ABSTRACT
The highly conserved TORC1 complex controls cell growth in response to nutrients, especially amino acids. The EGO complex activates TORC1 in response to glutamine and leucine. Here, we demonstrate that the I-BAR domain-containing protein Ivy1 colocalizes with Gtr1 and Gtr2, a heterodimer of small GTPases that are part of the EGO complex. Ivy1 is a negative regulator of Gtr-induced TORC1 activation, and is contained within puncta associated with the vacuolar membrane in cells grown in nutrient-rich medium or after brief nitrogen starvation. Addition of glutamine to nitrogen-starved cells leads to dissipation of Ivy1 puncta and redistribution of Ivy1 throughout the vacuolar membrane. Continued stimulation with glutamine results in concentration of Ivy1 within vacuolar membrane invaginations and its spatial separation from the EGO complex components Gtr1 and Gtr2. Disruption of vacuolar membrane invagination is associated with persistent mislocalization of Ivy1 across the vacuolar membrane and inhibition of TORC1 activity. Together, our findings illustrate a novel negative-feedback pathway that is exerted by Ivy1 on Gtr-dependent TORC1 signaling and provide insight into a potential molecular mechanism underlying TORC1 activation by vacuolar membrane remodeling.
KEY WORDS: EGO complex, Gtr1, Gtr2, Ivy1, TORC1, Microautophagy, Vps1
Summary: The I-BAR domain-containing protein Ivy1 colocalizes with the EGO complex components Gtr1 and Gtr2 and regulates their activation.
INTRODUCTION
TORC1 is a multiprotein complex that couples availability of nutrients to an appropriate growth response. The EGO complex relays signals of amino acid availability, specifically glutamine and leucine, to TORC1 (Binda et al., 2009; Stracka et al., 2014; Péli-Gulli et al., 2015; Tanigawa and Maeda, 2017; Varlakhanova et al., 2017). It consists of a heterodimer of small GTPases, Gtr1 and Gtr2, and a vacuole-associated ternary complex consisting of Meh1 (Ego1), Ego2 and Slm4 (Ego3) (Nicastro et al., 2017). The EGO complex regulates TORC1 activity through direct physical interaction and also modulates the vacuolar distribution of Tor1, one of two alternate TORC1 kinase subunits (Kira et al., 2016). Activity of the EGO complex depends on the nucleotide status of Gtr1 and Gtr2 – active EGO complex contains Gtr1GTP and Gtr2GDP. Gtr1GTP interacts with Tco89 and the active Gtr heterodimer interacts with Kog1, both of which are subunits of TORC1 (Binda et al., 2009; Sekiguchi et al., 2014). A complex network of accessory factors that regulate and modulate the nucleotide status of Gtr1 and Gtr2 has been characterized. For example, the Lst4–Lst7 complex acts as a GTPase-activating protein (GAP) for Gtr2, which leads to the GDP-bound active state of Gtr2 (Péli-Gulli et al., 2017, 2015). Likewise, Vam6 is a nucleotide exchange factor (GEF) for Gtr1, which would regenerate active Gtr1 (Binda et al., 2009). The GTPase activity of Gtr1 is also regulated by the SEA complex, consisting of Iml1, Npr2 and Npr3, that acts as a GTPase-activating protein for Gtr1, hence, decreasing activation of TORC1 (Panchaud et al., 2013b).
Recently, Ivy1 has been demonstrated to be an interactor with the EGO complex on vacuoles (Numrich et al., 2015). However, its function in regulation of the EGO complex and/or TORC1 remains unclear. Ivy1 is an effector of Ypt7, a small GTPase involved in vacuolar fusion, localizes to vacuoles and is a regulator of vacuolar membrane homeostasis. Ivy1 has also been shown to bind and inhibit the Fab1 complex, which generates phosphatidylinositol 3,5-bisphosphate [PI(3,5)P2], which, in turn, regulates vacuolar dynamics (Malia et al., 2018). A key structural feature of Ivy1 is its inverse BAR (I-BAR) domain (Itoh et al., 2016). In other proteins, I-BAR domains have been shown to generate negative membrane curvature (Mattila et al., 2007; Millard et al., 2005). In accordance, Ivy1 has been shown to accumulate at invaginations in the vacuolar membrane and has been proposed to be a marker for microautophagy (Numrich et al., 2015). When cells enter stationary phase, Ivy1 and Gtr2 both distribute into sterol-rich microdomains in the vacuolar membrane (Toulmay and Prinz, 2013), which might be due to a recently reported direct interaction between Ivy1 and Gtr2 (Numrich et al., 2015). Such sterol-rich microdomains mediate stationary-phase lipophagy, a process that morphologically resembles microautophagy (Wang et al., 2014).
In this work, we demonstrate that glutamine- and leucine-dependent activation of TORC1 following nitrogen starvation results in a remarkable redistribution of Ivy1 throughout the vacuolar membrane. We further show that, in the cell, Ivy1 is in close proximity to both Gtr1 and Gtr2. Ivy1 inhibits Gtr-mediated activation of TORC1 and does so by inhibiting Gtr activation. Glutamine-induced redistribution of Ivy1 is temporary, as continued stimulation with glutamine leads to sequestration of Ivy1 within vacuolar membrane invaginations. This separates the Ivy1 pool from both Gtr1 and Gtr2. Disruption of vacuolar membrane invagination prevents Ivy1 sequestration away from Gtr1 and Gtr2, and results in inhibition of Gtr-dependent regulation of TORC1. We, therefore, propose that Ivy1 is part of a homeostatic mechanism that fine-tunes the EGO-complex–TORC1 signaling axis in response to amino acid availability.
RESULTS
Ivy1 redistributes on the vacuolar membrane in response to glutamine and leucine
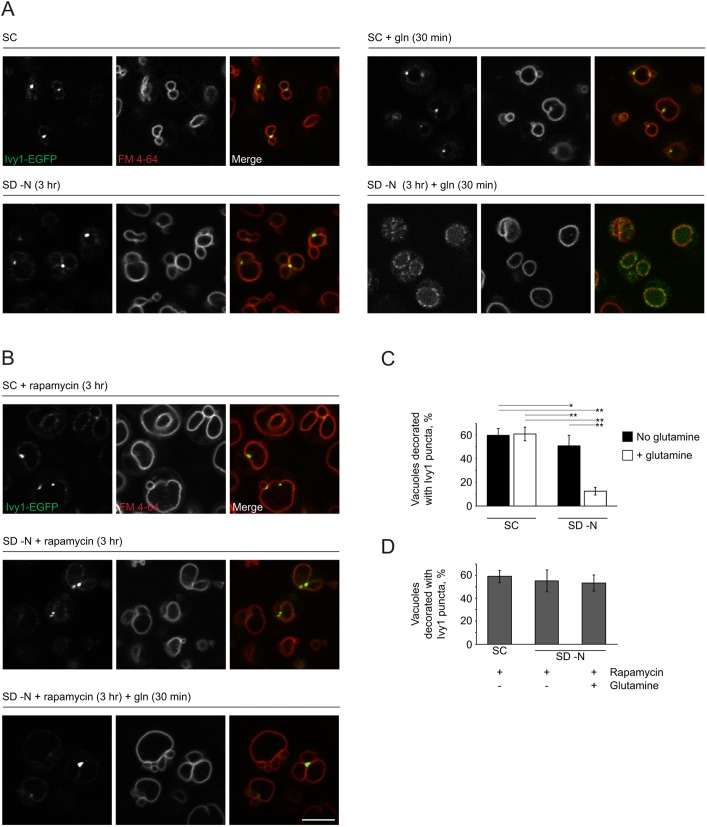
Ivy1 is an I-BAR domain-containing protein that associates with the vacuolar membrane and interacts with components of the EGO complex (Numrich et al., 2015). Previous work has demonstrated that Ivy1 has a punctate distribution in yeast grown in nutrient-rich medium. When cells enter stationary phase, Ivy1 accumulates at sterol-rich lipid microdomains at the vacuolar surface, together with Gtr2 (Toulmay and Prinz, 2013). These observations suggest that Ivy1 localization is nutrient-dependent and that it has a functional link to the EGO complex. Since the main function of the EGO complex is to transfer signals of amino acid availability – specifically glutamine and leucine – to TORC1, we examined whether localization of Ivy1 is sensitive to glutamine. In nutrient-rich medium, Ivy accumulated in puncta, often at junctions between vacuolar lobes (Fig. 1A,C) (59.7±5.8% of vacuoles were associated with at least one punctum of Ivy1-EGFP). Ivy1-EGFP remained punctate after nitrogen starvation (3 h), with a slight but significant reduction in the percentage of vacuoles associated with at least one punctum of Ivy1-EGFP (50.7±5.7%, P<0.01). Strikingly, stimulation of cells with glutamine (3 mM, 30 min) after nitrogen starvation (3 h) resulted in a redistribution of Ivy1 throughout the vacuolar membrane (12.7±3.2% of vacuoles remained associated with at least one punctum of Ivy1; compared to starved cells, P=9.9×10−13). Ivy1 localization at the vacuole was not uniform or homogeneous and appeared to be segregated into microdomains. By contrast, addition of glutamine (3 mM, 30 min) to cells grown in nutrient-rich (SC) medium did not change the punctate distribution of Ivy1 (60.1±9.2% of vacuoles were associated with at least one punctum of Ivy1: compared to cells grown in SC medium, P=0.77). This suggests that nitrogen starvation is a prerequisite for Ivy1 redistribution in response to glutamine. We observed a similar redistribution of Ivy1 in response to leucine treatment after nitrogen starvation (Fig. S1A) (14.2±3.4% of vacuoles were associated with at least one punctum of Ivy1). Redistribution of Ivy1 was dependent on TORC1 activity as addition of rapamycin, a pharmacological co-inhibitor of TORC1 (together with FKBP), prevented this redistribution (Fig. 1B,D). These findings demonstrate that Ivy1 redistributed only after nitrogen starvation and reactivation of TORC1 by glutamine. To confirm this interpretation, we used cells lacking both Gtr1 and Gtr2, which are unable to reactivate TORC1 (Binda et al., 2009; Dubouloz et al., 2005; Péli-Gulli et al., 2015; Varlakhanova et al., 2017). Ivy1 remained largely punctate after refeeding of nitrogen-starved Δgtr1 Δgtr2 cells with glutamine (3 mM, 30 min) (W303A SD −N +glutamine: 12.67±3.2% of vacuoles associated with Ivy1 puncta; Δgtr1 Δgtr2 SD −N+glutamine: 32.11±4.9% of vacuoles associated with puncta; t=10.43; P=8.4×10−9) (Fig. S1B,C). Therefore, Ivy1 redistrubution depends on glutamine- or leucine-dependent TORC1 reactivation after nitrogen starvation.
