Abstract
Evaluating potential adverse effects of complex chemical mixtures in the environment is challenging. One way to address that challenge is through more integrated analysis of chemical monitoring and biological effects data. In the present study, water samples from five locations near two municipal wastewater treatment plants in the St. Croix River basin, on the border of MN and WI, USA, were analyzed for 127 organic contaminants. Known chemical-gene interactions were used to develop site-specific knowledge assembly models (KAMs) and formulate hypotheses concerning possible biological effects associated with chemicals detected in water samples from each location. Additionally, hepatic gene expression data were collected for fathead minnows (Pimephales promelas) exposed in situ, for 12 d, at each location. Expression data from oligonucleotide microarrays were analyzed to identify functional annotation terms enriched among the differentially-expressed probes. The general nature of many of the terms made hypothesis formulation on the basis of the transcriptome-level response alone difficult. However, integrated analysis of the transcriptome data in the context of the site-specific KAMs allowed for evaluation of the likelihood of specific chemicals contributing to observed biological responses. Thirteen chemicals (atrazine, carbamazepine, metformin, thiabendazole, diazepam, cholesterol, p-cresol, phenytoin, omeprazole, ethyromycin, 17β-estradiol, cimetidine, and estrone), for which there was statistically significant concordance between occurrence at a site and expected biological response as represented in the KAM, were identified. While not definitive, the approach provides a line of evidence for evaluating potential cause-effect relationships between components of a complex mixture of contaminants and biological effects data, which can inform subsequent monitoring and investigation.
Keywords: Contaminants, Chemical mixtures, Chemical-gene interactions, Comparative Toxicogenomics Database, Adverse Outcome Pathway
Capsule:
Reverse causal reasoning and a knowledge assembly model were used to infer potential biological effects and chemicals driving those effects at 5 locations in the St. Croix River basin, USA.
For Table of Contents Only
1. Introduction
Evaluating the potential human health and ecological risks associated with exposure to complex chemical mixtures in the ambient environment is one of the central challenges of chemical safety assessment and environmental protection. To assess these risks and take appropriate management actions, there are a number of important questions that need to be addressed through research and/or monitoring efforts. These include: (1) what contaminants are present at a site and what is the potential for exposure to those contaminants; (2) what hazards may be associated with exposure to those contaminants; (3) what evidence exists that these hazards are occurring in exposed populations; (4) which contaminant(s) are most likely causing the effects observed; and (5) what is(are) their source(s).
Environmental monitoring has historically relied heavily upon targeted instrumental analysis for chemicals of known or potential concern. While chemical monitoring is well suited to identify and characterize targeted chemicals, it provides little or no information about potential biological effects. Chemical monitoring can be effective as a basis for environmental risk assessment when the hazards associated with detected chemicals are well characterized in terms of potency, effect concentration(s), and/or mode(s) of action, as is the case for many “legacy contaminants” such as PCBs and organochlorine pesticides. However, there are tens of thousands of chemicals for which little or no relevant toxicology data are available (Judson et al., 2009). In the case of these “contaminants of emerging concern” (CECs), chemical monitoring data alone are generally insufficient to support site-specific risk assessment and management.
Effects-based monitoring approaches can provide a useful complement to chemical monitoring. They allow for a direct measurement of biological effects which, if properly anchored to adverse outcomes, can be used to address hazards that may be associated with exposure of extant organisms (Altenburger et al., 2015; Brack et al., 2015; Ekman et al., 2013; Schroeder et al., 2016). Because effects-based monitoring tools measure the integrated biological activity of an entire mixture, they are capable of detecting exposure to chemicals, which investigators may not know to measure, or may not have the analytical methods to detect (Connon et al., 2012; Ekman et al., 2013). While many effects-based monitoring approaches provide a relatively narrow scope of characterization (Altenburger et al., 2015), more open-ended or unsupervised approaches can be employed to cover and evaluate a broader spectrum of biological effects. These include, for example, omics measurements performed on exposed organisms (Berninger et al., 2014; Garcia-Reyero et al., 2008, 2009, 2011; Martinović -Weigelt et al., 2014; Skelton et al., 2014), as well as batteries of pathway-based in vitro assays (Escher et al., 2014; Schroeder et al., 2016).
Despite their strengths, effects-based methods have important limitations. Notably, they rarely provide insights into which chemicals are causing the observed biological responses unless coupled with detailed and often costly and time-consuming bioassay-directed fractionation. Without the ability to connect specific chemicals, or at least chemical classes, to a particular effect, it is difficult to determine appropriate management actions. Consequently, approaches that integrate chemical monitoring with biological effects data may be useful to address the questions outlined above and to evaluate risks associated with specific chemicals present in the environment.
Combination of statistical and knowledge-based approaches to data integration can offer efficient means to generate additional lines of evidence that can inform subsequent research, monitoring, or decision-making as appropriate. Existing computational approaches can be used to build network models based on a priori knowledge about chemical exposures and biological effects which can allow for integration of these two types of monitoring data (Chindelevitch et al., 2012; Hoeng et al., 2012). For example, Reverse Causal Reasoning, a reverse engineering algorithm, has been used to identify chemicals that provide statistically significant explanations for differential measurements in a molecular profiling data set (Catlett et al., 2013). For this approach, a priori knowledge is first used to generate a large network of potential cause and effect relationships, i.e., a Knowledge Assembly Model (KAM). Smaller networks, termed hypotheses (HYPs), are derived from the KAM. For each HYP, the upstream node represents an experimental perturbation such as exposure to a chemical and the downstream nodes represent biological effects, such as a change in mRNA abundance. The edges in the networks specify an “increased”, “decreased”, or “ambiguous” relationship between chemicals and biological effects. These networks can then be evaluated for richness, which refers to the number of significantly increased or decreased downstream nodes relative to the entire population of nodes, and concordance, which refers to the consistency of the observed state, such as an increase or decrease in mRNA abundance, with the direction of change expected in response to the upstream node (Martin et al., 2012; Laifenfeld et al., 2014).
A KAM can be derived from a knowledge base that provides the cause and effect relationships necessary to develop the network. A number of publically available online resources have assembled, curated, and organized information about chemical-gene and chemical-protein interactions into computationally-accessible databases (Schroeder et al., 2016). For example, both the Search Tool for Interactions of Chemicals (STITCH; Kuhn et al., 2012) and the Comparative Toxicogenomics Database (CTD; Davis et al., 2013) provide information about the impacts of chemicals on biological responses utilizing experimental data from controlled laboratory studies.
Despite unavoidable limitations in terms of chemical and taxonomic coverage (e.g., heavy mammalian bias) and general lack of data for dose-, time-, target-dependency, and route of exposure for many of the chemicals, these sources nonetheless provide a knowledge base suitable for building qualitative KAMs that can be used as a tool for integrated analysis of chemical monitoring and biological effects data. For example, when only chemical monitoring data are available, the KAMs could be a useful first step for identifying contaminants of concern and hypothesizing the potential downstream biological impacts (i.e., perturbed genes or pathways; Schroeder et al., 2016). When both chemical and biological data are available for a site, HYPs derived from KAMs can support statistically guided inference concerning which chemicals are potentially associated with the observed biological responses (Martin et al., 2012). For example, studies utilizing this approach have identified biological effect signatures due to 2-butoxyethanol exposure (Laifenfeld et al., 2010) and drug-induced damage in the liver (Laifenfeld et al., 2014). Thus, KAMs have the potential to identify possible biological effects associated with a particular chemical exposure or, conversely, potential chemical causes associated with a given biological effect.
