Knockout of the peroxisomal MALATE DEHYDROGENASE2 drastically alters photosynthesis and the metabolism of fatty acids and starch upon nitrogen deprivation and high light exposure in the green microalga Chlamydomonas reinhardtii.
Abstract
Plants and algae must tightly coordinate photosynthetic electron transport and metabolic activities given that they often face fluctuating light and nutrient conditions. The exchange of metabolites and signaling molecules between organelles is thought to be central to this regulation but evidence for this is still fragmentary. Here, we show that knocking out the peroxisome-located MALATE DEHYDROGENASE2 (MDH2) of Chlamydomonas reinhardtii results in dramatic alterations not only in peroxisomal fatty acid breakdown but also in chloroplast starch metabolism and photosynthesis. mdh2 mutants accumulated 50% more storage lipid and 2-fold more starch than the wild type during nitrogen deprivation. In parallel, mdh2 showed increased photosystem II yield and photosynthetic CO2 fixation. Metabolite analyses revealed a >60% reduction in malate, together with increased levels of NADPH and H2O2 in mdh2. Similar phenotypes were found upon high light exposure. Furthermore, based on the lack of starch accumulation in a knockout mutant of the H2O2-producing peroxisomal ACYL-COA OXIDASE2 and on the effects of H2O2 supplementation, we propose that peroxisome-derived H2O2 acts as a regulator of chloroplast metabolism. We conclude that peroxisomal MDH2 helps photoautotrophs cope with nitrogen scarcity and high light by transmitting the redox state of the peroxisome to the chloroplast by means of malate shuttle- and H2O2-based redox signaling.
INTRODUCTION
Photoautotrophs convert light energy into reducing equivalents (NADPH) and phosphorylating power (ATP), which are used to drive the metabolic reactions of CO2 assimilation. Coordinating photosynthetic electron transport activities to downstream metabolic needs is essential for cell survival and growth because excess production of reducing power may result in an overreduction of the photosynthetic electron transport chain, which may lead to photooxidative damage (Niyogi, 2000). Multiple strategies have therefore evolved to facilitate fine-tuning of photosynthesis and allow plants and algae to rapidly acclimate to natural environments where nutrient, light, and temperature can change frequently (Saroussi et al., 2017). Several chloroplast-located alternative electron pathways (notably, cyclic electron flow, the water-to-water cycle, O2 photoreduction processes, and chlororespiration) have been identified to play roles in dissipation of photoreductant and/or reequilibration of the NADPH/ATP ratio (Peltier et al., 2010; Curien et al., 2016; Saroussi et al., 2017). Recently, it has been shown that chloroplast redox balance can also be achieved through export of excess reducing equivalents to mitochondria in green algae (Dang et al., 2014) and diatoms (Bailleul et al., 2015) yet the detailed molecular mechanisms remain to be elucidated. Moreover, alterations of mitochondrial metabolism have also been shown to influence photosynthetic performance in plants and algae (Sweetlove et al., 2006; Nunes-Nesi et al., 2011; Massoz et al., 2015; Larosa et al., 2018). Thus current knowledge on chloroplast redox poise is centered on the dissipation of excess reducing equivalents through chloroplast-based processes or through collaboration between chloroplast and mitochondria.
Alongside chloroplasts and mitochondria, peroxisomes are a further subcellular compartment involved in energetic metabolism. The peroxisome was originally defined as an organelle that carries out oxidative reactions leading to production of H2O2, a reactive oxygen species (ROS) that can cause oxidative damage if present in excess (Erickson et al., 2015; Dietz et al., 2016). In plant peroxisomes, the major sources of H2O2 are photorespiration and β-oxidation of fatty acids (FAs). However, in algae such as Chlamydomonas reinhardtii that possess a CO2 concentrating mechanism, photorespiration is negligible and does not produce H2O2 due to the absence of glyoxylate oxidase (Aboelmy and Peterhansel, 2014; Hagemann et al., 2016). Algal FA β-oxidation, which has recently been demonstrated to be the major pathway for FA catabolism in Chlamydomonas (Kong et al., 2017), is therefore the major contributor to H2O2 formation in peroxisomes. H2O2 is produced at the first step of β-oxidation, which is catalyzed by acyl-CoA oxidase (ACX). H2O2 can also be produced in chloroplasts through the Mehler reaction as a “safety valve” to photochemical reactions and has been shown to play a signaling role if present at sublethal levels (Dietz et al., 2016). However, it remains to be established whether peroxisome-derived H2O2 can also play a signaling role.
In addition to H2O2 and its end-product acetyl-CoA, FA β-oxidation generates one molecule of NADH (1:1:1) through the 3-hydoxyacyl-CoA dehydrogenase activity of the multifunctional protein (MFP-DH). Studies in yeasts and plants show that reoxidation of peroxisomal NADH must occur inside the organelle because the peroxisomal membrane is impermeable to NAD+ (van Roermund et al., 1995). NADH is an essential electron donor for numerous biochemical reactions occurring in different cellular compartments; therefore, NADH trafficking and homeostasis requires tight regulation and coordination (Mettler and Beevers, 1980; Scheibe, 2004). By catalyzing the reversible conversion of NADH to NAD+ via reduction of oxaloacetate to malate, which can be shuttled across subcellular membranes, malate dehydrogenases (MDHs) play a key role in intracellular trafficking of reducing equivalents (Mettler and Beevers, 1980; Scheibe, 2004). MDHs are ubiquitous enzymes, and each subcellular compartment usually contains at least one isoform (Mettler and Beevers, 1980; Scheibe, 2004). With the exception of the major chloroplast isoform which is NADP+-dependent (EC1.1.1.82) and so far the best characterized (Musrati et al., 1998; Miginiac-Maslow and Lancelin, 2002; Lemaire et al., 2005), all the other MDHs require NAD+ as a cofactor (EC1.1.1.37). Despite their potential importance in cellular redox homeostasis, none of the algal NAD+-dependent MDH has been studied. The Chlamydomonas genome encodes four putative NAD+-dependent MDHs (Merchant et al., 2007), and MDH2 has been localized to peroxisomes (Hayashi and Shinozaki, 2012; Lauersen et al., 2016; Kong et al., 2017).
In this study, through isolation and characterization of two Chlamydomonas mutants deficient in peroxisomal NAD+-dependent MALATE DEHYDROGENASE2 (MDH2), we show that MDH2 plays a key role in the reverse coupling of the redox/H2O2 signal from the peroxisome to the chloroplast. The mdh2 mutant cells are compromised in triacylglycerol (TAG) breakdown and display increased TAG and starch accumulation during both photoautotrophic nitrogen (N) deprivation and high light (HL) exposure. We propose that the increased carbon storage is a result of more active photosynthesis and increased levels of NADPH and H2O2. MDH2 thus connects peroxisomal FA catabolism to synthesis of starch and lipid reserves and photosynthetic electron transport in the chloroplast by transmitting the redox state of the peroxisome to the chloroplast. Therefore, this study provides unanticipated evidence for the role of peroxisomes in helping photoautotrophs to cope with N scarcity and HL, which are widespread phenomena in many terrestrial and aquatic environments.
RESULTS
Mutants Knocked out for MDH2 Are Compromised in Lipid Catabolism
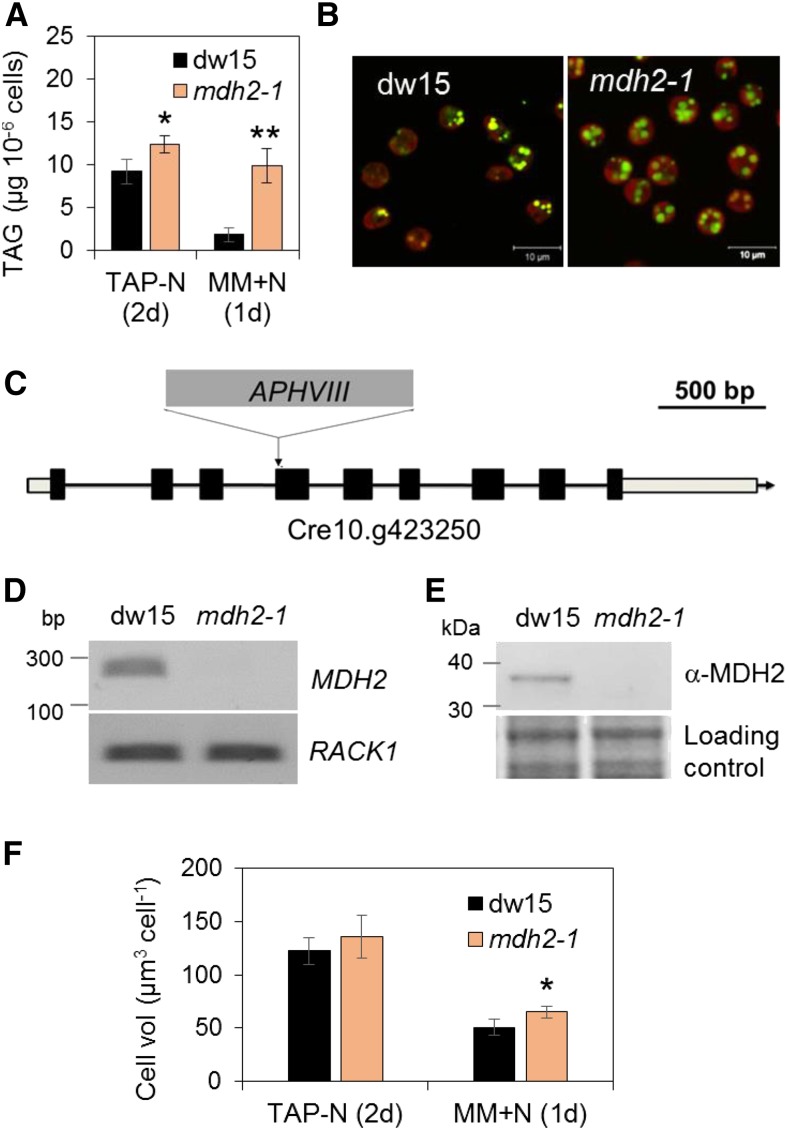
In Chlamydomonas, significant levels of TAG accumulate during N deprivation, and upon N resupply, TAGs are degraded to release carbon and energy contained in acyl groups to support cell division and growth (Liu and Benning, 2013; Schmollinger et al., 2014; Li-Beisson et al., 2015). To investigate the pathways and regulatory mechanisms of TAG homeostasis, a library of insertional mutants (∼4500) was screened for alterations in TAG content based on Nile red staining and flow cytometry (Cagnon et al., 2013). Among 80 mutants isolated, one mutant (Lb9G9) was found to be severely defective in TAG breakdown during the recovery phase after a period of mixotrophic N deprivation (Figure 1A). One day after N resupply while cells kept in the dark, the mutant cells retained ∼80% of the TAG amount accumulated before N resupply, whereas the parental strain (dw15) kept only ∼20% of the TAG accumulated. The observation that the mutant cells were 20% bigger than the wild type during the remobilization phase could not explain the fivefold higher TAG present in the mutant cells compared with the wild type at that stage (Figure 1F). The defect in TAG degradation could also be visualized by stronger BODIPY staining under confocal microscopy (Figure 1B). The experimental setup used in this study is depicted in Supplemental Figure 1A.
Figure 1.
Isolation of the mdh2-1 (Lb9G9) Mutant Impaired in TAG Breakdown during N Recovery.
(A) TAG content 2 d after N deprivation and 1 d after N resupply in mixotrophically grown cells.
(B) Visualization of LDs in cells upon N resupply (1 d) by staining with BODIPY.
(C) The insertion site of the cassette APHVIII in mdh2-1.
(D) RT-PCR analysis.
(E) Immunoblot analysis using anti-MDH2 antibodies.
(F) Cell volume determination.
Cells were first grown mixotrophically in N-deprived conditions. After N resupply, cells were kept in the dark in MM medium (1 d) then were stained with BODIPY, and pseudocolors were used: lipid droplet in green and chlorophyll in red. The housekeeping gene used for RT-PCR is RACK1. For immunoblots, samples were loaded at equal total protein amount and stained by Coomassie blue. RACK1, Receptor of activated protein C kinase 1. Values are the mean of biological replicates (i.e., independent shaking flask cultures; n = 5, sd). Biological replicates refer to cells that were grown in independent flasks. Asterisks indicate statistically significant changes compared with the parental line dw15 by paired-sample Student’s t test (*P ≤ 0.05 and **P ≤ 0.01).
The antibiotic resistance gene APHVIII was found to be inserted in the 4th exon of the Cre10.g423250 locus (Figure 1C), which codes for an isoform of NAD+-dependent MDH (MDH2/MDY2) (Merchant et al., 2007). The mutant was therefore designated as mdh2-1. DNA gel blot analyses revealed a single insertion in mdh2-1 (Supplemental Figure 2). RT-PCR performed on total RNAs isolated from exponentially grown cells showed the absence of MDH2 transcripts in mdh2-1 (Figure 1D). A polyclonal antibody raised against a synthetic peptide recognized in its parental strain (dw15) one polypeptide corresponding in size to the predicted molecular mass of MDH2 (∼36 kD), which was absent in mdh2-1 (Figure 1E). Taken together, these data suggest that mdh2-1 is a null mutant. Activity measurements performed on Chlamydomonas total protein extracts showed an ∼24% reduction in the activity of NAD+-dependent MDH in mdh2-1 in comparison to dw15 during optimal growth (Supplemental Figure 3). The residual NAD+-dependent MDH activity is likely due to existence of at least three other NAD+-requiring MDH isoforms in Chlamydomonas (Merchant et al., 2007) (Supplemental Table 1).
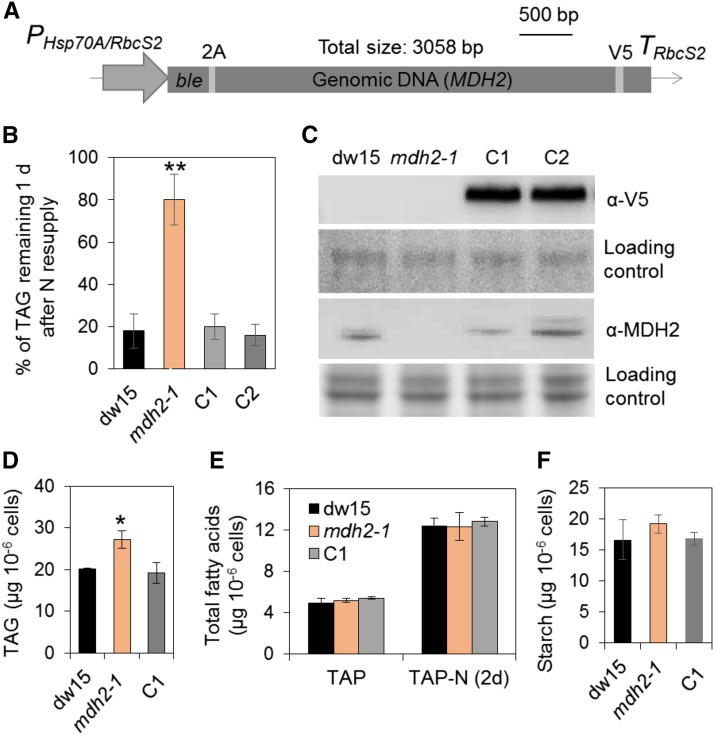
To confirm that the mdh2-1 mutant phenotype is due to a defect in the expression of MDH2, the mutant was complemented by transformation with a construct carrying a genomic copy of the full-length gene cloned in frame with the epitope tag V5 (Figure 2A). After screening ∼100 antibiotic resistant clones, we obtained two independent complemented lines that showed restoration in their capacity to remobilize oil (Figure 2B). The presence of the expressed protein MDH2 in these complemented lines was confirmed by immunoblot analysis using anti-V5 as well as anti-MDH2 antibodies (Figure 2C).
Figure 2.
Genetic Complementation of mdh2-1 Mutant.
(A) The construct used for genetic transformation.
(B) Restoration of TAG levels to that of the wild type in the complemented lines.
(C) Immunoblot detection of MDH2 in complemented lines using anti-V5 and anti-MDH2 antibodies.
(D) TAG analysis during mixotrophic N deprivation.
(E) Total FA quantification.
(F) Starch analysis during mixotrophic N deprivation.
TAG and starch contents were determined in N-deprived cells (2–3 d) and in N-resupplied cells (1 d). Two independent transformants (C1 and C2) were analyzed, and each line with three biological replicates (i.e., independent shaking flask cultures) and two technical replicates (i.e., different sampling from the same flask). Values are the mean of biological replicates (n = 3, sd). Asterisks indicate statistically significant changes compared with the control strains (dw15 and C1) by paired-sample Student’s t test (*P ≤ 0.05 and **P ≤ 0.01). Cells were grown in mixotrophic conditions under constant light. PHsp70A/RbcS2, the heat shock protein 70A and Rubisco small subunit promoter; ble, antibiotic resistance marker gene; TRbcS2, RbcS2 terminator; 2A, FMDV 2A self-cleaving peptide. For immunoblots, samples were loaded at equal total protein amounts. Loading controls were stained by Ponceau red for the upper panel and Coomassie blue for the lower panel.
