Abstract
Interactions between emerging nascent polypeptide chains and the ribosome can modulate cotranslational protein folding. However, it has remained unclear how such interactions can affect the binding of nascent chains to their cellular targets. We thus investigated on the ribosome the interaction between two intrinsically disordered proteins of opposite charge, ACTR and NCBD, which form a high-affinity complex in a coupled folding-and-binding reaction. Using fluorescence correlation spectroscopy and arrest-peptide-mediated force measurements in vitro and in vivo, we find that the ACTR-NCBD complex can form cotranslationally but only with ACTR as the nascent chain and NCBD free in solution, not vice versa. We show that this surprising asymmetry in behavior is caused by pronounced charge interactions: attraction of the positively charged nascent chain of NCBD to the negatively charged ribosomal surface competes with complex formation and prevents ACTR binding. In contrast, the negatively charged nascent ACTR is repelled by the ribosomal surface and thus remains available for productively binding its partner. Electrostatic interactions may thus be more important for cotranslational folding and binding than previously thought.
Introduction
Proteins can fold (1, 2, 3) and bind to cellular partners (4, 5) cotranslationally (6), as shown by studies performed by cryo-electron microscopy (EM) (7, 8), NMR (9), Förster-resonance energy transfer (FRET) (10), and optical tweezers (11, 12). The ribosome tunnel can accommodate an unstructured nascent chain of ∼30 residues, and secondary and tertiary structure have been reported to form in the tunnel, especially in the region of the vestibule where the tunnel widens to ∼20 Å (7, 13, 14, 15, 16, 17). Given the large negative charge of the ribosome (18), electrostatic interactions are expected to play an important role in cotranslational folding processes, including the modulation of translation rates. Negatively charged residues in nascent unstructured peptides can help overcoming translational stalling (19), whereas positively charged residues within the nascent chain can produce transiently arrested species (20) or lead to slowed translation (12, 20, 21).
Yet, it has remained largely unexplored whether the cotranslational binding of nascent chains to cellular partners can be affected by electrostatic interactions with the ribosome. Intrinsically disordered proteins (IDPs) (22), a large class of proteins that do not form tertiary structure under physiological conditions in the absence of a binding partner (23, 24), tend to be rich in charged amino acids (20, 25) and may thus be particularly susceptible to interactions with the ribosomal surface (26).
Here, we study the coupled folding and binding on the ribosome of two IDPs, the nuclear coactivator binding domain (NCBD) of CREB-bindingv protein and the activation domain from the p160 transcriptional coactivator, ACTR (Fig. S1). These proteins have opposite net charge and, upon binding, fold into a complex with high affinity in the low nanomolar range (23, 27) (Fig. 1 A). NCBD in isolation adopts a helical conformation similar to that in the complex (27) but lacks a cooperatively folded core (23). ACTR, instead, is largely unstructured and retains limited helicity in isolation (28). Importantly, formation of the complex is favored by electrostatics (29, 30).
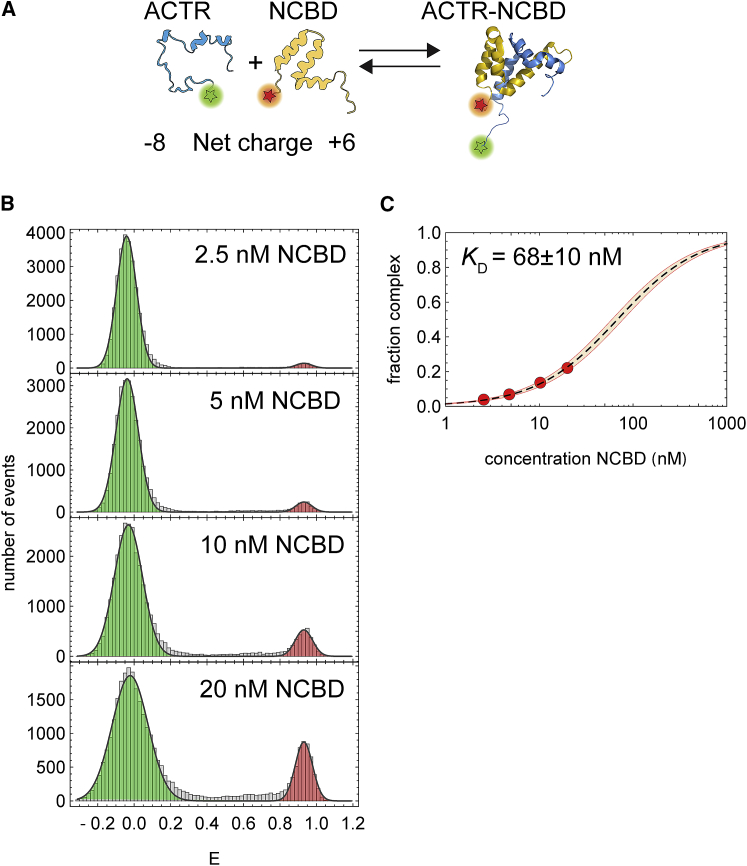
Figure 1.
Affinity of the ACTR-NCBD complex in the presence of the PURE system. (A) Schematic depiction of fluorescently labeled ACTR and NCBD and their complex formation (Protein Data Bank: 1KBH) (23). (B) An excess of NCBD labeled with CF680R was titrated to 50 pM ACTR labeled with Atto 532. Formation of the ACTR-NCBD complex can be monitored by the appearance of a high-transfer efficiency peak (red) while the low-transfer efficiency peak corresponding to free ACTR (green) decreases. (C) The fraction of ACTR-NCBD complex was calculated from the transfer efficiency histograms in (B) (by dividing the number of events with E > 0.4 by the total number of events) and fitted with a binding isotherm (dashed line). The shaded area corresponds to the 99% confidence interval of the fitted dissociation constant KD. NCBD could not be titrated to higher concentrations because of increased fluorescence background due to direct excitation of the acceptor fluorophore. To see this figure in color, go online.
We find that NCBD free in solution binds to ribosome-nascent chain complexes (RNCs) of ACTR, and its affinity for ACTR increases with increasing length of the nascent polypeptide chain separating ACTR and the peptidyl-transferase center (PTC). Arrest-peptide-mediated force measurements (7, 11) show that the formation of the complex exerts a force on the nascent chain and takes place in the vicinity of the exit tunnel, when ACTR is still partially embedded within the tunnel. Surprisingly, however, ACTR free in solution does not bind to NCBD emerging from the tunnel, and under the same conditions, we cannot detect any force applied on the nascent chain. We suggest that the attractive interaction between the NCBD nascent chain and the ribosomal surface interferes with ACTR binding, whereas the electrostatic repulsion between ACTR and the ribosome exit tunnel allows the formation of the ACTR-NCBD complex on the ribosome.
Materials and Methods
DNA manipulation and cloning
DNA sequences of ACTR and NCBD were polymerase chain reaction (PCR)-amplified using primers bearing 5′-SapI overhangs, after which the PCR products were purified from agarose gels and subsequently cloned into the p7XLNC3GH plasmids containing SecM sequences of increasing lengths followed by a C-terminal GFP, as previously described (31). Plasmids were sequence verified and further used for in vitro translation or to transform E. coli BL21 (DE3) cells. These plasmids were further used as templates for creating the sequence HA-ACTR-SecM Mannheimia succiniciproducens (supI), using forward and reverse primers that introduced the HA and the M. succiniciproducens (supI) sequence, respectively. PCR products were purified and cloned into a p7XC3H vector (32). Plasmids were sequence verified and used for in vitro translation. Mutant sequences of ACTR and NCBD were purchased from Integrated DNA Technologies (Coralville, IA) and directly cloned into the p7XC3H vector. All enzymes that were used for cloning were obtained from New England Biolabs (Ipswich, MA). All cloning procedures were based on fragment-exchange cloning (32).
