Abstract
The interaction between C-X-C chemokine receptor type 4 (CXCR4) and its cognate ligand C-X-C motif chemokine ligand 12 (CXCL12) plays a critical role in regulating hematopoietic stem cell activation and subsequent cellular mobilization. Extensive studies of these genes have been conducted in mammals, but much less is known about the expression and function of CXCR4 and CXCL12 in non-mammalian vertebrates. In the present study, we identify simultaneous expression of CXCR4 and CXCL12 orthologs in the epigonal organ (the primary hematopoietic tissue) of the little skate, Leucoraja erinacea. Genetic and phylogenetic analyses were functionally supported by significant mobilization of leukocytes following administration of Plerixafor, a CXCR4 antagonist and clinically important drug. Our results provide evidence that, as in humans, Plerixafor disrupts CXCR4/CXCL12 binding in the little skate, facilitating release of leukocytes into the bloodstream. Our study illustrates the value of the little skate as a model organism, particularly in studies of hematopoiesis and potentially for preclinical research on hematological and vascular disorders.
Keywords: C-X-C chemokine ligand 12, C-X-C chemokine receptor type 4, elasmobranch, mobilization, Plerixafor/AMD3100
INTRODUCTION
Mammalian bone marrow is composed of both endosteal (bone) and vascular (blood vessel) niches (38). Dormant hematopoietic stem cells (HSCs) are molecularly tethered to the endosteum before activation. Following activation, HSCs migrate to the marrow’s vasculature and undergo asymmetric division, whereby some daughter cells remain capable of self-renewal whereas others form multipotent progenitor cells that differentiate into the myeloid and lymphoid blood cell lineages (17, 21, 28). The production and differentiation of HSCs (i.e., hematopoiesis) is followed by mobilization of leukocytes into the bloodstream. One critical receptor/ligand pair that regulates this process is C-X-C chemokine receptor type 4 (CXCR4), which is expressed on HSCs, and its cognate ligand C-X-C motif chemokine ligand 12 [CXCL12; also known as stromal cell-derived factor 1 (SDF-1)] (47, 52). In mammals, CXCL12 chemotactically maintains homeostasis by attracting and retaining HSCs in the bone marrow (38, 52). Understanding the mechanisms of HSC activation and leukocyte mobilization offers important implications for hematological and vascular diseases and has been of great interest to scientists and physicians for decades (22, 32, 50).
The bulk of relevant studies have focused primarily on surgical bone marrow transplantation procedures. More recently, however, many preclinical and clinical studies have shifted attention toward using small molecules to activate HSCs and mobilize HSC progenitor cells from the bone marrow into the bloodstream. This less invasive approach, referred to as hematopoietic cell transplantation, has immense potential for improved treatment of pervasive diseases, but relevant methodologies still need to be optimized (6, 31, 41). One noteworthy mobilizing agent, Plerixafor (also known as AMD3100), is an antagonistic bicyclam molecule that preferentially binds to CXCR4 and prevents the subsequent binding of CXCL12 (5). Plerixafor-mediated disruption of molecular tethering results in a cascade of effects, including HSC activation, cellular differentiation, and mobilization. The release of multipotent progenitor cells into the peripheral blood occurs over a shorter period than that seen with other endogenous mobilizing agents, such as cytokines (16, 49). Healthy human subjects injected with Plerixafor demonstrated significant mobilization of CD34+ hematopoietic progenitor cells from the marrow to the peripheral blood as early as 3 h postinjection (6, 10, 31). Additional studies have shown mobilization of hematopoietic stem and progenitor cells, as well as endothelial cells, following Plerixafor administration; these studies illustrate the importance of Plerixafor in hematological and vascular disease research (16, 22, 60).
Researchers have largely utilized murine (e.g., see Refs. 25 and 52) and zebrafish (e.g., see Refs. 12 and 20) models in studies attempting to elucidate, mimic, and manipulate the processes that govern human HSC activation and mobilization. HSCs in murine models reside inside the protective bone matrix, which makes successfully accessing them (i.e., without depreciating the quality of the fragile and complex bone marrow) for harvest or experimental manipulation a challenge (8). In zebrafish, HSCs reside in the kidney marrow, but studies are limited due to the marrow size and location (19). Chondrichthyans (specifically the elasmobranchs: sharks, skates, and rays) offer a unique opportunity to enhance mechanistic studies of mobilization and overcome some of the aforementioned challenges. Cartilaginous fishes may complement current models because they lack endosteal compartmentalization but show remarkable conservation of cellular pathways and hematopoietic transcription factors (1, 29, 62).
In elasmobranchs, one of the primary organs that governs hematopoiesis is the epigonal organ (EO) (35). This organ, which forms a complex with the gonads, is responsible for the production and differentiation of leukocytes from HSCs (1, 33, 34). The EO also supplies immune cells of the various lymphoid and myeloid lineages to the body (1, 36).
One noteworthy elasmobranch model is the little skate, Leucoraja erinacea. Early studies of the EO in the little skate were constrained to morphological analyses, due primarily to a deficit of genomic and transcriptomic resources as well as a lack of cell surface markers for elasmobranch immune cells (1, 18, 34, 36). Genomic, transcriptomic, and proteomic sequence data are readily available for many sarcopterygii (i.e., lobe-finned) and actinopterygii (i.e., ray-finned) fishes, but equivalent information is comparatively limited for elasmobranchs (59). Given this deficit, the North East Bioinformatics Collaborative (NEBC) began sequencing and annotating the little skate genome in 2010 (26, 56, 59).
In the present study, we utilized novel genomic and transcriptomic information compiled by the NEBC to look for CXCR4 and CXCL12 expression in the little skate EO. Given that the EO is responsible for HSC activity in elasmobranchs, we hypothesized that both genes would be expressed. We also investigated whether Plerixafor administration to the little skate would facilitate leukocyte mobilization. The overall objectives of our study were to demonstrate the efficacy of novel little skate transcriptomic and genomic databases and to highlight the utility of the little skate as a model organism, particularly in studies of hematopoiesis.
MATERIALS AND METHODS
Experimental animals.
All animal procedures were approved by the Institutional Animal Care and Use Committees (IACUCs) of Salem State University (SSU; Salem, MA) and Endicott College (Beverly, MA). In collaboration with the Massachusetts Department of Marine Fisheries (Capt. Jim Ford, F/V Lisa Ann II/III, Newburyport, MA), a total of 23 little skates were collected by trawl fishing from northeastern Massachusetts waters between January 2013 and June 2015. All animals were euthanized humanely by way of cerebral/spinal pithing per IACUC protocol no. 04014-R. Fourteen little skates were dedicated to genetic analyses and nine were utilized for serological assessments of leukocyte mobilization following Plerixafor injection. The animals used for serological assessments were housed in recirculating seawater tanks at the Mount Desert Island Biological Laboratory (MDIBL; Mount Desert Island, ME) and the SSU Cat Cove Marine Laboratory.
RNA isolation and cDNA synthesis.
Following euthanasia, EO tissue samples from 14 little skates were isolated via microdissection and stored at −80°C in RNAlater (Thermo Fisher Scientific, Waltham, MA). RNA was isolated from the EO samples using an RNeasy Mini Kit (74104: Qiagen, Hilden, Germany). The protocol was optimized to remove excess salts through two additional washes with wash buffer RPE coupled with 5-min incubations at room temperature. Following the initial RNA elution step, the eluate was returned to the spin column, incubated for 10 min at room temperature, and centrifuged. Only tissues that yielded at least 500 ng of RNA were utilized for cDNA synthesis using the Superscript III First-Strand Synthesis System (18080051; Invitrogen, Carlsbad, CA) and oligo(dT) primers. A NanoDrop 1000 spectrophotometer (Thermo Fisher Scientific) was used to assess purity (via the 260/280- and 260/230-nm absorbance ratios) and concentration of RNA and cDNA.
PCR and sequencing.
PCR primers for CXCL12 and CXCR4 were designed using novel little skate genomic and transcriptomic data sets and SkateBase (56, 59). Basic Local Alignment Search Tool searches using human CXCL12 and CXCR4 amino acid sequences identified partial cDNA sequence for both genes in the SkateBase little skate transcriptome database. The retrieved 390-base pair (bp) CXCL12 partial cDNA (contig 25291) and 1,634-bp CXCR4 partial cDNA (contig 73076) were used as templates to design the little skate primers for PCR and sequencing. The PrimerQuest tool from Integrated DNA Technologies was used to generate primers. Primer sequences (listed 5′ to 3′) were as follows: CXCL12 forward GGAAAGCCGTCCTCGATAATAA, CXCL12 reverse ATCATGGCAGCGTCTACATC; CXCR4 forward CTTGCCGTGTATGTACTCTCTC, CXCR4 reverse ATGCCTCGGTGATGGTTATC.
To determine whether CXCL12 and CXCR4 were expressed in the little skate EO samples, synthesized cDNA was amplified via PCR using the exACTGene Complete PCR Kit (Thermo Fisher Scientific). The reaction mixture consisted of 33 µl of nuclease-free water, 5 µl of 10× PCR buffer A (1.5 mM MgCl2), 1 µl of PCR nucleotide mix (200 µM each dNTP), 2 µl of forward primer (1.0 µM), 2 µl of reverse primer (1.0 µM), 0.25 µl of Taq DNA polymerase (1.25 U), and 6.75 µl of template cDNA (5–10 ng/µl) for a total volume of 50 µl. The cycling (30 cycles) conditions for the PCR reactions were as follows: 94°C for 30 s, 50–65°C for 30 s, and 72°C for 1 min. A thermal cycler (Bio-Rad, Hercules, CA) with temperature gradient capabilities was used to test a range of annealing temperatures. The primers were validated by the presence of a single band of expected length when the PCR products were electrophoresed in 1.5% agarose gel. A negative control (no template) reaction was performed for each primer pair.