Fig. 1.
Changes in Ivy1 vacuolar distribution in response to glutamine. (A) W303A cells expressing Ivy1-EGFP were grown in SC medium. For nitrogen starvation, cells were transferred to SD −N for 3 h. Glutamine (gln, 3 mM) was added for 30 min, where indicated. Vacuoles were labeled with FM 4-64 (10 µM in the appropriate medium) for 1 h prior to imaging. (B) Cells were treated with rapamycin (200 ng/ml in the appropriate medium) for 3 h. Vacuoles were stained and cells were imaged as in A. (C) Quantification of the results shown in A. For each condition and each field of cells, Z-stacks were taken and the number of puncta associated with the vacuole in each stack was determined. Shown is mean±s.d. Two-way ANOVA was conducted to determine the effects of growth medium (SC or SD −N) and absence or presence of glutamine supplementation on the percentage of vacuoles decorated with Ivy1 puncta. There was a significant interaction term (F1,31=62, hence P<0.0001). Each main effect was also significant: medium (F1,31=111.08, hence P<0.0001) and absence or presence of glutamine supplementation (F1,31=224.75, hence P<0.0001). Selected pairs of values significant at the 0.05 (*) and 0.01 (**) levels are indicated. (D) Quantification of the results shown in B. No significant differences were observed among any of the assessed conditions. Scale bar: 5 µm (for A and B).
Ivy1 localizes in close proximity to Gtr1 and Gtr2
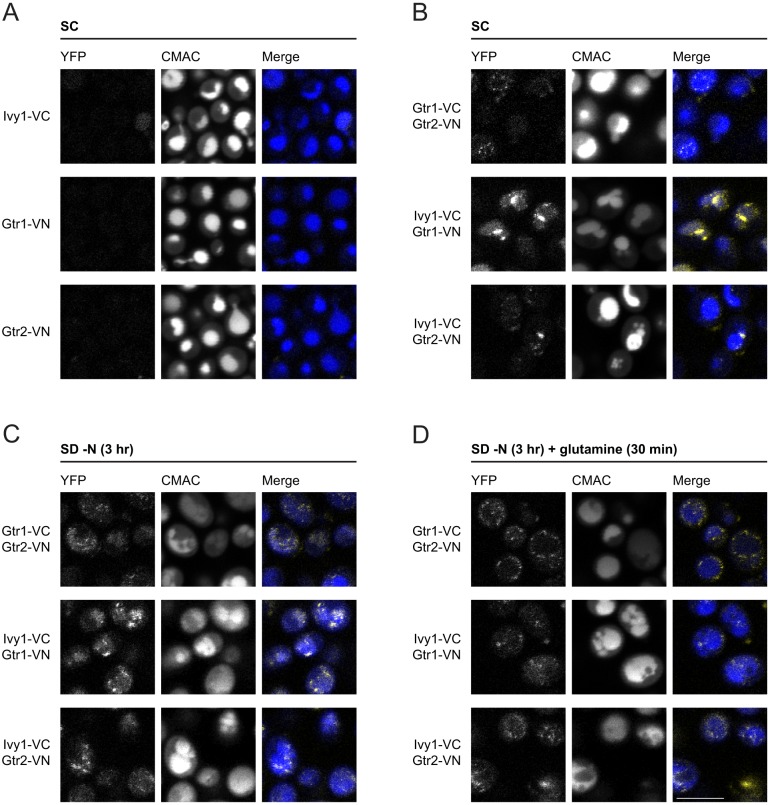
To assess the proximity of Ivy1 to both Gtr1 and Gtr2, we used bimolecular fluorescence complementation (hereafter, split YFP). In this approach, proteins that are potentially close enough to each other in order to interact in the cell are tagged with the N-terminal or C-terminal half of YFP. If the proteins are, indeed, close enough, the two YFP fragments will complement each other and YFP fluorescence will be detectable (Sung and Huh, 2007). When cells expressed Gtr1-VC or Gtr2-VN, no YFP fluorescence was detected (Fig. 2A). However, YFP fluorescence was clearly observed in cells co-expressing Gtr1-VC and Gtr2-VN (Fig. 2B). Importantly, we also detected YFP fluorescence in cells co-expressing Ivy1-VC and either Gtr1-VN or Gtr2-VN (Fig. 2B,C), but the signal was much more punctate. This is in agreement with the observed localization of Ivy1-GFP (Fig. 1A). When cells that co-express Ivy1-VC with either Gtr1-VN or Gtr2-VN were treated with glutamine following nitrogen starvation (3 h), YFP fluorescence remained but appeared much more dispersed (Fig. 2D), again in agreement with the observed localization of Ivy1-EGFP. Of note, an alternative tagging regime, where VC-Ivy1 was co-expressed with either Gtr1-VN or Gtr2-VN, did not produce YFP fluorescence (Fig. S2A). Our findings suggest that Ivy1 and Gtr1 or Gtr2 are in close-enough proximity in the cell to permit bimolecular functional complementation, but this interaction is preserved after glutamine-mediated redistribution of Ivy1.
Fig. 2.
Ivy1 localizes in close proximity to Gtr1 and Gtr2. (A) Controls for the split YFP assay. W303A cells expressing genomically integrated Ivy1, Gtr1 or Gtr2 C-terminally tagged with either the N-terminal (VN) or C-terminal (VC) fragments of YFP. Vacuolar lumens were labeled with CMAC. Cells were grown in SC medium. (B) YFP fluorescence was detected in W303A/α cells co-expressing appropriate pairs of proteins tagged with complementary fragments of YFP. Cells were grown and stained with CMAC as in A. (C,D) As in B but cells were grown in either SD −N for 3 h (C) or SD −N for 3 h followed by glutamine refeeding (3 mM) for 30 min prior to visualization (D). Scale bar: 5 µm (for A-D).
Next, we performed co-immunoprecipitation experiments with purified Ivy1 and the Gtr1-Gtr2 complex to determine whether there is a direct physical interaction. Unfortunately, purified Ivy1 robustly sedimented with the antibody-conjugated beads, even after extensive pre-clarification (Fig. S2B). No further enrichment of Ivy1 in the presence of Gtr1 and Gtr2 was observed. Hence, a direct interaction of Gtr1 and/or Gtr2 and Ivy1 is unlikely, at least under the conditions used. Ivy1 might, therefore, interact with other components of the EGO complex to regulate Gtr activation or function, or it might act more indirectly through regulation of other Gtr interaction partners or through modification of lipid microenvironments surrounding Gtrs.
Ivy1 localizes to vacuolar membrane invaginations
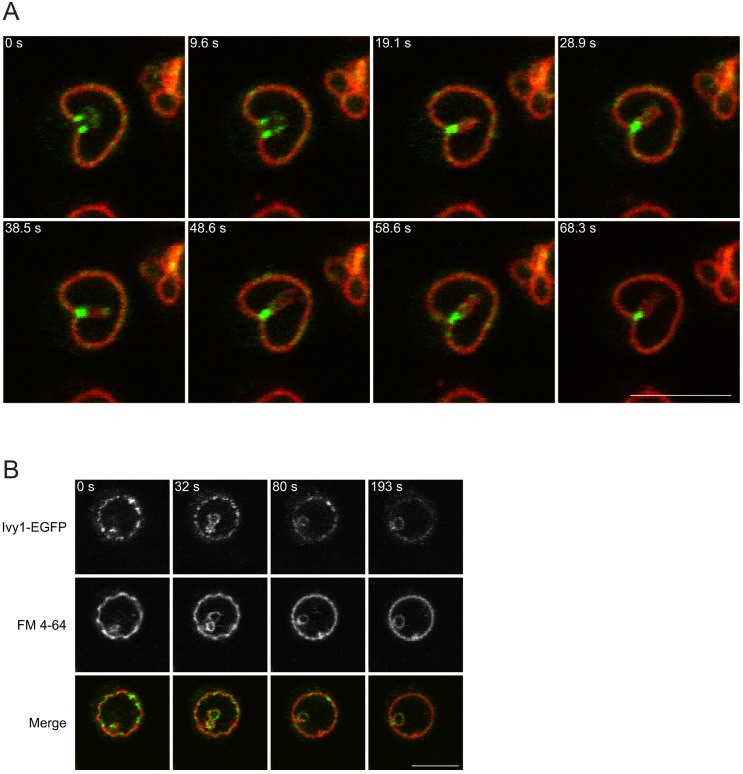
Continued stimulation of nitrogen-starved W303A cells with glutamine (1 h) led to the appearance of vacuolar membrane invaginations decorated with Ivy1 (Fig. 3A) (82.8±8.1% of invaginations were decorated). We also observed an apparent reorganization of vacuolar membrane microdomains marked by Ivy1. Ivy1-marked microdomains, characterized by a slight negative curvature, appeared to dissipate as Ivy1 was removed from them (Fig. 3B). Together, this suggests that the role of the vacuolar membrane invaginations is to reorganize the lipid microdomains and to sequester Ivy1 within the constrained region of the invagination.
Fig. 3.