In vitro translation and in-gel fluorescence analysis
In vitro translation of ACTR-SecM-GFP and NCBD-SecM-GFP constructs (Fig. 3) and the fusion constructs N-NCBD-ACTR and N-ACTR-NCBD (Fig. 4 C) were carried out using the Protein Synthesis Using Recombinant Elements (PURE) system (New England Biolabs), as described before (31). For experiments in the presence of binding partner, 8 μM of purified labeled ACTR or purified labeled NCBD was added before starting the reaction at 37°C. In-gel fluorescence analysis was carried out essentially as described previously (31). In-gel fluorescence was recorded using a LAS-3000 Fuji imager (GE Healthcare, Chicago, IL), and images were recorded for 8 s. For the ACTR-SecM constructs, in-gel fluorescence bands obtained from three gels were further quantified and used to determine means and standard deviations, whereas for NCBD-SecM constructs, values were quantified from one gel. In vitro translation of the HA-ACTR-SecM M. succiniciproducens (supI) constructs used for fluorescence correlation spectroscopy (FCS) analysis was carried out for 20 min at 37°C (Fig. S2) using the PURE system following the manufacturers’ instructions, after which samples were frozen in liquid nitrogen and stored at −80°C until further FCS analysis (Fig. 2) or Western blot analysis (Fig. S2). Samples were separated by sodium dodecyl sulfate/polyacrylamide gel electrophoresis at neutral pH (4–12% Bis-Tris protein gels (NuPAGE; Thermo Fischer Scientific, Waltham, MA)), followed by Western blotting using an antibody against the HA epitope (Merck, Kenilworth, NJ), according to the manufacturers’ instructions. In vitro translation of the HA-NCBD-SecM M. succiniciproducens (supI) constructs used for FCS analysis was carried out for 25, 40, or 50 min at 37°C in triplicates, after which samples were frozen in liquid nitrogen and stored at −80°C until being analyzed by Western blotting or FCS analysis, as described above.
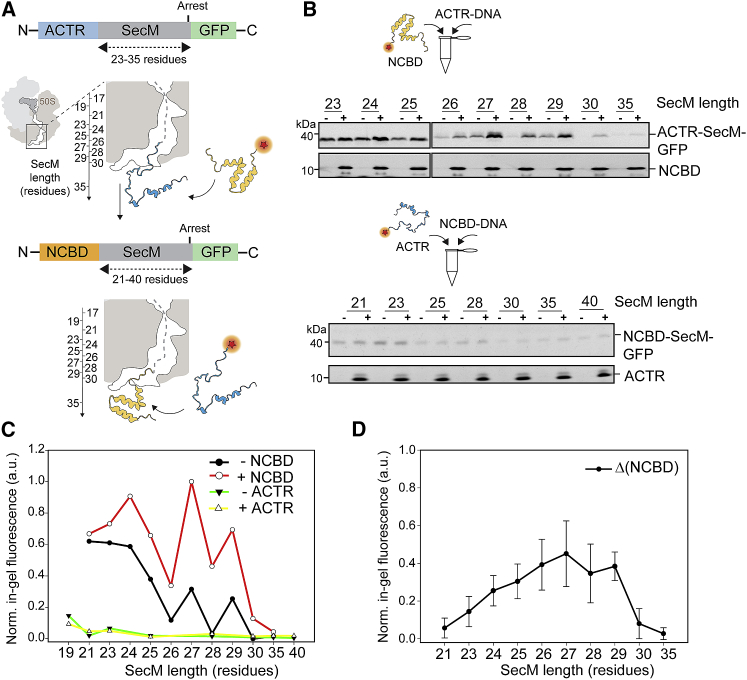
Figure 3.
Arrest-peptide-mediated force measurements upon formation of the ACTR-NCBD complex in vitro. (A) A schematic representation of the constructs used. ACTR (in light blue) is N-terminally fused to SecM sequences of increasing length (21–35 residues). Binding of fluorescently labeled NCBD creates force on the nascent chain that allows SecM stalling to be overcome and translation to continue to the C-terminal GFP. Similarly, NCBD was fused to SecM of increasing length (21–40 residues). (B) ACTR-SecM-GFP and NCBD-SecM-GFP constructs were translated in vitro, in the absence or presence of ∼8-μM CF680R-NCBD or ∼8-μM CF680R-ACTR, respectively. In-gel fluorescence recorded in the green (NCBD-SecM-GFP or ACTR-SecM-GFP) and red (CF680R-ACTR or CF680R-NCBD) channels is shown. (C) Quantification of the green fluorescent band intensities from the gels shown in (B) is depicted. (D) Difference between the expression of the ACTR-SecM-GFP constructs in the presence (+NCBD) and absence (-NCBD) of labeled NCBD, from experiments performed in triplicate, except from the construct ACTR-SecM-27, which was calculated from two experiments, is shown. To see this figure in color, go online. a.u., arbitrary units; Norm., normalized.
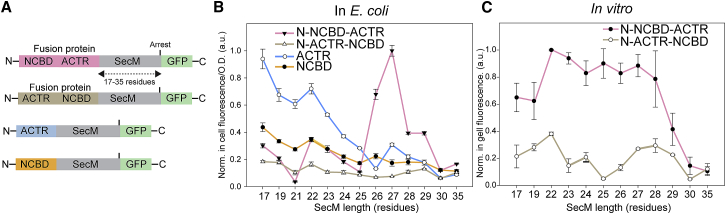
Figure 4.
Arrest-peptide-mediated force measurements of the fusion constructs N-NCBD-ACTR and N-ACTR-NCBD in E. coli and in vitro. (A) Schematic of the constructs used to express the fusion constructs of ACTR and NCBD in E. coli and ACTR and NCBD as individual nascent chains. (B) Force profiles of constructs shown in (A) obtained in vivo. Experiments were performed in triplicate using independent biological samples, after which in-cell fluorescence was quantified and normalized by cell density, and averages and SDs were calculated. Data are normalized relative to the construct NCBD-ACTR-SecM27-GFP, which displayed the highest fluorescence. (C) Force profiles of the constructs N-NCBD-ACTR and N-ACTR-NCBD expressed in vitro. Experiments were performed a minimum of three times. Fluorescent intensities were obtained from in-gel fluorescence, from which bands were quantified, and means and SDs were calculated. Values were normalized to a value of 1 for the highest fluorescence of the construct N-NCBD-ACTR-SecM(L = 22)-GFP. To see this figure in color, go online. a.u., arbitrary units; O.D., optical density.
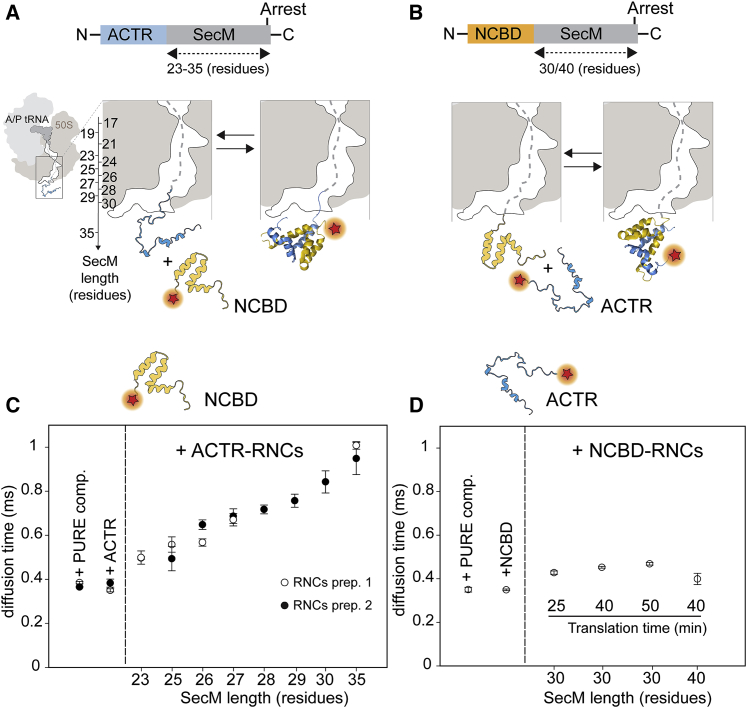
Figure 2.
