Chloroplast protein BFA1 is comprised primarily of two β-barrels and acts as a chaperone orchestrating the early steps of the CF1 assembly pathway via specific interaction with the CF1β, γ, and ε subunits.
Abstract
F-type ATP synthases produce nearly all of the ATP found in cells. The catalytic module F1 commonly comprises an α3β3 hexamer surrounding a γ/ε stalk. However, it is unclear how these subunits assemble to form a catalytic motor. In this work, we identified and characterized a chloroplast protein that interacts with the CF1β, γ, and ε subunits of the chloroplast ATP synthase and is required for assembly of its F1 module. We named this protein BIOGENESIS FACTOR REQUIRED FOR ATP SYNTHASE1 (BFA1) and determined its crystal structure at 2.8-Å resolution. BFA1 is comprised primarily of two interacting β-barrels that are oriented nearly perpendicularly to each other. The contact region between BFA1 and the CF1β and γ subunits was further mapped by yeast two-hybrid assays. An in silico molecular docking analysis was performed and revealed close fitting contact sites without steric conflicts between BFA1 and CF1β/γ. We propose that BFA1 acts mainly as a scaffold protein promoting the association of a CF1α/β heterodimer with CF1γ. The subsequent assembly of other CF1α/β heterodimers may shift the position of the CF1γ subunit to complete assembly of the CF1 module. This CF1 assembly process is likely to be valid for other F-type ATP synthases, as their structures are highly conserved.
INTRODUCTION
F-type ATPases, also called ATP synthases, are ubiquitous protein complexes found in energy-transducing membranes from bacteria, mitochondria, and chloroplasts. Utilizing the transmembrane proton gradient formed by respiratory or photosynthetic electron transport chains, these enzymes provide most of the cellular ATP, the universal energy currency required by living cells (Yoshida et al., 2001; Junge and Nelson, 2015). The overall architecture of ATP synthase is conserved in a variety of species and comprises two rotary motors, F1 and Fo (von Ballmoos et al., 2009; Hahn et al., 2018). The F1 module is a hydrophilic unit that catalyses ATP formation or hydrolysis and commonly consists of five subunits with the stoichiometry α3β3γ1ε1δ1. Whereas the γ/ε stalk is inserted into the α3β3 hexamer, subunit δ serves as clamp to fix the positions of F1 and Fo, the membrane sector of ATP synthase. The composition of the Fo subcomplex varies greatly between species, but generally it consists of hydrophobic subunits that form a transmembrane proton translocating unit (von Ballmoos et al., 2009). The rotational catalytic mechanism of ATP synthase is conserved between species; an efflux of protons through the Fo motor drives the rotation of the c-ring, which is connected to the γ/ε subunits of the F1 subcomplex. Because the α3β3 hexamer is fixed in place by the peripheral stalk, the relative rotation of γ/ε inside the α3β3 hexamer induces conformational changes in the catalytic sites of α3β3, resulting in the production of ATP from ADP and inorganic phosphate (Junge and Nelson, 2015).
Given the importance of the F-type ATP synthases in living cells and their potential usage in the development of molecular motors for nanomedicine in synthetic biology (Ahmad and Cox, 2014), the functional mechanisms and assembly pathway of this complex have been widely investigated. Although ATP formation in ATP synthase has been well explained by the binding change mechanism (Cross, 2000), the steps for the assembly of the complex remain elusive. The assembly of mitochondrial and bacterial F1Fo-ATP synthases is thought to involve the stepwise formation of several transient intermediates (Ackerman and Tzagoloff, 2005; Rak et al., 2011). For example, the mitochondrial c-ring and stator are independently assembled in the inner mitochondrial membrane, whereas the soluble F1 subcomplex is assembled in the matrix. The F1 module then binds to the c-ring, and the stator is integrated to form the functional complex. The assembly of the structurally similar chloroplast ATP synthase is thought to occur via a similar process (Rühle and Leister, 2015).
Depending upon cellular needs, the F1 module can catalyze either ATP synthesis or hydrolysis; therefore, understanding the mechanism of its assembly is of particular importance. In mitochondria, the assembly of the F1 module is facilitated by Atp11p and Atp12p, which interact with unassembled β and α subunits, respectively (Wang et al., 2000; Wang and Ackerman, 2000). However, the underlying molecular mechanisms are not well understood. In chloroplasts, in vitro reconstitution studies suggest that the formation of the α/β dimer, which involves several chaperones, including Cpn60, Cpn24, and Hsp70, is the first step of CF1 assembly (Chen and Jagendorf, 1994). An analysis of Arabidopsis thaliana mutants revealed that the chloroplast stromal proteins PAB (PROTEIN IN CHLOROPLAST ATPASE BIOGENESIS) and BFA3 (also known as CONSERVED IN THE GREEN LINEAGE AND DIATOMS11) are involved in the assembly of the CF1 subcomplex by specifically interacting with CF1γ and CF1β, respectively (Mao et al., 2015; Grahl et al., 2016; Zhang et al., 2016). However, because the whole assembly process of the CF1 module is still unclear, we isolated mutants that accumulated reduced amounts of chloroplast ATP synthase, with the goal of identifying proteins required for its assembly.
Here, we report the identification and characterization of the chloroplast protein BIOGENESIS FACTOR REQUIRED FOR ATP SYNTHASE1 (BFA1), which promotes the assembly of the CF1 module of chloroplast ATP synthase by interacting with several CF1 subunits. The crystal structure of BFA1 has been resolved at 2.8 Å and the BFA1-CF1β/γ interacting region has been mapped. Based on these results, we propose a model for the assembly of the α3β3γ subcomplex in which BFA1 interacts directly with the β and γ subunits.
RESULTS
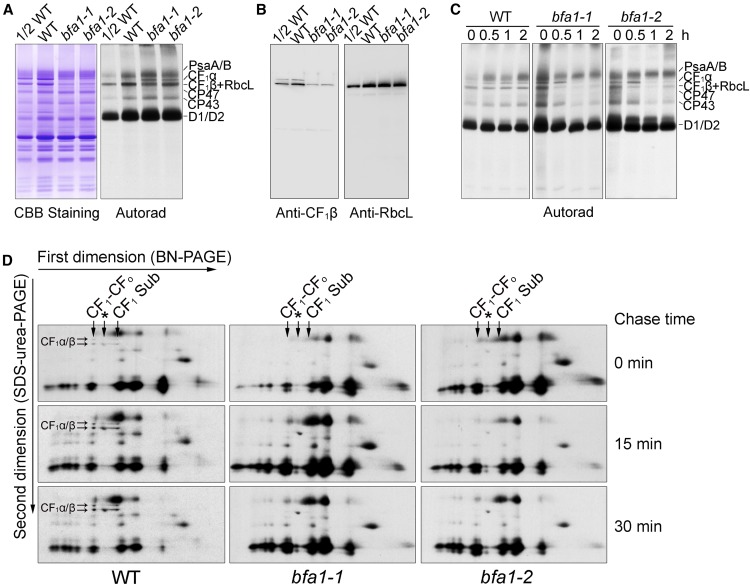
Chloroplast ATP Synthase Accumulation Is Specifically Reduced in bfa1
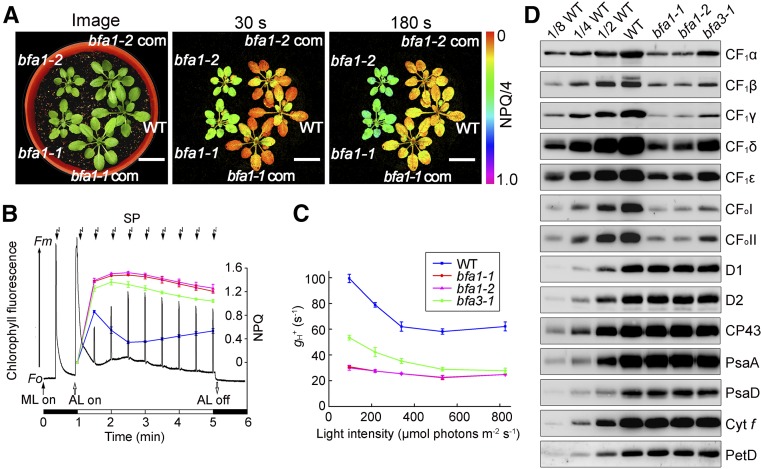
Nonphotochemical quenching of chlorophyll fluorescence (NPQ) is a photoprotective process employed by photosynthetic organisms to dissipate excess absorbed light energy into heat (Niyogi, 1999). The main component of NPQ, qE, is strictly dependent on the level of protons in the thylakoid lumen, which in turn is mainly controlled by photosynthetic electron transport activity and chloroplast ATP synthase. In the mutants with reduced ATP synthase activity, proton efflux from thylakoid lumen is less efficient, resulting in a high level of NPQ (Zhang et al., 2016). The bfa1-1 mutant was isolated on the basis of its high-NPQ phenotype upon illumination, as previously described (Figure 1A; Zhang et al., 2016). The bfa1-2 line was obtained from the NASC (European Arabidopsis Stock Centre). In both mutants, NPQ can be rapidly induced to ∼1.0 within 30 s of illumination (Figures 1A and 1B). Because chloroplast ATP synthase is activated by light, the protons that accumulate in the thylakoid lumen flow out efficiently through the ATP synthase in the wild type and NPQ is rapidly relaxed within 2 min (Figure 1B). By contrast, NPQ was only slightly relaxed in the two bfa1 mutants, as well as in our previously characterized mutant bfa3-1 (Figure 1B; Zhang et al., 2016), and this can be explained by their low chloroplast ATP synthase activities. The conductivity of the thylakoid membrane to protons, gH+, is often used to determine the activity of chloroplast ATP synthase (Cruz et al., 2001), and we found that gH+ was significantly reduced to ∼30% of wild-type levels in bfa1 under various light conditions (95, 216, 339, 531, and 825 μmol photons m–2 s–1) (Figure 1C), confirming the reduced activity of the chloroplast ATP synthase in this mutant.
Figure 1.
Accumulation of Chloroplast ATP Synthase Is Reduced in bfa1.
(A) Growth and high-NPQ phenotypes of the bfa1 mutants. An image of 4-week-old plants is shown in the left panel. Dark-adapted plants were illuminated at 80 μmol photons m–2 s–1, and the images of NPQ were captured following illumination for 30 s or 180 s. bfa1-1 com and bfa1-2 com are bfa1 mutants complemented with the wild-type (WT) BFA1 genomic sequence. Bars = 2 cm.
(B) Analysis of NPQ induction upon illumination with actinic light (AL; 80 μmol photons m–2 s–1). The black trace indicates a typical trace of chlorophyll fluorescence in wild-type plants. NPQ levels (colored lines) were calculated during saturating light pulses (SP; 3000 μmol photons m−2 s−1 for 0.8 s, black arrows at top). ML, measuring light; F0, minimal chlorophyll fluorescence; Fm, maximum chlorophyll fluorescence. Means ± sd (n = 3 biological replicates from independent plants).
(C) Analysis of the H+ conductivity through ATP synthase (gH+). gH+ was measured according to the fast decay kinetics of the electrochromic shift signal recorded in detached leaves following illumination at various light intensities. Means ± sd (n = 3 biological replicates from independent plants).
(D) Immunoblot analysis of the thylakoid proteins. Thylakoid protein extracts from wild-type, bfa1-1, bfa1-2, and bfa3-1 plants were loaded on an equal chlorophyll basis.
Immunoblot analysis revealed that the steady state levels of the chloroplast ATP synthase subunits (CF1α-CF1ε, CFoI, and CFoII) in bfa1 were reduced to approximately one-eighth of the wild-type level, but the amounts of other protein complexes such as photosystem II (PSII) (D1, D2, and CP43), photosystem I (PSI) (PsaA and PsaD), and cytochrome b6f (Cyt b6f) (Cyt f and PetD) were not affected (Figure 1D). Blue native (BN) and subsequent SDS-urea-PAGE analyses were performed to demonstrate that the retained subunits in bfa1 can assemble into the intact ATP synthase and the CF1 subcomplex containing α, β, γ, and ε subunits but lacking δ (marked with CF1 Sub in Supplemental Figure 1). The existence of the thylakoid-bound CF1 subcomplex lacking CF1δ was also reported earlier (Takabayashi et al., 2013). Consistent with the higher gH+ in bfa3 than in bfa1, bfa3 also produced more ATP synthase than bfa1 (Figure 1D). These results indicate that the reduced ATP synthase activity in bfa1 is due to its reduced amount of ATP synthase subunits.
The bfa1 seedlings were much smaller than those of the wild type following germination for 28 d under an irradiance of 50 µmol photons m–2 s–1 (Figure 1A). Like the phenotypes of bfa3 and other mutants defective in the accumulation of chloroplast ATP synthase (Rott et al., 2011; Zoschke et al., 2012; Rühle et al., 2014; Fristedt et al., 2015; Zhang et al., 2016), the electron transport rate (ETR) through PSII in bfa1 was lower than that of the wild type under light intensities higher than 100 µmol photons m–2 s–1 (Supplemental Figure 2A). Compared with the wild-type plants, bfa1 also had higher NPQ and 1-qL, which were also observed in bfa3-1 (Supplemental Figures 2B and 2C). The value of 1-qL reflects the reduction state of plastoquinone pool in thylakoids, and a high value of 1-qL indicates that the PSII acceptor side is more reduced in bfa1 compared with wild-type plants. The larger ∆pH also induced the higher donor-side limitation of PSI (Supplemental Figure 2D; Zhang et al., 2016). These photosynthetic properties can be explained by the fact that the decreased accumulation of chloroplast ATP synthase results in a higher accumulation of protons in the thylakoid lumen, leading to further restriction of the linear electron transport at the Cyt b6f complex via a mechanism termed photosynthetic control. This regulates photosynthetic electron transport in order to match the energy requirement for CO2 fixation in the chloroplast stroma (Foyer et al., 1990; Kanazawa and Kramer, 2002).
