Some proteins are put to work in more than one job, in a form of evolutionary improvisation. They are often described as “moonlighting” proteins, referring to the practice of people taking a second job, typically after dark, to help pay the bills. A classic example of a moonlighting protein is the enzyme lactate dehydrogenase, which was adapted to serve as a crystallin protein in the eye lens of some vertebrates (Wistow et al., 1987).
Schreier et al. (2018) now provide a striking example of a moonlighting function for plastidial NAD+-dependent malate dehydrogenase (pdNAD-MDH) in Arabidopsis thaliana. The authors report that pdNAD-MDH is essential for plastid development, not as a functional metabolic enzyme but, rather, as a protein constituent of a large AAA+ (ATPase associated with various cellular activities) protease complex localized to the chloroplast envelope inner membrane and essential for protein processing during plastid biogenesis.
In addition to pdNAD-MDH, plastids contain NADP+-dependent MDH (pdNADP-MDH). Redox-regulated pdNADP-MDH is active in the light and is considered to function in the “malate valve” that exports excess reducing equivalents from the chloroplast as malate (Scheibe, 2004). However, Arabidopsis mutants lacking pdNADP-MDH develop normally with only slightly reduced growth rate (Heyno et al., 2014). These observations raised the possibility that pdNAD-MDH might function in the malate valve mechanism, at least in the absence of pdNADP-MDH, by generating malate for chloroplast export.
Strikingly, Arabidopsis pdNAD-MDH knockout mutants are embryo-lethal, and a line with lowered pdNAD-MDH from gene silencing with an artificial microRNA had poor growth, pale leaves, disorganized chloroplasts, and low nighttime respiration (Beeler et al., 2014). In order to understand the essential function of this enzyme, Schreier et al. (2018) adopted several approaches.
First. they showed that embryo-specific biosynthesis of pdNAD-MDH using the ABI3 gene promoter rescued the pdnad-mdh mutant, confirming that pdNAD-MDH is essential during embryogenesis and seed development. However, the resulting seedlings produced white leaves and failed to develop properly. Similarly, inducible silencing of the pdNAD-MDH gene in adult plants led to the formation of white leaves. Electron microscopy visualization of plastids and analysis of galactolipids, carotenoids, and protochlorophyllide established that chloroplast development is impaired in the absence of pdNAD-MDH.
A possible explanation considered was that NAD+-dependent MDH activity is required in the plastid for chloroplast biogenesis. However, when Arabidopsis NAD+-dependent MDH enzymes from the peroxisome, mitochondrion, or cytosol were targeted to the plastid, they failed to complement the pdnad-mdh mutant. Even more surprisingly, enzymatically inactive variants of pdNAD-MDH produced by site-directed mutagenesis fully complemented the pdnad-mdh mutant. Therefore, the protein, but not its NAD+-dependent MDH enzyme activity, is required for plastid development.
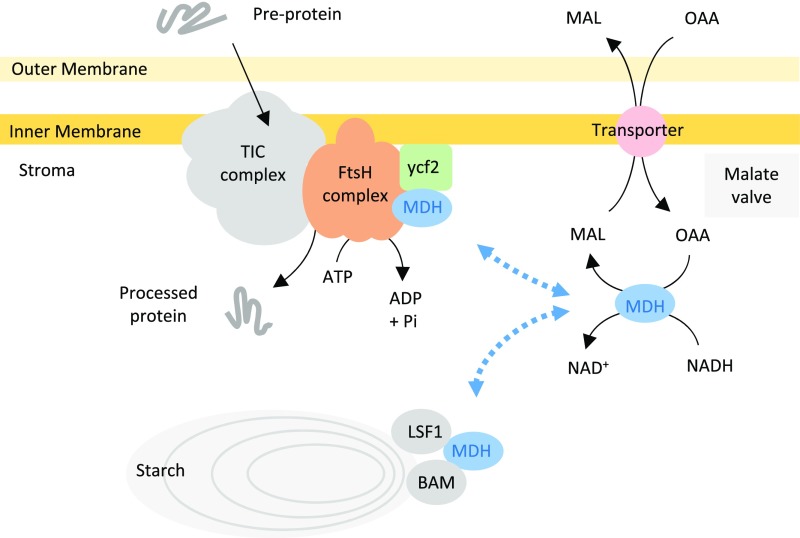
Next, they found that some of the most abundant peptides associated with pdNAD-MDH, identified by immunoprecipitation and LC-MS/MS, were derived from hypothetical chloroplast open reading frame 2 (Ycf2) and filamentous temperature sensitive (Fts) proteases—proteins known to be essential for chloroplast biogenesis. Functional interaction of pdNAD-MDH with FtsH12 was established with both enzymatically active and inactive forms of the enzyme. A knockout mutant of FstH12 was shown to be embryo lethal, and miRNA silencing of FtsH12 produced plants resembling those in which pdNAD-MDH is silenced. Moreover, FtsH12 protein was lacking in pdNAD-MDH-silenced plants, suggesting that pdNAD-MDH is required to stabilize FtsH12.
