Abstract
The tri-snRNP 27K protein is a component of the human U4/U6-U5 tri-snRNP and contains an N-terminal phosphorylated RS domain. In a forward genetic screen in C. elegans, we previously identified a dominant mutation, M141T, in the highly-conserved C-terminal region of this protein. The mutant allele promotes changes in cryptic 5′ splice site choice. To better understand the function of this poorly characterized splicing factor, we performed high-throughput mRNA sequencing analysis on worms containing this dominant mutation. Comparison of alternative splice site usage between the mutant and wild-type strains led to the identification of 26 native genes whose splicing changes in the presence of the snrp-27 mutation. The changes in splicing are specific to alternative 5′ splice sites. Analysis of new alleles suggests that snrp-27 is an essential gene for worm viability. We performed a novel directed-mutation experiment in which we used the CRISPR-cas9 system to randomly generate mutations specifically at M141 of SNRP-27. We identified eight amino acid substitutions at this position that are viable, and three that are homozygous lethal. All viable substitutions at M141 led to varying degrees of changes in alternative 5′ splicing of native targets. We hypothesize a role for this SR-related factor in maintaining the position of the 5′ splice site as U1snRNA trades interactions at the 5′ end of the intron with U6snRNA and PRP8 as the catalytic site is assembled.
Keywords: RS domain, cryptic splice sites, pre-mRNA splicing, spliceosome
INTRODUCTION
The two trans-esterification reaction steps of precursor messenger RNA (pre-mRNA) splicing are performed by the large ribonucleoprotein machine called the spliceosome. The spliceosome assembles onto the pre-mRNA at each intron through an ordered binding of its UsnRNP subunits. Early in this assembly, U1 snRNP recognizes the 5′ splice site through base-pairing interactions with the 5′ end of U1 snRNA (E complex). U2 snRNP is next recruited and identifies the branch point sequence via RNA–RNA interactions with U2snRNA (A complex). Next, the U4/U6-U5 tri-snRNP is recruited to the spliceosome (B complex) and a dramatic series of RNA-RNA and RNA–protein rearrangements, including the removal of U1 and U4 snRNAs, leads to formation of the catalytic complex (C complex), which carries out the first step of the splicing reaction (for a review, see Fica and Nagai 2017). In this first step of splicing, the 5′ splice site is catalytically committed by cleavage at the exon/intron junction and simultaneous formation of the intron lariat. As U1 leaves during B complex formation, U6 forms base-pairing interactions near the 5′ end of the intron prior to catalysis, and these interactions appear to be stabilized by PRP8 (Galej et al. 2016). The rearrangements in the spliceosome at the different steps are catalyzed and proofread by a series of ATPase helicases (Koodathingal and Staley 2013). In the past three years, there has been an explosion of new information about the structures of the different spliceosomal complexes brought about by advances in cryo-electron microscopy (for a review, see Fica and Nagai 2017; Shi 2017). This has led to major advances in our understanding of the complex RNA–protein, protein–protein, and RNA–RNA interactions of the spliceosome. It is a very exciting time in the field, yet there remain challenges to understanding the splicing mechanism, including understanding the dynamic rearrangements of the spliceosome as it transitions from one complex to the next, which may be best served by continued genetic and biochemical studies (Mayerle and Guthrie 2017).
The formation of spliceosomal complexes and their rearrangements are guided by many proteins that interact with the pre-mRNA and/or associate with the spliceosomal subunits. These include the SR family of proteins, characterized by the presence of N-terminal RNA recognition motif(s), and a C-terminal RS domain, containing phosphoserine-arginine dipeptide repeats (Shepard and Hertel 2009). SR proteins assemble onto the pre-mRNA and guide spliceosomal components, including the U1 and U2 snRNPs, to it (Howard and Sanford 2015). Phosphorylated RS domains are additionally present in protein components of the snRNPs: U1 snRNP-specific 70 kDa protein (SNRNP70); U5 snRNP 100 kDa protein (DDX23/PRP28) (Teigelkamp et al. 1997); and three tri-snRNP-specific proteins: 27K (SNRNP27) (Fetzer et al. 1997), 65K (USP39), and 110K (SART1) (Makarova et al. 2001). Unlike the canonical SR proteins with their C-terminal RS domains, these spliceosome-associated SR-related proteins have either an N-terminal RS domain (DDX23, SNRNP27, USP39, and SART1) or an internal RS domain (SNRNP70). SR proteins can interact with each other through their RS domains. This was evidenced initially by the ability to precipitate all the family members as a group by the addition of Mg2+ ions to cellular extracts (Zahler et al. 1992). SR proteins can regulate alternative splicing through differential recruitment of U1snRNP to different splice sites (Zahler and Roth 1995). U1 snRNP recruitment to the 5′ splice site at E complex formation is mediated by phosphorylation-dependent interactions between the RS and RRM domains of exon-bound SR proteins and the U1-70K RS domain (Xiao and Manley 1998; Cho et al. 2011). RS domains are highly disordered in their unphosphorylated state and are proposed to assume a rigid arch-like structure when phosphorylated (Xiang et al. 2013). The RS domains are strikingly absent from proteins in Saccharomyces cerevisiae. There are no homologs of classical SR proteins in S. cerevisiae, and an SNRNP27 homolog is also absent. For RS domain-containing proteins for which clear homologs are present in S. cerevisiae (i.e., SNRNP70 and PRP28), the RS domain is absent from the S. cerevisiae homolog.
We previously identified the C. elegans homolog of SNRNP27, snrp-27, in a genetic screen for phenotypic suppressors of uncoordination for the unc-73(e936) allele, which contains a G→U substitution at the first nucleotide position of intron 15 (Dassah et al. 2009). Cryptic 5′ splice sites are defined as sites that are never used by the splicing machinery unless an authentic nearby 5′ splice site is altered by mutation. When that occurs, nearby cryptic sites, usually defined by GU dinucleotides, become substrates for splicing but with a 2/3 chance of being out-of-frame. A dominant mutation in SNRP-27, M141T, in the highly conserved C-terminal region (a region with no detectable domain homology with other proteins) led to alteration of cryptic 5′ splice site usage in the unc-73(e936) allele, increasing the amount of in-frame splicing of unc-73(e936) with no apparent effects on worm viability. There is a very limited literature about this protein and its homologs. Human SNRNP27 was first identified as a gene of unknown function disrupted by HTLV-1 provirus insertion in a human T-cell line (then named RY-1) (Nakamura et al. 1994). The association of the human homolog with splicing was demonstrated when it was shown to be a component of the U4/U6-U5 tri-snRNP that is phosphorylated by a spliceosome-associated kinase (Fetzer et al. 1997). Even though it is detected in human tri-snRNPs prepared for cryo-EM structural analysis, its density has not been mapped in the proposed structure, likely due to the low complexity and potential unstructured nature of the protein (Agafonov et al. 2016). The Drosophila homolog, Yantar, was identified in a genetic deletion screen of lethal recessive mutants for factors that regulate hemocyte development; loss of Yantar is pupal lethal but leads to overproduction of hemocytes in larvae (Sinenko et al. 2004). In a reverse genetic screen for factors that affect splicing efficiency in Schizosaccharaomyces pombe, deletion of the fission yeast homolog SPCC162.01c led to both the accumulation of unspliced forms of some messages and increased splicing efficiency for others (Larson et al. 2016). SNRNP27′s high enrichment in arginine and lysine residues leaves a very limited number of peptides that can be observed in mass spectrometry proteomic experiments after tryptic digest. This likely leads to underreporting of the presence of this protein because of low confidence due to the lack of multiple distinct peptide fragments that can be assigned to it.
