Abstract
The genus Brachypodium represents a model system that is advancing our knowledge of the biology of grasses, including small grains, in the postgenomics era. The most widely used species, Brachypodium distachyon, is a C3 plant that is distributed worldwide. B. distachyon has a small genome, short life cycle, and small stature and is amenable to genetic transformation. Due to the intensive and thoughtful development of this grass as a model organism, it is well-suited for laboratory and field experimentation. The intent of this review is to introduce this model system genus and describe some key outcomes of nearly a decade of research since the first draft genome sequence of the flagship species, B. distachyon, was completed. We discuss characteristics and features of B. distachyon and its congeners that make the genus a valuable model system for studies in ecology, evolution, genetics, and genomics in the grasses, review current hot topics in Brachypodium research, and highlight the potential for future analysis using this system in the coming years.
INTRODUCTION: DEVELOPING BRACHYPODIUM AS A MODEL GENUS
Advances in biology have both been based on and benefited from research on model organisms. Arabidopsis thaliana, of course, has been transformative as a model system for plant biology research, with its short life cycle, ease of genetic manipulation, small diploid genome, and simple cultivation in the laboratory. In the past two decades, several experimental plants, including rice (Oryza sativa); the N2-fixing legumes Lotus japonicus and Medicago truncatula; Populus species; Nicotiana benthamiana; and Mimulus guttatus (also known as Erythranthe guttata) and Nicotiana attenuata (for population biology and evolutionary studies), have spearheaded current advances in plant biology. However, as cogently pointed out by Leonelli and Ankeny (2013), experimental organisms and model organisms differ, although both are essential for advances in biology. Specifically, model organisms are vital resources for studies in large-scale biology, ecology, evolution, genetics, and cell biology, with a plethora of diverse lines (wild, inbreds, and mutants) and infrastructure (databases, seeds, and so on) readily available. In addition, there is a culture of sharing such resources, which exhibit a short life cycle, are easily and inexpensively cultivated and are readily manipulated in the laboratory using standard molecular biology techniques (Table 1). By contrast, experimental organisms are used to solve a specific question, excel as a tool, or are interesting organisms or objects of scientific curiosity (Leonelli and Ankeny, 2013). Based on these criteria, Arabidopsis (for dicots) and Brachypodium (for monocots) can be defined as model organisms for plant biology.
Table 1. Key Features of Purple False Brome B. distachyon (Brachypodium, Brachypodieae, Pooideae, Poaceae, and Poales) as a Model Organism for Plant Biology.
| Key Features |
|---|
| Diploida |
| Annualb |
| Self-fertile |
| Short seed-to-seed life cycle (∼8 weeks) |
| Wild speciesc |
| Ease of outcrossing for genetics |
| Ease of growth under laboratory conditions |
| Broadly susceptible to plant microbesd |
| Evolutionary taxonomy of Brachypodium speciese |
| Small genome sizef |
| Synteny with major small grains (wheat, maize, millet, rice, barley) |
| Completed genome, transcriptomeg |
| Inbred linesg |
| Mutants (indels, SNPs, T-DNA insertions, CRISPR amenable)h |
| Community resources (seed, tools, genomic databases)h |
Key features of other Brachypodium species, both diploid and allopolyploid, are discussed in the text. See Table 2 for additional features of specific annual Brachypodium species.
Three annual species, B. distachyon, B. stacei, and B. hybridum, have been sequenced. B. sylvaticum, a perennial diploid species, has been sequenced, inbred lines have been developed, and it is amenable to Agrobacterium-mediated transformation (Steinwand et al., 2013). Other perennials are discussed in the text.
B. distachyon is found in its native Mediterranean region (Catalán et al., 2016a), and its close congener B. hybridum is found worldwide.
See Table 4 for a list of microbes and citations.
For more information, refer to updated taxonomic reports of annual (Catalán et al., 2016a) and of annual and perennial (Catalán et al., 2016b) Brachypodium species.
The 272-Mb genome of B. distachyon comprises five chromosomes. Genomes of other members of the genus Brachypodium are discussed in the text.
To date, 54 inbred lines of B. distachyon have been sequenced (Gordon et al., 2017), and several other have been characterized genomically.
See Table 3 for resources with pertinent links and citations.
The genus Brachypodium has come to the forefront for research on grasses (Poaceae). The development of its temperate flagship species, Brachypodium distachyon, as a model organism has been described previously (Brutnell et al., 2015; Kellogg, 2015b; Lyons and Scholthof, 2015, 2016; Vogel, 2016) and is briefly summarized below. B. distachyon was accepted at a remarkably rapid rate as a dynamic research tool that offers a plethora of opportunities for comparative work—with Arabidopsis as the model dicot—and fundamental advances in grass biology. Similar to Arabidopsis, building B. distachyon as a model grass species (Lyons and Scholthof, 2015; Vogel, 2016) has required the development of cultivation and transformation methods, plus the genetic and genomic tools deemed essential for 21st century plant biology studies. In addition, Brachypodium research has been greatly accelerated by next-generation sequencing (NGS) technologies and the remarkable efforts of community-based projects supported by the U.S. Department of Energy Joint Genome Institute (DOE-JGI) and the USDA.
Model organisms for laboratory research have primarily been used to dissect specific aspects of plant biology following a reductionist approach. With the rise of B. distachyon, scientists have found many opportunities to work across the spectrum of basic-translational-applied research, as elaborated below. For instance, B. distachyon has been rapidly subsumed into fundamental research on plant development, plant-microbe interactions, abiotic stress, evolutionary biology, systems biology, ecology research, and for the development of new tools and concepts toward improving other temperate C3 grasses—such as wheat (Triticum aestivum) and barley (Hordeum vulgare)—that are crucial small grain crops used worldwide for the production of food, forage, and feed. Today, B. distachyon provides an ideal and robust platform for discovery-based research in each of these aspects of plant biology (Table 1). Additionally, Brachypodium species have maintained their wildness, providing a treasure-trove of resources for plant ecology researchers to study the plants in situ.
Here, we describe key features of B. distachyon and other congeners, including descriptions of their origin, morphology, distribution, ecology, and genetic characteristics and provide examples of how they have been and can be used as genetic tools to understand and solve problems in plant biology.
BRACHYPODIUM SYSTEMATICS, EVOLUTION, SPECIATION, AND MORPHOLOGY
For more than a century, B. distachyon (Palisot de Beauvois, 1812) was considered the single annual representative species of the genus Brachypodium (Schippmann, 1991), and for more than three decades, three cytotypes of 2n=10, 20, and 30 chromosomes were recognized within the species, although they were thought to be diploid, tetraploid, and hexaploid individuals of an ascendant autopolyploid series with x=5 (Talavera, 1978). It was not until recently, however, that the accrued phenotypic, cytogenetic, and molecular phylogenetic evidence demonstrated that the three cytotypes corresponded to three independent species: two diploids, B. distachyon (2n=2x=10, x=5) and B. stacei (2n=2x=20, x=10), and their derived allotetraploid B. hybridum (2n=4x=30, x=10+5) (Catalán et al., 2012; Table 2). Despite having twice the number of chromosomes, the genome size of B. stacei (0.564 pg/2C; reference Brassica rapa Goldball 0.97pg/2C) is roughly similar to that of B. distachyon (0.631 pg/2C; reference Brassica rapa Goldball 0.97 pg/2C), whereas the genome size of B. hybridum corresponds to the sum of the two progenitor genomes (1.265 pg/2C; reference Lycopersicum esculentum Stupicke 1.96pg/2C) (Catalán et al., 2012). Molecular evolutionary data indicate that B. stacei is the oldest diploid lineage within the genus Brachypodium, splitting from the common ancestor ∼10 million years ago (Mya; 106 years). This was followed by the divergence of the B. distachyon lineage (∼7 Mya), which preceded the split of a clade of recent perennial lineages (core perennial clade; ∼3 Mya). The allotetraploid B. hybridum species originated ∼1 Mya (Catalán et al., 2012). The divergence estimations have been confirmed through nested-dating analysis within a (grass) family-wide phylogenomic analysis, based on plastome data that also dated the origin of the parental B. distachyon species to ∼1 Mya, concurrent with that of its descendant hybrid (Sancho et al., 2018).
Table 2. Key Features of B. distachyon, B. stacei, and B. hybridum.
| Trait | B. distachyon | B. stacei | B. hyridum |
|---|---|---|---|
| Ploidy | Diploid | Diploid | Allotetraploid |
| Annual | Yesa | Yes | Yes |
| Originb | ∼7 Mya (lineage) ∼1 Mya (species) | ∼10 Mya (lineage)c | ∼1 Mya (species)c |
| Chromosome | 2n=2x=10, x=5 | 2n=2x=20, x=10 | 2n=4x=30, x=10+5 |
| Genome size | 0.631 pg/2C 272 Mbp | 0.564 pg/2C 234 Mbp | 1.265 pg/2C 509 Mb |
| Occasional short rhizomes | No | Yes | Yes |
| Leaf blade color | Bright green | Pale green | Dark green |
| Leaf blade shape | Straight | Curled | Straight |
| Leaf softness | Brittle | Soft | Brittle |
| Leaf hairiness | Scarcely hairy or glabrous | Densely hairy | Scarcely hairy or glabrous |
| Vernalizationd | Yes | No | No |
Annual, although there are other Brachypodium species that are perennials.
Origin indicates million years ago (Mya; 106 years) since the split from a common ancestor. Lineage indicates split from the stem ancestor, and species indicates split from the crown ancestor.
B. hybridum is the derived allotetraploid of B. distachyon × B. stacei.
Vernalization is required to induce flowering. Most, but not all, B. distachyon lines require vernalization (Vogel et al., 2009).
Analysis of maternally inherited plastid genes support the recurrent origin of allotetraploid B. hybridum from bidirectional crosses of its parents, followed by whole-genome duplication of the unfertile interspecific hybrid (López-Alvarez et al., 2012). These authors also showed that most of the studied circum-Mediterranean B. hybridum populations were derived from a maternal B. stacei parent, whereas only relatively few western-Mediterranean populations were derived from a maternal B. distachyon parent.
Phylogenomic studies of B. distachyon based on 54 resequenced ecotypes showed a main split of two intraspecific lineages characterized by their flowering time. These included the extremely delayed flowering (EDF+) versus non-extremely delayed flowering (non-EDF+) lineages and their respective coevolving molecular variants of genes known to regulate vernalization (e.g., VRN1 and VRN2) and flowering (e.g., CO2, FTL9, FTL13, PHYC, and PPD1). However, none of those traits coevolved with latitude (Gordon et al., 2017). Both lineages branched off approximately half a million years ago (Sancho et al., 2018): The first clade contained lines distributed across the Mediterranean region, and the second clade showed the divergence of two geographically constrained eastern Mediterranean (Turkey and other countries, T+) and western Mediterranean (Spain and other countries, S+) groups. Comparative genomics of nuclear and plastome data further identified four introgressions (two of them also plastomic) and nine chloroplast capture events between the three groups, suggesting that flowering time variation is the main factor driving rapid intraspecific divergence in B. distachyon, although it is counterbalanced by repeated introgression between previously isolated lineages (Gordon et al., 2017; Sancho et al., 2018), thus maintaining the biological reproductive concept of the species.
PERENNIAL BRACHYPODIUM SPECIES
Besides the three most intensively investigated annual species, the genus Brachypodium also contains ∼17 perennial species distributed worldwide (Schippmann, 1991; Catalán et al., 2016b) (Figure 1). The 20 recognized Brachypodium taxa are characterized by their typical subsessile spikelet and exclusive forms of embryo development, seed storage proteins, polysaccharides and globulins, stem and leaf fructosans, small genome sizes, and large disploidy (i. e., species showing different chromosome base numbers) (Catalán et al., 2016b). The Brachypodium taxa belong to the monotypic tribe Brachypodieae and are evolutionarily placed in an intermediate position between the ancestral basal pooids and the recently evolved clade of core pooid lineages, including the economically important Triticeae + Bromeae and Poaceae (Sancho et al., 2018).
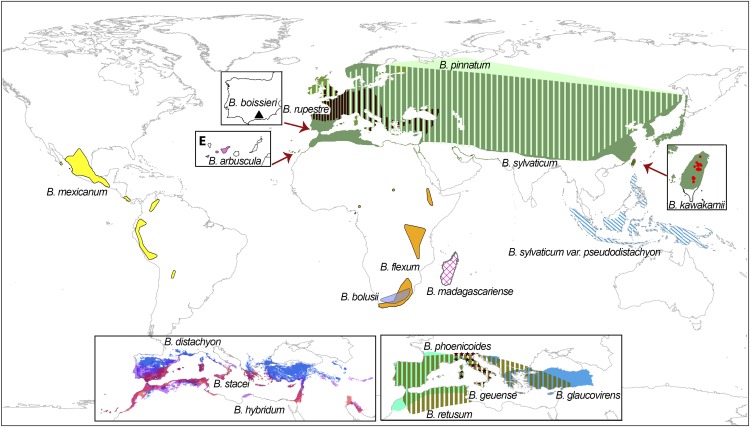
Figure 1.
Worldwide Geographic Distribution of 18 Brachypodium Taxa.
The 18 worldwide species of Brachypodium taxa (B. arbuscula, pink; B. boissieri, black triangle; B. bolusii, violet; B. distachyon, dark blue; B. flexum, orange; B. genuense, dark-brown star; B. glaucovirens, light blue; B. hybridum, purple; B. kawakamii, magenta dot; B. madagascariense, open pink square; B. mexicanum, yellow; B. phoenicoides, aquamarine; B. pinnatum, light green; B. retusum, pale brown; B. rupestre, dark brown; B. stacei, red; B. sylvaticum, sage green; B. sylvaticum var pseudodistachyon, light-blue diagonal line). (Image adapted from Catalán et al. [2016b], Figure 2, page 15, copyright 2016, Springer.)
Perennial Brachypodium species vary widely both in phenotype and origin. They range from the short-rhizomatose, self-fertile American allotetraploid B. mexicanum, a species closely related to the oldest B. stacei lineage and biologically and genomically similar to the annual species, to the strong-rhizomatose, outcrossing, and recently evolved Eurasian and African diploid and allopolyploid species of the core-perennial clade. This clade includes some of the widely distributed palaearctic species, such as the diploids B. pinnatum and B. sylvaticum, together with other more restricted endemic species (Catalán et al., 2016b). Two Mediterranean high ploidy-level allopolyploids, B. retusum and B. boissieri, characterized by their branched woody stems and short inrolled leaves, have inherited ancestral, intermediately evolved and recent genomes, whereas core perennial allotetraploids B. phoenicoides, B. pinnatum 4x and B. rupestre 4x, characterized by their nonbranched stems and long flat leaves, have only inherited recently evolved genomes (Catalán et al., 2016b; Díaz-Pérez et al., 2018).
The diploid B. sylvaticum, the best-known perennial species of the genus, was recently selected as a model plant for perenniality (Gordon et al., 2016). Genomic and transcriptomic resources are available for B. sylvaticum, including its reference genome (B. sylvaticum Ain1) and a second resequenced line (B. sylvaticum Sin1) (see Phytozome). This plant is predominantly self-fertile (94.6%), with a small, compact genome (340 Mb) distributed on nine chromosomes that can be easily transformed (Steinwand et al., 2013). Though it belongs to the core perennial clade of predominantly robust strong-rhizomatose outbreeding species, B. sylvaticum shows a slender habit and rhizomes and a selfing reproductive system; however, the species, like the other perennials, is an overwintering plant, characterized by its hairy indumentum, soft leaves, nodding panicle, and long awned lemma (Catalán et al., 2016b). Its distribution covers the largest native Old World geographical range of any Brachypodium species, ranging from the Canary Islands (West) to Japan and New Guinea (East) and from Scandinavia and Siberia (North) to northern Africa and Malaysia (South), although some of the East Asian and Malaysian populations may correspond to different microtaxa (Catalán et al., 2016b).
