Glycosyltransferases and acyltransferases in corms of the ornamental plant montbretia are involved in a complex flavonoid biosynthetic system relevant to human health.
Abstract
Plant specialized metabolism serves as a rich resource of biologically active molecules for drug discovery. The acylated flavonol glycoside montbretin A (MbA) and its precursor myricetin 3-O-(6’-O-caffeoyl)-glucosyl rhamnoside (mini-MbA) are potent inhibitors of human pancreatic α-amylase and are being developed as drug candidates to treat type-2 diabetes. MbA occurs in corms of the ornamental plant montbretia (Crocosmia x crocosmiiflora), but a system for large-scale MbA production is currently unavailable. Biosynthesis of MbA from the flavonol myricetin and MbA accumulation occur during early stages of corm development. We established myricetin 3-O-rhamnoside (MR), myricetin 3-O-glucosyl rhamnoside (MRG), and mini-MbA as the first three intermediates of MbA biosynthesis. Contrasting the transcriptomes of young and old corms revealed differentially expressed UDP-sugar-dependent glycosyltransferases (UGTs) and BAHD-acyltransferases (BAHD-ATs). UGT77B2 and UGT709G2 catalyze the consecutive glycosylation of myricetin to produce MR and of MR to give MRG, respectively. In addition, two BAHD-ATs, CcAT1 and CcAT2, catalyze the acylation of MRG to complete the formation of mini-MbA. Transcript profiles of UGT77B2, UGT709G2, CcAT1, and CcAT2 during corm development matched the metabolite profile of MbA accumulation. Expression of these enzymes in wild tobacco (Nicotiana benthamiana) resulted in the formation of a surrogate mini-MbA, validating the potential for metabolic engineering of mini-MbA in a heterologous plant system.
INTRODUCTION
Plants produce more than 200,000 different specialized metabolites, traditionally referred to as secondary metabolites (Dixon and Strack, 2003). These metabolites have functions beyond growth and development, such as in interactions of plants with their environment (Hartmann, 2008). For millennia, humans have been exploring the chemical diversity of plants for medicinal activities. Consequently, more than half of the drugs currently used in western medicine are based on specialized metabolites found in nature, and about a quarter of all prescription drugs contain plant-derived compounds (Pan et al., 2013).
One of the major health challenges of the 21st century is the increasing occurrence of type-2 diabetes mellitus, a chronic disease that affects over 320 million people worldwide (http://www.who.int). Type-2 diabetes is characterized by the body’s inefficient use of insulin and hyperglycemia, which may cause long-term damage to different organs of the human body. The regulation of blood glucose levels in patients diagnosed with type-2 diabetes can be achieved, among other treatments, by inhibiting the human enzymes involved in starch degradation, such as the pancreatic α-amylase (HPA), which cleaves starch into oligosaccharides, and the gut α-glucosidases, which degrade those oligosaccharides, as well as dietary oligosaccharides such as sucrose, to glucose. Drugs currently on the market under the trade names Acarbose, Miglitol, and Voglibose inhibit the gut α-glucosidases and have the positive effect of reducing sugar uptake. However, they also result in the movement of oligo- and disaccharides to the lower bowel where they result in osmotic imbalance and can be rapidly fermented by the gut microbiome, causing undesirable side effects such as abdominal pain, flatulence, or diarrhea. These side effects can lead to patient noncompliance (Scheen, 1997, 2003; Scott and Spencer, 2000).
The use of a specific inhibitor of the amylase but not the α-glucosidases should bypass these problems, thereby minimizing the described side effects of existing drugs in the treatment of type-2 diabetes. Relatively recently, Tarling et al. (2008) screened a library of 30,000 extracts from terrestrial and marine sources for inhibitors of HPA and identified montbretin A (MbA) isolated from montbretia (Crocosmia x crocosmiiflora) corms as a highly specific and potent HPA inhibitor. MbA selectively inhibits HPA with a Ki of 8.1 nM but does not affect the gut α-glucosidases (Tarling et al., 2008). Animal studies with Zucker diabetic fatty rats demonstrated the biological activity of MbA as an effective blood-glucose-lowering molecule (Yuen et al., 2016).
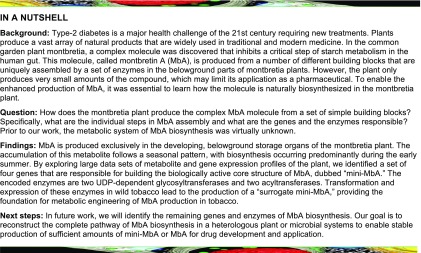
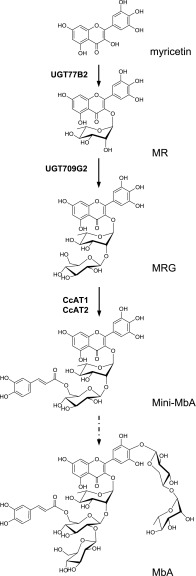
MbA is a complex acylated flavonol glycoside (Figure 1) (Asada et al., 1988; Tarling et al., 2008). It contains a myricetin core, which is decorated with two carbohydrate chains, a trisaccharide chain (β-d-glucosyl 1,2-β-d-glucosyl 1,2-α-l-rhamnoside) at O3 of the benzopyrone and a disaccharide chain (α-l-rhamnosyl 1,4-β-d-xyloside) at C4 of the phenyl moiety. The second glucose of the trisaccharide chain is linked at C6 to a caffeoyl moiety. In a series of structure-activity assays, the core active structure of MbA was identified as myricetin 3-O-(6’-O-caffeoyl)-β-d-glucosyl 1,2-α-l-rhamnoside (MRG-Caff) (Williams et al., 2015). This structure, dubbed mini-MbA (Williams et al., 2015), has a Ki of 93 nM with HPA and is a much less complex molecule yet still a highly potent HPA inhibitor.
Figure 1.
Structure of MbA and Schematic Structures of MbA and MbA-Related Compounds.
(A) The building blocks of MbA are labeled: myricetin, Rha, Glc, Xyl, and caffeic acid. Position of hydroxy groups of the myricetin core and the A, B, C designation of aromatic rings are shown. Dotted line marks the mini-MbA structure.
(B) Schematic of MbA, mini-MbA, MR, and MRG. M, myricetin (pink); R, rhamnose (yellow); G, glucose (green); X, xylose (orange); C, caffeic acid (blue).
The only known source of MbA is the ornamental plant montbretia (Asada et al., 1988), a perennial of the Iridaceae family native to southern and eastern Africa (Zurawik et al., 2015). Montbretia propagates by seed and through bulb-like belowground storage/reproductive organs, called corms, from which MbA was isolated. Research on montbretia has almost entirely focused on its improvement as an ornamental plant (Hannweg et al., 2013), and the MbA biosynthetic pathway is currently unknown.
Williams et al. (2015) reported the isolation of variable amounts of ∼0.3 to 3 mg MbA per g corm, depending on the montbretia cultivar. These amounts are sufficient for in vitro tests and initial animal and clinical trials but are likely too low for drug production. Chemical synthesis of MbA is not currently feasible due to its complex structure, while the scale of montbretia cultivation is limited to horticultural applications. Development of MbA as a drug for the treatment of type-2 diabetes would require reliable large-volume production, which may be achieved by farming of improved montbretia cultivars with higher levels of MbA or by metabolic engineering of MbA biosynthesis in a heterologous plant or microbial system. These strategies require knowledge of the genes and enzymes involved in MbA biosynthesis.
We are proposing a biosynthetic pathway for MbA that involves the formation of a set of five different building blocks, specifically the flavonol myricetin, the activated sugars UDP-rhamnose (UDP-Rha), UDP-glucose (UDP-Glc), UDP-xylose (UDP-Xyl), and the activated phenylpropanoid caffeoyl-CoA, followed by their stepwise assembly toward MbA. Transcriptome sequencing revealed the majority of the genes that constitute the three basic pathways that produce these five building blocks in montbretia, namely, the flavonol pathway, the nucleotide sugar pathway, and the phenylpropanoid pathway (SRP108844). The goal of current research is to discover the genes and enzymes that generate the unique glycosylation and acylation patterns in the biosynthesis of MbA, starting from the flavonol core myricetin. Enzymes that catalyze specific glycosylation and acylation reactions in plant specialized metabolism cannot be easily identified purely by predictive homology searches of sequence databases. The discovery of genes and enzymes with specific roles in specialized metabolism typically requires various functional characterization steps.
Glycosylation is a common modification of flavonoids, contributing to their stability and solubility as well as generating the complexity and diversity of flavonoids found in nature (Gachon et al., 2005; Yonekura-Sakakibara et al., 2012). Acylation also increases solubility and affects stability and transport (Bontpart et al., 2015). Glycosylation of plant specialized metabolites is typically catalyzed by UDP-sugar dependent glycosyltransferases (UGTs) belonging to family GT-1 of the CAZy-based glycosyltransferase classification (Caputi et al., 2012; Lombard et al., 2014). Plant UGTs utilize UDP-activated sugar donors, their sequences contain a highly conserved plant secondary product glycosylation (PSPG) motif, and they are encoded by members of some of the largest plant gene families. For example, Arabidopsis thaliana, sorghum (Sorghum bicolor), and poplar (Populus trichocarpa) contain 120, 180, and 223 sequences, respectively, annotated as UGTs (Ross et al., 2001; Tuskan et al., 2006; Paterson et al., 2009). Based on sequence phylogeny, plant UGTs fall into 16 clades (A–P) (Ross et al., 2001; Caputi et al., 2012). Several of these clades have been described to contain UGTs catalyzing the glycosylation of hydroxy groups of flavonoids at one or more of the 3-O-, 5-O-, 7-O-, 3ʹ-O-, 4ʹ-O-, or 5ʹ-O-positions (Caputi et al., 2012). Flavonoid glycosylation often starts with the 3-hydroxy group on the C-ring, and UGTs involved in flavonoid 3-O-glycosylation have been characterized in several plant species (Ford et al., 1998; Nakajima et al., 2001; Yamazaki et al., 2002; Jones et al., 2003; Li et al., 2015). In addition to UGTs catalyzing the direct glycosylation of the core flavonoid structure, side chain-elongating UGTs have also been described (Frydman et al., 2013). Enzyme of two classes can catalyze flavonoid acylation: BAHD-acyltransferases (BAHD-ATs) and serine carboxypeptidase-like acyltransferases (SCPL-ATs) (Milkowski and Strack, 2004; D’Auria, 2006). While BAHD-ATs are cytosolic and use acyl-CoA donors, SCLP-ATs are vacuolar and use 1-O-β-glucose esters as acyl donors (Bontpart et al., 2015). Over 60 BAHD-ATs have been characterized, while few SCPL-ATs have known functions (Bontpart et al., 2015). BAHD-ATs are grouped into five major phylogenetic clades (D’Auria, 2006). Clade I members function with flavonoids as acyl acceptors, but acyl donor specificity cannot be predicted (D’Auria, 2006; Bontpart et al., 2015).
Here, we describe insights into MbA biosynthesis based on metabolite and enzyme activity profiling, as well as the transcriptome-based discovery, cDNA cloning, and functional characterization of four enzymes that catalyze the first three steps in MbA biosynthesis and the complete biosynthesis of mini-MbA. Specifically, we describe two UGTs and two BAHD-ATs, a myricetin 3-O-rhamnosyltransferase, a myricetin 3-O-rhamnoside 1,2-glucosyltransferase, and two myricetin 3-O-glucosyl 1,2-rhamnoside 6’-O-caffeoyltransferases involved in the formation of myricetin 3-O-α-l-rhamnoside (MR), myricetin 3-O-β-d-glucosyl 1,2-α-l-rhamnoside (MRG), and mini-MbA (or MRG-Caff; Figure 1), respectively. This work enables the metabolic engineering of mini-MbA toward the production of an antidiabetic drug candidate.
RESULTS
MbA Accumulates in Developing Young Corms
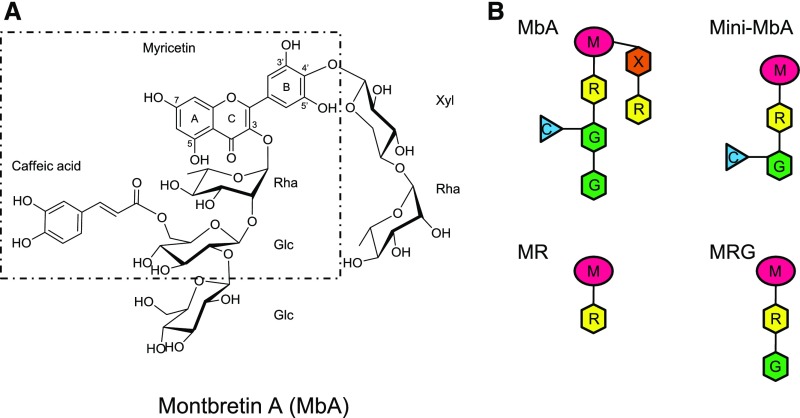
As a foundation for investigating MbA biosynthesis, we first monitored temporal patterns of MbA accumulation in corms over a 1-year growing season, February 2016 to February 2017 (Figure 2). Montbretia propagates vegetatively via corms (Figures 2A and 2B). In early spring, the original overwintering corm (old corm [oC]) produces multiple belowground shoots, called stolons. The stolon tip grows toward the surface, where it gives rise to the aboveground parts of the plant including leaves, stems, and flowers. Over the growing season, the base of the newly grown, aboveground shoot expands into a developing new corm (young corm [yC]). Over the 1-year growing season, yCs were observed first at the June 10th time point and continued to grow in diameter while the aboveground foliage, stem, and flowers developed. A single oC gave rise to between 1 and 10 yCs. We collected corms at 14 time points over the year. MbA was extracted from yC and oC and identified by LC-UV/MS (liquid chromatography-UV/mass spectrometry) based on retention time and fragmentation pattern in comparison to an authentic standard (Supplemental Figures 1A and 1B). Throughout the year, levels of MbA were relatively constant in oC, with an average of 2.8 ± 0.1 mg × g−1 fresh weight (FW) (Figure 2C; Supplemental Table 1). By comparison, yC had significantly higher levels of MbA from June to August relative to oC and showed a pattern of maximum accumulation during the summer. MbA levels in yC increased from 4.9 ± 0.2 mg × g−1 FW in early June to 6.1 ± 0.3 mg × g−1 FW in late June, remained steady in July and August, and decreased to 3.9 ± 0.3 mg × g−1 FW in September, more closely resembling the concentration of MbA in oC (Figure 2C; Supplemental Table 1). To validate that the observed differences in MbA levels between yC and oC and between the summer and fall time points for yC were not due to differences in water content, corms from a subset of time points were dried and MbA content and water loss were determined. Profiles of MbA accumulation showed the same pattern as observed for fresh material, and water loss did not differ between young and old corms (Supplemental Figure 2B and Supplemental Table 2).
Figure 2.
Time Course of MbA Accumulation in oCs and Developing yCs.
(A) Schematic of vegetative growth and corm development. Plant organs are labeled as oC, roots (r), stolon (st), yC, and shoot (s).
(B) Representative images of yC and oC at the June 10th and September 12th sampling time points. Bar = 1 cm.
(C) Corms were collected at 14 time points over a 1-year growing period (February 2016–February 2017). Levels of MbA in corm extracts are shown as the average with standard errors for four to six biological replicates for each time point. Different letters above the data points indicate significant differences between time points. Asterisks next to the sampling dates indicate statistically significant differences (P < 0.05) between yC and oC for that time point.
Once the aboveground parts of the plants had developed, we also analyzed roots, stolons, leaves, stems, flowers, and seedpods for MbA levels from late June until mid August (Supplemental Figure 2A). MbA was not detectable in leaves or roots, and only low levels of MbA were found in stolons, stems, flowers, and seedpods (Supplemental Table 3). Across these samples, flowers at the late June time point had the highest levels of MbA at 0.2 ± 0.3 mg × g−1 FW, which was <4% of the MbA levels in yC at the same time point. The identity of MbA was verified for each sample using an authentic standard and LC-UV/MS (Supplemental Figure 1).
