Abstract
Circulating microRNAs (c-miRNAs), plasma-based noncoding RNAs that control posttranscriptional gene expression, mediate processes that underlie phenotypical plasticity to exercise. The relationship and biological relevance between c-miRNA expression and variable dose exercise exposure remains uncertain. We hypothesized that certain c-miRNAs respond to changes in exercise intensity and/or duration in a dose-dependent fashion. Muscle release of such c-miRNAs may then deplete intracellular stores, thus facilitating gene reprogramming and exercise adaptation. To address these hypotheses, healthy men participated in variable intensity (n = 12, 30 × 1 min at 6, 7, and 8 miles/h, order randomized) and variable duration (n = 14, 7 × 1 mile/h for 30, 60, and 90 min, order randomized) treadmill-running protocols. Muscle-enriched c-miRNAs (i.e., miRNA-1 and miRNA-133a) and others with known relevance to exercise were measured before and after exercise. c-miRNA responses followed three profiles: 1) nonresponsive (miRNA-21 and miRNA-210), 2) responsive to exercise at some threshold but without dose dependence (miRNA-24 and miRNA-146a), and 3) responsive to exercise with dose dependence to increasing intensity (miRNA-1) or duration (miRNA-133a and miRNA-222). We also studied aerobic exercise-trained mice, comparing control, low-intensity (0.5 km/h), or high-intensity (1 km/h) treadmill-running protocols over 4 wk. In high- but not low-intensity-trained mice, we found increased plasma c-miR-133a along with decreased intracellular miRNA-133a and increased serum response factor, a known miR-133a target gene, in muscle. Characterization of c-miRNAs that are dose responsive to exercise in humans and mice supports the notion that they directly mediate physiological adaptation to exercise, potentially through depletion of intracellular stores of muscle-specific miRNAs.
NEW & NOTEWORTHY In this study of humans and mice, we define circulating microRNAs in plasma that are dose responsive to exercise. Our data support the notion that these microRNAs mediate physiological adaptation to exercise potentially through depletion of intracellular stores of muscle-specific microRNAs and releasing their inhibitory effects on target gene expression.
Keywords: aerobic exercise, cardiovascular biomarker, exercise intensity, fitness, microRNA
INTRODUCTION
Endurance exercise stimulates adaptive changes in skeletal and cardiac muscle, inducing enhanced mitochondrial function (31), muscle hypertrophy (5), and angiogenesis (26). The cellular and molecular mechanisms that control these and other adaptations are not fully understood. In humans, the release of circulating effectors in response to exercise may reflect molecular adaptive mechanisms. The responsiveness of cardiac and skeletal muscle tissue to bouts of exercise has been demonstrated convincingly through biomarker profiling of conventional circulating proteins, including creatine kinase, cardiac-specific troponin species (50), and cardiac natriuretic peptides (48). Although these canonical biomarkers have helped to establish exercise-induced responses to tissue injury and muscular endocrine function, they do not capture the full biology of exercise-induced muscle adaptation. Recently, nonprotein molecules have emerged as key potential mediators of this process.
MicroRNAs (miRNAs) are nonprotein coding ribonucleic acid molecules that control posttranscriptional gene expression and downstream cellular function by directly binding specific sequences in mRNA transcripts (1). miRNAs play critical roles in numerous biological processes, including those that relate to muscle physiology during development and disease (60) as well as exercise (38). miRNAs can be secreted as stabilized factors into the bloodstream, packaged either in microvesicles or as free RNA-protein complexes, after which they become circulating miRNAs (c-miRNAs) (19). Previous studies have demonstrated dynamic regulation of specific c-miRNAs during single bouts of aerobic exercise (3, 6, 20, 21, 32, 46, 57) as well as extended durations of aerobic exercise (2, 4, 17, 22, 40, 43, 44, 54). Emerging evidence supports a role of c-miRNAs as endocrine or paracrine messengers via their release and uptake by source and recipient tissues throughout the body (27), and their utility as biomarkers of cardiovascular health and disease is under active investigation. Complementary to that model of delivery of active miRNAs to recipient tissue, release of miRNAs may also represent an efficient method for alteration of gene expression in source tissue, as we have proposed in hypoxia (27), by removing specific inhibitory effects of miRNAs through rapid depletion of intracellular stores. This latter model would offer key advantages in facilitating time-sensitive molecular and physiological changes necessary in skeletal muscle in aerobic exercise, but this model, particularly in exercise physiology, has not been investigated in depth.
Endurance exercise exposure is quantified by three principal variables: intensity, duration, and frequency. The triple product of these factors can be used to quantify exercise load during athletics and to evaluate exercise dose as it relates to health promotion, disease management, or clinical outcomes. Prior studies from our group and others (44) have indicated that the c-miRNA responses to prolonged moderate-intensity aerobic exercise, such as marathon running (4, 40), differ substantially from those that occur during comparatively shorter and more intense bouts of exercise (3). These observations suggest discrete dose-response relationships between aerobic exercise and c-miRNA expression, but this concept has not yet been rigorously assessed. We hypothesized that expression of specific c-miRNAs with established roles in skeletal and muscle function would exhibit dose-response relationships with varying levels of exercise intensity and duration. We further postulated that such release of muscle-specific c-miRNAs reflects an active process in exercising muscle to deplete intracellular miRNA stores, thus removing the specific inhibitory effects of miRNAs on source tissue gene expression. To address these hypotheses, we conducted parallel studies in healthy human volunteers designed to characterize the c-miRNA response to variable levels of both exercise intensity and duration. We then pursued proof-of-concept studies in aerobic exercise-trained mice to determine whether some of those same muscle-specific c-miRNAs released during exercise are simultaneously depleted in source muscle tissue with concomitant alterations of known target genes recognized by those miRNAs.
METHODS
Study approval.
All animal experiments were approved by the University of Pittsburgh School of Medicine. All experimental procedures involving the use of human tissue and plasma were approved via the Partners Human Research Committee (2014P002381) and the University of Pittsburgh School of Medicine Institutional Review Board (PRO15110113). Ethical approval for this study conformed with the standards of the Declaration of Helsinki. Informed consent was obtained for blood sampling.
Human study design.
We conducted two prospective studies in sequential order designed to examine the impact of variable exercise dose on c-miRNA expression. The first study varied exercise intensity, and the second study varied exercise duration. Both studies used a prospective, repeated-measures design in which healthy young men (age range: 19–24 yr) participated in serial treadmill running sessions that controlled exercise intensity or duration. At the initiation of both studies, participant enrollment criteria were confirmed, comprehensive medical and athletic history were obtained, and maximal effort-limited cardiopulmonary exercise testing was performed as described below. Participants in the intensity study then participated in serial treadmill tests (constant distance: 5 miles) at 1-wk intervals at three intensities (6, 7, and 8 miles/h) and a final 5-mile test at maximal volitional speed, as determined by the participants. Participants in the duration study similarly participated in serial treadmill tests (constant speed: 7 miles/h) at 1-wk intervals at three durations (30, 60, and 90 min) in randomized order. Participants in the duration study also completed a peak effort 5-mile treadmill run to serve as a direct control to the identical data obtained during the intensity protocol. Blood samples were collected before and immediately after each exercise bout.
Participant recruitment.
Participants were recruited from the Harvard University undergraduate summer research program by the use of a posted paper flyer. To minimize the impact of hormonal fluctuation, only men were recruited for this study. Additional eligibility criteria included ages of 18−30 yr, no history of heart, liver, or kidney disease, no viral illnesses within 2 wk of study enrollment, and a habitual moderate running schedule of 20–30 miles/wk. To minimize the potential impact of any training effect, participants agreed not to escalate or reduce exercise training during the study period (i.e., to maintain weekly running of 20–30 miles) and to confine all exercise sessions aside from those dictated by the study protocol to <30 min. Participants received a complete verbal description of the study’s goals, methods, and risks and benefits of participation. Participants were provided with the written consent form for review and then provided with an opportunity to ask questions in private one-on-one sessions with a dedicated study investigator. At the time of enrollment, participants were instructed that they would be withdrawn from the study in the event of new-onset illness, musculoskeletal injury, and/or consumption of anti-inflammatory drugs (i.e., nonsteroidal anti-inflammatory drugs) during the study period. Participants maintained daily exercise diaries throughout the study period and were informed that performance of any strenuous exercise lasting longer than 30 min, including running or alternative endurance activity, weight lifting, and recreational or competitive sport participation, would result in removal from the study.
Baseline testing.