Gene Expression Defects in bfa1 Are Unlikely to Be Directly Responsible for Its Phenotype
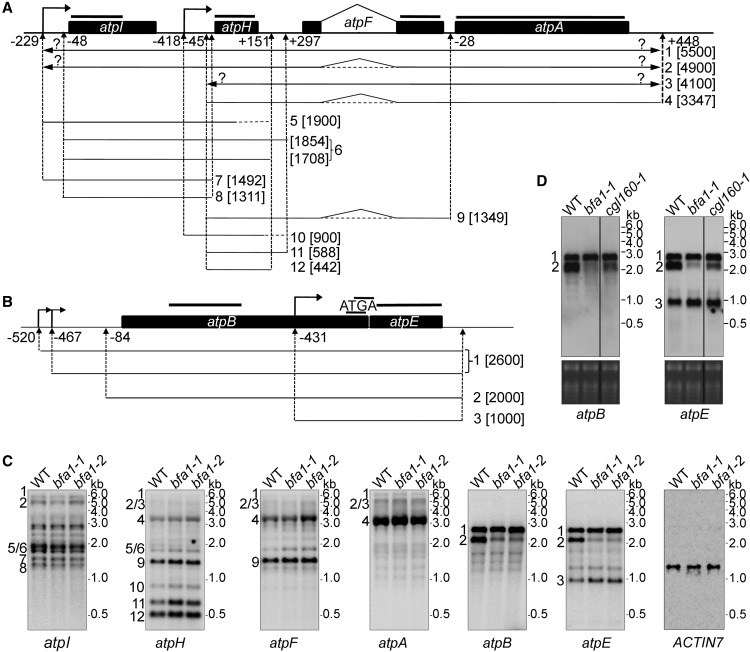
Because some subunits of chloroplast ATP synthase are encoded by the plastid genome, the reduced accumulation of ATP synthase in bfa1 may be caused by defects in the expression of plastid-encoded ATP synthase genes. To investigate this possibility, we performed RNA gel blot analyses and polysome association assays. For the large gene cluster atpI/H/F/A encoding the CFoIV, CFoIII, CFoI, and CF1α subunits, no obvious differences in expression patterns were observed between bfa1 and the wild type, except that the level of atpH and atpH/F transcripts was slightly higher in bfa1 than in the wild type (transcripts 9, 11, and 12, Figures 2A and 2C). For the small gene cluster atpB/E encoding the CF1β and CF1ε subunits, two major bands can be detected by the atpB probe in Arabidopsis (transcripts 1 and 2, Figures 2B and 2C; Schweer et al., 2006). The upper band corresponds to the primary dicistronic atpB/E transcripts that originate from the −520 and the −467 promoters. The lower band represents the processed dicistronic atpB/E transcript derived from the −520 and/or −467 promoter(s) and is generated by the activity of endo- and/or exonucleases that remove the 5′ region of the transcript (Supplemental Figure 3; Malik Ghulam et al., 2012). The monocistronic atpE transcript that starts at −431 from the atpE AUG can be detected by the atpE probe (transcript 3, Figure 2C; Schweer et al., 2006; Malik Ghulam et al., 2012).
Figure 2.
RNA Gel Blot Analysis of the Transcripts of Chloroplast-Encoded ATP Synthase Genes.
(A) and (B) Schematic representations of the large ATP synthase operon atpI/H/F/A (A) and the small ATP synthase operon atpB/E (B). Transcript maps were generated based on previously reported data (Pfalz et al., 2009; Malik Ghulam et al., 2012, 2013). Filled boxes represent coding regions of the ATP synthase genes. Hybridization probes used for the RNA gel blot analysis are indicated by black lines above the diagram. Question marks indicate remaining ambiguities. Transcription start sites are indicated with bent arrows. Upward arrows indicate the positions of 5′ or 3′ ends of the atp transcripts. The numbers adjacent to the upward arrows indicate the distance from the stop codon of upstream cistrons (+) or from the start codon of downstream cistrons (−). The numbers 1 to 12 in (A) and 1 to 3 in (B) represent the putative transcripts detected in Figure 2C. The size of each transcript is shown in the brackets.
(C) RNA gel blot analysis. Total leaf RNA was separated by denaturing agarose gel electrophoresis and transferred to nylon membranes. The RNA was probed with DIG-labeled DNA probes corresponding to the plastid atpA, atpF, atpH, atpI, atpB, and atpE transcripts. An ACTIN7 probe was used as a loading control. The positions of the RNA size markers (kb) are indicated on the right of the blots. The numbers to the left of the gels correspond to the corresponding transcripts illustrated above in (A) and (B).
(D) RNA gel blot analysis of atpB/E transcripts in the baf1-1 and cgl160-1 mutants. The RNA gel blot was prepared as in (C) and was then probed with DIG-labeled DNA probes corresponding to atpB and atpE transcripts. Ethidium bromide staining of the rRNAs before blotting, shown at the bottom of the blots, was used to verify equal loading. The numbers to the left of the gels correspond to the corresponding transcripts illustrated in (B).
Our results show that accumulation of the processed dicistronic atpB/E transcript was significantly reduced in bfa1, whereas the level of the primary dicistronic atpB/E transcripts was not affected and the monocistronic atpE transcript slightly overaccumulated in the bfa1 mutants (Figure 2C). Reduction of the processed dicistronic atpB/E transcript in bfa1 was in striking contrast to the bfa3 mutants in which the opposite pattern is observed (Zhang et al., 2016). We further investigated the level of processed dicistronic atpB/E transcript in the cgl160-1 mutant, in which assembly of the CFo subcomplex of chloroplast ATP synthase is impaired (Rühle et al., 2014; Fristedt et al., 2015). The accumulation of the processed dicistronic atpB/E transcript was also reduced in cgl160-1 (Figure 2D). In addition, we found that the ratio of the primary to the processed atpB/E transcripts in the ATP synthase mutant suppressor of variegation7 is altered in a similar way as in bfa1 and cgl160-1 (Zoschke et al., 2013). Taken together these results suggest that the level of processed dicistronic atpB/E transcript is highly sensitive to differences in accumulation of chloroplast ATP synthase subunits in various mutants affected in this complex. Moreover, our pulse-chase radiolabeling experiments showed that, even if synthesized at a reduced level in bfa1, CF1β cannot be properly assembled into ATP synthase (see below for more details). The reduction of processed dicistronic atpB/E transcript is thus unlikely to account for the low accumulation of chloroplast ATP synthase in the bfa1 and cgl160-1 mutants (Rühle et al., 2014; Fristedt et al., 2015).
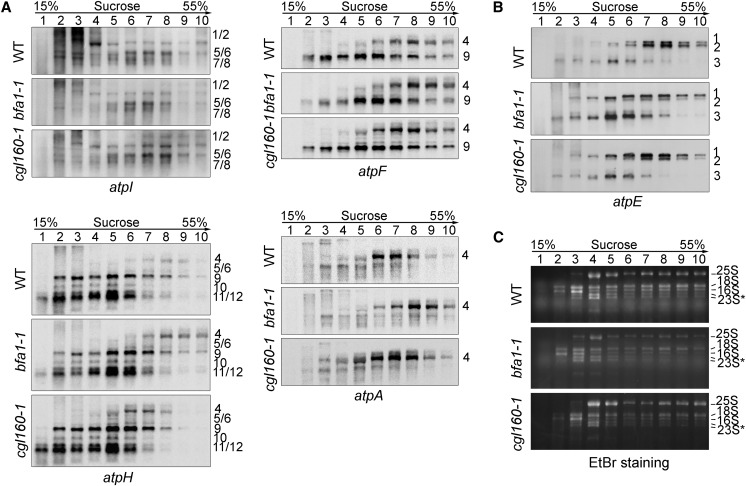
To investigate whether translation initiation of atp genes was affected in bfa1, we performed polysome association analyses (Figure 3). Our results show that a major shift to higher molecular weight fractions was detected for the large transcript atpH/F/A of bfa1-1, but not for the cgl160-1 transcript (transcript 4, Figure 3A). Moreover, a clear shift of primary dicistronic atpB/E transcript toward lower molecular weight fractions in the bfa1-1 mutant compared with the wild type was observed (transcript 1, Figure 3B). A similar polysome profile with this atpB/E transcript was also found in the cgl160-1 mutant (Figure 3B). Although the level of processed dicistronic atpB/E mRNA was reduced in bfa1, the polysome association pattern of this transcript was almost identical in bfa1-1, cgl160-1, and wild-type plants (transcript 2, main peak at fractions 7 to 9, Figure 3B). Similarly, the polysome profile of the monocistronic atpE transcript was not altered in the bfa1-1 and cgl160-1 mutants (transcript 3, main peak at 5 and 6 fractions, Figure 3B).
Figure 3.
Analysis of Polysome Association of the Plastid Transcripts Encoding ATP Synthase Subunits.
Total leaf extracts from wild-type, bfa1-1, and cgl160 plants were fractionated by sucrose density gradient centrifugation (15–55%) under native conditions. After centrifugation, the sucrose gradients were divided into 10 fractions of equal volume for RNA isolation and analysis, as noted above the gels. The RNAs were blotted and probed with DIG-labeled DNA probes corresponding to the plastid atpA, atpF, atpH, and atpI (A) as well as atpE (B) transcripts. rRNAs detected by ethidium bromide staining were used as fractionation and loading controls (C). 23S*, two breakdown products of the chloroplast 23S rRNA. Numbers to the right of the gel indicate sedimentation coefficients of the major RNAs.
Because CGL160 has been shown to be required for assembly of chloroplast ATP synthase, defects of atpB/E mRNA accumulation and polysome association in the cgl160-1 mutant are very likely secondary effects of the inefficient assembly of ATP synthase. These findings led us to investigate whether BFA1 is involved in the assembly of chloroplast ATP synthase, as CGL160.
Chloroplast ATP Synthase Assembly Is Impaired in bfa1
We used in vivo protein pulse labeling to test whether the synthesis and turnover rate of the chloroplast ATP synthase subunits was affected in bfa1. To reduce the complexity of protein labeling, cytosolic translation inhibitor cycloheximide was used to block the synthesis of nuclear-encoded proteins, and proteins encoded by the chloroplast genome were radiolabeled with [35S]Met (Figure 4). Almost no differences were observed in the synthesis of thylakoid proteins such as the PSI (PsaA/B) and PSII (CP47, CP43, D2, and pD1/D1) subunits in the thylakoid membrane of bfa1 and wild-type plants after labeling for 20 min. However, the rate of synthesis of CF1α was significantly higher in bfa1 compared with the wild type (Figure 4A), and this is consistent with the enhanced polysome association of the atpH/F/A transcript (transcript 4, Figure 3A). In contrast, the level of the protein band below CF1α in bfa1 was ∼50% lower in comparison with the wild type (Figure 4A). Immunoblot analysis showed that, besides the CF1β subunit, the large subunit of Rubisco (RbcL) was also present in this region (Figure 4B; Supplemental Figure 1), indicating that the band below CF1α contained both CF1β and RbcL. The RbcL contamination might have been due to the mild thylakoid isolation procedure used. Given that the levels of RbcL in bfa1 and the wild type are comparable (Figure 4B), reduction in the radiolabeled band corresponding to the CF1β and RbcL subunits in the bfa1 mutants indicated that the synthesis of CF1β in bfa1 was reduced compared with the wild type (Figure 4A), although we could not clearly determine to what extent. The lower rate of synthesis of CF1β may result from the reduced level of processed dicistronic atpB/E transcript as well as from the reduced association of the primary dicistronic atpB/E transcript with polysomes (Figures 2C and 3B).
Figure 4.
Assembly of the Chloroplast ATP Synthase Is Less Efficient in bfa1 Mutants.
(A) In vivo pulse labeling of the thylakoid proteins of the wild type (WT) and the bfa1 mutants. Primary leaves of 12-d-old plants were labeled with [35S]Met in the presence of cycloheximide, and the thylakoid proteins were isolated and separated using SDS-urea-PAGE. The gels were stained with Coomassie blue (left panel), and labeled thylakoid proteins were detected by autoradiography (right panel).
(B) Immunoblot analysis of the thylakoid proteins used in (A) probed with antibodies against CF1β and RbcL.
(C) Pulse and pulse-chase labeling of the thylakoid proteins in wild-type and bfa1 plants. After labeling for 20 min, the leaves were further incubated with unlabeled Met for the indicated durations (0 to 2 h). Labeled thylakoid proteins were detected by autoradiography.
(D) Analysis of the assembly of the thylakoid protein complexes. After pulse labeling for 20 min (indicated as 0 min) and a subsequent chase of 15 and 30 min, thylakoid proteins were separated by 2D BN/SDS-urea-PAGE and labeled proteins were detected using autoradiography. Arrows indicate intact CFo-CF1 ATP synthase, the CF1 subcomplex containing CF1α/β/γ/ε but lacking CF1δ (CF1 Sub), and the Rubisco complex (asterisks). The positions of CF1α and CF1β are indicated by horizontal arrows in the panels of wild-type samples.
