Abstract
Comprised of telomeric TTAGGG repeats and shelterin, telomeres ensure that the natural ends of chromosomes remain impervious to the DNA damage response. Telomeres carry a long constitutive 3′ overhang that can bind replication protein A (RPA) and activate the ATR Ser/Thr kinase (ATR), which induces cell cycle arrest. A single-stranded (ss) TTAGGG repeat–binding protein in mouse shelterin, POT1a, has been proposed to repress ATR signaling by preventing RPA binding. Repression of ATR at telomeres requires tethering of POT1a to the other shelterin subunits situated on the double-stranded (ds) telomeric DNA. The simplest model of ATR repression, the “tethered exclusion model,” suggests that the only critical features of POT1a are its connection to shelterin and its binding to ss telomeric DNA. In agreement with the model, we show here that a shelterin-tethered variant of RPA70 (lacking the ATR recruitment domain) can repress ATR signaling at telomeres that lack POT1a. However, arguing against the tethered exclusion model, the nearly identical POT1b subunit of shelterin has been shown to be much less proficient than POT1a in repression of ATR. We now show that POT1b has the intrinsic ability to fully repress ATR but is prevented from doing so when bound to Ctc1, Stn1, Ten1 (CST), the complex needed for telomere end processing. These results establish that shelterin represses ATR with a tethered ssDNA-binding domain that excludes RPA from the 3′ overhang and also reveal an unexpected effect of CST on the ability of POT1b to repress ATR.
Keywords: telomere; shelterin; DNA binding protein; DNA damage response; checkpoint control; ATR signaling; CST (Ctc1, Stn1, Ten1); POT1; RPA; single-stranded DNA; POT1a; POT1b; 3′ overhang
Introduction
One of the many tasks of the telomeric shelterin complex is to repress the ATR-dependent DNA damage response at the natural ends of chromosomes (reviewed in Refs. 1–3). The main mode of ATR3 activation involves the binding of the abundant trimeric RPA (RPA70, RPA32, and RPA14) complex to ssDNA, recruitment of ATR through an interaction between RPA70 and the ATR interacting partner ATRIP, loading of the 9-1-1 (Rad9–Hus1–Rad1) complex on the end of the duplex, and activation of ATR by 9-1-1–bound TopBP1 (reviewed in Ref.4). A second pathway involves activation of ATR by RPA-bound ETAA1 and is independent of 9-1-1 and TopBP1 (5, 6).
The two DNA structures required for TopBP1-dependent ATR activation are present at telomeres, which carry a 9-1-1–loading site and a 3′ overhang of TTAGGG repeats, sufficiently long to bind one or more RPA trimers. Although telomeres often occur in the t-loop configuration (7, 8), this structure is unlikely to guard against ATR signaling because the base of the t-loop retains the critical features required for loading of RPA and 9-1-1 (reviewed in Ref. 3). Rather, ATR repression is achieved by the POT1 subunit of shelterin (9), which binds to the ss TTAGGG repeats (10). Deletion of POT1 leads to activation of ATR at most telomeres in a manner that depends on RPA and TopBP1 (11). Super-resolution STORM imaging showed that this ATR activation takes place despite the presence of t-loops (8).
POT1 proteins bind the sequence 5′-TTAGGGTTAG-3′ either at a DNA end or at an internal position using two oligosaccharide/oligonucleotide binding (OB) folds in their N-terminal half (10, 12, 13). The interaction of POT1 with ss TTAGGG repeats is neither sufficient nor necessary to target the protein to telomeres (14–17). Accumulation of POT1 at telomeres requires its association with shelterin, which is mediated by the binding of the shelterin subunit TPP1 to the C-terminal half of POT1 (15, 16, 18). TPP1 binds to the TIN2 shelterin subunit, which in turn interacts with the duplex telomeric DNA-binding proteins TRF1 and TRF2, thus anchoring POT1 on the duplex telomeric DNA.
As a result of a gene duplication event, rodent telomeres contain two closely related POT1 proteins, POT1a and POT1b, which have diverged in function. Both POT1a and POT1b can repress ATR kinase signaling in G1 but repression of ATR in S/G2 requires POT1a (11, 19, 20). In S phase, POT1b, but not POT1a, governs the postreplicative generation of the telomeric 3′ overhang (21–23). In part, POT1b executes this function through the recruitment of CST (Ctc1, Stn1, Ten1) (reviewed in Ref. 24), an RPA-like complex that binds ssDNA and interacts with polymerase α/primase (polα/primase) to mediate fill-in synthesis that restores the correct overhang length (25).
Telomeres lacking POT1 proteins accumulate RPA, which is not detectable at functional telomeres (11, 26). This observation suggested a simple competition model whereby the binding of POT1 to the ssDNA prevents RPA-dependent ATR activation (9, 11, 26–28). As RPA is ∼200-fold more abundant than POT1a and POT1b and has the same subnanomolar affinity for ss telomeric DNA (29), it is unlikely that simple competition could explain the repression of ATR. Indeed, POT1 is not able to compete with RPA for ss telomeric DNA in vitro (26). It was therefore proposed that the repression of ATR requires the local tethering of the POT1 proteins to the rest of shelterin, thus increasing the local concentration of POT1. Consistent with this tethering model, repression of ATR requires the association of the POT1 proteins with TPP1 and the TPP1- and TIN2-mediated interaction with TRF1 and/or TRF2. ATR activation throughout the cell cycle is observed when telomeres lack either TPP1 or TIN2 (17, 29), or when telomeres contain alleles of TPP1 or TIN2 that do not recruit POT1a/b (30).
In its simplest form, the RPA exclusion model predicts that any shelterin-tethered protein with the ability to bind ss TTAGGG repeats can repress ATR signaling at telomeres. However, this prediction is not met in the context of POT1b, which can repress ATR signaling in G1 but not in S phase. POT1b and POT1a are equally abundant at telomeres, bind TPP1, and have indistinguishable DNA-binding features (17, 19, 29, 31). Thus, the current knowledge of POT1b argues against the idea that any shelterin-tethered ss TTAGGG repeat–binding protein can exclude RPA from telomeres and thus repress ATR signaling.
There are alternative models for how POT1 represses ATR signaling. The ability of human POT1 to protect telomeric G4 DNA from unfolding by RPA has been proposed to repress ATR signaling (32). It is also possible that the greater affinity of POT1 for the telomeric 3′ end (when ending in TTAG-3′) (13) protects telomeres by providing POT1 with a competitive advantage over RPA. In another proposal, an S/G2-specific interplay between hnRNPA1, an RNA-binding protein with high affinity for ss TTAGGG repeats, and the telomeric long noncoding RNA TERRA serves an intermediary function in removing RPA from telomeric DNA and then allowing POT1 to bind (26). Finally, it is possible that the POT1 proteins interfere with 9-1-1 loading. Although POT1a/b do not recognize the telomeric ds-ss junction in vitro (19), it is not excluded that the junction is bound by POT1 in the context of the whole shelterin complex.
