ABSTRACT
Cancer cells thrive when challenged with proteotoxic stress by inducing components of the protein folding, proteasome, autophagy and unfolded protein response (UPR) pathways. Consequently, specific molecular chaperones have been validated as targets for anti-cancer therapies. For example, inhibition of Hsp70 family proteins (hereafter Hsp70) in rhabdomyosarcoma triggers UPR induction and apoptosis. To define how these cancer cells respond to compromised proteostasis, we compared rhabdomyosarcoma cells that were sensitive (RMS13) or resistant (RMS13-R) to the Hsp70 inhibitor MAL3-101. We discovered that endoplasmic reticulum-associated degradation (ERAD) and autophagy were activated in RMS13-R cells, suggesting that resistant cells overcome Hsp70 ablation by increasing misfolded protein degradation. Indeed, RMS13-R cells degraded ERAD substrates more rapidly than RMS cells and induced the autophagy pathway. Surprisingly, inhibition of the proteasome or ERAD had no effect on RMS13-R cell survival, but silencing of select autophagy components or treatment with autophagy inhibitors restored MAL3-101 sensitivity and led to apoptosis. These data indicate a route through which cancer cells overcome a chaperone-based therapy, define how cells can adapt to Hsp70 inhibition, and demonstrate the value of combined chaperone and autophagy-based therapies.
This article has an associated First Person interview with the first author of the paper.
KEY WORDS: Hsp70, Protein degradation, Autophagy, Cancer, Apoptosis, ERAD, Ubiquitin-proteasome pathway
Summary: We demonstrate that cancer cells can compensate for Hsp70 inhibition by inducing autophagy, emphasizing the role of autophagy/chaperone-based therapies.
INTRODUCTION
Protein homeostasis (‘proteostasis’) is regulated to ensure that protein quality control pathways are employed only when needed. In some cell types, such as cancer cells, proteostasis pathways are differentially regulated to ensure survival (Benbrook and Long, 2012; Kim et al., 2013; McConkey, 2017; Wang and Kaufman, 2014). Notably, cancer cells thrive under conditions of nutrient and ATP depletion, hypoxia and exposure to cytotoxic agents (Mei et al., 2013; Rubiolo et al., 2014; Yan et al., 2015). Many cancer cells are also aneuploid, which leads to imbalanced levels of protein complexes that must be resolved by degradative pathways (Weaver and Cleveland, 2005; Williams and Amon, 2009). Finally, owing to their accelerated growth rate – and because some driver oncogenes are secreted or are membrane proteins – cancer cells contend with high levels of proteins that enter the secretory pathway (Deshaies, 2014). As a result, cancer cell adaptation requires that levels of molecular chaperones and the endoplasmic reticulum (ER) unfolded protein response (UPR) pathway are differentially regulated (Lorin et al., 2013; Mei et al., 2013; Rubiolo et al., 2014; Sannino and Brodsky, 2017; Vandewynckel et al., 2013; Yan et al., 2015). Nevertheless, the relative contributions of these potential pro-survival pathways in most cancers have not been investigated.
The ER plays a central role in proteostasis because it governs the synthesis and folding of secreted and membrane proteins, which constitute one-third of the proteome, as well as redox homeostasis and lipid biogenesis (Anelli et al., 2015; Ellgaard and Helenius, 2003; Rothman and Schekman, 2011; Sitia and Braakman, 2003; Song et al., 2017). When these processes are compromised, the UPR is induced. In mammals, the UPR is regulated by three stress sensors, known as inositol-required enzyme 1 (IRE1; also known as ERN1), PKR ER-resident kinase (PERK, also known as EIF2AK3) and activating transcription factor 6 (ATF6) that reside in the ER membrane, detect ER stress, and restore proteostasis or induce apoptotic cell death if a stress response cannot be rectified (Bi et al., 2005; Halterman et al., 2010; Ron and Walter, 2007; Yamamoto et al., 2004). The UPR is induced in many cancers, but factors that increase cellular fitness – such as the Hsp70 family (hereafter Hsp70) of molecular chaperones – are also induced (Garrido et al., 2006; Guo et al., 2005; Mosser and Morimoto, 2004; Sliutz et al., 1996; Yang et al., 2012). These data suggest that the UPR and Hsp70 might be therapeutically targeted in cancer (Clarke and Cook, 2015; Hazari et al., 2016; Ojha and Amaravadi, 2017; Sabnis et al., 2016; Shajahan et al., 2009).
Even though stress response pathways are upregulated, cancer cells still accumulate protein aggregates and unfolded proteins to a higher degree than normal cells (Clarke et al., 2011; Kim et al., 2015; Schonthal, 2012a,b). Consequently, the efficacy of select protein degradation pathways is also enhanced, such as the ubiquitin-proteasome and autophagy pathways (Amaravadi et al., 2011; Goldberg, 2003; Kim et al., 2013; Ma et al., 2014). These degradative processes decrease the burden of unfolded proteins and recycle amino acids, which promote protein synthesis and cell survival (Levine and Kroemer, 2008; Mizushima et al., 2008; Mizushima et al., 2002). In some transformed cells, proteasome inhibition, which represents a first line therapy in multiple myeloma, induces autophagy (Liu et al., 2013, 2016; Rapino et al., 2014; Wojcik, 2013). UPR induction also augments autophagy (B'Chir et al., 2013; Clarke et al., 2014). These data further highlight the interrelationship between distinct proteostatic pathways.
Another critical proteostatic pathway is ER-associated degradation (ERAD), which, with a few exceptions (Erzurumlu and Ballar, 2017; Singh et al., 2015), has not been measured in tumor-derived cells. ERAD reduces ER stress by identifying and destroying misfolded, improperly processed and orphaned subunits of oligomeric proteins for proteasome-mediated degradation (Haas et al., 1982; Olzmann et al., 2013; Raasi and Wolf, 2007; Vembar and Brodsky, 2008; Ye et al., 2001). Early work indicated that UPR induction increases the expression of ERAD components (Casagrande et al., 2000; Travers et al., 2000; Yoshida et al., 2001a,b), and proteasome inhibitors act by slowing the ERAD of misfolded immunoglobulins in multiple myeloma, which induces the UPR and decreases cell viability (Bianchi et al., 2009). Unfortunately, FDA-approved proteasome inhibitors, such as bortezomib (Velcade) and carfilzomib (Kyprolis), have minimal effects on solid tumors, suggesting cancer cell adaptation or a reduced dependence on the proteasome pathway (Blaney et al., 2004; Chen et al., 2011; Deshaies, 2014).
Another emerging target is the cytosolic Hsp70 molecular chaperone, which is upregulated in many cancers and inhibits multiple steps in the apoptotic pathway (Garrido et al., 2006; Santarosa et al., 1997; Uozaki et al., 2000). Inhibition of Hsp70 or the ER luminal homolog, BiP (also known as GRP78 and HSPA5), kills colorectal carcinomas, breast cancers, leukemia cells, glioblastomas and ovarian cancer cells (Cerezo et al., 2016; Guo et al., 2005; Kawiak et al., 2017; Lee, 2001, 2007; Ni et al., 2009; Powers et al., 2008, 2009; Sabnis et al., 2016). Furthermore, Hsp70 promotes chemotherapeutic resistance, and Hsp70 induction predicts metastasis in several cancers (Brodsky and Chiosis, 2006; Calderwood and Gong, 2016; Nanbu et al., 1998; Patury et al., 2009; Powers et al., 2009). These data highlight the potential of Hsp70-based therapies, but the compensatory resistance mechanisms that limit efficacy of Hsp70 inhibition are unexplored.
We recently reported that rhabdomyosarcoma (RMS) cells exhibit a heightened sensitivity to Hsp70 inhibition. RMS is a soft-tissue sarcoma that mainly affects children with an average 5-year survival of ∼60% (Ward et al., 2014). Chemotherapy, radiotherapy and surgery represent the only existing treatments for this cancer (Olanich and Barr, 2013; Shern et al., 2014; Ward et al., 2014; Weigel et al., 2016). Our prior study employed a small molecule inhibitor, MAL3-101, that binds an allosteric site on the Hsp70 ATPase domain and specifically blocks the stimulatory effect of the Hsp40 (also known as DNAJB) family of cochaperones (Fewell et al., 2004; Sabnis et al., 2016; Wisen et al., 2010). We reported that MAL3-101 treatment induces the PERK branch of the UPR pathway, thereby increasing expression of CHOP (also known as DDIT3) and triggering apoptosis (Sabnis et al., 2016). To define the MAL3-101 mechanism of action, and to delineate whether other compensatory pathways lessen its efficacy, we isolated an isogenic MAL3-101-resistant RMS cell line.
Here, we report that Hsp70 inhibitor-resistant RMS cells possess more efficient protein degradation networks, which prevent MAL3-101-dependent apoptosis. Nevertheless, we could re-sensitize the MAL3-101-resistant RMS cells by chemically inhibiting one of several steps in the autophagy pathway or by silencing a gene encoding an protein that is required for autophagy and is regulated in a PERK- and UPR-dependent manner. In contrast, inhibition of ERAD or the ubiquitin-proteasome pathway had no effect on survival. These findings delineate distinct roles of compensatory proteostatic networks in a cancer model, and show that combined Hsp70 and autophagy-targeted therapeutics might be used to treat rhabdomyosarcoma.
RESULTS
ERAD efficiency is higher in Hsp70 inhibitor resistant rhabdomyosarcoma cells
Rhabdomyosarcoma (RMS) cell lines are hypersensitive to Hsp70 inhibition compared to other cancer cells (Sabnis et al., 2016). To determine why RMS cells are sensitive to MAL3-101, a dose escalation strategy was used to isolate a MAL3-101-resistant line, denoted RMS13-R, from the parental cell line, RMS13. Interestingly, the UPR could still be activated in the RMS13-R-resistant cells, but CHOP was no longer induced after MAL3-101 treatment. In addition, the RMS13-R cells remained sensitive to conventional chemotherapeutics (Sabnis et al., 2016; see below), suggesting that acquired resistance to Hsp70 inhibition results from an increase in compensatory proteostatic pathways.
Hsp70 coordinates the folding, degradation, transport and assembly of proteins, and acts at multiple checkpoints to inhibit apoptosis (Evans et al., 2010; Goloudina et al., 2012; Morishima et al., 2000; Nanbu et al., 1998; Powers et al., 2010; Rodina et al., 2014; Young et al., 2004). To define which pathway(s) might be altered and circumvent Hsp70 inhibition in the resistant cells, we first asked whether proteasome activity was higher in RMS13-R relative to RMS13 cells. Total ubiquitylated protein levels in the RMS13 and RMS13-R cells were measured in the presence or absence of the proteasome inhibitor MG132. The steady-state levels of ubiquitylated proteins were similar in the two cell lines, either with or without MG132 (Fig. 1A). Second, to test whether the proteasome was more active in RMS13-R cells, we quantified the signal produced by the proteasome-dependent degradation of the fluorogenic substrate Suc-LLVY-7-amino-4-methylcoumarin (Stein et al., 1996), but no difference in proteasome activity was detected (Fig. S1A). These data suggest that another pathway is activated to offset the consequences of compromised Hsp70 activity in the resistant cells.
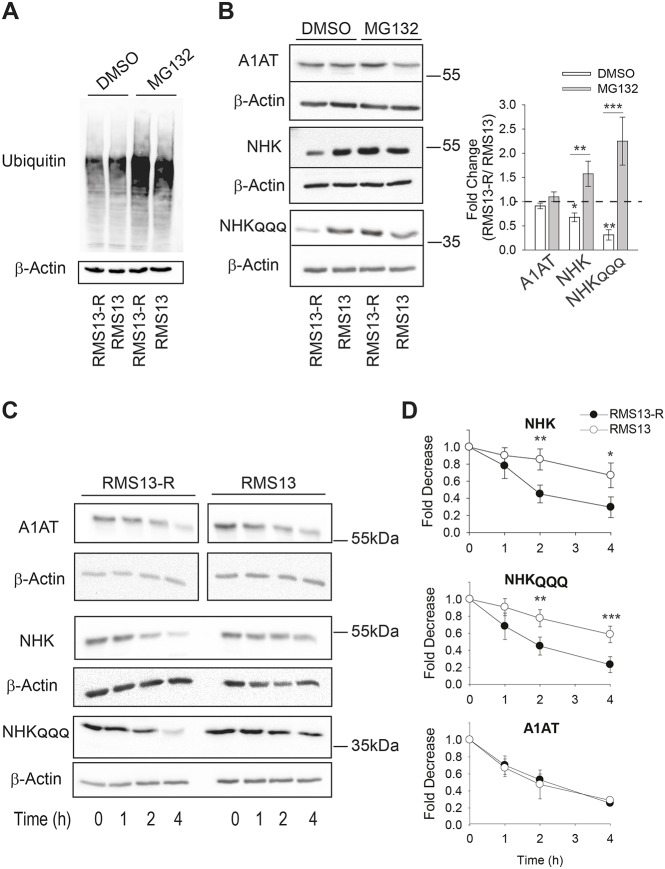
Fig. 1.
RMS13-R cells exhibit heightened levels of ERAD. (A) The MAL3-101-resistant rhabdomyosarcoma cell line (RMS13-R) and the parental MAL3-101-sensitive cell line (RMS13) were treated with DMSO or 20 µM MG132 for 4 h followed by an immunoblot analysis to visualize the total ubiquitylated protein. In the presence of MG132, there was a 2.5-fold increase in ubiquitylated protein levels. (B) Relative ERAD efficiency was measured by analyzing the steady-state levels of the NHK and NHKQQQ mutants in the presence or absence of 20 µM MG132 for 4 h. A1AT was used as a negative control. The graph on the right shows the ratio between the levels of each protein in the RMS13-R and RMS13 cells after treatment with DMSO (in white) and MG132 (in gray). The mean±s.d. of eight different experiments are plotted. (C,D) Both NHK and NHKQQQ are degraded faster in RMS13-R cells. The RMS13-R and RMS13 cells were treated with cycloheximide after a 3-h MG132 pre-treatment, aliquots were removed at the indicated times and lysates were resolved by SDS-PAGE and immunoblotted to detect A1AT abundance. Graphs represent the mean±s.d. of four independent experiments. *P<0.05, **P<0.005, ***P<0.0001.
Hsp70 identifies and degrades misfolded proteins (Daugaard et al., 2007; Goloudina et al., 2012; Morishima et al., 2000; Young et al., 2004). Because MAL3-101 induces the UPR (Sabnis et al., 2016), we reasoned that ERAD might be activated in RMS13-R cells, thus lessening the unfolded protein burden. To test this hypothesis, RMS13 and RMS13-R cells were transfected with vectors to express wild-type α1 anti-trypsin (A1AT), which folds efficiently and is mostly secreted (Marcus and Perlmutter, 2000; Perlmutter, 2011), and a C-terminally truncated variant of this protein known as null Hong Kong (NHK) as well as an NHK variant that cannot be glycosylated, denoted NHKQQQ, both of which are destroyed by ERAD (Hosokawa et al., 2003, 2006, 2007; Zhong et al., 2015). As shown in Fig. 1B, steady-state levels of NHK and NHKQQQ were lower in RMS13-R cells than in RMS13 cells, whereas the amount of A1AT was unchanged between the two lines (Fig. 1B). In addition, A1AT secretion was similar in the two cell lines, indicating that A1AT secretion is unaltered (Fig. S1B). As a positive control, we inhibited A1AT lysosomal degradation using chloroquine (CQ), a drug that impairs lysosomal acidification and protease activity (Ahlberg et al., 1985). CQ increased the amount of secreted A1AT to a comparable extent in both cell lines (Fig. S1B), suggesting that some of the protein is degraded in the lysosome. To determine whether the lower levels of both NHK and NHKQQQ in RMS13-R cells were due to increased degradation, the proteasome was inhibited. MG132 treatment significantly increased the levels of NHK and NHKQQQ in RMS13-R cells to a greater extent than A1AT, where no significant increase was observed (Fig. 1B).
We next conducted cycloheximide chase assays to measure the rate of substrate degradation. To build-up the substrate pool, cells were pre-treated with MG132 and then, after the proteasome inhibitor was washed-out, cycloheximide was added and protein levels were assessed over time. As anticipated, NHK and NHKQQQ were degraded faster in RMS13-R versus RMS13 cells, while no significant difference in the degradation rate was detected for A1AT (Fig. 1C,D). We also tested the degradation rate of misfolded membrane proteins targeted for ERAD. CD3δ and GluR1, which are, respectively, an orphaned subunit of the T cell receptor and an AMPA-type glutamate receptor subunit (Christianson et al., 2011; Frenkel et al., 2003; Goo et al., 2015), were also destroyed faster in RMS13-R cells (Fig. S1C). To confirm that enhanced degradation in RMS13-R cells was due to proteasome and not lysosomal degradation, cycloheximide was added in the presence of MG132 and the NHKQQQ protein levels were assessed over time (Fig. S1D). No significant difference in the initial levels or rate of degradation was detected. We conclude that ERAD is more efficient or that there is an increase in the number of ERAD-requiring complexes in RMS cells that are MAL3-101 resistant.