MR and MRG Are Likely Intermediates in MbA Biosynthesis
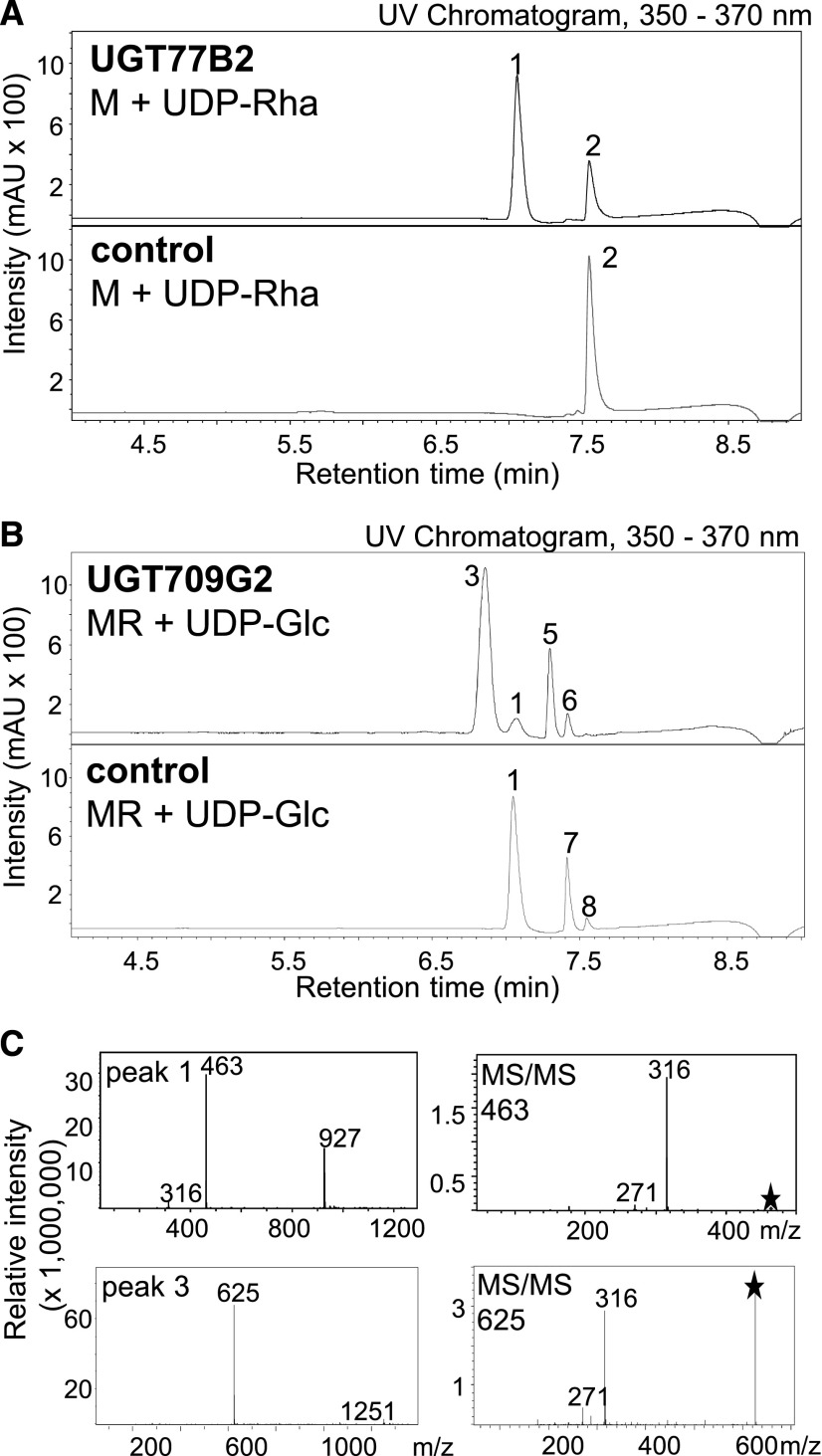
Given the multiple glycosylations of MbA (Figure 1), the sequence of the assembly of this molecule from its building blocks is not obvious. To support predictions of a potential biosynthetic route, we performed metabolite screens for theoretical intermediates in the modification of myricetin toward MbA in yC and oC. While myricetin could not be detected, LC-MS analysis revealed the presence of its 3-hydroxy rhamnosylated form, MR (m/z 463) (Figure 3A, peak 1), in corm extracts. MR was identified by comparison of retention time and fragmentation pattern to an authentic standard (Figure 3A). The MS/MS fragmentation of the mother ion (m/z 463) of the MR identified in corm extracts showed the expected difference in the relative abundance of the aglycone ion [Y]– compared with the radical aglycon ion [Y-H]–⋅, with the latter (m/z 316) being more dominant and indicative of glycosylation at the 3-hydroxy group (Griesser et al., 2008) (Figure 3B). Over the year-long time-course analysis, MR displayed a similar pattern of accumulation in yC as MbA, with the highest levels in late June to August (Supplemental Figure 3A and Supplemental Table 1).
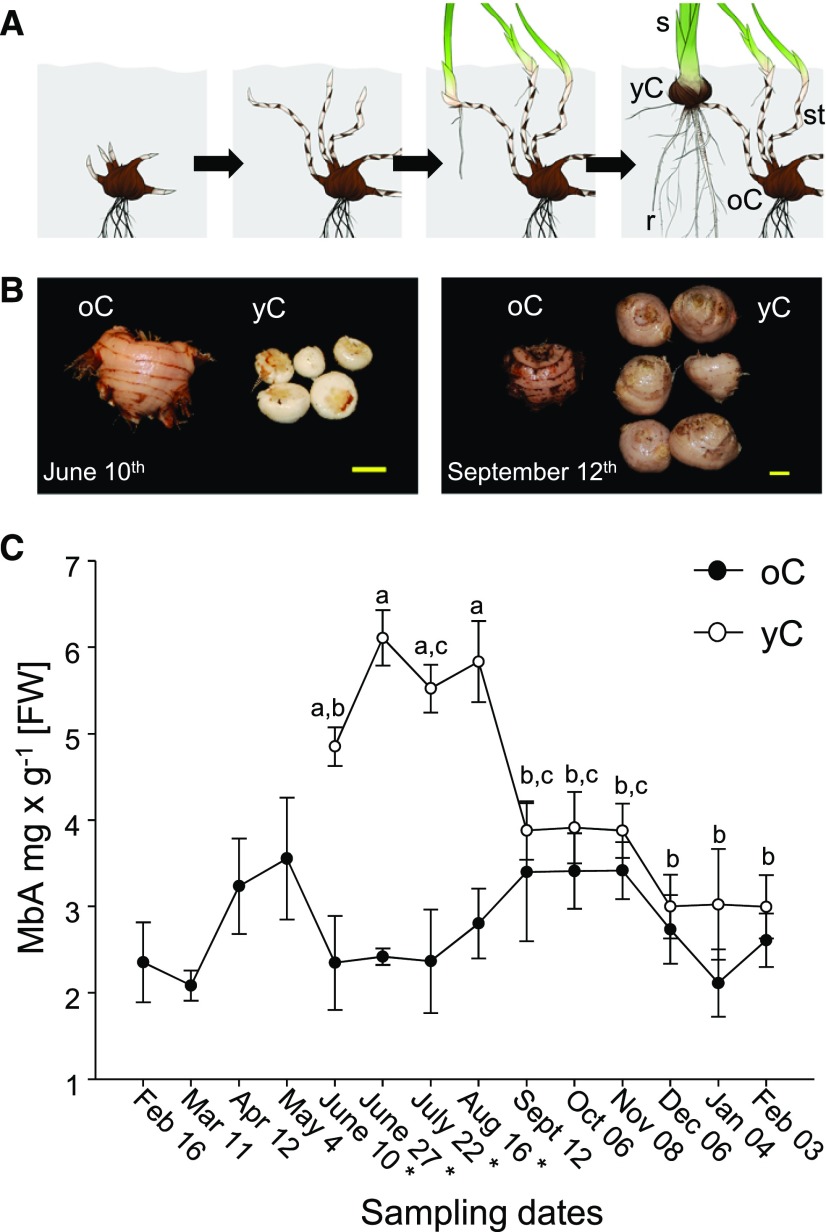
Figure 3.
Potential Intermediates in MbA Biosynthesis.
(A) Detection of a metabolite with m/z 463 in yCs and its formation in enzyme activity assays with yC total protein extracts. Peak 1 matches the retention time of an authentic standard of MR.
(B) MR was identified by comparison of the MS/MS fragmentation pattern with that of an authentic standard.
(C) Detection of metabolites with m/z 625 in yC and their formation in enzyme activity assays with yC total protein extracts.
(D) MS/MS fragmentation of m/z 625 was used for disaccharide identification.
(E) Formation of m/z 787 in enzyme activity assays with yC total protein extracts.
(F) Mini-MbA was identified by comparison of MS/MS fragmentation pattern with that of an authentic standard.
All samples were analyzed using LC-MS in negative-ionization mode and extracted ion chromatograms (EIC; [A], [C], and [E]) or MS/MS fragmentation patterns ([B], [D], and [F]) are shown. Asterisks indicate mother ion for MS/MS fragmentation. Peak 1, MR; peak 2, unknown; peak 3, tentatively identified as MGR; peak 4, MRG; peaks 5 to 7, tentatively identified as myricetin 3-O-rhamnoside-O-glucoside; 8, mini-MbA.
In addition, we detected two metabolite peaks (peak 3 and peak 4) in yC extracts (Figure 3C) with m/z 625 in the LC-MS analysis. The MS/MS spectra for peak 3 and peak 4 were nearly identical and showed a prominent m/z 316 for myricetin, but not ions indicative of monoglycosylated myricetin (m/z 479 and 463) (Figure 3D), suggesting a disaccharide (Ablajan et al., 2006; Griesser et al., 2008). The LC-MS spectra for peaks 3 and 4 resembled the mass and expected MS/MS fragmentation pattern of MRG or myricetin 3-O-rhamnosyl glucoside (MGR) (Figure 3D). Peak 4 was subsequently identified by NMR as MRG (see below), which led to the tentative annotation of peak 3 as MGR. Over the year-long time-course analysis, peak 4 resembled the accumulation pattern of MbA and MR in corms (Supplemental Figure 3B and Supplemental Table 1), while peak 3 did not show the similar high abundance in the early summer (Supplemental Figure 3C and Supplemental Table 1).
Another metabolite with m/z 1065 was detected in the LC-MS analysis of corm extracts (Supplemental Figure 1C). This compound matched the retention time and fragmentation pattern of an authentic standard for myricetin 3-O-glucosyl glucosyl rhamnoside 4’-O-rhamnosyl xyloside (MbA-C), which corresponds to MbA without the caffeoyl moiety (Supplemental Figure 1C). Over the year-long time-course analysis, the accumulation pattern of m/z 1065 did not reflect that of MbA or MR, and there was no difference in the abundance of m/z 1065 between yC and oC (Supplemental Figure 3D and Supplemental Table 1). No additional peaks for potential intermediates in MbA biosynthesis could be detected.
UGT Activities in Corm Protein Extracts Catalyze the Formation of MR and MRG
To verify the conversion of myricetin to MR and MRG as the first two steps in MbA biosynthesis and the involvement of UGTs in MR and MRG formation, we tested for enzyme activities in total protein extracts from yC and oC collected at the June 10th time point. As substrates, we used myricetin and a UDP-rhamnose preparation (UDP-RhaP) produced with Arabidopsis UDP-Rha synthase, or MR and UDP-Glc. Samples were analyzed using LC-MS.
Protein extracts from yC converted myricetin and UDP-RhaP into MR (peak 1, m/z 463) (Figure 3A). Assays with protein extracts from yC produced almost 20-fold more MR (peak 1) than protein extracts derived from the same amount of oC (Supplemental Figure 4A). The same assays of yC with myricetin and UDP-RhaP gave two additional products (m/z 625), which matched the retention times, masses, and fragmentations of peak 3 (MGR) and peak 4 (MRG) detected in yC metabolite extracts (Figures 3C and 3D). These additional products may be due to remaining UDP-Glc in the UDP-RhaP preparation (Supplemental Figure 5).
Protein extracts from yC incubated with MR and UDP-Glc resulted in a dominant m/z 625 product (peak 4) in the LC-MS analysis, matching the retention time, mass, and fragmentation pattern of MRG (Figures 3C and 3D). In addition, three smaller m/z 625 diglycoside products, peak 5, peak 6, and peak 7, were formed. Their MS/MS fragmentation showed daughter ions m/z 479 and m/z 463, which suggest that the glucose was linked directly to hydroxy groups of the flavonol core of MR, but not to the rhamnosyl side chain (Griesser et al., 2008) (Figure 3D). Assays with protein extracts from yC produced 250 times more MRG (peak 4) at the June 10th time point than protein extracts derived from the same amount of oC of the same time point (Supplemental Figures 4B and 4D). At the later sampling time points, protein extracts from yC showed a decrease in product formation (especially of m/z 625 peak 4), but product formation remained higher compared with oC protein extracts (Supplemental Figure 4D).
To assess the identity of m/z 625 peak 3 and peak 4, we performed a set of comparative assays with yC protein extracts. They included assays with myricetin, UDP- RhaP, and UDP-Glc and assays with myricetin 3-O-glucoside (MG) and UDP-RhaP (Supplemental Figure 6). Only assays with myricetin, UDP-RhaP, and UDP-Glc produced peak 4 in addition to a lower abundance of peak 3. By contrast, assays with MG and UDP-RhaP produced m/z 625 peak 3 but not peak 4. These results support the identification of peak 4 as MRG, the predicted intermediate in MbA biosynthesis. Peak 3 is likely MGR, which would not be an intermediate in MbA biosynthesis.
As MbA possesses a xylose at the 4’-hydroxy group, we also tested for alternative UGT activities in the conversion of myricetin or MR with UDP-Xyl as a sugar donor. When yC protein extracts were incubated with myricetin and UDP-Xyl, we detected the formation of three low-abundance m/z 449 peaks, representing putative myricetin xylosides (Supplemental Figure 7A). The peak area of MR formed when UDP-Rha was used as a sugar donor was over 200-fold higher compared with the peak areas of any of the m/z 449 products. Incubation of yC protein extracts with MR and UDP-Xyl led to the formation of an m/z 595 peak (Supplemental Figure 7B). The peak area of MRG formed when UDP-Glc was used as a sugar donor was over 10-fold greater compared with that of the m/z 595 product. The fragmentation of the mother ion (m/z 595) into the daughter ion m/z 316 indicated the attachment of Xyl to the Rha of MR (Supplemental Figures 7B and 7C); however, myricetin xylosyl rhamnoside is not a potential intermediate in MbA biosynthesis. Conversion of myricetin or MR with UDP-Xyl in yC protein extracts appears to be a background UGT activity that is likely not specific for MbA biosynthesis.
BAHD-AT Activity in Corm Protein Extracts Catalyzes the Formation of Mini-MbA
Next, we assessed the formation of mini-MbA as the third step in MbA biosynthesis and its possible formation from MRG by acyl-CoA-dependent BAHD-AT activity (D’Auria, 2006). Protein extracts from yC were incubated with MRG and the acyl donor caffeoyl-CoA (Caff-CoA), which resulted in the detection of an m/z 787 product peak that matched the retention time and fragmentation pattern of mini-MbA detected by LC-MS (Figures 3E and 3F). The mother ion m/z 787 fragmented into m/z 625 (MRG; loss of the caffeoyl group with its weaker ester linkage) and m/z 316 (myricetin; additional loss of the disaccharide group). The acylation activity that converted MRG to mini-MbA was over 15-fold greater in yC extracts compared with oC extracts (Supplemental Figure 4C). We also tested for alternative reactions that could conceivably represent the third step in MbA biosynthesis. To this end, corm protein extracts were incubated with MRG and the sugar donors UDP-Glc or UDP-Xyl. Glucosylation of MRG (m/z 625) was observed when UDP-Glc was incubated with yC or oC extracts, showing the formation of an m/z 787 peak (Supplemental Figure 8A). The MS/MS fragmentation pattern of m/z 787 showed the prominent daughter ion m/z 315 for myricetin, as well as ions for mono- and diglycosylated myricetin (m/z 478 corresponding to MG; m/z 625 corresponding to MRG) (Supplemental Figure 8C). These patterns indicated the attachment of the additional glucose moiety to the flavonol core in contrast to its attachment to the terminal glucose moiety of MRG, as would be required for MbA biosynthesis (Ablajan et al., 2006; Griesser et al., 2008). No glycosylation activity was observed when yC or oC protein extracts were incubated with UDP-Xyl and MRG (Supplemental Figure 8B). These results support the formation of mini-MbA from MRG as the third step in MbA biosynthesis catalyzed by a caffeoyl-CoA-dependent BAHD-AT.
Differential Transcriptome Expression across the Large UGT Gene Family in yC and oC
To enable the discovery of UGT genes involved in MbA biosynthesis, we established a montbretia corm reference transcriptome. Based on the temporal profiles of MbA accumulation and UGT activities over the time course of corm development (Figure 2; Supplemental Figure 4), we selected corms of the June 10th time point for RNA isolation and transcriptome sequencing of yC and oC. Sequencing on the Illumina HiSeq platform generated ∼400 million paired-end (PE) reads, which were assembled using Trinity into 171,070 nonredundant (NR) contigs with an average length of 691 bp, encoding 54,266 predicted NR peptides with an average length of 308 amino acids. Using reciprocal BLASTP searches against Arabidopsis and maize (Zea mays) UGTs, which represent all of the known UGT families A to P, we identified 159 different putative UGTs in the montbretia combined yC and oC transcriptome.
