Abstract
The process of translation is characterized by irregularities in the local decoding rates of specific mRNA codons. This includes the occurrences of long pauses that can take place when ribosomes decode certain peptide sequences, encounter strong RNA secondary structures, or decode “hungry” codons. Examples are known where such pausing or stalling is used for regulating protein synthesis. This can be achieved at the level of translation via direct alteration of ribosome progression through mRNA or by altering mRNA stability via NoGo decay. Ribosome pausing has also been implicated in the cotranslational folding of proteins. Ribosome profiling data often are used for inferring the locations of ribosome pauses. However, no dedicated online software is available for this purpose. Here we present PausePred (https://pausepred.ucc.ie/), which can be used to infer ribosome pauses from ribosome profiling (Ribo-seq) data. Peaks of ribosome footprint density are scored based on their magnitude relative to the background density within the surrounding area. The scoring allows the comparison of peaks across the transcriptome or genome. In addition to the score, PausePred reports the coordinates of the pause, the footprint density at the pause site, and the surrounding nucleotide sequence. The pauses can be visualized in the context of Ribo-seq and RNA-seq density plots generated for specific transcripts or genomic regions with the Rfeet tool. PausePred does not require input on the location of protein coding ORFs (although gene annotations can be optionally supplied). As a result, it can be used universally and its output does not depend on ever evolving annotations.
Keywords: ribosome pause, translation, Ribo-seq, ribosome profiling, ribosome stalling
INTRODUCTION
The progression of ribosomes along mRNA is often accompanied by the presence of strong pauses at specific locations (Richter and Coller 2015). Among the known causes of such site-specific pauses are strong RNA secondary structures whose unwinding is required for ribosome progression (Somogyi et al. 1993; Tholstrup et al. 2012) and specific nascent peptides interacting with the ribosome (Tenson and Ehrenberg 2002). Such peptide-mediated pauses are known to function as part of translation regulators sensing specific molecules as has been demonstrated for the fungal arginine attenuator peptide (Fang et al. 2004), erythromycin resistance genes in bacteria (Arenz et al. 2014) and in the polyamine responsive translation of the AMD1 mRNA in mammals (Law et al. 2001). Ribosome pausing was recently found to occur at AUG-Stop ORFs in response to boron (Tanaka et al. 2016). Ribosome pausing is also known to affect the stability of mRNA via NoGo decay (Doma and Parker 2006). Differences in local decoding rates have been implicated in cotranslational protein folding (Tsai et al. 2008).
Ribosome profiling (Ribo-seq) (Ingolia et al. 2009) provides a snapshot of ribosome locations across all mRNAs by capturing mRNA fragments protected by ribosomes (Michel and Baranov 2013; Ingolia 2016; Andreev et al. 2017). The detection of abnormally high peaks of ribosome footprint density can aid in the identification of ribosome pauses (Ingolia et al. 2011; Woolstenhulme et al. 2015), and features associated with these pauses (Sabi and Tuller 2017). Since the invention of ribosome profiling, numerous bioinformatics methods have been developed for the analysis of differential mRNA translation (Olshen et al. 2013; Xiao et al. 2016; Oertlin et al. 2017; Zhong et al. 2017), identification of translated open reading frames (ORFs) (Michel et al. 2012; Fields et al. 2015; Calviello et al. 2016; Chun et al. 2016; Raj et al. 2016; Malone et al. 2017; Ndah et al. 2017), characterization of global determinants of local decoding rates (O'Connor et al. 2016) as well as general use platforms (Chung et al. 2015; Crappé et al. 2015; Legendre et al. 2015; Dunn and Weissman 2016; Michel et al. 2016; Carja et al. 2017) and databases (Michel et al. 2014, 2015, 2018; Wan and Qian 2014; Olexiouk et al. 2016; Xie et al. 2016; Liu et al. 2017); see Calviello and Ohler (2017) for a recent review. While many ribosome profiling studies involved the identification of pauses, no dedicated software for pause detection and analysis is currently available. Further, most custom scripts detect pauses relative to the average density of the protein coding ORF. As a result, the predictions made with such scripts change when the annotations are updated.
To fill this gap, we have developed the PausePred tool for the identification of pauses by analyzing ribosome footprints aligned to transcriptome or genome reference sequences. PausePred extracts sequences surrounding high density peaks which can be readily used for exploring sequence motifs enriched in the vicinity of the pause sites. PausePred can also be used for analyzing changes in pause strength in response to different experimental conditions. The pauses can be visually examined using the Rfeet tool that we have developed to plot Ribo-seq and RNA-seq read density profiles.
RESULTS
PausePred
PausePred takes the number of reads r mapped to each position i within a sliding window of length n (step = n/2) and normalizes it over the average density within the window. The average of these values across overlapping windows is used as the pause score, Si:
| (1) |
Pause scores exceeding a certain threshold are selected for further analysis. The default threshold is 20, which is a highly permissive score that typically reports pauses on 1%–3% of coordinates; see Figure 1A. Highly atypical pauses would normally have a higher score than that. However, in certain cases, a smaller threshold could be more appropriate, e.g., when data sets are small and there is a need to increase the total number of data points as in the example used for the PausePred illustration. Due to differences in ribosome footprint coverage, the statistical significance of the pauses with the same score may differ. To take this into account, PausePred carries out a z-score transformation similar to what has been used for differential gene expression analysis (Andreev et al. 2015), i.e., pauses are grouped into bins of 300 based on footprint coverage within the corresponding windows and z-scores are calculated based on the parameters of the pause score distribution within each bin.