Pulse-chase labeling was performed to investigate the fate of the newly synthesized proteins in the thylakoid membrane. Following a 20-min labeling period (indicated as 0 in Figures 4C and 4D), the proteins were chased with unlabeled Met for various durations. After a 2-h chase, the CF1α and CF1β/RbcL signals was unchanged in the wild-type thylakoids. However, the level of labeled CF1α and CF1β/RbcL was drastically reduced already after a 1-h chase in the bfa1 mutants (Figure 4C). Newly synthesized RbcL protein was efficiently assembled into Rubisco complex and stably accumulated in thylakoids (Figure 4D). Thus, reduction of the levels of CF1α and CF1β/RbcL signals during the chase indicates that the newly synthesized CF1α/β subunits in thylakoids are unstable in the absence of BFA1.
We separated labeled thylakoid proteins by 2D BN/SDS-PAGE in order to further analyze the kinetics of ATP synthase assembly. After 20 min of pulse labeling, the newly synthesized CF1α/β subunits were incorporated into the intact CF1-CFo ATP synthase and the CF1 Sub subcomplex (Figure 4D). As expected, the RbcL subunit was assembled into the Rubisco complex in both wild-type and bfa1 plants (Figure 4D). The level of newly assembled complexes containing CF1α/β subunits in the wild type progressively increased during the chase period (Figure 4D). However, in the bfa1 mutants, the signal of newly synthesized CF1α and CF1β was undetectable in the intact CF1-CFo ATP synthase, the CF1 subcomplex, and at the position of the free proteins throughout the pulse and pulse-chase period (Figure 4D). This indicated that assembly of the chloroplast ATP synthase is impaired in the bfa1 mutants. CF1α subunits were synthesized at a higher rate in bfa1 compared with the wild type, as shown in 1D/SDS-PAGE (Figures 4A and 4C, chase time 0), but they could not be detected in the 2D BN/SDS-PAGE (Figure 4D). One possible reason is that unassembled CF1α and CF1β are lost during the 12,000g centrifugation after solubilization of thylakoids with dodecyl-β-d-maltoside in our experimental procedure for BN-PAGE. This is consistent with the observation that unassembled CF1α and CF1β tend to aggregate in vivo. In wild-type plants, newly synthesized CF1α and CF1β are assembled progressively during the chase, resulting in a gradual increase of the signal intensities for assembled ATP synthase in the 2D BN/SDS-PAGE. However, assembled ATP synthase was not detected in bfa1 even after a 30-min chase (Figure 4D), indicating that assembly of the ATP synthase is impaired in bfa1.
In our previous report, we found that a substantial amount of the CF1 subcomplex is present in the chloroplast stroma (Zhang et al., 2016). This subcomplex, containing all five CF1 subunits (Zhang et al., 2016), either may have been detached from thylakoids during sample preparation or may represent a free complex in the chloroplast stroma. Its molecular identity and physiological function will need further investigation. To distinguish this complex from the CF1 subcomplex detected in the thylakoids, we designated this putative CF1 subcomplex in the chloroplast stroma CF1 SubII (Supplemental Figure 4). To investigate whether the assembly of this putative CF1 subcomplex was affected in bfa1 mutants, total soluble protein was separated using clear native (CN)-PAGE and 2D CN/SDS-PAGE after pulse-chase labeling in the presence of cycloheximide. After a 20-min pulse labeling of the wild type, the newly synthesized CF1α/β subunits were efficiently incorporated into CF1 SubII. During a subsequent 1-h chase, CF1 SubII was stable and gradually increased in abundance in the wild type (Supplemental Figure 4). In contrast, the level of newly assembled CF1 SubII in bfa1 was significantly lower than in the wild type after pulse labeling for 20 min and after the subsequent 1-h chase (Supplemental Figure 4). These results support a key role of BFA1 in the assembly of the CF1 subcomplex of chloroplast ATP synthase.
The rate of CF1β synthesis in the bfa1 mutants was reduced compared with the rate in wild-type plants (Figure 4A), and the newly synthesized CF1β subunit was not efficiently assembled into either the intact ATP synthase or into the CF1 subcomplex (Figure 4D; Supplemental Figure 4). Instead, the synthesized CF1β subunit was degraded quickly in the thylakoids (Figure 4C). These observations imply that the low accumulation of chloroplast ATP synthase in bfa1 results from defects in the assembly of its CF1 module rather than from the lower accumulation of processed atpB/E transcripts and the altered polysome profile of the primary atpB/E transcript that is observed in both bfa1 and cgl160 mutants (Figures 2 and 3). Thus, it is very likely that the absence of BFA1 affects the assembly and accumulation of the ATP synthase CF1 subcomplex in chloroplasts. However, as described earlier (Schöttler et al., 2015), chloroplast ATP synthase is highly stable under standard growth conditions. Thus, we could not determine the turnover (or degradation) rate of newly assembled chloroplast ATP synthase in our short time pulse chase labeling experiments (20-min pulse and 30-min chase) aimed at following the kinetics of assembly of the ATP synthase complex. It is unclear whether the stability of ATP synthase is affected in the absence of BFA1.
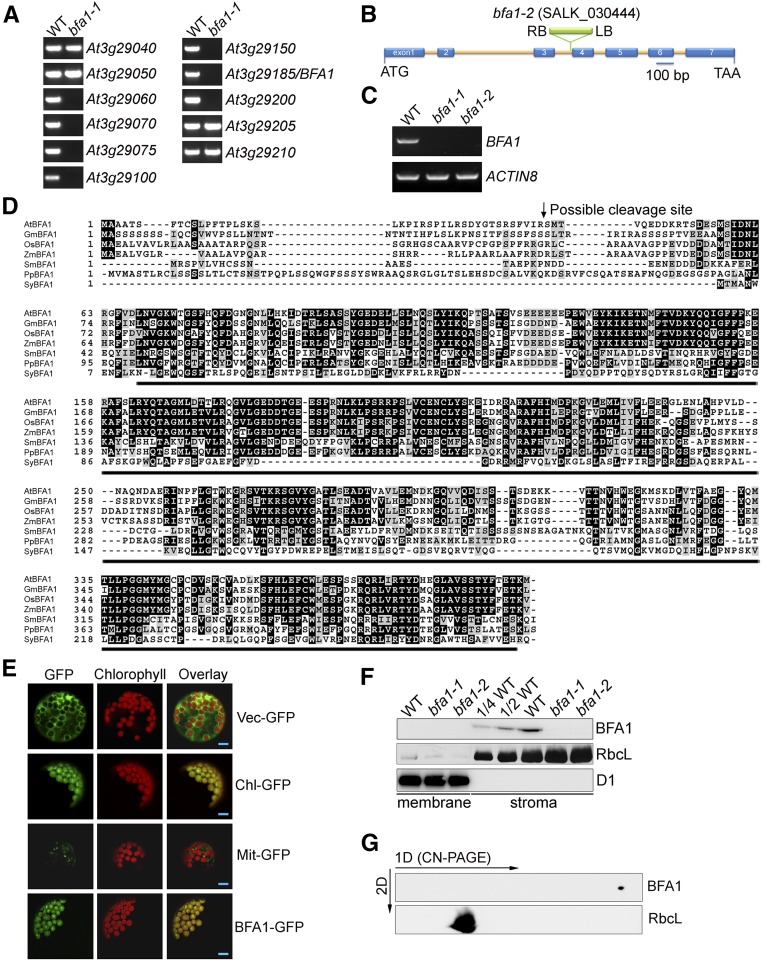
BFA1 Is a Chloroplast Stromal Protein with Unknown Function
Map-based cloning was performed to identify the mutation in bfa1-1, which was obtained from a large collection of T-DNA insertion mutants (Zhang et al., 2016). A large fragment of ∼120 kb on chromosome 3 (from At3g29060 to At3g29200) appears to be absent in bfa1-1 (Figure 5A; Supplemental Table 1). Among the 27 genes, only three genes encode proteins predicted to be in the chloroplast (Supplemental Table 1). We selected the At3g29185 gene for further analysis because its product has no known functional motif. We obtained the knockout allele bfa1-2, which carries a T-DNA insertion in the third intron of At3g29185 (Figure 5B). As expected, neither bfa1-1 nor bfa1-2 produced At3g29185 transcripts (Figure 5C) and the mutants have similar phenotypes (Figure 1). Moreover, addition of the wild-type At3g29185 gene into the bfa1-1 and bfa1-2 mutants fully complemented their high-NPQ phenotypes (Figure 1A), confirming that the absence of At3g29185 in bfa1 is responsible for the impaired assembly of the CF1 subcomplex in chloroplasts.
Figure 5.
Characterization of the bfa1 Mutants and BFA1.
(A) PCR amplification of the indicated genes in the bfa1-1 mutant. The results suggest that a large fragment from At3g29060 to At3g29200 is absent in bfa1-1.
(B) Structure of BFA1 and the position of the T-DNA insertion in the bfa1-2 mutant. The seven exons are indicated by blue rectangles and the introns are indicated by lines.
(C) RT-PCR analysis of the BFA1 transcripts. Arabidopsis ACTIN8 was used as the reference RNA.
(D) Sequence alignment of BFA1 and several homologs. AtBFA1 (AT3G29185, Arabidopsis), GmBFA1 (XP_003554683.1, Glycine max), OsBFA1 (XP_015630963.1, Oryza sativa), ZmBFA1 (NP_001143673.1, Zea mays), SmBFA1 (XP_002966592.1, Selaginella moellendorffii), PpBFA1 (XP_024367774.1, Physcomitrella patens), and SyBFA1 (WP_010874065.1, Synechocystis sp PCC 6803). Protein sequences above the black lines represent a putative DUF3598 domain. The arrow shows the possible cleavage site of the chloroplast transit peptide in Arabidopsis BFA1.
(E) Subcellular localization of BFA1. Arabidopsis protoplasts were transformed with constructs expressing GFP (Vec-GFP), RbcS-GFP (Chl-GFP, chloroplast localization control showing the signal of the Rubisco small subunit), FRO1-GFP (Mit-GFP, mitochondrial localization control showing the signal of FROSTBITE1), and BFA1-GFP. The GFP and chlorophyll signals were imaged using confocal microscopy. Bars = 10 μm.
(F) Immunodetection of BFA1 in chloroplasts. Intact chloroplasts isolated from the wild type and bfa1 mutants were separated into membrane (left three lanes) and stromal (right five lanes) fractions, and their proteins were immunoblotted with antibodies against BFA1, RbcL (stromal marker), and D1 (thylakoid marker).
(G) CN-PAGE and immunoblot analysis of BFA1. Chloroplast stromal proteins isolated from the wild type were separated using CN-PAGE and subsequent 2D SDS-PAGE. Stromal proteins were transferred onto a nitrocellulose membrane and immunodetected using antibodies against BFA1 and RbcL.
BFA1 encodes a 396-amino acid protein with a domain of unknown function known as DUF3598 that is found in bacteria and eukaryotes (Lakshmi et al., 2015). A BLAST search revealed that orthologs of BFA1 are found in land plants and cyanobacteria, but no homolog was found in the green alga Chlamydomonas reinhardtii (Figure 5D). To determine the subcellular localization of BFA1, we transiently expressed a BFA1-GFP fusion protein in Arabidopsis protoplasts and used confocal microscopy to observe that the GFP signal was present in chloroplasts (Figure 5E). Moreover, immunoblot analysis showed that BFA1 was present in the stromal fraction of wild-type plants, but that the BFA1 signal was absent from the bfa1 mutants as well as from the membrane fraction of wild-type plants (Figure 5F). These results demonstrate that BFA1 is a chloroplast stromal protein.