Recently, we used a KAM based on information in the CTD to predict the biological impacts of chemicals on field-exposed fish when chemical and biological data were not obtained simultaneously (Martinović-Weigelt et al., 2014). The objective of the present study was to further demonstrate the potential utility of knowledge-based approaches. Specifically, we used knowledge from the CTD to develop chemical-gene interaction network models (i.e., KAMs) and applied them, not only to predict potential biological effects of chemicals but also to identify the chemicals in environmental samples that may be associated with observed biological responses (Figure 1). To achieve this we first measured contaminant concentrations in water collected at five locations associated with two wastewater treatment plants (WWTPs) as well as relative hepatic mRNA transcript abundance in fish exposed in situ at each location. The CTD was used to identify genes whose expression had been previously reported to be affected by one or more of the detected chemicals. A KAM was developed for each location to generate location-specific hypotheses about the potential impacts of the chemicals detected in the environmental samples on gene expression. Reverse Causal Reasoning and the KAMs were then used to statistically evaluate HYPs as a means to identify which chemicals in the environmental samples were potentially contributing to the gene expression responses observed in the fish. The present study illustrates how KAMs can be used to integrate exposure and effects data and provide a line of evidence that can help address key questions important to environmental assessments concerning complex mixtures of emerging contaminants.
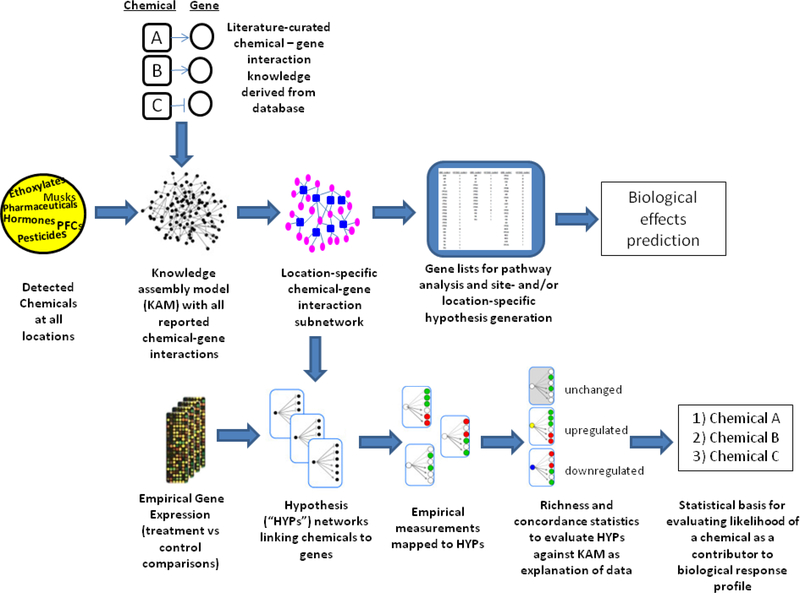
Figure 1 –
Workflow diagram showing the construction of the knowledge assembly model (KAM) for location-specific hypothesis generation and biological effects prediction or statistical, weight-of-evidence, based approach for evaluating which chemicals are likely associated with empirical gene expression results based on richness and concordance in relation to prior knowledge. (Modified from 14,15).
2. Materials and Methods
2.1. Site Characterization and Targeted Chemical Analyses
The present study focused on two sites near WWTPs in the St. Croix River basin in Minnesota and Wisconsin (Supplementary Figure S1). Both WWTPs treat influent that is 90 to 100 percent domestic, have biological phosphorus removal and ammonia reduction, and are located in mixed land use areas (Supplementary Table S1). The WWTP near North Branch, MN serves a population of 10,000 people and discharges an average of 0.62 million gallons of treated effluent daily into the North Branch of the Sunrise River (stream N). The WWTP near Chisago, MN serves a population of 11,000 and discharges every 2.5 h (1.1 million gallons per day) into a small stream (stream C) with no flow upstream of the WWTP. Upstream (US), effluent (EFF), and downstream (DS) locations were established at stream N. At stream C, only EFF and DS locations were sampled, as there was no flow upstream of the WWTP during the study period. Downstream sampling locations were assigned to areas where effluent was completely mixed with stream water as indicated by a return to uniform specific conductance across the stream channel.
Water samples were collected during the summer of 2012. Effluent samples were collected in 1L solvent-rinsed glass amber bottles by multiple vertical dips using a weighted sampler. In-stream samples were collected using Teflon bottles with a depth-integrating sampler at 5 to 10 equally spaced points across the channel to ensure samples were representative of the entire stream cross-section. All samples from each location were composited before dispensing into sample bottles and shipping for analysis. Water samples were analyzed at the U.S. Geological Survey National Water Quality Laboratory for 137 analytes including 69 wastewater-indicator compounds, 48 pharmaceuticals, and 20 natural or synthetic steroid hormones (for analytical methods and complete analyte list see Supplementary text and Table S2).
To visualize chemical occurrence patterns, hierarchical clustering was conducted (Pearson correlation with average linkage clustering) using Multi Experiment Viewer (MeV v 4.8; Saeed et al., 2003). Chemicals with concentrations below the method detection limit were considered to not be present and those chemical concentrations that were above the detection limit were considered detected analytes. For KAM development and analyses, only chemical “presence” or “absence” was considered; concentrations were not.
2.2. Development of Chemical-gene Interaction Knowledge Assembly Model (KAM)
A KAM focused on chemical-gene interactions was developed from information available in the CTD (http://ctdbase.org; Davis et al., 2013). The CTD was queried in June 2014 to identify genes whose expression was reported to be affected by the chemicals detected in water monitoring (Supplementary Table S2). A batch query was performed using Chemical Abstract Service (CAS) numbers for each chemical detected at a location. For the current analysis, gene transcripts whose expression was affected in any manner (i.e., reported as effects, up, or down regulated) by chemical exposure were included in the KAM. The chemical, gene symbol, species the interaction was reported in, and the literature reference were downloaded and compiled. Any transcript whose expression was reported to be affected due to co-treatment with another chemical not detected in the samples was removed (i.e., mixture data were not used). Because the CTD can report the same chemical-gene interactions, but from different species, redundant interactions were identified and removed to prevent over-representation of a single chemical-gene interaction within the KAM.
A location-specific sub-network was developed from the KAM to extract chemical-gene interaction information specific to each location. The KAM and location-specific chemical-gene interaction model was visualized using Cytoscape v2.8.3 (Smoot et al.,2011). The gene symbol, degree number for each gene, chemical(s) interacting with the gene, and the directionality of the gene expression was extracted for all of the genes present in the sub-network. Each KAM-derived gene list represents a set of testable hypotheses concerning which sites or locations would have the greatest biological effects and which genes and pathways are likely to be altered following in situ exposure at a given location.