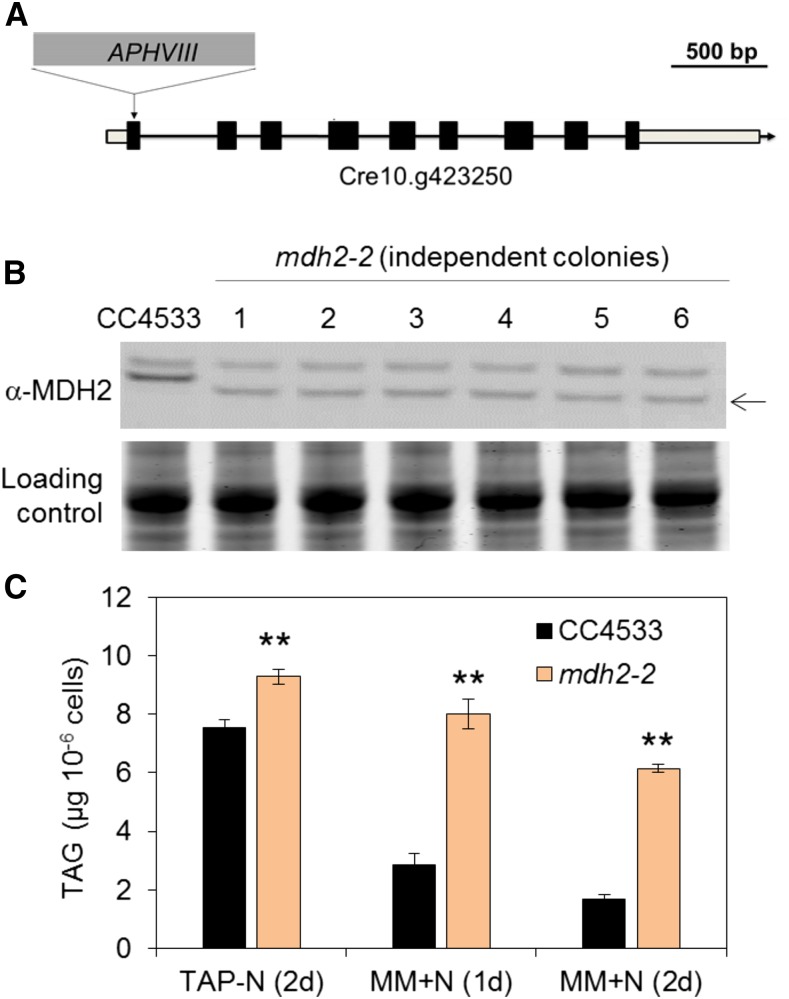
In parallel to the genetic complementation experiment, we also identified a second allele of the same locus (Cre10.g423250) in the CC4533 background from the Chlamydomonas Mutant Library (CLiP; https://www.chlamylibrary.org) (Li et al., 2016). This mutant, which we named mdh2-2, harbored an insertion of the antibiotic resistance gene in the first exon of the coding sequence (Figure 3A). Immunoblot analysis revealed the absence of the full-length protein and the presence of a smaller hybridizing signal likely due to the formation of a truncated protein (Figure 3B). Oil content analyses showed a similar oil remobilization phenotype (upon N resupply) in mdh2-2 (Figure 3C), as was observed in mdh2-1.
Figure 3.
mdh2-2 Mutant Characterization.
(A) The insertion site of the cassette APHVIII in mdh2-2.
(B) Immunoblot analysis using anti-MDH2 antibodies.
(C) TAG content analysis.
Cells were grown in mixotrophic conditions under constant light. TAG contents were determined in N-deprived cells (2 d) and in N-resupplied cells (1d and 2d). Values are the mean of biological replicates (i.e., independent shaking flask cultures; n = 3, sd). Asterisks indicate statistically significant changes compared with parental strain (CC4533) by paired-sample Student’s t test (**P ≤ 0.01). Note: We have screened six independent lines, and all of them possessed a hybridizing signal just below the expected size, suggesting the likely formation of a truncated protein (black arrow). For immunoblot, samples were loaded at equal total protein amounts. Loading controls were stained by Coomassie blue.
To summarize, based on genetic complementation and isolation of an independent allelic mutant, we conclude that the defect in oil remobilization observed in mdh2-1 and mdh2-2 results from the absence of a functional MDH2 protein. MDH2 has been previously shown to be located in peroxisomes of Chlamydomonas (Hayashi and Shinozaki, 2012; Kong et al., 2017) and is expected to play a role in FA β-oxidation based on its function in plants (Pracharoenwattana et al., 2010). In peroxisomes, in addition to FA β-oxidation, the glyoxylate cycle also requires the activity of MDH. Defects in enzymes of glyoxylate cycle often result in strains unable to grow in the dark when acetate is supplied as the only carbon source (Plancke et al., 2014). It was observed here that mdh2-1 and mdh2-2 mutants grew normally in the dark either in liquid culture or on agar plates (Supplemental Figures 4A and 4B), ruling out that MDH2 is required for acetate utilization. Moreover, the mutants grew as well as dw15 when cultivated photoautotrophically in the presence of CO2-enriched air, but reached a lower cell density at stationary phase when cultivated mixotrophically (Supplemental Figures 4C and 4D).
mdh2 Mutants Overaccumulate TAG and Starch during Photoautotrophic N Deprivation (Supplemented with 2% CO2 in the Air)
Increasing evidence suggests the occurrence of lipid remobilization simultaneous to synthesis (Miller et al., 2010). For example, knocking down lipid catabolism has been demonstrated to lead to strains that overaccumulate TAGs (Trentacoste et al., 2013; Kong et al., 2017). Disruption in the first step of FA β-oxidation (i.e., ACX2) in Chlamydomonas resulted in a mutant strain (acx2) that accumulated 20% more TAG than its parental strain (dw15) during mixotrophic N deprivation (Kong et al., 2017). Furthermore, in this study, a ∼20% increase was detected under mixotrophic culture conditions in the two mutants in which MDH2 was knocked out (Figures 1A, 2D, and 3C), although total fatty acids remained similar between the three genotypes (dw15, mdh2-1, and C1) (Figure 2E).
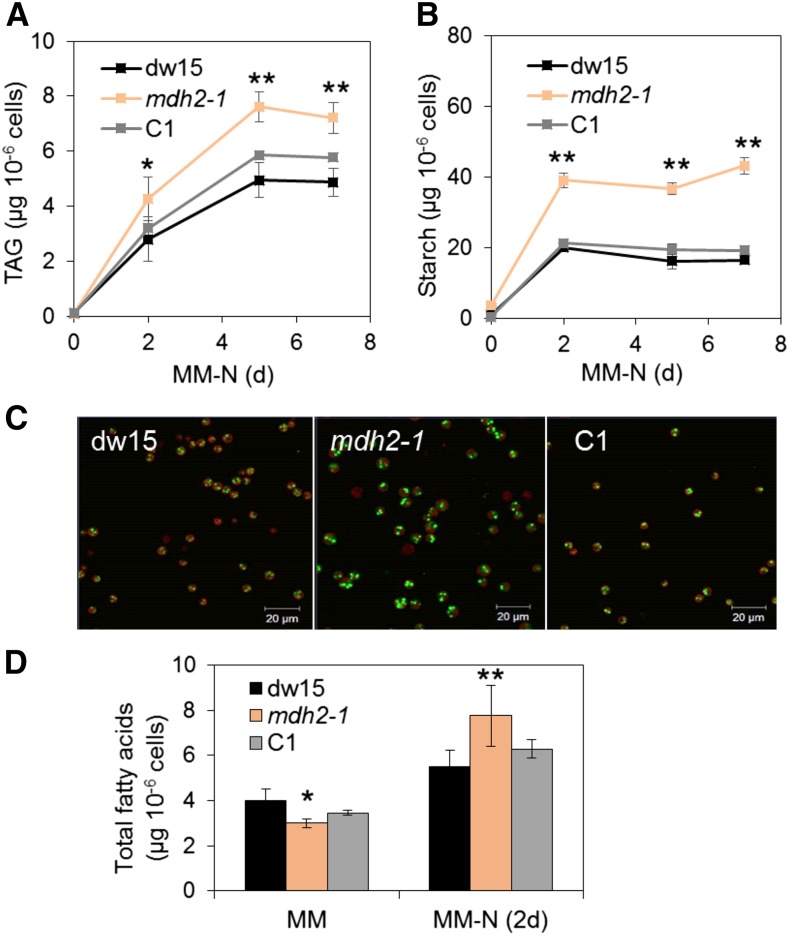
Based on immunoblot analysis, we observed that photoautotrophically grown wild-type cells made twice as much MDH2 protein than those grown mixotrophically (Supplemental Figure 5). Unless otherwise specified, cells were grown throughout this work under photoautotrophic conditions using air supplemented with 2% CO2 (Supplemental Figure 1B). To study the role of MDH2 under photoautotrophic condition, we followed TAG accumulation in N-starved cells cultivated under this condition. Interestingly, we found a much more dramatic increase in TAG when cultivated photoautotrophically, i.e., the mutant cells accumulated >50% more TAGs than dw15 following 2 d of N deprivation (Figure 4A). This is considerably greater than the 20% increase in mixotrophically grown N-deprived cells (Figures 1A and 2D). Accordingly, an increased number and size of lipid droplets (LDs) was observed in mdh2-1 by confocal microscopy in comparison to the control strains (Figure 4C). In addition, although mdh2-1 mutant cells were smaller during photoautotrophic N-sufficient growth (i.e., before the onset of N starvation), they grew bigger than the wild type after 2-d N starvation (Supplemental Figure 6A). To determine whether observed increases in TAG and starch content in mdh2-1 mutant result from metabolic effects or from an increase in cell size, we also plotted TAG and starch content on cellular volume basis (Supplemental Figures 6B and 6C). Despite the changes being less dramatic compared with the per cell basis, TAG and starch content per cellular volume were still 30% higher in the mdh2-1 mutant as compared with its control strains (dw15 and C1). Similar phenotypes (i.e., increase in TAG and LDs) were also observed in mdh2-2 in comparison to its parental strain (CC4533) on a cell basis (Figures 5A and 5D) or based on cellular volume (Supplemental Figures 7A and 7B).
Figure 4.
The mdh2-1 Mutant Overaccumulates TAG and Starch during Photoautotrophic N Deprivation.
(A) TAG content.
(B) Starch content.
(C) Confocal microscopy image of LDs in N-deprived cells (2 d).
(D) Total FA content.
Cells were cultivated under constant light in photoautotrophic conditions with additional supply of 2% CO2 in the air. Values are the mean of biological replicates (i.e., independent shaking flask cultures; n = 6, sd). Cells were stained with BODIPY, and pseudocolors were used: lipid droplet in green and chlorophyll in red. Asterisks represent statistically significant difference from both control strains (the parental line dw15 and C1) by paired-sample Student’s t test (*P ≤ 0.05 and ** P ≤ 0.01). C1, one representative complemented line.
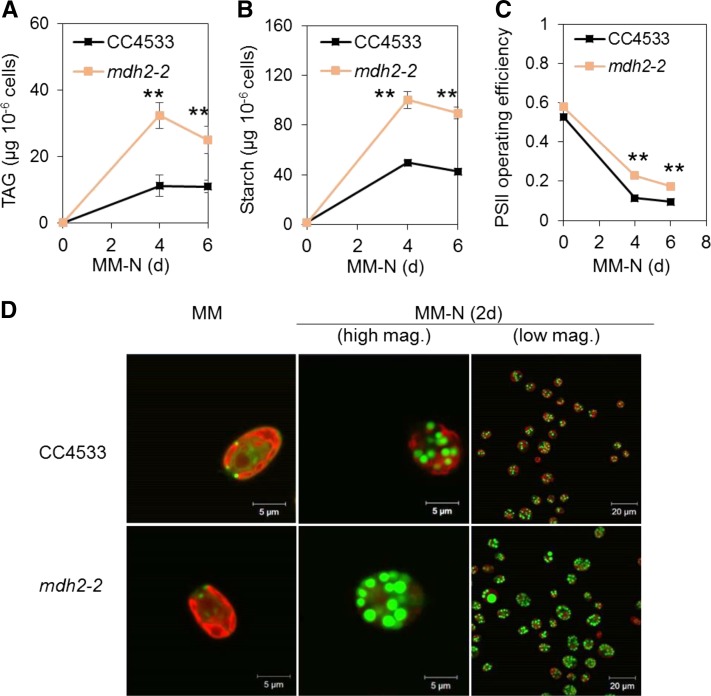
Figure 5.
mdh2-2 Overaccumulates TAG and Starch, and Possesses Higher Photosynthetic Activity during Photoautotrophic N Deprivation.
(A) TAG content.
(B) Starch content.
(C) PSII operating efficiency.
(D) LD imaging after cells being stained by BODIPY.
Cells were cultivated under constant light in photoautotrophic conditions with additional supply of CO2 at 2% in the air. Values are the mean of biological replicates (i.e., independent shaking flask cultures; n = 6, sd). Actinic light (200 µmol m−2 s−1) supplied by a red LED source was used for PSII yield measurement. Asterisks indicate statistically significant changes compared with the parental strain (CC4533) by paired-sample Student’s t test (**P ≤ 0.01). Cells were stained with BODIPY, and pseudocolors were used: lipid droplet in green and chlorophyll in red. mag., magnification.
Gas chromatography-mass spectrometry (GC-MS) analysis of total cellular FAs as their methyl esters (FAMEs) showed that there was a net increase in total cellular FAs in mdh2-1 following 2 d of N deprivation (Figure 4D), but interestingly there was no difference found when cells were cultivated under mixotrophic conditions (Figure 2E). It is worth noting here that prior to N deprivation, slightly smaller amounts of total FAs were present in the mdh2-1 mutant mostly due to its smaller cell size compared with the wild type. With the exception of an increase in 18:2(9,12), no significant change occurred in FA composition in comparison to the control strains (dw15 and C1) 2 d after photoautotrophic N starvation (Supplemental Figure 8). That said, during N-sufficient growth, the mdh2-1 mutant contains relatively higher amount of 16:0 and slightly lower amount of 18:3(9,12,15) than the control strains (dw15 and C1). Chlorophyll content in all strains was reduced upon N starvation, and the reduction was similar between all three strains (Supplemental Figures 6D and 6E).
Metabolic changes in mdh2 mutants were not limited to storage lipids. It was also found that the mutants accumulated twofold more starch than control strains (dw15 and C1) following 2 d of N deprivation (Figures 4B and 5B). By contrast, during mixotrophic N deprivation, starch accumulated to similar amounts in dw15 and the mdh2 mutants (Figure 2F). We therefore conclude that during photoautotrophic N starvation, higher TAG and starch accumulation observed in the mdh2 mutants are the consequence of two cumulative events, a metabolic effect and an increase in the cell size.
mdh2 Mutants Maintain a Higher Photosynthetic Activity during Photoautotrophic N Deprivation
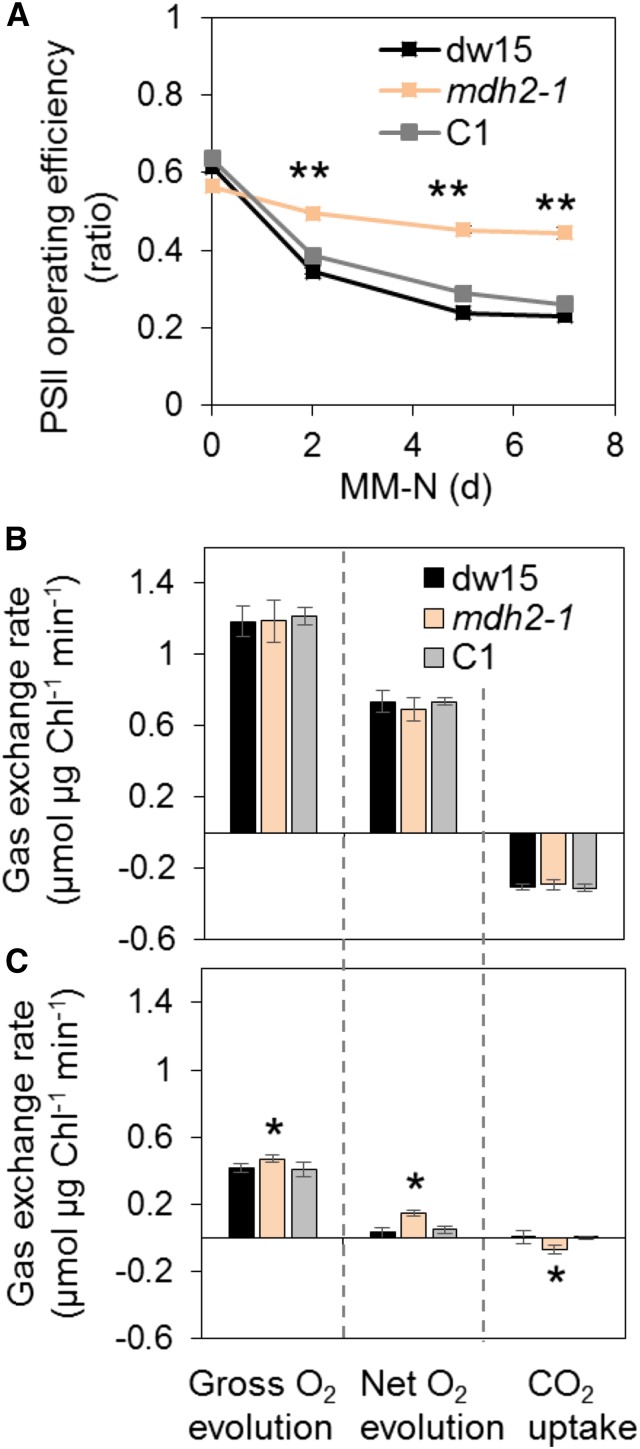
While the overaccumulation of TAG in mdh2 may not be surprising given the involvement of MDH2 in lipid catabolism, the effect on starch accumulation was more puzzling. To gain insight into the overaccumulation of storage compounds in mdh2 mutants, we measured photosynthetic parameters under photoautotrophic conditions during optimal growth and N deprivation. Photosynthetic performances (measured here as the photosystem II [PSII] operating efficiency) decreased in wild-type cells during N deprivation as previously reported (Figure 6A) (Saroussi et al., 2016; Schulz-Raffelt et al., 2016), but interestingly, the reduction in PSII yield was stronger in their respective parental lines than in the two mutants (Figures 5C and 6A). No significant difference in PSII yield could be observed when cells were cultivated in the presence of acetate (TAP condition) (Supplemental Figure 9). Measurement of O2 evolution and CO2 uptake using membrane inlet mass spectrometry (MIMS) demonstrated that gross and net photosynthetic O2 evolution and net CO2 uptake rates, although similar in N-replete conditions (Figure 6B), were significantly higher in the mdh2-1 mutant than in the control lines following 2 d of N deprivation (dw15 and C1) (Figure 6C).
Figure 6.
mdh2-1 Displays Higher Photosynthetic Yield during Photoautotrophic N Deprivation.
(A) Measurement of PSII operating efficiency.
(B) and (C) Measurements of O2 production and net CO2 fixation using MIMS in N-replete (B) and in N-deprived cells (C).
Cells were cultivated under constant light in photoautotrophic conditions with additional supply of 2% CO2 in the air. Values are the mean of biological replicates (i.e., independent shaking flask cultures; n = 3, sd). Actinic light (200 µmol m−2 s−1) supplied by a red LED was used for PSII yield measurement, whereas for MIMS analyses, light was supplied by a green LED source. Asterisks indicate statistically significant difference from both control strains (dw15 and C1) by paired-sample Student’s t test (*P ≤ 0.05 and **P ≤ 0.01). C1, one representative complemented line.
