Abstract
RNA 5′ cap structures comprising the metabolic effector nicotinamide adenine dinucleotide (NAD) have been identified in diverse organisms. Here we report a simple, two-step procedure to detect and quantitate NAD-capped RNA, termed “NAD-capQ.” By use of NAD-capQ we quantitate NAD-capped RNA levels in Escherichia coli, Saccharomyces cerevisiae, and human cells, and we measure increases in NAD-capped RNA levels in cells from all three organisms harboring disruptions in their respective “deNADding” enzymes. We further show that NAD-capped RNA levels in human cells respond to changes in cellular NAD concentrations, indicating that NAD capping provides a mechanism for human cells to directly sense and respond to alterations in NAD metabolism. Our findings establish NAD-capQ as a versatile, rapid, and accessible methodology to detect and quantitate 5′-NAD caps on endogenous RNA in any organism.
Keywords: NAD cap, NAD-capQ, mRNA deNADding, mRNA decapping
INTRODUCTION
The chemical nature of the 5′ end of RNA is a key determinant of all aspects of mRNA metabolism–synthesis, nucleo-cytoplasmic transport, translation, and turnover (Muthukrishnan et al. 1975; Furuichi et al. 1977; Konarska et al. 1984; Drummond et al. 1985). Thus, cellular processes that alter the chemical nature of the 5′ end of RNA provide a layer of “epitranscriptomic” regulation. One well-characterized RNA 5′ end modification is the 7-methylguanylate (m7G) “cap” structure that is added to many eukaryotic messenger RNAs after transcription initiation by a capping complex that binds RNA polymerase II (RNAP II) and interacts with nascent RNA (Shuman 1995). m7G capping protects eukaryotic mRNA from exonuclease digestion, and enables binding of proteins that facilitate mRNA translation and RNA processing.
Recently, a new form of RNA 5′ capping was identified (Jäschke et al. 2016; Kiledjian 2018) on bacterial (Chen et al. 2009; Cahová et al. 2015), yeast (Walters et al. 2017), and human (Jiao et al. 2017) RNA species. In this form of RNA 5′ capping, nicotinamide adenine dinucleotide (NAD), a critical redox cofactor present in all organisms, is used as a noncanonical initiating nucleotide (NCIN) by RNAP to generate the NAD cap (Bird et al. 2016). NCIN-mediated NAD capping has been demonstrated for bacterial RNAP and eukaryotic RNAP II (Malygin and Shemyakin 1979; Bird et al. 2016; Julius and Yuzenkova 2017), suggesting NAD capping is likely a shared property of all RNAPs. Moreover, mammalian cells appear to also harbor an additional mode of NAD capping that occurs post-transcriptionally (Kiledjian 2018). Thus, in contrast to m7G capping, which occurs only in organisms carrying specialized capping enzymes, NAD-capping, which can be added by RNAP, likely occurs in all organisms. Utilization of NAD as an RNA cap might provide a direct mechanism by which alterations in cellular NAD metabolism can be linked to alterations in gene expression. Studies of NAD capping may represent a new frontier for understanding the interplay between cellular metabolism and RNA biology.
Two methods for detection of NAD-capped RNAs in cells have been reported. The first method involves enrichment of small RNA products by size-exclusion chromatography, enzymatic RNA hydrolysis of the isolated small RNA fraction to completion, and detection of free NAD generated upon hydrolysis by comparative high-resolution LC/MS (Chen et al. 2009). The second method, termed NAD-captureSeq (Cahová et al. 2015; Winz et al. 2017), involves chemoenzymatic modification of the terminal nicotinamide to an alkyne moiety amendable to click-chemistry-mediated biotinylation, isolation of biotinylated RNA by a streptavidin matrix, and analysis of the isolated RNA by high-throughput sequencing. Both of these methods are labor-intensive, time consuming, and require specialized laboratory equipment and reagents. We therefore sought to develop an alternative method for detection of NAD-capped RNA that would overcome these limitations.