Autophagy inhibition re-sensitizes RMS13-R cells to MAL3-101
Because ERAD efficiency was elevated in the MAL3-101 resistant cells, we reasoned that they might exhibit greater sensitivity to an ERAD inhibitor. After recognition in the ER and ubiquitylation at the ER membrane, ERAD substrates are retrotranslocated to the cytosol and degraded by the proteasome (Bagola et al., 2011; Raasi and Wolf, 2007; Smith et al., 2011; Vembar and Brodsky, 2008; Weissman, 1997). Retrotranslocation is linked to ATP-dependent substrate transport by the AAA+ ATPase p97 (also known as VCP) (Bays et al., 2001; Jarosch et al., 2002; Raasi and Wolf, 2007; Rabinovich et al., 2002; Ye et al., 2001). Therefore, we used CB-5083, which targets the D2 ATPase domain in p97 (Zhou et al., 2015). This domain was recently shown to be involved in substrate unfolding during retrotranslocation (Bodnar and Rapoport, 2017a,b). In addition, CB-5083 kills multiple myeloma, colon and pancreatic cancer cells (Anderson et al., 2015; Le Moigne et al., 2017). Surprisingly, we found that the RMS13 and RMS13-R cells displayed identical CB-5083 sensitivities (Fig. 2A; IC50 of ∼0.85 µM).
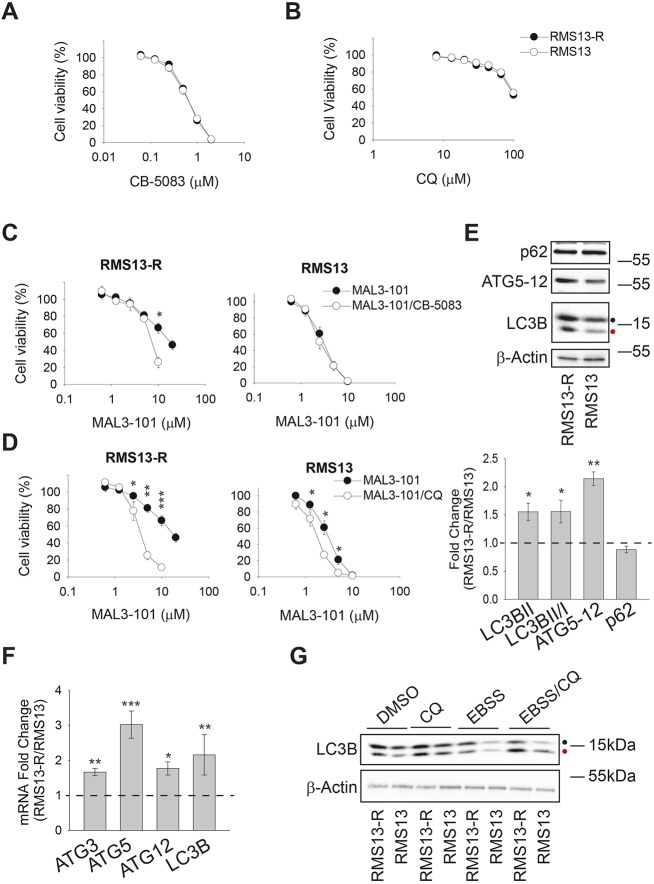
Fig. 2.
The autophagy pathway is required for RMS13-R resistance to Hsp70 inhibition. (A,B) RMS13 and RMS13-R cells were seeded into 96-well plates and treated with increasing doses of CB-5083, a p97 inhibitor, or chloroquine (CQ), an autophagy inhibitor for 72 h. Viability was measured using the CellTiter-Glo assay and the mean±s.e.m. of three independent experiments was plotted. (C,D) Cells were treated with increasing doses of MAL3-101 in the presence (white) or absence (black) of a sub-lethal dose of CB-5083 (0.5 µM) or CQ (40 µM). Cell viability was detected using the CellTiter-Glo assay after 72 h, and data represent the of three independent experiments. (E) Top, steady state levels of select autophagy related proteins in RMS13-R and RMS13 cells were analyzed by immunoblotting. A black and a red dot, respectively, indicate the soluble (LC3BI) and the autophagosome-associated isoform (LC3BII) of the autophagy marker LC3B. Bottom, the ratio of each protein in RMS13-R and RMS13 cells is depicted as the mean± s.e.m. (n=10). (F) The mean±s.d. fold change between RMS13-R and RMS13 cells corresponding to autophagy-related transcripts, as determined by quantitative PCR is plotted (n=5). (G) The induction of LC3B was analyzed by immunoblotting lysates from RMS13-R and RMS13 cells after a 4 h treatment with 50 µM CQ in complete medium or under starvation conditions (EBSS). *P<0.05, **P<0.005, ***P<0.0001.
Autophagy can also contribute to the clearance of ubiquitylated misfolded proteins, alleviating excess demands on the protein folding machinery and promoting cell survival (Levine and Klionsky, 2004; Liu et al., 2013, 2016; Rapino et al., 2014; Wojcik, 2013). Autophagy can also serve as a back-up pathway for the disposal of select ERAD substrates (Ishida et al., 2009; Kruse et al., 2006). Therefore, we asked whether RMS13 and RMS13-R cells exhibit different sensitivities to autophagy inhibitors. First, RMS13-R and RMS13 cells were treated with CQ, which not only impairs lysosomal acidification but also blocks the final step in the autophagy pathway (Ahlberg et al., 1985; Mizushima and Klionsky, 2007; Mizushima et al., 2011). As shown in Fig. 2B, the RMS13-R and RMS13 cell lines were equally resistant to CQ (IC50 >100 µM). Second, we tested two other compounds that inhibit different steps in the autophagy pathway: bafilomycin, a lysosomal proton-pump inhibitor that compromises autophagosome-lysosome fusion, and tubacin, an HDAC6 inhibitor that perturbs both autophagolysosome formation and the recruitment of ubiquitylated proteins to nascent autophagosomes (Bennett et al., 2005; Hubbert et al., 2002; Kawaguchi et al., 2003; Manic et al., 2014; Mizushima et al., 2011; Pandey et al., 2007; Tanida et al., 2005). The two lines again exhibited identical sensitivities (Fig. S2A; IC50 of ∼3.5 nM for bafilomycin and 4.5 µM for tubacin), suggesting that RMS cells initiate mechanisms that compensate for the loss of ERAD and autophagy.
Based on the interplay between protein degradation pathways and chaperone-dependent stress responses (Benbrook and Long, 2012; Guerriero and Brodsky, 2012; Kouroku et al., 2007; Liebl and Hoppe, 2016; Powers et al., 2009; Song et al., 2017; Yan et al., 2015; Yousefi et al., 2006), we next asked whether partial inhibition of one or both degradative pathways would re-sensitize RMS13-R cells to MAL3-101, highlighting relevant resistance mechanisms. We tested MAL3-101 sensitivity in the presence of sub-maximal concentrations of either CB-5083 (0.5 µM) or CQ (40 µM) in RMS13-R and RMS13 cells. As shown in Fig. 2C, CB-5083 modestly increased the sensitivity of RMS13-R cells to MAL3-101, but only in the presence of 10 µM MAL3-101, and no increase in RMS13 sensitivity was apparent under the same conditions. In contrast, CQ significantly re-sensitized RMS13-R cells to MAL3-101 (Fig. 2D). A stronger effect on RMS13-R cells was observed when we titrated MAL3-101 in presence of bafilomycin and tubacin (Fig. S2B,C). Thus, autophagy compensates for the toxic effects of Hsp70 inhibition in RMS cells.
To confirm these data, two other RMS13 clones resistant to MAL3-101 (clone 4B and clone 4E) were analyzed. As expected, both lines were similarly resistant to CQ (IC50 >100 µM, Fig. S2D) and were less sensitive to MAL3-101 (IC50 6.4 µM and 6.5 µM, respectively). We then tested whether partial autophagy inhibition achieved by adding 40 µM CQ increased sensitivity to MAL3-101. As hypothesized, CQ lowered the IC50s to 3.5 µM and 3.1 µM (Fig. S2E), confirming autophagy as a compensatory mechanism to attenuate the effect of Hsp70 inhibition. We then observed that cell lines derived from osteosarcomas, another mesenchymal malignancy of childhood and adolescence, were MAL3-101 sensitive (Sabnis et al., 2016). The effects of MAL3-101 as well as CQ were tested in U2OS and SaOS2 osteosarcoma cell lines, and in contrast to the RMS cells, both were sensitive to MAL3-101 and CQ (IC50 2 µM and 1.3 µM for MAL3-101, and 7 µM and 20 µM for CQ, respectively; data not shown).
Based on the protective effects of the autophagy pathway, we reasoned that this pathway might be upregulated more profoundly in the MAL3-101-resistant RMS cells. To test this hypothesis, the abundance of autophagy markers, including LC3B (also known as MAP1LC3B), ATG5–ATG12 and p62 (also known as SQSTM1), was measured (Kabeya et al., 2000; Pyo et al., 2005). Immunoblot analysis demonstrated greater amounts of ATG5–ATG12 in the RMS13-R line (Fig. 2E), suggesting that the ATG12 conjugation system, which is involved in the elongation phase of autophagy, is induced (Mizushima et al., 2011; Stromhaug et al., 2004). In line with this observation, more LC3BII – the lipidated LC3B isoform associated with autophagosomal membranes (Barth et al., 2010; Kabeya et al., 2000; McLeland et al., 2011) – accumulated in RMS13-R cells, and the conversion of LC3BI, the soluble LC3B isoform, into LC3BII was enhanced (Fig. 2E; LC3BI and LC3BII are highlighted by a black and a red dot, respectively). In contrast, the level of p62, an autophagy receptor, was unchanged (Bjørkøy et al., 2005; Ichimura et al., 2000; Katsuragi et al., 2015; Komatsu et al., 2007). The transcription of select autophagy-related genes (ATG3, ATG5, and ATG12) and the gene encoding LC3B were also higher in RMS13-R cells (Fig. 2F). ATG3 was included in this analysis as it acts in the LC3II conjugation system during elongation (Bernard et al., 2015; Ichimura et al., 2000; McEwan and Dikic, 2011). It is also important to note that the magnitude of these effects is in accordance with the increased levels of autophagy-associated factors observed in related studies (B'Chir et al., 2013; Fullgrabe et al., 2016; Klionsky et al., 2012).
We then asked whether autophagy pathway activation was also higher in RMS13-R cells under conditions of nutrient deprivation (i.e. when pathway induction should be maximal). First, autophagy inhibition after addition of CQ led to the accumulation of LC3BII in both RMS13 and RMS13-R cell lines, as expected; however, LC3BII levels, which serves as a read-out of autophagy induction, were higher in RMS13-R cells (Fig. 2G, ‘CQ’, red dot; Fig. S2F). These results were confirmed when bafilomycin was used (Fig. S2F). In addition, short-term starvation in serum-depleted (EBSS) medium decreased LC3BII abundance in both cell lines, consistent with the fact that autophagic flux was activated. In this case, the levels of LC3BII and the LC3BII:LC3BI ratio decreased more significantly in RMS13-R cells than in RMS13 cells (Fig. 2G; Fig. S2F). These data indicate that RMS13-R cells mount a greater autophagic response. Moreover, combined nutrient deprivation and CQ addition established that RMS13-R cells respond to nutrient deprivation and that autophagic flux is also higher in these cells (Fig. 2G, ‘EBSS/CQ’). Finally, we quantified the levels of select transcripts after starvation and found that the RMS13-R line harbored greater amounts of autophagy-associated messages (Fig. S2G). Together, these observations suggest that enhanced levels of basal autophagy mediate RMS13-R survival in the presence of an Hsp70 inhibitor.
RMS13-R cells adapt to Hsp70 inhibition by inducing autophagy
In contrast to the chronic conditions under which the RMS13-R cells were initially selected (Sabnis et al., 2016), we were curious as to whether RMS13-R cells mount a more-profound immediate autophagic response when challenged with MAL3-101. Positive results from this experiment would suggest that the proteostatic machinery had been rewired to minimize both short- and long-term stress as a result of compromised chaperone activity. Therefore, we next analyzed the transcriptional response of RMS13-R and RMS13 cells during a 6-h MAL3-101 treatment. RMS13-R cells significantly induced the expression of messages encoding proteins that act during phagosome elongation compared to RMS13 cells (Fig. 3A). We also found that the magnitude of induction in the RMS13-R cells in response to MAL3-101 and MG132 was similar (Fig. S3A), suggesting that autophagy-mediated degradation overcomes compromised proteasome activity to maintain proteostasis. In contrast, tubacin decreased the levels of the transcripts, perhaps consistent with the fact that this compound inhibits autophagy progression.
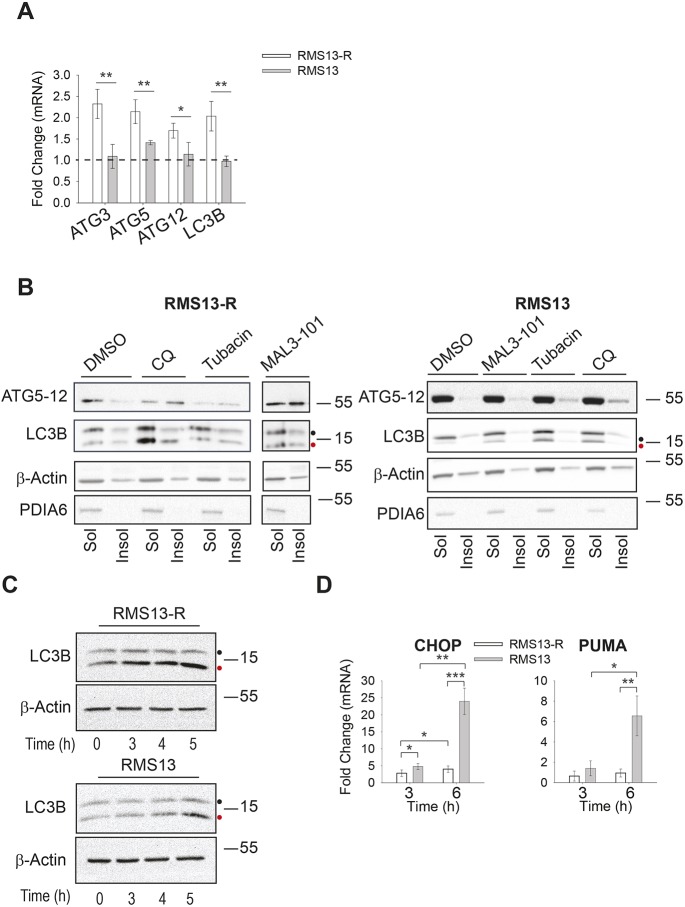
Fig. 3.
Hsp70 inhibition induces autophagy in RMS13-R cells. (A) The mean±s.d. relative level of autophagy gene expression was detected by qPCR in the presence of DMSO or 7.5 µM MAL3-101 for 6 h in RMS13-R (white) and RMS13 (gray) cells (n=4). *P<0.05, **P<0.005. (B) The amount of the indicated soluble (Sol) and membrane-associated (Insol) proteins was examined by immunoblotting after treating cells with 50 µM CQ, 11 µM tubacin or 7.5 µM MAL3-101 for 6 h. PDIA6, a soluble ER-resident chaperone, was used as a control. (C) RMS13-R and RMS13 cells were treated for the indicated times with 7.5 µM MAL3-101 and CQ was added during the last hour of the treatment to test autophagic flux. Therefore, time ‘0’ indicates a 4 h treatment with DMSO plus a 1 h treatment with CQ. Aliquots of cell lysates were resolved by SDS-PAGE and immunoblotted for LC3B as a marker for autophagic flux. LC3BI is indicated by a black dot and the autophagosome-associated form LC3BII is highlighted with a red dot. (D) RMS13-R or RMS13 cells were treated for 3 or 6 h with 7.5 µM MAL3-101 and the expression of the pro-apoptotic CHOP and PUMA genes was measure by qPCR relative to the DMSO control as the mean±s.d. (n=3). *P<0.05, **P<0.001, ***P<0.0001.
To measure whether enhanced autophagy in the presence of MAL3-101 could also be detected at the protein level, both detergent-insoluble (membrane associated; Insol) and soluble (Sol) proteins were isolated after DMSO or MAL3-101 treatment for 6 h. Previous work has shown that NP40 insoluble fractions contain components of the phagosome as well as autophagy receptor-protein aggregates, and LC3B and p62 abundance correlated with autophagy induction (Milan et al., 2015; Tanida et al., 2008). As a control, cells were treated with CQ and tubacin. Both ATG5-12 and LC3BII were enriched in the insoluble fraction of RMS13-R cells incubated with MAL3-101 compared to RMS13 cells under the same conditions, and LC3BI to LC3BII conversion was also higher in the resistant cells (Fig. 3B; Fig. S3B, compare black bars). Consistent with the fact that CQ blocks the final step in autophagy, the amount of LC3BII and the ATG5–ATG12 complex in the insoluble fraction of MAL3-101-treated RMS13-R cells was lower compared to the amount detected in CQ-treated cells. This result would be anticipated if Hsp70 inhibition induces autophagy, since LC3BII is degraded as the autophagolysosome is formed. This result also strongly suggests that Hsp70 inhibition directly induces autophagy rather than simply favoring the accumulation of autophagy-related markers. Furthermore, even though neither autophagy marker was significantly enriched in the membrane-associated fraction from RMS13 cells after MAL3-101 treatment, the use of CQ confirmed that RMS13 cells remain autophagy competent because LC3BII and ATG5–ATG12 accumulate (Fig. 3B; Fig. S3B, compare white and grey bars). Nevertheless, the accumulation of the markers after CQ treatment is greater in the resistant cell line compared to the sensitive one, reinforcing our hypothesis that autophagy is hyperactivated in the resistant cancer cells.