Baseline data acquisition was performed at the Massachusetts General Hospital’s Cardiovascular Performance Program Human Physiology Laboratory. Demographic, basic anthropometric, medical historical, and athletic participation data were obtained, including participant height, body mass, resting vital signs, active and past medication and supplement use, personal/family medical historical data, dietary restrictions, and athletic training and competition history, including average minutes of physical activity per day and per week. Next, each participant underwent a maximal, effort-limited cardiopulmonary exercise test with continuous heart rate (HR) monitoring and measurement of metabolic gas exchange. HR was recorded using a wireless 12-lead system (Mortara, San Francisco, CA). Breath-by-breath gas exchange data were collected using a commercially available metabolic cart (Ultima CardiO2, Medgraphics, St. Paul, MN). We used a maximal effort limited treadmill protocol (Woodway Pro, Woodway USA, Waukesha, WI) to determine maximal oxygen consumption (V̇o2max). After 3 min of resting data collection, participants ran at 5 miles/h (1% grade) for 10 min to ensure adequate musculoskeletal warmup, after which treadmill speed was increased by 1 mile/h every minute until the participant reached volitional exhaustion. Oxygen consumption (V̇o2) data were smoothed using a five-breath rolling mean with the highest and lowest values in each seven breaths removed, and peak oxygen consumption (V̇o2peak) was defined as the highest five-breath mean value during exercise (7). The highest 30-s average V̇o2 was recorded as V̇o2max. Peak effort testing was confirmed by the following criteria: 1) plateau of V̇o2 despite increasing workload, 2) a final respiratory exchange ratio of >1.1, and 3) a HR of ≥85% of the age-predicted maximum HR (30). The ventilatory threshold was determined by the modified V-slope method (7), and HR at the ventilatory threshold was recorded.
Variable dose exercise protocols.
Participants in both the variable intensity and variable duration studies were randomly assigned to one of three protocol groups that dictated their initial treadmill speed or study duration, respectively. Participants abstained from all exercise above and beyond activities of daily living for ≥48 h before each exercise session to minimize the impact of biochemical signal summation. Participants arrived for exercise sessions after an overnight fast, during which they were permitted to ingest only water. Exercise sessions for both the variable intensity and variable duration studies were completed on four consecutive Tuesdays with a consistent start time of 9 AM EST in Harvard University’s Malkin Athletic Center. Ambient room temperature and humidity were kept constant across study visits at 69–72°F and 20–30%, respectively. Participants in the variable intensity study completed three constant-distance (5-mile) treadmill runs at 6, 7, and 8 miles/h and a fourth and final timed 5-mile treadmill run at peak effort. Participants in the variable duration study completed three constant speed (7 miles/h) treadmill runs lasting 30, 60, and 90 min. Continuous HR recording using a commercially available chest strap (Polar T-31 Coded Transmitter) was performed during all exercise testing, with HR values recorded at 1-mile intervals for both protocols.
Blood sampling.
Venous blood was collected in conjunction with each study visit. Specifically, blood samples were obtained immediately before and immediately after treadmill running. All blood was obtained from a superficial upper extremity vein using standard phlebotomy techniques by the same trained phlebotomist. Samples were drawn into standard anticoagulant EDTA-treated vacutainer tubes (BD, Franklin Lakes, NJ) and spun at 2,700–2,800 relative centrifugal force in a Medilite centrifuge (Thermo Scientific, Waltham, MA) for 12 min to separate plasma. Plasma aliquots (400 μl) were frozen and stored at −80° C for subsequent analysis.
Candidate miRNA and mRNA target gene selection.
We chose to measure seven c-miRNAs (Table 1) known to play a role in processes inherent to physiological exercise adaptation. These processes include angiogenesis [miRNA (miR)-210 (24) and miR-222 (36, 45, 51)], inflammation [miR-21 (29, 55) and miR-146a (52)], skeletal and cardiac contractility [miR-21 (16, 53) and miR-133a (15)], and hypertrophy [miR-1 (15), miR-133a (12), miR-21 (56), and miR-24 (56)], and hypoxic adaptation [miR-21 (35), miR-146a (28), and miR-210 (13, 14)]. A known target gene of miR-133a (15) and regulator of skeletal muscle phenotype, serum response factor (SRF), was also chosen for analysis.
Table 1.
Candidate microRNAs that are involved in processes inherent to physiological exercise adaptation
| MicroRNA | Selection of Targets | Biological Function | Enriched Tissue |
|---|---|---|---|
| miR-1 and miR-133a | HDAC4, SRF, CCND2, CASP9 | Proliferation, apoptosis muscle contractility, cardiac hypertrophy | Muscle/heart |
| miR-210 | ISCU, EFNA3, E2F3 | Mitochondrial metabolism, angiogenesis, proliferation, survival, hypoxic adaptation | Ubiquitous (hypoxia dependent) |
| miR-222 | KIT (c-kit), CDKN1B (p27/KIP) | Angiogenesis, proliferation | Endothelium |
| miR-21, miR-24, and miR-146a | PDCD4, RECK, PTEN, TIMP3, SPRY2, TPM1, RHOB, MYC, KCNJ2 IRAK1, TRAF6 | Apoptosis, proliferation, inflammation, hypoxic adaptation, muscle contractility, cardiac hypertrophy, apoptosis, protection against myocardial ischemia, inflammation, hypoxic adaptation | Heart/endothelium |
HDAC4, Histone deacetylase 4; SRF, serum response factor; CCND2, cyclin D2; CASP9, caspase 9; ISCU, iron-sulfur cluster assembly enzyme; EFNA3, ephrin A3; E2F3, E2F transcription factor 3; CDKN1B, cyclin-dependent kinase inhibitor 1B; PDCD4, programmed cell death 4; RECK, reversion-inducing cysteine-rich protein with Kazal motifs; PTEN, phosphatase and tensin homolog; TIMP3, tissue inhibitor of metalloproteinase 3; SPRY2, Sprouty RTK signaling antagonist 2; TPM1, tropomyosin 1; RHOB, Ras homolog family member B; MYC, Myc protooncogene; KCNJ2, potassium voltage-gated channel subfamily J member 2; IRAK1, interleukin-1 receptor-associated kinase 1; TRAF6, TNF receptor-associated factor 6.
Mouse exercise protocol.
An aerobic exercise protocol in mice was based on prior reports designed to elicit a functional “training” effect on exercise performance (25) and mimic at least a modest degree of physiological left ventricular hypertrophy that occurs in highly trained athletes (23). Three mouse cohorts [n = 6 mice/cohort, 12-wk-old male littermate mice (C57BL6), The Jackson laboratory, Bar Harbor, ME] were divided evenly into either a control (baseline activity), low-intensity (0.5 km/h), or high-intensity (1 km/h) exercise training group. Exercise training was performed on an Exer-3/6 open treadmill with manual incline (Columbus Instruments, Columbus, OH), and divided lanes allowed six mice to run simultaneously. Manual stimulation (tapping the tail) and shock grid were implemented to encourage running. For 1 wk before exercise training initiation, mice in all groups were acclimated to treadmill conditions, allowing the mice to sit on the treadmill for 3 min, followed by 15 min of exercise at gradual increases of speed from 6 to 9 m/min. For low-intensity and high-intensity training groups, mice then ran for 1 h/day for 5 days/wk (Monday to Friday) over the course of 4 wk.
Rodent echocardiography.
Serial rodent transthoracic echocardiography was performed at both preexercise (time = 0 days) and postexercise training (time = 28 days) time points, as previously described (10). Briefly, echocardiography was performed using a 15- to 45-MHz transthoracic transducer and a VisualSonics Vevo770 system. Inhaled isoflurane anesthesia was used at 2% in 100% O2 during positioning and hair removal and then decreased to isoflurane 0.8% during imaging. Digital echocardiograms were analyzed offline for quantitative analysis, as previously described (8–10).
Rodent tissue and plasma harvesting.
Upon completion of the exercise protocol, each mouse was euthanized followed by harvesting of mouse plasma and major organs, including right gastrocnemius skeletal muscle, as we have previously described (8–10). Organs were flash frozen in liquid N2. Plasma samples were aliquoted and frozen and stored at −80° C for subsequent analysis.
Plasma RNA extraction.
Plasma samples were thawed and centrifuged at 12,700 rpm for 15 min to remove the remaining cellular debris. Supernatant was aliquoted into 150-μl volumes. All 12 samples from 1 individual were analyzed concurrently to minimize possible variability due to plasma handling. All samples were confirmed to have spectrophotometric absorption (414 nm, absorbance peak of free hemoglobin) far below 0.3, as previously described, as a cutoff for maximal specificity to rule out substantial hemolysis (49). In addition, the researcher performing the extraction and quantification was blinded to the randomization of each participant, thereby minimizing observation bias. To quantitatively normalize c-miRNA plasma levels, 2 μl of 0.002 μM cel-miR-67 mimic (ThermoFisher Scientific, Waltham, MA) was added to each 150 μl volume of plasma, as we have previously described (41). Notably, total RNA was extracted using a MicroRNA Extraction Kit (Benevbio, Mission Viejo, CA). Extracted RNA was subsequently reverse transcribed to generate cDNA (MicroRNA Assay Kit, Life Technologies), which corresponds to levels of mature c-miRNA molecules. Importantly, to ensure the accuracy and precision of this protocol, we compared the quantitative results of this single spike in miRNA protocol to other recently described operating procedures (44), using a mixture of two separate spike-in control miRNAs as well as an independent spike in control miRNA during cDNA synthesis. Over 10 separate and randomly selected plasma samples collected across both variable intensity and variable duration exercise, the quantitative fold change of candidate miRNAs did not differ significantly when we compared calculations derived from single versus multiple spikes in protocols. As such, we elected to use a single spike in control miRNA protocol to remain consistent with our prior reported findings across other exercise modalities (3, 4, 40).
