Abstract
ADP-ribosylation of proteins plays key roles in multiple biological processes, including DNA damage repair. Recent evidence suggests that serine is an important acceptor for ADP-ribosylation, and that serine ADP-ribosylation is hydrolyzed by ADP-ribosylhydrolase 3 (ARH3 or ADPRHL2). However, the structural details in ARH3-mediated hydrolysis remain elusive. Here, we determined the structure of ARH3 in a complex with ADP-ribose (ADPR). Our analyses revealed a group of acidic residues in ARH3 that keep two Mg2+ ions at the catalytic center for hydrolysis of Ser-linked ADP-ribosyl group. In particular, dynamic conformational changes involving Glu41 were observed in the catalytic center. Our observations suggest that Mg2+ ions together with Glu41 and water351 are likely to mediate the cleavage of the glycosidic bond in the serine–ADPR substrate. Moreover, we found that ADPR is buried in a groove and forms multiple hydrogen bonds with the main chain and side chains of ARH3 residues. On the basis of these structural findings, we used site-directed mutagenesis to examine the functional roles of key residues in the catalytic pocket of ARH3 in mediating the hydrolysis of ADP-ribosyl from serine and DNA damage repair. Moreover, we noted that ADPR recognition is essential for the recruitment of ARH3 to DNA lesions. Taken together, our study provides structural and functional insights into the molecular mechanism by which ARH3 hydrolyzes the ADP-ribosyl group from serine and contributes to DNA damage repair.
Keywords: ADP-ribosylation, protein crystallization, structural biology, crystal structure, DNA damage, DNA damage response, DNA repair
Introduction
ADP-ribosylation is a unique posttranslational modification that regulates a number of biological processes, such as DNA damage repair and gene transcription (1–3). This posttranslational modification is catalyzed by poly(ADP-ribose) polymerase (PARP)4 family enzymes (1, 2, 4). To date, there are 17 PARPs that have been identified in human cells (5). PARPs use NAD+ as the donor and covalently links the ADP-ribose to reside at the side chain of amino acid residues, such as Asp, Glu, Arg, and Cys (3). ADP-ribosylation exists as either polymer form or monomer form, namely poly(ADP-ribosyl)ation (PARylation) or mono(ADP-ribosyl)ation (MARylation) (6–10).
Recently, Ser has been shown as one of the major acceptors for ADP-ribosylation (11, 12). In response to H2O2 treatment, PARP1, the founding member of the PARP family, acts together with it co-factor HPF1 to mediate Ser-linked ADP-ribosylation (12, 13). The detailed analysis indicates that Ser-linked ADP-ribosylation is likely to be MARylation (14). Although the functional significance of Ser-linked ADP-ribosylation remains elusive, MS analysis suggests that Ser-linked ADP-ribosylation is one of the major forms of ADP-ribosylation catalyzed by PARP1 in vivo (11, 12).
In addition to ADP-ribosylation, hydrolyzation of ADP-ribose also plays key roles in numerous biological processes (9, 15–17). In particular, ADPR hydrolyzation has been shown to participate in DNA damage repair (18, 19). To date, more than 10 ADP-ribose hydrolases have been identified (18, 20). Among these enzymes, PARG is the most potent ADP-ribose hydrolase to degrade PARylation (21). Several others show amino acid-specific deMARylation activities. For example, both Macro D1/D2 and TARG1 mediate ADPR hydrolyzation on Asp or Glu; whereas ARH1 catalyzes deMARylation on Arg (18, 22). Thus, these enzymes coordinate together for the complicated ADP-ribose degradation on different amino acid residues.
Recently, it has been shown that ARH3 specifically mediates deMARylation on Ser during DNA damage repair (14, 20). ARH3 has been shown to localize in nucleus, cytosol, and mitochondria (23). Previous studies have shown that it hydrolyzes poly(ADP-ribose) (PAR) as well as O-acetyl-ADP-ribose, a byproduct generated during Sirtuin-mediated deacetylation (23). However, compared with PARG, ARH3 shows a strong deMARylation activity on PARP1 (14). Two recent studies suggest that Ser-linked ADP-ribosylation is one of the major substrates of ARH3 (14, 20). Thus, ARH3 may act as an amino acid-specific deMARylation enzyme in cells.
The crystal structure of apo-ARH3 was determined by Mueller-Dieckmann et al. (24) who have shown that ARH3 is a monomeric molecule with 19 α-helices. A possible substrate-binding mode was proposed using computer modeling, which was further validated by mutagenesis analysis (24). However, the crystal structure of the ARH3 and ligand complex has not yet been reported. Here, using X-ray crystallography, we determined the detailed structure of ARH3 in the complex with ADP-ribose (ADPR). Combining with biochemistry and cell biology assays, our results demonstrate that the enzymatic activity of ARH3 plays an important role in DNA damage repair.
Results
Overall structure of the ARH3-ADPR complex
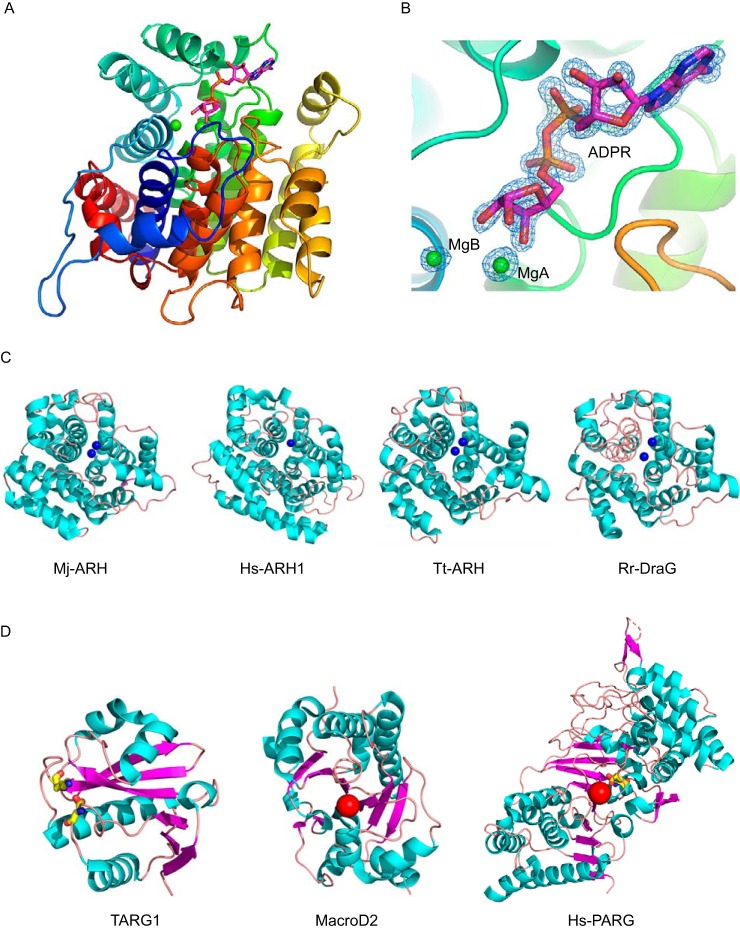
To understand the molecular mechanism by which ARH3 mediates ADP-ribosylhydrolyzation, the crystal structure of the ARH3-ADPR complex was determined at 1.58-Å resolution by X-ray diffraction. The final model contains one ARH3 molecule in the asymmetric unit (Fig. 1A). The ARH3 monomer binds to one ADPR molecule and two Mg2+ ions according to the electron density map (Fig. 1B).
