Abstract
Eukaryotic transfer RNAs (tRNA) contain on average 13 modifications that perform a wide range of roles in translation and in the generation of tRNA fragments that regulate gene expression. Queuosine (Q) modification occurs in the wobble anticodon position of tRNAs for amino acids His, Asn, Tyr, and Asp. In eukaryotes, Q modification is fully dependent on diet or on gut microbiome in multicellular organisms. Despite decades of study, cellular roles of Q modification remain to be fully elucidated. Here we show that in human cells, Q modification specifically protects its cognate tRNAHis and tRNAAsn against cleavage by ribonucleases. We generated cell lines that contain completely depleted or fully Q-modified tRNAs. Using these resources, we found that Q modification significantly reduces angiogenin cleavage of its cognate tRNAs in vitro. Q modification does not change the cellular abundance of the cognate full-length tRNAs, but alters the cellular content of their fragments in vivo in the absence and presence of stress. Our results provide a new biological aspect of Q modification and a mechanism of how Q modification alters small RNA pools in human cells.
Keywords: queuosine modification, tRNA, tRNA fragment
INTRODUCTION
Transfer RNAs (tRNAs) are essential for protein synthesis. Among all RNA species, tRNAs undergo the greatest number of chemically diverse modifications. Eukaryotic tRNAs contain on average over 13 modifications. A great number of modifications are found at position 34, the wobble anticodon nucleotide (Phizicky and Hopper 2010; El Yacoubi et al. 2012). Growing evidence indicates that tRNA modifications are critical for many aspects of tRNA functions such as folding, stability, and decoding (Kirchner and Ignatova 2015). tRNA modifications also play important roles in human diseases (Torres et al. 2014).
Queuosine is a hypermodified 7-deaza-guanosine that occurs at the wobble anticodon position 34 of four tRNAs for amino acids His, Asn, Tyr, and Asp with 5′GUN anticodons (recently reviewed in Fergus et al. 2015). Queuosine (Q) is the nucleoside; the modified base is designated as queuine. Bacteria synthesize queuine through a multistep biosynthesis pathway that results in the wobble Q34 base in these four tRNAs. Eukaryotes do not synthesize queuine, but rely on dietary sources and/or the gut microbes to obtain queuine, which is exchanged for the G34 base in the same four tRNAs by an enzyme encoded in the genomes. In humans, queuine produced by the gut microbiome is taken up by all cells and incorporated into the wobble anticodon nucleotide by the heterodimeric enzyme of QTRT1/QTRT2 (formerly QTRTD1, Supplemental Fig. S1). Thus, Q modification in human cells can depend on microbiome activity. The physiological requirements for the tRNA Q modifications in eukaryotes have been documented for decades, linking its relationship to many biological processes such as development, cancer, and tyrosine biosynthesis (Fergus et al. 2015). The role of Q modification in cells was suggested to be related to codon-biased translation of discrete mRNA transcripts given that Q modification is located in the wobble anticodon position critical for codon recognition (Yasukawa et al. 2001; Zaborske et al. 2014; Endres et al. 2015). Using the Turnip yellow mosaic virus coat protein mRNA as a reporter in Xenopus oocyte, Q-modified tRNAHis was found to decode the His codons CAU/CAC equally, while unmodified tRNAHis (G34) preferred CAC over CAU (Meier et al. 1985). Whether this result can be applied transcriptome-wide, however, is unknown. Q modification may also tune translational fidelity which has led to genome-wide reprogramming of coding sequences in Drosophilids (Zaborske et al. 2014).
A recent study in the fission yeast Schizosaccharomyces pombe showed that Q modification directly affects the activity of Dnmt2-dependent m5C38 modification in tRNAAsp, suggesting an unanticipated role of Q modification in RNA metabolism (Muller et al. 2015; Ehrenhofer-Murray 2017). Dnmt2 modifies m5C38 in three tRNA isoacceptors among which only tRNAAsp contains Q modification. Previous work in Drosophila found that m5C38 methylation by Dnmt2 protects cognate tRNAs against stress-induced cleavage and affects the biogenesis of tRNA-derived RNA fragments, which are functional small RNAs, especially in response to stress conditions (Lee et al. 2009; Yamasaki et al. 2009; Schaefer et al. 2010; Goodarzi et al. 2015).
tRNA fragments have recently received considerable attention due to their effects in the regulation of gene expression (Raina and Ibba 2014; Goodarzi et al. 2015; Kumar et al. 2016). tRNA fragments can be generated by multiple cellular ribonucleases; angiogenin among the best characterized (Yamasaki et al. 2009; Emara et al. 2010; Ivanov et al. 2011). Angiogenin is a stress-activated member of the RNase A superfamily. This human protein cleaves tRNA molecules within anticodon loops, leading to the production of tRNA-derived stress-induced fragments which are components of the cellular stress response program (Fu et al. 2009; Yamasaki et al. 2009; Ivanov et al. 2011).
Here, we investigated the role of Q modification in tRNA fragment generation in human cells. We generated cell lines fully depleted of Q modification and the same cells with fully Q-modified tRNAs. Using these resources we show that in vitro, Q modification protects cognate tRNAs against angiogenin cleavage in their anticodon loops. We applied high-throughput sequencing of full-length tRNAs and of tRNA fragments and found that although Q modification did not alter the levels of full-length tRNA, Q-modified cognate tRNA fragments were present in substantially higher levels in cells without Q modification. We also show that Q modification protects against cognate tRNA cleavage in vivo upon oxidative stress, thus altering the cellular tRNA fragment pools. Our study suggests a previously uncharacterized link between Q modification and small RNA metabolism in human cells.