To confirm that MAL3-101 induced autophagy in RMS13-R cells, we acutely inhibited autophagy for 1 h with CQ after Hsp70 activity had been compromised with MAL3-101 for different times (Fig. 3C; Fig. S3C,D). Again, RMS13-R cells exhibited an elevated autophagic flux, as measured by greater amounts of LC3B-II and increased conversion of LC3BI into LC3BII. This was evident in the absence of MAL3-101 (i.e. time 0) but became more apparent with longer incubation periods with the Hsp70 inhibitor. LC3BII accumulation was also quantified as a ratio between CQ-treated and untreated samples to confirm that LC3BII accumulation was not due to impaired lysosomal degradation (Fig. S3D). As expected, LC3BII accumulates to a greater degree in RMS13-R cells and Hsp70 inhibition favors LC3BII accumulation in RMS13-R cells (Fig. S3D). This effect became even more profound over time after MAL3-101 addition.
We also collected RNA from cells treated for 3 or 6 h with MAL3-101 and analyzed levels of ATG3, ATG5 and LC3B mRNA. Quantitative PCR analysis showed that these messages rose only in the RMS13-R line (Fig. S3E). In agreement with our previous data (Sabnis et al., 2016), MAL3-101 treatment also induced CHOP in RMS13 cells but not in the RMS13-R line (Fig. 3D). Because CHOP triggers apoptotic cell death downstream of UPR induction (Marciniak et al., 2004), we quantified transcript levels corresponding to the pro-apoptotic factor, PUMA (also known as BBC3), which is transcriptionally activated by ATF4–CHOP (Galehdar et al., 2010; Matus et al., 2013). Like CHOP, PUMA transcript levels also rose after MAL3-101 treatment (Fig. 3D) in the sensitive but not in the resistant cell line.
Next, we confirmed these results by western blotting. First, downstream induction of apoptosis in the MAL3-101-sensitive cells was clearly evident, as quantified by cleavage of caspase-8, caspase-3 and PARP (Fig. 4A,B). In contrast, LC3BII abundance and LC3BI to LC3BII conversion increased over time in RMS13-R cells, but no significant changes were detected in RMS13 cells under similar conditions. Thus, there is a mutually exclusive relationship between induction of CHOP and apoptosis (in the sensitive cells) and autophagy (in the resistant cells) when the Hsp70 molecular chaperone is inhibited.
Fig. 4.
Hsp70 inhibition induces autophagy but not apoptosis in RMS13-R cells. (A) RMS13-R and RMS13 cells were treated with 7.5 µM MAL3-101 for the indicated times and lysates were prepared and immunoblotted for LC3B, a marker for autophagic flux, and for indicators of apoptosis (PARP, cleaved caspase-8, cleaved caspase-3 and CHOP). (B) The corresponding fold increase of the indicated apoptotic and autophagy associated markers in RMS13-R (black) and RMS13 (white) cells relative to the DMSO control are plotted as the mean±s.e.m. (n=8 for LC3B, CHOP, and cleaved PARP; n=3 for cleaved caspase-3; n=2 for cleaved caspase-8). *P<0.05, **P<0.005, ***P<0.0001.
Higher steady-state levels of basal and MAL3-101-induced autophagy in RMS13-R cells should be reflected by an accumulation of autophagic vesicles. To test this hypothesis, we incubated RMS13-R and RMS13 cells in the presence or absence of MAL3-101 and performed thin section electron microscopy. A 4 h time point was chosen, since both autophagy and apoptosis were apparent in both cell lines (see above and data not shown). The cells were also treated with CQ, which served as a positive control. RMS13-R cells possessed both a greater number and larger size of vacuolar structures (Fig. 5, arrowhead), even in the presence of DMSO. After Hsp70 inhibition, dense material was present in the nuclei (‘N’) of RMS13 cells, consistent with an apoptotic response. In contrast, the nuclei in the RMS13-R line after MAL3-101 treatment exhibited wild-type morphology (data not shown) but the cells instead possessed electron-dense and empty autophagic structures. A small number of vacuolar structures also accumulated in CQ-treated RMS13 cells. However, this phenomenon was more striking in RMS13-R cells, further establishing that RMS13-R cells coopt the autophagy pathway to adapt to compromised Hsp70 function.
Fig. 5.
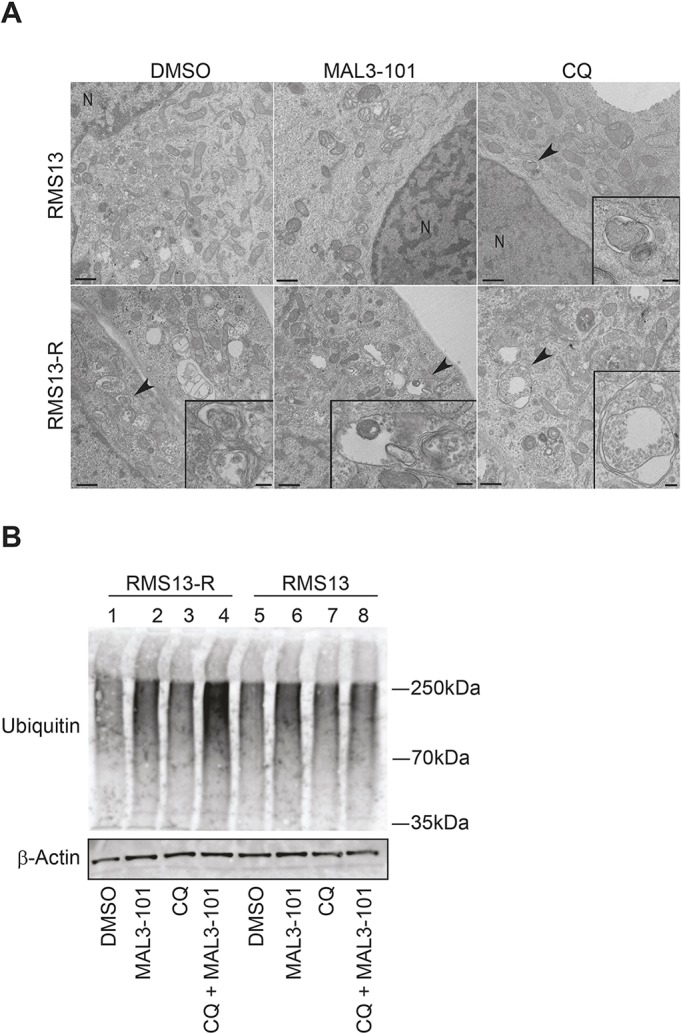
RMS13-R cells show evidence of constitutive autophagy. (A) Electron microscopy analysis was conducted on RMS13 and RMS13-R cells treated with DMSO, 7.5 µM MAL3-101, or 50 µM CQ for 4 h. Insets (areas indicated by black arrowheads) under high magnification are shown in the bottom right and provide evidence of autophagosomes or autophagic structures. Of note, heterochromatin in the nucleus (indicated by ‘N’) as well as disrupted mitochondrial morphology are visible upon MAL3-101 treatment in RMS13 cells. Scale bars: 200 nm. (B) RMS13-R and RMS13 cells were treated with DMSO, MAL3-101, CQ or MAL3-101/CQ for 4-h, lysates were prepared, and cellular protein was immunoblotted for total ubiquitylated proteins. When CQ was administrated together with MAL3-101 to RMS13-R cells, ubiquitylated protein content increased ∼3-fold compared to the DMSO control.
To investigate whether the combined addition of MAL3-101 and CQ led to an accumulation of unfolded proteins, we prepared lysates and performed an immunoblot analysis. Total ubiquitylated protein levels in the RMS13 and RMS13-R cells were also measured in the presence or absence of MAL3-101 and/or CQ (Fig. 5B). While steady-state levels of ubiquitylated proteins were similar in the two cell lines (lanes 1 and 5), Hsp70 inhibition slightly enhanced the accumulation of polyubiquitylated protein (compare lane 1 to 2 and 5 to 6). However, only when both Hsp70 and autophagy were inhibited was the accumulation of polyubiquitylated proteins substantially higher (lane 4), especially in RMS13-R cells. These results are consistent with autophagy playing a compensatory role to reduce toxic protein accumulation in the absence of Hsp70 function. In contrast, polyubiquitylated protein levels were unchanged in the presence of MAL3-101 and CQ in RMS13 cells (compare lanes 5 to 8).
Genetic inhibition of autophagy induces RMS13-R cell death in the presence of MAL3-101
To establish that autophagy inhibition sensitizes resistant cells to an Hsp70 inhibitor (Fig. 2), we silenced ATG5, which was induced at both the mRNA and protein levels in response to MAL3-101 (Fig. 3; Fig. S3). ATG5 facilitates elongation, and together with ATG12 and ATG16L, helps link LC3BII to the growing autophagosomal membrane (Cuervo, 2004; Mizushima and Klionsky, 2007; Mizushima et al., 2011). Two different oligonucleotides directed against ATG5 were transfected into RMS13-R and RMS13 cells, and knockdown efficiency was measured after 48 and 72 h (Fig. S4A; data not shown). We found that ∼25% of the ATG5–ATG12 complex remained after 72 h, but Hsp72 (also known as HSPA1A) – the major stress-inducible Hsp70 – was unaffected (data not shown). We also found that the ATG5 transcript was present at ∼20% of the control (scrambled siRNA; Fig. S4B). Previous work demonstrated that LC3BII abundance decreased after ATG5 knockdown or knockout due to reduced LC3BI to LC3BII conversion (Mizushima et al., 2001; Nishiyama et al., 2007). Therefore, we similarly measured LC3BI to LC3BII conversion as well as LC3BII abundance. In accordance with prior results, ATG5 silencing decreased LC3BI conversion and steady-state levels of LC3BII in both cell lines (Fig. S4A).
Next, we investigated whether ATG5 knockdown re-sensitized RMS13-R cells to MAL3-101. We first confirmed that the viability of RMS13-R cells was only modestly affected by MAL3-101, whereas the RMS13 cells were significantly more sensitive when both cells were treated with the control siRNA (Fig. 6A). Each cell line was then treated with the ATG5 siRNA (or the control), incubated with increasing doses of MAL3-101 for 24 h, and viability was measured. In agreement with data obtained using small-molecule autophagy inhibitors (Fig. 2D; Fig. S2B,C), knockdown with two different oligonucleotides re-sensitized the resistant line to MAL3-101, but a more modest effect was observed in the sensitive cells (Fig. 6B).
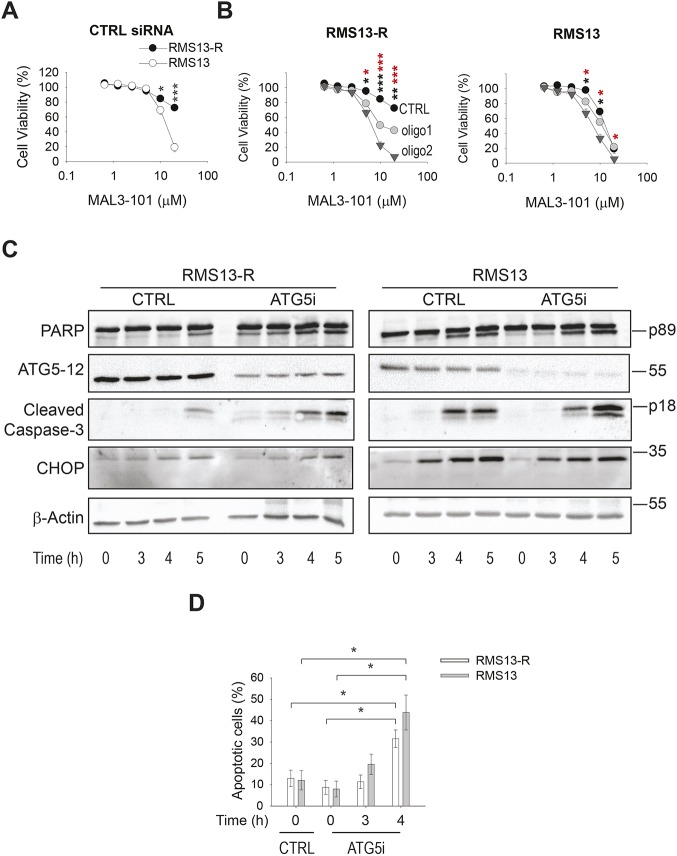
Fig. 6.
Autophagy inhibition induces MAL3-101-dependent cell death in RMS13-R cells. (A) RMS13-R and RMS13 cells were transfected with a control siRNA (CTRL), seeded into a 96-well plate, and treated with increasing doses of MAL3-101 for 24 h. Viability was assessed 72 h after transfection with the CellTiter-Glo assay. The mean±s.e.m. of three independent experiments are shown. (B) RMS13-R and RMS13 cells were transfected with a control siRNA (black) or with 2 different siRNA oligonucleotides directed against ATG5 (gray circles and inverted triangles). The mean±s.e.m. of three independent experiments are shown. Black asterisks correspond to the statistical significance between the control and oligonucleotide 1, and red asterisks represent the statistical significance between the control and oligonucleotide 2. (C) RMS13-R and RMS13 cells were transfected with a control siRNA or a mixture of the oligonucleotide 1 and oligonucleotide 2 ATG5 siRNAs and 70 h after transfection the cells were treated with 7.5 µM MAL3-101 for the indicated times. Lysates from cells prepared at each time point were immunoblotted for cleaved PARP, ATG5-12, cleaved caspase-3, and CHOP to monitor apoptotic induction. (D) RMS13 (gray) and RMS13-R (white) cells were transfected with control or a mixture of siRNA oligonucleotides 1 and 2 directed against ATG5, and 70 h after transfection the cells were treated with 7.5 µM MAL3-101 or with DMSO for the indicated times. Cells from each time point were stained for annexin-V and PI to monitor apoptosis. The sum of annexin-V-positive and annexin-V and PI double-positive cells is represented in the graph as the percentage of apoptotic cells. The mean±the range of the data are shown for two independent experiments. *P<0.05, **P<0.005, ***P<0.0001.
To determine whether apoptosis was induced, cells were transfected with the control siRNA or a mixture of the two ATG5-directed oligonucleotides, and, at 70 h post-transfection 7.5 µM MAL3-101 was added. As shown in Fig. 6C, cleaved PARP and caspase-3 accumulated to a greater extent in RMS13-R cells in the silenced cells (ATG5i), consistent with the fact that diminished autophagy re-sensitizes the resistant line to MAL3-101 (left panels). Also as anticipated, RMS13 cells exhibited a similar timecourse of PARP and caspase-3 cleavage after MAL3-101 addition, regardless of whether ATG5 had been silenced (right panels). Interestingly, there was no increased accumulation of CHOP in the ATG5-silenced RMS13-R cells compared to the RMS13 cells, suggesting that ATG5 knockdown is insufficient to increase CHOP expression, or possibly that induction of the CHOP protein (but not the mRNA; see Fig. 3D) requires a longer time frame after MAL3-101 addition. In fact, translation of the CHOP message is regulated via its 5′UTR, which contains an upstream open reading frame (uORF) that limits ribosome binding and inhibits expression of the downstream ORF (Chen et al., 2010; Jousse et al., 2001).
To confirm that an apoptotic response had been initiated, we then performed annexin-V–propidium iodide (PI) staining (Koopman et al., 1994; Vermes et al., 1995) and analyzed the percentage of apoptotic cells in the presence or absence of ATG5 knockdown and in the presence or absence of MAL3-101 (Fig. 6D). Although ATG5 silencing had no effect on viability in either cell line, time-dependent Hsp70 inhibition under conditions of impaired autophagy (ATG5i) increased the percentage of apoptotic cells in RMS13 and RMS13-R cells. In contrast, Hsp70 inhibition had no effect on cell viability in RMS13-R cells treated with control oligonucleotides (data not shown). These data confirm that RMS cancer cells are significantly more susceptible to Hsp70 inhibition when the autophagy pathway is compromised, and that combined application of Hsp70 and autophagy inhibitors synergistically compromises cell viability.
Finally, we examined whether MAL3-101 induced autophagy-associated genes in an animal model, as shown in the RMS13-R and (to a lesser extent) in the RMS13 cells (Fig. S3). To this end, 55 mg/kg body weight of MAL3-101 was introduced intraperitoneally into mice and whole liver was isolated after 4, 8 and 24 h. Western blot analysis showed increased LC3BII and conversion of LC3BI into LC3BII in the MAL3-101-treated animals compared to those treated with the vehicle, especially at the 4-h time point (Fig. S5A,B). After 24 h, the autophagic response returned to the baseline. Based on a quantitative (q)PCR analysis, increased ATG5, ATG3 and LC3B transcripts were also detected at these time points (data not shown). These data are also consistent with the observed pharmacokinetics of MAL3-101, which demonstrate early peak concentrations that are sustained over 8 h and fall to pre-dose levels by 24 h (data not shown). Because we previously reported that RMS cells respond to MAL3-101 treatment by upregulating BiP and CHOP (Sabnis et al., 2016), we then measured the levels of BiP and CHOP mRNA (Fig. S5C). Messages encoding both proteins accumulated 8 h after MAL3-101 injection, thereby indicating a comparable cellular response to MAL3-101 treatment between RMS cell lines and in vivo. To confirm the synergistic effect of autophagy inhibition with MAL3-101 in another cancer model, a colony formation assay was next performed with RMS13-R and RMS13 cells (Fig. S5D), and we discovered that MAL3-101-treated RMS13 cells form fewer colonies than RMS13-R cells. When MAL3-101 was added in combination with CQ, the number of RMS13-R colonies declined dramatically, confirming the compensatory role of autophagy in RMS13-R cell line survival.