Figure 1.
Overall structure of the ARH3-ADPR complex. A, rainbow-colored scheme representation of the ARH3-ADPR complex. ARH3 exhibits a compact all α-helical-fold. The ADPR molecule is displayed in sticks, with carbons in magenta. MgA and MgB are in green spheres. B, 2Fo − Fc map for the ADPR, MgA, and MgB contoured at 1.0 σ. C, comparison of the similar structural folding of ARH3 orthologous. Cyan, α-helix; salmon, loop. Mj-ARH, M. jannaschii ARH MJ1187 (PDB code 1T5J); Hs-ARH1, H. sapiens ARH1 (PDB code 3HFW); Tt-ARH, T. thermophilus ARH (PDB code 2YZV); and Rr-DraG, R. rubrum DraG (PDB code 2WOC). The Mg2+ ion in the active site is displayed in the blue sphere. D, different folding of macrodomain ADPR hydrolases. Cyan, α-helix; salmon, loop; and magenta, β-strand. Catalytic residue is in yellow stick, and the catalytic water is shown in the red sphere. From left to right: TARG1 (PDB code 4J5R), MacroD2 (PDB code 4IQY), and PARG (PDB code 4B1H).
The overall structure of ARH3 in the complex shows a compact all α-helical-fold (Fig. 1A). Notably, earlier studies on the 3D structure of apo-ARH3 show two crystal forms that are conformationally different, especially in the loop between η1 and α2 (η1-α2 loop, residues Glu41–Asp47) around the active pocket (24). Structural comparison shows that the corresponding η1-α2 loop in the ARH3-ADPR complex is more similar to that of the orthorhombic crystal form (PDB code 2FOZ) (Fig. S1).
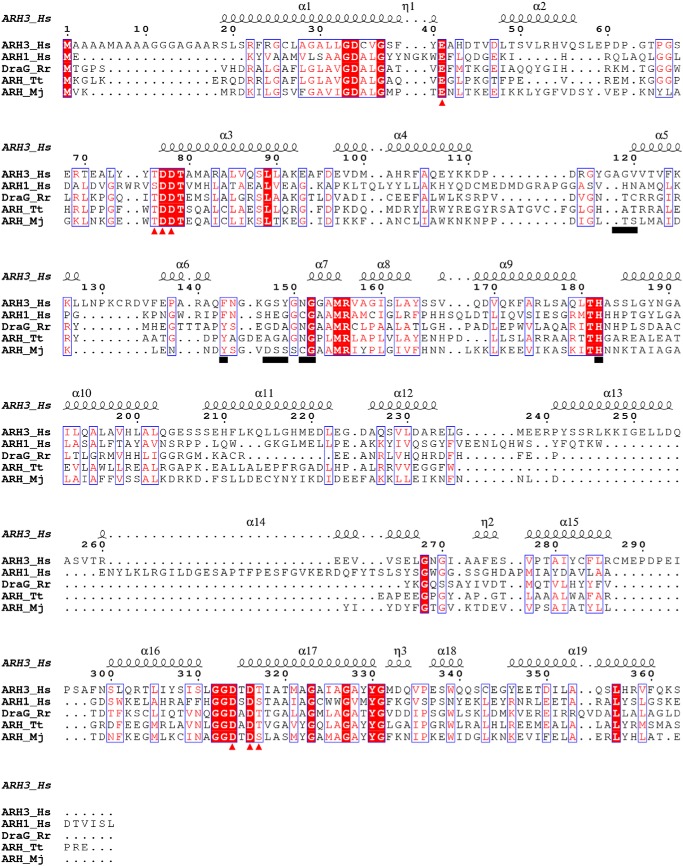
Structural alignment using the DALI program reveals that ARH3 shares a similar structural fold with several other ADP-ribosylglycohydrolases (ARH) despite their relatively low sequence identity (Fig. 2). These ARHs include ARH MJ1187 from Methanococcus jannaschii (PDB code 1T5J), ARH1 from Homo sapiens (PDB code 3HFW), putative ARH from Thermus thermophilus (PDB ID code 2YZV), and mono-ADP-ribosylhydrolase DraG from Rhodospirillum rubrum (PDB code 2WOC), giving a root mean square deviation of 2.4 ± 0.2 Å for all equivalent Ca atoms (Fig. 1C). These ARHs share common structural characteristics representing a compact all α-helical-fold, which is remarkably distinguished from that of macro domain ADPR hydrolases including PARG (PDB code 4B1H), MacroD2 (PDB code 4IQY), and TARG1 (PDB code 4J5R). The core structure of these macro domain enzymes contains a three-layered α-β-α sandwich, exhibiting the typical macro domain-fold (Fig. 1D).
Figure 2.
Sequence alignment of ARH3_Hs, ARH1_Hs, DraG_Rr, ARH_Tt, and ARH_Mj is generated via T-Coffee and the results were displayed using ESPript. The residues contributed to the ADPR binding are underlined. The Mg2+ coordinating residues are marked in red triangles. Hs, H. sapiens; Rr, R. rubrum; Tt, T. thermophiles; Mj, M. janaschii.
The catalytic site of ARH3
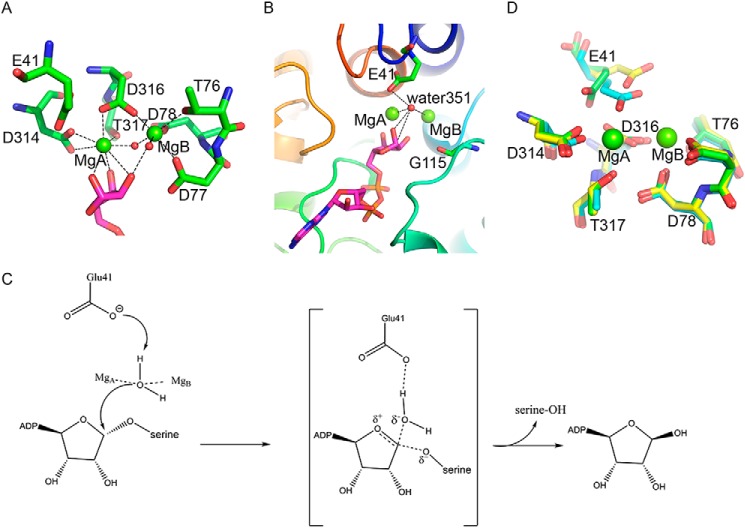
The ADPR-binding cavity of ARH3 unambiguously contains a binuclear Mg2+ site according to 2Fo − Fc electron density map, termed MgA and MgB, respectively (Fig. 1B). The distance between MgA and MgB is 3.8 Å. Both MgA and MgB have octahedral coordination. Structural comparison between apo-ARH3 (PDB codes 2FOZ and 2FP0) and the ARH3-ADPR complex reveals that both Mg2+ ions are shifted about 0.6 Å toward the distal ribose of ADPR, enabling the hydroxyl groups of the distal ribose to coordinate the two Mg2+ ions. MgA is coordinated with Asp314, Asp316, Thr317, three hydroxyl groups of the distal ribose and water269. Meanwhile, MgB is coordinated with the 2′-hydroxyl group of the distal ribose, Thr76, Asp77, Asp78, Asp316, and two water molecules (water269 and water85) (Fig. 3A). The sequence alignment and structural comparison revealed that the Mg2+ coordinating acidic residues Asp77, Asp78, Asp314, and Asp316 are fully conserved in homologous ARH enzymes (Fig. 2).
