Abstract
Genomic sequencing has undergone massive expansion in the past 10 yr, from a rarely used research tool into an approach that has broad applications in a clinical setting. From rare disease to cancer, genomics is transforming our knowledge of biology. The transition from targeted gene sequencing, to whole exome sequencing, to whole genome sequencing has only been made possible due to rapid advancements in technologies and informatics that have plummeted the cost per base of DNA sequencing and analysis. The tools of genomics have resolved the etiology of disease for previously undiagnosable conditions, identified cancer driver gene variants, and have impacted the understanding of pathophysiology for many diseases. However, this expansion of use has also highlighted research’s current voids in knowledge. The lack of precise animal models for gene-to-function association, lack of tools for analysis of genomic structural changes, skew in populations used for genetic studies, publication biases, and the “Unknown Proteome” all contribute to voids needing filled for genomics to work in a fast-paced clinical setting. The future will hold the tools to fill in these voids, with new data sets and the continual development of new technologies allowing for expansion of genomic medicine, ushering in the days to come for precision medicine. In this review we highlight these and other points in hopes of advancing and guiding precision medicine into the future for optimal success.
Keywords: clinical sequencing, ethics, GWAS, VUS, whole genome sequencing
THE PAST
Road to Personalized Medicine
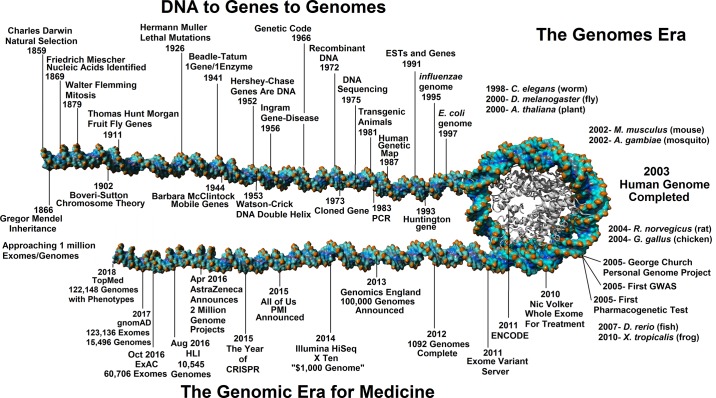
Many books and reviews have highlighted past achievements in understanding what DNA and genes are, how to sequence this DNA, and finally into the genomes era of the late 1990s and early 2000s, all culminating into what is now the “Genomics Era for Medicine” (Fig. 1). From the rediscovery of Mendel defining modes of trait inheritance, the Morgan laboratory defining trait linkage and mutations, to the identification of DNA as the genetic material (5), genetics grew as a field of biology in the 1900s. The development of sequencing technologies in the mid to late 1970s (97) opened up the door to sequence the first genes (78) and genomes (96). The beginning years of sequencing for clinical impact were mostly based on targeted gene approaches, with power to resolve only common variants within disease patients. Among these first gene variants to disease associations, found using genetics, were the polymorphisms of beta-globin genes for sickle cell disease in 1978 (59), the F508- of CFTR in cystic fibrosis in 1989 (60), CAG repeat length in the HTT gene in Huntington’s disease in 1993 (71), and the 1990 discovery of BRCA1 variants for breast cancer risk (45). Shortly following the completion of the first human genome in the early 2000s (51, 114), projects were proposed to take genomics into the medical field for personalized healthcare, such as the Personal Genome Project (19), with hopes of identifying genetic events for disease etiology in a broad range of diseases. In 2005, publications started solidifying genomics into an understanding of disease at a genome level through the development of genome-wide association studies (GWAS), with the first published study addressing the genomics of age-related macular degeneration (62).
Fig. 1.
Timeline for the discovery of DNA/genes into the genomic era for medicine.
Population Genetics and GWAS
One of the initial GWAS studies contained 116,204 single nucleotide polymorphisms (SNPs) and only looked at 96 cases with 50 controls, at the time a ground-breaking paper in Science (62). GWAS continued to advance from that initial point, and several reviews of those first few years have previously been published (18, 115). As the cost of performing the genotyping assays of GWAS decreased, in combination with increased density of variants on the assays, larger cohorts were collected for other indications such as neurological (81), cardiovascular (2), metabolic (108), renal (63), hepatic (15), and reproductive (85). Studies have grown to include more than 100,000 individuals, such as the body mass index association studies in 249,796 European individuals (103) and the 126,559 individuals studied for educational attainment (94).
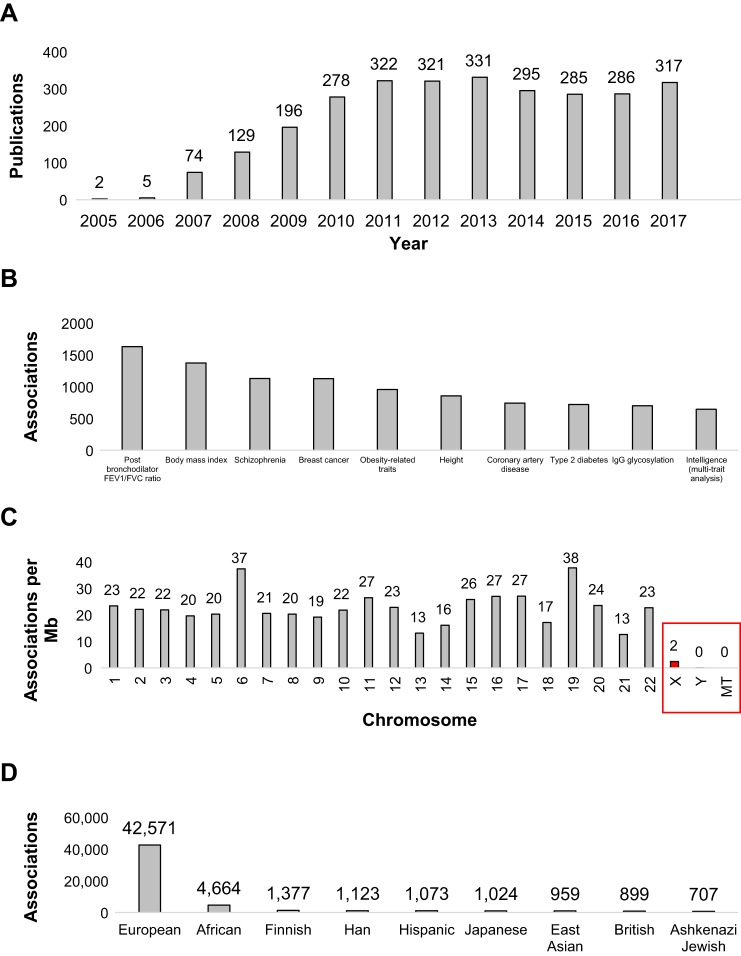
As of the writing this article, nearly 70,000 genetic associations have been curated from ~3,000 publications within the National Human Genome Research Institute-European Bioinformatics Institute GWAS database (116) consisting of ~2,500 separate publication-measured traits (https://www.ebi.ac.uk/gwas/, accessed on March 23, 2018). Following the initial growth phase of GWAS from 2005 to 2010, there has been a stable ~300 papers per year with quality GWAS data (Fig. 2A). These studies have revealed the most associations for traits such as asthma, body mass/obesity/height, schizophrenia, breast cancer, coronary artery disease, and Type 2 diabetes (Fig. 2B). Several chromosomes (chr) such as chr6 and chr19 return a higher than average phenotypic activity from GWAS relative to the chromosome size (Fig. 2C). Surprisingly the sex chr and mitochondrial genome are poorly associated with disease in GWAS, due not to biological association but to limited assessment within genome studies. The X-chromosome in males is highly associated with hemizygous disease risk and in females with random X-inactivation, resulting in ~50% of cells with hemizygous loss of function mutations (68, 90). In addition there is suggestions these sex chromosomes, particularly the Y-chromosome, have high phenotypic impact within high-throughput animal phenotyping studies in genetic crosses (88, 89).
Fig. 2.
Current statistics from the National Human Genome Research Institute-European Bioinformatics Institute (EBI/NHGRI) database. The EBI/NHGRI genome-wide association study (GWAS) catalog (https://www.ebi.ac.uk/gwas/docs/file-downloads, downloaded 3/23/2018) was assessed for publications per year (A), traits with the most associations (B), most associations per chromosome Mb (C), and associations per ethnicity (D).
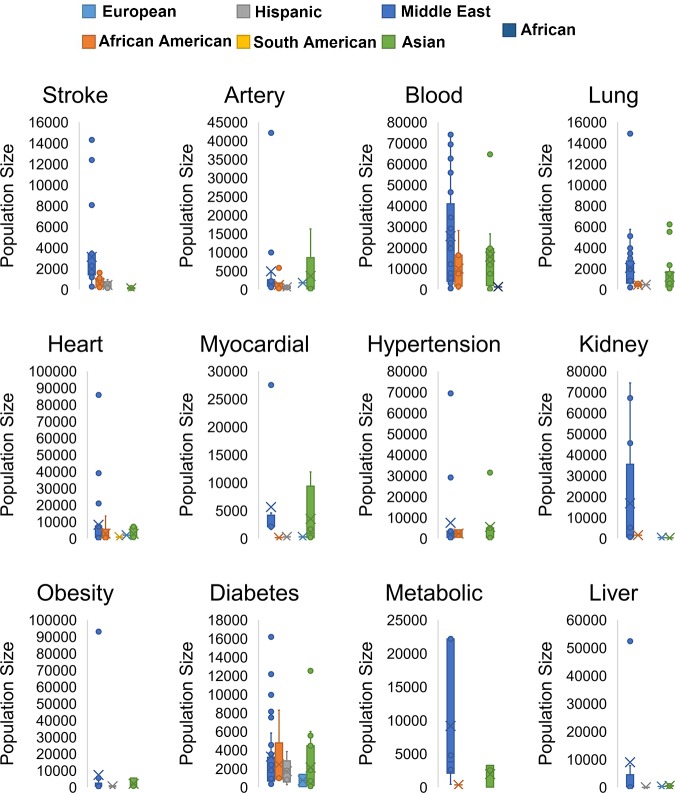
As the GWAS database continues to increase in size, it contains an overwhelming number of associations for individuals of European ancestry, at an astonishing ~63% (42,571/67,857 as of March 2018) of all associations (Fig. 2D), far exceeding the actual global population composition. This is a result of many disease areas having predominantly larger European populations within the studies, therefore increasing statistical power for the one ethnicity (Fig. 3). Addressing this lack of diversity to ensure just distribution of benefits from genomic technologies is one of the leading ELSI (Ethical Legal and Social Implications of Genetics) concerns for the future of genomic research and clinical practice (20). This represents a new iteration of a long-existing problem in biomedical research, resulting in the National Institutes of Health (NIH) Revitalization Act of 1993 mandating inclusion of minorities in all NIH-funded research (16). In the past, not only minorities but women and children have been underrepresented in clinical trials, resulting in the need to incentivize inclusion of more representative populations through the 1997 Food and Drug Administration Modernization Act and the 2002 Best Pharmaceuticals for Children Act (74). In short, convenience and ready availability are threatening to leave some populations out as the beneficiaries of scientific advancement; and at least one policy working group found that skewed representation in which higher socioeconomic populations were overrepresented made assessing the risks of genomic testing difficult, as higher socioeconomic populations may be immune or less vulnerable to many common concerns about the risks of testing (such as employment or insurance concerns) (21).
Fig. 3.
Population structure in current GWAS studies. Each search term was queried against the GWAS studies on November 1, 2016 from the EBI/NHGRI GWAS catalog. Populations were extracted and plotted as box and whisker plots. Caucasian/European = cyan, African American = red, Hispanic = gray, South American = yellow, Middle Eastern = Blue, Asian = green, African = dark blue.
In the past few years, GWAS approaches have expanded into Epigenome-wide association studies (EWAS) (14, 91), an approach where the methylation of cytosines in the DNA are studied at a genome-wide level for disease association. These experiments have expanded to such a point that meta-analyses are now being performed on the generated data (69), much like those done for GWAS (117). Moreover, as medical health record information is more reliable to be data-mined and correlated to genomic information, phenome-wide association studies (PheWAS) (25) have been increasing in potential utility for genomic medicine (24, 48, 84). Even as the development and expansion of these GWAS, EWAS, and PheWAS tools increase and impact medicine, they still require extensive ability to segregate phenotypes, test causality of genotype-to-phenotype, and are underpowered for rare variants. Arguments can be made for both rare and common variants in disease etiology (39); thus, many of these genomic tools are underpowered for full clinical utility. Other genomic tools such as animal models of disease and the expansion of whole exome/genome sequencing are beginning to overcome these limitations.
Genotype to Phenotype: from Animal Models to Cells for Human Health
Human genetic studies have many limitations, mainly resulting from the difficulty in deciphering one variant among a landscape of thousands of other unique variants; therefore, connecting a genotype to a phenotype with high confidence is very difficult in humans. Important questions arise once a variant is identified, such as does the variant alter the function of the gene, does the functional change result in disease or another phenotype, and is that associated disease or phenotype relevant to the specific clinical condition in the tested individual? These questions are hard to answer with current scientific tools within the index subject. Thus, the dissection of genetic elements that contribute to traits and diseases has always needed models within other organisms. Very early in genetics, animal models served this very function, with the fruit fly (Drosophila melanogaster) serving as one of the first and most successful examples of genotype-to-phenotype relationships (105). This included groundbreaking work by the Morgan laboratory mapping trait inheritance (79), Sturtevant defining trait linkage mapping (106), Muller defining mutation due to environmental exposures (80), studies of homology between different genera (107), and finally having complete genomes from 12 of the Drosophila species (104). Yet the fly is still very distantly related to humans, with 7 chr composing ~149 Mb with 30,443 proteins (https://www.ncbi.nlm.nih.gov/genome/?term=txid7227[orgn]) compared with the humans’ 23 chr pairs (22 autosome pairs and 2 sex chromosomes) with ~2,991 Mb and 79,074 proteins (https://www.ncbi.nlm.nih.gov/genome/?term=homo+sapien). The rat and mouse genomes are more similar to humans and thus provide a more reliable testing model organism for genotype-to-phenotype relationships. While primate models are even more similar to human, they have disadvantages ranging from increased time of study, expense, and ethical concerns for performing experiments. For the rat we have provided a table of the major milestones for genetic understanding and community resources that have relevance to understanding genotype-phenotype relationships (Table 1), many of which can also be found within the mouse community as well.
Table 1.
