Abstract
Evidence shows that proteins secreted from skeletal muscle influence a broad range of metabolic signaling pathways. We previously reported that essential polyunsaturated fatty acids (PUFA) improved whole-body glucose homeostasis in obese Zucker rats; however, the mechanisms underlying these benefits remain enigmatic. While PUFA and obesity influence skeletal muscle function, their effects on the secretome are unknown. The aim of this work was to determine if improvements in whole-body glucose homeostasis in obese Zucker rats fed diets supplemented with either linoleic acid (LA) or alpha-linolenic acid (ALA) for 12 wk are related to changes in the skeletal muscle secretome. Secreted proteins were identified with a predictive bioinformatic analysis of microarray gene expression from red tibialis anterior skeletal muscle. Approximately 130 genes were differentially expressed (false discovery rate = 0.05) in obese rats compared with lean controls. The expression of 15 genes encoding secreted proteins was differentially regulated in obese controls, obese LA-supplemented, and obese ALA-supplemented rats compared with lean controls. Five secreted proteins (Col3a1, Col15a1, Pdgfd, Lyz2, and Angptl4) were differentially regulated by LA and ALA. Most notably, ALA supplementation reduced Angptl4 gene expression compared with obese control and obese-LA supplemented rats and reduced circulating ANGPTL4 serum concentrations. ALA also influenced Angptl4 gene expression and ANGPTL4 secretion from differentiated rat L6 myotubes. Altogether, the present data indicate that obesity has a greater global impact on skeletal muscle gene expression than either essential PUFA; however, LA and ALA may exert their metabolic benefits in part by regulating the skeletal muscle secretome.
Keywords: alpha-linolenic acid, linoleic acid, microarray, obesity, skeletal muscle
BACKGROUND
Skeletal muscle is a key metabolic tissue with a critical role in whole-body fat oxidation and insulin-stimulated glucose clearance (11, 16). With obesity, skeletal muscle function is compromised leading to reduced fat oxidation (25) and glucose uptake (59). Consequently, skeletal muscle dysfunction is strongly implicated in the development of obesity-associated complications such as insulin resistance and dyslipidemia. While adipose tissue is well recognized to be an important endocrine organ through the secretion of adipokines (i.e., proteins secreted from adipose tissue), considerably less is known about the skeletal muscle secretome. Nevertheless, a growing body of evidence demonstrates that skeletal muscle is able to secrete proteins, termed myokines, that participate in tissue cross talk and play key roles in health and disease (14, 19, 58). Thus, investigating factors that regulate the skeletal muscle secretome promises to improve our understanding of the interaction between skeletal muscle and other tissues in the body, as well as identify new therapeutic targets.
In the secretion media of cultured primary human skeletal muscle cells, over 300 distinct proteins have been identified by a combination of microarray, proteomic, and bioinformatic tools (22). Proteins secreted from skeletal muscle have been proposed to regulate numerous metabolic signaling pathways in a paracrine and endocrine manner (2), including myogenesis (33), glucose uptake in myocytes (32), pancreatic β-cell function and glucose-stimulated insulin secretion (37), and lipid uptake in adipocytes and hepatocytes (47). Although the secretome associated with skeletal muscle dysfunction is less characterized than that of healthy skeletal muscle, many of the past studies investigating gene expression in obese and insulin-resistant skeletal muscle identified genes that encode secreted proteins (4, 23, 28, 38, 41, 48, 50, 51, 61), intimating that the skeletal muscle secretome may be altered with obesity and insulin resistance. Due to the involvement of the skeletal muscle secretome in the regulation of whole-body lipid and glucose homeostasis, a better understanding of how the expression and secretion of proteins are regulated, as well as how they are altered with metabolic disease, is needed.
Interestingly, there is some evidence in the literature that fatty acids that influence glucose tolerance can regulate myokine expression differently. For example, differentiated C2C12 myotubes treated with palmitate showed reduced expression of two myokines: irisin (Fndc5) and myonectin (60). In contrast, a combination of oleic acid and linoleic acid did not alter irisin expression in differentiated C2C12 myotubes (26). Recently, a small clinical trial revealed that supplementation with omega-3 fats improved glycemic status in diabetic patients and increased circulating irisin levels (5); however, it is unknown if the increase in irisin originated from skeletal muscle specifically. To date, no studies have examined how the essential dietary fats, linoleic acid (LA) and alpha-linolenic acid (ALA), regulate the skeletal muscle secretome.
We previously reported that essential polyunsaturated fatty acids (PUFA) improved whole-body glucose homeostasis in obese Zucker rats (34, 35); however, the mechanisms underlying these benefits remain to be elucidated. The goal of the present study was to investigate how obesity-associated insulin resistance with and without LA or ALA supplementation influenced the expression of genes encoding skeletal muscle secreted proteins. We hypothesized that essential PUFA supplementation would counter changes in the skeletal muscle secretome seen with obesity-associated insulin resistance.
METHODS
Animals
Housing, care, and sample collection.
Five-week-old male lean (n = 12) and obese (n = 36) Zucker rats were purchased from Charles River Laboratories. Animals were housed in a temperature-regulated room on a 12:12 h light-dark cycle with water available ad libitum. Lean and obese control animals were given unrestricted access to a control diet, while obese animals fed PUFA-supplemented diets were pair-fed to match for caloric intake of obese controls. After 12 wk, animals were anesthetized with a 60 mg/kg of pentobarbital sodium injection. Nonfasted blood was collected by cardiac puncture and serum was isolated by centrifugation at 4°C for 15 min at 1,500 g. Red tibialis anterior (TA) skeletal muscle was collected, flash-frozen in liquid nitrogen, and stored at −80°C until analysis. This study was approved by the University of Guelph Animal Care Committee.
Diets and feeding.
All diets were purchased from Research Diets (New Brunswick, NJ). Daily food consumption of obese rats fed the control diet (no. AIN-93G; 20% protein, 64% carbohydrate, and 16% fat) was recorded by weight to pair-feed rats given LA-supplemented (AIN-93G + 10% safflower oil; 20% protein, 54% carbohydrate, and 26% fat) and ALA-supplemented (AIN-93G + 10% flaxseed oil; 20% protein, 54% carbohydrate, and 26% fat) diets to ensure caloric consumption was similar across diets. Diet composition is provided in Supplemental Table S1. (The online version of this article contains supplemental material.) Fatty acid composition of the diets was confirmed by gas chromatography, as previously reported (35).
RNA extraction.
Total RNA was extracted from TA with Trizol and the Qiagen RNeasy Mini Kit (Mississauga, ON, Canada), according to manufacturer’s instructions. The quantity of total RNA was determined with a NanoDrop 2.0 (Fisher Scientific, Waltham, MA), and quality was assessed with an Agilent 2100 BioAnalyzer (Agilent Technologies, Santa Clara, CA). All samples had an RNA integrity number > 9.0. Total RNA was stored at −80°C until analysis.
Microarray analysis.
A total of eight microarrays per group were run for each of the following groups: lean control (LC), obese control (OC), obese-LA (O-LA) and obese-ALA (O-ALA). For each sample, 100 ng of total RNA was prepared for hybridization to Affymetrix Rat Gene 2.1 ST array strips, according to manufacturer’s instructions (Affymetrix, Fremont, CA). Microarray CEL files were loaded into R and analyzed with the oligo package (12). Quality assessment was performed on the raw data. and all microarrays were deemed high quality. The arrays were preprocessed by the robust multiarray average method (27). Probe sets were filtered to remove those without an annotation, leaving 20,258 transcript clusters corresponding to 19,540 unique Entrez GeneIDs. For genes represented by more than one probe set, the corresponding expression values were averaged so that each GeneID had only a single value for each sample. To determine which genes were differentially expressed among the four treatments, a one-way analysis of variance (one-way ANOVA) with a false discovery rate (FDR) correction of 5% was used. For genes identified as differentially expressed in the one-way ANOVA, a post hoc pairwise Student’s t-test analysis with an FDR correction of 5% was subsequently used. KEGG pathway analyses were performed with DAVID 6.8 Bioinformatic software (17). A pathway was considered significant if it had a Bonferroni adjusted P value of < 0.05. Microarray data have been deposited in the National Center for Biotechnology information Gene Expression Omnibus (GEO) and can be downloaded through GEO accession number GSE104567.
