High-confidence assemblies enabled comparative genomics analyses of the centromeric regions of Oryza genomes and provided insight into centromere evolution.
Abstract
Centromeres are dynamic chromosomal regions, and the genetic and epigenetic environment of the centromere is often regarded as oppressive to protein-coding genes. Here, we used comparative genomic and phylogenomic approaches to study the evolution of centromeres and centromere-linked genes in the genus Oryza. We report a 12.4-Mb high-quality BAC-based pericentromeric assembly for Oryza brachyantha, which diverged from cultivated rice (Oryza sativa) ∼15 million years ago. The synteny analyses reveal seven medium (>50 kb) pericentric inversions in O. sativa and 10 in O. brachyantha. Of these inversions, three resulted in centromere movement (Chr1, Chr7, and Chr9). Additionally, we identified a potential centromere-repositioning event, in which the ancestral centromere on chromosome 12 in O. brachyantha jumped ∼400 kb away, possibly mediated by a duplicated transposition event (>28 kb). More strikingly, we observed an excess of syntenic gene loss at and near the centromeric regions (P < 2.2 × 10−16). Most (33/47) of the missing genes moved to other genomic regions; therefore such excess could be explained by the selective loss of the copy in or near centromeric regions after gene duplication. The pattern of gene loss immediately adjacent to centromeric regions suggests centromere chromatin dynamics (e.g., spreading or microrepositioning) may drive such gene loss.
INTRODUCTION
Centromeres in higher eukaryotes are typically composed of tandemly arranged repeats (Henikoff et al., 2001). The flanking pericentromeric regions often exhibit complex DNA structures such as the interspersed satellite DNA tracts and the large segmental duplications described in animal pericentromeres (Bailey and Eichler, 2006) or the Ty3/gypsy retrotransposons that colonize at plant pericentromeres (Neumann et al., 2011). Due to this complex and repetitive nature, the pericentromeric and centromeric regions are vastly underrepresented in whole genome assemblies, thus encumbering the pursuit of in-depth evolutionary and genomic analyses within such regions. Targeted efforts are necessary to confer the completeness and accuracy of the centromeric and pericentromeric assembly toward a reliable genomic analysis as shown in some important model genomes (Nagaki et al., 2004; She et al., 2004; Zhang et al., 2004; Hoskins et al., 2007; Wolfgruber et al., 2016).
Through evolution and speciation, the physical position of a centromere can change; a phenomenon most frequently described in conjunction with (and likely driven by) larger chromosomal rearrangements. However, there are reports of occurrences of novel centromere establishment, known as centromere repositioning, which occurs in the absence of any obvious casual chromosomal rearrangements (Montefalcone et al., 1999). Centromere repositioning was used to explain the emergence of evolutionarily new centromeres (Ventura et al., 2001). Evolutionarily new centromeres are centromeres that form de novo at a new site and have been documented, mostly through cytological observations, in both animals (Rocchi et al., 2012) and plants (Han et al., 2009). However, the mechanism underlying their occurrence is not well understood. Several hypotheses have been proposed, including those driven solely through epigenetic changes (Ferreri et al., 2005; Ventura et al., 2007), while others point to latent centromeres (Ventura et al., 2004) and chromosomal rearrangements (Ventura et al., 2003). Important insights have been recently obtained using ChIP-seq (chromatin immunoprecipitation followed by sequencing) analyses. Centromeres from rice (Oryza sativa; Yan et al., 2008), potato (Solanum tuberosum; Gong et al., 2012), and maize (Zea mays; Wolfgruber et al., 2009), as well as in horse (Equus caballus; Wade et al., 2009; Piras et al., 2010) and chicken (Gallus gallus domesticus; Shang et al., 2010), were precisely delineated with regard to size, organization, and centromeric boundaries, by mapping reads from ChIP assays using antibodies against CenH3 (a H3 histone variation, assembled at functional centromeres) to the reference genomes. These results demonstrated that centromere size in higher eukaryotes can vary substantially in length from several kilobases in chicken, to multiple megabases in maize. ChIP-seq analysis in a maize population (Gent et al., 2015) revealed that centromere positions vary even among individuals and this diversity appears to arise from genetic variation rather than epigenetic “drift,” thus offering an alternative explanation for centromere repositioning and emphasizing the role of DNA elements on centromere specification. This is further evidenced by the finding that genetic variation in centromeric satellite repeats on human chromosome 17 influence centromere location (Aldrup-MacDonald et al., 2016).
The genomic characterization of many neocentromeres and evolutionarily new centromeres indicated centromeres may have evolved from euchromatic regions that originally hosted protein coding genes (Nagaki et al., 2004; Marshall et al., 2008; Fu et al., 2013). However, centromeres seem to harbor an environment of both genetic and epigenetic components that hamper the functional responsibilities of coding genes. On one hand, both the surrounding pericentromeric heterochromatin (Elgin and Reuter, 2013) and centromeric chromatin (CenH3 nucleosomes) (Allshire et al., 1995; Shang et al., 2013) were found to be incompatible with gene transcription. Genes uncovered at the centromeres of many plant species, including rice (Yan et al., 2008), maize (Zhao et al., 2016), and potato (Gong et al., 2012), as well as in some human neocentromeres (Marshall et al., 2008), are generally found in CenH3-depleted subdomains that exhibit euchromatic-like histone modifications. On the other hand, genes that are closely linked to centromeres may suffer a high risk of disruption from local structural arrangements due to frequent double-strand DNA breaks occurring in or near centromeres (Wolfgruber et al., 2016). Thus, genes within or near centromeres are more prone to be affected by position-effect variegation (Elgin and Reuter, 2013) that occurs when a euchromatic gene is juxtaposed with heterochromatin and centromeric chromatin via structural rearrangements. Recently, Schneider et al. (2016) found that selection for centromere-linked genes lead to centromere selection during domestication in just a few thousand years. The existence of active genes within or near centromeric regions appears to act as a barrier to prevent centromeric chromatin expansion by inhibiting CenH3 loading. This is probably due to the deleterious effect of silencing a gene by centromeric components, especially when the gene is essential (Fukagawa and Earnshaw, 2014). Taken together, these results suggested an antagonistic relationship between centromere environment and gene survival.
Here, we used comparative genomic and phylogenomic approaches to study centromere evolution in the genus Oryza, with a focus on centromere movement and the fate of centromere-linked genes. We previously reported the whole genome assembly of Oryza brachyantha, which diverged from rice (O. sativa) ∼15 million years ago (Mya; Chen et al., 2013). In this study, we generated, de novo, a 12.4-Mb high-quality BAC-based assembly of pericentromeric/centromeric regions of O. brachyantha. This assembly was independently validated by large single-molecule derived optical maps and hence provided a high degree of confidence in the following analyses. Through interrogating available Oryza genomes (Stein et al., 2018) as well as closely related grass genomes (as outgroups), we were able to perform a comprehensive syntenic and phylogenomic analyses of the centromeric regions on all rice chromosomes. Our results provide a striking insight into centromere evolution, including conservation of centromere location, potential mechanisms underlying centromere repositioning, and the extent to which natural selection may have contributed to gene exclusion from centromeric regions.
RESULTS
Improvement of the O. brachyantha Pericentromeric/Centromeric Sequence
In order to improve on the accuracy and completeness of the pericentromeric and centromeric sequence of O. brachyantha and therefore bolster interspecies genomic comparisons, we de novo sequenced 126 BAC clones (Supplemental Data Set 1) from such regions using a BAC-by-BAC strategy with 454 pyrosequencing (see more details in Methods). A total of 12.4 Mb of nonredundant BAC-based assembly for 10 centromeres was generated (Supplemental Table 1). We also confirmed the assembly of the other two centromeric regions by BAC end sequence alignments and manual inspection. In this genome assembly, the pericentromeric regions on all chromosomes except 6 and 10 extend into the CentO-F centromeric satellites (Supplemental Table 1), demonstrating that the sequence and structure of these pericentromeric regions are well covered. Furthermore, we compared our assembly to a separate build that was generated using optical mapping (BioNano Genomics Irys system) (Lam et al., 2012) to validate the contiguity and completeness of our BAC-based assembly. In comparison to the in silico Nt.BspQI recognition site maps, no obvious discrepancies were identified, demonstrating the high quality of the BAC-based assembly (Supplemental Figure 1).
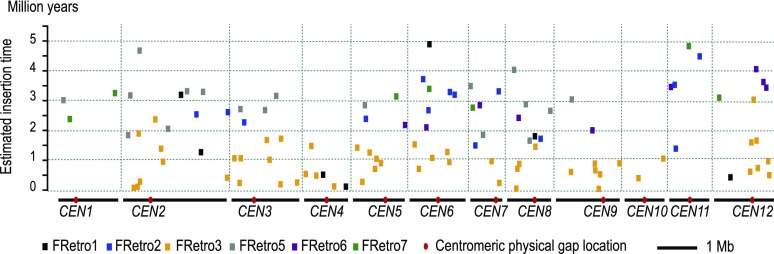
In addition to contiguity, the sequence completeness is also significantly improved compared with the whole-genome shotgun assembly thereby allowing us to generate a high-quality annotation of transposable elements in the O. brachyantha pericentromeric and centromeric regions. Approximately 45.6% and 19.7% of the BAC assemblies are composed of retrotransposons and DNA transposons, respectively (Supplemental Table 2). This is slightly different from the centromeric and pericentromeric regions of O. sativa, in which 60.3% and 8.2% are made up of retroelements and DNA transposons, respectively (Supplemental Table 3). Six LTR-retrotransposon families that have intact elements were identified from the O. brachyantha pericentromeric and centromeric regions. Among them, centromeric retrotransposon FRetro3 (Gao et al., 2009) is the most abundant family having 45 intact elements and 252 solo-LTRs (Supplemental Data Set 2). Previous analysis of the Cen8 BAC sequence from O. brachyantha revealed that the amplification of FRetro3 only occurred within the last few million years (Gao et al., 2009). We found that nearly all FRetro3 elements were inserted into the O. brachyantha genome within the last two million years suggesting a more recent insertion than all other families identified (Figure 1). The overall ratio of solo-LTRs to intact elements in the O. brachyantha pericentromeric and centromeric regions is ∼3.5:1, which is significantly higher than previously described in O. sativa (∼0.9:1; Ma and Bennetzen, 2006), suggesting a considerable proportion of the O. brachyantha pericentromeric sequence has been removed due to unequal recombination events.
Figure 1.
Estimated Insertion Times for Retroelements Identified from the O. brachyantha Pericentromeric Regions.