After the gels were analyzed and probable EO expression of CXCL12 (n = 11) and CXCR4 (n = 14) was identified, two EO samples that expressed both genes were chosen for sequencing. The ExoSAP-IT Cleanup Kit (Affymetrix, Santa Clara, CA) was used to remove excess primers and nucleotides from the PCR amplicon products, and sequencing was carried out by the MDIBL using a 3130xl Genetic Analyzer (Applied Biosystems, Foster City, CA). For both genes, the resultant sequence data was incomplete. The full-length consensus nucleotide sequences were inferred by using additional sequence data to add to the beginning (CXCL12 and CXCR4) and end (CXCR4) of the genes.
Phylogenetic analysis of CXCL12 and CXCR4.
Protein sequences for little skate CXCL12 and CXCR4 were obtained by translating the consensus nucleotide sequences using the NCBI Open Reading Frame (ORF) Finder (58). Transmembrane domains were predicted using TMHMM Server version 2.0 (24), and putative N-glycosylation site [N-X-(S/T)] was predicted using NetNGlyc 1.0 Server (http://www.cbs.dtu.dk/services/NetNGlyc/).
Chemokine ligand and receptor multiple sequence alignments and phylogenetic trees were constructed to verify the identity of the proposed CXCL12 and CXCR4 little skate orthologs. The chemokine ligand analysis included sequences from the CXCL8, CXCL11, CXCL12, CXCL13, and CXCL14 gene families. The chemokine receptor analysis included sequences from the CXCR1/2, CXCR3, CXCR4, CXCR5, and CXCR6 gene families. For all gene families, Uniprot and Ensembl were searched for little skate orthologs, as well as orthologs from the following vertebrate species, with their two-letter designations for phylogenetic analyses: human [Homo sapiens (Hs)], mouse [Mus musculus (Mm)], chicken [Gallus gallus (Gg)], zebrafish [Danio rerio (Dr)], pufferfish [Takifugu rubripes (Tr)], catshark [Scyliorhinus canicular (Sc)], elephant shark [Callorhincus milii (Cm)], coelacanth [Latimeria chalumnae (Lc)], and lamprey [Petromyzon marinus, (Pm)]. These species were chosen to put the little skate sequences within an appropriate evolutionary context. If a gene was not found in Uniprot or Ensembl, SkateBase and NCBI were searched. Several sequences were retrieved from Zou et al. (62) as well. Sequence accession information is located in Supplemental Table S1 (Supplemental Material for this article can be found on the AJP-Regulatory, Integrative and Comparative Physiology web site). Alignments were created using the Jalview software (57) and MuscleWS alignment program (13) and then manually curated.
Phylogenetic and molecular evolutionary analyses were conducted in MEGA version 6 (54). The MEGA Model Selection program was used to identify the best-fitting amino acid substitution model under the Bayesian Information Criterion (BIC). The complete-deletion option was used for gap sites, and the branch swap filter was set to “weak” to create a more exhaustive optimization with respect to branch lengths and improvements in log likelihood. The evolutionary history was then inferred through the Maximum Likelihood method based on the best model with 10,000 bootstraps. Initial trees for the heuristic search were obtained by applying the Neighbor-Joining method to a matrix of pairwise distances estimated using a JTT model (23). The tree with the highest log likelihood was chosen as the consensus tree and edited in FigTree version 1.4.3 (45).
Mobilization of leukocytes using Plerixafor.
In the 9 days leading up to Plerixafor (Selleckchem, Houston, TX) injection, blood was drawn from nine little skates every third day to acquire baseline total cell counts. Before each blood collection, the little skates were anesthetized in 20 liters of sea water containing 100 mg/l of Tricaine methanesulfonate (E10521: Sigma-Aldrich, St. Louis, MO) for 5 to 7 min. After anesthesia, whole blood (2–4 ml) was collected from the caudal vein of each little skate using a 22-g sterile needle and deposited into 10-ml sodium-heparinized tubes (026896: Thermo Fisher Scientific).
On the tenth day of the experiment, six little skates were given an intraperitoneal injection of Plerixafor dissolved in elasmobranch Ringer solution (0.3 M NaCl, 2.7 mM MgSO4, 5.2 mM KCl, 5 mM CaCl2, 0.37 M urea, and 15 mM Tris, pH 7.4) at the dose recommended by an IACUC veterinarian (160 μg/kg) (33). The remaining three little skates (designated as controls) received equal-volume injections of elasmobranch Ringer solution (i.e., placebo). After injections, whole blood was collected from three Plerixafor-injected little skates at 2 h postinjection, three control little skates at 4 h postinjection, and the remaining three Plerixafor-injected little skates at 6 h postinjection. Control animals were euthanized at 4 h postinjection because this time point was directly between the two experimental time points. This schedule reduced the number of control animals needed while also facilitating assessment of discernible mobilization caused by Plerixafor. Given that it was unclear how the little skate would metabolize Plerixafor, time points were chosen based on the drug’s terminal half-life of approximately 3 h and response times from human and murine studies (6, 10, 31, 41).
Serological assessment of leukocyte mobilization.
For all nine little skates, two blood smears per blood draw were prepared using 20 µl of whole blood and slides coated in poly-l-lysine. The slides were dried, fixed in methanol, and stained with Wrights-Giemsa (Sigma-Aldrich, Flinn Scientific, Batavia, IL). Each slide was analyzed at ×400 magnification using bright-field microscopy. Using ImageJ, 100 cells were quantified and typed (i.e., leukocyte or erythrocyte) within three randomly selected areas of each blood smear.
Statistical analyses.
To determine whether there was evidence of significant leukocyte mobilization following Plerixafor injection, an analysis of covariance (ANCOVA) controlling for baseline (i.e., preinjection) leukocyte percentages was used to compare the two experimental groups with each other and with the control group. For each group, the baseline leukocyte percentage was calculated as the averaged leukocyte percentage of the three little skates for the three blood draws before injection. Similarly, the postinjection leukocyte percentage was calculated as the averaged leukocyte percentage of the three little skates in each group for the single blood draw postinjection. Mean differences between baseline and postinjection leukocyte percentages were calculated by comparing the averaged values for each group both pre- and postinjection. Two planned orthogonal comparisons were performed to determine whether there was a significant difference between the percentage of leukocytes in experimental little skates 2 h postinjection compared with 6 h postinjection as well as between leukocyte percentages in experimental versus control little skates. Differences between means were considered significant if P < 0.05.
RESULTS
Sequencing of CXCL12 and CXCR4 genes.
One PCR product was obtained from the CXCL12 primers and, when sequenced, was found to contain the partial little skate CXCL12 cDNA. By coupling the partial cDNA sequence (covering 213 bp) with additional SkateBase sequence data, the full-length little skate CXCL12 ORF was deduced. No sequence was retrieved for the 3′- and 5′-untranslated regions (UTRs), and no N-glycosylation sites were predicted. The inferred full-length little skate CXCL12 ORF consists of 279 bp, coding for a predicted protein of 92 amino acids (Supplemental Fig. S1). Based on the human CXCL12 sequence, both a predicted receptor activation motif and receptor and heparin binding site were identified in the little skate CXCL12 sequence.
For CXCR4, three PCR products were obtained from the primers and, when sequenced, were found to contain the partial little skate CXCR4 cDNA. The partial cDNA sequence (covering 759 bp) was coupled with SkateBase sequence data, new paired-end RNA-sequencing data from stage 29 little skate embryos (King BL, unpublished data), and RNA-sequencing data from little skate ampullary cells, ampullary canals, body skin, and liver (4) to deduce the full-length little skate CXCR4 cDNA (Supplemental Fig. S2). The inferred full-length CXCR4 cDNA consists of 1,610 bp with an ORF of 1,086 bp, which codes for a predicted protein of 361 amino acids. The cDNA has a 5′-UTR of 55 nucleotides and a 3′-UTR of 469 nucleotides. The 5′-UTR contains an in-frame (frame 2) stop codon upstream of the true ORF. The 3′-UTR contains two mRNA instability motifs (ATTTA) and a polyadenylation signal (AATAAA). Six potential N-glycosylation sites were identified, with three localized to the extracellular NH2 terminus and one in each of the following cytoplasmic domains: intracellular loop (ICL) 2, ICL3, and the COOH-terminus. The inferred little skate CXCR4 has a seven-transmembrane domain structure.
Multiple sequence alignments.
In total, 38 sequences were included in the chemokine ligand alignment (Supplemental Fig. S3), and 50 sequences were included in the chemokine receptor alignment (Supplemental Fig. S4). Sequence accession information is located in Supplemental Table S1.