2.3. In Situ Fish Exposure
Fish exposures were conducted concurrently in summer of 2012. Sexually mature, 7-month old, laboratory-reared fathead minnows were transported to the field sites in identical, aerated containers holding well-water from the Saint Cloud State University’s Aquatic Facility. Once at the assigned exposure site, 20 randomly selected male fish (two replicates of 10 males per treatment) were moved into mini-mobile environmental monitoring units (MMUs; Kolok et al.,2012). The MMUs were continually supplied with a flow of water from: 1) a well (Control treatment), 2) stream C-EFF, 3) stream C-DS, 4) stream N-US, 5) stream N-EFF or 6) stream N-DS. Flow rates were approximately 200 mL/min per chamber with additional water being directed into a jacket surrounding the fish exposure chambers to buffer against water temperature changes associated with fluctuations in air temperature. As a result, water temperature in the MMUs closely mimicked the stream conditions of 23.4 (mean) ± 2.1 (SD) °C during the time of exposure with variations in water temperature reflecting diurnal temperature cycling (measured by automated temperature loggers). Water was continually aerated, and fish were fed daily with frozen brine shrimp. After 12 d of exposure, fish were transported (3 h transit) to the University of St. Thomas (UST) in identical, aerated containers containing site-specific water. Tissue collection started immediately upon arrival at UST. Fish were anesthetized using buffered MS-222, and morphometric measurements (mass, length, secondary sex characteristics assessment) were conducted. Liver and testes were excised, weighed, and stored appropriately (see below) for either microarray or histopathology analyses. All procedures involving live fish were reviewed and approved by the SCSU and UST Institutional Animal Care and Use Committees.
2.4. Plasma vitellogenin analysis
Circulating plasma vitellogenin was measured following previously published protocols (Dammann et al., 2011); competitive ELISA method that incorporates a species-specific polyclonal anti-vitellogenin antibody and purified vitellogenin standard was used.
2.5. Morphological endpoints
Whole body weights were measured for each fish (0.01g precision, Acculab Vicon, Edgewood, NY). Gonads and livers from each fish were excised and immediately weighed (0.001g precision, Mettler Toledo AG245, Columbus, OH). Liver and whole body weights were used to calculate the hepatosomatic index (HSI = liver weight/whole body weight × 100). Gonad and whole body weights were used to calculate the gonadosomatic index (GSI = gonad weight/whole body weight × 100). Body weight and total length was used to calculate the body condition factor (BCF = body weight/total length3), a measure of the overall metabolic condition of the fish (Fulton, 1904). Development of male-specific secondary sex characteristics (tubercles and dorsal pad) was scored using a qualitative scale 0 (no pad; no tubercles) to 3 (sharp, prominent tubercles; dorsal pad wide and thick forming a sharp nape behind head); this approach has been widely published/deployed by others (e.g., Danylchuk and Tonn, 2001).
2.6. Histopathology
After a 1-week fixation period in 10% neutral buffered formalin, tissue samples were dehydrated through a series of ethanol and xylene baths in a Leica automated tissue processor TP 050 (Leica, Wetzlar, Germany) and embedded in paraffin using a Thermo Scientific Microm EC 350–1 embedding station (Waltham, MA). Embedded tissues were sectioned at approximately 1/3 and 2/3 of the depth of the gonads (resulting in tissue slices ~ 100 μm apart) using a Reichert-Jung cassette microtome (Leica, Wetzlar, Germany; 4 μm sections). At least 6 sections from each organ (gonad, liver) were stained using standard hematoxylin and eosin techniques (Carson, 1997) in a Leica Autostainer XL similar to methods used in other histopathological studies (Kidd et al., 2007; Barber et al., 2011). Histological sections were assessed by an experienced histologist (HLS) and ranked on a semi-quantitative scale (0–4) for vacuolization of the liver (0 no vacuoles visible; 1 <5% of total area; 2 vacuoles small but throughout image <25% of area; 3 broad presence of large vacuoles 25%−50% of area; 4 >50% of area vacuolated) and the presence or absence of eosinic staining proteinaceous fluid. The developmental stage of the testis was also ranked on a semi-quantitative scale (0–4; Writer et al., 2010). Randomly selected slides were ranked for a second time to determine between analysis variance, which was found to be less than 1%.
2.7. Statistical analyses vitellogenin, morphological, histological endpoints
Effects of the exposure on plasma vitellogenin, and morphological and histopathological endpoints were evaluated using Kruskal-Wallis ANOVA; STATISTICA 10 (StatSoft Inc., Tulsa, OK, USA). If results were significant (p < 0.05) post hoc comparison of mean ranks as implemented in STATISTICA (Siegel and Castellan, 1988) was conducted.
2.8. RNA Extraction and Microarray Analysis
Total RNA was isolated from male liver samples (n = 6–7) using commercial extraction kits (RNeasy, Qiagen, Valencia, CA, USA). Microfluidic gel electrophoresis was used to assess RNA quality (Agilent 2100 Bioanalyzer, Agilent, Wilmington, DE, USA). Quantity was determined using a Nanodrop ND-1000 spectrophotometer (Nanodrop Technologies, Wilmington, DE, USA). Total RNA was stored at −80°C until analyzed.
A custom fathead minnow 60,000 gene array (GPL15775; Garcia-Reyero et al.,2014) was purchased from Agilent Technologies (Palo Alto, CA, USA). One μg of total RNA was used for all hybridizations. Probe labeling, amplification, and hybridization were performed using kits following the manufacturer’s protocols (Quick Amp Labeling Kit and One-Color Microarray Hybridization Protocol, version 6.5; Agilent) and scanned with a high-resolution microarray scanner (Agilent). Data were resolved from microarray images using Agilent Feature Extraction software version 10.7 (Agilent). Raw microarray data were deposited at the Gene Expression Omnibus Web site (GSE81263; http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE81263).
Microarray data were imported into GeneSpring GX 12.6. (SAS Institute Inc., Cary, NC, USA). Data for all treatments and control were subjected to the default normalization procedure for Agilent one-color microarray data (Agilent). GeneSpring’s Guided Workflow default quality control settings (e.g., expression values, flags, filter parameters) were used. Genes that were differentially expressed (DEGs) between controls and treated fish were identified by one-way ANOVA (p < 0.05) followed by post-hoc comparisons of each treatment to control (Tukey’s test, p < 0.05).
2.9. Functional Analysis
Biological pathways statistically enriched in gene lists derived from microarray DEGs or genes represented in the location-specific KAM were identified using GeneCodis3 (Tabas-Madrid et al., 2012). Pathway identifications were based on Gene Ontogeny (GO) Slim biological process annotations and pathways in the Kyoto Encyclopedia of Genes and Genomes (KEGG). Zebrafish-specific processes and pathways were used for these analyses.