RESULTS AND DISCUSSION
Generation of 0Q and 100Q cell lines
In order to investigate the role of tRNA Q modifications, cells devoid of Q modifications are required. As a nonessential modification, Q depleted cells may be obtained either by knocking out the QTRT1 gene or by queuine depletion in the culture medium (Rakovich et al. 2011). We went with the queuine depletion approach as QTRT1 knockout may alter the physiological response of the cell. We found that HEK293T cells had considerable amounts of Q modifications in the cognate tRNAHis and tRNAAsn when cultured with regular fetal bovine serum, ranging from 10%–60% (not shown). This result was consistent with previous reports that cells could obtain tRNA Q-modification by taking up the small molecule queuine from regular FBS used in cell culture, whereas using dialyzed FBS avoided this issue (Gunduz and Katze 1984; Zallot et al. 2014). We generated Q-depleted HEK293T cells by growing them in dialyzed FBS through continuous passages until the Q modification became undetectable, while Q modification in tRNAHis and tRNAAsn was fully restored by adding queuine to the medium (Fig. 1A). Q modifications in tRNAAsp and tRNATyr were not detected by our 3-acrylamidophenylboronic acid (APB) gel-based method due to further glycosylation of these Q-modified tRNAs that eliminated the cis-diol in the Q base responsible for the shift of the modified tRNA in the APB gel (Kasai et al. 1976). We performed HPLC of the completely nuclease-digested, purified tRNATyr and tRNAAsp followed by mass spectrometry to identify the presence of galactose-Q (galQ) in tRNATyr and mannose-Q (manQ) in tRNAAsp in tRNAs isolated from 100Q cells (Supplemental Fig. S2). We identified galQ and manQ from both UV absorbance profile and the mass of the modified nucleotide 5′ monophosphate.
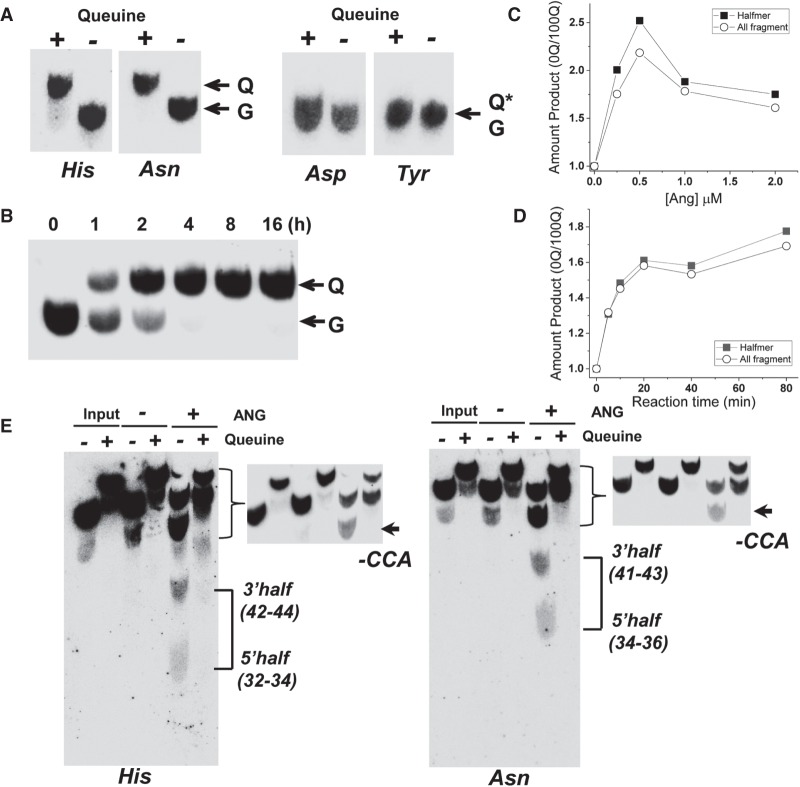
FIGURE 1.
Q modification inhibits angiogenin cleavage in vitro. (A) Northern blot analysis of total RNAs isolated from 0Q, 100Q HEK293T cells probed against tRNAHis, tRNAAsn, tRNAAsp, and tRNATyr separated by APB-containing gels. Q, Q*, and G indicate tRNA with Q34, glycosylated Q34 and G34, respectively. (B) Appearance of Q modification for tRNAAsn starting from the addition of queuine to HEK293T 0Q cells. Q and G indicate tRNA with Q34 and G34, respectively. (C,D) Comparative amount of angiogenin cleavage products upon varying the angiogenin concentration (C) or reaction time (D) of total tRNA isolated from 0Q and 100Q HEK293T cells. tRNAs were 5′ 32P-labeled, so the products can be identified by size. Comparison includes either only tRNA halfmer products corresponding to cleavage in the anticodon loop or all fragments derived from cleavage anywhere in the tRNA body. (E) Northern blot analysis of tRNA cleavage by angiogenin separated by APB-containing gels using the tRNAHis probe on the left and tRNAAsn probe on the right. Northern blot detected both 5′ and 3′ cleavage product in the anticodon loop; these halfmers in the 0Q sample are indicated by connecting lines on the right, and the size of the products is shown in parentheses. The product near the full-length likely corresponds to angiogenin cleavage of the 3′CCA tail; these are indicated by an arrow and -CCA on the right. A high contrast image of this portion is also shown for better visualization. Quantitation of these products is shown in Supplemental Table S1. Q-containing tRNA fragments are shifted in the 100Q sample.
We measured the time course for HEK293T cells to fully restore Q modification in tRNAAsn (Fig. 1B). tRNAAsn became fully modified after ∼8 h in the presence of queuine. We designated the completely Q-depleted cells as 0Q cells and fully Q-modified cells as 100Q cells hereafter. We focused on tRNAHis and tRNAAsn in our subsequent studies since for these cognate tRNAs, Q modification levels and the resulting properties could be directly assessed.