Prediction of secreted proteins.
The list of all differentially expressed GeneIDs was run through the Gene database in Batch Entrez (https://www.ncbi.nlm.nih.gov/sites/batchentrez) and subsequently through the RefSeq Proteins option of the Protein related database to retrieve corresponding protein IDs. The FASTA format of the resulting list of proteins was downloaded, and predicted protein sequences were removed, leaving only known protein sequences. This final list of protein FASTA sequences was then run through three signal peptide-prediction tools: Signal-BLAST, SignalP 4.1, and TOPCONS2. Signal-BLAST uses sequence alignment to compare a sequence of interest with reference data comprising curated secreted and nonsecreted proteins (21). SignalP 4.1 distinguishes signal peptide sequences from similar NH2-terminal transmembrane sequences (43). The sensitive setting was used to reduce the number of false negatives at the expense of receiving more false positives. TOPCONS2 predicts both signal peptide and transmembrane sequences (54). A final list of “potential secreted proteins” was established and corresponded to genes found to contain a signal peptide by Signal-BLAST, SignalP, and/or TOPCONS2. Each protein was then searched individually in the UniProt database (6) to confirm its secretion. In instances when little information was available in UniProt for rat proteins, we searched for their human orthologs.
Myotube Cell Culture
Chemicals and cell culture reagents.
Dulbecco’s modified Eagle’s medium (DMEM; catalog #SH3024301) and penicillin streptomycin (catalog #SV30010) were purchased from HyClone laboratories (Logan, UT). Fetal bovine serum and fatty acid-free bovine serum albumin (BSA) were purchased from Sigma Aldrich (St. Louis, MO). Horse serum was purchased from Fisher Scientific (Waltham, MA). Rat L6 myoblasts were purchased from ATCC (catalog #CRL-1458; Manassas, VA). LA and ALA were purchased from Cayman Chemical (Ann Arbor, MI).
Rat L6 myotube cell culture.
L6 rat myoblasts were seeded in six-well plates at a density of ~200,000 cells per well and incubated at 5% CO2 and 100% humidity at 37°C. Cells were maintained in DMEM supplemented with 10% FBS and 1% penicillin streptomycin for 4 days. Media were changed every 48 h. At 90% confluence, cells were incubated with differentiation medium comprised of DMEM, 2% horse serum and 1% penicillin-streptomycin. On day 8, media were switched to 1% horse serum before PUFA treatment. Differentiated L6 myotube cells were treated for 24 h with 2% BSA (control) or 50, 100, or 200 µM of LA or ALA.
RNA extraction.
Cells were lysed with RLT buffer, collected using cell scrapers (Fisher Scientific, Ottawa, ON, Canada), and passed through a 27 G × 1/2 inch needle before RNA was extracted using the RNeasy mini-kit (Qiagen).
Targeted Analysis of Secreted Proteins
Quantitative reverse-transcription PCR.
Primers were designed using the Roche online Universal Probe Library Assay Design Centre and sequence specificity was confirmed using NCBI Primer-BLAST. Total RNA (1 µg) from either L6 myotubes or TA was used to synthesize cDNA with the High-Capacity Reverse Transcription kit (Life Technologies, Burlington, ON, Canada). Analyses were performed with a Bio-Rad CFX-96 system using 0.025 µg cDNA and SsoFast Evagreen Supermix (Bio-Rad Laboratories, Mississauga, ON, Canada), as previously described (49). We used Rn18s as a housekeeping gene because it was not differentially expressed in our microarray data set and had Ct values with a coefficient of variance (%CV) <3% for both rat samples and L6 myoblasts. Data were analyzed with a one-way or two-way ANOVA followed by a post hoc least significant difference (LSD) test to assess pairwise comparisons.
ELISA.
Angiopoietin Like 4 protein (ANGTPL4) levels were measured in L6 secretion media and rat serum by ELISA (#E02A0507, BlueGene Biotech, Shanghai, China), according to manufacturer’s instructions. ANGPTL4 kit sensitivity was 0.1 ng/ml, as per the manufacturer’s product details. ELISA data were analyzed by one-way ANOVA with a post hoc LSD test.
RESULTS
Metabolic Response to Essential PUFA Diets in Obese Rats
Complete details regarding the metabolic and physiologic responses in LC, OC, O-LA, and O-ALA rats has been previously reported in detail (34, 35) but are summarized in Table 1. Briefly, OC, O-LA and O-ALA rats showed increased body weight compared with LC rats, with no differences seen between the three obese groups. OC rats showed impaired glucose tolerance compared with LC rats. However, obese rats pair-fed diets supplemented with LA or ALA showed improved glucose tolerance and fasting glucose compared with OC rats, with ALA-supplementation having the greatest benefits. Concomitant with improvements in glycemic control, TA fatty acid composition analyses showed that O-LA rats had higher total n-6 PUFA compared with OC and O-ALA rats, while O-ALA rats had higher n-3 PUFA compared with OC and O-LA rats.
Table 1.
Anthropometric measures and TA muscle PUFA concentrations of lean and obese Zucker rats fed either control or PUFA-supplemented diets
| Parameter | LC | OC | O-LA | O-ALA |
|---|---|---|---|---|
| Body weight, g | 418 ± 6a | 527 ± 7b | 542 ± 11b | 542 ± 4b |
| Fasting serum glucose, mM | 5.7 ± 0.9a | 14.2 ± 1.0b | 10 ± 1.3c | 9.4 ± 1.1c |
| Intraperitoneal glucose tolerance test area, under the curve | 975 ± 68a | 1,582 ± 111b | 1,381 ± 83a,b | 1,065 ± 152a |
| Intraperitoneal insulin tolerance test area, under the curve | 58 ± 6a | 107 ± 14b | 80 ± 14a,b | 62 ± 6a |
| Fasting serum free fatty acids, μM | 259 ± 22a | 847 ± 170b | 700 ± 65b | 957 ± 174b |
| Total n-3 PUFA in TA muscle, μmol/g dry mass | 11.2 ± 2.4a | 21.2 ± 1.5b | 7.68 ± 1.4a | 50.9 ± 5.1c |
| Total n-6 PUFA in TA muscle, μmol/g dry mass | 36.8 ± 7.2a | 77.1 ± 3.7b | 108.7 ± 10.6c | 61.9 ± 4.2b |
Data are expressed as means ± SE. Within a parameter, values that share letters are not significantly different from each other (P < 0.05). n = 8 animals per diet group. TA, tibialis anterior; PUFA, polyunsaturated fatty acids; LC, lean control; OC, obese control; O-LA, obese linoleic acid supplemented; O-ALA, obese alpha-linoleic acid supplemented.