In this paper, we further characterize the role of the dominant SNRP-27 mutation in splicing. We perform high-throughput mRNA sequencing analysis from the mutant strains and show that it activates alternative 5′ splice sites in 26 native genes; no other types of alternative splicing changes are detected. We confirmed these changes experimentally for 13 targets and characterize a likely role for this factor in 5′ splice site choice. Worms containing a CRISPRcas9-induced apparent null allele of this gene are inviable, with heterozygotes showing no apparent defects. We used a novel approach to randomize amino acid M141 and recovered viable worms with eight different substitutions at this position. We also identified three other substitutions that were homozygous lethal. All of the viable mutants led to changes in alternative 5′ splice site choice for the native targets tested, indicating that the highly conserved M141 is optimized for protein function. We propose that SNRP-27 functions during the handoff of the 5′ splice site from U1snRNA to U6snRNA and PRP8 to promote splicing fidelity.
RESULTS AND DISCUSSION
High-throughput mRNA-seq analysis of SNRP-27 M141T mutant strains
We have previously described the genetic screen that resulted in the isolation of snrp-27(az26) (Dassah et al. 2009). In that approach, we identified a suppressor allele of snrp-27 that altered the cryptic splicing of unc-73(e936), resulting in a twofold increase in in-frame messages leading to improved coordination and movement. In order to determine whether the snrp-27(az26) allele also promotes changes in splicing of native genes, we compared high-throughput mRNA sequencing results of strains CB936 (unc-73(e936)I) versus SZ118 (unc-73(e936) snrp-27(az26)I). mRNA was isolated from young adult animals, cDNA prepared and high-throughput sequencing performed to obtain strand-specific 100 nucleotide (nt) paired-end reads. Two independent libraries from two independent RNA isolations were prepared from each strain and the resulting sequences were mapped to the C. elegans genome using STAR (Dobin et al. 2013).
We next set out to identify native genes whose alternative splicing is changed in the presence of the snrp-27(az26) allele. We identified alternative splicing events and used MISO (Katz et al. 2010) to do four pairwise comparisons of the events in these libraries against each other (CB936 library 1 versus SZ118 library 1; CB936 library 1 versus SZ118 library 2; CB936 library 2 versus SZ118 library 1; CB936 library 2 versus SZ118 library 2) for all the de novo identified alternative splicing events in three categories (alternative 5′ splice site, alternative 3′ splice site, alternative cassette exon). Those events with a change in percent spliced in (delta PSI) value of ≥0.20 in all four library comparisons were further examined. There were 22 alternative 3′ splice site events that met these criteria, but upon examination of individual reads supporting these events using Sashimi and display of library BAM tracks in the UCSC Genome Browser, only one was a potential library-specific alternative 3′ splicing event. However, when tested directly by 32P RT-PCR analysis (data not shown), it showed no difference in alternative splicing between the strains. There were many alternative splicing events falsely flagged by our pipeline leading to a requirement that the flagged events be examined individually. False flagging was due to (i) pairwise comparison by MISO of two very minor isoforms when a third majority isoform was found sharing the same splice site, (ii) low coverage of reads for both isoforms of an alternatively spliced region such that a small number of changes led to large changes in percentages, (iii) complex splicing in a region that led to a false conclusion for a specific splicing event, or (iv) reads with alignment to multiple adjacent regions. Three alternative cassette exon events were flagged in this analysis as well, but upon examination none appeared to be true alternative splicing events dependent on snrp-27 mutation. Therefore, we could find no strong evidence for alternative 3′ splice site or cassette exons whose usage is misregulated in the presence of the SNRP-27 M141T mutation. In contrast, 61 alternative 5′ splice site events showed changes in all four comparisons. We determined that 26 of the 61 alternative 5′ splice site events have strong supportive evidence of alternative splicing differences between these strains. These are summarized in Figure 1. As snrp-27(az26) was identified as an allele that could alter cryptic 5′ splice site usage, this connection to alternative 5′ splice site usage of native C. elegans genes warrants further analysis to understand the nature of the SNRP-27 function.
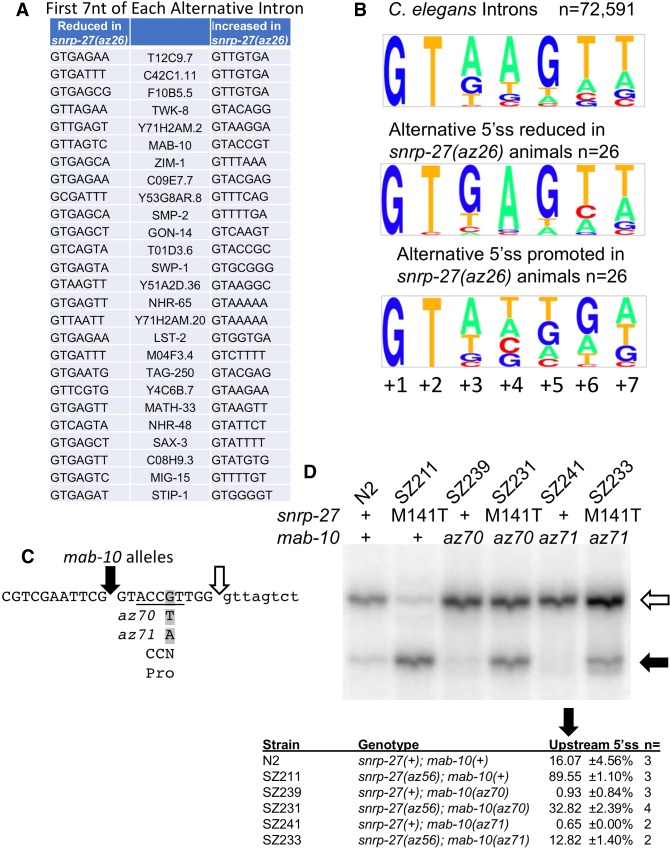
FIGURE 1.
Alternative 5′ splice sites of native genes whose usage is changed as suggested by mRNA-seq analysis. For each of the 26 genes, the DNA sequence of the alternative 5′ splice site region is indicated. Lower case indicates intron sequences, while upper case indicates sequences that can be exons. The splicing regions are ordered by the increasing length of the distance between the two 5′ splice sites in two sets. The top set (whose names are in bold) was confirmed by 32P RT-PCR (Fig. 2). Also indicated is whether the alternative splicing has been previously annotated by Wormbase and the chromosomal location of the corresponding region in C. elegans genome release WS190/ce6. The average delta PSI of the four pairwise library comparisons and standard deviation are indicated. Black triangles indicate the 5′ splice site promoted by the snrp-27(az26) mutation. White triangles indicate the alternative 5′ splice site whose usage decreases in the snrp-27(az26) strains. Inverted triangles indicate 3′ splice site location for introns in which the 3′ splice site fits on the line. Lines ending in “…” indicate that the intron is longer than the sequence shown. Note that for ZIM-1 there are two alternative 3′ splice sites separated by 9 nt and both are indicated.
