Abstract
Deletions of the 1p region appear as a pejorative prognostic factor in multiple myeloma patients (especially 1p22 and 1p32 deletions) but there’s a lack of data on the real impact of 1p abnormalities on an important and homogeneous group of patients. To address this issue we studied by fluorescence in situ hybridization (FISH) the incidence and prognostic impact of 1p22 and 1p32 deletions in 1195 patients from the IFM (Institut Francophone du Myélome) cell collection. Chromosome 1p deletions were present in 23.3% of the patients (271): 15.1% (176) for 1p22 and 7.3% (85) for 1p32 regions. In univariate analyses, 1p22 and 1p32 appeared as negative prognostic factors for PFS: 1p22: 19.8 months vs 33.6 months (p<0.001) and 1p32: 14.4 months vs 33.6 months p(<0.001); and OS: 1p22: 44.2 months vs 96.8 months (p=0.002) and 1p32: 26.7 months vs 96.8 months (p<0.001). In multivariate analyses 1p22 and 1p32 deletions still appear as independent negative prognostic factors for PFS and OS. In conclusion our data show that 1p22 and 1p32 deletions are major negative prognostic factors for PFS and OS for patients with MM. We thus suggest that 1p32 deletion should be tested for all patients at diagnosis
Keywords: deletion 1p22, deletion 1p32, multiple myeloma, prognosis
Keywords: myeloma, cytogenetics, prognosis
Introduction
Cytogenetic findings are the most important prognostic factors for patients with multiple myeloma (MM) with the International Staging System (ISS) based on evaluation of serum levels of β2-microglobulin (β2m) and albumin1. Classically, routine evaluation consists of FISH analysis of deletion 17p, t(4;14) and t(14;16)2. Others have mixed those two classifications (ISS + FISH) to improve the prognostic classification of MM patients3. However many others abnormalities occur in Multiple Myeloma such as gains of chromosomes 3, 5, 7, 9, 11, 15, 19 and 21 (hyperdiploidy) in 50 to 60% of patients4, structural abnormalities such as del(13) in approximately 50% of patients5, del(16q) (20% of patients)6, del(17p) (10% of patients)7, gain of 1q observed in approximately 30% of patients8 or translocations involving immunoglobulin heavy chain locus t(4;14)(p16.3;q32) (15% of patients)9 and t(14;16)(q32;q23) (3% of patients)10. New technologies such as SNP-based mapping array provides a deeper knowledge of diversity and heterogeneity of cytogenetic abnormalities in MM11–12; almost all chromosomes may be involved with a frequency and prognostic impact on outcome specific to each.
Current molecular classification based only on del(17p), t(4;14) and t(14;16) is clearly inadequate to explain the heterogeneity of MM patients and to perform a tailored evaluation of outcome. Thus, some cytogenetic abnormalities could be targeted in routine, individually or in combination, for a better assessment of patient outcome. Deletions of 1p are frequent events in MM patients (18 to 38%) and seem to impact outcome of patients 13–15. However, a study on a large cohort of newly diagnosed uniformlytreated MM patients is still missing. This study sought to evaluate the incidence and prognostic significance of presence of deletions 1p22 and 1p32 by FISH analysis on a large homogeneous cohort of 1195 patients from the IFM cell collection.
Patients and methods
We studied 1195 patients with newly diagnosed MM, younger than 66 years of age. They were all treated by induction therapy: VAD (vincristine-adriamycin-dexamethasone) or bortezomib based induction followed by high-dose melphalan autologous stem cell transplantation in IFM centers. No patient received maintenance therapy. All patients signed an informed consent form for research purposes, validated by the local Ethics committee.