Disploidy is a major feature of Brachypodium, a genus that contains diploid species with x=10, 9, 8, and 5 chromosomes and allopolyploid species with different combinations of chromosome base numbers (Catalán et al., 2016b). Phylogenetic and comparative chromosome painting data have been used to propose evolutionary hypotheses on descendant versus descendant-ascendant disploidy series along the Brachypodium tree (Betekhtin et al., 2014) and secondary origins for the allopolyploids (Catalán et al., 2016b). The advent of the sequenced reference genomes facilitated the reconstruction of the path of nested chromosome fusions that support the descendant disploidy hypothesis. Interspecific breeding barriers between Brachypodium species (Khan and Stace, 1999) are fully congruent with the Brachypodium phylogeny and explain the reproductive isolation of the early diverging B. stacei type (B. hybridum) and B. mexicanum. These barriers also explain the crossability of the intermediately evolved B. distachyon with the core perennial species and the highly fertile descendants of all attempted interspecific crosses between recently evolved core perennial taxa (Khan and Stace, 1999; Catalán et al., 2016b).
ECOLOGY OF BRACHYPODIUM
The ecology of the ∼20 Brachypodium species varies drastically depending on their geographical distributions and adaptation to different climates and habitats. Among these, the three annual species and the perennials B. retusum and B. boissieri have adapted to xeric Mediterranean conditions. The Canarian endemic species B. arbuscula grows in more humid places, while the endemic South African species B. bolusii and Taiwanese B. kawakamii thrive in alpine vegetation belts. The tropical African B. flexum and Malagasy B. madagascariense grow in the Afromontane forests, and the American B. mexicanum is found in xeric to humid neotropical habitats. The western Mediterranean B. phoenicoides is adapted to mesic to dry places with humid soils, and the predominantly Eurasian B. pinnatum, B. rupestre, and B. sylvaticum species grow in mesic to humid open grasslands and forests (Catalán et al., 2016b). Two Brachypodium species have been confirmed as invasive species. B. hybridum has successfully and predominantly colonized other Mediterranean-type ecoregions (California, South Africa, South America, and southern Australia), while B. sylvaticum is spreading in humid, forested regions of western North America and Australia (Catalán et al., 2016b).
Detailed ecological studies have been conducted using the three annual circum-Mediterranean species of the B. distachyon complex. Environmental niche modeling analysis indicated that, overall, B. distachyon grows in higher, cooler, and wetter places north of 33°; B. stacei in lower, warmer, and drier places south of 40° 30’; and B. hybridum in places with intermediate ecological features and across latitudinal boundaries but also overlapping with those of its parents, more often with those of B. stacei (López-Alvarez et al., 2015; Catalán et al., 2016b). This concurs with the finding that most B. distachyon lines require vernalization treatment in order to flower, whereas B. stacei and B. hybridum lines do not (Vogel et al., 2009). Additionally, B. stacei grows in shady habitats, whereas B. distachyon and B. hybridum occur in open habitats (López-Alvarez et al., 2015; Catalán et al., 2016a). Paleoenvironmental modeling data support the notion that the Mediterranean basin and adjacent areas serve as long-term refugia for B. stacei and B. distachyon, and some of them serve as potential hybrid zones, which could have favored the recurrent origins of B. hybridum since the late Pleistocene.
Niche similarity tests showed evidence of niche conservatism for B. hybridum and each of its parents; the allotetraploid shares niche occupancy with its progenitors but is reproductively isolated from both. Also, B. hybridum has the largest niche overlap with its parental niches but a similar distribution range and niche breadth, indicating that the hybrid does not outcompete its parents in their native ranges (López-Alvarez et al., 2015; Catalán et al., 2016b). Conversely, B. hybridum is the only species of the complex that has successfully colonized other non-native world regions. This suggests that the allotetraploid has greater ecological tolerance compared with the diploids, which could be associated with the boost in diversifying selection due to increasing genomic and epigenomic expression, as well as rapid shifts in physiological and adaptive traits such as photoperiod and weediness (Bakker et al., 2009; Catalán et al., 2016b).
Field analyses demonstrated that environmental aridity gradients in Spain affect the predominant northern and southern Mediterranean distributions of the less efficient water user B. distachyon and the more efficient water users B. hybridum and B. stacei (water use efficiency), respectively, under water-restricted growing conditions (Manzaneda et al., 2012; Martínez et al., 2018). Under drought conditions, B. hybridum individuals behave as drought escapists, maintaining higher photosynthesis and stomatal conductance and showing earlier flowering times to cope with water stress than the less adapted B. distachyon individuals (Manzaneda et al., 2015). Translocation experiments in mixed southern Spanish B. distachyon–B. hybridum populations have demonstrated the superior capability of the allotetraploid in colonizing densely occupied competitive habitats and the balance of intra/interspecies competition favoring the establishment of B. hybridum over B. distachyon populations under natural field conditions at the rear-edge distribution of the diploid B. distachyon parent (Rey et al., 2017). By contrast, field analyses in southern Mediterranean Israel microsites have revealed the predominant presence of allotetraploid B. hybridum over its diploid parents, especially the more frequent B. stacei, along a large-scale latitudinal range. However, the distribution of B. hybridum was not correlated with an aridity cline, although clustered patterns suggested that the distributions of B. stacei and B. hybridum were not random (Bareither et al., 2017). Ongoing ecogenomic studies of B. distachyon–B. hybridum populations and B. stacei–B. hybridum populations in Spain and Israel will further help to decipher the potential drivers of the ecological success of parental diploid and allotetraploid populations in different microenvironments.
GENETICS AND GENOMICS
B. distachyon has some obvious advantages over rice, its closest genetic competitor, as a grass model system, including its smaller habit, ease of cultivation in the laboratory, and shorter seed-to-seed life cycle (Figure 2). B. distachyon also has a smaller genome (∼272 Mb) (International Brachypodium Initiative, 2010) than rice (∼430 Mb) (Sasaki and Antonio, 2004), although genome size may become less important with the rapid advances in NGS technologies. Importantly, the genus Brachypodium is evolutionarily closer to several major cereal crops than rice, such as wheat, rye (Secale cereale), and barley, which makes it a better suited model system to study cereal biology (Draper et al., 2001; Brkljacic et al., 2011; Catalán et al., 2016b).
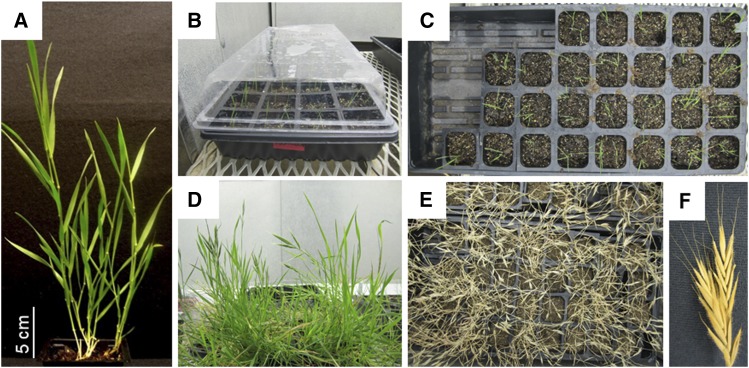
Figure 2.
Brachypodium Life Cycle and Propagation.
(A) Mature B. distachyon (Bd21-3) at ∼6 weeks old. Bar is an approximate estimate.
(B) Seven-day cold-treated (vernalized) Bd21-3 seeds are germinated in small 36-cell sheets in trays to accommodate several plants in a limited space. Typically, trays are covered until seeds germinate and seedlings are established approximately 1 week after planting.
(C) Uncovered trays containing two to three plants per pot in the early vegetative phase (∼2 weeks after planting).
(D) Mature Bd21-3 plants with inflorescence (∼8 weeks after planting).
(E) Senesced plants, ready for harvest ∼12 weeks after planting.
(F) Close-up of dried inflorescence (panicle). The seeds can be separated from the panicle, desiccated, and stored at 4°C.
B. distachyon and rice, both C3 plants, have less complex genomes compared with other grasses with larger genomes. For instance, comparative genomics analyses of B. distachyon, rice, sorghum (Sorghum bicolor; 730 Mb), and goat grass (Aegilops tauschii; 4020 Mb) revealed that sorghum and goat grass genes are distributed in clusters (gene insulae) in the genome. The gene insulae contain an average of 3.2 genes/cluster, with noncoding regions separating the clusters. The genes of rice and B. distachyon, on the other hand, are more uniformly distributed, with short intergenic distances. These differences highlight the complex effects of genome expansion on gene distribution and spacing in grasses (Gottlieb et al., 2013), and they underscore the simplicity of the B. distachyon genome.
The fully annotated reference genome sequences of two commonly used B. distachyon accessions (Bd21 and Bd21-3) are publicly available through Phytozome (https://phytozome.jgi.doe.gov/pz/portal.html). The accession Bd21 assembly was completed in 2010, and versions (v) 2.1 and 3.1 are currently available through Phytozome. The Bd21 assembly was found to comprise ∼272 Mb with an N50 of 6.4 Mb during the 2.1 version update and encodes 31,694 protein-coding loci. The recently completed Bd21 v3.1 update focused on inspecting smaller sized regions, which were verified by clone-based shotgun sequencing using Sanger and Illumina sequencing. This increased the assembly size by 1.43 Mb and filled several gaps that existed in the previous versions. The chromosomal genome assembly of accession Bd21-3 was completed using MECAT and refined using ARROW; this assembly is also ∼272 Mb in size. Users can also browse a Phytozome database of more than 800,000 single-nucleotide polymorphism (SNP) mutations in this newly released Bd21-3 assembly.
In addition to accessions Bd21 and Bd21-3, de novo assemblies of 54 diverse B. distachyon inbred accessions were completed to enable identification of the full gamut of genes (or pan-genome) of B. distachyon (Gordon et al., 2017). The availability of the B. distachyon pan-genome (https://brachypan.jgi.doe.gov)—with its annotated genomes, transcriptomes and transposons—opens new avenues to uncover key physiological and adaptive processes in grasses. The B. distachyon pan-genome is based on 54 fully sequenced genome assemblies of geographically diverse ecotypes, 36 of which were also analyzed at the transcriptome level (Gordon et al., 2017). From this, 61,155 pan-genome clusters were classified as core (present in all lines), softcore (95–98%), shell (5–94%), and cloud (2–5%) genes, comprising nearly twice the number of genes present in any individual genome. The study showed that the core genes were enriched for essential biological functions (e.g., glycolysis) and were constrained by purifying selection, whereas shell genes were enriched for potentially beneficial functions (e.g., defense, development, and gene regulation), displayed higher evolutionary rates, were located closer to and were more functionally affected by transposable elements, and were less syntenic with orthologous genes in other grasses (Gordon et al., 2017). Shell genes contribute substantially to phenotypic variation and influence population evolutionary history within B. distachyon, as demonstrated for the three phylogenetic groups detected in the study (EDF+, T+, S+), which are characterized by different flowering time traits and their molecular regulators associated with different types of core and shell ingroup genes (Gordon et al., 2017).
Reference genomes of additional Brachypodium species, B. stacei, B. sylvaticum, and B. hybridum, were also recently completed and are publicly available at Phytozome. Although sequencing and annotation of the mitochondrial genome is yet to be completed for most species, the plastid genomes of B. distachyon (54 lines), B. stacei (1 line), and B. hybridum (2 lines) have been sequenced (Sancho et al., 2018). Plastome lengths varied from 134,991 to 135,214 bp in B. distachyon and between 136,326 and 136,330 bp in B. stacei and B. hybridum, as the two allotetraploid lines were derived from maternal B. stacei-type parents. The B. distachyon plastomes contained 133 genes, 76 of which were protein-coding genes (including seven duplicated genes), 20 nonredundant tRNAs (out of a total 38), four rRNAs in both inverted repeats, four pseudogenes (trnI, rps12a, trnT, and trnI), and two hypothetical open reading frames (ycf). The B. stacei and B. hybridum plastomes showed the same general features as the B. distachyon plastomes, except for a 1161-bp insertion between psaI and rbcL in the large single-copy region that corresponds to a coding sequence fragment annotated as pseudogene rpl23, and a deletion of an rps19 copy between psbA and trnH in the IRb repeat (Sancho et al., 2018). Together, these Brachypodium spp genomes are invaluable resources for grass evolutionary biology, polyploidy, and speciation studies.
AVAILABLE TOOLS, RESOURCES, AND DATABASES
A growing suite of tools, resources, and databases exist for the scientific community interested in embracing Brachypodium spp for their research. The DOE-JGI has spearheaded sequencing of additional Brachypodium species and accessions, as described above, to complement the Bd21 genome. Through the efforts of the International Brachypodium Initiative, the community aims to allow free access to genetics and genomics resources among Brachypodium research groups. To this end, a B. distachyon gene expression atlas was recently completed, which maps gene expression in major organs at different developmental stages (Sibout et al., 2017). The gene atlas is a new, invaluable tool to study gene function and to identify metabolic pathways. Another gene atlas project is underway, funded by the JGI Community Science Program (CSP), which will allow genome-wide identification of conserved and unique gene expression responses that occur during diverse B. distachyon-microbe interactions, including pathogenic and beneficial interactions (CSP503406; https://jgi.doe.gov/csp-2018-mandadi-gene-atlases-grass-microbe-interactions/).
Several functional genetics tools and resources are currently available for Brachypodium research at the molecular, cytogenetic, and biochemical levels (Table 3). These include a diverse collection of B. distachyon accessions; T-DNA and EMS mutants; and BAC, EST, and yeast two-hybrid (Y2H) libraries. Many of these are publicly available for scientific research.
Table 3. Genetic and Genomic Resources for Brachypodium.
More than 800,000 SNP mutants are available for B. distachyon Bd21-3.
Sodium azide mutagenesis of Bd21-3 (Dalmais et al., 2013; de Bang et al., 2018).
Phytozome has resources for B. distachyon lines Bd21 and Bd21-3, plus three additional species of Brachypodium (B. hybridum, B. stacei, and B. sylvaticum).
Brachypan represents the sequence, assembly, and annotation of the genomes of 54 B. distachyon accessions (Gordon et al., 2017).
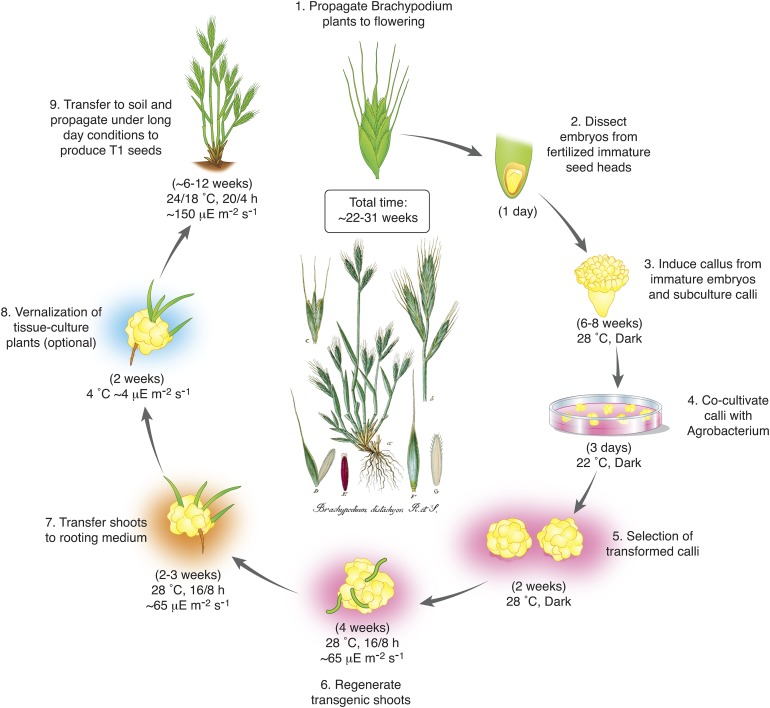
Well-established Agrobacterium tumefaciens-based and biolistic-based transformation methodologies exist to transform B. distachyon using mature (Bablak et al., 1995; Vogel et al., 2006; Sogutmaz Ozdemir and Budak, 2018) and immature (Bablak et al., 1995; Draper et al., 2001; Christiansen et al., 2005; Păcurar et al., 2008; Vain et al., 2008; Vogel and Hill, 2008) embryos. Agrobacterium-based transformation of B. distachyon Bd21-3 can be successfully completed in ∼22 to 31 weeks (Figure 3). Typically, immature embryos are dissected from developing seeds of vernalized Bd21-3 plants grown to maturity. To produce sufficient embryogenic callus for transformation, subculturing is performed twice on callus initiation medium at 28°C in the dark, with transfer to fresh medium every 2 weeks. Optimal calli that are well structured and yellow can be generated within ∼6 to 8 weeks. These calli are then cocultured with Agrobacterium (OD600 = 0.6) containing the desired construct at 22°C in the dark for 3 d. The calli are then transferred to selection medium containing the bactericide Timentin (300 mg/L) and an appropriate antibiotic for the selectable marker (e.g., hygromycin B). After 2 weeks on selection medium at 28°C in the dark, healthy transformed calli are transferred to regeneration medium with the appropriate selection agents. Shoots typically emerge from the calli in ∼4 weeks when incubated in a growth chamber with a 16/8-h light/dark cycle (28°C, ∼65 μEm m−2 s−1). For rooting, the shoots are transferred to MS medium with sucrose. Within ∼2 to 3 weeks, the plants are ready for vernalization (to promote flowering) by cold treatment at 4°C for 2 weeks under continuous low light (∼4 μEm m−2 s−1), or the plants can be transferred to soil and propagated under long-day diurnal conditions (20/4 h, 24/18°C light/dark, ∼150 μEm m−2 s−1). T1 seeds can be harvested from vernalized plants in ∼6 to 12 weeks. Bragg et al. (2015) have developed a comprehensive transformation protocol with detailed information about nutrient media compositions.