We subjected 13 of the alternative 5′ splicing events affected by the snrp-27(az26) mutation to experimental confirmation by doing reverse-transcription followed by PCR with 32P 5′ end-labeled primers. With these alternative 5′ splice sites in close proximity to each other (several are separated by only 3 nt), the resolution provided by separation of 32P-labeled PCR products on denaturing polyacrylamide gels is required to visualize the individual products. Six of the 13 alternative splicing events have 5′ splice sites that are not separated by a multiple of 3 nt. This would imply that their use would result in alternative messages with different reading frames downstream from the alternative splicing event and may result in the formation of a premature termination codon (PTC). Alternative splicing was determined for five of these in a nonsense-mediated decay mutant background. Figure 2 shows representative 32P RT-PCR analyses of these 13 alternative splicing events. For simplicity, 32P RT-PCR results for only CB936 and SZ118 (strains with wild-type snrp-27 and M141T mutant snrp-27) are shown for all examples. For five of the alternative 5′ splice sites not separated by a multiple of 3 nt, alternative splicing for the substrate in a nonsense-mediated decay-defective strain, CB4354 with a mutation in smg-3, is included.
FIGURE 2.
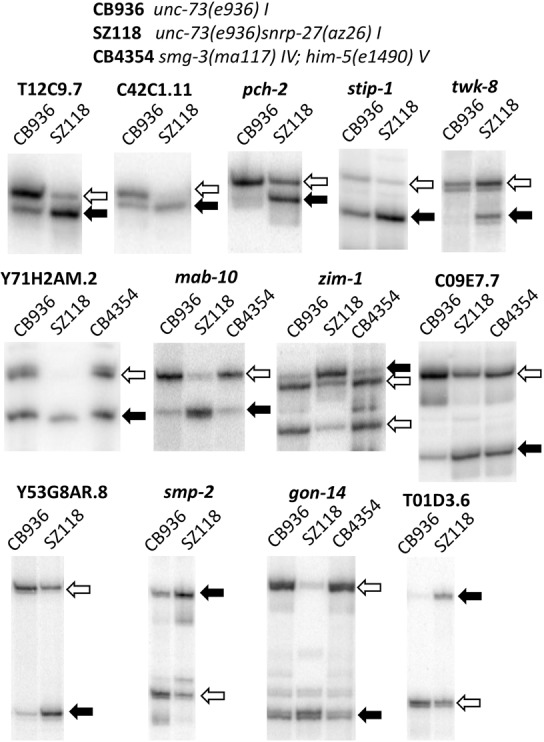
Confirmation of predicted az26-induced alternative splicing by 32P RT-PCR. Sample 32P RT-PCR reactions for each alternatively spliced region noted in bold in Figure 1. Genotypes of strains that were the source of RNA are indicated at the top of the figure. Alternative splicing events not separated by a multiple of 3 nt also show splicing for an NMD mutant strain. Black arrows indicate the 5′ splice site whose usage is promoted in the snrp-27(az26) strains, and the white arrows indicate its alternative splicing partner whose usage is reduced in the snrp-27(az26) strains.
The alternative splicing changes identified by high-throughput sequencing analysis are strongly supported by the individual RT-PCR reactions. The snrp-27(az26) allele promotes usage of the upstream 5′ splice site for the three alternative 5′ splice sites separated by 3 nt. These are interesting because the recognition sites for U1 snRNA and U6 snRNA are overlapping for these substrates, and the beginning intron sequence for the upstream snrp-27(az26)-promoted 5′ splice site, GTTGTGA, is identical for all three substrates. The other alternative splicing events have 5′ splice site sequences separated by 5 to 36 nt, and in these cases, either the upstream 5′ splice site or the downstream 5′ splice site can become preferentially used in the snrp-27(az26) mutant background. The 13 events tested had an average MISO delta PSI from the sequencing analysis of between 0.300 and 0.695, with stip-1 the weakest one tested. stip-1 had a confirmed change in alternative splicing in the snrp-27(az26) mutant, going from 75% usage of the upstream 5′ splice site in snrp-27(+) strains to 95% usage of that site in the snrp-27(az26) strains (an experimental delta PSI of 0.20, quantitation data not shown).
PTC-containing messages can be substrates for degradation by the nonsense-mediated decay (NMD) pathway (Maquat 2002). C. elegans is unique among model organism metazoans in that it is viable in the absence of NMD function (Pulak and Anderson 1993). In C. elegans, the smg genes (suppressors with morphogenetic effects on genitalia) are required for nonsense-mediated decay (Hodgkin et al. 1989). In order to confirm that we are seeing changes in 5′ splice site choice as opposed to changes in message stability in the snrp-27(az26) background, we isolated total RNA from smg-1(r869), smg-2(e2008) and smg-3(ma117) NMD-defective worms and tested 12 events for alternative 5′ splicing changes. In four of the five alternative splicing events tested that are not separated by a multiple of 3 nt, the alternative 5′ splice site usage was identical between wild-type and NMD mutant worms (Y71H2AM.2, mab-10, zim-1, and gon-14), indicating that neither alternative transcript is a substrate for the NMD pathways (only the smg-3 mutant is shown in Fig. 2 for simplicity). However, for the intron in C09E7.7, the NMD mutant backgrounds show higher usage of the upstream splice site relative to the CB936 strain. This indicates that usage of the upstream 5′ splice site in this case triggers NMD decay. It is notable that in the snrp-27(az26) mutant background (center lane–SZ118) there is also an increase in the steady-state level of transcripts that use the upstream 5′ splice site. Since those transcripts decay more rapidly via NMD than those that use the downstream 5′ splice site, it indicates snrp-27(az26) promotes an even stronger usage of the upstream 5′ splice site than is evidenced by just the relative steady state levels of the transcripts in that lane. Therefore, the changes observed in relative abundance of the different isoforms between CB936 and SZ118 are at the level of alternative splice site choice, not NMD-regulated message stability.
One of the alternative splicing events, in an intron in the zim-1 gene, is interesting in that it involves alternative 5′ splice sites separated by 11 nt and alternative 3′ splice sites separated by 9 nt. The upstream 5′ splice site is strongly preferred in the snrp-27(+) background and shows splicing to both alternative 3′ splice sites at relatively equal levels. Both 3′ splice sites have relatively strong consensus sequences, so from previous work in our laboratory we might expect their alternative usage to not be regulated in a tissue-specific manner (Ragle et al. 2015). In contrast, the downstream 5′ splice site is strongly preferred in the snrp-27(az26) background, and it splices exclusively to the downstream 3′ splice site. This specificity in alternative 3′ ss usage correlated with the downstream 5′ splice site is likely due to the fact that the distance between the downstream 5′ splice site and the upstream 3′ splice site, 33 nt, is below the minimal threshold for intron size in this organism (Lim and Burge 2001), so that the downstream 5′ splice site can only be paired with the downstream 3′ splice site.