Obesity Affects Global Skeletal Muscle Gene Expression More Than Essential PUFA
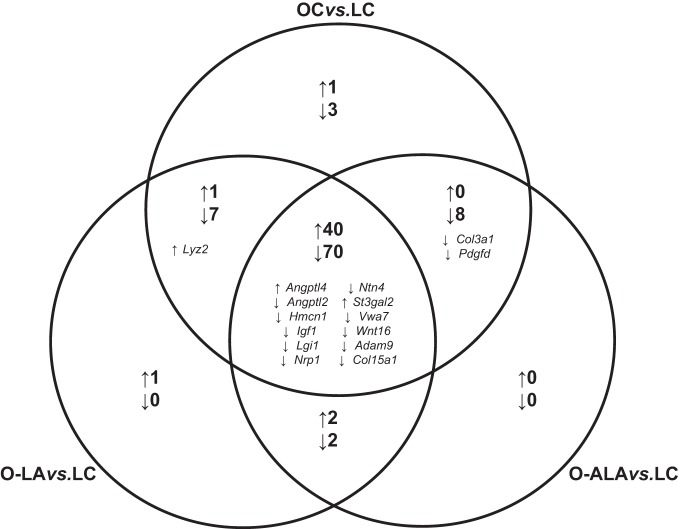
Microarray analysis identified 135 differentially expressed genes in TA among the four experimental groups (Supplemental Table S2). Post hoc pairwise comparisons revealed that 130 genes were differentially expressed between OC and LC rats (90 downregulated, 40 upregulated), 123 genes were differentially expressed between O-LA and LC rats (81 downregulated, 42 upregulated), and 121 genes were differentially expressed between O-ALA and LC rats (82 downregulated, 39 upregulated). As illustrated in Fig. 1, over 80% of the differentially expressed genes were directionally concordant between the three pairwise comparisons. Subsequent DAVID pathway analysis revealed that two KEGG pathways were significantly over-represented in the list of differentially expressed genes. Specifically, OC (P = 0.00082), O-LA (P = 0.0011), and O-ALA groups (P = 0.00062) all showed an upregulation of genes associated with the PPAR signaling pathway (Acadl, Angptl4, Cpt1a, Fabp4, Cd36) compared with LC. Additionally, O-LA (P = 0.049) and O-ALA (P = 0.047) showed a downregulation of genes associated with the hypertrophic cardiomyopathy pathway (Igf1, Prkag2, Tpm2, Tpm3, Tnnc1) compared with LC rats. While OC animals also showed a downregulation in this pathway, this change was of borderline significance (P = 0.085).
Fig. 1.
Venn diagram for differentially expressed genes in obese groups vs. lean control. The number of differentially expressed genes identified from the microarray analysis that were in common between different pairwise comparisons is shown in boldface, as are the specific myokine genes identified from our bioinformatic analysis. OC vs. LC (obese vs. lean control rats), O-ALA vs. LC (obese alpha-linoleic acid supplemented vs. lean control rats), and O-LA vs. LC (obese linoleic acid supplemented vs. lean control rats). ↑, Upregulated genes; ↓, downregulated genes; n = 8 per group.
We were also interested if supplementation with either LA or ALA caused differences in global TA gene expression when compared with OC. This secondary analysis revealed that 30 genes were differentially regulated between O-LA and OC (6 downregulated, 24 upregulated) and 26 genes were differentially regulated between O-ALA and OC (4 downregulated, 22 upregulated). Of these, O-LA and O-ALA normalized the expression of 11 genes and 10 genes, respectively, back to LC levels (Supplemental Table S2). Four of these genes (Gpm6b, Dok4, Asb3, Mid1ip1) were normalized by both LA and ALA in a directionally concordant manner.
Expression of Secreted Proteins was Altered with Obesity and PUFA
Our primary objective was to identify genes encoding secreted proteins that were differentially expressed with obesity and/or PUFA supplementation. The 135 differentially expressed genes identified in microarray data corresponded to 133 known proteins. From this list of 133 proteins, our bioinformatic analysis identified 28 with predicted secretory signals. From this subset, 18 genes corresponded to proteins with evidence of secretion according to UniProt, four of which were different isoforms of insulin-like growth factor 1 (Igf1). This resulted in a final number of 15 unique secreted proteins (Table 2). As shown in Fig. 1, 12 of the 15 secreted proteins were differentially expressed in all three obese groups compared with LC in a directionally concordant manner. We validated all microarray gene expression changes by quantitative reverse-transcription PCR (qRT-PCR), with the results for the OC vs. LC pairwise comparison shown in Table 3. All genes except for angiopoietin-like 2 (Angptl2) were confirmed as significantly differentially expressed in the OC vs. LC pairwise comparison.
Table 2.
Proteins encoded by differentially expressed genes predicted by at least one method to contain a signal peptide sequence
| Signal Peptide Prediction Method |
|||||||
|---|---|---|---|---|---|---|---|
| Gene Symbol | Gene Name | Entrez Gene ID | RefSeq Protein ID | Signal-BLAST | TOPCONS2 | SignalP 4.1 | UniProt Evidence of Secretion |
| Acacb | Acetyl-CoA carboxylase beta | 116719 | NP_446374.1 | ● | ● | ||
| Acadl | Acyl-CoA dehydrogenase, long chain | 25287 | NP_036951.1 | ● | |||
| Adam9 | ADAM metallopeptidase domain 9 | 290834 | NP_001014772.1 | ● | ● | ● | ● |
| Angptl2 | Angiopoietin-like 2 | 171100 | NP_598253.1 | ● | ● | ● | ● |
| Angptl4 | Angiopoietin-like 4 | 362850 | NP_954546.1 | ● | ● | ● | ● |
| Bcam | Basal cell adhesion molecule | 78958 | NP_113940.1 | ● | ● | ● | |
| Casq | Calsequestrin 2 | 29209 | NP_058827.3 | ● | ● | ● | |
| Cd34 | CD34 molecule | 305081 | NP_001100672.1 | ● | ● | ● | |
| Col15a1 | Collagen type XV alpha 1 chain | 298069 | NP_001094005.1 | ● | ● | ● | ● |
| Col3a1 | Collagen type III alpha 1 chain | 84032 | NP_114474.1 | ● | ● | ● | ● |
| Fbxl4 | F-box and leucine-rich repeat protein 4 | 313101 | NP_001101389.1 | ● | |||
| Gpihbp1 | Glycosylphosphatidylinositol anchored high density lipoprotein binding protein 1 | 300027 | NP_001124019.1 | ● | ● | ● | |
| Gscsh | Glycine cleavage system protein H | 171133 | NP_598282.2 | ● | |||
| Hmcn1 | Hemicentin 1 | 289094 | NP_001258221.1 | ● | ● | ● | ● |
| Igf1 (isoform a) | Insulin-like growth factor 1 | 24482 | NP_001075946.2 | ● | ● | ● | ● |
| Igf1 (isoform b) | Insulin-like growth factor 1 | 24482 | NP_849197.3 | ● | ● | ● | ● |
| Igf1 (isoform c) | Insulin-like growth factor 1 | 24482 | NP_001075947.1 | ● | ● | ● | ● |
| Igf1 (isoform d) | Insulin-like growth factor 1 | 24482 | NP_001075948.1 | ● | ● | ● | ● |
| Lgi1 | Leucine-rich, glioma inactivated 1 | 252892 | NP_665712.1 | ● | ● | ● | ● |
| Lyz2 | Lysozyme 2 | 25211 | NP_036903.2 | ● | ● | ● | ● |
| Nrp1 | Neuropilin 1 | 246331 | NP_659566.1 | ● | ● | ● | ● |
| Ntn4 | Netrin 4 | 299737 | NP_001100250.1 | ● | ● | ● | ● |
| Pdgfd | Platelet derived growth factor D | 66018 | NP_076452.1 | ● | ● | ● | ● |
| Serpinh1 | Serpin family H member 1 | 29345 | NP_058869.1 | ● | ● | ● | |
| St3gal2 | ST3 beta-galactoside alpha-2,3-sialyltransferase 2 | 64442 | NP_113883.2 | ● | ● | ● | |
| Tm2d2 | TM2 domain containing 2 | 290833 | NP_001017444.1 | ● | ● | ||
| Vwa7 | Von Willebrand factor A domain containing 7 | 309611 | NP_997664.1 | ● | ● | ● | ● |
| Wnt16 | Wnt family member 16 | 500047 | NP_001102693.1 | ● | ● | ● | ● |
| Total | 23 | 24 | 28 | 18 | |||
SignalP 4.1 was used with sensitive settings as opposed to the default settings, which reduced the number of false negatives. Default settings were used for Signal-BLAST and TOPCONS2. Genes in boldface (n = 18, 15 unique) were those with evidence of secretion as per UniProt, used for microarray validation, and investigated for potentially being affected by polyunsaturated fatty acid treatment.
Table 3.
