Abstract
MscL is a bacterial mechanosensitive channel that serves as a cellular emergency release valve, protecting the cell from lysis upon a drop in external osmolarity. The channel has an extremely large pore (30Å) and can be purified and reconstituted into artificial membranes. Moreover, MscL has been modified to open in response to alternative external stimuli including changes in pH. These properties suggest this channel’s potential as a triggered “nanopore” for localized release of vesicular contents such as MRI contrast agents and drugs. Toward this end, we have engineered several variants of pH-triggered MscL nanovalves. We have reconstituted them into stealth vesicles previously been shown to evade normal in vivo clearance and passively accumulate in inflamed and malignant tissues. These vesicles were loaded with Gd-DOTA, an MRI contrast reagent, and the resulting nanodevices tested for their ability to release Gd-DOTA as evidenced by enhancement of the longitudinal relaxation rate (R1) of the bulk water proton spins. We have identified nanovalves that are responsive to physiological pH changes, but differ in sensitivity and efficacy, thus giving an array of nanovalves that could potentially be useful in different settings. These triggered nanodevices may be useful in delivering both diagnostic and therapeutic agents.
Keywords: MscL, nanovalve, pH sensor, drug targeting, theranostics
1. Introduction.
MscL serves as a biological emergency release valve allowing the bacterial cell to survive acute decreases in osmotic environment.[1] Briefly, when bacteria encounter an environment that is of much lower osmolarity than that to which they have equilibrated, the cell swells, thus causing membrane tension. Mechanosensitive channels, such as MscL, open in response to this tension and jettison many cytoplasmic solutes, thus relieving the increased cell turgor.[1] The MscL channel structure has been well studied; it is a homopentamer with each subunit consisting of a bit over 100 amino acids and two transmembrane domains for most orthologues, with known crystal structures in the closed and partially open states,[2–4] as well as models for the opening of the channel and open channel structure.[5, 6]
The MscL channels are extremely tractable and tolerant of significant changes that can influence the channel’s sensing and gating properties. The channels are easily reconstituted in lipid bilayers, and when opened their pore size is around 30Å in diameter; it is the largest gated channel pore known.[7] Thus far, either by design or serendipity, modified MscL channels have been shown to be sensitive to diverse stimuli including redox, light and pH.[8–12]
The detection and treatment of damaged and cancerous tissues is critically important for favorable medical outcomes. One of the most commonly used clinical imaging modalities is magnetic resonance imaging (MRI), which provides exquisite anatomical images of soft tissues with excellent spatial resolution. A Gd-based contrast agent is often used to enhance image contrast. However, intravenously injected conventional small molecular weight Gd-based MRI agents have a short distribution half-life of about 5 min because they rapidly leak from the blood pool into the interstitium due to their low molecular weight. Previous studies have shown that liposome encapsulated contrast agents accumulate in cancerous and inflammatory tissues by passive targeting and in addition, in stealth liposomal systems, the presence of PEGylated lipids enable the liposomal MRI agents to escape clearance by the reticular endoplasmic system[13]. [14–16] Unfortunately, relaxivity quenching due to the slow diffusion rate of water molecules across the lipid bilayer is an issue.[17] What is needed is an efficient way to release the payload, whether it be an MRI contrast or even an anti-cancer drug.
The ability to reconstitute the large gated MscL pores into vesicles, and to change their sensory modality to pH and other effectors make a modified MscL attractive as a triggered nanovalve for liposomal-based imaging agents and drug delivery systems. In past studies a pH-sensitive MscL was generated by modifying a glycine residue within the pore region, at position 22, in each of the 5 subunits.[11, 12] The G22 residue was first mutated to Cys, then the Cys residues were functionalized with sulfhydryl-reactive pH-modulators whose protonation induced channel opening. However, the resulting triggered nanovalve is extremely inefficient. The choice of modifying G22 is largely historical, and subsequent screening and characterization of modifications in the region have suggested that this may not be the only or most optimal site to be engineered to change MscL modality.[18–21] The goal of this present work is to explore how various MscL mutants, engineered by varying the placement and number of modified sites of MscL, influence and potentially improve upon the efficacy of a liposome encapsulated pH-responsive MRI agent; several of the engineered MscL-based triggered channels, or “nanovalves described here are vastly superior to any previously reported or studied and thus may serve in multiple forms of pH-sensitive nanodevices.
2. Results.
2.1. MscL nanopores modified at multiple sites show faster kinetic responses and improved release of MRI agent upon pH decreases relative to MscL nanopores with a single alteration.
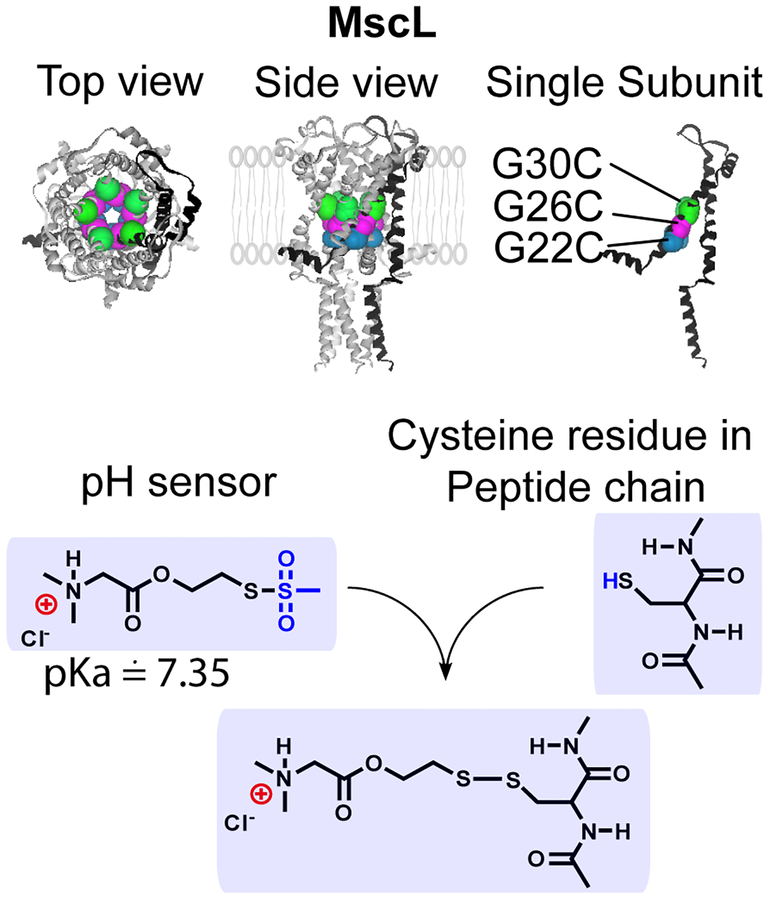
Modifications, especially adding charged residues within the constriction of the MscL pore, have previously been shown to increase channel gating.[18, 19, 21–25] This finding has led to studies in which researchers have changed the modality of the MscL channel by modifying residues in the pore.[8, 11, 12, 20, 26–28] In the vast majority of these studies, the modification site was G22. Using electrophysiological approaches and a calcein dye flux assay, we have tested several other possible sites for channel modification and have recently found an alternative site, G26, to be superior in many aspects including sensitivity and effectiveness.[19–21] We therefore directly compared the pH-responsiveness when these residues, G22, G26, as well as another site shown to have promise, G30C[19]. These residues were individually modified to contain a pH sensitive moiety, dimethylglycine, and assembled into a liposomal responsive device appropriate for MR analysis. Stealth vesicles, loaded with the macrocyclic magnetic resonance imaging contrast agent Gd-DOTA and containing the triggered nanovalves within the lipid membrane were prepared, as described in Materials and Methods. Figure 1 shows the modified residues transposed onto a model for the Escherichia coli (E. coli) channel[29] derived from the M. tuberculosis MscL structure,[3]. These residues were mutated to cysteines (G22C, G26C and G30C) and reacted with N, N-dimethylglycine 2-bromoethyl ester, a thiol selective reagent, to covalently modify the channel (Figure 1). The pH response of the Gd-DOTA loaded proteoliposomes containing labeled MscL were characterized by measuring the increase in the longitudinal relaxation rate (R1) of the bulk water protons upon decreasing the pH from 7.4 to 5.5. We have also tested the pH response of proteoliposomes containing non-labeled MscL, or no protein at all, and found that neither responded to changes in pH; for simplicity, the no protein vesicles were used as the control for the experiments presented. As shown in Figure 2a, labeling the nanovalve at G26 improved the pH response and have faster kinetics relative to vesicles containing the nanovalves labeled at G22, demonstrating that labeling at the G26 site has advantages over the G22 modified nanopore. Labeling at the single site G30C did not yield any pH response (not shown). However, even labeling at the G26 site yielded less than 50% pH-responsive release of the total possible liposomal R1 signal, which was determined by detergent lysis at the end of the experiment.