Analysis of the sequence context of SNRP-27 M141T-promoted alternative splicing events
We performed an analysis of the sequence context of the 5′ splice site intronic regions of the alternative isoforms. We focused on the first 7 nt of the 5′ end of the introns defined by the alternative 5′ splice sites as these sequences have overlapping interactions with U1snRNA and U6snRNA. SNRNP27 was isolated by its biochemical association with purified tri-snRNP complexes (Fetzer et al. 1997). As the tri-snRNP enters the spliceosome to form the B and then the Bact complexes, the interaction of U1snRNA with the 5′ splice site is exchanged for U6snRNA interactions (Ares and Weiser 1995). Figure 3A,B shows the sequences of the splice sites and a Pictogram display (http://genes.mit.edu/pictogram.html) of the first 7 nt of these introns. A consensus Pictogram from 72,591 randomly chosen 5′ splice sites is shown at the top of Figure 3B and it matches the published Pictogram consensus for the C. elegans 5′ splice site (Lim and Burge 2001). Next is the Pictogram for the 26 alternative 5′ splice sites whose usage is reduced in the snrp-27(az26) background. They are a fairly good match to the consensus 5′ splice site sequence, with the exception that the +3 position of these 26 introns tend to exclude A residues (only 1/26 has an A at the +3 position), while the consensus 5′ splice site finds an A residue at 58% of total C. elegans introns (P-value from a hypergeometric test for nucleotide frequency of A at position +3 in the 5′ splice sites whose usage is reduced in the snrp-27 mutants relative to all 111,187 annotated 5′ splice sites is 7.4 × 10−9). In the case of the 26 alternative 5′ splice sites promoted in the snrp-27(az26) background, the nucleotides at positions +4, +5, and +6 of these introns differ markedly from the C. elegans 5′ splice site consensus sequence and that of their alternative splicing counterparts (lower part of Fig. 3B). Intronic positions +4 and +5 of the snrp-27(az26)-promoted splice sites show no base preference [P-value for frequency of A at position +4 of the snrp-27(az26)-promoted splice sites relative to all introns is 5.0 × 10−5; P-value for frequency of G at position +5 of the snrp-27(az26)-promoted splice sites relative to all introns is 2.7 × 10−5]. In contrast, the alternative splice sites whose usage is reduced in the snrp-27(az26) background have a strong preference for A at +4 and G at +5 that matches the consensus 5′ splice site for all introns. Another difference is at the +6 position where G is rare in both the total intron pool (only 8.03% of total introns have a G at this position) and the alternative 5′ splice sites whose usage is reduced in the snrp-27(az26) background (1/26 have a G at the +6). However, G is enriched at the +6 position (14/26 introns) in the snrp-27(az26)-promoted alternative 5′ splice sites [P-value for enrichment of G at +6 of the snrp-27(az26)-promoted splice sites relative to total introns is 6.0 × 10−10]. The consensus sequence for the first 6 nt of the intron, GUAAGU, has direct base-pairing interactions with U1 snRNA. Positions +4, +5, and +6 of the intron have been shown in genetics experiments and cryoEM structures to interact with U6 snRNA in the active spliceosome, indicating that these sequences have two important interactions in spliceosome assembly (Fica and Nagai 2017; Shi 2017). We looked at the nucleotide composition at positions +4, +5, and +6 for 110,339 annotated C. elegans introns beginning with GU. Of these 5′ splice sites, only 30,054 (27.2%) have less than two of three nucleotides at positions +4, +5, +6 match the AGU consensus. For the snrp-27(az26)-promoted 5′ splice sites, 21/26 (80.8%) have less than two of the three nucleotides in this region match the consensus. In contrast, only 1/26 of their alternative 5′ splice site partners have less than two of the three nucleotides match the consensus. The differences in nucleotide composition of the snrp-27 M141T-promoted alternative 5′ splice sites, especially at positions +4, +5, and +6 of the intron, suggest a model in which the mutation in snrp-27 relaxes the specificity by which U6 assembles with the pre-mRNA into an active site for splicing.
FIGURE 3.
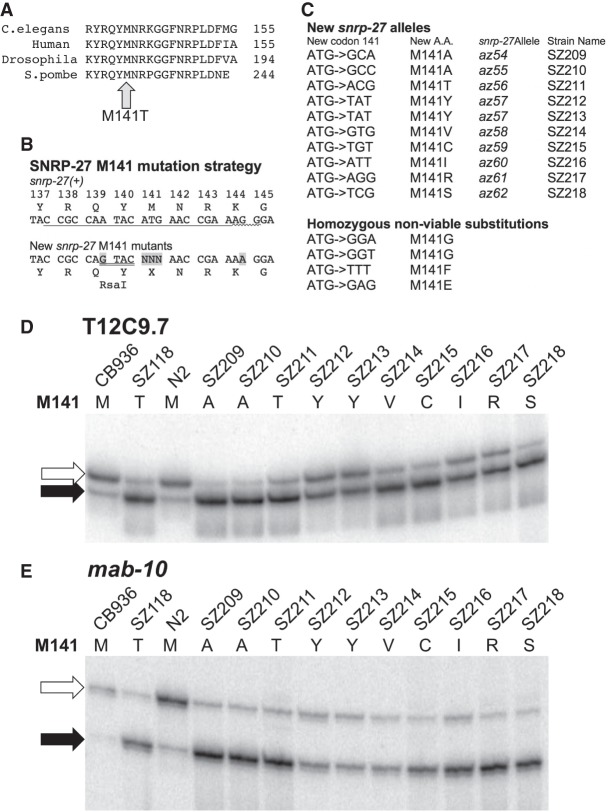
Comparison of SNRP-27 M141T-sensitive alternative 5′ splice site sequences. (A) Sequence of the first 7 nt of the intron for the alternative 5′ splice sites used in each indicated gene. The 5′ splice site that is promoted in the snrp-27(az26) animals is indicated in the right column, and its partner alternative 5′ splice site whose usage is reduced in snrp-27(az26) is indicated in the left column. (B) Pictograms (http://genes.mit.edu/pictogram.html) of the consensus sequence for the first 7 nt of the splice sites for total C. elegans introns (72,591 randomly chosen introns) (top), the alternative 5′ splice site whose usage is reduced in the snrp-27(az26) strain (middle), and the alternative 5′ splice site whose usage is promoted in the snrp-27(az26) strain (bottom). (C) Two new alleles of mab-10, az70, and az71, generated by CRISPRcas9 alteration. Both new point mutation alleles alter the intronic +6G of the upstream 5′ splice site (highlighted in gray). Black arrow indicates the upstream 5′ splice site, and the open arrow indicates the downstream 5′ splice site. The substitutions are translationally silent as they alter the wobble position of a proline codon. (D) 32P RT-PCR products for the alternatively spliced region of mab-10 separated on a 6% polyacrylamide denaturing gel. The black arrow corresponds to the product of the upstream 5′ splice site and the open arrow to the products of the downstream alternative 5′ splice site. The indicated genotypes of snrp-27 and mab-10 corresponding to each lane are shown above. The az56 allele of snrp-27 (M141T) is described in Figure 4C and encodes the same protein as the az26 allele, and the mutant mab-10 alleles (az70 and az71) are described in Figure 3C. Below the gel is a table indicating the relative fraction of upstream 5′ splice site usage of mab-10 determined over several independent experiments similar to the one in this figure. Quantitation was done using ImageQuant software (GE Life Sciences) from a PhosphorImager image of the gel.