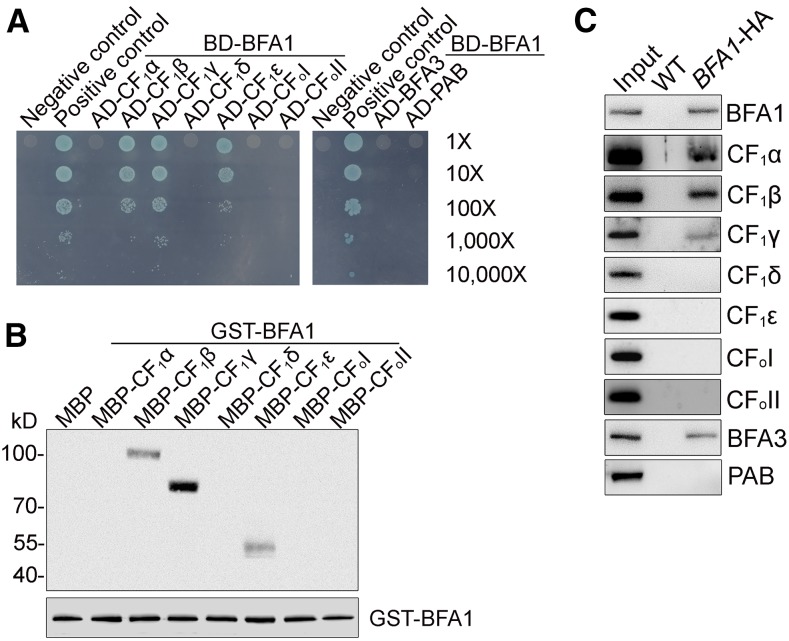
BFA1 Interacts with Several Chloroplast ATP Synthase CF1 Subunits
A 2D CN/SDS-PAGE and subsequent immunoblot analysis of stromal protein complexes showed that BFA1 is present in a free form and does not form a stable complex with other protein/complexes (Figure 5G). Yeast two-hybrid assays were performed to verify whether BFA1 promotes CF1 assembly by transiently and directly interacting with one or more of its subunits. As shown in Figure 6A, the mature BFA1 protein was found to interact with CF1β, CF1γ, and CF1ε, but not with CF1α and CF1δ. Moreover, no interaction was detected between BFA1 and the extramembranous parts of CFoI or CFoII, the two CFo subunits that interact with the α3β3 hexamer (Figure 6A). In addition, no interaction between BFA1 and two ATP synthase assembly factors, BFA3 and PAB, was detected with this assay (Figure 6A). In vitro pull-down assays further confirmed the interactions between BFA1 and CF1β, CF1γ, or CF1ε (Figure 6B). We also performed affinity chromatography using bfa1/BFA1-HA (hemagglutinin) plants, which fully complemented the phenotype of the bfa1-1 mutant. To copurify CF1 subunits with BFA1 from intact chloroplasts, chemical cross-linking with DSP (dithiobis [succinimidyl propionate]) was required, consistent with the detection of BFA1 as a free protein in the chloroplast stroma by CN-PAGE (Figure 5G). In addition to CF1β and CF1γ, CF1α and another assembly factor, BFA3, were also copurified with BFA1 following the DSP cross-linking and affinity purification using the HA matrix (Figure 6C). In contrast, chloroplast ATP synthase assembly factor PAB could not be copurified with BFA1 (Figure 6C), suggesting that PAB is involved in a different process from BFA1 and BFA3. Surprisingly, CF1ε, which was shown to interact with BFA1 in the yeast two-hybrid and pull-down assays, could not be purified with BFA1-HA (Figure 6C). Our failure to copurify CF1ε with BFA1 might be related to the different conformations of CF1ε or to shielding of CF1ε in the putative BFA1-CF1β/γ/ε complex.
Figure 6.
BFA1 Interacts with Several CF1 Subunits.
(A) Yeast two-hybrid assays using BFA1. The mature BFA1 protein was fused to the GAL4 DNA binding domain (BD). The five CF1 subunits (CF1α to CF1ε), the extramembranous parts of CFoI and CFoII, as well as the mature proteins BFA3 and PAB were fused to the GAL4 activation domain (AD). Cotransformation of pGBKT7-53 with pGADT7-T and pGBKT7-Lam with pGADT7-T were used as positive and negative controls, respectively. Numbers at the right represent dilutions from 1× to 10,000×.
(B) Pull-down analysis of BFA1. GST-tagged BFA1 was used as bait, and the indicated maltose binding protein (MBP)-tagged ATP synthase subunits were used as prey. The bound proteins were immunodetected with antibodies against MBP (upper panel) and BFA1 (lower panel), respectively.
(C) Coimmunoprecipitation assay of BFA1. Intact chloroplasts isolated from the wild-type and bfa1/BFA1-HA plants were cross-linked with DSP and then solubilized with 1.2% Triton X-100 for 30 min. The bound proteins were immunodetected using antibodies directed against selected proteins, as noted to the right of the images.
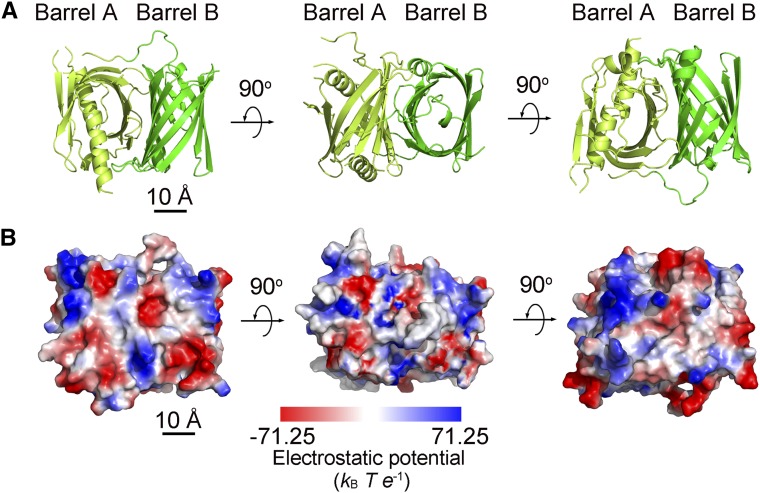
Crystal Structure of BFA1
To gain insights into the molecular mechanisms underlying the action of BFA1, we resolved its crystal structure at 2.8 Å (Figure 7; Supplemental Table 2 and Supplemental File 1). The mature BFA1 protein without the signal peptide (amino acids 42–396) was expressed in, and purified from, Escherichia coli. Size-exclusion chromatography analysis showed that the majority of BFA1 was present in a monomeric form during the purification (Supplemental Figure 5). A clear electron density in the structure of BFA1 allowed for the structural modeling of 344 residues (amino acids 53–396), although the first 11 N-terminal residues were missing from the model. The overall architecture of BFA1, with dimensions of 49 Å × 27 Å × 44 Å, consists primarily of two distinct β-barrels, designated A and B (Figure 7). While β-barrel A (Ser53–Val247) consists of nine β-sheets and two α-helices to form an atypical β-barrel, β-barrel B (Leu248–Met396) is composed of ten β-sheets and one α-helix and forms a typical β-barrel. The two β-barrels closely interact and are oriented at ∼90° relative to each other (Figure 7).
Figure 7.
Crystal Structure of BFA1.
(A) Overall structure of BFA1 in ribbon representation. The N-terminal β-barrel (Barrel A) is in light green, and the C-terminal β-barrel (Barrel B) is darker green. Bar = 10 Å.
(B) Electrostatic potential surface of BFA1 as viewed in (A). Positively and negatively charged patches are colored in blue and red, respectively. Three views are presented with a 90° rotation along the horizontal axis. The bar at the bottom shows the color scale of the electrostatic potential surface, which was generated by PyMOL software. Bar = 10 Å.
Identification of the Contact Regions between BFA1 and Its Partners
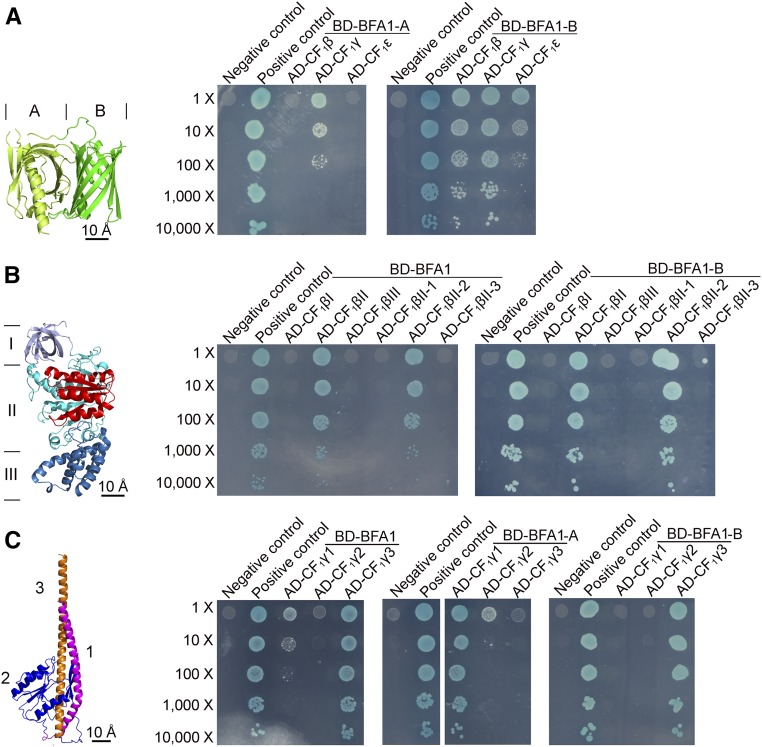
To investigate how BFA1 interacts with its partners, we attempted to determine the crystal structure of the BFA1-CF1β/γ/ε complex. We were able to purify the heterocomplexes of BFA1-CF1β, BFA1-CF1γ, and BFA1-CF1ε, but we were unable to crystallize these complexes. Therefore, we used yeast two-hybrid assays to determine the contact regions between BFA1 and its partners (Figure 8). We investigated which of the two BFA1 β-barrels is responsible for its interaction with CF1β, γ, and ε and found that β-barrel A interacted with CF1γ, whereas β-barrel B strongly interacts with CF1β, γ, and ε (Figure 8A).
Figure 8.
Determination of the Interaction Regions between BFA1 and CF1β/γ Subunits.
(A) Yeast two-hybrid assays of the two BFA1 barrels and the CF1β/γ/ε subunits. The BFA1 protein contains N- and C-terminal β-barrels, designated BFA1-A and BFA1-B, respectively (left). Each barrel was fused to the bait vector and tested for its interaction with the CF1β/γ/ε subunits as described for Figure 6A. Bar = 10 Å.
(B) Analysis of the BFA1 binding region in CF1β. CF1β was divided into three major domains (CF1βI, Met1–Asn96; CF1βII, Pro97–Pro377; CF1βIII, Arg378–Lys497) and the second domain was further separated into three subregions (CF1βII-1, Pro97–Gly165; CF1βII-2, Gly166–Ile275; CF1βII-3, Phe276–Pro377). All of the CF1β variants were fused to the prey vectors, while the mature BFA1 and the BFA1-B barrel were fused to the bait vectors. The interacting region in CF1β (CF1βII-2) is colored red (left). Bar = 10 Å.
(C) Analysis of the interaction region between CF1γ and BFA1. CF1γ was divided into three parts: the N-terminal helix (CF1γ1, Ala51–Val114), the central region (CF1γ2, Asp115–Pro297), and the C-terminal helix (CF1γ3, Val298–Val373), which are colored magenta, blue, and orange, respectively (left). Interactions between each of the CF1γ variants and intact BFA1, BFA1-A, and BFA1-B were analyzed. Bar = 10 Å.
Next, we divided the CF1 subunits into segments according to their crystal structures. The β subunit could be divided into three major domains (CF1βI–III), with CF1βII further divided into three subregions according to their positions relative to CF1α (CF1βII-1, -2, or -3 in Figure 8B) (Zhang et al., 2016). BFA1-B was shown to interact only with the CF1βII-2 subregion, which associates with the adjacent CF1α to form the catalytic site (Figure 8B). For the CF1γ subunit, both the N- and C-terminal ends comprised a long α-helix with a coiled-coil domain inserted into the α3β3 hexamer. We divided the γ subunit into three domains: the N-terminal helix (CF1γ1), the middle section (CF1γ2), and the C-terminal helix (CF1γ3) (Figure 8C). Yeast two-hybrid assays revealed that intact BFA1 interacts with the N- and C-terminal helices (CF1γ1 and CF1γ3), but not with the central domain, CF1γ2. Further analyses showed that BFA1-A and BFA1-B interact with CF1γ1 and CF1γ3, respectively (Figure 8C). The small subunit CF1ε can be divided into an N-terminal β-sandwich and a C-terminal helical hairpin domain (Uhlin et al., 1997). The C-terminal helical hairpin contains only 48 amino acids, and the removal of part or all of the C-terminal helical hairpin induced self-activation of the prey. We were therefore unable to determine the interaction region between BFA1 and CF1ε using yeast two-hybrid assays. In summary, our results demonstrated that the β-barrel B, but not A, of BFA1 interacted with the CF1βII-2 subregion, whereas BFA1 β-barrels B and A interacted with the N- and C-terminal helices of CF1γ, respectively.
Molecular Docking Analysis of BFA1 and Its Interacting Partners
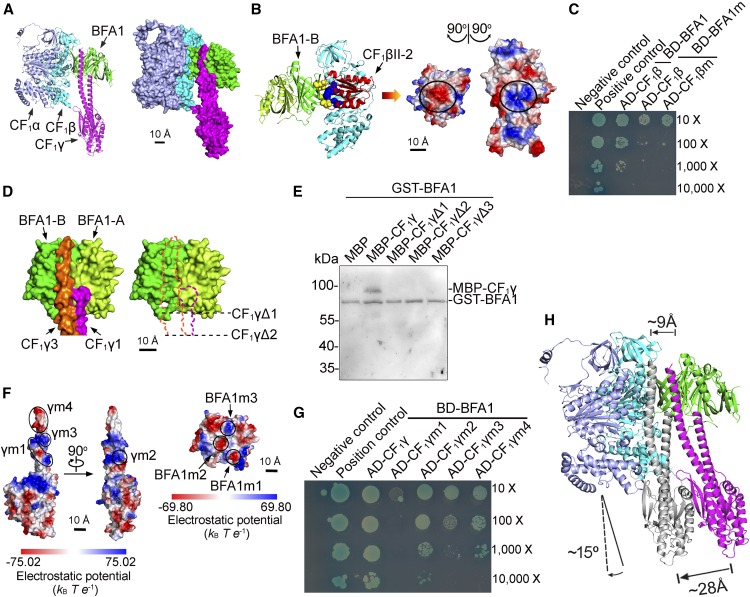
The availability of the BFA1 and CF1 subunit homolog crystal structures, as well as the identification of the regions of interaction between BFA1 and its CF1 partners, enabled us to investigate their interaction using in silico docking (Figure 9). The structure of the spinach (Spinacia oleracea) chloroplast ATP synthase, which is closely related to its Arabidopsis ortholog, has recently been resolved by cryo-electron microscopy at resolution of 2.9 Å (CF1) to 3.4 Å (CFo) (Hahn et al., 2018). We found that both the CF1β and CF1γ subunits could dock on the surface of BFA1 without steric conflicts, and their relative positions were similar to their final positions in the intact ATP synthase (Figure 9A). The region with the best fit for docking with the surface of the BFA1-B barrel was located in the CF1βII domain, which was oriented toward the inside of the α3β3 hexamer, close to the catalytic site, and adjacent to a CF1α subunit (Figure 9B). In the center of this contact region, the CF1β subunit had a domain enriched with positively charged amino acids, whereas the similarly sized corresponding region in BFA1 contained negatively charged residues (Figure 9B).