Figure 1: Labeling of MscL channel at specific sites.
Closed structure of the E. coli MscL based on the M. tuberculosis MscL crystal structure. The side view of MscL shows its location in lipid bilayer. For clarification, one single subunit is highlighted in black in the homopentameric structure.
Figure 2: Comparison of pH sensitive R1 release mediated by different labeled MscL nanovalves.
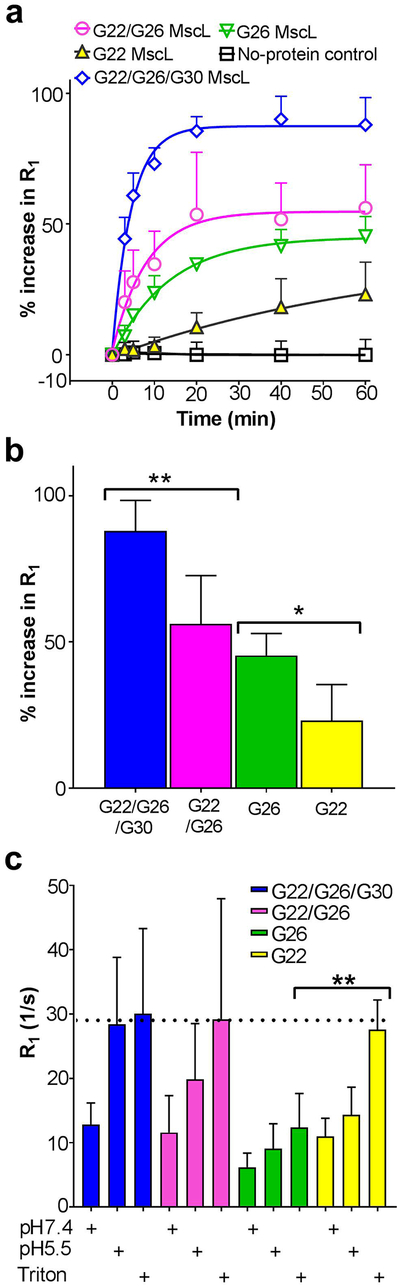
(a) kinetics R1 change at different time point was normalized to the Triton X-100 induced maximal change and expressed in percentage. Non-linear fitting of the data was analyzed by one phase decay equation in Graphpad Prism software. τ value for G22/G26/G30 (blue diamonds), G22/G26C (magenta circles), G26 (green triangles) and G22 (yellow triangles) nanovalves were 4.52 min (n = 5), 8.09 min (n = 5), 13.46 min (n = 3) and 67.42 min (n = 4), respectively. Control liposomes (black squares) without MscL showed no apparent change upon decrease in pH (n = 9); (b) Percentage increase at the 1 hour time point “**”: p < 0.01, t = 3.64, df = 8; “*”: p < 0.05, t = 2.78, df = 6, Student t-test. (c) Absolute R1 relaxation rates at the 1 hour time point. The dashed line is for comparison of maximum possible signals, as determined by proteoliposome lysis. “**”, p < 0.01 student t test.
In an attempt to increase the efficacy of the system, we generated double and triple mutants of MscL for pH sensor placements, thus allowing labeling at more sites within each subunit. Two of the most promising were one double (G22C+G26C) and one triple (G22C+G26C+G30C) combination. Figure 2a and b compare the kinetics and extent of the pH-dependent R1 responses of the vesicles containing the G22/G26 and G22/G26/G30 MscL modified nanopore relative to the nanopore modified at the single sites. Upon pH stimulus, both the vesicles containing the double-labeled and triple-labeled nanopore showed greater efficacy as determined by the observed relaxation rate expressed as the percentage of maximum possible R1 signal, as well as faster kinetics. The triple labeled nanopore showed an efficacy achieving nearly 100% of the maximal signal upon pH stimulus. This nanodevice also displayed the fastest kinetics with a τ of 4.5 min (n = 5) compared with vesicles containing nanopores labeled at G22/G26 (τ = 8.1 min) (n = 5), or only at the single sites G26 (τ = 13.5 min) (n = 3) and G22 (τ = 67.4 min) (n = 4). Figure 2c shows the absolute R1 values at pH 7.4 and 5.5 for each kind of liposomes at the 1 hour time point, demonstrating the stability and pH responsiveness of the proteoliposomes within this time frame. Note that the maximal absolute signal (highest R1 relaxation rate) for each type of proteoliposome observed after lysing the liposomes with Triton X-100, which destroys the lipid bilayer thus making the water exchange on the Gd-DOTA unimpeded, were similar for G22/26/30, G22/G26 and G22 MscL containing proteoliposomes, but reduced for the G26 MscL containing liposomes. However, the loading of Gd-DOTA (expressed as relative increase in R1 after lysis) for G26 is similar to the other proteoliposomes, suggesting a consistent decrease in the number of viable proteoliposomes for G26, perhaps due to sporadic openings of the G26 nanovalves upon proteoliposome synthesis (Supplemental Figure 2).