• Cell sorting and FISH analysis
Upon receipt, bone marrow plasma cells were sorted using nanobeads and an anti-CD138 antibody (RoboSep, Stem Cell Technologies). After immunomagnetic sorting, the plasma cell suspension purity was checked, and only samples with at least 90% of plasma cells were kept. Cells were then fixed in Carnoy’s fixative. Deletions of 1p22 and 1p32 were evaluated using probes RP11–242C15 and RP11–100D3 for 1p22 and RP11–762I3 for 1p32. Hybridizations were performed according to previously published techniques.15 For analysis, at least 100 plasma cells with correct signals were scored using a Zeiss epifluorescence microscope. Abnormality was considered positive if present in at least 30% of analyzed plasma cells except for del(17p) where the threshold is at 60%.16
• Statistics
Statistical analysis were performed by the Unité de Soutien Méthodologique à la Recherche Clinique (CHU Toulouse). Group comparison was made using Chi2 and Wilcoxon test. Median follow-up was assessed using reverse Kaplan-Meier method17. Overall Survival (OS) and Progression Free Survival (PFS) were calculated from the date of diagnosis using the Kaplan-Meier method, with the log rank test used to compare survival curves. First a stratified univariate Cox model was performed to estimate hazard ratios (HR) and 95% confidence intervals (95% CI). Stratification by clinical trials was done to correct for the different therapeutic schemas. The multivariate Cox model was finally adjusted on all prognostic variables (age, beta2 microglobulin, induction therapy, cytogenetic variables). First order interactions between cytogenetic variables and between induction therapy and cytogenetic variables have been explored. The proportionality assumptions have been checked with Cox-Snell residuals and log-log plots. For the overall survival analysis, we observed a violation of the proportional hazards assumption for del1p22 so, an interaction term with a function of time was added to model the evolvement of the HR over time. Tests were two sided and p values lower than 0.05 were considered significant. All analyses were conducted using stata® Version 11.0.
Results
-Patients characteristics
We analyzed a large cohort of 1195 patients from 21 IFM centers treated either by VAD (64.3%) or bortezomib-based (35.7%) induction followed by high dose melphalan (200 mg/m2) and autologous stem cell transplantation. Median age at diagnosis was 57.7 years (IQR [52.2 – 62.0]) with 56.3% men and 43.7% women. Overall 33.6% were ISS1, 37.4% ISS2 and 29.0% ISS3;
-cytogenetic findings
We observed a usual distribution for classical cytogenetic abnormalities in our cohort with 128 patients (12.5%) with t(4;14), 93 patients (9.5%) with a del(17p) in more than 60% of the plasma cells, and 529 (48.5%) with a deletion of chromosome 13. Information on adverse cytogenetic was missing in 170, 219 and 105 patients for t(4;14), del(17p) and deletion of chromosome 13 respectively.
-Incidence of 1p deletion
Deletions 1p were present in 23.3% of our cohort (261 patients), 15.1% (n=176) had deletion 1p22 and 7.3% (n=85) had deletion 1p32. Thirty-one patients were not evaluable for del(1p22) and 29 for de(1p32). Deletion 1p22 was associated with higher β2m (p<0.07), t(4;14) (p<0.001), del(17p) (p<0.001), deletion of chromosome 13 (p<0.001) and ISS status at diagnosis (p=0.01) (Table 1). Similarly, deletion 1p32 was associated with t(4;14) (p=0.001), del(17p) (p=0.002), deletion of chromosome 13 (p=0.001) and ISS status (p=0.03). So, deletions 1p are a frequent event for MM patients, with an incidence comparable to cytogenetic abnormalities of clinical interest: t(4;14) : 12.5% and del(17p): 9.5% versus respectively 15.1% and 7.3% for deletion 1p22 and 1p32. Thus, those deletions are not marginal events in MM patients, especially if we compare in the literature with t(14;16), another translocation of clinical interest to evaluate the prognosis of patients, observed in approximately 3% of the patients9.
Table 1.