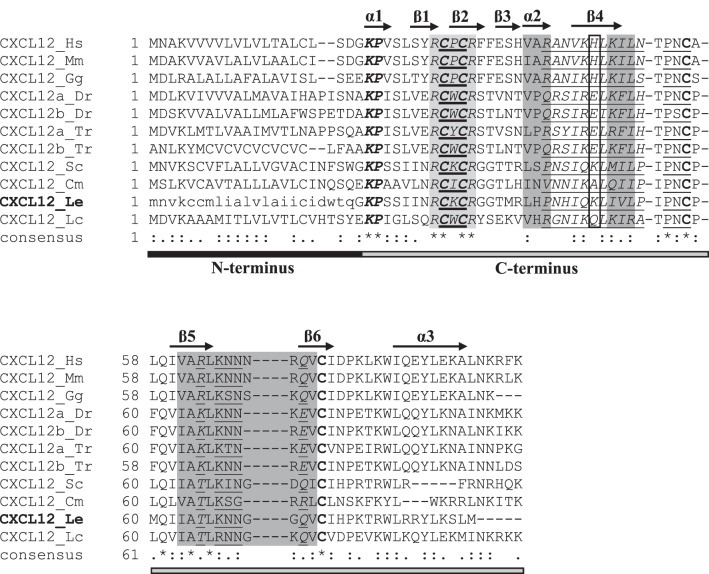
In the chemokine ligand alignment, all sequences showed uniform conservation of the C-X-C motif as well as uniform conservation of cysteines at positions 58 and 79 (of the consensus sequence). The proposed little skate CXCL12 sequence shared the most similarities with other CXCL12 genes. A closer look at the CXCL12 subset of the chemokine ligand alignment revealed areas of high amino acid conservation across the eleven sequences examined (Fig. 1). Sequence conservation suggests that the secondary structure of little skate CXCL12 mirrors that seen in mammalian versions of the gene, with three α-helices, six β-strands, and two turns.
Fig. 1.
Annotated multiple alignment of vertebrate ligand C-X-C motif chemokine ligand 12 (CXCL12; subset of the vertebrate chemokine ligand multiple alignment; see Supplemental Fig. S3). The little skate sequence (CXCL12_Le) is in boldface. The portion of the little skate sequence retrieved from sequencing is written in capital letters, whereas the inferred portion is written in lowercase letters. The characteristic C-X-C motif is denoted by a boldfaced underline. Sequences involved in receptor binding are highlighted in dark gray, whereas an amino acid involved in dimerization is boxed. Sites responsible for heparin binding are italicized. The receptor activation motif (KP) is bolded and italicized. The receptor and heparin binding site (RCXCR) is highlighted in light gray and italicized. Uniformly conserved cysteines are in boldface. Individual β-strands (β1–6) and α-helices (α1–3), inferred from the human CXCL12 sequence, are denoted by arrows above the alignment. Sequences responsible for turns in the secondary protein structure in human CXCL12 are underlined. For the consensus sequence, identical amino acids are denoted by an asterisk (*), whereas those with low similarity (i.e., 50%) are denoted by a period (.) and those with high similarity (i.e., 70%) are denoted by colon (:). Under the consensus sequence, the NH2 terminus and COOH terminus are shown as black and gray bars, respectively. The sequence accession numbers are described in Supplemental Table S1, and species abbreviations are as follows: Cm, elephant shark (Callorhincus milii); Dr, zebrafish (Danio rerio); Gg, chicken (Gallus gallus); Hs, human (Homo sapiens); Mm, mouse (Mus musculus); Lc, coelacanth (Latimeria chalumnae); Le, little skate (Leucoraja erinacea); Sc, catshark (Scyliorhinus canicula); Tr, pufferfish (Takifugu rubripes).
In general, CXCL12 sequence similarity in the α-helix and β-strand regions followed class lines, resulting in groupings of cartilaginous fishes (i.e., little skate, catshark, elephant shark), ray-finned fishes (i.e., zebrafish, pufferfish), lobe-finned fishes (i.e., coelacanth), birds (i.e., chicken), and mammals (i.e., mouse, human). Throughout this article, birds and mammals will be referenced by their overarching clade, amniotes, and ray-finned and lobe-finned fish will be combined into the osteichthyes (i.e., bony fish) superclass. The sequences included in the CXCL12 portion of the alignment showed uniform conservation of four bonding cysteines, with disulfide bonds resulting from cysteine 1 (C1) linking with C3 and C2 linking with C4. Cartilaginous fishes have an additional cysteine at position 6 that is absent in the other vertebrates examined, whereas the little skate alone possesses a cysteine at position 5. The predicted little skate CXCL12 receptor activation motif (KP) and receptor and heparin binding site (RCXCR) were uniformly conserved across all CXCL12 sequences.
Cartilaginous fishes (excluding elephant shark) and amniotes have a positive amino acid at position 48 (K and H, respectively). Bony fishes, however, have a negative (E; ray-finned fishes) or hydrophilic (Q; lobe-finned fishes) amino acid at position 48. For the three regions involved in heparin binding, the overall hydrophobicity pattern is relatively conserved even when substitutions occur. In general, the little skate CXCL12 sequence is most similar to the other cartilaginous fish sequences, with clear clustering of all 12 sequences along class lines.
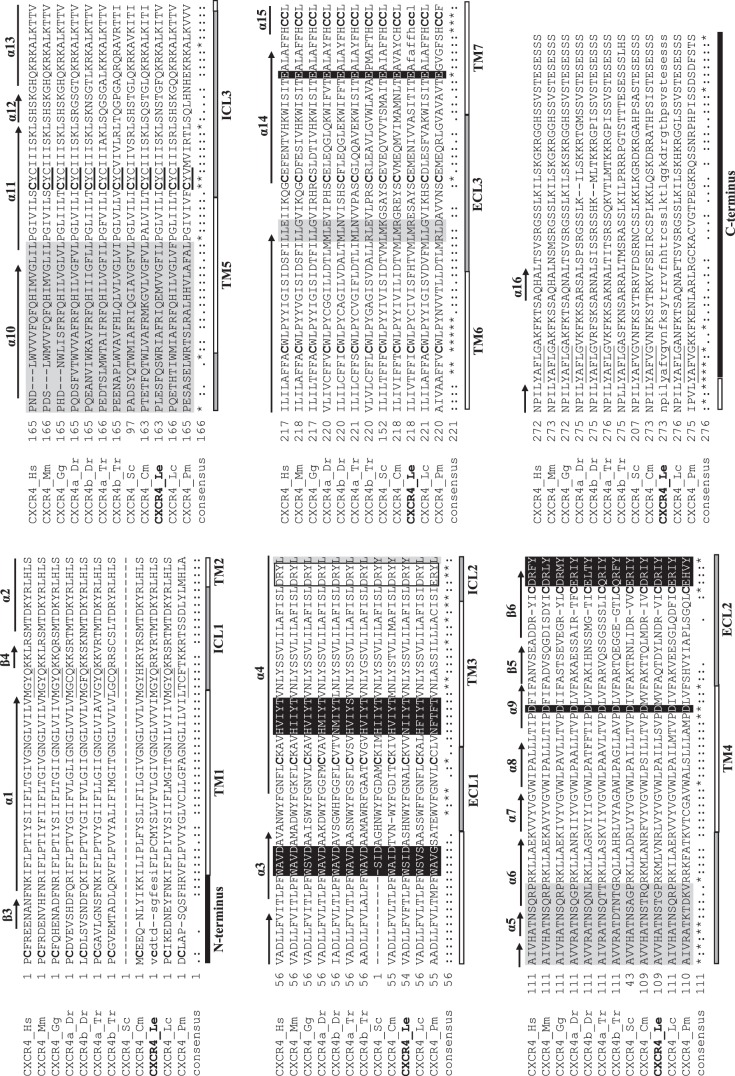
The chemokine receptor alignment shows uniform conservation of cysteines at positions 84, 161, and 196 (of the consensus sequence) as well as uniform conservation of tryptophan (position 135), phenylalanine (position 226), and proline (position 232) residues. Once again, the proposed little skate CXCR4 sequence shared the most similarities with other CXCR4 genes. Within the CXCR4 subset of the chemokine receptor alignment, the characteristic C-X-C motif is conserved in all sequences, except for lamprey (Fig. 2). Transmembrane domain prediction confirmed that the little skate CXCR4 has seven transmembrane domains [3 ICLs and 3 extracellular loops (ECLs)], an extracellular NH2 terminus, and an intracellular COOH terminus. Widespread sequence conservation further suggests that the secondary structure of little skate CXCR4 mirrors that seen in mammalian versions of the gene, with 16 α-helices, six β-strands, and two turns.
Fig. 2.
Annotated multiple alignment of vertebrate chemokine receptor type 4 (CXCR4; subset of the vertebrate chemokine ligand multiple alignment; see Supplemental Fig. S4). The little skate sequence (CXCR4_Le) is in boldface. The portion of the little skate sequence retrieved from sequencing is written in capital letters, whereas the inferred portion is written in lowercase letters. The characteristic C-X-C motif is denoted by a boldfaced underline. Sequences involved in chemokine binding are highlighted in black with white letters, whereas sequences involved in dimerization are highlighted in light gray. The boxed sequence (DRY for all species except lamprey) denotes an area that is critical for signaling. Uniformly conserved cysteines are in boldface. Sequences responsible for turns in the secondary protein structure in human CXCR4 are underlined. Individual β-strands (β3–6) and α-helices (α1–16) inferred from the human CXCR4 sequence are denoted by arrows above the alignment. For the consensus sequence, identical amino acids are denoted by an asterisk (*), whereas those with low similarity (i.e., 50%) are denoted by a period (.), and those with high similarity (i.e., 70%) are denoted by a colon (:). Under the consensus sequence, the NH2 terminus and COOH terminus are shown as black bars, transmembrane domains (TM1–7) as white bars, and extracellular loops (ECL1–3) and intracellular loops (ICL1–3) as gray bars. The sequence accession numbers are described in Supplemental Table S1, and species abbreviations are as follows: Cm, elephant shark (Callorhincus milii); Dr, zebrafish (Danio rerio); Gg, chicken (Gallus gallus); Hs, human (Homo sapiens); Mm, mouse (Mus musculus); Lc, coelacanth (Latimeria chalumnae); Le, little skate (Leucoraja erinacea); Pm, lamprey (Petromyzon marinus); Sc, catshark (Scyliorhinus canicula); Tr, pufferfish (Takifugu rubripes).