We tested whether the unusual abundance of G at the +6 position of the snrp-27(az26)-promoted 5′ splice sites was an important feature for this recognition. To do this, we generated two new alleles of the mab-10 gene using the CRISPRcas9 system. These new alleles result in a point mutation to the sixth position of the upstream snrp-27(az26)-promoted intron; +6G→T in the az70 allele and +6G→A in the az71 allele (Fig. 3C). The +6 position of this 5′ splice site in mab-10 was chosen for analysis because it fit two criteria; mutation of this position resulted in no change in protein coding sequence as it is the wobble position of a proline codon, and this point mutation disrupted a HpyCH4III restriction site, leading to ease of screening and recovery of single-point mutants by PCR across this region followed by HpyCH4III digestion. In the snrp-27(+) background, these two new alleles resulted in less relative usage of the mab-10 upstream 5′ splice site, from ∼16% in the wild-type N2 strain to ∼1% in the two mab-10 mutant alleles (Fig. 3D). When combined with snrp-27 M141T mutant allele az56, there was an increase in upstream 5′ splice site usage, ∼33% for az70 and ∼13% for az71. For both alleles, the snrp-27 mutant was able to increase the usage of the upstream 5′ splice site even without a G at the +6 position of the intron. However, the relative increase in upstream 5′ splice site usage in the presence of the snrp-27 mutation was different for the two mab-10 mutant alleles relative to each other as well as relative to mab-10(+). The upstream mab-10 alternative 5′ splice site also has noncanonical C nucleotides at positions +4 and +5. These data indicate that the snrp-27 mutant increases the relative usage of this unusual upstream 5′ splice site, and that the presence of an intronic +6G is not essential for this to occur but may contribute to this recognition.
Generation of new SNRP-27 M141 mutants and analysis of their role in splicing
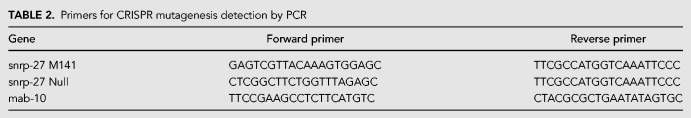
The SNRP-27 protein sequence is highly conserved in evolution. Figure 4A shows the alignment of the terminal 20aa of this protein from, C. elegans, human, Drosophila melanogaster, and Schizosaccharomyces pombe. There is striking conservation of the amino acid sequence of the protein in this 20-residue stretch at the C terminus. There is no homolog of this protein in Saccharomyces cerevisiae, and even a BLAST search (Altschul et al. 1990) with just this 20 amino acid conserved region yielded no potential homologs. The az26 M141T mutation is located within this region. In order to better understand the role of M141 in SNRP-27 function, we performed a novel genetic experiment in which we used the CRISPR-cas9 system coupled with oligonucleotide-directed recombination repair to specifically randomize the amino acid at position 141. Figure 4B outlines this repair strategy. We recovered 10 independent strains with nine different sequence alleles leading to eight different amino acid possibilities at position 141, including the original M141T substitution (Fig. 4C). These strains all grow vigorously like wild-type, with the exception of the two M141A strains, which grow slightly slower and have fewer progeny.
FIGURE 4.
Testing additional SNRP-27 M141 mutations in alternative 5′ splice site choice. (A) Alignment of the C-terminal region of tri-snRNP 27K proteins. The highly conserved terminal 20aa are aligned. Number at the right of each line indicates the position of the C-terminal amino acid in each homolog. The position of the C. elegans M141T substitution in snrp-27(az26) is also indicated. (B) CRISPR-cas9 strategy for generating amino acid substitutions at M141. The top part shows the native SNRP-27 amino acid sequence for this region. The underlined part is the crRNA used in the experiment, and the squiggly underline is the PAM sequence. The up arrow indicates the site of CRISPR-cas9 cleavage guided by the crRNA. The lower part of the figure shows the sequence of the central region of the repair guide oligonucleotide. Nucleotides highlighted in gray are changes from wild-type. The highlighted NNN are randomized during repair oligonucleotide synthesis and will create a range of potential amino acid substitutions at M141. (C) Sequences of the new snrp-27 alleles with amino acid substitutions at M141. The first column shows the new codon generated by the strategy outlined in part B of this figure. The second column shows the amino acid substitution at M141. The third column is the new allele name for this sequence, and the fourth column is the new strain name. In addition to the new codon 141 sequence, all new alleles had the two silent mutations highlighted in part B of this figure. The lower part of this section indicates the nucleotide and amino acid sequences of new alleles of M141 that were homozygous lethal. (D) Alternative 5′ splice site usage for T12C9.7 in the presence of different alleles of snrp-27. Sequence of the alternatively spliced region of T12C9.7 is found in Figure 1, and sequences of the different snrp-27 alleles are found in Figure 4C. The amino acid substitution at position 141 in each lane is also indicated. Black arrow indicates the alternative 5′ splice site promoted in the snrp-27(az26) background, and white arrow indicates its partner alternative 5′ splice site. (E) Alternative 5′ splice site usage for mab-10 in the presence of different alleles of snrp-27. Description is the same as in part D of this figure.
We next set out to test whether these new snrp-27 M141 mutations promote changes in splicing. This is simplified by the fact that we have identified native genes whose alternative splicing changes in the presence of the snrp-27(az26) mutation (Fig. 1). Figure 4D and E show the results of alternative 5′ splicing for T12C9.7 and mab-10 in the different strains listed in Figure 4C. For both alternative splicing events, the downstream 5′ splice site is strongly preferred in the wild-type snrp-27(+) backgrounds (CB936 (unc-73(e936) I) and N2) while any substitution to M141 leads to increased usage of the upstream 5′ splice site. This ranges from ∼50% in the M141Y substitution strains to almost 100% in the M141A substitution strains, which also showed a mild growth defect. These results suggest that methionine at position 141 is optimized for SNRP-27 function in 5′ splice site choice, and that substitutions to this highly conserved position weaken this specificity. While this experiment did not saturate for every possible amino acid substitution at M141, we were able to generate and test a broad range of new alleles with different classes of amino acid side chains.
In addition to the viable substitutions at M141, we identified four different lines that were heterozygous for changes at M141, but for which the mutant allele could not be made homozygous (Fig. 4C). These alleles appeared completely normal as heterozygotes, indicating that these were recessive mutations, but were homozygous lethal. We sequenced PCR products across the mutant genomic region from heterozygous individuals, and by reading the electropherograms, we were able to deduce both the wild-type sequence as well as the sequence of the amino acid substitution in the mutant (Fig. 4C). M141G, M141F, and M141E were all homozygous lethal substitutions in this protein. This indicates that while several amino acid substitutions may lead to a functional protein with weakened ability to promote 5′ splice site specificity, other amino acid substitutions lead to a complete interference in protein function.