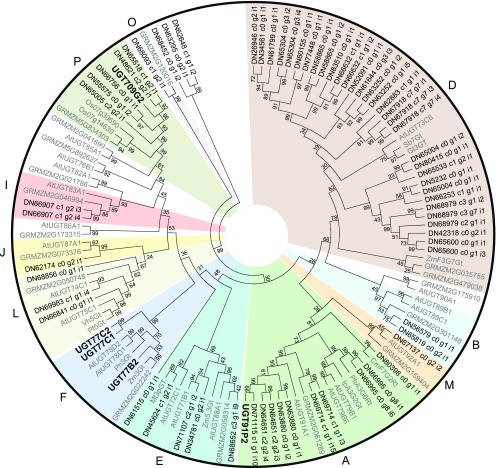
The predicted montbretia UGT protein sequences clustered with 14 of the 16 different known UGT families (Supplemental Figure 9) (Ross et al., 2001; Caputi et al., 2012). Only two UGT families, H and N, were not represented in the montbretia corm transcriptome. The largest number of montbretia UGTs were identified as members of family D, comprising 35% of corm-expressed UGTs (56 members). Of the 159 montbretia UGTs, 147 were differentially expressed (DE) in yC versus oC, and 70 had at least 2-fold higher transcript abundance in yC compared with oC (Supplemental Data Set 1). These 70 UGTs clustered into 11 of the 16 UGT families (Figure 4).
Figure 4.
Phylogeny of Montbretia UGTs.
Predicted amino acid sequences of montbretia UGTs with at least 2-fold higher transcript abundance in the yC transcriptome compared with the oC transcriptome were aligned with selected UGTs from other plant species using MUSCLE. A neighbor-joining tree was constructed using MEGA6. Alignments and phylogeny were used to cluster montbretia UGTs with known UGT clades. Montbretia UGTs characterized in this work are in bold; all other montbretia UGTs are labeled with their DN contig numbers. Clades are labeled with letters outside the circle. Bootstrap values are shown beside each node. Accession numbers can be found in Supplemental Table 6. At, Arabidopsis thaliana; Cm, Citrus maxima; Gt, Gentiana triflora; Ip, Ipomoea purpurea; Mt, Medicago truncatula; Os, Oryza sativa; Pf, Perilla frutescens; Ph, Petunia hybrid; Sb Scutellaria baicalensis; Vh, Verbena hybrid; Zm, Zea mays; GRMZM, UGT sequences from maize sequence database.
Since metabolite profiling and enzyme activity assays with yC and oC showed that UGTs of MR and MRG formation are most active in yC, we focused on the 70 UGTs with preferential expression in yC for further characterization. Based on the results from enzyme assays with whole protein extracts from corms and metabolite profiling, we hypothesized the formation of MR as the first step in the conversion of myricetin in MbA biosynthesis. UGTs that glycosylate the 3-hydroxy position of flavonoids have been described for other plant species to cluster in clade F of family 1 UGTs (Yamazaki et al., 2002; Di et al., 2015). The phylogeny of the DE 70 UGTs with higher expression in yC identified three montbretia UGTs, UGT77B2 (contig DN63814_c1_g1_i), UGT77C1 (contig DN63602_c0_g1_i1), and UGT77C2 (contig DN63602_c0_g1_i2) in the F-clade (Figure 4). Transcripts of UGT77B2, UGT77C1, and UGT77C2 were 35-fold, 10-fold, and 8-fold, respectively, more abundant in yC compared with oC at the June 10th time point (Supplemental Data Set 1).
UGT77B2 Catalyzes the Formation of MR
The full-length open reading frame for UGT77B2, which encodes a protein of 456 amino acids, was amplified from cDNA from yC and oC of the June 10th time point. The open reading frame sequence matched transcriptome contig DN63814_c1_g1_i1 with 99.8% identity. Contigs DN63602_c0_g1_i1 and DN63602_c0_g1_i2 appeared to be truncated. DN63602_c0_g1_i1 contained an early stop codon and encoded a protein of 390 amino acids missing the C-terminal region including the sugar binding motif. DN63602_c0_g1_i2 encoded a protein of 254 amino acids missing the N-terminal part. Full-length cDNAs for these contigs could not be identified, and thus they were not further investigated.
To test UGT77B2 for rhamnosyltransferase activity, the cDNA was expressed in Escherichia coli, protein expression was verified using protein gel blot analysis (Supplemental Figure 10), and recombinant protein assayed using myricetin and UDP-RhaP followed by LC-UV/MS analysis of the products. In the initial screen with nonpurified, heterologously expressed protein, UGT77B2 showed the formation of a single product peak with m/z 463 identified as MR based on matching retention time and fragmentation pattern with an authentic standard (Figure 5A). Ni-affinity-purified UGT77B2 protein catalyzed the same reaction (Supplemental Figure 11).
Figure 5.
Enzyme Activity of UGT77B2 and UGT709G2.
UGTs were heterologously expressed in E. coli and protein extracts were assayed for activity with myricetin and UDP-Rha for UGT77B2 (A) or with MR and UDP-Glc for UGT709G2 (B). As controls, assays were performed with protein extracts of E. coli transformed with empty vector. Products were analyzed using LC-MS/UV. UV detection was monitored at 350 to 370 nm. The mass spectrum and MS/MS fragmentation patterns of the reaction products are shown (C). Asterisks indicate mother ion for MS/MS fragmentation. Peak 1, MR; peak 2, myricetin; peak 3, MRG. Peaks 5 to 8 were tentatively identified as peak 5, quercetin O-rhamnoside (m/z 447); peak 6, kaempferol O-rhamnoside (m/z 431); peak 7, quercetin O-glucosyl rhamnoside (m/z 609); peak 8, kaempferol O-glucosyl rhamnoside (m/z 593). Asterisks indicate mother ion for MS/MS fragmentation.
In addition, purified UGT77B2 was also active with myricetin and UDP-Glc or UDP-Xyl, leading to the formation of, respectively, m/z 479 identified as MG based on comparison with an authentic standard and m/z 449 tentatively identified as myricetin 3-O-xyloside based on fragmentation pattern (Supplemental Figure 12). UGT77B2 was ∼45-fold and 50-fold more efficient with UDP-Rha compared with UDP-Glc and UDP-Xyl, respectively, as sugar donors (Supplemental Figure 12C). We also tested specificity of UGT77B2 toward different flavonoid and phenolic substrates, specifically myricetin, quercetin, naringenin, dihydrokaempferol, dihydroquercetin, dihydromyricetin, epicatechin, MR, quercetin 4’-O-glucoside, caffeic acid, salicin, and trichlorophenol. Of these substrates, UGT77B2 converted the flavonols myricetin, kaempferol, and quercetin into the corresponding rhamnosides MR, quercetin O-rhamnoside, and kaempferol O-rhamnoside (Supplemental Figure 13). UGT77B2 did not appear to be active with flavan(onol)s, 4’-hydroxy or 3-hydroxy glycosylated flavonols, or simpler phenolics (Supplemental Table 4). To determine the relative activity of UGT77B2 with the three different flavonols, relative turnover rates were determined using purified UDP-Rha and myricetin, quercetin, or kaempferol. The turnover rates for these three substrates were similar, with 2.4-fold and 3.6-fold higher turnover rates for kaempferol compared with quercetin and myricetin, respectively (Supplemental Figure 13B).
UGT709G2 Catalyzes the Formation of MRG
Enzymes catalyzing sequential reactions in the biosynthesis of plant specialized metabolites may be coexpressed, and such coexpression can be explored for gene discovery in a biosynthetic pathway (Zerbe et al., 2014; Celedon et al., 2016). We used Haystack coexpression analysis with UGT77B2 as a bait across the transcriptomes of montbretia flowers, stems, leaves, stolons, and corms to screen for candidate UGTs that may catalyze the second glycosylation step in MbA biosynthesis, specifically, the glycosylation of MR to form MRG. Two different transcripts, DN65518_c1_g1_i2 and DN71115_c1_g1_i4, encoded apparent full-length UGTs and showed strong correlation (R ≥ 0.9) with UGT77B2 expression (Supplemental Data Set 2A), as well as a ≥10× higher transcript abundance in yC compared with oC (Supplemental Data Set 1).
We cloned the full-length cDNAs of UGT709G2 and UGT91P2 corresponding to contigs DN65518_c1_g1_i2 (100% identity at the translated amino acid level) and DN71115_c1_g1_i4 (97.9% amino acid identity), respectively, from cDNA from yC and oC of the June 10th time point. UGT709G2 clustered into the P-clade, and DN71115 into the A-clade of family 1 UGTs (Figure 4). Both UGTs were heterologously expressed in E. coli, and protein expression was verified using protein gel blot analysis (Supplemental Figure 10). Total protein extracts were assayed for enzyme activity with MR and UDP-Glc as substrates, followed by LC-UV/MS analysis of the products. Only UGT709G2 showed glycosyltransferase activity in these assays, with the formation of a single m/z 625 product peak (Figure 5B). LC-MS/MS of m/z 625 showed the direct fragmentation into myricetin (m/z 316), suggesting that the sugars rhamnose and glucose are attached in the m/z 625 product as a disaccharide. The fragmentation pattern and retention time of the m/z 625 product of UGT709G2 matched the fragmentation pattern and retention time of peak 4 m/z 625 detected in corm extracts, and it was tentatively identified as MRG (Supplemental Figure 14). Purification of the UGT709G2 product m/z 625 and structural elucidation via NMR spectroscopy confirmed its identity as MRG (Supplemental Figure 15). Ni-affinity-purified UGT709G2 showed a sigmoidal profile of product formation over time and appeared to be affected by substrate inhibition at low substrate concentration (Supplemental Figure 16).
We tested purified UGT709G2 for substrate specificity with UDP-Glc as a sugar donor and different flavonoids and phenolics as sugar acceptors, specifically, myricetin, quercetin, MR, quercetin 3-O-glucoside, myricetin 3-O-glucoside, epicatechin, quercetin sophoroside, quercetin 4’-O-glucoside, arbutin, salicin, caffeic acid, and trichlorophenol (Supplemental Table 4). In addition to producing MRG with MR and UDP-Glc, UGT709G2 also glucosylated myricetin 3-O-glucoside and quercetin 3-O-glucoside (Supplemental Figure 17B). UGT709G2 was not active with any of the other acceptor substrates tested (Supplemental Table 4). In addition to UDP-Glc, purified UGT709G2 also accepted UDP-Xyl, producing myricetin 3-O-xylosyl rhamnoside from MR. The ∼50-fold higher turnover rate of UGT709G2 with MR and UDP-Glc compared with MR and UDP-Xyl suggested a substrate preference of UGT709G2 for UDP-Glc. UDP-Rha was not accepted as a sugar donor (Supplemental Figures 17A and 17D). The product peak m/z 609, which was formed with UDP-Rha and MR as substrates, fragmented into m/z 301, likely representing quercetin O-glucosyl rhamnoside (QRG) (Supplemental Figures 17A and 17C).
Identification of CcAT Candidates by Transcriptome Expression Analysis and Coexpression with UGT77B2 and UGT709G2
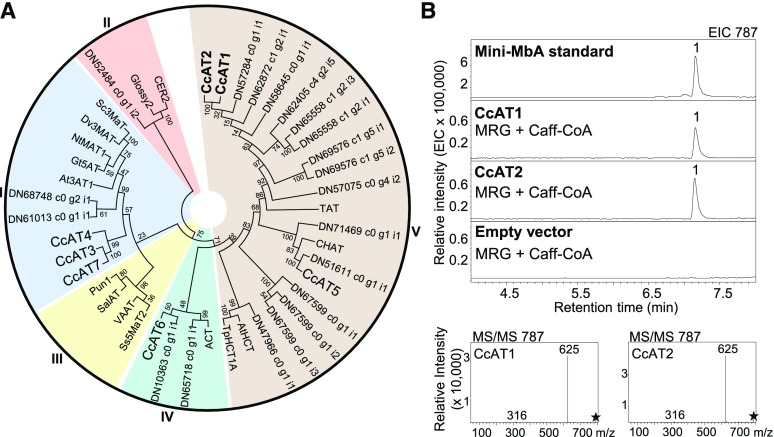
Using reciprocal BLASTP searches with BAHD-ATs from other plant species (Supplemental Table 5), we identified 59 different putative BAHD-ATs in the combined yC and oC transcriptome (Supplemental Figure 18). Of these, 50 were DE in yC versus oC and 27 had at least 2-fold higher transcript abundance in yC (Supplemental Data Set 3). These 27 sequences clustered into four of the five known BAHD-AT clades, with the majority falling into clade V (Figure 6A) (D’Auria, 2006). To reduce the list of target BAHD-ATs for functional characterization, we performed Haystack coexpression analysis across the flower, stem, leaf, stolon, and corm transcriptomes using UGT77B2 and UGT709G2 as baits. Seven different BAHD-AT transcripts, DN66658_c0_g1_i2 (CcAT1), DN66658_c0_g1_i1 (CcAT2), DN69556_c0_g2_i1 (CcAT3), DN63150_c0_g1_i1 (CcAT4), DN29265_c0_g1_i1 (CcAT5), DN37908_c0_g1_i1 (CcAT6), and DN61381_c0_g1_i1 (CcAT7), showed strong correlation (R ≥ 0.9) of expression with UGT77B2 and UGT709G2 (Supplemental Data Set 4A), as well as a ≥10-fold higher transcript abundance in yC compared with oC (Supplemental Data Set 3).
Figure 6.
Phylogeny of Montbretia ATs and Enzyme Activity of CcAT1 and CcAT2.
(A) Predicted amino acid sequences of montbretia ATs with at least 2-fold higher transcript abundance in the yC transcriptome compared with the oC transcriptome were aligned with selected BAHD-ATs from other plant species using MUSCLE. A neighbor joining tree was constructed using MEGA6. Alignments and phylogeny were used to cluster montbretia ATs with known AT clades. Montbretia ATs functionally characterized in this work are in bold; all other montbretia ATs are labeled with their DN contig numbers. Clades are labeled outside the circle. Bootstrap values are shown beside each node. Accession numbers are provided in Supplemental Table 5.
(B) ATs were heterologously expressed in E. coli and purified proteins were assayed for activity with MRG and Caff-CoA. As controls, assays were performed with purified protein of E. coli transformed with empty vector. Products were analyzed using LC-MS. The extracted ion chromatogram and MS/MS fragmentation patterns of the reaction products are shown. Peak 1, mini-MbA. Asterisks indicate mother ion for MS/MS fragmentation. At, Arabidopsis thaliana; Ca, Capsicum annum; Dv, Dahlia variabilis; Fv, Fragaria vesca; Gt, Gentiana triflora, Hv, Hordeum vulgare; Nt, Nicotiana tabacum; Pc, Pericallis cruenta; Ps, Papaver somniferum; Ss, Salvia splendens; Tc, Taxus cuspidate; Tp, Trifolium pretense.
CcAT1 and CcAT2 Catalyze the Formation of Mini-MbA
We cloned the full-length cDNAs of CcAT1 – CcAT7 from yC and oC of the June 10th time point. Predicted proteins CcAT1, CcAT2, and CcAT5 clustered into clade V; CcAT3, CcAT4, and CcAT7 clustered into clade I; and CcAT6 clustered into clade IV (Figure 6A). CcAT1 and CcAT2 shared 96.3% amino acid sequence identity. We expressed all seven ATs in E. coli and verified protein expression by protein gel blot analysis (Supplemental Figure 10). Total protein extracts were assayed for enzyme activity with MRG and Caff-CoA as substrates, followed by LC-MS analysis of the products. Only CcAT1 and CcAT2 showed acyltransferase activity in these assays (Supplemental Figure 19A), resulting in a single product peak with m/z 787, which was identified as mini-MbA by comparison of retention time and fragmentation pattern with an authentic standard (Supplemental Figure 19; Figure 6B). No activity with MRG and Caff-CoA was detected for CcAT3 to CcAT7 or the empty vector control (Supplemental Figure 19A).