Formation of the ACTR-NCBD complex on the ribosome monitored by FCS. (A and B) Schematic representations show the coupled folding and binding reaction of ACTR and NCBD on the ribosome. The ribosome is depicted as stalled with SecM (dashed gray line), ACTR is depicted in light blue, and NCBD is depicted in yellow. (C) Diffusion times of CF680R-NCBD upon binding to ACTR-RNCs as obtained by FCS measurements are shown. The diffusion time of labeled NCBD was first monitored in the presence of 25% PURE system and with 50 nM free unlabeled ACTR. Binding of labeled NCBD to two ACTR-RNC preparations obtained from different PURE system kits are reported, indicated as “RNC prep. 1” and “RNC prep. 2.” RNC preparation 1 did not include ACTR-RNCs at L = 28, 29, and 30. (D) FCS measurements of labeled ACTR upon binding to NCBD-RNCs are shown. FCS of CF680R-ACTR in the presence of 25% untranslated PURE system (+PURE components), 25% PURE system with 50 nM unlabeled NCBD, and NCBD-RNCs at SecM lengths of 30 and 40 residues obtained at increasing translation times (Fig. S6) are shown. Experiments were performed three times, and averages and SDs were calculated (except for NCBD-RNCs-30 translated for 25 min and NCBD-RNCs-40 translated for 40 min, which were performed twice). To see this figure in color, go online.
Expression in E. coli and in-cell fluorescence
Fusion constructs N-ACTR-NCBD and N-NCBD-ACTR were obtained by overlapping PCR. PCR products were purified and cloned using fragment-exchange cloning (32) into p7XLNC3GH plasmids, containing the SecM sequence and a C-terminal GFP, as described previously (31). Expression in E. coli BL21 (DE3) was carried out in 96-well plates, and subsequent in-cell fluorescence quantification was carried out as described previously (31).
Expression and purification of ACTR and NCBD
Cysteine variants of ACTR and NCBD (Fig. S1; Table S1) were generated by site-directed mutagenesis and coexpressed from a pET-47b (+) vector. The expression construct contained an N-terminal His6 tag cleavable with human rhinovirus 3C protease. Proteins were expressed in E. coli C41 (DE3) for 1 h after induction with 1 mM IPTG. The harvested cells were lysed by sonication and the His6-tagged protein was enriched via immobilized-metal affinity chromatography on Ni-IDA resin (ABT). The His6-tag was then cleaved off with human rhinovirus 3C protease and separated from the protein by another round of immobilized-metal affinity chromatography. Finally, ACTR and NCBD were separated via reversed-phase high-performance liquid chromatography (HPLC) on a C18 column (Reprosil Gold 200, Dr. Maisch; Ammerbuch, Germany) with a water/0.1% trifluoroacetic acid-acetonitrile gradient. The purified protein was lyophilized.
Labeling of ACTR and NCBD
Lyophilized protein was dissolved under a nitrogen atmosphere to a concentration of 200 μM in 100 mM potassium phosphate buffer (pH 7.0) (labeling buffer). For labeling with CF680R maleimide (Biotium, Hayward, CA), the protein was incubated for 3 h at room temperature with an equimolar amount of dye. For labeling with Atto 532, the protein was treated analogously with a 0.7-fold molar ratio of dye. Labeled protein was separated from unlabeled protein with HPLC as described above, and the appropriate fraction was lyophilized. The correct mass of all labeled proteins was confirmed by electrospray ionization mass spectrometry.
Single-molecule instrumentation and measurements
Single-molecule fluorescence data were recorded using a custom-built confocal microscope in a configuration described previously (33). The sample was excited with a continuous-wave solid-state laser (Oxxius LaserBoxx, Lannion, France) at a radiant flux of 50 μW (measured at the back aperture of the objective). Fluorescence was spatially selected with a 50-μm pinhole, split according to polarization and wavelength, and detected on four avalanche photodiodes. Data for transfer efficiency histograms were recorded for 30 min at sample concentrations of 50 pM donor-labeled ACTR in PURE system supplied with increasing concentrations of acceptor-labeled NCBD. The measurement buffer additionally contained 0.01% Tween 20 to prevent surface adhesion of the sample.
The arrival time of every photon was recorded; photons from all detection channels were binned at 0.75 ms, and all bins with >70 photons were retained as bursts. Identified bursts were corrected for background, the quantum yield of the dyes, the detection efficiency of the detectors, cross talk, and direct excitation of the acceptor (34). The bursts were histogrammed by the transfer efficiency (E) calculated from the corrected numbers of photons detected in the donor (nD) and acceptor (nA) channels, . The fraction of ACTR-NCBD complex was determined by dividing the number of bursts with E > 0.4 by the total number of detected bursts. The dissociation constant, KD, was then determined by fitting a binding isotherm (fraction bound = ) to the measured fractions of ACTR-NCBD complex, with cNCBD being the concentration of NCBD added. Note that this treatment is only valid under the following three conditions: 1) the contribution of fluorescent impurities to the donor-only peak is negligible compared to the contribution of donor-labeled ACTR molecules, 2) there is no significant fraction of ACTR-NCBD complex formed with an inactive acceptor dye, and 3) direct excitation of unbound acceptor-labeled NCBD does not lead to the detection of additional bursts. The first two conditions were addressed by reversed-phase HPLC in combination with fluorescence detection, which confirmed that the purity of the labeled protein was >95%. Furthermore, the minimal number of photons in a fluorescence burst was set to 70, a threshold in which no bursts were observed in the PURE system without added ACTR during the same time of measurement. This high threshold also eliminates all bursts that might arise from direct excitation of acceptor-labeled molecules, as reflected by the similarity in the number of detected bursts in all four measurements (in order of increasing NCBD concentration: 28,267, 26,938, 30,496, and 30,667). Taken together, our estimate of KD is thus likely to present an upper limit because additional contributions to the donor-only peak are difficult to exclude completely.