Figure 3.
Overview of the Brachypodium Agrobacterium-Based Transformation Pipeline.
For a detailed protocol with nutrient medium composition, please refer to Bragg et al. (2015). Inset: public domain botanical illustration from Sturm (1843).
The transformation efficiency (TE), defined as the percentage of cocultivated calli that successfully produce transgenic plants, is highly dependent on several factors including the B. distachyon accession (e.g., Bd21 and Bd21-3), embryo stage, callus morphology, Agrobacterium strain, as well as the promoters and selectable markers used in the binary vector (Bragg et al., 2015). When using immature embryos as the starting material, TEs of 50 to 70% can be achieved (Bragg et al., 2015). Typically, immature embryos <0.3 mm in size often produce the best embryogenic callus. It is also very important to only select and subculture compact, structured, yellow callus. Both Bd21 and Bd21-3 are amenable to transformation. However, Bd21-3 produces more structured and more yellow callus than Bd21, which is easily distinguished from inferior callus, resulting in higher TEs (Bragg et al., 2015). Therefore, Bd21-3 was used as the preferred background to develop the T-DNA knockout collection (Bragg et al., 2012; Hsia et al., 2017). Although multiple Agrobacterium strains and selection agents have been employed for Brachypodium transformation, Agrobacterium strain AGL1 and hygromycin selection are the materials of choice (Bragg et al., 2015). Commonly used monocot promoters, such as the maize (Zea mays) ubiquitin promoter, also work well in B. distachyon.
Rhizobium rhizogenes (formerly Agrobacterium rhizogenes, strain 18r12v) was recently evaluated for use with B. distachyon and B. sylvaticum, using paromomycin as a selectable marker (Collier et al., 2016). For B. distachyon, the TE using R. rhizogenes was comparable to or slightly greater (up to 60%) than that of Agrobacterium. However, the TE of B. sylvaticum was lower (∼4–6%) irrespective of the strain or selection marker used, indicating species-specific recalcitrance within Brachypodium spp. A new transformation strategy using selective overexpression of two maize morphogenic regulators, Baby boom (Bbm) and maize Wuschel2 (Wus2), dramatically enhanced the TE of several highly recalcitrant maize, sorghum, sugarcane (Saccharum officinarum), and rice lines, using either immature embryos or mature leaf (somatic) tissues as explants (Lowe et al., 2016). In this context, it may be possible to further enhance Brachypodium spp TE by combining the current transformation procedures with the Bbm and Wus2 system for functional genetic studies.
Using Agrobacterium-based transformation, multiple libraries of insertional T-DNA mutants have been generated. BrachyTAG (John Innes Center, Norwich, UK) contains ∼4117 T-DNA lines in the Bd21 background (Thole et al., 2010), but the collection is currently not accessible. The second T-DNA collection, which is accessible to users, was generated by the USDA and the DOE-JGI (https://jgi.doe.gov/our-science/science-programs/plant-genomics/Brachypodium/Brachypodium-t-dna-collection/) and consists of 23,649 T-DNA lines derived from the Bd21-3 accession, ∼7145 of which were described previously (Bragg et al., 2012). Recently, using an Illumina-based NGS approach, ∼21,165 T-DNA lines from this collection were sequenced and indexed (Hsia et al., 2017). This analysis identified ∼31% of the B. distachyon genes that were tagged by the T-DNAs, either in the gene body or within 500 bp of the gene locus.
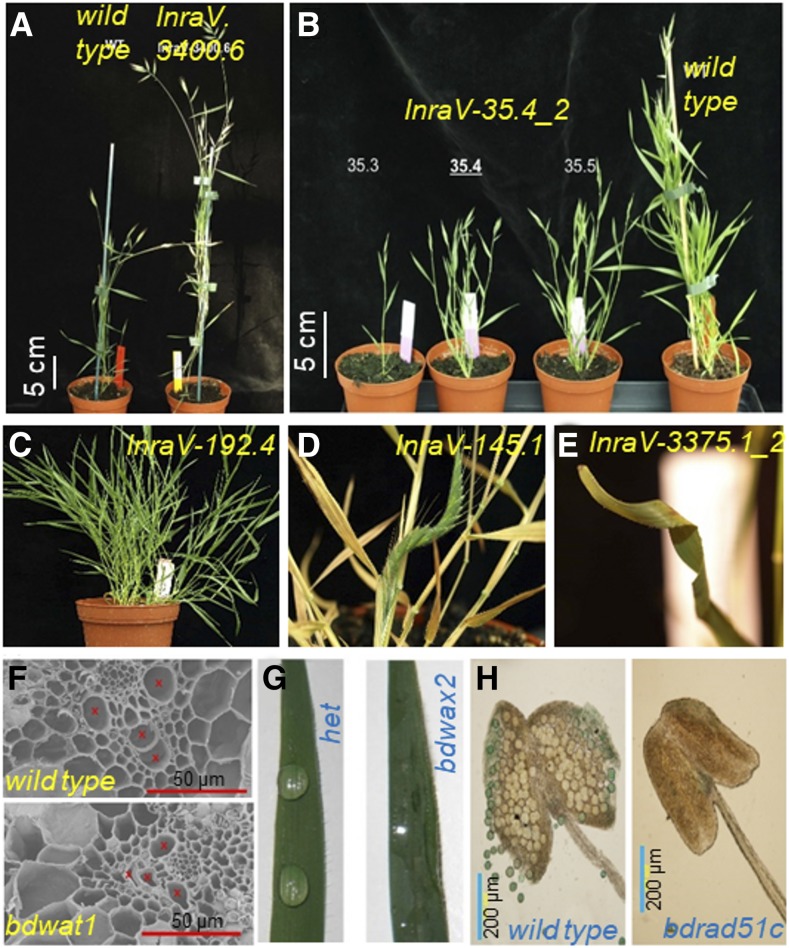
In addition to the T-DNA mutant collections, Tnt1 retrotransposon lines (Gill et al., 2018) and chemical mutagen lines (Dalmais et al., 2013) are available for functional genetic studies. For instance, ∼6000 sodium azide (NaN3) and EMS-mutagenized M2 families in the Bd21-3 background are available as part of a BRACHYTIL collection (http://urgv.evry.inra.fr/UTILLdb), with a mutation rate of ∼1 mutation per ∼354 kb (Dalmais et al., 2013). These mutants are generated and maintained by the Genomic Resources Center for B. distachyon at INRA (Versailles-Grignon; https://www-ijpb.versailles.inra.fr/en/pave/equipes/paroi-secondaire/index.htm). Together, the B. distachyon mutant collections are a valuable resource for the research community to investigate various topics in monocot biology. Researchers can find a range of phenotypes in these collections. For instance, several mutants in BRACHYTIL and the T-DNA collection display morphological, anatomical, and physiological defects reflecting perturbations in plant growth, development, flowering time pathways, cell wall, wax, and reproductive pathways (Figure 4).
Figure 4.
B. distachyon Mutant Resources and Selected Phenotypes.
Mutants from the BRACHYTIL sodium azide collection displaying abnormalities such as tall (A), dwarf (B), high tillering (C), and curved panicle and leaf phenotypes ([D] and [E]). (Images: R. Sibout, INRA, adapted from Dalmais et al. [2013], Figure 2, page 3, copyright 2013, PLoS ONE.) Mutants from Brachy T-DNA collection showing anatomical and physiological defects in cell wall (F), wax (G), and reproductive tissues (H). (Images: J. Vogel, DOE-JGI, adapted from Hsia et al. [2017], Figures 3D, 4C, and 5C, pages 365–366, copyright 2017, The Plant Journal.) Bars are approximate estimates.
In addition to making these mutant lines available to the Brachypodium community, researchers at DOE-JGI, USDA, and INRA are spearheading an international collaborative project, with the goal of sequencing ∼2000 interesting mutant lines. Several participants have initiated screening of the mutant collections for various traits of interest (https://docs.google.com/spreadsheets/d/16p85FUkyHFGYygEkzBtcz2Se-scieclbIJsUd31xPCY/edit#gid=0). Currently, ∼859 user-selected mutant lines have been sequenced, and ∼804,508 putative SNP mutations and ∼5540 putative small deletions were identified (J. Vogel, personal communication). The mutants are indexed on the Bd21-3 (v1.1) reference genome sequence available through Phytozome.
In addition to the genetic resources, two Y2H libraries are available for Brachypodium researchers to conduct protein-protein interaction studies (Cao et al., 2011). These libraries represent genes/proteins expressed in short- and long-day grown shoots collected over 24 h, as well as from shoots and roots treated with eight hormones (e.g., salicylic acid, methyl jasmonate, and so on) collected 24 h after treatment. Both Y2H libraries have very good coverage of the B. distachyon genome, as represented by ∼5.5 × 107 clones of ∼1500 bp. As a proof-of-concept, an ortholog of one of the Arabidopsis nuclear factor-Y proteins (NF-YC) in B. distachyon (Bradi3g05270) was used as a bait to identify BdNF-YB and its chaperonin containing the T-complex polypeptide-1 (CCT) protein interactors from both Y2H libraries, along with several novel interactors, demonstrating the usefulness of the Y2H libraries (Cao et al., 2011). The compact genome, small chromosome base number (x=5), and availability of a genomic BAC library of B. distachyon make it amenable for cytogenetic studies across the genus. Recently, cytogenetic maps of Brachypodium spp were developed using fluorescence in situ hybridization (FISH), chromosome painting, and chromosome barcoding (Hasterok et al., 2015; Idziak-Helmcke and Betekhtin, 2018; and references therein). Together, these cytogenetic and biochemical tools, databases, and resources are of tremendous value for Brachypodium gene discovery and genetics pertaining to diverse biological processes.
HOT TOPICS IN BRACHYPODIUM RESEARCH
Given the availability of myriad community resources and the amenability of Brachypodium species for grass functional genetics, the genus has rapidly become a model for understanding diverse grass traits including, but not limited to, cell wall biology, vernalization, evolutionary biology, root biology, host-microbe/microbiota interactions, and responses to abiotic stresses. Here, we detail some of the hot topics in Brachypodium research today and potentially the coming years.
Cell Wall and Saccharification
The composition and control of cell wall formation have primarily been studied in the eudicot model Arabidopsis. However, grasses are an evolutionarily separate group, and an appropriate model is needed to better study their cell walls (Coomey and Hazen, 2016). Grass cell wall carbohydrates predominantly include cellulose, β-glucans [(1,3;1,4)-β-d-glucans], and arabinoxylans. At the metabolic level, B. distachyon cell wall contents resemble those of field grasses (Bouvier d’Yvoire et al., 2013), making it an ideal model for studying cell wall biosynthesis. Furthermore, because of the similarity of B. distachyon vascular bundle anatomy to that of larger C3 grasses, it is a good model to study vascular system composition and development—with implications toward increasing biomass and biofuel production in feedstock grasses (Matos et al., 2013). Cell wall composition depends on tissue type and developmental stage. In B. distachyon, variations in cell wall components (cellulose, hemicellulose, and lignin) have been studied in different organs at the seedling, elongating stem and mature stages. For instance, lignin levels are lowest in the leaf and sheath and higher in the stem, with levels increasing as the plant ages (Rancour et al., 2012; Matos et al., 2013).
Cellulose, the main cell wall component in grasses, is the major target of saccharification in biofuel production. To enhance this process, several groups have focused on understanding and modifying lignin contents and quality (Bouvier d’Yvoire et al., 2013; Dalmais et al., 2013; Trabucco et al., 2013). Marriott et al. (2014) screened a B. distachyon mutant population and identified 12 lines with reduced recalcitrance to lignin hydrolyzing enzymes, named saccharification 1-12 (sac1-12). These lines show improved saccharification, with no major negative effects on plant size or mechanical strength. In another genetic screen, the spaghetti1 (spa1) mutant was identified, which exhibited pleiotrophic phenotypes such as brittle and floppy stems. At the metabolic level, the spa1 mutant contains reduced levels of crystalline cellulose but increased levels of xylan and lignins. Surprisingly, the mutants showed a 2-fold increase in saccharification compared with the wild type, despite the higher lignin content in the cell wall (Timpano et al., 2015). The authors suggest that perhaps the higher friability of the spa1 biomass and the lower cellulose crystallinity could have enhanced its enzymatic digestibility. Another study used RNAi tools to knock down B. distachyon UDP-arabinopyranose mutase activity encoded by BdRGP1, BdRGP2, and BdRGP3, which resulted in altered cell wall composition, such as reduced levels of arabinose sugars, ferulic acid, and p-coumarates (Rancour et al., 2015). The BdRGP1 RNAi lines also showed approximately 2-fold increases in released carbohydrate levels after enzymatic digestion (Rancour et al., 2015).
The mechanics of cellulose biosynthesis have been challenging to study, in part because the enzymes responsible become inactive when purified from the plant tissue. Recently, an indirect approach, based on studies with Arabidopsis, was used to help address this issue in B. distachyon (Liu et al., 2017). A fluorescent protein tag on the N-end of a cellulose synthase catalytic subunit (CESA) was used to monitor cellulose synthesis motility in B. distachyon. This fluorescence microscopy strategy also demonstrated that B. distachyon has a CESA motility practically indistinguishable from that of Arabidopsis (Liu et al., 2017).
At the transcriptional level, the regulation of cell wall formation has also been primarily described in Arabidopsis, while in grasses, these pathways have been less defined. However, with the use of B. distachyon as a model, an equivalent outline of cell wall transcriptional regulation in grasses is being established (Handakumbura and Hazen, 2012). As in Arabidopsis, there are several nascent polypeptide-associated complex (NAC) proteins in B. distachyon that regulate secondary wall biosynthesis. Of these, eight SECONDARY WALL NACs (SWNs) have been identified by phylogenetic analysis (Valdivia et al., 2013). The first six members of this family (BdSWN1-6) are orthologous to VASCULAR NAC DOMAIN transcription factors (Ohashi-Ito et al., 2010; Yamaguchi et al., 2011), while BdSWN7 and 8 are orthologous to the SND, NST1, and NST2 transcription factors in Arabidopsis (Mitsuda et al., 2005).
The mechanisms of cell wall assembly and remodeling during grain formation and the functions of cell wall proteins (CWPs) during this important process are poorly understood in grasses (Francin-Allami et al., 2016). Studies in B. distachyon, taking advantage of its similarities in grain development to that of other grasses, are helping to elucidate these pathways. A liquid chromatography-tandem mass spectrometry analysis of purified CWPs during three developmental stages in B. distachyon grain formation has helped increase our knowledge of the cell wall proteome in grasses. For instance, this study led to the identification of 111 new proteins. Many of these CWPs are likely involved in cell wall remodeling during grain formation (Francin-Allami et al., 2016). Of crucial importance for agriculture and crop research, the first comprehensive study of starch granule development in B. distachyon (Bd21) was performed and compared with Chinese Spring wheat and Aegylops peregrina. The results showed that starch granule size and development are highly similar between Bd21 and wheat, with most of the starch synthesis genes in B. distachyon being more similar to those in wheat than in rice (Chen et al., 2014). In summary, B. distachyon has proven to be an excellent model for studying grass cell wall biology, providing information that could be used to enhance food, biomass, and bioenergy-related traits in field grasses.