Here we report that, consistent with the tethering model, ATR signaling can be partially repressed by the DNA-binding domain of RPA70 when it is tethered to shelterin. Furthermore, we solve the POT1b conundrum by showing that POT1b, like POT1a, can repress ATR signaling in S phase but that its interaction with CST prevents it from doing so. The results provide evidence for the idea that ATR repression involves RPA exclusion by a shelterin-tethered ssDNA-binding protein with no other specialized features.
Results
Engineering a synthetic shelterin-tethered ATR repressor
We sought to test the idea that the exclusion of RPA by POT1 is based on the tethering of the ssDNA-binding domain of POT1 to the duplex telomeric DNA rather than POT1-specific DNA-binding features (e.g. binding to G4 DNA, binding to the 3′ end, or binding to junctions) (Fig. 1A). We argued that the most stringent test of this tethering model would involve creating a synthetic telomeric ATR repressor based on the DNA-binding domain of RPA70. RPA has the same affinity for ss TTAGGG repeats as POT1 (29) but a different DNA-binding mode: Whereas POT1 binds DNA with two OB-folds, RPA uses three OB-folds (OB-A, -B, and -C) in RPA70 (Fig. 1B) and one OB-fold in RPA32 (33). The C-terminal OB-fold (OB-C) of RPA70 interacts with RPA32, which in turn binds to RPA14.
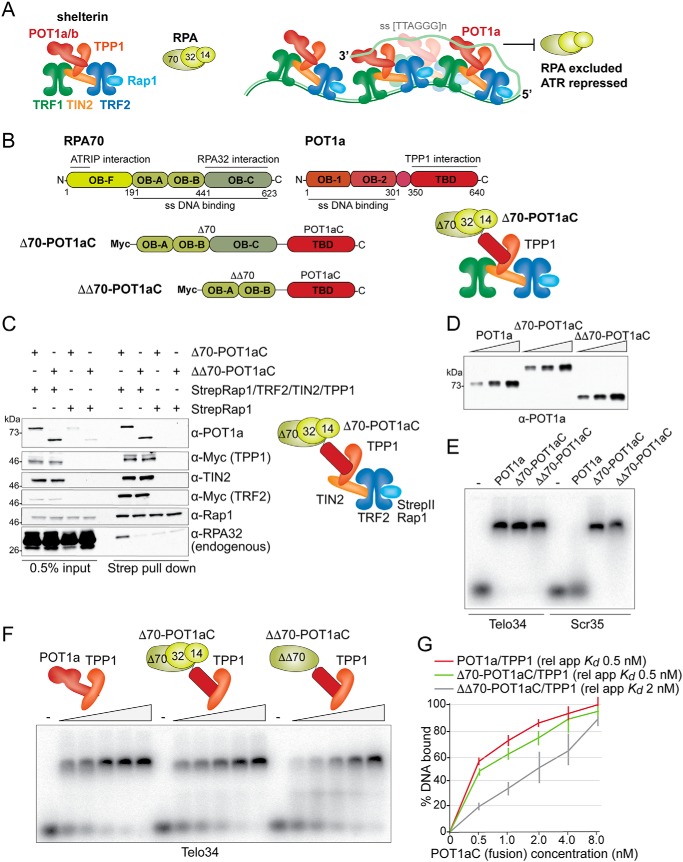
Figure 1.
Characterization of the RPA-POT1aC fusion proteins. A, schematic (left) of the mouse shelterin complex and RPA and model (right) for how the binding of POT1a to shelterin and ss TTAGGG repeats excludes RPA from telomeres and thereby prevents ATR activation. B, schematic of mouse RPA70, mouse POT1a and the two fusion proteins Δ70-POT1aC and ΔΔ70-POT1aC, indicating the amino acids and functions of the relevant domains. Right, schematic of mouse TPP1-tethered Δ70-POT1aC and RPA32 and RPA14. C, immunoblots showing co-IPs from transiently transfected 293T cells of Δ70-POT1aC or ΔΔ70-POT1aC with either Strep-tagged Rap1 or Strep-tagged Rap1 together with TRF2, TIN2, and TPP1. Antibodies to the expressed proteins and the endogenous RPA32 are indicated. D, example of immunoblotting to determine the relative protein concentration of Strep-tagged TPP1/POT1a or TPP1/RPA-fusion proteins isolated from transfected 293T cells. E, EMSA showing binding of TPP1/POT1a, TPP1/Δ70-POT1aC, and TPP1/ΔΔ70-POT1aC to telomeric and nontelomeric ssDNA. Telo34, 34 nts of single-stranded telomeric DNA; Scr35, 35 nts of single-stranded nontelomeric DNA (see “Experimental Procedures” for probe sequences). F, representative EMSA to determine the apparent relative Kds of TPP1/POT1a, TPP1/Δ70-POT1aC, and TPP1/ΔΔ70-POT1aC for Telo34. G, quantification of the affinity of TPP1/POT1a, TPP1/Δ70-POT1aC, and TPP1/ΔΔ70-POT1aC as shown in E from three independent experiments. Error bars represent S.D.s.
An obvious complication in creating an RPA-based ATR repressor is that tethering of WT RPA to telomeres would permanently recruit and likely activate the ATR kinase. The interaction of RPA70 with the ATRIP/ATR complex is mediated by its fourth (N-terminal) OB-fold (OB-F), which does not bind to DNA (34). We therefore based our construct on a version of RPA70 that lacks OB-fold OB-F (referred to as Δ70) (Fig. 1B). This part of RPA70 was fused to the C terminus of POT1a (referred to as POT1aC), which is the part of POT1a that interacts with TPP1 and ensures association with shelterin. The resulting chimera is referred to as Δ70-POT1aC.
Δ70-POT1aC and a second chimera, ΔΔ70-POT1aC, which also lacks the C-terminal RPA70 OB-fold where RPA70 interacts with RPA32, were co-expressed with TPP1, TIN2, TRF2, and StrepII-tagged Rap1 in 293T cells (Fig. 1C). Immunoblotting of proteins associated with StrepII-Rap1 showed that both chimeras had the ability to form a complex with TPP1/TIN2/TRF2/Rap1. As expected, Δ70-POT1aC showed an interaction with RPA32, whereas ΔΔ70-POT1aC did not.