Progress of genomics in the rat to create a model organism for studying human disease and available resources for the community
| Data Type | Reference | Year | Accomplishment |
|---|---|---|---|
| Sequencing | (38) | 2004 | Brown Norway (BN) rat sequenced: combining bacterial artificial chromosome, end sequencing, whole-genome shot gun, BAC fingerprinting mapping |
| Sequencing | (44) | 2008 | copy number variation found in inbred and outbred strains: computational and array-based comparative analysis of hybridization signals |
| Genome modification | (37) | 2009 | first use of Zinc-finger nucleases to begin humanizing the rat strains |
| Sequencing | (3) | 2010 | spontaneously hypertensive rat (SHR) sequenced: Illumina paired-end |
| Sequencing | (100) | 2012 | genomic variation in 2 founders (SHR/Olapcv and BN-Lx/Cub) of recombinant inbred panel and liver RNA-Seq data, paired-end library, SOLiD and Illumina |
| Sequencing | (42) | 2013 | DA and F344 strains sequenced: Illumina paired-end read technology |
| Sequencing | (92) | 2013 | 8 founders of the rat heterogeneous stock (46) sequenced: SOLiD |
| Sequencing | (4) | 2013 | comparative analysis of genome sequences for 25 inbred strains and substrains |
| Genome modification | (67) | 2013 | 1st use of CRISPR-Cas9 in rats |
| Expression | (122) | 2014 | generation of RNA-Seq library from the F344 strain for 11 tissues at 4 time points, in both males and females |
| Sequencing | (7) | 2014 | sequencing of the SHR Y-chromosome known to be involved in blood pressure regulation (31): first Y-chromosome sequence added in the Rnor6.0 genome release |
| Sequencing | (49) | 2015 | comprehensive inventory of 40 sequenced rat strains and substrains |
| Sequencing and phenotyping | (89) | 2016 | sequence analysis and consomic phenotyping in multiple strains of rats for all nuclear chromosomes including the Y-chromosome for the 1st time |
| Resources | https://rgd.mcw.edu/ | major source for rat genomic, genetic, phenotype, strain data, quantitative trait loci, genetic markers, correct nomenclature, functional annotations for rat, human, mouse derived data, pathway analysis (99) | |
| Resources | http://www.ensembl.org/Rattus_norvegicus/index.html | Ensembl Rat Browser | |
| Resources | https://rgd.mcw.edu/rgdweb/models/gerrc.html | source for models of gene knockout, knock-in, conditional expression, transgenic and Cre-recombinase in rat. | |
| Resources | http://www.rrrc.us/ | long-term preservation and distribution of rat models from USA | |
| Resources | http://www.anim.med.kyoto-u.ac.jp/NBR/ | long-term preservation and distribution of rat models from Japan | |
| Resources | http://pga.mcw.edu | physiological characterization of 2 panels of consomic rat strains (SS×BN, FHH×BN) and multiple inbred strains for cardiovascular, pulmonary, renal, vascular, and blood systems |
In the mouse (101) and rat (23), entire chromosomes (consomic) can be introgressed from strains of animals that have disease or particular phenotypes relative to “healthy” control animals, or vice versa, to determine the chr that contribute to disease. Combining the generation of every chromosome cross between two strains with the measuring of hundreds of phenotypes one can suggest chr with the highest impact on phenotypic traits (64, 73). For example, two complete rat consomic panels were developed by introgressing the Brown Norway (BN) to Fawn-Hooded Hypertensive (FHH) or the Salt Sensitive (SS) rat genome background. Surprisingly, per DNA base, the Y-chr contributes to a vastly elevated number of phenotypes relative to all other chr and also has twice the elevated rate of variation between strains (89). As the sex chr where already mentioned to be incredibly underpowered in GWAS (Fig. 2), animal models can have power to explain human phenotypes contributed by sex chr (88) that current techniques used in humans struggle to illuminate these roles of the sex chr. For example, animal models can be used in explaining the recently reported impact of the loss of Y-chromosome (LOY) as a major disease contributor (30, 35). To resolve the impact of specific regions of chr on phenotype, researchers utilize F1 crosses, heterogeneous stock breeding, and congenic mapping, all of which allow the association of regions/genes on chr with phenotype. This information can then be used in the human to map and test variants that may translate across organisms.
The development of technologies such as Zn fingers, TALENs, and now CRISPR/Cas9 has revolutionized our ability to characterize clinical variants found from whole genome sequencing in human (29, 33). As these technologies can be applied to nearly any species, particularly those that have had their genomes sequenced, it is possible to study a gene or variant in diverse species such as the fruit fly, Caenorhabditis elegans, zebrafish, mouse and rat. An easier approach, and what most groups have used to date, for characterization is either the deletion or insertion of an entire gene of interest. Yet these are based on all or none impacts, limiting the scope for potential gain of function or gene dosage contribution to disease. Utilizing these genome editing technologies, one of the growing approaches in animals is the generation of humanized species, such that the model organism is susceptible to human disease when it normally is not; therefore, opening the door for studies such as viral/bacterial infections and human tumor growth (26, 28, 32, 54). With more advanced humanized species, ethical concerns raised in the past from chimaera research return, and the NIH has only recently sought public feedback concerning plans to lift a moratorium forbidding federal funding, which is currently pending review by ethics panels (50, 58). More recently groups have begun to insert variants using donor sequence after cutting the genome with CRISPR/Cas9, thus generating single variants of interest. While these approaches are still difficult, they should continue to grow in the future and become more common practice for variant-to-phenotype associations.
While animal studies have significant scientific promise, they do result in multiple practical issues in addition to the ethical issues identified above. Primarily, their genomes are not the same as the human genome, and not all traits are shared between species. Therefore, many groups have moved into human cell culture characterization, using either isolated disease tissue from a patient, primary or stable cell lines, or generating organoids in culture. Moreover, human cells can be manipulated in many ways to define the biology of disease progression. Tumor biopsies can be sequenced and grown in culture, or they can be delivered to humanized models of mice and rats to study drug responses based on genetic variability of the specific tumor (43, 112). Germline cells can be obtained, sequenced, and then converted into inducible pluripotent stem cells (83, 109, 121). These cells can be manipulated with genome editing tools such as CRISPR/Cas9 (124) and then differentiated to many linages including neural (27), liver (102), fat (87), and heart (36, 125) to characterize phenotypic changes with the hope of potentially being used to treat disease. The question remains whether current research tools and costs can keep pace with the ever-growing number of human variants identified from clinical sequencing, or whether new methods are necessary.
Clinical Sequencing and Impact on Patient Treatment
Oncology and cancer research has long been at the forefront of using genomics in understanding disease etiology, including large-scale projects like COSMIC (34) and The Cancer Genome Atlas (9, 11). As mentioned earlier, sequencing initially was used for identifying common variants in conditions like cystic fibrosis, and GWAS was used to reach significance in large populations. With knowledge of the entire genome, including multiple ethnicities from the 1000 Genomes Project (1), new approaches [whole exome sequencing (WES)] were designed to sequence just those regions of the genome that code for proteins (~1% of the genome), opening the door for identifying potential pathogenic variants without spending the vast resources needed for the entire genome (6, 17). Some of the first large-scale sequencing projects, such as those done in cancer and the National Heart, Lung, and Blood Institute Gene Ontology (NHLBI GO) Exome Sequencing Project (111), began to highlight the full spectrum of variation using whole exome sequencing, showing a large number of rare variants that might influence disease. Around this same time, studies began to be published that showed discovery of rare diseases caused by rare variants (55). Among these first cases was a young boy whose life was saved through the identification of a C203Y variant in XIAP causing his inflammatory bowel disease. The variant was linked to a phenotype that primarily was an immunologic defect associated with a X-linked inhibitor of apoptosis deficiency. Based on this finding the clinicians caring for this patient applied a bone marrow transplant to prevent the development of life-threatening hemophagocytic lymphohistiocytosis. Here, WES not only provided the diagnosis but also suggested the previously unconsidered treatment option of a bone marrow transplant, bringing precision genomic medicine into the public spotlight for the first time (118).
With WES becoming more established in the clinic, the number of patients benefitting from these tools has rapidly grown (40, 120). It is estimated that the use of WES allows ~25% of rare disease cases to reach a molecular diagnosis (56, 119). With the robust yield and the continual decrease in cost of sequencing, many groups have begun the transition to whole genome sequencing (WGS) from exome (123). WGS can resolve protein-coding variants at a similar or higher rate as WES (72, 98) while also allowing for better coverage of the exons and identification of variants that might alter gene expression, gene splicing, and chromosome replication. With a limited knowledge of noncoding regions of the genome, investigators still interpret many variants with ambiguity, but with future research the role for noncoding regions may be better understood. Sequencing an entire genome just once allows for additional bioinformatics analysis to interpret future knowledge that whole exome does not provide, allowing for reassessment of variants instead of resequencing of the patient with new technology. Genome sequencing is still in its infancy, and identification of variants that are rare (such as n = 1 cases) does create issues that arise and warrant caution in reporting, with defined standards now becoming available (41). Constraints also exist as only some health insurance providers have started reimbursing the ordering institution for WES but few do for WGS.
THE PRESENT
Current Cost of Sequencing
As sequencing continues to become more routine (61), the larger numbers of sequenced individuals will further increase the utility and promise of genomics (57). In 2014, the threshold of a $1,000 cost for sequencing an entire whole genome (47), i.e., the raw cost to generate millions of short reads of ~100 bases for a genome, was crossed. Yet this is still a bit deceiving. Taking those 100 base reads and combining them into a genome by alignment to a reference and sorting through the hundreds of thousands of variants, many which are unique, means the actual cost depends on the depth of sequencing, the tools for computational analysis, and person time to analyze the data. All of this cannot be done for just $1,000. Sequencing a genome with lower depth, by decreasing the average of how many reads align to any site of the genome, reduces the cost but also decreases the confidence of variant interpretation, particularly for heterozygous variation. The choice of sequencing at low depth (~10×), average depth (30–40×), or high depth (~90×) should be made based on the circumstance to optimize cost/benefit.
While sequencing for most of the 2000s was performed as research grade, an increasing number of companies and academic institutes have begun offering certified sequencing through the College of American Pathologists (CAP) and Clinical Laboratory Improvement Amendments (CLIA) for clinical use. One of the biggest obstacles for genomic medicine to scale is the growing of sequencing infrastructure capacity to meet the clinical demand. Larger companies and academic institutes have expanded their sequencing capacities in the USA such that hundreds of thousands of genomes can be sequenced each year. The cost of bioinformatics for the growing amount of sequencing is also a hurdle, but as more genomes are completed and more advanced analytical pipelines are developed, these costs will decline. It should be noted that clinical-based sequencing must be ordered by a qualified clinician, but as the population continues to accept and embrace these approaches, there will be pressure on other physicians to learn and adopt these genomic tools to make them more common practice in patient care.
Assembly of Large Genomic Data Sets for Disease and Controls
With the decrease in sequencing cost, many individuals have now been sequenced, mostly at the research level. Large-scale projects have begun to publish these data sets, giving researchers a larger knowledge of variation within human populations. The genome Aggregation Database (gnomAD), updated in 2017, has grown to contain 123,136 exomes and 15,496 genomes from multiple ethnicities (66), providing a searchable format for genes and variants within each population (http://gnomad.broadinstitute.org/). NIH has invested heavily in the past few years in sequencing, moving projects like Trans-Omics for Precision Medicine (TopMed) into databases with >100,000 whole genome-sequenced individuals, many with detailed phenotype and clinical records (10). The Human Longevity, Inc. database (HLI) has made its first 10,545 30–40× whole genomes sequenced publicly available through the Open Search By Human Longevity, Inc. (110). With companies such as HLI announcing they intend to generate one million genomes or more, it is uncertain how much of this information will be released to the public for research in the future vs. being held within private siloes. The larger the public genomic data sets grow, especially with integration of other data, the more power will be available to interpret each individual genome. Sharing of information is vital for precision genomics medicine to reach its full utility.
The generation of genomic data also brings with it many ethical and legal questions. Unlike most traditional medical interventions, genomic testing often has direct implications for individuals and populations beyond the individual tested (75). Recent controversies raised in genomic research involving the Havasupai Tribe in Arizona illustrate the myriad indirect effects of such research, including potential stigmatization and threats to sacred cultural values and beliefs (77). Significant thought must be given to potentially unintended consequences that result from testing and the proper scope of information that should be obtained (Should we attempt to identify “the gay gene”? What sex- or ethnicity-correlated genetic dispositions might be useful or harmful? Can genetics be used for designer babies or preimplantation screening? Do we create stratifications in costs for health care based on the genetics of ethnicity? Etc.) (76).
Large-scale clinical sequencing projects are also extending to pharmacogenomics, with companies such as AstraZeneca launching a 2 million genomes project for their own clinical trials. They aim to incorporate genomics into clinical trials and revolutionize pharmacogenomics, with many additional companies and studies following in their footsteps. While the potential benefit of these efforts are great, they do come with additional ethical considerations that must not be ignored, particularly issues of patient data protection, oversight of pharmaceutical companies use/patenting/marketing of the data, stratification of groups, and differences in costs for treatments based on genomic data (8, 53, 70).
From Variant to Function
As the number of genomes continue to rise, interpreting the millions of variants that will be found for potential clinical impact becomes even more essential. In Fig. 4, we provide a sample workflow (our group utilizes) for analysis of variants, particularly for variants that appear promising but have not been associated with the disease of interest. Taking known disease causing variants from databases such as ClinVar (65) and filtering against a variant list from any genome is relatively simple, allowing a clear connection from variant to disease. However, when variants are rare or fall within a gene previously not identified with disease, additional tools are needed. For rare diseases, trio sequencing (i.e., the sequencing of mom and dad along with child/proband) can be used to resolve variants that are found in an affected child. For tumors, normal tissue and the tumor can both be sequenced to identify the somatic mutations that may give rise to cancers. These analyses, in combination with the level of conservation of a site throughout vertebrate evolution, can help filter impactful variants. Tools created to combine many aspects of evolution and protein changes can precompute every single possible variant that could be found, therefore decreasing computational costs when analyzing a single genome (52). Yet frequently these approaches identify genes not previously established for disease. These variants are referred to as variants of uncertain significance (VUS) or within genes of uncertain disease significance (GUDS) and are not clinically actionable (93). With additional experimentation, however, these variants can be reclassified as likely pathogenic (93). One approach is to identify additional patients with similar phenotypes and nominated genes using tools of the Global Alliance for Genomics and Health (GA4GH), such as Matchmaker Exchange (86), connecting individuals across the globe that share variation within genes. Tools such as MyGene2 (https://www.mygene2.org) even expand this idea directly into the patient’s hands, connecting researchers, clinicians, and patients on the basis of genes and phenotypes. Identifying a large number of similar patients combined with some basic biochemical, molecular, and animal experimentation can further resolve a variant’s role in disease etiology (Fig. 4).
Fig. 4.
Sample workflow for clinical sequencing and interpretation of variants.
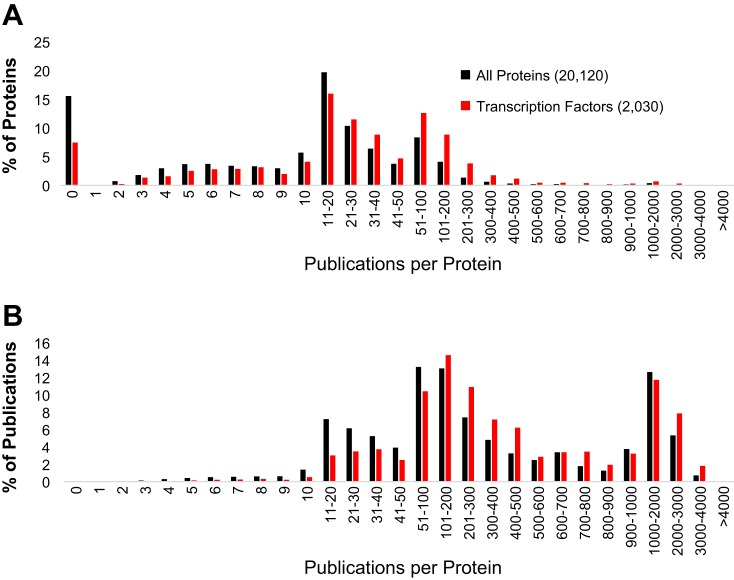
To speed up these analyses, detailed knowledge of every gene within the genome provides tools to interpret variants with higher resolution at a faster pace; however, we are currently far from understanding every gene. For example, querying annotated publications for the 20,120 high-confidence proteins or 2,030 transcription factors from the UniProt database (Fig. 5) shows a clear skew to our knowledge of proteins throughout the genome. A few proteins represent a very large amount of the curated work within UniProt. Around 15% of human proteins currently have 0 publications curated, which we refer to as the “Unknown Proteome.” Data sets such as the Human Protein Atlas (113) show the expression profile for the genes of the Unknown Proteome is 8% ubiquitously expressed and 17% tissue specific (primarily from the testis). Of the ubiquitously expressed genes, most of these are found in a wide range of vertebrate species, suggesting they greatly affect cell function and are highly conserved, yet we have minimal knowledge about their biology. This highlights the fact that many variants falling within the poorly defined regions of the genome will be missed from current bioinformatics, not because of limitations of bioinformatics but because of limited studies of these genes.