The α-helices show remarkable sequence conservation across species, although lamprey (the taxonomically oldest species and only jawless fish) often diverges from the other vertebrates. Notably, amniotes lack three amino acids in α10 compared with the other sequences. Consensus across the first three β-strands is minimal (β1 and β2 were manually trimmed from the alignment), likely because β1–3 are in the more variable NH2 terminus of the protein.
Across the 12 CXCR4 sequences examined, eight cysteine residues are conserved (with 1 in the NH2 terminus, 1 in each of the 3 ECLs, 1 in ICL3, 1 in TM6, and 2 in TM7). Of these eight cysteines, four are known to create two disulfide bonds, linking the NH2 terminus to ECL3 and ECL1 to ECL2 (9). A general trend in the sequences examined suggests a loss of CXCR4 cysteines over evolutionary time; the taxonomically oldest species, lamprey, has 15 cysteines, whereas the taxonomically youngest, human, has nine (Table 1). This trend was also apparent (albeit to a lesser extent) in CXCL12 cysteine counts; cartilaginous fishes have six or seven cysteines, whereas bony fishes and amniotes only have four or five (with the exception of the pufferfish B sequence) (Table 1).
Table 1.
CXCR4 and CXCL12 protein cysteine counts for selected vertebrates
| Species | CXCR4 Cysteines | CXCL12 Cysteines |
|---|---|---|
| Pm | 15 | – |
| Le | 12 | 7 |
| Cm | 11 | 6 |
| Sc | 9 | 6 |
| Tr A | 11 | 4 |
| Tr B | 12 | 10 |
| Dr A | 12 | 4 |
| Dr B | 11 | 4 |
| Lc | 9 | 5 |
| Gg | 9 | 4 |
| Mm | 9 | 5 |
| Hs | 9 | 5 |
CXCL12, C-X-C motif chemokine ligand 12; CXCR4, chemokine receptor type 4. Cm, elephant shark (Callorhincus milii); Dr, zebrafish (Danio rerio); Gg, chicken (Gallus gallus); Hs, human (Homo sapiens); Mm, mouse (Mus musculus); Lc, coelacanth (Latimeria chalumnae); Le, little skate (Leucoraja erinacea); Pm, lamprey (Petromyzon marinus); Sc, catshark (Scyliorhinus canicula); Tr, pufferfish (Takifugu rubripes).
The regions responsible for chemokine binding and dimerization are well conserved across all sequences examined, with the exception of the aforementioned dimerization sequence in α10. A DRY motif (ERY for lamprey), key for proper CXCR4 signaling, was present in ICL2 in all sequences. High CXCR4 sequence similarity was typically seen within classes (with little skate CXCR4 most similar to catshark and elephant shark CXCR4), and between-class similarity generally mirrored taxonomy.
Phylogenetic trees.
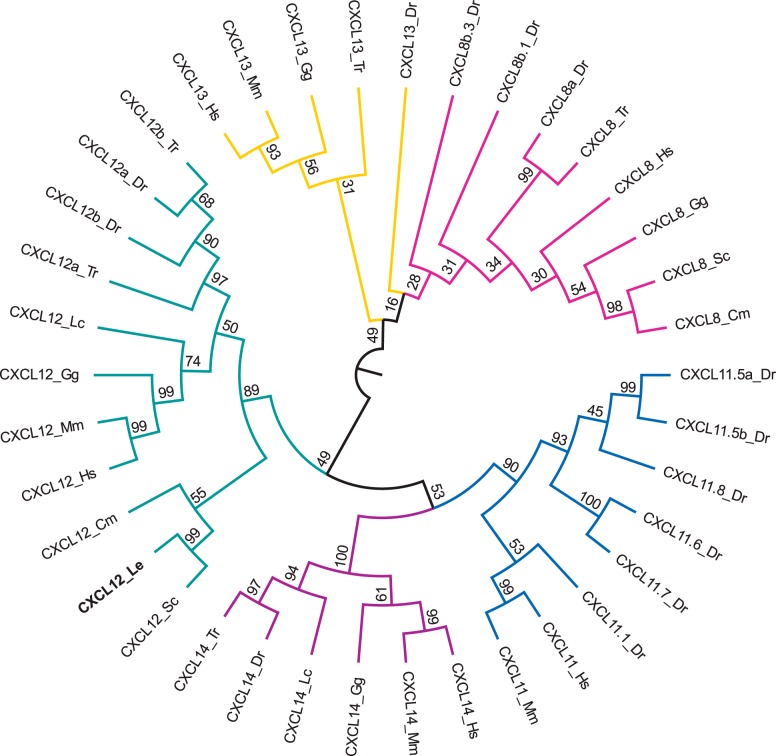
The model that best explained the evolutionary history of the chemokine ligand sequences was LG + G + I (BIC = 5,545.3), and the tree with the highest log likelihood (−2,520.9) was chosen as the consensus tree (Fig. 3) (30). In the ligand tree, the little skate CXCL12 gene grouped within the CXCL12 clade (branch support: 89%) and specifically within the cartilaginous fish branch (branch support: 55%). Little skate CXCL12 was most closely clustered with catshark CXCL12 (branch support: 99%).
Fig. 3.
Phylogenetic tree of vertebrate chemokine ligands from the C-X-C motif chemokine ligand (CXCL) 8 (pink), CXCL11 (blue), CXCL12 (green), CXCL13 (yellow), and CXCL14 (purple) gene families. The tree was constructed in the MEGA6 program (54) using a MuscleWS multiple alignment. The evolutionary history was inferred by using the maximum likelihood method based on the LG model (30) with invariable sites (+I; 6.1275% sites). A discrete γ-distribution was used to model evolutionary rate differences among sites (+G; five categories, parameter = 3.6495). Node values represent %bootstrap confidence derived from 10,000 replicates. The little skate CXCL12 sequence (CXCL12_Le) is in boldface. The analysis included 38 amino acid sequences. All positions containing gaps or missing data were eliminated, resulting in a total of 65 positions in the final data set. The sequence accession numbers are described in Supplemental Table S1, and species abbreviations are as follows: Cm, elephant shark (Callorhincus milii); Dr, zebrafish (Danio rerio); Gg, chicken (Gallus gallus); Hs, human (Homo sapiens); Mm, mouse (Mus musculus); Lc, coelacanth (Latimeria chalumnae); Le, little skate (Leucoraja erinacea); Sc, catshark (Scyliorhinus canicula); Tr, pufferfish (Takifugu rubripes).
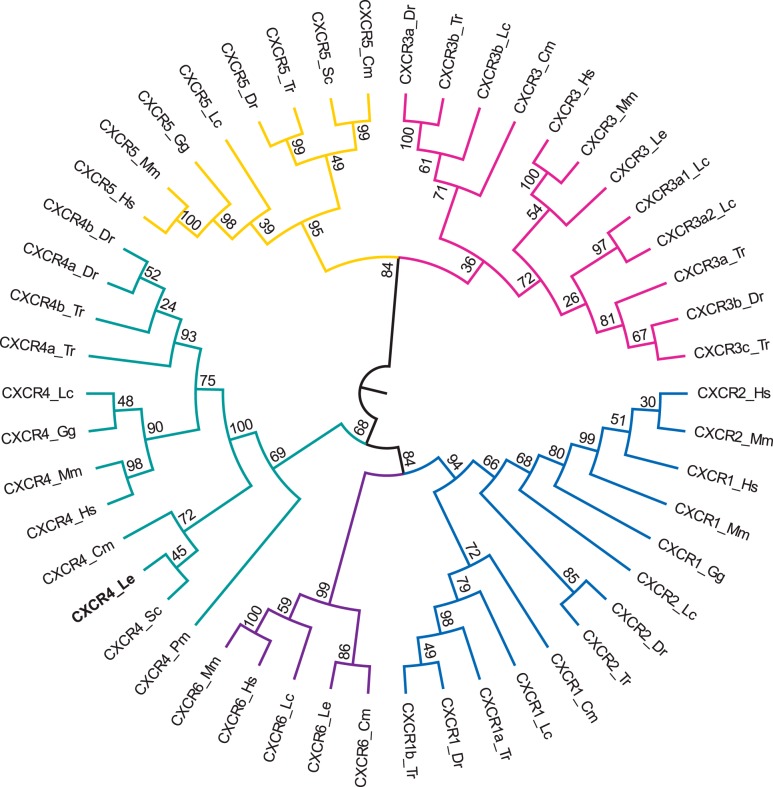
The best model for the chemokine receptor analysis was JTT + G + F (BIC = 31,1158.2) and the tree with the highest log likelihood (−15,025.4) was chosen as the consensus tree (Fig. 4) (23). In the receptor tree, the little skate CXCR4 gene grouped within the CXCR4 clade (branch support: 69%) and specifically within the cartilaginous fish branch (branch support: 72%). Little skate CXCR4 was, once again, most closely clustered with catshark CXCR4 (branch support: 45%).
Fig. 4.