Tissue RNA extraction.
Mouse skeletal muscle was homogenized in Qiazol (Qiagen, Hilden, Germany) using a Polytron PT 1200 E homogenizer (Kinematica, Bohemia, NY). Total RNA from gastrocnemius muscle was extracted via the miRNeasy Mini Kit (Qiagen, Hilden, Germany).
Quantification of miRNA and target gene expression in plasma and tissue.
Candidate c-miRNA and target mRNA levels were quantified using quantitative (“real time”) RT-PCR. As previously described (3), mRNAs were reverse transcribed using the Multiscript RT kit (ThermoFisher Scientific) to generate cDNA. miRNAs were reverse transcribed using TaqMan MicroRNA Assays (ThermoFisher Scientific). Generated cDNA was amplified using labeled Taqman probe and primer sets and an Applied Biosystems QuantStudio 6 Flex Real-Time PCR System, and the fold change was calculated using the following formula: , where CT is threshold cycle (3). For plasma miRNAs, ΔCT was calculated by subtracting the CT values of each c-miRNA in a sample from the CT value of the exogenously added cel-miR-67 in that sample. For human plasma, to determine the ΔΔCT, these ΔCT values were subsequently compared with each athlete’s own resting value that week (normalized to fold change of 1). For mouse plasma, ΔCT values were compared with the average level of the control mouse cohort (normalized to fold change of 1). For miRNA quantitation in mouse tissue, the mouse housekeeping small RNA sno-RNA-55 (ThermoFisher Scientific) was used for normalization. For mRNA quantitation in mouse tissue, the mouse β-actin transcript was used (ThermoFisher Scientific).
Statistical analysis.
Normality of distribution for all variables was assessed using the Shapiro-Wilk test. For human studies, participant characteristics, exercise testing data, and c-miRNA data are reported as means ± SE. The significance of HRs during exercise and c-miRNA changes across time points were assessed using mixed linear modeling, with V̇o2peak as a fixed effect and subject identification as a random effect. This regression technique was chosen to account for the correlated error inherent in our repeated-measures study design. Akaike’s information criterion tool was used to select optimal covariance structures for each model. Post hoc pairwise comparisons of variables between study visits were made using least-squares means derived from the mixed-effects models performed with the Bonferroni correction. Data analysis was performed using SPSS software version 23 (IBM). For mouse studies, data are also reported as means ± SE. Shapiro-Wilk testing was used to determine normality of data distribution. For normally distributed data, paired comparisons were performed by Student’s t-test, whereas multiple comparison testing was performed by one-way ANOVA and Bonferroni post hoc tests. For non-normally distributed data, Kruskal-Wallis testing combined with Dunn’s multiple comparisons testing were used. P values of <0.05 were considered significant.
RESULTS
Baseline participant characteristics.
In parallel exercise studies in healthy human volunteers, all enrolled participants completed all phases of their designated protocols and were included in all final analyses. Baseline data describing participants in both the variable intensity (n = 12) and variable duration (n = 14) protocols are shown in Table 2. The variable intensity cohort contained a higher percentage of Caucasian participants (50% vs. 33%), but both cohorts were otherwise similarly matched for age, height, weight, V̇o2peak, and peak exercise HR. Similar to a previous study with healthy individuals (41), all candidate c-miRNAs were detectable in circulating plasma at baseline and displayed ranges of variability (Fig. 1) consistent with prior studies of healthy male volunteers (3), thus indicating an adequate baseline for comparison with subsequent dose-dependent changes in response to exercise.
Table 2.
Baseline participant characteristics
| Clinical Parameter | Variable Intensity Cohort | Variable Duration Cohort |
|---|---|---|
| Number of participants | 12 | 14 |
| Age, yr | 21 ± 1 | 22 ± 3 |
| Height, m | 1.8 ± 0.1 | 1.78 ± 0.1 |
| Body mass, kg | 74 ± 8 | 70 ± 10 |
| Body mass index, kg/m2 | 23 ± 2 | 22 ± 3 |
| Peak O2 consumption, ml/min | 62 ± 5 | 66 ± 6 |
| Peak heart rate, beats/min | 196 ± 7 | 193 ± 6 |
Values are means ± SE.
Fig. 1.
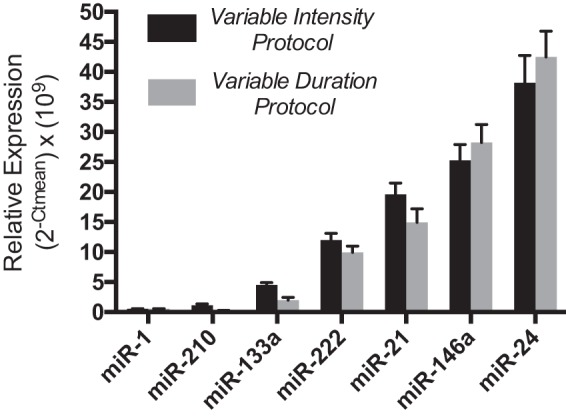
Baseline expression of circulating microRNAs (c-miRNAs) in plasma. At resting conditions before each weekly running session, c-miRNA levels in plasma (150 μl/sample) were measured by quantitative RT-PCR in volunteers participating in the fixed distance/variable intensity (n = 12; black bars) and fixed intensity/variable duration (n = 14; gray bars) portions of the study. Values are reported as relative levels based on the following formula: ( × 109), where Ct is threshold cycle. Data are presented as statistical means, and error bars reflect SEs.
Exercise physiology during variable intensity and duration exercise.
For the variable intensity protocol, exercise HRs increased significantly in parallel with increasing intensity during fixed distance 5-mile treadmill runs at 6 miles/h (76 ± 3% peak HR), 7 miles/h (81 ± 2% peak HR), and 8 miles/h (86 ± 2% peak HR) and maximal effort (92 ± 1% peak HR, P < 0.001). In contrast, exercise HRs during the variable duration/fixed intensity (7 miles/h) treadmill running protocol were unchanged across durations of 30 min (84 ± 3% peak HR), 60 min (85 ± 2% peak HR), and 90 min (87 ± 2% peak HR, P = not significant).
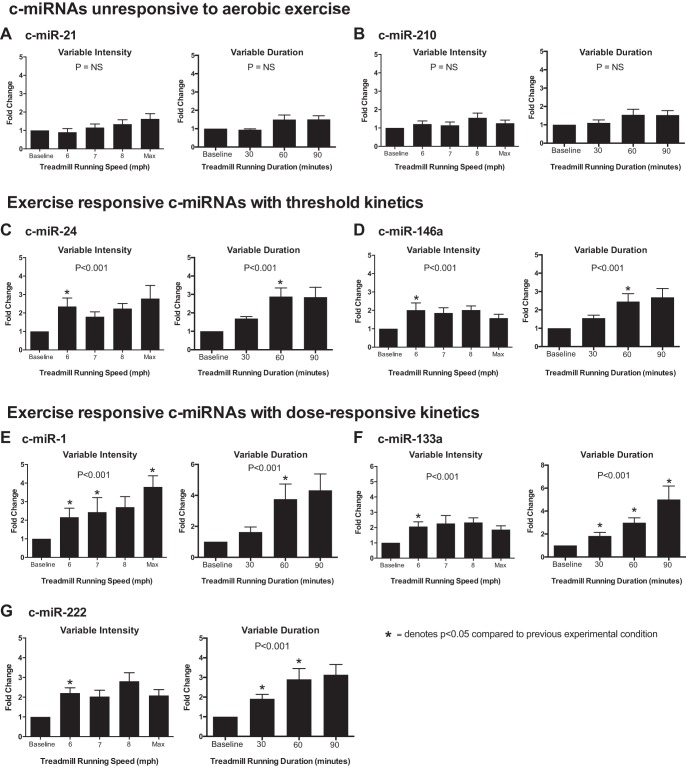
Differential responses of c-miRNAs to variable intensity and variable duration aerobic exercise.
Baseline concentrations of candidate c-miRNA are shown in Fig. 1. Candidate c-miRNA responses immediately postexercise were reported as fold changes during variable intensity and variable duration treadmill running (Fig. 2). The c-miRNA response showed three basic profiles: 1) nonresponsive to treadmill running regardless of intensity or duration (miR-21 and miR-210; Fig. 2, A and B), 2) responsive to treadmill running with threshold kinetics but no significant dose responsiveness (miR-24 and miR-146a, Fig. 2, C–E), and 3) responsive to treadmill running and significantly dose responsive to either increasing intensity (miR-1; Fig. 2E) or duration (miR-133a and miR-222; Fig. 2, F and G). Specifically, exercise unresponsive c-miR-21 and c-miR-210 displayed no significant differences between resting levels and any speed or duration during either the variable intensity or variable volume protocols. In contrast, c-miRNAs in the second category (c-miR-24 and c-miR-146a) were each upregulated similarly compared with baseline levels during all phases of the variable intensity protocol. These c-miRNAs were also upregulated significantly during the volume protocol, and both required 60 min of running for significant upregulation, with no additional augmentation after 90 min of running.