CcAT1 and CcAT2 activity was verified using Ni-purified proteins (Figure 6B). We also tested the acyl acceptor specificity of CcAT1 and CcAT2 toward myricetin, MG, MR, quercetin 3-O-sophoroside (QGG), rutin (QRG), salicin, and arbutin with Caff-CoA as the acyl donor (Supplemental Table 4). CcAT1 and CcAT2 produced an m/z 787 peak when QGG and Caff-CoA were used as substrates (Supplemental Figure 20A). The m/z 787 peak fragmented into m/z 625 (QGG) and m/z 300 (Q), and the product was tentatively identified as quercetin 3-O-(6’-O-caffeoyl)-glucosyl glucoside (Supplemental Figure 20B). No activity was observed for any of the other substrates tested (Supplemental Table 4). Acyl donor specificity of CcAT1 and CcAT2 was tested using MRG as the acyl acceptor and coumaroyl-CoA (Cou-CoA), feruloyl-CoA (Fe-CoA), acetyl-CoA (Ac-CoA), or malonyl-CoA (Ma-CoA) as the acyl donor (Supplemental Figure 21). Both enzymes accepted Cou-CoA, Fe-CoA, and Ac-CoA and produced m/z 771, m/z 801, and m/z 667, respectively, with MRG as the acyl acceptor (Supplemental Figures 21A and 21C). All three product peaks fragmented into expected daughter ions m/z 625 (MRG) and m/z 316 (M) (Supplemental Figure 21E). CcAT1 and CcAT2 had the highest turnover with Caff-CoA and Cou-CoA (Supplemental Figures 21B and 21D). The turnover with Fe-CoA was over 4 times less compared with Caff-CoA for both enzymes. Ac-CoA turnover was ∼9-fold and 3-fold less for CcAT1 and CcAT2, respectively, compared with Caff-CoA. No activity was observed with Ma-CoA as the acyl donor.
Transcript Profiles of UGT77B2, UGT709G2, CcAT1, and CcAT2 Match MbA Accumulation Profiles in yC and oC
We measured the profiles of transcript abundance of UGT77B2, UGT709G2, CcAT1, and CcAT2 during corm development in yC and oC from June 10th to October 6th using qRT-PCR (Figure 7). Putative serin-incorporator (MEP) and a putative zinc-finger protein (ZF) were used as references for quantitative transcript analysis (Supplemental Figure 22). UGT77B2, UGT709G2, CcAT1, and CcAT2 showed similar transcript expression patterns (Figure 7). In general, the transcript abundance of all genes was low and did not significantly change across all time points in oC and was significantly higher in yC at all time points. Transcript abundance was highest in June, with over 60-fold higher transcript levels for UGT77B2, UGT709G2, CcAT1, and CcAT2 at the June 27th time point compared with oC of the same time point. Transcript abundance of all genes significantly dropped from June toward August in yC. The temporal patterns of UGT77B2, UGT709G2, CcAT1, and CcAT2 transcript abundance (Figure 7) matched the MbA accumulation profiles in yC and oC (Figure 2C).
Figure 7.
Transcript Abundance of UGT77B2, UGT709G2, CcAT1, and CcAT2 in yC and oC.
RNA was isolated from yC and oC harvested at different time points of corm development. Transcript abundance was determined by qRT-PCR for UGT77B2 (A), UGT709G2 (B), CcAT1 (C), and CcAT2 (D). Means and standard errors are shown (n = 3). Different letters above the data points indicate significant differences between harvest points. Asterisks indicate statistically significant differences (P < 0.05) between yC and oC for all time points.
Reconstitution of Surrogate Mini-MbA Biosynthesis in Nicotiana benthamiana
To validate the functions of UGT77B2, UGT709G2, CcAT1, and CcAT2, we expressed the cDNAs in wild tobacco (N. benthamiana). In a first set of experiments, we tested the functions of UGT77B2 and UGT709G2, and then extended the experiments by additional coexpression of CcAT1 or CcAT2. In the first set of experiments, we used Agrobacterium tumefaciens transformed with plasmids carrying the promoter-UGT constructs 35Spro:UGT77B2 and 35Spro:UGT709G2 or the gene for the eGFP to infiltrate N. benthamiana leaves. Samples of leaves that expressed 35Spro:eGFP, 35Spro:UGT77B2, or 35Spro:UGT77B2 + 35Spro:UGT709G2 were collected 4 d after infiltration, and methanol/water extracts were analyzed by LC-MS. Myricetin glycosides were not detected in any of the samples expressing the montbretia UGTs, perhaps due to lack of access to myricetin as a substrate. We therefore performed experiments in which we supplied myricetin to leaves expressing 35Spro:eGFP or 35Spro:UGT77B2 or 35Spro:UGT77B2 + 35Spro:UGT709G2. For this purpose, transformed leaf discs were placed in a myricetin solution, followed by methanol/water extraction and LC-MS analysis. When myricetin was added as a substrate, MR (m/z 463 peak 2) was produced in N. benthamiana leaves expressing UGT77B2 (Supplemental Figure 23). In addition, we detected peak 4 with m/z 625 and a MS/MS daughter ion m/z 463 indicative of a monoglycosylated myricetin, which suggested that peak 4 represents MR with an additional glucose group on the flavonol ring. When UGT709G2 and UGT77B2 were coexpressed in tobacco leaves and myricetin was provided, myricetin was converted into MRG (m/z 625, peak 5). The lack of detection of an MR peak in the UGT709G2 and UGT77B2 coexpression assays demonstrated efficient conversion of MR into MRG. None of the above-mentioned product peaks were observed in the controls expressing eGFP. We detected two additional m/z 463 peaks, peak 1 and peak 3, which fragmented into m/z 301, indicative of quercetin O-glucosides. These peaks were also present in the eGFP control and were not affected by montbretia UGTs. No other myricetin disaccharides could be detected in these assays. We also screened for other myricetin glycosides; no myricetin xyloside was detected and only small amounts of myricetin 3-O-glucoside (m/z 479) were detected in all samples (Supplemental Figure 24).
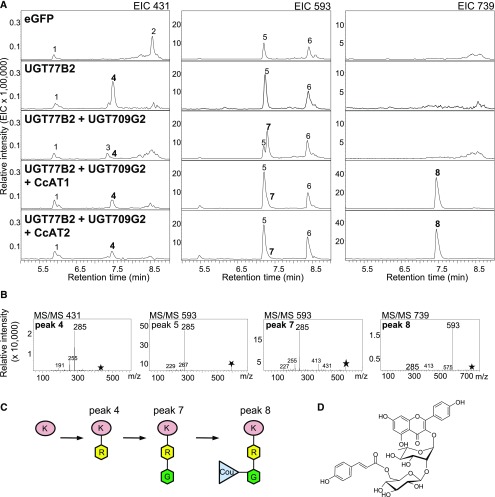
Tobacco leaves have been reported to contain various kaempferol glycosides (Chen et al., 2012), suggesting that kaempferol may be available as a flavonol substrate. We tested leaves that expressed 35Spro:eGFP, 35Spro:UGT77B2, or 35Spro:UGT77B2 + 35Spro:UGT709G2 for kaempferol glycosides (Figure 8). Leaves expressing UGT77B2 produced a unique m/z 431 peak 4 with the fragmentation pattern expected for kaempferol 3-O-rhamnoside. This peak was absent in leaves expressing eGFP, and only traces were found in leaves expressing both UGT77B2 and UGT709G2. Leaves expressing UGT77B2 and UGT709G2 together produced two peaks with the mass and fragmentation patterns expected for kaempferol 3-O-glucosyl rhamnoside (KRG). The fragmentation of the mother ion m/z 593 into m/z 285 indicated kaempferol as the core flavonol, and the absence of monoglycosylated daughter ions suggested a disaccharide side chain. The first m/z 593 peak (peak 5) was also present in eGFP-expressing leaves and likely represents kaempferol 3-O-rhamnosyl glucoside (KGR), which is known to be produced in tobacco (Chen et al., 2012). The other m/z 593 peak (peak 7) was specific to samples coexpressing the two montbretia UGTs. The identity of this peak is likely KRG, which is also supported by the depletion of KR (m/z 431) in these samples.
Figure 8.
N. benthamiana Transiently Expressing Montbretia UGTs and ATs Produce Kaempferol-Derived Surrogate Mini-MbA.
Tobacco leaves were infiltrated with Agrobacterium transformed with plasmids carrying the promoter-UGT constructs 35Spro:UGT77B2, 35Spro:UGT709G2, 35Spro:CcAT1, and 35Spro:CcAT2 or the gene for eGFP. Leaves were collected at day 4 after infiltration. Metabolites were extracted with methanol/water, analyzed by LC-MS, and tentatively identified based on their fragmentation patterns. The extracted ion chromatograms (EIC) (A) and MS/MS fragmentation patterns (B) are shown. Proposed pathway in transiently transformed N. benthamiana (C) and surrogate mini-MbA structure (D). Asterisks indicate mother ion for MS/MS fragmentation. Peaks 1 to 3, unknown; peak 4, kaempferol 3-O-rhamnoside (KR); peak 5, kaempferol disaccharide; peak 6, unknown; peak 7, KRG; 8, surrogate mini-MbA or KRG-Cou. K, kaempferol; R, rhamnose; G, glucose; Cou, coumaric acid.
In a second set of experiments, we transiently coexpressed UGT77B2 and UGT709G2 in combination with CcAT1 or CcAT2 in N. benthamiana (35Spro:UGT77B2 + 35Spro:UGT709G2 + 35Spro:CcAT1 or 35Spro:UGT77B2 + 35Spro:UGT709G2 + 35Spro:CcAT1). Small peaks for KR (m/z 431, peak 4) and KRG (m/z 593, peak 7) were detected in leaves coexpressing both UGTs and CcAT1 or CcAT2, suggesting depletion of the kaempferol glycosides in the presence of the CcATs (Figure 8). We screened for products with m/z 755, corresponding to the addition of a caffeoyl-group to KRG, and detected two peaks (Supplemental Figure 25). One of these peaks (peak 1; Supplemental Figure 25) was also present in controls coexpressing eGFP with UGT77B2 + UGT709G2, independent of CcAT expression. A second, much smaller m/z 755 peak (peak 2; Supplemental Figure 25) was only present in leaves coexpressing the two UGTs with CcAT1 or CcAT2 but was not detected in controls with eGFP or UGTs alone. This second m/z 755 peak showed daughter ions of m/z 609 (indicative of QRG) and m/z 300 (Q), as well as m/z 593 (KRG) and m/z 285 (K), suggesting that peak 2 represents the coelution of two compounds tentatively identified as quercetin 3-O-(6’-O-coumaroyl)-glucosyl rhamnoside (QRG-Cou) and kaempferol 3-O-(6’-O-caffeoyl)-glucosyl rhamnoside (KRG-Caff). Given the central role of coumaroyl-CoA in plant phenylpropanoid metabolism (Vogt, 2010), we also screened samples for metabolites of m/z 739, which would correspond to the addition of a coumaroyl group to KRG. Only samples expressing UGT77B2, UGT709G2, and CcAT1 or CcAT2 produced an m/z 739 peak (Figure 8). This peak fragmented into the daughter ions m/z 593 (KRG) and m/z 285 (K) and was tentatively identified as kaempferol 3-O-(6’-O-coumaroyl)-glucosyl rhamnoside (KRG-Cou) (Figure 8B). KRG-Cou appeared to be ∼10 times more abundant than KRG-Caff or QGG-Cou.
By transiently coexpressing UGT77B2, UGT709G2, and CcAT1 or UGT77B2, UGT709G2, and CcAT2 in N. benthamiana, we achieved the formation of 50.1 ± 11.4 µg × g−1 (FW) or 57.2 ± 3.8 µg × g−1 (FW) KRG-Cou, respectively. KRG-Cou is effectively a surrogate for mini-MbA (MRG-Caff) produced by the first three enzymes of the MbA biosynthetic pathways in transiently transformed N. benthamiana, where kaempferol may be more accessible to UGT77B2 than myricetin, and where Cou-CoA may be more accessible to CcAT1 and CcAT2 than Caff-CoA. We did not observe other glycosides of the flavonols kaempferol, quercetin, or myricetin that were specific to conditions of transient expression of UGT and AT. However, we cannot exclude further conversion of such compounds by endogenous N. benthamiana enzymes.
DISCUSSION
Type-2 diabetes is a major global health problem affecting over 320 million people. The specialized plant metabolite MbA, found in corms of the ornamental plant montbretia, is a promising novel drug candidate for the treatment of type-2 diabetes. MbA selectively inhibits HPA with a very high potency (Ki = 8 nM) (Tarling et al., 2008; Yuen et al., 2016). While MbA is a complex acylated flavonol glycoside (Figure 1), the simpler precursor mini-MbA, which is also a strong HPA inhibitor (Ki = 93 nM) (Williams et al., 2015), can be biosynthesized from the flavonol myricetin by a set of two UGT-catalyzed reactions and an additional acyltransferase reaction. We report the transcriptome- and metabolome-based discovery of two UGTs, a myricetin 3-O-rhamnosyltransferase and a myricetin 3-O-rhamnoside 1,2-glucosyltransferase, and two BAHD-ATs, myricetin 3-O-glucosyl rhamnoside 6’-O-caffeoyltransferases, from montbretia. These enzymes produce mini-MbA en route to MbA and may provide a foundation for improved production of MbA or mini-MbA through plant or microbial metabolic engineering.
Assembly of MbA Involves the Formation of MR, MRG, and Mini-MbA
The sequence of reactions by which MbA is assembled cannot be predicted from its structure alone. Using metabolite profiling, enzyme assays, and characterization of cloned UGTs and ATs, we resolved the first three steps in the assembly of MbA: the rhamnosylation of myricetin to yield MR, followed by the glucosylation of MR to form the disaccharide MRG, and next the acylation of MRG to form mini-MbA (Figure 9). A similar pattern of glycosylation occurs in the biosynthesis of lobelinin in edging lobelia (Lobelia erinus) (Hsu et al., 2017), where stepwise assembly of a disaccharide at the 3-hydroxy group of the anthocyanin core precedes the glycosylation on other positions of the flavonoid ring. Glycosylation of the 3-O-position is a common primary modification of flavonoids, which increases the stability and solubility of the molecule (Nakajima et al., 2001; Jones et al., 2003). In the biosynthesis of lobelinin in L. erinus, acylation of the disaccharide occurs prior to further glycosylations. By contrast, in anthocyanin biosynthesis in japanese iris (Iris ensata), acylation occurs after 5-O-glycosylation (Yabuya et al., 2002). The third step in MbA biosynthesis is the formation of the mini-MbA molecule. The biological activity of both mini-MbA and MbA as potent HPA inhibitors has been attributed to internal π-stacking interactions between the myricetin and caffeic acid building blocks, preorganizing the phenolic hydroxy groups for multiple hydrogen bonds with conserved HPA active site residues (Williams et al., 2015). With the identification of UGT77B2, UGT709G2, CcAT1, and CcAT2, the full set of enzymes required to produce mini-MbA is now available.
Figure 9.
Role of UGT77B2, UGT709G2, CcAT1, and CcAT2 in MbA Biosynthesis.
UGT77B2 converts myricetin and UDP-Rha into MR, which is then converted by UGT709G2 with UDP-Glc into MRG. CcAT1 and CcAT2 catalyze the acylation on the C6 position of the Glc of MRG to yield mini-MbA. Solid black arrows indicate pathway steps described in this work; dotted arrows indicate the proposed downstream biosynthesis.
MbA and Mini-MbA Biosynthesis Is Affected by Substrate Availability, Enzyme Expression, and Enzyme Specificity of UGTs and ATs
The montbretia corm transcriptome revealed a large number of different UGTs and BAHD-ATs, consistent with the large number of UGT and BAHD-AT genes in the genomes of other plant species (Li et al., 2001, 2014; Tuominen et al., 2011; Caputi et al., 2012). Using information about spatial and temporal patterns of MbA accumulation, enzyme activities, as well as the identification of intermediates in MbA biosynthesis, we discovered two UGTs, UGT77B2 and UGT709G2, and two BAHD-ATs, CcAT1 and CcAT2, that catalyze the biosynthesis of mini-MbA and the first three steps in the assembly of MbA. For the discovery of UGT77B2, a myricetin 3-O-rhamnosyltransferase that clusters in clade F of family 1 UGTs (Figure 4), we relied on the assumption that regio-specificity of UGTs in flavonoid biosynthesis is a conserved feature that occurred early in the evolution of family 1 UGTs. As a consequence, UGTs cluster across the gene family by regio-specificity rather than by species and sugar donor specificity (Li et al., 2001; Noguchi et al., 2009; Hsu et al., 2017). Other known UGT members of the F-clade mediate the transfer of UDP-sugars, mostly UDP-Glc, onto the 3-hydroxy groups of different flavonoids (Miller et al., 1999; Jones et al., 2003; Griesser et al., 2008; Sui et al., 2011; Tohge et al., 2015; Sun et al., 2016). Notably, UGTs of the F-clade appear to be missing in the genomes of the monocotyledonous reference plant systems maize and rice (Oryza sativa) (Caputi et al., 2012), and only one other flavonol 3-O-rhamnosyltransferase, UGT78D1 from Arabidopsis, has been characterized previously (Jones et al., 2003). Like AtUGT78D1, montbretia UGT77B2 can use different flavonols as sugar acceptors and showed high specificity toward UDP-Rha as a sugar donor. Sugar donor specificity is thought to be determined by the last amino acid residue of the PSPG-motif (Supplemental Figure 26). A glutamine or histidine in this position is important for glucosyl or galactosyl transfer activity, respectively (Kubo et al., 2004; Ono et al., 2010). AtUGT78D1 contains an asparagine at this position (Jones et al., 2003; Caputi et al., 2012), while montbretia UGT77B2 has a glutamine in this position, despite its characterized preference for UDP-Rha.