Differentially expressed genes identified by microarray and validated by qRT-PCR
| Entrez Gene ID | Gene Symbol | Microarray | qRT-PCR |
|---|---|---|---|
| 290834 | Adam9 | −1.22 | −1.58* |
| 171100 | Angptl2 | −1.40 | −1.27 |
| 362850 | Angptl4 | 6.54 | 17.13* |
| 298069 | Col15a1 | −1.86 | −5.05* |
| 84032 | Col3a1 | −1.75 | −2.67* |
| 289094 | Hmcn1 | −1.19 | −2.80* |
| 24482 | Igf1 | −1.71 | −2.04* |
| 252892 | Lgi1 | −2.77 | −7.24* |
| 25211 | Lyz2 | 2.00 | 1.77* |
| 246331 | Nrp1 | −1.48 | −1.53* |
| 299737 | Ntn4 | −1.41 | −4.41* |
| 66018 | Pdgfd | −1.37 | −2.17* |
| 64442 | St3gal2 | 1.23 | 1.64* |
| 309611 | Vwa7 | −1.33 | −1.78* |
| 500047 | Wnt16 | −1.71 | −1.69* |
Results correspond to fold changes for genes encoding secreted proteins that differed significantly between obese control (OC) vs. lean control (LC) rats (n = 8 per diet group).
Statistically significant quantitative (q)RT-PCR fold change (P < 0.05).
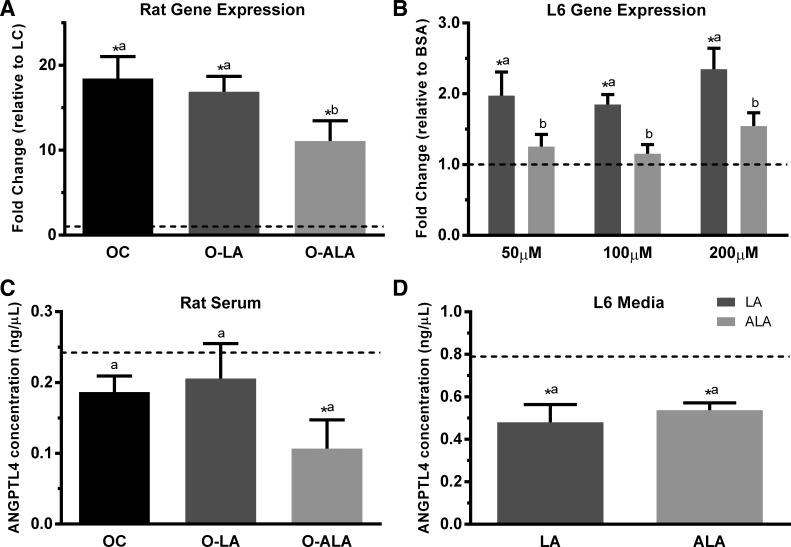
Interestingly, qRT-PCR revealed subtle differences in secreted protein gene expression between the three obese groups and the LC group that were not reflected in microarray data (Fig. 2). Collagen type III alpha-1 chain (Col3a1), Col15a1, and platelet-derived growth factor D (Pdgfd) were downregulated in the three obese groups compared with LC; however, the magnitude of the reduction differed. For the three aforementioned genes, the reduction observed in OC and O-ALA groups was significantly greater than that observed in O-LA (Fig. 2, A–C). Lysozyme (Lyz2) expression was similarly upregulated in OC and O-LA groups compared with LC animals, with no difference observed between O-ALA and LC groups (Fig. 2D). Gene expression for the myokine angiopoietin-like 4 (Angptl4) was increased in all three obese groups, but the increase observed in O-ALA was significantly less than that seen in OC and O-LA (Fig. 3A).
Fig. 2.
Expression of genes encoding skeletal muscle secreted proteins. Values from quantitative RT-PCR of tibialis anterior tissue samples are displayed as mean fold changes (±SE) in comparison to the lean control (LC), which was arbitrarily set to 1.0 and is represented with a dotted line. Statistical significance was determined by one-way ANOVA followed by a post hoc least significant difference test. *Statistical differences (P < 0.05) compared with LC. Bars sharing letters are not statistically different from one another. n = 8 per group.
Fig. 3.
Differential regulation of Angptl4 gene expression and protein secretion. Gene expression for Angptl4 in rat tibialis anterior (A) and differentiated L6 myotube cells (B) are reported as mean fold change (± SE), and ANGPTL4 protein levels in serum (C) and L6 myotube secretion media (D) are reported as ng/µl. The LC (A, C) and the BSA control (B, D) are represented with a dotted line, and were arbitrarily set to 1 for gene expression (A, B). Statistical significance was determined by one-way ANOVA (A, C, and D) or two-way ANOVA (B), followed by a post hoc least significant difference test. Bars sharing letters are not statistically significant from one another. *Statistical difference (P < 0.05) compared with LC (A, C) or BSA control (B, D). Rat work, n = 8 per group. L6 work, n = 3–6 per condition.
PUFA Regulation of Secreted Protein Expression in Cultured L6 Myotubes
Since our qRT-PCR analyses revealed that gene expression for five secreted proteins (Col3a1, Col15a1, Pdgfd, Lyz2, and Angptl4) was potentially differentially regulated by PUFA in TA, we subsequently investigated whether these genes were differentially expressed in a classical rat myotube cell model treated with pure LA or ALA. Pdgfd expression levels were below the limit of detection in L6 myotubes, and we found no evidence that Col3a1 and Col15a1 were differentially regulated with either ALA or LA treatment. Lyz2 expression was significantly upregulated with the highest dose of ALA (200 µM), with only upward trends seen with the 50 and 100 µM doses (data not shown). Angptl4 gene expression was significantly upregulated with all three doses of LA in comparison to ALA and the BSA control cells (Fig. 3B).
ANGPTL4 Protein Secretion is Regulated by ALA
We next examined if the differential gene expression observed for Angptl4 (Fig. 3A) was similarly reflected in circulating serum levels. As shown in Fig. 3C, serum levels of ANGPTL4 were similar between LC, OC, and O-LA groups, but significantly lower in O-ALA. In differentiated rat L6 myotubes, we found that only LA appeared to change Angptl4 gene expression (Fig. 3B); however, we saw that both LA and ALA (200 µM) reduced the levels of ANGPTL4 protein in L6 secretion media (Fig. 3D).
DISCUSSION
The current study investigated the effects of diets supplemented with either ALA or LA on the skeletal muscle secretome in a rodent model of obesity. Using a predictive bioinformatic approach, our study shows that the expression of genes encoding secreted proteins was primarily regulated by obesity, with essential PUFA having a comparatively smaller influence. We identified 15 genes encoding secreted proteins as differentially expressed in skeletal muscle of obese rats compared with lean controls, of which five (Col3a1, Col15a1, Pdgfd, Lyz2, and Angptl4) were found to be potentially differentially regulated by PUFA. Fatty acid regulation of secreted proteins was subsequently examined in the commonly used rat L6 myotube cell model. We also showed that ANGPTL4 protein levels in circulation and differentiated L6 myotube secretion media were regulated by essential PUFA. Together, our results suggest that the skeletal muscle secretome is altered with obesity, and that PUFA supplementation may regulate specific secreted proteins to improve metabolic health.
The predictive bioinformatic approach used in the current study is a validated method to identify secreted proteins. Indeed, the combination of global gene expression profiling and bioinformatic predictive algorithms has been successfully applied to both adipocytes (24, 36) and skeletal muscle (13, 22, 31). However, we acknowledge that our bioinformatic approach to study secreted proteins would miss any potential myokines that are encoded by genes lacking a signal sequence for the secretory pathway. Nevertheless, past studies using a similar bioinformatic approach have highlighted the vast secretory capacity of skeletal muscle. Previous investigations of myokine regulation by lifestyle factors have predominantly focused on exercise (42); thus the present study constitutes the first investigation of essential PUFA regulation of the skeletal muscle secretome.