To determine the DNA-binding features of the chimeric proteins, they were co-expressed with StrepII-tagged TPP1 and isolated on Strep-Tactin beads (Fig. 1, D–G; Fig. S1). TPP1 promotes the binding of POT1 to telomeric DNA but does not bind DNA by itself. The concentration of the partially purified TPP1/chimera heterodimers was adjusted based on immunoblotting (Fig. 1D) with an antibody to the C terminus of POT1a, and the complexes were analyzed in a gel-shift assay with labeled 34 or 35 nt probes (Fig. 1, E–G). As expected from previous work (29), the RPA70-containing complexes bound to ss TTAGGG repeat as well as a scrambled sequence probe, whereas the POT1a/TPP1 heterodimer only bound the TTAGGG repeat probe (Fig. 1E). Titration experiments showed that Δ70-POT1aC/TPP1 had the same affinity (apparent relative Kd ∼0.5 nm) for TTAGGG repeats as POT1a/TPP1 (Fig. 1, F and G). Consistent with the absence of the third RPA70 OB-fold, the ΔΔ70-POT1aC/TPP1 heterodimer showed a lower affinity (apparent relative Kd ∼2 nm) for ss TTAGGG repeats.
Repression of ATR by Δ70-POT1aC
To test the ability of the chimeric proteins to prevent the activation of ATR at telomeres (Fig. 2A), they were expressed in SV40-LT immortalized POT1aF/F MEFs from which POT1a can be deleted with the Cre recombinase (22) (Fig. 2B). As controls, the POT1aC fragment and the Δ70 mutant of RPA70 were expressed individually. Expression vectors were chosen to yield moderate expression levels of the chimeric proteins, because an excess of the mutant forms of RPA70 (Δ70 and ΔΔ70) might interfere with the essential functions of RPA in DNA replication and ATR signaling. Immunoblotting with RPA70 and POT1a antibodies showed that although all exogenous proteins were overexpressed compared with the endogenous POT1a, their abundance was far less than the endogenous RPA70 (Fig. 2B), making it unlikely that that the RPA variants would have a deleterious effect. Indeed, cells expressing the mutant RPA70 alleles showed no overt proliferation defect and had comparable S phase indices (Fig. S2, A and B). As expected, all exogenous proteins that carried the C terminus of POT1a localized to telomeres (Fig. 2, C and D).
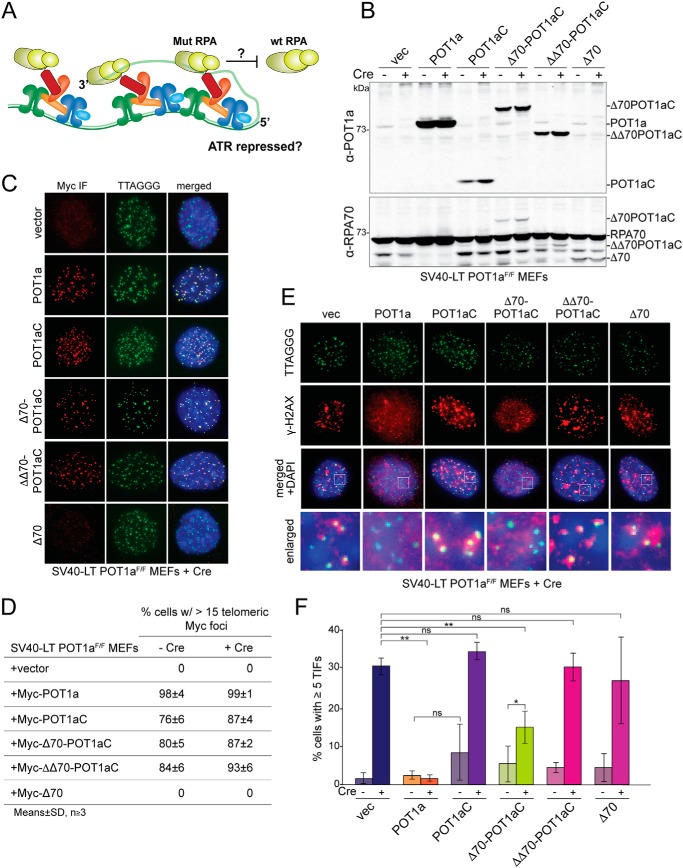
Figure 2.
A shelterin-tethered RPA70 mutant can repress ATR signaling. A, schematic of the experimental setup. Shelterin-tethered Δ70-POT1aC or (ΔΔ70-POT1aC) is tested for its ability to replace the role of POT1a in ATR repression. B, immunoblot for POT1a and RPA70 in SV40-LT immortalized POT1aF/F MEFs expressing the indicated proteins before and after Cre treatment (96 h time point). The endogenous POT1a is lost upon Cre treatment and the introduced proteins are overexpressed compared with endogenous POT1a (top). In contrast, the expressed RPA70 proteins are much less abundant than the endogenous RPA70 (bottom). The bands running below RPA70 are assumed to be degradation products. C, representative IF images showing the localization of the indicated exogenous Myc-tagged proteins in POT1aF/F cells after deletion of the endogenous POT1a with Cre (96 h). IF was performed with an α-Myc antibody (red) together with FISH for telomeric DNA (green). D, quantification of the telomeric localization (as in panel C) of the indicated proteins in POT1aF/F MEFs with and without Cre treatment (96 h). Averages of three independent experiments and S.D.s. E, representative IF images of the DNA damage response at telomeres of Cre-treated POT1aF/F cells expressing the indicated proteins (96 h). γH2AX foci that co-localize with telomeres (TIFs) were detected with α-γH2AX antibody (red) combined with FISH for telomeric DNA (green). F, quantification of the TIF response as shown in E in the indicated POT1aF/F cells with and without Cre treatment. Data represent averages from three to five independent experiments with S.D.s. p values were based on a two-tailed Student's t test. **, p ≤ 0.01; *, p ≤ 0.05; ns, not significant.
Deletion of POT1a showed the expected induction of a DNA damage response at telomeres. In absence of POT1a, ∼30% of the cells showed accumulation of γ-H2AX and 53BP1 at telomeres (Fig. 2, E and F; Fig. S2, C and D). This percentage of cells showing a DNA damage response is consistent with prior reports that showed that POT1a is required for the repression in S/G2 whereas in G1, POT1a is redundant with POT1b (11, 22).
As expected, this S/G2 ATR signaling was fully repressed by complementation of the cells with WT POT1a, but not by POT1aC (Fig. 2, E and F). Remarkably, Δ70-POT1aC showed a significant reduction in ATR signaling at telomeres, resulting in only 15% of cells with detectable γ-H2AX foci at their telomeres. As expected, the untethered Δ70 protein was unable to repress ATR signaling at telomeres (Fig. 2, E and F). Cell cycle analysis indicated that the changes in ATR signaling were not because of changes in S phase index (Fig. S2, A and B) and quantitative analysis of the ss TTAGGG repeats at telomeres (Fig. S2E) showed that the chimeric protein did not reduce the telomeric overhang signal, which could have confounded the interpretation of the reduced ATR signaling. In fact, overexpression of both POT1a and Δ70-POT1aC induced a slight increase in the telomeric overhang signal, presumably because they compete with POT1b for TPP1 interaction.