PERK is activated upon Hsp70 inhibition
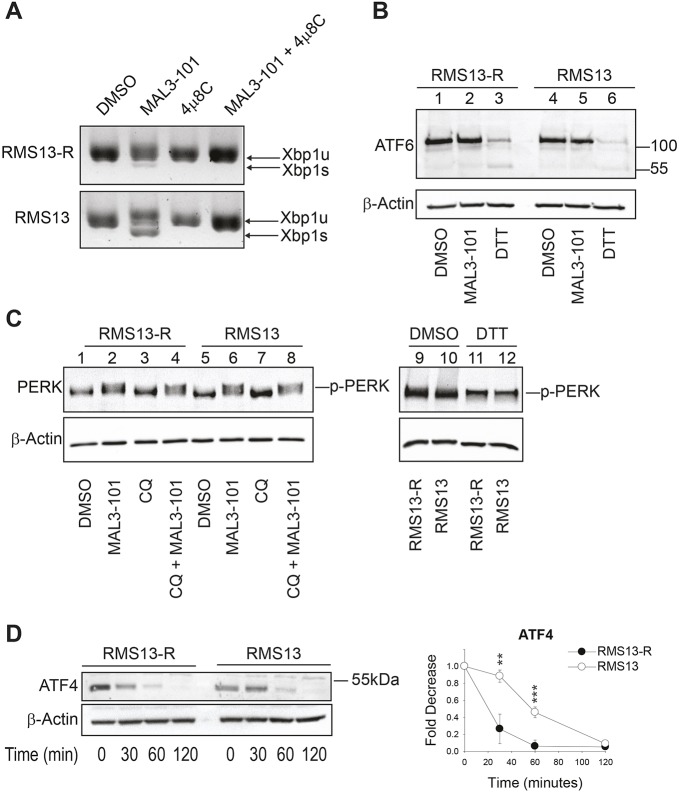
A consequence of unfolded protein accumulation is the induction of one or more of the three UPR branches (Bakunts et al., 2017; Walter and Ron, 2011). To dissect the contribution of each UPR branch when Hsp70 was inhibited, we first monitored XbpI splicing (as indicated by the presence of the spliced form, denoted XbpIs) as a readout of IRE1 endonuclease activity (Walter and Ron, 2011; Yoshida et al., 2001a) (Fig. 7A). The appearance of a higher mobility band after MAL3-101 treatment indicated a modest but clear induction of IRE1 in RMS13 but not RMS13-R cells. As a control, XbpIs accumulation was blocked in the presence of the IRE1 inhibitor 4µ8C (Stewart et al., 2017). Second, we monitored ATF6 cleavage in the presence of MAL3-101 in RMS13-R and RMS13 cells. Dithiothreitol (DTT), a compound that reduces disulfide bonds, was used as a positive control (Walter and Ron, 2011). As shown in Fig. 7B, the ATF6 cleaved isoform was absent in the presence of MAL3-101 in both the cell lines, indicating that ATF6 does not participate in the MAL3-101-driven cellular response. Next, we confirmed that PERK was induced by MAL3-101 in RMS13-R and RMS13 cells. PERK activation was detected both when cells were treated with MAL3-101 alone or in combination with CQ (Fig. 7C, compare lanes 1 to 2 and 1 to 4 for RMS13-R, and lanes 5 to 6 and 5 to 8 for RMS13).
Fig. 7.
The PERK branch of the UPR is activated in response to MAL3-101. (A) RMS13-R and RMS13 cells were treated with MAL3-101 for 4 h in the presence or absence of the IRE1 inhibitor 4µ8C and the accumulation of the unspliced and spliced XbpI (XbpIu and XbpIs) mRNAs were monitored by PCR. (B) RMS13-R and RMS13 cells were treated with MAL3-101 to detect the full-length and cleaved forms of ATF6. A 1 h DTT treatment was used as a positive control. (C) RMS13-R and RMS13 cells were treated with the indicated compounds or with DMSO for 4 h and PERK phosphorylation (p-PERK) was monitored by immunoblotting. p-PERK accumulation is detected upon MAL3-101 or MAL3-101/CQ treatment. A 1 h DTT treatment was used as a positive control. (D) ATF4 is degraded more rapidly in RMS13-R cells. The RMS13-R and RMS13 cells were pre-treated with MAL3-101 for 3 h and then cycloheximide was added for the indicated times. Lysates were resolved by SDS-PAGE and immunoblotted for ATF4. Graphs represent the mean± s.d. of three independent experiments. **P<0.005, ***P<0.0001.
If MAL3-101 induces the same stress response (i.e. PERK induction) in MAL3-101-sensitive and resistant cells, the final outcomes are unique: autophagy is activated in RMS13-R cells and CHOP-mediated cell death arises in RMS13 cells. Interestingly, ATF4 alone, or in combination with CHOP, has been reported to induce transcription of ATG mRNA, which then activates autophagy (B'Chir et al., 2013; Clarke et al., 2014; Rouschop et al., 2010; Rzymski et al., 2009, 2010). Thus, to determine whether ATF4 is differentially regulated, we measured ATF4 stability after a 3-h MAL3-101 treatment in a cycloheximide chase assay (Fig. 7D). We noted that ATF4 was degraded more efficiently in RMS13-R cells, suggesting that heightened ATF4 turnover may favor the induction of ATG genes rather than the activation of an apoptotic response.
DISCUSSION
Understanding how cancer cells evolve and escape death is critical to identify new methods to prevent therapeutic resistance, metastasis and relapse. Here, we analyzed the pro-survival roles of two cellular protein degradation pathways – ERAD and autophagy – in a cancer model. To define how these components of the cellular proteostasis network limit proteotoxicity, we examined RMS cells that were either sensitive (RMS13) or resistant (RMS13-R) to the specific Hsp70 inhibitor MAL3-101, which has been widely used in other cancer models (Adam et al., 2014; Braunstein et al., 2011; Rodina et al., 2007; Sabnis et al., 2016). We also studied how these cells respond to Hsp70 inhibition since this chaperone prevents apoptosis (Daugaard et al., 2007; Garrido et al., 2006; Guo et al., 2005; Mosser and Morimoto, 2004; Powers et al., 2008; Sliutz et al., 1996; Yang et al., 2012; Young et al., 2004) and may limit the efficacy of inhibitors targeting the Hsp90 family in clinical trials (Gabai et al., 2005; Guo et al., 2005; McCollum et al., 2006; Powers et al., 2008; Voellmy and Boellmann, 2007). Discovering potential Hsp70-based therapies, as well as defining the effect of Hsp70 inhibition on cancer cell proteostasis and survival are critical undertakings.
We previously showed that MAL3-101 induces the PERK branch of the UPR, thus triggering CHOP-mediated apoptosis (Sabnis et al., 2016). We now report that PERK is primarily induced in both RMS13-R and RMS13 cells (Fig. 7C), suggesting a crucial role of PERK and its downstream effectors in the MAL3-101-derived cellular response. We also present evidence that two protein degradation networks, ERAD and autophagy, are more active in resistant cells. RMS13-R cells exhibit a heightened ability to degrade both soluble and integral membrane ERAD substrates. To our knowledge, this is the first study in which ERAD efficiency has been addressed by specifically measuring this pathway in different cancer cells. The compensatory network that leads to increased ERAD in the RMS13-R cells is not clear, but a dissection of the factors and mechanisms that augment ERAD efficiency in RMS13-R cells is in progress.
Not only is ERAD activity higher in Hsp70 inhibitor-resistant cells, but autophagy is also more efficient. This conclusion is supported by the findings showing increased levels of the mRNAs and proteins linked to autophagosome elongation, and LC3BII conjugation to the autophagosomal membrane as well as by electron microscopy. The pro-survival activity of the autophagy pathway in select cancers has been established (Clarke et al., 2012; Hambright and Ghosh, 2017; Jiang and Mizushima, 2014; Levine and Kroemer, 2008; Maycotte and Thorburn, 2014), and, consistent with our results, rhabdomyosarcomas also coopt this pro-survival pathway (Peron et al., 2012; Rapino et al., 2014). Surprisingly, inhibition of autophagy, but not ERAD, re-sensitized RMS13-R cells to MAL3-101. We suggest that RMS13-R cells become refractory to the proteotoxic stress that arises after Hsp70 inhibition because autophagy is induced. By analogy, some cancer cells compensate for proteasome inhibition by inducing autophagy (Liu et al., 2013, 2016; Rapino et al., 2014; Wojcik, 2013). Our data support ongoing efforts to develop autophagy inhibitors as adjuncts to therapy in a variety of cancers (Carew et al., 2017; Cufi et al., 2013; Gómez et al., 2015; Jain et al., 2013; Maycotte and Thorburn, 2014).
The negligible effect of proteasome and ERAD inhibition in the RMS model is consistent with the inability of proteasome inhibitors to kill solid tumors (Chen et al., 2011; Deshaies, 2014; Le Moigne et al., 2017), yet these results also seem to contradict the role of protein degradation pathways in cancer cell survival (Mei et al., 2013; Rubiolo et al., 2014; Vandewynckel et al., 2013; Yan et al., 2015). In general, cancer cells survive stressful conditions by upregulating protein degradation networks (Clarke et al., 2011; Goldberg, 2003; Kim et al., 2013, 2015; Ma et al., 2014; Schonthal, 2012a,b). What is less clear – and can only be determined empirically at this point – is an understanding of which protein degradation pathway within the network is critical for cell survival.
Based on our results, we propose a model (Fig. 8) where Hsp70 inhibition increases protein unfolding, favoring accumulation of proteasome and autophagy substrates that trigger PERK induction. We reason that ATF4 and CHOP, both of which are activated by PERK, induce ATG5 and LC3B transcription when Hsp70 is inhibited and increase autophagy efficiency. In line with this model, ATF4 degradation is faster after acute MAL3-101 treatment in the resistant cells, suggesting that ATF4 stability regulates the life-and-death ‘decision’ after MAL3-101-dependent PERK induction (Fig. 7D). When autophagy fails to compensate for Hsp70 inhibition, unfolded proteins and ATF4 and CHOP accumulation induce cancer cell death via the apoptotic pathway. In summary, our increased understanding of how cancer cells compensate for Hsp70 inhibition demonstrates how different degradation pathways are integrated in cancer cells and almost certainly many other cell types. Further chemical optimization of Hsp70 inhibitors will also augment progress on the development of combination therapies that include autophagy inhibitors for the treatment of a range of cancers, including rhabdomyosarcomas.
Fig. 8.
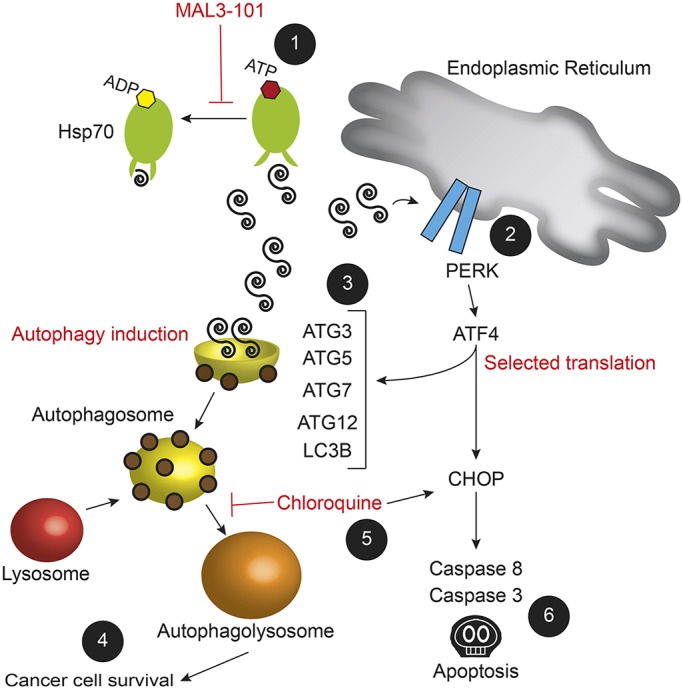
A model for cancer cell adaptation to Hsp70 inhibition. MAL3-101 treatment of RMS13 cells inhibits Hsp70 activity and induces (1) the accumulation of unfolded proteins that (2) lead to PERK-mediated transcription of ATF4 and CHOP, which (6) results in cell death (Sabnis et al., 2016). In contrast, (3) higher steady state levels of autophagy and perhaps an ATF4-mediated increase in ATG gene expression upon MAL3-101 treatment in RMS13-R cells (4) protects against cell death and favors cancer cell survival. If, however, autophagy is impaired [for example by CQ (5)], (6) Hsp70 inhibition is again toxic to MAL3-101-resistant cancer cells, which results in apoptosis.
MATERIALS AND METHODS
Cell culture, chemicals and transfection
RMS13 cells were purchased from ATCC and authenticated, and RMS13-R cells were generated by clonal derivation from the parental RMS13 cells after culture in escalating doses of MAL3-101 (Sabnis et al., 2016). In brief, to derive the MAL3-101-resistant cell lines, RMS13 cells were plated at 200,000 cells/well in a in six-well plate and treated with progressively increasing doses of MAL3-101 starting from 500 nM up to 10 μM, with interval splitting to avoid over-confluence. Once a resistant polyclonal population of cells was obtained, the cells were trypsinized and resuspended at 5000 cells/30 ml medium in a 15-cm dish with 3 μM MAL3-101. After 2 weeks, single-cell colonies were isolated using autoclaved vacuum grease and glass cloning cylinders (Sigma-Aldrich, Saint Louis, MO), trypsinized and expanded. Resistance to MAL3-101 in these single-cell clonal populations was then reconfirmed using CellTiter-Glo assays, as described above. Both RMS13 and the resistant clones were grown at 37°C and 5% CO2 in RPMI-1640 medium (GE Healthcare Hyclone, Logan, UT) supplemented with 10% FBS (GE Healthcare Hyclone), 1× penicillin/streptomycin (Gibco, Thermo Fisher Scientific, Waltham, MA), 10 mM HEPES pH 7.5 (MP Biomedicals, Solon, OH), 0.25% glucose (TEKNOVA, Hollister, CA) and 1 mM sodium pyruvate (Thermo Fisher Scientific, Waltham, MA) (complete medium). To induce autophagy, cells were starved for 4 h in Earle's balanced salt solution (EBSS, Thermo Fisher Scientific) containing 1% FBS, after any residual complete medium was removed with PBS.
MAL3-101 (Fewell et al., 2004), tubacin (Sigma-Aldrich, Saint Louis, MO), bafilomycin, MG132 and CB-5082 (Selleck Chemicals, Houston, TX) were dissolved in DMSO at 20 mM, except bafilomycin, which was prepared at 1 mM. Chloroquine (CQ; Sigma-Aldrich) was dissolved in sterile water at 100 mM. All compounds were stored at −80°C, except MG132, which was kept at −20°C. Prior to use, the desired amount of each compound was added to pre-warmed medium, mixed thoroughly, and added directly onto cells.
To transfect plasmids encoding ERAD substrates (Christianson et al., 2011), cells were seeded at a density of 200,000 cells/well in six-well plates, and after 24 h a total of 2 µg of the indicated plasmid was introduced using the FuGENE6 reagent (Promega, Madison, WI) according to the manufacturer's instructions. Cells were treated with the indicated compounds or directly processed for RNA or protein extraction (see below) 24 h after transfection. In experiments in which ATG5 was silenced, cells were seeded at a density of 250,000 in a six-well plate, and after 24 h two different siRNA oligonucleotides against ATG5 (siGENOME9474, D-004374-03 and D-004374-05, Dharmacon, Lafayette, CO) or a control siRNA (BLOCK-iT™ Alexa Fluor® Red Fluorescent Control, Invitrogen, Thermo Fisher Scientific) were transfected using Lipofectamine RNAiMAX (Invitrogen, Thermo Fisher Scientific) at 20 nM according to the manufacturer's instructions. After 72 h, cells were harvested and seeded for cell viability assays or collected for RNA and protein extraction to detect knockdown efficiency, as described below. The time frame and concentration of siRNA-mediated silencing were selected after conducting timecourse experiments in which ATG5–ATG12 and Hsp70 protein abundance was analyzed by immunoblotting (data not shown).
Cell viability assays
Cells were plated at a density of 3000 cells/100 μl in 96-well clear-bottomed plates (Greiner bio-one, NC). After 72 h of the indicated treatment, the cells were lysed and incubated with the CellTiter-Glo reagent (Promega), and luminescence was read on a Bio-Rad ChemiDoc XRS+ with the associated Image Lab software (Bio-Rad, Thermo Fisher Scientific). Under conditions in which cells were treated with more than one compound (e.g. Fig. 2C,D; Fig. S2B,C,E), the cells were incubated with increasing doses of MAL3-101 in the presence of 0.5 µM CB-5083, 40 µM CQ, 2.75 µM tubacin or 1.5 nM bafilomycin. These doses were selected so that no greater than 30% cell death was observed in the RMS13 and RMS13-R cell lines 72 h after treatment at these concentrations. Viability was quantified after normalizing the values obtained upon CB-5083, CQ, tubacin, or bafilomycin treatment alone at the indicated concentrations. When viability was measured after ATG5 knockdown, MAL3-101 sensitivity was measured 24 h after treatment and 72 h after siRNA transfection.