Phylogenetic tree of vertebrate chemokine receptors from the chemokine receptor type (CXCR) 1/2 (blue), CXCR3 (pink), CXCR4 (green), CXCR5 (yellow), and CXCR6 (purple) gene families. The tree was constructed in the MEGA6 program (54) using a MuscleWS multiple alignment. The evolutionary history was inferred by using the maximum likelihood method based on the JTT mixture-based model (23) with frequencies (+F). A discrete γ-distribution was used to model evolutionary rate differences among sites (+G; 5 categories, parameter = 2.3322). Node values represent %bootstrap confidence derived from 10,000 replicates. The little skate CXCR4 sequence (CXCR4_Le) is in boldface. The analysis included 50 amino acid sequences. All positions containing gaps and missing data were eliminated, resulting in a total of 166 positions in the final data set. The sequence accession numbers are described in Supplementary Table S1, and species abbreviations are as follows: Cm, elephant shark (Callorhincus milii); Dr, zebrafish (Danio rerio); Gg, chicken (Gallus gallus); Hs, human (Homo sapiens); Mm, mouse (Mus musculus); Lc, coelacanth (Latimeria chalumnae); Le, little skate (Leucoraja erinacea); Pm, lamprey (Petromyzon marinus); Sc, catshark (Scyliorhinus canicula); Tr, pufferfish (Takifugu rubripes).
Whole blood analysis pre- and postinjection.
In the control group, the mean leukocyte percentage was 24 ± 1% at baseline and 20 ± 1% at 4 h postinjection of placebo solution, resulting in a mean decrease (i.e., difference in the baseline and postinjection means) of 4.0 ± 0.3% (Fig. 5). In the first experimental group, mean leukocyte percentage was 24 ± 1% at baseline and 57 ± 12% at 2 h postinjection of Plerixafor, resulting in a mean increase of 32 ± 13% (Fig. 5). In the second experimental group, mean leukocyte percentage was 29 ± 2% at baseline and 51 ± 17% at 6 h postinjection of Plerixafor, resulting in a mean increase of 21 ± 17% (Fig. 5).
Fig. 5.
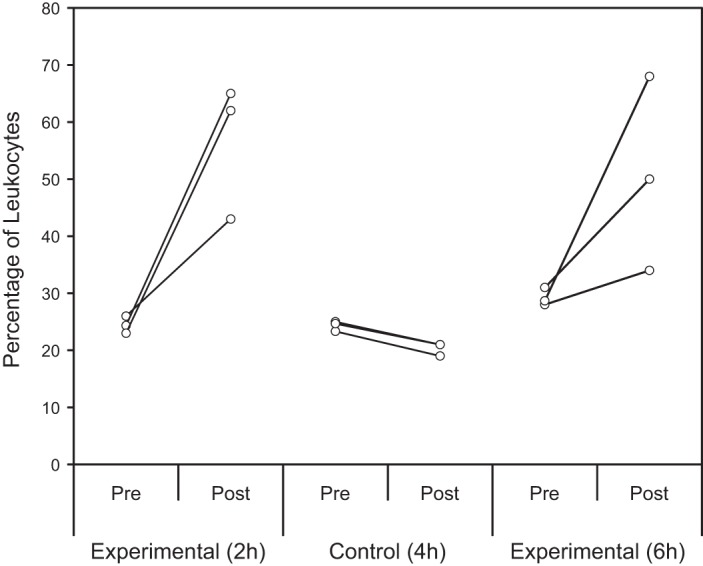
%Leukocytes pre- and post-Plerixafor injection. For all animals, the preinjection percentage was calculated by averaging the leukocyte percentages from the 3 blood draws before injection. The postinjection percentage was calculated by taking the leukocyte percentage from a single blood draw at 3 different end points: 4 h postinjection for the control group (n = 3), 2 h postinjection for 1 experimental group (n = 3), and 6 h postinjection for the other experimental group (n = 3). An ANCOVA comparing the 3 groups on postinjection leukocyte percentages after controlling for baseline (preinjection) leukocyte percentages was statistically significant [F(2,5) = 6.33, P = 0.04], with a large effect size (partial η2 = 0.72). Planned orthogonal comparisons found no significant difference in the no. of mobilized leukocytes between the 2 experimental groups postinjection (P = 0.93), but the experimental animals had significantly higher leukocyte percentages than control animals postinjection (P = 0.04).
An analysis of covariance (ANCOVA) comparing the three little skate groups on postinjection leukocyte percentages after controlling for baseline (preinjection) leukocytes was statistically significant [F(2,5) = 6.33, P = 0.04], with a large effect size (partial η2 = 0.72). A planned orthogonal contrast between experimental leukocyte percentages at 2 and 6 h postinjection found no significant difference (P = 0.93), allowing results from the two experimental groups to be pooled together. An additional planned orthogonal contrast between postinjection control and pooled experimental leukocyte percentages showed that the experimental animals had significantly higher leukocyte percentages than the controls (P = 0.04).
DISCUSSION
Characterization of little skate CXCR4.
Given the dearth of invertebrate chemokine receptor discoveries to date, coupled with the presence of at least six chemokine receptors in an early vertebrate (the lamprey P. marinus, a member of the agnathan superclass), genetic and phylogenetic evidence suggests that chemokine receptors first appeared in the genome of a vertebrate lineage ancestor (3, 42, 44). Indeed, CXCR4 is widely considered to be a primordial chemokine receptor due to its strong sequence conservation throughout evolution and across species; this makes it an ideal launching point for studies of chemokine activity and immune regulation in nonmammalian vertebrates (61).
The identity of the proposed little skate CXCR4 sequence was confirmed through comparison with 49 other vertebrate chemokine receptor sequences, as evidenced by the little skate CXCR4 gene clustering with other CXCR4 genes in the chemokine receptor phylogenetic tree. The CXCR4 subset of the tree has three overarching clusters, with cartilaginous fishes separated from the other species and lamprey forming an outgroup. Little skate CXCR4 is most similar to catshark (78% identities, 90% positives) and elephant shark (68% identities, 81% positives) CXCR4, and all sequences clustered according to class. The two mammalian CXCR4 sequences (human and mouse) were most similar to each other and also shared many similarities with the chicken and coelacanth sequences. This clustering is logical, given that lobe-finned fishes share a more recent common ancestor with tetrapods than with ray-finned or cartilaginous fishes (53). The ray-finned fish sequences clustered together as well.
More in-depth annotation of the little skate CXCR4 gene compared with the other CXCR4 genes suggests that many of the structurally and functionally important features are conserved across species. Little skate CXCR4 appears to have the expected seven-transmembrane domain configuration with a characteristic DRY signaling motif in ICL2 (40). Overall, the residues comprising each α-helix are well conserved across the 12 CXCR4 sequences included in our study, with the exception of three amino acids missing from the α10 region of amniotes but present in all other sequences. The α10 helix, as well as flanking amino acids, is involved in dimerization in human CXCR4; additional functional experiments are needed to assess the same residues in nonmammalian vertebrates to determine whether dimerization still occurred before the deletion. The β-strands show slightly more variability across species, but overall hydrophobicity patterns are typically conserved even when substitutions are made.
In mammalian CXCR4, four cysteine residues form two disulfide bonds, linking the NH2 terminus to ECL3 and ECL1 to ECL2. In addition to these four bond-forming cysteines, four other cysteines were conserved across species. A general trend in the sequences examined suggests a loss of cysteines in both CXCR4 and CXCL12 over evolutionary time (Table 1); disulfide bond prediction using CYSPRED (14) suggests that the additional CXCR4 cysteines are nonbonding. These findings diverge from previous work showing that cysteine representation is positively correlated with organism complexity and that evolution tends to favor an increase, rather than decrease, in protein cysteines over time (37). One possible explanation for the alternative trend seen here is chemokine receptor/ligand coevolution, which is discussed later.
In human CXCR4, five different sequence regions are involved in chemokine binding. As was a trend throughout this analysis, the lamprey CXCR4 sequence showed the greatest divergence from the other sequences. The five chemokine binding regions showed remarkable sequence conservation, with the overall hydrophobicity pattern of residues typically maintained. The same trend was seen when the three sequence regions involved in dimerization were examined (with the exception of the region encompassing α10, discussed above).
Characterization of little skate CXCL12.
Chemokines are characterized by the presence of four conserved cysteine residues that form two disulfide bonds; these four cysteines are conserved in all 38 sequences included in the chemokine ligand analysis. Considered a primordial chemokine, CXCL12 has been identified as far back in the evolutionary tree as agnathans (3). The little skate CXCL12 gene clustered with other CXCL12 genes in the chemokine ligand phylogenetic tree. The CXCL12 subset of the tree has two overarching clusters, with cartilaginous fishes separated from the other species. Little skate CXCL12 is most similar to catshark (70% identities, 85% positives) and elephant shark (46% identities, 81% positives) CXCL12. Like CXCR4, CXCL12 clustering follows class lines, with amniote and coelacanth sequences separated from ray-finned fish sequences. Disulfide bond prediction using CYSPRED (14) found that, aside from the aforementioned four cysteine residues, all cysteines in the CXCL12 genes are nonbonding.
Previous mutagenesis studies found that CXCL12 has several sequence regions responsible for heparin binding (48). The oligomeric state of CXCL12 is modulated by heparin (as well as pH and multivalent anions), with the monomeric form required for full chemotactic activity and the dimeric form acting as a partial CXCR4 agonist (39, 55). The primary role of heparin and heparin derivatives appears to be inhibition of CXCL12-mediated chemotaxis of CXCR4 (39). General conservation of the overall hydrophobicity pattern of residues in CXCL12 heparin binding areas suggests that function may be conserved as well. Future analyses are required to test this theory and draw functional conclusions about CXCL12/heparin binding and the subsequent effect on CXCR4 in the little skate EO.
Sequence regions that play crucial roles in the CXCL12/CXCR4 interaction, such as the receptor activation motif and receptor and heparin binding site, are uniformly conserved in all 11 CXCL12 sequences included in our study. Cartilaginous fishes do show some additional variation compared with the other vertebrates in three additional sequences involved in receptor binding, which is likely a reflection of chemokine receptor/ligand coevolution. Cartilaginous fishes (excluding elephant shark) and amniotes have a positive amino acid at position 48 (K and H, respectively), whereas ray-finned fishes have a negative amino acid and lobe-finned fishes have a hydrophilic amino acid. Given that the H in human CXCL12 is responsible for dimer formation, it is likely that the K in cartilaginous fish CXCL12 plays a similar functional role, whereas dimer formation in bony fishes may be facilitated by a different residue.