In contrast to Δ70-POT1aC, ΔΔ70-POT1aC was incapable of blocking ATR signaling at telomeres. We also tested the ability of Escherichia coli SSB to repress ATR signaling when tethered to shelterin (Fig. S3). The SSB-POT1aC chimera, which binds telomeric DNA with an apparent relative Kd of ∼4 nm (Fig. S3, A–D), localized to telomeres but did not repress the DNA damage response (Fig. S3, E–H). We do not know whether the higher relative Kd of ΔΔ70-POT1aC and SSB-POT1aC is the cause of the lack of protection by these chimeras. In principle, in the context of tethered chimeras, changes in the relative Kd should not have a great impact. Possibly, the chimeras are defective in other aspects (e.g. exact positioning of the ssDNA-binding domains within the shelterin complex).
CST binding interferes with the repression of ATR by POT1b
It remains to be explained why POT1b is incapable of repressing ATR in S/G2 although it can do so in G1. POT1b interacts with CST, allowing appropriate fill-in synthesis of the telomere terminus during S/G2 (21). We therefore asked whether the interaction of POT1b with CST interferes with its ability to repress ATR. Prior work had shown that a POT1b allele with mutations in two sites of the protein (Fig. 3A) does not bind to CST and is deficient in regulating the postreplicative processing of the telomere terminus (21). Notably, expression of this mutant version of POT1b fully restored the ATR repression in POT1a-deficient cells. The WT POT1b, even when overexpressed, did not have this effect (Fig. 3, B–D).
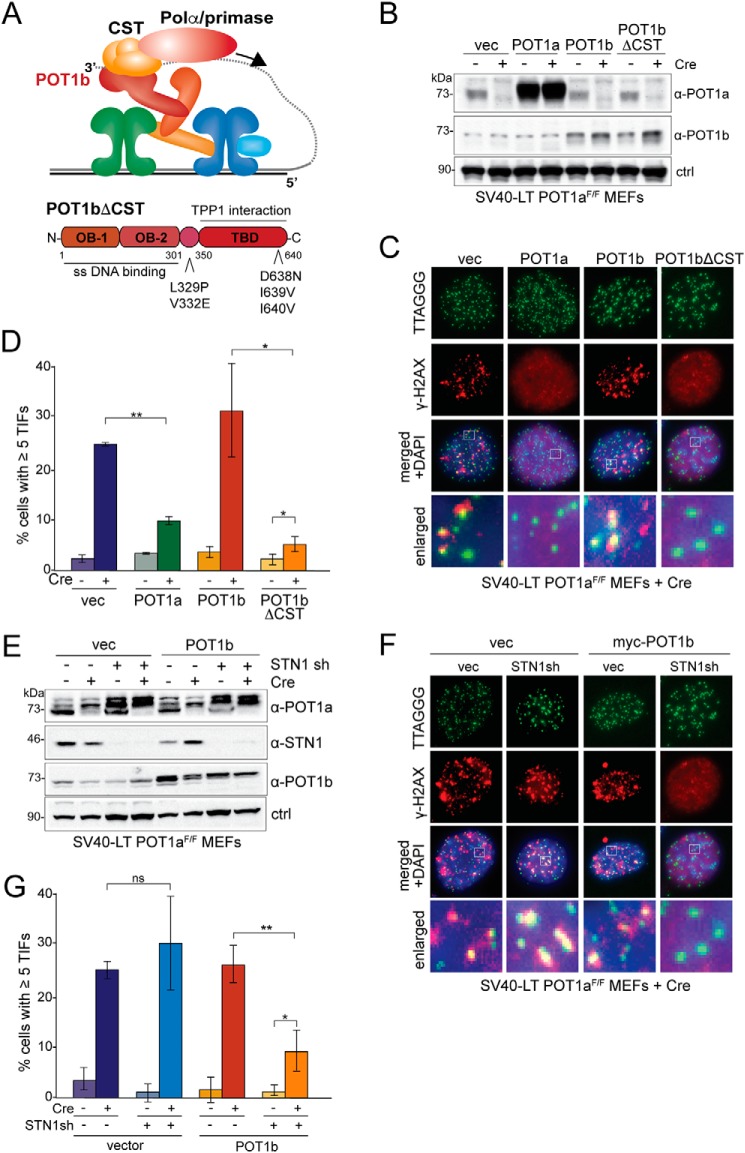
Figure 3.
ATR repression by POT1b is limited by its interaction with CST. A, schematic of the role of POT1b in recruiting CST and polymerase α/primase to telomeres (top). Schematic of POT1b domains and the mutations that disrupt the POT1b-CST interaction (bottom). B, immunoblot for POT1b in POT1aF/F MEFs expressing the indicated proteins before and after treatment with Cre (96 h). ctrl, nonspecific band used as loading control. C, representative IF-FISH images showing the DNA damage response at telomeres in Cre-treated POT1aF/F MEFs expressing POT1a, POT1b, or POT1bΔCST (96 h). γH2AX foci that co-localize with telomeres (TIFs) were detected using α-γH2AX antibody (red) and a telomeric FISH probe (green). D, quantification of the TIF response as shown in C of POT1aF/F MEFs before and after Cre treatment. Bar graphs represent averages from three experiments and S.D.s. p values were derived from a two-tailed Student's t test. p values: **, p ≤ 0.01; *, p ≤ 0.05. E, immunoblot for POT1a, POT1b, and Stn1 in POT1aF/F MEFs (with and without 96 h Cre treatment) expressing exogenous POT1b and/or Stn1 shRNA (indicated with +) or a scrambled sh (indicated with −). Asterisks indicate nonspecific bands. ctrl, nonspecific band used as loading control. F, representative images of IF-FISH to monitor the DNA damage response at telomeres in the indicated Cre-treated (96 h) POT1aF/F MEFs with and without POT1b and expressing Stn1sh or a scrambled control (ctrl sh). γH2AX foci that co-localize with telomeres (TIFs) were detected with IF with an α-γH2AX antibody (red) and FISH with a telomeric PNA probe (green). G, quantification of TIF response in the indicated POT1aF/F MEFs (as in F). Bar graphs represent averages from three independent experiments and S.D.s. p values: **, p < 0.001; *, p < 0.05; ns, not significant.
