NSI is an active chloroplast Lys acetyltransferase required for state transitions in Arabidopsis independently of LHCII phosphorylation.
Abstract
The amount of light energy received by the photosynthetic reaction centers photosystem II (PSII) and photosystem I (PSI) is balanced through state transitions. Reversible phosphorylation of a light-harvesting antenna trimer (L-LHCII) orchestrates the association between L-LHCII and the photosystems, thus adjusting the amount of excitation energy received by the reaction centers. In this study, we identified the enzyme NUCLEAR SHUTTLE INTERACTING (NSI; AT1G32070) as an active lysine acetyltransferase in the chloroplasts of Arabidopsis thaliana. Intriguingly, nsi knockout mutant plants were defective in state transitions, even though they had a similar LHCII phosphorylation pattern as the wild type. Accordingly, nsi plants were not able to accumulate the PSI-LHCII state transition complex, even though the LHCII docking site of PSI and the overall amounts of photosynthetic protein complexes remained unchanged. Instead, the nsi mutants showed a decreased Lys acetylation status of specific photosynthetic proteins including PSI, PSII, and LHCII subunits. Our work demonstrates that the chloroplast acetyltransferase NSI is needed for the dynamic reorganization of thylakoid protein complexes during photosynthetic state transitions.
INTRODUCTION
Light quality and quantity regulate photosynthetic light harvesting through dynamic reorganization of thylakoid membranes and the embedded protein complexes. A pool of LHCII trimers (L-LHCII) can function as an antenna either for photosystem II (PSII) or photosystem I (PSI; Galka et al., 2012) and thereby adjust the amount of excitation energy received by the two photosystems (Allen, 1992). Changes in the association of L-LCHII between the reaction centers are referred to as state transitions, and they are regulated in a light-dependent manner via the reversible phosphorylation of L-LHCII subunits LHCB1 and LHCB2 (Bonaventura and Myers, 1969; Murata, 1969; Pietrzykowska et al., 2014). Upon illumination that leads to plastoquinone (PQ) pool reduction, LHCB1 and LHCB2 are phosphorylated by the STN7 kinase (Depège et al., 2003; Bellafiore et al., 2005), which results in increased absorbance of PSI (state 2). By contrast, illumination favoring PQ oxidation leads to dephosphorylation of LHCII by the PPH1/TAP38 phosphatase (Pribil et al., 2010; Shapiguzov et al., 2010) and energy distribution toward PSII (state 1). Specifically, phosphorylation of LHCB2 is required for the attachment of L-trimers to PSI via the PSAH subunit (Lunde et al., 2000; Crepin and Caffarri, 2015; Longoni et al., 2015), which results in the formation of a PSI-LHCII complex in the nonstacked regions of the thylakoid membrane (Kouřil et al., 2005). Additionally, some L-trimers appear to interact with PSI via the LHCA proteins (Benson et al., 2015). In C3 plants, state transitions have an important role in protecting PSI from photodamage under fluctuating light intensity (Grieco et al., 2012).
While phosphorylation is the best-studied posttranslational modification regulating protein function, recent progress in enrichment techniques and high precision mass spectrometry have provided evidence that other modification types, such as the reversible acetylation of lysine (Lys) residues, are abundant on chloroplast proteins as well (Finkemeier et al., 2011; Wu et al., 2011; Hartl et al., 2017; Schmidt et al., 2017). Lys acetylation was originally identified as a regulator of gene expression in the nucleus, where histone proteins undergo extensive acetylation/deacetylation by histone acetyltransferases and deacetylases. However, the acetylation machinery and the functional significance of Lys acetylation in chloroplasts have remained largely unknown. To gain insight into the role of Lys acetylation in the regulation of chloroplast function, we studied the Arabidopsis thaliana enzyme NSI (NUCLEAR SHUTTLE INTERACTING; ATNSI; SNAT; AT1G32070). Based on its amino acid sequence, NSI is predicted to contain an acetyltransferase domain and a chloroplast targeting transit peptide, which makes it a putative chloroplast acetyltransferase enzyme.
In this study, we have employed quantitative mass spectrometry and in vitro Lys acetyltransferase assays to investigate the role of NSI as a Lys acetyltransferase in Arabidopsis. The results showed that NSI is an active chloroplast-localized Lys acetyltransferase that affects the acetylation status of several chloroplast proteins. Since some of the affected proteins were found to be involved in the light reactions of photosynthesis, we further characterized the photosynthetic properties of two Arabidopsis knockout lines lacking NSI (nsi-1 and nsi-2). We found that the nsi mutants were not able to undergo state transitions in response to changes in illumination, even though the plants had wild-type levels of LHCII phosphorylation and the LHCII docking site on PSI was not impaired. In light of our results, we propose that NSI is critical for the dynamic rearrangements of thylakoid membranes (i.e., state transitions). Possible mechanistic explanations will be discussed, but the exact mechanism for NSI action will remain an interesting topic for future research.
RESULTS
NSI Is a Chloroplast-Localized Lys Acetyltransferase
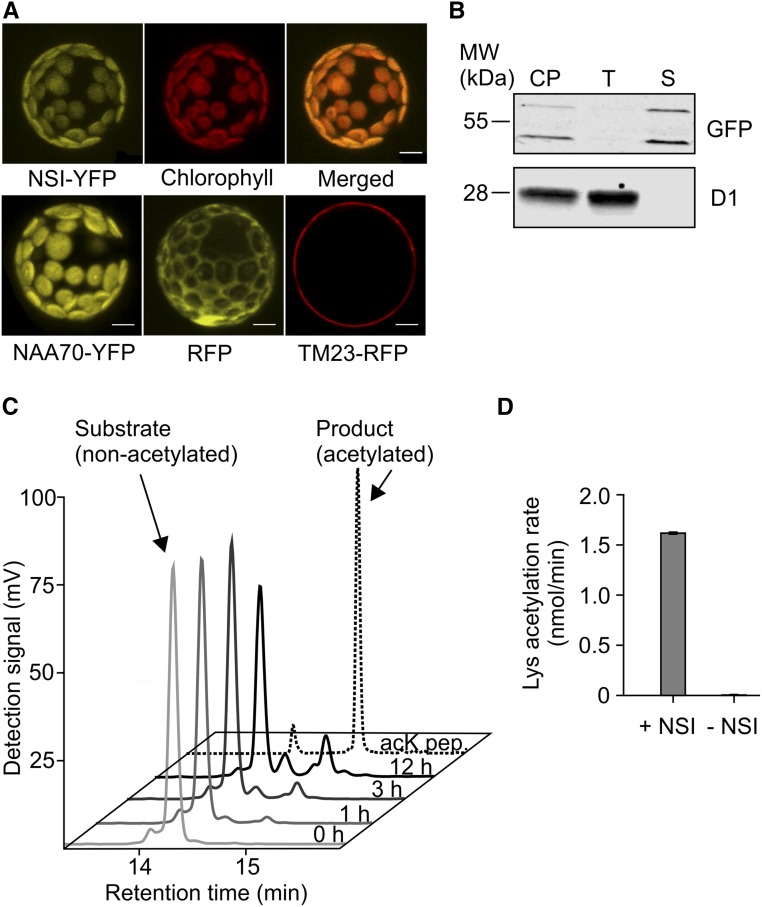
NSI localization was studied with a transient overexpression of NSI-YFP fusion protein in Arabidopsis protoplasts. The fusion protein clearly colocalized with chlorophyll autofluorescence, while no signal was detected in other compartments such as the nucleus (Figure 1). Immunoblotting of chloroplast fractions isolated from transgenic Arabidopsis lines expressing NSI-YFP further revealed that the majority of NSI is present in the soluble chloroplast fraction (Figure 1B). This observed plastid localization corroborates earlier results by Lee et al. (2014). To test whether NSI is able to function as a Lys acetyltransferase, an N-terminally His6-tagged recombinant protein lacking the predicted transit peptide (57 amino acids) was produced, and the Lys acetylation activity of the purified recombinant protein was examined on a general Lys acetyltransferase peptide substrate. NSI was indeed able to acetylate the ε-amino group of Lys residues using acetyl-CoA as a substrate (Figures 1C and 1D), indicating that NSI is an active chloroplast Lys acetyltransferase.
Figure 1.
Localization and Lys Acetylation Activity of NSI.
(A) Confocal microscopy image of Arabidopsis protoplast transiently expressing NSI-YFP (35S:NSI-YFP) fusion protein. Upper left panel shows the YFP signal, middle panel chlorophyll fluorescence of the same protoplast, and right panel is a merged image of the two. The lower panel shows the control lines: NAA70-YFP (left) was used as a chloroplast control marker, RFP (middle) as a cytoplasmic control, and TM23-RFP (right) as a plasma membrane control. Bar = 10 µm.
(B) Immunoblot detection of chloroplast protein fractions isolated from transgenic plants expressing NSI-YFP (35S:NSI-YFP) and separated on 12% acrylamide gel. GFP antibody was used for the detection of NSI-YFP and D1 antibody as a thylakoid membrane marker. NSI-YFP was detected as two bands, which may represent the preprotein (molecular weight based on mobility = 61.0 kD; expected molecular weight = 56.5 kD) and processed mature protein (molecular weight based on mobility = 49.0 kD; expected molecular weight = 49.9 kD). Ten micrograms of protein was loaded per sample. CP, chloroplasts; T, thylakoid fraction; S, soluble fraction.
(C) HPLC analysis of a general lysine acetyltransferase substrate and its acetylated product after conversion by His6-NSI for 1, 3, or 12 h. Identities of nonacetylated (0 h) and acetylated (acK pep.) standard peptides were confirmed by MS.
(D) Lysine acetylation rate of a peptide substrate by 10 µM His6-NSI (n = 3 technical replicates, ±sd).
To examine the role of NSI in Lys acetylation of chloroplast proteins in vivo, we characterized two nsi knockout mutant lines: nsi-1 and nsi-2 (Figure 2). Even though the visual phenotype of the nsi plants resembled that of the wild type (Figure 2B) and they accumulated an equal amount of chlorophyll, the chlorophyll a/b ratio of the mutants was slightly lower compared with the wild type (Table 1). Additionally, our quantitative mass spectrometry (MS) analysis of the Lys acetylomes showed a decreased Lys acetylation level of several plastid proteins in both mutants as compared with the wild type (Figure 2C; Supplemental Data Set 1). Some of these proteins, including PSBP-1 (AT1G06680), PSAH-1/2 (AT3G16140; AT1G52230), and LHCB1.4 (AT2G34430) (Figure 2; Supplemental Data Set 1) are involved in the light reactions of photosynthesis. In particular, Lys acetylation on K88 of PSBP-1 was more than 12-fold downregulated in both nsi mutants (LIMMA P value ≤ 0.00018). Interestingly, some thylakoid proteins, such as LHCB6 (AT1G15820) and the ATPase β-subunit (ATCG00480), had slightly increased Lys acetylation levels in the mutants, suggesting that Lys acetylation in chloroplasts also occurs independently of NSI and that there is an interplay between the acetylation of different proteins. The decreased Lys acetylation status of the identified chloroplast proteins was not caused by changes in protein abundance, as the quantities of these proteins were similar between nsi and wild-type plants (Figure 2D; Supplemental Data Set 2). In addition, it has to be pointed out that NSI might control additional acetylation sites that cannot be detected with the trypsin-based digestion method, since peptide fragments might be generated that are either too big or too small for detection.