Figure 9.
Molecular Docking Analysis of the BFA1-CF1β/γ Interaction.
(A) Ribbon (left) and surface (right) representation of the CF1β/γ-BFA1 complex. BFA1, CF1α, CF1β, and CF1γ are shown in green, light blue, cyan, and magenta, respectively. Bar = 10 Å.
(B) Ribbon representation of the CF1β-BFA1 complex (left) and the electrostatic potential interface of CF1β and BFA1 (right). CF1βII-2 is shown in red, as in Figure 8B. Acidic residues (Glu-291, Asp-294, Asp-301, and Glu-318) in BFA1 and basic residues (Lys-178, Arg-205, Arg-207, and Arg-277) in CF1β are shown as yellow and blue spheres, respectively. The circles indicate the interacting patches. Bar = 10 Å.
(C) Yeast two-hybrid assay of the interaction between mutated CF1β and BFA1. BFA1m, BFA1 protein in which the acidic amino acids (Glu-291, Asp-294, Asp-301, and Glu-318) were changed to Lys. CF1βm, CF1β subunit in which the basic amino acids (Lys-178, Arg-205, Arg-207, and Arg-277) were changed to Glu.
(D) Surface representation of the CF1γ-BFA1 complex. The N-terminal β-barrel of BFA1 (Barrel A) is colored light green, and the C-terminal β-barrel (Barrel B) is a darker green. The N-terminal (CF1γ1) and C-terminal helices (CF1γ3) of CF1γ are in magenta and orange, respectively. The black lines show the position of the deletion of the N- and C-terminal regions in the CF1γ subunit (CF1γΔ1 and CF1γΔ2), which were used in the pull-down assay in (E). Bar = 10 Å.
(E) In vitro pull-down assay of the truncated CF1γ subunit and BFA1. CF1γΔ1, CF1γΔ2, and CF1γΔ3 contain amino acids 69 to 342, 77 to 334, and 91 to 315 of CF1γ, respectively.
(F) Electrostatic potential surface of CF1γ and BFA1. Positively and negatively charged regions are colored blue and red, respectively. The color scale of the electrostatic potential surface is shown below. Basic amino acids in γm1 (Arg-54, Arg-57, Arg-59, Lys-64, and Lys-68), γm2 (Lys-343 and Lys-344), γm3 (Arg-352, Lys-353, Arg-354, and Lys-357), and BFA1m3 (Lys-272 and Lys-156) were changed to Glu. Acidic amino acids in γm4 (Glu-361, Glu-364, Ala-367, Gly-368, Ala-369, and Asn-370), BFA1m1 (Asp-213, Glu-236, and Glu-237), and BFA1m2 (Glu-157 and Glu-367) were changed to Lys. Bars = 10 Å.
(G) Yeast two-hybrid assay of the interaction between mutated CF1γ and BFA1. CF1γm1, CF1γm2, CF1γm3, and CF1γm4 are the mutated CF1γ subunits indicated in (F).
(H) The position of the CF1γ subunit in the CF1α/β/γ-BFA1 assembly intermediate (magenta) and in the intact CF1 sector (gray). According to this structure model, the CF1γ subunit will move ∼9 to 28 Å and rotate ∼15° during the subsequent assembly of other CF1α/β subunits to reach its final position in the intact CF1 subcomplex. The structure of CF1βDP (PDB ID 6FKF) was used as a basis for refinement in this model.
We mutated the charged amino acids in these corresponding BFA1 and CF1β sections to determine their contribution to the strength of the interaction using a yeast model. When the acidic amino acids (Glu-291, Asp-294, Asp-301, and Glu-318) in the BFA1 region were mutated to Lys (BFA1m), the growth rate of the yeast harboring BFA1m and CF1β was significantly lower than that of yeast cells transformed with the native BFA1 and CF1β protein (Figure 9C). However, the interaction was not regained when the basic amino acids (Lys-178, Arg-205, Arg-207, and Arg-277) in the CF1β interaction region were changed to Glu (CF1βm; Figure 9C). Instead, the growth rate of the yeast harboring BFA1m and CF1βm was further decreased. These results imply that steric constraints (i.e., the conformations of these charged clusters in BFA1 and CF1β) are also important for the interaction. Taken together, our results indicate that BFA1 interacts with CF1β via these two corresponding regions and that electrostatic interactions as well as steric constraints seem to be essential for their interaction.
Three different conformations of CF1α/β dimers can be found in the structure of chloroplast ATP synthase. Whereas two of the catalytic β subunits (βDP and βTP) contain Mg-ADP, the nucleotide binding domain of the third β subunit (βempty) is unoccupied with nucleotide (Hahn et al., 2018). To investigate whether the interaction between CF1β and BFA1 is dependent on nucleotide binding, we analyzed structural differences among three CF1β subunits with different conformations, concentrating on the interaction region of CF1β-BFA1. We found that the structure of βDP is similar to that of βTP (Supplemental Figure 6A). However, when the N terminus of βDP and βempty were lined up, the region of βDP from the last α-helices of CF1βII-2 to the C terminus rotates clockwise ∼30° compared with the structure of βempty (Supplemental Figure 6A), resulting in a slight structural change of the CF1β-BFA1 contact region of CF1β (Supplemental Figure 6B). Due to the limitation of the docking analyses, we could not determine which conformation of CF1β is more prone to interact with BFA1. To resolve this issue experimentally, we performed a pull-down analysis of BFA1 and CF1β in the absence or presence of ADP/ATP. The results show that addition of ADP and ATP in the reactions does not influence the copurification of BFA1 and CF1β (Supplemental Figure 6C). Thus, BFA1 and CF1β interaction is unlikely to depend on nucleotide binding.
Although the secondary structure of CF1α is very similar to that of CF1β (Supplemental Figures 6A and 6B), their sequences differ. The surface charge of CF1α in the corresponding CF1β-BFA1 contact region is also different from that of CF1β (Supplemental Figure 6B). It is possible that steric constraints as well as the surface charge determine why BFA1 interacts with CF1β but not with CF1α.
We also found that the surface of BFA1-B and BFA1-A formed a distinct groove facing the inside of the α3β3 hexamer (Figure 9D) and that the surface of the C-terminal helices of the CF1γ subunit fit this groove. The surface of the N-terminal helices of the CF1γ subunit fits into a small adjacent groove in the BFA1-A domain (Figure 9D). This interaction model is consistent with our yeast two-hybrid assays (Figure 8C), in which the N- and C-terminal helices of the CF1γ subunit were found to interact with BFA1-A and BFA1-B, respectively. To further support this model, we performed a yeast two-hybrid assay using a truncated CF1γ subunit in which the contacting regions for BFA1 binding in the N and C termini were removed (Figure 9D). However, strong self-activation of the prey (yeast transformed with the truncated CF1γ subunit, CF1γΔ1) prevented its use for the assay. We therefore used an in vitro pull-down assay. As expected, the truncated CF1γ subunits did not copurify with BFA1 (Figure 9E), confirming the interaction region between CF1γ and BFA1.
In our proposed CF1γ-BFA1 contact region, there are several positively and negatively charged patches on CF1γ and BFA1, respectively (Figure 9F). To determine whether these charged patches are essential for the interaction of CF1γ-BFA1, we mutated the charged amino acids in these CF1γ parts and then investigated their interaction with BFA1 using the yeast two-hybrid system. Whereas yeast harboring CF1γm1 and BFA1 cannot grow, the growth rate of yeast transformed with CF1γm3/m4 and BFA1 was lower than that of yeast transformed with wild-type BFA1 and CF1γ (Figure 9G). By contrast, mutation in the m2 region (CF1γm2), which localizes to the side facing the contact α-helix of CF1γ, slightly decreased the growth rate of yeast (Figure 9G). These results indicate that electrostatic interactions are important for the interaction between BFA1 and CF1γ. We also mutated amino acids in the charged patches of BFA1 (BAF1m1, BFA1m2, and BFA1m3, respectively). However, yeast transformed with mutated BFA1 exhibited self-activation to various extents and we were therefore unable to determine the contribution of charged patches in the interaction of BFA1-CF1γ (Supplemental Figure 7).
The CF1ε subunit is a small polypeptide that binds to the central domain of CF1γ and plays a regulatory role in an ATP-dependent manner in E. coli (Yagi et al., 2007). Two different states of ε (contracted and extended) were found in the E. coli F1 complex, and only the former was detected in chloroplast ATP synthase (Hahn et al., 2018). Our docking analysis showed that, when in an extended state, the C-terminal helical hairpin domain of F1ε barely reached the bottom of BFA1 in our proposed BFA1-CF1β/γ complex (Supplemental Figure 8A). Therefore, the putative contact region was very small, making the docking analysis difficult to perform. By contrast, when CF1ε is in a contracted state, it associates with the CF1γ central domain and is far from the BFA1 protein in our proposed BFA1-CF1β/γ complex (Supplemental Figure 8B). It is currently unclear how BFA1 participates in assembling CF1ε into the intact chloroplast ATP synthase. According to our proposed BFA1-CF1β/γ model, it is likely that the CF1ε subunit, together with the CF1γ subunit, is incorporated into the CF1 sector in an extended state.
DISCUSSION
Our results show that the loss of BFA1 significantly affects the assembly of CF1. In the absence of BFA1, newly synthesized CF1α and CF1β subunits are rapidly degraded instead of being assembled into intact chloroplast ATP synthase in the thylakoids (Figures 4C and 4D). Pulse-labeling analyses of stromal complexes further narrowed down the function of BFA1 to the assembly of the CF1 module (Supplemental Figure 4). Although the level of processed atpB/E transcript was significantly reduced and polysome association with primary atpB/E mRNA was slightly affected in bfa1 (Figures 2 and 3), this is unlikely to be the primary cause of the observed phenotype. Similar properties were also observed for the cgl160 mutant, in which assembly of the CFo subcomplex is impaired (Figures 2D and 3). In contrast, the bfa3 mutant accumulates higher levels of processed atpB/E transcript compared with wild-type plants (Zhang et al., 2016). These observations indicate that the level of processed dicistronic atpB/E transcript is affected when ATP synthase assembly is compromised. Broad defects in ATP synthase gene expression were also observed in other Arabidopsis and maize (Zea mays) ATP synthase mutants (Zoschke et al., 2012, 2013; Rühle et al., 2014; Fristedt et al., 2015; Grahl et al., 2016; Zhang et al., 2016). The concomitant effect on chloroplast RNA metabolism and gene expression, and in particular the lower accumulation of specific transcripts, is probably an indirect consequence of perturbed assembly steps of chloroplast ATP synthase. However, we cannot fully rule out an additional role of BFA1 in gene expression besides its major function as an assembly factor.
BFA1 is conserved in land plants and cyanobacteria but is absent from the Chlorophyta (Figure 5D), suggesting that BFA1 represents an ancient ATP synthase assembly factor present in photosynthetic prokaryotes. As evolution progressed, BFA1 appears to have been retained in land plants but lost from green algae. Chlorophytes may have acquired a different protein with a structure similar to that of BFA1. In contrast, two other CF1 assembly factors, BFA3 and PAB, are conserved in land plants and chlorophytes but absent from cyanobacteria (Mao et al., 2015; Grahl et al., 2016; Zhang et al., 2016), indicating that BFA3 and PAB were acquired by photosynthetic eukaryotes during evolution to optimize the assembly of CF1. Because ATP synthase is conserved among the phototrophs, the acquisition of these assembly factors in land plants may have facilitated their adaptation to the chloroplast environment or to specific amino acid changes in the ATP synthase subunits (Zhang et al., 2016).
BFA1 consists mainly of two interacting β-barrels, A and B, which are oriented at roughly 90° relative to each other. A typical β-barrel contains a specific number of β-sheets arranged in an antiparallel fashion to form a closed structure with a hydrophobic core. The β-barrel is commonly embedded in the membrane to mediate the transmembrane transport of ions, small molecules, and proteins and is present in porins and translocases (Jap and Walian, 1996; Schleiff and Soll, 2005). Besides the β-barrels, two α-helices and one helix domain are present near the ends of the A and B β-barrels in BFA1, respectively (Figure 7). These helices likely block the entrance of the β-barrel cores in BFA1, excluding the possibility that this protein participates in the transport of small molecules unless these helices are not blocked under specific physiological conditions or modified by some other unknown factors.