Association between deletions 1p and other factors
| Characteristics | No deletion N=904 |
Deletion 1p22 N=176 |
P* | Deletion 1p32 N=85 |
P* | |
|---|---|---|---|---|---|---|
| Beta2 microglobulin | Median (IQR) | 3.2 (2.3–5.1) | 3.5 (2.5–6.4) | 0.07 Wilcoxon | 3.8 (2.7–7.2) | 0.02 Wilcoxon |
| ISS N(%) | 1 | 239 (35.7) | 36 (29.0) | 0.01 | 13 (20.6) | 0.03 |
| 2 | 254 (37.9) | 39(31.5) | Chi2 | 26(41.3) | Chi2 | |
| 3 | 177(26.4) | 49 (39.5) | 24(38.1) | |||
| t(4;14) N(%) | No | 706(91.2) | 116(74.8) | <0.001 | 54(78.3) | 0.001 |
| Yes | 68(8.8) | 39 (25.2) | Chi2 | 15(21.7) | Chi2 | |
| Deletion 17 p N(%) | No | 686 (93.0) | 119(82.6) | <0.001 | 62 (82.7) | 0.002 |
| Yes | 52(7.1) | 25(17.4) | Chi2 | 13(17.3) | Chi2 | |
| Deletion 13 N(%) | No | 463 (56.3) | 53(33.3) | <0.001 | 29 (36.3) | 0.001 |
| Yes | 359 (43.7) | 106(66.7) | Chi2 | 51 (63.8) | Chi2 | |
| Bortezomib induction N(%) | No | 589 (65.2) | 107(60.8) | 0.27 | 48 (56.5) | 0.11 |
| Yes | 315(34.9) | 69 (39.2) | Chi2 | 37 (43.5) | Chi2 |
The comparisons are made versus the patients with no deletion 1p.
Survival analysis
The median follow-up was 81.3 months [35.3–105.9]
-Prognostic impact of 1p deletions on PFS
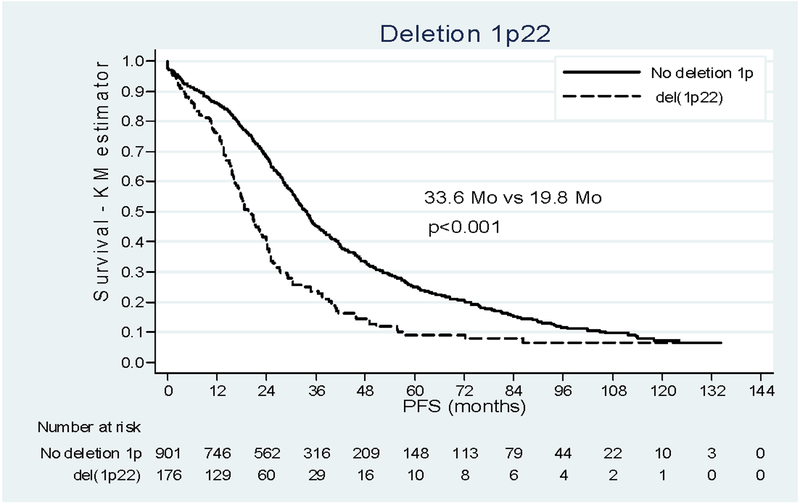
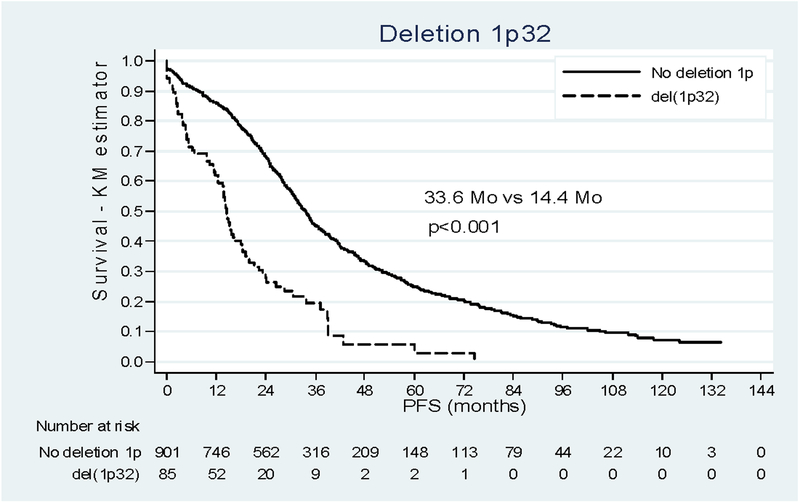
Survival analysis of the whole cohort showed a median duration of PFS of 30.2 months (16.3 – 54.0). Del(1p22) impacted negatively the outcome of patients, with a median PFS of 19.8 months [12.5 – 34.6] versus 33.6 months [20.3 – 60.1] when deletion 1p was absent. Del(1p32) also dramatically reduced the median PFS of patients from 33.6 months to 14.4 months [4.8 – 26.4] p<0.001 (Figure 1). Restricting the analysis to the patients with complete information on prognostic variables (N=679, Table 2) we found the negative incidence on prognosis already described15 for t(4;14), del(17p) and beta 2 microglobulin. Adjusting for these confounders, age and the type of induction therapy, both del(1p22) and del(1p32) remained independently associated with progression (HRa 1.56 (1.21–2.01); HRa 2.84 (2.02–3.98)). No interaction was found between del(1p22) or del(1p32) and others prognostic factors.