Ivy1 concentrates in vacuolar membrane invaginations. (A) W303A cells expressing Ivy1-EGFP were grown in SD −N for 3 h. Glutamine (3 mM) was added for 1 h. Vacuoles were labeled with FM 4-64 for 1 h prior to imaging. Images were collected at the indicated time points. (B) W303A cells expressing Ivy1-EGFP were grown and treated as in A. A time-lapse was taken and selected frames at various time points (indicated), showing the formation of a microautophagic invagination with simultaneous redistribution of Ivy1-EGFP across the vacuolar membrane, are shown. Scale bars: 5 µm.
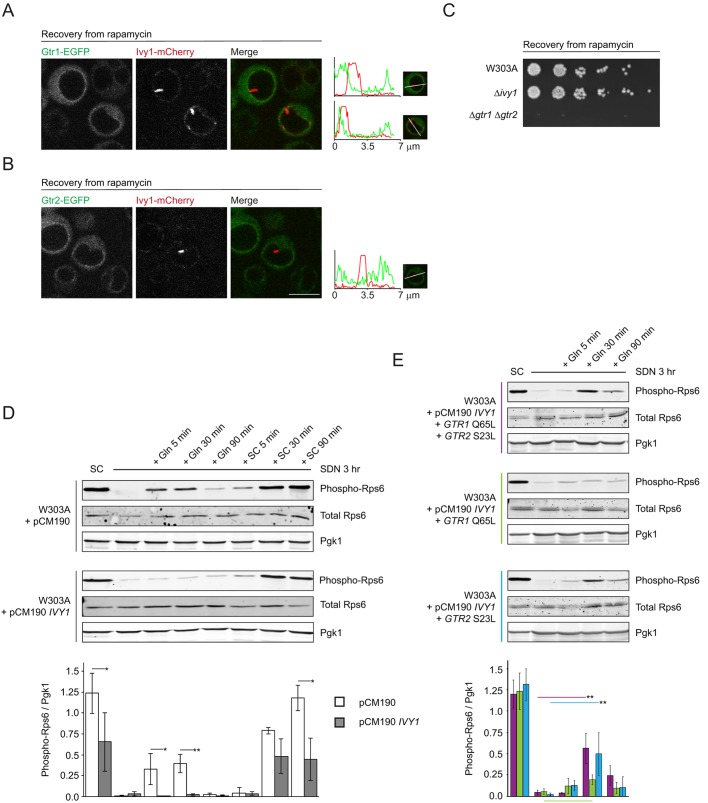
Since the vacuolar membrane invaginations appeared to sequester Ivy1 away from the remainder of the vacuolar membrane, we evaluated the localization of Ivy1, Gtr1 and Gtr2. Previous studies reported induction of vacuolar membrane invaginations, described as microautophagy, by treating cells with rapamycin and following their recovery (Dubouloz et al., 2005). We observed that, during recovery from rapamycin, Ivy1 relocalized into microautophagic invaginations (Fig. 4A,B) (88.7±9.3% of invaginations were decorated). These invaginations lacked both Gtr1 and Gtr2 (Fig. 4A,B), suggesting that vacuolar membrane invaginations that form during recovery from rapamycin sequester Ivy1 away from Gtr1 and Gtr2.
Fig. 4.
Ivy1 inhibits glutamine-stimulated TORC1 activation. (A,B) Cells expressing Ivy1-mCherry and Gtr1-EGFP (A) or Gtr2-EGFP (B) were visualized using confocal microscopy. Where indicated, cells were treated with rapamycin (200 ng/ml) for 3 h, washed and recovered in YPD for 1 h prior to visualization. Representative cells are shown. Line scans show fluorescence intensity profiles along the indicated path for the EGFP and mCherry signals. The maxima for mCherry and EGFP fluorescence along the path were equalized. (C) W303A, Δivy1 and Δgtr1 Δgtr2 cells were treated with 200 ng/ml rapamycin in YPD for 5 h at 30°C. After washing, cells were plated on YPD and were incubated at 30°C for 2 days. The left-most spot in each case corresponds to 2 µl of a culture with OD600 0.5. Spots to the right of this correspond to 2 µl of sequential 5-fold dilutions. (D) Phosphorylation levels of Rps6 were evaluated under the indicated conditions. Untreated cells were grown in SC medium. Cells were nitrogen-starved by incubation in SD −N for 3 h. For stimulation, cells were treated with SD −N supplemented with either glutamine (Gln, 3 mM) or were re-fed with complete SC medium and were incubated for the indicated times prior to lysis and processing. Pgk1 and total Rps6 were used as loading control. Representative blots are shown. The quantification of the 3–4 replicates of the blots is shown below. Shown are the means of the ratios of phosphorylated Rps6 (phospho-Rps6) to Pgk1 (mean±s.d.) for each condition, in the control case (pCM190, white bars) or with Ivy1 overexpression (pCM190 IVY1, gray bars). Significant differences between pairs of measurements (pCM190 and pCM190 IVY1) are indicated (*P<0.05, **P<0.01). (E) W303A cells expressing the indicated combinations of constructs were nitrogen-starved by incubation in SD −N for 3 h. Where indicated, cells were treated with SD −N supplemented with Gln (3 mM) and were incubated for the indicated times prior to lysis and processing as in D. Representative blots are shown. The quantification of 3 replicates for each condition is depicted below the blots. Shown are the means of the ratios of phosphorylated Rps6 (phospho-Rps6) to Pgk1 (mean±s.d.) under each condition, color-coded according to the combination of constructs expressed in the cells as indicated to the left of the corresponding representative blot (maroon: W303A+pCM190 IVY1+GTR1 Q65L+GTR2 S23L; green: W303A+pCM190 IVY1+GTR1 Q65L; blue: W303A+pCM190 IVY1+GTR2 S23L). For each combination, the means of the untreated, treated and recovery measurements were determined to be significantly heterogeneous one-way ANOVA (maroon: F4,10=48.09, P=1.7×10−6; green: F4,10=59.98, P=5.95×10−7; blue: F4,10=49.82, P=1.74×10−8). The most-relevant significantly different pairs of means, as assessed using the post-hoc Tukey HSD test, are indicated by the appropriate colored bar (**P<0.01). The most-relevant pair of means showing no significant difference is shown with a color-coded bar below the chart. Scale bar: 5 µm (for A and B).
Ivy1 is distributed throughout the vacuolar surface but is enriched in lipid microdomains when cells enter stationary phase or after prolonged periods of starvation (Toulmay and Prinz, 2013). We confirmed this observation by starving a saturated yeast culture for 7 h in nitrogen-free medium. Ivy1 distributed throughout the vacuolar membrane (data not shown). Refeeding of these starved cells for 1 h with complete medium resulted in some cells developing vacuolar membrane invaginations. Where this occurred, Ivy1 was invariably found within the invagination and was separated from Gtr2 (Fig. S3A).
Although Ivy1 was enriched in vacuolar invaginations, it was not required for their formation (Numrich et al., 2015). Vacuoles of cells in which IVY1 had been deleted (Δivy1 cells) still developed intravacuolar invaginations during recovery from exposure to rapamycin (Fig. S3B). Cells that are known to be defective in formation of vacuolar membrane invaginations (microautophagy), such as Δgtr1 Δgtr2, are impaired in recovery from exposure to rapamycin (Dubouloz et al., 2005) (Fig. 4C). Cells lacking Ivy1, however, recovered as efficiently as W303A cells (Fig. 4C). These results demonstrate that Ivy1 is not essential for microautophagy.
The EGO complex communicates glutamine and leucine availability to TORC1. The distribution of Ivy1 in response to treatment with glutamine and leucine, and subsequent separation of Ivy1 from Gtr1 and Gtr2 during microautophagy, suggests that it plays a role in Gtr-dependent regulation of TORC1. We, therefore, evaluated activation of TORC1 in response to glutamine in W303A cells, in cells lacking Ivy1 and cells overexpressing Ivy1, by using a tetracycline-regulatable promoter system (Garí et al., 1997). We used phosphorylation of Rps6 as a well-characterized readout of TORC1 activity (Gonzalez et al., 2015; Varlakhanova et al., 2017). In W303A cells, nitrogen starvation (3 h) decreased Rps6 phosphorylation, a direct result of TORC1 inhibition. Stimulation with glutamine after nitrogen starvation resulted in TORC1 activation, as previously reported (Varlakhanova et al., 2017). Deletion of IVY1 did not result in significant changes in TORC1 activation (Fig. S4A). In cells overexpressing Ivy, however, incubation with glutamine did not result in Rps6 phosphorylation and, by extension, stimulation of TORC1 activity (Fig. 4D). Similarly, leucine-dependent TORC1 activation, as assessed by Rps6 phosphorylation, was also inhibited when Ivy1 was overexpressed (Fig. S4B). The defect in TORC1 activation was glutamine- or leucine-specific, as TORC1 could still be activated by refeeding cells that overexpress Ivy1 with SC medium, to an extent similar to that observed in untreated cells (Fig. 4D).
We hypothesized that, due to the proximity of Ivy1 to Gtrs, Ivy1 inhibits TORC1 activation by disrupting Gtr activation. To test this, we overexpressed Ivy1 in cells expressing Gtr mutants that are constitutively active, either alone or simultaneously. Gtr1 Q65L and Gtr2 S23L are predicted to be GTP- and GDP-locked, respectively, and are, thus, constitutively active (Gao and Kaiser, 2006; Nicastro et al., 2017). Whereas expression of Gtr1 Q65L did not result in rescue of TORC1 reactivation by glutamine, expression of either Gtr2 S23L alone or of both Gtr1 Q65L and Gtr2 S23L together resulted in a significant recovery of TORC1 reactivation (Fig. 4E). This suggests that Ivy1 inhibits activation of Gtrs, particularly Gtr2. Of note, when Ivy1 was overexpressed, it localized not only to puncta but also throughout the vacuolar membrane (Fig. S4C). As previously reported, overexpression of Ivy1 results in spherical vacuoles that appear to be resistant to fragmentation (Malia et al., 2018), probably due to Ivy1 regulating PI(3,5)P2 levels. Expression of activated forms of the Gtrs did not alter this vacuolar phenotype associated with Ivy1 overexpression (Fig. S4D). Thus, active Gtrs did not simply restore TORC1 activation by reversing the vacuolar phenotype associated with Ivy1 overexpression. Our results, therefore, indicate that Ivy1 is a negative regulator of Gtr-dependent TORC1 activation.