A total of 107 intact and partially truncated LTR-retrotransposon elements from six families were identified and used for estimation of the insertion time. Different families are represented by different colors. The most abundant element, FRetro3, depicted as yellow rectangles, was found to be inserted into the O. brachyantha centromeric regions within the last two million years.
Comparison of local size variations in the centromeric regions between O. sativa and O. brachyantha revealed that nearly all centromeric regions in O. sativa have been expanded relative to the orthologous centromeric regions of O. brachyantha, and sizes increased from 0.05 in CEN3 to 1.07-fold in CEN4 (Supplemental Table 4). The gene content in the pericentromeric regions is generally low; for genes conserved between O. sativa and O. brachyantha, the density is ∼2.6 per 100 kb in O. brachyantha, compared with ∼1.86 in O. sativa (Supplemental Table 4).
ChIP-Seq with CenH3 in O. brachyantha
To determine which sequences/regions harbor the functional centromeres in O. brachyantha, we performed ChIP-seq analysis using antibodies targeting centromeric histones (CenH3). We obtained a total of 295,118 high-quality 454 reads with an average length of 250 bp. The mapping strategy applied has been previously described in O. sativa (Yan et al., 2008). To avoid mapping bias due to an incomplete CentO-F assembly, we first filtered the reads containing CentO-F sequence. The subsequence mapping results revealed no obvious continuous genomic regions that were enriched for ChIP-seq reads (Supplemental Figure 2), indicating the functional centromeric regions in O. brachyantha were embedded exclusively in highly repetitive sequences, rather than in unique or low copy sequences (Yan et al., 2008; Wade et al., 2009). Surprisingly, we found some genomic regions, including a region from the short arm of chromosome 9 and the pericentromeric regions of chromosomes 6 and 11, which were significantly enriched in our read counts. Sequence analysis of these regions revealed they are 45S rRNA genes, suggesting that 45S rRNA clusters may contribute to the centromeric landscape in O. brachyantha. To test this hypothesis, we performed a classification analysis of the 454 reads in ChIP-seq data set and a control genomic data set also generated using 454 sequencing. The ChIP-seq data set contained 112,177 (∼38%) and 11,684 (∼4%) reads from CentO-F (a more detailed analysis of CentO-F is provided in Supplemental Text 1) and FRetro3, respectively, based on the RepeatMasker annotation (Supplemental Table 5). In contrast, we only observed ∼1.6% and ∼2.5% in the genomic data set, indicating CentO-F and Fretro3 are indeed centromere-associated as shown in previous findings (Lee et al., 2005). As expected, we found that 10,863 reads (∼3.7%) were derived from 45S rRNA genes in the ChIP-seq data set (Supplemental Table 5) compared with only 1.3% in the genomic data set, suggesting some functional centromeric regions may contain 45S rRNA genes in O. brachyantha. We found an ∼600-kb 45S rRNA gene cluster immediately adjacent to the centromeric region of chromosome 9 (Supplemental Figure 3); thus, this cluster most likely contribute to the enrichment of 45S rRNA gene sequence in the ChIP-seq data set.
Centromere Synteny
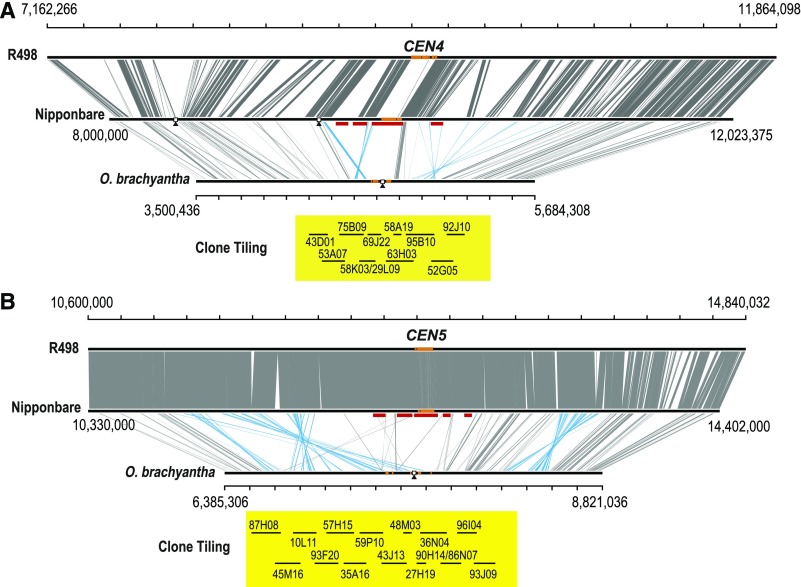
We generated a comparative sequence map between O. sativa and O. brachyantha to infer centromere synteny. We used two genome assemblies for O. sativa: the japonica rice Nipponbare (MSU7) (Kawahara et al., 2013) and the indica rice Shuhui 498 (R498) (Du et al., 2017). Nipponbare was assembled using a traditional map-based, clone-by-clone approach, and R498 was assembled with the integration of single-molecule sequencing and mapping data set, a genetic map, and fosmid sequence tags. Approximately 2 Mb of homologous region flanking each centromere (Supplemental Table 6) was selected from a whole-genome alignment between Nipponbare and O. brachyantha, including ∼5000 syntenic markers (Supplemental Table 7). The synteny map revealed extensive structural rearrangements in centromeric and pericentromeric regions between these two Oryza genomes (Figure 2; Supplemental Figure 4). Next, R498 was aligned to Nipponbare to ascertain whether significant discrepancies exist in the pericentromeric/centromeric regions between the genome assemblies of these two subspecies, which indeed verified the structural rearrangements identified between Nipponbare and O. brachyantha. Shifting of the centromere positions on chromosomes 1, 7 and 9 can each be resolved through a single inversion event. The differences in centromere position on chromosomes 3, 4, 5, 6, and 8 however appear to have arisen from a series of complex local arrangements. The centromere position is conserved on chromosomes 2, 10, and 11 between these two species. We found that only the centromere on chromosome 12 appears to have repositioned considerably, which likely resulted from a centromere repositioning event.
Figure 2.
Comparative Analysis of Syntenic and Conserved Sequences at the Centromeric and Pericentromeric Regions of Chromosomes 4 and 5 between O. sativa and O. brachyantha.
Approximate 4-Mb region with functional centromeric domains embedded for Nipponbare (middle) assembly and the corresponding homologous regions from R498 (up) and O. brachyantha (bottom) are compared. Syntenic and conserved sequences are connected by lines, and sky blue indicates inversions. The remaining sequence gaps in R498 and Nipponbare, and the physical map gaps in O. brachyantha centromeric regions are depicted with black triangles. The centromere-specific satellite repeats (CentO in O. sativa and CentO-F in O. brachyantha) are represented by orange rectangles. CenH3 domains previously defined for Nipponbare are depicted as red rectangles below the middle panel. The assembly of O. brachyantha was based on the combination of the improved BAC-based sequences and the draft genome assembly of the V1.4 (Chen et al., 2013) with extensive manually inspection. The centromere positions of O. brachyantha are defined as the only physical gap for each centromeric region based on the CentO-F distribution and CenH3 ChIP-seq analysis. Clone selected for sequencing are shown below in yellow box. See 10 other centromeric regions in Supplemental Figure 4.
To infer the species of origin in which the inversions occurred, we performed an additional gene-based synteny analysis that included at least outgroup species from Leersia perrieri (Stein et al., 2018), a sister genus to Oryza, Brachypodium distachyon, Setaria italica (foxtail millet), and Sorghum bicolor (Supplemental Figure 5). The homologous regions corresponding to the reference rice genome for the distantly grass species are provided in Supplemental Data Set 3. In total, we were able to trace the origin of 17 pericentromeric inversions, with sizes ranging from ∼50 to ∼1700 kb (Table 1, Figure 3; Supplemental Figures 6 to 13), 10 of which are specific to the O. brachyantha lineage and seven to O. sativa. A pericentric inversion specific to O. sativa has previously been reported to cause the physical shift of Cen8 (Ma et al., 2007), which is inconsistent with our result. We found both L. perrieri and B. distachyon share a relatively similar gene order to O. sativa; therefore, the inversion is presumably specific to O. brachyantha (Supplemental Figure 6) and appears to have no direct effect on the centromere position. We caution that synteny analysis within complex genomic regions such as centromeres can be ambiguous if pursued with only a few available syntenic markers.
Table 1. The Major Pericentric Inversions between Rice and O. brachyantha.
| Name | Size (kb)a | Specificb | Synteny Maps | |
|---|---|---|---|---|
| 1 | PeriCen1 | ∼150 | OB | Supplemental Figure 6A |
| 2 | PeriCen2 | ∼1727 | OB | Supplemental Figure 7 |
| 3 | PeriCen3 | ∼800 | OS | Supplemental Figure 8 |
| 4 | PeriCen4 | ∼630 | OS | Supplemental Figure 9 |
| 5 | PeriCen5_1 | ∼820 | OB | Supplemental Figure 10 |
| 6 | PeriCen5_2 | ∼140 | OB | Supplemental Figure 10 |
| 7 | PeriCen6 | ∼424 | OS | Supplemental Figure 11 |
| 8 | PeriCen7_1 | ∼50 | OB | Figure 3A |
| 9 | PeriCen7_2 | ∼421 | OS | Figure 3A |
| 10 | PeriCen7_3 | ∼501 | OB | Figure 3A |
| 11 | PeriCen8_1 | ∼750 | OB | Supplemental Figure 6B |
| 12 | PeriCen9_1 | ∼155 | OS | Figure 3B |
| 13 | PeriCen9_2 | ∼315 | OB | Figure 3B |
| 14 | PeriCen10_1 | ∼250 | OS | Supplemental Figure 12 |
| 15 | PeriCen10_2 | ∼335 | OB | Supplemental Figure 12 |
| 16 | PeriCen11_1 | ∼110 | OB | Supplemental Figure 13 |
| 17 | PeriCen11_2 | ∼120 | OB | Supplemental Figure 13 |
The size of each pericentric inversion is determined by the length of the inverted segment in the species in which the inversion occurred.
OB, O. brachyantha; OS, O. sativa.