Figure 1:
Impact of deletion 1p22 and 1p32 on PFS. (a) Patients deleted for 1p22 (dashed line) versus patient not deleted for 1p (solid line). (b) Patients deleted for 1p32 (dashed line) versus patient not deleted for 1p (solid line).
Table 2:
Progression Free Survival
| Univariate analysis | Multivariate analysis | |||
|---|---|---|---|---|
| Variable | HR [IC95%] | P | HRa [IC95%] | P |
| Bortezomib Induction | ||||
| No (415 / 352) | 1 | 1 | ||
| Yes (264 / 158) | 0.65 [0.43–0.97] | 0.04 | 0.53 [0.35 ; 0.80] | 0.002 |
| Beta2 microglobulin (679 / 510) | 1.03 [1.02–1.04] | <0.001 | 1.03 [1.01 ; 1.04] | 0.001 |
| Deletion 1p | ||||
| No deletion 1p (532 / 388) | 1 | 1 | ||
| Del(1p22) (97 / 80) | 1.94 [1.52–2.47] | <0.001 | 1.56 [1.20 ; 2.01] | 0.001 |
| Del(1p32) (50 / 42) | 3.21 [2.32–4.44] | <0.001 | 2.84 [2.02 ; 3.98] | <0.001 |
| t(4;14) | ||||
| No (601 / 458) | 1 | 1 | ||
| Yes (78 / 52) | 2.28 [1.76–2.95] | <0.001 | 1.85 [1.40 ; 2.44] | <0.001 |
| Deletion 17p | ||||
| No (617 / 458) | 1 | 1 | ||
| Yes (62 / 52) | 2.12 [1.59–2.83] | <0.001 | 1.88 [1.39 ; 2.54] | <0.001 |
-Prognostic impact of 1p deletions on OS
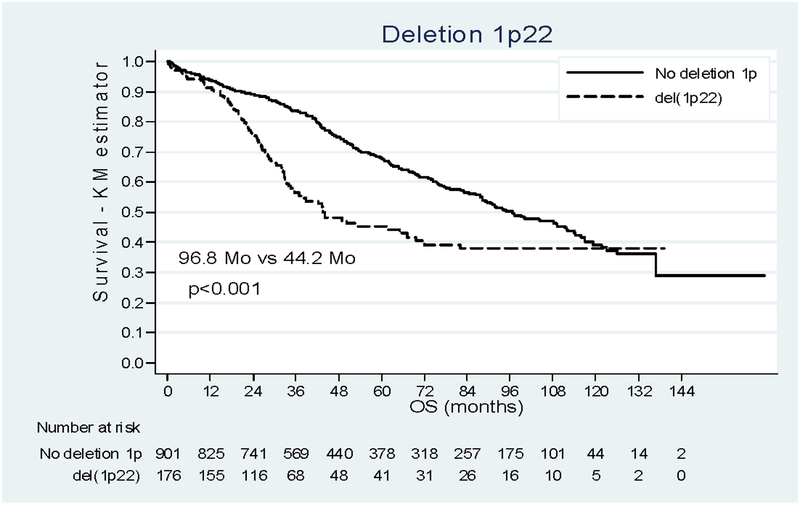
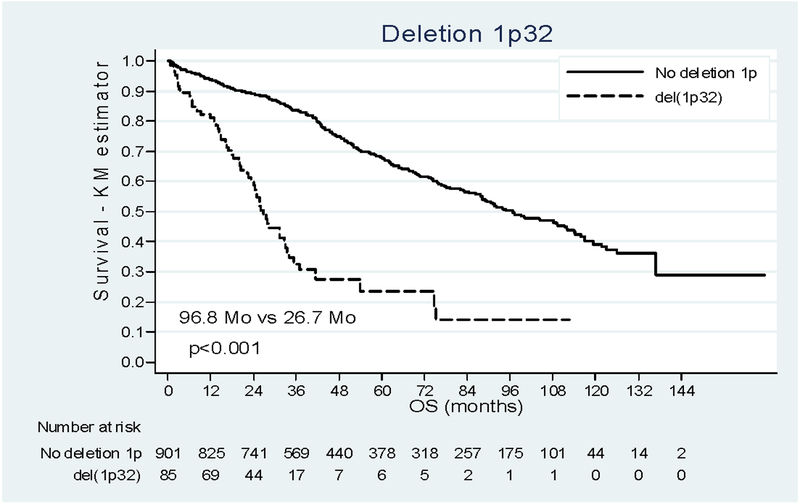
Survival analysis of the whole cohort showed a median duration of OS of 88.2 months [38.7 - Not Reached (NR)]. For OS, the impact of 1p deletions was more contrasted (Figure 2). The median OS was 96.8 months [47.6 - NR] in patients without del(1p) and significantly shorter 44.2 months [25.0 - NR] in patients with del(1p22). In the multivariate analysis (N=679, Table 3), del(1p22) appears as a negative prognostic factor and was found to decrease over time (del(1p22) by time interaction=0.002). The adjusted HR was, 1.82 [1.17–2.84] at diagnosis and it was equal to 1.61 [1.09–2.38], 1.10 [0.72–1.67], 0.58 [0.25–1.35] at 24, 48 and 72 months respectively. After 72 months, only one