The OC rats used in the current study displayed whole-body and skeletal muscle insulin resistance, as previously reported (35). We identified 130 genes to be differentially expressed between OC and LC animals. This number aligns with findings from past microarray studies in which anywhere between 85 and 195 differentially expressed genes were identified in skeletal muscle from Type 2 diabetic individuals compared with healthy controls (39, 51, 61). Related to the 130 differentially expressed genes, only two KEGG pathways reached statistical significance in the DAVID pathway analysis: PPAR signaling (up in OC) and hypertrophic cardiomyopathy (down in OC). Given our interest in secreted proteins, it is notable that the myokine Angptl4 was associated with the PPAR signaling pathway. Most genes encoding secreted proteins (12 of 15) were similarly regulated in all three obese groups compared with LC, with 10 genes downregulated and two genes upregulated. Hmcn1, Wnt16, Pdgfd, Vwa7, St3gal2, and Lgi1 correspond to potentially novel components of the skeletal muscle secretome. In contrast, the remaining nine genes have been previously reported in the literature to be secreted from muscle: Angptl2, Angptl4, Col3a1, Igf1, and Nrp1 in insulin-resistant C2C12 mouse skeletal muscle cells (18); Col15a1 and the human homolog of Lyz2 in exercise-induced humans (13, 45); and Ntn4 and Adam9 in a computational analysis of human skeletal muscle gene expression (8). qRT-PCR confirmed that five genes encoding secreted proteins were differentially regulated by ALA and LA in obese rats. Supplementation with LA regulated Col3a1, Col15a1, and Pdgfd, whereas Lyz2 and Angptl4 were altered with ALA. Overall, the minor differences in gene expression observed between the OC, O-LA, and O-ALA groups suggest that obesity has a dominant effect on gene expression in rat TA compared with essential PUFA. We acknowledge the caveat that these findings may be specific to the obese Zucker rat model; therefore, further corroboration is necessary in other models of obesity.
Col3a1 and Col15a1 are members of the collagen family and have roles in extracellular matrix (ECM) remodeling. Some reports have shown that skeletal muscle Col3a1 gene expression is upregulated with lipid oversupply (46) and weight gain (53) in humans, while others have reported a downregulation in insulin-resistant rat L6 myotubes (62) and human skeletal muscle biopsies (39). It is difficult to interpret the reduction in Col3a1 expression observed in OC rats given the aforementioned contrasting reports; however, it is notable that LA supplementation (and not ALA) reversed OC-induced reductions in Col3a1 expression. In comparison with Col3a1, less is known about Col15a1 in skeletal muscle. Eklund et al. (20) developed a global Col15a1−/− mouse and reported that these mice showed evidence of muscular disease and were more susceptible to exercise-induced muscle injury. Thus, it is possible that the reduction in Col15a1 expression seen in OC rats may compromise skeletal muscle stability. Although LA supplementation partially reversed OC-induced reductions in Col15a1 expression, levels were not returned to that of LC rats. Moreover, it is notable that neither LA nor ALA altered the expression of either collagen isoform in cultured rat L6 cells, suggesting that LA may have a more pronounced influence on Col3a1 and Col15a1 expression in an insulin-resistant context (i.e., obese Zucker rats) compared with an insulin-sensitive one (i.e., L6 myotubes).
We also found that OC animals experienced a downregulation in Pdgfd gene expression. Recently, Borkham-Kamphorst et al. (7) reported that PDGFD affects ECM remodeling by regulating matrix metalloprotease activity. This is intriguing given the aforementioned changes in various collagen isoforms, which reinforces a potential regulation of skeletal muscle ECM remodeling. Moreover, past studies showed that Pdgfd may also regulate monocyte migration (55, 57). In the current study, LA supplementation (and not ALA) reversed OC-induced reductions in Pdgfd expression. The reduction in Pdgfd expression in OC rats is unexpected given that obese, insulin-resistant skeletal muscle is typically characterized by increased macrophage infiltration (59). Although we did not study immune cell content in skeletal muscle, lower Pdgfd expression suggests a potential reduction in monocyte recruitment. We were unable to detect Pdgfd in rat L6 cells, which implies that myotubes may not be the cell type within skeletal muscle that expresses this myokine. Future studies should consider investigating if LA and ALA alter immune cell infiltration in skeletal muscle.
Lysozyme was previously identified as a putative myokine that is induced following acute exercise in humans (13, 45). Interestingly, lysozyme was detected on plasma extracellular vesicle membranes and shown to influence epithelial cell migration and immune signaling (1). We found that Lyz2 gene expression was upregulated in the OC and O-LA groups, but unchanged in O-ALA rats (compared with LC). Additionally, Lyz2 gene expression was upregulated in L6 myotubes treated with ALA. Although we saw different effects of ALA on Lyz2 gene expression in the two experimental models, our data suggest that the manner in which ALA influences Lyz2 expression may vary with metabolic status.
ANGPTL4 is an important inhibitor of lipoprotein lipase (LPL) activity and influences the clearance of circulating triacylglycerol-rich lipoproteins. In comparison with the four previous secreted proteins, considerably more is known about the regulation of the myokine ANGPTL4. For example, skeletal muscle Angptl4 gene expression is increased with fatty acids and exercise (45, 52). Additionally, human plasma levels of ANGPTL4 are increased with long-term fasting (30) and are differentially affected by dietary fats (10). We observed an increase in skeletal muscle Angptl4 gene expression in obese rats compared with LC rats, which aligns with findings in ob/ob mice (56). Interestingly, ALA partially attenuated this increase, which may correspond to a potential improvement in skeletal muscle LPL activity. The increase in skeletal muscle Angptl4 gene expression was not mirrored in serum ANGPTL4 levels in obese rats, with OC and O-LA rats showing no difference in comparison with LC rats. However, a significant reduction in serum ANGPTL4 levels was seen in O-ALA rats compared with LC. Overall, this suggests there may be differential regulation of ANGPTL4 by ALA at the levels of gene expression, protein translation, and secretion that requires further elucidation. Interpreting changes in ANGPTL4 blood levels is challenging because this gene is expressed and secreted by numerous tissues, including white adipose tissue, liver, and skeletal muscle (30). This latter point is particularly salient given our previous report that ALA supplementation in these rats increased levels of this fatty acid in adipose tissue and liver to a greater extent than in skeletal muscle (35). In rat L6 myotubes, LA-treatment increased Angptl4 gene expression, which aligns with the increase observed in skeletal muscle from O-LA rats. In contrast, ALA treatment had no effect on Angptl4 gene expression in rat L6 myotubes. However, both LA and ALA caused reductions in ANGPTL4 levels in rat L6 myotube secretion media. Collectively, our findings in L6 myotubes are difficult to reconcile with our in vivo data but suggest that PUFA may regulate myocyte Angptl4 expression and secretion in a differential manner.
Our findings concerning the relationship between obesity and the skeletal muscle secretome adds valuable information to a relatively understudied area of investigation. However, a brief discussion of discrepancies with past research is warranted to better appreciate the challenges associated with myokine discovery. One of the most widely studied myokines is irisin, which is encoded by the Fndc5 gene (44). Irisin was first reported to promote browning in white adipose tissue in response to exercise (9); however, its association with obesity and diabetes remains unclear due to discrepant findings (40). In the current study, Fndc5 was not differentially expressed as per our criteria; however, we did observe a reduction (−1.26-fold; P = 0.0005) in OC compared with LC rats when multiple testing was not accounted for (data not shown). Additionally, other studies have shown reductions in blood levels of various myokines (e.g., IL-6, IL-8, IL-13, and myostatin) in humans with insulin resistance and extreme obesity (3, 23, 28, 29, 38, 48). However, we did not detect changes in skeletal muscle gene expression related to any of these putative myokines. Overall, there are several possible explanations for apparent discrepancies between our findings and those reported in the literature. First, it is possible that myokines are not always regulated at the level of gene expression (58); thus our bioinformatic approach using microarray data would miss any changes in protein levels and/or secretion. Furthermore, myokines that do not contain a secretory signal peptide would not be identified with our approach. Second, many circulating proteins are derived from more than one tissue. Therefore, it is possible that previous findings have not conclusively demonstrated that changes in circulating proteins are specifically related to altered expression in the skeletal muscle secretome. Third, the rats in our study were fed diets for 12 wk, as opposed to receiving an acute intervention. While many secreted proteins are reported to respond to acute exercise (15), insulin (62) and inflammatory stimulation (62), it is plausible that the chronic feeding model used in the present study is capturing changes in the skeletal muscle secretome that are different from those noted in acute models. We also recognize that changes in the expression of secreted proteins in the O-LA and O-ALA groups could be attributed to lower carbohydrate content in the essential PUFA-supplemented diets compared with the OC diet. Fourth, the skeletal muscle secretome is not solely determined by myocytes, but all resident cell types (e.g., myocytes, macrophages, endothelial cells, etc.); thus it is not surprising to see differences between rat skeletal muscle and cultured L6 cells. Finally, we cannot exclude the possibility that differences in PUFA regulation of the secretome may exist between different skeletal muscles due to varying fiber composition; therefore, further studies using different muscles are warranted. Despite these various caveats, our transcriptomic approach using TA skeletal muscle provides new insights regarding the potential regulation of the skeletal muscle secretome by dietary LA and ALA.