In calculating test statistics and enrichment significance, all unique genes present in the CTD were included in the analyses. Gene symbols from all genes represented on the microarray were used as input for the MyVenn function in the CTD to identify genes present in the CTD. There were 23,887 non-redundant and annotated genes present on the microarray and 14,375 (60%) of the microarray genes were represented in the CTD. These 14,375 genes were used as the reference gene list for calculating enrichment. Enrichment was determined using a hypergeometric test with p-value correction with a false discovery rate. Pathway enrichment was considered significant following the software default parameters of at least three genes associated with a pathway and p < 0.05.
2.10. Statistical Analyses for Inferring Chemicals Associated with Biological Responses
Because the contaminants in environmental water samples occur as complex mixtures and multiple chemicals are capable of influencing the expression of a single gene transcript, statistical analyses were used to evaluate each HYP to identify the chemical(s) possibly contributing to the observed mRNA state changes. For each HYP, two independent scores were calculated, an enrichment score (also referred to as richness) and an activation score (also referred to as concordance; Catlett et al., 2013; Pollard et al., 2005; Krämer et al., 2014). The null hypothesis tested for each HYP is that globally observed biological responses cannot be explained by chemical(s) present in a complex environmental mixture, while the alternative hypothesis is that observed biological responses can be explained by chemical(s) present. Richness is the probability that the number of observed state changes downstream of a HYP could have occurred by chance alone and was calculated using a hypergeometric probability distribution. Richness does not account for the direction of change in the mRNA expression, so ambiguous (e.g., “affects”, as opposed to “up-regulates” or “down-regulates” expression) state changes are included in the calculation. Richness was calculated using the phyper function in R (R Core Team, 2014).
Concordance uses the mechanistic information about the gene regulation (up-regulation versus down-regulation) to calculate which chemicals are likely contributing to a particular biological response. Concordance was calculated using a binomial z-score to determine the accuracy of KAM-derived HYPs in predicting gene expression changes observed in field-exposed fish. The z-score is based on a probability of occurrence of 0.5, because the gene expression state change can only be up-regulated or down-regulated. Ambiguous state changes were not included in the calculation. The cumulative binomial probability was calculated using the pbinom function in R (R Core Team, 2014). A HYP was considered statistically (although not necessarily biologically) significant if both richness and concordance met the probability cutoff of less than 0.1 (Catlett et al., 2013).
3. Results and Discussion
3.1. Chemical Characterization of Sites
There were significant differences in the number of chemicals detected in water samples collected from each stream and among locations along a stream (Figure 2; Supplementary Table S2). For example, 46 analytes were detected in stream N among all three locations; four in water from the N-US location, 45 in the N-EFF, and 11 in water from the N-DS location. Chemicals detected at the N-US location and N-EFF appear to be contributing to the overall chemical composition downstream, although with significant in-stream dilution or degradation, as only 25% of the chemicals detected in the effluent were detected downstream. The agricultural pesticide metolachlor was the only chemical that was detected at N-US and N-DS but not in N-EFF.
Figure 2 –
Hierarchical clustering (Euclidean distance, complete linkage) of upstream (UP), effluent (EFF), and downstream (DS) locations for the North Branch (Stream N) and effluent and downstream locations for the Chisago (Stream C) WWTPs based on chemical composition. Chemicals detected at each location are indicated in yellow and chemicals below the method detection limit are indicated in black.
Greater numbers of chemicals were detected in stream C than stream N, with 85 and 79 chemicals measured in water from the C-EFF and C-DS locations, respectively (Figure 2; Supplementary Table S2). Seventy chemicals were detected at both locations, indicating the effluent is a major contributor to the chemicals detected downstream (as one might expect given that C-EFF constituted the headwater of stream C). Among the 46 analytes detected in water from stream N, there were only three which were not detected in water collected from either C-EFF or C-DS: phenanthrene, methotrexate, and bis(2-ethylhexyl) phthalate.
3.2. Bio-Effects Prediction using Knowledge Assembly Models
One of the main challenges associated with analytical monitoring of contaminants of emerging concern is inferring the types(s) of hazards that may be associated with chemical exposure at a site. Effects-based monitoring approaches can complement chemical monitoring, but there remains a question about what endpoint(s), biological pathway(s), taxa, etc. those effects-based approaches should focus on. Knowledge in the literature, summarized in a computationally accessible manner in on-line sources of chemical-gene interaction data, represents one potential source that can be mined to develop hypotheses to help focus subsequent effects-based monitoring (Schroeder et al., 2016).
In the present study, chemical-gene interaction networks developed from the KAM were used to hypothesize the types of biological responses (i.e., genes and pathways) that may occur in fish exposed at each location (i.e., addressing question 2 – what hazards may be associated with exposure to those contaminants). Note, these hypotheses were based only on consideration of the presence or absence of a given chemical, not consideration of its concentration relative to biological potency. Thus, this analysis should be considered a worst case hazard prediction, not a risk-based prediction. Further, the HYPs were neither species nor target tissue-specific. They considered all effects (chemical-gene interactions) reported in the CTD without regard to the species or target organ those effects were measured in. Consequently, the approach is meant to prioritize effects for further hypothesis-based testing, not for definitive prediction of outcomes nor predictive risk assessment.
Genes that could potentially be affected based on chemical-gene interactions from the CTD and the chemicals detected at the three stream N locations (Supplementary Table S3) were subjected to functional enrichment analyses (Supplementary Table S4). Considering enriched biological process gene ontology (GO) terms, there were six that were commonly enriched at all three stream N locations: translation, response to stress, transport, lipid metabolic process, carbohydrate metabolic process, and embryo development. Five of the terms do not directly translate into a well-defined endpoint for effects-based monitoring. However, embryo development does. Based on the results, one recommendation for follow-up investigation may be to test water samples or extracts in a fish embryo development assay, to evaluate whether environmentally-relevant concentrations of the mixture of chemicals found at the stream N sites may adversely affect fish development. “Growth” was the only GO term common to N-EFF and N-DS. Consequently, another endpoint to target for follow-up investigation might be to compare growth of larval fish held in water from N-US versus N-DS.
Considering the enriched KEGG pathways, there were 10 terms common to all N stream sites. Among those, steroid hormone biosynthesis, p53 signaling pathway, and glutathione metabolism suggest endpoints for future effects-based monitoring work. Measures of plasma steroid concentrations or ex vivo steroid production could be recommended as endpoints for future in situ exposures with fish, along with glutathione-related biomarkers. While p53 regulates cell cycle and is widely recognized for its function as a tumor suppressor, cancer is generally not a major endpoint of concern from an ecological risk assessment standpoint. Investigation of a tumor promotion effect may be of lesser interest, unless there was other anecdotal evidence suggesting tumor occurrence in native fish. Finally, considering the KEGG pathways common to N-EFF and N-DS, two-PPAR (peroxisome proliferator-activated receptors) signaling pathway and VEGF (vascular endothelial growth factor) signaling pathway-are specific enough to suggest potential endpoints of interest. Examination of embryo development and growth would be relevant relative to a potential VEGF signaling effect, while follow-up with an in vitro assay designed to characterize PPAR-mediated potency may be an effective first step for considering the potential for PPAR-related effects. For example, there are over 10 commercially-available in vitro assays for PPAR-mediated activity that are currently employed in US EPA’s ToxCast program (actor.epa.gov/dashboard) that could be used to screen water or extracts from the N-DS or N-EFF locations.