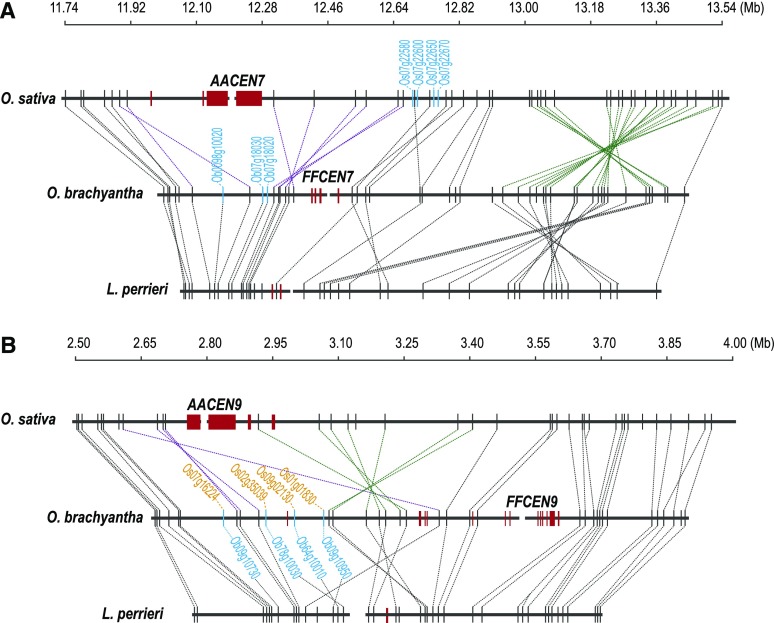
Figure 3.
Syntenic Gene Loss/Movements at the Cen7 and Cen9 Regions.
Red rectangles represent centromere satellite repeats; homologous genes are connected by dashed lines.
(A) Synteny map of the Cen7 region. Light-blue genes indicate genes that have been lost between O. sativa and O. brachyantha. Three inversions were shown to have occurred in the Cen7 region, two (in purple) specific to O. sativa with one leading to the centromere movement. The one remaining inversion (in green) occurred in O. brachyantha.
(B) Synteny map of the Cen9 region. Two inversions occurred between O. sativa and O. brachyantha. One hemicentric inversion occurred specifically in O. brachyantha (green lines), which may lead to centromere movement. The other inversion occurred specifically in O. sativa (purple lines). Four genes (in light blue color) were found to be lost in the centromeric region of O. sativa, but still retains in the inactive centromeric region of O. brachyantha, their homologs (in orange) present at other genomic regions.
Our syntenic data demonstrates that the centromere movement involving chromosome 1 in O. brachyantha (Supplemental Figure 6) and chromosome 7 in O. sativa (Figure 3A) are unique to their respective species and each was caused by a single pericentric inversion. Our data also reveal that the centromere movement on chromosome 9 is specific to O. brachyantha (Figure 3B). We detected a hemicentric inversion (one break in the active centromere and a second break on the chromosome arm) (Lamb et al., 2007) within the O. brachyantha Cen9 region. However, the new centromere is not located exactly at the distal breakpoint region; rather, it moved across a genomic segment of ∼120 kb, distally. Therefore, the centromere movement on O. brachyantha chromosome 9 might have involved a micro-repositioning event, in addition to the inversion. We also observed hemicentric inversions in the Cen5 and Cen10 regions of O. brachyantha (Supplemental Figures 10 and 12).
We were unable to resolve the discrepant centromere synteny observed in Chr3, Chr5, and Chr6 due to extreme syntenic decay within these centromeric regions. Although extensive chromosomal rearrangements have occurred within the centromeric regions between O. brachyantha and O. sativa, the centromeric synteny (approximate centromere position) is preserved with the exception of a few “subtle” changes (<500 kb). Our results suggest that the shift in centromere position between O. sativa and O. brachyantha can be explained by small local inversions and repositioning.
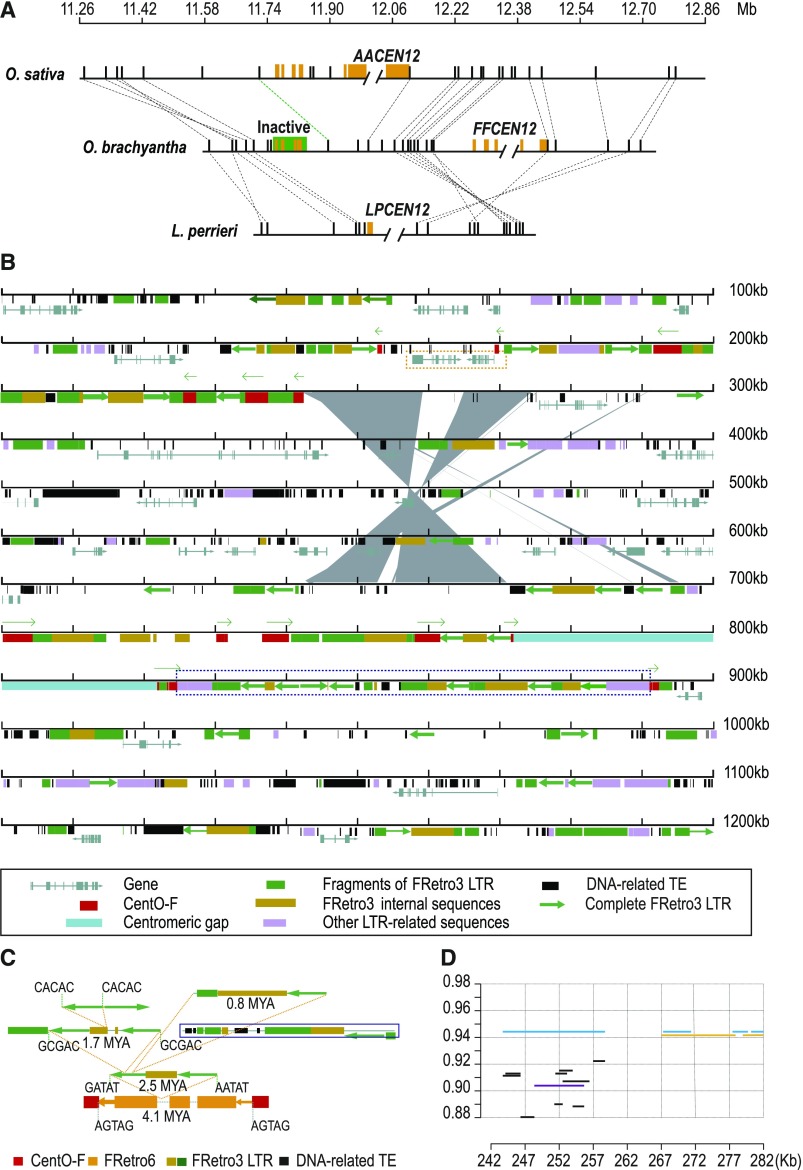
Evolution of Cen12
Centromere movement on chromosome 12 can either be explained via two successive inversions or as the result of a repositioning event. To elucidate the evolutionary origin of this event, we performed a gene-based synteny analysis of Cen12 regions, using L. perrieri as the outgroup (Figure 4A). We verified the sequence contigs from L. perrieri using BAC-end sequence alignments (only uniquely mapped sequences were used) (Supplemental Figure 14). The ancestral centromere position of chromosome 12 can be determined by the outgroup species, L. perrieri, in which the centromere position was shown to be syntenic with O. sativa. This is supported by the fact that the corresponding region of ancestral centromere position in O. brachyantha still retains a small cluster of residual centromeric satellite repeats (Figure 4A). Therefore, we conclude that the centromere of O. brachyantha has likely changed from its ancestral position. The new centromere location separated by ∼400 kb from the ancestral position. An inversion involving only one gene might have caused the centromere movement between the centromere of O. sativa and the inactive centromere of O. brachyantha (Figure 4A). However, due to the lack of syntenic information from L. perrieri, we were unable to assign this inversion and its associated centromere movement to a specific lineage.
Figure 4.
Evolutionarily New Centromere on O. brachyantha Chromosome 12 Probably Resulted from a Duplicated Transposition.
(A) Gene synteny map revealed centromere on O. brachyantha chromosome 12 has repositioned. Double slashes indicate the positions of centromeric gaps, representing the potential centromere positions in each species. Orange rectangles depict centromeric satellite repeats. Orthologous genes (black rectangles) are connected by dashed lines. The predicted ancestral centromere retains a small cluster of residual centromeric satellite repeats (shaded in green). The dashed line in green indicates an inversion containing only one gene has occurred between O. sativa and O. brachyantha.
(B) Annotation map of the 1.2-Mb region of O. brachyantha Cen12. Gene models, transposable elements (see detailed figure legend in the black box below), CentO-F clusters (red squares, with directions indicated by green arrows above), and a segmental duplication event (gray polygon) are depicted in the map. The size of duplicated segment is approximate 28 kb. The paralogous segments are exactly adjacent to the CentO-F sequence in the ancestral centromere position. Two genes in the dashed orange box were inserted into the ancestral centromeric position in O. brachyantha.
(C) Reconstructing the nested insertion history of LTR-retrotransposons at the region within CentO-F cluster at the distal edge of centromeric gap (highlighted with dashed blue box in Figure 4B). A FRetro6 element inserted into the CentO-F cluster occurred at 4.1 Mya. Three FRetro3 insertion events occurred after then, ranging in age from 2.5 to 0.8 Mya. Sequences that can’t be inferred are shown in dashed black box, but their insertion times should be younger than 2.5 Mya.
(D) Pattern of pericentromeric segmental duplications at O. brachyantha Cen12 region. The horizontal axis indicates the selected segment from the O. brachyantha Cen12 region (see coordinate in Figure 4B). This segment traces genomic regions from three chromosomes (black lines, Chr4; purple lines, Chr9; orange lines, Chr1; also see Supplemental Table 8). The light-blue lines indicate the local paralogous segment within the Cen12 region. The vertical axis indicates sequence identity. Sequence identity of these two paralogous segments is of the highest value, ∼0.945.
To investigate whether specific genomic features are associated with this centromere movement, we performed a detailed sequence analysis of the ∼1.2 Mb Cen12 region in O. brachyantha. Interestingly, we identified an inverted segmental duplication of ∼28 kb in the Cen12 region (Figure 4B). The duplicated segments were located precisely at the ancestral and new centromeric positions. The paralogous segments showed a high degree (∼94.5%) of sequence similarity (Figure 4D), suggesting that the duplication occurred recently. The calculation of nucleotide substitutions per site between these two paralogous sequences allow us to estimate an approximate divergent time of ∼1.54 Mya, using the formula: T = K/2r, where K is the average number of substitutions per aligned site and r is the average substitution rate of 1.3 × 10−8 (Ma and Bennetzen, 2004). It should be noted that since genomic regions near centromeres always have a higher mutation rate (Ossowski et al., 2010), the divergent time may still be overestimated. This duplicated segment is immediately adjacent to the CentO-F cluster, indicating at least a portion, if not all, of the ancestral CentO-F sequences are involved in this segmental duplication, consistent with the inverted orientation of CentO-F clusters between the new and old centromeric loci. Since segmental duplications have been proposed to play a role in the new centromere seeding (Cardone et al., 2007) and local segmental duplications facilitate centromere expansion in rice (Ma and Jackson, 2006), we suggest that Cen12 movement in O. brachyantha was more likely as the result of centromere repositioning triggered by segmental duplications.