Flowering, Vernalization, and Cold Acclimation
An aspect of cold adaptation among temperate grasses and cereals is the requirement to undergo vernalization—a biological process that ensures that the transition to flowering will not occur before winter. Prolonged cold exposure in winter, followed by a gradual increase in daylength during spring and summer, is typically necessary for vernalization. Much of our knowledge of the underlying mechanisms of this phenomenon in monocots is based on genetic studies of “spring” and “winter” genotypes of barley and wheat, and even extrapolations from Arabidopsis. Briefly, three key genes regulate this process: VERNALIZATION1 (VRN1), VRN2, and FLOWERING LOCUS T (FT) (Ream et al., 2014; Woods et al., 2017). VRN1 is upregulated by exposure to gradual cold temperatures, thus initiating a positive feedback loop involving the activation of FT that ensures that the flowering signal is maintained in the spring, while VRN2 is the flowering repressor. Orthologs of the three vernalization genes were recently identified and characterized in B. distachyon. The vernalization mechanisms in B. distachyon appear to be conserved compared with wheat and barley. For instance, in a manner similar to wheat and barley, VRN1 and FT are positive regulators of flowering in B. distachyon. Overexpression of VRN1 and FT results in early flowering in B. distachyon, while loss of function in VRN1 and FT delays this process (Lv et al., 2014; Ream et al., 2014; Woods et al., 2016).
For proper flowering induction, VRN1 must also be repressed before cold exposure, but the mechanisms of this repression have not been studied. Recently, forward-genetic screening of B. distachyon mutants identified another protein, REPRESSOR OF VERNALIZATION1 (RVR1), that is required to downregulate VRN1 before the onset of cold (Woods et al., 2017). RVR1 encodes a protein with bromo-adjacent homology and transcriptional elongation factor S-II domain. RVR1 also appears to be conserved among other grasses and is likely required for the vernalization requirement in other pooid grasses (Woods et al., 2017). The function of VRN2 in B. distachyon as a flowering repressor was also recently investigated. Unlike in wheat and barley, the expression of VRN2 is unaltered by cold and does not correlate with the levels of VRN1 and FT. However, genetic studies utilizing an artificial microRNA to downregulate VRN2 (amiVRN2) revealed early flowering phenotypes and enhanced VRN1 and FT expression in amiVRN2 lines (Woods et al., 2016). By contrast, overexpression of VRN2 resulted in delayed flowering and reduced expression of VRN1 and FT, suggesting that VRN2 is the flowering repressor in B. distachyon (Woods et al., 2016).
Cold and freezing temperatures can severely limit grain yields and cereal productivity. As a protective mechanism, vernalization helps to delay the transition from the vegetative to reproductive phase and thus protects the floral meristem from freezing temperatures. Additionally, “hardy” cereals adapt to freezing temperatures by precisely sensing their environment and modulating multiple physiological, biochemical, and molecular parameters, resulting in a freezing tolerance (Thomashow, 1999; Dhillon et al., 2010). Some of these changes include—but are not limited to—the upregulation of cold-regulated (COR) genes, accumulation of carbohydrates, organic acids, proline, and enzymes, and modification of the cell wall, lipid membranes, and photosynthetic apparatus.
B. distachyon was recently employed to evaluate the responses of temperate cereals to cold and freezing temperatures. The freezing tolerance and cold-hardiness of seven B. distachyon accessions were evaluated using a strategy that included comparisons at the phenological, metabolic, and molecular levels (Colton-Gagnon et al., 2014). This study demonstrated that B. distachyon accessions acclimate to the cold in a manner similar to winter cereals and that they can also become freezing tolerant. For instance, when subjected to low and freezing temperatures throughout the plant, B. distachyon “winter” genotypes (Bd1-1, Bd18-1, and Bd29-1) showed the expected accumulation of osmoprotectants such as proline and sugars, as well as activated COR gene expression. Surprisingly, accessions that were thought of as “spring” genotypes (Bd2-3, Bd3-1, Bd21, and Bd30-1) also exhibited responses typical of “winter” genotypes, thus indicating that these spring genotypes are actually “facultative” genotypes that are tolerant of low temperatures, yet do not demand vernalization (von Zitzewitz et al., 2005; Colton-Gagnon et al., 2014). Nevertheless, this study demonstrated the existence of molecular and metabolic wiring that is needed for cold adaptation and freezing tolerance in B. distachyon. It will be interesting to extend these experiments using (wild and laboratory) perennial and annual species of Brachypodium. Taken together, these studies exemplify the value of Brachypodium as a model system to study flowering, vernalization, and cold-acclimation dynamics relevant to agronomic grasses and cereals.
Perenniality and Polyploidy
Plant perenniality is an outstanding issue in biological research aimed at identifying the mechanisms causing the allocation of resources and the subsequent life strategy (Fjellheim et al., 2014). As it pertains to applied science, with possible environmental benefits, some perennial grasses have been developed to produce cellulosic biofuels (e.g., Miscanthus and switchgrass), and other grasses (e.g., intermediate wheatgrass) are being studied with the goal of developing perennial cereal crops (Gordon et al., 2016). The perennial-annual transition (or its converse) has occurred several times across the evolution of the Poaceae (Kellogg, 2015a). This suggests that either an overarching mechanism controls the life cycle or that more than one mechanism directs perenniality-annuality, if perenniality is considered to be a syndrome (Fjellheim et al., 2014). Some authors have identified “rhizomatosity” genes responsible for the perennial cycle in wild species of Oryza and Sorghum (e. g., Rh2 and Rh3) and suggested that convergent mutations have resulted in the evolution of the respective annual species (Hu et al., 2003, 2011).
The genetic control of the life cycle transition likely shares molecular components with the developmental networks that control the floral transition in annual grasses; some of these mechanisms and the associated genes (vernalization, photoperiod, and flowering) have been identified in B. distachyon and in several cereals (Higgins et al., 2010; Fjellheim et al., 2014; Woods et al., 2016) but have not yet been analyzed in perennial grasses. Two evolutionary switches from more ancestral annual lineages to more recently evolved perennial lineages (B. stacei → B. mexicanum and B. distachyon → core perennial clade) have occurred along the Brachypodium phylogeny (Catalán et al., 2016a), framing an exceptional evolutionary scenario for the ongoing investigation of annuality/perenniality switches in the model genus (JGI CSP503006; https://jgi.doe.gov/brachypodium-model-grass-genus-bioenergy/). The construction of the B. sylvaticum pan-genome will allow for comparative pan-genomic studies between this perennial species and the annual B. distachyon.
Allopolyploidy is recognized as a major evolutionary event in angiosperms, which are all considered descendants of paleopolyploid ancestors, and with some subsequently diploidized lineages accumulating more recent mergings (hybridization) and genome doublings in meso- and neopolyploid species (Soltis and Soltis, 2016). This is especially evident in the grass family, where allopolyploids account for 70 to 80% of all species (Stebbins, 1949; Kellogg, 2015a), including allopolyploid wheats, a primary cereal for human consumption (Marcussen et al., 2014). Despite the enormous biological and economic importance of allopolyploid grasses, relatively few studies have investigated the regulation and functional expression of homoeologous genes in the allopolyploids, mostly due to the complexity, large genome sizes, and abundance of repetitive DNA of their subgenomes (Pfeifer et al., 2014; Gordon et al., 2016).
Brachypodium again serves as an optimal model genus, in this case to explore the evolution and functional outcomes of allopolyploidy (Catalán et al., 2014; Gordon et al., 2016). Of the ∼20 species in this genus, half are diploids and half are allopolyploids, with the latter showing differentially inherited (more ancestral to more recently evolved) compact subgenomes that are phylogenetically and syntenically close to those of the wheats and other allopolyploid crop and forage grasses.
The three annual Brachypodium species were selected as a tractable grass polyploid system to investigate the effects of subgenome dominance and origin in the phenotypic, physiological, and adaptive responses of the allotetraploid hybrids (Catalán et al., 2016a; Gordon et al., 2016; Takahagi et al., 2018) (Table 2). These species are the allotetraploid B. hybridum and its diploid parents B. stacei and B. distachyon, which have repeatedly and bidirectionally crossed during the last million years in the wild. Furthermore, the recent development of a synthetic fertile allotetraploid hybrid (♀B. distachyon Bd3-1 × ♂B. stacei Bsta5 and genome doubling with colchicine) that phenotypically resembles the wild B. hybridum (Dinh Thi et al., 2016) will allow for a comprehensive comparative functional genomic and epigenomic analysis of ancestral versus recently produced allotetraploids, the estimation of the heterosis effects, and the analysis of mechanisms that allow the neopolyploid to stabilize and speciate soon after “allopolyploid shock” (drastic subgenomic changes). This integrative genomic approach to studying polyploidy and flowering is the basis of an ongoing JGI-CSP project (CSP503504; https://jgi.doe.gov/csp-2018-chalhoub-shaping-brachypodium-polyploid-model/).
Abiotic Stress
The study of plant abiotic stress tolerance mechanisms is of great importance to agriculture, perhaps allowing breeders to develop superior crops that can withstand climate change and environmental extremes. Brachypodium is a useful model to study the genetics and genomics of abiotic stress responses that affect major cereals and bioenergy grasses. As discussed in earlier sections, B. distachyon and its congeners thrive in a wide range of temperate environments, thus providing researchers the opportunity to use this genetic diversity to study adaptation traits to local climates and abiotic stressors. B. distachyon abiotic stress responses also closely reflect those of wheat (Triticum), barley (Hordeum), and rye (Secale), owing to similar adaptation to temperate climates and common C3 photosynthetic mechanisms (Des Marais and Juenger, 2016).
With the development of high-throughput phenotyping and imaging technologies, screening for complex traits related to stress-tolerance has advanced in the past decade (Fahlgren et al., 2015). Several drought-tolerant wild B. distachyon accessions were recently identified by screening leaf temperature using thermal imaging techniques (Ruíz et al., 2016). The wild accessions were collected from different geographical regions with contrasting rainfall and temperature patterns in the Iberian Peninsula. The drought-adapted genotypes had higher leaf temperatures under drought stress, consistent with increased stomatal closure and decreased evapotranspiration (Jones, 1999), compared with lines not adapted to arid conditions. This study also supports the existence of stomatal-based mechanisms governing natural variation to drought adaptation among wild B. distachyon genotypes. Further molecular and genetic characterization of these drought-adapted and sensitive accessions will undoubtedly help decipher the underlying mechanisms.
B. distachyon exposed to four major abiotic stresses (heat, high salinity, drought, and cold stress) exhibited transcriptome changes in 22 gene modules, 10 of which belong to defined biological processes (Priest et al., 2014). Gene coexpression network analysis further revealed that drought and salinity trigger synergistic coexpression profiles, while heat and cold trigger antagonistic coexpression profiles. Interestingly, drought and salinity stress upregulated the expression of gene modules with unique Gene Ontology categories, while cold and heat stress activated transcription factors and putative genes involved in protein folding and stability, respectively. These stress-associated gene networks are important additions to the outstanding resources available to Brachypodium researchers for the dissection of abiotic stress signaling processes and mechanisms (Priest et al., 2014).
Priest et al. (2014) also determined that desiccation stress downregulates B. distachyon genes (∼5000 genes over the 24-h sample collection period), many of which are involved in regulating the cell cycle and DNA replication, suggesting a gradual shutdown of cellular growth during dehydration stress (Priest et al., 2014). Yet, at the anatomical level, the responses of B. distachyon to desiccation appear to contradict those of Arabidopsis and other cereals. While most plants, including Arabidopsis, show reduced cell division and cell expansion under dehydration stress, B. distachyon showed no changes in cell number compared with the unstressed controls. The reduced leaf size in B. distachyon under dehydration stress appears to primarily result from decreased cell expansion (Verelst et al., 2013). These results point toward some unique aspects of B. distachyon with regards to the cellular and anatomical responses to dehydration stress.
A growing number of studies have used B. distachyon as a model system for genetic analysis of abiotic stress responses. Among the cellular processes, protein ubiquitination is a key process that plays an important role in plant adaptation to abiotic stress. Overexpression of the wheat Ta-Ub gene in tobacco was found to confer resistance to drought (Guo et al., 2008) and heat (Tian et al., 2014) stresses, demonstrating the role of Ta-UB in plant stress tolerance in dicots. However, little is known about its role in monocots. Kang et al. (2016) recently demonstrated that overexpression of Ta-Ub2 under the control of the stress-inducible RD29A promoter conferred transgenic B. distachyon plants with enhanced drought tolerance, enhanced water retention, and increased expression of reactive oxygen species-scavenging related genes. However, constitutive expression of Ta-Ub2 driven by the CaMV35S promoter had negative effects on growth and development in B. distachyon, suggesting that stress-associated gene expression needs to be carefully regulated to avoid deleterious consequences to plant growth and yield. Sun et al. (2015) recently showed that B. distachyon WRKY36 (a group IIe WRKY) positively regulates the drought tolerance response in transgenic Nicotiana tabacum (tobacco). WRKY36 expression is induced during drought stress in B. distachyon, and overexpression of BdWRKY36 in tobacco enhanced its drought tolerance, as characterized by reduced ion leakage and reactive oxygen species accumulation. Together, these results indicate that dicots and monocots share conserved stress tolerance mechanisms involving Ta-Ub2 and WRKY36.
Biotic Stress
Plant diseases and pests are economically devastating to growers, causing annual crop yield losses of 40 to 100%. Much of our knowledge of the plant immune system is based on studies using the dicot model plant Arabidopsis and several other plants with developed genetic systems (e.g., M. truncatula, N. attenuata, rice, maize, and tomato [Solanum lycopersicum]), as well as N. benthamiana. Grass-microbe interaction studies have been hindered by the lack of a tractable monocot model system. Of course, rice and maize have been invaluable tools for plant biologists, but they lack several key features of model systems (Table 1). Recently, as we have noted, Brachypodium has risen to model status for the investigation of diverse grass-microbe and grass-insect interactions, supporting the life cycles of a remarkable diversity of fungi, oomycetes, viruses, bacteria, insects, as well as other invertebrates (Mandadi and Scholthof, 2013)((Table 4).