RESULTS
NAD cap detection and quantitation (NAD-capQ)
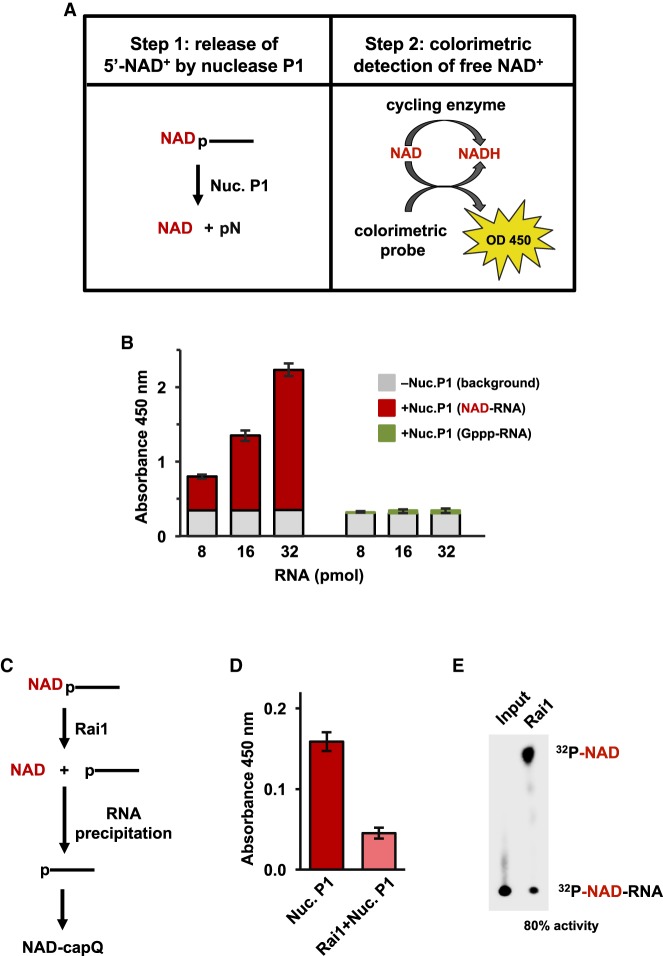
To develop a simple and rapid method to detect and quantitate levels of NAD caps in cells, we built on the approach developed by Liu and coworkers (Chen et al. 2009) by replacing the mass spectrometry analysis with a versatile colorimetric assay. Our method, termed NAD cap detection and quantitation (NAD-capQ), is a two-step procedure consisting of enzymatic treatment of RNA with nuclease P1 to cleave the phosphodiester bonds within RNA and release 5′-NAD, followed by quantitation of released 5′-NAD by a commercially available colorimetric assay (Fig. 1A). The colorimetric assay is based on an enzymatic cycling reaction in which NAD is reduced to NADH, which further reacts with the probe to produce a product that absorbs light at a wavelength of 450 nm, which can be monitored by spectrophotometry (Fig. 1A).
FIGURE 1.
NAD-cap detection and quantitation assay. (A) Steps in NAD-capQ. (B) NAD-capQ analysis of RNA generated in vitro. (Red) NAD-RNA; (green) Gppp-RNA; (gray) background signal detected without nuclease P1. Error bars represent ±SD, n ≥ 3. (C) Schematic of 5′-NAD cap removal from NAD-capped RNA using pretreatment with Rai1. (D) NAD-capQ analysis with or without pretreatment of NAD-RNA with Rai1. (Dark red) Background corrected absorbance without Rai1; (light red) background corrected absorbance with Rai1. Significance as in B. (E) Thin-layer chromatography analysis revealing 80% efficiency of Rai1 hydrolysis of 32P-5′ end labeled NAD-capped RNA spiked into 50 µg total RNA.