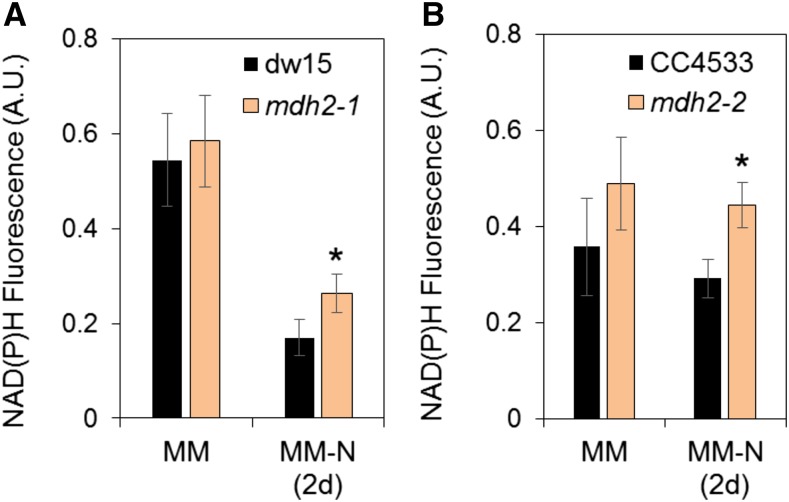
In addition to the increased rates of O2 production and CO2 fixation during N starvation, we also measured the production of NADPH upon the transition from dark to light by following changes in NAD(P)H fluorescence (Roach et al., 2015). This analysis revealed that while no significant difference between the strains was observed in N-replete conditions, both mdh2 mutants displayed a higher level of NADPH fluorescence under illumination in comparison to their corresponding parental strain following 2 d of photoautotrophic N deprivation (Figures 7A and 7B; Supplemental Figure 10). We conclude from these measurements that photosynthesis (measured as PSII efficiency, photosynthetic gas exchange rates, or NADPH accumulation) is less affected by N deprivation in mdh2 mutants than in the control lines.
Figure 7.
Measurement of NAD(P)H Fluorescence during Photoautotrophic N Starvation.
(A) NADPH fluorescence in mdh2-1 and its parental strain dw15.
(B) NADPH fluorescence in mdh2-2 and its parental strain CC4533.
Cells were grown to a constant OD under continuous light in photoautotrophic condition (+2% CO2 supplemented in the air), then NADPH fluorescence was measured before (MM) and after N deprivation (MM-N 2 d). Cells were kept in the dark for 1 min before an actinic light exposure (at 70 µmol photons m−2 m−1 provided by a red LED). Values are the mean of the NAD(P)H fluorescence level obtained across the 20-s light exposure (i.e., independent shaking flask cultures; n = 3, sd). Original NAD(P)H fluorescence traces are shown in Supplemental Figure 10. Asterisks indicate significant difference compared with their respective parental strains by paired-sample Student’s t test (*P ≤ 0.05). A.U., arbitrary unit.
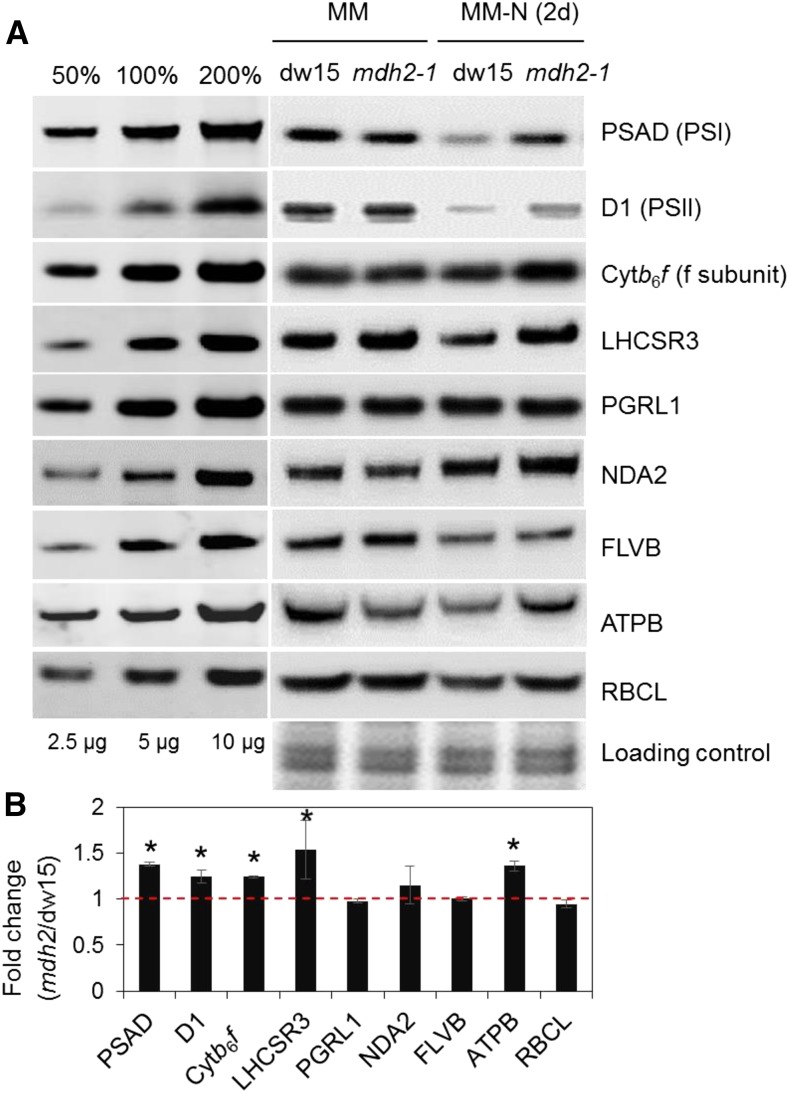
To better understand the higher photosynthetic activities in mdh2 mutants, we next determined the levels of representative photosynthetic proteins by immuno-analysis. While photosystem I (PSI) and PSII amounts probed by PSAD and D1 subunits, respectively, strongly decreased following 2 d of N deprivation, the decrease was less pronounced in the mdh2-1 mutant (Figure 8). Similar effects were observed on the light-harvesting complex stress-related 3 protein (LHCSR3), on the cytochrome b6f complex (probed by “the cytochrome f subunit”), and on the ATPase complex (probed by the ATPB subunit) albeit to a lesser extent than the photosystem proteins (Figure 8). Levels of other proteins, including the chloroplast type II NADPH dehydrogenase (NDA2), proton gradient regulation like 1 (PGRL1), flavodiiron protein (FLVB), and Rubisco large subunit (RBCL1) remained essentially unchanged. We conclude from this experiment that the general decrease in components of the photosynthetic machinery occurring during N deprivation in the control strains, and particularly in PSII and PSI amounts, is less pronounced in mdh2, which may at least partly explain the maintenance of higher photosynthetic performance in the mutant during N deprivation.
Figure 8.
Immunoblot Analyses of Photosystem Proteins during Photoautotrophic N Deprivation.
(A) Representative images of immunoblot analysis.
(B) Quantification of signal intensities from cells being N-starved for 2 d.
Proteins were collected from cells before (MM) and after photoautotrophic (+2% CO2 supplemented in the air) N deprivation (MM-N, 2d) under constant light. Representative images of immunoblot analysis are shown. Signals for (B) were averaged from three biological replicates (i.e., independent shaking flask cultures) for N-starved cells (n = 3; sd). Asterisks indicate significant difference from the parental strain dw15 by paired-sample Student’s t test (*P ≤ 0.05). Samples were loaded at equal total protein amounts and stained using Coomassie blue.
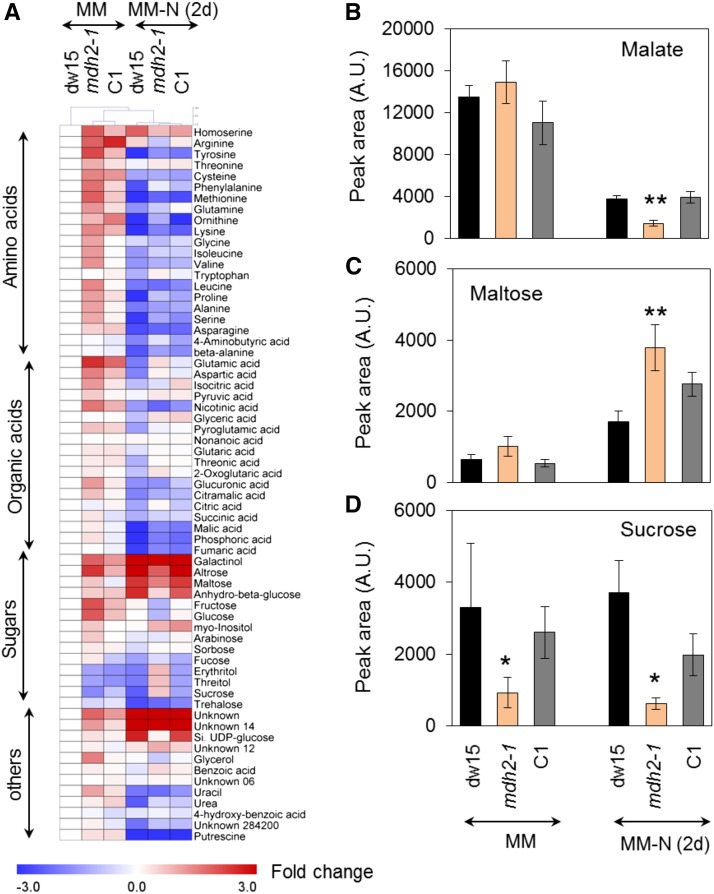
Metabolomics Analyses Reveal Reduction of Malate and Readjustment of Primary Metabolism in mdh2 during Photoautotrophic N Starvation
To determine if major changes in primary carbon metabolism occurred in mdh2 mutants, we performed metabolomics using a GC-MS-based method (Lisec et al., 2006). A total of 60 metabolites including organic acids, free amino acids, sugars, and their derivatives were quantified. In comparison to N-replete condition, over 60% of the total polar metabolites decreased in their cellular amount in N-deprived cells (Figure 9A; Supplemental Data Set 1). Furthermore, most organic acids and free amino acids were significantly more reduced in the mdh2-1 mutant than control strains (dw15 and C1) following 2 d of N deprivation, whereas the changes in sugars were more varied. No significant difference in the total content of malate, the product of MDH, was detected prior to N deprivation between mdh2-1 and control strains. In all strains, the malate content was reduced as a response to N deprivation, but the extent of reduction in mdh2-1 was far greater than that in control strains (dw15 and C1) (Figure 9B). A >60% reduction in intracellular malate content was observed in mdh2-1 in comparison to control strains (dw15 and C1) following 2 d of N deprivation. It is worth noting that, maltose, a breakdown product of starch degradation, was increased by 200% following 2 d of N deprivation in mdh2-1 compared with its control strains (Figure 9C). In addition, we observed a ∼80% decrease in sucrose content in the mdh2-1 mutant compared with its control strains following 2 d of N deprivation (Figure 9D).
Figure 9.
Metabolomics Analysis of Polar Metabolites in Photoautotrophically Grown Cells before and 2 d after N Deprivation.
(A) A heat map view of metabolic changes.
(B) Intracellular malate content.
(C) Intracellular maltose content.
(D) Intracellular sucrose content.
Cells were cultivated under constant light in photoautotrophic conditions with additional supply of 2% CO2 in the air. Values are the mean of biological replicates (i.e., independent shaking flask cultures; n = 8, sd). Asterisks indicate significant difference from control strains by paired-sample Student’s t test (*P ≤ 0.05 and **P ≤ 0.01). A.U., arbitrary unit.
The observed metabolic shift during N deprivation is in clear contrast to cells grown under optimal conditions, where most metabolites were found to be present in higher quantities in mdh2-1 than in their control strains (dw15 and C1) (Figure 9A, Supplemental Data Set 1). For example, with the exception of the two nonproteogenic amino acids (β-alanine and ornithine), which did not change significantly, most other free amino acids were present at levels that are 200 to 500% higher in mdh2-1 during N-replete growth. Yet, following 2 d of N deprivation, most free amino acid levels were present at much lower amount in the mdh2-1 mutant compared with its control strains (Figure 9A). Taken together, these results suggest that during response to N deficiency, greater metabolic adjustment occurs in the mutant compared with its parental strain.
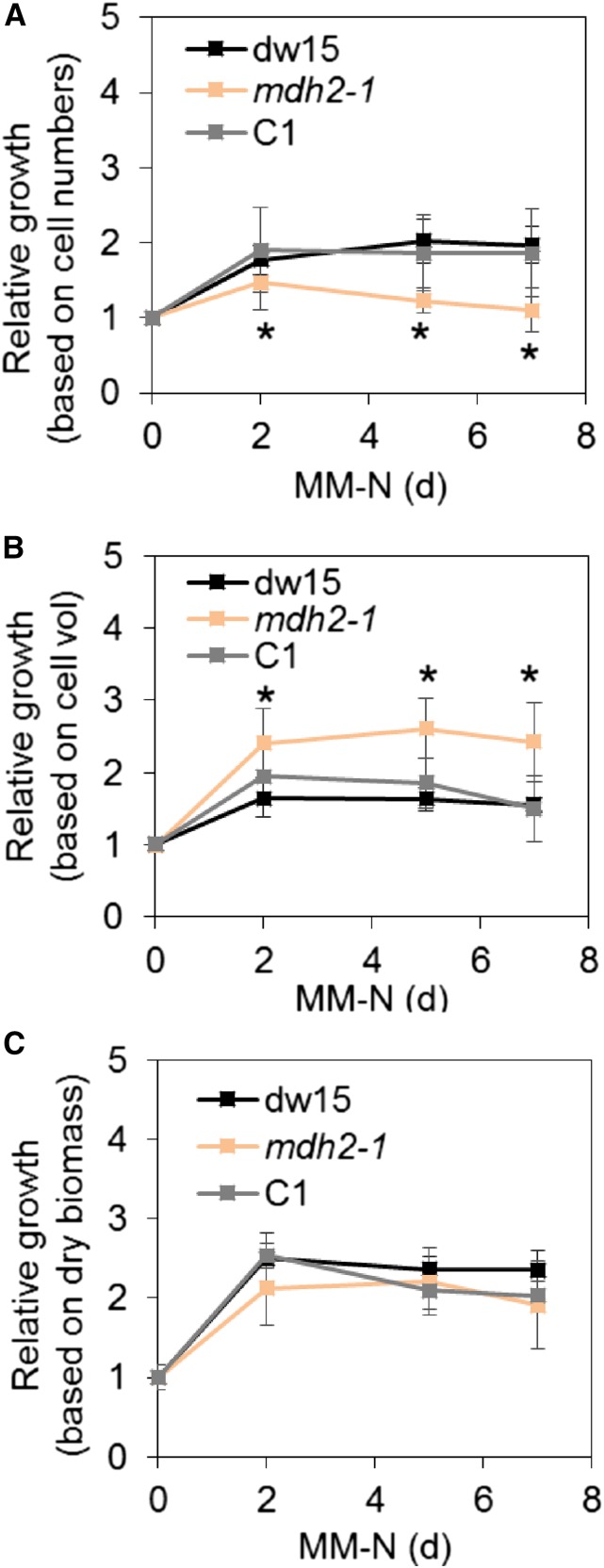
The mdh2-1 Mutant Is Delayed in Cell Division during N Deprivation but Overall Biomass Productivity Is Not Affected
Upon N starvation, the mdh2-1 mutant showed delayed cell division (1.5-fold increase in cell concentration) compared with the control strains (dw15 and C1), which typically show a doubling of the cell concentration 1 d after the onset of N deprivation under photoautotrophy (Figure 10A). However, on a cellular volume basis, growth was higher in the mdh2-1 mutant than in control strains (Figure 10B). Interestingly, there was no significant difference found when growth was evaluated on a dry biomass basis (Figure 10C). The increase in cell volume in the mdh2-1 mutant could be partly due to an impairment in cell division, due to the accumulation of carbon reserves, and partly due to the considerable increases in sugars and organic acids (Figure 9), some of which are known to alter cellular osmotic potential (Centeno et al., 2011; Araújo et al., 2012). The lack of an increase in dry biomass, in contrast to the increase in total cell volume, supports the latter hypothesis. Taken together, in spite of a slight retardation in cell division, there is no compromise in overall biomass productivity, possibly because the biomass of the mdh2-1 mutant is enriched in oil and starch (Figure 4).
Figure 10.
Cell Growth during Photoautotrophic N Deprivation.
(A) Relative growth based on cell number per milliliter of culture.
(B) Relative growth based on cell volume per milliliter of culture.
(C) Relative growth based on dry biomass per milliliter of culture.
Cells were cultivated under constant light in photoautotrophic N starvation conditions with additional supply of CO2 at 2% in the air. Cell growth was monitored every day using a Coulter counter. Then, cell concentration was normalized to that before N deprivation (which is set as 1). Values are the mean of biological replicates (i.e., independent shaking flask cultures; n = 11, sd). Asterisks indicate statistically significant difference from control strains by paired-sample Student’s t test (*P ≤ 0.05). Cell vol, cellular volume.
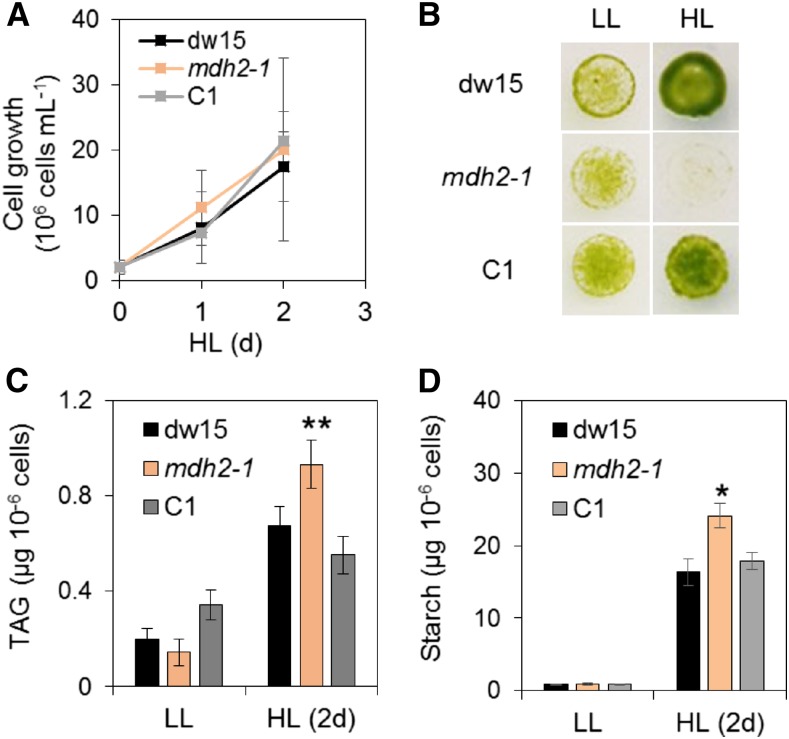
mdh2 Overaccumulates Starch upon HL Exposure and during Diurnal Growth
To determine if MDH2 has a function in other situations beyond N deprivation, we examined mutant behavior under HL, a condition frequently encountered by many algae. We observed that although cells grew at a similar rate in liquid cultures (Figure 11A), mdh2-1 is more sensitive to HL (500 µmol m−2 s−1) during photoautotrophic growth on agar plates than the control strains (dw15 and C1) visible 6 d after being exposed to HL, while the growth was similar under low light (LL; 50 µmol m−2 s−1) (Figure 11B). Such a difference in growth performance observed between solid and liquid cultures may be surprising but not unprecedented and have indeed also been observed in the pgrl1 mutant (Dang et al., 2014). Multiple physiological parameters could account for this difference among which the shading effect encountered quickly in liquid cultures as a result of cell doubling could explain a lack of growth difference between mdh2-1 and the wild type (i.e., dw15). Another reason might be due to the length of incubation, i.e., 2 d in liquid cultures versus 5 to 7 d in solid agar plate. Higher amounts of TAG and starch accumulated in all strains exposed to HL, which was consistent with previous observations in Chlamydomonas wild-type strains (Goold et al., 2016), yet the increase in TAG and starch content was even greater in mdh2-1 (Figures 11C and 11D).