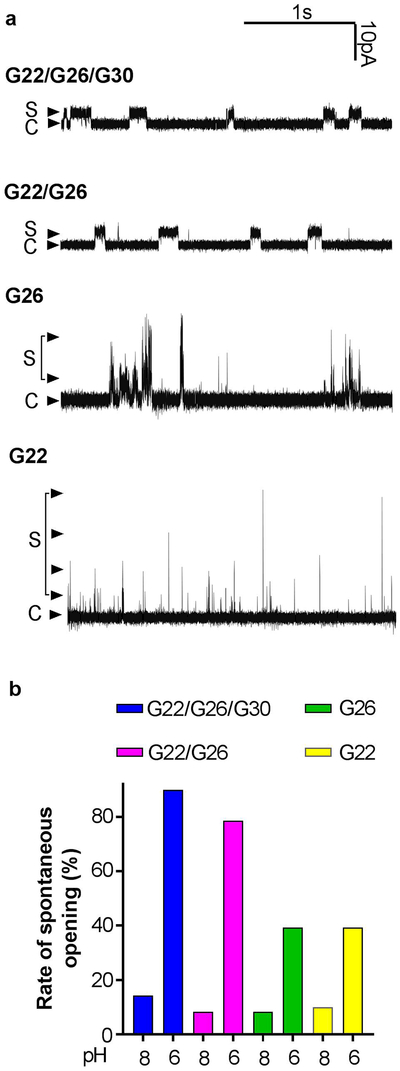
To determine the properties of individual pH-induced opening events of the labeled MscL nanopores, we reconstituted the nanopores into azolectin lipids and formed unilamellar blisters suitable for electrophysiological recording.[30, 31] As we had anticipated, and as shown in Figure 3a, all three modified channels showed spontaneous openings at low pH (6.0) (no activity was observed in blisters with no reconstituted protein). However, differences in gating of the three labeled channels were noted: the triple labeled G22/G26/G30 and double labeled G22/G26 nanopores have small squared openings with a small conductance of about 200 pS, while nanopores labeled at G22 and at G26 appear to have larger but extremely short, spike-like openings, making conductance impossible to accurately measure. In addition to short open dwell times, labeled G26 MscL also has slightly longer small openings at a conductance similar to that of the labeled G22/G26 nanopore. Note that unlabeled MscL channels under these conditions, when gated by membrane tension, has a conductance of greater than 3 nS, and often have short but square openings (the bulk of the channels have open dwell times of a few milliseconds).[10, 23, 32] The reasons for these differences in channel conductance and kinetics, or even whether they reflect the function of the nanopores when inserted within the stealth liposomes that contain a lipid mixture not conducive to patch clamp, is unclear. Figure 3b is an analysis of multiple patches that shows that all four labeled channels have higher spontaneous opening at pH 6.0 (rate of 39 to 90%; n = 7 to 19) relative to pH 8.0 (rate of 8 to 14%; n = 10 to 29). The rate of spontaneous opening for the triple labeled and double-labeled MscL at pH 6.0 are similar, but much higher than that for those only labeled at single sites. In sum, the results of single channel recordings are consistent with the pH sensitive responsiveness of Gd-DOTA loaded liposomes.
Figure 3: Single channel recordings of pH sensor labeled MscL channels.
(a): Patch clamp recordings. “C” and “S” represent the closed, sub-opening state of the channels. (b): Percentage of recordings that show the spontaneous openings of labeled channels in pH 8.0 and pH 6.0 patch buffer.
2.2. pH Response of the double and triple-labeled MscL nanopores.
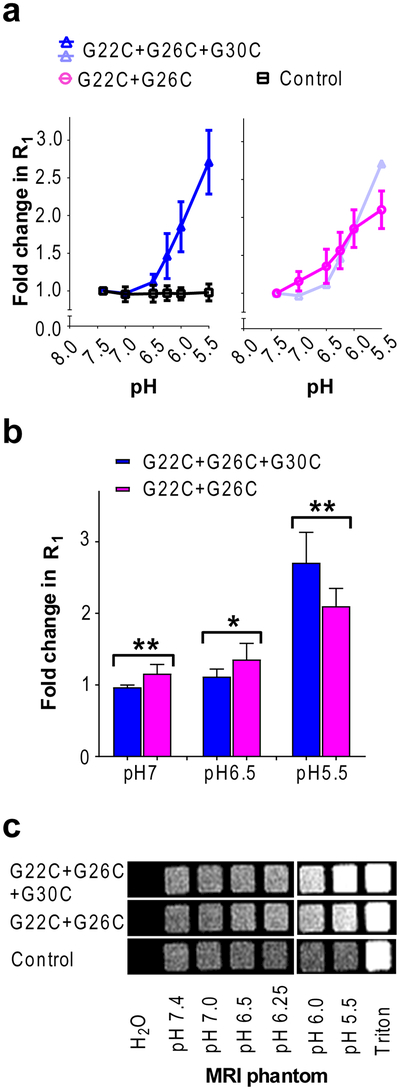
While the double and triple-labeled MscL nanovalve have the higher response to a low pH (5.5) relative to the single mutants, it is important to test if these nanovalves give a graded response, and if they easily detect pH values associated with pathophysiological conditions such as cancer and inflammation (pH 6.0–6.5).[33],[34] The samples for this experiment were prepared by distributing liposomal elution equally into 6 vials that were pelleted by ultracentrifugation, and ultimately the relaxation rate measured at 6 different pH values. As shown in Figure 4a, as expected for the G22/G26/G30 modified nanovalve, a 2.7 ± 0.4 fold increase in R1 was observed (n = 7), while the G22/G26 nanovalve only had 2.1 ± 0.2 fold increase at pH 5.5 (n = 7); no-protein control vesicles showed no signal changes (n = 10). Thus, the triple modified nanovalve had greater efficacy. However, as shown in Figure 4a right panel and Figure 4b, careful analyses revealed that the double-labeled nanovalve is more sensitive to small changes in pH. The G22/G26 nanovalve has significantly higher MR signal than the triple-labeled MscL at pH 7.0 and 6.5, while the triple-labeled nanovalve has significantly higher R1 signal than the double-labeled MscL at pH 5.5. Both channels show similar R1 enhancement at pH 6.25 and 6.0, which is consistent with our electrophysiological recording showing similar spontaneous opening rates for both channels at pH 6.0. We also tried several triple mutant channels that were modified at positions G22 and G26 and one other site near the pore lining region of MscL including I24, S34 and A38, and tested their pH sensitive responsiveness. As shown in Supplemental Figure 3, none of them showed a superior pH response compared to G22/G26 and G22/G26/G30 nanovalves in the range of pH tested.
Figure 4: pH dependent R1 increase and MR contrast enhancement of Gd-DOTA liposomes reconstituted with modified nanopores.
(a). The R1 relaxation rates at 1.4 T and 25 °C. (b). Comparison of R1 for the double and triple labeled mutant. “**”, p < 0.01, t = 3.83, df = 7; “*” p < 0.05, t = 2.48, df = 10; “**”, p < 0.01, t = 3.28, df = 12; student t test. (c). Spin echo T1 weighted MR image (at 1 T) of phantoms. Imaging parameters: TR = 200 ms, TE = 13.1 ms, data matrix = 128 × 128, FOV = 70 mm, slice thickness = 1 mm.