Figure 3.
Structural analysis of the catalytic site of ARH3. A, the binuclear Mg2+ site. The Mg2+ ions are in green spheres, and the coordinating water molecules are in red spheres. The coordinating residues are displayed in green sticks. The distal ribose of ADPR is shown in magenta sticks. The hydrogen bonds are indicated by black dashed lines. B, positions of Glu41 and the catalytic water351 in the ARH3-ADPR complex. C, the deduced ADPR hydrolyzation mediated by ARH3. D, coordinating residues in the ARH3-ADPR complex (green) is superimposed with apo-ARH3 orthorhombic (PDB code 2FOZ, cyan) and monoclinic crystal (PDB code 2FP0, yellow) forms.
In addition to the abovementioned coordinating residues, we notice that Glu41 is also involved in the Mg2+ coordination in the apo-ARH3 (PDB code 2FOZ) as the hydroxyl groups from the distal ribose of ADPR are absence in the apo-ARH3. However, in the ARH3-ADPR complex, the carboxyl side chain of Glu41 forms a hydrogen bond with water351 located at the catalytic center, with a close distance of 3.0 Å. Another hydrogen bond forms between water351 and the 1′-hydroxyl group of the distal ribose of ADPR, with a close distance of 2.8 Å. Moreover, the distance between MgA and water351 is 2.5 Å, and the distance between MgB and water351 is 3.4 Å. Thus, a hydrogen bond network is formed among Glu41, water351, 1′-hydroxyl group of the distal ribose, and MgA and MgB (Fig. 3B).
Moreover, water351 lies close to the C1′ atom of the distal ribose with a distance of 3.7 Å (Fig. 3B). Because the 1′-hydroxyl group of the distal ribose of ADPR is covalently linked to the side chain of serine via a glycosidic bond, it is likely that Glu41 functions as the catalytic residue to activate water351, which in turn carries out a nucleophilic attack on the C1′ atom of the distal ribose for the cleavage of the scissile glycosidic bond (Fig. 3C). This hydrolyzation mechanism mimics the canonical catalytic mechanism of glycosidases, in which an acidic residue is used as a proton acceptor to activate a water molecule, and then cleave the scissile glycosidic bond via nucleophilic attack (25).
Both Mg2+ ions are involved in the positioning of water351 in the catalytic center through hydrogen-bonding interaction. In contrast, in MacroD2, both the α-phosphate and distal ribose of ADPR contribute to hold the catalytic water (26). In PARG, the stabilizing factors for the catalytic water are Glu755, Asp737, and the diphosphate moiety of ADPR (27). Thus, the mechanism of ARH3-dependent ADPR hydrolyzation is distinct from macro domain enzymes.
Collectively, this structural analysis suggests that water351 is the catalytic water molecule that exists in the center of the catalytic pocket of ARH3, and Glu41 works as a key catalytic residue in the ADPR hydrolyzation reaction, which is consistent with earlier reports showing that Glu41 is required for catalysis (14, 28). Thus, our analysis of the catalytic center of ARH3 may reveal the detailed molecular mechanism by which ARH3 mediates the ADPR hydrolyzation.
It has been reported that the conformation of Glu41 in the ARH3 orthorhombic crystal form (PDB code 2FOZ) is obviously different from that of the monoclinic crystal form (PDB code 2FP0) (Fig. 3D). In the orthorhombic crystal form, Glu41 is involved in the Mg2+ coordination, whereas Glu41 moves about 1.8 Å toward to the protein surface and does not coordinate with Mg2+ in the monoclinic crystal form (24). Notably, Glu41 undergoes a significant conformational change in the ADPR-bound ARH3, the side chain of Glu41 is shifted 2.1 Å compared with that in the apo-ARH3 orthorhombic crystal form (PDB code 2FOZ), and is pushed away from the MgA (Fig. 3D). Taken together, three conformations have been observed for Glu41, indicating that Glu41 is in the dynamic status at the acidic catalytic center. This conformational diversity of Glu41 is in good agreement with its catalytic roles, which was also confirmed by the ADPR hydrolyzation assays (see below).
The structure of the ADPR binding cavity
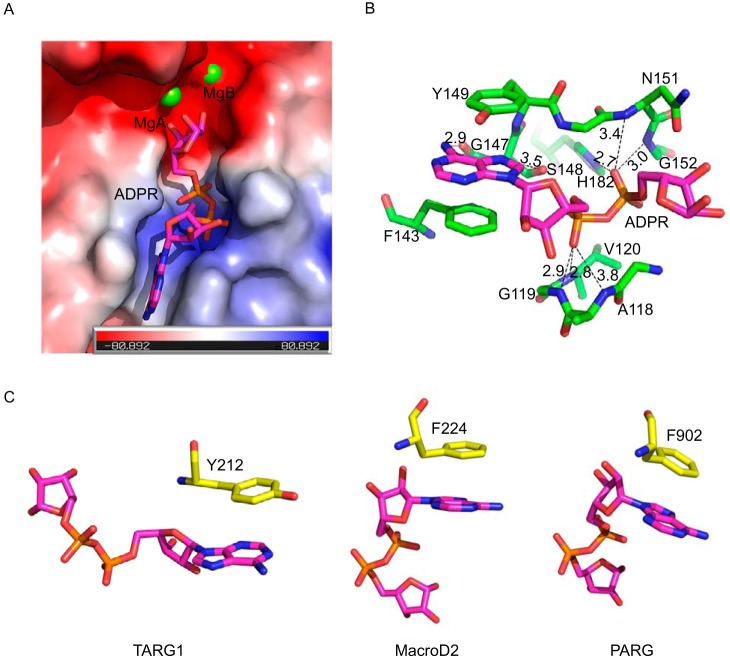
The electron density map reveals that an ADPR molecule binds to a deep cavity that is mainly encircled by loops (Fig. 4A). A number of residues are responsible for the binding and stabilizing ADPR via main chain and side chain contacts (Fig. 4B). The adenine ring is stacked by Tyr149 and Phe143 through the π–π interaction (Fig. 4B). This stacking interaction between the adenine ring and aromatic residues is common in ADP-ribose hydrolases. For example, Tyr212 contributes to the stacking interaction with the adenine ring of ADPR in DraG (29), and the corresponding stacking residues are Phe224 and Phe902 in MacroD2 and PARG, respectively (26, 27) (Fig. 4C). The double stacking interaction in the complex of ARH3-ADPR may clamp the adenine ring tightly and makes the adenine ring in a rigid conformation compared with the single stacking interaction. In addition to the π–π interaction, the adenine ring hydrogen bonds to the main chain of Gly147 and Ser148, which further contributes to the selectivity of the substrate in the catalytic pocket of ARH3 (Fig. 4B).