To confirm the findings with POT1bΔCST, we employed shRNA-mediated knockdown of Stn1. Consistent with the results with the POT1bΔCST mutant, partial knockdown of Stn1 restored the ability of overexpressed POT1b to repress ATR signaling at telomeres. In contrast, knockdown of Stn1 did not restore the ability of the endogenous POT1b to protect telomeres from ATR activation (Fig. 3, E–G). This is most likely because of the residual Stn1 interacting with endogenous POT1b, whereas the residual level of Stn1 is insufficient to bind all overexpressed POT1b. As CST is essential for cell viability, we are unable to test whether complete removal of Stn1 will restore ATR repression by endogenous POT1b.
Discussion
Because mammalian telomeres carry a 3′ protrusion of ssDNA, ATR signaling has to be repressed throughout the cell cycle. Given the abundance of telomere ends in the nucleus, a system that fails at as few as 1% of the telomeres could lead to induction of cell cycle arrest and senescence or apoptosis. Our data clarify how this stringent ATR repression is achieved by the POT1 subunits of the telomeric shelterin complex.
The shelterin-tethered chimera of RPA70 and the C terminus of POT1a (Δ70-POTaC) is remarkably proficient in repressing ATR signaling at telomeres, although not as effective as WT POT1a. The reduction in ATR signaling at telomeres containing Δ70-POT1aC suggests that the ability of POT1a to repress ATR signaling is primarily determined by its ability to bind to ssDNA telomeric DNA. Because the DNA-binding domain of POT1a can be replaced with the OB-folds of RPA70, we consider it unlikely that any other feature of POT1-DNA interaction beyond its binding to ssDNA is involved in the control of ATR signaling. The preference of POT1 for a TTAG-3′ telomere end appears to be largely irrelevant in this context. Indeed, TTAG-3′ ends only make up 40% of the telomere ends in human cells (35), yet all telomeres are shielded from the ATR pathway. Similarly, the ability of POT1 to protect G4 DNA from unfolding by RPA (32) is unlikely to be the primary determinant of ATR repression. Finally, although POT1 does not show a preference for its site next to the ds-ss junction in vitro (19), it could be argued that bound to shelterin, POT1 might have the ability to recognize the junction and prevent 9-1-1 loading. This hypothetical attribute is also rendered unlikely to be relevant to ATR repression based on the results with the shelterin-tethered chimeric RPA70 protein.
The inability of POT1b to repress ATR signaling in S/G2 has presented a conundrum because POT1a and POT1b have identical TPP1- and DNA-binding features. Furthermore, it can be inferred that POT1b resides at telomeres in S/G2 because it governs telomere end processing during this cell cycle stage. Our data indicate that the interaction of POT1b with the CST and perhaps the polymerase α/primase complex prevents POT1b from acting as an ATR repressor. Only when the interaction of POT1b with CST is abrogated, is POT1b proficient in the repression of ATR in S/G2. This is an unexpected result because both POT1b and CST bind ss TTAGGG repeats. One possibility is that the recently noted ability of CST to unfold G4 structures (36) leads to a greater ability of RPA to gain access to TTAGGG repeat–binding sites.
The single POT1 subunit in human shelterin has been implicated in the repression of ATR signaling (9, 20). Interestingly, the recruitment of CST to human telomeres is mediated by TPP1, not POT1 (37). Human POT1 does play a role in how CST functions at telomeres (38, 39) and may therefore have a (transient) interaction with CST, but the recruitment of CST primarily involves binding to TPP1. This arrangement may avoid the complication of CST interfering with the ability of human POT1 protein to repress ATR signaling.
Experimental procedures
Cell lines, expression constructs, and introduction of shRNAs
SV40 large T antigen (SV40LT) immortalized POT1aF/F MEFs were reported previously (22). For expression of Myc-tagged proteins the following constructs were used: pWZL-Myc (vector), Myc-POT1a (pWZL-Myc-POT1a), Myc-POT1aC (pWZL-Myc-POT1aC) containing POT1a aa 149–640 with an C-terminal Myc tag, Myc-Δ70-POT1aC (pWZL-Myc-Δ70-POT1aC) containing mouse RPA70 aa 191–623 fused to the N terminus of POT1aC, Myc-ΔΔ70-POT1aC (pWZL-Myc-ΔΔ70-POT1aC) containing mouse RPA70 aa 191–331 fused to the N terminus of POT1aC, Myc-Δ70 (pWZL-Myc-Δ70) containing RPA70 aa 191–623, Myc-SSB-POT1aC (pWZL-Myc-SSB-POT1aC) containing full-length E. coli SSB fused to POT1aC, Myc-POT1b (pWZL-Myc-POT1b), and Myc-POT1bΔCST (pWZL-Myc-POT1bΔCST) (21). For each retroviral construct, 20 μg DNA was transfected into Phoenix packaging cells using CaPO4 co-precipitation. Medium was changed 12 and 24 h after transfection, and the retroviral supernatant was used for four infections of the MEFs at 12-h intervals. Cells were subjected to selection in 135 μg/ml hygromycin for 3 days. To delete endogenous POT1a from POT1aF/F MEFs, cells were subjected to three infections at 12-h intervals with pMMP Hit&Run Cre retrovirus derived from transfected Phoenix cells as described previously (40). Time point 0 was set at 12 h after the first Hit&Run Cre infection. For Stn1 knockdown, the Stn1 shRNA (5′-GATCCTGTGTTTCTAGCCTTT-3′) (21) in a pLKO.1 lentiviral vector (Open Biosystems) was produced in Phoenix-ECO cells as described above and introduced with three infections (6-h intervals). Infected cells were selected for 2.5 days in 2 μg/ml puromycin.
Co-immunoprecipitation
4 × 106 HEK 293T cells were plated in a 15-cm dish 24 h prior to transfection using CaPO4 co-precipitation of 3 μg of pQE-StrepII-Rap1, pcDNA3.1-TRF2, pLPC-TIN2, pLPC-Myc-TPP1, pWZL-Myc-Δ70-POT1aC, pWZL-Myc-ΔΔ70-POT1aC as indicated. Medium was changed 8 h after transfection and cells were harvested 28 h later. Cells were resuspended in lysis buffer (60,000 cells/μl) (50 mm NaH2PO4, pH 8.0, 50 mm NaCl, 10 mm DTT, 0.5% Tween, 1× cOmplete Protease Inhibitors (Roche), 0.5 mm PMSF, and 8 μg/ml avidin and incubated on a rotator at 4 °C for 10 min. After centrifugation at 15,000 × g at 4 °C for 10 min, the supernatant was incubated with StrepII beads (Qiagen; 1 × 106 cells equivalent per μl beads) at 4 °C for 1 h. Beads were washed three times in 500 μl lysis buffer and proteins were eluted with 20 μl of 2× Laemmli buffer (100 mm Tris-HCl, pH 6.8, 200 mm DTT, 3% SDS, 20% glycerol, 0.05% bromphenol blue). Samples were heated to 95 °C for 5 min and analyzed by separation on 8% SDS-PAGE.