Figure 4c shows MR imaging of samples of G22/G26 and G22/G26/G30 modified nanopores that were prepared by a dilution of the original solutions. The MR images recorded at 1 T also demonstrated that as pH decreases the MRI signal intensity increases for the double and triple labeled MscL nanopore-containing proteoliposomes with the triple labeled MscL liposomes exhibiting highest signal intensity at pH 5.5. Again, no-protein control liposomes showed no apparent increase. After lysis of the liposomes by Triton X-100 treatment, both control and test proteoliposomes increased their signal intensity to a similar level, demonstrating that all sets of vesicles contained similar amounts of Gd-DOTA. The r1 relaxivity (the paramagnetic contribution to relaxation rate normalized to 1 mM of Gd) estimated from the relaxation rates and the total amount of Gd in the G22/G26/G30 samples (2.21 mM by ICP-OES) was 1.8 mM−1s−1 at pH 7.4 where the channel is closed, reflecting the decrease in relaxivity as a result of limited diffusibility of water molecules across the lipid bilayer.[13] At pH 5.5 where the channel is fully open, the r1 increased after about an hour to about 4.6 mM−1s−1 as the Gd-DOTA was released from the liposomes. After lysis of liposomes the r1 was around 5 mM−1s−1 at 1.4 T, 25 °C, which agrees well with relaxivity data reported for unencapuslated Gd-DOTA[35].
In sum, the data show that the G22/G26 and G22/G26/G30 modified nanovalves have superior sensitivity and efficacy over single-modified nanovalves in the physiological range of pH tested, with G22/G26 being more sensitive at smaller pH changes, and G22/G26/G30 having greater efficacy.
2.3. Reversible gating of the MscL nanopore.
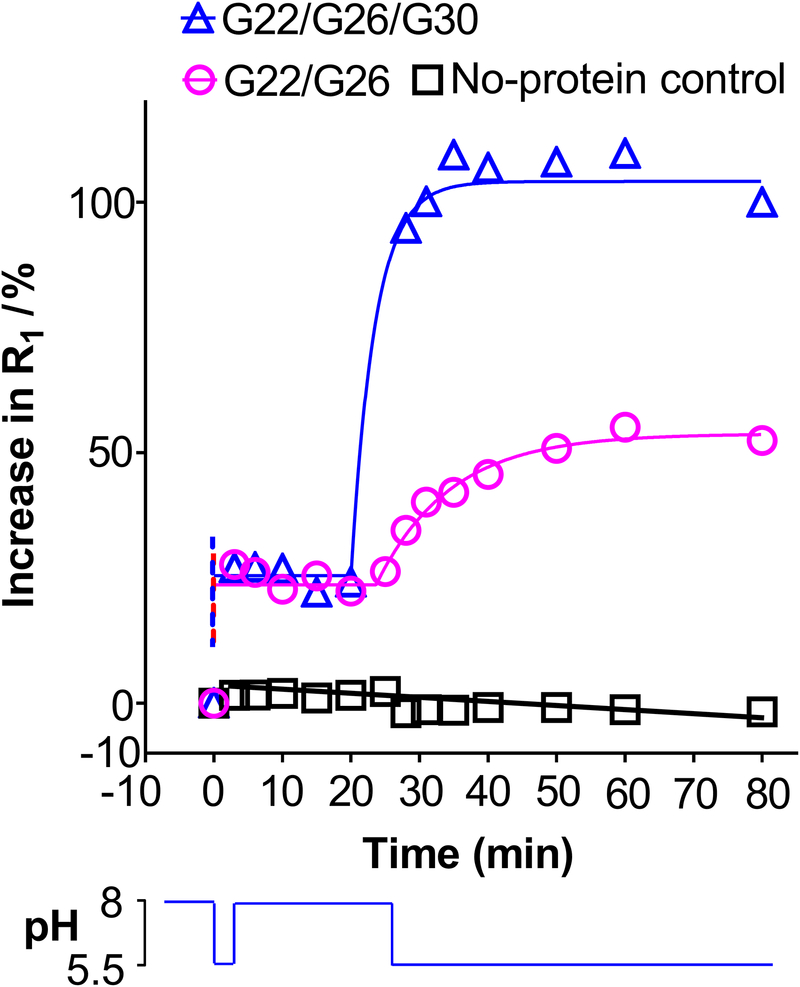
For a pH sensitive nanovalve, reversible gating that could further enhance specific release only at the targeted tissue is highly desirable – if the devise becomes dislodged from the inflamed or cancerous area, the devise should shut off. Theoretically, the pH-sensitive nanovalves we engineered should be reversible: able to be opened at lower pH by electrostatic repulsion, but closed if the pH increases, then opened again when a shift to a lower pH is again detected. To test this, the R1 value of the double and triple labeled nanopore-containing proteoliposome samples were first measured at pH 8.0. The pH of the liposome solution was then decreased to 5.5, where they were kept for 3 min before switching back to pH 8.0. As shown in Figure 5, the R1 of the proteoliposome preparation jumped to a new level after this brief pH shift, but remained at that level as long as the pH of the solution remained at pH 8.0. After a second pH decrease to 5.5, the R1 of the proteoliposome preparation further increased with a τ value of 3.38 min and 10.21 min for the triple and double labeled nanovalve, respectively, which is similar to that observed during first time pH treatment as shown in Figure 2. These data demonstrate that labeled MscL nanovalves could reversibly respond to pH changes: opening at lower pH, then closing when the pH is increased, but able to efficiently re-open upon a second decrease in the pH. In addition, the observation that switching the pH of the solution back to 8.0 did not recover the R1 back to the initial level implies that although the pore can open and close reversibly, Gd-DOTA is irreversibly released from proteoliposomes upon opening of the very large MscL nanopore.
Figure 5: Reversible responses of MscL nanopore toward pH change.
R1 change of Gd-DOTA liposomes at each time point was normalized by that after lysing the vesicles completely by Triton X-100 treatment. The bottom panel shows the pH transient decrease, increase and subsequent decrease that led to the release in the top panel. This is a representative of 7 experiments.
3. Discussion.
Core-encapsulated liposomal MRI contrast agents have been previously used for MR angiography as well as tumor imaging.[36] Although they can be made “stealth” and show ‘passive targeting’ in tumors and damaged tissues, their major limitation has been the relaxivity quenching due to the limited diffusion rate of water molecules across the lipid bilayer.[36] We have worked under the assumption that the incorporation of a small molecule-permeable nanovalve into the lipid bilayer would allow release of the encapsulated MR agent, leading to enhanced water permeation, and therefore overcome this limitation. We have thus engineered a biologically derived nanovalve derived from the bacterial MscL channel; to add an additional level of specificity, we have designed the nanovalve to be responsive to the low pH often observed in cancerous and inflamed tissues. Here we demonstrate that our newly designed nanovalve far exceeds previous attempts [11, 37] at engineering a triggered nanovalve from the MscL channel.