Using a combination of biochemical analyses with structural approaches, we identified contact sites between BFA1 and the CF1β and CF1γ subunits (Figures 6 to 9). In particular, a negatively charged sequence in the center of the BFA1-B barrel was found to correspond to a positively charged region on the CF1β subunit, indicating electrostatic interactions between BFA1 and CF1β (Figure 9B). The same analysis also revealed corresponding sequences between the BFA1 surface inside the α3β3 hexamer and the surface of the coiled-coil domain formed by the N and C termini of the CF1γ subunit (Figure 9D). It is therefore clear that BFA1 interacts closely with some of the CF1 subunits, suggesting that it might act as a chaperone.
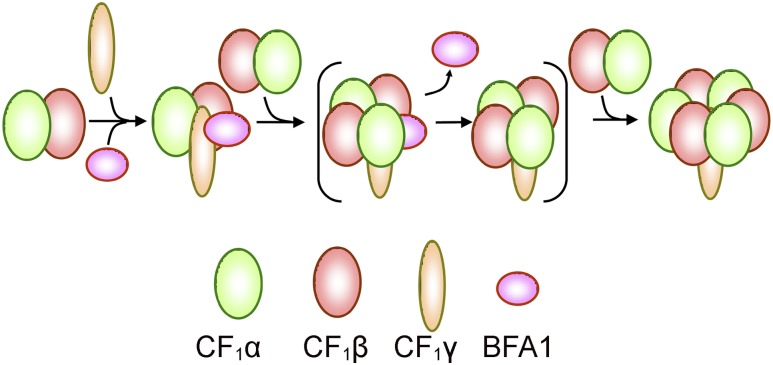
A particularly interesting finding is that, in the BFA1-CF1β/γ model, the CF1γ subunit does not occupy its final position in the mature CF1 complex. Rather, it has to shift from its position in the precomplex to the center of the α3β3 hexamer in the mature complex, requiring an 9 to 28 Å shift and a rotation of ∼15° (Figure 9H). We postulate that, upon binding of the CF1α/β heterodimers around CF1γ, CF1γ undergoes a conformational change and/or a position change, which in turn destabilizes the interaction between BFA1 and CF1β/γ and triggers the release of BFA1 from the assembly intermediate (Figure 10).
Figure 10.
Putative Model of BFA1-Assisted Assembly of the Chloroplast ATP Synthase CF1 Subcomplex.
After binding to the CF1α/β heterodimer, the BFA1 protein further associates with the CF1γ subunit. It is also possible that BFA1 acts at an earlier step by interacting first with a single β subunit followed by the association of subunits α and γ. In the next assembly step, another CF1α/β heterodimer is added into the intermediate complex around the CF1γ subunit. The association of the second CF1α/β pushes the CF1γ into the appropriate position and simultaneously triggers the release of BFA1. In the final step, the last CF1α/β heterodimer replaces BFA1 to form the intact CF1 subcomplex. The mechanism of assembly of the ε/δ subunit is unclear and is not described in this model, although yeast two-hybrid and pull-down assays demonstrate that BFA1 interacts with CF1ε (Figures 6A and 6B), implying that BFA1 may also be involved in the assembly of CF1ε into the α3β3γ complex.
Another assembly factor, BFA3, is required for the assembly of the CF1 subcomplex of the chloroplast ATP synthase, via its interaction with the CF1β subunit (Grahl et al., 2016; Zhang et al., 2016). Although our yeast two-hybrid assay did not detect their direct interaction, BFA3 can be copurified with BFA1 after DSP cross-linking (Figure 6C), implying that they might promote CF1 assembly in a cooperative manner. Our results show that both BFA1 and BFA3 interact with CF1β at the catalytic site adjacent to CF1α (Figure 8B; Zhang et al., 2016). In the BFA1-CF1β/γ complex, BFA1 interacts with CF1β in a region mainly oriented toward the inside of the α3β3 hexamer (Figure 9B). In a previous study, we mapped the critical residues of CF1β for BFA3 interaction and found that BFA3 associates with CF1β in the region oriented toward the outside of the α3β3 hexamer (Supplemental Figure 9; Zhang et al., 2016). Thus, BFA1 and BFA3 assembly factors bind to different sites of CF1β and thus can act simultaneously on the BFA1-CF1β/γ assembly intermediate and are probably involved in closely related processes. In contrast, another ATP synthase assembly factor, PAB, cannot be copurified with BFA1 and yeast two-hybrid assays did not detect any interaction between these two proteins (Figures 6A and 6C). These data suggest that PAB acts independently from BFA1/BFA3 in the assembly of chloroplast ATP synthase. It was postulated that PAB is an assembly chaperone that functions downstream of chaperonin 60 and assists in the integration of CF1γ into the CF1 subcomplex (Mao et al. (2015).
Two assembly models for the chloroplast ATP synthase CF1 subcomplex have previously been presented. Based on in vitro reconstitution studies (Gao et al., 1995), the first step of CF1 assembly following formation of the α/β heterodimer was proposed to be the formation of the α3β3 hexamer from three α/β heterodimers. After the insertion of the γ subunit into the α3β3 hexamer and the binding of the ε and δ subunits, the CF1 subcomplex was complete (Strotmann et al., 1998; Rühle and Leister, 2015). An alternative pathway is that one α/β heterodimer binds around the coiled-coil N/C terminus of the γ subunit. Afterwards two other α/β heterodimers are recruited to form the α3β3γ complex (Wollman et al., 1999; Choquet and Vallon, 2000; Hippler et al., 2002).
Our proposed BFA1-CF1β/γ assembly intermediate clearly supports the latter model for the assembly of the CF1 subcomplex. Although BFA1 does not directly interact with CF1α, this subunit can be copurified with BFA1 in vivo (Figure 6C), indicating that BFA1 interacts with the CF1α/β heterodimers. This is also consistent with early findings that CF1α/β heterodimer formation is a prerequisite for assembly of the CF1 sector (Wollman et al., 1999; Choquet and Vallon, 2000; Hippler et al., 2002; Rühle and Leister, 2015). Although we could not demonstrate the details of the following assembly steps, it is reasonable to propose that the binding of other CF1α/β heterodimers around CF1γ promotes the release of BFA1 and the formation of the α3β3γ complex (Figure 10). Because the F1 sector of ATP synthase is highly conserved among species, our proposed assembly model for α3β3γ may be also valid for other ATP synthases, which may involve additional species-specific assembly factors.
In summary, we have identified and characterized BFA1, a novel assembly factor of the chloroplast CF1 ATP synthase. We have determined the crystal structure of BFA1 and have identified the CF1 subunits and their domains that interact with BFA1. Based on these results plus a comparative analysis of the assembly of CF1 in wild-type and mutant plants lacking BFA1 and a molecular docking analysis, we propose a model for the assembly of CF1 in which BFA1 acts as a chaperone that orchestrates the early steps of the CF1 assembly pathway.
METHODS
Plant Materials and Growth Conditions
Arabidopsis thaliana plants were grown in soil in a glasshouse at 23°C, with a 16-h photoperiod and an irradiance of 50 μmol photons m–2 s–1. The bfa1-2 and cgl160-1 mutants were obtained from the European Arabidopsis Stock Centre (NASC), and bfa1-1 was identified in a screen of a T-DNA insertion mutant population using a Closed FluorCam FC 800-C (Photon System Instruments) (Zhang et al., 2016). To complement the bfa1 mutant, genomic DNA fragments of BFA1 were cloned into the pCAMBIA1301 vector (Hajdukiewicz et al., 1994). To produce bfa1/BFA1-HA plants, the cDNA of BFA1 was fused with the sequence encoding the HA tag and cloned into the pBI121 vector under the control of the cauliflower mosaic virus 35S promoter (Chen et al., 2003). The vectors were transferred into Agrobacterium tumefaciens C58C for transformation of Arabidopsis (Clough and Bent, 1998).
Analysis of Photosynthetic Properties
Chlorophyll fluorescence was measured using a MINI-PAM fluorometer (Walz). Chlorophyll fluorescence was monitored as described previously (Shikanai et al., 1999). The minimum fluorescence yield (Fo) of dark-adapted leaves was measured under the measuring light (650 nm, 0.1 μmol photons m−2 s−1). A saturating pulse of white light (3000 μmol photons m−2 s−1 for 0.8 s) was applied to determine the maximum fluorescence yield in the dark (Fm). Then, the leaves were illuminated with actinic light (80 μmol photons m−2 s−1) for 4 min. During illumination, the maximum chlorophyll fluorescence in the light (Fm′) was measured every 30 s by applying saturating pulses. Fv and NPQ were calculated as Fm – Fo and (Fm – Fm′)/Fm′, respectively. For determination of the light intensity-dependent photosynthetic parameters NPQ, ETR, and 1-qL, the steady state level of chlorophyll fluorescence (Fs) and Fm′ was recorded after AL illumination (20, 45, 90, 210, 300, 450, 720, and 1100 μmol photons m−2 s−1) for 2 min. ETR was calculated as Y(II) × PAR × 0.5 × 0.84. Y(II) was calculated as (Fm′ – Fs)/Fm’. 1-qL was calculated as 1 – {Y(II)/(1 –Y(II))} × [(1 – Fv/Fm)/(Fv/Fm)] × (NPQ + 1) (Miyake et al., 2009; Yamamoto et al., 2011). The oxidation state of the donor side of PSI was determined using a Dual-PAM-100 (Walz) and calculated automatically using the Dual-PAM-100 software (Zhang et al., 2016). The proton conductivity of the thylakoids (gH+) was measured at various light intensities using a Dual-PAM-100 equipped with a P515/535 emitter-detector module (Walz) (Schreiber and Klughammer, 2008). The fast phase (between 0 and 500 ms after a 10-min illumination) of the dark-interval relaxation kinetics measured at various light intensities (95, 216, 339, 531, and 825 μmol photons m−2 s−1) was fitted with a single exponential decay function. The reciprocal value of the halftime of the electrochromic shift decay represents thylakoid conductivity through chloroplast ATP synthase (gH+) and was used to estimate ATP synthase activity (Rott et al., 2011).
Isolation of Chloroplast Stromal Proteins and Thylakoid Membranes
Intact chloroplasts were isolated in isolation buffer (0.33 M sorbitol and 20 mM HEPES/KOH, pH 7.6) then osmotically ruptured in 20 mM HEPES/KOH (pH 7.6) (Munekage et al., 2002). Chloroplast stromal proteins were separated from the thylakoid membranes using centrifugation, as described previously (Zhang et al., 2016). The concentration of stromal proteins was determined using a Protein Assay Kit (Bio-Rad Laboratories). The chlorophyll contents of the thylakoid membrane were determined as described previously (Porra et al., 1989). For the immunoblot analysis, the proteins were separated on an SDS-PAGE gel containing 6 M urea and then transferred to nitrocellulose membranes (GE Healthcare). The membranes were incubated with various antibodies against the representative subunits of thylakoid protein complexes (PhytoAB), and the signal was captured with a LuminoGraph WSE-6100 (ATTO Technology). Rabbit CF1α (PHY0311), CF1β (PHY0312), CF1γ (PHY0313), CF1ε (PHY0314), CF1δ (PHY0315), CFoI (PHY0316), CFoII (PHY0170S), Cyt f (PHY0321), and PetD (PHY0354) antibodies were used at 1:1000 dilutions. Antibodies D1 (PHY0057), D2 (PHY0060), and PsaA (PHY0342) were used at 1:2000 dilutions. CP43 (PHY0318), PsaD (PHY0343), RbcL (PHY0346), and MBP (PHY0329) antibodies were used at 1:5000 dilutions.
RNA Gel Blot and Polysome Association Analysis
Total RNA was isolated from the leaves of 4-week-old Arabidopsis plants using the Trizol reagent (Thermo Fisher Scientific). A total of 5 μg (for blotting with atpA, atpB, atpE, and ACTIN7 probes) or 2.5 μg (for blotting with atpI, atpH, and atpF probes) RNA per genotype was separated using formamide denaturing agarose gel electrophoresis (Barkan, 1998) and then transferred onto a nylon membrane (GE Healthcare). An RNA gel blot analysis was performed using digoxigenin (DIG)-labeled nucleic acid probes (DIG Easy Hyb system; Roche). The polysome association assays were performed following the protocol described by Barkan (1988), with minor modifications. Briefly, the leaf total extracts were fractionated in 15 to 55% sucrose gradients by centrifugation at 45,000 rpm (246,000g) at 4°C for 65 min in a SW Ti55 rotor. After centrifugation, 10 fractions of 500 μL were collected from the sucrose gradients. The total RNA was purified from each fraction and an equal proportion of RNA was used for the RNA gel blot analysis. The rRNAs were detected by ethidium bromide staining, and the signals of the RNA gel blot were visualized with a LuminoGraph WSE-6100 (ATTO).
BFA1 Antibody Production
The cDNA sequence encoding the mature BFA1 protein (lacking the N-terminal 41 amino acid residues) was cloned into the pET-28a expression vector (Merck Millipore) and the recombinant protein was expressed in Escherichia coli BL21 (DE3) LysS strain (Novagen) treated with 1 mM IPTG. The protein was purified under denaturing conditions using Ni-NTA agarose (Qiagen) and used for immunoinjection into rabbits to produce polyclonal antibodies (PhytoAB). The BFA1 antibody was used at a 1:1000 dilution.