Q modification and tRNA cleavage by angiogenin in vitro
The ribonuclease angiogenin primarily targets tRNA anticodon loops for cleavage in vivo (Anderson and Ivanov 2014; Ivanov et al. 2014). We reasoned that the Q modification in the wobble anticodon position may directly affect angiogenin cleavage of the cognate, Q-modified tRNAs. Total tRNAs from 0Q and 100Q cells were 5′ 32P-labeled and gel purified, and incubated with recombinant human angiogenin protein either at varying concentration or incubation time (Fig. 1C,D). We quantified the fraction of products for tRNA fragments of all sizes from angiogenin cleavage throughout the tRNA body and those corresponding to tRNA halfmers which were derived from cleavage in the anticodon loop, and compared the amount of products from tRNAs isolated from 0Q over 100Q cells. We found that the amount of both all and halfmer cleavage products was always lower using total tRNAs from 100Q cells compared to those from 0Q cells, either at varying concentration of angiogenin from 0.25–2 µM or over a time course from 5–80 min. To specifically examine the cleavage products for the Q-modified tRNAHis and tRNAAsn, we performed northern blot analysis for these tRNAs (Fig. 1E). We found three products for both tRNAs. The two shorter ones correspond to the 5′ and 3′ halfmers of the angiogenin cleavage in the anticodon loop. Both halfmer products were stable in vitro and detected by the same northern probe we used here. We found an approximately twofold decrease in the halfmer products for 100Q tRNAs versus 0Q tRNAs for tRNAHis and an approximately ninefold decrease for tRNAAsn (Supplemental Table S1). The longest product close to the full-length tRNA corresponds to the known, in vitro angiogenin cleavage of 3′CCA of the tRNAs reported previously (Czech et al. 2013). The amount of this product was increased by ∼1.5-fold for 100Q tRNAs over those for 0Q tRNAs which was in the opposite direction as the halfmer products (Supplemental Table S1). These results indicate that Q modification can directly protect cognate tRNAHis and tRNAAsn from angiogenin cleavage in the anticodon loop in vitro.
Q modification and full-length tRNA abundance and tRNA fragments in vivo
To examine the cellular effects of Q modification on tRNA, we performed demethylase tRNA-seq (DM-tRNA-seq [Zheng et al. 2015]) to quantitatively compare the levels of all full-length tRNAs. We previously developed the DM-tRNA-seq method that removes many Watson–Crick face base methylations prior to cDNA synthesis by a processive, thermophilic group II intron reverse transcriptase to enable accurate quantitation of full-length tRNAs. We found by comparative analysis that 0Q and 100Q cells have very similar tRNA expression profiles (Fig. 2A), indicating that Q modification did not change the levels of full-length tRNAs.
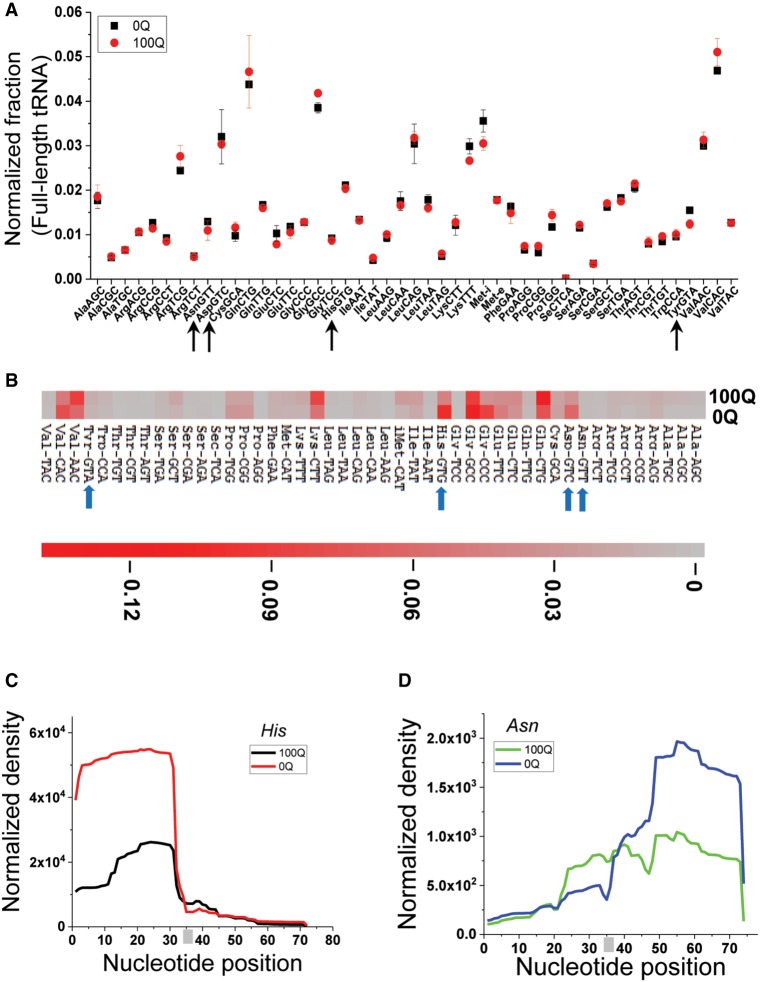
FIGURE 2.
Q modification and tRNA fragments in vivo by RNA-seq. (A) Comparison of the tRNA isoacceptor family abundance of 0Q and 100Q HEK293T cells by DM-tRNA-seq. Arrows indicate the four Q-modified cognate tRNAs. The total number of assigned tRNA reads is set to 1 in each sample, and the fraction of each tRNA isoacceptor is shown. (B) Heatmap of tRNA fragment abundance at the isoacceptor family level. The amount of all tRNA fragment products of all sizes in each sample are added together and set as 1; shown are the fractions of individual tRNA isoacceptors. Arrows indicate the four Q-modified cognate tRNAs. The scale bar shows the fraction tRNA fragments in the heatmap. (C,D) tRNAHis and tRNAAsn fragment coverage from 0Q and 100Q HEK293T cells. The anticodons (GTG for tRNAHis, and GTT for tRNAAsn) are indicated by a gray bar.