Soon after its discovery, using random mutagenesis we identified several single-site mutations within the MscL channel that inhibited cell growth when expressed in bacteria; the mutated channels were found to be more sensitive to stimuli, or even open spontaneously.[23] Most of these mutations were within the first transmembrane domain, TM1, and were substitutions to more hydrophilic residues. One of the sites identified in this screen was G22. Based on these findings, we mutated G22 within the TM1 domain to all other amino acids and correlated hydrophilicity of the amino acid side chains to the severity of the channel phenotype.[25] These findings were subsequently confirmed and extended by post-translational chemical modification of the MscL G22C cysteine with charged methanesulfonate compounds, MTSET+([2-(trimethylammonium) ethyl]methanethiosulfonate bromide) and MTSES− (Sodium (2-sulfonatoethyl)methanethiosulfonate), which also increased channel sensitivity.[24]
Chemical modifications of G22C were later used to introduce molecules into MscL that would generate charges upon UV light or low pH treatment, thus changing the channel modality to these stimuli. Hence, these modifications lead to channels that opened and released the dye calcein when stimulated.[11, 12] Other studies, however, have suggested that G22 is not the only, or even the best choice for channel modification for the engineering of a triggered nanopore. More extensive libraries in which almost every single residue in the protein was mutated to cysteine, combined with a rapid in vivo screening using charged MTS reagents, including MTSET+ revealed several additional candidate sites for modification,[18, 19, 21] with two of them, G26 and G30 confirmed by electrophysiological studies.[19–21] Our more recent study, demonstrated that engineering the G26 site for modifications leading to UV sensitive gating may indeed have advantages over the G22 site.[20]
In the present study, we compare single MscL mutants (G22C, G26C and G30C) as well as double and triple mutants based on the G22 modified nanopore in an attempt to optimize the construction of a more efficient pH-sensitive “smart” liposomal MRI contrast agent. We have found many of the mutants were responsive to changes in pH after labeling with N,N-dimethylglycine as the pH modulator. The G26 modified nanopore showed a higher percentage release of payload and faster kinetics than that modified at G22, but the extent of the response for either was still far from fast and total release of contrast agent from the vesicle. By expanding our studies to include multiple labeling sites within the pore, we further improved the design and performance of the nanovalve. Using Gd-DOTA-loaded liposomes we have found that labeling at both sites, G22 and G26 leads not only to a greater response, but also faster kinetics when compared with either of the single mutants. Moreover, labeling at three sites, G22, G26 and G30 leads to a near 100% increase of R1 and the fastest kinetics upon pH decrease among all mutants tested. These data are also consistent with single channel recording of the mutants. Our pH titration experiments indicated that the double (G22/G26) and triple (G22/G26/G30) labeled nanopores responded in a pH range that covers the extracellular pH of many cancerous tissues (pH 6.0–6.5). There is also a difference in pH sensitivity of the nanopores labeled at more than one site. Basically, the double mutant has higher response at smaller pH shifts while the triple mutant has a higher response at the greater pH changes, which renders them useful in different conditions, and potentially in detection and treatment of different pathologies. The increase of R1 relaxation rate with decreasing pH is nicely illustrated by T1 weighted MR imaging of a phantom containing Gd-DOTA labeled proteoliposome samples at different pH values. Moreover, we have demonstrated that these labeled nanovalves could be reversibly gated when the pH of the liposome suspension is shifted alternatively between 8.0 and 5.5. This reversibility is a potential advantage over pH-sensitive liposomes that simply and irreversibly dissociate at a certain pH and release the cargo [38, 39].
A recent paper demonstrated that stealth liposomes with the G22C mutated MscL channel labeled with N.N-dimethylglycine as pH-modulator can be used for the delivery of a Gd-based MR contrast agent in a glioma rodent model.[37] Although the release of the MR agent from the liposomes in mildly acid extracellular environment was observed, this device had obvious limitations, especially when compared to our system. In particular, the pH response for this G22C modified system in the biologically relevant pH range was weaker than that of our G22/G26 MscL proteoliposomes reflecting the less efficient actuation of the G22C modified MscL nanopore. Perhaps more importantly, the observed T1 values for these proteoliposome preparations after lysis with Triton X-100 were reported to be quite long, around 2200 ms, only barely shorter than that of pure water. In contrast, our proteoliposome systems with G22, G22/G26 or G22/G26/G30 MscL nanovalves had significantly shorter T1 values (around 33 ms). To put these numbers into context, the T1 value of most living tissues is around 1000–1500 ms at 3 T.[40]
4. Conclusion.
The biologically derived MscL-based pH-triggered nanopore described here has all the major properties desired for releasing a vesicular load at low-pH environments. While stable at pH 7.4, vesicles containing these nanovalves shows not only faster kinetics but also greater efficacy of pH-dependent load release when compared to those previously reported. When loaded with a contrast agent, significant increases in R1 bulk water relaxation rates were observed when the pH dropped from 7.4 to 7.0, and progressive increases in R1 were also observed at pH values down to pH 5.5. The discovery of triggered nanovalves with slightly different sensitivities may also be useful for some specialized purposes. This study reflects a major step in designing a pH-responsive “smart” liposomal contrast agent. However, it is important to note that this devise is obviously not limited to releasing MRI contrast agents in low pH environments. An identical design could potentially be used as a targeted delivery device for drugs. Thus, by loading the vesicles with different molecules, (e. g. imaging and therapeutic agents) the system described here can be used as a theranostic nanodevice.
5. Experimental section
5.1. Reagents.
All solvents and reagents were purchased from commercial sources and used as received unless otherwise stated. The reagents for the chemical synthesis, MscL protein expression, purification and labeling was purchased from Sigma-Aldrich, Co., Roche Diagnostics, Fischer Scientific, Anatrace Inc., Maumee, OH, USA, Thermo Scientific, Rockford, IL, USA and were used without further purification. The elemental analyses were done at Galbraith Laboratories, Inc (Knoxville, TN, USA). MS-ESI spectra were recorded with a Q tof MS-XEVO instrument (Waters; Milford, MA, USA).
5.1.1. N, N-Dimethylglycine 2-bromoethyl ester
This compound was prepared as described in the literature.[11]
5.1.2. 1,4,7,10-Tetraazacyclodoecane-1,4,7,10-tetraacetic acid (DOTA) and the Gd(III) complex of DOTA.
DOTA was synthesized as described in the literature.[41] The Gd(III) complex was prepared as follows. DOTA (4.00 g, 8.5 mmols) was dissolved in water (50 mL) and Gd2O3 (2.00 g, 5.52 mmols) was added. The mixture was stirred and heated at 60 °C for one day. The pH was then adjusted to around 8 with NaOH and the mixture was stirred at 60 °C for 2 days. The solution was filtered and then neutralized by adding small amounts of DOTA as needed. The resulting solution was filtered and lyophilized to give quantitative yield of the complex with Gd% of 21.30% (ICP-OES). MS (ESI positive mode), m/z: Calculated for [M+3H]2+/2 280.5; Found, 280.5 (100 %). Calculated for [M+2H]+ 560; Found 560 (75 %).