Fig. 2.
Distinct profiles of circulating microRNAs (c-miRNA) responses after variable intensity and duration of exercise. Levels of c-miRNAs were measured before exercise and immediately postexercise. For each individual, the fold change of c-miRNA expression postexercise was calculated compared with the individual’s level before exercise. A and B: profile 1. c-miRNAs nonresponsive to treadmill running regardless of intensity or duration [miRNA (miR)-21 and miR-210] are shown. C and D: profile 2. c-miRNAs responsive to treadmill running with threshold kinetics but without a progressive dose response (miR-24 and miR-146a) are shown. E–G: profile 3. c-miRNAs responsive to treadmill running with a significant dose response to either increasing intensity (miR-1) or duration (miR-133a and miR-222) are shown. Data are expressed as means ± SE. P values denote the significance across repeated measures, as defined by mixed linear modeling. *Significant change (P < 0.05) compared with the prior experimental condition (i.e., speed or duration) using post hoc pairwise comparisons of variables and least-squares means derived from the mixed-effects models performed with the Bonferroni correction.
Finally, and of most interest, highly muscle-enriched c-miR-1 and c-miR-133a as well as c-miR-222 were all upregulated with distinct and differential dose responsiveness during either the variable intensity or variable duration protocols. Specifically, c-miR-1 demonstrated progressive responsiveness to increasing running speeds during the variable intensity protocol but was upregulated only during the variable volume protocol after 60 and 90 min of running, with no apparent dose effect. In contrast, c-miR-133a was upregulated by a similar magnitude after all stages of the variable intensity protocol but demonstrated strong dose-responsive upregulation during the variable volume protocol (comparison of c-miR-1, c-miR-133a, and the nonresponsive reference c-miR-21 is shown in Fig. 3). Similar to miR-133a, c-miR-222 was upregulated by a similar magnitude by 5-mile running regardless of intensity but displayed some dose responsiveness to variable exercise duration, with maximal upregulation occurring with 60 and 90 min of exercise.
Fig. 3.
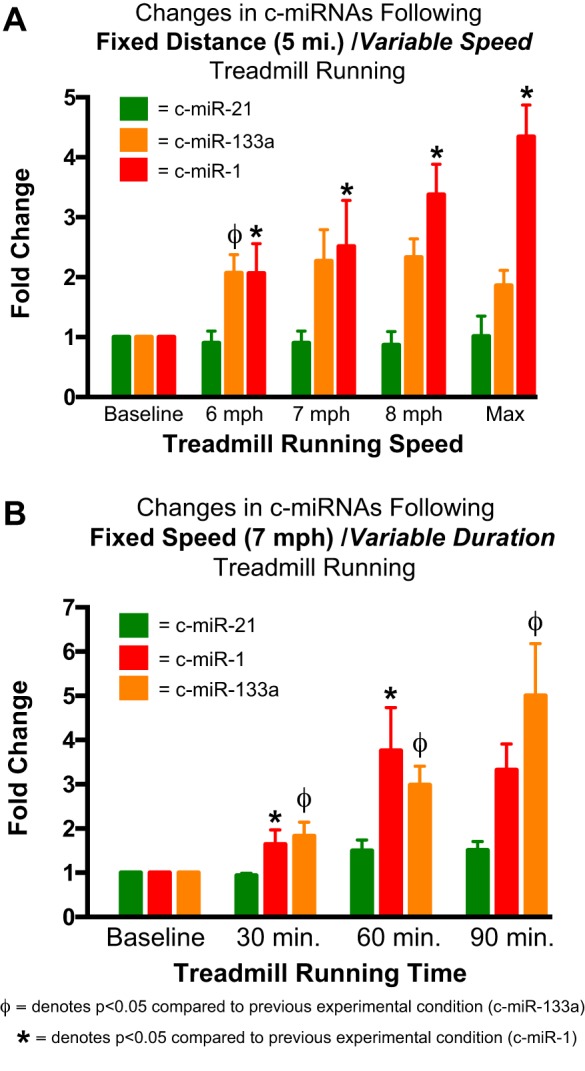
Comparison of the dose responsiveness of circulating microRNA (c-miR)-1 and c-miR-133a after variable intensity and duration of exercise. Changes in selected plasma concentrations of c-miRNAs (c-miR-1, c-miR-133a, and c-miR-21) are shown in response to fixed distance/variable intensity (A) and fixed intensity/variable duration (B) treadmill running among young healthy men. c-miR-21 demonstrated no significant response to exercise. c-miR-1 was upregulated by treadmill running under both conditions but demonstrated dose responsiveness to varying exercise intensity (A), whereas c-miR-133a was upregulated under both conditions but demonstrated dose responsiveness to varying exercise duration (B). Data represent fold changes of c-miRNA expression postexercise compared with each subject’s level before exercise. Data are expressed as means ± SE. * and ΦP < 0.05 compared with the previous experimental condition, as defined by mixed linear modeling with post hoc pairwise comparisons of variables and using least-squares means derived from the mixed-effects models performed with the Bonferroni correction.
High-intensity exercise training in mice increased plasma c-miR-133a, decreased intracellular miR-133a in skeletal muscle, and increased skeletal muscle expression of the known miR-133a target gene SRF.
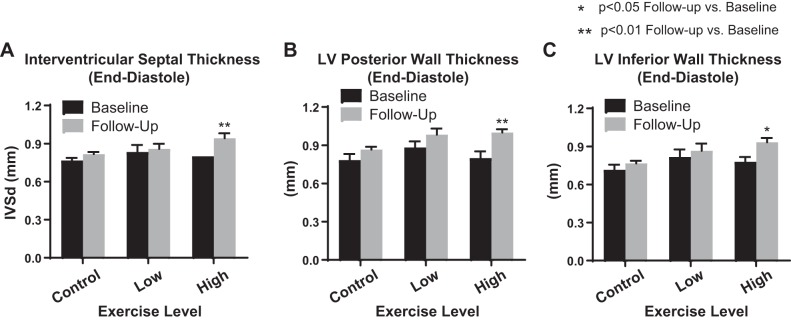
To determine whether the dose responsiveness of plasma miRNAs such as c-miR-133a may be functionally active in modulating gene expression in exercising muscle, we pursued proof-of-concept studies in aerobic exercise-trained mice to compare dynamic alterations of c-miR-133a with miRNA changes found in source muscle tissue, accompanied by its known target gene, SRF (15). To do so, an aerobic exercise training protocol in mice was performed to generate a functional “training” effect on exercise performance. Three mouse cohorts (n = 6 mice/cohort) were exposed to either a control (baseline activity), low-intensity (0.5 km/h), or high-intensity (1 km/h) running protocol on an open treadmill for 1 h/day for 5 consecutive weekdays over the course of 4 wk. As assessed by echocardiography at preexercise and postexercise training, interventricular septal thickness as well as left ventricular inferior/posterior wall thickness at end diastole were all increased in the high-intensity training group, consistent with the modest left ventricular hypertrophic response typically observed with this type of running protocol in mice (Fig. 4) (23). In that context, miR-133a expression levels in plasma were compared with intracellular skeletal muscle expression of this miRNA and its known target gene, SRF (Fig. 5). In contrast to miR-499, which displayed no significant response to this type of training, plasma c-miR-133a was specifically upregulated by high-intensity exercise training compared with control. A nonsignificant trend toward increased expression was observed in low-intensity exercise-trained mice. Conversely, high-intensity- but not low-intensity-trained mice displayed a decrease in intracellular miR-133a, but not miR-499, in skeletal muscle compared with control. Importantly, transcript levels of SRF were upregulated in response to high-intensity training, thus correlating with the notion of active gene regulatory reprogramming in exercising skeletal muscle, as intracellular miR-133a is released and depleted.
Fig. 4.
High-intensity exercise training promoted left ventricular (LV) hypertrophy in mice. A–C: echocardiography was performed pre- and postexercise training in mice to quantify interventricular septal thickness (IVSd) as well as LV inferior/posterior wall thickness at end diastole. Within the high-intensity training group (n = 6), IVSd was upregulated after exercise training compared with their own baseline values (A). Likewise, LV inferior and posterior wall diameters were upregulated after high-intensity training (B and C). However, neither the low-intensity training cohort (n = 6) nor the control cohort (n = 6) displayed significant alterations in LV thickness (A–C). Data presented are means ± SE. *P < 0.05 and **P < 0.01, significant change compared with each group’s own baseline mean (Student’s t-test).
Fig. 5.