death was observed and the Kaplan-Meier curves cannot be clearly interpreted since data beyond this point were sparse. For del(1p32), we observed a dramatic reduction of median OS : 26.7months [14.9–53.9]. In the multivariate analysis, the risk of death was 4 times higher among people with del(1p32) (p<0.001, HRa 4.07 [2.67–6.20]) after adjustment on age, treatment, β2m, del(17p), t(4;14) and induction treatment.
Figure 2:
Impact of deletion 1p22 and 1p32 on OS. (a) Patients deleted for 1p22 (dashed line) versus patient not deleted for 1p (solid line). (b) Patients deleted for 1p32 (dashed line) versus patient not deleted for 1p (solid line).
Table 3:
Overall Survival Analysis
| Univariate analysis | Multivariate analysis | |||
|---|---|---|---|---|
| Variable | HR [IC95%] | P | HRa [IC95%] | P |
| Bortezomib Induction | ||||
| No (415 / 208) | 1 | 1 | ||
| Yes (264 / 50) | 0.58 [0.31–1.05] | 0.07 | 0.52 [0.28 ; 0.95] | 0.03 |
| Beta2 microglobulin (679 / 258) | 1.04 [1.02–1.05] | <0.001 | 1.03 [1.01–1.05] | 0.001 |
| Deletion 1p | ||||
| No deletion (532 / 185) | 1 | 1 | ||
| Del1p22* (97 / 41) | 2.48* [1.62–3.80] | <0.001 | 1.82* [1.17 ; 2.84] | 0.008 |
| Del1p32 (50 / 32) | 5.17 [3.49–7.67] | <0.001 | 4.07 [2.67 ; 6.20] | <0.001 |
| t(4;14) | ||||
| No (601 / 213) | 1 | 1 | ||
| Yes (78 / 45) | 2.39 [1.73–3.30] | <0.001 | 2.02 [1.43 ; 2.85] | <0.001 |
| Deletion 17p | ||||
| No (617 / 214) | 1 | 1 | ||
| Yes (62 / 44) | 3.43 [2.48–4.76] | <0.001 | 2.99 [2.13 ; 4.20] | <0.001 |
The hazard ratio for 1p22 was not proportional over time. The estimates reported in this table are HR at diagnosis.
In this model, β2m, del(17p) , t(4;14) and induction treatment kept their independent prognostic value. The prognostic effect of del(1p22) and del(1p32) were similar across levels of the prognostic variables , as no interaction was found between del(1p22) or del(1p32) and these factors.