Figure 11.
HL Response of the mdh2-1 Mutant during Photoautotrophic Growth.
(A) Growth kinetics in liquid cultures under HL.
(B) Growth comparison on agar plates.
(C) TAG content on a per cell basis.
(D) Starch content on a per cell basis.
Cells were cultivated under constant light (either HL or LL) in photoautotrophic conditions with additional supply of 2% CO2 in the air. Light was provided by a cool LED white light. The same number of cells were inoculated on MM agar plate and kept under continuous light to monitor cell growth. Images were taken 6 d after cells being deposited. Values are the mean of biological replicates (i.e., independent shaking flask cultures; n = 4, sd). Asterisks indicate significant difference from control strains by paired-sample Student’s t test (*P ≤ 0.05 and **P ≤ 0.01). LL, 50 µmol m−2 s−1; HL, 500 µmol m−2 s−1.
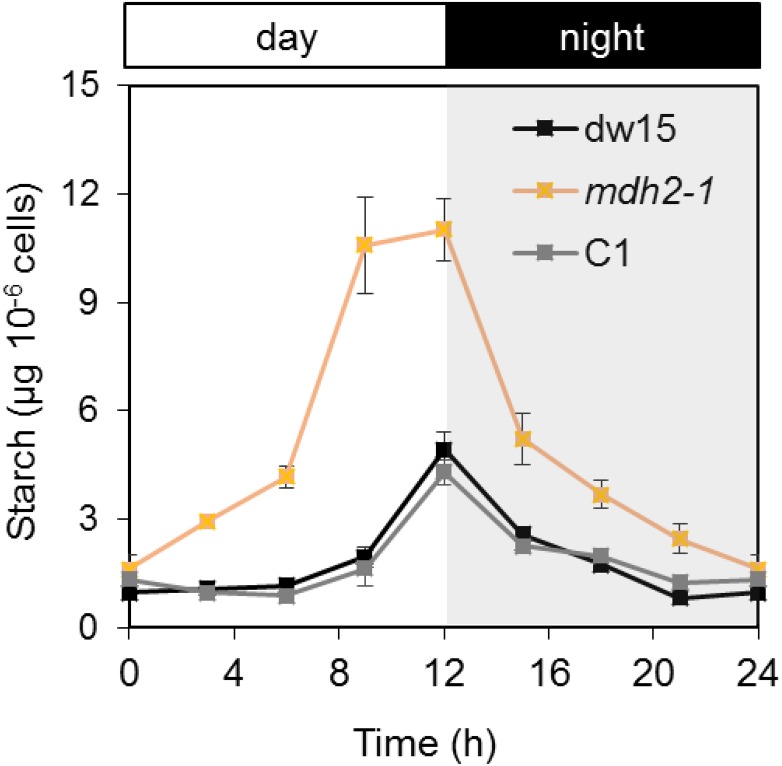
Changes in starch metabolism are not limited to N deprivation or HL. During diurnal growth, starch is accumulated during the day and consumed at night (Ball et al., 1990). Therefore, we also followed starch content in mdh2-1 and the control strains during a 12-h-light/12-h-dark cycle. We observed that starch accumulation followed the typical “bell shape” in all three strains, i.e., starch reached the highest level at the end of day and was degraded at night (Figure 12), but strikingly much higher amounts of starch (>300% increase at the end of day) were accumulated in the mutant. Intriguingly, at the end of night, the starch content returned to a similar level as found in the parental strain, which showed that mdh2-1 mutant degraded a greater amount of starch within the same time period, indicating a possible increase in catalytic activities of starch-degrading enzymes and implying that the circadian regulation of starch degradation is similar in Chlamydomonas as that previously reported in Arabidopsis thaliana (Smith and Stitt, 2007).
Figure 12.
Starch Accumulation during Photoautotrophic Diurnal Growth.
Cells were cultivated in a diurnal cycle of 12 h light/12 h dark in a controlled incubation chamber supplied with 2% CO2 in air. Light was supplied via fluorescent tubes at an intensity of 100 µmol m−2s−1. Values are the mean of biological replicates (i.e., independent shaking flask cultures; n = 4, sd). Shaded area refers to the night period.
mdh2-1 Produces Higher Amounts of H2O2 under N Starvation and HL as Well as during Diurnal Growth
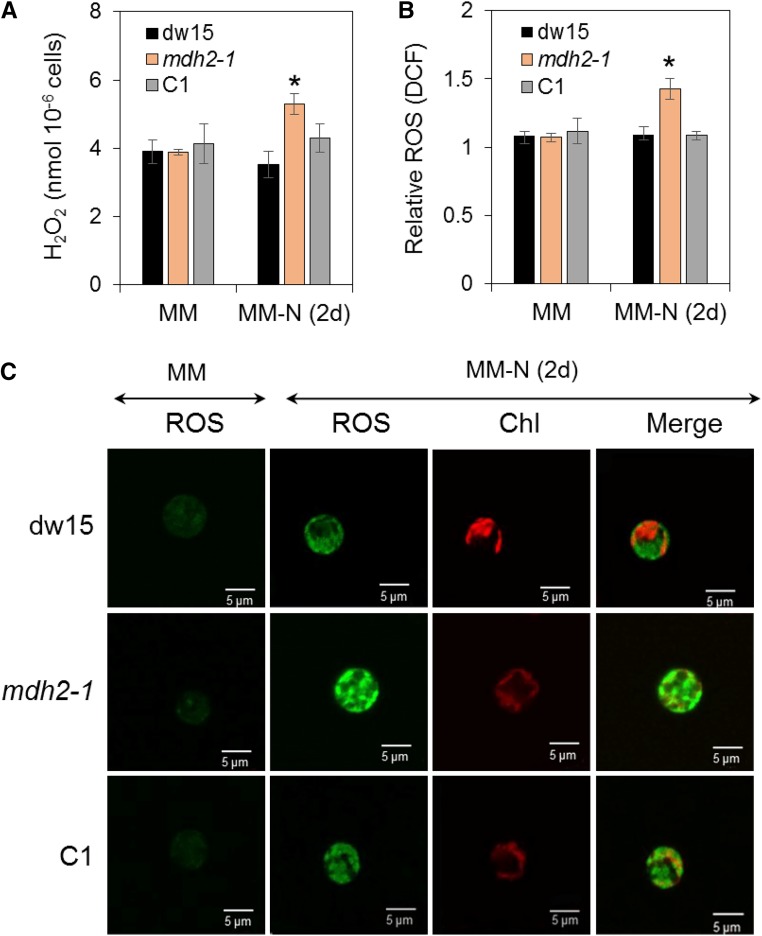
The absence of MDH2 is expected to hinder reoxidation of NADH produced by the 3-hydoxyacyl-CoA dehydrogenase activity of the β-oxidation spiral, therefore likely resulting in an overreduced peroxisome. The observed increases in de novo FA synthesis, photosynthetic activity, and starch accumulation suggested that the imbalance in peroxisomal redox status can be transmitted to chloroplast. H2O2 is a known cellular messenger. We therefore determined extracellular levels of H2O2 using the Amplex Red method (Allorent et al., 2013; Dang et al., 2014). The H2O2 level was 20% higher in mdh2 compared with control lines (dw15 and C1) following photoautotrophic N starvation (Figure 13A). The increase in the level of H2O2 in the mdh2-1 mutant triggered the production of higher amount of catalase 1 protein (CAT1) compared with wild-type strain during photoautotrophic N starvation (Supplemental Figure 11). To further investigate the overproduction of H2O2 by the mutant, we used the oxidant-sensing fluorescent probe 2',7'-dichlorodihydrofluorescein diacetate (H2DCFDA), which is converted to the green fluorescent dichlorofluorescein (DCF) upon exposure to conditions high in ROS. DCF fluorescence can be captured with a confocal microscope, and intensity of signal reflects the intracellular level of ROS including H2O2. Increased fluorescence was observed in all strains (dw15, mdh2, and C1) upon N starvation, conforming to previous observations on the increase in ROS level in N-deprived Chlorella sorokiniana (Zhang et al., 2013), Dunaliella salina (Yilancioglu et al., 2014), and Chlamydomonas (Du et al., 2018). Furthermore, the increase in ROS level upon N deprivation was even greater in mdh2 as observed by measurement of DCF fluorescence using a spectrofluorometer (Figure 13B) as well as under confocal imaging (Figure 13C). Nonuniform patches of green fluorescent spots were observed in cells of all genotypes, and although we could not be sure that they are present in the peroxisomes, it was clear that they are not limited to the chloroplasts. Furthermore, we also determined extracellular H2O2 levels in the parental strain and mutants upon HL exposure and during a diurnal growth. Whereas no difference was observed between strains under LL or in the dark, a 20 to 30% increase in H2O2 was observed in mdh2-1 compared with its control strains (dw15 and C1) after 1 d of HL exposure or during the light period (Supplemental Figure 12).
Figure 13.
Determination of H2O2 Level during Photoautotrophic N Deprivation.
(A) Extracellular H2O2 level determined by Amplex Red.
(B) Relative DCF fluorescence level.
(C) Intracellular ROS level determined using H2DCFDA staining.
Cells were cultivated under constant light in photoautotrophic conditions with additional supply of 2% CO2 in the air. Values are the mean of biological replicates (i.e., independent shaking flask cultures; n = 4, sd). Asterisks indicate significant difference compared with control strains (dw15 and C1) by paired-sample Student’s t test (*P < 0.05). C1, the complemented line.
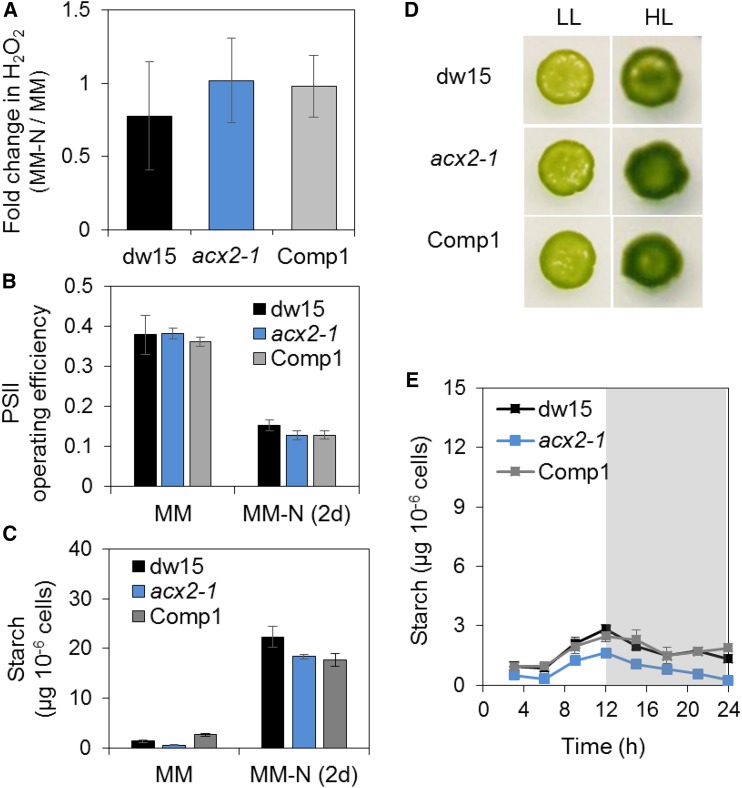
In plant/algal peroxisomes, H2O2 is produced by ACX, the initial step of β-oxidation (Graham, 2008; Kong et al., 2017). To identify the source of the observed increase in H2O2, we determined the level of ACX activity in cell-free protein extracts from mdh2-1 and its parental strain (dw15) using stearoyl-CoA (18:0) as a substrate given that it represents one of the major acyl-chain lengths in Chlamydomonas. ACX activity (18:0) was found to be almost twice higher in mdh2 than in dw15 following N-deprivation (2 d) (Supplemental Figure 13), consistent with the increased H2O2 production. To further explore a possible link between peroxisomal production of H2O2 by ACX and reserve formation, we also analyzed the acx2-1 mutant, which we have previously shown to be blocked in the H2O2-generating reaction of FA β-oxidation (Kong et al., 2017). We observed that under N deprivation, the acx2-1 mutant did not overaccumulate H2O2 and didn’t overproduce starch or show a difference in PSII operating efficiency from the control strains (Figures 14A to 14C). The lack of a reduction in H2O2 level in acx2-1 compared with its parental strain dw15 is not surprising given the occurrence of four other ACX isoforms in Chlamydomonas (Kong et al., 2017). Furthermore, we also examined the response of the acx2-1 mutant to HL exposure. The acx2-1 mutant grew as well as dw15 under HL (Figure 14D). Unlike the mdh2-1 mutant, where starch is overaccumulated during a diurnal growth period (+N), the acx2-1 mutant did not overaccumulate, or made even less, starch during the same period (Figure 14E). Overall, the observation on the increase of ACX (18:0) activity in mdh2-1 mutant together with the lack of starch overaccumulation in acx2 mutant therefore supported the idea that peroxisomal FA β-oxidation is a major source of H2O2 in mdh2.
Figure 14.
Effect of Silencing ACX2 on Starch Accumulation and HL Response.
(A) Fold change in H2O2 production in acx2-1.
(B) Operating PSII efficiency.
(C) Starch content before and after N deprivation (2 d).
(D) Light sensitivity test.
(E) Starch content during diurnal growth.
Cells were grown in liquid culture under constant light with additional supply of 2% CO2 and then the same number of cells was inoculated on MM agar plate and kept under continuous light (supplied by cool LED white light) to monitor cell growth at 25°C. Images were taken 7 d after cells being deposited. Actinic light (200 µmol m−2 s−1) supplied by a red LED source was used for PSII yield measurement. LL, 50 µmol m−2 s−1; HL, 500 µmol m−2 s−1. Shown are the parental strain dw15, acx2-1, and one complemented line (Comp1). Shaded area refers to the night period. Values are the mean of biological replicates (i.e., independent shaking flask cultures; n = 4, sd).
H2O2 Supplementation Results in TAG and Starch Overaccumulation in the mdh2-1 Mutant
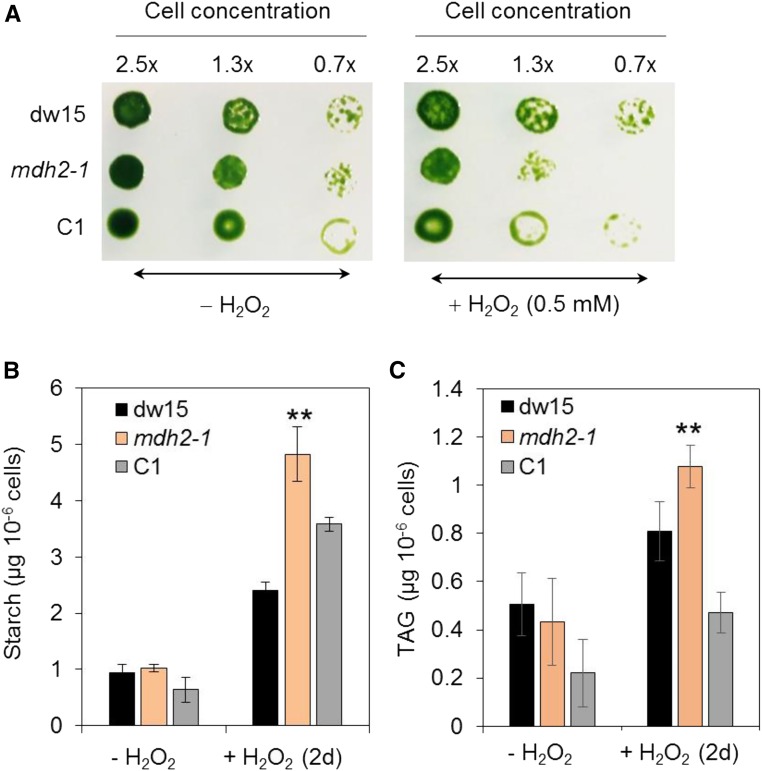
To further evaluate the idea that oil and starch over-accumulation observed in mdh2 may result from an increased H2O2, we determined TAG and starch contents in cells supplemented with H2O2. Considering the dual role of H2O2 on cell physiology and metabolism, we first tested the effect of various H2O2 concentrations on growth of the wild-type strain. We chose to use a final concentration of 0.5 mM H2O2 because at this concentration, cells were still growing and did not show any obvious deleterious effects seen with higher concentrations (Supplemental Figure 14A). Cell growth of all strains in the presence of H2O2 supplementation was similar in liquid cultures (Supplemental Figure 14B), whereas when cells were plated out in solid agar plate in serial dilutions, the mdh2-1 mutant showed increased sensitivity in growth compared with its parental line visible 7 d after being deposited (Figure 15A). This might be due to a cumulative effect of light and H2O2 supplementation encountered more severely in solid cultures.
Figure 15.
Effect of H2O2 Supplementation on TAG and Starch Content during Photoautotrophic Growth.
(A) Cell growth.
(B) Starch content per cell.
(C) TAG content per cell.