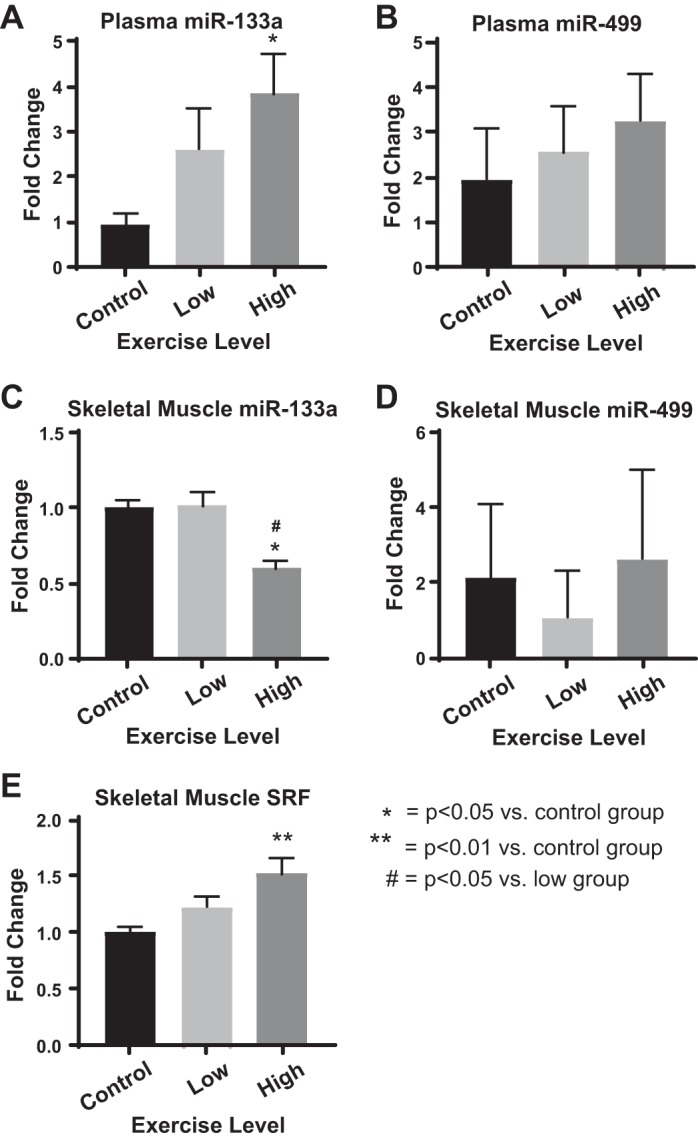
High-intensity training increased plasma miR-133a while promoting a decrease of intracellular skeletal muscle miR-133a and corresponding increase of the known skeletal muscle target gene of miR-133a, serum response factor (SRF). Expression levels in plasma and skeletal muscle of selected c-miRNAs (miR-133a and miR-499) and SRF mRNA transcript in skeletal muscle were assessed after aerobic exercise training (n = 6 mice/cohort) A: plasma circulating miRNA (c-miR)-133a expression was upregulated by high-intensity exercise training compared with control. A trend toward increased c-miR-133a after low-intensity exercise training was observed. B: in contrast, plasma c-miR-499 levels demonstrated no significant response to exercise training. C: intracellular miR-133a was downregulated in skeletal muscle by high- but not low-intensity exercise training compared with control. D: intracellular miR-499 demonstrated no significant response to training. E: correlating with decreased intracellular miR-133a, SRF, a validated target gene of miR-133a in skeletal muscle (15), was upregulated in response to the high-intensity training compared with control. Data are expressed as means ± SE. One-way ANOVA with Bonferroni post hoc testing was used for normally distributed data (A, C, and E). Kruskal-Wallis testing with Dunn’s multiple-comparison testing was used for nonnormally distributed data (B and D). *P > 0.05 and **P < 0.01 compared with control; #P < 0.05 compared with the low-intensity training cohort.
DISCUSSION
The popularity of aerobic and endurance exercise is steadily on the rise, as its importance in health and longevity becomes increasingly recognized. As recently summarized (59), habitual activity levels are a strong independent determinant of mortality, and physical activity guidelines exist to promote an optimal exercise dose (44a). However, the effects of different levels of intensity and duration are not understood, complicating the ability to evaluate the benefits and potential costs of different exercise doses. In addition, the molecular and consequent cardiac and muscle adaptations to specific exercise dose, as determined by the triple product of exercise intensity, duration, and frequency, have yet to be elucidated. In that context, although miRNAs are emerging as potential key mediators of exercise adaptation, specific thresholds of intensity and/or duration that are required for cardiac-specific and muscle-specific c-miRNA upregulation had not yet been rigorously assessed until now. The results from this study suggest that c-miRNAs demonstrate heterogeneous and molecule-specific responses to variable intensity and duration treadmill running. We identified specific c-miRNAs that display either threshold kinetics or progressive dose responsiveness to exercise intensity and duration, thereby supporting the notion that certain miRNAs may be exquisitely regulated to control physiological responses to exercise dose. Furthermore, by extending these human studies to mouse experimentation, we found that muscle release of one of these dose-responsive miRNAs, miR-133a, may reflect an active process to deplete intracellular miR-133a, thus removing its inhibitory effect and facilitating gene reprogramming and exercise adaptation. Thus, these findings represent an important step forward in revealing the crucial role of miRNA release in exercising muscle, thus facilitating the realistic development of these circulating molecules as biomarkers of cardiovascular adaptation and as pharmacological targets for modulating functional capacity.
Despite the recognition of a specific and dynamic regulatory program governing c-miRNAs in a variety of aerobic modalities (3, 4, 40, 46), dose-dependent c-miRNA expression has not previously been defined rigorously. A particularly intriguing finding in this study was the distinctly different responses of the tandem muscle-enriched miRNAs c-miR-1 and c-miR-133a, which are typically coregulated in a pairwise fashion as bicistronic transcripts but which we found to respond differently to variations in exercise intensity and duration. Specifically, c-miR-1 demonstrated progressive upregulation in response to increasing exercise intensity, whereas c-miR-133a demonstrated similar dose responsiveness to increasing exercise duration. The exact molecular explanation underlying these different regulatory expression profiles remains unclear. As previously reviewed (42), the myogenic factors myogenin and MyoD as well as the ERK1/2 signaling pathway are well-established positive regulators of both miRNAs. Since these regulatory molecules are differentially expressed in exercising muscle (37, 61), it is possible that these same molecules could play a role in the divergent expression of these miRNAs with modality of exercise dose in muscle and in plasma.
Furthermore, in light of these powerful miRNA-dependent actions, coupled with our observation in trained mice of higher extracellular and lower intracellular miR-133a along with a concomitant increase of miR-133a target gene SRF expression (Fig. 5), an attractive model of c-miRNA biology now emerges whereby the divergent c-miR-1 and c-miR-133a extracellular profiles likely reflect substantial functional differences in the intracellular levels of these molecules in source tissue in the context of exercise. Although microparticle release of c-miRNAs may allow for greater stability and uptake into recipient tissue, our data suggest that paracrine or endocrine messaging is not the only biological reason for c-miRNA release in exercise. Rather, we have proposed in other physiological conditions such as hypoxia (27) that rapid downregulation of intracellular miRNA content can be achieved by release of c-miRNAs, and this can serve as a key stimulus for fast “on/off” gene regulatory switches. Consistent with that notion, miR-1 and miR-133 may be released from exercising muscle tissue in a dose-dependent fashion, thus potentially promoting gradations of hypertrophic signaling and reprogramming dictated by quantitative declines of intracellular miRNA levels. This idea brings a new level of complexity to the known biology of these miRNAs, which carry critical roles in cardiac myogenesis and skeletal muscle development (42). Although they collaborate together early in myogenic development, later these miRNAs often have opposing and homeostatic molecular roles in differentiation and dedifferentiation of skeletal and cardiac muscle. Both miRNAs regulate distinct downstream target genes, but intracellular miR-133a (12) and miR-1 (47) both robustly suppress muscle and cardiac hypertrophy and are known to be downregulated in pathological contexts of hypertrophic disease. Intracellular downregulation of these miRNAs may have a similar role in modulating exercise-induced muscle hypertrophy, and the correlative increase of SRF muscle expression in trained mice (Fig. 5) supports that notion. Future work should be designed to interrogate this model further, specifically aimed at direct tracking of these molecules from the intracellular to extracellular space in exercising animal models and thus determining whether progressive alterations of intracellular miR-1 and miR-133 are dependent on exercise dose. If so, the progressive yet distinct alterations of these c-miRNAs may reflect a unique miR-dependent muscle remodeling program driven not just by exercise but more specifically by exercise intensity and duration with significantly different functional implications.
Similarly to c-miR-133a, c-miR-222 displayed dose responsiveness to variable exercise duration but not intensity. The functional consequences of these physiological profiles on exercise adaptation are not yet known. Notably, miR-222 was recently reported to control cardiac growth and proliferation in response to multiple forms of exercise via modulating the downstream cardiac genes homeodomain-interacting protein kinase 1 and homeobox-containing protein 1 (39). Interestingly, in its circulating form, c-miR-222 levels were found to differ among endurance- and resistance-trained athletes (58), increasing with endurance activity consistent with our findings (Fig. 3) but decreasing with strength training. c-miR-222 levels also were found to be higher in healthy adults with low versus high V̇o2max (11). Consequently, evidence is mounting to indicate the distinct activity of this c-miRNA in specific scenarios of exercise and other physiological states. Our findings implicating the context-specific dose responsiveness of c-miR-222 reinforce that notion and, coupled with the distinct cardiac actions of miR-222 already known in aerobic exercise (39), further promote the possibility of this molecule serving as a unique noninvasive marker of aerobic exercise dose.