Immunoblotting
Harvested MEFs (1 × 106 cells) were resuspended in 100 μl 2× Laemmli buffer, incubated with 250 units Benzonase (Sigma-Aldrich) on ice for 10 min, denatured at 95 °C for 5 min and separated on 8–16% SDS-PAGE (2 × 105 cell equivalent per lane). After immunoblotting, the membranes were blocked in TBS with Tween 20 with 2.5% nonfat dry milk and incubated with the following primary antibodies α-POT1a (no. 1221), α-Myc (9B11, Cell Signaling Technology), α-TIN2 (no. 1447), α-Rap1 (no. 1253), α-RPA32 (A300–244A, Bethyl), α-RPA70 (A300–241A, Bethyl), α-POT1b (no. 1223), α-STN1 (sc-376450, Santa Cruz Biotechnology), α-tubulin (GTU88, Abcam) in 1% nonfat dry milk. Immunoblots for POT1a were performed using the renaturation protocol described previously (14). Secondary antibodies were horseradish peroxidase–conjugated donkey α-mouse IgG (GE Healthcare) or donkey α-rabbit IgG (GE Healthcare). Immunoblots were developed with enhanced chemiluminescence (Amersham Biosciences).
Protein expression and isolation
4 × 106 HEK 293T cells were plated in a 15-cm dish 24 h prior to transfection using CaPO4 co-precipitation with 3 μg of pQE-StrepII-Myc-TPP1 (mouse), pLPC-Myc-POT1a, pLPC-Myc-Δ70-POT1aC, pLPC-Myc-ΔΔ70-POT1aC, and pLPC-Myc-SSB-POT1aC as indicated. Medium was changed 8 h after transfection and cells were harvested 28 h later. Subsequently, cells were pelleted, washed twice in PBS, resuspended in lysis buffer (60,000 cells/μl) (50 mm NaH2PO4, pH 8.0, 50 mm LiCl, 10 mm DTT, 0.5% Tween, 1× cOmplete Protease Inhibitors, 0.5 mm PMSF, and 8 μg/ml avidin) and incubated at 4 °C for 10 min on a rotator. After centrifugation at 15,000 × g at 4 °C for 10 min, the supernatant was incubated with Strep-Tactin beads (Qiagen; 1 × 106 cells equivalent per μl beads) and 250 units Benzonase (Sigma-Aldrich) for 1 h at 4 °C on a rotator. Beads were washed three times in lysis buffer and eluted twice in Elution buffer (50 mm NaH2PO4, pH 8.0, 50 mm LiCl, 1 mm DTT, 0.1% Tween, 10 mm biotin). Samples were aliquoted, snap-frozen, and stored at −80 °C.
Electrophoretic mobility shift assay
Labeling reactions and gel-shift assays were performed as described previously (19) with minor modifications. All oligonucleotides were obtained from Sigma. The probe sequences were as follows: Telo34 (5′-TTAGGGTTAGGGTTAGGGTTAGGGTTAGGGTTAG-3′) and Scr35 (5′-ATGCGACTCGAGCTAGATGATGTCTTCTGCAATCA-3′). Gel-shift reactions were performed in 10 μl reaction buffer (50 mm Hepes-KOH, pH 8.0, 50 mm LiCl, 0.1 mm DTT, 50 ng/μl β-casein) with 0.2 nm polynucleotide kinase end-labeled DNA probes. Proteins were added last and the mixture was incubated for 10 min on ice. Electrophoresis was performed in 0.8% agarose gels run in 0.5× Tris borate/EDTA, pH 8.3. Gels were run for 45 min at 100 V at room temperature, fixed in 20% methanol/10% acetic acid, dried on Whatman DE81 paper at 80 °C with vacuum and exposed to phosphorimager screens. The binding fractions were calculated taking all protein-DNA complexes into account (binding fraction (%)) = (proteins-DNA complex)/(free DNA probes) to derive apparent relative Kd values.
IF, IF-FISH, and TIF analysis
Cells grown on polylysine-covered coverslips were harvested at time point 96 h after Cre to analyze protein localizations and TIF response. Coverslips were incubated on ice, rinsed once with PBS containing Mg2+ and Ca2+, and soluble proteins were pre-extracted with ice cold Triton X-100 buffer (20 mm Hepes-KOH, pH 7.9, 50 mm NaCl, 3 mm MgCl2, 0.5% Triton X-100, 300 mm sucrose) for 30 s on ice. Coverslips were rinsed twice with cold PBS containing Mg2+ and Ca2+, transferred to room temperature, and fixed in 3% paraformaldehyde with 2% sucrose for 10 min. Coverslips were washed twice in PBS at room temperature for 5 min. IF was carried out as described previously (40–42) with the following primary antibodies: α-Myc (9B11, Cell Signaling Technology), α-γH2AX (no. 05–636, Millipore), and α-53BP1 (ab175933, Abcam). For IF-FISH staining, after the secondary antibody incubation and wash step, cells were fixed again with 2% paraformaldehyde for 5 min; dehydrated in 70, 95, and 100% ethanol for 5 min each and allowed to air dry. Hybridizing solution (10 mm Tris-HCl, pH 7.2, 70% formamide, 1 mg/ml blocking reagent (Roche), containing 100 nm fluorescent PNA probe (TelC-Alexa488 (PNA Bio Inc.)) was added to each coverslip, and the DNA was denatured by heating for 10 min at 80 °C on a heat block. After 2 h of incubation at room temperature in the dark, cells were washed twice with washing solution (70% formamide, 10 mm Tris-HCl, pH 7.2) for 15 min each and with PBS three times for 5 min each. DAPI (0.1 μg/ml) was added to the second PBS wash. Coverslips were sealed onto glass slides with embedding medium (ProLong Gold Antifade Reagent, Invitrogen). Digital images were captured on a Zeiss Axioplan II microscope with a Hamamatsu C4742–95 camera using Volocity software.
S phase index
Before harvesting, cells seeded on coverslips were incubated for 1.5 h in medium containing 10 μm BrdU and subsequently fixed with 2% PFA at room temperature for 10 min. Following an incubation in Nonidet P-40 buffer (0.5% Nonidet P-40 in PBS) at room temperature for 10 min, coverslips were washed three times in PBS for 5 min and then incubated in 4N HCl at room temperature for 10 min. Next, the coverslips were washed three times in PBS for 5 min and incubated with blocking solution (PBS, 0.1% BSA, 3% serum donkey, 0.1% Triton X-100, 1 mm EDTA, pH 8.0) at room temperature for 45 min before addition of α-BrdU antibody (B35130, Invitrogen). Cells were incubated with the antibody at 4 °C for overnight. Coverslips were washed three times in PBS for 5 min and incubated with secondary antibody at room temperature for 45 min. Coverslips were washed three times in PBS for 5 min, with the second PBS wash containing DAPI (0.15 μg/ml) to stain for DNA. Coverslips were sealed and processed as described for IF above.