Chemokine receptor/ligand coevolution.
Evidence strongly suggests that chemokine receptors and ligands coevolve to maintain functional interactions; genes encoding both the receptor and ligand must undergo parallel evolution with continuous adaptation to preserve the desired function while also allowing for novel functional pathways to be created (2, 15). Few chemokine receptor/ligand pairs demonstrate this phenomenon better than CXCR4 and CXCL12. In the teleost lineage, both genes have been duplicated but still maintain receptor/ligand specificity and binding ability (2, 43). This preservation of specificity suggests that the functional interaction between CXCR4 and CXCL12 has been preserved as well (2). Indeed, although this chemokine receptor/ligand pair carries out slightly different roles in different organisms, an overall trend of modulating immune responses and hematopoiesis prevails (11, 27, 46).
As noted above, cartilaginous fishes do show some variation in both receptor and ligand binding sequences compared with other vertebrates. Given the prevalence of chemokine receptor/ligand coevolution, these variations are not uncommon or unexpected. Detection of CXCR4 and CXCL12 in the primary hematopoietic organ of the little skate suggests that this chemokine receptor/ligand pair is carrying out its evolutionarily conserved function of immune regulation in elasmobranchs, but this function is likely not limited to the EO. Expression of both CXCR4 and CXCL12 has been documented preliminarily in the little skate in other immune-relevant organs, including the Leydig organ and spleen (7). The presence of both genes in these tissues opens the door for subsequent comparative assays of immune response in this elasmobranch model, which will clarify our understanding of CXCR4 and CXCL12 expression and functionality in all three immune organs.
Functional implications of leukocyte mobilization with Plerixafor.
Results from the serological portion of our study suggest that Plerixafor administration to little skates resulted in leukocyte mobilization. When compared with preinjection and control, blood smears from experimental animals showed an increase in leukocytes following Plerixafor injection. In a replication of this experiment, four additional little skates had significantly greater leukocyte percentages after receiving Plerixafor injections; once again, there was no significant difference in experimental animal leukocyte percentages 2 and 6 h postinjection (Ruth BA and Lutton BV, unpublished data). Our results suggest that, as in humans and murine models, Plerixafor-mediated leukocyte mobilization in the little skate is rapid and maintained at least 6 h postinjection (6, 31, 41).
Collectively, our results highlight the utility of the little skate as a preclinical model in studies of hematopoiesis and leukocyte mobilization. It is important to note, however, that the little skate is listed by the International Union for Conservation of Nature as nearly threatened. Given the depleted population levels of this species, it would be prudent to use specimens from captive breeding programs or other more abundant skate species (such as the clearnose skate, Raja eglanteria) in future studies until the conservation status of the little skate improves.
One limitation of the present study is an incomplete understanding of the cell types that express CXCR4 and CXCL12 in the little skate. Future studies attempting to detect CXCR4 and CXCL12 expression on leukocytes will clarify the elasmobranch role for these genes and will help verify whether the Plerixafor-mediated release of leukocytes seen in our study is due to inhibition of little skate CXCR4 and CXCL12 binding. Plerixafor can also mobilize circulating angiogenic cells (51). CXCL12 recruits CXCR4-expressing hemangiocytes (i.e., hematopoietic/endothelial progenitor cells) to ischemic tissues where these cells promote revascularization (22). Taken together, this suggests Plerixafor has the capacity to mobilize not just leukocytes for immune surveillance but also endothelial cells for blood vessel repair. Future research in our laboratory will focus on characterizing the cellular and molecular components behind angiogenesis in the little skate.
Perspectives and Significance
The cartilaginous fishes offer a unique perspective on the evolutionary mechanisms of hematopoiesis and immunity. These species are positioned at the point of divergence between animals that either lack or possess jaws, a backbone, and an immunoglobulin (Ig)-based adaptive immune system, among other physiological characteristics. Indeed, the acquisition of Ig-based adaptive immune cells and molecules by the ancestors of modern sharks is considered to mark a pivotal juncture in the coevolution between pathogens and their hosts and thus may offer insight into infectious diseases moving forward. Moreover, elasmobranchs do not possess several of the limitations observed in mammalian and teleost models for studies of hematopoiesis and are the only species known to possess primary reproductive organs in direct cellular and vascular contact with primary immune (i.e., hematopoietic) organs; importantly, neuroendocrine-immune interactions are well known to play a role in maintaining homeostasis. We believe that the little skate in particular will complement hematopoiesis studies in other models, as it is the first elasmobranch with both a developed embryonic cell line and sequenced genome. The NEBC selected the little skate for genome sequencing in 2010 over other elasmobranchs based on biomedical potential, experimental tractability, genome size, and phylogeny (as a member of the first group of gnathostomes) (26, 56, 59). Here, by demonstrating the presence of critical conserved hematopoietic regulatory genes, along with in vivo mobilization of cells using the human drug, Plerixafor, we further illustrate the utility of the little skate as a potential preclinical model that may open novel pathways for drug discovery, development, and testing in hematological and vascular diseases.
In conclusion, we have demonstrated simultaneous expression of a primordial chemokine receptor/ligand pair, CXCR4/CXCL12, in the primary hematopoietic tissue of the little skate. Chemokine receptor and ligand identities were determined through genetic analysis and verified through phylogenetic comparison. Administration of a CXCR4-antagonist, Plerixafor, resulted in significant mobilization of leukocytes into the bloodstream and was sustained for at least 6 h. We hypothesize that this mobilization was the direct result of Plerixafor-mediated disruption of CXCR4 and CXCL12 binding, but further studies are necessary to causatively and definitively assess this hypothesis. Our results not only illustrate the experimental tractability of the little skate but also demonstrate the potential preclinical relevance of this model organism in studies of hematopoiesis and leukocyte mobilization.
GRANTS
This work was financially supported by the National Science Foundation Research Experience for Undergraduates program through grants to T. A. Hersh (DBI-0453391) and J. C. Bruce (DBI-1005003), the James Slater Murphy, M.D., Fellowship Fund through a student fellowship to A. L. Dimond, the Maine Institutional Development Award (IDeA) Network of Biomedical Research Excellence (INBRE) through a scholarship to A. L. Dimond, and the Hancock County Scholars Program through a STEER student award to N. V. Lupica (1-R25-ES016254–01). B. L. King was supported by National Institute of General Medical Sciences (NIGMS) grants P20-GM-103423 and P20-GM-104318. Fellowship funds to B. V. Lutton from the MDIBL were provided for summer research between 2011 and 2017 via the Salisbury Cove Research Fund, the Dave Evans Fellowship Fund, the Leon Goldstein Fund, the John W. Boylan Fund, and the Terence C. Boylan Fellowship Fund. This research was supported by an IDeA from the NIGMS under grant no. P20-GM-103423.
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the authors.
AUTHOR CONTRIBUTIONS
T.A.H., A.L.D., B.A.R., N.V.L., J.C.B., B.L.K., and B.V.L. conceived and designed research; T.A.H., A.L.D., B.A.R., N.V.L., J.C.B., and B.V.L. performed experiments; T.A.H., A.L.D., B.A.R., N.V.L., J.C.B., J.M.K., B.L.K., and B.V.L. analyzed data; T.A.H., A.L.D., B.A.R., N.V.L., J.C.B., J.M.K., B.L.K., and B.V.L. interpreted results of experiments; T.A.H., A.L.D., J.M.K., and B.L.K. prepared figures; T.A.H., A.L.D., and B.V.L. drafted manuscript; T.A.H., A.L.D., B.A.R., N.V.L., J.C.B., J.M.K., B.L.K., and B.V.L. edited and revised manuscript; T.A.H., A.L.D., B.A.R., N.V.L., J.C.B., J.M.K., B.L.K., and B.V.L. approved final version of manuscript.
Supplemental Data
Figure S1: Deduced nucleotide and amino acid sequences of the little skate CXCL12 ORF - .docx (17 KB)
Figure S2: Deduced nucleotide and amino acid sequences of the little skate CXCR4 - .docx (23 KB)
Figure S3: Multiple alignment of vertebrate chemokine ligands from the CXCL8, CXCL11, CXCL12, CXCL13, and CXCL14 gene families - .docx (21 KB)
Figure S4: Multiple alignment of vertebrate chemokine receptors from the CXCR1/2, CXCR3, CXCR4, CXCR5, and CXCR6 gene families - .docx (42 KB)
ACKNOWLEDGMENTS
Specimens used in our study were maintained by staff, particularly Scott Weston and Dr. Joe Buttner, and students at the SSU Cat Cove Marine Laboratory as well as by Dr. James Sulikowski at the University of New England Biddeford Campus marine laboratory. We thank Dr. Dannie Durand and Pieter Spealman for phylogenetics assistance, Chris Smith for sequencing and instrumentation support, Dr. Jessica Kaufman for bioinformatics guidance, Capt. Jim Ford for little skate acquisition, and Dr. Raimon Duran-Struuck for Plerixafor dosage advice. This work enabled many high school and undergraduate student research experiences, and we accordingly thank former and current laboratory members for their efforts and assistance: Yuka Takemon, Morgan Bresnahan, Jesse Lupica, Eve Galen, Marina MacKinnon, and Juan Castaneda. Institutional support was provided by Endicott College, SSU, the MDIBL, and The Jackson Laboratory (Bar Harbor, ME).