Another notable feature of the O. brachyantha Cen12 region is that the genomic region (∼66 kb) immediately adjacent to CentO-F cluster of the new centromeric region is composed almost entirely of nested LTR-retrotransposons (Figure 4C). Therefore, LTR-retrotransposon accumulation may have contributed to the new centromere expansion.
Sequence analysis of the duplicated segments revealed a mosaic composition of genomic segments that originated from three chromosomes (Figure 4D; Supplemental Table 8). The structural and evolutionary patterns observed in pericentromeric segmental duplications is similar to those reported in humans (Horvath et al., 2000), suggesting an example of convergent evolution among pericentromeric regions between distant eukaryotic lineages.
An Excess of Syntenic Gene Losses near the O. sativa Centromeres
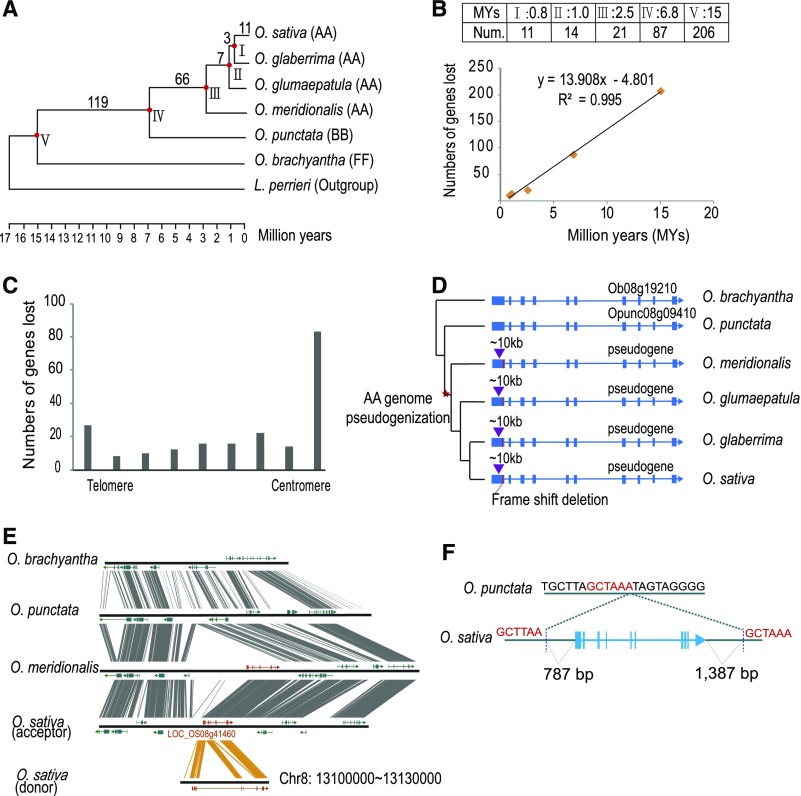
Gene loss is one of the major reasons for synteny decay in plant genomes. To characterize the rate and distribution of syntenic gene loss in O. sativa, we attempted to identify single or low-copy genes (identified as gene copies equal to or less than 3 based on OrthoMCL analysis) that are present in O. brachyantha and have syntenic orthologs in at least one outgroup species (L. perrieri and/or B. distachyon), yet are absent in the syntenic regions of O. sativa. Using intensive manual inspection, we identified 206 genes that had been lost from their original position in the O. sativa genome after its divergence from O. brachyantha, which accounted for ∼1% of the total syntenic gene pairs (206/20,467). We further assigned each gene loss event to the Oryza phylogenetic tree by including four additional Oryza genomes, i.e., O. glaberrima, O. glumaepatula, O. meridionalis, and O. punctata, which diverged from O. sativa 0.8 to 6.8 Mya (Stein et al., 2018) (Figure 5A). We found that the relative syntenic gene loss along the branches leading to O. sativa are proportional to the branch lengths (Figure 5B) with an exception of the branch between nodes II and III, suggesting that gene loss occurred at a fairly constant rate during Oryza genome evolution.
Figure 5.
Syntenic Gene Loss/Movements during Rice Genome Evolution.
(A) Placement of syntenic gene loss events along the branches leading to O. sativa in the Oryza species tree. Nodes (red circles) are labeled with Roman numerals. The length of branches (units, million years) is drawn to scale. The number of syntenic gene loss events is shown at each node. The phylogenetic inference and species divergence times are resolved and described in a recent report (Stein et al., 2018).
(B) Correlation of gene loss event and divergence time. The number of gene loss events was plotted against the divergence time of each node (see [A]). A strong correlation is found (R2 = 0.995), suggesting gene loss events occurred at a constant rate during the Oryza genome evolution.
(C) Syntenic gene loss events are enriched at centromeric or telomeric regions. Each chromosomal arm was divided into 9 bins, each with the same gene number. Gene loss events were calculated for each bin. An apparent enrichment of gene loss events in the bin closest to the centromere was observed.
(D) An example of pseudogenization event in the O. sativa Cen8 region. The purple inverted triangle indicates an insertion event, while the red line indicates a reading-frame shift mutation (2-bp deletion).
(E) LOC_OS08g41460 (MSU7 Chr8: 26,174,014–26,185,145) is highly homologous to Ob08g19210 and the pseudogene sequence in Cen8 region. Analysis of this locus reveals it was derived from an insertion event occurred before the divergence of O. sativa and O. meridionalis but after its divergence with O. punctata, implying this gene was a duplicated copy of the donor sequence. The donor gene at Cen8 became a pseudogene.
(F) Breakpoint analysis of the genomic segment containing locus LOC_OS08g41460 suggests the insertion was associated with the repair of double-strand breaks through nonhomologous end joining. A target site duplication of 6 bp was detected exactly at the insertion point.
We analyzed the distribution of these 206 genes along the 12 chromosomes in O. sativa. We divided each chromosome arm into nine bins from the telomere to the centromere, each bin with an identical gene number (Figure 5C). The gene loss events are enriched at the bins closest to centromeres and telomeres, a phenomenon that also been described in Drosophila melanogaster (Han and Hahn, 2012). Approximately a quarter of the gene loss events (47/206) fell into the regions immediately flanking each centromere with 10 syntenic genes on each side (Supplemental Figure 15 and Supplemental Data Set 4), which are generally within previously defined crossing-over suppressed pericentromeric regions (Yan et al., 2008; Tian et al., 2009). Therefore, centromeric or pericentromeric regions have experienced a significant excess of syntenic gene loss compared with other genomic regions. This is especially apparent in the centromeric regions of chromosome 6 and 9 other chromosomes with the exception of chromosomes 10 and 12 (Binomial test, P < 2.2 × 10−16; Table 2).
Table 2. Analysis of the Pattern of Gene Loss/Movements.
| Chr./Cen. | Expectationa | Observedb | P Value |
|---|---|---|---|
| 1 | 0.007 | 2/20 | 0.0086 |
| 2 | 0.008 | 3/20 | 5.2 × 10−4 |
| 3 | 0.007 | 5/20 | 2.4 × 10−7 |
| 4 | 0.0055 | 3/20 | 1.7 × 10−4 |
| 5 | 0.007 | 5/20 | 2.4 × 10−7 |
| 6 | 0.0095 | 9/20 | 9.6 × 10−14 |
| 7 | 0.012 | 5/20 | 3.3 × 10−6 |
| 8 | 0.018 | 3/20 | 0.0053 |
| 9 | 0.0145 | 4/20 | 1.8 × 10−4 |
| 10 | 0.014 | 1/20 | 0.2457 |
| 11 | 0.018 | 7/20 | 3.8 × 10−8 |
| 12 | 0.013 | 0/20 | 1 |
| All | 0.01 | 47/240 | <2.2 × 10−16 |
Based on the assumption that each gene has the equal frequency to be lost or moved; thus, the number can be approximately calculated as n = (observed total number of gene loss or movement for a given chromosome/total number of syntenic gene pairs between O. sativa and O. brachyantha in that chromosome).
The observed number of gene loss or movement for 20 genes (10 genes on each side of the centromere).
Most Genes Absent from Syntenic Regions near Centromeres Have Duplicated to Nonsyntenic Positions
Since the genes that were identified as missing at the pericentromeric and centromeric regions are of single or low-copy number and also evolutionarily conserved between distantly related grass species, they should be essential for Oryza genomes. Indeed, the functional annotation of these genes revealed that some of them encode important proteins, such as FAD binding and arabino-lactone oxidase domains, AMP binding domain, dolichol phosphate-mannose biosynthesis regulatory, AMP binding domain containing, glycosyltransferase, and so on (Supplemental Data Set 4). Thus, the loss of such proteins at the genomic level seems unlikely since it would otherwise affect gene dosage balance and/or result in lethality. Two circumstances provide reasonable explanations for the missing syntenic genes: These genes existed in two or more copies in the ancestral species when it diverged into O. sativa and O. brachyantha or, alternatively, the genes have simply moved to other genomic positions after the divergence of O. sativa and O. brachyantha. As we expect, among the 47 missing genes in syntenic positions near centromeres in O. sativa, 41 have homologs elsewhere in the genome (Supplemental Data Set 4). To investigate which circumstance lead to the gene loss, we sought to examine whether those nonsyntenic homologs (41 cases) arrived before or after the divergence of O. sativa and O. brachyantha. To achieve this goal, we performed synteny analyses that included five outgroup species, i.e., L. perrieri, B. distachyon, foxtail millet, sorghum, and maize. If a gene was present in a syntenic region with any of the outgroup genomes, it was regarded as an ancestral gene, whereas if it was absent in the syntenic regions from all outgroup species, it would be regarded as a derived gene (Supplemental Figure 16). Using these criteria, we are able to determine that at least 33 genes were derived loci in O. sativa, suggesting that genes moved here from centromeric regions after the split of the O. sativa and O. brachyantha lineages. Of these 33 genes, 32 are subject to purifying selection as shown that the nonsynonymous to synonymous nucleotide substitution rates are significantly less than one by comparing them with homologous genes in the centromeric regions in O. brachyantha (Supplemental Table 9). In addition, 31 genes show expression in seedlings (Zhang et al., 2012; Supplemental Table 9), 27 of which have an FPKM (fragments per kilobase of transcript per million mapped reads) value larger than one. These results indicate the set of relocated genes described retain function after settling into their new locus. Notably, we only found two duplication events that involved more than one gene, each with three and two genes, respectively, indicating that most of gene movement occurred independently (Supplemental Data Set 4).