The homozygous lethal snrp-27 alleles raised the question as to whether snrp-27 is an essential gene. This two-exon gene is the sixth gene of an eight-gene operon, so we were concerned that creating a large deletion of snrp-27 to generate a null allele might affect the function of surrounding genes. Instead we generated an allele by CRISPR-cas9 in which we inserted a stop codon at amino acid 64 and asked whether these worms could be viable. This mutation truncated the protein partway through the RS domain and lacked the highly conserved C-terminal region. This allele was viable as a heterozygote and behaved normally, however this allele could not be made homozygous. Occasionally we would recover a sick late larval stage animal that was homozygous for the putative null mutant but could not maintain it because it was sterile. Given that the heterozygous worms with this mutation do not show any phenotypes, we do not believe the lethality of this allele to be the result of an RS domain poison product; we think of it as a putative null allele. The inability to make this putative null allele homozygous, along with the results that the M141G, M141F and M141E alleles appear normal as heterozygotes but are homozygous lethal, point to snrp-27 as an essential gene. This is consistent with the pupal lethal phenotype of the Drosophila Yantar gene deletion (Sinenko et al. 2004).
Conclusions
The tri-snRNP 27K protein is a poorly understood splicing factor. It was initially identified as a component of purified tri-snRNPs that could undergo phosphorylation in its RS domain (Fetzer et al. 1997). Our subsequent genetic study identified a dominant mutation, M141T, in the C. elegans homolog snrp-27 that promotes changes in cryptic 5′ splice site usage (Dassah et al. 2009). In this paper we present data that the M141T mutation promotes changes in alternative 5′ splice site usage for a number of native genes. Many amino acid substitutions at M141 in the highly conserved C terminus are viable and also promote the alternative 5′ splice site usage that we identified in the M141T allele. However, a subset of M141 substitutions lead to homozygous lethality (Fig. 4C), indicating that there is an essential role for this highly conserved region in protein function.
The ability of the SNRP-27 M141 alleles to promote alternative 5′ splice site usage on native genes may be due to a rare combination of conditions that allow for slippage of the spliceosome between two juxtaposed 5′ splice sites as the spliceosome assembles. In every case, the snrp-27(az26) promoted 5′ splice site has usage that can be detected at least at a low level in wild-type animals (Fig. 2). Perhaps the combination of a weaker 5′ splice site whose usage is reduced in the snrp-27(az26) background (note the lack of an A residue at the +3 position of these 5′ splice sites) and an unusual juxtaposition of bases in the M141T mutant-promoted 5′ splice site (Fig. 3A,B) combine to give a region of “slipperiness” that is enhanced in the snrp-27 mutant, resulting in activation of the neighboring 5′ splice site. This is consistent with its role in cryptic 5′ splice site suppression of unc-73(e936) (Dassah et al. 2009). We noted that 14 of the 26 M141T-promoted alternative 5′ splice sites have a G at the +6 position of the intron. There is evidence for noncanonical base pairing of U1snRNA with 5′ splice sites involving bulged nucleotides in order to accommodate a G at the +6 position of the intron (Roca and Krainer 2009; Roca et al. 2012, 2013; Tan et al. 2016). However, in the case of the alternative 5′ splice sites we noted here, the preferences change as the result of mutations in the SNRP-27 protein associated with the tri-snRNP, not the initial U1snRNA interactions. We also note that the snrp-27(az26)-dependent change in alternative 5′ splice sites still occurs for the mab-10 event even when the +6G of the intron is changed to another nucleotide (Fig. 3C,D).
Based on our data, we propose a model for the role of the SNRP-27 M141 mutations in changes in alternative 5′ splice site usage. SNRP-27 is found associated with the U4/U6-U5 tri-snRNP (Fetzer et al. 1997), and interacts with the spliceosome after U1snRNA has established a base-pairing interaction with the 5′ splice site. As the tri-snRNP assembles into the spliceosome, U1 interactions with the 5′ end of the intron are replaced by interactions with U6 and PRP8 (Galej et al. 2016). Perhaps RS domain interactions between U1snRNP component U170K and SNRP-27 help to mediate the handoff of the 5′ splice site and ensure fidelity. For these unusual alternative 5′ splice sites whose relative usage changes in the presence of the SNRP-27 M141T mutant protein (Fig. 1), we hypothesize that this handoff becomes less efficient with the mutant SNRP-27, leading to a slippage event that allows U6 and PRP8 to assemble onto juxtaposed lower consensus alternative 5′ splice sites. While the new cryo-EM structural data have provided important new insights into the mechanism of splicing, it is clear that this is a dynamic process relying on multiple assembly steps. Our data strongly support the idea that there are additional factors not captured in these spliceosomal snapshots that play important roles in the fidelity of the assembly process.
MATERIALS AND METHODS
RNA isolation and high-throughput mRNA library production
RNA was isolated for library construction from the indicated strains from populations of worms enriched for young adults using TRIzol reagent (Invitrogen). mRNA libraries for high-throughput sequencing were prepared from 5 µg of total RNA after poly(A) selection using the NEXTflex Rapid Directional qRNA-Seq Kit from BioO Scientific. Paired directional 100 nt reads, eight libraries per lane were obtained from an Illumina HiSeq 4000 operated by the QB3 Vincent J. Coates Genomics Sequencing Laboratory at UC Berkeley. FASTQ files of raw reads from the high-throughput RNA-seq libraries and processed .gtf files of alternative splicing events in each library have been submitted to the NCBI Gene Expression Omnibus (GEO; http://www.ncbi.nlm.nih.gov/geo) under accession number GSE113275. Libraries were prepared from two independent RNA isolations for each strain. These resulting sequences were trimmed to 80 bases on each end and then mapped to the C. elegans genome using STAR (Dobin et al. 2013) and duplicates were removed. We used a custom script to identify all alternative 5′ splice site events and all alternative 3′ splice site events separated by 50 nt or less in these libraries. We also used AltEventFinder (Zhou et al. 2012) to identify cassette exon alternative splicing events in these libraries. We used MISO (Katz et al. 2010) to do four pairwise comparisons of these libraries against each other (CB936 library 1 versus SZ118 library 1; CB936 library 1 versus SZ118 library 2; CB936 library 2 versus SZ118 library 1; CB936 library 2 versus SZ118 library 2) for all the alternative splicing events in the three categories (alternative 5′ss, alternative 3′ss, alternative cassette exon). Those events with a change in percent spliced in (PSI) value of ≥0.20 in the comparisons were noted and events that met this threshold in all four library comparisons were further examined.
Calculation of significance of nucleotide frequency at specific positions in the snrp-27-affected alternative 5′ splice sites
We applied a hypergeometric test to determine the statistical significance of observed deviations in nucleotide frequencies at splice site positions. Briefly, we counted the number of times a nucleotide (e.g., “A”) occurred at a given position (e.g., “+3”) in a collection of 111,187 annotated 5′ splice sites, and in the subset of 26 affected pairs of alternative 5′ splice sites. We then applied a hypergeometric test (in Python, using scipy.stats.hypergeom.cdf) to determine P-values and whether the observed nucleotide counts in the subset of affected sites diverged significantly from the genome-wide frequency.