Ivy is mislocalized in cells lacking membrane invaginations
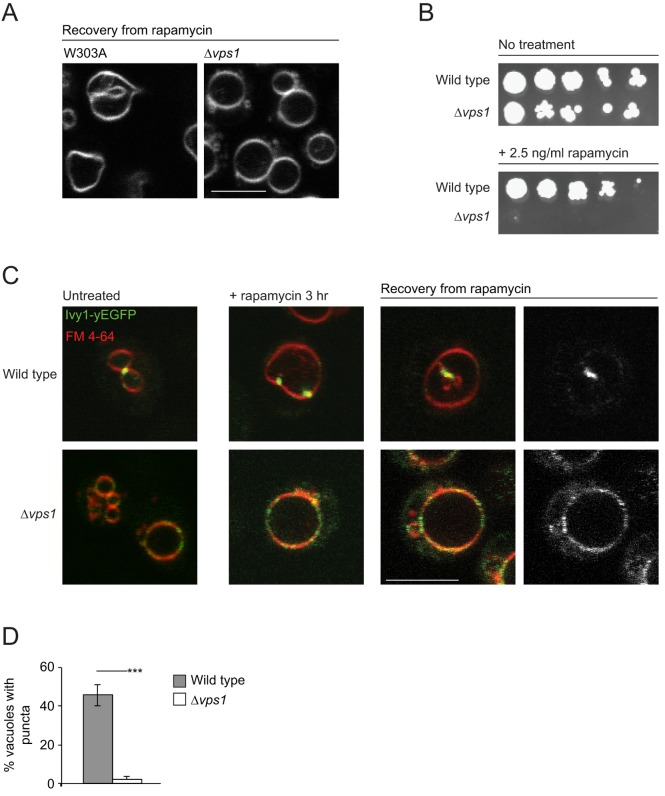
Since our results demonstrate that Ivy1 is sequestered into vacuolar invaginations, we next evaluated the consequences of disrupting the formation of vacuolar membrane invaginations on Ivy1 function in TORC signaling. To this end, we used cells that lack the dynamin-related protein Vps1. Vps1 has been shown to localize principally to the endosomal compartment where it regulates endosomal dynamics (Chi et al., 2014; Varlakhanova et al., 2018). Loss of Vps1 results in a Class F vacuolar morphology (Raymond et al., 1992) (Fig. S5A) and endosomal trafficking defects (Chi et al., 2014; Varlakhanova et al., 2018). Vacuoles in Δvps1 cells conspicuously lacked vacuolar membrane invaginations and intravacuolar vesicles, both in untreated cells and those recovering from exposure to rapamycin (Fig. 5A, Fig. S5A). Δvps1 cells also exhibited sensitivity to rapamycin, suggesting a defect in TORC1 signaling (Fig. 5B). Inhibition of TORC1 results in increased phosphorylation of eIF2α, a downstream target of TORC1 signaling (Cherkasova and Hinnebusch, 2003) and can, therefore, be used as an alternative readout of TORC1 activity. Rapamycin treatment resulted in enhanced phosphorylation of eIF2α in both W303A and Δvps1 cells, to comparable levels (Fig. S5B,C) (∼7.5-fold increase compared to levels in untreated W303A cells). During recovery from rapamycin exposure, a gradual decrease in eIF2α phosphorylation was observed in W303A cells, as TORC1 reactivated. By contrast, eIF2α remained phosphorylated in Δvps1 cells, even after 24 h of recovery (Fig. S5C, P=0.015), suggesting these cells fail to reactivate TORC1 during recovery.
Fig. 5.
Inhibition of vacuole invagination results in Ivy1 mislocalization on the vacuolar membrane. (A) W303A and Δvps1 cells were treated with rapamycin (200 ng/ml) for 3 h, followed by washing and recovery in YPD. Vacuoles were stained with FM 4-64 prior to visualization using confocal microscopy. Representative images are shown. (B) W303A and Δvps1 cells were plated in serial dilutions on YPD (top) or YPD supplemented with 2.5 ng/ml rapamycin (bottom) and were incubated at 30°C for 2 days. The left-most spot in each case corresponds to 2 µl of a culture with OD600 0.5. Spots to the right of this correspond to 2 µl of sequential 5-fold dilutions. (C) W303A or Δvps1 cells expressing Ivy1-EGFP were stained with FM 4-64 and were visualized using confocal microscopy. Where indicated, cells were treated with rapamycin (200 ng/ml) for 3 h, or were treated, washed and recovered in YPD for 1 h prior to visualization. Representative cells are shown. (D) Quantification of untreated vacuoles associated with Ivy1 puncta in W303A and Δvps1 cells. For untreated fields of W303A and Δvps1 cells, Z-stacks were obtained and the number of puncta associated with the vacuole in each stack was determined. The means of the ratios of vacuoles with Ivy1 puncta to total vacuoles were significantly different in W303A and Δvps1 cells (t=12.8; ***P<0.001; n=143 and 152 for W303A and Δvps1 vacuoles, respectively). Scale bars: 5 µm (A and C).
The vacuolar membrane remodeling during microautophagy is thought to compensate for membrane addition to the vacuolar membrane by macroautophagy (Dubouloz et al., 2005; Sattler and Mayer, 2000). EGO complex mutants are known to be defective in microautophagy and, hence, develop enlarged vacuoles after exposure to rapamycin (Dubouloz et al., 2005; Varlakhanova et al., 2017). If cells that lack Vps1 are, indeed, defective in microautophagy, they should exhibit enlarged vacuoles, such as EGO complex mutants, upon exposure to rapamycin. As expected, vacuoles of W303A cells increased in volume on rapamycin treatment (Chan and Marshall, 2014) (increase to 34.6 µm3 compared to 8.8 µm3 for untreated vacuoles, P=0.001). During recovery, the vacuolar volume returned to pre-exposure levels (10.6 µm3 after 48 h recovery, n.s.) (Fig. S5D). Vacuoles of Δvps1 cells were likewise enlarged upon treatment with rapamycin but never stopped expanding, even during recovery from rapamycin exposure, similar to what has been observed for EGO complex mutants (Fig. S5D) (19.3, 32.4 and 46.0 µm3, respectively, for untreated vacuoles, vacuoles treated with rapamycin for 3 h and vacuoles after 48 h recovery, P=9×10−10). Notably, during recovery, vacuoles from Δvps1 cells lacked any vacuolar membrane invaginations. These findings illustrate that Δvps1 cells are defective in formation of vacuolar membrane invaginations.
We next evaluated Ivy1 subcellular localization in cells lacking Vps1. In the absence of Vps1, Ivy1 was diffusely distributed through the vacuolar membrane, even in untreated cells (Fig. 5C). Since Ivy1 is known to interact with both PI3P and PI(3,5)P2, Δvps1 cells are likely to have an altered membrane lipid composition, which might be responsible for Ivy1 mislocalization in these cells. In W303A cells treated with rapamycin for 3 h, Ivy1 concentrated at nascent vacuolar invaginations (Fig. 5C). During recovery from rapamycin exposure, Ivy1 decorated the stalks of the vacuolar invaginations. By contrast, Ivy1 remained diffuse throughout the vacuolar membrane in Δvps1 cells, both during rapamycin exposure and subsequent recovery. During recovery from rapamycin, Ivy1 did not separate from Gtr1 and Gtr2 in Δvps1 cells (Fig. S6A and data not shown). Ivy1 also did not form any puncta. Hence, vacuolar membrane invaginations during and after rapamycin treatment could serve as a sink, to concentrate and accumulate Ivy1 and its potential interaction partners.