Fig. 5.
Curated protein publication bias with the UniProt database. A: number of genes binned into various number of publications as a percent of total publications. Shown in black are the values for all 20,120 validated human proteins of UniProt and in red are the values for 2,030 proteins annotated in UniProt as transcription factors. B: the same data from A but plotted as the total number of publications represented by each group.
While projects such as the Encyclopedia of DNA Elements (ENCODE) (31a) and RoadMap Epigenomics (95) have begun elucidating noncoding regions within the genome, significant progress must still be made. It is likely that many of the disease-causing variants remaining unidentified fall within these noncoding regions. From GWAS, it is suggested that the majority of variants identified for trait association are noncoding and likely expression quantitative trait loci (eQTLs) (82), meaning the variant does not alter the function of the protein but more likely changes the expression of the protein. GWAS has not resolved the causal variant for the majority of identified loci, so additional tools and detailed analysis of GWAS mechanism to disease will be needed to address rare variants. In addition, the field of EvoDevo hypothesizes that speciation events often happen through changes in gene regulation timing through alteration in the noncoding DNA (12, 13). Therefore, evolutionary conservation is also not able to resolve many potential impactful noncoding variants within the genome. Future tools are needed to resolve the role of noncoding variants at a cellular level if we want to fully understand all the genetic contribution to disease development and progression.
THE FUTURE
Where Can Sequencing Go?
As these large-scale projects that involve millions of genomes return results, we expect to gain a clearer view of what regions within the genome have variation and those that do not, including noncoding control regions. As we perform cell-level ENCODE-style experiments at higher resolution, we can also correlate these conserved sites with functional enhancers and promoters. With pharmaceutical companies analyzing large numbers of WES, we expect pharmacogenomics to expand continually. With the number of genomes being sequenced ever growing, and with integration of other data sets such as medical health records, imaging, expression profiles, and mobile health data (such as Fitbit), the field is moving into the big data age. Each individual could be represented by terabytes of information sitting in datacenters, which need to be secured for patient protection while being available for analysis for correlations among other patients by research centers. This level of data will require massive datacenters with fast data transfer speeds as well as high security to protect private patient information. Research databases will continue increasing in size, such that maintaining NIH-funded data sets in multiple locations will not be feasible, but cloud storage and computing must increase to disseminate the information into the hands of many scientists. If social media data can be leveraged in marketing and even politics, the power of medical and genomic information can be seen as even more of a risk. The genome by its nature is difficult to deidentify due to its uniqueness, making it more difficult to keep genotype-to-phenotype associations stored within datacenters and necessitating a delicate approach as genomic medicine advances. Technologies will continue advancing in the coming years as well, with technologies moving the cost of whole genome sequencing potentially to a few hundred dollars with shorter turn-around time, opening the possibility for a human to be routinely sequenced or even building multicellular human genomes, yielding even more data per individual.
Even as WGS becomes the standard method for researchers in the USA, GWAS studies need to continue around the world, particularly in areas such as Africa, Asia and South America where populations are underrepresented and understudied. From functional studies, many groups will continue to break down the linkage disequilibrium block of GWAS to resolve the functional units and define transcription factor networks involved in disease progression, which may unlock new tools for deciphering the role of rare variants in the noncoding portion of the genome. To decrease the time needed for classification of every protein-coding variant associated with disease, we need to develop fuller databases for protein knowledge. A larger focus and more resources need to be applied to characterizing poorly studied proteins if we want to understand the importance of clinical variants anywhere within the genome at a speed compatible with real-time clinical care.
Potential Issues and Conclusions for the Future
Moving genomic medicine into the science of individuals, known as n of 1 science, we must make clearer associations of variants that are either common or rare with changes and mechanisms of biology. As the databases of sequenced genomes continue to grow, we will better understand the dynamics of the human genome, the acceptability of variation at any one site within it, and the degree of structural changes in the genome. Yet resources need to be put into the functional outcome of genetic variants, developing cheaper and quicker tools for variant characterization. With continual identification of VUS and GUDS, time and money will be needed to test and reclassify variants. Much of the future for genomic medicine relies more on phenotype associations and knowledge of genes and the proteins those genes code and less on further advancements of sequencing technologies. As we try to understand the functional effects of the genome, a better understanding of the interactions with the metabolome and microbiome will further those goals in humans. The use of animal models, cell culture, microdevices mimicking organs, and even basic biochemistry of proteins will facilitate these advances. We must begin addressing our knowledge of proteins in the genome, starting with elimination of the Unknown Proteome. We need to start paying more attention to the sex chromosomes (X and Y) and also the mitochondria for phenotype association in humans, as animal models have suggested these regions to be drivers for phenotypic diversity. The role of noncoding variants need to be moved out of standard cell lines and human tissues and into individual cells of the human tissues to understand normal cellular control and the diversity of cell types within any human tissue. Any one variant may not act alone, further complicating our understanding of biology; therefore, we must also consider how to model and test the role of epistasis and gene modifiers within the genome. Above all, for genomic medicine to become a major factor in the future, we must remember that genomics, as powerful as it is, cannot explain all disease risk, including those risks that are passed from generation to generation. Lifestyle choices, environmental exposures, the microbiome, and cultural upbringing need to be merged with genetics to create a full view of diversity of human biology. These integrations will move our genomic understanding to phenotypic outcomes through precision medicine and revolutionize health care.
GRANTS
Supported by National Institute of Environmental Health Sciences Grant K01ES-025435 (to J. W. Prokop).
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the authors.
AUTHOR CONTRIBUTIONS
J.W.P. analyzed data; J.W.P. and J.L. prepared figures; J.W.P., T.M., K.S., S.M.B., C.B., S.R., E.A.W., and J.L. drafted manuscript; J.W.P., T.M., K.S., S.M.B., C.B., S.R., E.A.W., and J.L. edited and revised manuscript; J.W.P., T.M., K.S., S.M.B., C.B., S.R., E.A.W., and J.L. approved final version of manuscript.
REFERENCES
- 1.1000 Genomes Project Consortium, Abecasis GR, Auton A, Brooks LD, DePristo MA, Durbin RM, Handsaker RE, Kang HM, Marth GT, McVean GA. An integrated map of genetic variation from 1,092 human genomes. Nature 491: 56–65, 2012. doi: 10.1038/nature11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arking DE, Pulit SL, Crotti L, van der Harst P, Munroe PB, Koopmann TT, Sotoodehnia N, Rossin EJ, Morley M, Wang X, Johnson AD, Lundby A, Gudbjartsson DF, Noseworthy PA, Eijgelsheim M, Bradford Y, Tarasov KV, Dörr M, Müller-Nurasyid M, Lahtinen AM, Nolte IM, Smith AV, Bis JC, Isaacs A, Newhouse SJ, Evans DS, Post WS, Waggott D, Lyytikäinen L-P, Hicks AA, Eisele L, Ellinghaus D, Hayward C, Navarro P, Ulivi S, Tanaka T, Tester DJ, Chatel S, Gustafsson S, Kumari M, Morris RW, Naluai ÅT, Padmanabhan S, Kluttig A, Strohmer B, Panayiotou AG, Torres M, Knoflach M, Hubacek JA, Slowikowski K, Raychaudhuri S, Kumar RD, Harris TB, Launer LJ, Shuldiner AR, Alonso A, Bader JS, Ehret G, Huang H, Kao WHL, Strait JB, Macfarlane PW, Brown M, Caulfield MJ, Samani NJ, Kronenberg F, Willeit J, CARe Consortium, COGENT Consortium, Smith JG, Greiser KH, Meyer Zu Schwabedissen H, Werdan K, Carella M, Zelante L, Heckbert SR, Psaty BM, Rotter JI, Kolcic I, Polašek O, Wright AF, Griffin M, Daly MJ, DCCT/EDIC, Arnar DO, Hólm H, Thorsteinsdottir U, eMERGE Consortium; Denny JC, Roden DM, Zuvich RL, Emilsson V, Plump AS, Larson MG, O’Donnell CJ, Yin X, Bobbo M, D’Adamo AP, Iorio A, Sinagra G, Carracedo A, Cummings SR, Nalls MA, Jula A, Kontula KK, Marjamaa A, Oikarinen L, Perola M, Porthan K, Erbel R, Hoffmann P, Jöckel KH, Kälsch H, Nöthen MM, HRGEN Consortium, den Hoed M, Loos RJ, Thelle DS, Gieger C, Meitinger T, Perz S, Peters A, Prucha H, Sinner MF, Waldenberger M, de Boer RA, Franke L, van der Vleuten PA, Beckmann BM, Martens E, Bardai A, Hofman N, Wilde AA, Behr ER, Dalageorgou C, Giudicessi JR, Medeiros-Domingo A, Barc J, Kyndt F, Probst V, Ghidoni A, Insolia R, Hamilton RM, Scherer SW, Brandimarto J, Margulies K, Moravec CE, del Greco MF, Fuchsberger C, O’Connell JR, Lee WK, Watt GC, Campbell H, Wild SH, El Mokhtari NE, Frey N, Asselbergs FW, Mateo Leach I, Navis G, van den Berg MP, van Veldhuisen DJ, Kellis M, Krijthe BP, Franco OH, Hofman A, Kors JA, Uitterlinden AG, Witteman JC, Kedenko L, Lamina C, Oostra BA, Abecasis GR, Lakatta EG, Mulas A, Orrú M, Schlessinger D, Uda M, Markus MR, Völker U, Snieder H, Spector TD, Ärnlöv J, Lind L, Sundström J, Syvänen AC, Kivimaki M, Kähönen M, Mononen N, Raitakari OT, Viikari JS, Adamkova V, Kiechl S, Brion M, Nicolaides AN, Paulweber B, Haerting J, Dominiczak AF, Nyberg F, Whincup PH, Hingorani AD, Schott JJ, Bezzina CR, Ingelsson E, Ferrucci L, Gasparini P, Wilson JF, Rudan I, Franke A, Mühleisen TW, Pramstaller PP, Lehtimäki TJ, Paterson AD, Parsa A, Liu Y, van Duijn CM, Siscovick DS, Gudnason V, Jamshidi Y, Salomaa V, Felix SB, Sanna S, Ritchie MD, Stricker BH, Stefansson K, Boyer LA, Cappola TP, Olsen JV, Lage K, Schwartz PJ, Kääb S, Chakravarti A, Ackerman MJ, Pfeufer A, de Bakker PI, Newton-Cheh C. Genetic association study of QT interval highlights role for calcium signaling pathways in myocardial repolarization. Nat Genet 46: 826–836, 2014. doi: 10.1038/ng.3014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Atanur SS, Birol I, Guryev V, Hirst M, Hummel O, Morrissey C, Behmoaras J, Fernandez-Suarez XM, Johnson MD, McLaren WM, Patone G, Petretto E, Plessy C, Rockland KS, Rockland C, Saar K, Zhao Y, Carninci P, Flicek P, Kurtz T, Cuppen E, Pravenec M, Hubner N, Jones SJM, Birney E, Aitman TJ. The genome sequence of the spontaneously hypertensive rat: Analysis and functional significance. Genome Res 20: 791–803, 2010. doi: 10.1101/gr.103499.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Atanur SS, Diaz AG, Maratou K, Sarkis A, Rotival M, Game L, Tschannen MR, Kaisaki PJ, Otto GW, Ma MCJ, Keane TM, Hummel O, Saar K, Chen W, Guryev V, Gopalakrishnan K, Garrett MR, Joe B, Citterio L, Bianchi G, McBride M, Dominiczak A, Adams DJ, Serikawa T, Flicek P, Cuppen E, Hubner N, Petretto E, Gauguier D, Kwitek A, Jacob H, Aitman TJ. Genome sequencing reveals loci under artificial selection that underlie disease phenotypes in the laboratory rat. Cell 154: 691–703, 2013. doi: 10.1016/j.cell.2013.06.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Avery OT, Macleod CM, McCarty M. Studies on the Chemical Nature of the Substance Inducing Transformation of Pneumococcal Types: Induction of Transformation by a Desoxyribonucleic Acid Fraction Isolated from Pneumococcus Type III. J Exp Med 79: 137–158, 1944. doi: 10.1084/jem.79.2.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bamshad MJ, Ng SB, Bigham AW, Tabor HK, Emond MJ, Nickerson DA, Shendure J. Exome sequencing as a tool for Mendelian disease gene discovery. Nat Rev Genet 12: 745–755, 2011. doi: 10.1038/nrg3031. [DOI] [PubMed] [Google Scholar]