FIGURE 1.
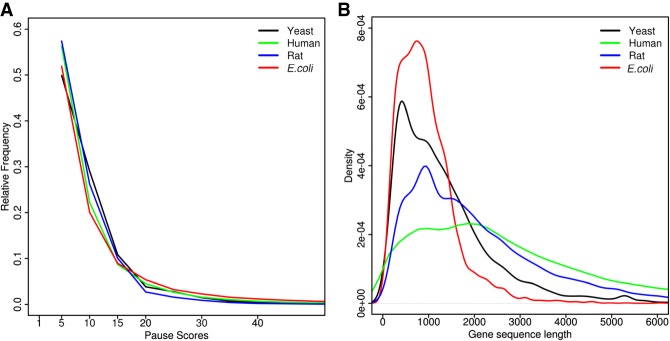
(A) The fraction of coordinates with a pause score greater than or equal to the score indicated on the x-axis (E. coli, SRA accession number SRR1734432 from Woolstenhulme et al. 2015; Yeast, control sample from Yerlikaya et al. 2016; Rat, SRA accession number SRR1557705 from Andreev et al. 2015; Human, SRA accession number SRR2433794 from Calviello et al. 2016). (B) Kernel density distributions of the lengths of protein coding ORFs in different organisms based on the transcriptome sequences downloaded from the following resources: Yeast: Saccharomyces genome database (Cherry et al. 1997), strain: S288C; E. coli: NCBI (NCBI Resource Coordinators 2017) assembly accession GCA_000005845.2; Human: Gencode (Harrow et al. 2012) release 25, GRCh37; Rat: Ensembl (Aken et al. 2016), Rnor_6.0 (GCA_000001895.4).
The user can specify the footprint read lengths to be considered (28–30 nt by default), the size of the window n for calculating the background density (the default is 1000 nt) and the offset value for inferring the position of the A-site from either the 5′ or 3′ end of the mapped reads.
The length of footprints varies depending on the organism or organelle used (Oh et al. 2011; Rooijers et al. 2013), as well as on parameters of the protocols (O'Connor et al. 2016) such as translation inhibitors (Ingolia et al. 2011; Lareau et al. 2014), nuclease digestion (Gerashchenko and Gladyshev 2017), and ribosome footprint size selection (Mohammad et al. 2016). The default window size of 1000 nt corresponds to the length of a typical protein coding region (Fig. 1B). When using PausePred for the detection of pauses specifically in small ORFs (e.g., in regulatory uORFs), it is advisable to reduce the length of the window size. Obviously, the presence of multiple pauses within the same window would reduce the pause score. Thus, it might be advisable to carry out analysis with smaller window sizes when such a possibility is a concern. The offset for inferring the A-site is an important parameter that is often data specific and should be determined based on the data. Also, due to different asymmetries in ribosome footprint length in bacteria (O'Connor et al. 2013), an offset from the 3′ end is likely to give more accurate predictions of the A-site position (Mohammad et al. 2016). With PausePred, the user can specify both the offset as well as the end from which it should be applied. The recently developed riboWaltz is an effective approach for determining read length specific A-site offsets (Lauria et al. 2017), and in conjunction with PausePred it can be used for precise pause localization using footprints of each specific length separately, since PausePred supports the use of multiple offsets for footprints of different lengths. Other tools also exist for offset detection including Plastid (Dunn and Weissman 2016), RiboProfiling (Popa et al. 2016), and RP-BP (Malone et al. 2017).
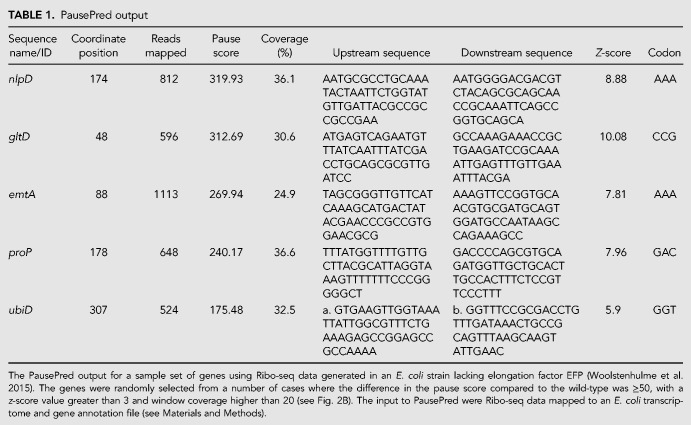
For each inferred pause, the sequence name/ID, the coordinate of the pause, the number of reads mapped, pause score, average coverage percentage of overlapping windows (where coverage is defined as the percentage of positions within a window having a minimum of one read mapped) and the z-score are output in a csv file. The flanking sequences are also provided (the default is 100 nt with the pause site corresponding to position 51) (Table 1). If an annotation file has been supplied, PausePred will provide codon level information related to the predicted pause (Table 1) (see Materials and Methods). We tested PausePred on ribosome profiling data from different organisms including E. coli (Woolstenhulme et al. 2015), yeast (Yerlikaya et al. 2016), rat (Andreev et al. 2015), and human (Calviello et al. 2016) (Fig. 1A).