Table 4. Brachypodium as a Model Organism for Investigations of Plant-Microbe and Plant-Invertebrate Interactions.
| Microbe or Invertebrate | References |
|---|---|
| MICROBES | |
| Viruses | |
| Bamboo mosaic virus (BaMV) | Liou et al. (2014) |
| Satellite RNA (BaMV satRNA) | Liou et al. (2014) |
| Barley stripe mosaic virus (BSMV) | Pacak et al. (2010); Yuan et al. (2011); Cui et al. (2012); Mandadi et al. (2014); Cheuk and Houde (2017); Wang et al. (2017) |
| Barley yellow dwarf virus (BYDV-GAV) | Tao et al. (2016) |
| Brome mosaic virus (BMV) | Mandadi et al. (2014) |
| Foxtail mosaic virus (FoMV) | Mandadi et al. (2014) |
| Maize dwarf mosaic virus | Rosenkranz (1978) |
| Maize mild mottle virus (MMMV) | Mandadi et al. (2014) |
| Panicum mosaic virus (PMV) | Mandadi and Scholthof (2012) |
| Satellite panicum mosaic virus (SPMV) | Mandadi and Scholthof (2012); Mandadi et al. (2015) |
| Satellite RNA (satRNA) | Pyle et al. (2017); Pyle and Scholthof (2018) |
| Sorghum yellow banding virus (SYBV) | Mandadi et al. (2014) |
| Sugarcane mosaic virus (SCMV) | Rosenkranz (1978) |
| Wheat steak mosaic virus (WSMV) | Mandadi et al. (2014) |
| Bacteria | |
| Agrobacterium tumefaciens, crown gall, transformationa | Vain et al. (2008); Steinwand et al. (2013); Bragg et al. (2015); Sogutmaz Ozdemir and Budak (2018) |
| Azospirillum brasilense, rhizobacteria, growth promoting | do Amaral et al. (2016) |
| Bacillus subtilis, endophyte | Gagné-Bourque et al. (2015) |
| Herbaspirillum seropedicae, rhizobacteria, growth promoting | do Amaral et al. (2016) |
| Rhizobium rhizogenes, biocontrol, transformationa | Collier et al. (2016) |
| Xanthomonas translucens, black chaff (wheat/barley) | Fitzgerald et al. (2015) |
| Fungi: Ascomycetes | |
| Alternaria sp, leaf spotb | Roy et al. (2011) |
| Ascochyta sp, leaf spotb | Roy et al. (2011) |
| Bipolaris sorokiniana | Falter and Voigt (2014) |
| Blumeria graminis, powdery mildew | Draper et al. (2001) |
| Claviceps purpurea, ergot | Kind et al. (2017) |
| Cochliobolus heterostrophus, southern corn leaf blight | Falter and Voigt (2014) |
| Cochliobolus sativus, root rot/leaf spot | Zhong et al. (2015) |
| Colletotrichum cereale, anthracnose (turf) | Sandoya and de Oliveira Buanafina (2014) |
| Epichloë sylvatica, endophyteb | Meijer and Leuchtmann (1999); Brem and Leuchtmann (2001) |
| Fusarium culmorum, head blight | Peraldi et al. (2014) |
| Fusarium graminearum, head blight | Peraldi et al. (2014) |
| Fusarium pseudograminearum, crown rot | Powell et al. (2017) |
| Gaeumannomyces graminis | Sandoya and de Oliveira Buanafina (2014) |
| Magnaporthe grisea, rice blast | Routledge et al. (2004) |
| Magnaporthe poae | Sandoya and de Oliveira Buanafina (2014) |
| Microdochium nivale, snow mold (turf) | Rioux et al. (2017) |
| Neotyphodium sp, endophyte | Brem and Leuchtmann (2001) |
| Oculimacula spp, eye spot (barley) | Peraldi et al. (2014) |
| Ophiosphaerella agrostis, dead spot (turf) | Sandoya and de Oliveira Buanafina (2014) |
| Ophiosphaerella korrae, necrotic ringspot (turf) | Sandoya and de Oliveira Buanafina (2014) |
| Pithomyces chartarum | Falter and Voigt (2014) |
| Pyrenophora erythrospila, leaf spotb | Halbritter et al. (2012) |
| Pyrenophora teres, net blotch | Brown et al. (1993) |
| Ramularia collo-cygni, Ramularia leaf spot (barley) | Peraldi et al. (2014) |
| Sclerotinia homeocarpa, dollar spot (turf) | Sandoya and de Oliveira Buanafina (2014); Rioux et al. (2017) |
| Stagonospora macropycnidia | Falter and Voigt (2014) |
| Stagonospora nodorum, glume blotchb | Halbritter et al. (2012) |
| Zymoseptoria tritici, Septoria tritici blotch | O’Driscoll et al. (2015) |
| Fungi: Basidiomycetes | |
| Puccinia brachypodii, leaf rusta | Barbieri et al. (2011) |
| Puccinia emaculata, switchgrass rust | Gill et al. (2015) |
| Puccinia graminis f. sp lolii | Figueroa et al. (2013) |
| Puccinia graminis f. sp phlei-pratensis | Figueroa et al. (2013) |
| Puccinia striiformis f. sp avenea, stripe rust (oat) | Ayliffe et al. (2013) |
| Puccinia striiformis f. sp bromi, stripe rust (brome grass) | Barbieri et al. (2011) |
| Puccinia striiformis f. sp hordei, stripe rust (barley) | Barbieri et al. (2011) |
| Puccinia striiformis f. sp tritici, stripe rust (wheat) | Barbieri et al. (2011); Ayliffe et al. (2013) |
| Puccinia triticina | Ayliffe et al. (2013) |
| Rhizoctonia solani, brown patch (turf) | Schneebeli et al. (2015); Rioux et al. (2017) |
| Ustilago bromivora, loose smut | Rabe et al. (2016) |
| Fungal-like organisms: oomycetes | |
| Pythium aphanidermatum | Sandoya and de Oliveira Buanafina (2014) |
| Mycrorhizal fungi | |
| Glomus candidum | Hong et al. (2012) |
| Glomus versiforme | Hong et al. (2012) |
| Gigaspora decipiens | Hong et al. (2012) |
| Gigaspora gigantea | Hong et al. (2012) |
| Rhizophagus irregularis (Glomus intraradices) | Hong et al. (2012); Jakobsen et al. (2016) |
| Rhizosphere | |
| Ascomycetes | |
| Chaetomium globosum | Kawasaki et al. (2016) |
| Emericellopsis mirabilis | Kawasaki et al. (2016) |
| Chitridomycetes | |
| Rhizophlyctis rosea | Kawasaki et al. (2016) |
| Actinobacteria | |
| Arthrobacter sp | Staley et al. (2017) |
| Alphaproteobacteria | Kawasaki et al. (2016) |
| Betaproteobacteria | |
| Herbaspirillum seropedicae, endophyte | do Amaral et al. (2016) |
| Achromobacter xylosoxidans | Staley et al. (2017) |
| Deltaproteobacteria | |
| Sorangium, cellulose degradation; fungicide and bactericide production; biocontrol agent | Kawasaki et al. (2016) |
| Gammaproteobacteria | |
| Stenotrophomonas | Tkacz et al. (2015) |
| Firmicutes | Kawasaki et al. (2016); Staley et al. (2017) |
| INVERTEBRATES | |
| Nematodes | |
| Heterodera avenae, cereal cyst nematodec | Kong et al. (2016) |
| Earthworms | |
| Apporectodea caliginosa, endogeic earthworm | Agapit et al. (2017) |
| Insects | |
| Diuraphis noxia, Russian wheat aphid | Azhaguvel et al. (2014); Sandoya and de Oliveira Buanafina (2014) |
| Laodelphax striatellus, small brown leafhopper | Zhou et al. (2016) |
| Schizaphis graminum, greenbug (aphid) | Azhaguvel et al. (2014); Tao et al. (2016) |
| Spodoptera frugiperda, Fall army worm | Brem and Leuchtmann (2001); Sandoya and de Oliveira Buanafina (2014) |
Within each grouping, microbes are listed in alphabetical order. Vernacular names and functions are indicated, as known.
For both B. distachyon and B. sylvaticum.
Host is B. sylvaticum.
On B. distachyon, H. avenae did not progress beyond the J2 stage.
B. distachyon is a host to several economically important viruses, including Barley stripe mosaic virus (BSMV), Brome mosaic virus, Panicum mosaic virus (PMV) and its satellites (satellite panicum mosaic virus and satellite RNAs [satRNAs]), Foxtail mosaic virus, Wheat streak mosaic virus, Sorghum yellow banding virus, and Maize mild mottle virus (Cui et al., 2012; Lee et al., 2012; Mandadi and Scholthof, 2012, 2015; Mandadi et al., 2014, 2015; Fitzgerald et al., 2015; Pyle et al., 2017; Irigoyen et al., 2018; Pyle and Scholthof, 2018).
In the first demonstration of the use of B. distachyon for pivotal investigations of Brachypodium-virus interactions, a set of geographically diverse accessions were screened for BSMV resistance. Through fine-mapping, BARLEY STRIPE MOSAIC VIRUS RESISTANCE1 was identified as a resistance (R) gene, opening the possibility of identifying similar R genes in barley and other small grains that are negatively affected by BSMV infection (Cui et al., 2012).
Using B. distachyon, we previously characterized genome-wide changes in transcriptome and defense signaling networks, splicing landscapes, as well as virulence determinants critical for PMV infection in grasses (Mandadi and Scholthof, 2012, 2015; Mandadi et al., 2015). Mandadi et al. (2014) identified the conserved and unique defense responses triggered by grass-virus interactions. For this, comparative analyses of B. distachyon and Setaria viridis (a C4 grass) immune responses to eight monocot-infecting viruses were performed (Table 4). These studies underscore the importance of B. distachyon as a model system to advance fundamental studies of grass-virus interactions (Mandadi et al., 2014).
B. distachyon has also been used to study of viral synergism and subviral agents, a largely understudied area of plant-virus interactions. B. distachyon supports the synergism between PMV and its satellite virus, satellite panicum mosaic virus, as well as replication of satRNAs. Pyle and colleagues recently demonstrated that a conserved PMV satRNA (satS) undergoes dynamic 3′-end modifications in B. distachyon and proso millet and attenuates PMV symptoms, resulting in less severe disease symptoms (Pyle et al., 2017; Pyle and Scholthof, 2018). These findings suggest that plant-virus interactions can be studied to understand the mechanisms by which the host machinery subverts the replicative ability of virus infections in grasses and to develop defense strategies to reduce virus infections in agronomic grass hosts.
Brachypodium is an excellent model to study the genetics and genomics of biotrophic, hemibiotrophic, and necrotrophic fungal pathogens of grasses such as Puccinia spp, Fusarium spp, Rhizoctonia spp, Colletotrichum spp, and Magnaporthe spp (Routledge et al., 2004; Ayliffe et al., 2013; Figueroa et al., 2013; Sandoya and de Oliveira Buanafina, 2014; Fitzgerald et al., 2015), as well as bacterial pathogens (Xanthomonas spp) (Fitzgerald et al., 2015). B. distachyon is also a host plant for several insect species, including aphids and the fall army worm (Spodoptera frugiperda) (Table 4). Aphids, in addition to causing extensive yield losses due to feeding on plant hosts, often transmit plant viruses. For example, Barley yellow dwarf virus GAV strain is transmitted by Schizaphis graminum. Using Brachypodium spp, it will now be possible to dissect the molecular genetics and biochemistry of aphid-virus and aphid-plant interactions (Tao et al., 2016). Similarly, the identification of B. distachyon as a host for two key pest species, fall army worm and Russian wheat aphid (Diuraphis noxia), will allow researchers to pursue studies related to plant-insect resistance mechanisms and to dissect the roles of green leafy volatiles in grass defense signaling (Sandoya and de Oliveira Buanafina, 2014). The results of these studies should be readily applicable to the development of field-based pest control measures.
Root Biology and the Microbiome: Holistic Studies to Guide Reductionist Laboratory Science
The study of root biology in Brachypodium is a relatively new field, but the features of this system make it an ideal model for temperate grass root biology, as well as comparative studies with dicot plants. Because B. distachyon is a wild grass, it also is an excellent choice for comparative studies of root traits that might have been modified or lost during crop domestication in field grasses and cereal crops. The natural variations in root morphology and physiology among B. distachyon accessions result in differential responses to nutrient deficiency or osmotic stress (Pacheco-Villalobos and Hardtke, 2012).
Angiosperms exhibit diverse root architecture; however, two distinct patterns can be found among monocot and dicot plants (Osmont et al., 2007). B. distachyon roots display a typical monocot system (Chochois et al., 2012; Pacheco-Villalobos and Hardtke, 2012), which starts with a single primary root, that persists throughout the plant life cycle. Typically, two coleoptile node roots emerge during the postembryonic stage, and several stem node roots can form, depending on nutrient availability (Ingram et al., 2012; Poiré et al., 2014). At the anatomical level, B. distachyon roots have one epidermis layer, five cortex layers that constitute the ground tissue, and a single endodermis layer (Watt et al., 2009; Pacheco-Villalobos and Hardtke, 2012). The stele is surrounded by a single pericycle layer containing the vascular tissues arranged in a radial pattern of alternating xylem and phloem poles. The ring of xylem and phloem poles surrounds the central metaxylem at the center of the root. This vasculature pattern is representative of most monocot root types, with slight variations (Watt et al., 2009; Pacheco-Villalobos and Hardtke, 2012).
At the genetic level, there appears to be some degree of conservation in gene function and signal transduction processes between Brachypodium and Arabidopsis, which might facilitate functional studies in Brachypodium. For example, the Arabidopsis BREVIS RADIX loss-of-function mutant, which displays shorter roots due to impaired cell growth in the elongation zone, is rescued by genetic transformation with members of the B. distachyon BREVIS RADIX-LIKE gene family (Mouchel et al., 2004; Beuchat et al., 2010). Another example is the Arabidopsis loss-of-function mutant for the brassinosteroid receptor BRASSINOSTEROID INSENSITIVE1, which displays reduced root growth. A mutation in the B. distachyon homolog also produces plants with reduced root size, although to a lesser degree (Thole et al., 2012; Goddard et al., 2014). Like Arabidopsis, B. distachyon also responds to the exogenous application of brassinosteroids; however, different root phenotypes are observed in ethylene and auxin signaling mutants (Stepanova et al., 2008; Won et al., 2011; Liang et al., 2012; Pacheco-Villalobos and Hardtke, 2012; Tao et al., 2016).
The anatomy of B. distachyon and wheat roots and their environmental and nutritional requirements are comparable (Watt et al., 2009; Chochois et al., 2012), but B. distachyon roots are much smaller and more convenient than wheat for laboratory research, since the adult root system can be contained in a small volume of soil without negatively affecting root physiology (Watt et al., 2009; Chochois et al., 2012). Furthermore, B. distachyon root exudates and rhizosphere microbial communities are comparable to those of wheat (Kawasaki et al., 2016) and include bacteria in the orders Burkholderiales, Sphingobacteriales, and Xanthomonadales and fungal communities belonging to the phyla Ascomycota, Chytridiomycota, and Basidiomycota (Kawasaki et al., 2016).
Arbuscular mycorrhiza (AM) symbiosis is the most common type of mycorrhizal association found in vascular plants. Unlike Arabidopsis, Brachypodium forms associations with AM fungi, thus allowing researchers to explore this interaction in a model system. For B. distachyon and many other plants, AM enhance phosphorous (P) and nitrogen (N) uptake from the soil and promote shoot growth. The B. distachyon-AM association results in distinct morphologies and functional associations unlike those formed by rice and M. truncatula roots—two model systems that have been widely used for AM symbiosis studies (Smith et al., 2003; Cui et al., 2012; Jakobsen et al., 2016). The AM associations in B. distachyon roots are exclusively intracellular (Paris-type), while AM associations in Medicago and rice are primarily intercellular (Arum-type) (Hong et al., 2012). Similarly, under P-limiting conditions, Medicago displayed a positive AM growth response, while B. distachyon, wheat, and barley showed neutral to negative growth (Jakobsen et al., 2016). Furthermore, although multiple AM fungi (e.g., Glomus candidum, G. intraradices, and G. versiformes) promote shoot growth and P uptake in B. distachyon, a few species (e.g., Gigaspora gigantea and G. decipiens) do not, suggesting a degree of functional diversity among AM associations with B. distachyon (Hong et al., 2012). In summary, Brachypodium is a suitable system to dissect grass-AM interactions and the associated morphological and functional diversity.
With its ∼20 species and well-defined evolutionary biology, wild Brachypodium species are being used to study complex host-microbe interactions. Of particular interest will be the investigation of naturally occurring interactions of microbes with Brachypodium strains aimed at defining the holobiont, which may have direct applications for increasing yields in grain crops as well as pasture grasses by promoting plant health. Similarly, investigating natural populations is key in determining the coevolution of host-microbe interactions in the identification of resistance genes and innate immune responses, which may be broadly beneficial for maintaining host homeostasis. For this rapidly advancing area of study, Brachypodium has become a key resource for understanding the broad implications of complex interactions between host genetics and the environment.
For example, within the broad topic of host microbiota, several studies have identified bacteria and fungi in the rhizosphere that can produce biopesticides (fungicides and bactericides), as well as biostimulants that abrogate plant dysbiosis (microbial imbalance). Thus far, three dozen microbes (viruses, fungi, and bacteria) have been approved by the European Union for crop application under organic or sustainable farm practices (Matyjaszczyk, 2015). Microbiota studies of B. distachyon have shown that Bacillus subtilis B26, which is vertically transmitted in the seed, reduces the impact of drought stress in subsequent generations of plants (Gagné-Bourque et al., 2015) (Table 4). From these initial studies, soil microbiota (much like human microbiota; Yong, 2016) will have specific species associated with plant health and others that are indicative of less fit plants (Ramírez-Puebla et al., 2013). For B. distachyon, increased fitness is associated with bacterial volatiles from B. subtilis GB03, resulting in increased total biomass (Delaplace et al., 2015). These findings provide a means by which new strategies and applications may be developed to improve grasses, reduce fertilizer and water inputs, and improve soil health.
OUTLOOK: EMBRACING AND ENHANCING THE BRACHYPODIUM MODEL SYSTEM
For Brachypodium to succeed as a model system for plant biology akin to Arabidopsis (Lyons and Scholthof, 2015, 2016), a dedicated stock center for the centralized maintenance and distribution of Brachypodium spp seeds and reagents is vital. Currently, the burden of this task is taken on by only a few individual laboratories around the world. Although the Brachypodium research community is ready to share this responsibility, many laboratories do not have the required resources. Moreover, when investigators retire, lose funding, or move into new areas of study, the germplasm and other resources are oftentimes no longer available to the community. A dedicated Brachypodium seed/reagent stock center with long-term financial support would alleviate these concerns.