- 7.Bellott DW, Hughes JF, Skaletsky H, Brown LG, Pyntikova T, Cho T-J, Koutseva N, Zaghlul S, Graves T, Rock S, Kremitzki C, Fulton RS, Dugan S, Ding Y, Morton D, Khan Z, Lewis L, Buhay C, Wang Q, Watt J, Holder M, Lee S, Nazareth L, Alföldi J, Rozen S, Muzny DM, Warren WC, Gibbs RA, Wilson RK, Page DC. Mammalian Y chromosomes retain widely expressed dosage-sensitive regulators. Nature 508: 494–499, 2014. [Erratum in Nature 514: 126, 2014] 10.1038/nature13206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Breckenridge A, Lindpaintner K, Lipton P, McLeod H, Rothstein M, Wallace H. Pharmacogenetics: ethical problems and solutions. Nat Rev Genet 5: 676–680, 2004. doi: 10.1038/nrg1431. [DOI] [PubMed] [Google Scholar]
- 9.Brennan CW, Verhaak RGW, McKenna A, Campos B, Noushmehr H, Salama SR, Zheng S, Chakravarty D, Sanborn JZ, Berman SH, Beroukhim R, Bernard B, Wu C-J, Genovese G, Shmulevich I, Barnholtz-Sloan J, Zou L, Vegesna R, Shukla SA, Ciriello G, Yung WK, Zhang W, Sougnez C, Mikkelsen T, Aldape K, Bigner DD, Van Meir EG, Prados M, Sloan A, Black KL, Eschbacher J, Finocchiaro G, Friedman W, Andrews DW, Guha A, Iacocca M, O’Neill BP, Foltz G, Myers J, Weisenberger DJ, Penny R, Kucherlapati R, Perou CM, Hayes DN, Gibbs R, Marra M, Mills GB, Lander E, Spellman P, Wilson R, Sander C, Weinstein J, Meyerson M, Gabriel S, Laird PW, Haussler D, Getz G, Chin L, Benz C, Barnholtz-Sloan J, Barrett W, Ostrom Q, Wolinsky Y, Black KL, Bose B, Boulos PT, Boulos M, Brown J, Czerinski C, Eppley M, Iacocca M, Kempista T, Kitko T, Koyfman Y, Rabeno B, Rastogi P, Sugarman M, Swanson P, Yalamanchii K, Otey IP, Liu YS, Xiao Y, Auman JT, Chen P-C, Hadjipanayis A, Lee E, Lee S, Park PJ, Seidman J, Yang L, Kucherlapati R, Kalkanis S, Mikkelsen T, Poisson LM, Raghunathan A, Scarpace L, Bernard B, Bressler R, Eakin A, Iype L, Kreisberg RB, Leinonen K, Reynolds S, Rovira H, Thorsson V, Shmulevich I, Annala MJ, Penny R, Paulauskis J, Curley E, Hatfield M, Mallery D, Morris S, Shelton T, Shelton C, Sherman M, Yena P, Cuppini L, DiMeco F, Eoli M, Finocchiaro G, Maderna E, Pollo B, Saini M, Balu S, Hoadley KA, Li L, Miller CR, Shi Y, Topal MD, Wu J, Dunn G, Giannini C, O’Neill BP, Aksoy BA, Antipin Y, Borsu L, Berman SH, Brennan CW, Cerami E, Chakravarty D, Ciriello G, Gao J, Gross B, Jacobsen A, Ladanyi M, Lash A, Liang Y, Reva B, Sander C, Schultz N, Shen R, Socci ND, Viale A, Ferguson ML, Chen Q-R, Demchok JA, Dillon LAL, Shaw KRM, Sheth M, Tarnuzzer R, Wang Z, Yang L, Davidsen T, Guyer MS, Ozenberger BA, Sofia HJ, Bergsten J, Eckman J, Harr J, Myers J, Smith C, Tucker K, Winemiller C, Zach LA, Ljubimova JY, Eley G, Ayala B, Jensen MA, Kahn A, Pihl TD, Pot DA, Wan Y, Eschbacher J, Foltz G, Hansen N, Hothi P, Lin B, Shah N, Yoon J, Lau C, Berens M, Ardlie K, Beroukhim R, Carter SL, Cherniack AD, Noble M, Cho J, Cibulskis K, DiCara D, Frazer S, Gabriel SB, Gehlenborg N, Gentry J, Heiman D, Kim J, Jing R, Lander ES, Lawrence M, Lin P, Mallard W, Meyerson M, Onofrio RC, Saksena G, Schumacher S, Sougnez C, Stojanov P, Tabak B, Voet D, Zhang H, Zou L, Getz G, Dees NN, Ding L, Fulton LL, Fulton RS, Kanchi K-L, Mardis ER, Wilson RK, Baylin SB, Andrews DW, Harshyne L, Cohen ML, Devine K, Sloan AE, VandenBerg SR, Berger MS, Prados M, Carlin D, Craft B, Ellrott K, Goldman M, Goldstein T, Grifford M, Haussler D, Ma S, Ng S, Salama SR, Sanborn JZ, Stuart J, Swatloski T, Waltman P, Zhu J, Foss R, Frentzen B, Friedman W, McTiernan R, Yachnis A, Hayes DN, Perou CM, Zheng S, Vegesna R, Mao Y, Akbani R, Aldape K, Bogler O, Fuller GN, Liu W, Liu Y, Lu Y, Mills G, Protopopov A, Ren X, Sun Y, Wu C-J, Yung WKA, Zhang W, Zhang J, Chen K, Weinstein JN, Chin L, Verhaak RGW, Noushmehr H, Weisenberger DJ, Bootwalla MS, Lai PH, Triche TJ Jr, Van Den Berg DJ, Laird PW, Gutmann DH, Lehman NL, VanMeir EG, Brat D, Olson JJ, Mastrogianakis GM, Devi NS, Zhang Z, Bigner D, Lipp E, McLendon R; TCGA Research Network . The somatic genomic landscape of glioblastoma. Cell 155: 462–477, 2013. [Erratum in Cell 157: 753, 2014] 10.1016/j.cell.2013.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brody JA, Morrison AC, Bis JC, O’Connell JR, Brown MR, Huffman JE, Ames DC, Carroll A, Conomos MP, Gabriel S, Gibbs RA, Gogarten SM, Gupta N, Jaquish CE, Johnson AD, Lewis JP, Liu X, Manning AK, Papanicolaou GJ, Pitsillides AN, Rice KM, Salerno W, Sitlani CM, Smith NL, Heckbert SR, Laurie CC, Mitchell BD, Vasan RS, Rich SS, Rotter JI, Wilson JG, Boerwinkle E, Psaty BM, Cupples LA; NHLBI Trans-Omics for Precision Medicine (TOPMed) Consortium; Cohorts for Heart and Aging Research in Genomic Epidemiology (CHARGE) Consortium; TOPMed Hematology and Hemostasis Working Group; CHARGE Analysis and Bioinformatics Working Group . Analysis commons, a team approach to discovery in a big-data environment for genetic epidemiology. Nat Genet 49: 1560–1563, 2017. doi: 10.1038/ng.3968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cancer Genome Atlas Research Network, Weinstein JN, Collisson EA, Mills GB, Shaw KR, Ozenberger BA, Ellrott K, Shmulevich I, Sander C, Stuart JM;. The Cancer Genome Atlas Pan-Cancer analysis project. Nat Genet 45: 1113–1120, 2013. doi: 10.1038/ng.2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Carroll SB. Endless forms: the evolution of gene regulation and morphological diversity. Cell 101: 577–580, 2000. doi: 10.1016/S0092-8674(00)80868-5. [DOI] [PubMed] [Google Scholar]
- 13.Carroll SB. Evo-devo and an expanding evolutionary synthesis: a genetic theory of morphological evolution. Cell 134: 25–36, 2008. doi: 10.1016/j.cell.2008.06.030. [DOI] [PubMed] [Google Scholar]
- 14.Chambers JC, Loh M, Lehne B, Drong A, Kriebel J, Motta V, Wahl S, Elliott HR, Rota F, Scott WR, Zhang W, Tan S-T, Campanella G, Chadeau-Hyam M, Yengo L, Richmond RC, Adamowicz-Brice M, Afzal U, Bozaoglu K, Mok ZY, Ng HK, Pattou F, Prokisch H, Rozario MA, Tarantini L, Abbott J, Ala-Korpela M, Albetti B, Ammerpohl O, Bertazzi PA, Blancher C, Caiazzo R, Danesh J, Gaunt TR, de Lusignan S, Gieger C, Illig T, Jha S, Jones S, Jowett J, Kangas AJ, Kasturiratne A, Kato N, Kotea N, Kowlessur S, Pitkäniemi J, Punjabi P, Saleheen D, Schafmayer C, Soininen P, Tai E-S, Thorand B, Tuomilehto J, Wickremasinghe AR, Kyrtopoulos SA, Aitman TJ, Herder C, Hampe J, Cauchi S, Relton CL, Froguel P, Soong R, Vineis P, Jarvelin M-R, Scott J, Grallert H, Bollati V, Elliott P, McCarthy MI, Kooner JS. Epigenome-wide association of DNA methylation markers in peripheral blood from Indian Asians and Europeans with incident type 2 diabetes: a nested case-control study. Lancet Diabetes Endocrinol 3: 526–534, 2015. doi: 10.1016/S2213-8587(15)00127-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chambers JC, Zhang W, Sehmi J, Li X, Wass MN, Van der Harst P, Holm H, Sanna S, Kavousi M, Baumeister SE, Coin LJ, Deng G, Gieger C, Heard-Costa NL, Hottenga J-J, Kühnel B, Kumar V, Lagou V, Liang L, Luan J, Vidal PM, Mateo Leach I, O’Reilly PF, Peden JF, Rahmioglu N, Soininen P, Speliotes EK, Yuan X, Thorleifsson G, Alizadeh BZ, Atwood LD, Borecki IB, Brown MJ, Charoen P, Cucca F, Das D, de Geus EJC, Dixon AL, Döring A, Ehret G, Eyjolfsson GI, Farrall M, Forouhi NG, Friedrich N, Goessling W, Gudbjartsson DF, Harris TB, Hartikainen A-L, Heath S, Hirschfield GM, Hofman A, Homuth G, Hyppönen E, Janssen HLA, Johnson T, Kangas AJ, Kema IP, Kühn JP, Lai S, Lathrop M, Lerch MM, Li Y, Liang TJ, Lin J-P, Loos RJF, Martin NG, Moffatt MF, Montgomery GW, Munroe PB, Musunuru K, Nakamura Y, O’Donnell CJ, Olafsson I, Penninx BW, Pouta A, Prins BP, Prokopenko I, Puls R, Ruokonen A, Savolainen MJ, Schlessinger D, Schouten JNL, Seedorf U, Sen-Chowdhry S, Siminovitch KA, Smit JH, Spector TD, Tan W, Teslovich TM, Tukiainen T, Uitterlinden AG, Van der Klauw MM, Vasan RS, Wallace C, Wallaschofski H, Wichmann H-E, Willemsen G, Würtz P, Xu C, Yerges-Armstrong LM, Alcohol Genome-wide Association (AlcGen) Consortium, Diabetes Genetics Replication and Meta-analyses (DIAGRAM+) Study, Genetic Investigation of Anthropometric Traits (GIANT) Consortium, Global Lipids Genetics Consortium, Genetics of Liver Disease (GOLD) Consortium, International Consortium for Blood Pressure (ICBP-GWAS); Meta-analyses of Glucose and Insulin-Related Traits Consortium (MAGIC), Abecasis GR, Ahmadi KR, Boomsma DI, Caulfield M, Cookson WO, van Duijn CM, Froguel P, Matsuda K, McCarthy MI, Meisinger C, Mooser V, Pietiläinen KH, Schumann G, Snieder H, Sternberg MJ, Stolk RP, Thomas HC, Thorsteinsdottir U, Uda M, Waeber G, Wareham NJ, Waterworth DM, Watkins H, Whitfield JB, Witteman JC, Wolffenbuttel BH, Fox CS, Ala-Korpela M, Stefansson K, Vollenweider P, Völzke H, Schadt EE, Scott J, Järvelin MR, Elliott P, Kooner JS. Genome-wide association study identifies loci influencing concentrations of liver enzymes in plasma. Nat Genet 43: 1131–1138, 2011. doi: 10.1038/ng.970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen MS Jr, Lara PN, Dang JHT, Paterniti DA, Kelly K. Twenty years post-NIH Revitalization Act: enhancing minority participation in clinical trials (EMPaCT): laying the groundwork for improving minority clinical trial accrual: renewing the case for enhancing minority participation in cancer clinical trials. Cancer 120, Suppl 7: 1091–1096, 2014. doi: 10.1002/cncr.28575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Choi M, Scholl UI, Ji W, Liu T, Tikhonova IR, Zumbo P, Nayir A, Bakkaloğlu A, Ozen S, Sanjad S, Nelson-Williams C, Farhi A, Mane S, Lifton RP. Genetic diagnosis by whole exome capture and massively parallel DNA sequencing. Proc Natl Acad Sci USA 106: 19096–19101, 2009. doi: 10.1073/pnas.0910672106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chung CC, Magalhaes WCS, Gonzalez-Bosquet J, Chanock SJ. Genome-wide association studies in cancer–current and future directions. Carcinogenesis 31: 111–120, 2010. doi: 10.1093/carcin/bgp273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Church GM. The personal genome project. Mol Syst Biol 1: 2005.0030, 2005. doi: 10.1038/msb4100040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cohn EG, Henderson GE, Appelbaum PS. Distributive justice, diversity, and inclusion in precision medicine: what will success look like? Genet Med 19: 157–159, 2017. doi: 10.1038/gim.2016.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.COMMITTEE ON BIOETHICS; COMMITTEE ON GENETICS, AND; AMERICAN COLLEGE OF MEDICAL GENETICS AND; GENOMICS SOCIAL; ETHICAL; LEGAL ISSUES COMMITTEE . Ethical and policy issues in genetic testing and screening of children. Pediatrics 131: 620–622, 2013. doi: 10.1542/peds.2012-3680. [DOI] [PubMed] [Google Scholar]
- 23.Cowley AW Jr, Liang M, Roman RJ, Greene AS, Jacob HJ. Consomic rat model systems for physiological genomics. Acta Physiol Scand 181: 585–592, 2004. doi: 10.1111/j.1365-201X.2004.01334.x. [DOI] [PubMed] [Google Scholar]
- 24.Denny JC, Bastarache L, Ritchie MD, Carroll RJ, Zink R, Mosley JD, Field JR, Pulley JM, Ramirez AH, Bowton E, Basford MA, Carrell DS, Peissig PL, Kho AN, Pacheco JA, Rasmussen LV, Crosslin DR, Crane PK, Pathak J, Bielinski SJ, Pendergrass SA, Xu H, Hindorff LA, Li R, Manolio TA, Chute CG, Chisholm RL, Larson EB, Jarvik GP, Brilliant MH, McCarty CA, Kullo IJ, Haines JL, Crawford DC, Masys DR, Roden DM. Systematic comparison of phenome-wide association study of electronic medical record data and genome-wide association study data. Nat Biotechnol 31: 1102–1110, 2013. doi: 10.1038/nbt.2749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Denny JC, Ritchie MD, Basford MA, Pulley JM, Bastarache L, Brown-Gentry K, Wang D, Masys DR, Roden DM, Crawford DC. PheWAS: demonstrating the feasibility of a phenome-wide scan to discover gene-disease associations. Bioinformatics 26: 1205–1210, 2010. doi: 10.1093/bioinformatics/btq126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Difilippantonio S, Celeste A, Fernandez-Capetillo O, Chen H-T, Reina San Martin B, Van Laethem F, Yang Y-P, Petukhova GV, Eckhaus M, Feigenbaum L, Manova K, Kruhlak M, Camerini-Otero RD, Sharan S, Nussenzweig M, Nussenzweig A. Role of Nbs1 in the activation of the Atm kinase revealed in humanized mouse models. Nat Cell Biol 7: 675–685, 2005. doi: 10.1038/ncb1270. [DOI] [PubMed] [Google Scholar]