In conclusion, we report a genomics-based model for predicting the secretion of proteins from skeletal muscle and show that LA and ALA may differentially regulate the skeletal muscle secretome. Moreover, ALA-induced reductions in ANGPTL4 blood levels may provide a partial explanation for the greater improvements in whole-body glucose tolerance previously reported in ALA-supplemented rats (34).
GRANTS
This work was supported by a grant from the Ontario Ministry of Agriculture, Food and Rural Affairs. A. Rajna was supported by an Undergraduate Research Award scholarship from The Natural Sciences and Engineering Research Council of Canada.
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the authors.
AUTHOR CONTRIBUTIONS
A.R., H.G., O.S., and S.M. performed experiments; A.R., H.G., and S.M. analyzed data; A.R., H.G., and D.M.M. interpreted results of experiments; A.R. prepared figures; A.R. and H.G. drafted manuscript; A.R., H.G., O.S., S.M., G.P.H., and D.M.M. edited and revised manuscript; A.R., H.G., O.S., S.M., G.P.H., and D.M.M. approved final version of manuscript; G.P.H. and D.M.M. conceived and designed research.
Supplemental Data
Table S1: Supplemental Table S1: Experimental diet composition; Table S2: Obesity induced dysregulation of skeletal muscle gene expression is partially restored by PUFA supplementation. - .docx (39 KB)
REFERENCES
- 1.Abey SK, Yuana Y, Joseph PV, Kenea ND, Fourie NH, Sherwin LB, Gonye GE, Smyser PA, Stempinski ES, Boulineaux CM, Weaver KR, Bleck CK, Henderson WA. Lysozyme association with circulating RNA, extracellular vesicles, and chronic stress. BBA Clin 7: 23–35, 2016. doi: 10.1016/j.bbacli.2016.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ahima RS, Park HK. Connecting Myokines and Metabolism. Endocrinol Metab (Seoul) 30: 235–245, 2015. doi: 10.3803/EnM.2015.30.3.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Al-Shukaili A, Al-Ghafri S, Al-Marhoobi S, Al-Abri S, Al-Lawati J, Al-Maskari M. Analysis of inflammatory mediators in type 2 diabetes patients. Int J Endocrinol 2013: 976810, 2013. doi: 10.1155/2013/976810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Alibegovic AC, Sonne MP, Højbjerre L, Bork-Jensen J, Jacobsen S, Nilsson E, Faerch K, Hiscock N, Mortensen B, Friedrichsen M, Stallknecht B, Dela F, Vaag A. Insulin resistance induced by physical inactivity is associated with multiple transcriptional changes in skeletal muscle in young men. Am J Physiol Endocrinol Metab 299: E752–E763, 2010. doi: 10.1152/ajpendo.00590.2009. [DOI] [PubMed] [Google Scholar]
- 5.Ansari S, Djalali M, Mohammadzadeh Honarvar N, Mazaherioun M, Zarei M, Agh F, Gholampour Z, Javanbakht MH. The Effect of n-3 Polyunsaturated Fatty Acids Supplementation on Serum Irisin in Patients with Type 2 Diabetes: A Randomized, Double-Blind, Placebo-Controlled Trial. Int J Endocrinol Metab 15: e40614, 2017. doi: 10.5812/ijem.40614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Apweiler R, Bairoch A, Wu CH, Barker WC, Boeckmann B, Ferro S, Gasteiger E, Huang H, Lopez R, Magrane M, Martin MJ, Natale DA, O’Donovan C, Redaschi N, Yeh LS. UniProt: the Universal Protein knowledgebase. Nucleic Acids Res 32: D115–D119, 2004. doi: 10.1093/nar/gkh131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Borkham-Kamphorst E, Alexi P, Tihaa L, Haas U, Weiskirchen R. Platelet-derived growth factor-D modulates extracellular matrix homeostasis and remodeling through TIMP-1 induction and attenuation of MMP-2 and MMP-9 gelatinase activities. Biochem Biophys Res Commun 457: 307–313, 2015. doi: 10.1016/j.bbrc.2014.12.106. [DOI] [PubMed] [Google Scholar]
- 8.Bortoluzzi S, Scannapieco P, Cestaro A, Danieli GA, Schiaffino S. Computational reconstruction of the human skeletal muscle secretome. Proteins 62: 776–792, 2006. doi: 10.1002/prot.20803. [DOI] [PubMed] [Google Scholar]
- 9.Boström P, Wu J, Jedrychowski MP, Korde A, Ye L, Lo JC, Rasbach KA, Boström EA, Choi JH, Long JZ, Kajimura S, Zingaretti MC, Vind BF, Tu H, Cinti S, Højlund K, Gygi SP, Spiegelman BM. A PGC1-α-dependent myokine that drives brown-fat-like development of white fat and thermogenesis. Nature 481: 463–468, 2012. doi: 10.1038/nature10777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brands M, Sauerwein HP, Ackermans MT, Kersten S, Serlie MJ. Omega-3 long-chain fatty acids strongly induce angiopoietin-like 4 in humans. J Lipid Res 54: 615–621, 2013. doi: 10.1194/jlr.M030049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bruce CR, Hoy AJ, Turner N, Watt MJ, Allen TL, Carpenter K, Cooney GJ, Febbraio MA, Kraegen EW. Overexpression of carnitine palmitoyltransferase-1 in skeletal muscle is sufficient to enhance fatty acid oxidation and improve high-fat diet-induced insulin resistance. Diabetes 58: 550–558, 2009. doi: 10.2337/db08-1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Carvalho BS, Irizarry RA. A framework for oligonucleotide microarray preprocessing. Bioinformatics 26: 2363–2367, 2010. doi: 10.1093/bioinformatics/btq431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Catoire M, Mensink M, Kalkhoven E, Schrauwen P, Kersten S. Identification of human exercise-induced myokines using secretome analysis. Physiol Genomics 46: 256–267, 2014. doi: 10.1152/physiolgenomics.00174.2013. [DOI] [PubMed] [Google Scholar]
- 14.Ciaraldi TP, Ryan AJ, Mudaliar SR, Henry RR. Altered Myokine Secretion Is an Intrinsic Property of Skeletal Muscle in Type 2 Diabetes. PLoS One 11: e0158209, 2016. doi: 10.1371/journal.pone.0158209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Covington JD, Tam CS, Bajpeyi S, Galgani JE, Noland RC, Smith SR, Redman LM, Ravussin E. Myokine Expression in Muscle and Myotubes in Response to Exercise Stimulation. Med Sci Sports Exerc 48: 384–390, 2016. doi: 10.1249/MSS.0000000000000787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.DeFronzo RA, Jacot E, Jequier E, Maeder E, Wahren J, Felber JP. The effect of insulin on the disposal of intravenous glucose. Results from indirect calorimetry and hepatic and femoral venous catheterization. Diabetes 30: 1000–1007, 1981. doi: 10.2337/diab.30.12.1000. [DOI] [PubMed] [Google Scholar]
- 17.Dennis G Jr, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, Lempicki RA. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol 4: P3, 2003. doi: 10.1186/gb-2003-4-5-p3. [DOI] [PubMed] [Google Scholar]
- 18.Deshmukh AS, Cox J, Jensen LJ, Meissner F, Mann M. Secretome Analysis of Lipid-Induced Insulin Resistance in Skeletal Muscle Cells by a Combined Experimental and Bioinformatics Workflow. J Proteome Res 14: 4885–4895, 2015. doi: 10.1021/acs.jproteome.5b00720. [DOI] [PubMed] [Google Scholar]