Another avenue for the improvement of resources is data deposition, access, and mining tools (Table 3). To enable uniformity in data deposition and the mining of valuable data, the community needs an integrated Brachypodium data resource platform that enables data mining in a manner similar to Araport (Krishnakumar et al., 2015) and PlaNET (Ruprecht et al., 2017). This latter resource, PlaNET, does host some B. distachyon data sets, but it is limited to the visualization and analysis of coregulated gene networks. Another, and more well-developed example, is the N. attenuata data hub (NaDH); N. attenuata, like Brachypodium, spans the wild plant-laboratory divide and has a platform for genetic, transcriptomic, and metabolomics resources (Brockmöller et al., 2017).
A unique aspect of the Brachypodium scientific community, compared with that of Arabidopsis, is their interest in the ecology and evolution of grasses in temperate climates. With the recent developments in plant microbiome research, Brachypodium is poised to serve several additional roles, including long-term ecology studies (Hæggström and Skytén, 1996), evolution of host-microbe interactions, and speciation in nature (Smiley et al., 2016). Long-term field studies of Brachypodium-microbe interactions, soil fertility, and insect and nematode ecology could prove to be valuable additions to the resources used for applied, fundamental, and translational research on perennial and annual C3 grasses and small grains.
Lastly, grant funding is a major bottleneck for the Brachypodium research community. Future outcomes are, of course, difficult to predict, but one thing is certain: Progress is predicated on the continued development and sharing of community resources, which requires long-term financial support from federal agencies, private institutions, and educational institutions (particularly colleges of agriculture at land-grant universities) for research, seed bank, and collaborative efforts. With the advent of NGS and phenotyping resources and the emphasis on translational and applied research in crop plants, with a few exceptions, many agencies and programs have directed their limited resources away from fundamental research and model systems to crop plants. Although it is important to ultimately translate basic research findings to field crops, something that has been extremely successful for rice and maize, vast knowledge gaps in our understanding of plant biology persist, more so in monocots. Because of this, many researchers (new and established) are currently faced with the challenge of supporting their basic research programs and highly discouraged from initiating new projects leveraging model plants. If we want to meaningfully, and sustainably, carry out our research endeavors as a scientific enterprise, we need to balance the current grant funding landscape to support fundamental research in model plants, such as Brachypodium spp, that will have direct, positive outcomes in our understanding of allopolyploidy, speciation, host-microbe interactions, ecology, and improvements of (wild and forage) grasses and key cereal crops.
Note from the Authors
To share your knowledge and to learn more about Brachypodium, international collaborative research projects, and community goals—from the landscape to the laboratory—please join us in Spain in 2019 for the 4th International Brachypodium meeting (https://sites.google.com/view/Brachypodium/home).
We apologize for not covering any other research topics or studies due to space constraints.
Dive Curated Terms
The following phenotypic, genotypic, and functional terms are of significance to the work described in this paper:
Acknowledgments
We appreciate the critical comments provided by Herman Scholthof and Will Cody. This project was supported in part by the Agriculture and Food Research Initiative competitive grants program (2016-67013-24738) from the USDA National Institute of Food and Agriculture (K.-B.G.S. and K.K.M); the Joint Genome Institute CSP503006 (P.C.), CSP503406 (K.K.M.), and CSP503504 projects (P.C.); the Spanish Ministry of Economy and Competitiveness CGL2016-79790-P grant (P.C.); and the cofunded Spanish Aragon Government and European Social Fund Bioflora research group (P.C.).
AUTHOR CONTRIBUTIONS
All authors contributed to the planning, writing, and editing of this review article.
Footnotes
Articles can be viewed without a subscription.
References
- Agapit C., Gigon A., Puga-Freitas R., Zeller B., Blouin M. (2017). Plant-earthworm interactions: influence of age and proportion of casts in the soil on plant growth, morphology and nitrogen uptake. Plant Soil 424: 49–61. [Google Scholar]
- Ayliffe M., Singh D., Park R., Moscou M., Pryor T. (2013). Infection of Brachypodium distachyon with selected grass rust pathogens. Mol. Plant Microbe Interact. 26: 946–957. [DOI] [PubMed] [Google Scholar]
- Azhaguvel P., Mornhinweg D., Vidya-Saraswathi D., Rudd J.C., Chekhovskiy K., Saha M., Close T.J., Dahleen L.S., Weng Y. (2014). Molecular mapping of greenbug (Schizaphis graminum) resistance gene Rsg1 in barley. Plant Breed. 133: 227–233. [Google Scholar]
- Bablak P., Draper J., Davey M., Lynch P. (1995). Plant regeneration and micropropagation of Brachypodium distachyon. Plant Cell Tissue Organ Cult. 42: 97–107. [Google Scholar]
- Bakker E.G., Montgomery B., Nguyen T., Eide K., Chang J., Mockler T.C., Liston A., Seabloom E.W., Borer E.T. (2009). Strong population structure characterizes weediness gene evolution in the invasive grass species Brachypodium distachyon. Mol. Ecol. 18: 2588–2601. [DOI] [PubMed] [Google Scholar]
- Barbieri M., Marcel T.C., Niks R.E. (2011). Host status of false brome grass to the leaf rust fungus Puccinia brachypodii and the stripe rust fungus P. striiformis. Plant Dis. 95: 1339–1345. [DOI] [PubMed] [Google Scholar]
- Bareither N., Scheffel A., Metz J. (2017). Distribution of polyploid plants in the common annual Brachypodium distachyon (s.l.) in Israel is not linearly correlated with aridity. Isr. J. Plant Sci. 64: 83–92. [Google Scholar]
- Betekhtin A., Jenkins G., Hasterok R. (2014). Reconstructing the evolution of Brachypodium genomes using comparative chromosome painting. PLoS One 9: e115108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beuchat J., Scacchi E., Tarkowska D., Ragni L., Strnad M., Hardtke C.S. (2010). BRX promotes Arabidopsis shoot growth. New Phytol. 188: 23–29. [DOI] [PubMed] [Google Scholar]
- Bouvier d’Yvoire M., et al. (2013). Disrupting the cinnamyl alcohol dehydrogenase 1 gene (BdCAD1) leads to altered lignification and improved saccharification in Brachypodium distachyon. Plant J. 73: 496–508. [DOI] [PubMed] [Google Scholar]
- Bragg J.N., Wu J., Gordon S.P., Guttman M.E., Thilmony R., Lazo G.R., Gu Y.Q., Vogel J.P. (2012). Generation and characterization of the Western Regional Research Center Brachypodium T-DNA insertional mutant collection. PLoS One 7: e41916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bragg J.N., Anderton A., Nieu R., Vogel J.P. (2015). Brachypodium distachyon In Agrobacterium Protocols, Vol. 1, Wang K., ed (New York: Springer; ), pp. 17–33. [Google Scholar]
- Brem D., Leuchtmann A. (2001). Epichloë grass endophytes increase herbivore resistance in the woodland grass Brachypodium sylvaticum. Oecologia 126: 522–530. [DOI] [PubMed] [Google Scholar]
- Brkljacic J., et al. (2011). Brachypodium as a model for the grasses: today and the future. Plant Physiol. 157: 3–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brockmöller T., Ling Z., Li D., Gaquerel E., Baldwin I.T., Xu S. (2017). Nicotiana attenuata Data Hub (NaDH): an integrative platform for exploring genomic, transcriptomic and metabolomic data in wild tobacco. BMC Genomics 18: 79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown M.P., Steffenson B.J., Webster R.K. (1993). Host range of Pyrenophora teres f. teres isolates from California. Plant Dis. 77: 942–947. [Google Scholar]
- Brutnell T.P., Bennetzen J.L., Vogel J.P. (2015). Brachypodium distachyon and Setaria viridis: Model genetic systems for the grasses. Annu. Rev. Plant Biol. 66: 465–485. [DOI] [PubMed] [Google Scholar]
- Cao S., Siriwardana C.L., Kumimoto R.W., Holt B.F. III (2011). Construction of high quality Gateway™ entry libraries and their application to yeast two-hybrid for the monocot model plant Brachypodium distachyon. BMC Biotechnol. 11: 53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catalán P., Müller J., Hasterok R., Jenkins G., Mur L.A.J., Langdon T., Betekhtin A., Siwinska D., Pimentel M., López-Alvarez D. (2012). Evolution and taxonomic split of the model grass Brachypodium distachyon. Ann. Bot. 109: 385–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catalán P., Chalhoub B., Chochois V., Garvin D.F., Hasterok R., Manzaneda A.J., Mur L.A.J., Pecchioni N., Rasmussen S.K., Vogel J.P., Voxeur A. (2014). Update on the genomics and basic biology of Brachypodium: International Brachypodium Initiative (IBI). Trends Plant Sci. 19: 414–418. [DOI] [PubMed] [Google Scholar]
- Catalán P., López-Álvarez D., Bellosta C., Villar L. (2016a). Updated taxonomic descriptions, iconography, and habitat preferences of Brachypodium distachyon, B. stacei, and B. hybridum (Poaceae). An. Jardin Botanico Madr. 1979 73: e028. [Google Scholar]
- Catalán P., López-Álvarez D., Díaz-Pérez A., Sancho R., López-Herránz M.L. (2016b). Phylogeny and evolution of the genus Brachypodium. In Genetics and Genomics of Brachypodium, Vogel J.P., ed (Switzerland: Springer; ), pp. 9–38. [Google Scholar]
- Chen G., Zhu J., Zhou J., Subburaj S., Zhang M., Han C., Hao P., Li X., Yan Y. (2014). Dynamic development of starch granules and the regulation of starch biosynthesis in Brachypodium distachyon: comparison with common wheat and Aegilops peregrina. BMC Plant Biol. 14: 198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheuk A., Houde M. (2017). A rapid and efficient method for uniform gene expression using the barley stripe mosaic virus. Plant Methods 13: 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chochois V., Vogel J.P., Watt M. (2012). Application of Brachypodium to the genetic improvement of wheat roots. J. Exp. Bot. 63: 3467–3474. [DOI] [PubMed] [Google Scholar]
- Christiansen P., Andersen C.H., Didion T., Folling M., Nielsen K.K. (2005). A rapid and efficient transformation protocol for the grass Brachypodium distachyon. Plant Cell Rep. 23: 751–758. [DOI] [PubMed] [Google Scholar]
- Collier R., Bragg J., Hernandez B.T., Vogel J.P., Thilmony R. (2016). Use of Agrobacterium rhizogenes strain 18r12v and paromomycin selection for transformation of Brachypodium distachyon and Brachypodium sylvaticum. Front. Plant Sci. 7: 716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colton-Gagnon K., Ali-Benali M.A., Mayer B.F., Dionne R., Bertrand A., Do Carmo S., Charron J.-B. (2014). Comparative analysis of the cold acclimation and freezing tolerance capacities of seven diploid Brachypodium distachyon accessions. Ann. Bot. 113: 681–693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coomey J.H., Hazen S.P. (2016). Brachypodium distachyon as a model species to understand grass cell walls. In Genetics and Genomics of Brachypodium, Vogel J.P., ed (Switzerland: Springer; ), pp. 197–217. [Google Scholar]
- Cui Y., et al. (2012). Fine mapping of the Bsr1 barley stripe mosaic virus resistance gene in the model grass Brachypodium distachyon. PLoS One 7: e38333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalmais M., et al. (2013). A TILLING platform for functional genomics in Brachypodium distachyon. PLoS One 8: e65503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Bang L., Torp A.M., Rasmussen S.K. (2018). TILLING in Brachypodium distachyon. In Brachypodium Genomics: Methods and Protocols, Sablok G., Budak H., Ralph P.J., eds (New York: Springer; ), pp. 173–186. [DOI] [PubMed] [Google Scholar]
- Delaplace P., Delory B.M., Baudson C., Mendaluk-Saunier de Cazenave M., Spaepen S., Varin S., Brostaux Y., du Jardin P. (2015). Influence of rhizobacterial volatiles on the root system architecture and the production and allocation of biomass in the model grass Brachypodium distachyon (L.) P. Beauv. BMC Plant Biol. 15: 195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Des Marais D.L., Juenger T.E. (2016). Brachypodium and the abiotic environment. In Genetics and Genomics of Brachypodium, Vogel J., ed (Switzerland: Springer; ), pp. 291–311. [Google Scholar]
- Dhillon T., Pearce S.P., Stockinger E.J., Distelfeld A., Li C., Knox A.K., Vashegyi I., Vágújfalvi A., Galiba G., Dubcovsky J. (2010). Regulation of freezing tolerance and flowering in temperate cereals: the VRN-1 connection. Plant Physiol. 153: 1846–1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Díaz-Pérez A., López-Álvarez D., Sancho R., Catalan P. (2018). Reconstructing the origins and the biogeography of species’ genomes in the highly reticulate allopolyploid-rich model grass genus Brachypodium using minimum evolution, coalescence and maximum likelihood approaches. Mol. Phylogenet. Evol. 127: 256–271. [DOI] [PubMed] [Google Scholar]
- Dinh Thi V.H., Coriton O., Le Clainche I., Arnaud D., Gordon S.P., Linc G., Catalán P., Hasterok R., Vogel J.P., Jahier J., Chalhoub B. (2016). Recreating stable Brachypodium hybridum allotetraploids by uniting the divergent genomes of B. distachyon and B. stacei. PLoS One 11: e0167171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- do Amaral F.P., Pankievicz V.C.S., Arisi A.C.M., de Souza E.M., Pedrosa F., Stacey G. (2016). Differential growth responses of Brachypodium distachyon genotypes to inoculation with plant growth promoting rhizobacteria. Plant Mol. Biol. 90: 689–697. [DOI] [PubMed] [Google Scholar]
- Draper J., Mur L.A.J., Jenkins G., Ghosh-Biswas G.C., Bablak P., Hasterok R., Routledge A.P.M. (2001). Brachypodium distachyon. A new model system for functional genomics in grasses. Plant Physiol. 127: 1539–1555. [PMC free article] [PubMed] [Google Scholar]
- Fahlgren N., Gehan M.A., Baxter I. (2015). Lights, camera, action: high-throughput plant phenotyping is ready for a close-up. Curr. Opin. Plant Biol. 24: 93–99. [DOI] [PubMed] [Google Scholar]
- Falter C., Voigt C.A. (2014). Comparative cellular analysis of pathogenic fungi with a disease incidence in Brachypodium distachyon and Miscanthus x giganteus. BioEnergy Res. 7: 958–973. [Google Scholar]
- Figueroa M., Alderman S., Garvin D.F., Pfender W.F. (2013). Infection of Brachypodium distachyon by formae speciales of Puccinia graminis: early infection events and host-pathogen incompatibility. PLoS One 8: e56857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzgerald T.L., et al. (2015). Brachypodium as an emerging model for cereal-pathogen interactions. Ann. Bot. 115: 717–731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fjellheim S., Boden S., Trevaskis B. (2014). The role of seasonal flowering responses in adaptation of grasses to temperate climates. Front. Plant Sci. 5: 431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francin-Allami M., Lollier V., Pavlovic M., San Clemente H., Rogniaux H., Jamet E., Guillon F., Larré C. (2016). Understanding the remodelling of cell walls during Brachypodium distachyon grain development through a sub-cellular quantitative proteomic approach. Proteomes 4: 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gagné-Bourque F., Mayer B.F., Charron J.-B., Vali H., Bertrand A., Jabaji S. (2015). Accelerated growth rate and increased drought stress resilience of the model grass Brachypodium distachyon colonized by Bacillus subtilis B26. PLoS One 10: e0130456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gill U.S., Uppalapati S.R., Nakashima J., Mysore K.S. (2015). Characterization of Brachypodium distachyon as a nonhost model against switchgrass rust pathogen Puccinia emaculata. BMC Plant Biol. 15: 113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gill U.S., Serrani-Yarce J.C., Lee H.-K., Mysore K.S. (2018). Tissue culture (somatic embryogenesis)-induced Tnt1 retrotransposon-based mutagenesis in Brachypodium distachyon. Methods Mol. Biol. 1667: 57–63. [DOI] [PubMed] [Google Scholar]