Most of the missing genes have moved to other genomic regions, thus mitigating the effect of gene dosage imbalance; however, mechanistic action of these movements is poorly understood. In our previous study, we reported that the breakpoint sequence analysis suggested nonhomologous end joining through the repair of double-strand breaks is the major mechanism account for creating the non-collinear genes in the rice genome (Chen et al., 2013). In this study, most of the derived genes (32/33) moved from centromeric regions prior to the divergence of the AA genome species (Supplemental Data Set 4), accounting for ∼2.5 million years of divergence. During such an evolutionary time period, sequence features necessary for a mechanistic characterization are sparse, especially within the rapidly evolved centromeric regions, resulting in only six cases that we were able identify degenerated sequences of parental genes in the ancestral loci and propose a mechanism. In the cases within the rice Cen8 region, two genes (homologs of O. brachyantha Ob08g19210 and Ob08g19230) were lost, while remnants of homolog of Ob08g19210 were observed in the corresponding syntenic region of O. sativa Cen8. Its orthologous gene is syntenically conserved at the Cen8 region in O. brachyantha (Ob08g19210), O. punctata (Opunc08g09410), and L. perrieri, but in all AA species investigated, the first exon of this gene (sequence detected also conserved at syntenic region in AA species) was disrupted by an insertion of ∼10 kb of sequence and a reading frame shift arising by a 2-bp deletion (Figure 5D). Thus, the pseudogenization event occurred before the radiation of the AA species. We found a highly homologous gene, Os08g41460, located on the long arm of chromosome 8, which may be the derived copy (acceptor). The derived copy appears to have originated from an insertion event (Figure 5E) by the double-strand break repair through nonhomologous ending joining (Figure 5F).
If gene duplication is indeed the major mechanism responsible for genes moving out of centromeric regions, we expect there should be considerable and recent gene duplications in centromeric gene regions. An example of this can be observed in the rice Cen4 region, in which a segment from the core region of this centromere was duplicated to the long arm of chromosome 4. This event was demonstrated to have occurred only after the divergence of O. sativa ssp japonica and O. sativa ssp indica (Supplemental Figure 17), suggesting it is a very recent event. The segment contains four annotated genes, including Os04g17650, Os04g17660, Os04g17680, and Os04g17700. In the derived loci, three genes were retained (Os04g24410, Os04g24430, and one unannotated gene model). Thus, gene movements in centromeric regions are still ongoing.
Gene Gain at the Genomic Regions near the O. sativa Centromeres
To investigate the extent of gene gain near centromeres in O. sativa, we focused on 240 genes, which are the most centromere-proximal (10 on each side of each centromere) and with high confidence, i.e., supported by full-length cDNAs, expression evidence (Supplemental Data Set 5), or evolutionary conservation. Of these 240 genes, 146 are shared with O. brachyantha and 14 were lost from the syntenic positions in O. brachyantha, leaving the remaining 80 candidates of gene gain. Since most of them (64/80) have no homologs in O. brachyantha or any other outgroup species, most likely, they have originated after the divergence of O. sativa and O. brachyantha. By introducing a number of intermediate species (Figure 5A), we found 59/64 are specific to the AA lineage. These results are consistent with the recent report (Stein et al., 2018) that the AA-type genomes experienced a higher rate of new gene emergence and new genes are more likely to be found near centromeres over low-recombination regions.
Despite of a striking number of gene gains near the O. sativa centromeres, most are likely new genes and not present before the divergence of O. sativa and O. brachyantha. For those genes that existed before the divergence of O. sativa and O. brachyantha, only 10 can be unambiguously traced from other genomic regions of origin. Notably, during the evolutionary period from the O. sativa-O. brachyantha split to the O. sativa-O. punctata split, only four genes moved in, compared with 24 genes moved out in the same evolutionary duration. This result indicates that gene translocations to O. sativa centromeres are less frequently than moving out, at least during the evolutionary period before it divergent from O. punctata.
Reciprocal Syntenic Gene Loss between O. sativa and O. brachyantha Centromeric Regions Provides Evidence for Antagonism between Gene and Centromere
We identified 45 syntenic gene losses (Supplemental Data Set 6) in the pericentromeric and centromeric regions (genome-wide survey is excluded due to the draft-quality of genomic sequence) of O. brachyantha after its split with O. sativa. Similarly, most of these missing syntenic genes can be traced to their original location within regions immediately adjacent to O. brachyantha centromeres (Figure 3; Supplemental Figures 7 to 13). By combining gene loss events with centromere movements between O. sativa and O. brachyantha, we found an apparent link between the occurrence of gene loss and the position of the functional centromere. For example, the position of O. sativa Cen7 has moved via an inversion (Figure 3A), resulting in a newly defined centromeric region that lost three genes (orthologs of O. brachyantha Ob07g18020, Ob07g18030, and Ob0098g10020). In contrast, the ancient centromeric region retained 4 genes (Os07g22580, Os07g22600, Os07g22650, and Os07g22670) whose orthologs in the corresponding syntenic region of O. brachyantha were lost. This reciprocal gene loss pattern suggests that centromeric environments are not conducive to functional gene activity. Similar trends can be observed in other centromeric regions as well, such as Cen9 (Figure 3B), Cen3 (Supplemental Figure 8), Cen11 (Supplemental Figure 13), and Cen6 (Supplemental Figure 11). These observations provide evidence for the proposed antagonistic relationship between genes and centromeres, and also reveal that selective drive generally pushes genes out of centromeric and pericentromeric regions.
DISCUSSION
Our study presents a sequence-level comparative genomic analysis of pericentromeric/centromeric regions in a relatively complex eukaryotic genome. The high-quality data generated here bolster our ability to accurately identify structural variations and hence permit reliably tracking the evolutionary history within complex pericentromeric and centromeric regions, which in turn allow us to relate each centromere movement to specific chromosomal rearrangements. We also showed that gene loss near centromeres may have accompanied the evolution of centromeres.
Conservation of Centromere Location
Although O. sativa and O. brachyantha separated each other for more than 15 million years and left <10% sequence conserved at the flanking regions of centromeres, our comparative analysis revealed that centromere synteny between these two Oryza genomes is still well maintained with the exception of a few subtle changes. This conservation can even extend to more distantly related grass species, such as B. distachyon, sorghum, maize, and wheat (Triticum aestivum; Qi et al., 2009, 2010; Wang and Bennetzen, 2012). Therefore, from a genomic perspective, the location of the plant centromeres as observed here is unlikely to change substantially, supporting the view that chromosomal location may be an evolutionarily conserved primary determinant for functional centromere identity (Thakur and Sanyal, 2013). This viewpoint could be further supported by recent observations that centromere positions between Z. mays and some distantly related species, e.g., Zea diploperennis, Zea luxurians, and Tripsacum dactyloides are stable within a constraint genomic region although extensive variations existed in CentC content (Gent et al., 2017).
Genomic Variations and Centromere Location
We documented many pericentric inversions in O. sativa and O. brachyantha, and some of the events that gave rise to centromere movements can be explained via evolutionary analysis. It should be noted that the overall inversion number can’t be accurately measured due to the remarkable divergence of sequences and therefore we anticipate the exact number to be larger than what is reported here. Several hemicentric inversions were found in centromeric regions, suggesting such a phenomenon is common in plant centromeric regions (Lamb et al., 2007; Schneider et al., 2016; Wolfgruber et al., 2016). The frequency of detected hemicentric inversions is indeed puzzling given the propensity of these types of inversions to disrupt centromere integrity.
Centromeric chromatin (CenH3-containing nucleosomes) can spread over the flanking noncentromeric regions (Lam et al., 2006; Müller and Almouzni, 2017). In contrast to the dynamic genomic architecture of centromeres, the physical size of a centromere may be a critical element in maintaining a robust epigenetic inheritance (Bodor et al., 2014). For example, in grass species, centromere sizes are positively correlated with genome sizes (Zhang and Dawe, 2012) and are found to be uniform within a given species (Wang et al., 2014b). Centromere expansion may be the key factor in determining whether a genomic variation is tolerated within a centromeric region. Hemicentric inversions can split centromeric chromatin and this centromeric chromatin can then spread into the flanking regions of the breakpoint in order to restore the optimal size of the centromere. The abundance of repetitive sequences around centromeres might be beneficial, as they can provide a mechanism to buffer genomic sequence from variation and exchanges within centromeric regions, thereby stabilizing the region (Fukagawa and Earnshaw, 2014). While some regional centromeres may cover hundreds of kilobases to several megabases along the chromosome (Wolfgruber et al., 2009), any degree of genomic variation within centromeres, both small (e.g., single-nucleotide polymorphisms and indels) and large (e.g., copy number variations and inversions), may have the potential to reshape centromeric chromatin.
Genes Escaping from Genomic Regions near/within Functional Centromeres during Evolution
Genomic positional bias in gene loss patterns have been observed in several evolutionary events, such as during the diploidization that follows whole-genome duplication (Langham et al., 2004; Thomas et al., 2006) and during the evolution of sexual chromosomes from autosomes (Cortez et al., 2014; Bergero et al., 2015). In this study, we observed an excess of syntenic gene loss at the centromeric regions of Oryza genomes, and this bias is generally resulted from selective loss of parental gene copies in the centromeric regions after gene duplications. Two major explanations are proposed to address this pattern, i.e., why duplicated genes in centromeric and pericentromeric regions are more likely to be pseudogenized or lost. First, meiotic recombination is known to be severely repressed or even absent near centromeres; thus, natural selection against deleterious gene mutations would be reduced, a scenario similar to the degeneration of Y chromosome (Charlesworth and Charlesworth, 2000). Our results reveal that most of the relocated genes currently reside within recombination-repressed regions. In addition, deleterious variants are indeed found enriched in regions of low recombination in the rice genome (Liu et al., 2017). Second and more likely, our result revealed genes immediately adjacent to functional centromeric regions are more likely to be lost, which can be explained by the antagonistic relationship between centromeric chromatin (CenH3) and active genes (Wolfgruber et al., 2016). The encapsulation of CenH3 across an active gene may result in the silencing of that gene and is thereby deleterious, especially when the gene is essential. Therefore, genes located close to the centromere may act as a barrier to prevent the spread of centromeric chromatin (CenH3) (Lomiento et al., 2008; Alonso et al., 2010), analogous to the boundary elements for maintaining a local balance of heterochromatin and euchromatin (Wang et al., 2014a). However, if the gene has paralogs (e.g., as a result of gene duplication) in other genomic regions, the barrier function will become weakened or completely lost because redundant genes can compensate the function of this gene. The spread of CenH3 encompassing this gene will have greatly reduced effect on genome function. Once this gene is silenced by CenH3 encapsulation, it would likely accumulate mutations due to the relaxation of functional constraints and eventually become pseudogenized or lost. In addition, gene loss at centromeric regions may be beneficial for centromere establishment and maintenance. Genomic variations are frequently found within centromeric regions; therefore, the expansion of CenH3 may provide a proactive mechanism to maintain optimal centromere size and organization. In this regard, selective loss of a duplicated gene within centromeric regions could provide a sequence resource for use during CenH3 expansion. This would be especially crucial to new centromeres because they are relatively small and successful expansion is thought to play a key role on their final survival and maturity.