To examine the effect of Q modification on tRNA fragment profiles, we gel purified small RNAs in the size range of 20–60 nucleotides from 0Q and 100Q HEK293T cells. To reduce the interference of Watson–Crick face methylations for the quantitative comparison among the tRNA fragments, we again treated this small RNA pool with the AlkB enzymes, which can efficiently remove m1A, m1G, m3C and in part m22G (Clark et al. 2016). Removal of these modifications should be more efficient for tRNA fragments than full-length tRNA due to the loss of tRNA structure in tRNA fragments (Cozen et al. 2015). We found that HEK293T cells already contained a considerable amount of tRNA fragments in the absence of stress (Fig. 2B), on the order of ∼1% relative to total full-length tRNAs (see below). At this time we do not know which and how much of these fragments were derived from angiogenin specifically. Since angiogenin is inhibited by its protein inhibitor RNH1 in the absence of stress (Yamasaki et al. 2009), it is conceivable that many fragments were produced by other ribonucleases in cells. This may explain that in the comparative analysis of individual tRNA fragments within each sample, fragments from several tRNAs that do not contain Q-modification (GlnCTG, LysCTT, ValAAC) were present at higher levels in 100Q than in 0Q cells (Fig. 2B).
On the other hand, Q-modified cognate tRNAs consistently showed higher fragment levels in 0Q cells versus 100Q cells. Among these, tRNAHis and tRNAAsp fragments were present at higher levels than tRNAAsn and tRNATyr, and the levels for both were also substantially higher in 0Q cells (Fig. 2B; Supplemental Fig. S3A). We further analyzed the tRNA fragment pattern of the Q-modified cognate tRNAs (Fig. 2C; Supplemental Fig. S3). Unlike in vitro conditions where both 5′ and 3′ tRNA halfmer fragments are stable (Fig. 1E), only one of the two products are generally stable in cells. In 0Q cells, tRNAHis fragments were primarily derived from cleavage near the 5′ end of the anticodon loop, with the major product encompassing the 5′ end to nucleotides 30–31, consistent with those observed in other cell lines in previous studies (Honda et al. 2015, 2017). The same 5′ fragments were significantly reduced in 100Q cells which also contained slightly more fragments encompassing the middle portion of the tRNA (Fig. 2C; Supplemental Fig. S3B). tRNAAsn fragments were present at a lower level than tRNAHis, and they were enriched from cleavage near the 3′ end of the anticodon loop (Fig. 2D; Supplemental Fig. S3C).
One question remained whether the differential amount of tRNA fragments among Q-modified cognate tRNAs may also be derived from another modification in the same tRNA that is dependent on the Q-modification. One precedent for this is the tRNAAsp in S. pombe where Q34 influences the m5C38 modification in the anticodon loop; m5C38 is known to reduce tRNA fragment generation for several tRNAs in cells (Schaefer et al. 2010). Ideally one would quantitatively compare the fractions of all other modifications in the Q-modified cognate tRNAs from 0Q and 100Q cells. This approach however is beyond the scope of this work; therefore we adapted our DM-tRNA-seq approach where several Watson–Crick modifications can be quantitatively compared between two samples using mutation and stop signatures generated by the RT enzyme during cDNA synthesis (Supplemental Fig. S4; Clark et al. 2016). In tRNAHis, two modifications can be measured this way that include the m1A58 in the T loop, and a newly found m1G37 (Clark et al. 2016) in the anticodon loop. In tRNAAsn, the four measurable modifications include m1A58, m22G26 between D and anticodon stems, acp3U in the D loop, and m1G9 between acceptor and D stems. In all cases, the mutation plus stop fractions at these positions are identical or very similar in 0Q and 100Q tRNAs, suggesting that Q modification does not significantly change the patterns of other modifications in these cognate tRNAs.
Q modification and tRNA cleavage under stress and angiogenin treatment in vivo
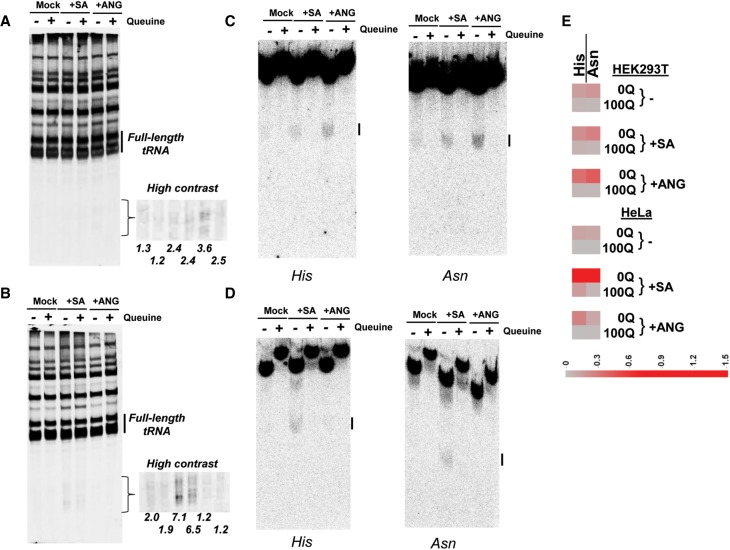
To examine the role of Q modification in generating tRNA fragments in cells under stress, we treated HEK293T and HeLa cells with sodium arsenite which was known to induce tRNA cleavage by ribonucleases (Yamasaki et al. 2009; Ivanov et al. 2011), or directly treated cells with recombinant human angiogenin protein (Fig. 3). In HEK293T cells (Fig. 3A), we found that small RNAs in the size range corresponding to tRNA halfmers increased in the arsenite stress and upon angiogenin treatment relative to the amount of total full-length tRNA. In HeLa cells devoid of Q modifications (Supplemental Fig. S5), we found that the amount of these small RNAs increased substantially under the arsenite stress, but not upon the addition of angiogenin (Fig. 3B). The cell line dependent result may be derived from differential ribonuclease activities in these cells. Northern blots of these same samples (Fig. 3C,D) showed that tRNA halfmer fragments derived specifically from the Q-modified cognate tRNAHis and tRNAAsn increased under all conditions. Importantly, the amount of these tRNA fragments was always higher in 0Q cells compared to 100Q cells (Fig. 3E).