- Goddard R., Peraldi A., Ridout C., Nicholson P. (2014). Enhanced disease resistance caused by BRI1 mutation is conserved between Brachypodium distachyon and barley (Hordeum vulgare). Mol. Plant Microbe Interact. 27: 1095–1106. [DOI] [PubMed] [Google Scholar]
- Gordon S.P., et al. (2017). Extensive gene content variation in the Brachypodium distachyon pan-genome correlates with population structure. Nat. Commun. 8: 2184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon S.P., Liu L., Vogel J.P. (2016). The genus Brachypodium as a model for perenniality and polyploidy. In Genetics and Genomics of Brachypodium, Vogel J.P., ed (Switzerland: Springer; ), pp. 313–325. [Google Scholar]
- Gottlieb A., et al. (2013). Insular organization of gene space in grass genomes. PLoS One 8: e54101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Q., Zhang J., Gao Q., Xing S., Li F., Wang W. (2008). Drought tolerance through overexpression of monoubiquitin in transgenic tobacco. J. Plant Physiol. 165: 1745–1755. [DOI] [PubMed] [Google Scholar]
- Hæggström C.-A., Skytén R. (1996). Flowering and individual survival of a population of the grass Brachypodium sylvaticum in Nåtö, Åland Islands, SW Finland. Ann. Bot. Fennici 33: 1–10. [Google Scholar]
- Halbritter A.H., Carroll G.C., Güsewell S., Roy B.A. (2012). Testing assumptions of the enemy release hypothesis: generalist versus specialist enemies of the grass Brachypodium sylvaticum. Mycologia 104: 34–44. [DOI] [PubMed] [Google Scholar]
- Handakumbura P.P., Hazen S.P. (2012). Transcriptional regulation of grass secondary cell wall biosynthesis: playing catch-up with Arabidopsis thaliana. Front. Plant Sci. 3: 74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasterok R., Marasek A., Donnison I.S., Armstead I., Thomas A., King I.P., Wolny E., Idziak D., Draper J., Jenkins G. (2006). Alignment of the genomes of Brachypodium distachyon and temperate cereals and grasses using bacterial artificial chromosome landing with fluorescence in situ hybridization. Genetics 173: 349–362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasterok R., Betekhtin A., Borowska-Zuchowska N., Braszewska-Zalewska A., Idziak-Helmcke D., Robaszkiewicz E., Wolny E. (2015). Molecular cytogenetics in the genus Brachypodium. In Genetics and Genomics of Brachypodium, Vogel J.P., ed (Switzerland: Springer; ), pp. 39–54. [Google Scholar]
- Higgins J.A., Bailey P.C., Laurie D.A. (2010). Comparative genomics of flowering time pathways using Brachypodium distachyon as a model for the temperate grasses. PLoS One 5: e10065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong J.J., Park Y.-S., Bravo A., Bhattarai K.K., Daniels D.A., Harrison M.J. (2012). Diversity of morphology and function in arbuscular mycorrhizal symbioses in Brachypodium distachyon. Planta 236: 851–865. [DOI] [PubMed] [Google Scholar]
- Hsia M.M., et al. (2017). Sequencing and functional validation of the JGI Brachypodium distachyon T-DNA collection. Plant J. 91: 361–370. [DOI] [PubMed] [Google Scholar]
- Hu F.Y., Tao D.Y., Sacks E., Fu B.Y., Xu P., Li J., Yang Y., McNally K., Khush G.S., Paterson A.H., Li Z.K. (2003). Convergent evolution of perenniality in rice and sorghum. Proc. Natl. Acad. Sci. USA 100: 4050–4054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu F., Wang D., Zhao X., Zhang T., Sun H., Zhu L., Zhang F., Li L., Li Q., Tao D., Fu B., Li Z. (2011). Identification of rhizome-specific genes by genome-wide differential expression analysis in Oryza longistaminata. BMC Plant Biol. 11: 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Idziak D., Betekhtin A., Wolny E., Lesniewska K., Wright J., Febrer M., Bevan M.W., Jenkins G., Hasterok R. (2011). Painting the chromosomes of Brachypodium: current status and future prospects. Chromosoma 120: 469–479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Idziak-Helmcke D., Betekhtin A. (2018). Methods for cytogenetic chromosome barcoding and chromosome painting in Brachypodium distachyon and its relative species. Methods Mol. Biol. 1667: 1–19. [DOI] [PubMed] [Google Scholar]
- Ingram P.A., Zhu J., Shariff A., Davis I.W., Benfey P.N., Elich T. (2012). High-throughput imaging and analysis of root system architecture in Brachypodium distachyon under differential nutrient availability. Philos. Trans. R. Soc. Lond. B Biol. Sci. 367: 1559–1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- International Brachypodium Initiative (2010). Genome sequencing and analysis of the model grass Brachypodium distachyon. Nature 463: 763–768. [DOI] [PubMed] [Google Scholar]
- Irigoyen S., Bedre R.H., Scholthof K.-B.G., Mandadi K.K. (2018). Genomic approaches to analyze alternative splicing, a key regulator of transcriptome and proteome diversity in Brachypodium distachyon. Methods Mol. Biol. 1667: 73–85. [DOI] [PubMed] [Google Scholar]
- Jakobsen I., Smith S.E., Smith F.A., Watts-Williams S.J., Clausen S.S., Grønlund M. (2016). Plant growth responses to elevated atmospheric CO2 are increased by phosphorus sufficiency but not by arbuscular mycorrhizas. J. Exp. Bot. 67: 6173–6186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones H.G. (1999). Use of thermography for quantitative studies of spatial and temporal variation of stomatal conductance over leaf surfaces. Plant Cell Environ. 22: 1043–1055. [Google Scholar]
- Kang H., Zhang M., Zhou S., Guo Q., Chen F., Wu J., Wang W. (2016). Overexpression of wheat ubiquitin gene, Ta-Ub2, improves abiotic stress tolerance of Brachypodium distachyon. Plant Sci. 248: 102–115. [DOI] [PubMed] [Google Scholar]
- Kawasaki A., Donn S., Ryan P.R., Mathesius U., Devilla R., Jones A., Watt M. (2016). Microbiome and exudates of the root and rhizosphere of Brachypodium distachyon, a model for wheat. PLoS One 11: e0164533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellogg E.A. (2015a). Flowering Plants. Monocots. Poaceae. (Switzerland: Springer; ). [Google Scholar]
- Kellogg E.A. (2015b). Brachypodium distachyon as a genetic model system. Annu. Rev. Genet. 49: 1–20. [DOI] [PubMed] [Google Scholar]
- Khan M.A., Stace C.A. (1999). Breeding relationships in the genus Brachypodium (Poaceae: Pooideae). Nord. J. Bot. 19: 257–269. [Google Scholar]
- Kind S., Schurack S., Hinsch J., Tudzynski P. (2017). Brachypodium distachyon as alternative model host system for the ergot fungus Claviceps purpurea. Mol. Plant Pathol. 19: 1005–1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong L.A., Wu D.Q., Cui J.K., Huang W.K., Peng H., Peng D.L. (2016). Testing and modelling the potential of three diploid plants in Poaceae as a new pathosystem to investigate the interactions between cereal hosts and cereal cyst nematode (Heterodera avenae). Plant Pathol. 65: 682–688. [Google Scholar]
- Krishnakumar V., et al. (2015). Araport: the Arabidopsis information portal. Nucleic Acids Res. 43: D1003–D1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee M.Y., et al. (2012). Brachypodium distachyon line Bd3-1 resistance is elicited by the barley stripe mosaic virus triple gene block 1 movement protein. J. Gen. Virol. 93: 2729–2739. [DOI] [PubMed] [Google Scholar]
- Leonelli S., Ankeny R.A. (2013). What makes a model organism? Endeavour 37: 209–212. [DOI] [PubMed] [Google Scholar]
- Liang X., Wang H., Mao L., Hu Y., Dong T., Zhang Y., Wang X., Bi Y. (2012). Involvement of COP1 in ethylene- and light-regulated hypocotyl elongation. Planta 236: 1791–1802. [DOI] [PubMed] [Google Scholar]
- Liou M.-R., Huang Y.-W., Hu C.-C., Lin N.-S., Hsu Y.-H. (2014). A dual gene-silencing vector system for monocot and dicot plants. Plant Biotechnol. J. 12: 330–343. [DOI] [PubMed] [Google Scholar]
- Liu D., Zehfroosh N., Hancock B.L., Hines K., Fang W., Kilfoil M., Learned-Miller E., Sanguinet K.A., Goldner L.S., Baskin T.I. (2017). Imaging cellulose synthase motility during primary cell wall synthesis in the grass Brachypodium distachyon. Sci. Rep. 7: 15111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- López-Alvarez D., López-Herranz M.L., Betekhtin A., Catalán P. (2012). A DNA barcoding method to discriminate between the model plant Brachypodium distachyon and its close relatives B. stacei and B. hybridum (Poaceae). PLoS One 7: e51058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- López-Alvarez D., Manzaneda A.J., Rey P.J., Giraldo P., Benavente E., Allainguillaume J., Mur L., Caicedo A.L., Hazen S.P., Breiman A., Ezrati S., Catalán P. (2015). Environmental niche variation and evolutionary diversification of the Brachypodium distachyon grass complex species in their native circum-Mediterranean range. Am. J. Bot. 102: 1073–1088. [DOI] [PubMed] [Google Scholar]
- Lowe K., et al. (2016). Morphogenic regulators Baby boom and Wuschel improve monocot transformation. Plant Cell 28: 1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lv B., Nitcher R., Han X., Wang S., Ni F., Li K., Pearce S., Wu J., Dubcovsky J., Fu D. (2014). Characterization of FLOWERING LOCUS T1 (FT1) gene in Brachypodium and wheat. PLoS One 9: e94171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyons C.W., Scholthof K.-B.G. (2015). Watching grass grow: The emergence of Brachypodium distachyon as a model for the Poaceae. In New Perspectives on the History of Life Sciences and Agriculture, Phillips D. and Kingsland S., eds (Switzerland: Springer; ), pp. 479–501. [Google Scholar]
- Lyons C.W., Scholthof K.-B.G. (2016). Brachypodium as an Arabidopsis for the grasses: Are we there yet? In Genetics and Genomics of Brachypodium, Vogel J.P., ed (Switzerland: Springer; ), pp. 327–341. [Google Scholar]
- Mandadi K.K., Scholthof K.-B.G. (2012). Characterization of a viral synergism in the monocot Brachypodium distachyon reveals distinctly altered host molecular processes associated with disease. Plant Physiol. 160: 1432–1452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandadi K.K., Scholthof K.-B.G. (2013). Plant immune responses against viruses: how does a virus cause disease? Plant Cell 25: 1489–1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandadi K.K., Scholthof K.-B.G. (2015). Genome-wide analysis of alternative splicing landscapes modulated during plant-virus interactions in Brachypodium distachyon. Plant Cell 27: 71–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandadi K.K., Pyle J.D., Scholthof K.-B.G. (2014). Comparative analysis of antiviral responses in Brachypodium distachyon and Setaria viridis reveals conserved and unique outcomes among C3 and C4 plant defenses. Mol. Plant Microbe Interact. 27: 1277–1290. [DOI] [PubMed] [Google Scholar]
- Mandadi K.K., Pyle J.D., Scholthof K.-B.G. (2015). Characterization of SCL33 splicing patterns during diverse virus infections in Brachypodium distachyon. Plant Signal. Behav. 10: e1042641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manzaneda A.J., Rey P.J., Bastida J.M., Weiss-Lehman C., Raskin E., Mitchell-Olds T. (2012). Environmental aridity is associated with cytotype segregation and polyploidy occurrence in Brachypodium distachyon (Poaceae). New Phytol. 193: 797–805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manzaneda A.J., Rey P.J., Anderson J.T., Raskin E., Weiss-Lehman C., Mitchell-Olds T. (2015). Natural variation, differentiation, and genetic trade-offs of ecophysiological traits in response to water limitation in Brachypodium distachyon and its descendent allotetraploid B. hybridum (Poaceae). Evolution 69: 2689–2704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcussen T., Sandve S.R., Heier L., Spannagl M., Pfeifer M., Jakobsen K.S., Wulff B.B.H., Steuernagel B., Mayer K.F.X., Olsen O.-A.; International Wheat Genome Sequencing Consortium (2014). Ancient hybridizations among the ancestral genomes of bread wheat. Science 345: 1250092. [DOI] [PubMed] [Google Scholar]
- Marriott P.E., Sibout R., Lapierre C., Fangel J.U., Willats W.G.T., Hofte H., Gómez L.D., McQueen-Mason S.J. (2014). Range of cell-wall alterations enhance saccharification in Brachypodium distachyon mutants. Proc. Natl. Acad. Sci. USA 111: 14601–14606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez L.M., Fernández-Ocaña A., Rey P.J., Salido T., Amil-Ruiz F., Manzaneda A.J. (2018). Variation in functional responses to water stress and differentiation between natural allopolyploid populations in the Brachypodium distachyon species complex. Ann. Bot. 121: 1369–1382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matos D.A., Whitney I.P., Harrington M.J., Hazen S.P. (2013). Cell walls and the developmental anatomy of the Brachypodium distachyon stem internode. PLoS One 8: e80640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matyjaszczyk E. (2015). Products containing microorganisms as a tool in integrated pest management and the rules of their market placement in the European Union. Pest Manag. Sci. 71: 1201–1206. [DOI] [PubMed] [Google Scholar]
- Meijer G., Leuchtmann A. (1999). Multistrain infections of the grass Brachypodium sylvaticum by its fungal endophyte Epichloë sylvatica. New Phytol. 141: 355–368. [DOI] [PubMed] [Google Scholar]
- Mitsuda N., Seki M., Shinozaki K., Ohme-Takagi M. (2005). The NAC transcription factors NST1 and NST2 of Arabidopsis regulate secondary wall thickenings and are required for anther dehiscence. Plant Cell 17: 2993–3006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouchel C.F., Briggs G.C., Hardtke C.S. (2004). Natural genetic variation in Arabidopsis identifies BREVIS RADIX, a novel regulator of cell proliferation and elongation in the root. Genes Dev. 18: 700–714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Driscoll A., Doohan F., Mullins E. (2015). Exploring the utility of Brachypodium distachyon as a model pathosystem for the wheat pathogen Zymoseptoria tritici. BMC Res. Notes 8: 132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohashi-Ito K., Oda Y., Fukuda H. (2010). Arabidopsis VASCULAR-RELATED NAC-DOMAIN6 directly regulates the genes that govern programmed cell death and secondary wall formation during xylem differentiation. Plant Cell 22: 3461–3473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osmont K.S., Sibout R., Hardtke C.S. (2007). Hidden branches: developments in root system architecture. Annu. Rev. Plant Biol. 58: 93–113. [DOI] [PubMed] [Google Scholar]
- Pacak A., Geisler K., Jørgensen B., Barciszewska-Pacak M., Nilsson L., Nielsen T.H., Johansen E., Grønlund M., Jakobsen I., Albrechtsen M. (2010). Investigations of barley stripe mosaic virus as a gene silencing vector in barley roots and in Brachypodium distachyon and oat. Plant Methods 6: 26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pacheco-Villalobos D., Hardtke C.S. (2012). Natural genetic variation of root system architecture from Arabidopsis to Brachypodium: towards adaptive value. Philos. Trans. R. Soc. Lond. B Biol. Sci. 367: 1552–1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Păcurar D.I., Thordal-Christensen H., Nielsen K.K., Lenk I. (2008). A high-throughput Agrobacterium-mediated transformation system for the grass model species Brachypodium distachyon L. Transgenic Res. 17: 965–975. [DOI] [PubMed] [Google Scholar]