Selective forces contributing to biased gene movement have been documented in animal sex chromosome evolution. For example, genes moving out of the mammalian X chromosome are thought to be driven by natural selection to attain male germline function (Emerson et al., 2004). Such forces have also been reported in the X chromosome in Drosophila (Betrán et al., 2002; Vibranovski et al., 2009) and the Z chromosome in silkworm (Wang et al., 2012). In our study, we reveal a potential novel selective force where CenH3 chromatin dynamic spreading or repositioning can drive gene movement or gene relocation in the Oryza genomes.
The observation of abundant new genes at pericentromeric regions appears paradoxical to our conclusion. Several assumptions may help to explain this paradox. First, although new genes can generate at a high rate within the pericentromeric regions as driving by the rapid evolutionary turnover in such regions (She et al., 2004), most would be rapidly lost and a small set will be retained by selection (Carvunis et al., 2012; Neme and Tautz, 2013). New genes have relatively shorter exons and lower expressions, even with relaxed selective constrains comparing to old genes (Stein et al., 2018), suggesting their “dispensable” role to the genome. Our result that most new genes are specific to AA lineage (∼2.5 million years), few were found to be emerged within the duration (∼8.2 million years) between the O. sativa-O. punctata split and the O. sativa-O. brachyantha split, supports the assertion that only a small set of new genes can survive over time if the rate of new gene emergence is constant during evolution. Second, centromere maturity level can affect the rate of new gene creation and/or survival. While centromeres have not been completely evolved to the repeat-based centromeres, their immediate pericentromeric regions are more resistant to accept new genes. However, once centromeres are gradually becoming mature by acquiring sufficient satellite repeats within their core, the expanded flanking pericentromeric regions would become less sensitive to new gene creation because the new created genes are already far away from the centromeric chromatin. Therefore, the distinct extend of new genes observed before and after O. sativa diverged from O. punctata may reflect this effect.
METHODS
BAC Selection, Sequencing, Assembling, and Verification
The BAC library, BAC-end sequences, and fingerprinted contig data of Oryza brachyantha were obtained from the Arizona Genomics Institute (http://www.omap.org/resources.html) (Ammiraju et al., 2006; Kim et al., 2008). BAC clones selected for sequencing were identified by alignments of BAC-end sequences onto pseudomolecules representing the pericentromeric regions of O. brachyantha. The pseudomolecules were constructed by manually ordering and orienting the raw scaffolds that were used to generated the whole genome assembly (Chen et al., 2013). Tiling-path BAC physical maps were also constructed for each centromeric region (covering at least 2 Mb on each side).
We sequenced a total of 126 BAC clones using the Roche 454 platform. These BAC clones represent 21 contigs, including 18 pericentromeric regions, two regions from chromosome 2 that were sequenced to verify the inversion breakpoints, and one region from the telomere on the short arm of chromosome 9 (Supplemental Data Set 1). In addition, we sequenced clone 45C05 to extend the centromeric region of chromosome 8. Several clones for the O. brachyantha centromere 8 have already been sequenced using the standard Sanger methods (Gao et al., 2009). We sequenced 56 clones using a pooled BAC clone strategy (each pool contained two to three clones), and independently sequenced 71 clones due to their repetitive nature (Supplemental Data Set 1).
All BAC sequences were assembled with Newbler (version 2.6) using the default parameters, followed by intensive manual correction. We determined overlaps between neighboring clones using BLASTN and constructed the final pseudomolecules with careful inspection and verification of each overlap.
We compared our BAC-based assemblies to an optical physical genome map to validate accuracy and completeness. We produced the genome map using the BioNano Genomics Irys system (Lam et al., 2012). In total, we collected 39 GB of data (>100 kb) representing ∼130× genome coverage with a N50 length of 154 kb (Supplemental Table 10). The assembly has 244 maps with a N50 of 1.33 Mb that spans 255 Mb.
ChIP-Seq Analysis
ChIP experiments performed as previously described (Yan et al., 2008). ChIP DNA was subjected to 454 sequencing. Reads contain CentO-F sequences were first filtered using BLASTN and RepeatMasker (www.repeatmasker.org). The rest reads were mapped onto the improved O. brachyantha genome assembly using LASTZ (Schwartz et al., 2003) with coverage of 90% and identity of 95% (Supplemental Methods). We plotted the number of CenH3 ChIP-seq reads along individual chromosomes in widow size of 20 kb, spaced every 10 kb.
Identification of LTR-Retrotransposon and Timing of Its Integration
LTR-retrotransposons were identified using LTR_STRUC (McCarthy and McDonald, 2003) with default settings and RepeatMasker with a custom TE library for O. brachyantha (Supplemental Methods). The integration times of full LTR-retrotransposon elements were estimated as previously described (Yang et al., 2012).
Sequence-Based Synteny Map
Genome sequences of Oryza sativa ssp japonica (IRGSP1.0), O. sativa ssp indica R498, and O. brachyantha were soft masked using RepeatMasker (www.repeatmasker.org) with independent custom TE libraries. Chromosome one-to-one alignments were performed with LASTZ, with parameters set as: K = 2200, L = 6000, Y = 3400, E = 30, H = 0, O = 400, and T = 1. The resultant alignment blocks were further parsed with the Chain/Net package (Kent et al., 2003) as described previously (Chen et al., 2013). A custom Perl script (https://github.com/yiliao1022) was used to identify syntenic blocks, requiring neighboring blocks to be no more than five blocks apart. Manual inspection was conducted if necessary. Syntenic blocks identified for 2 Mb on each side of centromeres were extracted and used for constructing the synteny map.
Gene Synteny Map
We used syntenic and orthologous genes as markers to construct the synteny map between O. sativa ssp japonica and O. brachyantha at each centromeric region, using appropriate species as outgroups, such as Leersia perrieri, Brachypodium distachyon, Setaria italica, Sorghum bicolor, and/or Zea mays (Supplemental Methods).
Centromere Satellite Repeats for L. perrieri
Approximate 2-Mb sequences from the pseudomolecules of L. perrieri (Stein et al., 2018) corresponding to the homologous regions of rice centromeres were searched by Tandem Repeat Finder (Benson, 1999). The most abundant tandem repeat sequences found were the satellite repeats with a unit size of 180 bp, which were exclusively mapped at the homologous regions of rice centromere.
Sequence Annotation for O. brachyantha Cen12 Region
We annotated genes, transposable elements, and inter/intrasegmental duplications in the O. brachyantha Cen12 region (∼1.2 Mb). Gene models identified using the MAKER v2.31 annotation engine (http://www.yandell-lab.org/software/maker.html), incorporating homology, expression evidence, and ab initio gene prediction methods. TE annotation is described as the above. Segmental duplications refer to paralogous sequences with coverage >1 kb and identity >90%. More details are provided in Supplemental Methods.
Syntenic Gene Losses
In this study, syntenic gene loss refers to a gene that has been lost in a syntenic position between O. sativa and O. brachyantha. The gene may be either completely lost from the genome or moved to other genomic locations. To identify syntenic gene losses in O. sativa after its divergence from O. brachyantha, we searched genes that are syntenically conserved between O. brachyantha and at least one outgroup species, L. perrieri and/or B. distachyon, but absent at the corresponding syntenic region in O. sativa. To filter false positives that may result from a missing annotation or sequence gap, we conducted a three-step process as previously described (Schnable et al., 2012). To place gene loss events on the branches of the Oryza phylogenic tree leading to O. sativa, we also assessed these events in four internal branch species, including O. glaberrima, O. glumaepatula, O. meridionalis, and O. punctata, which represent an evolutionary gradient.
Accession Numbers
The CenH3 ChIP-seq 454 reads have been deposited in GenBank under accession number SRX4224850. The monomer sequence of centromeric satellite repeat of L. perrieri has been deposited in EMBL under accession number LS9738733.1.The assembly of BAC clones are available at https://de.cyverse.org/dl/d/A07342B7-6E3D-485D-8605-867C93509F74/Obrac_Centromere_BAC_seq.fasta.
Supplemental Data
Supplemental Figure 1. Alignment of the in silico reference sequence motif map for the pericentromeric regions (upper panel) and the map of the same regions produced by genome mapping (lower panel).
Supplemental Figure 2. Plot of CenH3 ChIP-seq reads along the O. brachyantha chromosomes.
Supplemental Figure 3. BioNano genome maps aligned to chromosome 9.
Supplemental Figure 4. Comparative sequence analysis of the centromeres and flanking regions between O. sativa and O. brachyantha.
Supplemental Figure 5. Phylogenetic tree and divergence times of some grass species
Supplemental Figure 6. Synteny map of orthologous genes at Cen1 and Cen8 regions.
Supplemental Figure 7. Gene synteny at the centromeric regions of chromosome 2 between O. sativa and O. brachyantha and the homologous region (scaffold_1: 16.6–21.6 Mb) from S. italica (outgroup).
Supplemental Figure 8. Gene synteny at the centromeric regions of chromosome 3 in rice (O. sativa) and O. brachyantha and the homologous regions from S. italica (outgroup 1) and S. bicolor (outgroup 2).
Supplemental Figure 9. Gene synteny at the centromeric region of chromosome 4 in rice (O. sativa) and O. brachyantha and the homologous regions from B. distachyon (Bd5: 51.80–68.45 Mb) and S. bicolor (Chromosome_6: 6.68–10.11 Mb).
Supplemental Figure 10. Gene synteny at the centromeric region of chromosome 5 in rice (O. sativa), O. brachyantha, and L. perrieri, and the homologous region from S. italica (scaffold3: 28.7–34.4 Mb).
Supplemental Figure 11. Gene synteny at the centromeric region of chromosome 6 in rice (O. sativa) and O. brachyantha, as well as the homologous region from S. italica (scaffold_4: 18.50–22.87 Mb).