5.2. MscL protein expression, purification and labeling.
The E. coli MscL mutants, G22C, G26C, G22C+G26C and G22C+G26C+G30C with a 6-histidine tag at c-terminus of protein were generated using the Mega Primer method as described previously.[32] Both G26C and G22C+G26C mutant MscL were also truncated at their c-terminal end from residue 110 to 136 which has been shown to improve protein expression while maintain same level of channel function.[42–44] Mutants were inserted within the Pet21a vector plasmid, transformed in E. coli strain PB116, which is a λDE3 lysogenization (Novagen, San Diego, CA, USA) of PB106,[42] inoculated and selected on Lennox Broth (LB) (Fisher Scientific, Fair Lawn, NY, USA) agar plate supplemented with ampicillin (Fisher Scientific, Fair Lawn, NJ, USA). Single colony on agar plate was then picked and inoculated in LB media containing ampicillin (100 μg/ml) in a shaker-incubator at 37 °C and rotated at 250 cycles/min overnight, which was followed by subculture after 100-fold dilution the next day. When OD 600 nm of the culture reached 0.6 – 0.8, expression of protein was induced by the addition of isopropyl-β-d-thiogalactopyranoside (IPTG, 1 mM) (Anatrace Inc., Maumee, OH, USA) for 2 h. Bacteria were then pelleted by centrifugation at 4993 g for 30 min and stored at −80 °C. For MscL purification, cell pellets were suspended in Tris buffer (10 mM, pH 8.0) that was supplemented with dithiothreitol (DTT, 5 mM) (Fisher Scientific, Fair Lawn, NJ, USA), deoxyribonuclease I (DNAse, 0.5 μg/mL) (Sigma-Aldrich, Co., St Louis, MO, USA), protease inhibitor cocktail (Roche Diagnostics, Indianapolis, IN, USA) and lysozyme (1 mg/ml, Sigma-Aldrich, Co.,St Louis, MO,USA). The suspension was incubated at room temperature for 30 min before being passed through French press twice at 16K PSI in cold room. After ultracentrifugation at 70 kg for 30 min, the cell membrane containing pellet was homogenized in Tris buffer (10 mM, pH 8.0) with 2-mercaptoethanol (β-ME, 1 mM) (Sigma-Aldrich, Co., St Louis, MO, USA). In order to extract membrane proteins the homogenate was added with Triton X-100 (2 %) for 30 min at room temperature, which was followed by one hour incubation with equilibrated Ni-NTA beads (Thermo Scientific, Rockford, IL, USA) in the presence of imidazole (15 mM, Sigma-Aldrich, Co., St Louis, MO, USA) to allow the binding of the histidine tagged MscL protein to the beads. The Ni-NTA beads were then washed by Tris buffer (10 mM, pH 8.0) containing Triton X-100 (0.05%) (protein buffer) thoroughly to remove non-specifically bound protein. The bead bound MscL protein was labeled at room temperature for 45 min with N, N-dimethylglycine 2-bromoethyl ester, which was freshly dissolved in protein buffer with the labeling molecule to protein ratio in mg/mg to be 10 for single mutant, 20 for double mutant MscL and 30 for triple mutant MscL. The bead suspension was then transferred into an elution column to allow flow through of labeling solution. And the remaining beads were washed 3 times with protein buffer to completely remove free labeling molecules and labeling byproduct. Protein buffer containing imidazole (250 mM) was then used to elute protein from the Ni-NTA beads. The eluted protein solution was dialyzed against protein buffer overnight at 4 °C to remove imidazole before concentration measurement using BCA kit (Thermo Scientific, Rockford, IL, USA). The protein was concentrated to 2 mg/ml by centrifugal filter tube (Merck Millipore Ltd., IRL) and then divided into aliquots, flash frozen in liquid nitrogen before stored at −80 °C freezer, which could remain stable for months.
5.3. Liposomal preparation
lipid mixtures composed of 1,2-dioleoyl-sn-glycero-3-phosphocholine (70% in M), cholesterol (20% in M) and 1,2-distearoyl-sn-glycero-3-phosphoethanolamine-N-[methoxy(polyethylene glycol)-2000] (10% in M) (Avanti Polar Lipids Inc., Alabaster, AL, USA) in chloroform were dried under argon while rotating in a glass tube to form a thin lipid layer on glass wall, which was further dried under argon for 40 min before rehydrating with Gd-DOTA solution (420 mM) made in Tris buffer (10 mM, pH 8.0). After thorough vortex the liposome suspension was incubated at 37 °C with shaking for 1 h, which was then divided into aliquots and frozen in liquid nitrogen before storing at −80°C.
5.4. Protein reconstitution
For protein reconstitution, liposomes stored at −80 °C were melted at 60 °C for 15 min, added with Triton X-100 (Anatrace Inc.) before protein solution was introduced. Carefully titration of detergent lipid ratio and protein lipid ratio were done for each batch of liposomes and protein. Protein reconstitution mixture was incubated at 60 °C for 40 min before calcein disodium (25 mM final concentration, Sigma Aldrich, St. Louis, MO, USA), a fluorescent dye indicator, was added and incubated at room temperature for 20 min. The detergent was removed by adding Bio-beads™ SM-2 (6.25 mg wet weight per ul of 10% Triton X-100) (Bio-Rad Laboratories, inc, USA) in an overnight incubation at 4 °C. Free calcein and Gd-DOTA was then removed by passage through a G-50 fine Sephadex column (GE Healthcare Inc., Piscataway, NJ, USA) washed with vesicle buffer (10 mM Tris (Fisher Scientific, Fair Lawn, NJ, USA), 600 mM sucrose (Sigma-Aldrich, Steinheim, Germany), 300 mM NaCl (Sigma-aldrich, Steinheim, Germany) and 1 mM EDTA (Sigma-Aldrich, St Louis, MO, USA), pH 8.0). After liposomal portion of the elution was collected, vesicles were pelleted by ultracentrifugation at 264,597 g and stored on ice before relaxivity rate (R1) measurement and MRI. Although the modified MscL protein could be frozen for long periods of time, all reported experiments were performed the same day that the liposomes were generated. Storage of loaded liposomes for 24 to 48 hours often lead to decrease in the cargo load of more than 50%, as determined by measurements after Triton X-100 lysis, suggesting a slow leakage of Gd-DOTA. Improving the stability and storage of the loaded vesicles is an active area of study.
5.5. Determination of the optimal Gd-DOTA concentration for liposome preparation.
After reconstitution the final concentration of Gd-DOTA inside proteoliposomes is calculated to be 280 mM and we have observed that the R1 of the liposomal solution is enhanced by 4.05 ± 0.19 fold after lysing the liposomes by 0.5% Triton X-100 treatment, which indicates a good loading of Gd-DOTA inside liposomes. As the Gd-DOTA concentration decrease, the signal difference after Triton X-100 treatment decreased, which is consistent with literature [45] and suggest that the relaxivity quenching is inversely proportional to the water permeability of the liposome bilayer and directly proportional to the concentration of the Gd(III) inside the liposome. We did not further increase the concentration of Gd-DOTA considering the future use of the vesicles in animal experiments where they have to be compatible with normal range of plasma osmolarity.