Figure 2.
Characterization of the nsi Knockout Lines and Quantitative Lys Acetylome Analysis.
(A) The left panel represents the gene model of NSI based on TAIR10. Positions of T-DNA insertions in each line are marked with arrows. Sand colored boxes represent 5′- and 3′-UTR regions, orange boxes exons, and black lines introns. The right panel shows the absence of NSI mRNA verified with end-point RT-PCR. ACTIN2 was used as a control of cDNA quality.
(B) Phenotypes of 5-week-old wild type and nsi mutant lines grown in short day (8 h light/16 h dark), PPFD 100 µmol m−2 s−1, 50% humidity, and 23°C.
(C) and (D) Volcano plots representing quantitative Lys acetylome (C) and proteome (D) analyses of the nsi knockout lines (nsi-1 and nsi-2) compared with the wild type. Sums indicate numbers of quantified Lys acetylation sites and proteins, respectively. For statistical analyses, nsi-1 and nsi-2 were treated as group (defect in NSI) and tested against the wild type. Values had to be present in at least six out of the eight biological replicates. All replicate values are listed in the Supplemental Data Sets1 and 2. Green (plastid localization) and blue (nonplastid localization) circles illustrate significant data points with log2 fold changes ≥0.5 or ≤−0.5 and false discovery rate corrected P value ≤ 0.05 (LIMMA). Proteins involved in state transitions have been marked with text in the figure. 1, KEA1/2 K168/K170 (AT1G01790.1/AT4G00630.2); 2, unknown protein K62 (AT2G05310.1); 3, FER1 K134 (AT5G01600.1); 4, LHCB6 K220 (AT1G15820.1); 5, plastid-lipid associated protein PAP K225 (AT3G26070.1); 6, ATPF K119 (ATCG00130.1); 7, SOUL heme binding family protein K320 (AT5G20140.1); 8, SBPase K307 (AT3G55800.1); 9, ENH1 K233 (AT5G17170.1); 10, PSBH (ATCG00710.1); 11, ARM repeat superfamily protein (AT5G48120.1); 12, FAD6 (AT4G30950.1).
Table 1. Chlorophyll Content and State Transition Parameters of the Wild Type, nsi, and stn7.
| Plant Line | Chl a+b (µg/mg)a | Chl a/bb | Fv/Fmc,d | qTe,f | qSg,h |
|---|---|---|---|---|---|
| Wild type | 1.32 ± 0.11 (n = 27) | 3.50 ± 0.14 | 0.79 ± 0.04 (n = 4) | 0.10 ± 0.01 | 0.80 ± 0.02 |
| nsi-1 | 1.34 ± 0.12 (n = 27) | 3.44 ± 0.13 | 0.81 ± 0.01 (n = 4) | 0.01 ± 0.01 | 0.23 ± 0.03 |
| nsi-2 | 1.27 ± 0.11 (n = 27) | 3.40 ± 0.13 | 0.81 ± 0.02 (n = 4) | 0.01 ± 0.01 | 0.25 ± 0.03 |
| stn7 | 1.26 ± 0.11 (n = 14) | 3.36 ± 0.10 | 0.82 ± 0.01 (n = 4) | −0.01 ± 0.01 | 0.08 ± 0.04 |
Differences between genotypes were tested with ANOVA (Supplemental File 1). All data were normally distributed, apart from the stn7 qT parameter, and variances were homogenous. Multiple comparisons were done with Tukey HSD. Averages ± sd are shown, and n is marked in parentheses.
ANOVA, P = 0.079.
ANOVA, P = 0.005. Multiple comparisons: wild type versus nsi-1, P = 0.322; wild type versus nsi-2, P = 0.025; wild type versus stn7, P = 0.009; nsi-1 versus nsi-2, P = 0.649; nsi-1 versus stn7, P = 0.264; nsi-2 versus stn7, P = 0.820.
ANOVA, P = 0.294.
Fv/Fm = (Fm-F0)/Fm.
ANOVA, P = 1.493×10−8. Multiple comparisons: wild type versus nsi-1, P = 1.168 × 10−7; wild type versus nsi-2, P = 1.852 × 10−7; wild type versus stn7, P = 2.151 × 10−8; nsi-1 versus nsi-2, P = 0.953; nsi-1 versus stn7, P = 0.216; nsi-2 versus stn7, P = 0.094.
qT = (Fm1-Fm2)/Fm1, where Fm1 and Fm2 are the maximum fluorescence yields after illumination causing state 1 and 2, respectively.
ANOVA, P = 1.385 × 10−12. Multiple comparisons: wild type versus nsi-1, P = 2.266 × 10−11; wild type versus nsi-2, P = 3.052 × 10−11; wild type versus stn7, P = 2.512 × 10−12; nsi-1 versus nsi-2, P = 0.894; nsi-1 versus stn7, P = 0.007 × 10–2; nsi-2 versus stn7, P = 0.027 × 10−3.
qS = (FS1′-FS2′)/(FS1′-FS2), where FS1' and FS2' are fluorescence yields in the beginning and at the end, respectively, of illumination causing state 2, and FS2 is fluorescence yield immediately after switching on illumination causing state 1.
nsi Knockout Plants Have a Defect in State Transitions and Are Not Able to Form the PSI-LHCII Complex in Response to Illumination
The decreased Lys acetylation status of PSII, PSI, and LHCII prompted us to study the photosynthetic properties and organization of thylakoid protein complexes in the nsi mutants. In line with the unaffected growth phenotype of the nsi mutants, the maximum quantum yield of PSII, represented as Fv/Fm, was similar to that of the wild type (Table 1). However, when we measured rapid light response curves of both chlorophyll fluorescence and P700 absorbance, a severe decrease in the yield of PSII was detected under low light intensities (Supplemental Data Set 3). This decrease was not due to an increase in NPQ, since the mutants showed similar or even slightly lower NPQ in low light intensities. However, the mutants seemed to have more closed PSII reaction centers, as shown by the decreased values of the coefficients of photochemical quenching (qP and qL) under these conditions (Supplemental Data Set 3), which can be a sign of excess excitation of PSII in these conditions. Intriguingly, in higher light intensities this phenotype was lost or even reversed (Supplemental Data Set 3).
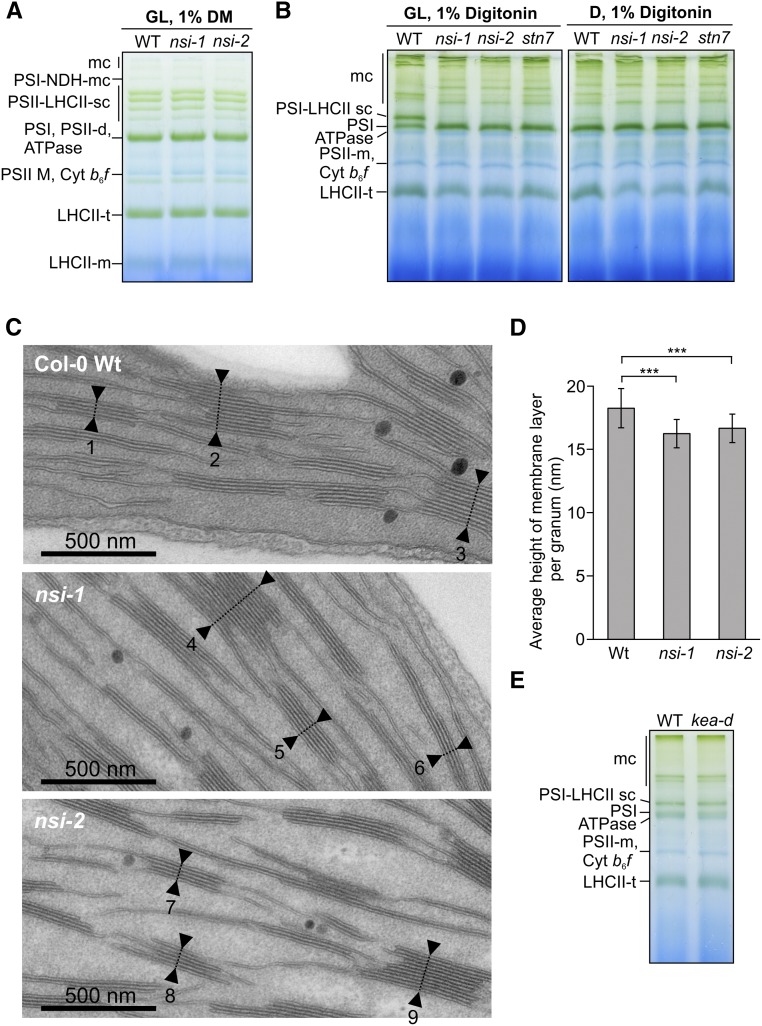
To study the status of the photosynthetic electron transfer machinery further, we extracted thylakoids from growth light (GL)-acclimated plants, solubilized the protein complexes with different detergents, and separated them with large-pore blue native gel electrophoresis (lpBN-PAGE). When thylakoids were solubilized with β-dodecylmaltoside (a detergent that solubilizes protein complexes from the whole thylakoid membrane but breaks down labile interactions), no differences in the abundances of any of the protein complexes between nsi and the wild type could be detected (Figure 3). However, analysis of the nsi thylakoids solubilized with digitonin (a detergent that preferentially solubilizes the PSI-rich stroma thylakoids and preserves supramolecular interactions of the thylakoid protein complexes) revealed that the nsi mutants lacked the PSI-LHCII complex in GL (Figure 3B) (Kouřil et al., 2005; Pesaresi et al., 2009; Suorsa et al., 2015). This complex is composed of PSI, LHCI, and LHCII subunits (Supplemental Data Set 4; Pesaresi et al., 2009; Suorsa et al., 2015) and accumulates in low to moderate light and upon transitioning from state 1 to state 2 (Kouřil et al., 2005; Pesaresi et al., 2009; Suorsa et al., 2015). In accordance with previous findings (Suorsa et al., 2015), the complex was disassembled in the dark (Figure 3B). Intriguingly, the megacomplex pattern of nsi closely resembled that of stn7 (Figure 3B) (Suorsa et al., 2015), which is the LHCII kinase mutant incapable of performing state transitions (Depège et al., 2003; Bellafiore et al., 2005).
Figure 3.
Organization of Thylakoid Protein Complexes of the Wild Type, nsi, and stn7 and Thylakoid Ultrastructure of the Wild Type and nsi.
(A) Large-pore blue native gel of thylakoid protein complexes from thylakoids that were isolated from GL (100 µmol photons m−2 s−1) adapted plants and solubilized with 1% β-dodecylmaltoside (DM). Representative image from experiment repeated with three biological replicates is shown. mc, megacomplex; sc, supercomplex; t, trimer; d, dimer; m, monomer.
(B) Large-pore blue native gels after digitonin solubilization of thylakoids isolated from plants after GL or D adaptation.
(C) Transmission electron microscopy (TEM) analysis of the nsi chloroplasts. TEM pictures of palisade mesophyll cells with chloroplasts in close-up view. Leaves of the two T-DNA insertion lines nsi-1 and nsi-2 and of wild-type Col-0 were prepared as thin section samples. Numbers and arrows display exemplary thylakoid stacks.