FIGURE 3.
Q modification protects cognate tRNA cleavage under arsenite stress and angiogenin treatment in cells. (A) Total small RNA fragment analysis of total cellular RNA isolated from 0Q and 100Q HEK293T cells by SYBR gold staining under conditions of no stress (Mock), plus sodium arsenite (SA), plus recombinant human angiogenin protein (ANG). Total RNA was analyzed on 10% denaturing PAGE containing 7 M urea. Full-length tRNA bands are indicated by a bar on the right. The size region corresponding to tRNA halfmers is also shown in higher contrast on the right. Percentages of total tRNA halfmer sized fragments relative to full-length tRNA are shown below each lane. (B) Same as A for HeLa cells. (C) Northern blot analyses of the same samples in A using tRNAHis and tRNAAsn probes separated by APB-containing gels. tRNA fragments corresponding to the halfmers are indicated by a bar on the right. (D) Same as C for HeLa cells. (E) Heatmap of tRNAHis and tRNAAsn halfmer fragment products for 0Q and 100Q cells. The scale bar shows the percent product under these conditions relative to full-length tRNA on the same northern blot.
Concluding remarks
Taken together, our results indicate that queuosine tRNA modification directly protects the cognate tRNAs against ribonuclease cleavage and alters the tRNA fragment derived small RNA pools in human cells. tRNA fragments are heterogeneous in size, composition, biogenesis, and function (Fu et al. 2009; Yamasaki et al. 2009; Phizicky and Hopper 2010) and play important roles in cell physiology and human health and disease (Anderson and Ivanov 2014; Goodarzi et al. 2015; Gapp and Miska 2016). Like the known function for many fragments from tRNAs not modified with Q, Q modification dependent tRNA fragments may also play a role in cellular stress response, although this biological hypothesis was not addressed here. Previous studies showed that tRNA Q modification levels could be lower in cancer cells and tRNA Q hypo-modification could affect cell proliferation and malignancy (Pathak et al. 2005; Vinayak and Pathak 2009). Future investigation will examine what roles the tRNA fragments derived from tRNAHis and tRNAAsn play under different cellular contexts.
In summary, we show here a previously unappreciated biological aspect of tRNA Q modification in human cells which complements its known roles in translational control through codon–anticodon interaction. The level and pattern of Q-mediated tRNA fragments may serve as markers for the status of cellular stress, nutrient deprivation and diseases.
MATERIALS AND METHODS
Cell culture and generation of Q-depleted cell lines
Human embryonic kidney HEK293T cells (CRL-11268) and human cervical cancer HeLa cells (CCL-2) from ATCC were grown in Dulbecco's modified Eagle's medium supplemented with 10% dialyzed fetal bovine serum (US origin, Life Technologies), and 1% penicillin–streptomycin. Cells were passaged at the confluency of 70%–90% for ∼10 passages to generate Q-depleted cell lines (designated as 0Q cells). Total RNA was isolated using the TRIzol method, and the tRNA queuosine modification level was determined by northern blot analysis with specific tRNA probes as described below. 0Q cell lines proliferated normally under standard culture conditions, and were stocked for subsequent experiments. To fully restore tRNA Q modification, Q-depleted cells were supplemented with queuine (Toronto Research Chemicals) at 1 µM for 24 h (designated as 100Q cells).
In vitro tRNA angiogenin cleavage assay
In vitro angiogenin cleavage assay was performed in accordance with a previous study (Czech et al. 2013). Briefly, to prepare the tRNA substrates, total tRNAs from 0Q and100Q HEK293T cells were 5′ 32P-labeled and gel purified. Nonradioactive, gel purified tRNAs (1 µg/µL) from corresponding 0Q and 100Q cells were spiked with respective 32P-labeled tRNAs in 30 mM TrisHCl, pH 7.4, 30 mM NaCl, heated at 90°C for 2 min and then incubated at room temperature for 3 min. MgCl2 was added to a final concentration of 2 mM, and the sample was further incubated for 5 min at 37°C. Recombinant human angiogenin (R&D systems) was added to a final concentration of 0–2 µM to the reaction and incubated at 37°C for the indicated period of time. The reactions were stopped by adding 2× urea loading buffer (9 M urea, 100 mM EDTA), and the full-length tRNAs and tRNA fragments were separated on a denaturing 10% PAGE gel followed by exposure to phosphorimaging plates. Band intensity of fraction products after angiogenin digestion was quantified using Imager software.
tRNA fragments generation under stress conditions and angiogenin treatment in vivo
0Q HEK293T cells and HeLa cells were grown in Dulbecco's modified Eagle's medium supplemented with 10% dialyzed fetal bovine serum and 1% penicillin–streptomycin. At the confluency of ∼50%, half of the cells were supplemented with or without queuine at 1 µM for 24 h to generate100Q cells. Subsequently, 0Q and 100Q cells were treated with or without stress conditions. For angiogenin (ANG) treatment, cells were treated with recombinant human angiogenin (R&D systems) at 1 µg/mL for 2 h at 37°C. For oxidative stress, cells were treated with 500 µM sodium arsenite (SA) for 2 h at 37°C. The stress conditions were the same for 0Q and 100Q HeLa cells as for HEK293T cells. Total RNAs were extracted by the mirVana miRNA Isolation Kit and separated on 10% denaturing PAGE, and stained by SYBR gold staining to visualize tRNA fragments. Q modification levels and tRNA fragments were detected by northern blot analysis as described below.