Figure 4.
Structural analysis of the ADPR-binding cavity in ARH3. A, electrostatic potential map of the binding cavity of ARH3. The ADPR molecule is shown in magenta sticks. MgA and MgB are in green spheres. B, the ADPR binding mode. The ADPR is in magenta sticks. Residues involved in the interaction with ADPR were shown in green stick. The hydrogen bonds are in black dashed lines. C, the stacking interaction between different ADPR hydrolases and the adenine ring of ADPR. From left to right: TARG1 (PDB code 4J5R), MacroD2 (PDB code 4IQY), and PARG (PDB code 4B1H). The stacking residues are in yellow stick.
Moreover, the adenosine ribose in ADPR forms a hydrogen bond with the main chain amino group of Gly119 (Fig. 4B). The diphosphate moiety in ADPR sits in the N terminus of two α-helices of ARH3, and it hydrogen bonds the main chain amino groups of Ala118, Gly119, Val120, Asn151, and Gly152, and the side chain of His182, generating a network of hydrogen bonds that comprise a large proportion of the interaction between ARH3 and ADPR (Fig. 4B). In addition, several water molecules are involved in this hydrogen bond network formation. Notably, there is no direct interaction between the distal ribose of ADPR and ARH3. Instead, the two catalytic Mg2+ ions interact with the distal ribose via coordination.
Different catalytic mechanisms of ADP-ribose hydrolases
Sequence alignment reveals that these catalytically related residues consisting of Glu41, Asp77, Asp78, Asp314, and Asp316 are not conserved between ARH family enzymes and macro domain ADP-ribose hydrolases, such as PARG, MacroD1/D2, and TARG1. Moreover, unlike ARHs, there is no binuclear metal-binding site in these macro domain enzymes. Thus, the catalytic mechanism of protein ADP-ribose hydrolases can be classified into two different groups, namely metal-dependent and metal-independent catalysis. ARHs, such as ARH3, belong to metal-dependent catalysis, utilizing two Mg2+ ions and acidic residues to complete the catalytic reaction, which might be highly conserved.
In contrast, the catalytic mechanism is not conserved in the macro domain ADP-ribose hydrolases, For example, Glu756 and a water molecule act together to catalyze the reaction in PARG (27), whereas the key catalytic factor in MacroD2 is an activated water (26). For TARG1, the Lys84/Asp125 catalytic dyad may contribute to the catalytic reaction (30).
Structural comparison of the binding pocket between ARH3 and PARG
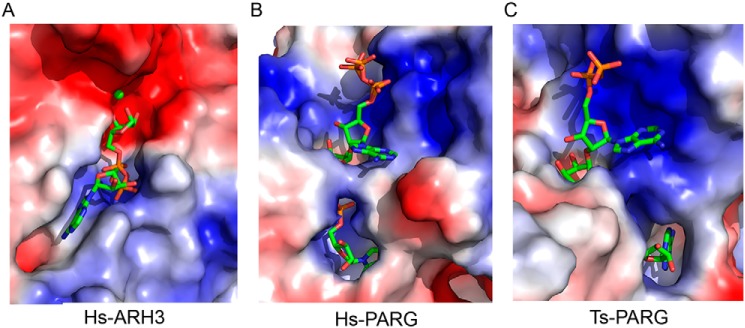
The electrostatic potential molecular surface shows that the binding pocket of ARH3 is divided into two subunits by the binuclear Mg2+ sites. The subunit accommodating ADPR is mainly composed of a basic region (positive charged), which has strong affinity for the diphosphate group of ADPR. The other subunit, the putative binding site for the remaining substrate groups, is entirely composed of an acidic region (negative charged) (Fig. 5A). Interestingly, the charge characteristic of the binding pocket in ARH3 is remarkably distinguished from that in PARG. The binding pocket of PARG, accommodating the ADPR dimer, is mostly composed of the basic region (PDB code 5A7R) (Fig. 5B). Moreover, the binding pocket in T. thermophila PARG (PDB code 4L2H) predominantly comprises the basic region (Fig. 5C). The electric charge differences in the substrate-binding pocket may be closely related to their divergent enzyme-degrading activities.
Figure 5.
Electrostatic surface potential comparison of the binding cavity between ARH3 and two PARGs. Blue, positive charged; red, negative charged; gray, neutral. A, Hs-ARH3, the ADPR are in green stick. B, Hs-PARG (PDB code 5A7R), the ADPR dimer is in green stick. C, Tt-PARG (PDB code 4L2H), the ADPR dimer is shown in green stick.
Key residue mutations in ARH3 abolish the enzymatic activity
To validate the in vitro structural analysis, we generated a series of mutations in the catalytic binding pocket of ARH3, including E41Q, D77N, S148A, Y149A, H182A, D314A, and T317A. Among these mutations, E41Q, D77N, D314A, and T317A occur at the catalytic site, whereas S148A, Y149A, and H182A exist in the ADPR recognition pocket. We generated the recombinant glutathione S-transferase-ARH3 mutants, and examined these proteins in the in vitro ADPR hydrolyzation assays. Compared with the WT ARH3, all these mutations abolished the enzymatic activity of ARH3, suggesting that all these residues are essential for the ARH3-dependent ADPR hydrolyzation (Fig. S2), which is also consistent with earlier studies (14, 24). Given Asp77, Asp314, and Thr317 are involved in Mg2+ coordination, our results suggest that Mg2+ is important in the catalytic process.