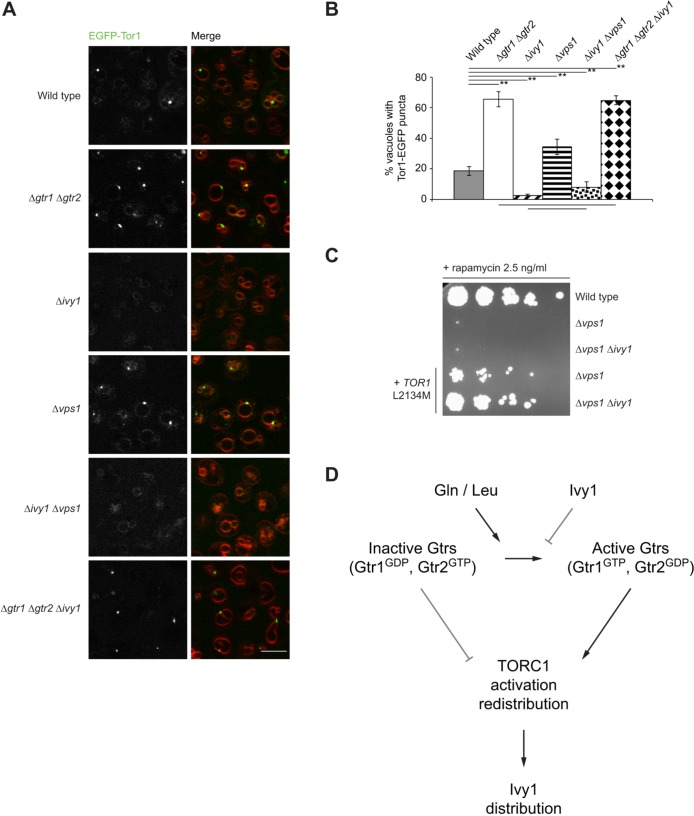
Our data suggest that Ivy1 regulates the activation status of Gtrs. The GTP/GDP status of Gtrs is known to affect the distribution of TORC1 on the vacuolar membrane (Kira et al., 2016; Prouteau et al., 2017). Specifically, it has been demonstrated that Gtr1GDP and Gtr2GTP favors formation of TORC1 perivacuolar puncta, whereas Gtr1GTP and Gtr2GDP promotes redistribution of TORC1 throughout the vacuolar membrane. We, therefore, used the distribution and localization of EGFP-tagged Tor1, one of the two alternative kinase subunits of TORC1, as an indirect and dynamic readout of Gtr status. EGFP-tagged Tor1 was functional, as assessed using rapamycin sensitivity assays in Δtor1 cells (Fig. S6B). It also localized, as expected, on vacuolar membranes in Δtor1 cells (Fig. S6C). Loss of both Gtr1 and Gtr2 resulted in accumulation of EGFP-Tor1 in puncta located on the vacuolar membrane (Fig. 6A) (66% of Δgtr1 Δgtr2 vacuoles were associated with Tor1 puncta compared to 19% of W303A vacuoles, P=0.001) (Kira et al., 2016). Loss of Ivy1 resulted in a significant drop in vacuoles with Tor1 puncta (Fig. 6A,B) (2.5% of vacuoles in Δivy1 cells compared to 19% of W303A vacuoles, P=0.001). This phenotype mimics the effect of expression of active Gtr1 and Gtr2 on Tor1 localization (Kira et al., 2016; Prouteau et al., 2017). More vacuoles exhibited Tor1 puncta in Δvps1 cells (Fig. 6A,B) (34% of vacuoles in Δvps1 cells compared to 19% in W303A cells, P=0.001). We hypothesize that this is a consequence of Ivy1 being distributed throughout the vacuolar membrane in these cells. Indeed, simultaneous deletion of Vps1 and Ivy1 significantly reduced the percentage of vacuoles with puncta (Fig. 6A) (8% compared to 19% in W303A cells, P=0.01). Taken together, these observations suggest that Ivy1 regulates Gtr-dependent Tor1 localization.
Fig. 6.
Ivy1 regulates Tor1 localization. (A) Cells, as indicated, expressing EGFP-Tor1 were stained with FM 4-64 and visualized using confocal microscopy. Representative fields are shown. Scale bar: 5 µm. (B) Quantification of Tor1 puncta on vacuoles. The means of the ratios of vacuoles with Tor1 puncta to total vacuoles for the indicated strains were significantly heterogeneous (one-way ANOVA, F5,18=227.83, P=1.3×10−15). Pairs of means not significantly different from one another (Tukey HSD post-hoc test) are indicated by horizontal lines below the chart (P>0.05). Selected pairs of means that are significantly different from one another are indicated by horizontal lines above the graph (**P<0.01). 260–480 vacuoles were quantified for each strain. (C) Cells expressing Tor1 L2134M as indicated were plated on YPD supplemented with 2.5 ng/ml rapamycin and incubated at 30°C for 3 days. The left-most spot in each case corresponds to 2 µl of a culture with OD600 0.5. Spots to the right of this correspond to 2 µl of sequential 5-fold dilutions. (D) Model for the regulation of TORC1 signaling by Ivy1. Glutamine or leucine activate Gtrs that regulate TORC1 activation and distribution. Reactivated TORC1 results in redistribution of Ivy1 on the vacuolar membrane. Ivy1, in turn suppresses activation of Gtrs.
Importantly, loss of Ivy1 did not alter Tor1 distribution in cells lacking both Gtr1 and Gtr2, suggesting that Ivy1 effects on Tor1 localization depend on the presence of Gtr1 and/or Gtr2 (Fig. 6A,B) (66% of vacuoles were associated with TORC1 puncta in Δgtr1 Δgtr2 cells compared to 65% in Δgtr1 Δgtr2 Δivy1 cells, n.s.). These data support our hypothesis that the effects Ivy1 has on Tor1 localization are mediated through its effects on Gtrs.
To determine whether rapamycin sensitivity of Δvps1 cells is mediated by mislocalized Ivy1, we evaluated sensitivity to rapamycin of Δivy1 Δvps1 cells. The double deletion cells remained sensitive to rapamycin (Fig. 6C), suggesting that the defect associated with the loss of Vps1 is not simply due to mislocalization of Ivy1. Loss of Ivy1 also did not rescue the defect in TORC1 activation by glutamine in Δvps1 cells (Fig. S6D). In Δvps1 cells, expression of Tor1 L2134M, a hyperactive Tor1 mutant whose activation is independent of Gtr1 and Gtr2 (Kingsbury et al., 2014; Takahara and Maeda, 2012) resulted in only partial growth on plates containing rapamycin. As Tor1 localization is changed in Δvps1 cells compared to W303A cells, this might account for the inability of Tor1 to activate its downstream effectors. Simultaneous deletion of Ivy1 and Vps1 leads to a redistribution of Tor1 throughout the vacuolar membrane. These cells, when expressing Tor1 L2134M, grew at a rate comparable to that of W303A cells on rapamycin (Fig. 6C). Thus, the TORC1 defect observed in Δvps1 cells can be partially attributed to mislocalization of Tor1 due to Ivy1 accumulation or distribution on the vacuolar membrane.
DISCUSSION
In this work, we show that Ivy1 localizes in close proximity to the EGO complex components Gtr1 and Gtr2, suggesting a functional link between Ivy1 and TORC1 signaling. The EGO complex mediates glutamine- and leucine-dependent activation of TORC1 (Binda et al., 2009; Péli-Gulli et al., 2015; Varlakhanova et al., 2017). We show that Ivy1 inhibits Gtr-dependent stimulation of TORC1. Furthermore, glutamine- or leucine-mediated stimulation of TORC1, after nitrogen starvation, results in a temporary redistribution of Ivy1 throughout the vacuolar membrane. Ivy1 is then sequestered into vacuolar membrane invaginations, morphologically described as microautophagic invaginations. This process physically separates Ivy1 from Gtr1 and Gtr2. Disruption of Ivy1 sequestration, by abolishing invaginations of the vacuolar membrane, leads to chronic dispersal of Ivy1 across the vacuolar surface and is associated with an inhibition of Gtr function on TORC1 activity and localization. We, therefore, propose that, after TORC1 is reactivated by Gtrs in response to amino acids, Ivy1 functions to repress Gtr activation.
How might Ivy1 regulate Gtr activation? Gtr1 and Gtr2 assemble into a constitutive heterodimer. The EGO complex is active when Gtr1 is GTP-bound and Gtr2 is GDP-bound, respectively, and inactive in the reverse configuration. Our data demonstrate that Ivy1 primarily affects Gtr2 activation. Therefore, Ivy1 may play a role in activation of Gtr2 through the Lst4–Lst7 complex, which functions as a GAP for Gtr2 (Péli-Gulli et al., 2017, 2015). In the absence of amino acids, Lst4–Lst7 is recruited to the vacuolar membrane. Upon amino acid refeeding, Lst4–Lst7 stimulates Gtr2 GTPase activity, resulting in TORC1 reactivation. Active TORC1 phosphorylates Lst4–Lst7, which results in its release from the vacuolar membrane. Future work will need to assess whether Ivy1 redistribution throughout the vacuolar membrane on refeeding with glutamine or leucine affects Lst4–Lst7 vacuolar localization or function. Alternatively, the GEF for Gtr1 is Vam6, also known as Vps39 (Binda et al., 2009). Vacuolar localization of Vam6 depends on the small GTPase Ypt7, which is a factor required for homotypic vacuolar fusion (Zick and Wickner, 2016). Ivy1 interacts with Ypt7 (Lazar et al., 2002) and, thus, may physically compete for binding to Ypt7 with Vam6, thereby disrupting Gtr1 and EGO complex activation.
We did not detect a direct physical interaction between Ivy1 and Gtr1 and/or Gtr2, suggesting that the effect of Ivy1 on Gtr activation is indirect. A recent report demonstrates that Ivy1 directly binds to, and inhibits, the multi-component Fab1 complex, which consists of Fab1 kinase and several accessory factors (Malia et al., 2018). The Fab1 complex generates and is the sole source of PI(3,5)P2 in the vacuolar membrane. We, therefore, hypothesize that Ivy1, by regulating the local lipid microenvironment surrounding the Gtrs, influences the activation of Gtrs. This provides an exciting potential link between PI(3,5)P2 – which has previously been shown to regulate TORC1 signaling (Jin et al., 2014) – and Gtr activation.
In exponentially growing cells, Ivy1 is punctate (Fig. 1A) (Numrich et al., 2015), where the puncta are associated with the vacuolar membrane. Once cells reach stationary phase, Ivy1 expression has been shown to be upregulated and its localization becomes dispersed across the vacuolar membrane (Numrich et al., 2015). In these cells, Ivy1 accumulates in sterol-rich lipid microdomains together with Gtr2 (Toulmay and Prinz, 2013). The functional significance of Ivy1 and Gtr2 segregation remains unclear. Since stationary phase is an adaptation to prolonged stress and/or starvation, it is tempting to speculate that, on the basis of our data, Ivy1 mediates a long-term suppression of Gtr2 activation.
Our data show that Ivy1 is a negative regulator of Gtr2 activation, which suggests that cells that lack Ivy1 have enhanced levels of activated Gtr2 (Gtr2GDP). Previous work has demonstrated that expression of constitutively active Gtr2 (Gtr2 S23L) is, by itself, not sufficient to enhance TORC1 activation (Panchaud et al., 2013a), which is consistent with our findings that loss of Ivy1 does not alter TORC1 reactivation by glutamine (Fig. S4A).