Determination of Q modification levels in tRNAHis and tRNAAsn using 3-acrylamidophenylboronic acid (APB) gel and northern blot analysis
This method was based on the protocol developed previously (Igloi and Kossel 1985; Zaborske et al. 2014). Total RNA was deacylated by incubating in 100 mM Tris-HCl (pH 9.0) for 30 min at 37°C. Deacylated RNA samples were combined with an equal volume of denaturing 2× gel loading buffer containing 9 M urea, 100 mM EDTA. Samples were loaded onto denaturing 10% polyacrylamide gels containing 5% 3-Acrylamidophenylboronic acid (Frontier Scientific), and gel electrophoresis was run at 4°C in TAE buffer. RNA was transferred onto Hybond-XL membrane (GE Healthcare) using a vacuum gel dryer for 2 h at 80°C. The membrane was then washed twice for 30 min each in the hybridization buffer (20 mM phosphate, pH 7.2, 300 mM NaCl, 1% SDS), followed by hybridization with 5′ 32P-labeled oligonucleotide probes of four Q-containing tRNAs in the hybridization buffer for 16 h at 60°C. Membranes were washed twice for 30 min each in a solution containing 20 mM phosphate, pH 7.2, 300 mM NaCl, 2 mM EDTA, and 0.1% SDS and exposed to phosphorimaging plates. Band intensity was quantified using Imager software. Oligonucleotide probe sequences were: tRNAHis: 5′TGCCGTGACTCGGATTCGAACCGAGGTTGCTGCGGCCACAACGCAGAGTACTAACCACTATACGATCACGGC; tRNAAsn: 5′CGTCCCTGGGTGGGCTCGAACCACCAACCTTTCGGTTAACAGCCGAACGCGCTAACCGATTGCGCCACAGAGAC; tRNAAsp: 5′CTCCCCGTCGGGGAATCGAACCCCGGTCTCCCGCGTGACAGGCGGGGATACTCACCACTATACTAACGAGGA; tRNATyr: 5′TCCTTCGAGCCGGASTCGAACCAGCGACCTAAGGATCTACAGTCCTCCGCTCTACCARCTGAGCTATCGAAGG.
Identification of glycosylated Q-tRNA modifications by HPLC and mass spectrometry
Total RNA extracted from 100Q cells was used to purify tRNAAsp and tRNATyr which are known to be further modified to mannose-Q and galactose-Q, respectively. 160 μg of total RNA was hybridized with 160 pmole 5′biotin-labeled oligos of the same sequences as the northern blot probes above for tRNAAsp and tRNATyr in 2X SSC at 16°C for 16 h, followed by incubation with Dynabeads M-280 streptavidin beads (Invitrogen) at room temperature for 1 h. The RNAs were released from beads by DNase I digestion at 37°C for 30 min, followed by acid phenol extraction (pH 4.5) and ethanol precipitation. RNAs were dissolved in water and the concentration was determined by UV absorbance using Nanodrop.
Purified tRNAAsp or tRNATyr were digested in 45 µL containing 11 mM Tris-HCl, pH 7.5, 1.1 mM EDTA, pH 8.0, and 89 ng/µL RNase A and 22 ng/µL nuclease T1 at 37°C for 1 h. 160 µl water was added, the mixture spun at highest speed for 10 min and transferred to a HPLC vial. A total of 200 µl was injected, and the HPLC gradient was 0%–15% acetonitrile in 0.05 M TEAA in 30 min. All peaks except for those of mononucleotide 3′-phosphates were collected. After speedvac, the RNA fragments were subjected to digestion with 0.044 U/µL nuclease P1 (Sigma) in 25 mM NaCl, 2.5 mM ZnCl2, pH 5.5 at 37°C for 2 h to generate the corresponding 5′-phosphate. The reaction mixture was diluted to 500 µL and spun at highest speed for 10 min. The supernatant was transferred to a new vial and injected to HPLC eluting with 0%–12% acetonitrile in 0.05 M TEAA in 30 min while collecting each peak. Absorbance spectra for each peak was taken and compared to the known absorbance profile of Q-modified nucleotides (Costa et al. 2004). After speedvac, the pellet was dissolved in 10 µL water, and 2 µL were mixed with THAP matrix followed by MALDI MS analysis.
Full-length tRNA sequencing
High-throughput tRNA sequencing of 0Q and 100Q HEK293T cells was performed using the demethylase-tRNA-seq (DM-tRNA-seq) method according to the published protocol (Zheng et al. 2015; Evans et al. 2017).
tRNA fragment sequencing
tRNA fragment sequencing was adapted from a previously reported method with the added step of AlkB demethylase treatment to remove Watson–Crick face methyl modifications (Zheng et al. 2015; Evans et al. 2017). Total RNAs were extracted from 0Q and 100Q HEK293T cells with the mirVana miRNA Isolation Kit. 5′ and 3′ tRNA fragments in the size range of 20–60 nt were gel-size selected, and RNAs were eluted by incubation with crush and soak buffer (50 mM KOAc/200 mM KCl, pH 7.0) at 4°C overnight. The eluted RNA was dissolved in water. Demethylation reaction was performed under previously optimized conditions (Zheng et al. 2015; Evans et al. 2017). A total of 100 μl of reaction mixture containing 0.5 μg of isolated small RNA (∼40 pmole) was treated with 2× molar ratio of wild-type AlkB enzyme (80 pmole) and 4× molar ratio of D135S mutant (160 pmole). The reaction buffer contained 300 mM KCl, 2 mM MgCl2, 50 μM of (NH4)2Fe(SO4)2·6H2O, 300 μM 2-ketoglutarate (2-KG), 2 mM L-ascorbic acid, 50 μg/mL BSA, 50 mM MES, pH 5.0. The reaction was incubated for 2 h at room temperature and quenched by the addition of 5 mM EDTA final concentration. After phenol–chloroform extraction, 1/10 volume of 3 M NaOAc/HOAc, pH 4.8 was added and RNAs were recovered by ethanol precipitation.