32P RT-PCR
Starting with 4 µg of total mixed stage RNA, reverse transcription was performed with Avian Myeloblastosis Virus reverse transcriptase (Promega) with a primer specific for the downstream exon (see Tables 1 and 2) in 25 µL reaction mixtures. This was followed by polymerase chain reactions containing 1.3 µL of the reverse transcription product amplified with Taq polymerase using the same reverse primer as in the reverse transcription reaction along with a forward facing primer that anneals to the upstream exon. The PCR mixtures were spiked with a 32P 5′ end-labeled reverse primer at 2% of the concentration of unlabeled reverse primer. After 25 extension cycles, PCR products were recovered by phenol:CHCl3 extraction and ethanol precipitation. The ethanol precipitates were resuspended in formamide with dyes and 2 µL were separated on a 40 cm long 6% polyacrylamide urea denaturing gel in TBE buffer. Products were visualized with a Molecular Dynamics Typhoon PhosphorImager, and quantifications of band intensities were performed using ImageQuant software.
TABLE 1.
Primers for 32P RT-PCR
TABLE 2.
Primers for CRISPR mutagenesis detection by PCR
CRISPR homologous recombination repair oligonucleotides
snrp-27 M141 mutagenesis
CGTGGATGGCTGCGTGAACATCAAGAAACCTCGCAGATACCGCCAgTACNNNAACCGAAAaGGAGGATTCAATCGTCCACTTGATTTTATGGGATAATTTA
Notes: The Met141 codon is replaced with three Ns (underlined) in which all nucleotides are randomly incorporated at these positions. The lower case “g” (underlined) is a silent mutation at the wobble position of Q139. It creates a GTAC RsaI restriction site which is used for detection of the altered snrp-27 coding region. The underlined lower-case “a” is a silent mutation at the wobble position of K144 that will disrupt the PAM sequence.
snrp-27 null allele mutagenesis
GTCGAAGCAGAAGTCCGCGCGATAAACGAGATCGGCGGGAAAGAAGTCGACTCACGTGAT CGAAAAGAACGTGATCGTGAACGACAAAAAAAGGATCGTGA
Notes: The underlined C is a 1 nt insertion that changes reading frame at amino acid 64 resulting in a stop codon two codons downstream. It also creates a new SalI restriction site, GTCGAC that is useful for the detection and screening of altered alleles. The underlined T disrupts the PAM sequence and becomes the T of the out-of-frame TGA stop codon.
mab-10 5′ss mutagenesis
TCATCGTCTCCGTCACTGAATCTGAACACGACATCGTCGAATTCG GTACCHTTGG GTTAGTCTTTGTCGCTGGGCGTTTGGTAGCACACGCATGTCCATAA
Notes: The mutagenesis will disrupt an HpyCH4III site ACNGT (underlined) which is useful for detection of altered genes. The H with an underline is a substitution of a G nucleotide with A, C, or T randomly generated at this position during oligo synthesis.
CRISPR crRNAs
AltR crRNAs were synthesized by Integrated DNA Technology with the following crRNA guide sequences
snrp-27 M141 mutagenesis
CCGCCAAUACAUGAACCGAA
snrp-27 null allele
GGCGGGAAAGAAGUCGAUCA
mab-10
AUCGUCGAAUUCGGUACCGU
CRISPR Cas9 mutant production
CRISPRCas9-directed mutagenesis was performed by micro-injecting Cas9 ribonucleoprotein complexes and homologous recombination repair oligonucleotides into the syncytial gonad of adult hermaphrodites (Paix et al. 2015). In addition to the crRNA and repair oligonucleotides for target genes snrp-27 and mab-10, we used co-CRISPR of dpy-10 II to the dominant roller cn64 allele for snrp-27 I mutagenesis or co-CRISPR of unc-58 X to the dominant paralysis e665 allele for mab-10 II mutagenesis using previously described crRNA and repair oligonucleotide sequences (Arribere et al. 2014). We prepared RNP complexes by incubating crRNAs and tracrRNA in 10 µl mixtures containing 500 pmols of tracrRNA, 450 pmols of target gene crRNA, and 50 pmols of co-CRISPR target crRNA (dpy-10 or unc-58). All RNAs in the mixture were synthesized by IDT using their proprietary Alt-R 5′ and 3′ end modifications to increase stability and had been resuspended at 100 µM concentration in IDT-supplied Nuclease-Free Duplex buffer prior to mixing. The crRNA/tracrRNA mixtures were heated to 95°C for 2 min followed by a 5 min incubation at room temperature. Five microliters of annealed RNA mixture were mixed with 5 µL of 40 µM S. pyogenes Cas9-NLS protein (in 20 mM HEPES-KOH pH 7.5, 150 mM KCl, 10% glycerol, 1 mM DTT), which was purchased from the QB3 MacroLab core facility at UC Berkeley. The cas9/crRNA/tracrRNA mixture was incubated for 5min at room temperature to form RNP complexes. Then, 1 µL of 40 µM co-CRISPR repair oligo [either for dpy-10(cn64) or unc-58(e665)] and 2 µL of 40 µM target gene repair oligo were added to make the injection mixture. The mixture was centrifuged at 13,000g for 5 min to clear any insoluble contaminants prior to filling an injection needle with the supernatant. In a typical experiment, 50 young adult C. elegans hermaphrodites were injected in their syncytial gonad with this mixture [Bristol N2 wild-type worms in all cases with the exception of the mab-10 mutagenesis experiment in which SZ211 snrp-27(az56) worms were initially injected]. The P0 injected adults were allowed to recover for 4 h and each was placed on its own NGM agar plate. F1 progeny were screened for the dominant co-injection phenotype, and these were put onto individual plates and allowed to lay embryos. Then the adult was removed and DNA extracted from the single worm, and subject to PCR with primers spanning the targeted region of interest. Restriction digests were performed on the PCR products allowing us to identify F1s heterozygous for the mutation. F2 progeny of these heterozygous F1s that were wild-type in movement (had segregated away the unlinked dominant co-CRISPR allele) were followed by this same method in order to identify homozygous mutants in the target gene. The PCR products of the homozygous alleles were then subjected to Sanger sequencing methods to identify the new alleles of the target genes.
ACKNOWLEDGMENTS
This research was supported by the National Science Foundation MCB-1613867 to A.M.Z. L.E.R. was a fellow of the UCSC PREP research program funded by the National Institute of General Medical Sciences, National Institutes of Health, R25GM104552. We thank Manny Ares and Naomi Tesfuzigta for the idea of an oligonucleotide-directed homologous repair genetic experiment to randomize a single site. We thank Joshua Arribere for valuable help and guidance with statistical analysis. We are grateful to Zhouli Ni and Sam Guoping Gu for their assistance in establishing the C. elegans CRISPR system in our laboratory. Thanks to Jeremy Sanford, Melissa Jurica, Alec Barrett, Jessie Lopez, Ken Osterhoudt, Sidney Owen, and Megan Mayerle for many helpful discussions.
Footnotes
Article is online at http://www.rnajournal.org/cgi/doi/10.1261/rna.066878.118.