Present address of T. A. Hersh: Dalhousie University, Life Sciences Centre, 1355 Oxford St., Halifax, NS, B3H 4R2, Canada.
Present address of A. L. Dimond: Regis College, School of Nursing, 235 Wellesley St., Weston, MA 02493.
Present address of B. A. Ruth: LeMoyne College, Dept. of Physician Assistant Studies, 1419 Salt Springs Rd., Syracuse, NY 13214.
Present address of N. V. Lupica: Brown University, Warren Alpert Medical School, 222 Richmond St., Providence, RI 02903.
Present address of J. C. Bruce: Katahdin Analytical Services, 600 Technology Way, Scarborough, ME 04074.
REFERENCES
- 1.Anderson MK, Pant R, Miracle AL, Sun X, Luer CA, Walsh CJ, Telfer JC, Litman GW, Rothenberg EV. Evolutionary origins of lymphocytes: ensembles of T cell and B cell transcriptional regulators in a cartilaginous fish. J Immunol 172: 5851–5860, 2004. doi: 10.4049/jimmunol.172.10.5851. [DOI] [PubMed] [Google Scholar]
- 2.Bajoghli B. Evolution and function of chemokine receptors in the immune system of lower vertebrates. Eur J Immunol 43: 1686–1692, 2013. doi: 10.1002/eji.201343557. [DOI] [PubMed] [Google Scholar]
- 3.Bajoghli B, Aghaallaei N, Hess I, Rode I, Netuschil N, Tay BH, Venkatesh B, Yu JK, Kaltenbach SL, Holland ND, Diekhoff D, Happe C, Schorpp M, Boehm T. Evolution of genetic networks underlying the emergence of thymopoiesis in vertebrates. Cell 138: 186–197, 2009. doi: 10.1016/j.cell.2009.04.017. [DOI] [PubMed] [Google Scholar]
- 4.Bellono NW, Leitch DB, Julius D. Molecular basis of ancestral vertebrate electroreception. Nature 543: 391–396, 2017. doi: 10.1038/nature21401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bilgin YM, de Greef GE. Plerixafor for stem cell mobilization: the current status. Curr Opin Hematol 23: 67–71, 2016. doi: 10.1097/MOH.0000000000000200. [DOI] [PubMed] [Google Scholar]
- 6.Broxmeyer HE, Orschell CM, Clapp DW, Hangoc G, Cooper S, Plett PA, Liles WC, Li X, Graham-Evans B, Campbell TB, Calandra G, Bridger G, Dale DC, Srour EF. Rapid mobilization of murine and human hematopoietic stem and progenitor cells with AMD3100, a CXCR4 antagonist. J Exp Med 201: 1307–1318, 2005. doi: 10.1084/jem.20041385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bruce JC, Galen E, King B, Lutton BV. Identification and quantification of gene expression during angiogenesis in the skate, Leucoraja erinacea. MDIBL Bulletin 54: 6–7, 2015. [Google Scholar]
- 8.Callis G, Sterchi D. Decalcification of bone: literature review and practical study of various decalcifying agents, methods, and their effects on bone histology. J Histotechnol 21: 49–58, 1998. doi: 10.1179/his.1998.21.1.49. [DOI] [Google Scholar]
- 9.Chabot DJ, Zhang PF, Quinnan GV, Broder CC. Mutagenesis of CXCR4 identifies important domains for human immunodeficiency virus type 1 X4 isolate envelope-mediated membrane fusion and virus entry and reveals cryptic coreceptor activity for R5 isolates. J Virol 73: 6598–6609, 1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Devine SM, Flomenberg N, Vesole DH, Liesveld J, Weisdorf D, Badel K, Calandra G, DiPersio JF. Rapid mobilization of CD34+ cells following administration of the CXCR4 antagonist AMD3100 to patients with multiple myeloma and non-Hodgkin’s lymphoma. J Clin Oncol 22: 1095–1102, 2004. doi: 10.1200/JCO.2004.07.131. [DOI] [PubMed] [Google Scholar]
- 11.Doitsidou M, Reichman-Fried M, Stebler J, Köprunner M, Dörries J, Meyer D, Esguerra CV, Leung T, Raz E. Guidance of primordial germ cell migration by the chemokine SDF-1. Cell 111: 647–659, 2002. doi: 10.1016/S0092-8674(02)01135-2. [DOI] [PubMed] [Google Scholar]
- 12.Eckfeldt CE, Mendenhall EM, Flynn CM, Wang TF, Pickart MA, Grindle SM, Ekker SC, Verfaillie CM. Functional analysis of human hematopoietic stem cell gene expression using zebrafish. PLoS Biol 3: e254, 2005. doi: 10.1371/journal.pbio.0030254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32: 1792–1797, 2004. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fariselli P, Riccobelli P, Casadio R. Role of evolutionary information in predicting the disulfide-bonding state of cysteine in proteins. Proteins 36: 340–346, 1999. doi:. [DOI] [PubMed] [Google Scholar]
- 15.Goh CS, Bogan AA, Joachimiak M, Walther D, Cohen FE. Co-evolution of proteins with their interaction partners. J Mol Biol 299: 283–293, 2000. doi: 10.1006/jmbi.2000.3732. [DOI] [PubMed] [Google Scholar]
- 16.Hartmann T, Hübel K, Monsef I, Engert A, Skoetz N. Additional plerixafor to granulocyte colony-stimulating factors for haematopoietic stem cell mobilisation for autologous transplantation in people with malignant lymphoma or multiple myeloma. Cochrane Database Syst Rev 10: CD010615, 2015. doi: 10.1002/14651858.CD010615.pub2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.He S, Nakada D, Morrison SJ. Mechanisms of stem cell self-renewal. Annu Rev Cell Dev Biol 25: 377–406, 2009. doi: 10.1146/annurev.cellbio.042308.113248. [DOI] [PubMed] [Google Scholar]
- 18.Honma Y, Okabe K, Chiba A. Comparative histology of the Leydig and epigonal organs in some elasmobranchs. Jpn J Ichthyol 31: 47–54, 1984. doi: 10.11369/jji1950.31.47. [DOI] [Google Scholar]
- 19.Ivanovski O, Kulkeaw K, Nakagawa M, Sasaki T, Mizuochi C, Horio Y, Ishitani T, Sugiyama D. Characterization of kidney marrow in zebrafish (Danio rerio) by using a new surgical technique. Prilozi 30: 71–80, 2009. [PubMed] [Google Scholar]
- 20.Jagannathan-Bogdan M, Zon LI. Hematopoiesis. Development 140: 2463–2467, 2013. doi: 10.1242/dev.083147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Januschke J, Näthke I. Stem cell decisions: a twist of fate or a niche market? Semin Cell Dev Biol 34: 116–123, 2014. doi: 10.1016/j.semcdb.2014.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jin DK, Shido K, Kopp HG, Petit I, Shmelkov SV, Young LM, Hooper AT, Amano H, Avecilla ST, Heissig B, Hattori K, Zhang F, Hicklin DJ, Wu Y, Zhu Z, Dunn A, Salari H, Werb Z, Hackett NR, Crystal RG, Lyden D, Rafii S. Cytokine-mediated deployment of SDF-1 induces revascularization through recruitment of CXCR4+ hemangiocytes. Nat Med 12: 557–567, 2006. [Erratum in Nat Med 12: 978, 2006.] 10.1038/nm1400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jones DT, Taylor WR, Thornton JM. The rapid generation of mutation data matrices from protein sequences. Comput Appl Biosci 8: 275–282, 1992. doi: 10.1093/bioinformatics/8.3.275. [DOI] [PubMed] [Google Scholar]
- 24.Kahsay RY, Gao G, Liao L. An improved hidden Markov model for transmembrane protein detection and topology prediction and its applications to complete genomes. Bioinformatics 21: 1853–1858, 2005. doi: 10.1093/bioinformatics/bti303. [DOI] [PubMed] [Google Scholar]
- 25.Kiel MJ, Iwashita T, Yilmaz ÖH, Morrison SJ. Spatial differences in hematopoiesis but not in stem cells indicate a lack of regional patterning in definitive hematopoietic stem cells. Dev Biol 283: 29–39, 2005. doi: 10.1016/j.ydbio.2005.03.037. [DOI] [PubMed] [Google Scholar]
- 26.King BL, Gillis JA, Carlisle HR, Dahn RD. A natural deletion of the HoxC cluster in elasmobranch fishes. Science 334: 1517, 2011. doi: 10.1126/science.1210912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Knaut H, Werz C, Geisler R, Nüsslein-Volhard C; Tübingen 2000 Screen Consortium . A zebrafish homologue of the chemokine receptor Cxcr4 is a germ-cell guidance receptor. Nature 421: 279–282, 2003. doi: 10.1038/nature01338. [DOI] [PubMed] [Google Scholar]
- 28.Knoblich JA. Mechanisms of asymmetric stem cell division. Cell 132: 583–597, 2008. doi: 10.1016/j.cell.2008.02.007. [DOI] [PubMed] [Google Scholar]
- 29.Kuroda N, Uinuk-ool TS, Sato A, Samonte IE, Figueroa F, Mayer WE, Klein J. Identification of chemokines and a chemokine receptor in cichlid fish, shark, and lamprey. Immunogenetics 54: 884–895, 2003. doi: 10.1007/s00251-002-031-z. [DOI] [PubMed] [Google Scholar]
- 30.Le SQ, Gascuel O. An improved general amino acid replacement matrix. Mol Biol Evol 25: 1307–1320, 2008. doi: 10.1093/molbev/msn067. [DOI] [PubMed] [Google Scholar]
- 31.Liles WC, Broxmeyer HE, Rodger E, Wood B, Hübel K, Cooper S, Hangoc G, Bridger GJ, Henson GW, Calandra G, Dale DC. Mobilization of hematopoietic progenitor cells in healthy volunteers by AMD3100, a CXCR4 antagonist. Blood 102: 2728–2730, 2003. doi: 10.1182/blood-2003-02-0663. [DOI] [PubMed] [Google Scholar]