(D) Average heights per granum membrane layer ± sd for the two nsi knockout lines in comparison to the wild type Col-0 (calculated from [C]). Seven hundred thylakoid stacks per plant line displayed in 70 TEM pictures from seven independent biological replicates were analyzed (***P ≤ 0.001 using two-tailed Student’s t test).
(E) Large-pore blue native gel of GL adapted wild-type and kea1 kea2 double knockout (kea-d) thylakoids solubilized with 1% digitonin.
Moreover, a liquid chromatography-tandem mass spectrometry (LC-MS/MS) analysis was performed on the PSI-LHCII complex and the LHCII trimers separated by the lpBN-PAGE to search for acetylation sites within the complexes. However, for this analysis, no immunoaffinity enrichment step for Lys-acetylated peptides could be included due to the low amount of peptides retrieved from the gel bands. First of all, the quantitative proteome analysis of the LHCII trimer bands showed that the overall protein composition was not different between wild type, stn7, and both nsi mutants (Supplemental Data Sets 4A and 4C). Since the PSI-LHCII complex was not present in the nsi mutants and in stn7 (Figure 3B), this complex was analyzed only in the wild type. Within this complex, a few lysine-acetylated proteins were identified, including PSAH (K99), LHCB1.2 (K44), and LHCB1.5 (K41) (Supplemental Data Set 4B). A few acetylation sites were also detected in the LHC trimers. Interestingly, acetylation on the LHCB2.2 protein (Lys-42 and Lys-120) was not detected in nsi-1 and only detected in one replicate of nsi-2, while it was detected in two to three replicates of both stn7 and the wild type (Supplemental Data Set 4D). Unfortunately, the Lys acetylation sites of PSAH1/2 (K138) and LHCB1.4 (K40), which were identified in the full acetylome analysis (Figure 2C), were not detected in this analysis, most likely due to the missing immunoaffinity enrichment step. Furthermore, the thylakoid preparation was done under native conditions, which makes it difficult to maintain the in vivo acetylation status of all proteins.
Because of the observed changes in thylakoid protein organization, we also studied the structure of nsi chloroplasts with transmission electron microscopy. In line with the fact that nsi plants have no severe visual phenotype, they showed no major differences in plastid ultrastructure compared with the wild type (Figure 3C). However, the grana stacks of the knockout plants showed more compact packing than the wild type (Figure 3D).
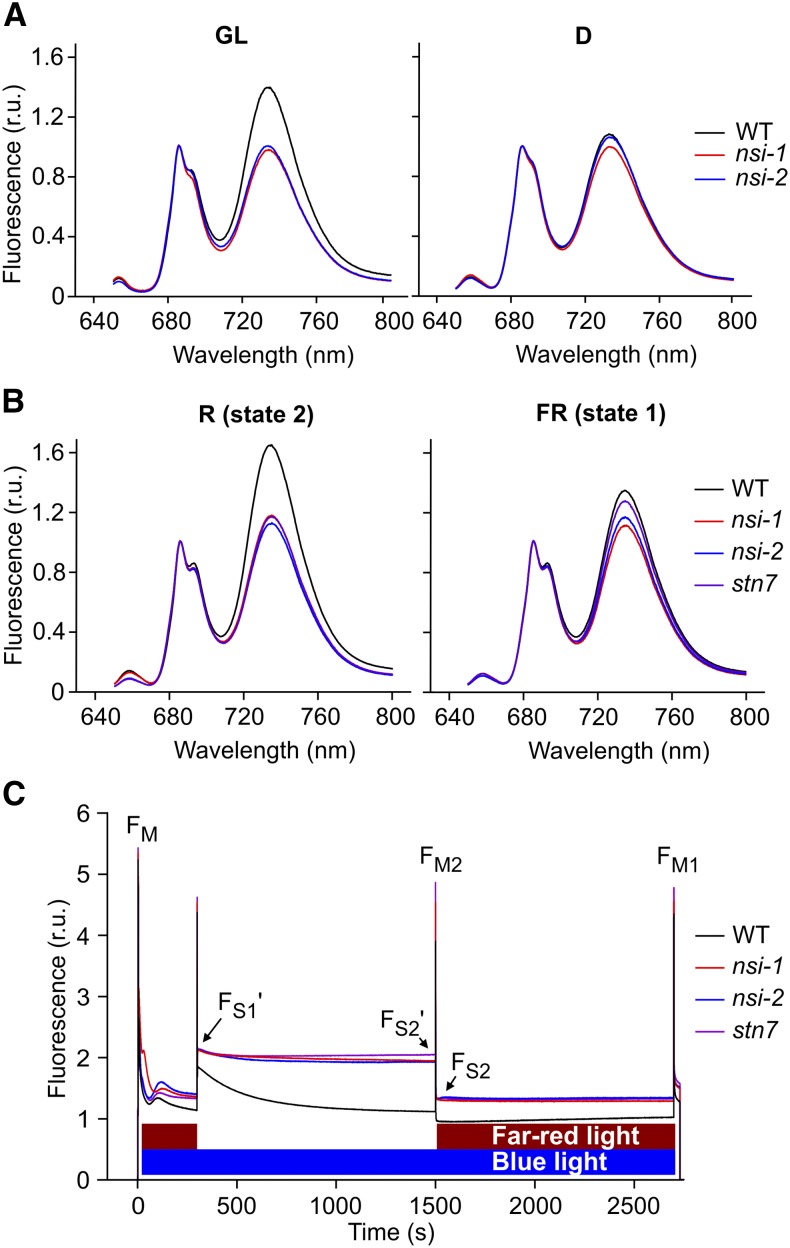
Due to the absence of the PSI-LHCII state transition complex in nsi, we examined the effect of NSI knockout on state transitions further. First, we measured 77K fluorescence emission spectra of wild-type and nsi thylakoids. Thylakoids were extracted in the middle of the light period (GL, 100 µmol photons m−2 s−1) as well as at the end of the dark period (D). In GL samples, the fluorescence emission peak originating from PSI (735 nm) was clearly lower in the nsi mutants than in the wild type, whereas the emission spectra from D was similar between all lines (Figure 4). These results suggest that the nsi mutant lines are defective in light-dependent adjustment of the excitation energy distribution between PSII and PSI, i.e., state transitions. To confirm this conclusion, we further investigated the behavior of nsi plants by treating them for 1 h in red (R; 660 nm; excites preferentially PSII) or far-red (FR; 735 nm; excites preferentially PSI) light to induce state 2 and state 1, respectively, and included the stn7 mutant line as a control. Thereafter, thylakoids were isolated and 77K fluorescence emission spectra were measured. Red light treatment induced state 2 in wild-type plants, while no such effect could be detected in nsi or stn7 (Figure 4B). Moreover, in vivo fluorescence measurements with a pulse amplitude modulation fluorometer confirmed that state transitions in all mutant lines were very weak and significantly different from the wild type (Figure 4C, Table 1).
Figure 4.
State Transitions in Wild Type, nsi, and stn7 under Different Light Treatments.
(A) 77K fluorescence emission spectra from thylakoids isolated from GL- and D-adapted plants.
(B) 77K fluorescence emission spectra from thylakoids isolated from R (660 nm) or FR light (735 nm) treated plants. Spectra in (A) and (B) were normalized to 685 nm and present an average of three biological replicates. Fluorescence emission around 685 to 695 nm originates from PSII and fluorescence emission around 735 nm from PSI.
(C) Representative graphs of state transition measurements with a pulse amplitude modulation fluorometer.
KEA1 and/or KEA2 were among the most drastically downregulated Lys-acetylated targets detected in nsi (Figure 2C). KEA1 and KEA2 are chloroplast envelope K+/H+ antiporters, which are essential for chloroplast pH and osmoregulation (Kunz et al., 2014). Because kea1 kea2 double knockout mutants showed impaired partitioning of the proton motive force across the thylakoid membrane, the function of the KEA transporters appears to be linked to the photosynthetic light reactions (Kunz et al., 2014). Therefore, we studied state transitions in the kea1 kea2 plants. However, no defects were detected in state transitions (qT = 0.10 ± 0.01, n = 12) or in the accumulation of the PSI-LHCII complex in the kea1 kea2 plants (Figure 3E), suggesting that inactivation of the KEA transporters does not result in impaired state transitions.
LHCII Phosphorylation and the PSI Docking Site for LHCII Are Not Impaired in nsi
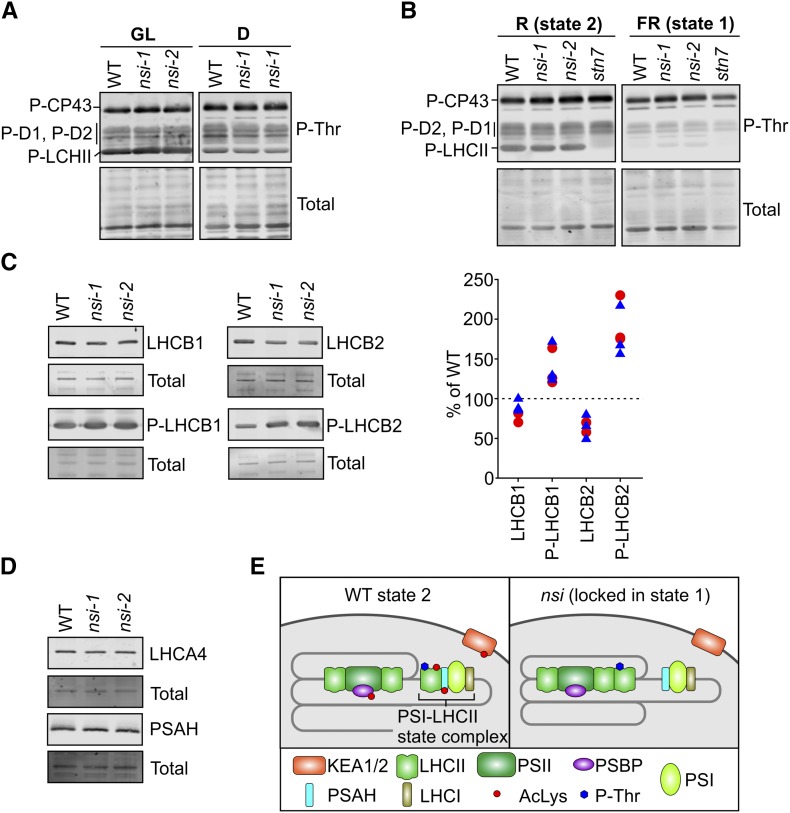
Since phosphorylation of LHCII, and especially that of the LHCB2 subunit of the L-LHCII trimers, has been shown to be the main determinant of state transitions (Leoni et al., 2013; Pietrzykowska et al., 2014; Crepin and Caffarri, 2015; Longoni et al., 2015), we analyzed the phosphorylation status of thylakoid proteins of nsi mutants by immunoblotting with a phospho-threonine (P-Thr) antibody. No differences were detected between the nsi mutants and the wild type in the overall phosphorylation status of thylakoid proteins isolated from GL or darkness (Figure 5). In addition, after red (state 2) light treatment LHCII was found to be phosphorylated in nsi and wild-type plants, whereas, as expected, no phosphorylation was seen in stn7 (Figure 5B). Far-red light treatment led to LHCII dephosphorylation in all lines (Figure 5B). To analyze LHCII phosphorylation in more detail, we immunoblotted thylakoids isolated from GL-adapted plants with (phospho) LHCB1 and LHCB2 antibodies. Figure 5C shows that phosphorylation levels of both of these proteins, but especially that of LHCB2, were even higher in nsi than in the wild type, even if the total amounts appeared similar or slightly lower in the signal from the immunoblot. However, no differences in LHCB1 and LHCB2 abundances between the wild type and mutants were detected by quantitative mass spectrometry (Supplemental Data Set 2), which suggests that there was less efficient binding of antibody to the hyperphosphorylated LHCB1/2. Because L-LHCII has been shown to interact with PSI through the PSAH subunit (Lunde et al., 2000; Crepin and Caffarri, 2015; Longoni et al., 2015) or through the LHCA antenna (Benson et al., 2015), we also checked nsi for the abundance of these proteins. Both proteins were present similar in abundance in the mutants compared with the wild type (Figure 5D). Taken together, we conclude that the inability of nsi plants to undergo state transitions is not due to impaired LHCII phosphorylation or defects in the PSI docking sites for L-LHCII.