In the case of stream C, despite the overall detection of more chemicals and a corresponding increase in the number of chemical-gene interactions represented in the site-specific KAMs, the number of enriched functional annotation terms was not markedly higher than for stream N (Supplementary Table S5). Most of the enriched biological process GO terms relate to fundamental cellular processes such as cell cycle and cell division, synthesis, process and transport of various classes of biomolecules (Supplementary Table S6), and thus do not readily suggest specific endpoints for monitoring. However, enrichment of the embryo development term suggests, once again, that testing of water samples or extracts in a fish embryo assay may be worthwhile. Relative to KEGG pathways, there was a single enriched term associated with both the C-EFF and C-DS sites, nucleotide excision repair (Supplementary Table S6). Impairment of nucleotide excision repair can result in the formation of bulky DNA adducts and may lead to increased mutation rates (Memisoglu and Samson, 2000). Consequently, measures of DNA damage in fish or other organisms exposed at the site would be a reasonable follow-up. However, it should be noted that even if altered gene expression associated with this pathway were to occur, it may not manifest as impairment of DNA repair. Rather it may simply signal adaptive activation of such repair pathways as organisms deal with the additional DNA damage associated with exposure to some of the contaminants present at the site.
While high accuracy should not be expected, knowledge captured in the CTD provided a reasonable basis for formulating testable hypotheses that could be used to focus further investigation. As the case study illustrates, annotation at the level of pathways and biological processes often does not reveal highly specific biological interpretations. For example, it is unclear what apical outcomes may be related to predicted effects on the ribosome pathway. Nevertheless, some of the pathway annotations provide adequate specificity to hypothesize apical effects and target further analyses. For example, the suggestion that steroid hormone biosynthesis would be affected in fish exposed in stream N, could guide efforts to quantify circulating steroid concentrations in fish from those sites and assess reproductive toxicity. Similarly, embryo development signatures at each site in streams N and C suggest that embryo toxicity testing may be warranted. As more biological process or KEGG pathway terms are linked to specific adverse outcomes (i.e., through development of adverse outcome pathways; Ankley et al., 2010; Schroeder et al., 2016), developing testable hypotheses based on prior knowledge found in sources like CTD should become increasingly useful for predicting biological effects.
3.3. Unsupervised Biological Effects Surveillance Using Microarray-Based Gene Expression Profiling
The biological effects prediction approach described above employs analytical characterization of the chemicals present at a site, along with prior knowledge concerning reported chemical-gene interactions as a basis for hypothesis formulation. Although useful if chemical monitoring data are the only source available for characterizing a site, there are numerous limitations. For example, one can only infer effects for chemicals detected at the site. However, it is widely recognized that analytical monitoring approaches only detect a small fraction of the overall exposome (Tang et al.,2013). Likewise, even for the chemicals detected, the scope and scale of data curated into sources like the CTD is generally insufficient to tailor the hypotheses to the species, target tissues, concentration ranges, and routes of exposure relevant to a given field study. For example, in the present study when KAM building was limited to fish-specific data, the networks generated were too sparse to support useful hypothesis generation (not shown). Consequently, when resources and capacity allow, unsupervised biological effects surveillance approaches can be a very useful complement to the analytical chemistry and knowledge-driven biological effects prediction approaches (Ekman et al., 2013; Schroeder et al., 2016; Tang et al., 2013).
In the present study, the approach used for unsupervised biological effects surveillance was examination of hepatic transcriptome responses in fathead minnows exposed in situ. However, the use of a DNA microarray-based approach allowed for screening of effects on a broad range of biological pathways within the liver, a key organ that can serve as both a target site in terms of toxicity and as a site of detoxification with regard to xenobiotic metabolism. It should be noted that unlike CTD-based biological effects prediction or biological effects surveillance employing a battery of high throughput in vitro assays (Schroeder et al., 2016), hepatic transcriptome responses to in situ exposure are not necessarily specific to effects of chemicals. Other environmental variables such as water-quality parameters, dietary influences, etc. also can influence biological response profiles. Although other environmental variables were controlled to the extent possible through the use of MMUs, it cannot be assumed that all transcriptional differences between fish exposed to control well water and stream water were driven by chemicals.
In the case of stream N, the total numbers of genes differentially expressed compared to well water-exposed controls ranged from 4369 for males exposed to water from N-US (Supplementary Table S7) to 3561 for those exposed to N-EFF (Supplementary Table S8), with N-DS showing an intermediate effect at 3918 differentially expressed genes (Supplementary Table S9). In terms of numbers of DEGs, the trend was opposite of what might have been predicted based on total numbers of contaminants detected at each site (4, 45, 11 for N-US, N-EFF, and N-DS, respectively). Nearly 30% (1893) of the differentially-expressed probes were common to the N-US, N-EFF, and N-DS exposed fish. This suggests that the handful of contaminants detected at all three stream N locations (e.g., atrazine, p-cresol, and/or cholesterol) were influencing the transcriptome response. However, it is plausible that non-contaminant-related differences between the well-water controls and N-exposed fish and other contaminants not detected with the analytical techniques used were also contributing.
From a pathway analysis perspective, we were most interested in functional annotation terms associated with the genes enriched among those differentially expressed at the N-EFF and N-DS locations, but not N-US, because the effluent appeared to be a major source of contaminant introduction. No such enriched terms were observed (Figure 3; Supplementary Table S10). Therefore, there was not compelling evidence for an effluent contaminant-related, pathway-specific, effect on the hepatic transcriptome. Genes annotated as playing a functional role in embryo development and lipid metabolic processes were enriched among those differentially expressed in males exposed at the N-US and N-DS, but not N-EFF site. This differed from the site-specificity of the pathway-effects predicted based on the KAM. However, considering that the pathway analysis did not differentiate up-regulation from down-regulation, embryo exposures would still represent a useful follow-up investigation. None of the other enriched terms associated with alterations in the hepatic transcriptome of fish exposed at the N sites helped define endpoints or outcomes to monitor in a whole organism.