The enzymatic activity of ARH3 plays an important role in DNA damage repair
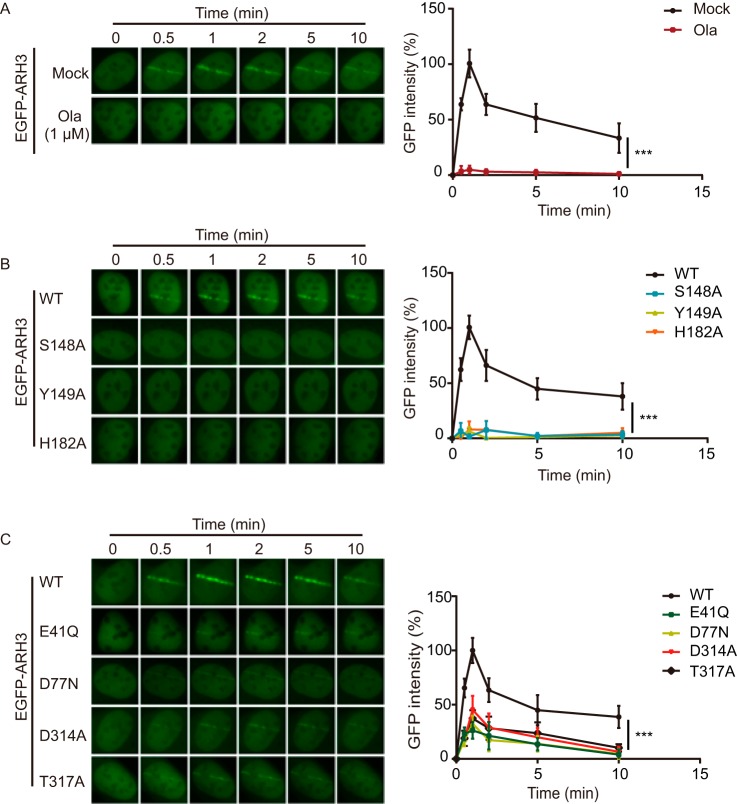
It has been shown that ARH3 is involved in DNA damage repair (23). Using a laser microirradiation assay, we examined the recruitment of ARH3 to DNA lesions. WT of ARH3 was recruited to the sites of DNA damage within 30 s, and this relocation process was suppressed by the PARP inhibitor olaparib (Fig. 6A), suggesting that the recruitment of ARH3 to DNA lesions is mediated by DNA damage-induced ADP-ribosylation. Compared with WT ARH3, S148A, Y149A, and H182A mutants failed to relocate to DNA lesions (Fig. 6B). Because S148A, Y149A, and H182A disrupted the ADPR-binding pocket, it suggests that ADPR recognition plays a key role for the recruitment of ARH3. Notably, E41Q, D77N, D314A, and T317A mutants were still recruited to DNA lesions, albeit at low intensity (Fig. 6C), suggesting that the catalytic site is not essential for the recruitment of ARH3. Instead, our results demonstrate that ADPR recognition is required for the recruitment of ARH3 to DNA lesions.
Figure 6.
The recruitment of ARH3 to DNA lesions is mediated by ADPR recognition. A, the recruitment of ARH3 is mediated by ADP-ribosylation. U2OS cells were expressing GFP-ARH3. The cells were treated with or without 1 μm olaparib. The recruitment kinetics was measured in 30 cells in three independent experiments. B, S148A, Y149A, and H182A mutations abolish the recruitment of ARH3. C, E41Q, D77N, D314A, and T317A mutations partially impair the recruitment of ARH3. The recruitment kinetics is measured in 30 cells in three independent experiments. Data are represented as mean ± S.D. as indicated from three independent experiments. ***, statistically significant (p < 0.001).
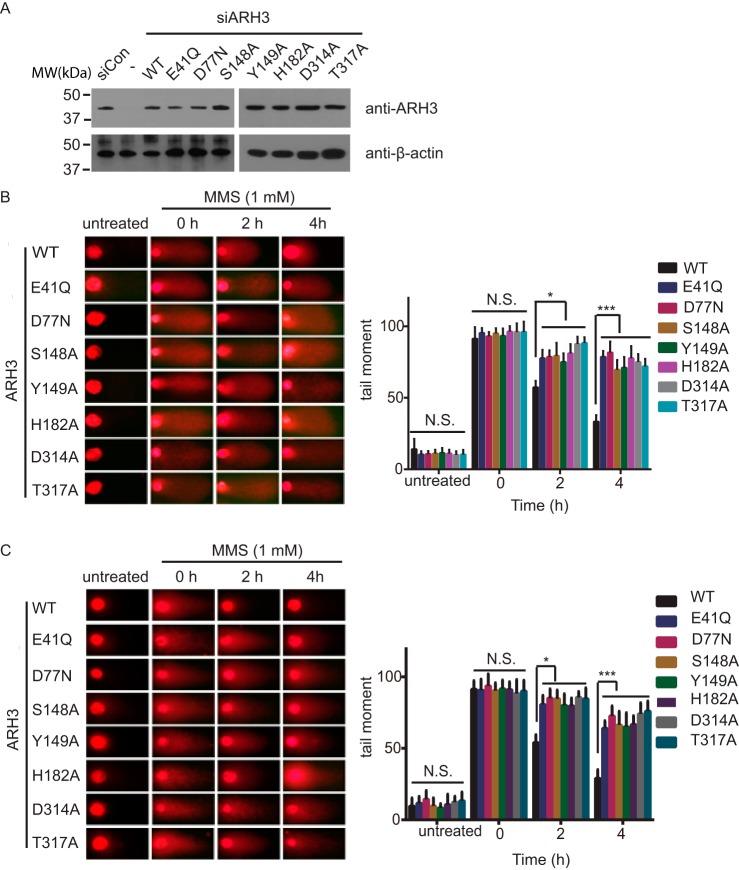
Next, we examined the roles of ARH3 in DNA damage repair. We knocked down the endogenous ARH3 with siRNA, and expressed WT ARH3 and its mutants in U2OS cells (Fig. 7A). The cells were treated with methyl methanesulfonate, a DNA damaging agent. We performed comet assays to examine the DNA damage repair kinetics. Under both alkaline and neutral conditions, the DNA damage repair was impaired when cells were expressing mutant ARH3 (Fig. 7, B and C), suggesting that ARH3-dependent ADPR hydrolyzation plays an important role in DNA damage repair.
Figure 7.
Enzymatic activity of ARH3 plays an important role for DNA damage repair. A, U2OS cells were expressing the ARH3 mutants. U2OS cells were treated with siRNA to knock down the endogenous ARH3, and the cells were expressing WT ARH3 or its mutants. Comet assays were performed under either alkaline (B) or neutral conditions (C). Comet tail moments were examined to indicate the DNA damage repair kinetics. Data are represented as mean ± S.D. as indicated from three independent experiments. *, statistically significant (p < 0.05); ***: statistically significant (p < 0.001). MMS, methyl methanesulfonate.
Discussion
In 2006, Mueller-Dieckmann et al. (24) determined the 3D structure of apo-ARH3, which are actually in two different crystal forms, including monoclinic and orthorhombic crystal forms. The significant conformational differences occur in η1-α2 loop around the active pocket, indicating that this region is very flexible. In our structure analysis, we show that the η1-α2 loop in the ARH3-ADPR complex is more close to that of orthorhombic crystal form (Fig. S1), suggesting that ligand binding stabilizes this region, which plays a key role for catalysis. Moreover, in the ARH3-ADPR complex, we observed that a potential nucleophilic water molecule (water351) is at the catalytic center (Fig. 3). Combining the conformation diversity of Glu41 and the canonical catalytic mechanism of glycosidases, we proposed a model of catalysis mediated by Glu41 (Fig. 3C). Our analysis on the ARH3-ADPR complex provides the structural evidence. Moreover, based on this unique conformational change surrounding Glu41, we examined the role of Glu41 and confirmed that loss of Glu41 abolished the enzymatic activity of ARH3 (Fig. S2), which is in agreement with Mueller-Dieckmann et al. (24). Because ARH3 mediates ADP-ribosylhydrolation in response to DNA damage, we have shown the first evidence that the E41Q mutation impaired ARH3-dependent DNA damage repair (Figs. 6 and 7). Taken together, our study demonstrates the catalytic mechanism by which ARH3 hydrolyzes ADP-ribosylation.