5.6. Preparation of proteoliposomes for patch clamp recording.
As described previously,[30, 31] Azolectin, also known as soy total lipid extract (Soy PC 20% Cat# 541601, Avanti Polar Lipids Inc., Alabaster, AL, USA) was dissolved in chloroform, dried under argon for 40 min to form lipid film, and then rehydrated into 20 mg/ml suspension with buffer A (10 mM Tris, 1mM EDTA, and 1mM EGTA, pH 8.0) at 45°C for at least 2 hours. After sonication of the liposome suspension MscL proteins were added with 1 to 250 protein-lipid ratio. Detergent was removed by dialysis with 3 × 1000 ml of buffer B (10 mM Tris, 100 mM NaCl, 0.2 mM EDTA, and 0.002% NaN3, pH 8.0) containing Bio-beads (Bio-Rad, Hercules, CA, USA). The resultant proteoliposomes were collected by a 20-minute air driven ultracentrifugation at 30 psi (AirfugeTM, Beckman Instruments Inc, Palo Alto, CA, USA). The pellet was re-suspended into buffer C (10 mM MOPS, 5% ethylene glycerol, pH 8.0) and desiccated overnight under vacuum at 4 °C.
5.7. Patch clamp recording
Patch clamp of liposomes was performed as described previously,[31] with modifications. The above mentioned desiccated liposomes were rehydrated in buffer D (5 mM Hepes, 10 mM KCl, and 2 mM MgCl plus 320 mM sucrose, pH 8.0) at a lipid concentration of 90 mg/ml. Patches were excised at room temperature in patch buffers (5 mM HEPES, 200 mM KCl, 90 mM MgCl2, 10 mM CaCl2) with pH adjusted to 8.0 and 6.0 respectively. Data were acquired at −20 mV at a sampling rate of 33 kHz and a 5 KHz low pass filter using an AxoPatch 200B amplifier and Digidata 1440A from Molecular Devices (Sunnyvale, CA, USA) and analyzed using pCLAMP10 software (Molecular Devices, Sunnyvale, CA, USA).
5.8. R1 measurements and MRI.
The relaxivity measurements were performed on a Bruker Minispec MQ 60 instrument operating at 1.4 T. An inversion recovery pulse sequence (180°-τ−90°) was used to collect the data for an array of the τ, and the data points were then fitted to an exponential recovery curve to get the actual T1 values. MR imaging was performed on an Aspect M2 horizontal bore small animal imaging system operating at 1 T. The T1w images of the phantoms were obtained using a standard spin echo sequence. Imaging parameters: TR = 200 ms, TE = 13.1 ms, data matrix = 128 × 128, FOV = 70 mm, slice thickness = 1 mm. For generation of all data except those in Figure 5, samples were re-suspended in modified vesicle buffer of pH 7.4, 7.0, 6.5, 6.25, 6.0 or 5.5 before R1 measurement or MRI. The modified vesicle buffer containing same components as vesicle buffer except Tris (50 mM) for buffering pH at 7.4 and 7.0, and MES (50 mM) (Sigma-Aldrich, Steinheim, Germany) for buffering pH at 6.5, 6.25, 6.0 and 5.5, respectively. The liposome re-suspension was put into melting point capillary for R1 measurement. For the reversibility experiment in Figure 5, HCl or NaOH (150 mM, less than 5 % of sample volume) was added to liposome to change pH to desired value which was monitored by pH measurement. After experiments were completed, samples treated with Triton X-100 (final contcentration of 0.5%) to lyse liposomes were sent for ICP-OES analysis of their Gd-DOTA concentration.
5.9. Transmission electron microscopy of liposomes
The electron microscopy of the liposomes was carried out on a FEI Tecnai G2 spirit transmission electron microscope. Sample (2.0 μL) was loaded to a glow-discharged carbon-coated copper grid (CF-400-Cu, Electron Microscopy Science, Hatfield, PA, USA). After 30 s incubation, the grid was blotted with a piece of Whatman No. 4 filter paper (Sigma-Aldrich, Co., St Louis, MO, USA) to remove excessive solution, then incubated with ammonium molybdate (6.0 %) (Sigma-Aldrich, Co., St Louis, MO, USA) adjusted pH with KOH to 6.4 for 30 s. After removing excessive staining solution, the stained grid was dried in air and then observed under FEI Tecnai G2 spirit transmission electron microscope (Hillsboro, OR, USA). Images were taken with a Gatan 2Kx2K multiport readout post column CCD (Pleasanton, CA, USA) at −2 to −3 μm defocus.
5.10. Reduction of the size of proteoliposomes by sonication
For in vivo application of the proteoliposomes, it is necessary to have a uniform size of liposomes. Under transmission electron microscopy (TEM) we found out that liposomes tended to cluster and fuse after the protein reconstitution procedure, which would not be ideal for in vivo use, making it difficult for the liposomes to permeate through capillaries to reach the target tissue. Therefore, 1 min sonication using sonic cleaner (50/60 Hz, 40 W, Fisher Scientific, USA) was applied to the liposomes after protein reconstitution in the presence of Gd-DOTA solution in order to reduce liposome size while maintaining the Gd-DOTA concentration inside the liposomes. As shown in Supplemental Figure 1a, the diameters of liposomes were successfully reduced to an average of 100 nm after sonication and appeared less aggregated and to be of more uniform in size. Since sonication procedure was not applied in experiments for all figures in this work, the pH responsiveness of the G22/G26 and G22/G26/G30 liposomes after sonication was tested, which showed that the relative low release of G22/G26 was significantly improved after sonication, presumably due to the removal of clustered liposomes, thus allowing more complete release of contrast agent (Supplemental Figure 1b).
Supplementary Material
Acknowledgements
This work was supported by Grant RP130362 from the Cancer Prevention & Research Institute of Texas (CPRIT; http://www.cprit.state.tx.us/), Grant I-1420 of the Welch Foundation, and Grants GM121780, GM061028 and P41-EB015908 from the National Institutes of Health. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health or other funding organizations.