We characterized UGT709G2 as myricetin 3-O-rhamnoside 1,2-glucosyltransferase, which mediates the first chain-elongating glycosylation in MbA biosynthesis. Other flavonoid or polyphenol chain-elongating UGTs have been shown to catalyze 1,6-glycosylations (Kroon et al., 1994; Noguchi et al., 2008; Masada et al., 2009; Frydman et al., 2013; Rojas Rodas et al., 2014, 2016; Li et al., 2016; Hsu et al., 2017) or 1,2-glycosylations (Frydman et al., 2004; Morita et al., 2005; Sawada et al., 2005; Montefiori et al., 2011; Yonekura-Sakakibara et al., 2012; Di et al., 2015). For example, UGT79B6 from Arabidopsis catalyzes the 1,2-glucosylation of flavonol 3-O-glucosides and is important for the production of kaempferol and quercetin 3-O-glucosyl 1,2-glucosides in pollen (Yonekura-Sakakibara et al., 2014). Like UGT79B6 and other branch-elongating UGTs, montbretia UGT709G2 is specific toward its sugar donor and accepts different 3-O-glycosylated flavonols (Yonekura-Sakakibara et al., 2012, 2014; Rojas Rodas et al., 2014, 2016; Hsu et al., 2017). To our knowledge, all previously characterized chain-elongating UGTs of plant origin are from eudicots and form a separate branch within clade A of family 1 UGTs (Frydman et al., 2013). By contrast, montbretia UGT709G2 falls into clade P (Figure 4). UGTs of the P-clade appear to be absent in Arabidopsis, while the P-clade has expanded in montbretia compared with other plants (Caputi et al., 2012). It is not known if other members of this clade besides montbretia UGT709G2 also function as branch elongating UGTs. Expression of UGT77B2 and UGT709G2 in N. benthamiana supported the conclusion that sugar donor specificity is an inherent feature of these UGTs, while acceptor specificity is more promiscuous, and the biosynthetic product of these enzymes may be determined by flavonol availability in planta (Jones et al., 2003).
We characterized CcAT1 and CcAT2 as myricetin 3-O-glucosyl 1,2-rhamnoside 6’-O-caffeoyl transferases. CcAT1 and CcAT2 cluster into BAHD-AT family clade V, which contains ATs that act on a range of substrates including terpenoids, medium-chain alcohols, and quinic acid (Tuominen et al., 2011). For example, the hexenol acetyltransferase, CHAT, from Arabidopsis is involved in the production of volatile esters (D’Auria et al., 2002), and the hydroxycinnamoyl-transferases from red clover (Trifolium pratense), TpHCT1A, is involved in the production of p-coumaroyl-shikimate/quinate esters (Sullivan, 2009). Interestingly, all BAHD-ATs characterized to acylate flavonoids (mainly anthocyanins) are members of clade I, except for the malonyltransferase SsMAT2 from scarlet sage (Salvia splendens) in clade III (Bontpart et al., 2015). CcAT1 and CcAT2 appear to be unusual members of clade V in that they use flavonoids as the acyl acceptor. Flavonoid BAHD-ATs have been described to be specific toward the position of the glucose and the hydroxy group on the glucose but not toward the hydroxylation pattern on the B-ring of flavonoids (Bontpart et al., 2015). For example, Arabidopsis At3AT1 and At3AT2 acylate the C6 hydroxy of the glucose in position 3 of anthocyanins (Luo et al., 2007). Our results suggest that CcAT1 and CcAT2 are specific toward the C6’-hydroxy of the second glucose of the flavonoid 3-hydroxy disaccharide chain. Characterized flavonoid BAHD-ATs have been shown to be aliphatic ATs, transferring the malonyl moiety or aromatic ATs transferring hydroxycinnamoyl moieties (Bontpart et al., 2015). For example, At3AT1 and At3AT2 exhibit affinity for p-coumaroyl, feruloyl-CoA, and caffeoyl-CoA, but not sinapoyl-CoA (Luo et al., 2007). CcAT1 and CcAT2 were most active with p-coumaroyl and caffeoyl-CoA but also accept acetyl-CoA and to a lesser extent feruloyl-CoA.
In addition to the most abundant montbretin, MbA, montbretia produces smaller amounts of montbretin B (MbB) and montbretin C (MbC) (Asada et al., 1988; Tarling et al., 2008). MbB has a coumaroyl moiety, and MbC has a feruloyl moiety, instead of the caffeoyl moiety in MbA. Compared with MbA, the potency of MbB and MbC to inhibit HPA is 1000-fold lower, which highlights the importance of the meta-hydroxy group of the caffeoyl moiety (Williams et al., 2015). The lack of absolute acyl donor specificity suggests that CcAT1 and CcAT2, together with UGT77B2 and UGT709G2, may also be involved in the formation of MbB and MbC. As with UGT77B2 and UGT709G2, the in planta products of CcAT1 and CcAT2 may be controlled by substrate availability, whereby the availability of caffeoyl-CoA, coumaroyl-CoA and feruloyl-CoA may be a key factor in determining the preferential formation of MbA, MbB, or MbC, respectively. This would be similar to the features of cocaine synthase from the coca plant (Erythroxylum coca), which can form cocaine or cinnamoylcocaine based on the availability of benzoyl-CoA or cinnamoyl-CoA, respectively (Schmidt et al., 2015).
Overall, the high regio-specificity of UGT77B2 and UGT709G2, as well as CcAT1 and CcAT2, support their function in MbA biosynthesis. This conclusion is also supported by the correlation of gene expression, metabolite accumulation, and protein activity data over the time course of corm development. Substrate availability and expression of the UGT and BAHD-ATs are likely critical factors for the control of MbA biosynthesis.
Montbretia Corms and Alternative Systems for the Production of MbA
Montbretia is the only known source of MbA (Asada et al., 1988; Tarling et al., 2008), which is produced in a mostly organ-specific fashion in montbretia corms. However, MbA accounts for only 0.06 to 0.69% of the corm fresh weight (or 0.07 to 1.67% of corm dry weight). The small size of montbretia corms and the low abundance of MbA limit the production of MbA using its natural source. In addition, the harvesting of corms, which serve as the plant’s belowground storage and vegetative reproductive organs, is destructive and would require the development of specialized farm operations for large-scale planting, harvesting, and propagation. Animal studies suggest that treatment of type-2 diabetes with MbA requires a daily dose of 7.5 mg per kg body weight for rats (Yuen et al., 2016). As an alternative to MbA production in montbretia, it may be possible to produce MbA, and more easily mini-MbA, in an engineered biological system such as microorganisms or plants.
Progress has been made in engineering flavonoid glycoside production in microbial systems such as E. coli and yeast (Saccharomyces cerevisiae) (Wang et al., 2011; Yang et al., 2014; Trantas et al., 2015). However, as these hosts do not possess endogenous flavonoid biosynthesis, the introduction of a large suite of genes and engineering of metabolite flux will be necessary for MbA production, in addition to the expression of the genes that assemble MbA from its building blocks. Indeed, few projects have achieved flavonoid glycoside production in microbial hosts without supplementation of precursors (Trantas et al., 2015). For example, Yang et al. (2014) reported the production of 58 mg × L−1 of kaempferol 3-O-rhamnoside, which is a considerably less complex molecule than MbA, from glucose by engineering 12 different genes in E. coli. Utilizing the endogenous phenylpropanoid and flavonoid metabolism of a plant host for metabolic engineering may accomplish MbA production by metabolic engineering of fewer genes. The use of a plant host may also be advantageous for proper subcellular localization of engineered MbA biosynthesis. Transient expression in N. benthamiana is particularly suitable for the expression of multiple genes. For example, Lau and Sattely (2015) achieved the production of an etoposide aglycone, the precursor for the chemotherapeutic etoposide, by transiently expressing ten genes in N. benthamiana (Lau and Sattely, 2015). Another example is the production of glucoraphanin, a cancer preventive glucosinolate, by transient expression of 13 genes (Mikkelsen et al., 2010). In this system, the reduced production of undesired side products and improved yield were accomplished via the expression of two additional genes (Crocoll et al., 2016). This study highlighted a possible problem of using heterologous plant systems for metabolic engineering of a complex plant metabolite: the potential formation of non-target products, which may result from promiscuous activities of the introduced enzymes reacting with non-target substrates or from the conversion of engineered pathway intermediates and products by endogenous enzymes. In this work, transient expression of UGT77B2, UGT709G2, and CcAT1 or CcAT2 in N. benthamiana with only endogenous substrates resulted in the production of KRG-Cou instead of MRG-Caff (mini-MbA). On an experimental scale, this problem can be overcome by feeding of the substrates myricetin and caffeoyl-CoA, which are expensive. For larger scale production, we will attempt in future work to convert pools of kaempferol into myricetin and to redirect coumaroyl-CoA formation toward caffeoyl-CoA. In addition, the overall levels of phenylpropanoid and flavonoid biosynthesis may have to be increased, which may be achieved by overexpressing transcription factors that control these pathways (Liu et al., 2015).
Other approaches to facilitate MbA or mini-MbA production may include the use of cell cultures, as shown for the production of Taxol (Zhong, 2002), or increasing MbA levels in montbretia through breeding, as demonstrated for the enhanced production of artemisinin in sweet wormwood (Artimisia annua) (Simonnet et al., 2008). All of these approaches require a detailed understanding of the complex MbA biosynthetic system, which we have begun to explore here, including knowledge of the genes, enzymes, and spatial and temporal patterns of biosynthesis and accumulation.
Spatial and Temporal Patterns of MbA Biosynthesis May Suggest a Role in Plant Defense
MbA biosynthesis occurs during yC development in the early summer, as demonstrated by (1) higher levels of MbA and MbA-intermediates in yC compared with oC, (2) greater total biosynthetic enzyme activity in yC compared with oC at the same time points in early summer, and (3) the transcript profiles of UGT77B2, UGT709G2, CcAT1, and CcAT2 that match the profiles of metabolite and enzyme activities. The window of time for MbA biosynthesis appears to be tightly correlated to yC development. Corms are also the site of starch accumulation. It is conceivable that the spatially restricted pattern of MbA accumulation in corms and the temporally restricted pattern of MbA biosynthesis in developing yC followed by retention of MbA in oC provide protection for this storage and reproductive organ against feeding mammals by interfering with a digestive enzyme involved in starch degradation.
METHODS
Plant Material
Montbretia (Crocosmia x crocosmiiflora) plants of the variety ‘Emily McKenzie’ were obtained from a collection curated by Gary Brayer (Richmond, British Columbia, Canada) in July 2010. Plants were vegetatively propagated from corms and maintained in 4-liter pots containing perennial potting mix under natural outdoor conditions on the University of British Columbia campus in a semishaded area. Plant maintenance included the annual removal of dead aboveground leaves and stems after the active growing season in November, coverage of pots for overwintering, and annual repotting in February whereby clusters of corms were separated and a single corm per pot was replanted. Nicotiana benthamiana plants were grown from seed in potting soil in a controlled environment chamber (day, 26°C; night, 22°C; 16-h/8-h light/dark cycle; 120 μmol light intensity; fluorescent lamps).
Plant Sampling
Between February 2016 and February 2017, corms of four to six biological replicates (individual plants grown in separate pots) were collected at least once a month. Corms that were newly developing during this growing season appeared in June and were defined as young corms (yCs) and collected separately from the original corm, which was defined as the old corm (oCs). During the summer months (late June, July, and August), additional parts of the plants (stems, leaves, flowers, seedpods, stolons, and roots) were collected. Plants were dissected with a razor blade, and the samples were flash frozen in liquid nitrogen and stored at −80°C until further use. In addition, corms, stem, leaves, flowers, stolons, and roots were sampled on July 29, 2013 from three to four biological replicates (individual plants) for RNA-seq.
Metabolite Extraction
Frozen plant material was ground in liquid nitrogen to a fine powder, of which 100 mg per sample was extracted with 1 mL 50% methanol/water (v/v) (2 h shaking at 21°C). After removing the supernatant, one reextraction step was performed and both supernatants were combined. After two extraction steps, <10% MbA remained in the material. Undiluted and 1:10 diluted samples were used for metabolite analyses, which were performed with four to six separate biological replicates for each time point and sample type. To assess the potential that differences in MbA levels may be caused by differences in the water content of corms, 100 mg powdered tissue of a subset of samples was dried for 3 d at 50°C, dry weight was determined, and the sample was extracted as described above for nondried samples. The yC and oC did not differ in water content, and extraction from dried or nondried samples did not affect the results of temporal metabolite profiling in yC and oC (Supplemental Figure 2B). Extracts were analyzed using LC-MS (described below).
Protein Extraction and UGT and BAHD-AT Activity Assays
Total protein extracts were prepared as described by Nagel et al. (2012) and Martin et al. (2002) with minor modifications. In brief, 500 mg of powdered corm sample was extracted with 2.5 mL of buffer (100 mM NaPi, pH 7.4, 5 mM ascorbic acid, 5 mM sodium bisulfite, 5 mM dithiothreitol, 1 mM EDTA, 10% [v/v] glycerol, 1% [w/v] PVP, 4% [w/v] PVPP, 4% [w/v] Amberlite XAD-4, and 0.1% [v/v] Tween) for 1 h at 4°C. Following centrifugation at 4300g for 30 min (4°C), the supernatant was desalted three times into assay buffer (10 mM Tris-HCl, pH 7.5, 1 mM DTT, and 10% [v/v] glycerol) using NAP-5 columns (GE-Healthcare). Desalted protein extract (75 µL) was used in a 150-μL total assay volume with 100 µM myricetin or 100 µM MR (Sigma-Aldrich) as flavonoid substrates and one of the following UDP sugars: 1 mM UDP-Glc (Sigma-Aldrich), 1 mM UDP-Xyl (CarboSource Services), or 50 μL UDP-RhaP (see below for UDP-Rha preparation). Additional assays were performed using 50 µM MRG and 120 µM caffeoyl-CoA (TransMIT), 1 mM UDP-Glc, or 1 mM UDP-Xyl. Assays were incubated at 21°C for 6 h in a Teflon-sealed, screw-capped 1-mL GC glass vial and stopped by placing the vial on ice after the addition of 100 μL methanol. After centrifugation at 4300g for 20 min (4°C), the supernatant was transferred into a fresh vial and assays were analyzed for product formation by LC-MS.
LC-MS Analysis
LC was performed on an Agilent 1100 HPLC (Agilent Technologies) with Agilent ZORBAX SB-C18 column (50 × 4.6 mm, 1.8 μm particle size) (Merck) using aqueous formic acid (0.2% v/v) (mobile phase A) and acetonitrile plus formic acid (0.2% v/v) (mobile phases B). The elution profile was: 0 to 0.5 min, 95% A; 0.5 to 5 min, 5 to 20% B in A; 5 to 7 min 90% B in A; and 7.1 to 10 min 95% A. The flow rate was 0.8 mL × min−1 at a column temperature of 50°C. LC was coupled to an Agilent MSD Trap XCT-Plus mass spectrometer equipped with an electrospray operated in negative ionization mode (capillary voltage, 4000 eV; temperature, 350°C; nebulizing gas, 60 p.s.i.; dry gas 12 L/min) and an Agilent 1100 diode array detector (detection 200–700 nm; J&M Analytik). MS/MS was used to monitor daughter ion formation. LC/MSD Trap Software 5.2 (Bruker Daltonik) was used for data acquisition and processing. Metabolites were quantified using an MbA standard curve. Enzyme products were quantified, in addition, using the UV spectrum (350–370 nm) and external standard curves of myricetin, quercetin, or kaempferol (Sigma-Aldrich). Compounds were tentatively identified using their molecular masses and specific fragmentation patterns. Authentic standards were available for MR (Sigma-Aldrich), myricetin 3-O-glucoside (Extrasynthese), MbA, MbA-C, and mini-MbA (Williams et al., 2015).