A final notable observation was the responsiveness to exercise threshold of certain miRNAs such as c-miR-24 and 146a to variable duration but not intensity examined in this study. Although other miRNAs such as c-miR-21 and c-miR-210 displayed no response to either variable intensity or duration under the same parameters in this study, the existence of an activation threshold for these miRNAs may be extrapolated, since previous studies have reported dynamic changes in the expression of these molecules in response to comparatively longer-duration endurance exercise (4, 27, 40), such as marathon running. Thus, in aggregate, although the threshold set point may differ among miRNAs, these results suggest that these and perhaps other c-miRNAs exhibit some level of threshold for activation dependent upon exercise duration. Given the additional insight that concomitant statin use, which is known to have myotoxic effects in certain individuals, can further increase release of certain miRNAs after a marathon run (40), a reasonable hypothesis contends that at least some component of the release of these miRNAs may be driven more by muscle and tissue strain or injury with long periods of exercise rather than more adaptive pathways that are activated with different kinetics during shorter bouts of exertion. Alternatively, since a number of c-miRNAs are released in microparticles (19), a threshold effect could be governed by the microparticle assembly time necessary for packaging specific miRNAs.
Several limitations of this study merit consideration. First, regarding the study design of both the human and mouse studies, we chose to focus on young healthy male subjects, and thus our data may have limited generalizability to other populations that differ by age and health status. Second, the relatively small variable intensity (n = 12) and variable duration (n = 14) protocols in humans were comprised of different cohorts but were otherwise well matched, a fact particularly evident when comparing baseline c-miRNA levels among these participants (Fig. 1). In addition, our longitudinal and repeated-measures study design coupled with randomization of exercise intensity and duration within each protocol should have minimized the impact of any confounding variables introduced by the use of different subjects in each protocol. Third, it is important to emphasize that the rodent exercise protocol in this study was chosen to assess more directly the notion of intracellular depletion of muscle-specific miR-133a in the context of high levels of c-miR-133a seen with a variety of exercise modalities. It was not selected as an exact surrogate for the more intricate human studies, and we acknowledge that the protocols of variable intensity of exercise training that we used in mice do not fully parallel the intensity and duration of short-term exercise doses used in the human subjects. Moreover, in contrast to these rodent experiments, our group did not previously find alterations of c-miR-133a expression after 90 days of rowing training in humans (3), a discrepancy that may further highlight a substantial difference in adaptation to specific exercise protocols and/or animal species. Consistent with that notion, it is known that forced treadmill exercise training in mice likely does not recapitulate all aspects of aerobic training in humans (23), and some training effects in mice have been less reproducible with forced treadmill running as compared with voluntary training (34). Yet despite those issues, the dose sensitivity of c-miR-133a seen in this study across humans and mice when using different exercise intensities in both short-term bouts and long-term training underscores the functional relevance of exercise dose dependence in miRNA biology.
Regarding more technical issues of miRNA analysis, in an attempt to achieve maximum measurement accuracy, we chose to study a limited number of candidate miRNAs rather than perform a more comprehensive high-throughput c-miRNA screen. As such, our findings set the stage for more complete profiling of c-miRNAs that display exercise dose responsiveness. Additionally, although we undertook substantial effort to ensure accuracy in c-miRNA quantitation, we acknowledge that there still is a lack of standardized methodology in this field (33, 44, 49). Although future work will be necessary to ensure comparability in general across independent studies, we are nonetheless confident in the accuracy of the results presented herein, given the consistency of c-miRNAs levels across different individuals and exercise modalities (Fig. 1), the specificity of c-miRNA alterations (i.e., detection of c-miRNAs that are either responsive or entirely unresponsive to exercise protocol; Fig. 2, A–D), and the detection of changes in muscle-specific c-miRNAs that are not subject to confounding issues of hemolysis (Fig. 2, E–G). Furthermore, it is important to note that although some c-miRNAs are known to be muscle specific, such as miR-1 and miR-133a, the exact tissue source of c-miRNA release is not definitively known; in the future, more intricate miRNA labeling experiments during exercise will be necessary to achieve that insight. Finally, in this proof-of-concept study, we did not couple c-miRNA expression directly to any of the fundamental phenotypic exercise adaptations in humans and anticipate this as an important future goal.
In conclusion, by studying exercise physiology across humans and mice, we demonstrate that c-miRNA expression in response to aerobic exercise varies across c-miRNA molecules and in some cases follows distinct dose-responsive profiles. Such dose-dependent release of c-miRNAs from muscle may result in the depletion of intracellular stores of muscle-specific miRNAs, thus releasing their inhibitory effects on target gene expression and facilitating muscle adaptation to exercise. This dose dependence offers greater insights into the precision and complexity of the regulatory processes inherent in c-miRNA expression in the plasma. Furthermore, the exact coupling of exercise dose and specific c-miRNA regulation suggests a critical need for future work designed to define their role as quantitative mediators and biomarkers of exercise-induced adaptation, exercise capacity, and overall health.
GRANTS
This work was supported by National Institutes of Health Grants (NIH) R01-HL-124021, HL-122596, HL-138437, and UH2-TR-002073 (to S. Y. Chan) and R01-HL-125869 (to A. L. Baggish), a UPMC Department of Pathology Klionsky Summer Research Fellowship (to A. E. Ramos), the Hintze Family Charitable Foundation (to D. E. Lieberman), and the American Heart Association Undergraduate Student Summer Fellowship Program (to C. Lo).
DISCLOSURES
S.Y.C. has served as a consultant for Actelion (Significant), Gilead, Pfizer, and Vivus (Modest). The authors declare no other conflicts of interest.
AUTHOR CONTRIBUTIONS
A.E.R., S.Y.C., and A.B. conceived and designed research; A.E.R., C.L., L.E., Y.-Y.T., Y.T., J.Z., M.S., J.G., M.G.B., and S.Y.C. performed experiments; A.E.R., C.L., L.E., Y.-Y.T., Y.T., J.Z., M.S., J.G., M.G.B., D.E.L., S.Y.C., and A.B. analyzed data; A.E.R., C.L., L.E., Y.-Y.T., Y.T., J.Z., M.S., J.G., M.G.B., D.E.L., S.Y.C., and A.B. interpreted results of experiments; A.E.R., L.E., Y.-Y.T., S.Y.C., and A.B. prepared figures; A.E.R., L.E., Y.-Y.T., D.E.L., S.Y.C., and A.B. drafted manuscript; A.E.R., C.L., L.E., Y.-Y.T., Y.T., J.Z., M.S., J.G., M.G.B., D.E.L., S.Y.C., and A.B. edited and revised manuscript; A.E.R., C.L., L.E., Y.-Y.T., Y.T., J.Z., M.S., J.G., M.G.B., D.E.L., S.Y.C., and A.B. approved final version of manuscript.
ACKNOWLEDGMENTS
We thank Q. Yu, V. Negi, M. Culley, and N. Kelly for fruitful discussions.