Discussion
Multiple myeloma is characterized by the heterogeneity of patients: clinical presentation, cytogenetic abnormalities, response to treatment, and survival. The larger heterogeneity is for survival, some patients living more than ten years whereas some die within few months, with an identical biological and clinical presentation. Thus, many studies try to determine prognostic factors to predict outcome of patients. The most currently used are the ISS classification1, and FISH analysis for t(4;14), deletion 17p and t(14;16)2,3. But many others have been described: other cytogenetic abnormalities, cytokine levels, bone involvement at diagnosis, and combinations of these factors.
Among chromosomal changes, deletions of the 1p region seem to play a significant role on the outcome of the patients. In a work by Walker et al12, the incidence of deletion 1p rose to 30% (24% in the work of Avet-Loiseau et al11). In 2007, Giralt14 et al. identified deletion 1p in 18% of patients and found an association with a significantly shorter PFS and OS. Boyd et al13 have published a study of deletions at 1p12, 1p22 and 1p32 where del(1p32) was present in 11.3% of patients and impaired survival (34.5 months versus not reached, p<0.001) and 1p22 did not affect survival although it was the most frequently deleted region (22.5%). Moreover, they described putative target genes in 1p32 region: CDKN2C and FAF1; and MTF2 and TMED5 for a vast region from 1p22.1 to 1p21.3.
By comparison, we screened 1195 patients under 65 years of age, all treated intensively by stem cell transplantation rescue with a long follow-up. Our findings show a slightly lower incidence than previously described, 15.1% (versus 22.5%) for del(1p22) and 7.3% (versus 11.3%) for del(1p32). Our survival data confirm that deletions 1p22 and 1p32 impaired outcome of patients both on PFS and OS. Our data also underline the link between deletions 1p and t(4;14), del(17p), monosomy 13 and ISS. Another point of importance is our adjustment for possible imbalance in induction with VAD or bortezomib, showing that the effect on outcome of deletions 1p was independent of induction treatment. For OS, the deviation from proportional hazards for del1p22 may reflect selection effects which can appear in long term survival data due to the early death of patients with poor prognostic factors and some omitted covariates in the model like the type of relapse therapy. Even if other authors have suggested that there was a decreased effect over time for prognostic factors among patients with other malignancies, additional studies are needed with a longer follow-up in a larger population of patients del(1p22) to clearly understand this result. If we compare the hazard ratio obtained in multivariate analysis, we observe that del(1p32) presents the highest hazard ratio both for PFS and OS. In consequence, del(1p32) appears as a major independent prognostic factor with a relapse and death risk clearly increased in regards to other classical cytogenetic abnormalities.
In conclusion, it is the first time that the prognostic impact of deletions 1p22 and 1p32 is clearly confirmed on a large homogenous cohort of young patients. Both deletions are frequent events and have a significant impact on PFS and OS as independent prognostic factors in the multivariate analysis. Considering its dramatic effect on outcome, del(1p32) should be tested for every patient at diagnosis, with t(4;14) and del(17p).
Acknowledgements:
This study is partially supported by a grant from the French Institut National du Cancer (INCa), and by a grant from the American NIH (PO1-155258).
Footnotes
Presented in abstract form at the 54th annual meeting of the American Society of Hematology, Atlanta, GA, December 11, 2012
We have no conflict of interest to disclose.