To validate NAD-capQ as a method for detection and quantitation of NAD caps, we generated NAD-capped RNA in vitro, incubated different amounts of the RNA with or without nuclease P1, carried out the enzymatic cycling reaction and measured the absorbance in each sample at 450 nm. The signal detected in samples treated with nuclease P1 was above background levels of absorbance detected in samples without nuclease P1 and increased proportionally to the amount of input NAD-capped RNA (Fig. 1B, red bars). In contrast, the signal detected in samples containing a guanosine-capped RNA (Gppp-RNA) was indistinguishable with or without nuclease P1 treatment (Fig. 1B, green bars), indicating that the signal detected in the analysis of NAD-capped RNA is derived from released NAD. To confirm the signal detected in samples containing NAD-capped RNA was derived from the 5′-NAD cap, RNA was treated with Schizosaccharomyces pombe Rai1, a “deNADding” enzyme that hydrolyzes NAD caps without degrading the RNA body (Jiao et al. 2017) to remove NAD caps prior to nuclease P1 digestion (Fig. 1C). Treatment of NAD-capped RNA with Rai1 preceding RNA isolation and nuclease P1 treatment resulted in an ∼80% decrease in the signal detected by NAD-capQ (Fig. 1D), which matched the ∼80% Rai1 efficiency detected by thin-layer chromatographic analysis using radiolabeled NAD-capped RNA (Fig. 1E). The results indicate the signal detected by NAD-capQ is derived from 5′-NAD caps. Collectively, the results presented in Figure 1 establish NAD-capQ as a method for the detection and quantitation of NAD-capped RNA generated in vitro.
Quantitation of NAD-capped RNA in diverse organisms by NAD-capQ
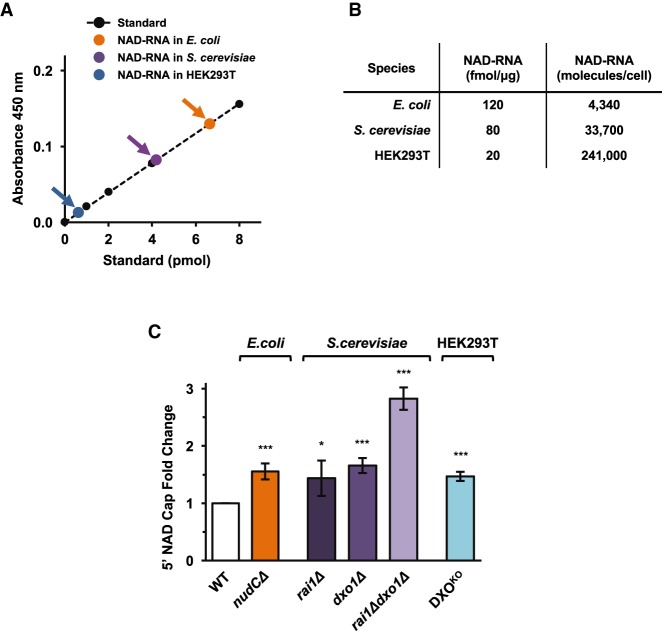
To establish the functionality of NAD-capQ on endogenous NAD-capped RNAs, we initially tested the method on RNA from E. coli. Plotting the colorimetric signal detected in the analysis of cellular RNA against a standard curve generated using NADH, ∼6 pmol NAD caps were detected per 50 µg of total E. coli RNA (Fig. 2A, orange data-point). Using an estimate of 60 fg RNA per E. coli cell, the value we measured by NAD-capQ corresponds to ∼4300 total molecules of NAD cap per cell (Fig. 2B), a value similar to that reported by Liu and coworkers using the LC/MS method (∼3300 NAD cap molecules per cell [Chen et al. 2009]). The corresponding values between NAD cap levels derived from the NAD-capQ assay and that obtained by mass spectrometry demonstrate the validity of NAD-capQ for NAD cap quantitation. We conclude that NAD-capQ enables detection and quantitation of NAD-capped RNA generated in vivo.
FIGURE 2.