Our findings illustrate that extended glutamine stimulation or refeeding of cells after prolonged starvation results in Ivy1 accumulating into vacuolar membrane invaginations (Fig. 3, Fig. S3A) and consequent separation of Ivy1 from the Gtrs. Such invaginations are morphologically identical to what has been described as microautophagic invaginations (Numrich et al., 2015). Microautophagy could, therefore, be a mechanism for rapid reorganization of vacuolar membrane microdomains and/or Ivy1 sequestration away from the Gtrs. Microautophagy has previously been demonstrated to contribute to the reactivation of TORC1 (De Virgilio and Loewith, 2006; Dubouloz et al., 2005). The EGO complex and TORC1 are both positive regulators of microautophagy. We, therefore, propose that microautophagic vacuolar membrane invaginations serve to remove Ivy1 from the Gtrs, thereby facilitating subsequent Gtr reactivation. In this scenario, amino acids stimulate Gtr activation, which, in turn, stimulates TORC1. Activated TORC1 redistributes Ivy1 throughout the vacuolar membrane, where it inactivates the Gtrs. Ivy1 is then sequestered and removed by concentration into microautophagic invaginations, which returns the system to its initial state (Fig. 6D).
Loss of Ivy1 does not prevent microautophagy (Fig. S3B) (Numrich et al., 2015) and does not result in rapamycin sensitivity (Fig. 4C). Ivy1 displays negative genetic interactions with many components of the vacuolar ATPase (V-ATPase) (Costanzo et al., 2016; Usaj et al., 2017). This suggests that Ivy1 and V-ATPase act independently, via parallel pathways, to affect the same fundamental process. Simultaneous deletion of Ivy1 and components of V-ATPase, such as Vma13 and Vma6, result in increased rapamycin sensitivity as well as a pronounced vacuolar phenotype: vacuoles display an enlarged surface area with many interconnected vacuolar invaginations (Numrich et al., 2015). Together, these observations suggest that Ivy1 and V-ATPase regulate microautophagy and, consequently, also TORC1 signaling. Ivy1 and the V-ATPase component Vph1 are sequestered into different lipid microdomains on the vacuolar membrane in stationary cells (Vph1 in lipid disordered, Ivy1 into sterol-rich lipid-ordered microdomains) (Toulmay and Prinz, 2013). Ivy1 contains an I-BAR domain that, in the context of mammalian IRSp53 is known to generate negative membrane curvature in vitro (Mattila et al., 2007). Some BAR domain- and I-BAR domain-containing proteins have been demonstrated to help generate and stabilize lipid microdomains by preventing diffusion of phosphoinositides (Saarikangas et al., 2009; Zhao et al., 2013). It would be of interest to determine whether Ivy1, alone or together with V-ATPase, promotes formation, scaffolding and persistence of such microdomains at the vacuolar surface, thus, impacting Gtr activation and TORC1 in a more indirect fashion. Scaffolding may in this case rely on self-assembly of Ivy1, as has been reported (Itoh et al., 2016). In support of this scenario is the finding that components of TORC1, Gtr1 and Iml1 accumulate in sterol-rich microdomains within the vacuolar membrane (Murley et al., 2017). Formation of microdomains leads to clustering of EGO complex components with its negative regulators, resulting in inhibition of TORC1 activity. Ivy1 might, therefore, regulate TORC1 and Gtrs activation by promoting microdomain formation and/or stability.
Taken together, we demonstrate that Ivy1 is a negative regulator of Gtr-dependent activation of TORC1. Gtr-dependent activation of TORC1 controls the vacuolar distribution of Ivy1. Microautophagic invaginations function to reorganize microdomains and to sequester Ivy1 away from the Gtrs.
MATERIALS AND METHODS
Yeast genetic manipulation and molecular biology
Strains used in this work are listed in Table S1. Gene deletions were generated in W303A/α diploids by homologous recombination and complete replacement of the target open reading frame using cassettes amplified from pFA6a-kanMX6, pFA6a-His3MX6 or pFA6-natMX4 flanked with sequence (30 bp) proximal to the coding sequence of the target gene (Goldstein and McCusker, 1999; Longtine et al., 1998). The ORF for VPH1 was C-terminally tagged with EGFP by PCR-mediated homologous recombination using cassettes amplified from pFA6a-link-yEGFP-Kan (Sheff and Thorn, 2004). ORFs for IVY1, GTR1 and GTR2 were N-terminally or C-terminally tagged with the N-terminal (residues 1–172) or C-terminal (residues 155–258) halves of YFP, as appropriate, by homologous recombination using cassettes amplified from pFA6a-VC-kanMX6, pFA6a-kanMX6-pGAL1-VC or pFA6a-VN-HIS3MX6. In all cases, diploids were sporulated by starvation in SPO. Following manual tetrad dissection, haploids were validated by colony PCR, microscopy and, in some cases, sequencing. All deletion strains were generated at 30°C with the exception of Δvps1::NAT, which was constructed at room temperature as Δvps1 cells are known to have a temperature-sensitive growth defect at 37°C (Rothman et al., 1990; Varlakhanova et al., 2018). Strains harboring more than one genomic modification were generated by mating and sporulation of appropriate parental strains, followed by extensive revalidation. For Bimolecular Functional Complementation, haploids of opposite mating type expressing the appropriate pairs of VN- and VC-tagged proteins were mated on YPD for 5 h at room temperature, after which diploids were selected by restreaking onto doubly selective plates.
Media
YPD (2% peptone, 1% yeast extract, 2% glucose, supplemented with L-tryptophan and adenine) was used for routine growth. Synthetic complete (SC; yeast nitrogen base, ammonium sulfate, 2% glucose, amino acids), synthetic defined (SD; yeast nitrogen base, ammonium sulfate, 2% glucose, appropriate amino acid dropout) or YPGal (YPD with galactose substituting for dextrose) media used prior to microscopy or to maintain plasmid selection as indicated. For sporulation, cells were successively cultured in YPA (2% potassium acetate, 2% peptone, 1% yeast extract) and SPO (1% potassium acetate, 0.1% yeast extract, 0.05% glucose). For nitrogen starvation, cells were grown in SD −N (0.17% yeast nitrogen base without amino acids and ammonium sulfate, 2% glucose). Where relevant, SC or SD −N medium was supplemented with 3 mM glutamine. For overexpression of Ivy1 or Ivy1-EGFP, cells harboring the appropriate pCM190 plasmid were cultured in medium lacking doxycycline for 12 h prior to experimentation. 2x TY (1.6% tryptone, 1% yeast extract, 0.5% NaCl) was used for growth of Escherichia coli for protein expression.
Cloning and plasmids
Plasmids in this work are listed in Table S2. S. cerevisiae IVY1, containing the IVY1 coding sequence, and upstream and downstream fragments of 500 and 146 bp, respectively, was amplified from genomic DNA prepared from W303A/α diploids using the Yeast DNA Extraction Kit (Thermo Fisher Scientific, Pittsburgh, PA) and was introduced into the target vector by Gibson assembly (Gibson et al., 2009). Coding, upstream and downstream sequences for TOR1, GTR1 and GTR2 were also amplified from W303A/α genomic DNA. TOR1 L2134M was generated as described (Varlakhanova et al., 2017). Fusion constructs of IVY1, TOR1, GTR1 and GTR2 with EGFP or mCherry, as appropriate, were made by PCR-mediated overlap extension (Heckman and Pease, 2007) followed by Gibson assembly. A PreScission-GTR1 fragment was cloned, in-frame with a His6 tag, into the first multiple cloning site (MCS), of pRSFDuet-1 by Gibson assembly. A GTR2-Myc fragment was cloned into the second MCS. The IVY1 coding sequence was cloned into pET-15b by Gibson assembly, generating an in-frame fusion with His6. All constructs were fully verified by sequencing. Primer and construct sequences are available on request.
Analysis of growth by serial dilution
Following overnight growth in YPD, target cells were diluted and regrown to mid-logarithmic phase in YPD at 30°C. Cells were then diluted to 0.5 OD600/ml and 5-fold serial dilutions were made in water. 2 µl of each dilution was spotted onto YPD or YPD+2.5 ng/ml rapamycin plates. Where relevant, cells were incubated for the indicated times with YPD supplemented with 200 ng/ml rapamycin at 30°C. After extensive washing, cells were resuspended in fresh YPD and recovered at 30°C for the indicated time prior to plating on YPD. Plates were then incubated at 30°C for 2 days prior to photographing.
Western blotting
Protein extracts for western blotting were obtained as described (Millen et al., 2009). Briefly, cells were lysed on ice by resuspension in 1 ml ice-cold H2O to which 150 µl of 1.85 M NaOH and 7.5% (v/v) β-mercaptoethanol were added. After 10 min incubation on ice, protein was precipitated by addition of 150 µl of 50% (w/v) trichloroacetic acid. Pellets were washed twice with acetone, resuspended in 150 µl 1× SDS-PAGE Buffer and incubated for 30 min at 30°C followed by 2 min at 95°C. Antibodies used were as follows: phospho-eIF2α (S51, 1:1000 dilution; Cell Signaling Technology, Danvers, MA), anti-EIF2S1 (ab94587, 1:1000 dilution; Abcam, Cambridge, MA), phospho-Rps6 (4858, 1:1000 dilution; Cell Signaling Technology), Rps6 (ab40820, 1:1000 dilution; Abcam) and anti-PGK1 (ab113687, 1:2000 dilution; Abcam). Labeled secondary antibodies were IRDye 680RD goat anti-rabbit antibody (926-68171; LI-COR, Lincoln, NE) and IRDye 680RD goat anti-mouse (926-68070; LI-COR). These were detected using the Odyssey system. Bands were integrated and quantified using the Fiji distribution of ImageJ (Schindelin et al., 2012).