Precipitated RNAs were deacylated in 20 µL 0.1 M Tris-HCl, pH 9.0 at 37°C for 30 min. Twenty microliters buffer containing 0.1 M Tris-HCl, pH 7.0, 20 mM MgCl2 and 2 U/µL T4 kinase (NEB) was added and the mixture was incubated at 37°C for another 30 min to remove the 3′ and 2′,3′ cyclic phosphates of the tRNA fragments (Honda et al. 2015, 2017). RNAs were extracted using acidic Phenol/CHCl3 (pH 4.5) with 0.3 M NaOAc/HOAc, pH 4.8, followed by ethanol precipitation. Directional ligation of adapters, cDNA generation, and PCR amplification were performed using the TruSeq Small RNA Sample Prep Kit (Illumina) according to the manufacturer's protocol. The amplified cDNAs were sequenced using Illumina HiSeq 2000 in the Genomics Core at the University of Chicago.
Sequencing data analysis
For full length tRNA sequencing, all libraries were sequenced on an Illumina HiSeq 1000 with paired-end mapping using read lengths of 100 bp. Standard quality control via FastQC was performed after sequencing and also after read processing. Reads were processed using Trimmomatic v0.32 to remove the standard Illumina adapter sequence followed by subsequent trimming using custom Python scripts to remove demultiplexing artifacts, primers, and trim the extended adapter. This second trimming step ensures that reads are not over-trimmed by Trimmomatic to ensure fidelity of the 3′ end of the raw reads. The resultant trimmed sequences were then aligned to the library using Bowtie 1.0 with sensitive options (-- k 1 –v 3 –best –strata). Sequencing reads were aligned to a modified tRNA hg19 genome file, containing nuclear-encoded tRNAs, mitochondrial-encoded tRNAs, and non-Homo sapiens tRNAs as standards.
For tRNA fragment sequencing, reads were mapped using bowtie version 1.0 on human tRNA sequences, provided by the genomic tRNA database (http://gtrnadb.ucsc.edu/genomes/eukaryota/Hsapi19/hg19-tRNAs.fa [Chan and Lowe 2016]). Only one copy of multiple identical tRNA sequences was retained for Bowtie's index files, for a total of n = 419 unique tRNA sequences. Pileup plots were derived from mapped reads using samtools version 0.1.19 (Li et al. 2009), and allowed to quantify tRNAs in the following manner: Reads that uniquely map to one tRNA were used to randomly assign reads that map to multiple tRNAs. The random assignment is such that it is proportional to the number of unique hits each tRNA has. For each sample, tRNA quantification was normalized with the help of the normalize.quantiles function of the “preprocessCore” R package (Bolstad et al. 2003).
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
DATA DEPOSITION
The full-length tRNA and tRNA fragment sequencing data have been deposited in NCBI database under GEO accession number GSE102570.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the Department of Defense/Congressionally Directed Medical Research Programs (DoD/CDMRP; BC160450 to T.P.) and the National Institutes of Health/National Institute of Diabetes and Digestive and Kidney Diseases (NIH/NIDDK; K01 DK111764 to X.Y.W.).
Footnotes
Article is online at http://www.rnajournal.org/cgi/doi/10.1261/rna.067033.118.
REFERENCES
- Anderson P, Ivanov P. 2014. tRNA fragments in human health and disease. FEBS Lett 588: 4297–4304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolstad BM, Irizarry RA, Astrand M, Speed TP. 2003. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics 19: 185–193. [DOI] [PubMed] [Google Scholar]
- Chan PP, Lowe TM. 2016. GtRNAdb 2.0: an expanded database of transfer RNA genes identified in complete and draft genomes. Nucleic Acids Res 44: D184–D189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark WC, Evans ME, Dominissini D, Zheng G, Pan T. 2016. tRNA base methylation identification and quantification via high-throughput sequencing. RNA 22: 1771–1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa A, Pais de Barros JP, Keith G, Baranowski W, Desgres J. 2004. Determination of queuosine derivatives by reverse-phase liquid chromatography for the hypomodification study of Q-bearing tRNAs from various mammal liver cells. J Chromatogr B Analyt Technol Biomed Life Sci 801: 237–247. [DOI] [PubMed] [Google Scholar]
- Cozen AE, Quartley E, Holmes AD, Hrabeta-Robinson E, Phizicky EM, Lowe TM. 2015. ARM-seq: alkB-facilitated RNA methylation sequencing reveals a complex landscape of modified tRNA fragments. Nat Methods 12: 879–884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czech A, Wende S, Morl M, Pan T, Ignatova Z. 2013. Reversible and rapid transfer-RNA deactivation as a mechanism of translational repression in stress. PLoS Genet 9: e1003767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrenhofer-Murray AE. 2017. Cross-talk between Dnmt2-dependent tRNA methylation and queuosine modification. Biomolecules 7: E14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Yacoubi B, Bailly M, de Crecy-Lagard V. 2012. Biosynthesis and function of posttranscriptional modifications of transfer RNAs. Annu Rev Genet 46: 69–95. [DOI] [PubMed] [Google Scholar]
- Emara MM, Ivanov P, Hickman T, Dawra N, Tisdale S, Kedersha N, Hu GF, Anderson P. 2010. Angiogenin-induced tRNA-derived stress-induced RNAs promote stress-induced stress granule assembly. J Biol Chem 285: 10959–10968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endres L, Dedon PC, Begley TJ. 2015. Codon-biased translation can be regulated by wobble-base tRNA modification systems during cellular stress responses. RNA Biol 12: 603–614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans ME, Clark WC, Zheng G, Pan T. 2017. Determination of tRNA aminoacylation levels by high-throughput sequencing. Nucleic Acids Res 45: e133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fergus C, Barnes D, Alqasem MA, Kelly VP. 2015. The queuine micronutrient: charting a course from microbe to man. Nutrients 7: 2897–2929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu H, Feng J, Liu Q, Sun F, Tie Y, Zhu J, Xing R, Sun Z, Zheng X. 2009. Stress induces tRNA cleavage by angiogenin in mammalian cells. FEBS Lett 583: 437–442. [DOI] [PubMed] [Google Scholar]