NAD-capQ detection and quantitation of RNA generated in vivo. (A) Quantitation of cellular NAD-capped RNA levels in E. coli, S. cerevisiae, or human HEK293T cells plotted on an NADH standard curve (orange data-point, E. coli; purple data-point, S. cerevisiae; blue data-point, human HEK293T; black data-points, NADH standard). n ≥ 3. (B) Values of NAD-RNA fmol/µg were calculated by dividing the calculated value of NAD-capped RNA detected by NAD-capQ in A by the amount of total cellular RNA analyzed. Quantitative values of NAD-capped RNA represented as RNA molecules/cell were calculated using total RNA concentrations per cell in E. coli (0.06 pg; Neidhardt et al. 1990), S. cerevisiae (0.7 pg; von der Haar 2008), and mammals (20 pg; Palazzo and Lee 2015), respectively. (C) Effects of disrupting deNADding enzymes on NAD-capped RNA levels in E. coli, S. cerevisiae, and human HEK293T cells. Graphs plot the fold-change in NAD-capped RNA values in the indicated mutant cells relative to the value observed in wild-type cells. (Error bars represent ±SD, n ≥ 3; [*] P < 0.05; [***] P < 0.001).
To assess the versatility of the NAD-capQ assay, we next used NAD-capQ to analyze levels of total NAD-capped RNA in yeast and human cells (Fig. 2A,B). We find that S. cerevisiae cells contain ∼4 pmol NAD caps per 50 µg of total RNA (Fig. 2A, purple data-point) while human HEK293T cells contain ∼1 pmol of NAD caps per 50 µg of total RNA (Fig. 2A, blue data-point). The results establish use of NAD-capQ to quantitate levels of NAD-capped RNA across a range of organisms, and suggests a hierarchal distribution of levels of NAD caps in bacteria, fungi, and human cells (Fig. 2A).
Disruption of “deNADding” enzymes alters NAD-capped RNA levels
To establish use of NAD-capQ to quantitate changes in levels of NAD-capped RNA in vivo, we measured levels of NAD-capped RNA in E. coli, S. cerevisiae and HEK293T cells harboring disruptions in their respective “deNADding” enzymes (Kiledjian 2018), and compared these values to levels of NAD-capped RNA in corresponding wild-type cells.
The E. coli NudC protein has been shown to function as a deNADding enzyme in vitro (Cahová et al. 2015). Furthermore, levels of NAD capping of the RNAI transcript in E. coli cells are higher in cells lacking NudC compared with wild-type cells (Cahová et al. 2015; Bird et al. 2016), providing evidence that NudC functions as a deNADding enzyme in vivo. NAD-capQ analysis of RNA isolated from wild-type E. coli cells or mutant cells carrying a disrupted nudC allele (ΔnudC) show that total levels of NAD caps are ∼1.6-fold higher in ΔnudC cells (Fig. 2C). The results provide further evidence that NudC functions as a deNADding enzyme in vivo, in E. coli.
We have recently demonstrated that mammalian and fungal noncanonical DXO/Rai1 decapping enzymes exhibit robust deNADding activity by removing the entire NAD moiety from the 5′ end of NAD-capped RNA (Jiao et al. 2017). To establish the effect of disruptions in DXO/Rai1 decapping enzymes on NAD cap levels in vivo, we first used NAD-capQ to measure NAD cap levels in wild-type S. cerevisiae or mutant strains harboring disruptions in rai1 (rai1Δ), dxo1 (dxo1Δ), or both (rai1Δdxo1Δ). Results show NAD cap levels are 1.4-fold and 1.7-fold higher in rai1Δ and dxo1Δ strains, respectively, relative to wild-type cells (Fig. 2C), indicating both proteins function as deNADding enzymes in vivo, in S. cerevisiae. Moreover, we find NAD cap levels are higher in cells lacking both enzymes (rai1Δdxo1Δ) than in cells harboring disruptions in either rai1 or dxo1 alone. These data suggest that Rai1p and Dxo1p each target a distinct set of NAD-capped RNAs in S. cerevisiae cells.
We next used NAD-capQ to measure NAD cap levels in human HEK293T cells that carried either the wild-type or homozygous mutant DXO alleles (DKOKO [Jiao et al. 2017]). Results show NAD cap levels are 1.5-fold higher in cells harboring a disruption in DXO (DKOKO) relative to cells carrying wild-type DXO alleles (Fig. 2C), providing further evidence that DXO functions as a deNADding enzyme in human cells. Together, the analysis of E. coli, S. cerevisiae and HEK293T cells harboring disruptions in their respective “deNADding” enzymes establishes NAD-capQ as a method for quantitative analysis of changes in levels of NAD capping in vivo that occur in response to alterations in the enzymatic processing of NAD caps.