- 27.Dimos JT, Rodolfa KT, Niakan KK, Weisenthal LM, Mitsumoto H, Chung W, Croft GF, Saphier G, Leibel R, Goland R, Wichterle H, Henderson CE, Eggan K. Induced pluripotent stem cells generated from patients with ALS can be differentiated into motor neurons. Science 321: 1218–1221, 2008. doi: 10.1126/science.1158799. [DOI] [PubMed] [Google Scholar]
- 28.Dorner M, Horwitz JA, Robbins JB, Barry WT, Feng Q, Mu K, Jones CT, Schoggins JW, Catanese MT, Burton DR, Law M, Rice CM, Ploss A. A genetically humanized mouse model for hepatitis C virus infection. Nature 474: 208–211, 2011. doi: 10.1038/nature10168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Doudna JA, Charpentier E. Genome editing. The new frontier of genome engineering with CRISPR-Cas9. Science 346: 1258096, 2014. doi: 10.1126/science.1258096. [DOI] [PubMed] [Google Scholar]
- 30.Dumanski JP, Rasi C, Lönn M, Davies H, Ingelsson M, Giedraitis V, Lannfelt L, Magnusson PKE, Lindgren CM, Morris AP, Cesarini D, Johannesson M, Tiensuu Janson E, Lind L, Pedersen NL, Ingelsson E, Forsberg LA. Mutagenesis. Smoking is associated with mosaic loss of chromosome Y. Science 347: 81–83, 2015. doi: 10.1126/science.1262092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ely DL, Turner ME. Hypertension in the spontaneously hypertensive rat is linked to the Y chromosome. Hypertension 16: 277–281, 1990. doi: 10.1161/01.HYP.16.3.277. [DOI] [PubMed] [Google Scholar]
- 31a.ENCODE Project Consortium The ENCODE (ENCyclopedia Of DNA Elements) Project. Science 306: 636–640, 2004. doi: 10.1126/science.1105136. [DOI] [PubMed] [Google Scholar]
- 32.Flister MJ, Endres BT, Rudemiller N, Sarkis AB, Santarriaga S, Roy I, Lemke A, Geurts AM, Moreno C, Ran S, Tsaih S-W, De Pons J, Carlson DF, Tan W, Fahrenkrug SC, Lazarova Z, Lazar J, North PE, LaViolette PS, Dwinell MB, Shull JD, Jacob HJ. CXM: a new tool for mapping breast cancer risk in the tumor microenvironment. Cancer Res 74: 6419–6429, 2014. doi: 10.1158/0008-5472.CAN-13-3212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Flister MJ, Prokop JW, Lazar J, Shimoyama M, Dwinell M, Geurts A; International Committee on Standardized Genetic Nomenclature for Mice; Rat Genome and Nomenclature Committee . 2015 Guidelines for Establishing Genetically Modified Rat Models for Cardiovascular Research. J Cardiovasc Transl Res 8: 269–277, 2015. doi: 10.1007/s12265-015-9626-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Forbes SA, Bindal N, Bamford S, Cole C, Kok CY, Beare D, Jia M, Shepherd R, Leung K, Menzies A, Teague JW, Campbell PJ, Stratton MR, Futreal PA. COSMIC: mining complete cancer genomes in the Catalogue of Somatic Mutations in Cancer. Nucleic Acids Res 39, Database: D945–D950, 2011. doi: 10.1093/nar/gkq929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Forsberg LA, Rasi C, Malmqvist N, Davies H, Pasupulati S, Pakalapati G, Sandgren J, Diaz de Ståhl T, Zaghlool A, Giedraitis V, Lannfelt L, Score J, Cross NCP, Absher D, Janson ET, Lindgren CM, Morris AP, Ingelsson E, Lind L, Dumanski JP. Mosaic loss of chromosome Y in peripheral blood is associated with shorter survival and higher risk of cancer. Nat Genet 46: 624–628, 2014. doi: 10.1038/ng.2966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gai H, Leung EL-H, Costantino PD, Aguila JR, Nguyen DM, Fink LM, Ward DC, Ma Y. Generation and characterization of functional cardiomyocytes using induced pluripotent stem cells derived from human fibroblasts. Cell Biol Int 33: 1184–1193, 2009. doi: 10.1016/j.cellbi.2009.08.008. [DOI] [PubMed] [Google Scholar]
- 37.Geurts AM, Cost GJ, Freyvert Y, Zeitler B, Miller JC, Choi VM, Jenkins SS, Wood A, Cui X, Meng X, Vincent A, Lam S, Michalkiewicz M, Schilling R, Foeckler J, Kalloway S, Weiler H, Ménoret S, Anegon I, Davis GD, Zhang L, Rebar EJ, Gregory PD, Urnov FD, Jacob HJ, Buelow R. Knockout rats via embryo microinjection of zinc-finger nucleases. Science 325: 433, 2009. doi: 10.1126/science.1172447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gibbs RA, Weinstock GM, Metzker ML, Muzny DM, Sodergren EJ, Scherer S, Scott G, Steffen D, Worley KC, Burch PE, Okwuonu G, Hines S, Lewis L, DeRamo C, Delgado O, Dugan-Rocha S, Miner G, Morgan M, Hawes A, Gill R, Celera, Holt RA, Adams MD, Amanatides PG, Baden-Tillson H, Barnstead M, Chin S, Evans CA, Ferriera S, Fosler C, Glodek A, Gu Z, Jennings D, Kraft CL, Nguyen T, Pfannkoch CM, Sitter C, Sutton GG, Venter JC, Woodage T, Smith D, Lee HM, Gustafson E, Cahill P, Kana A, Doucette-Stamm L, Weinstock K, Fechtel K, Weiss RB, Dunn DM, Green ED, Blakesley RW, Bouffard GG, De Jong PJ, Osoegawa K, Zhu B, Marra M, Schein J, Bosdet I, Fjell C, Jones S, Krzywinski M, Mathewson C, Siddiqui A, Wye N, McPherson J, Zhao S, Fraser CM, Shetty J, Shatsman S, Geer K, Chen Y, Abramzon S, Nierman WC, Havlak PH, Chen R, Durbin KJ, Egan A, Ren Y, Song XZ, Li B, Liu Y, Qin X, Cawley S, Worley KC, Cooney AJ, D’Souza LM, Martin K, Wu JQ, Gonzalez-Garay ML, Jackson AR, Kalafus KJ, McLeod MP, Milosavljevic A, Virk D, Volkov A, Wheeler DA, Zhang Z, Bailey JA, Eichler EE, Tuzun E, Birney E, Mongin E, Ureta-Vidal A, Woodwark C, Zdobnov E, Bork P, Suyama M, Torrents D, Alexandersson M, Trask BJ, Young JM, Huang H, Wang H, Xing H, Daniels S, Gietzen D, Schmidt J, Stevens K, Vitt U, Wingrove J, Camara F, Mar Albà M, Abril JF, Guigo R, Smit A, Dubchak I, Rubin EM, Couronne O, Poliakov A, Hübner N, Ganten D, Goesele C, Hummel O, Kreitler T, Lee YA, Monti J, Schulz H, Zimdahl H, Himmelbauer H, Lehrach H, Jacob HJ, Bromberg S, Gullings-Handley J, Jensen-Seaman MI, Kwitek AE, Lazar J, Pasko D, Tonellato PJ, Twigger S, Ponting CP, Duarte JM, Rice S, Goodstadt L, Beatson SA, Emes RD, Winter EE, Webber C, Brandt P, Nyakatura G, Adetobi M, Chiaromonte F, Elnitski L, Eswara P, Hardison RC, Hou M, Kolbe D, Makova K, Miller W, Nekrutenko A, Riemer C, Schwartz S, Taylor J, Yang S, Zhang Y, Lindpaintner K, Andrews TD, Caccamo M, Clamp M, Clarke L, Curwen V, Durbin R, Eyras E, Searle SM, Cooper GM, Batzoglou S, Brudno M, Sidow A, Stone EA, Venter JC, Payseur BA, Bourque G, López-Otín C, Puente XS, Chakrabarti K, Chatterji S, Dewey C, Pachter L, Bray N, Yap VB, Caspi A, Tesler G, Pevzner PA, Haussler D, Roskin KM, Baertsch R, Clawson H, Furey TS, Hinrichs AS, Karolchik D, Kent WJ, Rosenbloom KR, Trumbower H, Weirauch M, Cooper DN, Stenson PD, Ma B, Brent M, Arumugam M, Shteynberg D, Copley RR, Taylor MS, Riethman H, Mudunuri U, Peterson J, Guyer M, Felsenfeld A, Old S, Mockrin S, Collins F; Rat Genome Sequencing Project Consortium . Genome sequence of the Brown Norway rat yields insights into mammalian evolution. Nature 428: 493–521, 2004. doi: 10.1038/nature02426. [DOI] [PubMed] [Google Scholar]
- 39.Gibson G. Rare and common variants: twenty arguments. Nat Rev Genet 13: 135–145, 2012. doi: 10.1038/nrg3118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gilissen C, Hoischen A, Brunner HG, Veltman JA. Unlocking Mendelian disease using exome sequencing. Genome Biol 12: 228, 2011. doi: 10.1186/gb-2011-12-9-228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Goldstein DB, Allen A, Keebler J, Margulies EH, Petrou S, Petrovski S, Sunyaev S. Sequencing studies in human genetics: design and interpretation. Nat Rev Genet 14: 460–470, 2013. doi: 10.1038/nrg3455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Guo X, Brenner M, Zhang X, Laragione T, Tai S, Li Y, Bu J, Yin Y, Shah AA, Kwan K, Li Y, Jun W, Gulko PS. Whole-genome sequences of DA and F344 rats with different susceptibilities to arthritis, autoimmunity, inflammation and cancer. Genetics 194: 1017–1028, 2013. doi: 10.1534/genetics.113.153049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gura T. Systems for identifying new drugs are often faulty. Science 278: 1041–1042, 1997. doi: 10.1126/science.278.5340.1041. [DOI] [PubMed] [Google Scholar]
- 44.Guryev V, Saar K, Adamovic T, Verheul M, van Heesch SAAC, Cook S, Pravenec M, Aitman T, Jacob H, Shull JD, Hubner N, Cuppen E. Distribution and functional impact of DNA copy number variation in the rat. Nat Genet 40: 538–545, 2008. doi: 10.1038/ng.141. [DOI] [PubMed] [Google Scholar]
- 45.Hall JM, Lee MK, Newman B, Morrow JE, Anderson LA, Huey B, King MC. Linkage of early-onset familial breast cancer to chromosome 17q21. Science 250: 1684–1689, 1990. doi: 10.1126/science.2270482. [DOI] [PubMed] [Google Scholar]
- 46.Hansen C, Spuhler K. Development of the National Institutes of Health genetically heterogeneous rat stock. Alcohol Clin Exp Res 8: 477–479, 1984. doi: 10.1111/j.1530-0277.1984.tb05706.x. [DOI] [PubMed] [Google Scholar]
- 47.Hayden EC. Technology: the $1,000 genome. Nature 507: 294–295, 2014. doi: 10.1038/507294a. [DOI] [PubMed] [Google Scholar]
- 48.Hebbring SJ. The challenges, advantages and future of phenome-wide association studies. Immunology 141: 157–165, 2014. doi: 10.1111/imm.12195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hermsen R, de Ligt J, Spee W, Blokzijl F, Schäfer S, Adami E, Boymans S, Flink S, van Boxtel R, van der Weide RH, Aitman T, Hübner N, Simonis M, Tabakoff B, Guryev V, Cuppen E. Genomic landscape of rat strain and substrain variation. BMC Genomics 16: 357, 2015. doi: 10.1186/s12864-015-1594-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hyun I. Illusory fears must not stifle chimaera research. Nature 537: 281, 2016. doi: 10.1038/537281a. [DOI] [PubMed] [Google Scholar]
- 51.Human Genome Sequencing Consortium I; International Human Genome Sequencing Consortium . Finishing the euchromatic sequence of the human genome. Nature 431: 931–945, 2004. doi: 10.1038/nature03001. [DOI] [PubMed] [Google Scholar]
- 52.Ioannidis NM, Rothstein JH, Pejaver V, Middha S, McDonnell SK, Baheti S, Musolf A, Li Q, Holzinger E, Karyadi D, Cannon-Albright LA, Teerlink CC, Stanford JL, Isaacs WB, Xu J, Cooney KA, Lange EM, Schleutker J, Carpten JD, Powell IJ, Cussenot O, Cancel-Tassin G, Giles GG, MacInnis RJ, Maier C, Hsieh C-L, Wiklund F, Catalona WJ, Foulkes WD, Mandal D, Eeles RA, Kote-Jarai Z, Bustamante CD, Schaid DJ, Hastie T, Ostrander EA, Bailey-Wilson JE, Radivojac P, Thibodeau SN, Whittemore AS, Sieh W. REVEL: An Ensemble Method for Predicting the Pathogenicity of Rare Missense Variants. Am J Hum Genet 99: 877–885, 2016. doi: 10.1016/j.ajhg.2016.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Issa AM. Ethical considerations in clinical pharmacogenomics research. Trends Pharmacol Sci 21: 247–249, 2000. doi: 10.1016/S0165-6147(00)01493-0. [DOI] [PubMed] [Google Scholar]
- 54.Ito M, Kobayashi K, Nakahata T. NOD/Shi-scid IL2rgamma(null) (NOG) mice more appropriate for humanized mouse models. Curr Top Microbiol Immunol 324: 53–76, 2008. [DOI] [PubMed] [Google Scholar]
- 55.Jacob HJ. Next-generation sequencing for clinical diagnostics. N Engl J Med 369: 1557–1558, 2013. doi: 10.1056/NEJMe1310846. [DOI] [PubMed] [Google Scholar]
- 56.Jacob HJ, Abrams K, Bick DP, Brodie K, Dimmock DP, Farrell M, Geurts J, Harris J, Helbling D, Joers BJ, Kliegman R, Kowalski G, Lazar J, Margolis DA, North P, Northup J, Roquemore-Goins A, Scharer G, Shimoyama M, Strong K, Taylor B, Tsaih S-W, Tschannen MR, Veith RL, Wendt-Andrae J, Wilk B, Worthey EA. Genomics in clinical practice: lessons from the front lines. Sci Transl Med 5: 194cm5, 2013. doi: 10.1126/scitranslmed.3006468. [DOI] [PubMed] [Google Scholar]
- 57.Johansen Taber KA, Dickinson BD, Wilson M. The promise and challenges of next-generation genome sequencing for clinical care. JAMA Intern Med 174: 275–280, 2014. doi: 10.1001/jamainternmed.2013.12048. [DOI] [PubMed] [Google Scholar]
- 58.Kaiser J. BIOETHICS. NIH plans to fund human-animal chimera research. Science 353: 634–635, 2016. doi: 10.1126/science.353.6300.634. [DOI] [PubMed] [Google Scholar]
- 59.Kan YW, Dozy AM. Polymorphism of DNA sequence adjacent to human beta-globin structural gene: relationship to sickle mutation. Proc Natl Acad Sci USA 75: 5631–5635, 1978. doi: 10.1073/pnas.75.11.5631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kerem B, Rommens JM, Buchanan JA, Markiewicz D, Cox TK, Chakravarti A, Buchwald M, Tsui LC. Identification of the cystic fibrosis gene: genetic analysis. Science 245: 1073–1080, 1989. doi: 10.1126/science.2570460. [DOI] [PubMed] [Google Scholar]