- 19.Eckardt K, Görgens SW, Raschke S, Eckel J. Myokines in insulin resistance and type 2 diabetes. Diabetologia 57: 1087–1099, 2014. doi: 10.1007/s00125-014-3224-x. [DOI] [PubMed] [Google Scholar]
- 20.Eklund L, Piuhola J, Komulainen J, Sormunen R, Ongvarrasopone C, Fássler R, Muona A, Ilves M, Ruskoaho H, Takala TE, Pihlajaniemi T. Lack of type XV collagen causes a skeletal myopathy and cardiovascular defects in mice. Proc Natl Acad Sci USA 98: 1194–1199, 2001. doi: 10.1073/pnas.98.3.1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Frank K, Sippl MJ. High-performance signal peptide prediction based on sequence alignment techniques. Bioinformatics 24: 2172–2176, 2008. doi: 10.1093/bioinformatics/btn422. [DOI] [PubMed] [Google Scholar]
- 22.Hartwig S, Raschke S, Knebel B, Scheler M, Irmler M, Passlack W, Muller S, Hanisch FG, Franz T, Li X, Dicken HD, Eckardt K, Beckers J, de Angelis MH, Weigert C, Häring HU, Al-Hasani H, Ouwens DM, Eckel J, Kotzka J, Lehr S. Secretome profiling of primary human skeletal muscle cells. Biochim Biophys Acta 1844: 1011–1017, 2014. doi: 10.1016/j.bbapap.2013.08.004. [DOI] [PubMed] [Google Scholar]
- 23.Hittel DS, Berggren JR, Shearer J, Boyle K, Houmard JA. Increased secretion and expression of myostatin in skeletal muscle from extremely obese women. Diabetes 58: 30–38, 2009. doi: 10.2337/db08-0943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hoggard N, Cruickshank M, Moar KM, Bashir S, Mayer CD. Using gene expression to predict differences in the secretome of human omental vs. subcutaneous adipose tissue. Obesity (Silver Spring) 20: 1158–1167, 2012. doi: 10.1038/oby.2012.14. [DOI] [PubMed] [Google Scholar]
- 25.Houmard JA. Intramuscular lipid oxidation and obesity. Am J Physiol Regul Integr Comp Physiol 294: R1111–R1116, 2008. doi: 10.1152/ajpregu.00396.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huerta AE, Prieto-Hontoria PL, Fernández-Galilea M, Sáinz N, Cuervo M, Martínez JA, Moreno-Aliaga MJ. Circulating irisin and glucose metabolism in overweight/obese women: effects of α-lipoic acid and eicosapentaenoic acid. J Physiol Biochem 71: 547–558, 2015. doi: 10.1007/s13105-015-0400-5. [DOI] [PubMed] [Google Scholar]
- 27.Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, Speed TP. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics 4: 249–264, 2003. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- 28.Jiang LQ, Franck N, Egan B, Sjögren RJ, Katayama M, Duque-Guimaraes D, Arner P, Zierath JR, Krook A. Autocrine role of interleukin-13 on skeletal muscle glucose metabolism in type 2 diabetic patients involves microRNA let-7. Am J Physiol Endocrinol Metab 305: E1359–E1366, 2013. doi: 10.1152/ajpendo.00236.2013. [DOI] [PubMed] [Google Scholar]
- 29.Kern PA, Ranganathan S, Li C, Wood L, Ranganathan G. Adipose tissue tumor necrosis factor and interleukin-6 expression in human obesity and insulin resistance. Am J Physiol Endocrinol Metab 280: E745–E751, 2001. doi: 10.1152/ajpendo.2001.280.5.E745. [DOI] [PubMed] [Google Scholar]
- 30.Kersten S, Lichtenstein L, Steenbergen E, Mudde K, Hendriks HF, Hesselink MK, Schrauwen P, Müller M. Caloric restriction and exercise increase plasma ANGPTL4 levels in humans via elevated free fatty acids. Arterioscler Thromb Vasc Biol 29: 969–974, 2009. doi: 10.1161/ATVBAHA.108.182147. [DOI] [PubMed] [Google Scholar]
- 31.Le Bihan MC, Bigot A, Jensen SS, Dennis JL, Rogowska-Wrzesinska A, Lainé J, Gache V, Furling D, Jensen ON, Voit T, Mouly V, Coulton GR, Butler-Browne G. In-depth analysis of the secretome identifies three major independent secretory pathways in differentiating human myoblasts. J Proteomics 77: 344–356, 2012. doi: 10.1016/j.jprot.2012.09.008. [DOI] [PubMed] [Google Scholar]
- 32.Lee HJ, Lee JO, Lee YW, Kim SA, Park SH, Kim HS. Kalirin, a GEF for Rac1, plays an important role in FSTL-1-mediated glucose uptake in skeletal muscle cells. Cell Signal 29: 150–157, 2017. doi: 10.1016/j.cellsig.2016.10.013. [DOI] [PubMed] [Google Scholar]
- 33.Masuda S, Tanaka M, Inoue T, Ohue-Kitano R, Yamakage H, Muranaka K, Kusakabe T, Shimatsu A, Hasegawa K, Satoh-Asahara N. Chemokine (C-X-C motif) ligand 1 is a myokine induced by palmitate and is required for myogenesis in mouse satellite cells. Acta Physiol (Oxf) 222: 2018. doi: 10.1111/apha.12975. [DOI] [PubMed] [Google Scholar]
- 34.Matravadia S, Herbst EA, Jain SS, Mutch DM, Holloway GP. Both linoleic and α-linolenic acid prevent insulin resistance but have divergent impacts on skeletal muscle mitochondrial bioenergetics in obese Zucker rats. Am J Physiol Endocrinol Metab 307: E102–E114, 2014. doi: 10.1152/ajpendo.00032.2014. [DOI] [PubMed] [Google Scholar]
- 35.Matravadia S, Zabielski P, Chabowski A, Mutch DM, Holloway GP. LA and ALA prevent glucose intolerance in obese male rats without reducing reactive lipid content, but cause tissue-specific changes in fatty acid composition. Am J Physiol Regul Integr Comp Physiol 310: R619–R630, 2016. doi: 10.1152/ajpregu.00297.2015. [DOI] [PubMed] [Google Scholar]
- 36.Mutch DM, Rouault C, Keophiphath M, Lacasa D, Clément K. Using gene expression to predict the secretome of differentiating human preadipocytes. Int J Obes 33: 354–363, 2009. doi: 10.1038/ijo.2009.3. [DOI] [PubMed] [Google Scholar]
- 37.Natalicchio A, Marrano N, Biondi G, Spagnuolo R, Labarbuta R, Porreca I, Cignarelli A, Bugliani M, Marchetti P, Perrini S, Laviola L, Giorgino F. The Myokine Irisin Is Released in Response to Saturated Fatty Acids and Promotes Pancreatic β-Cell Survival and Insulin Secretion. Diabetes 66: 2849–2856, 2017. doi: 10.2337/db17-0002. [DOI] [PubMed] [Google Scholar]
- 38.Nielsen AR, Hojman P, Erikstrup C, Fischer CP, Plomgaard P, Mounier R, Mortensen OH, Broholm C, Taudorf S, Krogh-Madsen R, Lindegaard B, Petersen AM, Gehl J, Pedersen BK. Association between interleukin-15 and obesity: interleukin-15 as a potential regulator of fat mass. J Clin Endocrinol Metab 93: 4486–4493, 2008. doi: 10.1210/jc.2007-2561. [DOI] [PubMed] [Google Scholar]
- 39.Palsgaard J, Brøns C, Friedrichsen M, Dominguez H, Jensen M, Storgaard H, Spohr C, Torp-Pedersen C, Borup R, De Meyts P, Vaag A. Gene expression in skeletal muscle biopsies from people with type 2 diabetes and relatives: differential regulation of insulin signaling pathways. PLoS One 4: e6575, 2009. doi: 10.1371/journal.pone.0006575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Panati K, Suneetha Y, Narala VR. Irisin/FNDC5–an updated review. Eur Rev Med Pharmacol Sci 20: 689–697, 2016. [PubMed] [Google Scholar]