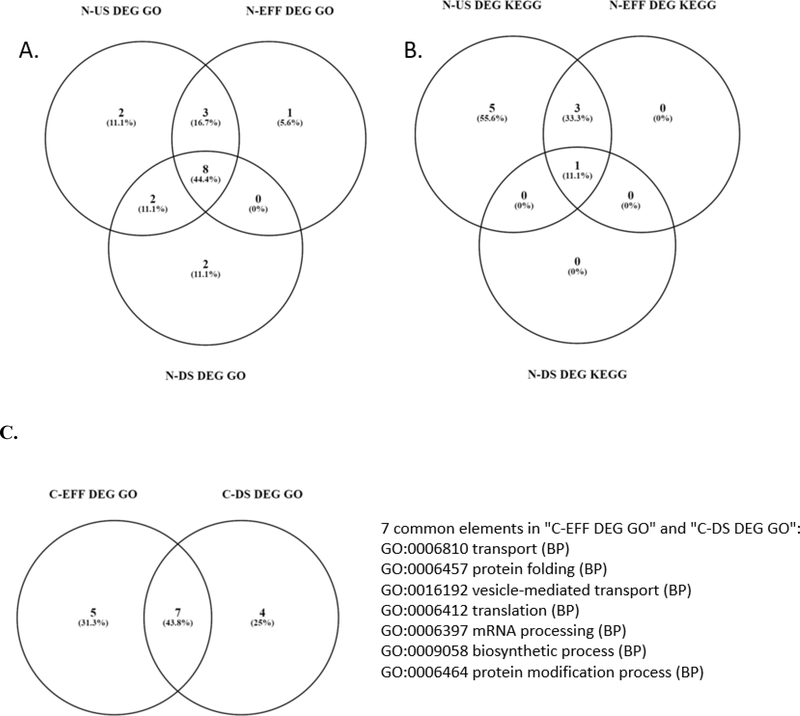
Figure 3 –
Venn diagrams showing the overlap in biological process (BP) gene ontology (GO) terms or Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway annotation terms identified as enriched among the differentially expressed genes identified for the hepatic transcriptome at each site. [A.] Numbers of overlapping biological process GO terms for N stream sites. [B.] Numbers of overlapping KEGG pathway annotation terms for N stream sites. [C.] Number of overlapping biological process GO terms for C stream sites, along with identification of the overlapping terms.
For the two stream C locations, the total numbers of DEGs were similar to that determined for the N stream locations, despite detection of significantly more chemicals. Numbers of DEGs ranged from 2983 for males exposed at the C-EFF location (Supplementary Table S11) to 3286 for those exposed at the C-DS location (Supplementary Table S12). In terms of enriched functional annotations, seven enriched biological process GO terms were common to hepatic transcriptome response to C-EFF and C-DS, relative to the well-water control (Supplementary Table S13; Figure 3C). Similarly, five of the enriched KEGG pathway annotations overlapped for C-EFF and C-DS. However, the overlapping functional annotations referred to very general cellular processes and did not lend much insight or suggest endpoints for future monitoring.
These results illustrate one of the challenges associated with the use of transcriptomics in organisms exposed in situ for biological effects surveillance. Although a rich database of putative DEGs are identified, functional annotation terms often lack the specificity to interpret what the responses may mean in the whole organism. Whereas it is possible to develop a large number of hypotheses by delving into the lists of DEGs in detail, objective selection of targets to focus on can be problematic when there are thousands of options to choose from.
Phenotypic anchoring offers one potential solution. When transcriptome-related responses can be linked to observed phenotypic changes, there is increased confidence in their interpretation. However, in the present study there were no notable morphological or histological changes associated with in situ exposure at the N and C stream locations (Supplementary Table S14). The only significant difference among exposures was reduced body condition factor in the fish exposed to water from C-DS compared to the well-water control. No effects on secondary sex characteristics, plasma vitellogenin, sperm, or liver histopathology were detected.
3.4. Identification of Chemicals Associated with Observed Transcriptomic Responses
The previous two analyses were based on location-specific chemical analyses alone or biological effects alone. However, when both analytical and biological effects data are available, KAM-based approaches can be used to statistically evaluate chemicals likely or unlikely to be causing the effects observed (i.e., addressing question 4 from the Introduction). This can be done using Reverse Causal Reasoning, which evaluates richness and concordance statistics for HYPs representing different chemical-biological interactions for each location (Table 1; Supplementary Tables S15, S16). For example, atrazine was detected at the N-US location. Based on the CTD, 303 RNAs were previously reported to be affected by atrazine exposure. Among those, 36 were identified as differentially expressed in the livers of fish exposed at the N-US location (Supplementary Figure S2). Twenty-three of those 36 DEGs showed a direction of change (up- or down-regulation) consistent with the state changes curated in CTD and subsequently represented in the KAM, providing significant richness and concordance of 1.01E-51 and 0.066, respectively (Table S15, Supporting Information). There were four HYPs with significant richness and concordance from the KAM for the N-EFF, including carbemazepine, metformin, thiabendazole, and diazepam (Table 1). The KAM for the N-DS location also identified four significant HYPs, including atrazine, carbemazepine, cholesterol, and p-cresol. The KAM for the C-EFF identified four significant HYPs (phenytoin, omeprazole, carbamazepine, erythromycin), whereas the KAM for the C-DS location identified five significant HYPs (17β-estradiol, cholesterol, cimetidine, erythromycin, and estrone; Table 1).
Table 1 —
Richness and concordance p-values for the significant HYPs (chemicals) identified at each location.
| Site | Location | HYP (Chemical) | Richness P-Value |
Concordance P-Value |
|---|---|---|---|---|
| North Branch | Upstream | Atrazine | 1.0E-51 | 0.066 |
| North Branch | Effluent | Carbamazepine | 1.7E-19 | 0.015 |
| North Branch | Effluent | Metformin | 3.9E-09 | 0.020 |
| North Branch | Effluent | Thiabendazole | 3.0E-07 | 0.031 |
| North Branch | Effluent | Diazepam | 0.0004 | 0.031 |
| North Branch | Downstream | Cholesterol | 3.1E-20 | 0.0009 |
| North Branch | Downstream | Atrazine | 3.6E-37 | 0.0063 |
| North Branch | Downstream | Carbamazepine | 1.8E-20 | 0.0064 |
| North Branch | Downstream | p-Cresol | 2.2E-07 | 0.027 |
| Chisago | Effluent | Phenytoin | 0.0057 | 0.0001 |
| Chisago | Effluent | Omeprazole | 2.6E-06 | 0.0078 |
| Chisago | Effluent | Carbamazepine | 4.5E-05 | 0.048 |
| Chisago | Effluent | Erythromycin | 0.0061 | 0.062 |
| Chisago | Downstream | 17-beta Estradiol | 0.0001 | 0.0088 |
| Chisago | Downstream | Cholesterol | 0.0029 | 0.011 |
| Chisago | Downstream | Cimetidine | 0.0009 | 0.015 |
| Chisago | Downstream | Erythromycin | 0.0033 | 0.062 |
| Chisago | Downstream | Estrone | 0.074 | 0.062 |
Among the chemicals detected at these locations, those with significant concordance and richness (p<0.1) of biological responses are arguably chemicals for which there is greater weight of evidence they may be directly contributing to responses in organisms exposed at the location. Although this does not mean with certainty that these chemicals are the cause of the responses observed, it does mean that based on existing knowledge, there is a quantifiable likelihood that these chemicals are contributors, which provides a basis for prioritizing them for further monitoring and/or investigation. The method still includes an inherent bias toward the detection of well-studied chemicals because they will have more documented HYPs upon which to base the concordance and richness calculations. Nonetheless, that bias is reflective of the current state of scientific knowledge and is similar to the bias an investigator may impose from simply reviewing available evidence in the literature. The real value in this statistical approach lies in the quantitative evaluation of the overall concordance with the existing literature. The approach utilizes weight-of-evidence for linking chemicals with observed biological response, and thus should have value for integrated environmental monitoring.