Cellular NAD levels can influence NAD capping
NAD plays a vital role in energy metabolism in eukaryotic cells by acting as an electron acceptor, becoming reduced through the metabolism of other molecules to form NADH to fuel oxidative phosphorylation. NAD and its metabolites also act as cofactors for a wide range of enzymes, such as sirtuins (Hall et al. 2013), linking cellular metabolism to changes in signaling and transcription events.
The demonstration that NAD functions as an NCIN suggests alterations in levels of free NAD will directly alter levels of NAD-capped RNA. To determine whether there is a correlation between cellular NAD levels and levels of NAD-capped RNA, we treated human HEK293T cells either with nicotinamide (NAM), which increases cellular levels of NAD (Hong et al. 2016) or FK866, a small molecule inhibitor of NAM phosphoribosyltransferase that reduces intracellular NAD (Khan et al. 2006; Wang et al. 2006; Fig. 3B), and analyzed levels of NAD caps by NAD-capQ. Results show that increasing the cellular levels of NAD leads to a corresponding increase in the levels of NAD-capped RNA, while decreasing the cellular levels of NAD leads to a corresponding decrease in the levels of NAD-capped RNA (Fig. 3C). The results indicate that cellular NAD levels and levels of NAD capped RNA are correlated in human cells, and establish NAD-capQ as a method to analyze the interplay between NAD levels and NAD capping in vivo.
FIGURE 3.

Cellular NAD concentrations can influence NAD cap levels. (A) The synthesis of NAD from NAM or NR. NAM is converted to NMN by the rate-limiting NAMPT, using PRPP as a cosubstrate. NMN is also the product of NR phosphorylation by NR kinases. NMN is converted to NAD by the NMNAT enzyme. FK866 is a small molecule inhibitor of NAMPT. (NAMPT) Nicotinamide phosphoribosyltransferase; (NMNAT) NMN adenylyltransferase; (NRK) NR kinase; (PRPP) phosphoribosyl pyrophosphate. (B) Quantitation of cellular total NAD following 48 h treatment with 5 nM FK866 or 5 mM NAM to decrease and increase cellular NAD levels, respectively. (C) Quantification of NAD-caps, detected with NAD-CapQ following 48 h treatment with 5 nM FK866 or 5 mM NAM. Mean values are plotted. Error bars represent ±SD. Statistical significance was calculated by one-way ANOVA with a Dunnett's multiple comparison post-test. P-values are denoted by asterisks. (*) P < 0.05; (**) P < 0.01; (***) P < 0.001.
DISCUSSION
Here we report NAD-capQ, a versatile, rapid and accessible methodology to detect and quantitate 5′-NAD caps on endogenous RNA (Fig. 1). We demonstrate the versatility of NAD-capQ by quantitating NAD-capped RNA levels in bacteria, yeast, and human cells (Fig. 2A,B), measuring increases in NAD-capped RNA levels in cells harboring disrupted “deNADding” enzymes (Fig. 2C), and establishing that levels of NAD-capped RNA are responsive to changes in levels of cellular NAD (Fig. 3).
In contrast to previously reported methods for analysis of NAD-capped RNA, NAD-capQ can be performed in a single day using common laboratory equipment. Strikingly, the results of our quantitative analysis of NAD-capped RNA in E. coli by NAD-capQ, in which a commercially available kit is used for the detection of NAD, matched results previously reported using LC/MS for detection of NAD (Fig. 2B; Chen et al. 2009). Thus, NAD-capQ is a simpler procedure for detection of NAD-capped RNA that provides sensitivity that matches previously reported methods for analysis of NAD-capped RNA.