Cells were cultivated under constant light in photoautotrophic conditions with additional supply of 2% CO2 in the air. Cells were collected before and 2 d after H2O2 addition (0.5 mM). A given number of cells was deposited on MM agar plates with or without addition of H2O2. Images were taken 7 d after cells being deposited. Values are the mean of biological replicates (i.e., independent shaking flask cultures; n = 4, sd). Asterisks indicate statistically significant difference from control strains (dw15 and C1) by paired-sample Student’s t test (**P ≤ 0.01).
We then compared the capacity of all three strains in accumulating starch and TAG following H2O2 supplementation. In the presence of exogenously added H2O2, it was found that starch increased >200%, whereas TAG increased by 50% in the wild type 2 d after H2O2 supplementation (Figures 15B and 15C). We further noticed that the increases in starch and TAG contents in the mdh2-1 mutant were much greater (>30% more) than those of control strains (dw15 and C1) (Figures 15B and 15C).
DISCUSSION
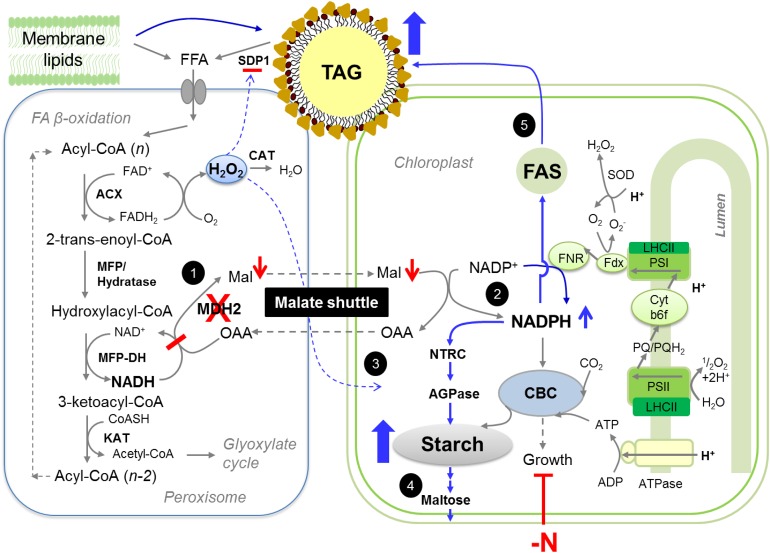
In all eukaryotic cells, biochemical reactions are compartmentalized in specific subcellular organelles rendering the coordination of metabolism across different compartments essential for cell functioning (Sweetlove and Fernie, 2013). The chloroplast, the major power house of photoautotrophs, is often found located in close proximity to peroxisomes (Hayashi and Shinozaki, 2012; Hu et al., 2012; Schwarz et al., 2017), but little is known about the interaction and exchange of information between them in algae. Here, via characterization of two knockout mutants of the peroxisomal MDH2 in the unicellular green alga Chlamydomonas, we provide evidence that (1) FA β-oxidation requires a functional MDH2; (2) lipid catabolism is connected to photosynthesis and chloroplast metabolism through MDH2; (3) the reverse coupling of redox/H2O2 from peroxisome to chloroplast is essential for cell division during N deprivation or HL exposure; and (4) MDH2 plays a role in modulating starch metabolism during diurnal growth. This study thus reveals regulation of chloroplast-based activities by factors derived from peroxisomes. We further identify malate and H2O2 as important players in this process. Below, we discuss and provide a working model explaining metabolic reorientations occurring in the absence of MDH2 in Chlamydomonas (Figure 16). The logics and reasoning in supporting this model are provided below.
Figure 16.
Tentative Model Explaining Redirection of Metabolism in the Absence of MDH2 during Photoautotrophic N Deprivation.
FA degradation starts with hydrolysis of TAGs and membrane lipids by lipases. FAs released enter the peroxisomes via an ABC transporter and subsequently are degraded to acetyl-CoAs by the core FA β-oxidation spiral, which consists of four enzymatic activities (ACX, MFP-hydratase, MFP-DH, and KAT). MDH2 plays a role in oxidation of NADH, which is generated by hydroxylacyl-CoA dehydrogenase (MFP-DH) at the third step of β-oxidation spiral. In the absence of MDH2, NADH is likely accumulated, thereby increasing the reduction state of peroxisome. This would result in two metabolic changes in peroxisome: a decrease in malate export and an increase in H2O2 level. These two metabolites would in turn significantly alter photosynthesis and chloroplast metabolism. Activated pathways in the mutant are indicated by blue arrows, whereas downregulated pathways are indicated by red arrows. Based on results obtained from this study and current literature, we propose a cascade of events leading to the observed phenotypes in mdh2 mutants: “High oil” phenotype (events 1, 2, and 5): A block in FA β-oxidation combined with increased de novo FA synthesis contributes to higher TAG accumulation. “High starch” phenotype (events 2 to 4): During N starvation, the more active CO2 fixation and photosynthesis in the mdh2-1 mutant provides more NADPH and carbon precursors for starch synthesis. The increased level of NADPH in chloroplast activates AGPase for starch synthesis and also activates several starch-degrading enzymes; therefore, it results in an increased carbon flux into and out of the starch route. Higher amounts of starch indicate high sink capacity, therefore sustaining photosynthesis. As a consequence, more NADPH is produced by photochemical reactions, and this further activates AGPase, together resulting in the 100 to 300% increase in starch content. “High H2O2” phenotype (event 3): The higher level of H2O2 in the mutant is supported by the 2-fold increase in the H2O2-generating reaction catalyzed by ACX activity. Once it is transmitted to chloroplast, H2O2 activates the starch synthesis pathway, as supported further by the observation of starch overaccumulation in wild-type cells supplemented with exogenous H2O2. “Sustained photosynthesis” (events 1 and 2): A blockage in FA β-oxidation at the step of MDH2 will result in less malate being produced in the peroxisome. Malate, a recognized electron carrier, can be transported to other compartments via the dicarboxylate transporter. When malate level is high, less NADP+ is available as electron acceptor at PSI, therefore downregulating photosynthesis. Conversely, in the absence of MDH2, the malate level is decreased, and more NADP+ is available to accept electrons from PSI. ACX, acyl-CoA oxidase; CAT, catalase; CBC, Calvin-Benson-Bassham cycle; CTS1, comatose 1; DH, dehydrogenase; FAD, flavin adenine dinucleotide; FFA, free fatty acid; Fdx, ferredoxin; FNR, ferredoxin-NADP+ reductase; KAT, ketoacyl-CoA thiolase; LHC, light-harvesting complex; Mal, malate; MFP, multifunctional protein; NTRC, NADP:thioredoxin reductase C; OAA, oxaloacetate; PQ, plastoquinone; SDP1, sugar dependent 1; SOD, superoxide dismutase.
MDH2 Is a Major Contributor to NAD+ Regeneration in Peroxisomal FA β-Oxidation
MDH2 has been localized to peroxisomes in Chlamydomonas by three independent studies (Hayashi and Shinozaki, 2012; Lauersen et al., 2016; Kong et al., 2017) and has further been observed that MDH2 colocalized with ACX2, the first enzyme of the β-oxidation cycle (Kong et al., 2017). The defect in TAG hydrolysis reported here together with a peroxisomal localization strongly supports the involvement of MDH2 in FA β-oxidation (Figure 16, event 1). Moreover, MDH2 showed the highest amino acid sequence similarity (>55% identity, BLAST) to the two Arabidopsis peroxisomal proteins AtpMDH1 and AtpMDH2 (Supplemental Table 1). Interestingly, the double Arabidopsis mutant (pmdh1 pmdh2) lost the capacity to remobilize TAG during seed germination (Pracharoenwattana et al., 2007). A defect in lipid catabolism has also been observed in a mutant of Saccharomyces cerevisiae deficient in the peroxisomal MDH3 (van Roermund et al., 1995). Taken together, these studies demonstrate that the metabolism of NADH generated via FA β-oxidation in peroxisomes is conserved between fungi, plants, and algae, and similar to the other two systems, the algal peroxisomal membranes are not permeable to NAD(H), thus prohibiting free exchange with cytoplasm where de novo NAD+ synthesis occurs (Noctor et al., 2006).
Oil hydrolysis is severely but not completely blocked in the mdh2 mutants (Figure 1). This suggests the possible occurrence of compensatory pathways in providing NAD+ for β-oxidation. Indeed, a number of other mechanisms controlling NAD+ homeostasis have been reported to operate in plant peroxisomes. Studies in Arabidopsis have shown that FA β-oxidation can be supported by peroxisomal hydroxypyruvate reductase (HPR) when AtpMDH1 and AtpMDH2 are absent (Pracharoenwattana et al., 2010). Plant peroxisomes are also known to contain high amounts of the ascorbate peroxidase (APX)/monodehydroascorbate reductase (MDAR1) electron transfer system, which has been reported to play a role in NADH reoxidation as well as in detoxifying H2O2 (Eastmond, 2007). In addition, an Arabidopsis peroxisomal NAD+ carrier (PXN) has been demonstrated to operate in the import of NAD+ into the peroxisomes from the cytoplasm (Bernhardt et al., 2012; van Roermund et al., 2016), and knockout mutants for PXN are defective in oil breakdown during seedling establishment. Genes homologous to HPR (Cre06.g295450), APX (Cre09.g401886), MDAR1 (Cre17.g712100), and PXN (Cre07.g353300) are encoded in the genome of Chlamydomonas (Merchant et al., 2007), and their potential involvement in lipid catabolism in algae is further supported by a recent transcriptomic study where their transcriptions were dysregulated in a mutant defective in oil hydrolysis following N resupply (i.e., the cht7 mutant, compromised in hydrolysis of TAG) (Tsai et al., 2014, 2018). That said, none of the algal proteins has yet been functionally characterized. Nevertheless, these alternative mechanisms in any case do not appear to play a major role, at least under the conditions tested in this study, since mdh2 mutants lost >80% of their capacity to remobilize TAG reserves.
The Role of Peroxisomal MDH2 in Fine-Tuning of Photosynthesis during Photoautotrophic N Deprivation
Chlamydomonas reduces its electron transport activities to adapt to a reduced metabolic need when N is depleted from the medium (Schmollinger et al., 2014; Park et al., 2015; Saroussi et al., 2016). Structural changes and transcriptomic responses during this adaptation have been reported (Schmollinger et al., 2014; Saroussi et al., 2016), but little is known concerning the underlying regulatory mechanisms. Findings in this study suggest that the peroxisomal MDH2 through participation in peroxisomal FA β-oxidation and NAD+ metabolism is, at least partly, involved in downregulating photosynthesis in N-deprived Chlamydomonas. This conclusion is evidenced by the fact that in the absence of MDH2, cells sustained a higher photosynthetic electron transport activity and generated higher amounts of NADPH during photoautotrophic N starvation than control strains (Figures 6, 7, and 16, event 2).
Two major metabolic changes could result in more sustained photosynthesis in mdh2, namely, a reduction in malate and an increase in starch. First, malate is a recognized electron carrier and can be transported across subcellular membranes by dicarboxylate transporters (Mettler and Beevers, 1980). In wild-type cells when malate level is high, less NADP+ is available as acceptor of electrons released from PSI; therefore, photosynthesis is downregulated to match to the reduced metabolic needs of the cell. By contrast, in the absence of MDH2, the malate level is lower and more NADP+ is available to accept electrons from PSI, thereby enhancing electron transport activities and producing more NADPH. A lower malate level in the chloroplast of mdh2 mutants may result from impaired shuttling of malate between peroxisomes and chloroplasts, such shuttling being possible thanks to a concerted functioning of both peroxisomal and plastidial MDHs. Although the redox-regulated plastidial MDH5 has been often reported to function as a malate valve exporting reducing power out of the chloroplast (Scheibe, 2004; Hebbelmann et al., 2012; Heyno et al., 2014), the enzymatic activity of NADP-MDH is highly reversible, and it is therefore conceivable that depending on the metabolic conditions (for instance, N availability and/or light intensity) and depending on the cellular redox poise the shuttle may work in one direction or in the other. Moreover, in addition to the redox-regulated NADP-MDH, plants and algal chloroplasts contain a NAD-MDH (MDH1) (Supplemental Table 1), which is not redox regulated and not involved in the functioning of the malate valve, but supports other metabolic functions (Beeler et al., 2014; Selinski and Scheibe, 2014).
Second, the higher photosynthetic activities of mdh2 (in comparison to the wild type) occurring during photoautotrophic N starvation could be due to increased sink strength since starch accumulates in mdh2 mutants (Figure 4), and starch is an excellent electron sink. When facing harsh conditions (nutrient deprivation or HL), the limiting factor for photosynthesis is the availability of electron acceptors. The observed increase in starch synthesis in mdh2 will consume a large amount of NADPH, therefore regenerating NADP+ for photochemical reactions, resulting in more sustained photosynthesis during photoautotrophic N starvation compared with the wild type. In addition, the chloroplast redox state has been reported to induce the expression of proteins of the photosynthetic apparatus (Oswald et al., 2001), which might explain the higher levels of these proteins observed in the mutant than the wild type during N starvation (Figure 8). The lower sucrose level present in the mdh2-1 mutant (Figure 9) could also be involved in sustaining photosynthesis, via relieving feedback regulation of photosynthesis, as has been reported previously in land plants (Pfannschmidt et al., 1999; Lobo et al., 2015). Together, the different metabolic adjustments occurring in mdh2 during photoautotrophic N starvation favor a higher photosynthetic electron flow and higher CO2 fixation into organic carbon as compared with the parental strains.
Malate, H2O2 and Relationships to Starch Metabolism during Photoautotrophic N Deprivation
In addition to lipids, we detected large amounts of starch in the mutants under N deprivation, HL, and during diurnal growth (Figures 4, 5, 11, and 15). In parallel, a reduction in malate and an increase in the levels of maltose, NADPH, and H2O2 was observed (Figures 7, 9, and 13). Together, these results point to a link between malate content, H2O2 level, NADP+ reduction state, and starch metabolism (Figure 16, event 3). A negative correlation between malate and starch content has been previously observed in potato tubers (Solanum tuberosum) and tomato fruits (Solanum lycopersicum) when mitochondrial malate metabolism is disturbed (Jenner et al., 2001; Centeno et al., 2011), and elevated photosynthesis has been reported following downregulation of the mitochondrial malate dehydrogenase in tomato (Nunes-Nesi et al., 2005). However, the identity of intracellular signal(s) involved in such a connection has not been established. Compared with previous reports, our work brings in two new aspects: (1) It provides an example in which chloroplast starch metabolism can be modulated by malate metabolism in peroxisomes; and (2) it provides evidence that diurnal starch metabolism can be modulated by a peroxisome-located protein.
Regulation of starch synthesis is known to occur at the level of the first and rate-limiting step catalyzed by ADP-glucose pyrophosphorylase (AGPase), which is under multiple regulations including principally allosteric and redox regulation (Geigenberger, 2011). Reductive activation of AGPase has been shown to occur in tomato as well as Chlamydomonas (Iglesias et al., 1994; Libessart et al., 1995; Buléon et al., 1997) and is mediated by a chloroplast-located NADPH-dependent thioredoxin reductase C (NTRC) (Michalska et al., 2009; Geigenberger, 2011; Lepistö et al., 2013; Naranjo et al., 2016; Thormählen et al., 2017). The increased reduction state of mdh2 chloroplasts (i.e., higher NADPH level) could activate AGPase via NTRC (Figure 16, event 3). In parallel, the higher CO2 fixation of mdh2 (Figure 6) results in an increased production of photosynthates through the CBC cycle; these two effects (enzyme activation and substrate availability) result in a boost of starch synthesis in mdh2 mutants.
Moreover, a positive correlation between H2O2 and starch amounts was observed, which is supported by three lines of evidence. (1) The H2O2 level was increased in mdh2 mutants together with enhanced starch accumulation. 2) When the H2O2 level remained similar to the wild type, i.e., in the case of the acx2-1 mutant, starch was not overaccumulated. (3) When wild-type cells were supplied with exogenous H2O2, starch was overaccumulated. This report thus establishes a positive correlation between starch amount and H2O2. The biochemical observation made here is supported by a recent H2O2-induced transcriptomic response study showing that mRNA abundance of several starch synthesis genes is increased in H2O2-supplemented cells (Blaby et al., 2015). Nevertheless, the underlying molecular mechanisms linking H2O2 and starch still need to be worked out.
Metabolomics analysis revealed that maltose and glucose, the major products of starch degradation, were dramatically increased in mdh2 in comparison to its parental strain. Moreover, despite the fact that more starch was made by the end of day in mdh2, the level of starch at the end of night period fell to the same level as those of control strains, indicating accelerated activities of starch-degrading enzymes. These two sets of data thus point to an increase in starch degradation in mdh2 (Figure 16, event 4). This could be explained by the elevated levels of NADPH present in mdh2 because several enzymes of the starch degradation pathway, notably, an α-glucan water dikinase, a β-amylase, and a phosphatase, have been shown to be under redox regulation (Mikkelsen et al., 2005; Sparla et al., 2006; Comparot-Moss et al., 2010; Geigenberger, 2011). The concomitant increase in synthesis and degradation of starch seems, at first sight, puzzling. Actually, increasing knowledge reveals the occurrence of starch degradation activities simultaneous to starch synthesis in the light, in specific cell types, or under particular environmental conditions (Sparla et al., 2006; Baslam et al., 2017; Daloso et al., 2017).
In parallel to energetic changes, the increase in CO2 assimilation in mdh2 results in higher production of precursors for the synthesis of carbon reserves i.e., starch and TAG. Together, the increased synthesis and turnover of starch in mdh2 led to the spectacular “high starch high maltose” phenotype. Adding to the already complex regulatory mechanisms of starch synthesis, our work provides a demonstration of the regulation of chloroplast starch synthesis and turnover by signal (and metabolite) derived from the peroxisomes.
Increased TAG Amount in N-Deprived mdh2
In this study, we found that significant amounts of TAGs are made in N-deprived mdh2 cells (Figures 1, 4, and 5). Upon N deprivation, a block in β-oxidation diverts otherwise toxic free FAs released from membrane lipids to make storage lipid. This is supported by two recent observations in Chlamydomonas, i.e., in acx2 mutants (Kong et al., 2017), as well as in the icl (isocitrate lyase) mutant that is blocked in glyoxylate cycle that functions downstream of β-oxidation allowing conversion of FAs to sugars (Plancke et al., 2014). However, this can only partly explain the increased TAG content in mdh2 under photoautotrophic N deprivation because we found that the increase in TAG content in mdh2 is much more dramatic under photoautotrophic N deprivation (50–100%) than under mixotrophic N deprivation (∼20%). The dramatic increase in TAG content in mdh2 under photoautotrophic conditions can partly be accredited to enhanced de novo FA synthesis, as suggested by the increase in total FA amount under photoautotrophy but not mixotrophy (Figures 2E and 4D). The difference in the photosynthetic responses during N starvation under mixo- versus photoautotrophic conditions supports an increased production of NADPH (and potentially total FA) only in photoautotrophically N-starved cells (Figure 4; Supplemental Figure 9). Thus, the additional increase in TAG content in photoautotrophically N-starved mdh2-1 cells is likely due to an increased de novo FA synthesis (Figure 16, event 5).