Analysis of cell surface annexin-V was performed by staining cells with the annexin-V apoptosis detection kit (eBioscience, Thermo Fisher Scientific) following the manufacturer's instructions. In brief, 48 h after siRNA transfection (see above) cells were seeded at a density of 200,000 cells in six-well plates and allowed to adhere overnight before performing the indicated treatments. After mild trypsinization, the cells were harvested and washed in PBS and then equilibrated in annexin-V-binding buffer. A cell resuspension of 150,000 cells in 100 µl was incubated with 5 µl of annexin-V conjugated to allophycocyanin (APC) for 15 min in the dark. After three washes with the binding buffer, cells were suspended in 400 µl annexin-V-binding buffer containing 5 µl propidium iodide (PI) and analyzed on an Accuri C6 FACS apparatus (BD-Biosciences, San Diego). Staining was used to discriminate annexin-V and PI-positive cells, respectively. Annexin-V and annexin-V and PI double-positive cells were summed and represented in the graph as percentage of apoptotic cells.
Immunoblot analysis
All antibodies used in this study were obtained from Cell Signaling Technology (Danvers, MA), unless indicated otherwise. To measure the levels of endogenous proteins, cells were plated at a density of 200,000 cells in six-well plates and allowed to adhere overnight before the indicated treatments. The supernatant/medium was collected, and after the cells were washed in PBS lysates were obtained after incubation in 1% SDS RIPA buffer (100 mM Tris-HCl pH 7.5, 1% NP40, 1% SDS, 300 mM NaCl and 0.5% sodium deoxycholate) supplemented with protease inhibitors (cOmplete Mini EDTA free tablets; Roche, Indianapolis, IN), 10 mM N-ethylmaleimide (NEM; Sigma-Aldrich) and 5 mM phenylmethanesulfonyl fluoride (PMSF; Sigma-Aldrich) for 15 min on ice. The lysate was then clarified by sonication and centrifugation at 13,000 g for 5 min at 4°C, and the amount of protein was quantified with a BCA assay kit (Thermo Fisher Scientific). Aliquots containing 25 µg of total protein were incubated at 95°C for 5 min, subjected to SDS-PAGE, and, after transfer, the blots were incubated with the following antibodies: anti-LC3B (D11, #3868S; at 1:1000), anti-ATG5 (D5F5 U, #12994S; at 1:1000), anti-p62/SQSTM1 (P0067, Sigma-Aldrich; at 1:2000) and anti-PERK (C33E10, #3192; at 1:1000) antibodies. When apoptotic marker proteins were examined, aliquots from the same lysates were instead heated to 75°C for 15 min prior to SDS-PAGE, and blots were incubated with anti-PARP (46D11, #9532S; at 1:1000), anti-CHOP (L63FZ, #2895S; at 1:1000), anti-cleaved caspase-3 (#9661S; at 1:500), and anti-cleaved caspase-8 (18C8, #9496; at 1:1000) antibodies. Anti-β-actin antibody was used as loading control (#4867L; at 1:2000). To measure LC3BII accumulation and LC3BI to LC3BII conversion in murine livers, 30 mg of tissue was sonicated on ice for 30 s in 1% SDS RIPA buffer supplemented with protease inhibitors (cOmplete Mini EDTA free tablets; Roche, Indianapolis, IN), 10 mM N-ethylmaleimide (NEM; Sigma-Aldrich), and 5 mM phenylmethanesulfonyl fluoride (PMSF; Sigma-Aldrich). After protein quantification, 40 µg of total protein was subjected to SDS-PAGE as described above.
To measure the levels of transfected HA-tagged proteins, 24 h after transfection (see above) the cells were treated for 4 h with 20 µM MG132 or an equivalent volume of DMSO. The cells were then washed in PBS and incubated in RIPA buffer (50 mM Tris-HCl pH 7.5, 1% NP40, 150 mM NaCl, 0.5% sodium deoxycholate and 0.1% SDS) supplemented with protease inhibitors (see above) and 10 mM NEM for 20 min on ice. Next, the samples were passed through a 20G syringe (BD/Fisher) five times and the mixture was centrifuged at 13,000 g for 15 min at 4°C. The supernatant fractions were collected and protein quantification was assessed as described above. Aliquots containing 20 μg were heated at 75°C for 15 min and subjected to SDS-PAGE. After blotting, rat monoclonal anti-HA antibody conjugated to horseradish peroxidase (HRP; 3F10; Roche) was used to detect the overexpressed protein of interest at a final dilution of 1:4000. Primary antibodies were visualized with HRP-conjugated anti-mouse-IgG secondary antibodies at 1:4000 for 2 h at room temperature.
To detect levels of protein ubiquitylation, 10 µg of total protein was subject to SDS-PAGE, and the resulting nitrocellulose membranes were incubated in boiling water for 1 h to expose antibody epitopes before the blots were blocked in a milk solution. An anti-ubiquitin antibody, P4D1 (Santa Cruz Biotechnology, Dallas, TX), was used at 1:1000, and HRP-conjugated anti-rabbit-IgG secondary antibodies (at 1:4000) were applied for 2 h at room temperature prior to imaging. In all cases, proteins were visualized using the SuperSignal Chemiluminescence kit (Thermo Scientific) and images were taken using a Bio-Rad ChemiDoc XRS+ with Image Lab software. Data were analyzed using ImageJ software.
To detect inactive and activated ATF6, cells were plated at a density of 200,000 in six-well plates and allowed to adhere overnight. Next, the cells were treated as indicated and 1 mM DTT was applied for 1 h as a positive control. All the treatments were performed in the presence of 5 µM MG132 to avoid ATF6 degradation (Horimoto et al., 2013). The supernatant/medium was then collected, and after the cells were washed in PBS containing 5 µM MG132, the cells were detached with trypsin, and after centrifugation the cell pellets were resuspended in 150 mM NaCl, 10 mM Tris-HCl pH 7.5, 10 µM MG132 and 2% SDS buffer, supplemented with protease inhibitors (cOmplete Mini EDTA free tablets; Roche, Indianapolis, IN), 10 mM N-ethylmaleimide (NEM; Sigma-Aldrich), and 5 mM phenylmethanesulfonyl fluoride (PMSF; Sigma-Aldrich). The lysate was then clarified by sonication and centrifugation at 13,000 g for 5 min at 4°C, and protein was quantified with the BCA assay kit as described above. 50 µg of total protein was incubated at 95°C for 5 min, subjected to SDS-PAGE using a 8% polyacrylamide gel (Acrylamide:Bis-acrylamide, 29:1, 40% Solution, OmniPur, Sigma-Millipore, Germany) and the resulting nitrocellulose filter was incubated in Tris-buffered saline with 0.1% Tween® 20 (TBST) containing 10% non-fat dry milk for 1 h at room temperature under gentle shaking. The nitrocellulose filter was then incubated with an aliquot of anti-ATF6 antibody (73-500, Bio Academia, Japan; at 1:1000) in presence of 5% non-fat dried milk powder for 1 h at room temperature. An anti-light chain-specific anti mouse IgG monoclonal antibody (115-035-174, Jackson ImmunoResearch, West Grove, PA) was used as the HRP-conjugated secondary antibody at 1:5000 in 1% non-fat milk TBST for 2 h at room temperature. Proteins were visualized using the SuperSignal Chemiluminescence kit (Thermo Scientific) and images were taken using a Bio-Rad ChemiDoc XRS+ with Image Lab software. Data were analyzed using ImageJ software.
Biochemical assays
To measure the stabilities of the indicated proteins, cycloheximide chase assays were performed 24 h after transfection on cells grown in six-well plates (see above). Where indicated cells were pre-incubated with 20 µM MG132 for 3 h prior to the addition of 50 µg/ml cycloheximide in the presence of fresh medium supplemented with MG132 or DMSO. Cells were harvested, lysed in RIPA buffer as indicated above, and the indicated HA-tagged protein substrates were detected after immunoblot analysis.
To measure ATF4 stability, cells were pre-treated with 7.5 µM MAL3-101 for 3 h prior to the addition of 50 µg/ml cycloheximide in fresh medium. Cells were harvested at the indicated times, lysed in RIPA buffer as indicated above, and 25 µg total protein was incubated at 37°C for 30 min and subjected to SDS-PAGE. The nitrocellulose filter was decorated with an anti-ATF4 antibody (D4B8, #11815; at 1:2000) and HRP-conjugated anti-rabbit-IgG secondary antibodies (at 1:4000) for 2 h at room temperature prior to imaging. Protein visualization and data analysis were performed as described above.
To differentiate soluble and membrane-associated proteins, cells were collected 6 h after the indicated treatments and lysed in 150 mM NaCl, 10 mM Tris-HCl pH 7.6 and 1% NP-40, which was supplemented with protease inhibitors (Roche), 10 mM NEM and 1 mM PMSF, for 15 min on ice. Soluble fractions were collected after centrifugation at 14,000 g for 20 min at 4°C, while the insoluble pellet fractions were resuspended in 150 mM NaCl, 10 mM Tris-HCl pH 7.5 and 2% SDS and sonicated to fragment the DNA (Milan et al., 2015). Aliquots of the soluble and insoluble fractions were resolved after SDS-PAGE and subject to immunoblot analysis, as indicated above, with anti-LC3B (D11, #3868S; at 1:1000), anti-ATG5 (D5F5U, #12994S; at 1:1000), anti-PDIA6 (#11432; Abcam, Cambridge, UK; at 1:2000) and anti-β-actin (#4867L; at 1:2000) antibodies.
Antitrypsin secretion assay
To measure A1AT secretion, cells were seeded at 200,000 cells/well in a six-well plate and the following day they were transfected with the indicated plasmid, as described above. At 24 h after transfection, cells were incubated with Opti-MEM (Gibco, Thermo Fisher Scientific) in the presence or absence of 50 µM CQ for 4 h. The secreted material was collected and centrifuged at 850 g for 4 min to remove any residual cells. Then, 10 mM NEM, 5 mM PMSF and a protease inhibitor cocktail (see above) were added to the secreted pool. The attached cells were treated as described above and lysed in RIPA buffer (50 mM Tris-HCl pH 7.5, 1% NP40, 150 mM NaCl, 0.5% sodium deoxycholate, 0.1% SDS) supplemented with protease inhibitors and 10 mM NEM for 20 min on ice. 20 µg total protein aliquots were loaded as intracellular material and a corresponding volume of secreted material was subjected to TCA precipitation overnight and then loaded onto the same gel. Rat monoclonal HRP-conjugated anti-HA antibody was used to detect the overexpressed protein of interest (see above).
Real-time quantitative PCR
Cells were seeded at 200,000 cells/well in six-well plates, grown overnight, and then treated for the indicated times with the indicated compounds. RNA was extracted using the RNeasy kit (Qiagen, Hilden, Germany) according to the manufacturer's instructions. cDNA was synthetized from 1 µg of the extracted RNA using MuLV reverse transcriptase (Thermo Fisher Scientific) and 40 ng were used for real-time qPCR using the StepOnePlus system (Applied Biosystems, Thermo Fisher Scientific). All primers are listed in Table S1. Each PCR was run on three or more biological replicates and with three technical replicates for each reaction. Primer efficiency was determined by serial dilution of the template and the relative expression ratios were calculated (Pfaffl, 2001). Primers amplifying β-actin were used as an internal control.
For the murine liver samples (see below for details of mouse handling), RNA was extracted from 20 mg of tissue after sonication using the RNeasy kit (Qiagen, Hilden, Germany), and cDNA was synthesized from 2 µg of the extracted RNA using MuLV reverse transcriptase (Thermo Fisher Scientific). A total of 20 ng of cDNA was used for real-time qPCR using the Quantstudio3 machine (Thermo Fisher Scientific). All primers are listed in Table S2. Each reaction was run on two different murine livers and with three technical replicates for each reaction. Primers directed against β-actin were used as an internal control.
XbpI splicing
RMS13 and RMS13-R cells were seeded at 200,000 cells/well in a six-well plate, grown overnight and, the following day, cells were pre-treated with 25 µM 4µ8C (Selleckchem) or vehicle for 1 h and then treated with either DMSO or 10 µM MAL3-101 for 6 h. RNA was extracted using the RNeasy kit as described above. cDNA was generated from 0.5 µg RNA using the RetroScript Kit (ThermoFisher Scientific). Diluted cDNA was used as a template for PCR amplification of unspliced and spliced Xbp1. DNA products were run on a 3% agarose gel, visualized with ethidium bromide, and an image was captured using a BioRad ChemiDoc XRS+ with Image Lab software.
Proteasome activity assays
Cellular lysates from RMS13 and RMS13-R cells were collected as described previously (Milan et al., 2015), and proteasome activity was assessed by monitoring the production of 7-amino-4-methylcoumarin (AMC) from the Suc-LLVY-AMC proteasome substrate (cat. no. I-1395; Bachem, Torrance, CA), which specifically detects the chymotrypsin-like activity of the proteasome (Stein et al., 1996). To this end, 10 µl aliquots of each lysate were incubated with 1 µl of the 5 mM stock solution of Suc-LLVY-AMC in 20 mM Tris-HCl, pH 7.5, 2 mM ATP, 2 mM MgCl2 and 0.2% bovine serum albumin in the presence or absence of 10 µM MG132 (Gleixner et al., 2017). The fluorescence of released AMC was measured at time 0 and after 30 min in a FluoroMax3 plate reader at an excitation wavelength of 380 nm and an emission wavelength of 460 nm (HORIBA Scientific Instrument & Systems, Kyoto, Japan). To calibrate the assay, a standard free fluorophore solution containing a range of AMC concentrations was used (VWR, Randor, PA). All measurements were performed in duplicate and values were normalized to protein content, as determined by the BCA protein assay kit according to the manufacturer's instructions (Thermo Fisher Scientific). Proteasome activity was calculated by: ((RFU2–RFU1)/(T2-T1))/[protein].
Transmission electron microscopy imaging
Cells were seeded at 200,000 cells/well in a six-well tissue culture plate. After 18 h, DMSO, 7.5 µM MAL3-101 or 40 µM CQ were added for 4 h. The cells were then washed twice with PBS, and fixed in 2.5% glutaraldehyde in 100 mM PBS (8 g/l NaCl, 0.2 g/l KCl, 1.15 g/l Na2HPO4•7H2O, 0.2 g/l KH2PO4, pH 7.4) for 1 h at room temperature. Monolayers were then washed in PBS three times and post-fixed in aqueous 1% osmium tetroxide and 1% Fe6CN3 for 1 h. Next, the fixed cells were washed three times in PBS and dehydrated through a 30–100% ethanol series with Polybed 812-embedding resin (Polysciences, Warrington, PA). The cell mixture was subsequently embedded by inverting Polybed 812-filled BEEM capsules on top of the cells. The blocks were cured overnight at 37°C, and then cured for 2 days at 65°C. Monolayers were pulled off the coverslips and sectioned en face. Ultrathin sections (60–70 nm) of the cells were obtained on a Riechart Ultracut E microtome, post-stained in 4% uranyl acetate for 10 min, and 1% lead citrate for 7 min. Sections were viewed on a JEOL JEM 1011 transmission electron microscope (JEOL, Peobody MA) at 80 kV. Images were taken using a side-mount AMT 2k digital camera (Advanced Microscopy Techniques, Danvers, MA).
Rodent studies
To create a MAL3-101 formulation for in vivo dosing, 250 mg of KolliphorHS 15 (Sigma Aldrich) was melted at 50°C and mixed with 150 mg ethanol and 100 mg dimethylacetamide (Sigma Aldrich). A total of 10.15 mg of MAL3-101 was added and the solution vortexed; 0.5 ml HPLC water was then added and the solution was vortexed for 10 min, centrifuged, and sterile-filtered to create a 1 mg/ml solution. Next, immunocompromised 6-week-old female nu/nu mice kept in the AAALAC-accredited barrier facility at UCSF were treated with 66 mg/kg body weight of MAL3-101 (or equal volume of vehicle) by intraperitoneal injection, then euthanized at 1, 4, 8 and 24 h. One lobe of the liver was harvested at the time of euthanasia and snap-frozen for immunoblotting analysis. Animal care was supervised by veterinary staff, and experiments were carried out under an IACUC-approved protocol.
Colony formation assay
50,000 RMS13 or RMS13-R cells were suspended in 0.4% agarose, 1× DMEM and 10% FBS, and plated over 0.6% agarose in DMEM with 10% FBS, in triplicate per treatment, in six-well plates. Once the agarose had set, 1 ml of RPMI-1640 with 10% FBS, 1× penicillin/streptomycin, and 40 µM CQ, 7.5 µM MAL3-101, a combination of the two, or DMSO, was gently pipetted onto the medium. Medium and drugs were refreshed twice weekly for 25 days. Plates were then washed with PBS and then fixed and stained with 0.05% Crystal Violet and 10% ethanol. After destaining with deionized water, plates were imaged using a 5× low power objective, and colonies >2 mm in diameter in each photomicrograph were counted using the Cell Counter plug-in for ImageJ. Colonies were counted for three low-power fields per condition.
Statistical analysis
IC50 concentrations from CellTiter-Glo assays were calculated as described previously (Sabnis et al., 2016) using SigmaPlot 11.0 (Systat Software, Inc.). GraphPad Prism was used to carry out a two-tailed Student's t-test (GraphPad Software, Inc.). In all experiments, P<0.05 was considered significant.