Figure 5.
Immunoblot Analysis of Thylakoid Protein Phosphorylation, L-LHCII Subunits, and PSI Docking Site for L-LHCII and Schematic Presentation of the Downregulated Lys Acetylation Sites in nsi.
(A) Phosphorylation of thylakoid proteins isolated from GL- or D-adapted plants. Proteins were separated on 15% acrylamide gels and immunoblotted with P-Thr antibody.
(B) Phosphorylation of thylakoid proteins isolated form R- or FR-treated plants. Proteins were separated on 12% acrylamide gels and immunoblotted with P-Thr antibody.
(C) Analysis of L-LHCII subunits from GL-adapted thylakoids. Proteins were separated on 12% acrylamide gels and immunoblotted using antibodies against LHCB1 and LHCB2 and their phosphoforms (P). Right panel shows quantification of LHCB1, LHCB2, and their phosphorylated forms in nsi mutants (nsi-1 is marked with red circles and nsi-2 with blue triangles). Protein amounts were quantified from the blots and calculated as a percentage of the wild type from the respective replicate.
(D) Immunoblot analysis of L-LHCII docking site on PSI. Proteins were isolated from GL-adapted plants, separated on 12% acrylamide gels, and immunoblotted with PSAH and LHCA4 antibodies. The lower panels (Total) show blots after staining with REVERT total protein stain to verify equal loading; representative blots are shown from three biological replicates ([A] to [D]).
(E) Comparison of chloroplast protein complexes between the wild type and nsi in state 2. Upon conditions favoring plastoquinone pool reduction, L-LHCII trimers are phosphorylated, which results in the interaction of L-LHCII with PSI, mediated by PSAH (state 2 in the wild type). In contrast to the wild type, P-LHCII is not able to interact with PSI in nsi under state 2 conditions. The phenotype may result from defects in Lys acetylation of (1) PSAH and LHCII, which may hinder the PSI-LHCII interaction; (2) PSBP and LHCII, which may result in a strong interaction between PSII and L-LHCII; or (3) proteins involved in chloroplast ion homeostasis (PSBP and KEA1/KEA2), which may be required for the dynamic reorganization of thylakoid protein complexes.
DISCUSSION
Despite the fact that Lys acetylation of various chloroplast proteins and the first plastid Lys deacetylase have been recently identified (Finkemeier et al., 2011; Wu et al., 2011; Hartl et al., 2017; Schmidt et al., 2017), the chloroplast Lys acetyltransferase enzymes have not been characterized to date. Nevertheless, Lys acetylation has been shown to have marked effects on chloroplast function, as it regulates the activities of Rubisco, Rubisco activase, and phosphoglycerate kinase enzymes (Finkemeier et al., 2011; Hartl et al., 2017) as well as the stability of ATP synthase (Schmidt et al., 2017). Here, we identified a plant organellar Lys acetyltransferase, NSI, which is localized in the chloroplast stroma (Figure 1). Loss of NSI affected the acetylation status of various chloroplast proteins, including PSAH-1/PSAH-2, LHCB1.4, PSBP-1, and KEA1/ KEA2 (Figure 2C; Supplemental Data Set 1). In addition, nsi plants were not able to carry out light-dependent reorganization of thylakoid protein complexes (i.e., state transitions) (Figures 3 and 4). Intriguingly, the nsi mutants closely resembled stn7, even though LHCII phosphorylation in nsi was not impaired (Figure 5). This finding suggests that NSI affects state transitions independently of LHCII phosphorylation.
In wild-type plants, state transitions occur as a response to plastoquinone pool reduction or oxidation (state 2 and state 1, respectively). Under state 2 conditions, the STN7 kinase is activated and phosphorylates the L-LHCII trimer composed of LHCB1 and LHCB2 subunits. The phosphorylated trimer then binds to PSI via the PSAH subunit (together with the nearby PSAO and PSAL subunits), increasing the absorption cross section of PSI (Lunde et al., 2000; Zhang and Scheller, 2004; Galka et al., 2012). Recently, the LHCA proteins, localized on the opposite site of PSI compared with PSAH, have been implicated in binding a pool of L-LHCII trimers (Benson et al., 2015). Lack of acetylation in one or more of these proteins might disturb direct protein-protein interactions, which are required for light energy transfer from LHCII to PSI. Indeed, we observed a small but consistent 1.2-fold decrease in Lys acetylation of PSAH-1/2 in nsi mutants. Additionally, a similar decrease in the acetylation status of LHCB1.4, an abundant subunit in the L-LHCII trimers (Galka et al., 2012), was detected in the nsi plants (Figure 2C). Since acetylation, in general, can affect the conformation of a protein and furthermore removes the charge of the Lys residue, adequate acetylation of one or more of these proteins may be necessary for a stable interaction between the thylakoid protein complexes (Figure 5E). It is noteworthy that the LHCII-PSI state transition complex has been shown to be composed of one L-LHCII attached to one PSI reaction center via the PSAH subunit (Kouřil et al., 2005) and that this complex was completely absent in nsi (Figure 3B). Indeed, the state transition phenotype in the PSAH cosuppression line was similar to nsi (Lunde et al., 2000). It should be noted, however, that the effect of NSI to state transitions might also be mediated via the formation of the PSII supercomplexes: Changes in the acetylation of PSII and LHCII subunits might result in permanent attachment of L-LHCII to PSII, which could prevent L-LHCII binding to PSI (Figure 5E).
As LHCII phosphorylation and an intact L-LHCII docking site of PSI have been shown to be indispensable for state transitions (Lunde et al., 2000; Depège et al., 2003; Zhang and Scheller, 2004; Bellafiore et al., 2005; Galka et al., 2012), practically all mutants defective in state transitions have problems either in the accumulation or phosphorylation of LHCII, or in the LHCII docking site of PSI. For instance, other mutations affecting the accumulation of LHCII, e.g., chlorina1-2 with impaired chlorophyll b biosynthesis and chaos deficient in cpSRP43, have an effect on state transitions, either due to the decreased level of LHCII phosphorylation in chlorina1-2 or the lack of L-LHCII in chaos (Wang and Grimm, 2016). The PSB33 protein, which provides stability to PSII-LHCII supercomplexes, is also indispensable for proper phosphorylation of the thylakoid proteins upon fluctuating light conditions and therefore affects accumulation of PSI-LHCII complexes and state transitions (Fristedt et al., 2015, 2017). The ics1 mutants with dysfunctional ISOCHORISMATE SYNTHASE1 protein, involved in the biosynthesis of phylloquinone and salicylic acid, possess higher PQ pool reduction levels and increase numbers of stacked thylakoids per granum compared with the wild type (Gawroński et al., 2013). Moreover, the decreased content of phylloquinone results in defective in state transitions in ics1 (Gawroński et al., 2013). However, as the level of LHCII phosphorylation in this mutant was not studied, speculation on the mechanistic details behind the defective state transitions is not possible (Gawroński et al., 2013).
The complex interactions between the chloroplasts and mitochondria in Chlamydomonas reinhardtii have been exemplified by examining the photosynthetic properties of mutants having defects in mitochondrial respiratory electron transfer (Cardol et al., 2003; Schönfeld et al., 2004). The respiration rates of the dum mutants were shown to correlate with the relative quantum yield of photosynthetic electron transfer, and the mutants were not able to perform transition to state 1, apparently because of an increased rate of nonphotochemical PQ pool reduction and persistent LHCII phosphorylation even under illumination with PSI light (Cardol et al., 2003). By contrast, the Chlamydomonas stm6 mutants showing reduced levels of cytochrome c oxidase and rotenone-insensitive external NADPH dehydrogenase activities were locked in state 1 (Schönfeld et al., 2004). The LHCII phosphorylation in stm6 was impaired even if the PQ pool was in a more reduced state compared with the wild type, possibly because of overreduction of stroma resulting in inactivation of the STN7 kinase (Rintamäki et al., 2000; Schönfeld et al., 2004). These examples show that even if state transitions in Chlamydomonas tune the ratio between linear and cyclic photosynthetic electron transfer, thus responding to the metabolic need for ATP (Wollman, 2001) rather than balancing energy distribution between the two photosystems (as in plants), the mechanistic background of state transitions is always based on reversible LHCII phosphorylation. LHCII phosphorylation, however, may be differently regulated; indeed, no such tight interplay between respiration and state transitions has been reported in higher plants compared with Chlamydomonas.
Thylakoid membrane architecture shows large rearrangements during state transitions, and unstacking of grana occurs upon transition to state 2 (Chuartzman et al., 2008). Interestingly, the grana in the nsi were more tightly packed than in the wild type (Figure 3C). On the other hand, as chloroplast ion content is known to be a key determinant of thylakoid membrane stacking, it can play an important role in state transitions in conjunction with LHCII phosphorylation (Kaňa and Govindjee, 2016). Indeed, one of the most drastically downregulated Lys acetylation sites in the nsi mutants was found in the PSBP-1 protein, where acetylation was more than 12-fold less abundant than in the wild type (Figure 2C). The PSBP-1 protein is a lumenal PSII subunit that stabilizes Ca2+ and Cl− binding in the oxygen evolving complex (Ifuku et al., 2008). Intriguingly, the altered Lys acetylation site of PSBP-1 is situated in the N-terminal domain (K88 of the preprotein; K11 in the mature protein), which is essential for ion binding and oxygen evolution (Ifuku et al., 2005, 2008). It has previously been shown that defects in the function of PSBP-1 affect thylakoid structure (Yi et al., 2009) and that a correct composition of the oxygen evolving complex influences state transition kinetics (Allahverdiyeva et al., 2013). Therefore, it is possible that lack of acetylation might alter ion binding by PSBP-1 in nsi, which could disrupt the protein complex reorganization required for state transitions.