FCS measurements were recorded on a MicroTime 200 confocal single-molecule instrument (PicoQuant, Berlin, Germany). ACTR or NCBD labeled with Biotium CF680R were measured at concentrations of 6–13 nM for 10 min in triplicate for each construct in buffer (50 mM sodium phosphate (pH 7.0), 10 mM MgAc2, 0.01% Tween) with the addition of 25% PURE system containing either empty ribosomes or ACTR/NCBD nascent chains. The sample was excited at 635 nm with a diode laser either operating in continuous wave mode or in pulsed mode at 20 MHz (LDH-D-C-635M; PicoQuant). In both cases, the radiant flux was 30 μW (measured at the back aperture of the objective). Fluorescence was separated from excitation light with a triple-band mirror (zt405/530/630rpc; Chroma, Irvine, CA), collected through a 100 μm pinhole, split according to polarization, and directed onto two detectors after filtering through LP647RU filters (Semrock, Rochester, NY). In a first step, the fluorescence intensity cross correlation, G(τ), between the detectors was fitted with an FCS model taking into account translational diffusion and triplet blinking to determine the diffusion time τD,free of the free protein in the PURE System through the confocal volume:
| (1) |
where N is the average number of labeled molecules in the confocal volume. The parameter s, the ratio of the lateral to the axial radii of the confocal volume, was set to 1/6, and the correlation time of triplet blinking, τT, was 10 μs. We obtained τD,free = 0.33 and 0.35 ms for ACTR and NCBD, respectively. We estimated the diffusion time of the 70S ribosome, τD,70S, through the confocal volume using the Stokes-Einstein equation and the resulting relation τD,70S/τD,free(ACTR) = RH,70S/RH,ACTR to be 1.81 ms. RH,70S and RH,ACTR are the previously determined hydrodynamic radii of the ribosome (12.6 nm) (35) and ACTR (2.3 nm) (36), respectively. We note that the diffusion times of ACTR and NCBD in the absence of the PURE system are 0.32 and 0.27 ms (experimental error due to variation in confocal volume size was ∼5%), indicating the absence of excluded volume effects of the PURE system on their translational diffusion but a slight retardation of NCBD diffusion by transient electrostatic interactions with the ribosome. Given the estimated macromolecular volume fraction of at most ∼1% in the PURE System (37), the absence of crowding effects is expected.
In a second step, the data sets in the presence of RNCs were fitted with a model taking into account a mixture of two species with different translational diffusion times, τD,free and τD,70S, corresponding to the free and RNC-bound labeled protein, and triplet blinking:
| (2) |
where the diffusion times for free ACTR or NCBD were set to the results obtained in the absence of RNCs, and the diffusion time of RNC-bound protein was set to τD,70S = 1.81 ms. The only free fit parameters were thus the average numbers of molecules, Nfree for unbound, and N70S for bound protein, as well as the triplet amplitude, cT. The mean fitted values for cT of the different data sets were 0.05 ± 0.01 for NCBD RNCs, 0.11 ± 0.01 for ACTR RNCs preparation (prep.) 1 and 0.1 ± 0.02 for ACTR RNCs prep. 2. We calculated the fractions of labeled proteins bound to the ribosome complex (see Fig. S5 B) from fbound = N70S/(Nfree + N70S) and the mean diffusion times (see Fig. 2) from .
Results
Coupled folding and binding of ACTR and NCBD in the presence of the PURE system
The coupled folding and binding of ACTR and NCBD is favored by electrostatics and thus strongly influenced by ionic strength (28, 30). To test the suitability of the PURE system (37) for producing RNCs of ACTR and NCBD and probing their interactions with the respective binding partner, we first investigated whether the free proteins (i.e., not attached to the ribosome) form a complex under the solution conditions of the PURE system. For intermolecular single-molecule FRET, ACTR was labeled with Atto 532 (Sigma-Aldrich, St. Louis, MO) as a donor dye, NCBD was labeled with Biotium CF680R as the acceptor dye (Fig. 1 A), and binding was investigated by confocal single-molecule fluorescence of freely diffusing molecules in the presence of all components and the buffer system of the PURE system.
From unbound ACTR molecules, only donor fluorescence is observed (green-shaded peak in Fig. 1 B). Upon the addition of acceptor-labeled NCBD and formation of the ACTR-NCBD complex, donor and acceptor dyes come into close proximity, leading to FRET, visible as a peak at high-transfer efficiency appearing at NCBD concentrations in the nanomolar range (red-shaded peak in Fig. 1 B). Note that the single-molecule approach allows us to determine the absolute fraction of the complex, so, in principle, a single measurement would suffice to determine the dissociation constant, KD. However, to increase the precision of the KD value, we titrated a constant concentration of donor-labeled ACTR (50 pM) with increasing amounts of acceptor-labeled NCBD until fluorescence background due to acceptor direct excitation became prohibitive, and determined the affinity by fitting a binding isotherm to the fraction of ACTR in complex with NCBD. We obtained a KD of 68 ± 10 nM (Fig. 1 C), demonstrating that the high affinity of the two binding partners (23) is not compromised by fluorescence labeling and the presence of the components of the PURE system.
Formation of the ACTR-NCBD complex on the ribosome monitored by FCS
Can the complex also form if one of the two binding partners is emerging from the ribosome? Single-molecule FRET experiments for detecting binding to RNCs were not feasible owing to the following combination of factors: 1) a small change in intramolecular transfer efficiencies upon binding for both FRET-labeled ACTR and NCBD at the salt concentrations present in the PURE system (33); 2) relatively low binding affinities, especially for FRET-labeled NCBD (38), which 3) require prohibitively high concentrations of RNCs, with a concomitant increase in fluorescence background upon donor excitation. We thus investigated complex formation with FCS via changes in translational diffusion times when labeled protein free in solution binds to unlabeled RNCs.
We produced RNCs of ACTR and NCBD (Fig. 2, A and B) using the C-terminal arrest sequence of the SecM signal peptide, which regulates the expression of the ATPase SecA in bacteria (39). Once translated, the C-terminal part of SecM induces translational stalling, with peptidyl transfer RNAs in the P/P state and hybrid A/P, P/E states, as shown by cryo-EM structures of the E. coli ribosome stalled by SecM (40). ACTR-RNCs were obtained by translation in vitro for 20 min at 37°C, so that a sufficient concentration of RNCs was produced for FCS measurements, but release of ACTR from the ribosome was minimal (Fig. S2 A). To further reduce nascent-chain release, we used the C-terminal variant (supI) of the M. succiniciproducens SecM sequence, which yields particularly strong stalling (7, 41, 42) (Fig. S2, B and C). A final concentration of RNCs of ∼35 nM was used for the FCS experiments, as quantified by Western blot analysis (Fig. S3).
ACTR-RNCs or NCBD-RNCs were presented with free NCBD or ACTR, respectively, labeled with CF680R to enable measurements of translational diffusion after excitation at 635 nm, in which little background fluorescence is detected that could bias the result. Binding of the labeled proteins to the unlabeled RNCs is expected to result in an increase in the translational diffusion time through the confocal volume. As reference values, we quantified the diffusion times of free ACTR and NCBD in the presence of the PURE system (Fig. 2, C and D). The diffusion times of fluorescently labeled NCBD in the presence of ACTR-RNCs showed a systematic increase with increasing length of the SecM sequence and thus the nascent chain (Fig. 2 A; Fig. S4 A), indicating a rising affinity of NCBD to the increasing part of ACTR exposed outside the ribosome tunnel. At a SecM length of L = 35 residues (Fig. 2 A), ACTR is expected to be completely outside the tunnel (40) and thus available for binding to NCBD with full affinity.
From the FCS data, we estimate ∼40% of the NCBD molecules to be bound for the longest SecM length under these conditions (Fig. S5 B), remarkably close to the ∼35% expected from the KD of 68 nM for free ACTR (Fig. 1 C) and an ACTR-RNC concentration of 35 nM. ACTR emerging from the ribosome thus reaches similar affinities to NCBD as the free protein. Surprisingly, a very different result was obtained when NCBD was tethered to the ribosome as a nascent chain and presented with free labeled ACTR: even with a tether length of 40 residues (Fig. 2 B), the diffusion time of ACTR was similar to that of free ACTR in the PURE system (Fig. 2 D; Fig. S5 A), indicating a lack of binding. Increasing the concentration of NCBD-RNCs by extending the translation times (Fig. 2 D; Fig. S5 B) did not change this result. The affinity of ACTR for NCBD thus seems to be much lower when NCBD is tethered to the ribosome.