The ability to rapidly quantitate NAD cap levels enabled us to readily compare NAD-capped RNA levels between several organisms (Fig. 2A,B). We find that the highest amount of total NAD-capped RNA per unit RNA was detected in E. coli, followed by S. cerevisiae and human HEK293T cells (Fig. 2A,B). The difference in NAD-capped RNA observed between S. cerevisiae and human cells is consistent with prior studies, indicating that intracellular NAD concentrations in S. cerevisiae are higher than in mammalian cells (∼1 to ∼2 mM for S. cerevisiae [Lin et al. 2004; Sporty et al. 2008] and ∼0.2–0.5 mM for mammalian cells [Cantó et al. 2015]).
The broad utility of NAD-capQ was evident from findings in S. cerevisiae where the Rai1 and Dxo1 proteins previously shown to possess deNADding activity in vitro, were shown to function as deNADding enzymes in cells. Whether the doubling of NAD cap levels in strains lacking both enzymes (rai1Δdxo1Δ) relative to either alone is due to an additive effect of both enzymes targeting the same NAD-capped RNAs or each enzyme targeting different subsets of NAD-capped RNAs is currently unknown. However, the presence of multiple deNADding enzymes and their potential to target distinct subsets of RNA would be analogous to the multiple canonical m7G-cap decapping enzymes that function on specific subsets of RNA substrates (Grudzien-Nogalska and Kiledjian 2017) demonstrating a common theme within decapping/deNADding enzymes.
NAD plays a vital role in energy metabolism in eukaryotic cells by acting as an electron acceptor, becoming reduced through the metabolism of other molecules to form NADH to fuel oxidative phosphorylation. NAD also acts as a cofactor for a wide range of enzymes, such as sirtuins (Hall et al. 2013), linking cellular metabolism to changes in signaling and transcription events. In addition, intracellular concentrations of NAD fluctuate along with the metabolic state of a cell and under cellular stress (Chen et al. 2008; Fulco et al. 2008; Cantó et al. 2009, 2010; Raynes et al. 2013). Our finding that, in human cells, alterations in NAD concentrations lead to corresponding alterations in NAD capping suggests NAD capping provides a mechanism by which human cells could directly sense and respond to alterations in NAD metabolism. The results presented here underscore the utility of NAD-capQ in providing a simple quantitative assay to assess the interplay between cellular metabolism and NAD capping.
Although the recent emphasis in the field has been on 5′-end NAD capped RNA (Jäschke et al. 2016; Kiledjian 2018), nucleotide metabolite modifications at the 5′ end of RNAs likely extend beyond NAD. In this regard, it has been shown that, in addition to NAD, RNAP can use several other nucleoside-containing metabolites as NCINs during transcription initiation, including desphosphocoenzyme A (dpCoA), FAD, UDP-Glucose, and UDP-N-acetylglucosamine (Malygin and Shemyakin 1979; Huang 2003; Bird et al. 2016; Julius and Yuzenkova 2017; Vvedenskaya et al. 2018). Furthermore, evidence for 5′-end dpCoA capped RNA has been reported in E. coli (Kowtoniuk et al. 2009). Thus, we anticipate that by incorporating a different colorimetric or fluorescent-based assay in step 2 of the NAD-capQ procedure (see Fig. 1A), it will be possible to develop assays to detect and quantitate RNA species that are capped with nucleoside-containing metabolites other than NAD.
MATERIALS AND METHODS
Bacteria growth and RNA extraction
E. coli MG1655 was obtained from the ATCC. MG1655 ΔnudC:Kan was previously reported (Bird et al. 2016). E. coli MG1655 or MG1655 ΔnudC:Kan cells were grown overnight and then back diluted 1:100 in 25 mL LB (10 g Bacto-tryptone, 5 g Bacto-yeast extract and 10 g NaCl per liter) containing 25 µg mL–1 kanamycin (only for the ΔnudC:Kan strain) in 125 mL flasks (Bellco). Cultures were shaken at 220 rpm at 37°C. When cell density reached an OD600 ∼ 3.4 (∼8 h), 5 mL of the cell suspension was centrifuged (1 min, 10,000g at room temperature) to collect cells, supernatants were removed, and cell pellets were rapidly frozen on dry ice. To extract total RNA, frozen pellets were resuspended in 1 ml of TRI Reagent solution (Molecular Research Center). Samples were incubated at 70°C for 10 min and then centrifuged (10 min, 21,000g, 4°C) to remove insoluble material. The supernatant was transferred to a fresh tube and 200 µL of chloroform was added, samples were mixed by vortexing, and then centrifuged (10 min, 21,000g, 4°C). The aqueous phase was transferred to a fresh tube, extracted with acid phenol:chloroform twice (Ambion), and RNA transcripts were recovered by ethanol precipitation, washed with 75% ethanol at 4°C and resuspended in RNase free water (Ambion).