Another strongly affected site was found in a peptide that is common to the KEA1 and KEA2 proteins. KEA1 and KEA2 are homologous K+/H+ antiporters localized in the chloroplast envelope (Kunz et al., 2014). Double knockout kea1 kea2 plants are severely affected in their growth and contain malformed, swollen chloroplasts, which underlines the importance of a proper ion balance in chloroplasts (Kunz et al., 2014). It is thus possible that the lack of acetylation in KEA1 and KEA2 proteins (Figure 2C) could cause the compact thylakoid stacking detected in nsi chloroplasts (Figures 3C and 3D) through altered ion content in the chloroplast, which in turn might lead to obstruction of protein complex reorganization (Figure 5E). If this is the case, acetylation is most likely required for the inactivation of KEA channels in the wild type, since state transitions in the kea1 kea2 mutant plants were fully functional. Whether acetylation of KEA1 and KEA2 is involved in channel inactivation, and thereby has an influence on ion homeostasis and state transitions, remains to be tested.
Our results reveal a new layer in the dynamic regulation of chloroplast light responses and implicate the chloroplast acetyltransferase NSI as a prerequisite for state transitions (Figure 5E). In addition to nsi and the docking site mutant psah, no other mutants have yet been described that have a wild-type-like growth phenotype, no defects in LHCII phosphorylation, but lack state transitions. This is an intriguing finding that correlates with the function of NSI as a Lys acetyltransferase and with a decreased acetylation status of a number of chloroplast proteins. Although the causal relationship between the LHCII phosphorylation and state transitions was described long ago (Bennett et al., 1980; Allen, 1992), numerous questions around the ways in which Lys acetylation could play a role have remained unanswered. For instance, what kind of effect(s) does the three-dimensional structure of the thylakoid network have on the state transitions, and how are changes in chloroplast ion content reflected in state transitions in vivo? Are components other than LHCB1, LHCB2, PSAH, and PSI subunits in the vicinity of PSAH required for state transitions? Are there still other, so far uncharacterized factors regulating state transitions? Whether the effect of NSI on state transitions is based directly on Lys acetylation and disturbed interaction between PSI and LHCII, on altered PSII-LHCII interaction, or on some other more indirect mechanism that affects thylakoid dynamics will be an important topic for further investigation.
METHODS
Plant Material
Arabidopsis thaliana (Col-0) was grown in 8 h light/16 h darkness at PPFD of 100 µmol m−2 s−1 (light source: Osram Powerstar HQI-BT 400W/D daylight), 50% humidity, and 23°C. Seed stocks for the nsi (At1g32070) T-DNA lines and the wild type were ordered from the NASC, and stn7 (SALK_073254) was from Mikko Tikkanen (Tikkanen et al., 2006). The two nsi T-DNA lines SALK_033944 and SALK_020577 (nsi-1 and nsi-2, respectively) were PCR screened according to Salk Institute Genomic Analysis Laboratory instructions and using the following primers: nsi-1, LP, 5′-AAGAAGTCCCCAGTAACAATCC and RP, 5′-CCGCCTTCTGTGTCAAATAAC; nsi-2, LP, 5′-CGAGCTGATTTACGTGGAAAG and RP, 5′-AGCTTATTGGTATGGCACGTG; BP for both lines was LBb1.3 (5′-ATTTTGCCGATTTCGGAAC). Absence of NSI mRNA was verified with end-point RT-PCR using gene-specific primers and ACTIN2 as a control (NSI_fw_1, 5′-GATTCATCAGAAGGCGGGGAT; NSI_rev_1, 5′-GATGCCTTCTGGATCAGCCT; ACTIN2_fw, 5′-GTGAACGATTCCTGGACCTGCCTC; ACTIN2_rev, 5′-GAGAGGTTACATGTTCACCACAAC). RNA was extracted using Agilent Plant RNA Isolation Mini Kit and treated with Ambion TURBO DNA-free DNase. cDNA was synthesized with Bio-Rad iScript cDNA synthesis kit.
Determination of Chlorophyll Content
Leaf discs were cut, weighed, and incubated overnight in 1 mL of dimethylformamide in darkness and room temperature. Chlorophyll content was calculated according to Inskeep and Bloom (1985).
Generation of Transgenic YFP Line
A transgenic Arabidopsis line expressing NSI with a C-terminal YFP-tag (35S:NSI-YFP) was generated via modified floral inoculation (Narusaka et al., 2010) using Agrobacterium tumefaciens GV3101:pMP90:pSoup. The coding sequence of NSI was amplified from Arabidopsis cDNA using NSI_fw_2, 5′-TATACCCGGGATGCTACTAATCCCA, and NSI_rev_2, 5′-TATAGGATCCCTTTGGGTACCAAAACATG. The PCR product was cloned into pGWR8-YFP (Rozhon et al., 2010), which was used for the transformation.
Fluorescence Microscopy
Detection of YFP fusion proteins was performed as previously described (Dinh et al., 2015). Signal of RFP was recorded using a 560- to 615-nm band-pass filter after excitation at 543 nm. Primers for expression of full-length NSI protein as an N-terminal fusion of YFP were: NSI_fw_3, 5′-GATCGGATCCATGCTACTAATCCCAATTTC, and NSI_rev_3, 5′-GATCGTCGACCTTTGGGTACCAAAACATGC. Localization of marker proteins in the chloroplast (NAA70-YFP), the cytosol (RFP), and the plasma membrane (TMD23-RFP) have been demonstrated earlier by Dinh et al. (2015) and Brandizzi et al. (2002), respectively.
Thylakoid Protein Extraction
Fresh Arabidopsis leaves were ground 3 × 2 s in cold buffer (300 mM sucrose, 50 mM HEPES-KOH, pH 7.6, 5 mM MgCl2, 1 mM Na-EDTA, 1.25% BSA, 22 mM ascorbate, and 10 mM NaF). Homogenate was filtered through Miracloth (Millipore), and the filtrate was centrifuged for 4 min, 4000g, 4°C to pellet chloroplasts and thylakoids. Chloroplasts were broken by resuspending the pellet to hypotonic lysis buffer (5 mM sucrose, 10 mM HEPES-KOH, pH 7.6, 5 mM MgCl2, 10 mM NaF, and Pierce protease inhibitor [Thermo Scientific]). The lysate was centrifuged at 18,000g for 5 min, 4°C, and the pellet (thylakoids) was resuspended in storage buffer (100 mM sucrose, 10 mM HEPES-KOH, pH 7.6, 10 mM MgCl2, and 10 mM NaF). Different biological replicates were prepared from plants grown at different times on separate trays. The chlorophyll concentration of thylakoids was determined as described (Porra et al., 1989).
Chloroplast Isolation and Fractionation
Fresh Arabidopsis rosettes were ground 3 × 2 s in cold buffer (330 mM sorbitol, 50 mM HEPES-KOH, pH 7.6, 1 mM MgCl2, 5 mM Na-EDTA, 0.1% BSA, and 5 mM ascorbate). Leaf homogenate was filtered through one layer of Miracloth (Millipore) and the filtrate centrifuged for 2 min, 2000g, 4°C. The pellet was gently resuspended into residual buffer. Chloroplast suspension was loaded on top of a Percoll step gradient (40%/80% Percoll in 330 mM sorbitol and 50 mM HEPES-KOH, pH 7.6) and centrifuged 6 min, 7000g, 4°C with mild acceleration and no breaks in a fixed angle rotor. Intact chloroplasts were collected from the gradient interface and washed twice (330 mM sorbitol, 50 mM HEPES-KOH, pH 7.6, and 2 mM Na-EDTA). Chloroplasts were pelleted 2 min, 10,000g, 4°C and resuspended into hypotonic lysis buffer (5 mM sucrose, 10 mM HEPES-KOH, pH 7.6, and 5 mM MgCl2) at a final concentration of 1 µg chlorophyll/μL buffer. Suspension was freeze-thawed with liquid nitrogen and fractions separated by centrifuging 10 min, 18,000g, 4°C. Supernatant was collected as the soluble fraction. The pellet (thylakoids) was washed and finally resuspended into storage buffer (100 mM sucrose, 10 mM HEPES-KOH, pH 7.6, and 10 mM MgCl2). Protein concentrations were determined with Bradford Protein Assay (Bio-Rad).
Immunoblotting of Thylakoids and Chloroplast Fractions
Proteins were solubilized with 2× Laemmli buffer (Laemmli, 1970) supplemented with 6 M urea and run on 12 or 15% acrylamide gels, as indicated, containing 6 M urea. The gels were blotted to Immobilon-FL membrane (Merck Millipore) in blotting buffer (39 mM glycine, 48 mM Tris, 1.3 mM SDS, and 20% methanol) using 1 mA/cm2 for 1 h with Hoefer TE77X semidry blotter. All blots were blocked using 5% BSA in TTBS (20 mM Tris-HCl, pH 7.5, 150 mM NaCl, and 0.05% Tween 20). Rabbit P-Thr antibody was purchased from New England Biolabs and used as a 1:3000 dilution with 0.5 µg of chlorophyll. Rabbit LHCA4 (AS01 008), LHCB1 (AS01 004), LHCB2 (AS01 003), P-LHCB1 (AS13 2704, lot 1310), P-LHCB2 (AS13 2705, lot 1310), and PSAH (AS06 105) antibodies were purchased from Agrisera and used as 1:5000 (LHCB1, LHCB2, and PSAH) or 1:10,000 (P-LHCB1 and P-LHCB2) dilutions (Leoni et al., 2013). Rabbit GFP antibody (SAB4301138, lot 492635538) was purchased from Sigma-Aldrich and used as a 1:5000 dilution. Rabbit D1 DE-loop antibody (Kettunen et al., 1996) was used as a 1:8000 dilution. LI-COR Goat anti-rabbit IRDye 800CW 2nd antibody was used for detection according to manufacturer’s instructions. Blots were imaged using LI-COR Odyssey CLx. Blots were stained with REVERT Total Protein Stain (LI-COR) to verify equal loading and transfer of proteins.
Native Electrophoresis
Large-pore blue native gels and samples were prepared as previously described (Järvi et al., 2011).
Heterologous Expression and Purification of Recombinant NSI Protein
NSI coding sequence, excluding that for the predicted transit peptide (57 N-terminal amino acid residues), was amplified from Arabidopsis cDNA with Phusion High Fidelity Polymerase (Thermo Fisher Scientific). The sequence was amplified using NSI_fw_4, 5′-CAAGGATCCTCTGGGTTTGTGAAG, and NSI_rev_4, 5′-GTACCCGGGCTACTTTGGGTACCA, primers. The NSI PCR product was cloned into pQE-30 vector (Qiagen). Protein was expressed in BL21(DE3)pLysS cells (Invitrogen) induced with 1 mM IPTG (Roth) for 15 h at 21°C. Cells were harvested, resuspended in buffer (100 mM Tris-HCl, pH 7.8, 150 mM NaCl, and protease inhibitor cocktail [Sigma–Aldrich]) and disrupted with a French press. After addition of 5 mM DTT and 10 units of lysozyme, recombinant NSI was purified from the soluble phase by Protino Ni-NTA affinity chromatography (Macherey-Nagel). Proteins were eluted with 500 mM imidazole and desalted on PD-10 gel filtration columns (GE Healthcare) using 50 mM Tris-HCl (pH 7.8), 150 mM NaCl, and 10% glycerol. The protein concentrations were determined with Pierce 660 nm protein assay (Thermo Scientific).