REFERENCES
- 1.Ambros V. The functions of animal microRNAs. Nature 431: 350–355, 2004. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 2.Aoi W, Ichikawa H, Mune K, Tanimura Y, Mizushima K, Naito Y, Yoshikawa T. Muscle-enriched microRNA miR-486 decreases in circulation in response to exercise in young men. Front Physiol 4: 80, 2013. doi: 10.3389/fphys.2013.00080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baggish AL, Hale A, Weiner RB, Lewis GD, Systrom D, Wang F, Wang TJ, Chan SY. Dynamic regulation of circulating microRNA during acute exhaustive exercise and sustained aerobic exercise training. J Physiol 589: 3983–3994, 2011. doi: 10.1113/jphysiol.2011.213363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baggish AL, Park J, Min PK, Isaacs S, Parker BA, Thompson PD, Troyanos C, D’Hemecourt P, Dyer S, Thiel M, Hale A, Chan SY. Rapid upregulation and clearance of distinct circulating microRNAs after prolonged aerobic exercise. J Appl Physiol 116: 522–531, 2014. doi: 10.1152/japplphysiol.01141.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baggish AL, Wang F, Weiner RB, Elinoff JM, Tournoux F, Boland A, Picard MH, Hutter AM Jr, Wood MJ. Training-specific changes in cardiac structure and function: a prospective and longitudinal assessment of competitive athletes. J Appl Physiol 104: 1121–1128, 2008. doi: 10.1152/japplphysiol.01170.2007. [DOI] [PubMed] [Google Scholar]
- 6.Banzet S, Chennaoui M, Girard O, Racinais S, Drogou C, Chalabi H, Koulmann N. Changes in circulating microRNAs levels with exercise modality. J Appl Physiol 115: 1237–1244, 2013. doi: 10.1152/japplphysiol.00075.2013. [DOI] [PubMed] [Google Scholar]
- 7.Beaver WL, Wasserman K, Whipp BJ. A new method for detecting anaerobic threshold by gas exchange. J Appl Physiol 60: 2020–2027, 1986. doi: 10.1152/jappl.1986.60.6.2020. [DOI] [PubMed] [Google Scholar]
- 8.Bertero T, Cottrill KA, Lu Y, Haeger CM, Dieffenbach P, Annis S, Hale A, Bhat B, Kaimal V, Zhang YY, Graham BB, Kumar R, Saggar R, Saggar R, Wallace WD, Ross DJ, Black SM, Fratz S, Fineman JR, Vargas SO, Haley KJ, Waxman AB, Chau BN, Fredenburgh LE, Chan SY. Matrix remodeling promotes pulmonary hypertension through feedback mechanoactivation of the YAP/TAZ-miR-130/301 circuit. Cell Rep 13: 1016–1032, 2015. doi: 10.1016/j.celrep.2015.09.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bertero T, Lu Y, Annis S, Hale A, Bhat B, Saggar R, Saggar R, Wallace WD, Ross DJ, Vargas SO, Graham BB, Kumar R, Black SM, Fratz S, Fineman JR, West JD, Haley KJ, Waxman AB, Chau BN, Cottrill KA, Chan SY. Systems-level regulation of microRNA networks by miR-130/301 promotes pulmonary hypertension. J Clin Invest 124: 3514–3528, 2014. doi: 10.1172/JCI74773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bertero T, Oldham WM, Cottrill KA, Pisano S, Vanderpool RR, Yu Q, Zhao J, Tai Y, Tang Y, Zhang YY, Rehman S, Sugahara M, Qi Z, Gorcsan J III, Vargas SO, Saggar R, Saggar R, Wallace WD, Ross DJ, Haley KJ, Waxman AB, Parikh VN, De Marco T, Hsue PY, Morris A, Simon MA, Norris KA, Gaggioli C, Loscalzo J, Fessel J, Chan SY. Vascular stiffness mechanoactivates YAP/TAZ-dependent glutaminolysis to drive pulmonary hypertension. J Clin Invest 126: 3313–3335, 2016. doi: 10.1172/JCI86387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bye A, Røsjø H, Aspenes ST, Condorelli G, Omland T, Wisløff U. Circulating microRNAs and aerobic fitness−the HUNT-Study. PLoS One 8: e57496, 2013. doi: 10.1371/journal.pone.0057496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Carè A, Catalucci D, Felicetti F, Bonci D, Addario A, Gallo P, Bang ML, Segnalini P, Gu Y, Dalton ND, Elia L, Latronico MV, Høydal M, Autore C, Russo MA, Dorn GW II, Ellingsen O, Ruiz-Lozano P, Peterson KL, Croce CM, Peschle C, Condorelli G. MicroRNA-133 controls cardiac hypertrophy. Nat Med 13: 613–618, 2007. doi: 10.1038/nm1582. [DOI] [PubMed] [Google Scholar]
- 13.Chan SY, Loscalzo J. MicroRNA-210: a unique and pleiotropic hypoxamir. Cell Cycle 9: 1072–1083, 2010. doi: 10.4161/cc.9.6.11006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chan SY, Zhang YY, Hemann C, Mahoney CE, Zweier JL, Loscalzo J. MicroRNA-210 controls mitochondrial metabolism during hypoxia by repressing the iron-sulfur cluster assembly proteins ISCU1/2. Cell Metab 10: 273–284, 2009. doi: 10.1016/j.cmet.2009.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen JF, Mandel EM, Thomson JM, Wu Q, Callis TE, Hammond SM, Conlon FL, Wang DZ. The role of microRNA-1 and microRNA-133 in skeletal muscle proliferation and differentiation. Nat Genet 38: 228–233, 2006. doi: 10.1038/ng1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cheng Y, Zhu P, Yang J, Liu X, Dong S, Wang X, Chun B, Zhuang J, Zhang C. Ischaemic preconditioning-regulated miR-21 protects heart against ischaemia/reperfusion injury via anti-apoptosis through its target PDCD4. Cardiovasc Res 87: 431–439, 2010. doi: 10.1093/cvr/cvq082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Clauss S, Wakili R, Hildebrand B, Kääb S, Hoster E, Klier I, Martens E, Hanley A, Hanssen H, Halle M, Nickel T. MicroRNAs as biomarkers for acute atrial remodeling in marathon runners (the miRathon Study−a sub-study of the Munich Marathon Study). PLoS One 11: e0148599, 2016. doi: 10.1371/journal.pone.0148599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Creemers EE, Tijsen AJ, Pinto YM. Circulating microRNAs: novel biomarkers and extracellular communicators in cardiovascular disease? Circ Res 110: 483–495, 2012. doi: 10.1161/CIRCRESAHA.111.247452. [DOI] [PubMed] [Google Scholar]
- 20.Cui SF, Wang C, Yin X, Tian D, Lu QJ, Zhang CY, Chen X, Ma JZ. Similar responses of circulating micrornas to acute high-intensity interval exercise and vigorous-intensity continuous exercise. Front Physiol 7: 102, 2016. doi: 10.3389/fphys.2016.00102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.de Gonzalo-Calvo D, Dávalos A, Montero A, García-González Á, Tyshkovska I, González-Medina A, Soares SM, Martínez-Camblor P, Casas-Agustench P, Rabadán M, Díaz-Martínez AE, Úbeda N, Iglesias-Gutiérrez E. Circulating inflammatory miRNA signature in response to different doses of aerobic exercise. J Appl Physiol 119: 124–134, 2015. doi: 10.1152/japplphysiol.00077.2015. [DOI] [PubMed] [Google Scholar]
- 22.Denham J, Prestes PR. Muscle-enriched microRNAs Isolated from whole blood are regulated by exercise and are potential biomarkers of cardiorespiratory fitness. Front Genet 7: 196, 2016. doi: 10.3389/fgene.2016.00196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Doevendans PA, Daemen MJ, de Muinck ED, Smits JF. Cardiovascular phenotyping in mice. Cardiovasc Res 39: 34–49, 1998. doi: 10.1016/S0008-6363(98)00073-X. [DOI] [PubMed] [Google Scholar]
- 24.Fasanaro P, D’Alessandra Y, Di Stefano V, Melchionna R, Romani S, Pompilio G, Capogrossi MC, Martelli F. MicroRNA-210 modulates endothelial cell response to hypoxia and inhibits the receptor tyrosine kinase ligand ephrin-A3. J Biol Chem 283: 15878–15883, 2008. doi: 10.1074/jbc.M800731200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fewell JG, Osinska H, Klevitsky R, Ng W, Sfyris G, Bahrehmand F, Robbins J. A treadmill exercise regimen for identifying cardiovascular phenotypes in transgenic mice. Am J Physiol Heart Circ Physiol 273: H1595–H1605, 1997. [DOI] [PubMed] [Google Scholar]
- 26.Gute D, Fraga C, Laughlin MH, Amann JF. Regional changes in capillary supply in skeletal muscle of high-intensity endurance-trained rats. J Appl Physiol 81: 619–626, 1996. doi: 10.1152/jappl.1996.81.2.619. [DOI] [PubMed] [Google Scholar]
- 27.Hale A, Lee C, Annis S, Min PK, Pande R, Creager MA, Julian CG, Moore LG, Mitsialis SA, Hwang SJ, Kourembanas S, Chan SY. An Argonaute 2 switch regulates circulating miR-210 to coordinate hypoxic adaptation across cells. Biochim Biophys Acta 1843: 2528–2542, 2014. doi: 10.1016/j.bbamcr.2014.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huang W, Tian SS, Hang PZ, Sun C, Guo J, Du ZM. Combination of microRNA-21 and microRNA-146a attenuates cardiac dysfunction and apoptosis during acute myocardial infarction in mice. Mol Ther Nucleic Acids 5: e296, 2016. doi: 10.1038/mtna.2016.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Iliopoulos D, Jaeger SA, Hirsch HA, Bulyk ML, Struhl K. STAT3 activation of miR-21 and miR-181b-1 via PTEN and CYLD are part of the epigenetic switch linking inflammation to cancer. Mol Cell 39: 493–506, 2010. doi: 10.1016/j.molcel.2010.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jones NL, Makrides L, Hitchcock C, Chypchar T, McCartney N. Normal standards for an incremental progressive cycle ergometer test. Am Rev Respir Dis 131: 700–708, 1985. [DOI] [PubMed] [Google Scholar]