References
- 1.Levina N; Totemeyer S; Stokes NR; Louis P; Jones MA; Booth IR, EMBO J. 1999, 18 (7), 1730–1737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Li J; Guo J; Ou X; Zhang M; Li Y; Liu Z, Proc Natl Acad Sci U S A 2015, 112 (34), 10726–31. DOI 10.1073/pnas.1503202112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chang G; Spencer RH; Lee AT; Barclay MT; Rees DC, Science 1998, 282, 2220–2226. [DOI] [PubMed] [Google Scholar]
- 4.Steinbacher S; Bass R; Strop P; Rees DC, Structures of the prokaryotic mechanosensitive channels MscL and MscS In Mechanosensitive Ion Channels (a volume in the Current Topics in Membranes series), Hamill OP, Ed. Elsievier Press: St. Louis, MO, 2007; Vol. 58, pp 1–20. [Google Scholar]
- 5.Iscla I; Blount P, Biophys J 2012, 103 (2), 169–74. DOI 10.1016/j.bpj.2012.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Perozo E; Cortes DM; Sompornpisut P; Kloda A; Martinac B, Nature 2002, 418 (6901), 942–8. [DOI] [PubMed] [Google Scholar]
- 7.Cruickshank CC; Minchin RF; Le Dain AC; Martinac B, Biophys. J 1997, 73 (4), 1925–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kocer A; Tauk L; Dejardin P, Biosens Bioelectron 2012, 38 (1), 1–10. DOI 10.1016/j.bios.2012.05.013. [DOI] [PubMed] [Google Scholar]
- 9.Blount P; Iscla I; Moe PC; Li Y, MscL: The bacterial mechanosensitive channel of large conductance In Mechanosensitive Ion Channels (a volume in the Current Topics in Membranes series), Hamill OP, Ed. Elsievier Press: St. Louis, MO, 2007; Vol. 58, pp 202–233. [Google Scholar]
- 10.Iscla I; Levin G; Wray R; Blount P, Biophys J 2007, 92 (4), 1224–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Koçer A; Walko M; Bulten E; Halza E; Feringa B; Meijberg W, Angew. Chem 2006, 45, 3126–3130. [DOI] [PubMed] [Google Scholar]
- 12.Koçer A; Walko M; Meijberg W; Feringa BL, Science 2005, 309 (5735), 755–8. [DOI] [PubMed] [Google Scholar]
- 13.Ayyagari AL; Zhang X; Ghaghada KB; Annapragada A; Hu X; Bellamkonda RV, Magn Reson Med 2006, 55 (5), 1023–9. DOI 10.1002/mrm.20846. [DOI] [PubMed] [Google Scholar]
- 14.Hashizume H; Baluk P; Morikawa S; McLean JW; Thurston G; Roberge S; Jain RK; McDonald DM, The American journal of pathology 2000, 156 (4), 1363–80. DOI 10.1016/s0002-9440(10)65006-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sercombe L; Veerati T; Moheimani F; Wu SY; Sood AK; Hua S, Frontiers in pharmacology 2015, 6, 286 DOI 10.3389/fphar.2015.00286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nehoff H; Parayath NN; Domanovitch L; Taurin S; Greish K, Int J Nanomedicine 2014, 9, 2539–55. DOI 10.2147/ijn.s47129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Delli Castelli D; Gianolio E; Geninatti Crich S; Terreno E; Aime S, Coordination Chemistry Reviews 2008, 252 (21–22), 2424–2443. [Google Scholar]
- 18.Bartlett JL; Levin G; Blount P, Proc Natl Acad Sci U S A 2004, 101, 10161–10165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bartlett JL; Li Y; Blount P, Biophys J 2006, 91 (10), 3684–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Iscla I; Eaton C; Parker J; Wray R; Kovacs Z; Blount P, Biosensors 2013, 3 (1), 171–84. DOI 10.3390/bios3010171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Iscla I; Wray R; Eaton C; Blount P, PloS one 2015, 10 (9), e0137994 DOI 10.1371/journal.pone.0137994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Batiza AF; Kuo MM; Yoshimura K; Kung C, Proc. Nat. Acad. Sci. USA 2002, 99 (8), 5643–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ou X; Blount P; Hoffman RJ; Kung C, Proc. Nat. Acad. Sci. USA 1998, 95 (19), 11471–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yoshimura K; Batiza A; Kung C, Biophys. J 2001, 80 (5), 2198–2206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yoshimura K; Batiza A; Schroeder M; Blount P; Kung C, Biophys. J 1999, 77, 1960–1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kocer A, J Liposome Res 2007, 17 (3–4), 219–25. DOI 10.1080/08982100701528203. [DOI] [PubMed] [Google Scholar]
- 27.Kocer A, Methods Mol Biol 2010, 605, 243–55. DOI 10.1007/978-1-60327-360-2_16. [DOI] [PubMed] [Google Scholar]
- 28.Kocer A; Walko M; Feringa BL, Nat Protoc 2007, 2 (6), 1426–37. DOI 10.1038/nprot.2007.196. [DOI] [PubMed] [Google Scholar]
- 29.Corry B; Hurst AC; Pal P; Nomura T; Rigby P; Martinac B, J Gen Physiol 2010, 136 (4), 483–94. DOI 10.1085/jgp.200910376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Moe P; Blount P, Biochemistry 2005, 44 (36), 12239–44. DOI 10.1021/bi0509649. [DOI] [PubMed] [Google Scholar]
- 31.Blount P; Sukharev SI; Moe PC; Martinac B; Kung C, Mechanosensitive channels of bacteria In Meth. Enzymol, Conn PM, Ed. Academic Press: San Diego, CA, 1999; Vol. 294, pp 458–482. [DOI] [PubMed] [Google Scholar]
- 32.Levin G; Blount P, Biophys J 2004, 86 (5), 2862–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tannock IF; Rotin D, Cancer Res 1989, 49 (16), 4373–84. [PubMed] [Google Scholar]
- 34.Punnia-Moorthy A, Journal of oral pathology 1987, 16 (1), 36–44. [DOI] [PubMed] [Google Scholar]
- 35.Aime S; Anelli PL; Botta M; Fedeli F; Grandi M; Paoli P; Uggeri F, Inorg Chem 1992, 31 (12), 2422–2428. DOI 10.1021/ic00038a023. [DOI] [Google Scholar]
- 36.Castelli DD; Gianolio E; Crich SG; Terreno E; Aime S, Coordination Chemistry Reviews 2008, 252 (21–22), 2424–2443. DOI 10.1016/j.ccr.2008.05.006. [DOI] [Google Scholar]
- 37.Pacheco-Torres J; Mukherjee N; Walko M; Lopez-Larrubia P; Ballesteros P; Cerdan S; Kocer A, Nanomedicine: nanotechnology, biology, and medicine 2015, 11 (6), 1345–54. DOI 10.1016/j.nano.2015.03.014. [DOI] [PubMed] [Google Scholar]
- 38.Lokling KE; Skurtveit R; Bjornerud A; Fossheim SL, Magn Reson Med 2004, 51 (4), 688–96. DOI 10.1002/mrm.20009. [DOI] [PubMed] [Google Scholar]
- 39.Lokling KE; Fossheim SL; Skurtveit R; Bjornerud A; Klaveness J, Magn Reson Imaging 2001, 19 (5), 731–8. [DOI] [PubMed] [Google Scholar]
- 40.Bojorquez JZ; Bricq S; Acquitter C; Brunotte F; Walker PM; Lalande A, Magnetic Resonance Imaging 2017, 35 (Supplement C), 69–80. DOI 10.1016/j.mri.2016.08.021. [DOI] [PubMed] [Google Scholar]
- 41.Desreux JF, Inorg Chem 1980, 19 (5), 1319–1324. DOI DOI 10.1021/ic50207a042. [DOI] [Google Scholar]
- 42.Blount P; Sukharev SI; Schroeder MJ; Nagle SK; Kung C, Proc. Nat. Acad. Sci. USA 1996, 93 (21), 11652–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Häse CC; Ledain AC; Martinac B, J. Membr. Biol 1997, 157 (1), 17–25. [DOI] [PubMed] [Google Scholar]
- 44.Andersson M; Okeyo G; Wilson D; Keizer H; Moe P; Blount P; Fine D; Dodabalapur A; Duran RS, Biosens Bioelectron 2008, 23 (6), 919–23. DOI S0956–5663(07)00409–5 [pii] 10.1016/j.bios.2007.09.014. [DOI] [PubMed] [Google Scholar]
- 45.Mulder WJ; Strijkers GJ; van Tilborg GA; Griffioen AW; Nicolay K, NMR Biomed 2006, 19 (1), 142–64. DOI 10.1002/nbm.1011. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.