Telomeric overhang analysis
Mouse telomeric DNA was analyzed on CHEF gels as described previously (21).
Author contributions
K. K. and T. d. L. conceptualization; K. K. and T. d. L. data curation; K. K. and T. d. L. formal analysis; K. K. and T. d. L. validation; K. K. and T. d. L. investigation; K. K. and T. d. L. visualization; K. K. and T. d. L. methodology; K. K. and T. d. L. writing-original draft; T. d. L. supervision; T. d. L. funding acquisition; T. d. L. project administration.
Supplementary Material
Acknowledgments
We thank Hiro Takai and Melissa Pamula for executing initial experiments and members of the de Lange lab for helpful discussion.
This work was supported by EMBO long-term fellowship EMBO ALTF-1044-2011 and Women & Science Fellowship Program at The Rockefeller University funds (to K. K.). This work was also supported by National Institutes of Health Grant AG016642 (to T. d. L.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
This article contains supporting Figs. S1–S3.
- ATR
- ATR Ser/Thr kinase
- RPA
- replication protein A
- ss
- single-stranded
- ds
- double-stranded
- OB
- oligosaccharide/oligonucleotide binding
- CST
- Ctc1, Stn1, Ten1
- MEF
- mouse embryonic fibroblasts
- aa
- amino acid
- SSB
- single-stranded DNA binding
- IF
- immunofluorescence
- FISH
- fluorescent in situ hybridization
- TIFs
- telomere dysfunction–induced foci.
References
- 1. Lazzerini-Denchi E., and Sfeir A. (2016) Stop pulling my strings—what telomeres taught us about the DNA damage response. Nat. Rev. Mol. Cell Biol. 17, 364–378 10.1038/nrm.2016.43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Palm W., and de Lange T. (2008) How shelterin protects mammalian telomeres. Annu. Rev. Genet. 42, 301–334 10.1146/annurev.genet.41.110306.130350 [DOI] [PubMed] [Google Scholar]
- 3. de Lange T. (2018) Shelterin-mediated telomere protection. Ann. Rev. Genetics, in press [DOI] [PubMed] [Google Scholar]
- 4. Ciccia A., and Elledge S. J. (2010) The DNA damage response: Making it safe to play with knives. Mol. Cell 40, 179–204 10.1016/j.molcel.2010.09.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bass T. E., Luzwick J. W., Kavanaugh G., Carroll C., Dungrawala H., Glick G. G., Feldkamp M. D., Putney R., Chazin W. J., and Cortez D. (2016) ETAA1 acts at stalled replication forks to maintain genome integrity. Nat. Cell Biol. 18, 1185–1195 10.1038/ncb3415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Haahr P., Hoffmann S., Tollenaere M. A., Ho T., Toledo L. I., Mann M., Bekker-Jensen S., Räschle M., and Mailand N. (2016) Activation of the ATR kinase by the RPA-binding protein ETAA1. Nat. Cell Biol. 18, 1196–1207 10.1038/ncb3422 [DOI] [PubMed] [Google Scholar]
- 7. Griffith J. D., Comeau L., Rosenfield S., Stansel R. M., Bianchi A., Moss H., and de Lange T. (1999) Mammalian telomeres end in a large duplex loop. Cell 97, 503–514 10.1016/S0092-8674(00)80760-6 [DOI] [PubMed] [Google Scholar]
- 8. Doksani Y., Wu J. Y., de Lange T., and Zhuang X. (2013) Super-resolution fluorescence imaging of telomeres reveals TRF2-dependent T-loop formation. Cell 155, 345–356 10.1016/j.cell.2013.09.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Denchi E. L., and de Lange T. (2007) Protection of telomeres through independent control of ATM and ATR by TRF2 and POT1. Nature 448, 1068–1071 10.1038/nature06065 [DOI] [PubMed] [Google Scholar]
- 10. Baumann P., and Cech T. R. (2001) Pot1, the putative telomere end-binding protein in fission yeast and humans. Science 292, 1171–1175 10.1126/science.1060036 [DOI] [PubMed] [Google Scholar]
- 11. Gong Y., and de Lange T. (2010) A Shld1-controlled POT1a provides support for repression of ATR signaling at telomeres through RPA exclusion. Mol. Cell 40, 377–387 10.1016/j.molcel.2010.10.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Loayza D., Parsons H., Donigian J., Hoke K., and de Lange T. (2004) DNA binding features of human POT1: A nonamer 5′-TAGGGTTAG-3′ minimal binding site, sequence specificity, and internal binding to multimeric sites. J. Biol. Chem. 279, 13241–13248 10.1074/jbc.M312309200 [DOI] [PubMed] [Google Scholar]
- 13. Lei M., Podell E. R., and Cech T. R. (2004) Structure of human POT1 bound to telomeric single-stranded DNA provides a model for chromosome end-protection. Nat. Struct. Mol. Biol. 11, 1223–1229 10.1038/nsmb867 [DOI] [PubMed] [Google Scholar]
- 14. Loayza D., and de Lange T. (2003) POT1 as a terminal transducer of TRF1 telomere length control. Nature 423, 1013–1018 10.1038/nature01688 [DOI] [PubMed] [Google Scholar]
- 15. Liu D., Safari A., O'Connor M. S., Chan D. W., Laegeler A., Qin J., and Songyang Z. (2004) PTOP interacts with POT1 and regulates its localization to telomeres. Nat. Cell Biol. 6, 673–680 10.1038/ncb1142 [DOI] [PubMed] [Google Scholar]
- 16. Ye J. Z., Hockemeyer D., Krutchinsky A. N., Loayza D., Hooper S. M., Chait B. T., and de Lange T. (2004) POT1-interacting protein PIP1: A telomere length regulator that recruits POT1 to the TIN2/TRF1 complex. Genes Dev. 18, 1649–1654 10.1101/gad.1215404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kibe T., Osawa G. A., Keegan C. E., and de Lange T. (2010) Telomere protection by TPP1 is mediated by POT1a and POT1b. Mol. Cell. Biol. 30, 1059–1066 10.1128/MCB.01498-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Rice C., Shastrula P. K., Kossenkov A. V., Hills R., Baird D. M., Showe L. C., Doukov T., Janicki S., and Skordalakes E. (2017) Structural and functional analysis of the human POT1-TPP1 telomeric complex. Nat. Commun. 8, 14928 10.1038/ncomms14928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Palm W., Hockemeyer D., Kibe T., and de Lange T. (2009) Functional dissection of human and mouse POT1 proteins. Mol. Cell. Biol. 29, 471–482 10.1128/MCB.01352-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hockemeyer D., Sfeir A. J., Shay J. W., Wright W. E., and de Lange T. (2005) POT1 protects telomeres from a transient DNA damage response and determines how human chromosomes end. EMBO J. 24, 2667–2678 10.1038/sj.emboj.7600733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Wu P., Takai H., and de Lange T. (2012) Telomeric 3′ overhangs derive from resection by Exo1 and Apollo and fill-in by POT1b-associated CST. Cell 150, 39–52 10.1016/j.cell.2012.05.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hockemeyer D., Daniels J. P., Takai H., and de Lange T. (2006) Recent expansion of the telomeric complex in rodents: Two distinct POT1 proteins protect mouse telomeres. Cell 126, 63–77 10.1016/j.cell.2006.04.044 [DOI] [PubMed] [Google Scholar]