- 61.Kingsmore SF, Saunders CJ. Deep sequencing of patient genomes for disease diagnosis: when will it become routine? Sci Transl Med 3: 87ps23, 2011. doi: 10.1126/scitranslmed.3002695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Klein RJ, Zeiss C, Chew EY, Tsai J-Y, Sackler RS, Haynes C, Henning AK, SanGiovanni JP, Mane SM, Mayne ST, Bracken MB, Ferris FL, Ott J, Barnstable C, Hoh J. Complement factor H polymorphism in age-related macular degeneration. Science 308: 385–389, 2005. doi: 10.1126/science.1109557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Köttgen A, Pattaro C, Böger CA, Fuchsberger C, Olden M, Glazer NL, Parsa A, Gao X, Yang Q, Smith AV, O’Connell JR, Li M, Schmidt H, Tanaka T, Isaacs A, Ketkar S, Hwang S-J, Johnson AD, Dehghan A, Teumer A, Paré G, Atkinson EJ, Zeller T, Lohman K, Cornelis MC, Probst-Hensch NM, Kronenberg F, Tönjes A, Hayward C, Aspelund T, Eiriksdottir G, Launer LJ, Harris TB, Rampersaud E, Mitchell BD, Arking DE, Boerwinkle E, Struchalin M, Cavalieri M, Singleton A, Giallauria F, Metter J, de Boer IH, Haritunians T, Lumley T, Siscovick D, Psaty BM, Zillikens MC, Oostra BA, Feitosa M, Province M, de Andrade M, Turner ST, Schillert A, Ziegler A, Wild PS, Schnabel RB, Wilde S, Munzel TF, Leak TS, Illig T, Klopp N, Meisinger C, Wichmann H-E, Koenig W, Zgaga L, Zemunik T, Kolcic I, Minelli C, Hu FB, Johansson A, Igl W, Zaboli G, Wild SH, Wright AF, Campbell H, Ellinghaus D, Schreiber S, Aulchenko YS, Felix JF, Rivadeneira F, Uitterlinden AG, Hofman A, Imboden M, Nitsch D, Brandstätter A, Kollerits B, Kedenko L, Mägi R, Stumvoll M, Kovacs P, Boban M, Campbell S, Endlich K, Völzke H, Kroemer HK, Nauck M, Völker U, Polasek O, Vitart V, Badola S, Parker AN, Ridker PM, Kardia SL, Blankenberg S, Liu Y, Curhan GC, Franke A, Rochat T, Paulweber B, Prokopenko I, Wang W, Gudnason V, Shuldiner AR, Coresh J, Schmidt R, Ferrucci L, Shlipak MG, van Duijn CM, Borecki I, Krämer BK, Rudan I, Gyllensten U, Wilson JF, Witteman JC, Pramstaller PP, Rettig R, Hastie N, Chasman DI, Kao WH, Heid IM, Fox CS. New loci associated with kidney function and chronic kidney disease. Nat Genet 42: 376–384, 2010. doi: 10.1038/ng.568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kwitek AE, Jacob HJ, Baker JE, Dwinell MR, Forster HV, Greene AS, Kunert MP, Lombard JH, Mattson DL, Pritchard KA Jr, Roman RJ, Tonellato PJ, Cowley AW Jr. BN phenome: detailed characterization of the cardiovascular, renal, and pulmonary systems of the sequenced rat. Physiol Genomics 25: 303–313, 2006. doi: 10.1152/physiolgenomics.00288.2005. [DOI] [PubMed] [Google Scholar]
- 65.Landrum MJ, Lee JM, Riley GR, Jang W, Rubinstein WS, Church DM, Maglott DR. ClinVar: public archive of relationships among sequence variation and human phenotype. Nucleic Acids Res 42: D980–D985, 2013. doi: 10.1093/nar/gkt1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lek M, Karczewski KJ, Minikel EV, Samocha KE, Banks E, Fennell T, O’Donnell-Luria AH, Ware JS, Hill AJ, Cummings BB, Tukiainen T, Birnbaum DP, Kosmicki JA, Duncan LE, Estrada K, Zhao F, Zou J, Pierce-Hoffman E, Berghout J, Cooper DN, Deflaux N, DePristo M, Do R, Flannick J, Fromer M, Gauthier L, Goldstein J, Gupta N, Howrigan D, Kiezun A, Kurki MI, Moonshine AL, Natarajan P, Orozco L, Peloso GM, Poplin R, Rivas MA, Ruano-Rubio V, Rose SA, Ruderfer DM, Shakir K, Stenson PD, Stevens C, Thomas BP, Tiao G, Tusie-Luna MT, Weisburd B, Won H-H, Yu D, Altshuler DM, Ardissino D, Boehnke M, Danesh J, Donnelly S, Elosua R, Florez JC, Gabriel SB, Getz G, Glatt SJ, Hultman CM, Kathiresan S, Laakso M, McCarroll S, McCarthy MI, McGovern D, McPherson R, Neale BM, Palotie A, Purcell SM, Saleheen D, Scharf JM, Sklar P, Sullivan PF, Tuomilehto J, Tsuang MT, Watkins HC, Wilson JG, Daly MJ, MacArthur DG; Exome Aggregation Consortium . Analysis of protein-coding genetic variation in 60,706 humans. Nature 536: 285–291, 2016. doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Li D, Qiu Z, Shao Y, Chen Y, Guan Y, Liu M, Li Y, Gao N, Wang L, Lu X, Zhao Y, Liu M. Heritable gene targeting in the mouse and rat using a CRISPR-Cas system. Nat Biotechnol 31: 681–683, 2013. doi: 10.1038/nbt.2661. [DOI] [PubMed] [Google Scholar]
- 68.Libert C, Dejager L, Pinheiro I. The X chromosome in immune functions: when a chromosome makes the difference. Nat Rev Immunol 10: 594–604, 2010. doi: 10.1038/nri2815. [DOI] [PubMed] [Google Scholar]
- 69.Ligthart S, Marzi C, Aslibekyan S, Mendelson MM, Conneely KN, Tanaka T, Colicino E, Waite LL, Joehanes R, Guan W, Brody JA, Elks C, Marioni R, Jhun MA, Agha G, Bressler J, Ward-Caviness CK, Chen BH, Huan T, Bakulski K, Salfati EL, Fiorito G, Wahl S, Schramm K, Sha J, Hernandez DG, Just AC, Smith JA, Sotoodehnia N, Pilling LC, Pankow JS, Tsao PS, Liu C, Zhao W, Guarrera S, Michopoulos VJ, Smith AK, Peters MJ, Melzer D, Vokonas P, Fornage M, Prokisch H, Bis JC, Chu AY, Herder C, Grallert H, Yao C, Shah S, McRae AF, Lin H, Horvath S, Fallin D, Hofman A, Wareham NJ, Wiggins KL, Feinberg AP, Starr JM, Visscher PM, Murabito JM, Kardia SL, Absher DM, Binder EB, Singleton AB, Bandinelli S, Peters A, Waldenberger M, Matullo G, Schwartz JD, Demerath EW, Uitterlinden AG, van Meurs JB, Franco OH, Chen YI, Levy D, Turner ST, Deary IJ, Ressler KJ, Dupuis J, Ferrucci L, Ong KK, Assimes TL, Boerwinkle E, Koenig W, Arnett DK, Baccarelli AA, Benjamin EJ, Dehghan A; WHI-EMPC Investigators; CHARGE epigenetics of Coronary Heart Disease . DNA methylation signatures of chronic low-grade inflammation are associated with complex diseases. Genome Biol 17: 255, 2016. doi: 10.1186/s13059-016-1119-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lipton P. Pharmacogenetics: the ethical issues. Pharmacogenomics J 3: 14–16, 2003. doi: 10.1038/sj.tpj.6500159. [DOI] [PubMed] [Google Scholar]
- 71.MacDonald ME, Ambrose CM, Duyao MP, Myers RH, Lin C, Srinidhi L, Barnes G, Taylor SA, James M, Groot N, MacFarlane H, Jenkins B, Anderson MA, Wexler NS, Gusella JF, Bates GP, Baxendale S, Hummerich H, Kirby S, North M, Youngman S, Mott R, Zehetner G, Sedlacek Z, Poustka A, Frischauf A-M, Lehrach H, Buckler AJ, Church D, Doucette-Stamm L, O’Donovan MC, Riba-Ramirez L, Shah M, Stanton VP, Strobel SA, Draths KM, Wales JL, Dervan P, Housman DE, Altherr M, Shiang R, Thompson L, Fielder T, Wasmuth JJ, Tagle D, Valdes J, Elmer L, Allard M, Castilla L, Swaroop M, Blanchard K, Collins FS, Snell R, Holloway T, Gillespie K, Datson N, Shaw D, Harper PS; The Huntington’s Disease Collaborative Research Group . A novel gene containing a trinucleotide repeat that is expanded and unstable on Huntington’s disease chromosomes. Cell 72: 971–983, 1993. doi: 10.1016/0092-8674(93)90585-E. [DOI] [PubMed] [Google Scholar]
- 72.Martin HC, Kim GE, Pagnamenta AT, Murakami Y, Carvill GL, Meyer E, Copley RR, Rimmer A, Barcia G, Fleming MR, Kronengold J, Brown MR, Hudspith KA, Broxholme J, Kanapin A, Cazier J-B, Kinoshita T, Nabbout R, Bentley D, McVean G, Heavin S, Zaiwalla Z, McShane T, Mefford HC, Shears D, Stewart H, Kurian MA, Scheffer IE, Blair E, Donnelly P, Kaczmarek LK, Taylor JC; WGS500 Consortium . Clinical whole-genome sequencing in severe early-onset epilepsy reveals new genes and improves molecular diagnosis. Hum Mol Genet 23: 3200–3211, 2014. doi: 10.1093/hmg/ddu030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Mattson DL, Dwinell MR, Greene AS, Kwitek AE, Roman RJ, Jacob HJ, Cowley AW Jr. Chromosome substitution reveals the genetic basis of Dahl salt-sensitive hypertension and renal disease. Am J Physiol Renal Physiol 295: F837–F842, 2008. doi: 10.1152/ajprenal.90341.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.May T. Expanding Bioshield: a call for caution. Am J Public Health 97, Suppl 1: S23–S25, 2007. doi: 10.2105/AJPH.2005.077453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.May T. Rethinking clinical risk for DNA sequencing. Am J Bioeth 12: 24–26, 2012. doi: 10.1080/15265161.2012.699152. [DOI] [PubMed] [Google Scholar]
- 76.May T, Zusevics KL, Derse A, Strong KA, Jeruzal J, La Pean Kirschner A, Farrell MH, Spellecy R. The limits of traditional approaches to informed consent for genomic medicine. HEC Forum 26: 185–202, 2014. doi: 10.1007/s10730-014-9247-3. [DOI] [PubMed] [Google Scholar]
- 77.Mello MM, Wolf LE. The Havasupai Indian tribe case–lessons for research involving stored biologic samples. N Engl J Med 363: 204–207, 2010. doi: 10.1056/NEJMp1005203. [DOI] [PubMed] [Google Scholar]
- 78.Min Jou W, Haegeman G, Ysebaert M, Fiers W. Nucleotide sequence of the gene coding for the bacteriophage MS2 coat protein. Nature 237: 82–88, 1972. doi: 10.1038/237082a0. [DOI] [PubMed] [Google Scholar]
- 79.Morgan TH, Bridges CB. Sex-linked Inheritance in Drosophila. Washington, DC: Carnegie Institution of Washington, 1916. [Google Scholar]
- 80.Muller HJ. Radiation and Genetics. Am Nat 64: 220–251, 1930. doi: 10.1086/280313. [DOI] [Google Scholar]
- 81.Network and Pathway Analysis Subgroup of Psychiatric Genomics Consortium Psychiatric genome-wide association study analyses implicate neuronal, immune and histone pathways. Nat Neurosci 18: 199–209, 2015. [Erratum in Nat Neurosci 18: 926, 2015] 10.1038/nn.3922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Nicolae DL, Gamazon E, Zhang W, Duan S, Dolan ME, Cox NJ. Trait-associated SNPs are more likely to be eQTLs: annotation to enhance discovery from GWAS. PLoS Genet 6: e1000888, 2010. doi: 10.1371/journal.pgen.1000888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Okita K, Ichisaka T, Yamanaka S. Generation of germline-competent induced pluripotent stem cells. Nature 448: 313–317, 2007. doi: 10.1038/nature05934. [DOI] [PubMed] [Google Scholar]
- 84.Pendergrass SA, Brown-Gentry K, Dudek SM, Torstenson ES, Ambite JL, Avery CL, Buyske S, Cai C, Fesinmeyer MD, Haiman C, Heiss G, Hindorff LA, Hsu C-N, Jackson RD, Kooperberg C, Le Marchand L, Lin Y, Matise TC, Moreland L, Monroe K, Reiner AP, Wallace R, Wilkens LR, Crawford DC, Ritchie MD. The use of phenome-wide association studies (PheWAS) for exploration of novel genotype-phenotype relationships and pleiotropy discovery. Genet Epidemiol 35: 410–422, 2011. doi: 10.1002/gepi.20589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Perry JRB, Day F, Elks CE, Sulem P, Thompson DJ, Ferreira T, He C, Chasman DI, Esko T, Thorleifsson G, Albrecht E, Ang WQ, Corre T, Cousminer DL, Feenstra B, Franceschini N, Ganna A, Johnson AD, Kjellqvist S, Lunetta KL, McMahon G, Nolte IM, Paternoster L, Porcu E, Smith AV, Stolk L, Teumer A, Tšernikova N, Tikkanen E, Ulivi S, Wagner EK, Amin N, Bierut LJ, Byrne EM, Hottenga J-J, Koller DL, Mangino M, Pers TH, Yerges-Armstrong LM, Zhao JH, Andrulis IL, Anton-Culver H, Atsma F, Bandinelli S, Beckmann MW, Benitez J, Blomqvist C, Bojesen SE, Bolla MK, Bonanni B, Brauch H, Brenner H, Buring JE, Chang-Claude J, Chanock S, Chen J, Chenevix-Trench G, Collée JM, Couch FJ, Couper D, Coveillo AD, Cox A, Czene K, D’adamo AP, Smith GD, De Vivo I, Demerath EW, Dennis J, Devilee P, Dieffenbach AK, Dunning AM, Eiriksdottir G, Eriksson JG, Fasching PA, Ferrucci L, Flesch-Janys D, Flyger H, Foroud T, Franke L, Garcia ME, García-Closas M, Geller F, de Geus EEJ, Giles GG, Gudbjartsson DF, Gudnason V, Guénel P, Guo S, Hall P, Hamann U, Haring R, Hartman CA, Heath AC, Hofman A, Hooning MJ, Hopper JL, Hu FB, Hunter DJ, Karasik D, Kiel DP, Knight JA, Kosma V-M, Kutalik Z, Lai S, Lambrechts D, Lindblom A, Mägi R, Magnusson PK, Mannermaa A, Martin NG, Masson G, McArdle PF, McArdle WL, Melbye M, Michailidou K, Mihailov E, Milani L, Milne RL, Nevanlinna H, Neven P, Nohr EA, Oldehinkel AJ, Oostra BA, Palotie A, Peacock M, Pedersen NL, Peterlongo P, Peto J, Pharoah PD, Postma DS, Pouta A, Pylkäs K, Radice P, Ring S, Rivadeneira F, Robino A, Rose LM, Rudolph A, Salomaa V, Sanna S, Schlessinger D, Schmidt MK, Southey MC, Sovio U, Stampfer MJ, Stöckl D, Storniolo AM, Timpson NJ, Tyrer J, Visser JA, Vollenweider P, Völzke H, Waeber G, Waldenberger M, Wallaschofski H, Wang Q, Willemsen G, Winqvist R, Wolffenbuttel BH, Wright MJ, Australian Ovarian Cancer Study, GENICA Network, kConFab, LifeLines Cohort Study, InterAct Consortium; Early Growth Genetics (EGG) Consortium, Boomsma DI, Econs MJ, Khaw KT, Loos RJ, McCarthy MI, Montgomery GW, Rice JP, Streeten EA, Thorsteinsdottir U, van Duijn CM, Alizadeh BZ, Bergmann S, Boerwinkle E, Boyd HA, Crisponi L, Gasparini P, Gieger C, Harris TB, Ingelsson E, Järvelin MR, Kraft P, Lawlor D, Metspalu A, Pennell CE, Ridker PM, Snieder H, Sørensen TI, Spector TD, Strachan DP, Uitterlinden AG, Wareham NJ, Widen E, Zygmunt M, Murray A, Easton DF, Stefansson K, Murabito JM, Ong KK. Parent-of-origin-specific allelic associations among 106 genomic loci for age at menarche. Nature 514: 92–97, 2014. doi: 10.1038/nature13545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Philippakis AA, Azzariti DR, Beltran S, Brookes AJ, Brownstein CA, Brudno M, Brunner HG, Buske OJ, Carey K, Doll C, Dumitriu S, Dyke SOM, den Dunnen JT, Firth HV, Gibbs RA, Girdea M, Gonzalez M, Haendel MA, Hamosh A, Holm IA, Huang L, Hurles ME, Hutton B, Krier JB, Misyura A, Mungall CJ, Paschall J, Paten B, Robinson PN, Schiettecatte F, Sobreira NL, Swaminathan GJ, Taschner PE, Terry SF, Washington NL, Züchner S, Boycott KM, Rehm HL. The Matchmaker Exchange: a platform for rare disease gene discovery. Hum Mutat 36: 915–921, 2015. doi: 10.1002/humu.22858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Pittenger MF, Mackay AM, Beck SC, Jaiswal RK, Douglas R, Mosca JD, Moorman MA, Simonetti DW, Craig S, Marshak DR. Multilineage potential of adult human mesenchymal stem cells. Science 284: 143–147, 1999. doi: 10.1126/science.284.5411.143. [DOI] [PubMed] [Google Scholar]