Protein purification and in vitro co-immunoprecipitation
Purification of the Gtr1-Gtr2 complex
The bicistronic expression construct for the Gtr1-Gtr2 complex was transformed into BL21 Codon Plus RIPL (Agilent Technologies, Santa Clara, CA). Cells were grown in 2×TY and protein was expressed overnight at 21°C by isopropyl β-D-1-thiogalactopyranoside (IPTG) induction. Cells were pelleted, washed and resuspended in TN buffer (20 mM TRIS/Cl pH 8.0, 150 mM NaCl, 1.93 mM β-mercaptoethanol) and lysed by homogenization using an Emulsiflex-C3 (Avestin, Ottawa, Canada). After clarification by centrifugation at 142,000 g for 45 min at 4°C, lysates were incubated with Ni-IDA beads (Macherey-Nagel, Bethlehem, PA). After extensive washing in TN buffer, the beads were loaded into a column and protein was eluted in TN buffer with a gradient to 250 mM of imidazole. Peak fractions were pooled and dialyzed against TN buffer to remove imidazole. The proteins were then passed over Q-sepharose and Superdex 200 16/60 columns in 20 mM Tris/Cl pH 8.0, 150 mM NaCl, 1.93 mM β-mercaptoethanol, after which the peak fractions were concentrated and flash frozen in liquid nitrogen and stored at −80°C until use.
Purification of Ivy1
The Ivy1 expression construct was transformed into SoluBL21 (Genlantis, San Diego, CA). Cells were grown in 2×TY at 37°C. Protein expression was induced by IPTG at mid-logarithmic phase and protein was expressed overnight at 21°C. Cells were pelleted and resuspended in TN buffer. After lysis by homogenization, the lysate was clarified by centrifugation at 142,000 g for 45 min at 4°C. The lysate was then incubated with Ni-IDA for 1 h at 4°C. After extensive washing with TN, the beads were packed into a column and protein was eluted with a gradient to 250 mM imidazole. Peak fractions were dialyzed into fresh TN buffer to remove imidazole. The protein was concentrated and flash frozen in liquid nitrogen for storage at −80°C. The identity of the protein was confirmed by mass spectrometry.
Co-immunoprecipitation
His6-Ivy1 was diluted in TN supplemented with HALT protease inhibitor cocktail (Thermo Fisher Scientific) and was pre-clarified by pre-incubation for 1 h at 4°C with 10 µl magnetic beads conjugated to a mouse anti-Myc monoclonal antibody (Cell Signaling Technology). The supernatant was removed and used, alone or in combination with purified Gtr1-Gtr2 complex, and incubated for 3 h at 4°C with fresh magnetic beads conjugated to a mouse anti-Myc monoclonal antibody. The beads were extensively washed with TN buffer supplemented with protease inhibitors. Protein was eluted from the beads by boiling in SDS sample buffer. Proteins were separated by SDS polyacrylamide gel electrophoresis and were detected by western blotting using an anti-His6 polyclonal antibody (ab137839, 1:1000 dilution; Abcam) or anti-Myc monoclonal (ab32, 1:1000 dilution; Abcam).
Preparation of yeast for microscopy
Cells were grown overnight in YPD or SD medium appropriately supplemented to maintain plasmid selection. Cells were diluted in YPD or SC medium and grown to mid-logarithmic phase. Vacuolar membranes were stained with 10 µM FM 4-64 (Thermo Fisher Scientific) for 45 min, followed by washing and incubation in YPD or SC medium without dye for 1 h. For rapamycin treatment, cells in YPD were treated for the indicated time with a final concentration of 200 ng/ml rapamycin (Thermo Fisher Scientific). For recovery from rapamycin exposure, cells were extensively washed and resuspended in fresh YPD and incubated as indicated. Cells were plated onto No. 1.5 glass-bottomed dishes (MatTek Corporation, Ashland, MA) previously treated with 15 µl 2 mg/ml concanavalin-A (Sigma-Aldrich, St. Louis, MO). For bimolecular functional complementation, cells were stained with 0.5 mM CMAC (Thermo Fisher Scientific) for 30 min at 30°C, washed once and resuspended in SC medium.
Confocal microscopy and image analysis
Confocal images were acquired on a Nikon A1 confocal microscope (Melville, NY), with a 100× Plan Apo 100× oil objective. NIS Elements microscope imaging software (Nikon) was used to control acquisition. For bimolecular functional complementation, samples were imaged using a 60×1.40 N.A. objective on a Nikon A1 confocal microscope equipped with a six-line laser launch with 405 nm, 457 nm, 488 nm, 514 nm, 561 nm and 640 nm lines. Samples were imaged utilizing 405 nm or 514 nm excitation and 460/50 or 535/30 barrier filters, respectively. Images were further processed using Fiji. Cells expressing genomically integrated Vph1-EGFP were used to measure vacuolar volumes. Z-series were deconvolved prior to image thresholding and segmentation. Volumes were obtained using NIS Elements microscope imaging software.
Supplementary Material
Acknowledgements
The authors thank Simon Watkins and Callen Wallace of the Center for Biologic Imaging (University of Pittsburgh School of Medicine) for expertise and technical support.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
Conceptualization: N.V.V., M.G.J.F.; Methodology: N.V.V., M.G.J.F.; Validation: N.V.V., M.G.J.F.; Formal analysis: N.V.V., M.G.J.F.; Investigation: N.V.V., B.A.T.; Writing - original draft: N.V.V., M.G.J.F.; Visualization: N.V.V., M.G.J.F.; Supervision: M.G.F.; Project administration: M.G.J.F.; Funding acquisition: M.G.J.F.
Funding
This work was supported by the National Institutes of Health [grant number: GM120102 (M.G.J.F)]. Deposited in PMC for release after 12 months.
Supplementary information
Supplementary information available online at http://jcs.biologists.org/lookup/doi/10.1242/jcs.218305.supplemental
References
- Binda M., Péli-Gulli M.-P., Bonfils G., Panchaud N., Urban J., Sturgill T. W., Loewith R. and De Virgilio C. (2009). The Vam6 GEF controls TORC1 by activating the EGO complex. Mol. Cell 35, 563-573. 10.1016/j.molcel.2009.06.033 [DOI] [PubMed] [Google Scholar]
- Chan Y.-H. M. and Marshall W. F. (2014). Organelle size scaling of the budding yeast vacuole is tuned by membrane trafficking rates. Biophys. J. 106, 1986-1996. 10.1016/j.bpj.2014.03.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherkasova V. A. and Hinnebusch A. G. (2003). Translational control by TOR and TAP42 through dephosphorylation of eIF2alpha kinase GCN2. Genes Dev. 17, 859-872. 10.1101/gad.1069003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi R. J., Liu J., West M., Wang J., Odorizzi G. and Burd C. G. (2014). Fission of SNX-BAR-coated endosomal retrograde transport carriers is promoted by the dynamin-related protein Vps1. J. Cell Biol. 204, 793-806. 10.1083/jcb.201309084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costanzo M., VanderSluis B., Koch E. N., Baryshnikova A., Pons C., Tan G., Wang W., Usaj M., Hanchard J., Lee S. D. et al. (2016). A global genetic interaction network maps a wiring diagram of cellular function. Science 353, aaf1420 10.1126/science.aaf1420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Virgilio C. and Loewith R. (2006). The TOR signalling network from yeast to man. Int. J. Biochem. Cell Biol. 38, 1476-1481. 10.1016/j.biocel.2006.02.013 [DOI] [PubMed] [Google Scholar]
- Dubouloz F., Deloche O., Wanke V., Cameroni E. and De Virgilio C. (2005). The TOR and EGO protein complexes orchestrate microautophagy in yeast. Mol. Cell 19, 15-26. 10.1016/j.molcel.2005.05.020 [DOI] [PubMed] [Google Scholar]
- Gao M. and Kaiser C. A. (2006). A conserved GTPase-containing complex is required for intracellular sorting of the general amino-acid permease in yeast. Nat. Cell Biol. 8, 657-667. 10.1038/ncb1419 [DOI] [PubMed] [Google Scholar]
- Garí E., Piedrafita L., Aldea M. and Herrero E. (1997). A set of vectors with a tetracycline-regulatable promoter system for modulated gene expression in Saccharomyces cerevisiae. Yeast 13, 837-848. [DOI] [PubMed] [Google Scholar]
- Gibson D. G., Young L., Chuang R.-Y., Venter J. C., Hutchison C. A. III and Smith H. O. (2009). Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat. Methods 6, 343-345. 10.1038/nmeth.1318 [DOI] [PubMed] [Google Scholar]
- Goldstein A. L. and McCusker J. H. (1999). Three new dominant drug resistance cassettes for gene disruption in Saccharomyces cerevisiae. Yeast 15, 1541-1553. [DOI] [PubMed] [Google Scholar]
- Gonzalez A., Shimobayashi M., Eisenberg T., Merle D. A., Pendl T., Hall M. N. and Moustafa T. (2015). TORC1 promotes phosphorylation of ribosomal protein S6 via the AGC kinase Ypk3 in Saccharomyces cerevisiae. PLoS ONE 10, e0120250 10.1371/journal.pone.0120250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heckman K. L. and Pease L. R. (2007). Gene splicing and mutagenesis by PCR-driven overlap extension. Nat. Protoc. 2, 924-932. 10.1038/nprot.2007.132 [DOI] [PubMed] [Google Scholar]
- Itoh Y., Kida K., Hanawa-Suetsugu K. and Suetsugu S. (2016). Yeast Ivy1p is a putative I-BAR-domain protein with pH-sensitive filament forming ability in vitro. Cell Struct. Funct. 41, 1-11. 10.1247/csf.15014 [DOI] [PubMed] [Google Scholar]