TABLE 1.
PausePred output
Illustration of PausePred usage
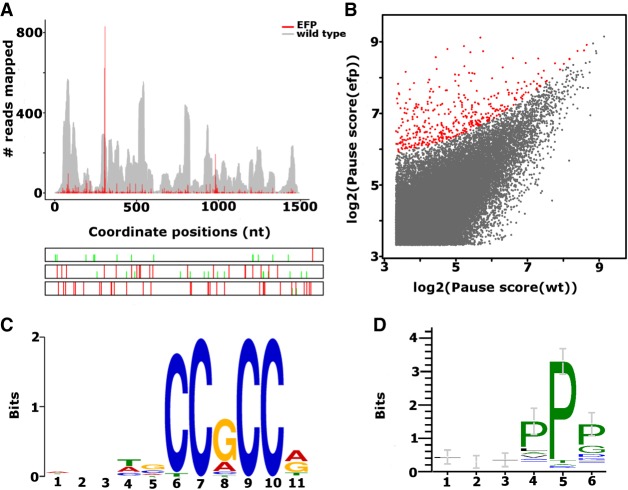
Here we illustrate PausePred predictions using the Ribo-seq data generated for the E. coli MG1655 strain and a mutant strain that lacks elongation factor EFP (Woolstenhulme et al. 2015). After processing the data (see Materials and Methods), the sorted BAM files were uploaded to the PausePred webtool (https://pausepred.ucc.ie/). To compare the EFP knockout strain with the wild-type, a pause score of 10, read lengths varying from 25–35, a 3′ offset of 12 nt for each read length and window coverage of 5 (a minimum of 5% of the positions within a window size of 1000 nt had at least one read mapped) were used for pause prediction in both the EFP mutant and the wild-type Ribo-seq samples. A lower threshold than the default pause score was used to increase the number of detected pauses. Table 1 provides the PausePred output for a subset of the detected pauses, and Figure 2A shows the Rfeet visualization (see Materials and Methods) of the pause detected in the Ribo-seq density profile of the ubiD gene. Of the predicted pauses, 399 candidates were found to have a difference in pause score of at least 50 compared to the corresponding position in the wild-type Ribo-seq alignments (highlighted in red in Fig. 2B).
FIGURE 2.
(A) Rfeet visualization of the ribosome density profile (red) for the ubiD gene containing a ribosome pause detected by PausePred at position 307 of the CDS in an E. coli strain lacking elongation factor EFP (Woolstenhulme et al. 2015). The wild-type strain is provided as a gray coverage profile, i.e., where the number of sequence reads aligning to each coordinate is displayed. The bottom panel shows the ORF organization with red lines for stop codons and green lines for AUG codons. (B) Comparison of pause scores predicted by PausePred in E. coli mutants lacking elongation factor EFP and the wild-type sample. The EFP pauses with a pause score ≥50 compared to WT are highlighted in red. (C) The sequence motif found to be overrepresented in 88 cases, using 15 nt upstream of the detected pauses in the filtered 399 cases (B, highlighted in red). The pause score of these locations is higher than in the wild-type by at least 50. (D) The enrichment of the PP motif in the 88 sequences found to contain a C-rich motif at the nucleotide level.
PausePred outputs the sequences flanking the pause sites (Table 1) which can be explored for pause-related motifs. We analyzed the 15 nucleotides (nt) upstream of the detected pauses (corresponding approximately to the five codons encoding the C-terminal part of the nascent peptide) in the 399 candidates for overrepresented motifs using Multiple EM for Motif Elicitation (MEME 4.11.2) (Bailey et al. 2009). The most significant motif was found in 88 of these sequences with an e-value of 2.6 × 10−49 (Fig. 2C). The last six nucleotides of this motif may reflect the enrichment of the polyproline motif reported by Woolstenhulme et al. (2015), to be associated with pauses in the EFP lacking E. coli mutants. The six amino acids upstream of the pause locations in the 88 sequences containing the C-rich motif were analyzed using Weblogo (Crooks et al. 2004) for a polyproline motif enrichment at the amino acid level by using the corresponding protein sequences obtained from Genbank (NCBI Resource Coordinators 2017) (assembly accession GCA_000005845.2) (Fig. 2D). Further, the PP motif was found to be 42.83 times more frequent in the 399 sequences containing the pauses than expected (P-value <0.0001 Fisher exact test).