Based on the computer modeling of ARH3 and ligand binding, Mueller-Dieckmann et al. (24) performed mutagenesis and characterized the catalytically associated amino acid residues. They mutated 9 individual residues, including Glu41, Asp77, Ser148, Tyr149, Asn151, His182, Glu275, Asp314, and Thr317, to examine the roles of these residues in the enzymatic activity of ARH3 (24). In our studies, we examined the structural analysis of the ARH3-ADPR complex and performed the mutagenesis analysis based on the detailed analysis of the ARH3-ADPR complex. We confirmed these specific residues that mediate the catalysis, which is in agreement with Mueller-Dieckmann et al. (24). Both Mueller-Dieckmann et al. (24) and us reveal that Ser148, Tyr149, and His182 are involved in ADPR recognition; Glu41, Asp77, Asp314, and Thr317 are catalytically related. Notably, in addition to the biochemical analysis, we further examined the roles of these residues in the ARH3-mediated DNA damage repair. Recent studies indicate that ARH3 plays a role in DNA damage repair. Using laser microirradiation assays, we have revealed that ARH3 is recruited by DNA lesions, which is mediated by DNA damage-induced poly(ADP-ribosyl)ation (Fig. 6A). We further analyzed ADPR recognition residues including Ser148, Tyr149, and His182, and found that these residues are required for the recruitment of ARH3 to DNA lesions (Fig. 6B). In contrast, catalytically related residues, including Glu41, Asp77, Asp314, and Thr317, although very important, are not essential for the recruitment of ARH3. Mutations of these residues only partially impair the recruitment. Furthermore, we performed comet assays to examine the roles of these residues in DNA damage repair. However, due to loss of the enzymatic activity, mutating catalytically related residues still abolished the ARH3-mediated DNA damage repair (Fig. 7). Thus, our detailed molecular biology analysis reveals the biological functions of two different groups of residues in ARH3. One group is for substrate recognition; the other group is for ADPR hydrolysis. Both groups of residues are important for ARH3-dependent DNA damage repair. In summary, our research work provides further insight on the catalytic mechanism and the structure-function analysis of ARH3.
Experimental procedures
Protein expression and purification
The ARH3 gene was amplified from genomic DNA of 293T cells and constructed into the modified pET-15b vector, containing an N-terminal hexahistidine tag and a cleavage site for human rhinovirus 3C (HRV 3C) protease. The N-terminal 18 residues of ARH3 were removed during the cloning process.
The recombinant plasmid of pET-15b-ARH3 was transformed into Escherichia coli BL21 (DE3) cells. The transforming cells were grown in liquid L broth media at 37 °C to A600 = 0.8–1.0, and then 0.12 mm isopropyl β-d-1-thiogalactopyranoside was added to induce recombinant protein expression at 16 °C overnight. Cells were harvested by centrifugation, resuspended in ice-cold lysis buffer (25 mm Tris-HCl, pH 8.0, 100 mm NaCl), and lysed through sonication. The cell lysate was then centrifuged at 35,000 × g for 45 min.
For protein purification, the supernatant containing the soluble ARH3 was loaded onto a nickel-chelating Sepharose (GE Healthcare) column pre-equilibrated with lysis buffer. After extensively washing with lysis buffer, ARH3 was eluted using elution buffer (25 mm Tris-HCl, pH 8.0, 100 mm NaCl, and 250 mm imidazole). The N-terminal His tag was removed by the HRV 3C protease. Then the eluent was further purified using an ion-exchange column Source 15Q (GE Healthcare) and Superdex 200 Increase column (GE Healthcare) successively. The purified protein was concentrated to 11 mg/ml in 10 mm Tris-HCl, pH 8.0, and 100 mm NaCl.
Crystallization
To screen for crystallization of the ARH3-ADPR complex, ARH3 and ADPR were mixed in the molar ratio of 1:3. Crystals were obtained using the sitting drop diffusion method at 293 K through mixing equal volumes of protein complex with reservoir solution consisting of 0.1 m MES, pH 6.0, 20% (w/v) PEG mme 2000. The best crystals were optimized by adding NDSB-201 up to the final concentration of 0.2 m.
Data collection and processing
To collect the X-ray data, the crystals were immersed in the cryoprotectant buffer containing 20% glycerol (v/v) plus reservoir buffer and then flash-cooled in a 100 K nitrogen stream. The X-ray diffraction data were collected on the beamline BL17u1 at Shanghai Synchrotron Radiation facility (SSRF). The crystal belongs to space group P21 with the unit cell dimensions a = 43.885 Å, b = 76.737 Å, c = 48.533 Å, and β = 103.598°. The data were processed using the HKL-3000 software suite (31).
Structure determination and refinement
The crystal structure of the ARH3-ADPR complex was solved through molecular replacement using Phaser (32). The structure of apo-ARH3 (PDB code 2FOZ) was used as the search model. The structure building was performed using the ARP/WARP software (33). The further refinement was carried out repeatedly using COOT (34) and PHENIX (35). The detailed statistics of data collection and structure refinement are summarized in Table 1. All the molecular graphics figures were prepared using PyMol.
Table 1.
Data collection and refinement statistics
| ARH3-ADPR | |
|---|---|
| Data collection | |
| Space group | P 21 |
| Cell dimensions | |
| a, b, c (Å) | 43.88, 76.74, 48.53 |
| α, β, γ (°) | 90.0, 103.0, 90.0 |
| Wavelength (Å) | 0.9791 |
| Resolution (Å) | 50–1.58 (1.64–1.58)a |
| Rmerge(%) | 6.1 (20.7) |
| 〈I/σ(I)〉 | 15.8 (5.0) |
| Completeness (%) | 96.4 (91.4) |
| Redundancy | 3.2 (3.2) |
| Refinement | |
| Resolution (Å) | 37.35–1.58 |
| No. reflections | 40,881 |
| Rwork / Rfree (%) | 15.74/17.99 |
| No. atoms | |
| Protein | 2,683 |
| ADPR | 36 |
| Mg2+ | 2 |
| Water | 262 |
| B-factors | |
| Protein | 13.20 |
| ADPR | 13.74 |
| Mg2+ | 15.09 |
| Water | 24.71 |
| Root mean square deviations | |
| Bond lengths (Å) | 0.006 |
| Bond angles (°) | 0.918 |
a Values in parentheses are for highest-resolution shell.
Plasmids and siRNA
Human ARH3 and its mutants were cloned into pEGFP-C1, pGEX-4T-1, and pCDNA3 vectors. Human PARP1 full-length cDNA was cloned to the pET-28a vector. ARH3 siRNA targeting sequence was 5′-auuggagagcuucuagaccaggcat-3′.
Cell culture
U2OS cells were maintained in Dulbecco's modified Eagle's medium with 10% fetal bovine serum and cultivated at 37 °C in 5% CO2 (v/v). U2OS cells were transfected with plasmids encoding ARH3 or its mutants.