Expression, Purification, and Crystallization of BFA1
The cDNA sequence encoding the mature BFA1 protein (amino acid residues 42–396) was amplified by PCR using the primers BFA1-pET28-TEV-F and BFA1-pET28-R (Supplemental Table 3) and was then cloned into the pET-28a(+) expression vector (Novagen). The nucleotides underlined in the primer sequence encode the tobacco etch virus (TEV) protease-cleavable sequence (ENLYFQS), which is used to remove the N-terminal His-tag from the recombinant BFA1 proteins after purification. The expression of the recombinant BFA1 protein was induced in the E. coli BL21 (DE3) cells by treatment at 16°C for 14 to 16 h in 0.2 mM IPTG. The cells were harvested and disrupted by sonication in a lysis buffer containing 20 mM Tris-HCl (pH 7.5), 150 mM NaCl, and 20 mM imidazole. After centrifugation at 12,000g for 20 min, the clear lysate was incubated for 30 min with Ni-NTA agarose (Qiagen) at 4°C. The recombinant protein was eluted in an elution buffer containing 20 mM Tris-HCl (pH 7.5), 150 mM NaCl, 5 mM DTT, and 200 mM imidazole and treated with His-tagged TEV protease (prepared in our laboratory) overnight at 4°C. A second round of incubation with Ni-NTA agarose was performed to remove the N-terminal His-tag, the His-tagged TEV protease, and any noncleaved His-tagged BFA1 recombinant protein. The flow-through was concentrated to 4 mL and then subjected to size-exclusion chromatography using a HiLoad 16/600 Superdex 200 pg column (GE Healthcare). The fractions corresponding to the BFA1 monomer were collected and concentrated by ultrafiltration to a concentration of 10 mg mL–1 in a buffer containing 20 mM Tris-HCl (pH 7.5), 150 mM NaCl, and 5 mM DTT.
For protein crystallization, both native and selenomethionine-substituted crystals of BFA1 were grown by the hanging-drop vapor diffusion method (McPherson, 1990) at 16°C in a mixture containing 1 to 2 μL of the protein sample and an equal volume of reservoir solution (0.1 M Bis-Tris, pH 6.3, 0.3 M MgCl2, and 28% [w/v] PEG3350). Before data collection, the crystals were harvested with nylon loops, cryo-protected in the crystallization solution supplemented with 30% (v/v) PEG3350, and shock-frozen in liquid nitrogen.
Data Collection and Structure Determination
X-ray diffraction data were collected at the Shanghai Synchrotron Radiation Facility beamline BL17U and processed using HKL-2000 software (HKL Research). The selenium positions were determined using the SHELXD program (Schneider and Sheldrick, 2002), and a preliminary model was built manually in Coot (Emsley and Cowtan, 2004). Further refinement was performed with phenix.refine and AutoBuild in PHENIX (Adams et al., 2010), and the model was manually adjusted in Coot according to the 2Fo-Fc and Fo-Fc electron density maps. The quality of the structure was assessed using PROCHECK (Laskowski et al., 1993), and the structure figures were created in PyMOL (Schrödinger). Data collection and structure refinement statistics are summarized in Supplemental Table 2.
Molecular Docking Analysis
The in silico docking of BFA1 to the β, γ, and ε subunits was performed using Coot (Emsley and Cowtan, 2004). For the molecular docking analyses, CF1α/β/γ/ε subcomplex structures from spinach (Spinacia oleracea; 6FKF; Hahn et al., 2018) and the structure of extended F1ε subunit from E. coli (5T4O; Supplemental Figure 8A; Sobti et al., 2016) were obtained from the Protein Data Bank (PDB). Three-dimensional models of the Arabidopsis CF1β/γ subunits (Figure 8) were built using the alignment mode in the SWISS-MODEL software (Arnold et al., 2006), and the 6FKF structure was used as a template (Hahn et al., 2018). The electrostatic potential at the CF1α/β/γ and BFA1 surface was calculated using PyMOL (Schrödinger).
In Vivo Labeling and Chasing of Chloroplast Proteins
Chloroplast protein labeling and chasing, subsequent separation using BN-PAGE and CN-PAGE, and the detection of the labeled proteins were performed as described by Zhang et al. (2016). Seeds were sterilized and sown on Murashige and Skoog medium containing 3% sucrose for 12 d. Primary leaves were detached and incubated in 45 μL of a buffer containing 1 mM KH2PO4 (pH 6.3), 0.1% (w/v) Tween 20, and 20 mg/mL cycloheximide for 30 min to block the synthesis of nuclear-encoded proteins. Then, 5 μL of [35S]Met (10.25 mCi/mL, NEG709A500UC; PerkinElmer) was added and incubated for 20 min under light of 80 μmol photons m–2 s–1. After labeling, the leaves were washed twice for protein isolation or further chased with a buffer containing 1 mM KH2PO4 (pH 6.3), 0.1% (w/v) Tween 20, 1 mM unlabeled Met, and 20 mg/mL cycloheximide for various times as indicated in the text. For membrane and soluble protein isolation following the in vivo labeling and chase, the labeled leaves were ground with a cold rod on ice in a buffer containing 20 mM HEPES/KOH (pH 7.6). Membrane proteins were pelleted by centrifugation at 10,000g for 5 min at 4°C. Soluble proteins were subjected to two additional rounds of centrifugation at 15,000g for 10 min at 4°C to remove trace amounts of membrane contaminants (Zhang et al., 2016).
Yeast Two-Hybrid Assays and Affinity Chromatography
Yeast two-hybrid assays were performed according to the Matchmaker Gold Yeast Two-hybrid System user manual (Clontech Laboratories). Cross-linking and affinity chromatography assays were performed as previously described (Zhang et al., 2016). Briefly, intact chloroplasts isolated from 4-week-old plants were suspended to a concentration of 0.5 mg chlorophyll/mL in a buffer containing 0.33 M sorbitol and 20 mM HEPES-KOH (pH 7.6). The cross-linker DSP was then added to a final concentration of 2.5 mM. After cross-linking for 30 min on ice, Tris-HCl (pH7.5) was added to stop the reaction. Chloroplasts were washed and then solubilized for 30 min at 4°C in a buffer containing 20 mM HEPES-KOH (pH 8.0), 1.2% Triton X-100, 200 mM NaCl, and 1 mM phenylmethylsulfonyl fluoride. Solubilized proteins were separated by centrifugation at 15,000g for 5 min at 4°C and mixed with Anti-HA Affinity Matrix (Roche) for 2 h at 4°C. The bound proteins were purified and immunodetected with antibodies as described above. Antibody against PAB was obtained from Lixin Zhang (Institute of Botany, Chinese Academy of Sciences) and used at a 1:1000 dilution.
Pull-Down Analysis
For the pull-down assays, the cDNA sequences encoding the mature BFA1 protein and ATP synthase subunits were subcloned into the pGEX-6P-1 (GE Healthcare) and the pMAL-c5x (New England Biolabs) expression vector, respectively, as described previously (Zhang et al., 2016). The fusion proteins were expressed in E. coli BL21 (DE3) LysS strain (Invitrogen) using 0.3 mM IPTG at 16°C overnight. GST-BFA1 and MBP fusion proteins were purified using glutathione-agarose 4B beads (GE Healthcare) and amylose resin (New England Biolabs), respectively. In the subsequent procedures, 5 µg of GST-BFA1 and MBP fusion proteins were mixed in a buffer containing 50 mM Tris-HCl (pH 7.5), 0.15 M NaCl, 1% Triton X-100, 5% glycerol, 1 mM DTT, and complete protease inhibitor cocktail (Roche) for 1 h at 4°C. Then, 25 μL of glutathione-agarose beads was added to purify the GST-BFA1 and its associated proteins. The bound proteins were immunodetected with antibodies against MBP and BFA1 (1:1000 dilutions), respectively (Zhang et al., 2016)
Subcellular Localization of GFP Proteins and Circular RT-PCR
For subcellular localization of BFA1-GFP, full length of BFA1 was amplified by RT-PCR with primers BFA1-GFP-F and BFA1-GFP-R (Supplemental Table 3) and then subcloned into the pBI221 vector (Zhong et al., 2013). The constructs expressing FRO1-GFP and RbcS-GFP, which are mitochondrial and chloroplast localization controls, respectively, were constructed as described previously (Cai et al., 2009). The constructs were introduced into Arabidopsis protoplasts via polyethylene glycol-mediated transformation (Mathur and Koncz, 1998), and GFP signals were observed by confocal scanning microscopy (LSM 510 Meta; Zeiss) as described by Zhang et al. (2016). For circular RT-PCR, total RNA was incubated with DNase I (Takara) and self-ligated for 2 h with T4 RNA ligase (New England Biolabs). After ligation, reverse transcription was performed using self-ligated RNA with M-MLV reverse transcriptase (Thermo). Then, 1/20th of the cDNA was used in a PCR amplification reaction and the DNA products were cloned into pMD-18T (TaKaRa) for sequencing.
Accession Numbers
Sequence data from this article can be found in the NCBI or Phytozome databases under the following accession numbers: At-BFA1 (AT3G29185, Arabidopsis), Gm-BFA1 (XP_003554683.1, Glycine max), Os-BFA1 (XP_015630963.1, Oryza sativa), Zm-BFA1 (NP_001143673.1, Zea mays), Sm-BFA1 (XP_002966592.1, Selaginella moellendorffii), Pp-BFA1 (XP_024367774.1, Physcomitrella patens), Sy-BFA1 (WP_010874065.1, Synechocystis sp PCC 6803), FRO1 (AT5G67590, Arabidopsis), RbcS (AT1G67090, Arabidopsis), ACTIN7 (AT5G09810, Arabidopsis), and ACTIN8 (AT1G49240, Arabidopsis). The coordinates and structure factors for At-BFA1 were deposited in the Protein Data Bank under accession code PDB ID 5YVF. Mutants used included bfa1-2 (SALK_030444), bfa3-1 (SALK_019326), and cgl160-1 (SALK_057229), which were obtained from the European Arabidopsis Stock Centre (NASC). The mutant pools of pSKI015 T-DNA insertion Arabidopsis lines (stock no. CS31400) used for screening bfa mutants were obtained from the ABRC.
Supplemental Data
Supplemental Figure 1. BN-PAGE and 2D SDS-urea-PAGE Analysis of the Thylakoid Protein Complexes in bfa1.
Supplemental Figure 2. Photosynthetic Properties of bfa1.
Supplemental Figure 3. Mapping of the Termini of the Primary atpB/E Transcripts.
Supplemental Figure 4. Analysis of the Assembly of the Putative CF1 Subcomplex (CF1 SubII) in Chloroplast Stroma.
Supplemental Figure 5. Elution Profile of BFA1 during Size-Exclusion Chromatography.
Supplemental Figure 6. Analysis of the Interaction between BFA1 and CF1α/β.
Supplemental Figure 7. Yeast Two-Hybrid Assay of the Interaction between Mutated BFA1 and CF1γ.
Supplemental Figure 8. Analysis of the Interaction between BFA1 and CF1ε.
Supplemental Figure 9. Prediction of the BFA3 Binding Site in the BFA1-CF1β/γ Model.
Supplemental Table 1. List of the Genes Deleted in the bfa1-1 Mutant.
Supplemental Table 2. Data Collection and Structure Refinement Statistics of Arabidopsis BFA1.
Supplemental Table 3. Primers Used in This Work.
Supplemental File 1. wwPDB X-ray Structure Validation Summary Report.
Dive Curated Terms
The following phenotypic, genotypic, and functional terms are of significance to the work described in this paper:
Acknowledgments
We thank Toshiharu Shikanai for the critical reading of the manuscript, Lixin Zhang for providing the PAB antibody, Hualing Mi for providing laboratory space, Cuimin Liu for the advice regarding the in silico docking analysis, and the NASC and ABRC for providing the mutant seeds. This work was supported by the National Natural Science Foundation of China Funds for Outstanding Youth (31322007), by the National Natural Science Foundation of China (31700202), by the Shanghai Engineering Research Center of Plant Germplasm Resources (17DZ2252700), by the China Postdoctoral Science Foundation (2017M621511), and by funds from Shanghai Normal University (SK201636).
AUTHOR CONTRIBUTIONS
L.Z. and L.P. conceived the study and designed the experiments. Z.D. and L.L. performed the crystallization experiments. H.P. and L.L. performed the in silico docking analysis. L.Z., Y.L., B.L., Q.Z., and W.L. performed all other experiments. All authors analyzed the data. L.Z., L.L., J.-D.R., and L.P. wrote the article. L.P. supervised the study.