References
- 1.Greipp PR, San Miguel J, Durie BG, Crowley JJ, Barlogie B, Blade J, et al. International Staging System for Multiple Myeloma. J Clin Oncol. 2005; 23:3412–3420. [DOI] [PubMed] [Google Scholar]
- 2.Fonseca R, Bergsagel PL, Drach J, Shaughnessy J, Gutierrez N, Stewart K, et al. International Myeloma Working Group molecular classification of multiple myeloma: spotlight review. Leukemia. 2009; 23:2210–2221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Avet-Loiseau H, Durie BG, Cavo M, Attal M, Gutierrez N, Haessler J, et al. Combining Fluorescent In Situ Hybridization (iFISH) data with ISS staging improves risk assessment in myeloma: an International Myeloma Working Group (IMWG) collaborative project. Leukemia. 2012. October 3 [Epub ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chng WJ, Kumar S, Vanwier S, Ahmann G, Price-Troska T, Henderson K, et al. Molecular dissection of hyperdiploid multiple myeloma by gene expression profiling. Cancer Res. 2007; 67:2982–2989. [DOI] [PubMed] [Google Scholar]
- 5.Facon T, Avet-Loiseau H, Guillerm G, Moreau P, Genevieve F, Zandecki M, et al. Chromosome 13 abnormalities identified by FISH analysis and serum beta2-microglobulin produce a very powerful myeloma staging system for patients receiving high dose therapy. Blood.2001; 97:1566–1571. [DOI] [PubMed] [Google Scholar]
- 6.Jenner MW, Leone PE, Walker BA, Ross FM, Johnson DC, Gonzalez D, et al. Gene mapping and expression analysis of 16q loss of heterozygosity identifies WWOX and CYLD as being important in clinical outcome in multiple myeloma. Blood. 2007; 110:3291–3300. [DOI] [PubMed] [Google Scholar]
- 7.Chang H, Qi C, Yi QL, Reece D, Stewart K. p53 gene deletion detected by fluorescence in situ hybridization is an adverse prognostic factor for patients with multiple myeloma following autologous stem cell transplantation. Blood. 2005; 105:358–360. [DOI] [PubMed] [Google Scholar]
- 8.Fonseca R, Van Wier SA, Chng WJ, Ketterling R, Lacy MQ, Dispenzieri A, et al. Prognostic value of chromosome 1q21 gain by fluorescent in situ hybridization and increase CKS1B expression in myeloma. Leukemia. 2006; 20:2034–2040. [DOI] [PubMed] [Google Scholar]
- 9.Moreau P, Facon T, Leleu X, Morineau N, Huyghe P, Harousseau JL, et al. Recurrent 14q32 translocations determine the prognosis of multiple myeloma, especially in patients receiving intensive chemotherapy. Blood. 2002; 100:1579–1583. [DOI] [PubMed] [Google Scholar]
- 10.Avet-Loiseau H, Malard F, Campion L, Magrangeas F, Sebban C, Lioure B, et al. Translocation t(14;16) and multiple myeloma: is it really an independent prognostic factor? Blood. 2011. ; 117(6):2009–11. [DOI] [PubMed] [Google Scholar]
- 11.Avet-Loiseau H, Li C, Magrangeas F, Gouraud W, Charbonnel C, Harousseau JL, et al. Prognostic significance of Copy-Number Alterations in Multiple Myeloma. J Clin Oncol. 2009; 27:4585–4590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Walker BA, Leone PE, Chiecchio L, Dickens NJ, Jenner MW, Boyd KD, et al. A compendium of myeloma-associated chromosomal copy number abnormalities and their prognostic value. Blood. 2010; 116(15):e56–65. [DOI] [PubMed] [Google Scholar]
- 13.Boyd K, Ross F, Walker BA, Wardell CP, Tapper WJ, Chiecchio L, et al. Mapping of Chromosome 1p Deletions in Myeloma Identifies FAM46C at 1p12 and CDKN2C at 1p32.3 as Being Genes in Regions Associated with Adverse Survival. Clin Cancer Res. 2011; 17:7776–7784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Qazilbash MH, Saliba RM, Ahmed B, Parikh G, Mendoza F, Ashraf N, et al. Deletion of the short arm of chromosome 1 (del 1p) is a strong predictor of poor outcome in myeloma patients undergoing an autotransplant. Biol Blood Marrow Transplant. 2007; 13(9):1066–72. [DOI] [PubMed] [Google Scholar]
- 15.Leone PE, Walker BA, Jenner MW, Chiecchio L, Dagrada G, Protheroe RK, et al. Deletions of CDKN2C in multiple myeloma: biological and clinical implications. Clin Cancer Res. 2008; 14(19):6033–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Avet-Loiseau H, Attal M, Moreau P, Charbonnel C, Garban F, Hulin C, et al. Genetic abnormalities and survival in multiple myeloma: the experience of the Intergroupe Francophone du Myelome. Blood. 2007; 109:3489–3495. [DOI] [PubMed] [Google Scholar]
- 17.Schemper M, Smith TL. A note on quantifying follow-up in studies of failure time. Control Clin Trials. 1996;17:343–6 [DOI] [PubMed] [Google Scholar]