4. Conclusions
The current study highlights the potential of KAMs to systematically utilize existing knowledge of chemical-biological interactions to help address key questions concerning the potential effects of chemical exposures in the environment. First, it illustrates how, in the absence of site-specific biological data, KAMs could be used to hypothesize biological effects that may be associated with chemicals detected at a field location. Second, it demonstrates how, when both chemistry and biological response data are available for a site, it is possible to use a KAM-based approach to evaluate involvement of specific chemicals in eliciting the observed biological responses.
Evaluation of chemicals detected at the five field locations along with knowledge concerning chemical-gene interactions curated in the CTD aided hypothesis formulation concerning possible biological effects, and associated identified assays or endpoints to potentially use in future location-specific monitoring. Specifically, follow-up investigations may want to examine effects on fish embryo development and larval growth. At a pathway-specific level, examination of steroid hormone concentrations and (or) production, as well as PPAR-related activity and glutathione status also may be useful. Direct analysis of the hepatic transcriptome responses following exposures to site water was not as fruitful for hypothesis formulation and endpoint selection, largely due to the rather broad and non-specific nature of many of the annotation terms identified via enrichment analyses and uncertainties as to whether the profiles were influenced primarily by chemical contaminants or other factors. Nonetheless, when the transcriptome data were analyzed in the context of the KAMs using reverse causal reasoning, a list of detected chemicals with some evidence to suggest they may be causing biological responses in fish caged at the site(s) were identified. Together these approaches identify a target set of both analytes and assays or endpoints to target in future studies. This type of hypothesis formulation represents an important step in initial environmental surveillance, which can be used to inform subsequent targeted investigation and monitoring (Ekman et al., 2013).
Supplementary Material
Acknowledgements
Support by the USGS and National Park Service (NPS), US EPA (Office of Research and Development’s Chemical Safety for Sustainability Research Program, and Region 5, Great Lakes National Program Office), and National Science Foundation (CBET-1336062 / 1336165 / 1336604). We thank Sarah Elliott, Jeffery Ziegeweid, and Brent Mason at USGS for their field support; and Byron Karns at NPS for logistical support. We thank Maya Peters, Jackie Heitzman, Abigail Lukowicz, Evan Eid, Kyle Stevens, Jenna Cavallin, Megan Hughes, Krysta Nelson, Rebecca Milsk, Travis Saari, and Eric Randolph for help with network development. We thank Lynn Escalon for microarray analysis. Any use of trade, product, or firm names is for descriptive purposes only and does not imply endorsement by the U.S. Government. The contents neither constitute, nor necessarily reflect, US EPA policy. Permission was granted by the Chief of Engineers to publish this information.
References
- Altenburger R, Ait-Aissa S, Antczak P, Backhaus T, Barceló D, Seiler TB, Brion F, Busch W, Chipman K, de Alda ML; de Aragão Umbuzeiro G, 2015. Future water quality monitoring – Adapting tools to deal with mixtures of pollutants in water resource management. Sci. Total Environ 512, 540–551. [DOI] [PubMed] [Google Scholar]
- Ankley GT, Bennett RS, Erickson RJ, Hoff DJ, Hornung MW, Johnson RD, Mount DR, Nichols JW, Russom CL, Schmieder PK, Serrano JA, Tietge JE, Villeneuve DL, 2010. Adverse outcome pathways: a conceptual framework to support ecotoxicology research and risk assessment. Environ. Toxicol. Chem 29, 730–741. [DOI] [PubMed] [Google Scholar]
- Barber LB, Brown GK, Nettesheim TG, Murphy EW, Bartell SE, Schoenfuss HL, 2011. Effects of organic contaminant mixtures on fish in a wastewater dominated urban stream. Sci. Total Environ 409, 4720–4728. [DOI] [PubMed] [Google Scholar]
- Berninger JP, Martinović-Weigelt D, Garcia-Reyero N, Escalon L, Perkins EJ, Ankley GT, Villeneuve DL, 2014. Using transcriptomic tools to evaluate biological effects across effluent gradients at a diverse set of study sites in Minnesota, USA. Environ. Sci. Technol 48, 2404–2412. [DOI] [PubMed] [Google Scholar]
- Brack W, Altenburger R, Schüürmann G, Krauss M, Herráez DL, van Gils J, Slobodnik J, Munthe J, Gawlik BM, van Wezel A, Schriks M, 2015. The SOLUTIONS project: challenges and responses for present and future emerging pollutants in land and water resources management. Sci. Total Environ 15, 503–522. [DOI] [PubMed] [Google Scholar]
- Carson FL, 1997. Histotechnology. ASCP Press, Chicago. [Google Scholar]
- Catlett NL, Bargnesi AJ, Ungerer S, Seagaran T, Ladd W, Elliston KO, Pratt D, 2013. Reverse causal reasoning: applying qualitative causal knowledge to the interpretation of high-throughput data. BMC Bioinformatics 14, 340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chindelevitch L, Ziemek D, Enavetallah A, Randhawa R, Sidders B, Brockel C, Huang ES, 2012. Causal reasoning on biological networks: interpreting transcriptional changes. Bioinformatics 28, 1114–1121. [DOI] [PubMed] [Google Scholar]
- Connon RE, Geist J, Werner I, 2012. Effect-based tools for monitoring and predicting the ecotoxicological effects of chemicals in the aquatic environment. Sensors (Basel) 12, 12741–12771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dammann AA, Shappell NW, Bartell SE, Schoenfuss HL, 2011. Comparing biological effects and potencies of estrone and 17β-estradiol in mature fathead minnows, Pimephales promelas. Aquat. Toxicol 105, 559–568. [DOI] [PubMed] [Google Scholar]
- Danylchuk AJ, Tonn WM, 2001. Effects of social structure on reproductive activity in male fathead minnows (Pimephales promelas). Behav. Ecol 12, 482–489. [Google Scholar]
- Davis AP, Murphy CG, Johnson R, Lay JM, Lennon-Hopkins K, Saraceni-Richards C, Sciaky D, King BL, Rosentein MC, Wiegers TC, Mattingly CJ, 2013. The Comparative Toxicogenomics Database: update 2013. Nucleic Acids Res. 2013, (Database issue), D1104–1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekman DR, Ankley GT, Blazer VS, Collette TW, Garcia-Reyero N, Iwanowicz LR, Jorgensen ZG, Lee KE, Mazik PM, Miller DH, Perkins EJ, Smith ET, Tietge JE, Villeneuve DL, 2013. Biological effects-based tools for monitoring impacted surface waters in the Great Lakes: a multiagency program in support of the Great Lakes Restoration Initiative. Environ. Practice 15, 409–426. [Google Scholar]