- 88.Prokop JW, Deschepper CF. Chromosome Y genetic variants: impact in animal models and on human disease. Physiol Genomics 47: 525–537, 2015. doi: 10.1152/physiolgenomics.00074.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Prokop JW, Tsaih S-W, Faber AB, Boehme S, Underwood AC, Troyer S, Playl L, Milsted A, Turner ME, Ely D, Martins AS, Tutaj M, Lazar J, Dwinell MR, Jacob HJ. The phenotypic impact of the male-specific region of chromosome-Y in inbred mating: the role of genetic variants and gene duplications in multiple inbred rat strains. Biol Sex Differ 7: 10, 2016. doi: 10.1186/s13293-016-0064-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Puck JM, Willard HF. X inactivation in females with X-linked disease. N Engl J Med 338: 325–328, 1998. doi: 10.1056/NEJM199801293380611. [DOI] [PubMed] [Google Scholar]
- 91.Rakyan VK, Down TA, Balding DJ, Beck S. Epigenome-wide association studies for common human diseases. Nat Rev Genet 12: 529–541, 2011. doi: 10.1038/nrg3000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Rat Genome Sequencing and Mapping Consortium; Baud A, Hermsen R, Guryev V, Stridh P, Graham D, McBride MW, Foroud T, Calderari S, Diez M, Ockinger J, Beyeen AD, Gillett A, Abdelmagid N, Guerreiro-Cacais AO, Jagodic M, Tuncel J, Norin U, Beattie E, Huynh N, Miller WH, Koller DL, Alam I, Falak S, Osborne-Pellegrin M, Martinez-Membrives E, Canete T, Blazquez G, Vicens-Costa E, Mont-Cardona C, Diaz-Moran S, Tobena A, Hummel O, Zelenika D, Saar K, Patone G, Bauerfeind A, Bihoreau MT, Heinig M, Lee YA, Rintisch C, Schulz H, Wheeler DA, Worley KC, Muzny DM, Gibbs RA, Lathrop M, Lansu N, Toonen P, Ruzius FP, de Bruijn E, Hauser H, Adams DJ, Keane T, Atanur SS, Aitman TJ, Flicek P, Malinauskas T, Jones EY, Ekman D, Lopez-Aumatell R, Dominiczak AF, Johannesson M, Holmdahl R, Olsson T, Gauguier D, Hubner N, Fernandez-Teruel A, Cuppen E, Mott R, Flint J. Combined sequence-based and genetic mapping analysis of complex traits in outbred rats. Nat Genet 45: 767–775, 2013. doi: 10.1038/ng.2644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, Grody WW, Hegde M, Lyon E, Spector E, Voelkerding K, Rehm HL; ACMG Laboratory Quality Assurance Committee . Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med 17: 405–424, 2015. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Rietveld CA, Medland SE, Derringer J, Yang J, Esko T, Martin NW, Westra H-J, Shakhbazov K, Abdellaoui A, Agrawal A, Albrecht E, Alizadeh BZ, Amin N, Barnard J, Baumeister SE, Benke KS, Bielak LF, Boatman JA, Boyle PA, Davies G, de Leeuw C, Eklund N, Evans DS, Ferhmann R, Fischer K, Gieger C, Gjessing HK, Hägg S, Harris JR, Hayward C, Holzapfel C, Ibrahim-Verbaas CA, Ingelsson E, Jacobsson B, Joshi PK, Jugessur A, Kaakinen M, Kanoni S, Karjalainen J, Kolcic I, Kristiansson K, Kutalik Z, Lahti J, Lee SH, Lin P, Lind PA, Liu Y, Lohman K, Loitfelder M, McMahon G, Vidal PM, Meirelles O, Milani L, Myhre R, Nuotio M-L, Oldmeadow CJ, Petrovic KE, Peyrot WJ, Polasek O, Quaye L, Reinmaa E, Rice JP, Rizzi TS, Schmidt H, Schmidt R, Smith AV, Smith JA, Tanaka T, Terracciano A, van der Loos MJHM, Vitart V, Völzke H, Wellmann J, Yu L, Zhao W, Allik J, Attia JR, Bandinelli S, Bastardot F, Beauchamp J, Bennett DA, Berger K, Bierut LJ, Boomsma DI, Bültmann U, Campbell H, Chabris CF, Cherkas L, Chung MK, Cucca F, de Andrade M, De Jager PL, De Neve JE, Deary IJ, Dedoussis GV, Deloukas P, Dimitriou M, Eiríksdóttir G, Elderson MF, Eriksson JG, Evans DM, Faul JD, Ferrucci L, Garcia ME, Grönberg H, Guðnason V, Hall P, Harris JM, Harris TB, Hastie ND, Heath AC, Hernandez DG, Hoffmann W, Hofman A, Holle R, Holliday EG, Hottenga JJ, Iacono WG, Illig T, Järvelin MR, Kähönen M, Kaprio J, Kirkpatrick RM, Kowgier M, Latvala A, Launer LJ, Lawlor DA, Lehtimäki T, Li J, Lichtenstein P, Lichtner P, Liewald DC, Madden PA, Magnusson PK, Mäkinen TE, Masala M, McGue M, Metspalu A, Mielck A, Miller MB, Montgomery GW, Mukherjee S, Nyholt DR, Oostra BA, Palmer LJ, Palotie A, Penninx BW, Perola M, Peyser PA, Preisig M, Räikkönen K, Raitakari OT, Realo A, Ring SM, Ripatti S, Rivadeneira F, Rudan I, Rustichini A, Salomaa V, Sarin AP, Schlessinger D, Scott RJ, Snieder H, St Pourcain B, Starr JM, Sul JH, Surakka I, Svento R, Teumer A, Tiemeier H, van Rooij FJ, Van Wagoner DR, Vartiainen E, Viikari J, Vollenweider P, Vonk JM, Waeber G, Weir DR, Wichmann HE, Widen E, Willemsen G, Wilson JF, Wright AF, Conley D, Davey-Smith G, Franke L, Groenen PJ, Hofman A, Johannesson M, Kardia SL, Krueger RF, Laibson D, Martin NG, Meyer MN, Posthuma D, Thurik AR, Timpson NJ, Uitterlinden AG, van Duijn CM, Visscher PM, Benjamin DJ, Cesarini D, Koellinger PD; LifeLines Cohort Study . GWAS of 126,559 individuals identifies genetic variants associated with educational attainment. Science 340: 1467–1471, 2013. doi: 10.1126/science.1235488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Roadmap Epigenomics Consortium, Kundaje A, Meuleman W, Ernst J, Bilenky M, Yen A, Heravi-Moussavi A, Kheradpour P, Zhang Z, Wang J, Ziller MJ, Amin V, Whitaker JW, Schultz MD, Ward LD, Sarkar A, Quon G, Sandstrom RS, Eaton ML, Wu Y-C, Pfenning AR, Wang X, Claussnitzer M, Liu Y, Coarfa C, Harris RA, Shoresh N, Epstein CB, Gjoneska E, Leung D, Xie W, Hawkins RD, Lister R, Hong C, Gascard P, Mungall AJ, Moore R, Chuah E, Tam A, Canfield TK, Hansen RS, Kaul R, Sabo PJ, Bansal MS, Carles A, Dixon JR, Farh K-H, Feizi S, Karlic R, Kim A-R, Kulkarni A, Li D, Lowdon R, Elliott G, Mercer TR, Neph SJ, Onuchic V, Polak P, Rajagopal N, Ray P, Sallari RC, Siebenthall KT, Sinnott-Armstrong NA, Stevens M, Thurman RE, Wu J, Zhang B, Zhou X, Beaudet AE, Boyer LA, De Jager PL, Farnham PJ, Fisher SJ, Haussler D, Jones SJM, Li W, Marra MA, McManus MT, Sunyaev S, Thomson JA, Tlsty TD, Tsai L-H, Wang W, Waterland RA, Zhang MQ, Chadwick LH, Bernstein BE, Costello JF, Ecker JR, Hirst M, Meissner A, Milosavljevic A, Ren B, Stamatoyannopoulos JA, Wang T, Kellis M. Integrative analysis of 111 reference human epigenomes. Nature 518: 317–330, 2015. doi: 10.1038/nature14248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Sanger F, Air GM, Barrell BG, Brown NL, Coulson AR, Fiddes CA, Hutchison CA, Slocombe PM, Smith M. Nucleotide sequence of bacteriophage phi X174 DNA. Nature 265: 687–695, 1977. doi: 10.1038/265687a0. [DOI] [PubMed] [Google Scholar]
- 97.Sanger F, Nicklen S, Coulson AR. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA 74: 5463–5467, 1977. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Saunders CJ, Miller NA, Soden SE, Dinwiddie DL, Noll A, Alnadi NA, Andraws N, Patterson ML, Krivohlavek LA, Fellis J, Humphray S, Saffrey P, Kingsbury Z, Weir JC, Betley J, Grocock RJ, Margulies EH, Farrow EG, Artman M, Safina NP, Petrikin JE, Hall KP, Kingsmore SF. Rapid whole-genome sequencing for genetic disease diagnosis in neonatal intensive care units. Sci Transl Med 4: 154ra135, 2012. doi: 10.1126/scitranslmed.3004041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Shimoyama M, Laulederkind SJF, De Pons J, Nigam R, Smith JR, Tutaj M, Petri V, Hayman GT, Wang S-J, Ghiasvand O, Thota J, Dwinell MR. Exploring human disease using the Rat Genome Database. Dis Model Mech 9: 1089–1095, 2016. doi: 10.1242/dmm.026021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Simonis M, Atanur SS, Linsen S, Guryev V, Ruzius F-P, Game L, Lansu N, de Bruijn E, van Heesch S, Jones SJM, Pravenec M, Aitman TJ, Cuppen E. Genetic basis of transcriptome differences between the founder strains of the rat HXB/BXH recombinant inbred panel. Genome Biol 13: r31, 2012. doi: 10.1186/gb-2012-13-4-r31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Singer JB, Hill AE, Burrage LC, Olszens KR, Song J, Justice M, O’Brien WE, Conti DV, Witte JS, Lander ES, Nadeau JH. Genetic dissection of complex traits with chromosome substitution strains of mice. Science 304: 445–448, 2004. doi: 10.1126/science.1093139. [DOI] [PubMed] [Google Scholar]
- 102.Si-Tayeb K, Noto FK, Nagaoka M, Li J, Battle MA, Duris C, North PE, Dalton S, Duncan SA. Highly efficient generation of human hepatocyte-like cells from induced pluripotent stem cells. Hepatology 51: 297–305, 2010. doi: 10.1002/hep.23354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Speliotes EK, Willer CJ, Berndt SI, Monda KL, Thorleifsson G, Jackson AU, Lango Allen H, Lindgren CM, Luan J, Mägi R, Randall JC, Vedantam S, Winkler TW, Qi L, Workalemahu T, Heid IM, Steinthorsdottir V, Stringham HM, Weedon MN, Wheeler E, Wood AR, Ferreira T, Weyant RJ, Segrè AV, Estrada K, Liang L, Nemesh J, Park J-H, Gustafsson S, Kilpeläinen TO, Yang J, Bouatia-Naji N, Esko T, Feitosa MF, Kutalik Z, Mangino M, Raychaudhuri S, Scherag A, Smith AV, Welch R, Zhao JH, Aben KK, Absher DM, Amin N, Dixon AL, Fisher E, Glazer NL, Goddard ME, Heard-Costa NL, Hoesel V, Hottenga J-J, Johansson A, Johnson T, Ketkar S, Lamina C, Li S, Moffatt MF, Myers RH, Narisu N, Perry JRB, Peters MJ, Preuss M, Ripatti S, Rivadeneira F, Sandholt C, Scott LJ, Timpson NJ, Tyrer JP, van Wingerden S, Watanabe RM, White CC, Wiklund F, Barlassina C, Chasman DI, Cooper MN, Jansson JO, Lawrence RW, Pellikka N, Prokopenko I, Shi J, Thiering E, Alavere H, Alibrandi MT, Almgren P, Arnold AM, Aspelund T, Atwood LD, Balkau B, Balmforth AJ, Bennett AJ, Ben-Shlomo Y, Bergman RN, Bergmann S, Biebermann H, Blakemore AI, Boes T, Bonnycastle LL, Bornstein SR, Brown MJ, Buchanan TA, Busonero F, Campbell H, Cappuccio FP, Cavalcanti-Proença C, Chen YD, Chen CM, Chines PS, Clarke R, Coin L, Connell J, Day IN, den Heijer M, Duan J, Ebrahim S, Elliott P, Elosua R, Eiriksdottir G, Erdos MR, Eriksson JG, Facheris MF, Felix SB, Fischer-Posovszky P, Folsom AR, Friedrich N, Freimer NB, Fu M, Gaget S, Gejman PV, Geus EJ, Gieger C, Gjesing AP, Goel A, Goyette P, Grallert H, Grässler J, Greenawalt DM, Groves CJ, Gudnason V, Guiducci C, Hartikainen AL, Hassanali N, Hall AS, Havulinna AS, Hayward C, Heath AC, Hengstenberg C, Hicks AA, Hinney A, Hofman A, Homuth G, Hui J, Igl W, Iribarren C, Isomaa B, Jacobs KB, Jarick I, Jewell E, John U, Jørgensen T, Jousilahti P, Jula A, Kaakinen M, Kajantie E, Kaplan LM, Kathiresan S, Kettunen J, Kinnunen L, Knowles JW, Kolcic I, König IR, Koskinen S, Kovacs P, Kuusisto J, Kraft P, Kvaløy K, Laitinen J, Lantieri O, Lanzani C, Launer LJ, Lecoeur C, Lehtimäki T, Lettre G, Liu J, Lokki ML, Lorentzon M, Luben RN, Ludwig B, Manunta P, Marek D, Marre M, Martin NG, McArdle WL, McCarthy A, McKnight B, Meitinger T, Melander O, Meyre D, Midthjell K, Montgomery GW, Morken MA, Morris AP, Mulic R, Ngwa JS, Nelis M, Neville MJ, Nyholt DR, O’Donnell CJ, O’Rahilly S, Ong KK, Oostra B, Paré G, Parker AN, Perola M, Pichler I, Pietiläinen KH, Platou CG, Polasek O, Pouta A, Rafelt S, Raitakari O, Rayner NW, Ridderstråle M, Rief W, Ruokonen A, Robertson NR, Rzehak P, Salomaa V, Sanders AR, Sandhu MS, Sanna S, Saramies J, Savolainen MJ, Scherag S, Schipf S, Schreiber S, Schunkert H, Silander K, Sinisalo J, Siscovick DS, Smit JH, Soranzo N, Sovio U, Stephens J, Surakka I, Swift AJ, Tammesoo ML, Tardif JC, Teder-Laving M, Teslovich TM, Thompson JR, Thomson B, Tönjes A, Tuomi T, van Meurs JB, van Ommen GJ, Vatin V, Viikari J, Visvikis-Siest S, Vitart V, Vogel CI, Voight