Asymmetry in complex affinity on the ribosome detected by arrest-peptide-mediated force measurements
If the affinity between ACTR and NCBD depends on which binding partner emerges from the ribosome exit tunnel, this asymmetry should also be detectable by arrest-peptide-mediated force measurements (7, 31, 42). SecM responds to molecular events that create force, such as the interactions of the nascent chain with the translocon (39, 42) and the cotranslational folding of proteins (7, 11, 12, 31). These forces can be detected by quantifying the translation efficiency of a reporter protein, such as GFP, placed after the arrest point of SecM. The amount of GFP produced as a function of the length of the nascent chain (L) is used to derive an arrest-peptide-mediated force profile, either in E. coli or in vitro using a reconstituted translation system (11, 31). This method, referred to as arrest-peptide-mediated force measurements (7, 8, 11, 12, 31, 42), can be used to determine at which point during translation cotranslational folding events take place as a function of the sequence separation of the protein under study from the PTC. Arrest-peptide-mediated force measurements of small domains generally revealed good agreement between in vitro and in vivo experiments (7, 11, 31).
We thus performed in vitro translation of ACTR or NCBD in the presence of ∼8 μM of the same fluorescently labeled NCBD or ACTR used for the FCS measurements (the ribosome concentration in the PURE system is ∼2 μM according to the manufacturer) and quantified the amount of GFP produced as a function of tether length (Fig. 3, A–D). As observed previously, there is a SecM-length-dependent variation in the resulting force profiles both with and without a binding partner present (Fig. 3, B and C), which likely arises from the sequence-specific stalling properties of SecM with different lengths (31). More importantly, however, in the case of ACTR-RNCs translated in the presence of labeled NCBD, a clear increase in the amount of GFP was observed compared to the situation without added NCBD, with a maximal effect in the range between L = 26 and L = 29 residues (Fig. 3, B–D). A steady increase in force exerted on the nascent chain, indirectly monitored via the amount of full-length-ACTR-SecM-GFP fusion protein produced, can be observed in Fig. 3 D. These values were obtained by subtracting the GFP fluorescence intensities obtained in the absence of NCBD as a reference from the reactions where NCBD was added.
This observation suggests that the ACTR-NCBD complex forms cotranslationally, in keeping with the FCS results, and exerts a force on the nascent chain. According to the cryo-EM structure of SecM (40) and the ADR1 domain fused to SecM (7), from which we can identify the position of SecM inside the ribosome exit tunnel and estimate the tether length required to span the tunnel as ∼30 residues, folding and binding of the ACTR-NCBD complex thus likely takes place while the ACTR sequence is still partially located in the vestibule of the exit tunnel. In contrast, when we inverted the experiment and produced NCBD-RNCs in the presence of labeled ACTR, no increase in GFP translation occurred in the presence of free ACTR (Fig. 3, B and C). In accordance with the FCS analysis (Fig. 2 D), the arrest-peptide-mediated force profiles thus confirm that formation of the ACTR-NCBD complex can only occur when ACTR emerges from the ribosomal exit tunnel.
To test whether this asymmetry in cotranslational folding and binding also occurs in vivo, we performed arrest-peptide-mediated force measurements in live E. coli cells (11, 31). We first determined the force profiles in vivo for ACTR and NCBD individually (Fig. 4, A and B) and performed expression in E. coli followed by in-cell fluorescence analysis to quantify the formation of GFP reporting on the force on the nascent chain. Similar to the trend observed in vitro (Fig. 3, B and C), ACTR showed high GFP translation at short tether lengths (L = 19–24), with a decrease to baseline when it was expected to have emerged completely from the tunnel (Fig. 4 B). NCBD instead showed much lower fluorescence values throughout, with only a slight increase for short tether lengths (Fig. 4 B). For investigating the effect of coupled folding and binding, because attaining a well-defined concentration of free protein compared to translating ribosomes is difficult in cells, we created a series of fusion constructs, in which ACTR was placed either at the N- or C-terminus of NCBD (Fig. 4, A and B). When expressing the fusion construct N-NCBD-ACTR, a clear force peak occurs at tether lengths between L = 26 and L = 29 residues. In contrast, the fusion construct N-ACTR-NCBD gave rise to a flat force profile without noticeable peaks (Fig. 4 B).
We then tested the same set of fusion constructs in vitro (Fig. 4 C) (i.e., in the PURE system). Importantly, the pronounced difference between the two fusion constructs is conserved: as in the in vivo results, N-NCBD-ACTR yields strongly increased GFP fluorescence, corresponding to large forces on the nascent chain, whereas N-ACTR-NCBD resulted in low fluorescence throughout, corresponding to small forces on the nascent chain (Fig. 4 C). Thus, the asymmetry in terms of the capability of generating force on the nascent chain was consistently observed in all tested conditions, both for the separate and the fused proteins. However, the force profile of N-NCBD-ACTR did not display the sharp peak between L = 26 and L = 29 residues obtained in vivo (Fig. 4 B) but instead a broad peak reminiscent of the ΔNCBD profile obtained for the separate proteins in vitro (Fig. 3 D). The difference in shape between the force profiles for N-NCBD-ACTR in vitro and in vivo is thus likely to originate from the lack of cellular components in the PURE system, in particular molecular chaperones, such as trigger factor, GroEL/GroES, or the DnaK/J/GrpE system, which are known to interact with nascent polypeptide chains in E. coli and have previously been shown to affect the forces exerted on the nascent polypeptide chain (43).
Charge interactions between ACTR and the ribosome create a force on the nascent chain
The FCS experiments in vitro (Fig. 2) and arrest-peptide-mediated force measurements in vitro and in vivo (Figs. 3 and 4) consistently indicate that the coupled folding and binding of ACTR and NCBD on the ribosome exhibits a pronounced asymmetry, and even the forces on the individual nascent chains are very different for ACTR and NCBD. Given that ACTR has a net charge of −8, whereas NCBD has a net charge of +6 (Fig. S1 A), a potential contribution to these effects originates from electrostatic interactions with the negatively charged surface of the ribosome and the exit tunnel (44).
To test this hypothesis, we altered the charge content of both IDPs (Fig. 5). In ACTR, we replaced Asp and Glu residues with Ala, whereas in NCBD, we exchanged the positively charged residues Arg and Lys and its C-terminal residue to Glu to mimic the charge distribution of ACTR (Fig. 5 C and D). The required mutations at the messenger RNA level are not expected to affect the translation rates, as predicted according to the E. coli transfer RNA abundance (45, 46). The changes in the sequence of ACTR and NCBD also do not introduce rare codons or sequences known to create translational stalling (47). Moreover, the first 50 bases near the starting Met, which are crucial for translation efficiency in E. coli (48), are equal among all ACTR and NCBD constructs.
Figure 5.