UDP-l-Rhamnose Preparation and Purification
As UDP-l-rhamnose was not commercially available when this study was performed, a preparation of this activated sugar (UDP-RhaP) was generated from UDP-Glc using the two active domains (RHM-D and RHM-ER) of the Arabidopsis thaliana UDP-l-rhamnose synthase (Rautengarten et al., 2014), kindly provided by Henrik Scheller (University of California, Berkeley). RHM-D and RHM-ER were individually expressed in Escherichia coli BL21(DE) (Invitrogen) following the protocol for expression and crude protein preparation as described below for UGTs, except that 1 mM IPTG was used to induce gene expression. Crude protein extract containing RHM-D (450 µL) was incubated with 2 mM UDP-Glc and 1 mM NAD+ in a total volume of 500 μL in assay buffer (see UGT expression below) for 24 h at 21°C. The enzyme was then inactivated and precipitated by incubation at 70°C for 10 min. After centrifugation, 450 μL RHM-ER crude protein extract and 1 mM NADPH was added to the supernatant to yield a total volume of 1 mL. The reaction was incubated for another 24 h at 21°C, followed by heat inactivation at 70°C for 10 min. After centrifugation, the supernatant (UDP-RhaP) was stored at −20°C until further use. All UDP-RhaP were analyzed by LC-MS to ensure the conversion of UDP-Glc (m/z 565) into UDP-Rha (m/z 549) (Supplemental Figure 4). Some of the UDP-RhaP was purified by HPLC using an Agilent Zorbax SB-Aq column (150 × 4.6 mm, 5 µm) with water (phase A) and acetonitrile (phase B). The elution profile was 0 to 5 min, 95% A; 5 to 10 min, 5 to 95% B in A; hold 2 min at 95% B; 12.1 to 15 min 95% A. The column temperature was 25°C. Purified UDP-Rha fractions were freeze-dried.
Purification and NMR Analysis of MRG
MRG was purified from enzyme assays by HPLC using an Agilent Zorbax SB-C18 column (150 × 4.6 mm, 5 µm) with water (phase A) and acetonitrile (phase B). The elution profile was changed to 0 to 0.5 min, 95% A; 0.5 to 8 min, 25% B in A; 8 to 12 min 95% B in A; and 12.1 to 15 min 95% A. The column temperature was 35°C. Purified MRG was freeze-dried and used for structure determination by NMR. All NMR spectra were acquired on a Bruker Avance 600 spectrometer with a 1H operating frequency of 600 MHz and equipped with a Bruker BioSpin TCI 1.7 mm MicroCryoProbe. The sample was dissolved in 13 μL of CD3OD and syringed into a 1-mm NMR tube. 1H chemical shifts were recorded with respect to the residual nondeuterated solvent signal from the NMR solvent, CD3OD. 13C chemical shifts were obtained from the HSQC and HMBC spectra. 1H, gradient COSY, ROESY, gradient HSQC, and gradient HMBC spectra were acquired for structure elucidation.
RNA Extraction
Plant material was ground into a fine powder under liquid nitrogen and total RNA extracted as described previously with minor modifications (Kolosova et al., 2004). In brief, 1 mL extraction buffer (200 mM Tris-HCl, pH 8.5, 1.5% [w/v] lithium dodecylsulfate, 300 mM LiCl, 10 mM disodium salt EDTA, 1% [w/v] sodium deoxycholate, 1% [w/v] Tergitol Nonidet P-40, 5 mM thiourea, 1 mM aurintricarboxylic acid, 10 mM DTT, and 2% [w/v] polyvinylpolypyrrolidone [PVPP]) was added to 100 mg plant material, the sample was flash-frozen in liquid nitrogen and thawed at RT, centrifuged at 20,000g for 10 min at 4°C, and one-thirtieth volume of 3.3 M sodium acetate (pH 6.1) and 0.1 volume 100% ethanol were added to the supernatant. After incubation on ice for 10 min, the centrifugation step was repeated and one-tenth volume of 3.3 M sodium acetate (pH 6.1) and 0.6 volume of ice-cold isopropanol were added to the supernatant, and then left at −80°C for at least 30 min. Samples were thawed at RT, centrifuged (20,000g, 20 min, 4°C), and the pellet was resuspended in 400 μL TE buffer (10 mM Tris-HCl, pH 8.0, and 1 mM EDTA) and 400 μL 5 M NaCl for at least 30 min on ice. Samples were mixed with 200 μL of 10% cetyltrimethylammonium bromide and incubated for 5 min at 65°C following two extractions with 1 mL chloroform/isoamylalcohol each time (24:1; v/v). A quarter volume of 10 M LiCl was added to the chloroform/isoamylalcohol extract and samples were incubated overnight at −20°C. RNA precipitates were collected by centrifugation (20,000g, 30 min, 4°C), and dissolved in 100 μL TE buffer for up to 1 h on ice, combined with 100 μL chilled isopropanol and 11 μL of 3.3 M sodium acetate, and RNA was precipitated for 30 min at −80°C. RNA precipitates were collected by centrifugation (20,000g, 30 min, 4°C) and washed with 100 μL 70% (v/v) ethanol. Centrifugation was repeated for 10 min to remove the remaining liquid. The RNA pellet was dried for 3 min at RT and resuspended in 25 μL DEPC-treated water on ice for 30 min. Total RNA concentration was determined using a NanoDrop 1000 (Thermo Fisher Scientific) and assessed for quality on an Agilent 2100 Bioanalyzer and Agilent RNA 6000 Nano Kit LabChips (Agilent Technologies). RNA was stored at −80°C until further use.
Transcriptome Sequencing and de Novo Assembly
RNA samples (RNA integrity number ≥ 9) prepared separately from yC and oC from the June 10, 2016 time point, each with two biological replicates, were sequenced at the McGill University and Génome Québec Innovation Centre (http://gqinnovationcenter.com). RNA-seq was performed on the Illumina HiSeq platform using 100-bp PE strand-specific libraries multiplexed on a single lane, generating ∼400 million PE reads. Sequence quality was assessed with FastQC (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/). Adapter sequences were trimmed with BBDuk of the BBTools software suite (v 36.62, sourceforge.net/projects/bbmap/). To improve assembly contiguity, overlapping PE reads were joined by BBMerge to generate longer single end reads. A second round of trimming, using BBDuk, was performed to remove low-quality base pairs in both merged and unmerged reads. A total of 274 million merged reads and 112 million unmerged PE reads were pooled and assembled de novo using Trinity (v 2.3.2) (Grabherr et al., 2011), resulting in 171,070 NR contigs with an average length of 691 bp. From the assembly, TransDecoder (version 3.0.0, https://transdecoder.github.io/) predicted 54,266 NR peptides with an average length of 308 amino acids. Additional RNA-seq data were produced from 16 RNA samples (three biological replicates for flowers, leafs, stems, and stolons and four replicates of corms; RNA integrity number ≥ 8) from plant material collected on July 29, 2013 to generate ∼75 Gb of sequencing data using the Illumina HiSeq 2000 platform with 150-bp PE sequencing at the McGill University and Génome Québec Innovation Centre.
Identification of UGT and BAHD-AT Transcripts
Putative montbretia UGT sequences were identified in the transcriptome assembly by performing a BLASTP search of the montbretia translated protein database using Arabidopsis and maize (Zea mays) UGTs from all currently known UGT-families (A–P) (for accession numbers, see Supplemental Table 6). Results were filtered using a reciprocal BLASTP search against the nonredundant protein database of NCBI. Montbretia UGT sequences and the Arabidopsis and maize UGTs were used to construct a phylogenetic tree to select representative montbretia UGTs from each family for a reciprocal BLASTP analysis. All putative montbretia UGTs were clustered at 98% amino acid identity, using CD-HIT (v 4.6.1) to collapse possible allelic variants (Fu et al., 2012). In addition, short sequences (≥250 amino acids) were removed and UGTs were manually assessed using alignments to join shorter sequences and to remove likely chimeric or misassembled genes, which resulted in a refined list of 159 montbretia UGTs (Supplemental Data Set 5). Montbretia BAHD-ATs were identified using the same pipeline as described for UGTs. BAHD-AT sequences used for BLASTP analysis are listed in Supplemental Table 5. A refined list of 59 montbretia-ATs was generated (Supplemental Data Set 6).
UGT and BAHD-AT Sequence Analysis and Phylogenetic Analysis
An amino acid alignment of montbretia UGTs and published UGTs from other plant species (Supplemental Table 6) was constructed using the MUSCLE algorithm (gap open, −2.9; gap extend, 0; hydrophobicity multiplier, 1.2; clustering method, upgmb) implemented in MEGA6 (Supplemental File 1; Tamura et al., 2011). Based on this alignment, a phylogenetic tree was produced using the neighbor-joining algorithm (Poisson model) in MEGA6. A bootstrap resampling analysis with 1000 replicates was performed to evaluate the topology of the phylogeny. An additional phylogeny was constructed using only UGTs that were at least 2-fold upregulated in yC compared with oC together with UGTs from other species (Supplemental File 2 and Supplemental Table 6). To associate montbretia UGTs with previously described UGT clades, we selected Arabidopsis and maize UGTs that represent clades A to P. In addition, we selected characterized UGTs that catalyze O-glycosylations and chain elongations. To visualize the PSPG-motif, an alignment was generated using BioEdit (http://www.mbio.ncsu.edu/bioedit/bioedit.html) and the ClustalW algorithm. A phylogenetic tree comprising the 59 montbretia BAHD-ATs and BAHD-ATs from other species (Supplemental File 3 and Supplemental Table 5), and a phylogeny of BAHD-ATs that were at least 2-fold upregulated in yC compared with oC together with BAHD-ATs from other species (Supplemental File 4), was constructed as described for UGTs.
Analysis of Differentially Expressed UGTs and Identification of Target UGTs
The refined UGT list was used to carry out a DE analysis between yC and oC on all predicted coding sequences using the voom/limma package in R with quantification results from Salmon (v 0.8) with numBootStrap = 100 (Law et al., 2014; Patro et al., 2017). Contigs with less than 100 counts per million were discarded. Expression of UGTs in other plant organs was assessed with RNA-seq data using Salmon with the same parameters as described above and using the refined UGT list from the yC and oC transcriptome assembly. Haystack (http://haystack.mocklerlab.org/) was used to test for correlations in expression patterns between UGT77B2 and other UGTs across yC, oC, and other plant organs (Supplemental Data Set 2). A correlation cutoff of R ≥ 0.9 was used; in addition, the results were filtered to meet criteria of at least 10-fold greater expression in yC compared with oC.
UGT cDNA Cloning and Heterologous Expression in E. coli
Target UGTs were amplified from cDNA prepared from yC from the June 10, 2016 time point and cloned into the pJET1.2/blunt vector (Thermo Fisher Scientific) for sequencing (Supplemental Table 7). Complete open reading frames of the target UGTs were cloned as BsaI or BspMI fragments into the pASK-IBA37 vector (IBA). The E. coli TOP10 strain (Invitrogen) was used for heterologous UGT expression. Cultures were grown at 21°C, induced at an OD600 = 0.5 with 200 µg/L anhydrotetracycline (Sigma-Aldrich), placed at 18°C, and grown for another 20 h. Cells were collected by centrifugation and disrupted by five freeze-and-thaw cycles in chilled extraction buffer (50 mM Tris-HCl, pH 7.5, 10 mM MgCl2, 5 mM DTT, 10% [v/v] glycerol, 1× Pierce protease inhibitor [EDTA-free; Thermo Fisher Scientific], 25 units Benzonase Nuclease [Merck], and 0.2 mg × mL−1 lysozyme). Cell fragments were removed by centrifugation at 14,000g, and the supernatant was desalted into assay buffer (10 mM Tris-HCl, pH 7.5, 1 mM DTT, and 10% [v/v] glycerol) using Econopac 10DG columns (Bio-Rad). For protein purification using Ni-NTA, a modified extraction buffer was used (50 mM Tris-HCl, pH 7.5, 10 mM MgCl2, 5 mM DTT, 2% [v/v] glycerol, 150 mM NaCl2, 20 mM imidazole, 1× Pierce protease inhibitor, 25 units Benzonase Nuclease, and 0.2 mg × mL−1 lysozyme), and the lysate was directly loaded onto a Ni-NTA agarose column (Qiagen). Protein was eluted with elution buffer (10 mM Tris-HCl, pH 7.5, 500 mM imidazole, 1 mM DTT, and 10% [v/v] glycerol) and desalted into assay buffer using Illustra NAP-5 Columns (GE Healthcare). Enzyme concentrations were determined using UV absorption at 280 nm. To ensure successful heterologous enzyme expression protein gel blot analysis was performed using, the His-Tag Antibody HRP Conjugate Kit (Novagene) according to the manufacturer’s instructions (Supplemental Figure 10).
Enzyme Assays with Recombinant UGTs
To test for UGT activity, initial enzyme assays were performed with 100 μL of the bacterial extract, 50 µM myricetin or 50 µM MR, and 1 mM UDP-Glc or UDP-Xyl or 50 μL of UDP-RhaP in a Teflon-sealed, screw-capped 1-mL GC glass vial. Unless stated otherwise, assays were performed in assay buffer in a final volume of 150 μL and incubated at 25°C. Assays were incubated for 6 h and stopped by placing on ice after the addition of an equal volume of methanol. To characterize UGTs and determine enzyme parameters, Ni-purified UGT77B2 or UGT709G2 was assayed with variations in incubation times and substrate concentrations. UGT77B2 was tested for substrate specificity using 2 µg of protein, 50 μL of UDP-RhaP and 50 µM of the different substrates, listed in Supplemental Table 4, and incubated for 1 h. To determine relative turnover rates for myricetin and different UDP-sugars, 5 µg purified UGT77B2 was assayed with 50 µM myricetin and 1 mM UDP-Glc or UDP-Xyl or HPLC-purified UDP-Rha for 15, 30, 45, 60, 120, 180, or 210 min. To determine relative turnover rates for different flavonol acceptors, 1.25 µg purified UGT77B2 enzyme was incubated with 50 µM flavonol (myricetin, kaempferol, or quercetin) and 1 mM UDP-Rha (HPLC-purified) for 15, 30, 45, 60, 120, and 210 min. UGT709G2 substrate specificity was tested by incubating 0.36 µg of purified protein with 1 mM UDP-Glc and 5 µM of different substrates (substrates are listed in Supplemental Table 4) for 2 h or 45 min for the different flavonol 3-O-glycoside substrates (MR, quercetin 3-O-glucoside, and myricetin 3-O-glucoside). Sugar specificity was tested using 0.26 µg purified UGT709G2 protein, 5 µM MR, and 1 mM UDP-Glc, UDP-Xyl, or UDP-Rha (HPLC purified) and 15, 30, 45, 60, 120, and 210 min incubation time. MRG formation was characterized using 0.18 µg purified UGT709G2 protein, 1 mM UDP-Glc, and different MR concentrations (1, 2, 3, 4, 5, and 10 μM) and incubation times (15, 30, 45, 60, 90, 120, 150, 180, and 240 min). Product quantification was done based on the UV (350–370 nm) signal of external standard curves of myricetin, quercetin, or kaempferol. Enzyme concentrations and incubation times were chosen so that the reaction velocity was constant during the incubation time period.
Analysis, Identification, and Characterization of BAHD-ATs
The refined BAHD-AT list was used to carry out a DE analysis between yC and oC and between other plant organs as described for UGTs. Haystack (http://haystack.mocklerlab.org/) was used to test for correlations in expression patterns between UGT77B2, UGT709G2, and BAHD-ATs across yC, oC, and other plant organs (Supplemental Data Set 4). A correlation cutoff of R ≥ 0.9 was used; in addition, results were filtered to meet criteria of at least 10-fold greater expression in yC compared with oC.
Target BAHD-ATs were amplified, cloned, heterologously expressed, and purified as described for UGTs. To test for BAHD-AT activity, initial enzyme assays were performed with 98 μL of the bacterial extract, 120 µM MRG, and 60 µM caffeoyl-CoA in a Teflon-sealed, screw-capped 1-mL GC glass vial. Unless stated otherwise, assays were performed in assay buffer in a final volume of 100 μL and incubated at 25°C. Assays were incubated for 6 h and stopped by placing on ice after the addition of an equal volume of methanol. To characterize BAHD-ATs and determine enzyme parameters, Ni-purified CcAT1 or CcAT2 was used. CcAT1 and CcAT2 were tested for acyl acceptor specificity using 1 µg of protein, 150 µM of caffeoyl-CoA, and 100 µM of the different substrates, listed in Supplemental Table 4, and incubated for 1 h. Acyl donor specificity for both enzymes was tested using 1 µg of protein, 100 µM MRG, and 150 µM of the different acyl donors: caffeoyl-CoA, coumaroyl-CoA (TransMIT), feruloyl-CoA (TransMIT), acetyl-CoA (Sigma-Aldrich), and malonyl-CoA (Sigma-Aldrich) and incubated for 1 h. For relative turnover rates for MRG and different acyl donors, 0.8 µg purified CcAT1 or CcAT2 was assayed with 25 µM MRG and 37.5 µM acyl donor for 10 min. Product quantification was done using an external standard curves for MbA.