REFERENCES
- Agafonov DE, Kastner B, Dybkov O, Hofele RV, Liu WT, Urlaub H, Lührmann R, Stark H. 2016. Molecular architecture of the human U4/U6.U5 tri-snRNP. Science 351: 1416–1420. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J Mol Biol 215: 403–410. [DOI] [PubMed] [Google Scholar]
- Ares M Jr, Weiser B. 1995. Rearrangement of snRNA structure during assembly and function of the spliceosome. Prog Nucleic Acid Res Mol Biol 50: 131–159. [DOI] [PubMed] [Google Scholar]
- Arribere JA, Bell RT, Fu BX, Artiles KL, Hartman PS, Fire AZ. 2014. Efficient marker-free recovery of custom genetic modifications with CRISPR/Cas9 in Caenorhabditis elegans. Genetics 198: 837–846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho S, Hoang A, Sinha R, Zhong XY, Fu XD, Krainer AR, Ghosh G. 2011. Interaction between the RNA binding domains of Ser-Arg splicing factor 1 and U1-70K snRNP protein determines early spliceosome assembly. Proc Natl Acad Sci 108: 8233–8238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dassah M, Patzek S, Hunt VM, Medina PE, Zahler AM. 2009. A genetic screen for suppressors of a mutated 5′ splice site identifies factors associated with later steps of spliceosome assembly. Genetics 182: 725–734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR. 2013. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29: 15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fetzer S, Lauber J, Will CL, Lührmann R. 1997. The [U4/U6.U5] tri-snRNP-specific 27K protein is a novel SR protein that can be phosphorylated by the snRNP-associated protein kinase. RNA 3: 344–355. [PMC free article] [PubMed] [Google Scholar]
- Fica SM, Nagai K. 2017. Cryo-electron microscopy snapshots of the spliceosome: structural insights into a dynamic ribonucleoprotein machine. Nat Struct Mol Biol 24: 791–799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galej WP, Wilkinson ME, Fica SM, Oubridge C, Newman AJ, Nagai K. 2016. Cryo-EM structure of the spliceosome immediately after branching. Nature 537: 197–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodgkin J, Papp A, Pulak R, Ambros V, Anderson P. 1989. A new kind of informational suppression in the nematode Caenorhabditis elegans. Genetics 123: 301–313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howard JM, Sanford JR. 2015. The RNAissance family: SR proteins as multifaceted regulators of gene expression. Wiley Interdiscip Rev RNA 6: 93–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katz Y, Wang ET, Airoldi EM, Burge CB. 2010. Analysis and design of RNA sequencing experiments for identifying isoform regulation. Nat Methods 7: 1009–1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koodathingal P, Staley JP. 2013. Splicing fidelity: DEAD/H-box ATPases as molecular clocks. RNA Biol 10: 1073–1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larson A, Fair BJ, Pleiss JA. 2016. Interconnections between RNA-processing pathways revealed by a sequencing-based genetic screen for pre-mRNA splicing mutants in fission yeast. G3 (Bethesda) 6: 1513–1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim LP, Burge CB. 2001. A computational analysis of sequence features involved in recognition of short introns. Proc Natl Acad Sci 98: 11193–11198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarova OV, Makarov EM, Lührmann R. 2001. The 65 and 110 kDa SR-related proteins of the U4/U6.U5 tri-snRNP are essential for the assembly of mature spliceosomes. EMBO J 20: 2553–2563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maquat LE. 2002. Nonsense-mediated mRNA decay. Curr Biol 12: 196–197. [DOI] [PubMed] [Google Scholar]
- Mayerle M, Guthrie C. 2017. Genetics and biochemistry remain essential in the structural era of the spliceosome. Methods 125: 3–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura Y, Moriuchi R, Nakayama D, Yamashita I, Higashiyama Y, Yamamoto T, Kusano Y, Hino S, Miyamoto T, Katamine S. 1994. Altered expression of a novel cellular gene as a consequence of integration of human T cell lymphotropic virus type 1. J Gen Virol 75(Pt 10): 2625–2633. [DOI] [PubMed] [Google Scholar]
- Paix A, Folkmann A, Rasoloson D, Seydoux G. 2015. High efficiency, homology-directed genome editing in Caenorhabditis elegans using CRISPR-Cas9 ribonucleoprotein complexes. Genetics 201: 47–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pulak R, Anderson P. 1993. mRNA surveillance by the Caenorhabditis elegans smg genes. Genes Dev 7: 1885–1897. [DOI] [PubMed] [Google Scholar]
- Ragle JM, Katzman S, Akers TF, Barberan-Soler S, Zahler AM. 2015. Coordinated tissue-specific regulation of adjacent alternative 3′ splice sites in C. elegans. Genome Res 25: 982–994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roca X, Krainer AR. 2009. Recognition of atypical 5′ splice sites by shifted base-pairing to U1 snRNA. Nat Struct Mol Biol 16: 176–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roca X, Akerman M, Gaus H, Berdeja A, Bennett CF, Krainer AR. 2012. Widespread recognition of 5′ splice sites by noncanonical base-pairing to U1 snRNA involving bulged nucleotides. Genes Dev 26: 1098–1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roca X, Krainer AR, Eperon IC. 2013. Pick one, but be quick: 5′ splice sites and the problems of too many choices. Genes Dev 27: 129–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shepard PJ, Hertel KJ. 2009. The SR protein family. Genome Biol 10: 242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y. 2017. Mechanistic insights into precursor messenger RNA splicing by the spliceosome. Nat Rev Mol Cell Biol 18: 655–670. [DOI] [PubMed] [Google Scholar]
- Sinenko SA, Kim EK, Wynn R, Manfruelli P, Ando I, Wharton KA, Perrimon N, Mathey-Prevot B. 2004. Yantar, a conserved arginine-rich protein is involved in Drosophila hemocyte development. Dev Biol 273: 48–62. [DOI] [PubMed] [Google Scholar]
- Tan J, Ho JX, Zhong Z, Luo S, Chen G, Roca X. 2016. Noncanonical registers and base pairs in human 5′ splice-site selection. Nucleic Acids Res 44: 3908–3921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teigelkamp S, Mundt C, Achsel T, Will CL, Luhrmann R. 1997. The human U5 snRNP-specific 100-kD protein is an RS domain-containing, putative RNA helicase with significant homology to the yeast splicing factor Prp28p. RNA 3: 1313–1326. [PMC free article] [PubMed] [Google Scholar]
- Xiang S, Gapsys V, Kim HY, Bessonov S, Hsiao HH, Möhlmann S, Klaukien V, Ficner R, Becker S, Urlaub H, et al. 2013. Phosphorylation drives a dynamic switch in serine/arginine-rich proteins. Structure 21: 2162–2174. [DOI] [PubMed] [Google Scholar]
- Xiao SH, Manley JL. 1998. Phosphorylation-dephosphorylation differentially affects activities of splicing factor ASF/SF2. EMBO J 17: 6359–6367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zahler AM, Roth MB. 1995. Distinct functions of SR proteins in recruitment of U1 small nuclear ribonucleoprotein to alternative 5′ splice sites. Proc Natl Acad Sci 92: 2642–2646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zahler AM, Lane WS, Stolk JA, Roth MB. 1992. SR proteins: a conserved family of pre-mRNA splicing factors. Genes Dev 6: 837–847. [DOI] [PubMed] [Google Scholar]
- Zhou A, Breese MR, Hao Y, Edenberg HJ, Li L, Skaar TC, Liu Y. 2012. Alt Event Finder: a tool for extracting alternative splicing events from RNA-seq data. BMC Genomics 13(Suppl 8): S10. [DOI] [PMC free article] [PubMed] [Google Scholar]