Lys Acetyltransferase Activity Assay
The acetyltransferase activity assay was performed by incubating His6-NSI (10 μM) and a general Lys acetyltransferase peptide substrate (50 µM) coupled to anthranilic acid at the N terminus at 30°C in reaction buffer (150 mM Na-phosphate, pH 7, and 50 mM NaCl) (Seidel et al., 2016). Reaction was started by addition of 50 µM acetyl-CoA. Twenty-microliter samples were collected from time points between 0 and 12 h, and the reaction was stopped by addition of 180 μL trifluoroacetic acid (TFA; final concentration 2%). Reaction products were analyzed by reversed phase HPLC chromatograph (Shimadzu) equipped with CBM-20A controller, two LC-20AD pumps, a DGU-20A degasser, an SPD-20A detector, and an SIL-20AC autosampler. Separation was achieved on a Hypersil GOLD column (4.6 mm × 250 mm, 5-µm particle size; Thermo Fisher Scientific). A gradient program consisting of solvent A (0.1% TFA [v/v] in distilled water) and solvent B (95% acetonitrile and 0.1% TFA [v/v] in distilled water) was applied at a flow rate of 1.0 mL/min as follows: 0 to 1 min, 5% B; 1 to 20 min, linear 5 to 100% B; 20 to 25 min, 100% B; 25 to 25.5 min, 100 to 5% B; 25.5 to 30 min, 5% B. One hundred microliters of sample solution was injected. The detector was set at 218 (peptide backbone) and 360 nm (anthranilic acid). All reaction rates were determined from three independent technical replicates. Reaction rates were calculated from the peak areas of the free Lys and Lys-acetylated peptides, which eluted at 14.34 and 14.85 min, respectively.
Protein Extraction, Peptide Dimethyl Labeling, and Lys-Acetylated Peptide Enrichment
Frozen leaf material was ground to fine powder in liquid nitrogen and extracted using a modified filter-assisted sample preparation protocol (Wiśniewski et al., 2009a) and treated as described (Wiśniewski et al., 2009b). Digested peptides were dimethyl labeled on C18 Sep-Pak plus short columns (Waters) as previously described (Boersema et al., 2009). Equal amounts of light and medium labeled peptides were pooled for each replicate and the solvent evaporated in a vacuum centrifuge. Fifteen micrograms of peptide mixture was stored for whole proteome analysis. Lys-acetylated peptide enrichment was performed as previously described (Hartl et al., 2015) with 8 mg peptide per combined sample (1 mg peptide/25 μL antibody slurry). After enrichment, the eluted peptides were desalted and fractionated in three steps using SDB Stagetips (Kulak et al., 2014) and evaporated in a vacuum centrifuge.
LC-MS/MS Data Acquisition
Dried peptides were dissolved in 2% ACN and 0.1% TFA for analysis. Samples were analyzed using an EASY-nLC 1200 (Thermo Fisher Scientific) coupled to a Q Exactive HF mass spectrometer (Thermo Fisher Scientific). Peptides were separated on 17-cm frit-less silica emitters (New Objective; 0.75-µm inner diameter), packed in-house with reversed-phase ReproSil-Pur C18 AQ 1.9 µm resin (Dr. Maisch). The column was kept at 50°C in a column oven throughout the run. The following parameters were used for whole-proteome analysis, and parameters for acetylome analysis are stated in brackets; if not stated separately, parameters are identical. Peptides were eluted for 115 (68) min using a segmented linear gradient of 0% to 98% solvent B (solvent A 0% ACN, 0.5% FA; solvent B 80% ACN, 0.5% FA) at a flow rate of 300 (250) nL/min. Mass spectra were acquired in data-dependent acquisition mode with a Top15 method. MS spectra were acquired in the Orbitrap analyzer with a mass range of 300 to 1759 m/z at a resolution of 60,000 (120,000) FWHM, maximum IT of 55 ms, and a target value of 3 × 106 ions. Precursors were selected with an isolation window of 1.3 (1.2) m/z. HCD fragmentation was performed at a normalized collision energy of 25. MS/MS spectra were acquired with a target value of 105 (5 × 104) ions at a resolution of 15,000 FWHM, maximum IT of 55 (150) ms, and a fixed first mass of m/z 100. Peptides with a charge of +1, >6, or with unassigned charge state were excluded from fragmentation for MS2, and dynamic exclusion for 30 s prevented repeated selection of precursors.
MS Data Analysis
Raw data were processed using MaxQuant software version 1.5.2.8 (http://www.maxquant.org/) (Cox and Mann, 2008; Tyanova et al., 2016a). MS/MS spectra were searched with the Andromeda search engine against the TAIR10 database (TAIR10_pep_20101214; ftp://ftp.arabidopsis.org/home/tair/Proteins/TAIR10_protein_lists/). Sequences of 248 common contaminant proteins and decoy sequences were automatically added during the search. Trypsin specificity was required and a maximum of two (proteome) or four missed cleavages (acetylome) was allowed. Minimal peptide length was set to seven amino acids. Carbamidomethylation of cysteine residues was set as fixed, oxidation of methionine and protein N-terminal acetylation as variable modifications. Acetylation of Lys was set as variable modification only for the antibody-enriched samples. Light and medium dimethylation of Lys and peptide N termini were set as labels. Peptide spectrum matches and proteins were retained if they were below a false discovery rate of 1%, and modified peptides were additionally filtered for a score ≥ 35 and a delta score of ≥ 6 to remove low quality identifications. Match between runs and requantify options were enabled. Downstream data analysis was performed using Perseus version 1.5.5.3 (Tyanova et al., 2016b). For proteome and acetylome, reverse hits and contaminants were removed, the site ratios log2 transformed, and flip-label ratios inverted. Plotting of the raw and the normalized site ratios confirmed that the automatic normalization procedure of MaxQuant worked reliably and normalized site ratios were used for all further analyses. For quantitative Lys acetylome analyses, sites were filtered for a localization probability of ≥0.75. The “expand site table” feature of Perseus was used to allow separate analysis of site ratios for multiply acetylated peptides occurring in different acetylation states. Technical replicates were averaged and sites as well as protein groups displaying less than two out of four ratios were removed. The resulting matrices for proteome and acetylome were exported, and significantly differential abundant protein groups and Lys acetylation sites were determined using the LIMMA package (Ritchie et al., 2015) in R 3.3.1 (R Core Team, 2016). Volcano plots were generated with R base graphics, plotting the nonadjusted P values versus the log2 fold change and marking data points below 5% false discovery rate (i.e., adjusted P values) when present.
Trypsin Digestion of Bands Excised from lpBN-PAGE and Data Analysis
Protein spots were excised from gels, trypsin-digested as described before (Morgan et al., 2008), and analyzed using LC-MS/MS. Raw data were processed using MaxQuant software version 1.5.2.8 (http://www.maxquant.org/) (Cox and Mann, 2008; Tyanova et al., 2016a). MS/MS spectra were searched with the Andromeda search engine against the Araport 11 database. Sequences of 248 common contaminant proteins and decoy sequences were automatically added during the search. Trypsin specificity was required and a maximum of two missed cleavages were allowed. Minimal peptide length was set to seven amino acids. Carbamidomethylation of cysteine residues was set as fixed and oxidation of methionine as variable modifications. Acetylation of Lys and phosphorylation (STY) were set as variable modification. Peptide spectrum matches and proteins were retained if they were below a false discovery rate of 1%, a score ≥35, and a delta score of ≥6 for modified peptides were required. Match between runs and iBAQ were enabled. Downstream data analysis was performed using Perseus version 1.6.1.3 (Tyanova et al., 2016b). Reverse hits and contaminants were removed, and peptide and iBAQ intensities were log2 transformed. Technical replicates were averaged and sites as well as protein groups identified in only one replicate of each genotype were removed. Data were analyzed from three independent biological replicates.
Fluorescence Measurements
77K fluorescence emission spectra were measured from thylakoids diluted with storage buffer to 0.33 µg/mL chlorophyll. Each spectrum was measured with QEPro spectrometer (Ocean Optics) from a 100-μL thylakoid batch using 3 s integration time and blue excitation light.
Rapid light response curves were measured with Dual-PAM-100 (Heinz Walz) equipped with DUAL-E emitter and DUAL-DR detector units, using a red measuring beam for fluorescence and red actinic light. Absorbance changes due to oxidation of the primary donor P700 of PSI were measured simultaneously with the same device at 830 nm. One leaf from an intact Arabidopsis rosette was used per biological replicate. Measurements were done after 20 min of dark incubation followed by determination of initial fluorescence F0 with the measuring beam alone, Fm with a saturating flash, and maximum P700 oxidation (Klughammer and Schreiber, 1994) by a saturating flash preceded by 10-s FR illumination. Thereafter, each measurement continued by 2-min illumination steps, with a saturating flash at the end of each step to allow the determination of parameters of PSII and PSI as follows. The quantum yield of PSII [Y(II); Genty et al., 1989], nonphotochemical quenching (NPQ; Demmig-Adams, 1990), photochemical quenching (qP [Schreiber et al., 1986] and qL [Kramer et al., 2004]), and the quantum yield of regulated and nonregulated nonphotochemical quenching [Y(NPQ) and Y(NO), respectively; Kramer et al., 2004] were calculated from the fluorescence data. The quantum yield of PSI [Y(I)] and the donor and acceptor side limitation of PSI [Y(ND) and Y(NA), respectively; Klughammer and Schreiber, 1994] were calculated from absorbance changes at 830 nm. One leaf from an intact rosette was used per biological replicate.
State transitions were measured using a Waltz PAM-101 fluorometer equipped with the FIP control software (Tyystjärvi and Karunen, 1990) using dark incubated (30 min) plants. First, F0 and Fm were measured from a dark-acclimated leaf, and the values were used to calculate Fv/Fm (Fv = Fm – F0). Then, the leaf was illuminated for 5 min with FR LED (Walz 102-FR; 53 µmol photons m−2 s−1) and blue LED (470 nm LED filtered through a 470-nm, 10-nm FWHM filter (Andover), 24 µmol m−2 s−1) to activate photosynthesis. The FR was turned off for 20 min to induce state 2, after which it was turned on again for 20 min to induce state 1. At the end of each illumination step, a saturating pulse was fired to obtain maximum fluorescence in state 1 (Fm1’) or state 2 (Fm2’). The state transition parameters qT and qS were calculated according to Ruban and Johnson (2009). One detached leaf from one Arabidopsis rosette was used per biological replicate. White-light saturating pulses (1 s, PPFD 2750 µmol m−2 s−1) and the measuring beam of the PAM-101 fluorometer were used.