Yeast growth and RNA extraction
The genotypes of the following Saccharomyces cerevisiae strains used in this study were described previously: ABD1 WT (Schwer et al. 2000), ABD1;rai1Δ, ABD1;ydr370C and ABD1;rai1Δ;ydr370CΔ (Jiao et al. 2010). Yeast strains were grown in minimal (synthetic defined) media at 30°C according to standard protocols. All strains were harvested for the experiments at the end of the exponential phase (OD600 = 0.9) and total RNAs were isolated with the acidic hot phenol method (Köhrer and Domdey 1991).
Mammalian cell culture and RNA extraction
Human embryonic kidney HEK293T cells were obtained from ATCC. Cells were maintained in DMEM medium (Thermo Fisher Scientific) supplemented with 10% fetal bovine serum (Atlanta Biologics), and antibiotics (100 units/mL penicillin and 100 µg/mL of streptomycin) under 5% CO2 at 37°C. The three different monoclonal HEK293T cell lines harboring CRISPR-Cas9n double-nick generated distinct homozygous deletions within the DXO gene genomic region corresponding to its catalytic site have been reported previously (Jiao et al. 2017). Total cellular RNAs were extracted with TRIzol Reagent according to the manufacturer's protocol (Thermo Fisher Scientific).
In vitro transcription of NAD-capped RNAs
For assays shown in Figure 1B and E, NAD-capped RNA, GpppA-RNA and 32P-NAD-capped RNA were generated by in vitro transcription as previously described (Jiao et al. 2017). Four-hundred nanograms of PCR generated ɸ2.5A-CA2-G16 linear template containing the T7 ɸ2.5 promoter (Coleman et al. 2004) that retained the first adenosine at the transcription start site and replaced all other adenosine with uracils, as well as harboring the G16 tract at the 3′ end (Jiao et al. 2017), was subjected to in vitro transcription. Reactions were in 50 µL reaction buffer (10 µL 5 × transcription buffer, 5 µL DTT [10 mM], 5 µL BSA [10 µg/µL], 5 µL rNTP Mix [5 mM], 2 µL T7 RNA polymerase [Promega], and 1 µL RNasin Ribonuclease Inhibitor [40 U/µL, Promega]). To generate 32P-NAD-cap labeled or unlabeled NAD-capped RNAs, rATP was replaced with 32P-NAD or NAD. For GpppA-capped RNA, 0.5 mM GpppA (Sigma-Aldrich) was used instead of rATP. Reactions were incubated 90 min at 37°C and then treated with 3 units of DNase RQ1 (Promega) for 20 min at 37°C. Samples were extracted with acidic phenol:chloroform and RNA recovered by ethanol precipitation.
Cellular total NAD detection
HEK293T cells (2 × 106) were seeded in 100 mm plates a day before the experiment. Actively growing cells were either subjected to 48 h treatment with 5 nM FK866 (Sigma-Aldrich) or 5 mM NAM (Sigma-Aldrich) to decrease and increase cellular NAD levels, respectively. To measure cellular total NAD levels, 1/5 of the cells were lysed in 400 µL of NAD/H Extraction Buffer (NAD/H Quantitation Kit, Sigma-Aldrich) and total proteins were extracted with two freeze/thaw cycles (20 min on dry-ice, 10 min at RT). Cell lysates were centrifuged at 13,000g at 4°C for 10 min and supernatant was taken to measure protein concentration. Ten micrograms of total protein was used to detect cellular NAD according to the manufacturer's protocol (NAD/H Quantitation Kit, Sigma).