BF, Waite LL, Wallaschofski H, Walters GB, Widen E, Wiegand S, Wild SH, Willemsen G, Witte DR, Witteman JC, Xu J, Zhang Q, Zgaga L, Ziegler A, Zitting P, Beilby JP, Farooqi IS, Hebebrand J, Huikuri HV, James AL, Kähönen M, Levinson DF, Macciardi F, Nieminen MS, Ohlsson C, Palmer LJ, Ridker PM, Stumvoll M, Beckmann JS, Boeing H, Boerwinkle E, Boomsma DI, Caulfield MJ, Chanock SJ, Collins FS, Cupples LA, Smith GD, Erdmann J, Froguel P, Grönberg H, Gyllensten U, Hall P, Hansen T, Harris TB, Hattersley AT, Hayes RB, Heinrich J, Hu FB, Hveem K, Illig T, Jarvelin MR, Kaprio J, Karpe F, Khaw KT, Kiemeney LA, Krude H, Laakso M, Lawlor DA, Metspalu A, Munroe PB, Ouwehand WH, Pedersen O, Penninx BW, Peters A, Pramstaller PP, Quertermous T, Reinehr T, Rissanen A, Rudan I, Samani NJ, Schwarz PE, Shuldiner AR, Spector TD, Tuomilehto J, Uda M, Uitterlinden A, Valle TT, Wabitsch M, Waeber G, Wareham NJ, Watkins H, Wilson JF, Wright AF, Zillikens MC, Chatterjee N, McCarroll SA, Purcell S, Schadt EE, Visscher PM, Assimes TL, Borecki IB, Deloukas P, Fox CS, Groop LC, Haritunians T, Hunter DJ, Kaplan RC, Mohlke KL, O’Connell JR, Peltonen L, Schlessinger D, Strachan DP, van Duijn CM, Wichmann HE, Frayling TM, Thorsteinsdottir U, Abecasis GR, Barroso I, Boehnke M, Stefansson K, North KE, McCarthy MI, Hirschhorn JN, Ingelsson E, Loos RJ, MAGIC; Procardis Consortium . Association analyses of 249,796 individuals reveal 18 new loci associated with body mass index. Nat Genet 42: 937–948, 2010. doi: 10.1038/ng.686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Stark A, Lin MF, Kheradpour P, Pedersen JS, Parts L, Carlson JW, Crosby MA, Rasmussen MD, Roy S, Deoras AN, Ruby JG, Brennecke J, Hodges E, Hinrichs AS, Caspi A, Paten B, Park SW, Han MV, Maeder ML, Polansky BJ, Robson BE, Aerts S, van Helden J, Hassan B, Gilbert DG, Eastman DA, Rice M, Weir M, Hahn MW, Park Y, Dewey CN, Pachter L, Kent WJ, Haussler D, Lai EC, Bartel DP, Hannon GJ, Kaufman TC, Eisen MB, Clark AG, Smith D, Celniker SE, Gelbart WM, Kellis M; Harvard FlyBase curators; Berkeley Drosophila Genome Project . Discovery of functional elements in 12 Drosophila genomes using evolutionary signatures. Nature 450: 219–232, 2007. doi: 10.1038/nature06340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Stephenson R, Metcalfe NH. Drosophila melanogaster: a fly through its history and current use. J R Coll Physicians Edinb 43: 70–75, 2013. doi: 10.4997/JRCPE.2013.116. [DOI] [PubMed] [Google Scholar]
- 106.Sturtevant AH. The behavior of the chromosomes as studied through linkage. Zeitschr Indukt Abstamm- Vererbungslehre 13: 234–287, 1915. [Google Scholar]
- 107.Sturtevant AH, Novitski E. The Homologies of the Chromosome Elements in the Genus Drosophila. Genetics 26: 517–541, 1941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Surakka I, Horikoshi M, Mägi R, Sarin A-P, Mahajan A, Lagou V, Marullo L, Ferreira T, Miraglio B, Timonen S, Kettunen J, Pirinen M, Karjalainen J, Thorleifsson G, Hägg S, Hottenga J-J, Isaacs A, Ladenvall C, Beekman M, Esko T, Ried JS, Nelson CP, Willenborg C, Gustafsson S, Westra H-J, Blades M, de Craen AJM, de Geus EJ, Deelen J, Grallert H, Hamsten A, Havulinna AS, Hengstenberg C, Houwing-Duistermaat JJ, Hyppönen E, Karssen LC, Lehtimäki T, Lyssenko V, Magnusson PKE, Mihailov E, Müller-Nurasyid M, Mpindi J-P, Pedersen NL, Penninx BWJH, Perola M, Pers TH, Peters A, Rung J, Smit JH, Steinthorsdottir V, Tobin MD, Tsernikova N, van Leeuwen EM, Viikari JS, Willems SM, Willemsen G, Schunkert H, Erdmann J, Samani NJ, Kaprio J, Lind L, Gieger C, Metspalu A, Slagboom PE, Groop L, van Duijn CM, Eriksson JG, Jula A, Salomaa V, Boomsma DI, Power C, Raitakari OT, Ingelsson E, Järvelin M-R, Thorsteinsdottir U, Franke L, Ikonen E, Kallioniemi O, Pietiäinen V, Lindgren CM, Stefansson K, Palotie A, McCarthy MI, Morris AP, Prokopenko I, Ripatti S; ENGAGE Consortium . The impact of low-frequency and rare variants on lipid levels. Nat Genet 47: 589–597, 2015. doi: 10.1038/ng.3300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, Yamanaka S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell 131: 861–872, 2007. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 110.Telenti A, Pierce LCT, Biggs WH, di Iulio J, Wong EHM, Fabani MM, Kirkness EF, Moustafa A, Shah N, Xie C, Brewerton SC, Bulsara N, Garner C, Metzker G, Sandoval E, Perkins BA, Och FJ, Turpaz Y, Venter JC. Deep sequencing of 10,000 human genomes. Proc Natl Acad Sci USA 113: 11901–11906, 2016. doi: 10.1073/pnas.1613365113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Tennessen JA, Bigham AW, O’Connor TD, Fu W, Kenny EE, Gravel S, McGee S, Do R, Liu X, Jun G, Kang HM, Jordan D, Leal SM, Gabriel S, Rieder MJ, Abecasis G, Altshuler D, Nickerson DA, Boerwinkle E, Sunyaev S, Bustamante CD, Bamshad MJ, Akey JM, Broad GO, Seattle GO; NHLBI Exome Sequencing Project . Evolution and functional impact of rare coding variation from deep sequencing of human exomes. Science 337: 64–69, 2012. doi: 10.1126/science.1219240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Tentler JJ, Tan AC, Weekes CD, Jimeno A, Leong S, Pitts TM, Arcaroli JJ, Messersmith WA, Eckhardt SG. Patient-derived tumour xenografts as models for oncology drug development. Nat Rev Clin Oncol 9: 338–350, 2012. doi: 10.1038/nrclinonc.2012.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Uhlen M, Oksvold P, Fagerberg L, Lundberg E, Jonasson K, Forsberg M, Zwahlen M, Kampf C, Wester K, Hober S, Wernerus H, Björling L, Ponten F. Towards a knowledge-based Human Protein Atlas. Nat Biotechnol 28: 1248–1250, 2010. doi: 10.1038/nbt1210-1248. [DOI] [PubMed] [Google Scholar]
- 114.Venter JC, Adams MD, Myers EW, Li PW, Mural RJ, Sutton GG, Smith HO, Yandell M, Evans CA, Holt RA, Gocayne JD, Amanatides P, Ballew RM, Huson DH, Wortman JR, Zhang Q, Kodira CD, Zheng XH, Chen L, Skupski M, Subramanian G, Thomas PD, Zhang J, Gabor Miklos GL, Nelson C, Broder S, Clark AG, Nadeau J, McKusick VA, Zinder N, Levine AJ, Roberts RJ, Simon M, Slayman C, Hunkapiller M, Bolanos R, Delcher A, Dew I, Fasulo D, Flanigan M, Florea L, Halpern A, Hannenhalli S, Kravitz S, Levy S, Mobarry C, Reinert K, Remington K, Abu-Threideh J, Beasley E, Biddick K, Bonazzi V, Brandon R, Cargill M, Chandramouliswaran I, Charlab R, Chaturvedi K, Deng Z, Di Francesco V, Dunn P, Eilbeck K, Evangelista C, Gabrielian AE, Gan W, Ge W, Gong F, Gu Z, Guan P, Heiman TJ, Higgins ME, Ji RR, Ke Z, Ketchum KA, Lai Z, Lei Y, Li Z, Li J, Liang Y, Lin X, Lu F, Merkulov GV, Milshina N, Moore HM, Naik AK, Narayan VA, Neelam B, Nusskern D, Rusch DB, Salzberg S, Shao W, Shue B, Sun J, Wang Z, Wang A, Wang X, Wang J, Wei M, Wides R, Xiao C, Yan C, Yao A, Ye J, Zhan M, Zhang W, Zhang H, Zhao Q, Zheng L, Zhong F, Zhong W, Zhu S, Zhao S, Gilbert D, Baumhueter S, Spier G, Carter C, Cravchik A, Woodage T, Ali F, An H, Awe A, Baldwin D, Baden H, Barnstead M, Barrow I, Beeson K, Busam D, Carver A, Center A, Cheng ML, Curry L, Danaher S, Davenport L, Desilets R, Dietz S, Dodson K, Doup L, Ferriera S, Garg N, Gluecksmann A, Hart B, Haynes J, Haynes C, Heiner C, Hladun S, Hostin D, Houck J, Howland T, Ibegwam C, Johnson J, Kalush F, Kline L, Koduru S, Love A, Mann F, May D, McCawley S, McIntosh T, McMullen I, Moy M, Moy L, Murphy B, Nelson K, Pfannkoch C, Pratts E, Puri V, Qureshi H, Reardon M, Rodriguez R, Rogers YH, Romblad D, Ruhfel B, Scott R, Sitter C, Smallwood M, Stewart E, Strong R, Suh E, Thomas R, Tint NN, Tse S, Vech C, Wang G, Wetter J, Williams S, Williams M, Windsor S, Winn-Deen E, Wolfe K, Zaveri J, Zaveri K, Abril JF, Guigó R, Campbell MJ, Sjolander KV, Karlak B, Kejariwal A, Mi H, Lazareva B, Hatton T, Narechania A, Diemer K, Muruganujan A, Guo N, Sato S, Bafna V, Istrail S, Lippert R, Schwartz R, Walenz B, Yooseph S, Allen D, Basu A, Baxendale J, Blick L, Caminha M, Carnes-Stine J, Caulk P, Chiang YH, Coyne M, Dahlke C, Mays A, Dombroski M, Donnelly M, Ely D, Esparham S, Fosler C, Gire H, Glanowski S, Glasser K, Glodek A, Gorokhov M, Graham K, Gropman B, Harris M, Heil J, Henderson S, Hoover J, Jennings D, Jordan C, Jordan J, Kasha J, Kagan L, Kraft C, Levitsky A, Lewis M, Liu X, Lopez J, Ma D, Majoros W, McDaniel J, Murphy S, Newman M, Nguyen T, Nguyen N, Nodell M, Pan S, Peck J, Peterson M, Rowe W, Sanders R, Scott J, Simpson M, Smith T, Sprague A, Stockwell T, Turner R, Venter E, Wang M, Wen M, Wu D, Wu M, Xia A, Zandieh A, Zhu X. The sequence of the human genome. Science 291: 1304–1351, 2001. doi: 10.1126/science.1058040. [DOI] [PubMed] [Google Scholar]
- 115.Visscher PM, Brown MA, McCarthy MI, Yang J. Five years of GWAS discovery. Am J Hum Genet 90: 7–24, 2012. doi: 10.1016/j.ajhg.2011.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Welter D, MacArthur J, Morales J, Burdett T, Hall P, Junkins H, Klemm A, Flicek P, Manolio T, Hindorff L, Parkinson H. The NHGRI GWAS Catalog, a curated resource of SNP-trait associations. Nucleic Acids Res 42, D1: D1001–D1006, 2014. doi: 10.1093/nar/gkt1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Willer CJ, Li Y, Abecasis GR. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics 26: 2190–2191, 2010. doi: 10.1093/bioinformatics/btq340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Worthey EA, Mayer AN, Syverson GD, Helbling D, Bonacci BB, Decker B, Serpe JM, Dasu T, Tschannen MR, Veith RL, Basehore MJ, Broeckel U, Tomita-Mitchell A, Arca MJ, Casper JT, Margolis DA, Bick DP, Hessner MJ, Routes JM, Verbsky JW, Jacob HJ, Dimmock DP. Making a definitive diagnosis: successful clinical application of whole exome sequencing in a child with intractable inflammatory bowel disease. Genet Med 13: 255–262, 2011. doi: 10.1097/GIM.0b013e3182088158. [DOI] [PubMed] [Google Scholar]
- 119.Yang Y, Muzny DM, Reid JG, Bainbridge MN, Willis A, Ward PA, Braxton A, Beuten J, Xia F, Niu Z, Hardison M, Person R, Bekheirnia MR, Leduc MS, Kirby A, Pham P, Scull J, Wang M, Ding Y, Plon SE, Lupski JR, Beaudet AL, Gibbs RA, Eng CM. Clinical whole-exome sequencing for the diagnosis of Mendelian disorders. N Engl J Med 369: 1502–1511, 2013. doi: 10.1056/NEJMoa1306555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Yang Y, Muzny DM, Xia F, Niu Z, Person R, Ding Y, Ward P, Braxton A, Wang M, Buhay C, Veeraraghavan N, Hawes A, Chiang T, Leduc M, Beuten J, Zhang J, He W, Scull J, Willis A, Landsverk M, Craigen WJ, Bekheirnia MR, Stray-Pedersen A, Liu P, Wen S, Alcaraz W, Cui H, Walkiewicz M, Reid J, Bainbridge M, Patel A, Boerwinkle E, Beaudet AL, Lupski JR, Plon SE, Gibbs RA, Eng CM. Molecular findings among patients referred for clinical whole-exome sequencing. JAMA 312: 1870–1879, 2014. doi: 10.1001/jama.2014.14601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Yu J, Vodyanik MA, Smuga-Otto K, Antosiewicz-Bourget J, Frane JL, Tian S, Nie J, Jonsdottir GA, Ruotti V, Stewart R, Slukvin II, Thomson JA. Induced pluripotent stem cell lines derived from human somatic cells. Science 318: 1917–1920, 2007. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
- 122.Yu Y, Fuscoe JC, Zhao C, Guo C, Jia M, Qing T, Bannon DI, Lancashire L, Bao W, Du T, Luo H, Su Z, Jones WD, Moland CL, Branham WS, Qian F, Ning B, Li Y, Hong H, Guo L, Mei N, Shi T, Wang KY, Wolfinger RD, Nikolsky Y, Walker SJ, Duerksen-Hughes P, Mason CE, Tong W, Thierry-Mieg J, Thierry-Mieg D, Shi L, Wang C. A rat RNA-Seq transcriptomic BodyMap across 11 organs and 4 developmental stages. Nat Commun 5: 3230, 2014. doi: 10.1038/ncomms4230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Yu Y, Wu BL, Wu J, Shen Y. Exome and whole-genome sequencing as clinical tests: a transformative practice in molecular diagnostics. Clin Chem 58: 1507–1509, 2012. doi: 10.1373/clinchem.2012.193128. [DOI] [PubMed] [Google Scholar]
- 124.Yusa K, Rashid ST, Strick-Marchand H, Varela I, Liu P-Q, Paschon DE, Miranda E, Ordóñez A, Hannan NRF, Rouhani FJ, Darche S, Alexander G, Marciniak SJ, Fusaki N, Hasegawa M, Holmes MC, Di Santo JP, Lomas DA, Bradley A, Vallier L. Targeted gene correction of α1-antitrypsin deficiency in induced pluripotent stem cells. Nature 478: 391–394, 2011. doi: 10.1038/nature10424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Zhang J, Wilson GF, Soerens AG, Koonce CH, Yu J, Palecek SP, Thomson JA, Kamp TJ. Functional cardiomyocytes derived from human induced pluripotent stem cells. Circ Res 104: e30–e41, 2009. doi: 10.1161/CIRCRESAHA.108.192237. [DOI] [PMC free article] [PubMed] [Google Scholar]