Supplemental Figure 12. Gene synteny at the centromeric region of chromosome 10 in rice (O. sativa), O. brachyantha, and L. perrieri (outgroup).
Supplemental Figure 13. Gene synteny at the centromeric region of chromosome 11 in rice (O. sativa), O. brachyantha, and L. perrieri, and the homologous region from B. distachyon (Bd4: 22.30–23.22 Mb).
Supplemental Figure 14. Two contigs from the L. perrieri centromeric regions of chromosome 12 are verified by BAC end sequence alignments.
Supplemental Figure 15. Our targeted regions (10 syntenic genes on each side of the centromere) generally overlap with crossing-over suppressed pericentromeric regions in rice.
Supplemental Figure 16. Examples illustrating the identification of an ancestral or derived gene locus in O. sativa.
Supplemental Figure 17. A segment containing multiple genes from rice Cen4 has duplicated to the long arm of chromosome 4.
Supplemental Figure 18. Chromosome-specific distribution of CentoF.
Supplemental Table 1. Statistics of BAC-based assembly.
Supplemental Table 2. O. brachyantha pericentromeric region repeat counts by repeat classes.
Supplemental Table 3. O. sativa pericentromeric/centromeric region* repeat counts by repeat classes.
Supplemental Table 4. Comparison of the conserved gene content and local size variation at the centromeric/pericentromeric regions between O. sativa and O. brachyantha.
Supplemental Table 5. Comparison of 454 reads between CenH3 ChIP data set and whole-genome shotgun data set.
Supplemental Table 6. Targeted regions for comparative mapping.
Supplemental Table 7. Number of syntenic blocks detected using LASTZ alignments between O. sativa and O. brachyantha.
Supplemental Table 8. Segmental duplications within the O. brachyantha Cen12 region.
Supplemental Table 9. Nonsynonymous to synonymous substitution rates (ka/ks) between O. sativa centromeric moved genes and their homologs in the centromeric regions of O. brachyantha, and the expression level of the moved genes in seedling.
Supplemental Table 10. Summary of Oryza brachyantha raw BioNano molecule data.
Supplemental Table 11. Average sequence similarity (%) of CentO-F monomers within and between O. brachyantha centromeres.
Supplemental Data Set 1. List of BAC clones sequenced for the O. brachyantha pericentromeric and/or centromeric regions.
Supplemental Data Set 2. Statistics of intact, truncated, and solo LTR-retrotransposons in O. brachyantha pericentromeric regions.
Supplemental Data Set 3. Major homologous/syntenic chromosomal segments of other grass species compared with O. sativa.
Supplemental Data Set 4. Genes identified that moved or were lost from the rice genome after its split with O. brachyantha.
Supplemental Data Set 5. Investigating of the most 240 centromere-proximal genes in O. sativa.
Supplemental Data Set 6. Genes identified that moved or were lost from the O. brachyantha pericentromeric regions after it split with O. sativa.
Supplemental Data Set 7. Gene synteny at 12 centromeric regions.
Supplemental Text 1. Centromeric satellite repeat sequence (CentO-F) in Oryza brachyantha.
Dive Curated Terms
The following phenotypic, genotypic, and functional terms are of significance to the work described in this paper:
Acknowledgments
This work was supported by grants from the National Natural Science Foundation of China (31571309, 31371284, and 31771409 to M.C., and 31401074 to B.L.) and by The State Key Laboratory of Plant Genomics.
AUTHOR CONTRIBUTIONS
Y.L. and M.C. conceived and carried out the experiments, analyzed the data, and wrote the article. X.Z., B.L., T.L., J.C., Z.B., M.W., and J.S. carried out the experiments and analyzed the data. J.G.W. and J.J. provided CenH3 ChIP-seq data and edited the article. R.A.W. provided unpublished Oryza genome assemblies and edited the article. All authors read and approved the final manuscript.
References
- Aldrup-MacDonald M.E., Kuo M.E., Sullivan L.L., Chew K., Sullivan B.A. (2016). Genomic variation within alpha satellite DNA influences centromere location on human chromosomes with metastable epialleles. Genome Res. 26: 1301–1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allshire R.C., Nimmo E.R., Ekwall K., Javerzat J.P., Cranston G. (1995). Mutations derepressing silent centromeric domains in fission yeast disrupt chromosome segregation. Genes Dev. 9: 218–233. [DOI] [PubMed] [Google Scholar]
- Alonso A, Hasson D, Cheung F, Warburton PE (2010). A paucity of heterochromatin at functional human neocentromeres. Epigenet. Chromatin 3: 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ammiraju J.S.S., et al. (2006). The Oryza bacterial artificial chromosome library resource: construction and analysis of 12 deep-coverage large-insert BAC libraries that represent the 10 genome types of the genus Oryza. Genome Res. 16: 140–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey J.A., Eichler E.E. (2006). Primate segmental duplications: crucibles of evolution, diversity and disease. Nat. Rev. Genet. 7: 898. [DOI] [PubMed] [Google Scholar]
- Benson G. (1999). Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Res. 27: 573–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergero R., Qiu S., Charlesworth D. (2015). Gene loss from a plant sex chromosome system. Curr. Biol. 25: 1234–1240. [DOI] [PubMed] [Google Scholar]
- Betrán E., Thornton K., Long M. (2002). Retroposed new genes out of the X in Drosophila. Genome Res. 12: 1854–1859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodor D.L., Mata J.F., Sergeev M., David A.F., Salimian K.J., Panchenko T., Cleveland D.W., Black B.E., Shah J.V., Jansen L.E.T. (2014). The quantitative architecture of centromeric chromatin. eLife 3: e02137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardone M.F., Lomiento M., Teti M.G., Misceo D., Roberto R., Capozzi O., D’Addabbo P., Ventura M., Rocchi M., Archidiacono N. (2007). Evolutionary history of chromosome 11 featuring four distinct centromere repositioning events in Catarrhini. Genomics 90: 35–43. [DOI] [PubMed] [Google Scholar]
- Carvunis A.R., et al. (2012). Proto-genes and de novo gene birth. Nature 487: 370–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth B., Charlesworth D. (2000). The degeneration of Y chromosomes. Philos. Trans. R. Soc. Lond. B Biol. Sci. 355: 1563–1572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J., et al. (2013). Whole-genome sequencing of Oryza brachyantha reveals mechanisms underlying Oryza genome evolution. Nat. Commun. 4: 1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortez D., Marin R., Toledo-Flores D., Froidevaux L., Liechti A., Waters P.D., Grützner F., Kaessmann H. (2014). Origins and functional evolution of Y chromosomes across mammals. Nature 508: 488–493. [DOI] [PubMed] [Google Scholar]
- Du H., et al. (2017). Sequencing and de novo assembly of a near complete indica rice genome. Nat. Commun. 8: 15324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elgin S.C., Reuter G. (2013). Position-effect variegation, heterochromatin formation, and gene silencing in Drosophila. Cold Spring Harb. Perspect. Biol. 5: a017780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emerson J.J., Kaessmann H., Betrán E., Long M. (2004). Extensive gene traffic on the mammalian X chromosome. Science 303: 537–540. [DOI] [PubMed] [Google Scholar]
- Ferreri G.C., Liscinsky D.M., Mack J.A., Eldridge M.D.B., O’Neill R.J. (2005). Retention of latent centromeres in the mammalian genome. J. Hered. 96: 217–224. [DOI] [PubMed] [Google Scholar]
- Fu S., Lv Z., Gao Z., Wu H., Pang J., Zhang B., Dong Q., Guo X., Wang X.J., Birchler J.A., Han F. (2013). De novo centromere formation on a chromosome fragment in maize. Proc. Natl. Acad. Sci. USA 110: 6033–6036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukagawa T., Earnshaw W.C. (2014). The centromere: chromatin foundation for the kinetochore machinery. Dev. Cell 30: 496–508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao D., et al. (2009). A lineage-specific centromere retrotransposon in Oryza brachyantha. Plant J. 60: 820–831. [DOI] [PubMed] [Google Scholar]
- Gent J.I., Wang K., Jiang J., Dawe R.K. (2015). Stable patterns of CENH3 occupancy through maize lineages containing genetically similar centromeres. Genetics 200: 1105–1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gent J.I., Wang N., Dawe R.K. (2017). Stable centromere positioning in diverse sequence contexts of complex and satellite centromeres of maize and wild relatives. Genome Biol. 18: 121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong Z., Wu Y., Koblízková A., Torres G.A., Wang K., Iovene M., Neumann P., Zhang W., Novák P., Buell C.R., Macas J., Jiang J. (2012). Repeatless and repeat-based centromeres in potato: implications for centromere evolution. Plant Cell 24: 3559–3574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han M.V., Hahn M.W. (2012). Inferring the history of interchromosomal gene transposition in Drosophila using n-dimensional parsimony. Genetics 190: 813–825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han Y., Zhang Z., Liu C., Liu J., Huang S., Jiang J., Jin W. (2009). Centromere repositioning in cucurbit species: implication of the genomic impact from centromere activation and inactivation. Proc. Natl. Acad. Sci. USA 106: 14937–14941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henikoff S., Ahmad K., Malik H.S. (2001). The centromere paradox: stable inheritance with rapidly evolving DNA. Science 293: 1098–1102. [DOI] [PubMed] [Google Scholar]
- Horvath J.E., Schwartz S., Eichler E.E. (2000). The mosaic structure of human pericentromeric DNA: a strategy for characterizing complex regions of the human genome. Genome Res. 10: 839–852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoskins R.A., et al. (2007). Sequence finishing and mapping of Drosophila melanogaster heterochromatin. Science 316: 1625–1628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawahara Y., et al. (2013). Improvement of the Oryza sativa Nipponbare reference genome using next generation sequence and optical map data. Rice (N.Y.) 6: 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent W.J., Baertsch R., Hinrichs A., Miller W., Haussler D. (2003). Evolution’s cauldron: duplication, deletion, and rearrangement in the mouse and human genomes. Proc. Natl. Acad. Sci. USA 100: 11484–11489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H., et al. (2008). Construction, alignment and analysis of twelve framework physical maps that represent the ten genome types of the genus Oryza. Genome Biol. 9: R45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam A.L., Boivin C.D., Bonney C.F., Rudd M.K., Sullivan B.A. (2006). Human centromeric chromatin is a dynamic chromosomal domain that can spread over noncentromeric DNA. Proc. Natl. Acad. Sci. USA 103: 4186–4191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam E.T., Hastie A., Lin C., Ehrlich D., Das S.K., Austin M.D., Deshpande P., Cao H., Nagarajan N., Xiao M., Kwok P.Y. (2012). Genome mapping on nanochannel arrays for structural variation analysis and sequence assembly. Nat. Biotechnol. 30: 771–776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamb J.C., Meyer J.M., Birchler J.A. (2007). A hemicentric inversion in the maize line knobless Tama flint created two sites of centromeric elements and moved the kinetochore-forming region. Chromosoma 116: 237–247. [DOI] [PubMed] [Google Scholar]