Supplementary Material
Acknowledgements
We thank Drs. John Christianson, Ineke Braakman, Taber Maskrey, Karen Arndt, Rehana Leak, Milena Vitale, Eelco Van Anken and Tia-Lynn Ashman for advice, reagents, and/or instrumentation.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
Conceptualization: S.S., A.J.S., P.W., S.C.W., T.G.B., J.L.B.; Methodology: S.S., C.J.G., A.J.S., D.B.S., C.T.W., S.C.W.; Validation: S.S., C.J.G., A.J.S.; Formal analysis: S.S., A.J.S., D.B.S., C.T.W., T.G.B., J.L.B.; Investigation: S.S., A.J.S., C.T.W.; Resources: A.J.S., D.B.S., P.W., S.C.W., T.G.B., J.L.B.; Data curation: C.J.G.; Writing - original draft: S.S., J.L.B.; Writing - review & editing: S.S., C.J.G., A.J.S., D.B.S., P.W., T.G.B., J.L.B.; Visualization: S.S., A.J.S., D.B.S., J.L.B.; Supervision: S.S., C.J.G., S.C.W., T.G.B.; Project administration: J.L.B.; Funding acquisition: T.G.B., J.L.B.
Funding
This work was support by National Institutes of Health (grants GM75061 and DK79307), a University of Pittsburgh Center for Precision and Translational Pharmacology grant, and by a Howard Hughes Medical Institute Collaborative Innovation Award. S.S. acknowledges the receipt of a long-term European Molecular Biology Organization (EMBO) post-doctoral fellowship (ALTF 823-2016). A.J.S. received support from the Frank A. Campini Foundation and a Damon Runyon-Sohn Fellowship (6P-13). This work was supported by the UCSF Helen Diller Family Comprehensive Cancer Center support grant (P30CA082103). Deposited in PMC for release after 12 months.
Supplementary information
Supplementary information available online at http://jcs.biologists.org/lookup/doi/10.1242/jcs.217760.supplemental
References
- Adam C., Baeurle A., Brodsky J. L., Wipf P., Schrama D., Becker J. C. and Houben R. (2014). The HSP70 modulator MAL3-101 inhibits Merkel cell carcinoma. PLoS One 9, e92041 10.1371/journal.pone.0092041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahlberg J., Berkenstam A., Henell F. and Glaumann H. (1985). Degradation of short and long lived proteins in isolated rat liver lysosomes. Effects of pH, temperature, and proteolytic inhibitors. J. Biol. Chem. 260, 5847-5854. [PubMed] [Google Scholar]
- Amaravadi R. K., Lippincott-Schwartz J., Yin X. M., Weiss W. A., Takebe N., Timmer W., DiPaola R. S., Lotze M. T. and White E. (2011). Principles and current strategies for targeting autophagy for cancer treatment. Clin. Cancer Res. 17, 654-666. 10.1158/1078-0432.CCR-10-2634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson D. J., Le Moigne R., Djakovic S., Kumar B., Rice J., Wong S., Wang J., Yao B., Valle E., Kiss von Soly S. et al. (2015). Targeting the AAA ATPase p97 as an Approach to Treat Cancer through Disruption of Protein Homeostasis. Cancer Cell 28, 653-665. 10.1016/j.ccell.2015.10.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anelli T., Sannino S. and Sitia R. (2015). Proteostasis and “redoxtasis” in the secretory pathway: tales of tails from ERp44 and immunoglobulins. Free Radic. Biol. Med. 83, 323-330. 10.1016/j.freeradbiomed.2015.02.020 [DOI] [PubMed] [Google Scholar]
- Bagola K., Mehnert M., Jarosch E. and Sommer T. (2011). Protein dislocation from the ER. Biochim. Biophys. Acta 1808, 925-936. 10.1016/j.bbamem.2010.06.025 [DOI] [PubMed] [Google Scholar]
- B'Chir W., Maurin A.-C., Carraro V., Averous J., Jousse C., Muranishi Y., Parry L., Stepien G., Fafournoux P. and Bruhat A. (2013). The eIF2alpha/ATF4 pathway is essential for stress-induced autophagy gene expression. Nucleic Acids Res. 41, 7683-7699. 10.1093/nar/gkt563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakunts A., Orsi A., Vitale M., Cattaneo A., Lari F., Tade L., Sitia R., Raimondi A., Bachi A. and van Anken E. (2017). Ratiometric sensing of BiP-client versus BiP levels by the unfolded protein response determines its signaling amplitude. Elife 6, e27518 10.7554/eLife.27518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barth S., Glick D. and Macleod K. F. (2010). Autophagy: assays and artifacts. J. Pathol. 221, 117-124. 10.1002/path.2694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bays N. W., Wilhovsky S. K., Goradia A., Hodgkiss-Harlow K. and Hampton R. Y. (2001). HRD4/NPL4 is required for the proteasomal processing of ubiquitinated ER proteins. Mol. Biol. Cell 12, 4114-4128. 10.1091/mbc.12.12.4114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benbrook D. M. and Long A. (2012). Integration of autophagy, proteasomal degradation, unfolded protein response and apoptosis. Exp. Oncol. 34, 286-297. [PubMed] [Google Scholar]
- Bennett E. J., Bence N. F., Jayakumar R. and Kopito R. R. (2005). Global impairment of the ubiquitin-proteasome system by nuclear or cytoplasmic protein aggregates precedes inclusion body formation. Mol. Cell 17, 351-365. 10.1016/j.molcel.2004.12.021 [DOI] [PubMed] [Google Scholar]
- Bernard A., Jin M., Xu Z. and Klionsky D. J. (2015). A large-scale analysis of autophagy-related gene expression identifies new regulators of autophagy. Autophagy 11, 2114-2122. 10.1080/15548627.2015.1099796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi M., Naczki C., Koritzinsky M., Fels D., Blais J., Hu N., Harding H., Novoa I., Varia M., Raleigh J. et al. (2005). ER stress-regulated translation increases tolerance to extreme hypoxia and promotes tumor growth. EMBO J. 24, 3470-3481. 10.1038/sj.emboj.7600777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bianchi G., Oliva L., Cascio P., Pengo N., Fontana F., Cerruti F., Orsi A., Pasqualetto E., Mezghrani A., Calbi V. et al. (2009). The proteasome load versus capacity balance determines apoptotic sensitivity of multiple myeloma cells to proteasome inhibition. Blood 113, 3040-3049. 10.1182/blood-2008-08-172734 [DOI] [PubMed] [Google Scholar]
- Bjørkøy G., Lamark T., Brech A., Outzen H., Perander M., Overvatn A., Stenmark H. and Johansen T. (2005). p62/SQSTM1 forms protein aggregates degraded by autophagy and has a protective effect on huntingtin-induced cell death. J. Cell Biol. 171, 603-614. 10.1083/jcb.200507002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blaney S. M., Bernstein M., Neville K., Ginsberg J., Kitchen B., Horton T., Berg S. L., Krailo M. and Adamson P. C. (2004). Phase I study of the proteasome inhibitor bortezomib in pediatric patients with refractory solid tumors: a Children's Oncology Group study (ADVL0015). J. Clin. Oncol. 22, 4804-4809. 10.1200/JCO.2004.12.185 [DOI] [PubMed] [Google Scholar]
- Bodnar N. and Rapoport T. (2017a). Toward an understanding of the Cdc48/p97 ATPase. F1000Res 6, 1318 10.12688/f1000research.11683.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodnar N. O. and Rapoport T. A. (2017b). Molecular Mechanism of Substrate Processing by the Cdc48 ATPase Complex. Cell 169, 722-735 e9. 10.1016/j.cell.2017.04.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braunstein M. J., Scott S. S., Scott C. M., Behrman S., Walter P., Wipf P., Coplan J. D., Chrico W., Joseph D., Brodsky J. L. et al. (2011). Antimyeloma effects of the heat shock protein 70 molecular chaperone inhibitor MAL3-101. J. Oncol. 2011, 232037 10.1155/2011/232037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodsky J. L. and Chiosis G. (2006). Hsp70 molecular chaperones: emerging roles in human disease and identification of small molecule modulators. Curr. Top. Med. Chem. 6, 1215-1225. 10.2174/156802606777811997 [DOI] [PubMed] [Google Scholar]
- Calderwood S. K. and Gong J. (2016). Heat shock proteins promote cancer: it's a protection racket. Trends Biochem. Sci. 41, 311-323. 10.1016/j.tibs.2016.01.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carew J. S., Espitia C. M., Zhao W., Han Y., Visconte V., Phillips J. and Nawrocki S. T. (2017). Disruption of autophagic degradation with ROC-325 antagonizes renal cell carcinoma pathogenesis. Clin. Cancer Res. 23, 2869-2879. 10.1158/1078-0432.CCR-16-1742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casagrande R., Stern P., Diehn M., Shamu C., Osario M., Zuniga M., Brown P. O. and Ploegh H. (2000). Degradation of proteins from the ER of S. cerevisiae requires an intact unfolded protein response pathway. Mol. Cell 5, 729-735. [DOI] [PubMed] [Google Scholar]
- Cerezo M., Lehraiki A., Millet A., Rouaud F., Plaisant M., Jaune E., Botton T., Ronco C., Abbe P., Amdouni H. et al. (2016). Compounds triggering ER stress exert anti-melanoma effects and overcome BRAF inhibitor resistance. Cancer Cell 30, 183 10.1016/j.ccell.2016.06.007 [DOI] [PubMed] [Google Scholar]
- Chen Y.-J., Tan B. C.-M., Cheng Y.-Y., Chen J.-S. and Lee S.-C. (2010). Differential regulation of CHOP translation by phosphorylated eIF4E under stress conditions. Nucleic Acids Res. 38, 764-777. 10.1093/nar/gkp1034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen D., Frezza M., Schmitt S., Kanwar J. and Dou Q. P. (2011). Bortezomib as the first proteasome inhibitor anticancer drug: current status and future perspectives. Curr. Cancer Drug Targets 11, 239-253. 10.2174/156800911794519752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christianson J. C., Olzmann J. A., Shaler T. A., Sowa M. E., Bennett E. J., Richter C. M., Tyler R. E., Greenblatt E. J., Harper J. W. and Kopito R. R. (2011). Defining human ERAD networks through an integrative mapping strategy. Nat. Cell Biol. 14, 93-105. 10.1038/ncb2383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke R. and Cook K. L. (2015). Unfolding the role of stress response signaling in endocrine resistant breast cancers. Front Oncol. 5, 140 10.3389/fonc.2015.00140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke R., Shajahan A. N., Wang Y., Tyson J. J., Riggins R. B., Weiner L. M., Bauman W. T., Xuan J., Zhang B., Facey C. et al. (2011). Endoplasmic reticulum stress, the unfolded protein response, and gene network modeling in antiestrogen resistant breast cancer. Horm. Mol. Biol. Clin. Investig. 5, 35-44. 10.1515/hmbci.2010.073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke R., Cook K. L., Hu R., Facey C. O., Tavassoly I., Schwartz J. L., Baumann W. T., Tyson J. J., Xuan J., Wang Y. et al. (2012). Endoplasmic reticulum stress, the unfolded protein response, autophagy, and the integrated regulation of breast cancer cell fate. Cancer Res. 72, 1321-1331. 10.1158/1538-7445.AM2012-1321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke H. J., Chambers J. E., Liniker E. and Marciniak S. J. (2014). Endoplasmic reticulum stress in malignancy. Cancer Cell 25, 563-573. 10.1016/j.ccr.2014.03.015 [DOI] [PubMed] [Google Scholar]
- Cuervo A. M. (2004). Autophagy: many paths to the same end. Mol. Cell. Biochem. 263, 55-72. 10.1023/B:MCBI.0000041848.57020.57 [DOI] [PubMed] [Google Scholar]
- Cufi S., Vazquez-Martin A., Oliveras-Ferraros C., Corominas-Faja B., Cuyàs E., López-Bonet E., Martin-Castillo B., Joven J. and Menendez J. A. (2013). The anti-malarial chloroquine overcomes primary resistance and restores sensitivity to trastuzumab in HER2-positive breast cancer. Sci. Rep. 3, 2469 10.1038/srep02469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daugaard M., Rohde M. and Jäättelä M. (2007). The heat shock protein 70 family: Highly homologous proteins with overlapping and distinct functions. FEBS Lett. 581, 3702-3710. 10.1016/j.febslet.2007.05.039 [DOI] [PubMed] [Google Scholar]
- Deshaies R. J. (2014). Proteotoxic crisis, the ubiquitin-proteasome system, and cancer therapy. BMC Biol. 12, 94 10.1186/s12915-014-0094-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellgaard L. and Helenius A. (2003). Quality control in the endoplasmic reticulum. Nat. Rev. Mol. Cell Biol. 4, 181-191. 10.1038/nrm1052 [DOI] [PubMed] [Google Scholar]
- Erzurumlu Y. and Ballar P. (2017). Androgen mediated regulation of endoplasmic reticulum-associated degradation and its effects on prostate cancer. Sci. Rep. 7, 40719 10.1038/srep40719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans C. G., Chang L. and Gestwicki J. E. (2010). Heat shock protein 70 (hsp70) as an emerging drug target. J. Med. Chem. 53, 4585-4602. 10.1021/jm100054f [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fewell S. W., Smith C. M., Lyon M. A., Dumitrescu T. P., Wipf P., Day B. W. and Brodsky J. L. (2004). Small molecule modulators of endogenous and co-chaperone-stimulated Hsp70 ATPase activity. J. Biol. Chem. 279, 51131-51140. 10.1074/jbc.M404857200 [DOI] [PubMed] [Google Scholar]
- Frenkel Z., Gregory W., Kornfeld S. and Lederkremer G. Z. (2003). Endoplasmic reticulum-associated degradation of mammalian glycoproteins involves sugar chain trimming to Man6-5GlcNAc2. J. Biol. Chem. 278, 34119-34124. 10.1074/jbc.M305929200 [DOI] [PubMed] [Google Scholar]
- Fullgrabe J., Ghislat G., Cho D. H. and Rubinsztein D. C. (2016). Transcriptional regulation of mammalian autophagy at a glance. J. Cell Sci. 129, 3059-3066. 10.1242/jcs.188920 [DOI] [PubMed] [Google Scholar]
- Gabai V. L., Budagova K. R. and Sherman M. Y. (2005). Increased expression of the major heat shock protein Hsp72 in human prostate carcinoma cells is dispensable for their viability but confers resistance to a variety of anticancer agents. Oncogene 24, 3328-3338. 10.1038/sj.onc.1208495 [DOI] [PubMed] [Google Scholar]
- Galehdar Z., Swan P., Fuerth B., Callaghan S. M., Park D. S. and Cregan S. P. (2010). Neuronal apoptosis induced by endoplasmic reticulum stress is regulated by ATF4-CHOP-mediated induction of the Bcl-2 homology 3-only member PUMA. J. Neurosci. 30, 16938-16948. 10.1523/JNEUROSCI.1598-10.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrido C., Brunet M., Didelot C., Zermati Y., Schmitt E. and Kroemer G. (2006). Heat shock proteins 27 and 70: anti-apoptotic proteins with tumorigenic properties. Cell Cycle 5, 2592-2601. 10.4161/cc.5.22.3448 [DOI] [PubMed] [Google Scholar]
- Gleixner A. M., Hutchison D. F., Sannino S., Bhatia T. N., Leak L. C., Flaherty P. T., Wipf P., Brodsky J. L. and Leak R. K. (2017). N-Acetyl-l-cysteine protects astrocytes against proteotoxicity without recourse to glutathione. Mol. Pharmacol. 92, 564-575. 10.1124/mol.117.109926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg A. L. (2003). Protein degradation and protection against misfolded or damaged proteins. Nature 426, 895-899. 10.1038/nature02263 [DOI] [PubMed] [Google Scholar]
- Goloudina A. R., Demidov O. N. and Garrido C. (2012). Inhibition of HSP70: a challenging anti-cancer strategy. Cancer Lett. 325, 117-124. 10.1016/j.canlet.2012.06.003 [DOI] [PubMed] [Google Scholar]
- Gómez V. E., Giovannetti E. and Peters G. J. (2015). Unraveling the complexity of autophagy: potential therapeutic applications in Pancreatic Ductal Adenocarcinoma. Semin. Cancer Biol. 35, 11-19. 10.1016/j.semcancer.2015.09.011 [DOI] [PubMed] [Google Scholar]
- Goo M. S., Scudder S. L. and Patrick G. N. (2015). Ubiquitin-dependent trafficking and turnover of ionotropic glutamate receptors. Front Mol Neurosci 8, 60 10.3389/fnmol.2015.00060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerriero C. J. and Brodsky J. L. (2012). The delicate balance between secreted protein folding and endoplasmic reticulum-associated degradation in human physiology. Physiol. Rev. 92, 537-576. 10.1152/physrev.00027.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo F., Rocha K., Bali P., Pranpat M., Fiskus W., Boyapalle S., Kumaraswamy S., Balasis M., Greedy B., Armitage E. S. et al. (2005). Abrogation of heat shock protein 70 induction as a strategy to increase antileukemia activity of heat shock protein 90 inhibitor 17-allylamino-demethoxy geldanamycin. Cancer Res. 65, 10536-10544. 10.1158/0008-5472.CAN-05-1799 [DOI] [PubMed] [Google Scholar]