FtsH12 can associate with the chloroplast inner envelope, and the immunoprecipitation experiments used to identify it and Ycf2 also identified components of the plastid protein import machinery. With this finding, and through analogy to the role of FtsH proteins in mitochondria, Schreier and colleagues speculate that the FtsH complex may act as the ATP-driven import motor, recently proposed by Nakai (2018), that functions with the Translocon of the Inner membrane of Chloroplasts (TIC) complex to draw proteins into the plastid (see figure).
Moonlighting functions of pdNAD-MDH. (Adapted from Screier et al. [2018], Figure 11.)
Intriguingly, the immunoprecipitation experiments also identified starch degradation enzymes, including Like SEX4 1, BETA-AMYLASE1 (BAM1), and BAM3. These observations suggest that pdNAD-MDH might have additional roles in plastid function.
These discoveries elevate pdNAD-MDH to a new level of importance in plant biology. First, they open the way to determining the functions of pdNAD-MDH and pdNADP-MDH in plastid redox metabolism. By expressing enzymatically inactive pdNAD-MDH protein in a pdnad-mdh mutant, it should be possible to cross this with an nadp-mdh mutant to create a double mutant lacking all plastid MDH activity. This will make it possible to determine the role of MDH activity in the malate valve and in other vital aspects of plastid metabolism including the Calvin cycle, starch metabolism, and fatty acid biosynthesis.
Second, this discovery identifies pdNAD-MDH as a good example of a moonlighting protein in plants. However, it also raises the question of the distinction between moonlighting and multitasking or coordinating proteins. Potentially, pdNAD-MDH operates simultaneously to coordinate redox status with plastid protein import and carbon metabolism. Thus, it could be argued that it is simply multitasking. Regardless of the semantics of such a distinction, pdNAD-MDH is now shown to be a pivotal protein in several aspects of plastid function.
Footnotes
Articles can be viewed without a subscription.
References
- Beeler S., Liu H.-C., Stadler M., Schreier T., Eicke S., Lue W.-L., Truernit E., Zeeman S.C., Chen J., Kötting O. (2014). Plastidial NAD-dependent malate dehydrogenase is critical for embryo development and heterotrophic metabolism in Arabidopsis. Plant Physiol. 164: 1175–1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heyno E., Innocenti G., Lemaire S.D., Issakidis-Bourguet E., Krieger-Liszkay A. (2014). Putative role of the malate valve enzyme NADP-malate dehydrogenase in H2O2 signalling in Arabidopsis. Philos. Trans. R. Soc. Lond. B Biol. Sci. 369: 20130228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakai M. (2018). New perspectives on chloroplast protein import. Plant Cell Physiol. 59: 1111–1119. [DOI] [PubMed] [Google Scholar]
- Scheibe R. (2004). Malate valves to balance cellular energy supply. Physiol. Plant. 120: 21–26. [DOI] [PubMed] [Google Scholar]
- Schreier T.B., Antoine C., Schläfli M., Galbier F., Stadler M., Demarsy E., Albertini D., Maier B.A., Kessler F., Hörtensteiner S., Zeeman S.C., Kötting O. (2018). Plastidial NAD-dependent malate dehydrogenase: a moonlighting protein involved in early chloroplast development through its interaction with an FtsH12-FtsHi protease complex. Plant Cell 30: 1745–1769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wistow G.J., Mulders J.W., de Jong W.W. (1987). The enzyme lactate dehydrogenase as a structural protein in avian and crocodilian lenses. Nature 326: 622–624. [DOI] [PubMed] [Google Scholar]