- 31.Kiessling KH, Pilström L, Karlsson J, Piehl K. Mitochondrial volume in skeletal muscle from young and old physically untrained and trained healthy men and from alcoholics. Clin Sci 44: 547–554, 1973. doi: 10.1042/cs0440547. [DOI] [PubMed] [Google Scholar]
- 32.Kilian Y, Wehmeier UF, Wahl P, Mester J, Hilberg T, Sperlich B. Acute response of circulating vascular regulating microRNAs during and after high-intensity and high-volume cycling in children. Front Physiol 7: 92, 2016. doi: 10.3389/fphys.2016.00092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kirschner MB, Edelman JJ, Kao SC, Vallely MP, van Zandwijk N, Reid G. The impact of hemolysis on cell-free microRNA biomarkers. Front Genet 4: 94, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Knab AM, Bowen RS, Moore-Harrison T, Hamilton AT, Turner MJ, Lightfoot JT. Repeatability of exercise behaviors in mice. Physiol Behav 98: 433–440, 2009. doi: 10.1016/j.physbeh.2009.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Krichevsky AM, Gabriely G. miR-21: a small multi-faceted RNA. J Cell Mol Med 13: 39–53, 2009. doi: 10.1111/j.1582-4934.2008.00556.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kuehbacher A, Urbich C, Zeiher AM, Dimmeler S. Role of Dicer and Drosha for endothelial microRNA expression and angiogenesis. Circ Res 101: 59–68, 2007. doi: 10.1161/CIRCRESAHA.107.153916. [DOI] [PubMed] [Google Scholar]
- 37.Legerlotz K, Smith HK. Role of MyoD in denervated, disused, and exercised muscle. Muscle Nerve 38: 1087–1100, 2008. doi: 10.1002/mus.21087. [DOI] [PubMed] [Google Scholar]
- 38.Liu X, Platt C, Rosenzweig A. The Role of microRNAs in the cardiac response to exercise. Cold Spring Harb Perspect Med 7: a029850, 2017. doi: 10.1101/cshperspect.a029850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu X, Xiao J, Zhu H, Wei X, Platt C, Damilano F, Xiao C, Bezzerides V, Boström P, Che L, Zhang C, Spiegelman BM, Rosenzweig A. miR-222 is necessary for exercise-induced cardiac growth and protects against pathological cardiac remodeling. Cell Metab 21: 584–595, 2015. doi: 10.1016/j.cmet.2015.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Min PK, Park J, Isaacs S, Taylor BA, Thompson PD, Troyanos C, D’Hemecourt P, Dyer S, Chan SY, Baggish AL. Influence of statins on distinct circulating microRNAs during prolonged aerobic exercise. J Appl Physiol 120: 711–720, 2016. doi: 10.1152/japplphysiol.00654.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mitchell PS, Parkin RK, Kroh EM, Fritz BR, Wyman SK, Pogosova-Agadjanyan EL, Peterson A, Noteboom J, O’Briant KC, Allen A, Lin DW, Urban N, Drescher CW, Knudsen BS, Stirewalt DL, Gentleman R, Vessella RL, Nelson PS, Martin DB, Tewari M. Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci USA 105: 10513–10518, 2008. doi: 10.1073/pnas.0804549105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mitchelson KR, Qin WY. Roles of the canonical myomiRs miR-1, -133 and -206 in cell development and disease. World J Biol Chem 6: 162–208, 2015. doi: 10.4331/wjbc.v6.i3.162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mooren FC, Viereck J, Krüger K, Thum T. Circulating microRNAs as potential biomarkers of aerobic exercise capacity. Am J Physiol Heart Circ Physiol 306: H557–H563, 2014. doi: 10.1152/ajpheart.00711.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nielsen S, Åkerström T, Rinnov A, Yfanti C, Scheele C, Pedersen BK, Laye MJ. The miRNA plasma signature in response to acute aerobic exercise and endurance training. PLoS One 9: e87308, 2014. doi: 10.1371/journal.pone.0087308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44a.Physical Activity Guidelines Advisory Committee Physical Activity Guidelines Advisory Committee Report. Washington, DC: United States Department of Health and Human Services, 2008. [Google Scholar]
- 45.Poliseno L, Tuccoli A, Mariani L, Evangelista M, Citti L, Woods K, Mercatanti A, Hammond S, Rainaldi G. MicroRNAs modulate the angiogenic properties of HUVECs. Blood 108: 3068–3071, 2006. doi: 10.1182/blood-2006-01-012369. [DOI] [PubMed] [Google Scholar]
- 46.Sawada S, Kon M, Wada S, Ushida T, Suzuki K, Akimoto T. Profiling of circulating microRNAs after a bout of acute resistance exercise in humans. PLoS One 8: e70823, 2013. doi: 10.1371/journal.pone.0070823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sayed D, Hong C, Chen IY, Lypowy J, Abdellatif M. MicroRNAs play an essential role in the development of cardiac hypertrophy. Circ Res 100: 416–424, 2007. doi: 10.1161/01.RES.0000257913.42552.23. [DOI] [PubMed] [Google Scholar]
- 48.Scharhag J, Herrmann M, Urhausen A, Haschke M, Herrmann W, Kindermann W. Independent elevations of N-terminal pro-brain natriuretic peptide and cardiac troponins in endurance athletes after prolonged strenuous exercise. Am Heart J 150: 1128–1134, 2005. doi: 10.1016/j.ahj.2005.01.051. [DOI] [PubMed] [Google Scholar]
- 49.Shah JS, Soon PS, Marsh DJ. Comparison of methodologies to detect low levels of hemolysis in serum for accurate assessment of serum microRNAs. PLoS One 11: e0153200, 2016. doi: 10.1371/journal.pone.0153200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shave R, Baggish A, George K, Wood M, Scharhag J, Whyte G, Gaze D, Thompson PD. Exercise-induced cardiac troponin elevation: evidence, mechanisms, and implications. J Am Coll Cardiol 56: 169–176, 2010. doi: 10.1016/j.jacc.2010.03.037. [DOI] [PubMed] [Google Scholar]
- 51.Suárez Y, Fernández-Hernando C, Pober JS, Sessa WC. Dicer dependent microRNAs regulate gene expression and functions in human endothelial cells. Circ Res 100: 1164–1173, 2007. doi: 10.1161/01.RES.0000265065.26744.17. [DOI] [PubMed] [Google Scholar]
- 52.Taganov KD, Boldin MP, Chang KJ, Baltimore D. NF-κB-dependent induction of microRNA miR-146, an inhibitor targeted to signaling proteins of innate immune responses. Proc Natl Acad Sci USA 103: 12481–12486, 2006. doi: 10.1073/pnas.0605298103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Thum T, Gross C, Fiedler J, Fischer T, Kissler S, Bussen M, Galuppo P, Just S, Rottbauer W, Frantz S, Castoldi M, Soutschek J, Koteliansky V, Rosenwald A, Basson MA, Licht JD, Pena JT, Rouhanifard SH, Muckenthaler MU, Tuschl T, Martin GR, Bauersachs J, Engelhardt S. MicroRNA-21 contributes to myocardial disease by stimulating MAP kinase signalling in fibroblasts. Nature 456: 980–984, 2008. doi: 10.1038/nature07511. [DOI] [PubMed] [Google Scholar]
- 54.Uhlemann M, Möbius-Winkler S, Fikenzer S, Adam J, Redlich M, Möhlenkamp S, Hilberg T, Schuler GC, Adams V. Circulating microRNA-126 increases after different forms of endurance exercise in healthy adults. Eur J Prev Cardiol 21: 484–491, 2014. doi: 10.1177/2047487312467902. [DOI] [PubMed] [Google Scholar]
- 55.Urbich C, Kuehbacher A, Dimmeler S. Role of microRNAs in vascular diseases, inflammation, and angiogenesis. Cardiovasc Res 79: 581–588, 2008. doi: 10.1093/cvr/cvn156. [DOI] [PubMed] [Google Scholar]
- 56.van Rooij E, Sutherland LB, Liu N, Williams AH, McAnally J, Gerard RD, Richardson JA, Olson EN. A signature pattern of stress-responsive microRNAs that can evoke cardiac hypertrophy and heart failure. Proc Natl Acad Sci USA 103: 18255–18260, 2006. doi: 10.1073/pnas.0608791103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wahl P, Wehmeier UF, Jansen FJ, Kilian Y, Bloch W, Werner N, Mester J, Hilberg T. Acute effects of different exercise protocols on the circulating vascular microRNAs -16, -21, and -126 in trained subjects. Front Physiol 7: 643, 2016. doi: 10.3389/fphys.2016.00643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wardle SL, Bailey ME, Kilikevicius A, Malkova D, Wilson RH, Venckunas T, Moran CN. Plasma microRNA levels differ between endurance and strength athletes. PLoS One 10: e0122107, 2015. doi: 10.1371/journal.pone.0122107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wasfy MM, Baggish AL. Exercise dose in clinical practice. Circulation 133: 2297–2313, 2016. doi: 10.1161/CIRCULATIONAHA.116.018093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Williams AH, Liu N, van Rooij E, Olson EN. MicroRNA control of muscle development and disease. Curr Opin Cell Biol 21: 461–469, 2009. doi: 10.1016/j.ceb.2009.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zanou N, Gailly P. Skeletal muscle hypertrophy and regeneration: interplay between the myogenic regulatory factors (MRFs) and insulin-like growth factors (IGFs) pathways. Cell Mol Life Sci 70: 4117–4130, 2013. doi: 10.1007/s00018-013-1330-4. [DOI] [PMC free article] [PubMed] [Google Scholar]