- Haas A. L., Warms J. V., Hershko A. and Rose I. A. (1982). Ubiquitin-activating enzyme. Mechanism and role in protein-ubiquitin conjugation. J. Biol. Chem. 257, 2543-2548. [PubMed] [Google Scholar]
- Halterman M. W., Gill M., DeJesus C., Ogihara M., Schor N. F. and Federoff H. J. (2010). The endoplasmic reticulum stress response factor CHOP-10 protects against hypoxia-induced neuronal death. J. Biol. Chem. 285, 21329-21340. 10.1074/jbc.M109.095299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hambright H. G. and Ghosh R. (2017). Autophagy: in the cROSshairs of cancer. Biochem. Pharmacol. 126, 13-22. 10.1016/j.bcp.2016.10.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazari Y. M., Bashir A., Haq E. U. and Fazili K. M. (2016). Emerging tale of UPR and cancer: an essentiality for malignancy. Tumour Biol. 37, 14381-14390. 10.1007/s13277-016-5343-0 [DOI] [PubMed] [Google Scholar]
- Horimoto S., Ninagawa S., Okada T., Koba H., Sugimoto T., Kamiya Y., Kato K., Takeda S. and Mori K. (2013). The unfolded protein response transducer ATF6 represents a novel transmembrane-type endoplasmic reticulum-associated degradation substrate requiring both mannose trimming and SEL1L protein. J. Biol. Chem. 288, 31517-31527. 10.1074/jbc.M113.476010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosokawa N., Tremblay L. O., You Z., Herscovics A., Wada I. and Nagata K. (2003). Enhancement of endoplasmic reticulum (ER) degradation of misfolded Null Hong Kong alpha1-antitrypsin by human ER mannosidase I. J. Biol. Chem. 278, 26287-26294. 10.1074/jbc.M303395200 [DOI] [PubMed] [Google Scholar]
- Hosokawa N., Wada I., Natsuka Y. and Nagata K. (2006). EDEM accelerates ERAD by preventing aberrant dimer formation of misfolded alpha1-antitrypsin. Genes Cells 11, 465-476. 10.1111/j.1365-2443.2006.00957.x [DOI] [PubMed] [Google Scholar]
- Hosokawa N., You Z., Tremblay L. O., Nagata K. and Herscovics A. (2007). Stimulation of ERAD of misfolded null Hong Kong alpha1-antitrypsin by Golgi alpha1,2-mannosidases. Biochem. Biophys. Res. Commun. 362, 626-632. 10.1016/j.bbrc.2007.08.057 [DOI] [PubMed] [Google Scholar]
- Hubbert C., Guardiola A., Shao R., Kawaguchi Y., Ito A., Nixon A., Yoshida M., Wang X.-F. and Yao T.-P. (2002). HDAC6 is a microtubule-associated deacetylase. Nature 417, 455-458. 10.1038/417455a [DOI] [PubMed] [Google Scholar]
- Ichimura Y., Kirisako T., Takao T., Satomi Y., Shimonishi Y., Ishihara N., Mizushima N., Tanida I., Kominami E., Ohsumi M. et al. (2000). A ubiquitin-like system mediates protein lipidation. Nature 408, 488-492. 10.1038/35044114 [DOI] [PubMed] [Google Scholar]
- Ishida Y., Yamamoto A., Kitamura A., Lamandé S. R., Yoshimori T., Bateman J. F., Kubota H. and Nagata K. (2009). Autophagic elimination of misfolded procollagen aggregates in the endoplasmic reticulum as a means of cell protection. Mol. Biol. Cell 20, 2744-2754. 10.1091/mbc.e08-11-1092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain K., Paranandi K. S., Sridharan S. and Basu A. (2013). Autophagy in breast cancer and its implications for therapy. Am. J. Cancer Res 3, 251-265. [PMC free article] [PubMed] [Google Scholar]
- Jarosch E., Taxis C., Volkwein C., Bordallo J., Finley D., Wolf D. H. and Sommer T. (2002). Protein dislocation from the ER requires polyubiquitination and the AAA-ATPase Cdc48. Nat. Cell Biol. 4, 134-139. 10.1038/ncb746 [DOI] [PubMed] [Google Scholar]
- Jiang P. and Mizushima N. (2014). Autophagy and human diseases. Cell Res. 24, 69-79. 10.1038/cr.2013.161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jousse C., Bruhat A., Carraro V., Urano F., Ferrara M., Ron D. and Fafournoux P. (2001). Inhibition of CHOP translation by a peptide encoded by an open reading frame localized in the chop 5'UTR. Nucleic Acids Res. 29, 4341-4351. 10.1093/nar/29.21.4341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabeya Y., Mizushima N., Ueno T., Yamamoto A., Kirisako T., Noda T., Kominami E., Ohsumi Y. and Yoshimori T. (2000). LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J. 19, 5720-5728. 10.1093/emboj/19.21.5720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katsuragi Y., Ichimura Y. and Komatsu M. (2015). p62/SQSTM1 functions as a signaling hub and an autophagy adaptor. FEBS J. 282, 4672-4678. 10.1111/febs.13540 [DOI] [PubMed] [Google Scholar]
- Kawaguchi Y., Kovacs J. J., McLaurin A., Vance J. M., Ito A. and Yao T.-P. (2003). The deacetylase HDAC6 regulates aggresome formation and cell viability in response to misfolded protein stress. Cell 115, 727-738. 10.1016/S0092-8674(03)00939-5 [DOI] [PubMed] [Google Scholar]
- Kawiak A., Domachowska A., Jaworska A. and Lojkowska E. (2017). Plumbagin sensitizes breast cancer cells to tamoxifen-induced cell death through GRP78 inhibition and Bik upregulation. Sci. Rep. 7, 43781 10.1038/srep43781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y. E., Hipp M. S., Bracher A., Hayer-Hartl M. and Hartl F. U. (2013). Molecular chaperone functions in protein folding and proteostasis. Annu. Rev. Biochem. 82, 323-355. 10.1146/annurev-biochem-060208-092442 [DOI] [PubMed] [Google Scholar]
- Kim H., Bhattacharya A. and Qi L. (2015). Endoplasmic reticulum quality control in cancer: Friend or foe. Semin. Cancer Biol. 33, 25-33. 10.1016/j.semcancer.2015.02.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klionsky D. J., Abdalla F. C., Abeliovich H., Abraham R. T., Acevedo-Arozena A., Adeli K., Agholme L., Agnello M., Agostinis P., Aguirre-Ghiso J. A. et al. (2012). Guidelines for the use and interpretation of assays for monitoring autophagy. Autophagy 8, 445-544. 10.4161/auto.19496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komatsu M., Waguri S., Koike M., Sou Y. S., Ueno T., Hara T., Mizushima N., Iwata J., Ezaki J., Murata S. et al. (2007). Homeostatic levels of p62 control cytoplasmic inclusion body formation in autophagy-deficient mice. Cell 131, 1149-1163. 10.1016/j.cell.2007.10.035 [DOI] [PubMed] [Google Scholar]
- Koopman G., Reutelingsperger C. P., Kuijten G. A., Keehnen R. M., Pals S. T. and van Oers M. H. (1994). Annexin V for flow cytometric detection of phosphatidylserine expression on B cells undergoing apoptosis. Blood 84, 1415-1420. [PubMed] [Google Scholar]
- Kouroku Y., Fujita E., Tanida I., Ueno T., Isoai A., Kumagai H., Ogawa S., Kaufman R. J., Kominami E. and Momoi T. (2007). ER stress (PERK/eIF2alpha phosphorylation) mediates the polyglutamine-induced LC3 conversion, an essential step for autophagy formation. Cell Death Differ. 14, 230-239. 10.1038/sj.cdd.4401984 [DOI] [PubMed] [Google Scholar]
- Kruse K. B., Brodsky J. L. and McCracken A. A. (2006). Autophagy: an ER protein quality control process. Autophagy 2, 135-137. 10.4161/auto.2.2.2388 [DOI] [PubMed] [Google Scholar]
- Le Moigne R., Aftab B. T., Djakovic S., Dhimolea E., Valle E., Murnane M., King E. M., Soriano F., Menon M. K., Wu Z. Y. et al. (2017). The p97 inhibitor CB-5083 is a unique disrupter of protein homeostasis in models of multiple myeloma. Mol. Cancer Ther. 16, 2375-2386. 10.1158/1535-7163.MCT-17-0233 [DOI] [PubMed] [Google Scholar]
- Lee A. S. (2001). The glucose-regulated proteins: stress induction and clinical applications. Trends Biochem. Sci. 26, 504-510. 10.1016/S0968-0004(01)01908-9 [DOI] [PubMed] [Google Scholar]
- Lee A. S. (2007). GRP78 induction in cancer: therapeutic and prognostic implications. Cancer Res. 67, 3496-3499. 10.1158/0008-5472.CAN-07-0325 [DOI] [PubMed] [Google Scholar]
- Levine B. and Klionsky D. J. (2004). Development by self-digestion: molecular mechanisms and biological functions of autophagy. Dev. Cell 6, 463-477. 10.1016/S1534-5807(04)00099-1 [DOI] [PubMed] [Google Scholar]
- Levine B. and Kroemer G. (2008). Autophagy in the pathogenesis of disease. Cell 132, 27-42. 10.1016/j.cell.2007.12.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liebl M. P. and Hoppe T. (2016). It's all about talking: two-way communication between proteasomal and lysosomal degradation pathways via ubiquitin. Am. J. Physiol. Cell Physiol. 311, C166-C178. 10.1152/ajpcell.00074.2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B.-Q., Du Z.-X., Zong Z.-H., Li C., Li N., Zhang Q., Kong D.-H. and Wang H.-Q. (2013). BAG3-dependent noncanonical autophagy induced by proteasome inhibition in HepG2 cells. Autophagy 9, 905-916. 10.4161/auto.24292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W. J., Ye L., Huang W. F., Guo L. J., Xu Z. G., Wu H. L., Yang C. and Liu H. F. (2016). p62 links the autophagy pathway and the ubiqutin-proteasome system upon ubiquitinated protein degradation. Cell. Mol. Biol. Lett. 21, 29 10.1186/s11658-016-0031-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorin S., Hamaï A., Mehrpour M. and Codogno P. (2013). Autophagy regulation and its role in cancer. Semin. Cancer Biol. 23, 361-379. 10.1016/j.semcancer.2013.06.007 [DOI] [PubMed] [Google Scholar]
- Ma X.-H., Piao S.-F., Dey S., McAfee Q., Karakousis G., Villanueva J., Hart L. S., Levi S., Hu J., Zhang G. et al. (2014). Targeting ER stress-induced autophagy overcomes BRAF inhibitor resistance in melanoma. J. Clin. Invest. 124, 1406-1417. 10.1172/JCI70454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manic G., Obrist F., Kroemer G., Vitale I. and Galluzzi L. (2014). Chloroquine and hydroxychloroquine for cancer therapy. Mol. Cell Oncol. 1, e29911 10.4161/mco.29911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marciniak S. J., Yun C. Y., Oyadomari S., Novoa I., Zhang Y., Jungreis R., Nagata K., Harding H. P. and Ron D. (2004). CHOP induces death by promoting protein synthesis and oxidation in the stressed endoplasmic reticulum. Genes Dev. 18, 3066-3077. 10.1101/gad.1250704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcus N. Y. and Perlmutter D. H. (2000). Glucosidase and mannosidase inhibitors mediate increased secretion of mutant alpha1 antitrypsin Z. J. Biol. Chem. 275, 1987-1992. 10.1074/jbc.275.3.1987 [DOI] [PubMed] [Google Scholar]
- Matus S., Lopez E., Valenzuela V., Nassif M. and Hetz C. (2013). Functional contribution of the transcription factor ATF4 to the pathogenesis of amyotrophic lateral sclerosis. PLoS One 8, e66672 10.1371/journal.pone.0066672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maycotte P. and Thorburn A. (2014). Targeting autophagy in breast cancer. World J. Clin. Oncol. 5, 224-240. 10.5306/wjco.v5.i3.224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCollum A. K., Teneyck C. J., Sauer B. M., Toft D. O. and Erlichman C. (2006). Up-regulation of heat shock protein 27 induces resistance to 17-allylamino-demethoxygeldanamycin through a glutathione-mediated mechanism. Cancer Res. 66, 10967-10975. 10.1158/0008-5472.CAN-06-1629 [DOI] [PubMed] [Google Scholar]
- McConkey D. J. (2017). The integrated stress response and proteotoxicity in cancer therapy. Biochem. Biophys. Res. Commun. 482, 450-453. 10.1016/j.bbrc.2016.11.047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McEwan D. G. and Dikic I. (2011). The Three Musketeers of Autophagy: phosphorylation, ubiquitylation and acetylation. Trends Cell Biol. 21, 195-201. 10.1016/j.tcb.2010.12.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLeland C. B., Rodriguez J. and Stern S. T. (2011). Autophagy monitoring assay: qualitative analysis of MAP LC3-I to II conversion by immunoblot. Methods Mol. Biol. 697, 199-206. 10.1007/978-1-60327-198-1_21 [DOI] [PubMed] [Google Scholar]
- Mei Y., Thompson M. D., Cohen R. A. and Tong X. (2013). Endoplasmic reticulum stress and related pathological processes. J. Pharmacol. Biomed. Anal. 1, 1000107. [PMC free article] [PubMed] [Google Scholar]
- Milan E., Perini T., Resnati M., Orfanelli U., Oliva L., Raimondi A., Cascio P., Bachi A., Marcatti M., Ciceri F. et al. (2015). A plastic SQSTM1/p62-dependent autophagic reserve maintains proteostasis and determines proteasome inhibitor susceptibility in multiple myeloma cells. Autophagy 11, 1161-1178. 10.1080/15548627.2015.1052928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizushima N. and Klionsky D. J. (2007). Protein turnover via autophagy: implications for metabolism. Annu. Rev. Nutr. 27, 19-40. 10.1146/annurev.nutr.27.061406.093749 [DOI] [PubMed] [Google Scholar]
- Mizushima N., Yamamoto A., Hatano M., Kobayashi Y., Kabeya Y., Suzuki K., Tokuhisa T., Ohsumi Y. and Yoshimori T. (2001). Dissection of autophagosome formation using Apg5-deficient mouse embryonic stem cells. J. Cell Biol. 152, 657-668. 10.1083/jcb.152.4.657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizushima N., Ohsumi Y. and Yoshimori T. (2002). Autophagosome formation in mammalian cells. Cell Struct. Funct. 27, 421-429. 10.1247/csf.27.421 [DOI] [PubMed] [Google Scholar]
- Mizushima N., Levine B., Cuervo A. M. and Klionsky D. J. (2008). Autophagy fights disease through cellular self-digestion. Nature 451, 1069-1075. 10.1038/nature06639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizushima N., Yoshimori T. and Ohsumi Y. (2011). The role of Atg proteins in autophagosome formation. Annu. Rev. Cell Dev. Biol. 27, 107-132. 10.1146/annurev-cellbio-092910-154005 [DOI] [PubMed] [Google Scholar]
- Morishima Y., Kanelakis K. C., Silverstein A. M., Dittmar K. D., Estrada L. and Pratt W. B. (2000). The Hsp organizer protein hop enhances the rate of but is not essential for glucocorticoid receptor folding by the multiprotein Hsp90-based chaperone system. J. Biol. Chem. 275, 6894-6900. 10.1074/jbc.275.10.6894 [DOI] [PubMed] [Google Scholar]
- Mosser D. D. and Morimoto R. I. (2004). Molecular chaperones and the stress of oncogenesis. Oncogene 23, 2907-2918. 10.1038/sj.onc.1207529 [DOI] [PubMed] [Google Scholar]
- Nanbu K., Konishi I., Mandai M., Kuroda H., Hamid A. A., Komatsu T. and Mori T. (1998). Prognostic significance of heat shock proteins HSP70 and HSP90 in endometrial carcinomas. Cancer Detect. Prev. 22, 549-555. 10.1046/j.1525-1500.1998.00069.x [DOI] [PubMed] [Google Scholar]
- Ni M., Zhou H., Wey S., Baumeister P. and Lee A. S. (2009). Regulation of PERK signaling and leukemic cell survival by a novel cytosolic isoform of the UPR regulator GRP78/BiP. PLoS One 4, e6868 10.1371/journal.pone.0006868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishiyama J., Miura E., Mizushima N., Watanabe M. and Yuzaki M. (2007). Aberrant membranes and double-membrane structures accumulate in the axons of Atg5-null Purkinje cells before neuronal death. Autophagy 3, 591-596. 10.4161/auto.4964 [DOI] [PubMed] [Google Scholar]
- Ojha R. and Amaravadi R. K. (2017). Targeting the unfolded protein response in cancer. Pharmacol. Res. 120, 258-266. 10.1016/j.phrs.2017.04.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olanich M. E. and Barr F. G. (2013). A call to ARMS: targeting the PAX3-FOXO1 gene in alveolar rhabdomyosarcoma. Expert Opin Ther. Targets 17, 607-623. 10.1517/14728222.2013.772136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olzmann J. A., Kopito R. R. and Christianson J. C. (2013). The mammalian endoplasmic reticulum-associated degradation system. Cold Spring Harb. Perspect. Biol. 5, a013185 10.1101/cshperspect.a013185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey U. B., Batlevi Y., Baehrecke E. H. and Taylor J. P. (2007). HDAC6 at the intersection of autophagy, the ubiquitin-proteasome system and neurodegeneration. Autophagy 3, 643-645. 10.4161/auto.5050 [DOI] [PubMed] [Google Scholar]