- Escher BI, Allinson M, Altenburger R, Bain PA, Balaguer P, Busch W, Crago J, Denslow ND, et al. , 2014. Benchmaking organic micropollutants in wastewater, recycled water and drinking water with in vitro bioassays. Environ. Sci. Technol 48, 1940–1956. [DOI] [PubMed] [Google Scholar]
- Fulton TW, 1904. The rate of growth of fishes. Fisheries Board of Scotland, Annual Report 1904. 22, 141–241. [Google Scholar]
- Garcia-Reyero N, Kennedy AJ, Escalon BL, Habib T, Laird JG, Rawat A, Wiseman S, Hecker M, Denslow N, Steevens JA, Perkins EJ, 2014. Differential effects and potential adverse outcomes of ionic silver and silver nanoparticles in vivo and in vitro. Environ. Sci. Technol 48, 4546–4555. [DOI] [PubMed] [Google Scholar]
- Garcia-Reyero N, Adelman I, Liu L, Denslow N, 2008. Gene expression profiles of fathead minnows exposed to surface waters above and below a sewage treatment plant in Minnesota. Mar. Environ. Res 66, 134–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Reyero N, Adelman IR, Martinovic D, Liu L, Denslow ND, 2009. Site-specific impacts on gene expression and behavior in fathead minnows (Pimephales promelas) exposed in situ to streams adjacent to sewage treatment plants. BMC Bioinformatics 10 (Suppl 11), S11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Reyero N, Lavelle CM, Escalon BL, Martinovic D, Kroll KJ, Sorensen PW, Denslow ND, 2011. Behavioral and genomic impacts of a wastewater effluent on the fathead minnow. Aquat. Toxicol 101, 38–48. [DOI] [PubMed] [Google Scholar]
- Hoeng J, Deehan R, Pratt D, Martin F, Sewer A, Thomson TM, Drubin DA, Waters CA, de Graaf D, Peitsch MC, 2012. A network-based approach to quantifying the impact of biologically active substances. Drug Discov. Today 17, 413–418. [DOI] [PubMed] [Google Scholar]
- Judson R, Richard A, Dix DJ, Houck K, Martin M, Kavlock R, Dellarco V, Henry T, Holderman T, Savre P, Tan S, Carpenter T, Smith E, 2009. The toxicity data landscape for environmental chemicals. Environ. Health Perspect 117, 685–695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kidd KJ, Blanchfield PJ, Mills KH, Palace VP, Evans RE, Lazorchak JM, Flick RW, 2007. Collapse of a fish population after exposure to a synthetic estrogen. PNAS 104, 8897–8901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolok AS, Miller JT, Schoenfuss HL, 2012. The mini mobile environmental monitoring unit: a novel bio-assessment tool. J. Environ. Monit 14, 202–208. [DOI] [PubMed] [Google Scholar]
- Krämer A, Green J, Pollard J, Tugendreich S, 2014. Causal analysis approaches in Ingenuity Pathway Analysis. Bioinformatics 30, 523–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn M, Szklarczyk D, Franceschini A, von Mering C, Jensen LJ, Bork P, 2012. STITCH 3: zooming in on protein-chemical interactions. Nucleic Acids Res. 40 (Database issue), D876–880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laifenfeld D, Gilchrist A, Drubin D, Jorge M, Eddy SF, Frushour BP, Ladd B, Obert LA, Gosink MM, Cook JC, Criswell K, Somps CJ, Koza-Taylor P, Lawton MP, 2010. The role of hypoxia in 2-butoxyethanol-induced hemangiosarcoma. Toxicol. Sci 113, 254–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laifenfeld D, Luing Q, Swiss R, Park J, Macoritto M, Will Y, Younis H, Lawton M, 2014. Utilization of reverse causal reasoning of hepatic gene expression in rats to identify molecular pathways of idiosyncratic drug-induced liver injury. Toxicol. Sci 137, 234–248. [DOI] [PubMed] [Google Scholar]
- Martin F, Thomson TM, Sewer A, Drubin DA, Mathis C, Weisensee D, Pratt D, Hoeng J, Peitsch MC, 2012. Assessment of network perturbation amplitudues by applying high-throughput data to causal biological networks. BMC Systems Biol. 6, 54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinović-Weigelt D, Mehinto AC, Ankley GT, Denslow ND, Barber LB, Lee KE, King RJ, Schoenfuss HL, Schroeder AL, Villeneuve DL, 2014. Transcriptomic effects-based monitoring for endocrine active chemicals: Assessing relative contribution of treated wastewater to downstream pollution. Environ. Sci Technol 48, 2385–2394. [DOI] [PubMed] [Google Scholar]
- Memisoglu A, Samson L, 2000. Base excision repair in yeast and mammals. Mutat. Res 451, 39–51. [DOI] [PubMed] [Google Scholar]
- Pollard J, Butte AJ, Hoberman S, Joshi M, Levy J, Pappo J, 2005. A computational model to define the molecular causes of type 2 diabetes mellitus. Diabetes Technol. Ther 7, 323–336. [DOI] [PubMed] [Google Scholar]
- R Core Team., 2014. R: A language and environment for statistical computing. (http://www.r-project.org).
- Saeed AI, Sharov V, White J, Li J, Liang W, Bhagabati N, Braisted J, Klapa M, Currier T, Thiagarajan M, Sturn A, Snuffin M, Rezantsev A, Popov D, Ryltsov A, Kostukovich E, Borisovsky I, Liu Z, Vinsavich A, Trush V, Quackenbush J, 2003. TM4: a free, open-source system for microarray data management and analysis. Biotechniques 34, 374–378. [DOI] [PubMed] [Google Scholar]
- Schroeder AL, Ankley GT, HKouck KA, Villeneuve DL, 2016. Environmental surveillance and monitoring—The next frontiers for high-throughput toxicology. Environ. Toxicol. Chem 35(3), 513–525. [DOI] [PubMed] [Google Scholar]
- Siegel S, Castellan NJ, 1988. Nonparametric statistics for behavioral sciences (2nd Ed.). McGraw-Hill, New York, NY., 213–214. [Google Scholar]
- Skelton DM, Ekman DR, Martinović-Weigelt D, Ankley GT, Villeneuve DL, Teng Q, Collette TW, 2014. Metabolomics for in situ environmental monitoring of surface waters impacted by contaminants from both point and nonpoint sources. Environ. Sci. Technol 48, 2395–2403. [DOI] [PubMed] [Google Scholar]
- Smoot ME, Ono K, Ruscheinski J, Wang PL, Ideker T, 2011. Cytoscape 2.8: new features for data integration and network visualization. Bioinformatics 27, 431–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabas-Madrid D, Nogales-Cadenas R, Pascual-Montano A, 2012. GeneCodis3: a non-redundant and modular enrichment analysis tool for functional genomics. Nucleic Acids Res. 40 (Web Server issue), W478–483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang JY, McCarty S, Glenn E, Neale PA, Warne MS, Escher BI, 2013. Mixture effects of organic micropollutants present in water: towards the development of effect-based water quality trigger values for baseline toxicity. Water Res. 47, 3300–3314. [DOI] [PubMed] [Google Scholar]
- Writer JH, Barber LB, Brown GK, Taylor HE, Kiesling RL, Ferrey ML, Jahns ND, Bartell SE, Schoenfuss HL, 2010. Anthropogenic tracers, endocrine disrupting chemicals, and endocrine disruption in Minnesota lakes. Sci. Total Environ 409, 100–111. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.