In mdh2, enhanced de novo FA synthesis can be attributed to the increased supply of carbon precursors as well as of the reducing equivalent NADPH. De novo FA synthesis occurs in the chloroplast of algae and plants and is catalyzed by fatty acid synthase (FAS), and this reaction requires a stoichiometric supply of carbon precursors, ATP and NAD(P)H (Ohlrogge and Browse, 1995; Li-Beisson et al., 2013). Enhanced CO2 assimilation in mdh2 will provide more organic carbon precursors fed into the FAS complex for FA synthesis. This conforms to the previous finding that carbon precursor supply for FA synthesis controls TAG accumulation in Chlamydomonas (Goodson et al., 2011; Fan et al., 2012).
The FAS complex is known to be activated by NADPH, which was present at higher levels in mdh2 as a result of enhanced photosynthetic activities and increased starch turnover. Indeed, a positive link between the level of NADPH and FA amount has been established in fungi, plants, and algae (Zhang et al., 2007; Xue et al., 2016). For instance, it has been observed that de novo FA synthesis can be enhanced via transgenic overexpression of an NADPH-producing malic enzyme in the diatom Phaeodactylum tricornutum and in the green alga Chlorella pyrenoidosa (Xue et al., 2015, 2016). By contrast, reducing the NADPH supply via inhibition of malic enzyme activities using sesamol in Haematococcus pluvialis, Nannochloropsis sp, and the filamentous fungus Mucor circinelloides led to reduction in total FAs (Wynn et al., 1997; Recht et al., 2012).
In addition, the increased TAG level in mdh2 could also be due to overproduction of H2O2. Indeed, a study in Arabidopsis has shown that H2O2 released from the peroxisomes defective in the APX/MDAR system inhibits the major TAG lipase SUGAR-DEPENDENT1, therefore resulting in impaired TAG hydrolysis during seed germination and postgerminative growth (Eastmond, 2007). Taken together, the dramatic increase in TAG content in photoautotrophic N-deprived mdh2 is a combinatorial result of downregulating FA β-oxidation and TAG lipolysis and enhancing their de novo synthesis due to increased availability of carbon precursors as well as reducing equivalent NADPH.
Conclusion
In summary, although the peroxisome was described >60 years ago in kidney (Rhodin, 1954), found later in plants (Beevers, 1979) and only fairly recently confirmed in Chlamydomonas (Hayashi and Shinozaki, 2012), the role of the peroxisomes as signaling organelle has just started to be recognized in mammals (Tripathi and Walker, 2016). We still know comparatively little about the role of peroxisomes in photoautotrophs beyond photorespiration in land plants. In addition to revealing previously unreported functions of peroxisome-chloroplast interactions in algal lipid metabolism (β-oxidation and de novo FA synthesis), this study uncovered an important function of the peroxisome in photoautotrophs in exerting control on photosynthesis and chloroplast metabolism. The proper exchange of information (malate/H2O2) from the peroxisome to the chloroplast is essential for cell growth especially when facing harsh environmental conditions. The peroxisomal H2O2 is likely the missing link between environmental stress, metabolism, and redox balance. This study also points out the importance of peroxisomes in the already complex landscape of chloroplast- and mitochondrion-based electron dissipation mechanisms. Findings from this study on the unicellular green model alga Chlamydomonas should be extendable to photosynthetic tissues of land plants where photosynthesis occurs alongside the formation of carbon reserves in the same cell. Our results therefore may suggest additional strategies for engineering plants and algae for improved biomass production and stress tolerance.
METHODS
Strains and Culture Conditions
The cell wall-less strain dw15.1 (CC4619 cw15, nit1, mt+; abbreviated as dw15 throughout this article), kindly provided by Christoph Benning, was used as the parental strain for mutant library generation (Li et al., 2012). The mutant mdh2-1 was isolated following the previously published protocol (Cagnon et al., 2013). The mdh2-2 mutant (LMJ.RY0402.208051) together with its background strain CC4533 was purchased from the CLiP collection at (https://www.chlamylibrary.org) (Li et al., 2016). Unless otherwise specified, cells were routinely cultivated in an incubation shaker (INFORS Multitron pro) maintained at 25°C, with 100 rpm shaking and constant illumination at 100 µmol m−2 s−1. Except the HL experiment (500 μmol m−2 s−1), which was provided by a cool LED white light, all other lightings used for cell cultivation in the INFORS were supplied by fluorescent tubes (Fluora Osram). Photoautotrophic growth refers to cells grown in MOPS-buffered minimal medium (MM) supplemented with 2% CO2 in the air (Harris, 2009; Schulz-Raffelt et al., 2016), whereas mixotrophic growth refers to cells cultivated in Tris-acetate-phosphate (TAP) medium (Harris, 2009). With the exception of TAG remobilization experiment (performed in the dark in air), all other cultures were grown in MM-MOPS supplemented with 2% CO2 in the air (Supplemental Figure 1). Cell concentration, cell size, and cellular volume were monitored with a Multisizer 3 Coulter counter (Beckman Coulter). For drop test, a series of diluted samples was either spotted on TAP plate for heterotrophic growth or spotted on MM agar plates and incubated for 5 to 10 d under either LL (50 μmol m−2 s−1) or HL (500 μmol m−2 s−1) supplied with a cool LED white light at 25°C. For growth test in the presence of H2O2, a given number of cells were spotted on MM agar plates with or without H2O2 supplementation (at a final concentration of 0.5 mM). For N deprivation, exponentially grown cells were centrifuged at 600g for 5 min, and cell pellets were washed twice with N-free media (TAP-N or MM-N) before being resuspended in N-free media for starvation experiments. Unless otherwise stated, biological replicates refer to cells grown in independent Erlenmeyer flasks.
Starch and Chlorophyll Quantification
Starch and chlorophyll were quantified from 1 mL of culture containing ∼5 million cells. Briefly, cell culture was centrifuged in a microfuge tube at 13,000g for 10 min. Pellet was resuspended into 1 mL of methanol, mixed vigorously, and stored frozen at −80°C before analyses. The supernatants were used to quantify chlorophyll photometrically (Lichtenthaler, 1987), and the remaining pellets were left at room temperature in a fume hood to evaporate the residual methanol. Pellets were then resuspended in 400 μL of distilled water and autoclaved for 15 min at 120°C to solubilize the starch polymer. Total starch was quantified using an enzymatic starch assay kit (Sigma-Aldrich; ref. SA-20) following the manufacturer’s instructions. Glucose converted from the starch was quantified using an automated YSI 2700 sugar analyzer (YSI Life Sciences) using a known amount of commercial glucose as a standard.
Biomass Determination
Biomass was determined by dry weight measurements. Briefly, cells in 10 mL of culture medium (N replete) and 5 mL of culture medium (N deplete) were collected on the preweighted glass fiber filter (VWR; ref. 611-0739) and dried overnight at 80°C in an incubator. The dried biomass was then gravimetrically measured.
FA and Lipid Analyses
FAs were analyzed by GC-MS after all acyl-lipids were converted to their methyl esters. TAG content was quantified after being separated from the bulk membrane lipids on a thin-layer chromatography plate. Unless otherwise stated, all methods related to lipid analyses were performed as previously described (Kong et al., 2017).
Insertion Site Identification by Restriction Enzyme Site-Directed Amplification-PCR
The position of the insertion of the cassette APHVIII in the genome of Chlamydomonas reinhardtii was determined according to the method of restriction enzyme site-directed amplification-PCR (González-Ballester et al., 2005; Kong et al., 2017). These and all other primer sequences used in this study are listed in Supplemental Table 2.
DNA Extraction, Gene Cloning, and Vector Construction for Complementation
Genomic DNA was isolated from exponentially grown cells by the CTAB method (Schroda et al., 2001). The genomic DNA coding for MDH2 was cloned using primers EcoRI-MDH2-F1 and XbaI-MDH2-R1. The PCR reaction was performed using the high fidelity KOD Hot Start DNA Polymerase (Merck Millipore). The amplified DNA fragment was cloned as an EcoRI-XbaI fragment into the pChlamy_4 vector (Life Technologies), which contains the ble gene conferring zeocin resistance (Stevens et al., 1996; Kong et al., 2015), generating pChlamy4-gMDH2. The target gene was cloned in frame with a V5 tag at its 3′ end, allowing screening complemented lines by anti-V5 (GKPIPNPLLGLDST) antibodies. The plasmids of pChlamy4-gMDH2 were transformed into the mdh2-1 mutant using electroporation, and independent zeocin (25 mg L−1) resistant clones were screened for TAG content by thin-layer chromatography (TLC). Briefly, a given number of cells (usually 20 million) was collected from each transformant grown to exponential phase, and total lipids were extracted following the method described (Kong et al., 2017). TAGs were separated out from other lipid classes by migration on a TLC plate in a solvent mixture of hexane/diethyl ether/acetic acid (17/3/0.2, v/v/v) as described (Siaut et al., 2011). TAG51:0 (17:0/17:0/17:0) (Sigma-Aldrich) was used as a standard to allow TAG quantification by densitometry. Detailed methods for TLC and quantification can be found in Siaut et al. (2011).
DNA Gel Blot Analysis
To determine the number of insertions in the mutant genome, genomic DNA was digested with NotI or StuI. The digested fragments were separated and blotted onto positively charged nylon membranes (GE Healthcare). The membrane containing DNA was then hybridized with a digoxigenin (DIG)-labeled probe made with a fragment of the APHVIII gene. The probe was made using a PCR DIG probe synthesis kit (Sigma-Aldrich) by PCR amplification with primers APHVIII-probe-F and APHVIII-probe-R. Imaging was done using the luminescent image analyzer G:Box Chemi XRQ system (Syngene).
RT-PCR
Total RNA was extracted as described before (Nguyen et al., 2013). The RNA was treated with DNase I (Ambion, Invitrogen) and then purified with Nucleospin RNA Clean Up (Macherey Nagel). First-strand cDNA was synthesized from 1 μg of the total RNA with the SuperScript VILO cDNA synthesis kit (Life Technologies) according to the manufacturer’s instructions. For RT-PCR, the cDNA fragment of MDH2 was amplified by PCR using gene-specific primers MDH2-F2 and MDH2-R2. The RACK1 (Cre06.g278222) gene was used as a housekeeping gene and amplified with primers RACK1-F1 and RACK1-R1.
Generation of Anti-MDH2 Antibodies
Antipeptide antibodies against MDH2 were made by immunizing rabbits with the synthetic peptide (PVSEYAYIRHPPRL). Synthesis of the peptide, rabbit immunization, and purification of antibodies were performed by Proteogenix SAS (Schiltigheim). Specificities of the antibodies were then tested on total proteins extracted from whole-cell Chlamydomonas parental lines (dw15 and CC4533) and the two mdh2 mutants.
Protein Extraction and Immunoblot Analysis
Exponentially grown cells were collected by centrifugation for 2 min at 1789g and resuspended in 1 mL lysis buffer (20 mM HEPES-KOH, pH 7.2, 10 mM KCl, 1 mM MgCl2, 154 mM NaCl, and 0.1× protease inhibitor cocktail; Sigma-Aldrich). Cells were then sonicated for 90 s with an alternating cycle of 1-s pulse/1-s pause. The homogenates were centrifuged at 14,000g at 4°C for 10 min, and the supernatant was used as total protein extracts. Protein concentrations were determined spectrophotometrically at 280 nm using a bicinchoninic acid protein assay kit (Bio-Rad). The loading controls were visualized either by Ponceau red staining or Coomassie Brilliant Blue staining as detailed in the legend. For immunoblots, a given amount of total proteins were separated on SDS-PAGE and then transferred to a nitrocellulose membrane using the semidry technique. Primary antibodies (at a dilution of 1/1000) included the specific polyclonal rabbit anti-V5 antibodies (Invitrogen; catalog no. PA1-993), the rabbit anti-MDH2 antibodies (this study), anti-PGRL1 (Tolleter et al., 2011), anti-NDA2 (Baltz et al., 2014), anti-FLVB (Chaux et al., 2017), and the anti-PSAD (catalog no. AS09-461), anti-PSBA (catalog no. AS01-016), anti-Cytb6f (subunit f; catalog no. AS06-119), anti-LHCSR3 (catalog no. AS14-2766), anti-ATPB (catalog no. AS05-085), anti-RBCL (catalog no. AS03-037), and anti-CAT1 (catalog no. AS15-2991) antibodies were purchased (Agrisera). Anti-rabbit horseradish peroxidase-conjugated antibodies (Sigma-Aldrich; catalog no. AQ132P) (1/20,000) were used as the secondary antibody. The detection was performed with the G:BOX Chemi XRQ system (Syngene) using ECL detection reagents (GE Healthcare). The images were captured using a CCD camera by GeneSys Image Acquisition Software (Syngene), and chemiluminescent fluorescence was quantitated by GeneTools Analysis Software (Syngene) according to the manufacturer’s instructions.
Total Protein Extraction and Enzymatic Activity Assays
Cells at the logarithm phase (around 20 million cells total) were collected and resuspended in extraction buffer (50 mM HEPES-KOH, 1 mM EDTA, 10% glycerol [v/v], 20 μL mL−1 protein protease inhibitor cocktail for plant cells [Sigma-Aldrich P9599], and 5 mM DTT at pH 7.5), and then lysed by sonication three times with a 10-s interval cycle. The concentrations of extracted protein were determined spectrophotometrically at 280 nm using a bicinchoninic acid protein assay kit (Bio-Rad) according to the manufacturer’s instructions.
MDH Activity Assay
The activity of MDH was determined photometrically by measuring the decrease in absorbance at 340 nm resulting from the oxidation of NADH to NAD+ as previously described (Mekhalfi et al., 2014). Enzyme assays were performed in a 1-cm path-length cuvette containing 2× assay buffer (90 mM KH2PO4-KOH, pH 7.4, 0.05% Triton X-100 [v/v], and 5 mM MgCl2) buffer with 0.2 mM NADH at 30°C. The reaction was initiated by addition of 3 mM oxaloacetate (Sigma-Aldrich), and the rate of NAD+ formation was monitored at 340 nm in a Kontron Uvikon 810 spectrophotometer (Thermo Fisher Scientific). All the solutions for enzyme activity assays were freshly prepared prior to use. The enzyme activities were calculated based on the molar extinction coefficient of NAD(P)H at 340 nm (ε340 = 6220 M−1 cm−1), and the enzymatic reaction rate was calculated using a linear regression. One unit of MDH activity is defined as 1 µmol of NADH oxidized per min per mg of protein.
ACX Activity Assay
Stearoyl-CoA (C18:0-CoA; Sigma-Aldrich) at 50 mM was used as the substrate for determining ACX activity in total cell-free extracts by following the production of H2O2, detected using the Amplex Red hydrogen peroxide/peroxidase assay kit (Invitrogen). A Xenius XC spectrofluorometer (SAFAS) was used to measure the fluorescence emission at 580 nm (excitation at 540 nm). Detailed protocols and quantification have been described previously (Kong et al., 2017).
LD Imaging
A confocal laser scanning microscope (TCS SP2; Leica) was used to observe LDs in cells first fixed in 0.25% glutaraldehyde and subsequently stained with BODIPY (4,4-difluoro-1,3,5,7-tetramethyl-4-bora-3a,4a-diaza-s-indacene; Thermo Fisher Scientific; D3921) at a final concentration of 10 µg mL−1. A 63× oil immersion objective was used for all imaging work, cells were excited with a 488-nm laser line, and emission was collected between 500 and 540 nm for BODIPY signal and between 650 and 714 nm for chlorophyll autofluorescence. Pseudo colors were applied using ZEN software (Carl Zeiss).
Determination of Extracellular H2O2 Levels
Chlamydomonas secrets H2O2 outside cell (Michelet et al., 2013). Extracellular H2O2 produced can be determined using the Amplex Red reagent (the Amplex Red hydrogen peroxide/peroxidase assay kit; Invitrogen). For this, 1.5 mL culture was centrifuged at 700g for 3 min in the dark. One milliliter of supernatant was mixed with the Amplex Red following the manufacturer’s instructions. The fluorescence emission spectra was read at 580 nm (with excitation at 540 nm) using a SAFAS Xenius XC fluorescence spectrophotometer. A known amount of commercial H2O2 (VWR chemicals) was first used to generate a standard curve. Concentration was calculated based on the standard curve and normalized by cell concentrations of the culture media at the time of sampling.
Intracellular ROS Level Imaging
Confocal microscopy images of cellular ROS were captured following the method described (Rastogi et al., 2010). Briefly, Chlamydomonas cells (∼20 million) were collected by centrifugation, washed once, and resuspended in 1× PBS buffer containing 5.0 μM of the oxidant-sensing fluorescent probe H2DCFDA (Thermo Fisher Scientific). After incubation at room temperature in the dark for 30 min with gentle shaking, the samples were washed three times with 1× PBS buffer. The cells were examined by a confocal laser scanning microscope (TCS SP2; Leica), for the detection of DCF fluorescence (excitation at 488 nm and emission 510 to 530 nm) and for chloroplast autofluorescence (excitation at 645 nm and emission at 685 to 720 nm). The DCF fluorescence of 2 million cells was also collected with a Cary Eclipse fluorescence spectrophotometer using excitation at 504 nm and emission at 524 nm.
Electron Transport Rate Measurement
Chlorophyll fluorescence was measured using a Dual Pulse Amplitude Modulated Fluorimeter (DUAL-PAM-100; Walz) as previously described (Schulz-Raffelt et al., 2016). Actinic light (supplied by a red LED light) increased stepwise (every 2 min) from 50 to 500 µmol photons m−2 s−1, and saturating flashes (10,000 µmol photons m−2 s−1, 200-ms duration) were applied to determine the PSII operating efficiency.