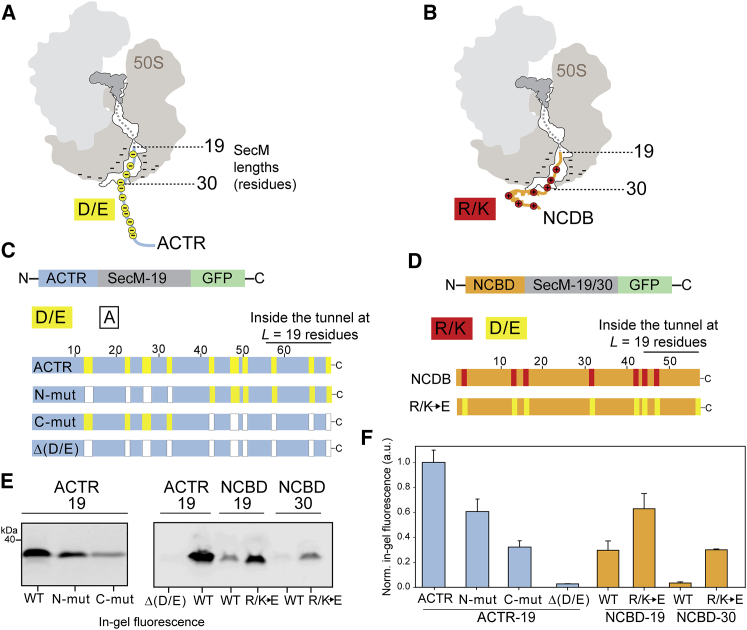
Arrest-peptide-mediated force measurements upon exchange of charged residues on ACTR and NCBD. (A and B) A schematic representation of ACTR and NCBD partially embedded within the ribosome tunnel is shown. Negative charges on ACTR are depicted as yellow circles, and positive charges on NCBD are depicted as red circles. Selected SecM lengths used to stall ACTR or NCBD in the ribosome exit tunnel are indicated. (C) A schematic representation of the constructs used for in vitro translation of ACTR-SecM-19 is shown. Wild-type ACTR and its variants with negatively charged residues replaced by Ala are indicated. Asp (D) and Glu (E) residues were replaced by Ala (A) in the N-terminal half of ACTR (N-mut), in the C-terminal half (C-mut), or throughout the sequence (Δ(D/E)). (D) Schematic representation of the NCBD-SecM (19/30 residues) constructs and NCBD variants in which all seven positively charged residues were replaced by Glu. (E) A representative in-gel fluorescence analysis upon translation in the PURE system is shown. (F) In vitro translation and in-gel fluorescence analysis were performed in triplicate, bands were quantified, and averages and SDs were calculated, normalized relative to the highest fluorescence intensity (ACTR wild type at L = 19). To see this figure in color, go online. a.u., arbitrary units; WT, wild-type.
The changes in charge content of the nascent chains had a pronounced effect on the arrest-peptide-mediated force measurements. For ACTR, the constructs were chosen to have a constant SecM length of 19 residues (Fig. 5 A) because ACTR confined within the ribosome tunnel at this tether length resulted in a high level of GFP fluorescence, indicating a release of SecM stalling (Fig. 4 B). We observed a reduction in fluorescence upon exchanging 6 of the 13 negatively charged residues in the N-terminal half of ACTR (N-mut, Fig. 5 C, E and F); the reduction in fluorescence was even more pronounced when 7 of the 13 negatively charged residues in the C-terminal half of ACTR were exchanged, 3 of which are likely to be still inside or near the exit tunnel (C-mut, Fig. 5, C, E, and F). Removal of all negative charges from ACTR, resulting in a net charge of +5, eliminated GFP release almost completely (Δ(D/E), Fig. 5, C, E, and F). This striking reduction in signal suggests that the Δ(D/E) variant should result in a correspondingly larger amount of arrested chains, which could indeed be detected by Western blot analysis (Fig. S6).
For NCBD, we studied the behavior at SecM lengths of L = 19 and L = 30 to have either part of NCBD still embedded in the tunnel (L = 19), as in the case of ACTR, or to have the protein completely outside the tunnel (L = 30) (Fig. 5 D). GFP translation increases substantially for both tether lengths upon introduction of the negatively charged residues, indicating increased chain release (Fig. 5 D–F). These results strongly support the hypothesis that positive charges in the nascent chain cause attractive interactions with the ribosomal surface in or near the exit tunnel, thus making the release of SecM less favorable. The same interactions are thus likely to make NCBD less available for complex formation with ACTR, as detected by FCS and the arrest-peptide-mediated force measurements in the presence of a binding partner.
Discussion
Using a combination of single-molecule FRET, FCS, and arrest-peptide-mediated force measurements, we investigated how ACTR and NCBD, IDPs of opposite net charge, mutually bind and fold in the vicinity of the ribosome exit tunnel. We find that the coupled folding and binding reaction can take place on the ribosome and produces sufficient force on the nascent chain to be detected by arrest-peptide-mediated force measurements in vitro and in vivo. However, folding and binding occur only if ACTR is the chain in direct proximity to the ribosomal exit tunnel. Based on the exchange of charged amino acid residues in ACTR and NCBD, we conclude that attractive charge interactions between NCBD and the ribosomal exit tunnel interfere with ACTR binding, whereas the lack of such attraction for ACTR keeps it available for binding NCBD. Our experiments thus illustrate the pronounced effect that charge interactions can have on the nascent chain.
We also observe a pronounced difference in force acting on the nascent chain depending on whether ACTR or NCBD is tethered to the ribosome, even in the absence of a binding partner. A nascent chain of ACTR generates stronger force when the chain is deeper in the tunnel (Fig. 3 F). At short SecM lengths, such as at L = 19, ∼10 residues of its C-terminus are expected to be inside the tunnel (assuming that the C-terminus of ACTR does not compact into a helical structure). Correspondingly, the release of SecM stalling becomes less efficient when ∼7 of the 13 negatively charged residues located in the C-terminal half of ACTR are replaced by alanine (Fig. 5 E). In contrast, NCBD is positively charged and does not produce a force on the stalling sequence while being extruded from the ribosome tunnel, as shown by our arrest-peptide-mediated force measurements (Fig. 4 B). Mutational analysis shows that by replacing the seven positively charged residues on NCBD by glutamic acid, the force increases, both when part of NCBD is still inside the tunnel (at L = 19 residues) and when NCBD is expected to be outside the tunnel (at L = 30 residues) (Fig. 5 F). Thus, the positively charged residues of NCBD are important for attenuating the transmission of force through the ribosome tunnel, presumably owing to an attractive force between the chain and the ribosome.
Our FCS experiments (Fig. 2 C) show no binding of free NCBD to the ribosome without nascent ACTR, indicating that the attractive interactions of the positively charged residues in nascent NCBD chains with the ribosome surface in or near the tunnel are not specific. However, owing to the high local charge density of the nascent chain in or at the exit tunnel, the contribution of electrostatic interactions to the force balance on the ribosome can apparently be substantial. However, the strength of the interaction seems to decay steeply with the distance of the positively charged chain segments from the exit tunnel, as suggested by the pronounced difference in forces on the nascent chain for the two permutants of the ACTR-NCBD fusion proteins in the experiments in vivo (Fig. 4 B).
When ACTR emerges from the tunnel and presents its negative charges to NCBD, complex formation can occur when ACTR is still partially inside the tunnel, at a distance from the PTC between L = 26 and L = 29 residues, as shown by the arrest-peptide-mediated force measurements (Figs. 3 and 4). Our FCS experiments indicate the nonequilibrium properties of this process: at tether lengths of 26 and 27 residues, where the force profile has its peak, barely any equilibrium binding of freely diffusing NCBD to ribosome-bound ACTR can be detected. However, binding must still occur to release SecM stalling. In contrast to the FCS experiment, which probes the average fraction bound, a single binding event can be sufficient to create force on the nascent chain and thus release SecM stalling. Single-molecule FRET experiments on doubly labeled ACTR and NCBD show that upon binding, the nascent chain of ACTR compacts (38). This compaction of ACTR upon binding to NCBD and its extrusion from the tunnel might contribute to creating the force peak that we observe at SecM lengths between L = 26 and L = 29 residues.
However, our results indicate that the cotranslational behavior of IDPs can be strongly influenced by negative and positive charges along the nascent polypeptide chain. Although the effect identified here is surprisingly strong, it is consistent with previous observations indicating charge interactions with a nascent IDP as monitored by an increase in mobility with increasingly negative charge density in the chain (26). It is known that peptides containing negatively charged residues are more likely to overcome translational arrest than peptides containing positively charged residues (19) and that translation slows down when the nascent chain contains positively charged residues located inside or in the vicinity of the ribosome exit tunnel, as these residues make transient interactions within the tunnel (12). Similarly, data from a ribosomal footprinting assay in Saccharomyces cerevisiae showed that positively charged residues slow down translation downstream of their incorporation into the polypeptide chain, suggesting a role for attractive interactions of these residues with the negatively charged surface of the ribosome exit tunnel (20). Finally, charge interactions have also been reported to modulate protein folding rates on the ribosome (21).
