMed] [Google Scholar]
- van Brunschot S. L., Bergervoet J. H. W., Pagendam D. E., de Weerdt M., Geering A. D. W., Drenth A., et al. (2014b). Development of a multiplexed bead-based suspension array for the detection and discrimination of Pospiviroid plant pathogens. PLoS One 9:e84743. 10.1371/journal.pone.0084743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verdin E., Wipf-Scheibel C., Gognalons P., Aller F., Jacquemond M., Tepfer M. (2017). Sequencing viral siRNAs to identify previously undescribed viruses and viroids in a panel of ornamental plant samples structured as a matrix of pools. Virus Res. 241 19–28. 10.1016/j.virusres.2017.05.019 [DOI] [PubMed] [Google Scholar]
- Vet J. A. M., Majithia A. R., Marras S. A. E., Tyagi S., Dube S., Poiesz B. J., et al. (1999). Multiplex detection of four pathogenic retroviruses using molecular beacons. Proc. Natl. Acad. Sci. U.S.A. 96 6394–6399. 10.1073/pnas.96.11.6394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villamor D. E. V., Mekuria T. A., Pillai S. S., Eastwell K. C. (2016). High-throughput sequencing identifies novel viruses in nectarine: insights to the etiology of stem pitting disease. Phytopathology 106 519–527. 10.1094/PHYTO-07-15-0168-R [DOI] [PubMed] [Google Scholar]
- Visser M., Burger J. T., Maree H. J. (2016). Targeted virus detection in next- generation sequencing data using an automated e-probe based approach. Virology 495 122–128. 10.1016/j.virol.2016.05.008 [DOI] [PubMed] [Google Scholar]
- Wei T., Pearson M. N., Blohm D., Nolte M., Armstrong K. (2009). Development of a short oligonucleotide microarray for the detection and identification of multiple potyviruses. J. Virol. Methods 162 109–118. 10.1016/j.jviromet.2009.07.024 [DOI] [PubMed] [Google Scholar]
- WenXing X., Ni H., QiuTing J., Farooq A. B. U., ZeQiong W., YanSu S., et al. (2009). Probe binding to host proteins: a cause for false positive signals in viroid detection by tissue hybridization. Virus Res. 145 26–30. 10.1016/j.virusres.2009.06.011 [DOI] [PubMed] [Google Scholar]
- Wu Q., Ding S.-W., Zhang Y., Zhu S. (2015). Identification of viruses and viroids by next-generation sequencing and homology-dependent and homology independent algorithms. Annu. Rev. Phytopathol. 53 425–444. 10.1146/annurev-phyto-080614-120030 [DOI] [PubMed] [Google Scholar]
- Wylie S. J., Luo H., Li H., Jones M. K. J. (2012). Multiple polyadenylated RNA viruses detected in pooled cultivated and wild plant samples. Arch. Virol. 157 271–284. 10.1007/s00705-011-1166-x [DOI] [PubMed] [Google Scholar]
- Zamora-Macorra E., Leobardo Ochoa-Martinez D., Valdovinos-Ponce G., Rojas-Martinez R., Ramirez-Rojas S., Sánchez-Navarro J. A., et al. (2015). Simultaneous detection of Clavibacter michiganensis subsp michiganensis, Pepino mosaic virus and Mexican papita viroid by non-radioactive molecular hybridization using a unique polyprobe. Eur. J. Plant Pathol. 143 779–787. 10.1007/s10658-015-0729-1 [DOI] [Google Scholar]
- Zhang Y., Yin J., Jiang D., Xin Y., Ding F., Deng Z., et al. (2013). A universal oligonucleotide microarray with a minimal number of probes for the detection and identification of viroids at the genus level. PLoS One 8:e64474. 10.1371/journal.pone.0064474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z., Peng S., Jiang D., Pan S., Wang H., Li S. (2012). Development of a polyprobe for the simultaneous detection of four grapevine viroids in grapevine plants. Eur. J. Plant Pathol. 132 9–16. 10.1007/s10658-011-9856-5 [DOI] [Google Scholar]
- Zhang Z., Qi S., Tang N., Zhang X., Chen S., Zhu P., et al. (2014). Discovery of replicating circular RNAs by RNA-seq and computational algorithms. PLoS Pathog. 10:e1004553. 10.1371/journal.ppat.1004553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X., Liu X., Ge B., Li M., Hong B. (2015). A multiplex RT-PCR for simultaneous detection and identification of five viruses and two viroids infecting chrysanthemum. Arch. Virol. 160 1145–1152. 10.1007/s00705-015-2360-z [DOI] [PubMed] [Google Scholar]
- Zheng L., Rodoni B. C., Gibbs M. J., Gibbs A. J. (2010). A novel pair of universal primers for the detection of potyviruses. Plant Pathol. 59 211–220. 10.1111/j.1365-3059.2009.02201.x [DOI] [Google Scholar]
- Zhu S., Zhang Y., Tiberini A., Barba M. (2017). “Detection and identification of viroids by microarrays,” in Viroids and Satellites, eds Hadidi A., Flores R., Randles J. W., Palukaitis P. (London: Elsevier Inc.), 393–412. [Google Scholar]
- Zipper H., Brunner H., Bernhagen J., Vitzthum F. (2004). Investigations on DNA intercalation and surface binding by SYBR Green I, its structure determination and methodological implications. Nucleic Acids Res. 32:e103. 10.1093/nar/gnh101 [DOI] [PMC free article] [PubMed] [Google Scholar]