- Gapp K, Miska EA. 2016. tRNA fragments: novel players in intergenerational inheritance. Cell Res 26: 395–396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodarzi H, Liu X, Nguyen HC, Zhang S, Fish L, Tavazoie SF. 2015. Endogenous tRNA-derived fragments suppress breast cancer progression via YBX1 displacement. Cell 161: 790–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunduz U, Katze JR. 1984. Queuine salvage in mammalian cells. Evidence that queuine is generated from queuosine 5′-phosphate. J Biol Chem 259:1110–1113. [PubMed] [Google Scholar]
- Honda S, Loher P, Shigematsu M, Palazzo JP, Suzuki R, Imoto I, Rigoutsos I, Kirino Y. 2015. Sex hormone-dependent tRNA halves enhance cell proliferation in breast and prostate cancers. Proc Natl Acad Sci 112: E3816–3825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honda S, Kawamura T, Loher P, Morichika K, Rigoutsos I, Kirino Y. 2017. The biogenesis pathway of tRNA-derived piRNAs in Bombyx germ cells. Nucleic Acids Res 45: 9108–9120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Igloi GL, Kossel H. 1985. Affinity electrophoresis for monitoring terminal phosphorylation and the presence of queuosine in RNA. Application of polyacrylamide containing a covalently bound boronic acid. Nucleic Acids Res 13: 6881–6898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov P, Emara MM, Villen J, Gygi SP, Anderson P. 2011. Angiogenin-induced tRNA fragments inhibit translation initiation. Mol Cell 43: 613–623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov P, O'Day E, Emara MM, Wagner G, Lieberman J, Anderson P. 2014. G-quadruplex structures contribute to the neuroprotective effects of angiogenin-induced tRNA fragments. Proc Natl Acad Sci 111: 18201–18206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasai H, Nakanishi K, Macfarlane RD, Torgerson DF, Ohashi Z, McCloskey JA, Gross HJ, Nishimura S. 1976. Letter: the structure of Q* nucleoside isolated from rabbit liver transfer ribonucleic acid. J Am Chem Soc 98: 5044–5046. [DOI] [PubMed] [Google Scholar]
- Kirchner S, Ignatova Z. 2015. Emerging roles of tRNA in adaptive translation, signalling dynamics and disease. Nat Rev Genet 16: 98–112. [DOI] [PubMed] [Google Scholar]
- Kumar P, Kuscu C, Dutta A. 2016. Biogenesis and function of transfer RNA-related fragments (tRFs). Trends Biochem Sci 41: 679–689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YS, Shibata Y, Malhotra A, Dutta A. 2009. A novel class of small RNAs: tRNA-derived RNA fragments (tRFs). Genes Dev 23: 2639–2649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R; Genome Project Data Processing Sungroup. 2009. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25: 2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meier F, Suter B, Grosjean H, Keith G, Kubli E. 1985. Queuosine modification of the wobble base in tRNAHis influences ‘in vivo’ decoding properties. EMBO J 4: 823–827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller M, Hartmann M, Schuster I, Bender S, Thuring KL, Helm M, Katze JR, Nellen W, Lyko F, Ehrenhofer-Murray AE. 2015. Dynamic modulation of Dnmt2-dependent tRNA methylation by the micronutrient queuine. Nucleic Acids Res 43: 10952–10962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pathak C, Jaiswal YK, Vinayak M. 2005. Hypomodification of transfer RNA in cancer with respect to queuosine. RNA Biol 2: 143–148. [DOI] [PubMed] [Google Scholar]
- Phizicky EM, Hopper AK. 2010. tRNA biology charges to the front. Genes Dev 24: 1832–1860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raina M, Ibba M. 2014. tRNAs as regulators of biological processes. Front Genet 5: 171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rakovich T, Boland C, Bernstein I, Chikwana VM, Iwata-Reuyl D, Kelly VP. 2011. Queuosine deficiency in eukaryotes compromises tyrosine production through increased tetrahydrobiopterin oxidation. J Biol Chem 286: 19354–19363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaefer M, Pollex T, Hanna K, Tuorto F, Meusburger M, Helm M, Lyko F. 2010. RNA methylation by Dnmt2 protects transfer RNAs against stress-induced cleavage. Genes Dev 24: 1590–1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres AG, Batlle E, Ribas de Pouplana, L. 2014. Role of tRNA modifications in human diseases. Trends Mol Med 20: 306–314. [DOI] [PubMed] [Google Scholar]
- Vinayak M, Pathak C. 2009. Queuosine modification of tRNA: its divergent role in cellular machinery. Biosci Rep 30: 135–148. [DOI] [PubMed] [Google Scholar]
- Yamasaki S, Ivanov P, Hu GF, Anderson P. 2009. Angiogenin cleaves tRNA and promotes stress-induced translational repression. J Cell Biol 185: 35–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasukawa T, Suzuki T, Ishii N, Ohta S, Watanabe K. 2001. Wobble modification defect in tRNA disturbs codon-anticodon interaction in a mitochondrial disease. EMBO J 20: 4794–4802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaborske JM, DuMont VL, Wallace EW, Pan T, Aquadro CF, Drummond DA. 2014. A nutrient-driven tRNA modification alters translational fidelity and genome-wide protein coding across an animal genus. PLoS Biol 12: e1002015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zallot R, Brochier-Armanet C, Gaston KW, Forouhar F, Limbach PA, Hunt JF, de Crecy-Lagard V. 2014. Plant, animal, and fungal micronutrient queuosine is salvaged by members of the DUF2419 protein family. ACS Chem Biol 9: 1812–1825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng G, Qin Y, Clark WC, Dai Q, Yi C, He C, Lambowitz AM, Pan T. 2015. Efficient and quantitative high-throughput tRNA sequencing. Nat Methods 12: 835–837. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.