References
- Ackerman S.H., Tzagoloff A. (2005). Function, structure, and biogenesis of mitochondrial ATP synthase. Prog. Nucleic Acid Res. Mol. Biol. 80: 95–133. [DOI] [PubMed] [Google Scholar]
- Adams P.D., et al. (2010). PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66: 213–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmad Z., Cox J.L. (2014). ATP synthase: the right size base model for nanomotors in nanomedicine. Sci. World J. 2014: 567398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold K., Bordoli L., Kopp J., Schwede T. (2006). The SWISS-MODEL workspace: a web-based environment for protein structure homology modelling. Bioinformatics 22: 195–201. [DOI] [PubMed] [Google Scholar]
- Barkan A. (1988). Proteins encoded by a complex chloroplast transcription unit are each translated from both monocistronic and polycistronic mRNAs. EMBO J. 7: 2637–2644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barkan A. (1998). Approaches to investigating nuclear genes that function in chloroplast biogenesis in land plants. Methods Enzymol. 297: 38–57. [Google Scholar]
- Cai W., Ji D., Peng L., Guo J., Ma J., Zou M., Lu C., Zhang L. (2009). LPA66 is required for editing psbF chloroplast transcripts in Arabidopsis. Plant Physiol. 150: 1260–1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen G.G., Jagendorf A.T. (1994). Chloroplast molecular chaperone-assisted refolding and reconstitution of an active multisubunit coupling factor CF1 core. Proc. Natl. Acad. Sci. USA 91: 11497–11501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen P.-Y., Wang C.-K., Soong S.-C., To K.-Y. (2003). Complete sequence of the binary vector pBI121 and its application in cloning T-DNA insertion from transgenic plants. Mol. Breed. 11: 287–293. [Google Scholar]
- Choquet Y., Vallon O. (2000). Synthesis, assembly and degradation of thylakoid membrane proteins. Biochimie 82: 615–634. [DOI] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. (1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743. [DOI] [PubMed] [Google Scholar]
- Cross R.L. (2000). The rotary binding change mechanism of ATP synthases. Biochim. Biophys. Acta 1458: 270–275. [DOI] [PubMed] [Google Scholar]
- Cruz J.A., Sacksteder C.A., Kanazawa A., Kramer D.M. (2001). Contribution of electric field (Delta psi) to steady-state transthylakoid proton motive force (pmf) in vitro and in vivo. control of pmf parsing into Delta psi and Delta pH by ionic strength. Biochemistry 40: 1226–1237. [DOI] [PubMed] [Google Scholar]
- Emsley P., Cowtan K. (2004). Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60: 2126–2132. [DOI] [PubMed] [Google Scholar]
- Foyer C., Furbank R., Harbinson J., Horton P. (1990). The mechanisms contributing to photosynthetic control of electron transport by carbon assimilation in leaves. Photosynth. Res. 25: 83–100. [DOI] [PubMed] [Google Scholar]
- Fristedt R., Martins N.F., Strenkert D., Clarke C.A., Suchoszek M., Thiele W., Schöttler M.A., Merchant S.S. (2015). The thylakoid membrane protein CGL160 supports CF1CF0 ATP synthase accumulation in Arabidopsis thaliana. PLoS One 10: e0121658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao F., Lipscomb B., Wu I., Richter M.L. (1995). In vitro assembly of the core catalytic complex of the chloroplast ATP synthase. J. Biol. Chem. 270: 9763–9769. [DOI] [PubMed] [Google Scholar]
- Grahl S., Reiter B., Gügel I.L., Vamvaka E., Gandini C., Jahns P., Soll J., Leister D., Rühle T. (2016). The Arabidopsis protein CGLD11 is required for chloroplast ATP synthase accumulation. Mol. Plant 9: 885–899. [DOI] [PubMed] [Google Scholar]
- Hahn A., Vonck J., Mills D.J., Meier T., Kühlbrandt W. (2018). Structure, mechanism, and regulation of the chloroplast ATP synthase. Science 360: eaat4318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajdukiewicz P., Svab Z., Maliga P. (1994). The small, versatile pPZP family of Agrobacterium binary vectors for plant transformation. Plant Mol. Biol. 25: 989–994. [DOI] [PubMed] [Google Scholar]
- Hippler M., Rimbault B., Takahashi Y. (2002). Photosynthetic complex assembly in Chlamydomonas reinhardtii. Protist 153: 197–220. [DOI] [PubMed] [Google Scholar]
- Jap B.K., Walian P.J. (1996). Structure and functional mechanism of porins. Physiol. Rev. 76: 1073–1088. [DOI] [PubMed] [Google Scholar]
- Junge W., Nelson N. (2015). ATP synthase. Annu. Rev. Biochem. 84: 631–657. [DOI] [PubMed] [Google Scholar]
- Kanazawa A., Kramer D.M. (2002). In vivo modulation of nonphotochemical exciton quenching (NPQ) by regulation of the chloroplast ATP synthase. Proc. Natl. Acad. Sci. USA 99: 12789–12794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lakshmi B., Mishra M., Srinivasan N., Archunan G. (2015). Structure-based phylogenetic analysis of the lipocalin superfamily. PLoS One 10: e0135507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laskowski R.A., MacArthur M.W., Moss D.S., Thornton J.M. (1993). PROCHECK: A program to check the stereochemical quality of protein structures. J. Appl. Cryst. 26: 283–291. [Google Scholar]
- Malik Ghulam M., Zghidi-Abouzid O., Lambert E., Lerbs-Mache S., Merendino L. (2012). Transcriptional organization of the large and the small ATP synthase operons, atpI/H/F/A and atpB/E, in Arabidopsis thaliana chloroplasts. Plant Mol. Biol. 79: 259–272. [DOI] [PubMed] [Google Scholar]
- Malik Ghulam M., Courtois F., Lerbs-Mache S., Merendino L. (2013). Complex processing patterns of mRNAs of the large ATP synthase operon in Arabidopsis chloroplasts. PLoS One 8: e78265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao J., Chi W., Ouyang M., He B., Chen F., Zhang L. (2015). PAB is an assembly chaperone that functions downstream of chaperonin 60 in the assembly of chloroplast ATP synthase coupling factor 1. Proc. Natl. Acad. Sci. USA 112: 4152–4157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathur J., Koncz C. (1998). PEG-mediated protoplast transformation with naked DNA. In Arabidopsis Protocols. Methods in Molecular Biology, Vol. 82, Martinez-Zapater J.M., Salinas J., eds (Totowa, NJ: Humana Press; ), pp. 267–276. [DOI] [PubMed] [Google Scholar]
- McPherson A. (1990). Current approaches to macromolecular crystallization. Eur. J. Biochem. 189: 1–23. [DOI] [PubMed] [Google Scholar]
- Miyake C., Amako K., Shiraishi N., Sugimoto T. (2009). Acclimation of tobacco leaves to high light intensity drives the plastoquinone oxidation system: relationship among the fraction of open PSII centers, nonphotochemical quenching of Chl fluorescence and the maximum quantum yield of PSII in the dark. Plant Cell Physiol. 50: 730–743. [DOI] [PubMed] [Google Scholar]
- Munekage Y., Hojo M., Meurer J., Endo T., Tasaka M., Shikanai T. (2002). PGR5 is involved in cyclic electron flow around photosystem I and is essential for photoprotection in Arabidopsis. Cell 110: 361–371. [DOI] [PubMed] [Google Scholar]
- Niyogi K.K. (1999). Photoprotection revisited: genetic and molecular approaches. Annu. Rev. Plant Physiol. Plant Mol. Biol. 50: 333–359. [DOI] [PubMed] [Google Scholar]
- Pfalz J., Bayraktar O.A., Prikryl J., Barkan A. (2009). Site-specific binding of a PPR protein defines and stabilizes 5′ and 3′ mRNA termini in chloroplasts. EMBO J. 28: 2042–2052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porra R.J., Thompson W.A., Kriedemann P.E. (1989). Determination of accurate extinction coefficients and simultaneous equations for assaying chlorophylls a and b extracted with four different solvents: Verification of the concentration of chlorophyll standards by atomic absorption spectrometry. Biochim. Biophys. Acta 975: 384–394. [Google Scholar]
- Rak M., Gokova S., Tzagoloff A. (2011). Modular assembly of yeast mitochondrial ATP synthase. EMBO J. 30: 920–930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rott M., Martins N.F., Thiele W., Lein W., Bock R., Kramer D.M., Schöttler M.A. (2011). ATP synthase repression in tobacco restricts photosynthetic electron transport, CO2 assimilation, and plant growth by overacidification of the thylakoid lumen. Plant Cell 23: 304–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rühle T., Leister D. (2015). Assembly of F1F0-ATP synthases. Biochim. Biophys. Acta 1847: 849–860. [DOI] [PubMed] [Google Scholar]
- Rühle T., Razeghi J.A., Vamvaka E., Viola S., Gandini C., Kleine T., Schünemann D., Barbato R., Jahns P., Leister D. (2014). The Arabidopsis protein CONSERVED ONLY IN THE GREEN LINEAGE160 promotes the assembly of the membranous part of the chloroplast ATP synthase. Plant Physiol. 165: 207–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schleiff E., Soll J. (2005). Membrane protein insertion: mixing eukaryotic and prokaryotic concepts. EMBO Rep. 6: 1023–1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider T.R., Sheldrick G.M. (2002). Substructure solution with SHELXD. Acta Crystallogr. D Biol. Crystallogr. 58: 1772–1779. [DOI] [PubMed] [Google Scholar]
- Schöttler M.A., Tóth S.Z., Boulouis A., Kahlau S. (2015). Photosynthetic complex stoichiometry dynamics in higher plants: biogenesis, function, and turnover of ATP synthase and the cytochrome b6f complex. J. Exp. Bot. 66: 2373–2400. [DOI] [PubMed] [Google Scholar]
- Schreiber U., Klughammer C. (2008). New accessory for the Dual-PAM-100: the P515/535 module and examples of its application. PAM Appl. Notes 1: 1–10. [Google Scholar]
- Schweer J., Loschelder H., Link G. (2006). A promoter switch that can rescue a plant sigma factor mutant. FEBS Lett. 580: 6617–6622. [DOI] [PubMed] [Google Scholar]
- Shikanai T., Munekage Y., Shimizu K., Endo T., Hashimoto T. (1999). Identification and characterization of Arabidopsis mutants with reduced quenching of chlorophyll fluorescence. Plant Cell Physiol. 40: 1134–1142. [DOI] [PubMed] [Google Scholar]
- Sobti M., Smits C., Wong A.S., Ishmukhametov R., Stock D., Sandin S., Stewart A.G. (2016). Cryo-EM structures of the autoinhibited E. coli ATP synthase in three rotational states. eLife 5: e21598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strotmann H., Shavit N., Leu S. (1998). Assembly and function of the chloroplast ATP synthase. In The Molecular Biology of Chloroplast and Mitochondria in Chlamydomonas, Rochaix J.D., Goldschmidt-Clermont M., and Merchant S., eds (Norwell, MA: Kluwer Academic Publishers; ), pp. 477–500. [Google Scholar]
- Takabayashi A., Kadoya R., Kuwano M., Kurihara K., Ito H., Tanaka R., Tanaka A. (2013). Protein co-migration database (PCoM-DB) for thylakoids and cells. Springerplus 2: 148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhlin U., Cox G.B., Guss J.M. (1997). Crystal structure of the ϵ subunit of the proton-translocating ATP synthase from Escherichia coli. Structure 5: 1219–1230. [DOI] [PubMed] [Google Scholar]
- von Ballmoos C., Wiedenmann A., Dimroth P. (2009). Essentials for ATP synthesis by F1F0 ATP synthases. Annu. Rev. Biochem. 78: 649–672. [DOI] [PubMed] [Google Scholar]
- Wang Z.G., Ackerman S.H. (2000). The assembly factor Atp11p binds to the beta-subunit of the mitochondrial F(1)-ATPase. J. Biol. Chem. 275: 5767–5772. [DOI] [PubMed] [Google Scholar]
- Wang Z.G., Sheluho D., Gatti D.L., Ackerman S.H. (2000). The alpha-subunit of the mitochondrial F(1) ATPase interacts directly with the assembly factor Atp12p. EMBO J. 19: 1486–1493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wollman F.A., Minai L., Nechushtai R. (1999). The biogenesis and assembly of photosynthetic proteins in thylakoid membranes1. Biochim. Biophys. Acta 1411: 21–85. [DOI] [PubMed] [Google Scholar]
- Yagi H., Kajiwara N., Tanaka H., Tsukihara T., Kato-Yamada Y., Yoshida M., Akutsu H. (2007). Structures of the thermophilic F1-ATPase ε subunit suggesting ATP-regulated arm motion of its C-terminal domain in F1. Proc. Natl. Acad. Sci. USA 104: 11233–11238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto H., Peng L., Fukao Y., Shikanai T. (2011). An Src homology 3 domain-like fold protein forms a ferredoxin binding site for the chloroplast NADH dehydrogenase-like complex in Arabidopsis. Plant Cell 23: 1480–1493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida M., Muneyuki E., Hisabori T. (2001). ATP synthase--a marvellous rotary engine of the cell. Nat. Rev. Mol. Cell Biol. 2: 669–677. [DOI] [PubMed] [Google Scholar]
- Zhang L., Duan Z., Zhang J., Peng L. (2016). BIOGENESIS FACTOR REQUIRED FOR ATP SYNTHASE 3 facilitates assembly of the chloroplast ATP synthase complex. Plant Physiol. 171: 1291–1306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong L., Zhou W., Wang H., Ding S., Lu Q., Wen X., Peng L., Zhang L., Lu C. (2013). Chloroplast small heat shock protein HSP21 interacts with plastid nucleoid protein pTAC5 and is essential for chloroplast development in Arabidopsis under heat stress. Plant Cell 25: 2925–2943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoschke R., Kroeger T., Belcher S., Schöttler M.A., Barkan A., Schmitz-Linneweber C. (2012). The pentatricopeptide repeat-SMR protein ATP4 promotes translation of the chloroplast atpB/E mRNA. Plant J. 72: 547–558. [DOI] [PubMed] [Google Scholar]
- Zoschke R., Qu Y., Zubo Y.O., Börner T., Schmitz-Linneweber C. (2013). Mutation of the pentatricopeptide repeat-SMR protein SVR7 impairs accumulation and translation of chloroplast ATP synthase subunits in Arabidopsis thaliana. J. Plant Res. 126: 403–414. [DOI] [PubMed] [Google Scholar]