Transient Expression of UGT77B2, UGT709G2, CcAT1, and CcAT2 in N. benthamiana
For expression in N. benthamiana, the coding regions of UGT77B2, UGT709G2, CcAT1, and CcAT2 were cloned into the pCAMBiA2300U vector. After sequence verification, pCAMBiA vectors carrying UGT77B2, UGT709G2, CcAT1, CcAT2, eGFP, and the pBIN:p19 were separately transferred into Agrobacterium tumefaciens strain C58pMP90. One mL overnight culture (220 rpm, 28°C) was used to inoculate 10 mL LB medium containing 50 µg × mL−1 kanamycin, 25 µg × mL−1 rifampicin and 25 µg × mL−1 gentamicin for overnight growth. The following day, the cultures were centrifuged (4000g, 5 min) and cells were resuspended in infiltration buffer (10 mM MES, pH 5.6, 10 mM MgCl2, and 100 µM acetosyringone) to a final OD600 of 0.5. After shaking for 3 h at RT, the following combinations of transformed Agrobacterium were prepared for leaf infiltration: (1) Agrobacterium 35Spro:UGT77B2 + Agrobacterium pBIN:p19; (2) Agrobacterium 35Spro:UGT77B2 + Agrobacterium 35Spro:UGT709G2 + Agrobacterium pBIN:p19; (3) Agrobacterium 35Spro:UGT77B2 + Agrobacterium 35Spro:UGT709G2 + Agrobacterium 35Spro:CcAT1 + Agrobacterium pBIN:p19, (4) Agrobacterium 35Spro:UGT77B2 + Agrobacterium 35Spro:UGT709G2 + Agrobacterium 35Spro:CcAT2 + Agrobacterium pBIN:p19, (5) Agrobacterium 35Spro:eGFP + AgrobacteriumpBIN:p19. Equal volumes of each line of transformed Agrobacterium were used to prepare the mixtures. The leaves of 4-week-old N. benthamiana plants were infiltrated with Agrobacterium solution using a 1-mL needle-free syringe to gently push the bacterial mixture into the abaxial surface. Infiltrated leaves were labeled with tape and harvested 4 d after infiltration. For substrate infiltration, leaf discs 1 cm in diameter were prepared from Agrobacterium-infiltrated N. benthamiana leaves using a cork borer and placed in a 24-well cell culture plate containing 800 μL of tap water with 500 µM myricetin (added as 4 μL of a 100 mM myricetin solution in DMSO). The remaining leaf material was directly frozen in liquid nitrogen and stored at −80°C until further analysis. After incubating leaf discs in a climate chamber (day, 26°C; night, 22°C; 16-h/8-h light/dark cycle) overnight, leaf discs were briefly flushed with water, dried, and flash-frozen in liquid nitrogen. Plant material was ground in liquid nitrogen into a fine powder, and 100 mg of tissue was extracted with 400 μL 50% (v/v) methanol for 2 h at RT. The extract was analyzed using LC-MS.
Reverse Transcription and qRT-PCR
cDNA synthesis was performed with 650 ng total RNA using a dsDNase and Maxima First Strand cDNA Synthesis Kit (Thermo Fisher Scientific) according to the manufacturer’s instructions. For qRT-PCR, the cDNA was diluted 1:5 with water. For the amplification of UGT and AT gene fragments ∼150 bp length, primer pairs were designed with a temperature ≥ 60°C, a GC content of 45 to 60%, and a primer length of 20 to 25 nucleotides (Supplemental Table 7). Primer specificity was confirmed by agarose gel electrophoresis, melting curve analysis, standard curve analysis, and by sequence verification of cloned PCR products. qRT-PCR was performed in duplicate on a Bio-Rad CFX96 instrument in optical 96-well plates using SsoFast EvaGreen Supermix (Bio-Rad) with the following PCR conditions: initial incubation at 95°C for 30 s followed by 40 cycles of amplification (95°C for 5 s, 60°C for 10 s). Fluorescence measurement was performed during the annealing and extension steps of each cycle. Data for the melting curves were recorded at the end of cycling from 55°C to 95°C. qRT-PCR analyses were performed with three biological replicates (corms from three different plants) and two technical replicates for each of the six different time points of yC and oC collections (June 10th, June 27th, July 22nd, August 16th, September 12th, and October 6th). The serine incorporator (MEP) and zinc-finger protein (ZF) genes were used as reference genes (Supplemental Table 7).
Statistical Analysis
To test for significant differences in MbA accumulation patterns (June 2016–February 2017) and UGT and AT gene expression in young and old corms at different time points, a two-way ANOVA was performed followed by a Tukey test using SigmaPlot 11.0 for Windows (Systat Software) (Supplemental Tables 8 and 9). Data for UGT77B2 and CcAT1/2 gene expression were transformed ^0.35 or ^0.25, respectively, in order to meet statistical requirements. MbA levels in old corms from February 2016–February 2017 were additionally analyzed using a one-way ANOVA (Supplemental Table 9).
Accession Numbers
Transcriptome libraries from old and young corms are available in the NCBI/GenBank Sequence Read Archive (SRP108844). UGT and BAHD-AT nucleotide sequences were deposited in GenBank with the accession numbers MG938542 (UGT77B2), MG938543 (UGT709G2), MH365462 (CcAT1), and MH365463 (CcAT2). Accession numbers for all other genes/proteins used in this work can be found in Supplemental Tables 5 and 6 and Supplemental Data Sets 5 and 6.
Supplemental Data
Supplemental Figure 1. MbA and MbA-C detection in corms.
Supplemental Figure 2. Images of montbretia and MbA levels in dried corms.
Supplemental Figure 3. Accumulation of potential intermediates in MbA biosynthesis in old corms and young corms over a 1-year time course.
Supplemental Figure 4. UGT and BAHD-AT activity in protein extracts of yC and oC.
Supplemental Figure 5. UDP-Rha produced by Arabidopsis thaliana RHM-D and RHM-ER.
Supplemental Figure 6. Product profile of total young corm protein assays with myricetin or myricetin 3-O-glucoside as substrates.
Supplemental Figure 7. Products of UGT enzyme activity assays with corm protein extracts and UDP-Xyl.
Supplemental Figure 8. Products of UGT enzyme activity assays with corm protein extracts using MRG and UDP-Glc or UDP-Xyl.
Supplemental Figure 9. Phylogeny of UGTs identified in the montbretia corm transcriptome.
Supplemental Figure 10. Protein gel blot showing UGT and AT enzyme expression.
Supplemental Figure 11. Formation of myricetin 3-O-rhamnoside in assays with purified UGT77B2.
Supplemental Figure 12. Activity of purified UGT77B2 with myricetin and different UDP-sugars.
Supplemental Figure 13. Activity of purified UGT77B2 with different flavonols.
Supplemental Figure 14. m/z 625 peak 4 detected in corm extracts and produced in assays with recombinant UGT709G2.
Supplemental Figure 15. Structural elucidation of MRG using NMR.
Supplemental Figure 16. Product formation catalyzed by UGT709G2 over time.
Supplemental Figure 17. Activity of UGT709G2 with different sugar donors and flavonol 3-O-glycoside acceptors.
Supplemental Figure 18. Phylogeny of BAHD-ATs identified in the montbretia corm transcriptome.
Supplemental Figure 19. Enzyme activity of BAHD-ATs.
Supplemental Figure 20. Activity of purified CcAT1 and CcAT2 with quercetin 3-O-sophoroside.
Supplemental Figure 21. Activity of CcAT1 and CcAT2 with different acyl donors.
Supplemental Figure 22. Cq values of housekeeping genes in montbretia corms.
Supplemental Figure 23. N. benthamiana leaves transiently expressing montbretia UGTs and supplemented with myricetin produce MR and MRG.
Supplemental Figure 24. Myricetin 3-O-glucoside in transformed N. benthamiana leave extracts.
Supplemental Figure 25. N. benthamiana leaves transiently expressing montbretia UGTs and ATs.
Supplemental Figure 26. Sequence alignment of montbretia UGTs with UGTs of other plant species.
Supplemental Table 1. Metabolite levels in old and young corms over a 1-year growing season.
Supplemental Table 2. MbA concentration in dried montbretia corm material.
Supplemental Table 3. MbA levels in different organs of montbretia plants.
Supplemental Table 4. Activity of UGT77B2, UGT709G2, CcAT1, and CcAT2 with different acceptors.
Supplemental Table 5. NCBI identification numbers of BAHD-AT sequences used in the phylogenetic analysis.
Supplemental Table 6. NCBI gene identification numbers of sequences used in the phylogenetic tree.
Supplemental Table 7. Oligonucleotides used in this study.
Supplemental Table 8. Statistical analysis of gene expression data.
Supplemental Table 9. Statistical analysis of MbA levels in montbretia corms.
Supplemental Data Set 1. Differential expression of UGT transcripts in montbretia corms.
Supplemental Data Set 2. Gene expression correlation analysis of UGTs.
Supplemental Data Set 3. Differential expression of BAHDP-AT transcripts in montbretia corms.
Supplemental Data Set 4. Gene expression correlation analysis of BAHD-ATs.
Supplemental Data Set 5. Protein sequences of 159 montbretia UGTs
Supplemental Data Set 6. Protein sequences of 59 BAHD-ATs.
Supplemental File 1. Alignment used to produce the phylogenetic tree shown in Supplemental Figure 9.
Supplemental File 2. Alignment used to produce the phylogenetic tree shown in Figure 4.
Supplemental File 3. Alignment used to produce the phylogenetic tree shown in Supplemental Figure 18.
Supplemental File 4. Alignment used to produce the phylogenetic tree shown in Figure 6.
Dive Curated Terms
The following phenotypic, genotypic, and functional terms are of significance to the work described in this paper:
Acknowledgments
We thank Gary Brayer for providing montbretia corms, Carol Ritland for project management, Michael Court and the UGT nomenclature committee for naming of montbretia UGTs, and Henrik Scheller for providing the Arabidopsis UDP-l-rhamnose synthase. The research was supported by funds from the GlycoNet NCE to S.G.W. and J.B., by the Natural Sciences and Engineering Research Council of Canada (NSERC) to J.B., and by the Canadian Institutes for Health Research to S.G.W. S.I. was supported by the Alexander von Humboldt foundation through a Feodor Lynen Research Fellowship. Se.J. was supported with an undergraduate fellowship from the UBC Genome Science and Technology Graduate Program and an NSERC Canada Graduate Scholarship-Master’s. C.R.R. was supported with scholarships from the British Columbia Epilepsy Society.
AUTHOR CONTRIBUTIONS
J.B. and S.G.W. conceived the project and secured the funding. S.I. and J.B. designed the research. S.I., Se.J., C.R.R., Sh.J., L.M., M.O.J., and R.W. carried out the experimental work. S.I., Se.J., C.R.R., M.M.S.Y., M.O.J., S.G.W. and J.B. analyzed data and interpreted results. S.I. and J.B. wrote the article. All authors read, edited, and approved the final manuscript.
Footnotes
Articles can be viewed without a subscription.
References
- Ablajan K., Abliz Z., Shang X.-Y., He J.-M., Zhang R.-P., Shi J.-G. (2006). Structural characterization of flavonol 3,7-di-O-glycosides and determination of the glycosylation position by using negative ion electrospray ionization tandem mass spectrometry. J. Mass Spectrom. 41: 352–360. [DOI] [PubMed] [Google Scholar]
- Asada Y., Hirayama Y., Furuya T. (1988). Acylated flavonols from Crocosmia-Crocosmiiflora. Phytochemistry 27: 1497–1501. [Google Scholar]
- Bontpart T., Cheynier V., Ageorges A., Terrier N. (2015). BAHD or SCPL acyltransferase? What a dilemma for acylation in the world of plant phenolic compounds. New Phytol. 208: 695–707. [DOI] [PubMed] [Google Scholar]
- Caputi L., Malnoy M., Goremykin V., Nikiforova S., Martens S. (2012). A genome-wide phylogenetic reconstruction of family 1 UDP-glycosyltransferases revealed the expansion of the family during the adaptation of plants to life on land. Plant J. 69: 1030–1042. [DOI] [PubMed] [Google Scholar]
- Celedon J.M., Chiang A., Yuen M.M.S., Diaz-Chavez M.L., Madilao L.L., Finnegan P.M., Barbour E.L., Bohlmann J. (2016). Heartwood-specific transcriptome and metabolite signatures of tropical sandalwood (Santalum album) reveal the final step of (Z)-santalol fragrance biosynthesis. Plant J. 86: 289–299. [DOI] [PubMed] [Google Scholar]
- Chen Y.K., Li X.S., Yang G.Y., Chen Z.Y., Hu Q.F., Miao M.M. (2012). Phenolic compounds from Nicotiana tabacum and their biological activities. J. Asian Nat. Prod. Res. 14: 450–456. [DOI] [PubMed] [Google Scholar]
- Crocoll C., Mirza N., Reichelt M., Gershenzon J., Halkier B.A. (2016). Optimization of engineered production of the glucoraphanin precursor dihomomethionine in Nicotiana benthamiana. Front. Bioeng. Biotechnol. 4: 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Auria J.C. (2006). Acyltransferases in plants: a good time to be BAHD. Curr. Opin. Plant Biol. 9: 331–340. [DOI] [PubMed] [Google Scholar]
- D’Auria J.C., Chen F., Pichersky E. (2002). Characterization of an acyltransferase capable of synthesizing benzylbenzoate and other volatile esters in flowers and damaged leaves of Clarkia breweri. Plant Physiol. 130: 466–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di S., Yan F., Rodas F.R., Rodriguez T.O., Murai Y., Iwashina T., Sugawara S., Mori T., Nakabayashi R., Yonekura-Sakakibara K., Saito K., Takahashi R. (2015). Linkage mapping, molecular cloning and functional analysis of soybean gene Fg3 encoding flavonol 3-O-glucoside/galactoside (1→2) glucosyltransferase. BMC Plant Biol. 15: 126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon R.A., Strack D. (2003). Phytochemistry meets genome analysis, and beyond. Phytochemistry 62: 815–816. [DOI] [PubMed] [Google Scholar]
- Ford C.M., Boss P.K., Hoj P.B. (1998). Cloning and characterization of Vitis vinifera UDP-glucose:flavonoid 3-O-glucosyltransferase, a homologue of the enzyme encoded by the maize Bronze-1 locus that may primarily serve to glucosylate anthocyanidins in vivo. J. Biol. Chem. 273: 9224–9233. [DOI] [PubMed] [Google Scholar]
- Frydman A., Weisshaus O., Bar-Peled M., Huhman D.V., Sumner L.W., Marin F.R., Lewinsohn E., Fluhr R., Gressel J., Eyal Y. (2004). Citrus fruit bitter flavors: isolation and functional characterization of the gene Cm1,2RhaT encoding a 1,2 rhamnosyltransferase, a key enzyme in the biosynthesis of the bitter flavonoids of citrus. Plant J. 40: 88–100. [DOI] [PubMed] [Google Scholar]
- Frydman A., Liberman R., Huhman D.V., Carmeli-Weissberg M., Sapir-Mir M., Ophir R., W Sumner L., Eyal Y. (2013). The molecular and enzymatic basis of bitter/non-bitter flavor of citrus fruit: evolution of branch-forming rhamnosyltransferases under domestication. Plant J. 73: 166–178. [DOI] [PubMed] [Google Scholar]
- Fu L., Niu B., Zhu Z., Wu S., Li W. (2012). CD-HIT: accelerated for clustering the next-generation sequencing data. Bioinformatics 28: 3150–3152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gachon C.M., Langlois-Meurinne M., Saindrenan P. (2005). Plant secondary metabolism glycosyltransferases: the emerging functional analysis. Trends Plant Sci. 10: 542–549. [DOI] [PubMed] [Google Scholar]
- Grabherr M.G., et al. (2011). Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 29: 644–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griesser M., Vitzthum F., Fink B., Bellido M.L., Raasch C., Munoz-Blanco J., Schwab W. (2008). Multi-substrate flavonol O-glucosyltransferases from strawberry (Fragaria x ananassa) achene and receptacle. J. Exp. Bot. 59: 2611–2625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannweg K., Sippel A., Bertling I. (2013). A simple and effective method for the micropropagation and in vitro induction of polyploidy and the effect on floral characteristics of the South African iris, Crocosmia aurea. S. Afr. J. Bot. 88: 367–372. [Google Scholar]