- Palisot de Beauvois A.M.F.J. (1812). Essai d’une Nouvelle Agrostographie ou Nouveaux Genres des Graminées. (Paris: Imprimerie de Fain; ). [Google Scholar]
- Peraldi A., Griffe L.L., Burt C., McGrann G.R.D., Nicholson P. (2014). Brachypodium distachyon exhibits compatible interactions with Oculimacula spp. and Ramularia collo-cygni, providing the first pathosystem model to study eyespot and ramularia leaf spot diseases. Plant Pathol. 63: 554–562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeifer M., Kugler K.G., Sandve S.R., Zhan B., Rudi H., Hvidsten T.R., Mayer K.F.X., Olsen O.-A.; International Wheat Genome Sequencing Consortium (2014). Genome interplay in the grain transcriptome of hexaploid bread wheat. Science 345: 1250091. [DOI] [PubMed] [Google Scholar]
- Poiré R., Chochois V., Sirault X.R., Vogel J.P., Watt M., Furbank R.T. (2014). Digital imaging approaches for phenotyping whole plant nitrogen and phosphorus response in Brachypodium distachyon. J. Integr. Plant Biol. 56: 781–796. [DOI] [PubMed] [Google Scholar]
- Powell J.J., Carere J., Sablok G., Fitzgerald T.L., Stiller J., Colgrave M.L., Gardiner D.M., Manners J.M., Vogel J.P., Henry R.J., Kazan K. (2017). Transcriptome analysis of Brachypodium during fungal pathogen infection reveals both shared and distinct defense responses with wheat. Sci. Rep. 7: 17212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Priest H.D., Fox S.E., Rowley E.R., Murray J.R., Michael T.P., Mockler T.C. (2014). Analysis of global gene expression in Brachypodium distachyon reveals extensive network plasticity in response to abiotic stress. PLoS One 9: e87499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pyle J.D., Scholthof K.-B.G. (2018). De novo generation of helper virus-satellite chimera RNAs results in disease attenuation and satellite sequence acquisition in a host-dependent manner. Virology 514: 182–191. [DOI] [PubMed] [Google Scholar]
- Pyle J.D., Monis J., Scholthof K.G. (2017). Complete nucleotide sequences and virion particle association of two satellite RNAs of panicum mosaic virus. Virus Res. 240: 87–93. [DOI] [PubMed] [Google Scholar]
- Rabe F., et al. (2016). A complete toolset for the study of Ustilago bromivora and Brachypodium sp. as a fungal-temperate grass pathosystem. eLife 5: e20522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramírez-Puebla S.T., Servín-Garcidueñas L.E., Jiménez-Marín B., Bolaños L.M., Rosenblueth M., Martínez J., Rogel M.A., Ormeño-Orrillo E., Martínez-Romero E. (2013). Gut and root microbiota commonalities. Appl. Environ. Microbiol. 79: 2–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rancour D.M., Marita J.M., Hatfield R.D. (2012). Cell wall composition throughout development for the model grass Brachypodium distachyon. Front. Plant Sci. 3: 266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rancour D.M., Hatfield R.D., Marita J.M., Rohr N.A., Schmitz R.J. (2015). Cell wall composition and digestibility alterations in Brachypodium distachyon achieved through reduced expression of the UDP-arabinopyranose mutase. Front. Plant Sci. 6: 446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ream T.S., Woods D.P., Schwartz C.J., Sanabria C.P., Mahoy J.A., Walters E.M., Kaeppler H.F., Amasino R.M. (2014). Interaction of photoperiod and vernalization determines flowering time of Brachypodium distachyon. Plant Physiol. 164: 694–709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rey P.J., Manzaneda A.J., Alcántara J.M. (2017). The interplay between aridity and competition determines colonization ability, exclusion and ecological segregation in the heteroploid Brachypodium distachyon species complex. New Phytol. 215: 85–96. [DOI] [PubMed] [Google Scholar]
- Rioux R.A., Van Ryzin B.J., Kerns J.P. (2017). Brachypodium: A potential model host for fungal pathogens of turfgrasses. Phytopathology 107: 749–757. [DOI] [PubMed] [Google Scholar]
- Rosenkranz E. (1978). Grasses native or adventive to the United States as new hosts of maize dwarf mosaic and sugarcane mosaic viruses. Phytopathology 68: 175–179. [Google Scholar]
- Routledge A.P.M., Shelley G., Smith J.V., Talbot N.J., Draper J., Mur L.A.J. (2004). Magnaporthe grisea interactions with the model grass Brachypodium distachyon closely resemble those with rice (Oryza sativa). Mol. Plant Pathol. 5: 253–265. [DOI] [PubMed] [Google Scholar]
- Roy B.A., Coulson T., Blaser W., Policha T., Stewart J.L., Blaisdell G.K., Güsewell S. (2011). Population regulation by enemies of the grass Brachypodium sylvaticum: demography in native and invaded ranges. Ecology 92: 665–675. [DOI] [PubMed] [Google Scholar]
- Ruíz M., Quemada M., García R.M., Carrillo J.M., Benavente E. (2016). Use of thermographic imaging to screen for drought-tolerant genotypes in Brachypodium distachyon. Crop Pasture Sci. 67: 99–108. [Google Scholar]
- Ruprecht C., Proost S., Hernandez-Coronado M., Ortiz-Ramirez C., Lang D., Rensing S.A., Becker J.D., Vandepoele K., Mutwil M. (2017). Phylogenomic analysis of gene co-expression networks reveals the evolution of functional modules. Plant J. 90: 447–465. [DOI] [PubMed] [Google Scholar]
- Sancho R., Cantalapiedra C.P., López-Alvarez D., Gordon S.P., Vogel J.P., Catalán P., Contreras-Moreira B. (2018). Comparative plastome genomics and phylogenomics of Brachypodium: flowering time signatures, introgression and recombination in recently diverged ecotypes. New Phytol. 218: 1631–1644. [DOI] [PubMed] [Google Scholar]
- Sandoya G.V., de Oliveira Buanafina M.M. (2014). Differential responses of Brachypodium distachyon genotypes to insect and fungal pathogens. Physiol. Mol. Plant Pathol. 85: 53–64. [Google Scholar]
- Sasaki T., Antonio B.A. (2004). Rice genome as a model system for cereals. In Cereal Genomics, Gupta P.K., Varshney R.K., eds (Dordrecht, The Netherlands: Kluwer; ), pp. 535–557. [Google Scholar]
- Schippmann U. (1991). Revision der europäischen Arten der Gattung Brachypodium Palisot de Beauvois (Poaceae). Boissiera 45: 1–250. [Google Scholar]
- Schneebeli K., Mathesius U., Watt M. (2015). Brachypodium distachyon is a pathosystem model for the study of the wheat disease rhizoctonia root rot. Plant Pathol. 64: 91–100. [Google Scholar]
- Sibout R., et al. (2017). Expression atlas and comparative coexpression network analyses reveal important genes involved in the formation of lignified cell wall in Brachypodium distachyon. New Phytol. 215: 1009–1025. [DOI] [PubMed] [Google Scholar]
- Smiley R.W., Machado S., Rhinhart K.E.L., Reardon C.L., Wuest S.B. (2016). Rapid quantification of soilborne pathogen communities in wheat-based long-term field experiments. Plant Dis. 100: 1692–1708. [DOI] [PubMed] [Google Scholar]
- Smith S.E., Smith F.A., Jakobsen I. (2003). Mycorrhizal fungi can dominate phosphate supply to plants irrespective of growth responses. Plant Physiol. 133: 16–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sogutmaz Ozdemir B., Budak H. (2018). Application of tissue culture and transformation techniques in model species Brachypodium distachyon. Methods Mol. Biol. 1667: 289–310. [DOI] [PubMed] [Google Scholar]
- Soltis P.S., Soltis D.E. (2016). Ancient WGD events as drivers of key innovations in angiosperms. Curr. Opin. Plant Biol. 30: 159–165. [DOI] [PubMed] [Google Scholar]
- Staley C., et al. (2017). Diurnal cycling of rhizosphere bacterial communities is associated with shifts in carbon metabolism. Microbiome 5: 65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stebbins G.L. (1949). The evolutionary significance of natural and artificial polyploids in the family Gramineae. Hereditas 35: 461–485. [Google Scholar]
- Steinwand M.A., Young H.A., Bragg J.N., Tobias C.M., Vogel J.P. (2013). Brachypodium sylvaticum, a model for perennial grasses: transformation and inbred line development. PLoS One 8: e75180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stepanova A.N., Robertson-Hoyt J., Yun J., Benavente L.M., Xie D.-Y., Doležal K., Schlereth A., Jürgens G., Alonso J.M. (2008). TAA1-mediated auxin biosynthesis is essential for hormone crosstalk and plant development. Cell 133: 177–191. [DOI] [PubMed] [Google Scholar]
- Sturm J. (1843). Deutschlands flora in abbildungen nach der natur Abt.1:Bd.19. Nurnberg. http://biodiversitylibrary.org/item/148126.
- Sun J., Hu W., Zhou R., Wang L., Wang X., Wang Q., Feng Z., Li Y., Qiu D., He G., Yang G. (2015). The Brachypodium distachyon BdWRKY36 gene confers tolerance to drought stress in transgenic tobacco plants. Plant Cell Rep. 34: 23–35. [DOI] [PubMed] [Google Scholar]
- Takahagi K., Inoue K., Shimizu M., Uehara-Yamaguchi Y., Onda Y., Mochida K. (2018). Homoeolog-specific activation of genes for heat acclimation in the allopolyploid grass Brachypodium hybridum. Gigascience 7: giy020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talavera S. (1978). Aportación al estudio cariológico de las gramíneas españolas. Lagascalia 7: 133–142. [Google Scholar]
- Tao Y., Nadege S.W., Huang C., Zhang P., Song S., Sun L., Wu Y. (2016). Brachypodium distachyon is a suitable host plant for study of Barley yellow dwarf virus. Virus Genes 52: 299–302. [DOI] [PubMed] [Google Scholar]
- Thole V., Worland B., Wright J., Bevan M.W., Vain P. (2010). Distribution and characterization of more than 1000 T-DNA tags in the genome of Brachypodium distachyon community standard line Bd21. Plant Biotechnol. J. 8: 734–747. [DOI] [PubMed] [Google Scholar]
- Thole V., Peraldi A., Worland B., Nicholson P., Doonan J.H., Vain P. (2012). T-DNA mutagenesis in Brachypodium distachyon. J. Exp. Bot. 63: 567–576. [DOI] [PubMed] [Google Scholar]
- Thomashow M.F. (1999). Plant cold acclimation: Freezing tolerance genes and regulatory mechanisms. Annu. Rev. Plant Physiol. Plant Mol. Biol. 50: 571–599. [DOI] [PubMed] [Google Scholar]
- Tian F., Gong J., Zhang J., Feng Y., Wang G., Guo Q., Wang W. (2014). Overexpression of monoubiquitin improves photosynthesis in transgenic tobacco plants following high temperature stress. Plant Sci. 226: 92–100. [DOI] [PubMed] [Google Scholar]
- Timpano H., Sibout R., Devaux M.-F., Alvarado C., Looten R., Falourd X., Pontoire B., Martin M., Legée F., Cézard L. (2015). Brachypodium cell wall mutant with enhanced saccharification potential despite increased lignin content. BioEnergy Res. 8: 53–67. [Google Scholar]
- Tkacz A., Cheema J., Chandra G., Grant A., Poole P.S. (2015). Stability and succession of the rhizosphere microbiota depends upon plant type and soil composition. ISME J. 9: 2349–2359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trabucco G.M., Matos D.A., Lee S.J., Saathoff A.J., Priest H.D., Mockler T.C., Sarath G., Hazen S.P. (2013). Functional characterization of cinnamyl alcohol dehydrogenase and caffeic acid O-methyltransferase in Brachypodium distachyon. BMC Biotechnol. 13: 61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vain P., Worland B., Thole V., McKenzie N., Alves S.C., Opanowicz M., Fish L.J., Bevan M.W., Snape J.W. (2008). Agrobacterium-mediated transformation of the temperate grass Brachypodium distachyon (genotype Bd21) for T-DNA insertional mutagenesis. Plant Biotechnol. J. 6: 236–245. [DOI] [PubMed] [Google Scholar]
- Valdivia E.R., Herrera M.T., Gianzo C., Fidalgo J., Revilla G., Zarra I., Sampedro J. (2013). Regulation of secondary wall synthesis and cell death by NAC transcription factors in the monocot Brachypodium distachyon. J. Exp. Bot. 64: 1333–1343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verelst W., Bertolini E., De Bodt S., Vandepoele K., Demeulenaere M., Pè M.E., Inzé D. (2013). Molecular and physiological analysis of growth-limiting drought stress in Brachypodium distachyon leaves. Mol. Plant 6: 311–322. [DOI] [PubMed] [Google Scholar]
- Vogel J.P. (2016). The rise of Brachypodium as a model system. In Genetics and Genomics of Brachypodium, Vogel J.P., ed (Switzerland: Springer; ), pp. 1–7. [Google Scholar]
- Vogel J., Hill T. (2008). High-efficiency Agrobacterium-mediated transformation of Brachypodium distachyon inbred line Bd21-3. Plant Cell Rep. 27: 471–478. [DOI] [PubMed] [Google Scholar]
- Vogel J.P., Garvin D.F., Leong O.M., Hayden D.M. (2006). Agrobacterium-mediated transformation and inbred line development in the model grass Brachypodium distachyon. Plant Cell Tissue Organ Cult. 84: 199–211. [Google Scholar]
- Vogel J.P., Tuna M., Budak H., Huo N., Gu Y.Q., Steinwand M.A. (2009). Development of SSR markers and analysis of diversity in Turkish populations of Brachypodium distachyon. BMC Plant Biol. 9: 88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Zitzewitz J., Szűcs P., Dubcovsky J., Yan L., Francia E., Pecchioni N., Casas A., Chen T.H.H., Hayes P.M., Skinner J.S. (2005). Molecular and structural characterization of barley vernalization genes. Plant Mol. Biol. 59: 449–467. [DOI] [PubMed] [Google Scholar]
- Wang G., Wang L., Cui Y., Yu M., Dang C., Wang H., Jin X., Yan L., Wu Q., Li D., Liu Z. (2017). RNA-seq analysis of Brachypodium distachyon responses to Barley stripe mosaic virus infection. Crop J. 5: 1–10. [Google Scholar]
- Watt M., Schneebeli K., Dong P., Wilson I.W. (2009). The shoot and root growth of Brachypodium and its potential as a model for wheat and other cereal crops. Funct. Plant Biol. 36: 960–969. [DOI] [PubMed] [Google Scholar]
- Wolny E., Hasterok R. (2009). Comparative cytogenetic analysis of the genomes of the model grass Brachypodium distachyon and its close relatives. Ann. Bot. 104: 873–881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Won C., Shen X., Mashiguchi K., Zheng Z., Dai X., Cheng Y., Kasahara H., Kamiya Y., Chory J., Zhao Y. (2011). Conversion of tryptophan to indole-3-acetic acid by TRYPTOPHAN AMINOTRANSFERASES OF ARABIDOPSIS and YUCCAs in Arabidopsis. Proc. Natl. Acad. Sci. USA 108: 18518–18523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woods D.P., McKeown M.A., Dong Y., Preston J.C., Amasino R.M. (2016). Evolution of VRN2/Ghd7-like genes in vernalization-mediated repression of grass flowering. Plant Physiol. 170: 2124–2135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woods D.P., Ream T.S., Bouché F., Lee J., Thrower N., Wilkerson C., Amasino R.M. (2017). Establishment of a vernalization requirement in Brachypodium distachyon requires REPRESSOR OF VERNALIZATION1. Proc. Natl. Acad. Sci. USA 114: 6623–6628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi M., Mitsuda N., Ohtani M., Ohme-Takagi M., Kato K., Demura T. (2011). Vascular-related NAC-domain 7 directly regulates a broad range of genes for xylem vessel differentiation. BMC Proc. 5 (suppl. 7): O37. [DOI] [PubMed] [Google Scholar]
- Yong E. (2016). I Contain Multitudes. (New York: HarperCollins; ). [Google Scholar]
- Yuan C., Li C., Yan L., Jackson A.O., Liu Z., Han C., Yu J., Li D. (2011). A high throughput barley stripe mosaic virus vector for virus induced gene silencing in monocots and dicots. PLoS One 6: e26468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong S., Ali S., Leng Y., Wang R., Garvin D.F. (2015). Brachypodium distachyon–Cochliobolus sativus pathosystem is a new model for studying plant–fungal interactions in cereal crops. Phytopathology 105: 482–489. [DOI] [PubMed] [Google Scholar]
- Zhou Y.-R., Li L.-Y., Li J.-M., Sun Z.-T., Xie L., Chen J.-P. (2016). Argonaute subfamily genes in the small brown planthopper, Laodelphax striatellus (Hemiptera: Delphacidae). Arch. Insect Biochem. Physiol. 91: 37–51. [DOI] [PubMed] [Google Scholar]