- 32.Liu T, Li X, You S, Bhuyan SS, Dong L. Effectiveness of AMD3100 in treatment of leukemia and solid tumors: from original discovery to use in current clinical practice. Exp Hematol Oncol 5: 19, 2016. doi: 10.1186/s40164-016-0050-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lutton BV, Callard IP. Effects of reproductive activity and sex hormones on apoptosis in the epigonal organ of the skate (Leucoraja erinacea). Gen Comp Endocrinol 154: 75–84, 2007. doi: 10.1016/j.ygcen.2007.06.014. [DOI] [PubMed] [Google Scholar]
- 34.Lutton BV, Callard IP. Influence of reproductive activity, sex steroids, and seasonality on epigonal organ cellular proliferation in the skate (Leucoraja erinacea). Gen Comp Endocrinol 155: 116–125, 2008. doi: 10.1016/j.ygcen.2007.03.011. [DOI] [PubMed] [Google Scholar]
- 35.McClusky LM, Sulikowski J. The epigonal organ and mature pole of the testis in the recreationally fished blue shark (Prionace glauca): histochemico-functional correlates. J Anat 225: 614–624, 2014. doi: 10.1111/joa.12242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Miracle AL, Anderson MK, Litman RT, Walsh CJ, Luer CA, Rothenberg EV, Litman GW. Complex expression patterns of lymphocyte-specific genes during the development of cartilaginous fish implicate unique lymphoid tissues in generating an immune repertoire. Int Immunol 13: 567–580, 2001. doi: 10.1093/intimm/13.4.567. [DOI] [PubMed] [Google Scholar]
- 37.Miseta A, Csutora P. Relationship between the occurrence of cysteine in proteins and the complexity of organisms. Mol Biol Evol 17: 1232–1239, 2000. doi: 10.1093/oxfordjournals.molbev.a026406. [DOI] [PubMed] [Google Scholar]
- 38.Morrison SJ, Scadden DT. The bone marrow niche for haematopoietic stem cells. Nature 505: 327–334, 2014. doi: 10.1038/nature12984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Murphy JW, Cho Y, Sachpatzidis A, Fan C, Hodsdon ME, Lolis E. Structural and functional basis of CXCL12 (stromal cell-derived factor-1 α) binding to heparin. J Biol Chem 282: 10018–10027, 2007. doi: 10.1074/jbc.M608796200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Murphy PM. Molecular piracy of chemokine receptors by herpesviruses. Infect Agents Dis 3: 137–154, 1994. [PubMed] [Google Scholar]
- 41.Nervi B, Ramirez P, Rettig MP, Uy GL, Holt MS, Ritchey JK, Prior JL, Piwnica-Worms D, Bridger G, Ley TJ, DiPersio JF. Chemosensitization of acute myeloid leukemia (AML) following mobilization by the CXCR4 antagonist AMD3100. Blood 113: 6206–6214, 2009. doi: 10.1182/blood-2008-06-162123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nomiyama H, Osada N, Yoshie O. A family tree of vertebrate chemokine receptors for a unified nomenclature. Dev Comp Immunol 35: 705–715, 2011. doi: 10.1016/j.dci.2011.01.019. [DOI] [PubMed] [Google Scholar]
- 43.Nomiyama H, Osada N, Yoshie O. Systematic classification of vertebrate chemokines based on conserved synteny and evolutionary history. Genes Cells 18: 1–16, 2013. doi: 10.1111/gtc.12013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nordström KJ, Fredriksson R, Schiöth HB. The amphioxus (Branchiostoma floridae) genome contains a highly diversified set of G protein-coupled receptors. BMC Evol Biol 8: 9, 2008. doi: 10.1186/1471-2148-8-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rambaut A. FigTree, a graphical viewer of phylogenetic trees (Online) http://tree.bio.ed.ac.uk/software/Figtree/ [14 Jan 2018].
- 46.Rankin SM. Chemokines and adult bone marrow stem cells. Immunol Lett 145: 47–54, 2012. doi: 10.1016/j.imlet.2012.04.009. [DOI] [PubMed] [Google Scholar]
- 47.Roland J, Murphy BJ, Ahr B, Robert-Hebmann V, Delauzun V, Nye KE, Devaux C, Biard-Piechaczyk M. Role of the intracellular domains of CXCR4 in SDF-1-mediated signaling. Blood 101: 399–406, 2003. doi: 10.1182/blood-2002-03-0978. [DOI] [PubMed] [Google Scholar]
- 48.Sadir R, Baleux F, Grosdidier A, Imberty A, Lortat-Jacob H. Characterization of the stromal cell-derived factor-1alpha-heparin complex. J Biol Chem 276: 8288–8296, 2001. doi: 10.1074/jbc.M008110200. [DOI] [PubMed] [Google Scholar]
- 49.Scala S. Molecular pathways: Targeting the CXCR-CXCL12 axis—untapped potential in the tumor microenvironment. Clin Cancer Res 21: 4278–4285, 2015. doi: 10.1158/1078-0432.CCR-14-0914. [DOI] [PubMed] [Google Scholar]
- 50.Shen ZH, Zeng DF, Ma YY, Zhang X, Zhang C, Kong PY. Are there any new insights for G-CSF and/or AMD3100 in chemotherapy of haematological malignants? Med Oncol 32: 262, 2015. doi: 10.1007/s12032-015-0705-9. [DOI] [PubMed] [Google Scholar]
- 51.Shepherd RM, Capoccia BJ, Devine SM, Dipersio J, Trinkaus KM, Ingram D, Link DC. Angiogenic cells can be rapidly mobilized and efficiently harvested from the blood following treatment with AMD3100. Blood 108: 3662–3667, 2006. doi: 10.1182/blood-2006-06-030577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sugiyama T, Kohara H, Noda M, Nagasawa T. Maintenance of the hematopoietic stem cell pool by CXCL12-CXCR4 chemokine signaling in bone marrow stromal cell niches. Immunity 25: 977–988, 2006. doi: 10.1016/j.immuni.2006.10.016. [DOI] [PubMed] [Google Scholar]
- 53.Swartz B. A marine stem-tetrapod from the Devonian of western North America. PLoS One 7: e33683, 2012. doi: 10.1371/journal.pone.0033683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol 30: 2725–2729, 2013. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Veldkamp CT, Peterson FC, Pelzek AJ, Volkman BF. The monomer-dimer equilibrium of stromal cell-derived factor-1 (CXCL 12) is altered by pH, phosphate, sulfate, and heparin. Protein Sci 14: 1071–1081, 2005. doi: 10.1110/ps.041219505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wang Q, Arighi CN, King BL, Polson SW, Vincent J, Chen C, Huang H, Kingham BF, Page ST, Rendino MF, Thomas WK, Udwary DW, Wu CH; North East Bioinformatics Collaborative Curation Team . Community annotation and bioinformatics workforce development in concert–Little Skate Genome Annotation Workshops and Jamborees. Database (Oxford) 2012: bar064, 2012. doi: 10.1093/database/bar064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Waterhouse AM, Procter JB, Martin DMA, Clamp M, Barton GJ. Jalview Version 2–a multiple sequence alignment editor and analysis workbench. Bioinformatics 25: 1189–1191, 2009. doi: 10.1093/bioinformatics/btp033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wheeler DL, Church DM, Federhen S, Lash AE, Madden TL, Pontius JU, Schuler GD, Schriml LM, Sequeira E, Tatusova TA, Wagner L. Database resources of the National Center for Biotechnology. Nucleic Acids Res 31: 28–33, 2003. doi: 10.1093/nar/gkg033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wyffels J, King BL, Vincent J, Chen C, Wu CH, Polson SW. SkateBase, an elasmobranch genome project and collection of molecular resources for chondrichthyan fishes. F1000Res 3: 191, 2014. doi: 10.12688/f1000research.4996.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Xiao Ling K, Peng L, Jian Feng Z, Wei C, Wei Yan Y, Nan S, Cheng Qi G, Zhi Wei W. Stromal derived factor-1/CXCR4 axis involved in bone marrow mesenchymal stem cells recruitment to injured liver. Stem Cells Int 2016: 8906945, 2016. doi: 10.1155/2016/8906945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zlotnik A, Yoshie O, Nomiyama H. The chemokine and chemokine receptor superfamilies and their molecular evolution. Genome Biol 7: 243, 2006. doi: 10.1186/gb-2006-7-12-243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zou J, Redmond AK, Qi Z, Dooley H, Secombes CJ. The CXC chemokine receptors of fish: Insights into CXCR evolution in the vertebrates. Gen Comp Endocrinol 215: 117–131, 2015. doi: 10.1016/j.ygcen.2015.01.004. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1: Deduced nucleotide and amino acid sequences of the little skate CXCL12 ORF - .docx (17 KB)
Figure S2: Deduced nucleotide and amino acid sequences of the little skate CXCR4 - .docx (23 KB)
Figure S3: Multiple alignment of vertebrate chemokine ligands from the CXCL8, CXCL11, CXCL12, CXCL13, and CXCL14 gene families - .docx (21 KB)
Figure S4: Multiple alignment of vertebrate chemokine receptors from the CXCR1/2, CXCR3, CXCR4, CXCR5, and CXCR6 gene families - .docx (42 KB)