Gas Exchange Measurement by MIMS
Gas exchanges were monitored inside a water-jacketed and thermo-regulated (25°C) measuring chamber (using a modified Hansatech electrode chamber filled with a 1.5-mL sample). The bottom of the chamber was sealed by a Teflon membrane (13-µm thickness) allowing dissolved gases to be directly introduced through a vacuum line into the ion source of the mass spectrometer (model Prima δB; Thermo Fisher Scientific) after passing through a −65°C cooled water trap. Cells of Chlamydomonas were grown to mid-log phase and then centrifuged at 450g for 4 min and resuspended in fresh high salt medium at pH 6.2 in a final concentration of 20 µg chlorophyll mL−1. The cell suspension was placed in the measuring chamber under constant stirring and bicarbonate (0.5 mM final concentration) was added to saturate CO2. Before gas exchange measurement, [18O]-enriched O2 (99% 18O2 isotope content; Euriso-Top) was bubbled at the top of the suspension until reaching approximately equal concentrations of 16O2 and 18O2. Gas abundances (N2, 16O2, 18O2, and CO2) were recorded adjusting the magnet current to the corresponding mass peaks (m/z = 28, 32, 36, and 44, respectively). The measuring chamber was then sealed and gas exchanges recorded for 2 min until light was provided with three green LEDs at 600 μmol photons m−2 s−1 for 10 min. Gas concentrations were calculated according to Cournac et al. (2002). O2 exchange rates were calculated using equations from Radmer and Kok (1976).
NAD(P)H Fluorescence Measurement
NAD(P)H fluorescence was measured as previously described (Kauny and Sétif, 2014; Roach et al., 2015). Briefly, cells (OD684 = 0.26) were centrifuged and resuspended in fresh medium, then dark-adapted for 1 min at room temperature inside the spectrophotometer before starting the measurement. Light-induced measurements were performed at room temperature using NADPH/9-AA module of the DUAL-PAM (Walz). NAD(P)H fluorescence was first measured in the dark, then the actinic light (light intensity 70 µmol photons m−2 m−1 provided by a red LED) was switched on for 20 s. After switching off actinic light, fluorescence was recorded again in the dark for 10 s. Thirty measurements were averaged to obtain a good signal-to-noise ratio for each replicate.
Polar Metabolite Analysis by GC-MS
Chlamydomonas cells were grown to mid log phase, and 60 million cells were collected quickly by centrifugation at 13,000g for 10 s at 4°C. Cell pellets were quickly frozen in liquid nitrogen until analysis. Metabolites extraction and derivatization using GC-MS were performed as described (Lisec et al., 2006). The GC-MS data were obtained using an Agilent 7683 series auto-sampler (Agilent Technologies), coupled to an Agilent 6890 gas chromatograph-Leco Pegasus two time-of-flight mass spectrometer (Leco). Identical chromatogram acquisition parameters were applied to those previously used (Cuadros-Inostroza et al., 2009). Chromatograms were exported from LECO CHROMATOF software (version 3.34) to R software. Ion extraction, peak detection, retention time alignment, and library searching were obtained using the Target Search package from Bioconductor (Cuadros-Inostroza et al., 2009).
Statistical Analysis
Student’s t test was performed on biological replicates using Excel software (2013 version).
Accession Numbers
Sequence data presented in this article can be found for Chlamydomonas genes in Phytozome (https://phytozome.jgi.doe.gov/pz/portal.html#!info?alias=Org_Creinhardtii) with gene identifications as follows: MDH1 (Cre03.g194850), MDH2 (Cre10.g423250), MDH3 (Cre02.g145800), MDH4 (Cre12.g483950), MDH5 (Cre09.g410700), RACK1 (g6364), FLVB (Cre16.g691800), Cytb6f (Cre16.g650100), PSAD (Cre05.g238332), LHCSR3 (Cre08.g367400), CAT1 (Cre09.g417150), NDA2 (Cre19.g750547), PGRL1 (Cre07.g340200), ATPB (Cre17.g698000), RBCL (Cre06.g298300), psbA (ChreCp.021), APX (Cre09.g401886), HPR (Cre06.g295450), MDAR1 (Cre17.g712100), PXN (Cre07.g353300), and ACX2 (Cre05.g232002). For proteins of Arabidopsis, protein IDs can be found in TAIR (www.arabidopsis.org).
Supplemental Data
Supplemental Figure 1. Major Experimental Setup Used in This Study.
Supplemental Figure 2. DNA Gel Blot of the APHVIII Gene Insertion Events in mdh2-1.
Supplemental Figure 3. NAD-Dependent Total MDH Activity Analyses during Mixotrophic Growth.
Supplemental Figure 4. Cell Growth under Heterotrophic, Mixotrophic, and Photoautotrophic Conditions.
Supplemental Figure 5. Comparison of MDH2 Protein Level in Mixotrophically and Photoautotrophically Grown Wild-Type (dw15) Cells.
Supplemental Figure 6. The mdh2-1 Mutant Overaccumulates TAG and Starch 2 d after N Starvation during Photoautotrophic Growth When Expressed Based on Cell Volume.
Supplemental Figure 7. The mdh2-2 Mutant Overaccumulates TAG and Starch 2 d after N Starvation during Photoautotrophic Growth (+2% CO2 Supplemented in the Air) When Expressed Based on Cell Volume.
Supplemental Figure 8. Fatty Acid Compositional Analysis by Gas Chromatography-Mass Spectrometry.
Supplemental Figure 9. PSII Operating Yield in mdh2-1 during Mixotrophic N Deprivation.
Supplemental Figure 10. Traces for Measurement of NAD(P)H Fluorescence in Photoautotrophically Grown Cells.
Supplemental Figure 11. The Level of Catalase 1 Protein Revealed by Immunoblot Using Anticatalase Antibodies.
Supplemental Figure 12. Measurement of Extracellular H2O2 Level in mdh2-1 and Control Strains.
Supplemental Figure 13. ACX (18:0) Activity Analysis.
Supplemental Figure 14. H2O2 Supplementation on Cell Growth.
Supplemental Table 1. Bioinformatic Comparison of the MDH Family in Chlamydomonas to That of Land Plant Arabidopsis thaliana.
Supplemental Table 2. Primer Sequences Used in This Study.
Supplemental Data Set 1. Polar Metabolite Profiling of the Wild Type, mdh2-1, and the Complementation Line C1 by GC-MS.
Dive Curated Terms
The following phenotypic, genotypic, and functional terms are of significance to the work described in this paper:
Acknowledgments
We thank John Ohlrogge for critical reading of the manuscript. We thank Isabelle Thé, Audrey Beyly, Stéphan Cuiné, and Pascaline Auroy for laboratory assistance. Adrien Burlacot is a recipient of a CEA (Irtelis) international PhD studentship. Yuanxue Liang acknowledges the China Scholarship Council for a PhD studentship. Work in the authors’ laboratory is supported by the French Agence Nationale pour la Recherche (ANR JCJC MUsCA) and by the A*MIDEX project funded by the “Investissements d’Avenir.” We acknowledge the European Union Regional Developing Fund, the Région Provence Alpes Côte d’Azur, the French Ministry of Research, and the CEA for funding the HelioBiotec platform. Support for the microscopy equipment was provided by the Région Provence Alpes Côte d’Azur, the Conseil Général des Bouches-du-Rhône, the French Ministry of Research, the CNRS, and the CEA. S.A. and A.R.F. thank the European Union for funding in the framework of the European Union 2020 TEAMING Project (SGA-CSA No 664621 and No 739582 under FPA No. 664620).
AUTHOR CONTRIBUTIONS
Y.L.-B. designed the project. F.K. performed all experiments except the ones detailed below. A.B. performed MIMS measurements with the supervision of G.P. Y.L. analyzed enzymatic activities and RT-PCR. B.L. supervised lipid analysis. S.A. and Y.B. performed metabolomics analysis with the supervision of A.R.F. A.K.-L. performed NAD(P)H measurement. Y.L.-B., F.B., G.P., and F.K. wrote the article with comments from A.R.F. and A.K.-L.
Footnotes
Articles can be viewed without a subscription.
References
- Aboelmy M.H., Peterhansel C. (2014). Enzymatic characterization of Chlamydomonas reinhardtii glycolate dehydrogenase and its nearest proteobacterial homologue. Plant Physiol. Biochem. 79: 25–30. [DOI] [PubMed] [Google Scholar]
- Allorent G., et al. (2013). A dual strategy to cope with high light in Chlamydomonas reinhardtii. Plant Cell 25: 545–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araújo W.L., Tohge T., Osorio S., Lohse M., Balbo I., Krahnert I., Sienkiewicz-Porzucek A., Usadel B., Nunes-Nesi A., Fernie A.R. (2012). Antisense inhibition of the 2-oxoglutarate dehydrogenase complex in tomato demonstrates its importance for plant respiration and during leaf senescence and fruit maturation. Plant Cell 24: 2328–2351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailleul B., et al. (2015). Energetic coupling between plastids and mitochondria drives CO2 assimilation in diatoms. Nature 524: 366–369. [DOI] [PubMed] [Google Scholar]
- Ball S., Dirick L., Decq A., Martiat J., Matagne R. (1990). Physiology of starch storage in the monocellular alga Chlamydomonas reinhardtii. Plant Sci. 66: 1–9. [Google Scholar]
- Baltz A., Dang K.-V., Beyly A., Auroy P., Richaud P., Cournac L., Peltier G. (2014). Plastidial expression of Type II NAD(P)H dehydrogenase increases the reducing state of plastoquinones and hydrogen photoproduction rate by the indirect pathway in Chlamydomonas reinhardtii. Plant Physiol. 165: 1344–1352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baslam M., Baroja-Fernández E., Ricarte-Bermejo A., Sánchez-López A.M., Aranjuelo I., Bahaji A., Muñoz F.J., Almagro G., Pujol P., Galarza R., Teixidor P., Pozueta-Romero J. (2017). Genetic and isotope ratio mass spectrometric evidence for the occurrence of starch degradation and cycling in illuminated Arabidopsis leaves. PLoS One 12: e0171245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beeler S., Liu H.C., Stadler M., Schreier T., Eicke S., Lue W.L., Truernit E., Zeeman S.C., Chen J., Kötting O. (2014). Plastidial NAD-dependent malate dehydrogenase is critical for embryo development and heterotrophic metabolism in Arabidopsis. Plant Physiol. 164: 1175–1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beevers H. (1979). Microbodies in higher-plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 30: 159–193. [Google Scholar]
- Bernhardt K., Wilkinson S., Weber A.P., Linka N. (2012). A peroxisomal carrier delivers NAD+ and contributes to optimal fatty acid degradation during storage oil mobilization. Plant J. 69: 1–13. [DOI] [PubMed] [Google Scholar]
- Blaby I.K., Blaby-Haas C.E., Pérez-Pérez M.E., Schmollinger S., Fitz-Gibbon S., Lemaire S.D., Merchant S.S. (2015). Genome-wide analysis on Chlamydomonas reinhardtii reveals the impact of hydrogen peroxide on protein stress responses and overlap with other stress transcriptomes. Plant J. 84: 974–988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buléon A., Gallant D.J., Bouchet B., Mouille G., D’Hulst C., Kossmann J., Ball S. (1997). Starches from A to C. Chlamydomonas reinhardtii as a model microbial system to investigate the biosynthesis of the plant amylopectin crystal. Plant Physiol. 115: 949–957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cagnon C., Mirabella B., Nguyen H.M., Beyly-Adriano A., Bouvet S., Cuiné S., Beisson F., Peltier G., Li-Beisson Y. (2013). Development of a forward genetic screen to isolate oil mutants in the green microalga Chlamydomonas reinhardtii. Biotechnol. Biofuels 6: 178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Centeno D.C., et al. (2011). Malate plays a crucial role in starch metabolism, ripening, and soluble solid content of tomato fruit and affects postharvest softening. Plant Cell 23: 162–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaux F., Burlacot A., Mekhalfi M., Auroy P., Blangy S., Richaud P., Peltier G. (2017). Flavodiiron proteins promote fast and transient O2 photoreduction in Chlamydomonas. Plant Physiol. 174: 1825–1836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comparot-Moss S., et al. (2010). A putative phosphatase, LSF1, is required for normal starch turnover in Arabidopsis leaves. Plant Physiol. 152: 685–697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cournac L., Latouche G., Cerovic Z., Redding K., Ravenel J., Peltier G. (2002). In vivo interactions between photosynthesis, mitorespiration, and chlororespiration in Chlamydomonas reinhardtii. Plant Physiol. 129: 1921–1928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuadros-Inostroza A., Caldana C., Redestig H., Kusano M., Lisec J., Peña-Cortés H., Willmitzer L., Hannah M.A. (2009). TargetSearch--a Bioconductor package for the efficient preprocessing of GC-MS metabolite profiling data. BMC Bioinformatics 10: 428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curien G., Flori S., Villanova V., Magneschi L., Giustini C., Forti G., Matringe M., Petroutsos D., Kuntz M., Finazzi G. (2016). The water to water cycles in microalgae. Plant Cell Physiol. 57: 1354–1363. [DOI] [PubMed] [Google Scholar]
- Daloso D.M., Medeiros D.B., Dos Anjos L., Yoshida T., Araújo W.L., Fernie A.R. (2017). Metabolism within the specialized guard cells of plants. New Phytol. 216: 1018–1033. [DOI] [PubMed] [Google Scholar]
- Dang K.-V., Plet J., Tolleter D., Jokel M., Cuiné S., Carrier P., Auroy P., Richaud P., Johnson X., Alric J., Allahverdiyeva Y., Peltier G. (2014). Combined increases in mitochondrial cooperation and oxygen photoreduction compensate for deficiency in cyclic electron flow in Chlamydomonas reinhardtii. Plant Cell 26: 3036–3050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietz K.-J., Turkan I., Krieger-Liszkay A. (2016). Redox- and reactive oxygen species-dependent signaling into and out of the photosynthesizing chloroplast. Plant Physiol. 171: 1541–1550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Z.Y., Lucker B.F., Zienkiewicz K., Miller T.E., Zienkiewicz A., Sears B.B., Kramer D.M., Benning C. (2018). Galactoglycerolipid lipase PGD1 is involved in thylakoid membrane remodeling in response to adverse environmental conditions in Chlamydomonas. Plant Cell 30: 447–465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eastmond P.J. (2007). MONODEHYROASCORBATE REDUCTASE4 is required for seed storage oil hydrolysis and postgerminative growth in Arabidopsis. Plant Cell 19: 1376–1387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erickson E., Wakao S., Niyogi K.K. (2015). Light stress and photoprotection in Chlamydomonas reinhardtii. Plant J. 82: 449–465. [DOI] [PubMed] [Google Scholar]
- Fan J., Yan C., Andre C., Shanklin J., Schwender J., Xu C. (2012). Oil accumulation is controlled by carbon precursor supply for fatty acid synthesis in Chlamydomonas reinhardtii. Plant Cell Physiol. 53: 1380–1390. [DOI] [PubMed] [Google Scholar]
- Geigenberger P. (2011). Regulation of starch biosynthesis in response to a fluctuating environment. Plant Physiol. 155: 1566–1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- González-Ballester D., de Montaigu A., Galván A., Fernández E. (2005). Restriction enzyme site-directed amplification PCR: a tool to identify regions flanking a marker DNA. Anal. Biochem. 340: 330–335. [DOI] [PubMed] [Google Scholar]
- Goodson C., Roth R., Wang Z.T., Goodenough U. (2011). Structural correlates of cytoplasmic and chloroplast lipid body synthesis in Chlamydomonas reinhardtii and stimulation of lipid body production with acetate boost. Eukaryot. Cell 10: 1592–1606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goold H.D., Cuiné S., Légeret B., Liang Y., Brugière S., Auroy P., Javot H., Tardif M., Jones B., Beisson F., Peltier G., Li-Beisson Y. (2016). Saturating light induces sustained accumulation of oil in plastidal lipid droplets in Chlamydomonas reinhardtii. Plant Physiol. 171: 2406–2417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham I.A. (2008). Seed storage oil mobilization. Annu. Rev. Plant Biol. 59: 115–142. [DOI] [PubMed] [Google Scholar]
- Hagemann M., Kern R., Maurino V.G., Hanson D.T., Weber A.P., Sage R.F., Bauwe H. (2016). Evolution of photorespiration from cyanobacteria to land plants, considering protein phylogenies and acquisition of carbon concentrating mechanisms. J. Exp. Bot. 67: 2963–2976. [DOI] [PubMed] [Google Scholar]
- Harris E.H. (2009). The Chlamydomonas Sourcebook: Introduction to Chlamydomonas and Its Laboratory Use. (Oxford, UK: Elsevier; ). [Google Scholar]
- Hayashi Y., Shinozaki A. (2012). Visualization of microbodies in Chlamydomonas reinhardtii. J. Plant Res. 125: 579–586. [DOI] [PubMed] [Google Scholar]
- Hebbelmann I., et al. (2012). Multiple strategies to prevent oxidative stress in Arabidopsis plants lacking the malate valve enzyme NADP-malate dehydrogenase. J. Exp. Bot. 63: 1445–1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heyno E., Innocenti G., Lemaire S.D., Issakidis-Bourguet E., Krieger-Liszkay A. (2014). Putative role of the malate valve enzyme NADP-malate dehydrogenase in H2O2 signalling in Arabidopsis. Philos. Trans. R. Soc. Lond. B Biol. Sci. 369: 20130228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J., Baker A., Bartel B., Linka N., Mullen R.T., Reumann S., Zolman B.K. (2012). Plant peroxisomes: biogenesis and function. Plant Cell 24: 2279–2303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iglesias A.A., Charng Y.Y., Ball S., Preiss J. (1994). Characterization of the kinetic, regulatory, and structural properties of ADP-glucose pyrophosphorylase from Chlamydomonas reinhardtii. Plant Physiol. 104: 1287–1294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenner H.L., Winning B.M., Millar A.H., Tomlinson K.L., Leaver C.J., Hill S.A. (2001). NAD malic enzyme and the control of carbohydrate metabolism in potato tubers. Plant Physiol. 126: 1139–1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kauny J., Sétif P. (2014). NADPH fluorescence in the cyanobacterium Synechocystis sp. PCC 6803: a versatile probe for in vivo measurements of rates, yields and pools. Biochim. Biophys. Acta 1837: 792–801. [DOI] [PubMed] [Google Scholar]