In view of the abundance of proteins with marked net charge, the type of effects and interactions we observed for ACTR and NCBD are likely to be no exception. It will thus be important to quantify both the repulsive and the attractive electrostatic forces between the nascent chain and the negative potential of the tunnel and the ribosomal surface in more detail and to compare them with those arising during folding (11). We note that even though we investigated here the interaction of the two eukaryotic proteins on prokaryotic ribosomes, the high negative surface charge is a universal property of ribosomes and is likely to result in similar interactions across all life forms.
Despite the consistent effect of charges on nascent chain-ribosome interactions and their coupled folding and binding in experiments both in vitro and in vivo, our results indicate the possibility of a pronounced modulation of these interactions by the cellular environment. Arrest-peptide-mediated force measurements for small protein domains have previously been shown to agree well in E. coli BL21(DE3) and the PURE system (31). The shapes of the force profiles for the N-NCBD-ACTR constructs, however, are very different in vivo and in vitro. ACTR and NCBD are largely disordered in the absence of their respective binding partner and might thus be recognized cotranslationally by cellular factors, especially molecular chaperones. Such interactions are likely to explain the difference in the shape of the force profiles we observe between the in vivo and in vitro experiments, in line with previous results that demonstrated that both trigger factor and GroEL reduce the force on the nascent polypeptide chain of DHFR tethered to the ribosome (43). Investigating such effects in more detail for ACTR/NCBD will be an interesting aspect of future work.
The experimental approach used in this study can, in principle, also be applied to study how multiprotein complexes assemble cotranslationally (4). For example, it could be possible to investigate whether the order in which proteins are synthesized (e.g., those encoded in bacterial operons) has been optimized during evolution to ensure that proteins emerging from the ribosome would preferentially interact with their binding partners rather than with the ribosome. Further work will be required to determine, on a larger scale, the cotranslational folding behavior of proteins with large negative or positive net charge and how electrostatic interactions with the ribosome affect their physiological functions or the binding of chaperones (43) and other cellular factors.
Acknowledgments
We thank Hagen Hofmann for his contribution to establishing the ACTR-NCBD system for single-molecule measurements in the group and Thomas Reinberg and the High-Throughput Facility of the Department of Biochemistry of the University of Zurich for providing an HA-tagged DARPin.
J.M. acknowledges support from the Novartis Foundation for Biomedical Research (Basel, Switzerland) and the Forschungskredit of the University of Zurich (FK-41101-04). This work was supported by the Swiss National Science Foundation (to B.S).
Editor: Elizabeth Komives.
Footnotes
Franziska Zosel's present address is Novo Nordisk A/S, Måløv, Denmark.
Six figures and one table are available at http://www.biophysj.org/biophysj/supplemental/S0006-3495(18)30928-7.
Contributor Information
Jacopo Marino, Email: jacopo.marino@psi.ch.
Benjamin Schuler, Email: schuler@bioc.uzh.ch.
Author Contributions
J.M. and B.S. designed and coordinated the project and wrote the manuscript with the help of all authors. J.M. performed arrest-peptide-mediated force measurements and produced samples for FCS analysis. K.J.B. and F.Z. expressed, purified, and labeled proteins. K.J.B. performed and analyzed FCS experiments. F.Z. performed and analyzed single-molecule FRET experiments. D.N. carried out FRET analysis. B.S. supervised the project.
Supporting Material
References
- 1.Jacobs W.M., Shakhnovich E.I. Evidence of evolutionary selection for cotranslational folding. Proc. Natl. Acad. Sci. USA. 2017;114:11434–11439. doi: 10.1073/pnas.1705772114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jacobson G.N., Clark P.L. Quality over quantity: optimizing co-translational protein folding with non-‘optimal’ synonymous codons. Curr. Opin. Struct. Biol. 2016;38:102–110. doi: 10.1016/j.sbi.2016.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hanes J., Plückthun A. In vitro selection and evolution of functional proteins by using ribosome display. Proc. Natl. Acad. Sci. USA. 1997;94:4937–4942. doi: 10.1073/pnas.94.10.4937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shieh Y.W., Minguez P., Bukau B. Operon structure and cotranslational subunit association direct protein assembly in bacteria. Science. 2015;350:678–680. doi: 10.1126/science.aac8171. [DOI] [PubMed] [Google Scholar]
- 5.Fedorov A.N., Baldwin T.O. Contribution of cotranslational folding to the rate of formation of native protein structure. Proc. Natl. Acad. Sci. USA. 1995;92:1227–1231. doi: 10.1073/pnas.92.4.1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Thommen M., Holtkamp W., Rodnina M.V. Co-translational protein folding: progress and methods. Curr. Opin. Struct. Biol. 2017;42:83–89. doi: 10.1016/j.sbi.2016.11.020. [DOI] [PubMed] [Google Scholar]
- 7.Nilsson O.B., Hedman R., von Heijne G. Cotranslational protein folding inside the ribosome exit tunnel. Cell Reports. 2015;12:1533–1540. doi: 10.1016/j.celrep.2015.07.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nilsson O.B., Nickson A.A., Clarke J. Cotranslational folding of spectrin domains via partially structured states. Nat. Struct. Mol. Biol. 2017;24:221–225. doi: 10.1038/nsmb.3355. [DOI] [PubMed] [Google Scholar]
- 9.Cabrita L.D., Cassaignau A.M.E., Christodoulou J. A structural ensemble of a ribosome-nascent chain complex during cotranslational protein folding. Nat. Struct. Mol. Biol. 2016;23:278–285. doi: 10.1038/nsmb.3182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Holtkamp W., Kokic G., Rodnina M.V. Cotranslational protein folding on the ribosome monitored in real time. Science. 2015;350:1104–1107. doi: 10.1126/science.aad0344. [DOI] [PubMed] [Google Scholar]
- 11.Goldman D.H., Kaiser C.M., Bustamante C. Ribosome. Mechanical force releases nascent chain-mediated ribosome arrest in vitro and in vivo. Science. 2015;348:457–460. doi: 10.1126/science.1261909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wruck F., Katranidis A., Hegner M. Translation and folding of single proteins in real time. Proc. Natl. Acad. Sci. USA. 2017;114:E4399–E4407. doi: 10.1073/pnas.1617873114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kramer G., Boehringer D., Bukau B. The ribosome as a platform for co-translational processing, folding and targeting of newly synthesized proteins. Nat. Struct. Mol. Biol. 2009;16:589–597. doi: 10.1038/nsmb.1614. [DOI] [PubMed] [Google Scholar]
- 14.Kelkar D.A., Khushoo A., Skach W.R. Kinetic analysis of ribosome-bound fluorescent proteins reveals an early, stable, cotranslational folding intermediate. J. Biol. Chem. 2012;287:2568–2578. doi: 10.1074/jbc.M111.318766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kosolapov A., Deutsch C. Tertiary interactions within the ribosomal exit tunnel. Nat. Struct. Mol. Biol. 2009;16:405–411. doi: 10.1038/nsmb.1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.O’Brien E.P., Hsu S.T., Dobson C.M. Transient tertiary structure formation within the ribosome exit port. J. Am. Chem. Soc. 2010;132:16928–16937. doi: 10.1021/ja106530y. [DOI] [PubMed] [Google Scholar]