Detection of previously reported pauses
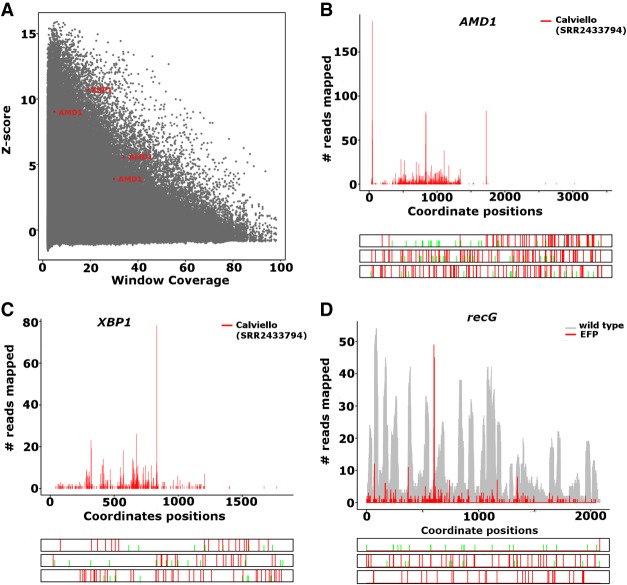
Ribosome pausing at an upstream ORF (MAGDIS) has been reported for the AMDl gene (Ruan et al. 1996) and recently (Yordanova et al. 2018), detected a ribosome stalling site in the 3′ leader of this gene. Using PausePred and ribosome profiling data generated for human from Calviello et al. (2016), we detected pauses at mRNA positions 47, 837, and 1725 with pause scores 114.98, 59.32, and 373.87 and z-scores of 5.55, 3.89, and 9.03, respectively (Fig. 3A). In Figure 3B we provide the Rfeet generated ribosome profile for the AMD1 gene showing the three detected pauses.
FIGURE 3.
(A) The z-score and coverage of all the pauses predicted in the Calviello et al. (2016) data. The pauses predicted in AMD1 and XBP1 are highlighted in red. (B) Ribosome profile and ORF architecture generated with Rfeet showing the footprint density for the human AMD1 gene (transcript ENST00000368885) using Calviello et al. (2016) data. Pauses were detected at positions 47, 837, and 1725 with a pause score of 114.98, 59.32, and 373.87; z-score value of 5.55, 3.89, and 9.03. (C) The footprint density for the XBP1 gene (transcript ENST00000403532) using Calviello et al. (2016) data. A pause was detected in the XBP1 gene at position 832 with pause score 194.6 and z-score value of 10.70 using PausePred. (D) The footprint density for the gene recG using Woolstenhulme et al. (2015) data. The recG gene was found to contain a pause at position 616 in the EFP lacking sample with a pause score of 246.05 and a z-score value of 6.9 using PausePred.
PausePred also detected the known pause in XBP1u (Ingolia et al. 2011; Yanagitani et al. 2011) at mRNA position 832 with a pause score of 194.6 and z-score value of 10.70 (Fig. 3A,C).
PausePred also identified many of the pauses predicted by Woolstenhulme et al. (2015). One such example is the pause at position 616 for the recG gene with a pause score 246.05 and z-score value of 6.9 (Fig. 3D).
DISCUSSION
A number of studies (Ingolia et al. 2011; Li et al. 2012; Woolstenhulme et al. 2015; Mohammad et al. 2016; Matsuo et al. 2017) have relied on in-house scripts to identify pause sites in Ribo-seq data. Such in-house scripts are often difficult to transfer to use in different settings due to differences in computer platforms, software dependencies and data specific reasons. Custom made scripts often require gene annotation information, i.e., GTF (general transfer format) or GFF (general feature format). As a result, the output of these scripts is not stable over time due to changes in annotations. The optional annotation independence of PausePred makes it an easy universal tool for consistent and reproducible analysis of ribosome pause locations. It also allows the detection of ribosome pauses outside of coding regions, e.g., a recently discovered stalling site in the stop codon readthrough extension of the human AMD1 mRNA; see Figure 3B (Yordanova et al. 2018).
PausePred provides users with a browser-based translational pause prediction tool. Coupled with the visualization functionality offered by Rfeet, these webtools avoid the need to download software or develop computer programs. For researchers comfortable with the command line, stand-alone versions of both tools are also available. To illustrate the utility of these tools, a downloadable example data set for both genome and transcriptome alignments is provided along with step-by-step help instructions including screen shots. In future, we plan to enhance the functionality of PausePred by incorporating differential pause analysis.
It is important to note that in its essence PausePred is a peak calling program that detects unusually high local density of footprints, but such density could arise due to reasons other than ribosome pauses. One source of high density is ambiguous alignments. Translation of mRNAs containing segments that share high sequence similarity would result in the alignment of all footprints generated from different mRNAs to the same locations elevating their local density. To avoid such artifacts, it is important to limit ambiguous mapping of sequencing reads or use an appropriate RNA-seq control that was generated with cDNA libraries of the same length, as it would produce high peaks of RNA-seq density for the same reason. The precise sequences of footprints vary depending on experimental protocol parameters; we discussed the sources of this variability elsewhere (O'Connor et al. 2016; Andreev et al. 2017). Therefore, we recommend to use multiple data sets for a reliable analysis to yield robust interpretations. A prominent example related to this is the claim that Shine–Dalgarno sequences induce ribosome pauses in bacteria (Li et al. 2012). This claim was challenged later on the basis that the original study did not sequence footprints of all lengths but focused only on long ribosome protected fragments (Mohammad et al. 2016). Longer footprints are associated with Shine–Dalgarno sequences (O'Connor et al. 2013) and therefore sequencing only long footprints results in elevated footprint density downstream from Shine–Dalgarno sequences (Mohammad et al. 2016).