- Jin N., Mao K., Jin Y., Tevzadze G., Kauffman E. J., Park S., Bridges D., Loewith R., Saltiel A. R., Klionsky D. J. et al. (2014). Roles for PI(3,5)P2 in nutrient sensing through TORC1. Mol. Biol. Cell 25, 1171-1185. 10.1091/mbc.e14-01-0021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kingsbury J. M., Sen N. D., Maeda T., Heitman J. and Cardenas M. E. (2014). Endolysosomal membrane trafficking complexes drive nutrient-dependent TORC1 signaling to control cell growth in Saccharomyces cerevisiae. Genetics 196, 1077-1089. 10.1534/genetics.114.161646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kira S., Kumano Y., Ukai H., Takeda E., Matsuura A. and Noda T. (2016). Dynamic relocation of the TORC1-Gtr1/2-Ego1/2/3 complex is regulated by Gtr1 and Gtr2. Mol. Biol. Cell 27, 382-396. 10.1091/mbc.e15-07-0470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazar T., Scheglmann D. and Gallwitz D. (2002). A novel phospholipid-binding protein from the yeast Saccharomyces cerevisiae with dual binding specificities for the transport GTPase Ypt7p and the Sec1-related Vps33p. Eur. J. Cell Biol. 81, 635-646. 10.1078/0171-9335-00290 [DOI] [PubMed] [Google Scholar]
- Longtine M. S., McKenzie A. III, Demarini D. J., Shah N. G., Wach A., Brachat A., Philippsen P. and Pringle J. R. (1998). Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14, 953-961. [DOI] [PubMed] [Google Scholar]
- Malia P. C., Numrich J., Nishimura T., Gonzalez Montoro A., Stefan C. J. and Ungermann C. (2018). Control of vacuole membrane homeostasis by a resident PI-3,5-kinase inhibitor. Proc. Natl. Acad. Sci. USA 115, 4684-4689. 10.1073/pnas.1722517115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattila P. K., Pykäläinen A., Saarikangas J., Paavilainen V. O., Vihinen H., Jokitalo E. and Lappalainen P. (2007). Missing-in-metastasis and IRSp53 deform PI(4,5)P2-rich membranes by an inverse BAR domain-like mechanism. J. Cell Biol. 176, 953-964. 10.1083/jcb.200609176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millard T. H., Bompard G., Heung M. Y., Dafforn T. R., Scott D. J., Machesky L. M. and Futterer K. (2005). Structural basis of filopodia formation induced by the IRSp53/MIM homology domain of human IRSp53. EMBO J. 24, 240-250. 10.1038/sj.emboj.7600535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millen J. I., Krick R., Prick T., Thumm M. and Goldfarb D. S. (2009). Measuring piecemeal microautophagy of the nucleus in Saccharomyces cerevisiae. Autophagy 5, 75-81. 10.4161/auto.5.1.7181 [DOI] [PubMed] [Google Scholar]
- Murley A., Yamada J., Niles B. J., Toulmay A., Prinz W. A., Powers T. and Nunnari J. (2017). Sterol transporters at membrane contact sites regulate TORC1 and TORC2 signaling. J. Cell Biol. 216, 2679-2689. 10.1083/jcb.201610032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicastro R., Sardu A., Panchaud N. and De Virgilio C. (2017). The architecture of the Rag GTPase signaling network. Biomolecules 7 10.3390/biom7030048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Numrich J., Péli-Gulli M. P., Arlt H., Sardu A., Griffith J., Levine T., Engelbrecht-Vandre S., Reggiori F., De Virgilio C. and Ungermann C. (2015). The I-BAR protein Ivy1 is an effector of the Rab7 GTPase Ypt7 involved in vacuole membrane homeostasis. J. Cell Sci. 128, 2278-2292. 10.1242/jcs.164905 [DOI] [PubMed] [Google Scholar]
- Panchaud N., Péli-Gulli M.-P. and De Virgilio C. (2013a). Amino acid deprivation inhibits TORC1 through a GTPase-activating protein complex for the Rag family GTPase Gtr1. Sci. Signal. 6, ra42 10.1126/scisignal.2004112 [DOI] [PubMed] [Google Scholar]
- Panchaud N., Péli-Gulli M.-P. and De Virgilio C. (2013b). SEACing the GAP that nEGOCiates TORC1 activation: evolutionary conservation of Rag GTPase regulation. Cell Cycle 12, 2948-2952. 10.4161/cc.26000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Péli-Gulli M. P., Sardu A., Panchaud N., Raucci S. and De Virgilio C. (2015). Amino acids stimulate TORC1 through Lst4-Lst7, a GTPase-Activating protein complex for the rag family GTPase Gtr2. Cell Rep 13, 1-7. 10.1016/j.celrep.2015.08.059 [DOI] [PubMed] [Google Scholar]
- Péli-Gulli M. P., Raucci S., Hu Z., Dengjel J. and De Virgilio C. (2017). Feedback inhibition of the Rag GTPase GAP complex Lst4-Lst7 safeguards TORC1 from hyperactivation by amino acid signals. Cell Rep 20, 281-288. 10.1016/j.celrep.2017.06.058 [DOI] [PubMed] [Google Scholar]
- Prouteau M., Desfosses A., Sieben C., Bourgoint C., Lydia Mozaffari N., Demurtas D., Mitra A. K., Guichard P., Manley S. and Loewith R. (2017). TORC1 organized in inhibited domains (TOROIDs) regulate TORC1 activity. Nature 550, 265-269. 10.1038/nature24021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raymond C. K., Howald-Stevenson I., Vater C. A. and Stevens T. H. (1992). Morphological classification of the yeast vacuolar protein sorting mutants: evidence for a prevacuolar compartment in class E vps mutants. Mol. Biol. Cell 3, 1389-1402. 10.1091/mbc.3.12.1389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothman J. H., Raymond C. K., Gilbert T., O'Hara P. J. and Stevens T. H. (1990). A putative GTP binding protein homologous to interferon-inducible Mx proteins performs an essential function in yeast protein sorting. Cell 61, 1063-1074. 10.1016/0092-8674(90)90070-U [DOI] [PubMed] [Google Scholar]
- Saarikangas J., Zhao H., Pykäläinen A., Laurinmäki P., Mattila P. K., Kinnunen P. K. J., Butcher S. J. and Lappalainen P. (2009). Molecular mechanisms of membrane deformation by I-BAR domain proteins. Curr. Biol. 19, 95-107. 10.1016/j.cub.2008.12.029 [DOI] [PubMed] [Google Scholar]
- Sattler T. and Mayer A. (2000). Cell-free reconstitution of microautophagic vacuole invagination and vesicle formation. J. Cell Biol. 151, 529-538. 10.1083/jcb.151.3.529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B. et al. (2012). Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676-682. 10.1038/nmeth.2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekiguchi T., Kamada Y., Furuno N., Funakoshi M. and Kobayashi H. (2014). Amino acid residues required for Gtr1p-Gtr2p complex formation and its interactions with the Ego1p-Ego3p complex and TORC1 components in yeast. Genes Cells 19, 449-463. 10.1111/gtc.12145 [DOI] [PubMed] [Google Scholar]
- Sheff M. A. and Thorn K. S. (2004). Optimized cassettes for fluorescent protein tagging in Saccharomyces cerevisiae. Yeast 21, 661-670. 10.1002/yea.1130 [DOI] [PubMed] [Google Scholar]
- Stracka D., Jozefczuk S., Rudroff F., Sauer U. and Hall M. N. (2014). Nitrogen source activates TOR (target of rapamycin) complex 1 via glutamine and independently of Gtr/Rag proteins. J. Biol. Chem. 289, 25010-25020. 10.1074/jbc.M114.574335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung M.-K. and Huh W.-K. (2007). Bimolecular fluorescence complementation analysis system for in vivo detection of protein-protein interaction in Saccharomyces cerevisiae. Yeast 24, 767-775. 10.1002/yea.1504 [DOI] [PubMed] [Google Scholar]
- Takahara T. and Maeda T. (2012). Transient sequestration of TORC1 into stress granules during heat stress. Mol. Cell 47, 242-252. 10.1016/j.molcel.2012.05.019 [DOI] [PubMed] [Google Scholar]
- Tanigawa M. and Maeda T. (2017). An in vitro TORC1 kinase assay that recapitulates the Gtr-independent glutamine-responsive TORC1 activation mechanism on yeast vacuoles. Mol. Cell. Biol. 37, e00075-17. 10.1128/MCB.00075-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toulmay A. and Prinz W. A. (2013). Direct imaging reveals stable, micrometer-scale lipid domains that segregate proteins in live cells. J. Cell Biol. 202, 35-44. 10.1083/jcb.201301039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Usaj M., Tan Y., Wang W., VanderSluis B., Zou A., Myers C. L., Costanzo M., Andrews B. and Boone C. (2017). TheCellMap.org: a web-accessible database for visualizing and mining the global yeast genetic interaction network. G3 (Bethesda) 7, 1539-1549. 10.1534/g3.117.040220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varlakhanova N. V., Mihalevic M. J., Bernstein K. A. and Ford M. G. J. (2017). Pib2 and the EGO complex are both required for activation of TORC1. J. Cell Sci. 130, 3878-3890. 10.1242/jcs.207910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varlakhanova N. V., Alvarez F. J. D., Brady T. M., Tornabene B. A., Hosford C. J., Chappie J. S., Zhang P. and Ford M. G. J. (2018). Structures of the fungal dynamin-1 related protein Vps1 reveal a unique, open helical architecture. J. Cell Biol. (In Press). 10.1083/jcb.201712021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C.-W., Miao Y.-H. and Chang Y.-S. (2014). A sterol-enriched vacuolar microdomain mediates stationary phase lipophagy in budding yeast. J. Cell Biol. 206, 357-366. 10.1083/jcb.201404115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao H., Michelot A., Koskela E. V., Tkach V., Stamou D., Drubin D. G. and Lappalainen P. (2013). Membrane-sculpting BAR domains generate stable lipid microdomains. Cell Rep 4, 1213-1223. 10.1016/j.celrep.2013.08.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zick M. and Wickner W. (2016). Improved reconstitution of yeast vacuole fusion with physiological SNARE concentrations reveals an asymmetric Rab(GTP) requirement. Mol. Biol. Cell 27, 2590-2597. 10.1091/mbc.e16-04-0230 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.