Sample Preparation for Transmission Electron Microscopy
Leaf discs (2 mm) from 6-week-old wild-type and mutant plants cultivated in 8 h light/16 h dark were excised using a biopsy punch, fixed with 2.5% (v/v) glutaraldehyde and 2% (w/v) paraformaldehyde in 0.05 M sodium cacodylate buffer (pH 6.9) for 3 h at room temperature, and then kept overnight at 4°C. Subsequently, samples were rinsed six times for 10 min in 0.05 M sodium cacodylate buffer (pH 6.9, rinses 3 and 4 supplemented with 0.05 M glycine) and postfixed in 1% osmium tetroxide in 0.05M sodium cacodylate (pH 6.9) supplemented with 0.15% potassium ferricyanide for 1 h on ice. After thorough rinsing in 0.05 M sodium cacodylate buffer (pH 6.9) and water, samples were further dehydrated with a series of ethanol, gradually transferred to acetone and embedded into Araldite 502/Embed 812 resin (EMS; catalog no. 13940) using the ultrarapid infiltration by centrifugation method revisited by McDonald (2014). Ultrathin (70–90 nm) sections were collected on nickel slot grids as described by Moran and Rowley (1987), stained with 0.1% potassium permanganate in 0.1 n H2SO4 (Sawaguchi et al., 2001), and examined with an Hitachi H-7650 TEM (Hitachi High-Technologies Europe) operating at 100 kV and fitted with an AMT XR41-M digital camera (Advanced Microscopy Techniques). Leaf samples of seven biological replicates per genotype were analyzed. For each of those leaf samples, 10 images at a magnification of 10,000 were taken from chloroplast areas from palisade parenchyma with a section orientation perpendicular to the majority of thylakoid membranes. In total, membrane layers and grana heights from 700 grana stacks per genotype were quantified.
Statistical Analyses
Experimental plant material was grown appropriately blocked for each experiment. Statistical analysis of chlorophyll content and fluorescence measurements was performed with IBM SPSS Statistics software. For the quantitative MS data, differential protein and peptide abundances from four independent biological replicates were tested with the LIMMA package (Ritchie et al., 2015) in R 3.3.1 (R Core Team, 2016). For statistical analysis of the membrane layers per grana height, data were analyzed in Microsoft Excel using a two-tailed Student’s t test assuming unequal variances.
Accession Numbers
The MS proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE (Vizcaíno et al., 2016) partner repository with the data set identifiers PXD007625 and PXD007630. NSI (At1g32070) T-DNA lines used in this work were nsi-1 (SALK_033944) and nsi-2 (SALK_020577), and STN7 (At1g68830) T-DNA line stn7 (SALK_073254).
Supplemental Data
Supplemental Data Set 1. Quantitative acetylome data analysis.
Supplemental Data Set 2. Quantitative proteome data analysis.
Supplemental Data Set 3. Fast light response curves of P700 absorbance and chlorophyll fluorescence.
Supplemental Data Set 4. Quantitative proteome data analysis of the PSI-LHCII, PSI, and LHCII trimer bands excised from the lpBN-PAGE.
Supplemental File 1. ANOVA tables.
Dive Curated Terms
The following phenotypic, genotypic, and functional terms are of significance to the work described in this paper:
Acknowledgments
We thank Taina Tyystjärvi and Daniel Gibbs for reading the manuscript and Hans-Henning Kunz for providing the kea seeds. This study was supported by the Academy of Finland (307335 “Centre of Excellence in Molecular Biology of Primary Producers” to P.M., M.M.K., A.I., M.G., and E.T. and 259075 for E.T.), the Doctoral Programme of Molecular Life Sciences at the University of Turku and The Finnish Concordia Fund to M.M.K., the “Professorinnenprogramm” of the University of Muenster to A.B. and I.F., the Max Planck Gesellschaft to I.L., U.N., and I.F., and the Deutsche Forschungsgemeinschaft (FI 1655/3-1, INST 211/744-1 FUGG) to I.F. Selected aspects of this work were supported by the German Research Foundation funds SFB 1036/TP13 and WI 3560/2-1 to M.W. This project was carried out within the ERA-CAPS Research Programme “KatNat.”
AUTHOR CONTRIBUTIONS
P.M., I.F., M.M.K., E.T., D.S., and M.W. designed the experiments. M.M.K., A.B., A.I., M.G., I.L., U.N., T.V.D., and J.S. performed research. M.M.K., P.M., I.F., I.L., A.B., and E.T. analyzed the data. M.M.K., P.M., and I.F. wrote the manuscript. All authors revised and approved the article.
Footnotes
Articles can be viewed without a subscription.
References
- Allahverdiyeva Y., et al. (2013). Arabidopsis plants lacking PsbQ and PsbR subunits of the oxygen-evolving complex show altered PSII super-complex organization and short-term adaptive mechanisms. Plant J. 75: 671–684. [DOI] [PubMed] [Google Scholar]
- Allen J.F. (1992). Protein phosphorylation in regulation of photosynthesis. Biochim. Biophys. Acta 1098: 275–335. [DOI] [PubMed] [Google Scholar]
- Bellafiore S., Barneche F., Peltier G., Rochaix J.-D. (2005). State transitions and light adaptation require chloroplast thylakoid protein kinase STN7. Nature 433: 892–895. [DOI] [PubMed] [Google Scholar]
- Bennett J., Steinback K.E., Arntzen C.J. (1980). Chloroplast phosphoproteins: regulation of excitation energy transfer by phosphorylation of thylakoid membrane polypeptides. Proc. Natl. Acad. Sci. USA 77: 5253–5257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benson S.L., Maheswaran P., Ware M.A., Hunter C.N., Horton P., Jansson S., Ruban A.V., Johnson M.P. (2015). An intact light harvesting complex I antenna system is required for complete state transitions in Arabidopsis. Nat. Plants 1: 15176. [DOI] [PubMed] [Google Scholar]
- Boersema P.J., Raijmakers R., Lemeer S., Mohammed S., Heck A.J.R. (2009). Multiplex peptide stable isotope dimethyl labeling for quantitative proteomics. Nat. Protoc. 4: 484–494. [DOI] [PubMed] [Google Scholar]
- Bonaventura C., Myers J. (1969). Fluorescence and oxygen evolution from Chlorella pyrenoidosa. Biochim. Biophys. Acta 189: 366–383. [DOI] [PubMed] [Google Scholar]
- Brandizzi F., Frangne N., Marc-Martin S., Hawes C., Neuhaus J.-M., Paris N. (2002). The destination for single-pass membrane proteins is influenced markedly by the length of the hydrophobic domain. Plant Cell 14: 1077–1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardol P., Gloire G., Havaux M., Remacle C., Matagne R., Franck F. (2003). Photosynthesis and state transitions in mitochondrial mutants of Chlamydomonas reinhardtii affected in respiration. Plant Physiol. 133: 2010–2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuartzman S.G., Nevo R., Shimoni E., Charuvi D., Kiss V., Ohad I., Brumfeld V., Reich Z. (2008). Thylakoid membrane remodeling during state transitions in Arabidopsis. Plant Cell 20: 1029–1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox J., Mann M. (2008). MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 26: 1367–1372. [DOI] [PubMed] [Google Scholar]
- Crepin A., Caffarri S. (2015). The specific localizations of phosphorylated Lhcb1 and Lhcb2 isoforms reveal the role of Lhcb2 in the formation of the PSI-LHCII supercomplex in Arabidopsis during state transitions. Biochim. Biophys. Acta 1847: 1539–1548. [DOI] [PubMed] [Google Scholar]
- Demmig-Adams B. (1990). Carotenoids and photoprotection in plants: A role for the xanthophyll zeaxanthin. Biochim. Biophys. Acta 1020: 1–24. [Google Scholar]
- Depège N., Bellafiore S., Rochaix J.-D. (2003). Role of chloroplast protein kinase Stt7 in LHCII phosphorylation and state transition in Chlamydomonas. Science 299: 1572–1575. [DOI] [PubMed] [Google Scholar]
- Dinh T.V., Bienvenut W.V., Linster E., Feldman-Salit A., Jung V.A., Meinnel T., Hell R., Giglione C., Wirtz M. (2015). Molecular identification and functional characterization of the first Nα-acetyltransferase in plastids by global acetylome profiling. Proteomics 15: 2426–2435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkemeier I., Laxa M., Miguet L., Howden A.J.M., Sweetlove L.J. (2011). Proteins of diverse function and subcellular location are lysine acetylated in Arabidopsis. Plant Physiol. 155: 1779–1790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fristedt R., Herdean A., Blaby-Haas C.E., Mamedov F., Merchant S.S., Last R.L., Lundin B. (2015). PHOTOSYSTEM II PROTEIN33, a protein conserved in the plastid lineage, is associated with the chloroplast thylakoid membrane and provides stability to photosystem II supercomplexes in Arabidopsis. Plant Physiol. 167: 481–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fristedt R., Trotta A., Suorsa M., Nilsson A.K., Croce R., Aro E.M., Lundin B. (2017). PSB33 sustains photosystem II D1 protein under fluctuating light conditions. J. Exp. Bot. 68: 4281–4293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galka P., Santabarbara S., Khuong T.T.H., Degand H., Morsomme P., Jennings R.C., Boekema E.J., Caffarri S. (2012). Functional analyses of the plant photosystem I-light-harvesting complex II supercomplex reveal that light-harvesting complex II loosely bound to photosystem II is a very efficient antenna for photosystem I in state II. Plant Cell 24: 2963–2978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gawroński P., Górecka M., Bederska M., Rusaczonek A., Ślesak I., Kruk J., Karpiński S. (2013). Isochorismate synthase 1 is required for thylakoid organization, optimal plastoquinone redox status, and state transitions in Arabidopsis thaliana. J. Exp. Bot. 64: 3669–3679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genty B., Briantais J., Baker N.R. (1989). The relationship between the quantum yield of photosynthetic electron transport and quenching of chlorophyll fluorescence. Biochim. Biophys. Acta 990: 87–92. [Google Scholar]
- Grieco M., Tikkanen M., Paakkarinen V., Kangasjärvi S., Aro E.-M. (2012). Steady-state phosphorylation of light-harvesting complex II proteins preserves photosystem I under fluctuating white light. Plant Physiol. 160: 1896–1910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartl M., et al. (2017). Lysine acetylome profiling uncovers novel histone deacetylase substrate proteins in Arabidopsis. Mol. Syst. Biol. 13: 949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartl M., König A.-C., Finkemeier I. (2015). Identification of lysine-acetylated mitochondrial proteins and their acetylation sites. In Plant Mitochondria: Methods and Protocols, Whelan J. and Murcha M., eds (New York: Humana Press; ), pp. 107–121. [DOI] [PubMed] [Google Scholar]
- Ifuku K., Nakatsu T., Shimamoto R., Yamamoto Y., Ishihara S., Kato H., Sato F. (2005). Structure and function of the PsbP protein of photosystem II from higher plants. Photosynth. Res. 84: 251–255. [DOI] [PubMed] [Google Scholar]
- Ifuku K., Ishihara S., Shimamoto R., Ido K., Sato F. (2008). Structure, function, and evolution of the PsbP protein family in higher plants. Photosynth. Res. 98: 427–437. [DOI] [PubMed] [Google Scholar]
- Inskeep W.P., Bloom P.R. (1985). Extinction coefficients of chlorophyll a and B in n,n-dimethylformamide and 80% acetone. Plant Physiol. 77: 483–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Järvi S., Suorsa M., Paakkarinen V., Aro E.-M. (2011). Optimized native gel systems for separation of thylakoid protein complexes: novel super- and mega-complexes. Biochem. J. 439: 207–214. [DOI] [PubMed] [Google Scholar]