Synthesis and purification of PAR
His-PARP1 was expressed in BL21 and purified by nickel-nitrilotriacetic acid affinity resin. PAR was synthesized in a 1-ml incubation buffer containing 100 mm Tris-HCl, pH 7.8, 10 mm MgCl2, 1 mm NAD+, 10 mm DTT, 50 μg of octameric oligonucleotide GGAATTCC, and 100 μg of recombinant PARP1. The mixture was incubated at 37 °C for 60 min and stopped by an addition of 20 ml of ice-cold 20% TCA. Oligo DNA was removed by DNase I. Following precipitation the pellet was washed with ice-cold 99.8% ethanol and stored at −20 °C.
ADPR hydrolyzation assay
Approximately 5 pmol of PAR and 1 pmol of each recombinant protein were incubated together in PBS. After incubation for 1 h at room temperature, heating at 90 °C for 8 min, 2-μl samples were dotted onto nitrocellulose membranes. After incubation for 20 min at 60 °C, dot blotting assays were performed with anti-PAR antibody.
Laser microirradiation and microscope image acquisition
Cells transfected with GFP-tagged corresponding plasmids were grown on 35-mm glass bottom dishes (NEST Biotechnology). Laser microirradiation was performed on OLYMPUS IX71 inverted fluorescence microscope coupled with the MicroPoint Laser Illumination and Ablation System (Photonic Instruments, Inc.). A 337.1-nm laser diode (3.4 milliwatts) transmits through a specific Dye Cell and then yields a 365-nm wavelength laser beam that is focused through X60 UPlanSApo/1.35 oil objective to yield a spot size of 0.5–1 μm. The pulse energy is 170 microjoules at 10 Hz. Images were taken by the same microscope with CellSens software (Olympus). The GPF fluorescence strips were recorded at the indicated time points and then analyzed with Image J software. 30 cells were analyzed from three independent experiments. Error bars represent the standard deviation.
Comet assay
Single-cell gel electrophoretic comet assay was performed under neutral conditions to detect DNA double strand breaks and alkaline conditions to detect single strand DNA breaks. U2OS cells with or without the indicated treatments were recovered in normal culture medium for the indicated time. Cells were collected and rinsed twice with ice-cold PBS; 2 × 104/ml cells were combined with 1% LMAgarose at 40 °C at a ratio of 1:3 (v/v) and immediately pipetted onto slides. For cellular lysis, the slides were immersed in the neutral lysis solution (2% Sarkosyl, 0.5 m EDTA, 0.5 mg/ml of proteinase K in pH 8.0) overnight at 37 °C in the dark, or alkaline lysis solution (1.2 m NaCl, 100 mm EDTA, 0.26 m NaOH, 0.1% Sarkosyl) followed by washing in the rinse buffer (90 mm Tris-HCl, pH 8.5, 90 mm boric acid, 2 mm EDTA) and alkaline wash buffer (0.03 m NaOH, 2 mm EDTA) for 30 min with two repeats. Then, the slides were subjected to electrophoresis at 20 V (0.6 V/cm) for 25 min and stained in 2.5 μg/ml of propidium iodide for 20 min. All images were taken with a fluorescence microscope and analyzed by Comet Assay IV software.
Author contributions
M. W., R. X., and Y. M. data curation; M. W., Z. Y., X. L., and X. Y. methodology; Z. Y., R. X., X. L., and X. Y. formal analysis; X. L. and X. Y. investigation; X. L. and X. Y. writing-original draft; X. L. project administration; X. Y. conceptualization; X. Y. supervision; X. Y. funding acquisition; X. Y. writing-review and editing.
Supplementary Material
Acknowledgment
We thank the beamline BL17u1 staff at the Shanghai Synchrotron Radiation facility for X-ray data collection experiments.
This work was supported by the National Natural Science Foundation of China Grants 81672794, 31570305, and 31572269, the “100 Talents Plan” of Hebei Province Grant E2016100014, Foundation of Hebei Educational committee Grant SLRC2017023, NCI, National Institutes of Health Grants CA130899, CA132755, and CA187209, NIGMS, National Institutes of Health Grant GM108467, and Leukemia and Lymphoma Society Grant 1325-15. The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
This article contains Figs. S1 and S2.
The atomic coordinates and structure factors (code 5ZQY) have been deposited in the Protein Data Bank (http://wwpdb.org/).
- PARP
- poly(ADP-ribose) polymerase
- PARylation
- poly(ADP-ribosyl)ation
- ADPR
- ADP-ribose
- MARylation
- mono(ADP-ribosyl)ation
- PAR
- poly(ADP-ribose)
- ARH
- ADP-ribosylglycohydrolases
- HRV 3C
- human rhinovirus 3C
- PDB
- Protein Data Bank.
References
- 1. Kraus W. L. (2015) PARPs and ADP-ribosylation: 50 years, and counting. Mol. Cell 58, 902–910 10.1016/j.molcel.2015.06.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bürkle A. (2005) Poly(ADP-ribose): the most elaborate metabolite of NAD. FEBS J. 272, 4576–4589 10.1111/j.1742-4658.2005.04864.x [DOI] [PubMed] [Google Scholar]
- 3. Hottiger M. O. (2015) Nuclear ADP-ribosylation and its role in chromatin plasticity, cell differentiation, and epigenetics. Annu. Rev. Biochem. 84, 227–263 10.1146/annurev-biochem-060614-034506 [DOI] [PubMed] [Google Scholar]
- 4. Luo X., and Kraus W. L. (2012) On PAR with PARP: cellular stress signaling through poly(ADP-ribose) and PARP-1. Genes Dev. 26, 417–432 10.1101/gad.183509.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Perina D., Mikoč A., Ahel J., Ćetković, HŽaja R., and Ahel I. (2014) Distribution of protein poly(ADP-ribosyl)ation systems across all domains of life. DNA Repair 23, 4–16 10.1016/j.dnarep.2014.05.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Hassa P. O., Haenni S. S., Elser M., and Hottiger M. O. (2006) Nuclear ADP-ribosylation reactions in mammalian cells: where are we today and where are we going? Microbiol. Mol. Biol. Rev. 70, 789–829 10.1128/MMBR.00040-05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Vyas S., Matic I., Uchima L., Rood J., Zaja R., Hay R. T., Ahel I., and Chang P. (2014) Family-wide analysis of poly(ADP-ribose) polymerase activity. Nat. Commun. 5, 4426 10.1038/ncomms5426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Leung A. K. (2014) Poly(ADP-ribose): an organizer of cellular architecture. J. Cell Biol. 205, 613–619 10.1083/jcb.201402114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Barkauskaite E., Jankevicius G., and Ahel I. (2015) Structures and mechanisms of enzymes employed in the synthesis and degradation of PARP-dependent protein ADP-ribosylation. Mol. Cell 58, 935–946 10.1016/j.molcel.2015.05.007 [DOI] [PubMed] [Google Scholar]