- Langham R.J., Walsh J., Dunn M., Ko C., Goff S.A., Freeling M. (2004). Genomic duplication, fractionation and the origin of regulatory novelty. Genetics 166: 935–945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H.R., Zhang W., Langdon T., Jin W., Yan H., Cheng Z., Jiang J. (2005). Chromatin immunoprecipitation cloning reveals rapid evolutionary patterns of centromeric DNA in Oryza species. Proc. Natl. Acad. Sci. USA 102: 11793–11798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q., Zhou Y., Morrell P.L., Gaut B.S. (2017). Deleterious variants in Asian rice and the potential cost of domestication. Mol. Biol. Evol. 34: 908–924. [DOI] [PubMed] [Google Scholar]
- Lomiento M., Jiang Z., D’Addabbo P., Eichler E.E., Rocchi M. (2008). Evolutionary-new centromeres preferentially emerge within gene deserts. Genome Biol. 9: R173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma J., Bennetzen J.L. (2004). Rapid recent growth and divergence of rice nuclear genomes. Proc. Natl. Acad. Sci. USA 101: 12404–12410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma J., Bennetzen J.L. (2006). Recombination, rearrangement, reshuffling, and divergence in a centromeric region of rice. Proc. Natl. Acad. Sci. USA 103: 383–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma J., Jackson S.A. (2006). Retrotransposon accumulation and satellite amplification mediated by segmental duplication facilitate centromere expansion in rice. Genome Res. 16: 251–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma J., Wing R.A., Bennetzen J.L., Jackson S.A. (2007). Evolutionary history and positional shift of a rice centromere. Genetics 177: 1217–1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall O.J., Chueh A.C., Wong L.H., Choo K.H.A. (2008). Neocentromeres: new insights into centromere structure, disease development, and karyotype evolution. Am. J. Hum. Genet. 82: 261–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy E.M., McDonald J.F. (2003). LTR_STRUC: a novel search and identification program for LTR retrotransposons. Bioinformatics 19: 362–367. [DOI] [PubMed] [Google Scholar]
- Montefalcone G., Tempesta S., Rocchi M., Archidiacono N. (1999). Centromere repositioning. Genome Res. 9: 1184–1188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller S., Almouzni G. (2017). Chromatin dynamics during the cell cycle at centromeres. Nat. Rev. Genet. 18: 192–208. [DOI] [PubMed] [Google Scholar]
- Nagaki K., Cheng Z., Ouyang S., Talbert P.B., Kim M., Jones K.M., Henikoff S., Buell C.R., Jiang J. (2004). Sequencing of a rice centromere uncovers active genes. Nat. Genet. 36: 138–145. [DOI] [PubMed] [Google Scholar]
- Neme R., Tautz D. (2013). Phylogenetic patterns of emergence of new genes support a model of frequent de novo evolution. BMC Genomics 14: 117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann P, Navratilova A, Koblizkova A, Kejnovsky E, Hribova E, Hobza R, Widmer A, Dolezel J, Macas J (2011). Plant centromeric retrotransposons: a structural and cytogenetic perspective. Mobile DNA 2: 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ossowski S., Schneeberger K., Lucas-Lledó J.I., Warthmann N., Clark R.M., Shaw R.G., Weigel D., Lynch M. (2010). The rate and molecular spectrum of spontaneous mutations in Arabidopsis thaliana. Science 327: 92–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piras F.M., Nergadze S.G., Magnani E., Bertoni L., Attolini C., Khoriauli L., Raimondi E., Giulotto E. (2010). Uncoupling of satellite DNA and centromeric function in the genus Equus. PLoS Genet. 6: e1000845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi L., Friebe B., Zhang P., Gill B.S. (2009). A molecular-cytogenetic method for locating genes to pericentromeric regions facilitates a genomewide comparison of synteny between the centromeric regions of wheat and rice. Genetics 183: 1235–1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi L., Friebe B., Wu J., Gu Y., Qian C., Gill B.S. (2010). The compact Brachypodium genome conserves centromeric regions of a common ancestor with wheat and rice. Funct. Integr. Genomics 10: 477–492. [DOI] [PubMed] [Google Scholar]
- Rocchi M., Archidiacono N., Schempp W., Capozzi O., Stanyon R. (2012). Centromere repositioning in mammals. Heredity (Edinb.) 108: 59–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnable J.C., Freeling M., Lyons E. (2012). Genome-wide analysis of syntenic gene deletion in the grasses. Genome Biol. Evol. 4: 265–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider K.L., Xie Z., Wolfgruber T.K., Presting G.G. (2016). Inbreeding drives maize centromere evolution. Proc. Natl. Acad. Sci. USA 113: E987–E996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz S., Kent W.J., Smit A., Zhang Z., Baertsch R., Hardison R.C., Haussler D., Miller W. (2003). Human-mouse alignments with BLASTZ. Genome Res. 13: 103–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang W.H., et al. (2013). Chromosome engineering allows the efficient isolation of vertebrate neocentromeres. Dev. Cell 24: 635–648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang W.H., Hori T., Toyoda A., Kato J., Popendorf K., Sakakibara Y., Fujiyama A., Fukagawa T. (2010). Chickens possess centromeres with both extended tandem repeats and short non-tandem-repetitive sequences. Genome Res. 20: 1219–1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- She X., et al. (2004). The structure and evolution of centromeric transition regions within the human genome. Nature 430: 857–864. [DOI] [PubMed] [Google Scholar]
- Stein J.C., et al. (2018). Genomes of 13 domesticated and wild rice relatives highlight genetic conservation, turnover and innovation across the genus Oryza. Nat. Genet. 50: 285–296. [DOI] [PubMed] [Google Scholar]
- Thakur J., Sanyal K. (2013). Efficient neocentromere formation is suppressed by gene conversion to maintain centromere function at native physical chromosomal loci in Candida albicans. Genome Res. 23: 638–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas B.C., Pedersen B., Freeling M. (2006). Following tetraploidy in an Arabidopsis ancestor, genes were removed preferentially from one homeolog leaving clusters enriched in dose-sensitive genes. Genome Res. 16: 934–946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian Z., Rizzon C., Du J., Zhu L., Bennetzen J.L., Jackson S.A., Gaut B.S., Ma J. (2009). Do genetic recombination and gene density shape the pattern of DNA elimination in rice long terminal repeat retrotransposons? Genome Res. 19: 2221–2230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ventura M., et al. (2004). Recurrent sites for new centromere seeding. Genome Res. 14: 1696–1703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ventura M., Archidiacono N., Rocchi M. (2001). Centromere emergence in evolution. Genome Res. 11: 595–599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ventura M., Mudge J.M., Palumbo V., Burn S., Blennow E., Pierluigi M., Giorda R., Zuffardi O., Archidiacono N., Jackson M.S., Rocchi M. (2003). Neocentromeres in 15q24-26 map to duplicons which flanked an ancestral centromere in 15q25. Genome Res. 13: 2059–2068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ventura M., Antonacci F., Cardone M.F., Stanyon R., D’Addabbo P., Cellamare A., Sprague L.J., Eichler E.E., Archidiacono N., Rocchi M. (2007). Evolutionary formation of new centromeres in macaque. Science 316: 243–246. [DOI] [PubMed] [Google Scholar]
- Vibranovski M.D., Zhang Y., Long M. (2009). General gene movement off the X chromosome in the Drosophila genus. Genome Res. 19: 897–903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wade C.M., et al.; Broad Institute Genome Sequencing Platform; Broad Institute Whole Genome Assembly Team (2009). Genome sequence, comparative analysis, and population genetics of the domestic horse. Science 326: 865–867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Bennetzen J.L. (2012). Centromere retention and loss during the descent of maize from a tetraploid ancestor. Proc. Natl. Acad. Sci. USA 109: 21004–21009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Long M., Vibranovski M.D. (2012). Retrogenes moved out of the z chromosome in the silkworm. J. Mol. Evol. 74: 113–126. [DOI] [PubMed] [Google Scholar]
- Wang J., Lawry S.T., Cohen A.L., Jia S. (2014a). Chromosome boundary elements and regulation of heterochromatin spreading. Cell. Mol. Life Sci. 71: 4841–4852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K., Wu Y., Zhang W., Dawe R.K., Jiang J. (2014b). Maize centromeres expand and adopt a uniform size in the genetic background of oat. Genome Res. 24: 107–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfgruber T.K., et al. (2009). Maize centromere structure and evolution: sequence analysis of centromeres 2 and 5 reveals dynamic Loci shaped primarily by retrotransposons. PLoS Genet. 5: e1000743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfgruber T.K., Nakashima M.M., Schneider K.L., Sharma A., Xie Z., Albert P.S., Xu R., Bilinski P., Dawe R.K., Ross-Ibarra J., Birchler J.A., Presting G.G. (2016). High quality maize centromere 10 sequence reveals evidence of frequent recombination events. Front. Plant Sci. 7: 308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan H., Talbert P.B., Lee H.R., Jett J., Henikoff S., Chen F., Jiang J. (2008). Intergenic locations of rice centromeric chromatin. PLoS Biol. 6: e286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L., Liu T., Li B., Sui Y., Chen J., Shi J., Wing R.A., Chen M. (2012). Comparative sequence analysis of the Ghd7 orthologous regions revealed movement of Ghd7 in the grass genomes. PLoS One 7: e50236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H., Dawe R.K. (2012). Total centromere size and genome size are strongly correlated in ten grass species. Chromosome Res. 20: 403–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W., Wu Y., Schnable J.C., Zeng Z., Freeling M., Crawford G.E., Jiang J. (2012). High-resolution mapping of open chromatin in the rice genome. Genome Res. 22: 151–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Huang Y., Zhang L., Li Y., Lu T., Lu Y., Feng Q., Zhao Q., Cheng Z., Xue Y., Wing R.A., Han B. (2004). Structural features of the rice chromosome 4 centromere. Nucleic Acids Res. 32: 2023–2030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao HN, Zhu XB, Wang K, Gent JI, Zhang WL, Dawe RK, Jiang JM. 2016. Gene expression and chromatin modifications associated with maize centromeres. G3 (Bethesda) 6: 183–192. [DOI] [PMC free article] [PubMed] [Google Scholar]