- Hartmann T. (2008). The lost origin of chemical ecology in the late 19th century. Proc. Natl. Acad. Sci. USA 105: 4541–4546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu Y.-H., Tagami T., Matsunaga K., Okuyama M., Suzuki T., Noda N., Suzuki M., Shimura H. (2017). Functional characterization of UDP-rhamnose-dependent rhamnosyltransferase involved in anthocyanin modification, a key enzyme determining blue coloration in Lobelia erinus. Plant J. 89: 325–337. [DOI] [PubMed] [Google Scholar]
- Jones P., Messner B., Nakajima J., Schäffner A.R., Saito K. (2003). UGT73C6 and UGT78D1, glycosyltransferases involved in flavonol glycoside biosynthesis in Arabidopsis thaliana. J. Biol. Chem. 278: 43910–43918. [DOI] [PubMed] [Google Scholar]
- Kolosova N., Miller B., Ralph S., Ellis B.E., Douglas C., Ritland K., Bohlmann J. (2004). Isolation of high-quality RNA from gymnosperm and angiosperm trees. Biotechniques 36: 821–824. [DOI] [PubMed] [Google Scholar]
- Kroon J., Souer E., de Graaff A., Xue Y., Mol J., Koes R. (1994). Cloning and structural analysis of the anthocyanin pigmentation locus Rt of Petunia hybrida: characterization of insertion sequences in two mutant alleles. Plant J. 5: 69–80. [DOI] [PubMed] [Google Scholar]
- Kubo A., Arai Y., Nagashima S., Yoshikawa T. (2004). Alteration of sugar donor specificities of plant glycosyltransferases by a single point mutation. Arch. Biochem. Biophys. 429: 198–203. [DOI] [PubMed] [Google Scholar]
- Lau W., Sattely E.S. (2015). Six enzymes from mayapple that complete the biosynthetic pathway to the etoposide aglycone. Science 349: 1224–1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law C.W., Chen Y., Shi W., Smyth G.K. (2014). voom: Precision weights unlock linear model analysis tools for RNA-seq read counts. Genome Biol. 15: R29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X.J., Zhang J.Q., Wu Z.C., Lai B., Huang X.M., Qin Y.H., Wang H.C., Hu G.B. (2015). Functional characterization of a glucosyltransferase gene, LcUFGT1, involved in the formation of cyanidin glucoside in the pericarp of Litchi chinensis. Physiol. Plant. 156: 139–149. [DOI] [PubMed] [Google Scholar]
- Li X.J., Lai B., Zhao J.T., Qin Y.H., He J.M., Huang X.M., Wang H.C., Hu G.B. (2016). Sequence differences in LcFGRT4 alleles are responsible for the diverse anthocyanin composition in the pericarp of Litchi chinensis. Mol. Breed. 36: 93. [Google Scholar]
- Li Y., Baldauf S., Lim E.K., Bowles D.J. (2001). Phylogenetic analysis of the UDP-glycosyltransferase multigene family of Arabidopsis thaliana. J. Biol. Chem. 276: 4338–4343. [DOI] [PubMed] [Google Scholar]
- Li Y., Li P., Wang Y., Dong R., Yu H., Hou B. (2014). Genome-wide identification and phylogenetic analysis of Family-1 UDP glycosyltransferases in maize (Zea mays). Planta 239: 1265–1279. [DOI] [PubMed] [Google Scholar]
- Liu J., Osbourn A., Ma P. (2015). MYB transcription factors as regulators of phenylpropanoid metabolism in plants. Mol. Plant 8: 689–708. [DOI] [PubMed] [Google Scholar]
- Lombard V., Golaconda Ramulu H., Drula E., Coutinho P.M., Henrissat B. (2014). The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res. 42: D490–D495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo J., et al. (2007). Convergent evolution in the BAHD family of acyl transferases: identification and characterization of anthocyanin acyl transferases from Arabidopsis thaliana. Plant J. 50: 678–695. [DOI] [PubMed] [Google Scholar]
- Martin D., Tholl D., Gershenzon J., Bohlmann J. (2002). Methyl jasmonate induces traumatic resin ducts, terpenoid resin biosynthesis, and terpenoid accumulation in developing xylem of Norway spruce stems. Plant Physiol. 129: 1003–1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masada S., Terasaka K., Oguchi Y., Okazaki S., Mizushima T., Mizukami H. (2009). Functional and structural characterization of a flavonoid glucoside 1,6-glucosyltransferase from Catharanthus roseus. Plant Cell Physiol. 50: 1401–1415. [DOI] [PubMed] [Google Scholar]
- Mikkelsen M.D., Olsen C.E., Halkier B.A. (2010). Production of the cancer-preventive glucoraphanin in tobacco. Mol. Plant 3: 751–759. [DOI] [PubMed] [Google Scholar]
- Milkowski C., Strack D. (2004). Serine carboxypeptidase-like acyltransferases. Phytochemistry 65: 517–524. [DOI] [PubMed] [Google Scholar]
- Miller K.D., Guyon V., Evans J.N.S., Shuttleworth W.A., Taylor L.P. (1999). Purification, cloning, and heterologous expression of a catalytically efficient flavonol 3-O-galactosyltransferase expressed in the male gametophyte of Petunia hybrida. J. Biol. Chem. 274: 34011–34019. [DOI] [PubMed] [Google Scholar]
- Montefiori M., Espley R.V., Stevenson D., Cooney J., Datson P.M., Saiz A., Atkinson R.G., Hellens R.P., Allan A.C. (2011). Identification and characterisation of F3GT1 and F3GGT1, two glycosyltransferases responsible for anthocyanin biosynthesis in red-fleshed kiwifruit (Actinidia chinensis). Plant J. 65: 106–118. [DOI] [PubMed] [Google Scholar]
- Morita Y., Hoshino A., Kikuchi Y., Okuhara H., Ono E., Tanaka Y., Fukui Y., Saito N., Nitasaka E., Noguchi H., Iida S. (2005). Japanese morning glory dusky mutants displaying reddish-brown or purplish-gray flowers are deficient in a novel glycosylation enzyme for anthocyanin biosynthesis, UDP-glucose:anthocyanidin 3-O-glucoside-2′'-O-glucosyltransferase, due to 4-bp insertions in the gene. Plant J. 42: 353–363. [DOI] [PubMed] [Google Scholar]
- Nagel R., Gershenzon J., Schmidt A. (2012). Nonradioactive assay for detecting isoprenyl diphosphate synthase activity in crude plant extracts using liquid chromatography coupled with tandem mass spectrometry. Anal. Biochem. 422: 33–38. [DOI] [PubMed] [Google Scholar]
- Nakajima J., Tanaka Y., Yamazaki M., Saito K. (2001). Reaction mechanism from leucoanthocyanidin to anthocyanidin 3-glucoside, a key reaction for coloring in anthocyanin biosynthesis. J. Biol. Chem. 276: 25797–25803. [DOI] [PubMed] [Google Scholar]
- Noguchi A., Fukui Y., Iuchi-Okada A., Kakutani S., Satake H., Iwashita T., Nakao M., Umezawa T., Ono E. (2008). Sequential glucosylation of a furofuran lignan, (+)-sesaminol, by Sesamum indicum UGT71A9 and UGT94D1 glucosyltransferases. Plant J. 54: 415–427. [DOI] [PubMed] [Google Scholar]
- Noguchi A., Horikawa M., Fukui Y., Fukuchi-Mizutani M., Iuchi-Okada A., Ishiguro M., Kiso Y., Nakayama T., Ono E. (2009). Local differentiation of sugar donor specificity of flavonoid glycosyltransferase in Lamiales. Plant Cell 21: 1556–1572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono E., Homma Y., Horikawa M., Kunikane-Doi S., Imai H., Takahashi S., Kawai Y., Ishiguro M., Fukui Y., Nakayama T. (2010). Functional differentiation of the glycosyltransferases that contribute to the chemical diversity of bioactive flavonol glycosides in grapevines (Vitis vinifera). Plant Cell 22: 2856–2871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan S.Y., Zhou S.F., Gao S.H., Yu Z.L., Zhang S.F., Tang M.K., Sun J.N., Ma D.L., Han Y.F., Fong W.F., Ko K.M. (2013). New perspectives on how to discover drugs from herbal medicines: CAM's outstanding contribution to modern therapeutics. eCAM 2013: 627375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paterson A.H., et al. (2009). The Sorghum bicolor genome and the diversification of grasses. Nature 457: 551–556. [DOI] [PubMed] [Google Scholar]
- Patro R., Duggal G., Love M.I., Irizarry R.A., Kingsford C. (2017). Salmon provides fast and bias-aware quantification of transcript expression. Nat. Methods 14: 417–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rautengarten C., et al. (2014). The Golgi localized bifunctional UDP-rhamnose/UDP-galactose transporter family of Arabidopsis. Proc. Natl. Acad. Sci. USA 111: 11563–11568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rojas Rodas F., Rodriguez T.O., Murai Y., Iwashina T., Sugawara S., Suzuki M., Nakabayashi R., Yonekura-Sakakibara K., Saito K., Kitajima J., Toda K., Takahashi R. (2014). Linkage mapping, molecular cloning and functional analysis of soybean gene Fg2 encoding flavonol 3-O-glucoside (1 → 6) rhamnosyltransferase. Plant Mol. Biol. 84: 287–300. [DOI] [PubMed] [Google Scholar]
- Rojas Rodas F., Di S., Murai Y., Iwashina T., Sugawara S., Mori T., Nakabayashi R., Yonekura-Sakakibara K., Saito K., Takahashi R. (2016). Cloning and characterization of soybean gene Fg1 encoding flavonol 3-O-glucoside/galactoside (1→6) glucosyltransferase. Plant Mol. Biol. 92: 445–456. [DOI] [PubMed] [Google Scholar]
- Ross J., Li Y., Lim E.K., Bowles D.J. (2001). Higher plant glycosyltransferases. Genome Biol. 2: 3004.1–3004.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawada S., Suzuki H., Ichimaida F., Yamaguchi M.A., Iwashita T., Fukui Y., Hemmi H., Nishino T., Nakayama T. (2005). UDP-glucuronic acid:anthocyanin glucuronosyltransferase from red daisy (Bellis perennis) flowers. Enzymology and phylogenetics of a novel glucuronosyltransferase involved in flower pigment biosynthesis. J. Biol. Chem. 280: 899–906. [DOI] [PubMed] [Google Scholar]
- Scheen A.J. (1997). Drug treatment of non-insulin-dependent diabetes mellitus in the 1990s. Achievements and future developments. Drugs 54: 355–368. [DOI] [PubMed] [Google Scholar]
- Scheen A.J. (2003). Is there a role for α-glucosidase inhibitors in the prevention of type 2 diabetes mellitus? Drugs 63: 933–951. [DOI] [PubMed] [Google Scholar]
- Schmidt G.W., Jirschitzka J., Porta T., Reichelt M., Luck K., Torre J.C.P., Dolke F., Varesio E., Hopfgartner G., Gershenzon J., D’Auria J.C. (2015). The last step in cocaine biosynthesis is catalyzed by a BAHD acyltransferase. Plant Physiol. 167: 89–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott L.J., Spencer C.M. (2000). Miglitol: a review of its therapeutic potential in type 2 diabetes mellitus. Drugs 59: 521–549. [DOI] [PubMed] [Google Scholar]
- Simonnet X., Quennoz M., Carlen C. (2008). New Artimisia annua hybrids with high artemisinin content. Acta Hortic. (769): 371–373. [Google Scholar]
- Sui X., Gao X., Ao M., Wang Q., Yang D., Wang M., Fu Y., Wang L. (2011). cDNA cloning and characterization of UDP-glucose: anthocyanidin 3-O-glucosyltransferase in Freesia hybrida. Plant Cell Rep. 30: 1209–1218. [DOI] [PubMed] [Google Scholar]
- Sullivan M. (2009). A novel red clover hydroxycinnamoyl transferase has enzymatic activities consistent with a role in phaselic acid biosynthesis. Plant Physiol. 150: 1866–1879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun W., Liang L., Meng X., Li Y., Gao F., Liu X., Wang S., Gao X., Wang L. (2016). Biochemical and molecular characterization of a flavonoid 3-O-glycosyltransferase responsible for anthocyanins and flavonols biosynthesis in Freesia hybrida. Front. Plant Sci. 7: 410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K., Peterson D., Peterson N., Stecher G., Nei M., Kumar S. (2011). MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28: 2731–2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarling C.A., Woods K., Zhang R., Brastianos H.C., Brayer G.D., Andersen R.J., Withers S.G. (2008). The search for novel human pancreatic alpha-amylase inhibitors: high-throughput screening of terrestrial and marine natural product extracts. ChemBioChem 9: 433–438. [DOI] [PubMed] [Google Scholar]
- Tohge T., Zhang Y., Peterek S., Matros A., Rallapalli G., Tandrón Y.A., Butelli E., Kallam K., Hertkorn N., Mock H.P., Martin C., Fernie A.R. (2015). Ectopic expression of snapdragon transcription factors facilitates the identification of genes encoding enzymes of anthocyanin decoration in tomato. Plant J. 83: 686–704. [DOI] [PubMed] [Google Scholar]
- Trantas E.A., Koffas M.A.G., Xu P., Ververidis F. (2015). When plants produce not enough or at all: metabolic engineering of flavonoids in microbial hosts. Front. Plant Sci. 6: 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuominen L.K., Johnson V.E., Tsai C.J. (2011). Differential phylogenetic expansions in BAHD acyltransferases across five angiosperm taxa and evidence of divergent expression among Populus paralogues. BMC Genomics 12: 236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuskan G.A., et al. (2006). The genome of black cottonwood, Populus trichocarpa (Torr. & Gray). Science 313: 1596–1604. [DOI] [PubMed] [Google Scholar]
- Vogt T. (2010). Phenylpropanoid biosynthesis. Mol. Plant 3: 2–20. [DOI] [PubMed] [Google Scholar]
- Wang Y., Chen S., Yu O. (2011). Metabolic engineering of flavonoids in plants and microorganisms. Appl. Microbiol. Biotechnol. 91: 949–956. [DOI] [PubMed] [Google Scholar]
- Williams L.K., et al. (2015). The amylase inhibitor montbretin A reveals a new glycosidase inhibition motif. Nat. Chem. Biol. 11: 691–696. [DOI] [PubMed] [Google Scholar]
- Yabuya T., Yamaguchi M.a., Imayama T., Katoh K., Ino I. (2002). Anthocyanin 5-O-glucosyltransferase in flowers of Iris ensata. Plant Sci. 162: 779–784. [DOI] [PubMed] [Google Scholar]
- Yamazaki M., Yamagishi E., Gong Z., Fukuchi-Mizutani M., Fukui Y., Tanaka Y., Kusumi T., Yamaguchi M., Saito K. (2002). Two flavonoid glucosyltransferases from Petunia hybrida: molecular cloning, biochemical properties and developmentally regulated expression. Plant Mol. Biol. 48: 401–411. [DOI] [PubMed] [Google Scholar]
- Yang S.M., Han S.H., Kim B.G., Ahn J.H. (2014). Production of kaempferol 3-O-rhamnoside from glucose using engineered Escherichia coli. J. Ind. Microbiol. Biotechnol. 41: 1311–1318. [DOI] [PubMed] [Google Scholar]
- Yonekura-Sakakibara K., et al. (2012). Two glycosyltransferases involved in anthocyanin modification delineated by transcriptome independent component analysis in Arabidopsis thaliana. Plant J. 69: 154–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yonekura-Sakakibara K., Nakabayashi R., Sugawara S., Tohge T., Ito T., Koyanagi M., Kitajima M., Takayama H., Saito K. (2014). A flavonoid 3-O-glucoside:2"-O-glucosyltransferase responsible for terminal modification of pollen-specific flavonols in Arabidopsis thaliana. Plant J. 79: 769–782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuen V.G., Coleman J., Withers S.G., Andersen R.J., Brayer G.D., Mustafa S., McNeill J.H. (2016). Glucose lowering effect of montbretin A in Zucker Diabetic Fatty rats. Mol. Cell. Biochem. 411: 373–381. [DOI] [PubMed] [Google Scholar]
- Zerbe P., Chiang A., Dullat H., O’Neil-Johnson M., Starks C., Hamberger B., Bohlmann J. (2014). Diterpene synthases of the biosynthetic system of medicinally active diterpenoids in Marrubium vulgare. Plant J. 79: 914–927. [DOI] [PubMed] [Google Scholar]
- Zhong J.J. (2002). Plant cell culture for production of paclitaxel and other taxanes. J. Biosci. Bioeng. 94: 591–599. [DOI] [PubMed] [Google Scholar]
- Zurawik P., Salachna P., Zurawik A., Dobrowolska A. (2015). Morphological traits, flowering and corm yield of Crocosmia x crocosmiiflora (Lemoine) N.E. cultivars are determined by planting time. Acta Sci. Pol. Hortorum Cultus 14: 97–108. [Google Scholar]