- Kaňa R., Govindjee (2016). Role of ions in the regulation of light-harvesting. Front. Plant Sci. 7: 1849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kettunen R., Tyystjärvi E., Aro E.-M. (1996). Degradation pattern of photosystem II reaction center protein D1 in intact leaves. The major photoinhibition-induced cleavage site in D1 polypeptide is located amino terminally of the DE loop. Plant Physiol. 111: 1183–1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klughammer C., Schreiber U. (1994). An improved method, using saturating light pulses, for the determination of photosystem I quantum yield via P700+-absorbance changes at 830 nm. Planta 192: 261–268. [Google Scholar]
- Kouřil R., Zygadlo A., Arteni A.A., de Wit C.D., Dekker J.P., Jensen P.E., Scheller H.V., Boekema E.J. (2005). Structural characterization of a complex of photosystem I and light-harvesting complex II of Arabidopsis thaliana. Biochemistry 44: 10935–10940. [DOI] [PubMed] [Google Scholar]
- Kramer D.M., Johnson G., Kiirats O., Edwards G.E. (2004). New fluorescence parameters for the determination of QA redox state and excitation energy fluxes. Photosynth. Res. 79: 209–218. [DOI] [PubMed] [Google Scholar]
- Kulak N.A., Pichler G., Paron I., Nagaraj N., Mann M. (2014). Minimal, encapsulated proteomic-sample processing applied to copy-number estimation in eukaryotic cells. Nat. Methods 11: 319–324. [DOI] [PubMed] [Google Scholar]
- Kunz H.H., Gierth M., Herdean A., Satoh-Cruz M., Kramer D.M., Spetea C., Schroeder J.I. (2014). Plastidial transporters KEA1, -2, and -3 are essential for chloroplast osmoregulation, integrity, and pH regulation in Arabidopsis. Proc. Natl. Acad. Sci. USA 111: 7480–7485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli U.K. (1970). Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227: 680–685. [DOI] [PubMed] [Google Scholar]
- Lee H.Y., Byeon Y., Lee K., Lee H.-J., Back K. (2014). Cloning of Arabidopsis serotonin N-acetyltransferase and its role with caffeic acid O-methyltransferase in the biosynthesis of melatonin in vitro despite their different subcellular localizations. J. Pineal Res. 57: 418–426. [DOI] [PubMed] [Google Scholar]
- Leoni C., Pietrzykowska M., Kiss A.Z., Suorsa M., Ceci L.R., Aro E.-M., Jansson S. (2013). Very rapid phosphorylation kinetics suggest a unique role for Lhcb2 during state transitions in Arabidopsis. Plant J. 76: 236–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longoni P., Douchi D., Cariti F., Fucile G., Goldschmidt-Clermont M. (2015). Phosphorylation of the light-harvesting complex II isoform Lhcb2 is central to state transitions. Plant Physiol. 169: 2874–2883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lunde C., Jensen P.E., Haldrup A., Knoetzel J., Scheller H.V. (2000). The PSI-H subunit of photosystem I is essential for state transitions in plant photosynthesis. Nature 408: 613–615. [DOI] [PubMed] [Google Scholar]
- McDonald K.L. (2014). Out with the old and in with the new: rapid specimen preparation procedures for electron microscopy of sectioned biological material. Protoplasma 251: 429–448. [DOI] [PubMed] [Google Scholar]
- Moran D.T., Rowley J.C. (1987). Biological specimen preparation for correlative light and electron microscopy. In Correlative Microscopy in Biology: Instrumentation and Methods, Hayat M.A., ed (New York: Academic Press; ), pp. 1–22. [Google Scholar]
- Morgan M.J., Lehmann M., Schwarzländer M., Baxter C.J., Sienkiewicz-Porzucek A., Williams T.C., Schauer N., Fernie A.R., Fricker M.D., Ratcliffe R.G., Sweetlove L.J., Finkemeier I. (2008). Decrease in manganese superoxide dismutase leads to reduced root growth and affects tricarboxylic acid cycle flux and mitochondrial redox homeostasis. Plant Physiol. 147: 101–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murata N. (1969). Control of excitation transfer in photosynthesis. I. Light-induced change of chlorophyll a fluorescence in Porphyridium cruentum. Biochim. Biophys. Acta 172: 242–251. [DOI] [PubMed] [Google Scholar]
- Narusaka M., Shiraishi T., Iwabuchi M., Narusaka Y. (2010). The floral inoculating protocol: A simplified Arabidopsis thaliana transformation method modified from floral dipping. Plant Biotechnol. 27: 349–351. [Google Scholar]
- Pesaresi P., Hertle A., Pribil M., Kleine T., Wagner R., Strissel H., Ihnatowicz A., Bonardi V., Scharfenberg M., Schneider A., Pfannschmidt T., Leister D. (2009). Arabidopsis STN7 kinase provides a link between short- and long-term photosynthetic acclimation. Plant Cell 21: 2402–2423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pietrzykowska M., Suorsa M., Semchonok D.A., Tikkanen M., Boekema E.J., Aro E.-M., Jansson S. (2014). The light-harvesting chlorophyll a/b binding proteins Lhcb1 and Lhcb2 play complementary roles during state transitions in Arabidopsis. Plant Cell 26: 3646–3660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porra R.J., Thompson W.A., Kriedemann P.E. (1989). Determination of accurate extinction coefficients and simultaneous equations for assaying chlorophylls a and b extracted with four different solvents: verification of the concentration of chlorophyll standards by atomic absorption spectroscopy. Biochim. Biophys. Acta 975: 384–394. [Google Scholar]
- Pribil M., Pesaresi P., Hertle A., Barbato R., Leister D. (2010). Role of plastid protein phosphatase TAP38 in LHCII dephosphorylation and thylakoid electron flow. PLoS Biol. 8: e1000288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team (2016). R: A Language and Environment for Statistical Computing. (Vienna, Austria: R Foundation for Statistical Computing), https://www.R-project.org/.
- Rintamäki E., Martinsuo P., Pursiheimo S., Aro E.M. (2000). Cooperative regulation of light-harvesting complex II phosphorylation via the plastoquinol and ferredoxin-thioredoxin system in chloroplasts. Proc. Natl. Acad. Sci. USA 97: 11644–11649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritchie M.E., Phipson B., Wu D., Hu Y., Law C.W., Shi W., Smyth G.K. (2015). limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 43: e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozhon W., Mayerhofer J., Petutschnig E., Fujioka S., Jonak C. (2010). ASKtheta, a group-III Arabidopsis GSK3, functions in the brassinosteroid signalling pathway. Plant J. 62: 215–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruban A.V., Johnson M.P. (2009). Dynamics of higher plant photosystem cross-section associated with state transitions. Photosynth. Res. 99: 173–183. [DOI] [PubMed] [Google Scholar]
- Sawaguchi A., Ide S., Goto Y., Kawano J., Oinuma T., Suganuma T. (2001). A simple contrast enhancement by potassium permanganate oxidation for Lowicryl K4M ultrathin sections prepared by high pressure freezing/freeze substitution. J. Microsc. 201: 77–83. [DOI] [PubMed] [Google Scholar]
- Schmidt C., Beilsten-Edmands V., Mohammed S., Robinson C.V. (2017). Acetylation and phosphorylation control both local and global stability of the chloroplast F1 ATP synthase. Sci. Rep. 7: 44068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schönfeld C., Wobbe L., Borgstädt R., Kienast A., Nixon P.J., Kruse O. (2004). The nucleus-encoded protein MOC1 is essential for mitochondrial light acclimation in Chlamydomonas reinhardtii. J. Biol. Chem. 279: 50366–50374. [DOI] [PubMed] [Google Scholar]
- Schreiber U., Schliwa U., Bilger W. (1986). Continuous recording of photochemical and non-photochemical chlorophyll fluorescence quenching with a new type of modulation fluorometer. Photosynth. Res. 10: 51–62. [DOI] [PubMed] [Google Scholar]
- Seidel J., Klockenbusch C., Schwarzer D. (2016). Investigating deformylase and deacylase activity of mammalian and bacterial sirtuins. ChemBioChem 17: 398–402. [DOI] [PubMed] [Google Scholar]
- Shapiguzov A., Ingelsson B., Samol I., Andres C., Kessler F., Rochaix J.-D., Vener A.V., Goldschmidt-Clermont M. (2010). The PPH1 phosphatase is specifically involved in LHCII dephosphorylation and state transitions in Arabidopsis. Proc. Natl. Acad. Sci. USA 107: 4782–4787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suorsa M., Rantala M., Mamedov F., Lespinasse M., Trotta A., Grieco M., Vuorio E., Tikkanen M., Järvi S., Aro E.-M. (2015). Light acclimation involves dynamic re-organization of the pigment-protein megacomplexes in non-appressed thylakoid domains. Plant J. 84: 360–373. [DOI] [PubMed] [Google Scholar]
- Tikkanen M., Piippo M., Suorsa M., Sirpiö S., Mulo P., Vainonen J., Vener A.V., Allahverdiyeva Y., Aro E.-M. (2006). State transitions revisited-a buffering system for dynamic low light acclimation of Arabidopsis. Plant Mol. Biol. 62: 779–793. [DOI] [PubMed] [Google Scholar]
- Tyanova S., Temu T., Cox J. (2016a). The MaxQuant computational platform for mass spectrometry-based shotgun proteomics. Nat. Protoc. 11: 2301–2319. [DOI] [PubMed] [Google Scholar]
- Tyanova S., Temu T., Sinitcyn P., Carlson A., Hein M.Y., Geiger T., Mann M., Cox J. (2016b). The Perseus computational platform for comprehensive analysis of (prote)omics data. Nat. Methods 13: 731–740. [DOI] [PubMed] [Google Scholar]
- Tyystjärvi E., Karunen J. (1990). A microcomputer program and fast analog to digital converter card for the analysis of fluorescence induction transients. Photosynth. Res. 26: 127–132. [DOI] [PubMed] [Google Scholar]
- Vizcaíno J.A., et al. (2016). 2016 update of the PRIDE database and its related tools. Nucleic Acids Res. 44: D447–D456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P., Grimm B. (2016). Comparative analysis of light-harvesting antennae and state transition in chlorina and cpSRP mutants. Plant Physiol. 172: 1519–1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiśniewski J.R., Zougman A., Mann M. (2009a). Combination of FASP and StageTip-based fractionation allows in-depth analysis of the hippocampal membrane proteome. J. Proteome Res. 8: 5674–5678. [DOI] [PubMed] [Google Scholar]
- Wiśniewski J.R., Zougman A., Nagaraj N., Mann M. (2009b). Universal sample preparation method for proteome analysis. Nat. Methods 6: 359–362. [DOI] [PubMed] [Google Scholar]
- Wollman F.A. (2001). State transitions reveal the dynamics and flexibility of the photosynthetic apparatus. EMBO J. 20: 3623–3630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X., Oh M.H., Schwarz E.M., Larue C.T., Sivaguru M., Imai B.S., Yau P.M., Ort D.R., Huber S.C. (2011). Lysine acetylation is a widespread protein modification for diverse proteins in Arabidopsis. Plant Physiol. 155: 1769–1778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi X., Hargett S.R., Frankel L.K., Bricker T.M. (2009). The PsbP protein, but not the PsbQ protein, is required for normal thylakoid architecture in Arabidopsis thaliana. FEBS Lett. 583: 2142–2147. [DOI] [PubMed] [Google Scholar]
- Zhang S., Scheller H.V. (2004). Light-harvesting complex II binds to several small subunits of photosystem I. J. Biol. Chem. 279: 3180–3187. [DOI] [PubMed] [Google Scholar]