- 10. Bütepage M., Eckei L., Verheugd P., and Lüscher B. (2015) Intracellular mono-ADP-ribosylation in signaling and disease. Cells 4, 569–595 10.3390/cells4040569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Leidecker O., Bonfiglio J. J., Colby T., Zhang Q., Atanassov I., Zaja R., Palazzo L., Stockum A., Ahel I., and Matic I. (2016) Serine is a new target residue for endogenous ADP-ribosylation on histones. Nat. Chem. Biol. 12, 998–1000 10.1038/nchembio.2180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Bonfiglio J. J., Fontana P., Zhang Q., Colby T., Gibbs-Seymour I., Atanassov I., Bartlett E., Zaja R., Ahel I., and Matic I. (2017) Serine ADP-ribosylation depends on HPF1. Mol. Cell 65, 932–940.e936 10.1016/j.molcel.2017.01.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Palazzo L., Leidecker O., Prokhorova E., Dauben H., Matic I., and Ahel I. (2018) Serine is the major residue for ADP-ribosylation upon DNA damage. eLife 7, e34334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Abplanalp J., Leutert M., Frugier E., Nowak K., Feurer R., Kato J., Kistemaker H. V. A., Filippov D. V., Moss J., Caflisch A., and Hottiger M. O. (2017) Proteomic analyses identify ARH3 as a serine mono-ADP-ribosylhydrolase. Nat. Commun. 8, 2055 10.1038/s41467-017-02253-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Gibson B. A., and Kraus W. L. (2012) New insights into the molecular and cellular functions of poly(ADP-ribose) and PARPs. Nat. Rev. Mol. Cell Biol. 13, 411–424 10.1038/nrm3376 [DOI] [PubMed] [Google Scholar]
- 16. Kasamatsu A., Nakao M., Smith B. C., Comstock L. R., Ono T., Kato J., Denu J. M., and Moss J. (2011) Hydrolysis of O-acetyl-ADP-ribose isomers by ADP-ribosylhydrolase 3. J. Biol. Chem. 286, 21110–21117 10.1074/jbc.M111.237636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Palazzo L., Mikoč A., and Ahel I. (2017) ADP-ribosylation: new facets of an ancient modification. FEBS J. 284, 2932–2946 10.1111/febs.14078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Liu C., Vyas A., Kassab M. A., Singh A. K., and Yu X. (2017) The role of poly ADP-ribosylation in the first wave of DNA damage response. Nucleic Acids Res. 45, 8129–8141 10.1093/nar/gkx565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Mashimo M., and Moss J. (2016) Functional role of ADP-ribosyl-acceptor hydrolase 3 in poly(ADP-ribose) polymerase-1 response to oxidative stress. Curr. Protein Peptide Sci. 17, 633–640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Fontana P., Bonfiglio J. J., Palazzo L., Bartlett E., Matic I., and Ahel I. (2017) Serine ADP-ribosylation reversal by the hydrolase ARH3. eLife 6, e28533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Slade D., Dunstan M. S., Barkauskaite E., Weston R., Lafite P., Dixon N., Ahel M., Leys D., and Ahel I. (2011) The structure and catalytic mechanism of a poly(ADP-ribose) glycohydrolase. Nature 477, 616–620 10.1038/nature10404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Moss J., Jacobson M. K., and Stanley S. J. (1985) Reversibility of arginine-specific mono(ADP-ribosyl)ation: identification in erythrocytes of an ADP-ribose-l-arginine cleavage enzyme. Proc. Natl. Acad. Sci. U.S.A. 82, 5603–5607 10.1073/pnas.82.17.5603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Mashimo M., Kato J., and Moss J. (2014) Structure and function of the ARH family of ADP-ribosyl-acceptor hydrolases. DNA Repair 23, 88–94 10.1016/j.dnarep.2014.03.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Mueller-Dieckmann C., Kernstock S., Lisurek M., von Kries J. P., Haag F., Weiss M. S., and Koch-Nolte F. (2006) The structure of human ADP-ribosylhydrolase 3 (ARH3) provides insights into the reversibility of protein ADP-ribosylation. Proc. Natl. Acad. Sci. U.S.A. 103, 15026–15031 10.1073/pnas.0606762103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Vasella A., Davies G. J., and Böhm M. (2002) Glycosidase mechanisms. Curr. Opin. Chem. Biol. 6, 619–629 10.1016/S1367-5931(02)00380-0 [DOI] [PubMed] [Google Scholar]
- 26. Jankevicius G., Hassler M., Golia B., Rybin V., Zacharias M., Timinszky G., and Ladurner A. G. (2013) A family of macrodomain proteins reverses cellular mono-ADP-ribosylation. Nat. Struct. Mol. Biol. 20, 508–514 10.1038/nsmb.2523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Tucker J. A., Bennett N., Brassington C., Durant S. T., Hassall G., Holdgate G., McAlister M., Nissink J. W., Truman C., and Watson M. (2012) Structures of the human poly (ADP-ribose) glycohydrolase catalytic domain confirm catalytic mechanism and explain inhibition by ADP-HPD derivatives. PloS One 7, e50889 10.1371/journal.pone.0050889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ono T., Kasamatsu A., Oka S., and Moss J. (2006) The 39-kDa poly(ADP-ribose) glycohydrolase ARH3 hydrolyzes O-acetyl-ADP-ribose, a product of the Sir2 family of acetyl-histone deacetylases. Proc. Natl. Acad. Sci. U.S.A. 103, 16687–16691 10.1073/pnas.0607911103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Berthold C. L., Wang H., Nordlund S., and Högbom M. (2009) Mechanism of ADP-ribosylation removal revealed by the structure and ligand complexes of the dimanganese mono-ADP-ribosylhydrolase DraG. Proc. Natl. Acad. Sci. U.S.A. 106, 14247–14252 10.1073/pnas.0905906106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Sharifi R., Morra R., Appel C. D., Tallis M., Chioza B., Jankevicius G., Simpson M. A., Matic I., Ozkan E., Golia B., Schellenberg M. J., Weston R., Williams J. G., Rossi M. N., Galehdari H., et al. (2013) Deficiency of terminal ADP-ribose protein glycohydrolase TARG1/C6orf130 in neurodegenerative disease. EMBO J. 32, 1225–1237 10.1038/emboj.2013.51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Otwinowski Z., and Minor W. (1997) Processing of x-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 10.1016/S0076-6879(97)76066-X [DOI] [PubMed] [Google Scholar]
- 32. McCoy A. J., Grosse-Kunstleve R. W., Adams P. D., Winn M. D., Storoni L. C., and Read R. J. (2007) Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 10.1107/S0021889807021206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Langer G., Cohen S. X., Lamzin V. S., and Perrakis A. (2008) Automated macromolecular model building for X-ray crystallography using ARP/wARP version 7. Nat. Protoc. 3, 1171–1179 10.1038/nprot.2008.91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Emsley P., Lohkamp B., Scott W. G., and Cowtan K. (2010) Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 10.1107/S0907444910007493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Adams P. D., Afonine P. V., Bunkóczi G., Chen V. B., Davis I. W., Echols N., Headd J. J., Hung L. W., Kapral G. J., Grosse-Kunstleve R. W., McCoy A. J., Moriarty N. W., Oeffner R., Read R. J., Richardson D. C., et al. (2010) PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 10.1107/S0907444909052925 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.