- 23. Hockemeyer D., Palm W., Wang R. C., Couto S. S., and de Lange T. (2008) Engineered telomere degradation models dyskeratosis congenita. Genes Dev. 22, 1773–1785 10.1101/gad.1679208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Price C. M., Boltz K. A., Chaiken M. F., Stewart J. A., Beilstein M. A., and Shippen D. E. (2010) Evolution of CST function in telomere maintenance. Cell Cycle 9, 3157–3165 10.4161/cc.9.15.12521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Diotti R., Kalan S., Matveyenko A., and Loayza D. (2015) DNA-directed polymerase subunits play a vital role in human telomeric overhang processing. Mol. Cancer Res. 13, 402–410 10.1158/1541-7786.MCR-14-0381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Flynn R. L., Centore R. C., O'Sullivan R. J., Rai R., Tse A., Songyang Z., Chang S., Karlseder J., and Zou L. (2011) TERRA and hnRNPA1 orchestrate an RPA-to-POT1 switch on telomeric single-stranded DNA. Nature 471, 532–536 10.1038/nature09772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Guo X., Deng Y., Lin Y., Cosme-Blanco W., Chan S., He H., Yuan G., Brown E. J., and Chang S. (2007) Dysfunctional telomeres activate an ATM-ATR-dependent DNA damage response to suppress tumorigenesis. EMBO J. 26, 4709–4719 10.1038/sj.emboj.7601893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Churikov D., and Price C. M. (2008) Pot1 and cell cycle progression cooperate in telomere length regulation. Nat. Struct. Mol. Biol. 15, 79–84 10.1038/nsmb1331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Takai K. K., Kibe T., Donigian J. R., Frescas D., and de Lange T. (2011) Telomere protection by TPP1/POT1 requires tethering to TIN2. Mol. Cell 44, 647–659 10.1016/j.molcel.2011.08.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Frescas D., and de Lange T. (2014) Binding of TPP1 to TIN2 is required for POT1a,b-mediated telomere protection. J. Biol. Chem. 289, 24180–24187 10.1074/jbc.M114.592592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Hockemeyer D., Palm W., Else T., Daniels J. P., Takai K. K., Ye J. Z., Keegan C. E., de Lange T., and Hammer G. D. (2007) Telomere protection by mammalian POT1 requires interaction with TPP1. Nat. Struct. Mol. Biol. 14, 754–761 10.1038/nsmb1270 [DOI] [PubMed] [Google Scholar]
- 32. Ray S., Bandaria J. N., Qureshi M. H., Yildiz A., and Balci H. (2014) G-quadruplex formation in telomeres enhances POT1/TPP1 protection against RPA binding. Proc. Natl. Acad. Sci. U.S.A. 111, 2990–2995 10.1073/pnas.1321436111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Fan J., and Pavletich N. P. (2012) Structure and conformational change of a replication protein A heterotrimer bound to ssDNA. Genes Dev. 26, 2337–2347 10.1101/gad.194787.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Xu X., Vaithiyalingam S., Glick G. G., Mordes D. A., Chazin W. J., and Cortez D. (2008) The basic cleft of RPA70N binds multiple checkpoint proteins, including RAD9, to regulate ATR signaling. Mol. Cell. Biol. 28, 7345–7353 10.1128/MCB.01079-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Sfeir A. J., Chai W., Shay J. W., and Wright W. E. (2005) Telomere-end processing the terminal nucleotides of human chromosomes. Mol. Cell 18, 131–138 10.1016/j.molcel.2005.02.035 [DOI] [PubMed] [Google Scholar]
- 36. Bhattacharjee A., Wang Y., Diao J., and Price C. M. (2017) Dynamic DNA binding, junction recognition and G4 melting activity underlie the telomeric and genome-wide roles of human CST. Nucleic Acids Res. 45, 12311–12324 10.1093/nar/gkx878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Wan M., Qin J., Songyang Z., and Liu D. (2009) OB fold-containing protein 1 (OBFC1), a human homolog of yeast Stn1, associates with TPP1 and is implicated in telomere length regulation. J. Biol. Chem. 284, 26725–26731 10.1074/jbc.M109.021105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Takai H., Jenkinson E., Kabir S., Babul-Hirji R., Najm-Tehrani N., Chitayat D. A., Crow Y. J., and de Lange T. (2016) A POT1 mutation implicates defective telomere end fill-in and telomere truncations in Coats plus. Genes Dev. 30, 812–826 10.1101/gad.276873.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Pinzaru A. M., Hom R. A., Beal A., Phillips A. F., Ni E., Cardozo T., Nair N., Choi J., Wuttke D. S., Sfeir A., and Denchi E. L. (2016) Telomere replication stress induced by POT1 inactivation accelerates tumorigenesis. Cell Rep. 15, 2170–2184 10.1016/j.celrep.2016.05.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Celli G. B., and de Lange T. (2005) DNA processing is not required for ATM-mediated telomere damage response after TRF2 deletion. Nat. Cell Biol. 7, 712–718 10.1038/ncb1275 [DOI] [PubMed] [Google Scholar]
- 41. Takai H., Smogorzewska A., and de Lange T. (2003) DNA damage foci at dysfunctional telomeres. Curr. Biol. 13, 1549–1556 10.1016/S0960-9822(03)00542-6 [DOI] [PubMed] [Google Scholar]
- 42. Dimitrova N., and de Lange T. (2006) MDC1 accelerates nonhomologous end-joining of dysfunctional telomeres. Genes Dev. 20, 3238–3243 10.1101/gad.1496606 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.