- Patury S., Miyata Y. and Gestwicki J. E. (2009). Pharmacological targeting of the Hsp70 chaperone. Curr. Top. Med. Chem. 9, 1337-1351. 10.2174/156802609789895674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perlmutter D. H. (2011). Alpha-1-antitrypsin deficiency: importance of proteasomal and autophagic degradative pathways in disposal of liver disease-associated protein aggregates. Annu. Rev. Med. 62, 333-345. 10.1146/annurev-med-042409-151920 [DOI] [PubMed] [Google Scholar]
- Peron M., Bonvini P. and Rosolen A. (2012). Effect of inhibition of the ubiquitin-proteasome system and Hsp90 on growth and survival of rhabdomyosarcoma cells in vitro. BMC Cancer 12, 233 10.1186/1471-2407-12-233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaffl M. W. (2001). A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29, e45 10.1093/nar/29.9.e45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powers M. V., Clarke P. A. and Workman P. (2008). Dual targeting of HSC70 and HSP72 inhibits HSP90 function and induces tumor-specific apoptosis. Cancer Cell 14, 250-262. 10.1016/j.ccr.2008.08.002 [DOI] [PubMed] [Google Scholar]
- Powers M. V., Clarke P. A. and Workman P. (2009). Death by chaperone: HSP90, HSP70 or both? Cell Cycle 8, 518-526. 10.4161/cc.8.4.7583 [DOI] [PubMed] [Google Scholar]
- Powers M. V., Jones K., Barillari C., Westwood I., van Montfort R. L. and Workman P. (2010). Targeting HSP70: the second potentially druggable heat shock protein and molecular chaperone? Cell Cycle 9, 1542-1550. 10.4161/cc.9.8.11204 [DOI] [PubMed] [Google Scholar]
- Pyo J.-O., Jang M.-H., Kwon Y.-K., Lee H.-J., Jun J.-I. L., Woo H.-N., Cho D.-H., Choi B. Y., Lee H., Kim J.-H. et al. (2005). Essential roles of Atg5 and FADD in autophagic cell death: dissection of autophagic cell death into vacuole formation and cell death. J. Biol. Chem. 280, 20722-20729. 10.1074/jbc.M413934200 [DOI] [PubMed] [Google Scholar]
- Raasi S. and Wolf D. H. (2007). Ubiquitin receptors and ERAD: a network of pathways to the proteasome. Semin. Cell Dev. Biol. 18, 780-791. 10.1016/j.semcdb.2007.09.008 [DOI] [PubMed] [Google Scholar]
- Rabinovich E., Kerem A., Frohlich K.-U., Diamant N. and Bar-Nun S. (2002). AAA-ATPase p97/Cdc48p, a cytosolic chaperone required for endoplasmic reticulum-associated protein degradation. Mol. Cell. Biol. 22, 626-634. 10.1128/MCB.22.2.626-634.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rapino F., Jung M. and Fulda S. (2014). BAG3 induction is required to mitigate proteotoxicity via selective autophagy following inhibition of constitutive protein degradation pathways. Oncogene 33, 1713-1724. 10.1038/onc.2013.110 [DOI] [PubMed] [Google Scholar]
- Rodina A., Vilenchik M., Moulick K., Aguirre J., Kim J., Chiang A., Litz J., Clement C. C., Kang Y., She Y. et al. (2007). Selective compounds define Hsp90 as a major inhibitor of apoptosis in small-cell lung cancer. Nat. Chem. Biol. 3, 498-507. 10.1038/nchembio.2007.10 [DOI] [PubMed] [Google Scholar]
- Rodina A., Taldone T., Kang Y., Patel P. D., Koren J. III, Yan P., DaGama Gomes E. M., Yang C., Patel M. R., Shrestha L. et al. (2014). Affinity purification probes of potential use to investigate the endogenous Hsp70 interactome in cancer. ACS Chem. Biol. 9, 1698-1705. 10.1021/cb500256u [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ron D. and Walter P. (2007). Signal integration in the endoplasmic reticulum unfolded protein response. Nat. Rev. Mol. Cell Biol. 8, 519-529. 10.1038/nrm2199 [DOI] [PubMed] [Google Scholar]
- Rothman J. E. and Schekman R. (2011). Molecular mechanism of protein folding in the cell. Cell 146, 851-854. 10.1016/j.cell.2011.08.041 [DOI] [PubMed] [Google Scholar]
- Rouschop K. M. A., van den Beucken T., Dubois L., Niessen H., Bussink J., Savelkouls K., Keulers T., Mujcic H., Landuyt W., Voncken J. W. et al. (2010). The unfolded protein response protects human tumor cells during hypoxia through regulation of the autophagy genes MAP1LC3B and ATG5. J. Clin. Invest. 120, 127-141. 10.1172/JCI40027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubiolo J. A., López-Alonso H., Martínez P., Millán A., Cagide E., Vieytes M. R., Vega F. V. and Botana L. M. (2014). Yessotoxin induces ER-stress followed by autophagic cell death in glioma cells mediated by mTOR and BNIP3. Cell. Signal. 26, 419-432. 10.1016/j.cellsig.2013.10.004 [DOI] [PubMed] [Google Scholar]
- Rzymski T., Milani M., Singleton D. C. and Harris A. L. (2009). Role of ATF4 in regulation of autophagy and resistance to drugs and hypoxia. Cell Cycle 8, 3838-3847. 10.4161/cc.8.23.10086 [DOI] [PubMed] [Google Scholar]
- Rzymski T., Milani M., Pike L., Buffa F., Mellor H. R., Winchester L., Pires I., Hammond E., Ragoussis I. and Harris A. L. (2010). Regulation of autophagy by ATF4 in response to severe hypoxia. Oncogene 29, 4424-4435. 10.1038/onc.2010.191 [DOI] [PubMed] [Google Scholar]
- Sabnis A. J., Guerriero C. J., Olivas V., Sayana A., Shue J., Flanagan J., Asthana S., Paton A. W., Paton J. C., Gestwicki J. E. et al. (2016). Combined chemical-genetic approach identifies cytosolic HSP70 dependence in rhabdomyosarcoma. Proc. Natl. Acad. Sci. USA 113, 9015-9020. 10.1073/pnas.1603883113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sannino S. and Brodsky J. L. (2017). Targeting protein quality control pathways in breast cancer. BMC Biol. 15, 109 10.1186/s12915-017-0449-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santarosa M., Favaro D., Quaia M. and Galligioni E. (1997). Expression of heat shock protein 72 in renal cell carcinoma: possible role and prognostic implications in cancer patients. Eur. J. Cancer 33, 873-877. 10.1016/S0959-8049(97)00002-6 [DOI] [PubMed] [Google Scholar]
- Schonthal A. H. (2012a). Endoplasmic reticulum stress: its role in disease and novel prospects for therapy. Scientifica (Cairo) 2012, 857516 10.6064/2012/857516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schonthal A. H. (2012b). Targeting endoplasmic reticulum stress for cancer therapy. Front Biosci. (Schol. Ed.) 4, 412-431. 10.2741/s276 [DOI] [PubMed] [Google Scholar]
- Shajahan A. N., Riggins R. B. and Clarke R. (2009). The role of X-box binding protein-1 in tumorigenicity. Drug News Perspect. 22, 241-246. 10.1358/dnp.2009.22.5.1378631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shern J. F., Chen L., Chmielecki J., Wei J. S., Patidar R., Rosenberg M., Ambrogio L., Auclair D., Wang J., Song Y. K. et al. (2014). Comprehensive genomic analysis of rhabdomyosarcoma reveals a landscape of alterations affecting a common genetic axis in fusion-positive and fusion-negative tumors. Cancer Discov. 4, 216-231. 10.1158/2159-8290.CD-13-0639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh N., Joshi R. and Komurov K. (2015). HER2-mTOR signaling-driven breast cancer cells require ER-associated degradation to survive. Sci. Signal. 8, ra52 10.1126/scisignal.aaa6922 [DOI] [PubMed] [Google Scholar]
- Sitia R. and Braakman I. (2003). Quality control in the endoplasmic reticulum protein factory. Nature 426, 891-894. 10.1038/nature02262 [DOI] [PubMed] [Google Scholar]
- Sliutz G., Karlseder J., Tempfer C., Orel L., Holzer G. and Simon M. M. (1996). Drug resistance against gemcitabine and topotecan mediated by constitutive hsp70 overexpression in vitro: implication of quercetin as sensitiser in chemotherapy. Br. J. Cancer 74, 172-177. 10.1038/bjc.1996.334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith M. H., Ploegh H. L. and Weissman J. S. (2011). Road to ruin: targeting proteins for degradation in the endoplasmic reticulum. Science 334, 1086-1090. 10.1126/science.1209235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song S., Tan J., Miao Y., Li M. and Zhang Q. (2017). Crosstalk of autophagy and apoptosis: Involvement of the dual role of autophagy under ER stress. J. Cell. Physiol. 232, 2977-2984. 10.1002/jcp.25785 [DOI] [PubMed] [Google Scholar]
- Stein R. L., Melandri F. and Dick L. (1996). Kinetic characterization of the chymotryptic activity of the 20S proteasome. Biochemistry 35, 3899-3908. 10.1021/bi952262x [DOI] [PubMed] [Google Scholar]
- Stewart C., Estrada A., Kim P., Wang D., Wei Y., Gentile C. and Pagliassotti M. (2017). Regulation of IRE1alpha by the small molecule inhibitor 4mu8c in hepatoma cells. Endoplasmic Reticulum Stress Dis. 4, 1-10. 10.1515/ersc-2017-0001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stromhaug P. E., Reggiori F., Guan J., Wang C. W. and Klionsky D. J. (2004). Atg21 is a phosphoinositide binding protein required for efficient lipidation and localization of Atg8 during uptake of aminopeptidase I by selective autophagy. Mol. Biol. Cell 15, 3553-3566. 10.1091/mbc.e04-02-0147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanida I., Minematsu-Ikeguchi N., Ueno T. and Kominami E. (2005). Lysosomal turnover, but not a cellular level, of endogenous LC3 is a marker for autophagy. Autophagy 1, 84-91. 10.4161/auto.1.2.1697 [DOI] [PubMed] [Google Scholar]
- Tanida I., Ueno T. and Kominami E. (2008). LC3 and Autophagy. Methods Mol. Biol. 445, 77-88. 10.1007/978-1-59745-157-4_4 [DOI] [PubMed] [Google Scholar]
- Travers K. J., Patil C. K., Wodicka L., Lockhart D. J., Weissman J. S. and Walter P. (2000). Functional and genomic analyses reveal an essential coordination between the unfolded protein response and ER-associated degradation. Cell 101, 249-258. 10.1016/S0092-8674(00)80835-1 [DOI] [PubMed] [Google Scholar]
- Uozaki H., Ishida T., Kakiuchi C., Horiuchi H., Gotoh T., Iijima T., Imamura T. and Machinami R. (2000). Expression of heat shock proteins in osteosarcoma and its relationship to prognosis. Pathol. Res. Pract. 196, 665-673. 10.1016/S0344-0338(00)80118-1 [DOI] [PubMed] [Google Scholar]
- Vandewynckel Y. P., Laukens D., Geerts A., Bogaerts E., Paridaens A., Verhelst X., Janssens S., Heindryckx F. and Van Vlierberghe H. (2013). The paradox of the unfolded protein response in cancer. Anticancer Res. 33, 4683-4694. [PubMed] [Google Scholar]
- Vembar S. S. and Brodsky J. L. (2008). One step at a time: endoplasmic reticulum-associated degradation. Nat. Rev. Mol. Cell Biol. 9, 944-957. 10.1038/nrm2546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vermes I., Haanen C., Steffens-Nakken H. and Reutelingsperger C. (1995). A novel assay for apoptosis. Flow cytometric detection of phosphatidylserine expression on early apoptotic cells using fluorescein labelled Annexin V. J. Immunol. Methods 184, 39-51. [DOI] [PubMed] [Google Scholar]
- Voellmy R. and Boellmann F. (2007). Chaperone regulation of the heat shock protein response. Adv. Exp. Med. Biol. 594, 89-99. 10.1007/978-0-387-39975-1_9 [DOI] [PubMed] [Google Scholar]
- Walter P. and Ron D. (2011). The unfolded protein response: from stress pathway to homeostatic regulation. Science 334, 1081-1086. 10.1126/science.1209038 [DOI] [PubMed] [Google Scholar]
- Wang M. and Kaufman R. J. (2014). The impact of the endoplasmic reticulum protein-folding environment on cancer development. Nat. Rev. Cancer 14, 581-597. 10.1038/nrc3800 [DOI] [PubMed] [Google Scholar]
- Ward E., DeSantis C., Robbins A., Kohler B. and Jemal A. (2014). Childhood and adolescent cancer statistics, 2014. CA Cancer J. Clin. 64, 83-103. 10.3322/caac.21219 [DOI] [PubMed] [Google Scholar]
- Weaver B. A. A. and Cleveland D. W. (2005). Decoding the links between mitosis, cancer, and chemotherapy: the mitotic checkpoint, adaptation, and cell death. Cancer Cell 8, 7-12. 10.1016/j.ccr.2005.06.011 [DOI] [PubMed] [Google Scholar]
- Weigel B. J., Lyden E., Anderson J. R., Meyer W. H., Parham D. M., Rodeberg D. A., Michalski J. M., Hawkins D. S. and Arndt C. A. (2016). Intensive multiagent therapy, including dose-compressed cycles of ifosfamide/etoposide and vincristine/doxorubicin/cyclophosphamide, irinotecan, and radiation, in patients with high-risk rhabdomyosarcoma: a report from the children's oncology group. J. Clin. Oncol. 34, 117-122. 10.1200/JCO.2015.63.4048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weissman A. M. (1997). Regulating protein degradation by ubiquitination. Immunol. Today 18, 189-198. 10.1016/S0167-5699(97)84666-X [DOI] [PubMed] [Google Scholar]
- Williams B. R. and Amon A. (2009). Aneuploidy: cancer's fatal flaw? Cancer Res. 69, 5289-5291. 10.1158/0008-5472.CAN-09-0944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wisén S., Bertelsen E. B., Thompson A. D., Patury S., Ung P., Chang L., Evans C. G., Walter G. M., Wipf P., Carlson H. A. et al. (2010). Binding of a small molecule at a protein-protein interface regulates the chaperone activity of hsp70-hsp40. ACS Chem. Biol. 5, 611-622. 10.1021/cb1000422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wojcik S. (2013). Crosstalk between autophagy and proteasome protein degradation systems: possible implications for cancer therapy. Folia Histochem. Cytobiol. 51, 249-264. 10.5603/FHC.2013.0036 [DOI] [PubMed] [Google Scholar]
- Yamamoto K., Yoshida H., Kokame K., Kaufman R. J. and Mori K. (2004). Differential contributions of ATF6 and XBP1 to the activation of endoplasmic reticulum stress-responsive cis-acting elements ERSE, UPRE and ERSE-II. J. Biochem. 136, 343-350. 10.1093/jb/mvh122 [DOI] [PubMed] [Google Scholar]
- Yan M. M., Ni J. D., Song D., Ding M. and Huang J. (2015). Interplay between unfolded protein response and autophagy promotes tumor drug resistance. Oncol. Lett. 10, 1959-1969. 10.3892/ol.2015.3508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X., Wang J., Zhou Y., Wang Y., Wang S. and Zhang W. (2012). Hsp70 promotes chemoresistance by blocking Bax mitochondrial translocation in ovarian cancer cells. Cancer Lett. 321, 137-143. 10.1016/j.canlet.2012.01.030 [DOI] [PubMed] [Google Scholar]
- Ye Y., Meyer H. H. and Rapoport T. A. (2001). The AAA ATPase Cdc48/p97 and its partners transport proteins from the ER into the cytosol. Nature 414, 652-656. 10.1038/414652a [DOI] [PubMed] [Google Scholar]
- Yoshida H., Matsui T., Yamamoto A., Okada T. and Mori K. (2001a). XBP1 mRNA is induced by ATF6 and spliced by IRE1 in response to ER stress to produce a highly active transcription factor. Cell 107, 881-891. 10.1016/S0092-8674(01)00611-0 [DOI] [PubMed] [Google Scholar]
- Yoshida H., Okada T., Haze K., Yanagi H., Yura T., Negishi M. and Mori K. (2001b). Endoplasmic reticulum stress-induced formation of transcription factor complex ERSF including NF-Y (CBF) and activating transcription factors 6alpha and 6beta that activates the mammalian unfolded protein response. Mol. Cell. Biol. 21, 1239-1248. 10.1128/MCB.21.4.1239-1248.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young J. C., Agashe V. R., Siegers K. and Hartl F. U. (2004). Pathways of chaperone-mediated protein folding in the cytosol. Nat. Rev. Mol. Cell Biol. 5, 781-791. 10.1038/nrm1492 [DOI] [PubMed] [Google Scholar]
- Yousefi S., Perozzo R., Schmid I., Ziemiecki A., Schaffner T., Scapozza L., Brunner T. and Simon H.-U. (2006). Calpain-mediated cleavage of Atg5 switches autophagy to apoptosis. Nat. Cell Biol. 8, 1124-1132. 10.1038/ncb1482 [DOI] [PubMed] [Google Scholar]
- Zhong Y., Shen H., Wang Y., Yang Y., Yang P. and Fang S. (2015). Identification of ERAD components essential for dislocation of the null Hong Kong variant of alpha-1-antitrypsin (NHK). Biochem. Biophys. Res. Commun. 458, 424-428. 10.1016/j.bbrc.2015.01.133 [DOI] [PubMed] [Google Scholar]
- Zhou H.-J., Wang J., Yao B., Wong S., Djakovic S., Kumar B., Rice J., Valle E., Soriano F., Menon M.-K. et al. (2015). Discovery of a first-in-class, potent, selective, and orally bioavailable inhibitor of the p97 AAA ATPase (CB-5083). J. Med. Chem. 58, 9480-9497. 10.1021/acs.jmedchem.5b01346 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.