PausePred and Rfeet are designed for specific purposes in ribosome profiling data analysis, pause detection and visualization of local footprint density, respectively, and cannot be used as stand-alone tools for ribosome profiling data analysis which is a multistep process that requires processing of raw reads, alignment to reference sequences, removal of rRNA and tRNA contaminations. There are existing platforms that have been developed for this purpose, e.g., RiboGalaxy (Michel et al. 2016) and Plastid (Dunn and Weissman 2016), that provide the tools for these steps as well as many additional utilities. We plan to integrate PausePred and Rfeet into RiboGalaxy, which should make them readily available for the users of RiboGalaxy.
MATERIALS AND METHODS
PausePred methodology
The PausePred and Rfeet tools are written in PERL and can be used via a web browser interface at https://pausepred.ucc.ie/. A standalone version is available for download on GitHub at https://github.com/romikasaini/Pausepred_offline. The web-based versions are served using the Apache2 web server. The website is designed using HTML and JAVA.
The PausePred workflow requires a reference sequence in FASTA format (genomic or transcriptomic) and Ribo-seq data alignments in sorted BAM format. A gene annotation file (a tab delimited text file with the first column specifying the name of the gene/ORF and second and third columns specifying the ORF coordinates) can also be optionally supplied. The coding frame information provided in the annotation file is used to detect the codon of the predicted pause.
The E. coli Ribo-seq data that we used to illustrate the utility of PausePred were downloaded from NCBI GEO accession number GSE64488 (Woolstenhulme et al. 2015). Raw reads need to be preprocessed to remove adapter sequences and rRNA contaminations followed by the alignment to reference sequences prior to using Pausepred. Raw reads were trimmed using Cutadapt (Martin 2011), and reads that mapped to rRNA and tRNA sequences were removed using the Bowtie short read alignment program (version 1.1.1) (Langmead et al. 2009) allowing a maximum of three mismatches. The E. coli transcriptome obtained from NCBI assembly accession GCA_000005845.2 and assembly name ASM584v2 was used as a reference sequence for the alignment of the remaining Ribo-seq reads also using Bowtie version 1.1.1. (Langmead et al. 2009), with no more than three mismatches allowed. The Bowtie SAM outputs were converted to BAM format and sorted using SAMtools (Li et al. 2009). The sorted BAM files were uploaded to the PausePred webtool (https://pausepred.ucc.ie/).
For determining a suitable fold change threshold (Fig. 1A), we used Ribo-seq data from E. coli, human, rat, and yeast. For the E. coli data, we used a 3′ offset of 12 nt as mentioned in Woolstenhulme et al. (2015). For human, we selected data from Calviello et al. (2016) (accession number SRR2433794). The BAM alignment file was generated using the same approach as for E. coli (see Materials and Methods). The offsets for the A-site positions were predicted by creating metagene plots as described in Andreev et al. (2015). An offset value of 17 was used for reads of length 28–30 and 18 for reads of length 31. For rat we used Ribo-seq data from Andreev et al. (2015) (accession number SRR1557705). The parameters used for this data set were the same as for the Calviello et al. (2016) human data set. For yeast we used a control sample from Yerlikaya et al. (2016) with a 5′ offset value of 15 nt for a read length range of 28–31 nt. A default window of 1000 nt was used and the fold change was set to 5.
Rfeet methodology
Rfeet takes a BAM alignment file (single end reads only) and a reference sequence FASTA file as inputs, and produces Ribo-seq and RNA-seq (if supplied) read density profiles for a sequence name/ID. The nucleotide sequences are fetched with bioperl modules to provide the ORF architectures for the mRNA (forward strand) or genomic region (forward and reverse strands) with color delineation for the strand orientation and the start (ATG) and stop (TAG, TAA, TGA) codons. The ribosome footprint density plots, i.e., ribosome profiles, display 5′ or 3′ footprint end counts or the inferred positions of the decoding center when an offset is used (Figs. 2A, 3B–D). The option to use a second BAM alignment file is available for comparing additional RNA-seq or Ribo-seq alignments. For a given profile region (Gene/Transcript/Chr:start-stop), Rfeet provides two plot type options. The “absolute plot” will show the raw counts of the mapped reads and is the default option. Absolute counts are also plotted for the second input file when a coverage plot is selected. A coverage plot provides the number of sequence reads that align to each coordinate. The “normalized plot” is generated by dividing the number of reads mapped on each position by the total number of reads mapped in a particular profile.
| (2) |
where r is the number of reads mapped to each position i across the profile region of length n.