- Kong F., Yamasaki T., Kurniasih S.D., Hou L., Li X., Ivanova N., Okada S., Ohama T. (2015). Robust expression of heterologous genes by selection marker fusion system in improved Chlamydomonas strains. J. Biosci. Bioeng. 120: 239–245. [DOI] [PubMed] [Google Scholar]
- Kong F., Liang Y., Légeret B., Beyly-Adriano A., Blangy S., Haslam R.P., Napier J.A., Beisson F., Peltier G., Li-Beisson Y. (2017). Chlamydomonas carries out fatty acid β-oxidation in ancestral peroxisomes using a bona fide acyl-CoA oxidase. Plant J. 90: 358–371. [DOI] [PubMed] [Google Scholar]
- Larosa V., Meneghesso A., La Rocca N., Steinbeck J., Hippler M., Szabò I., Morosinotto T. (2018). Mitochondria affect photosynthetic electron transport and photo-sensitivity in a green alga. Plant Physiol. 176: 2305–2314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauersen K.J., Willamme R., Coosemans N., Joris M., Kruse O., Remacle C. (2016). Peroxisomal microbodies are at the crossroads of acetate assimilation in the green microalga Chlamydomonas reinhardtii. Algal Res. 16: 266–274. [Google Scholar]
- Lemaire S.D., Quesada A., Merchan F., Corral J.M., Igeno M.I., Keryer E., Issakidis-Bourguet E., Hirasawa M., Knaff D.B., Miginiac-Maslow M. (2005). NADP-malate dehydrogenase from unicellular green alga Chlamydomonas reinhardtii. A first step toward redox regulation? Plant Physiol. 137: 514–521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lepistö A., Pakula E., Toivola J., Krieger-Liszkay A., Vignols F., Rintamäki E. (2013). Deletion of chloroplast NADPH-dependent thioredoxin reductase results in inability to regulate starch synthesis and causes stunted growth under short-day photoperiods. J. Exp. Bot. 64: 3843–3854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Moellering E.R., Liu B., Johnny C., Fedewa M., Sears B.B., Kuo M.-H., Benning C. (2012). A galactoglycerolipid lipase is required for triacylglycerol accumulation and survival following nitrogen deprivation in Chlamydomonas reinhardtii. Plant Cell 24: 4670–4686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Zhang R., Patena W., Gang S.S., Blum S.R., Ivanova N., Yue R., Robertson J.M., Lefebvre P.A., Fitz-Gibbon S.T., Grossman A.R., Jonikas M.C. (2016). An indexed, mapped mutant library enables reverse genetics studies of biological processes in Chlamydomonas reinhardtii. Plant Cell 28: 367–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li-Beisson Y., et al. (2013). Acyl-lipid metabolism. The Arabidopsis Book 11: e0161, doi/10.1199/tab.0161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li-Beisson Y., Beisson F., Riekhof W. (2015). Metabolism of acyl-lipids in Chlamydomonas reinhardtii. Plant J. 82: 504–522. [DOI] [PubMed] [Google Scholar]
- Libessart N., Maddelein M.L., Koornhuyse N., Decq A., Delrue B., Mouille G., D’Hulst C., Ball S. (1995). Storage, photosynthesis and growth - the conditional nature of mutations affecting starch synthesis and structure in Chlamydomonas. Plant Cell 7: 1117–1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lichtenthaler H. (1987). Chlorophylls and carotenoids - pigments of photosynthetic biomembranes. Methods Enzymol. 148: 350–382. [Google Scholar]
- Lisec J., Schauer N., Kopka J., Willmitzer L., Fernie A.R. (2006). Gas chromatography mass spectrometry-based metabolite profiling in plants. Nat. Protoc. 1: 387–396. [DOI] [PubMed] [Google Scholar]
- Liu B., Benning C. (2013). Lipid metabolism in microalgae distinguishes itself. Curr. Opin. Biotechnol. 24: 300–309. [DOI] [PubMed] [Google Scholar]
- Lobo A.K., de Oliveira Martins M., Lima Neto M.C., Machado E.C., Ribeiro R.V., Silveira J.A. (2015). Exogenous sucrose supply changes sugar metabolism and reduces photosynthesis of sugarcane through the down-regulation of Rubisco abundance and activity. J. Plant Physiol. 179: 113–121. [DOI] [PubMed] [Google Scholar]
- Massoz S., Larosa V., Horrion B., Matagne R.F., Remacle C., Cardol P. (2015). Isolation of Chlamydomonas reinhardtii mutants with altered mitochondrial respiration by chlorophyll fluorescence measurement. J. Biotechnol. 215: 27–34. [DOI] [PubMed] [Google Scholar]
- Mekhalfi M., Amara S., Robert S., Carrière F., Gontero B. (2014). Effect of environmental conditions on various enzyme activities and triacylglycerol contents in cultures of the freshwater diatom, Asterionella formosa (Bacillariophyceae). Biochimie 101: 21–30. [DOI] [PubMed] [Google Scholar]
- Merchant S.S., et al. (2007). The Chlamydomonas genome reveals the evolution of key animal and plant functions. Science 318: 245–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mettler I.J., Beevers H. (1980). Oxidation of NADH in glyoxysomes by a malate-aspartate shuttle. Plant Physiol. 66: 555–560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michalska J., Zauber H., Buchanan B.B., Cejudo F.J., Geigenberger P. (2009). NTRC links built-in thioredoxin to light and sucrose in regulating starch synthesis in chloroplasts and amyloplasts. Proc. Natl. Acad. Sci. USA 106: 9908–9913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michelet L., Roach T., Fischer B.B., Bedhomme M., Lemaire S.D., Krieger-Liszkay A. (2013). Down-regulation of catalase activity allows transient accumulation of a hydrogen peroxide signal in Chlamydomonas reinhardtii. Plant Cell Environ. 36: 1204–1213. [DOI] [PubMed] [Google Scholar]
- Miginiac-Maslow M., Lancelin J.M. (2002). Intrasteric inhibition in redox signalling: light activation of NADP-malate dehydrogenase. Photosynth. Res. 72: 1–12. [DOI] [PubMed] [Google Scholar]
- Mikkelsen R., Mutenda K.E., Mant A., Schürmann P., Blennow A. (2005). Alpha-glucan, water dikinase (GWD): a plastidic enzyme with redox-regulated and coordinated catalytic activity and binding affinity. Proc. Natl. Acad. Sci. USA 102: 1785–1790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller R., et al. (2010). Changes in transcript abundance in Chlamydomonas reinhardtii following nitrogen deprivation predict diversion of metabolism. Plant Physiol. 154: 1737–1752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musrati R.A., Kollárová M., Mernik N., Mikulásová D. (1998). Malate dehydrogenase: distribution, function and properties. Gen. Physiol. Biophys. 17: 193–210. [PubMed] [Google Scholar]
- Naranjo B., Mignée C., Krieger-Liszkay A., Hornero-Méndez D., Gallardo-Guerrero L., Cejudo F.J., Lindahl M. (2016). The chloroplast NADPH thioredoxin reductase C, NTRC, controls non-photochemical quenching of light energy and photosynthetic electron transport in Arabidopsis. Plant Cell Environ. 39: 804–822. [DOI] [PubMed] [Google Scholar]
- Nguyen H.M., Cuiné S., Beyly-Adriano A., Légeret B., Billon E., Auroy P., Beisson F., Peltier G., Li-Beisson Y. (2013). The green microalga Chlamydomonas reinhardtii has a single ω-3 fatty acid desaturase that localizes to the chloroplast and impacts both plastidic and extraplastidic membrane lipids. Plant Physiol. 163: 914–928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niyogi K.K. (2000). Safety valves for photosynthesis. Curr. Opin. Plant Biol. 3: 455–460. [DOI] [PubMed] [Google Scholar]
- Noctor G., Queval G., Gakière B. (2006). NAD(P) synthesis and pyridine nucleotide cycling in plants and their potential importance in stress conditions. J. Exp. Bot. 57: 1603–1620. [DOI] [PubMed] [Google Scholar]
- Nunes-Nesi A., Carrari F., Lytovchenko A., Smith A.M., Loureiro M.E., Ratcliffe R.G., Sweetlove L.J., Fernie A.R. (2005). Enhanced photosynthetic performance and growth as a consequence of decreasing mitochondrial malate dehydrogenase activity in transgenic tomato plants. Plant Physiol. 137: 611–622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunes-Nesi A., Araújo W.L., Fernie A.R. (2011). Targeting mitochondrial metabolism and machinery as a means to enhance photosynthesis. Plant Physiol. 155: 101–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohlrogge J., Browse J. (1995). Lipid biosynthesis. Plant Cell 7: 957–970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oswald O., Martin T., Dominy P.J., Graham I.A. (2001). Plastid redox state and sugars: interactive regulators of nuclear-encoded photosynthetic gene expression. Proc. Natl. Acad. Sci. USA 98: 2047–2052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J.-J., Wang H., Gargouri M., Deshpande R.R., Skepper J.N., Holguin F.O., Juergens M.T., Shachar-Hill Y., Hicks L.M., Gang D.R. (2015). The response of Chlamydomonas reinhardtii to nitrogen deprivation: a systems biology analysis. Plant J. 81: 611–624. [DOI] [PubMed] [Google Scholar]
- Peltier G., Tolleter D., Billon E., Cournac L. (2010). Auxiliary electron transport pathways in chloroplasts of microalgae. Photosynth. Res. 106: 19–31. [DOI] [PubMed] [Google Scholar]
- Pfannschmidt T., Nilsson A., Allen J.F. (1999). Photosynthetic control of chloroplast gene expression. Nature 397: 625. [Google Scholar]
- Plancke C., et al. (2014). Lack of isocitrate lyase in Chlamydomonas leads to changes in carbon metabolism and in the response to oxidative stress under mixotrophic growth. Plant J. 77: 404–417. [DOI] [PubMed] [Google Scholar]
- Pracharoenwattana I., Cornah J.E., Smith S.M. (2007). Arabidopsis peroxisomal malate dehydrogenase functions in beta-oxidation but not in the glyoxylate cycle. Plant J. 50: 381–390. [DOI] [PubMed] [Google Scholar]
- Pracharoenwattana I., Zhou W., Smith S.M. (2010). Fatty acid beta-oxidation in germinating Arabidopsis seeds is supported by peroxisomal hydroxypyruvate reductase when malate dehydrogenase is absent. Plant Mol. Biol. 72: 101–109. [DOI] [PubMed] [Google Scholar]
- Radmer R.J., Kok B. (1976). Photoreduction of O2 primes and replaces CO2 assimilation. Plant Physiol. 58: 336–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rastogi R.P., Singh S.P., Häder D.P., Sinha R.P. (2010). Detection of reactive oxygen species (ROS) by the oxidant-sensing probe 2′,7′-dichlorodihydrofluorescein diacetate in the cyanobacterium Anabaena variabilis PCC 7937. Biochem. Biophys. Res. Commun. 397: 603–607. [DOI] [PubMed] [Google Scholar]
- Recht L., Zarka A., Boussiba S. (2012). Patterns of carbohydrate and fatty acid changes under nitrogen starvation in the microalgae Haematococcus pluvialis and Nannochloropsis sp. Appl. Microbiol. Biotechnol. 94: 1495–1503. [DOI] [PubMed] [Google Scholar]
- Rhodin J.A.G. (1954). Correlation of Ultrastructural Organization and Function in Normal and Experimentally Changed Proximal Convoluted Tubule Cells of the Mouse Kidney: An Electron Microscopic Study. (Stockholm, Sweden: Karolinska Institutet; ). [Google Scholar]
- Roach T., Na C.S., Krieger-Liszkay A. (2015). High light-induced hydrogen peroxide production in Chlamydomonas reinhardtii is increased by high CO2 availability. Plant J. 81: 759–766. [DOI] [PubMed] [Google Scholar]
- Saroussi S.I., Wittkopp T.M., Grossmann E.R. (2016). The type II NADPH dehydrogenase facilitates cyclic electron flow, energy-dependent quenching, and chlororespiratory metabolism during acclimation of Chlamydomonas reinhardtii to nitrogen deprivation. Plant Physiol. 170: 1975–1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saroussi S., Sanz-Luque E., Kim R.G., Grossman A.R. (2017). Nutrient scavenging and energy management: acclimation responses in nitrogen and sulfur deprived Chlamydomonas. Curr. Opin. Plant Biol. 39: 114–122. [DOI] [PubMed] [Google Scholar]
- Scheibe R. (2004). Malate valves to balance cellular energy supply. Physiol. Plant. 120: 21–26. [DOI] [PubMed] [Google Scholar]
- Schmollinger S., et al. (2014). Nitrogen-sparing mechanisms in Chlamydomonas affect the transcriptome, the proteome, and photosynthetic metabolism. Plant Cell 26: 1410–1435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroda M., Vallon O., Whitelegge J.P., Beck C.F., Wollman F.-A. (2001). The chloroplastic GrpE homolog of Chlamydomonas: two isoforms generated by differential splicing. Plant Cell 13: 2823–2839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulz-Raffelt M., Chochois V., Auroy P., Cuiné S., Billon E., Dauvillée D., Li-Beisson Y., Peltier G. (2016). Hyper-accumulation of starch and oil in a Chlamydomonas mutant affected in a plant-specific DYRK kinase. Biotechnol. Biofuels 9: 55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz V., Andosch A., Geretschläger A., Affenzeller M., Lütz-Meindl U. (2017). Carbon starvation induces lipid degradation via autophagy in the model alga Micrasterias. J. Plant Physiol. 208: 115–127. [DOI] [PubMed] [Google Scholar]
- Selinski J., Scheibe R. (2014). Lack of malate valve capacities lead to improved N-assimilation and growth in transgenic A. thaliana plants. Plant Signal. Behav. 9: e29057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siaut M., Cuiné S., Cagnon C., Fessler B., Nguyen M., Carrier P., Beyly A., Beisson F., Triantaphylidès C., Li-Beisson Y., Peltier G. (2011). Oil accumulation in the model green alga Chlamydomonas reinhardtii: characterization, variability between common laboratory strains and relationship with starch reserves. BMC Biotechnol. 11: 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith A.M., Stitt M. (2007). Coordination of carbon supply and plant growth. Plant Cell Environ. 30: 1126–1149. [DOI] [PubMed] [Google Scholar]
- Sparla F., Costa A., Lo Schiavo F., Pupillo P., Trost P. (2006). Redox regulation of a novel plastid-targeted beta-amylase of Arabidopsis. Plant Physiol. 141: 840–850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens D.R., Rochaix J.D., Purton S. (1996). The bacterial phleomycin resistance gene ble as a dominant selectable marker in Chlamydomonas. Mol. Gen. Genet. 251: 23–30. [DOI] [PubMed] [Google Scholar]
- Sweetlove L.J., Fernie A.R. (2013). The spatial organization of metabolism within the plant cell. In Annual Review of Plant Biology, Vol. 64, Merchant S.S., ed (Palo Alto, CA: Annual Reviews; ), pp. 723–746. [DOI] [PubMed] [Google Scholar]
- Sweetlove L.J., Lytovchenko A., Morgan M., Nunes-Nesi A., Taylor N.L., Baxter C.J., Eickmeier I., Fernie A.R. (2006). Mitochondrial uncoupling protein is required for efficient photosynthesis. Proc. Natl. Acad. Sci. USA 103: 19587–19592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thormählen I., Zupok A., Rescher J., Leger J., Weissenberger S., Groysman J., Orwat A., Chatel-Innocenti G., Issakidis-Bourguet E., Armbruster U., Geigenberger P. (2017). Thioredoxins play a crucial role in dynamic acclimation of photosynthesis in fluctuating light. Mol. Plant 10: 168–182. [DOI] [PubMed] [Google Scholar]
- Tolleter D., et al. (2011). Control of hydrogen photoproduction by the proton gradient generated by cyclic electron flow in Chlamydomonas reinhardtii. Plant Cell 23: 2619–2630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trentacoste E.M., Shrestha R.P., Smith S.R., Glé C., Hartmann A.C., Hildebrand M., Gerwick W.H. (2013). Metabolic engineering of lipid catabolism increases microalgal lipid accumulation without compromising growth. Proc. Natl. Acad. Sci. USA 110: 19748–19753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tripathi D.N., Walker C.L. (2016). The peroxisome as a cell signaling organelle. Curr. Opin. Cell Biol. 39: 109–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai C.-H., Warakanont J., Takeuchi T., Sears B.B., Moellering E.R., Benning C. (2014). The protein Compromised Hydrolysis of Triacylglycerols 7 (CHT7) acts as a repressor of cellular quiescence in Chlamydomonas. Proc. Natl. Acad. Sci. USA 111: 15833–15838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai C.H., Uygun S., Roston R., Shiu S.H., Benning C. (2018). Recovery from N deprivation is a transcriptionally and functionally distinct state in Chlamydomonas. Plant Physiol. 176: 2007–2023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Roermund C.W.T., Elgersma Y., Singh N., Wanders R.J.A., Tabak H.F. (1995). The membrane of peroxisomes in Saccharomyces cerevisiae is impermeable to NAD(H) and acetyl-CoA under in vivo conditions. EMBO J. 14: 3480–3486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Roermund C.W., Schroers M.G., Wiese J., Facchinelli F., Kurz S. (2016). The peroxisomal NAD carrier from Arabidopsis imports NAD in exchange with AMP. Plant Physiol. 171: 2127–2139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wynn J.P., Kendrick A., Ratledge C. (1997). Sesamol as an inhibitor of growth and lipid metabolism in Mucor circinelloides via its action on malic enzyme. Lipids 32: 605–610. [DOI] [PubMed] [Google Scholar]
- Xue J., Niu Y.-F., Huang T., Yang W.-D., Liu J.-S., Li H.-Y. (2015). Genetic improvement of the microalga Phaeodactylum tricornutum for boosting neutral lipid accumulation. Metab. Eng. 27: 1–9. [DOI] [PubMed] [Google Scholar]
- Xue J., Wang L., Zhang L., Balamurugan S., Li D.-W., Zeng H., Yang W.-D., Liu J.-S., Li H.-Y. (2016). The pivotal role of malic enzyme in enhancing oil accumulation in green microalga Chlorella pyrenoidosa. Microb. Cell Fact. 15: 120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yilancioglu K., Cokol M., Pastirmaci I., Erman B., Cetiner S. (2014). Oxidative stress is a mediator for increased lipid accumulation in a newly isolated Dunaliella salina strain. PLoS One 9: e91957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Adams I.P., Ratledge C. (2007). Malic enzyme: the controlling activity for lipid production? Overexpression of malic enzyme in Mucor circinelloides leads to a 2.5-fold increase in lipid accumulation. Microbiology 153: 2013–2025. [DOI] [PubMed] [Google Scholar]
- Zhang Y.M., Chen H., He C.L., Wang Q. (2013). Nitrogen starvation induced oxidative stress in an oil-producing green alga Chlorella sorokiniana C3. PLoS One 8: e69225. [DOI] [PMC free article] [PubMed] [Google Scholar]