- 41.Patti ME, Butte AJ, Crunkhorn S, Cusi K, Berria R, Kashyap S, Miyazaki Y, Kohane I, Costello M, Saccone R, Landaker EJ, Goldfine AB, Mun E, DeFronzo R, Finlayson J, Kahn CR, Mandarino LJ. Coordinated reduction of genes of oxidative metabolism in humans with insulin resistance and diabetes: Potential role of PGC1 and NRF1. Proc Natl Acad Sci USA 100: 8466–8471, 2003. doi: 10.1073/pnas.1032913100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pedersen BK, Febbraio MA. Muscle as an endocrine organ: focus on muscle-derived interleukin-6. Physiol Rev 88: 1379–1406, 2008. doi: 10.1152/physrev.90100.2007. [DOI] [PubMed] [Google Scholar]
- 43.Petersen TN, Brunak S, von Heijne G, Nielsen H. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat Methods 8: 785–786, 2011. doi: 10.1038/nmeth.1701. [DOI] [PubMed] [Google Scholar]
- 44.Polyzos SA, Anastasilakis AD, Efstathiadou ZA, Makras P, Perakakis N, Kountouras J, Mantzoros CS. Irisin in metabolic diseases. Endocrine 59: 260–274, 2018. doi: 10.1007/s12020-017-1476-1. [DOI] [PubMed] [Google Scholar]
- 45.Pourteymour S, Eckardt K, Holen T, Langleite T, Lee S, Jensen J, Birkeland KI, Drevon CA, Hjorth M. Global mRNA sequencing of human skeletal muscle: Search for novel exercise-regulated myokines. Mol Metab 6: 352–365, 2017. doi: 10.1016/j.molmet.2017.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Richardson DK, Kashyap S, Bajaj M, Cusi K, Mandarino SJ, Finlayson J, DeFronzo RA, Jenkinson CP, Mandarino LJ. Lipid infusion decreases the expression of nuclear encoded mitochondrial genes and increases the expression of extracellular matrix genes in human skeletal muscle. J Biol Chem 280: 10290–10297, 2005. doi: 10.1074/jbc.M408985200. [DOI] [PubMed] [Google Scholar]
- 47.Seldin MM, Peterson JM, Byerly MS, Wei Z, Wong GW. Myonectin (CTRP15), a novel myokine that links skeletal muscle to systemic lipid homeostasis. J Biol Chem 287: 11968–11980, 2012. doi: 10.1074/jbc.M111.336834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shah R, Hinkle CC, Ferguson JF, Mehta NN, Li M, Qu L, Lu Y, Putt ME, Ahima RS, Reilly MP. Fractalkine is a novel human adipochemokine associated with type 2 diabetes. Diabetes 60: 1512–1518, 2011. doi: 10.2337/db10-0956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shaw B, Lambert S, Wong MH, Ralston JC, Stryjecki C, Mutch DM. Individual saturated and monounsaturated fatty acids trigger distinct transcriptional networks in differentiated 3T3-L1 preadipocytes. J Nutrigenet Nutrigenomics 6: 1–15, 2013. doi: 10.1159/000345913. [DOI] [PubMed] [Google Scholar]
- 50.Sparks LM, Xie H, Koza RA, Mynatt R, Hulver MW, Bray GA, Smith SR. A high-fat diet coordinately downregulates genes required for mitochondrial oxidative phosphorylation in skeletal muscle. Diabetes 54: 1926–1933, 2005. doi: 10.2337/diabetes.54.7.1926. [DOI] [PubMed] [Google Scholar]
- 51.Sreekumar R, Halvatsiotis P, Schimke JC, Nair KS. Gene expression profile in skeletal muscle of type 2 diabetes and the effect of insulin treatment. Diabetes 51: 1913–1920, 2002. doi: 10.2337/diabetes.51.6.1913. [DOI] [PubMed] [Google Scholar]
- 52.Staiger H, Haas C, Machann J, Werner R, Weisser M, Schick F, Machicao F, Stefan N, Fritsche A, Häring HU. Muscle-derived angiopoietin-like protein 4 is induced by fatty acids via peroxisome proliferator-activated receptor (PPAR)-delta and is of metabolic relevance in humans. Diabetes 58: 579–589, 2009. doi: 10.2337/db07-1438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tam CS, Covington JD, Bajpeyi S, Tchoukalova Y, Burk D, Johannsen DL, Zingaretti CM, Cinti S, Ravussin E. Weight gain reveals dramatic increases in skeletal muscle extracellular matrix remodeling. J Clin Endocrinol Metab 99: 1749–1757, 2014. doi: 10.1210/jc.2013-4381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tsirigos KD, Peters C, Shu N, Käll L, Elofsson A. The TOPCONS web server for consensus prediction of membrane protein topology and signal peptides. Nucleic Acids Res 43, W1: W401–W407, 2015. doi: 10.1093/nar/gkv485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Uutela M, Wirzenius M, Paavonen K, Rajantie I, He Y, Karpanen T, Lohela M, Wiig H, Salven P, Pajusola K, Eriksson U, Alitalo K. PDGF-D induces macrophage recruitment, increased interstitial pressure, and blood vessel maturation during angiogenesis. Blood 104: 3198–3204, 2004. doi: 10.1182/blood-2004-04-1485. [DOI] [PubMed] [Google Scholar]
- 56.Vienberg SG, Kleinridders A, Suzuki R, Kahn CR. Differential effects of angiopoietin-like 4 in brain and muscle on regulation of lipoprotein lipase activity. Mol Metab 4: 144–150, 2014. doi: 10.1016/j.molmet.2014.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wågsäter D, Zhu C, Björck HM, Eriksson P. Effects of PDGF-C and PDGF-D on monocyte migration and MMP-2 and MMP-9 expression. Atherosclerosis 202: 415–423, 2009. doi: 10.1016/j.atherosclerosis.2008.04.050. [DOI] [PubMed] [Google Scholar]
- 58.Whitham M, Febbraio MA. The ever-expanding myokinome: discovery challenges and therapeutic implications. Nat Rev Drug Discov 15: 719–729, 2016. doi: 10.1038/nrd.2016.153. [DOI] [PubMed] [Google Scholar]
- 59.Wu H, Ballantyne CM. Skeletal muscle inflammation and insulin resistance in obesity. J Clin Invest 127: 43–54, 2017. doi: 10.1172/JCI88880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yang M, Wei D, Mo C, Zhang J, Wang X, Han X, Wang Z, Xiao H. Saturated fatty acid palmitate-induced insulin resistance is accompanied with myotube loss and the impaired expression of health benefit myokine genes in C2C12 myotubes. Lipids Health Dis 12: 104, 2013. doi: 10.1186/1476-511X-12-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yang X, Pratley RE, Tokraks S, Bogardus C, Permana PA. Microarray profiling of skeletal muscle tissues from equally obese, non-diabetic insulin-sensitive and insulin-resistant Pima Indians. Diabetologia 45: 1584–1593, 2002. doi: 10.1007/s00125-002-0905-7. [DOI] [PubMed] [Google Scholar]
- 62.Yoon JH, Yea K, Kim J, Choi YS, Park S, Lee H, Lee CS, Suh PG, Ryu SH. Comparative proteomic analysis of the insulin-induced L6 myotube secretome. Proteomics 9: 51–60, 2009. doi: 10.1002/pmic.200800187. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1: Supplemental Table S1: Experimental diet composition; Table S2: Obesity induced dysregulation of skeletal muscle gene expression is partially restored by PUFA supplementation. - .docx (39 KB)