To generate individual plots for more than one transcript or a genomic region, a comma separated list of IDs should be used. Red and blue colors are used to display ribosome footprints aligned to positive and negative strands, respectively (for example figure, see the Rfeet help page).
COMPETING INTEREST STATEMENT
A.M.M. and P.V.B. are founders of Ribomaps Ltd., which offers ribosome profiling data generation and analysis as a service.
ACKNOWLEDGMENTS
We thank James P.A. Mullan for helping with the Apache2 server setup. We also thank Padraig Caomhain O'Flynn for testing PausePred. This work was supported by Science Foundation Ireland (grant 12/IA/1335 to P.V.B.).
Footnotes
Article is online at http://www.rnajournal.org/cgi/doi/10.1261/rna.065235.117.
REFERENCES
- Aken BL, Ayling S, Barrell D, Clarke L, Curwen V, Fairley S, Fernandez Banet J, Billis K, Garcia Giron C, Hourlier T, et al. 2016. The Ensembl gene annotation system. Database (Oxford) 2016: baw093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreev DE, O'Connor PB, Zhdanov AV, Dmitriev RI, Shatsky IN, Papkovsky DB, Baranov PV. 2015. Oxygen and glucose deprivation induces widespread alterations in mRNA translation within 20 minutes. Genome Biol 16: 90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreev DE, O'Connor PB, Loughran G, Dmitriev SE, Baranov PV, Shatsky IN. 2017. Insights into the mechanisms of eukaryotic translation gained with ribosome profiling. Nucleic Acids Res 45: 513–526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arenz S, Ramu H, Gupta P, Berninghausen O, Beckmann R, Vázquez-Laslop N, Mankin AS, Wilson DN. 2014. Molecular basis for erythromycin-dependent ribosome stalling during translation of the ErmBL leader peptide. Nat Commun 5: 3501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey TL, Boden M, Buske FA, Frith M, Grant CE, Clementi L, Ren J, Li WW, Noble WS. 2009. MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res 37: W202–W208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calviello L, Ohler U. 2017. Beyond read-counts: ribo-seq data analysis to understand the functions of the transcriptome. Trends Genet 33: 728–744. [DOI] [PubMed] [Google Scholar]
- Calviello L, Mukherjee N, Wyler E, Zauber H, Hirsekorn A, Selbach M, Landthaler M, Obermayer B, Ohler U. 2016. Detecting actively translated open reading frames in ribosome profiling data. Nat Methods 13: 165–170. [DOI] [PubMed] [Google Scholar]
- Carja O, Xing T, Plotkin JB, Shah P. 2017. riboviz: analysis and visualization of ribosome profiling datasets. bioRxiv 10.1101/100032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherry JM, Ball C, Weng S, Juvik G, Schmidt R, Adler C, Dunn B, Dwight S, Riles L, Mortimer RK, et al. 1997. Genetic and physical maps of Saccharomyces cerevisiae. Nature 387: 67–73. [PMC free article] [PubMed] [Google Scholar]
- Chun SY, Rodriguez CM, Todd PK, Mills RE. 2016. SPECtre: a spectral coherence-based classifier of actively translated transcripts from ribosome profiling sequence data. BMC Bioinformatics 17: 482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung BY, Hardcastle TJ, Jones JD, Irigoyen N, Firth AE, Baulcombe DC, Brierley I. 2015. The use of duplex-specific nuclease in ribosome profiling and a user-friendly software package for Ribo-seq data analysis. RNA 21: 1731–1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crappé J, Ndah E, Koch A, Steyaert S, Gawron D, De Keulenaer S, De Meester E, De Meyer T, Van Criekinge W, Van Damme P, et al. 2015. PROTEOFORMER: deep proteome coverage through ribosome profiling and MS integration. Nucleic Acids Res 43: e29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crooks GE, Hon G, Chandonia JM, Brenner SE. 2004. WebLogo: a sequence logo generator. Genome Res 14: 1188–1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doma MK, Parker R. 2006. Endonucleolytic cleavage of eukaryotic mRNAs with stalls in translation elongation. Nature 440: 561–564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn JG, Weissman JS. 2016. Plastid: nucleotide-resolution analysis of next-generation sequencing and genomics data. BMC Genomics 17: 958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang P, Spevak CC, Wu C, Sachs MS. 2004. A nascent polypeptide domain that can regulate translation elongation. Proc Natl Acad Sci 101: 4059–4064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fields AP, Rodriguez EH, Jovanovic M, Stern-Ginossar N, Haas BJ, Mertins P, Raychowdhury R, Hacohen N, Carr SA, Ingolia NT, et al. 2015. A regression-based analysis of ribosome-profiling data reveals a conserved complexity to mammalian translation. Mol Cell 60: 816–827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerashchenko MV, Gladyshev VN. 2017. Ribonuclease selection for ribosome profiling. Nucleic Acids Res 45: e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrow J, Frankish A, Gonzalez JM, Tapanari E, Diekhans M, Kokocinski F, Aken BL, Barrell D, Zadissa A, Searle S, et al. 2012. GENCODE: the reference human genome annotation for The ENCODE Project. Genome Res 22: 1760–1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingolia NT. 2016. Ribosome footprint profiling of translation throughout the genome. Cell 165: 22–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingolia NT, Ghaemmaghami S, Newman JR, Weissman JS. 2009. Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science 324: 218–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingolia NT, Lareau LF, Weissman JS. 2011. Ribosome profiling of mouse embryonic stem cells reveals the complexity and dynamics of mammalian proteomes. Cell 147: 789–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL. 2009. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10: R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lareau LF, Hite DH, Hogan GJ, Brown PO. 2014. Distinct stages of the translation elongation cycle revealed by sequencing ribosome-protected mRNA fragments. Elife 3: e01257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauria F, Tebaldi T, Bernabo P, Groen EJN, Gillingwater TH, Viero G. 2017. riboWaltz: optimization of ribosome P-site positioning in ribosome profiling data. bioRxiv 10.1101/169862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law GL, Raney A, Heusner C, Morris DR. 2001. Polyamine regulation of ribosome pausing at the upstream open reading frame of S-adenosylmethionine decarboxylase. J Biol Chem 276: 38036–38043. [DOI] [PubMed] [Google Scholar]
- Legendre R, Baudin-Baillieu A, Hatin I, Namy O. 2015. RiboTools: a Galaxy toolbox for qualitative ribosome profiling analysis. Bioinformatics 31: 2586–2588. [DOI] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, Genome Project Data Processing Subgroup. 2009. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25: 2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li GW, Oh E, Weissman JS. 2012. The anti-Shine-Dalgarno sequence drives translational pausing and codon choice in bacteria. Nature 484: 538–541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W, Xiang L, Zheng T, Jin J, Zhang G. 2017. TranslatomeDB: a comprehensive database and cloud-based analysis platform for translatome sequencing data. Nucleic Acids Res 46: D206–D212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malone B, Atanassov I, Aeschimann F, Li X, Grosshans H, Dieterich C. 2017. Bayesian prediction of RNA translation from ribosome profiling. Nucleic Acids Res 45: 2960–2972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin M. 2011. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnetjournal 17: 10–12. [Google Scholar]
- Matsuo Y, Ikeuchi K, Saeki Y, Iwasaki S, Schmidt C, Udagawa T, Sato F, Tsuchiya H, Becker T, Tanaka K, et al. 2017. Ubiquitination of stalled ribosome triggers ribosome-associated quality control. Nat Commun 8: 159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michel AM, Baranov PV. 2013. Ribosome profiling: a Hi-Def monitor for protein synthesis at the genome-wide scale. Wiley Interdiscip Rev RNA 4: 473–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michel AM, Choudhury KR, Firth AE, Ingolia NT, Atkins JF, Baranov PV. 2012. Observation of dually decoded regions of the human genome using ribosome profiling data. Genome Res 22: 2219–2229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michel AM, Fox G, M Kiran A, De Bo C, O'Connor PB, Heaphy SM, Mullan JP, Donohue CA, Higgins DG, Baranov PV. 2014. GWIPS-viz: development of a ribo-seq genome browser. Nucleic Acids Res 42: D859–D864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michel AM, Ahern AM, Donohue CA, Baranov PV. 2015. GWIPS-viz as a tool for exploring ribosome profiling evidence supporting the synthesis of alternative proteoforms. Proteomics 15: 2410–2416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michel AM, Mullan JP, Velayudhan V, O'Connor PB, Donohue CA, Baranov PV. 2016. RiboGalaxy: a browser based platform for the alignment, analysis and visualization of ribosome profiling data. RNA Biol 13: 316–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michel AM, Kiniry SJ, O'Connor PBF, Mullan JP, Baranov PV. 2018. GWIPS-viz: 2018 update. Nucleic Acids Res 46: D823–D830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohammad F, Woolstenhulme CJ, Green R, Buskirk AR. 2016. Clarifying the translational pausing landscape in bacteria by ribosome profiling. Cell Rep 14: 686–694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- NCBI Resource Coordinators. 2017. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res 45: D12–D17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ndah E, Jonckheere V, Giess A, Valen E, Menschaert G, Van Damme P. 2017. REPARATION: ribosome profiling assisted (re-)annotation of bacterial genomes. Nucleic Acid Res 45: e168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Connor PB, Li GW, Weissman JS, Atkins JF, Baranov PV. 2013. rRNA:mRNA pairing alters the length and the symmetry of mRNA-protected fragments in ribosome profiling experiments. Bioinformatics 29: 1488–1491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Connor PB, Andreev DE, Baranov PV. 2016. Comparative survey of the relative impact of mRNA features on local ribosome profiling read density. Nat Commun 7: 12915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oertlin C, Lorent J, Gandin V, Murie C, Masvidal L, Cargnello M, Furic L, Topisirovic I, Larsson O. 2017. Genome-wide analysis of differential translation and differential translational buffering using anota2seq. bioRxiv 10.1101/106922. [DOI] [Google Scholar]