- 17.O’Brien E.P., Christodoulou J., Dobson C.M. New scenarios of protein folding can occur on the ribosome. J. Am. Chem. Soc. 2011;133:513–526. doi: 10.1021/ja107863z. [DOI] [PubMed] [Google Scholar]
- 18.Ban N., Nissen P., Steitz T.A. The complete atomic structure of the large ribosomal subunit at 2.4 A resolution. Science. 2000;289:905–920. doi: 10.1126/science.289.5481.905. [DOI] [PubMed] [Google Scholar]
- 19.Lu J., Deutsch C. Electrostatics in the ribosomal tunnel modulate chain elongation rates. J. Mol. Biol. 2008;384:73–86. doi: 10.1016/j.jmb.2008.08.089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Charneski C.A., Hurst L.D. Positively charged residues are the major determinants of ribosomal velocity. PLoS Biol. 2013;11:e1001508. doi: 10.1371/journal.pbio.1001508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kaiser C.M., Goldman D.H., Bustamante C. The ribosome modulates nascent protein folding. Science. 2011;334:1723–1727. doi: 10.1126/science.1209740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Habchi J., Tompa P., Uversky V.N. Introducing protein intrinsic disorder. Chem. Rev. 2014;114:6561–6588. doi: 10.1021/cr400514h. [DOI] [PubMed] [Google Scholar]
- 23.Demarest S.J., Martinez-Yamout M., Wright P.E. Mutual synergistic folding in recruitment of CBP/p300 by p160 nuclear receptor coactivators. Nature. 2002;415:549–553. doi: 10.1038/415549a. [DOI] [PubMed] [Google Scholar]
- 24.Wright P.E., Dyson H.J. Linking folding and binding. Curr. Opin. Struct. Biol. 2009;19:31–38. doi: 10.1016/j.sbi.2008.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.van der Lee R., Buljan M., Babu M.M. Classification of intrinsically disordered regions and proteins. Chem. Rev. 2014;114:6589–6631. doi: 10.1021/cr400525m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Knight A.M., Culviner P.H., Cavagnero S. Electrostatic effect of the ribosomal surface on nascent polypeptide dynamics. ACS Chem. Biol. 2013;8:1195–1204. doi: 10.1021/cb400030n. [DOI] [PubMed] [Google Scholar]
- 27.Kjaergaard M., Teilum K., Poulsen F.M. Conformational selection in the molten globule state of the nuclear coactivator binding domain of CBP. Proc. Natl. Acad. Sci. USA. 2010;107:12535–12540. doi: 10.1073/pnas.1001693107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dogan J., Schmidt T., Jemth P. Fast association and slow transitions in the interaction between two intrinsically disordered protein domains. J. Biol. Chem. 2012;287:34316–34324. doi: 10.1074/jbc.M112.399436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Iešmantavičius V., Jensen M.R., Kjaergaard M. Modulation of the intrinsic helix propensity of an intrinsically disordered protein reveals long-range helix-helix interactions. J. Am. Chem. Soc. 2013;135:10155–10163. doi: 10.1021/ja4045532. [DOI] [PubMed] [Google Scholar]
- 30.Dogan J., Jonasson J., Jemth P. Binding rate constants reveal distinct features of disordered protein domains. Biochemistry. 2015;54:4741–4750. doi: 10.1021/acs.biochem.5b00520. [DOI] [PubMed] [Google Scholar]
- 31.Marino J., von Heijne G., Beckmann R. Small protein domains fold inside the ribosome exit tunnel. FEBS Lett. 2016;590:655–660. doi: 10.1002/1873-3468.12098. [DOI] [PubMed] [Google Scholar]
- 32.Geertsma E.R. FX cloning: a simple and robust high-throughput cloning method for protein expression. Methods Mol. Biol. 2014;1116:153–164. doi: 10.1007/978-1-62703-764-8_11. [DOI] [PubMed] [Google Scholar]
- 33.Dingfelder F., Wunderlich B., Schuler B. Rapid microfluidic double-jump mixing device for single-molecule spectroscopy. J. Am. Chem. Soc. 2017;139:6062–6065. doi: 10.1021/jacs.7b02357. [DOI] [PubMed] [Google Scholar]
- 34.Schuler B. Application of single molecule Förster resonance energy transfer to protein folding. Methods Mol. Biol. 2007;350:115–138. doi: 10.1385/1-59745-189-4:115. [DOI] [PubMed] [Google Scholar]
- 35.Koppel D.E. Study of Escherichia coli ribosomes by intensity fluctuation spectroscopy of scattered laser light. Biochemistry. 1974;13:2712–2719. doi: 10.1021/bi00710a009. [DOI] [PubMed] [Google Scholar]
- 36.Borgia A., Zheng W., Schuler B. Consistent view of polypeptide chain expansion in chemical denaturants from multiple experimental methods. J. Am. Chem. Soc. 2016;138:11714–11726. doi: 10.1021/jacs.6b05917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shimizu Y., Kanamori T., Ueda T. Protein synthesis by pure translation systems. Methods. 2005;36:299–304. doi: 10.1016/j.ymeth.2005.04.006. [DOI] [PubMed] [Google Scholar]
- 38.Zijlstra N., Dingfelder F., Schuler B. Rapid microfluidic dilution for single-molecule spectroscopy of low-affinity biomolecular complexes. Angew. Chem. Int. Ed. Engl. 2017;56:7126–7129. doi: 10.1002/anie.201702439. [DOI] [PubMed] [Google Scholar]
- 39.Oliver D., Norman J., Sarker S. Regulation of Escherichia coli secA by cellular protein secretion proficiency requires an intact gene X signal sequence and an active translocon. J. Bacteriol. 1998;180:5240–5242. doi: 10.1128/jb.180.19.5240-5242.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang J., Pan X., Sui S.F. Mechanisms of ribosome stalling by SecM at multiple elongation steps. eLife. 2015;4:e09684. doi: 10.7554/eLife.09684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yap M.N., Bernstein H.D. The plasticity of a translation arrest motif yields insights into nascent polypeptide recognition inside the ribosome tunnel. Mol. Cell. 2009;34:201–211. doi: 10.1016/j.molcel.2009.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ismail N., Hedman R., von Heijne G. A biphasic pulling force acts on transmembrane helices during translocon-mediated membrane integration. Nat. Struct. Mol. Biol. 2012;19:1018–1022. doi: 10.1038/nsmb.2376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nilsson O.B., Müller-Lucks A., von Heijne G. Trigger factor reduces the force exerted on the nascent chain by a cotranslationally folding protein. J. Mol. Biol. 2016;428:1356–1364. doi: 10.1016/j.jmb.2016.02.014. [DOI] [PubMed] [Google Scholar]
- 44.Lu J., Kobertz W.R., Deutsch C. Mapping the electrostatic potential within the ribosomal exit tunnel. J. Mol. Biol. 2007;371:1378–1391. doi: 10.1016/j.jmb.2007.06.038. [DOI] [PubMed] [Google Scholar]
- 45.Zhang G., Hubalewska M., Ignatova Z. Transient ribosomal attenuation coordinates protein synthesis and co-translational folding. Nat. Struct. Mol. Biol. 2009;16:274–280. doi: 10.1038/nsmb.1554. [DOI] [PubMed] [Google Scholar]
- 46.Zhang G., Ignatova Z. Generic algorithm to predict the speed of translational elongation: implications for protein biogenesis. PLoS One. 2009;4:e5036. doi: 10.1371/journal.pone.0005036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Woolstenhulme C.J., Parajuli S., Buskirk A.R. Nascent peptides that block protein synthesis in bacteria. Proc. Natl. Acad. Sci. USA. 2013;110:E878–E887. doi: 10.1073/pnas.1219536110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kudla G., Murray A.W., Plotkin J.B. Coding-sequence determinants of gene expression in Escherichia coli. Science. 2009;324:255–258. doi: 10.1126/science.1170160. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.