- Oh E, Becker AH, Sandikci A, Huber D, Chaba R, Gloge F, Nichols RJ, Typas A, Gross CA, Kramer G, et al. 2011. Selective ribosome profiling reveals the cotranslational chaperone action of trigger factor in vivo. Cell 147: 1295–1308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olexiouk V, Crappé J, Verbruggen S, Verhegen K, Martens L, Menschaert G. 2016. sORFs.org: a repository of small ORFs identified by ribosome profiling. Nucleic Acids Res 44: D324–D329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olshen AB, Hsieh AC, Stumpf CR, Olshen RA, Ruggero D, Taylor BS. 2013. Assessing gene-level translational control from ribosome profiling. Bioinformatics 29: 2995–3002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popa A, Lebrigand K, Paquet A, Nottet N, Robbe-Sermesant K, Waldmann R, Barbry P. 2016. RiboProfiling: a Bioconductor package for standard Ribo-seq pipeline processing. F1000Res 5: 1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raj A, Wang SH, Shim H, Harpak A, Li YI, Engelmann B, Stephens M, Gilad Y, Pritchard JK. 2016. Thousands of novel translated open reading frames in humans inferred by ribosome footprint profiling. Elife 5: e13328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter JD, Coller J. 2015. Pausing on polyribosomes: make way for elongation in translational control. Cell 163: 292–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rooijers K, Loayza-Puch F, Nijtmans LG, Agami R. 2013. Ribosome profiling reveals features of normal and disease-associated mitochondrial translation. Nat Commun 4: 2886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruan H, Shantz LM, Pegg AE, Morris DR. 1996. The upstream open reading frame of the mRNA encoding S-adenosylmethionine decarboxylase is a polyamine-responsive translational control element. J Biol Chem 271: 29576–29582. [DOI] [PubMed] [Google Scholar]
- Sabi R, Tuller T. 2017. Computational analysis of nascent peptides that induce ribosome stalling and their proteomic distribution in Saccharomyces cerevisiae. RNA 23: 983–994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somogyi P, Jenner AJ, Brierley I, Inglis SC. 1993. Ribosomal pausing during translation of an RNA pseudoknot. Mol Cell Biol 13: 6931–6940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka M, Sotta N, Yamazumi Y, Yamashita Y, Miwa K, Murota K, Chiba Y, Hirai MY, Akiyama T, Onouchi H, et al. 2016. The minimum open reading frame, AUG-Stop, induces boron-dependent ribosome stalling and mRNA degradation. Plant Cell 28: 2830–2849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenson T, Ehrenberg M. 2002. Regulatory nascent peptides in the ribosomal tunnel. Cell 108: 591–594. [DOI] [PubMed] [Google Scholar]
- Tholstrup J, Oddershede LB, Sorensen MA. 2012. mRNA pseudoknot structures can act as ribosomal roadblocks. Nucleic Acids Res 40: 303–313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai CJ, Sauna ZE, Kimchi-Sarfaty C, Ambudkar SV, Gottesman MM, Nussinov R. 2008. Synonymous mutations and ribosome stalling can lead to altered folding pathways and distinct minima. J Mol Biol 383: 281–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan J, Qian SB. 2014. TISdb: a database for alternative translation initiation in mammalian cells. Nucleic Acids Res 42: D845–850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woolstenhulme CJ, Guydosh NR, Green R, Buskirk AR. 2015. High-precision analysis of translational pausing by ribosome profiling in bacteria lacking EFP. Cell Rep 11: 13–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao Z, Zou Q, Liu Y, Yang X. 2016. Genome-wide assessment of differential translations with ribosome profiling data. Nat Commun 7: 11194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie SQ, Nie P, Wang Y, Wang H, Li H, Yang Z, Liu Y, Ren J, Xie Z. 2016. RPFdb: a database for genome wide information of translated mRNA generated from ribosome profiling. Nucleic Acids Res 44: D254–D258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanagitani K, Kimata Y, Kadokura H, Kohno K. 2011. Translational pausing ensures membrane targeting and cytoplasmic splicing of XBP1u mRNA. Science 331: 586–589. [DOI] [PubMed] [Google Scholar]
- Yerlikaya S, Meusburger M, Kumari R, Huber A, Anrather D, Costanzo M, Boone C, Ammerer G, Baranov PV, Loewith R. 2016. TORC1 and TORC2 work together to regulate ribosomal protein S6 phosphorylation in Saccharomyces cerevisiae. Mol Biol Cell 27: 397–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yordanova MM, Loughran G, Zhdanov AV, Mariotti M, Kiniry SJ, O'Connor PB, Andreev DE, Tzani I, Saffert P, Michel AM, et al. 2018. AMD1 mRNA employs ribosome stalling as a mechanism for molecular memory formation. Nature 553: 356–360. [DOI] [PubMed] [Google Scholar]
- Zhong Y, Karaletsos T, Drewe P, Sreedharan VT, Kuo D, Singh K, Wendel HG, Ratsch G. 2017. RiboDiff: detecting changes of mRNA translation efficiency from ribosome footprints. Bioinformatics 33: 139–141. [DOI] [PMC free article] [PubMed] [Google Scholar]