NAD-cap detection and quantitation (NAD-capQ)
To carry out NAD-capQ, it was necessary to minimize copurifying free NAD in the RNA preparation. To remove residual free NAD, purified RNAs were dissolved in 10 mM Tris-HCl (pH 7.5) containing 2 M urea. Samples were incubated 2 min at 65°C and immediately precipitated with isopropanol in the presence of 2 M (final concentration) ammonium acetate. Fifty micrograms of total RNA was subjected to NAD-cap Detection and Quantitation (NAD-capQ) assay. In the first step, RNA was digested with 1 U of Nuclease P1 (Sigma-Aldrich) in 20 µL reaction containing 10 mM Tris (pH 7.0), 20 µM ZnCl2 at 37°C for 20 min to release intact NAD/NADH. The control samples were prepared by incubating 50 µg of RNA under the same reaction conditions, except instead of adding nuclease P1, an equivalent volume of 10% glycerol (which is what was used to dissolve the enzyme) was added to correct for any potential deviation in background absorbance. Similarly, NADH standard was prepared in the same buffer condition as that for the nuclease P1 reaction. After at least a 2 h incubation at room temperature, 30 µL of NAD/NADH Extraction Buffer (NAD/NADH Quantitation Kit, Sigma-Aldrich) was added to each sample. In the second step, 50 µL samples were used in a colorimetric assay to measure absorbance at 450 nm according to the manufacturer's protocol (NAD/NADH Quantitation Kit, Sigma-Aldrich). The signal corrected for the background absorbance is proportional to concentrations of NAD and NADH within a sample. Although NAD/NADH Quantitation Kit from Sigma-Aldrich was used in this study, a number of similar kits are available from other manufacturers and could likely be substituted.
Rai1 digestion
S. pombe Rai1 was prepared from E. coli strain BL21(DE3) Rosetta using culture and induction procedures, immobilized-metal-ion affinity chromatography on Ni-NTA agarose, and size exclusion chromatography using Sephacryl S-300 as in Xiang et al. (2009). Fifty micrograms of total RNA isolated from cells were spiked with synthetic 32P-5′-end labeled NAD-capped RNA and incubated with 4 µg of recombinant SpRai1 in 20 µL reaction buffer (10 mM Tris-HCl [pH 7.5], 100 mM KOAc, 2 mM MgCl2, 2 mM MnCl2) for 1 h at 37°C. The reactions were stopped by the addition of 30 mM EDTA and RNAs were extracted with acid phenol:chloroform. Urea (2 M final concentration) was added, samples were heated at 65°C for 2 min and total RNA was recovered by isopropanol precipitation in the presence of ammonium acetate (2 M final concentration). DeNADding products were resolved by PEI-cellulose TLC plates (Sigma-Aldrich) and developed in 0.5 M LiCl2 in a TLC chamber at room temperature as previously described (Liu et al. 2008). Reaction products were visualized and quantitated with a Molecular Dynamics PhosphorImager (Storm860) with ImageQuant-5 software.
Statistical analysis
All statistical parameters are listed in the figure legends and represented as error bars in the figures. Statistical significance level was calculated by one-way ANOVA with a Dunnett's multiple comparison post-test. P < 0.05 is used to claim statistical significances, which are denoted as follows: (*) P < 0.05; (**) P < 0.01; (***) P < 0.001.
ACKNOWLEDGMENTS
This work was supported by National Institutes of Health (NIH) grant GM118059 to B.E.N. and GM126488 to M.K.
Author contributions: M.K. and E.G.N. designed the experiments. E.G.N. carried out all experiments unless otherwise indicated. J.B. provided the RNA isolated from bacterial strains. E.G.N., M.K., and B.E.N wrote the manuscript.
Footnotes
Article is online at http://www.rnajournal.org/cgi/doi/10.1261/rna.067686.118.
REFERENCES
- Bird JG, Zhang Y, Tian Y, Panova N, Barvik I, Greene L, Liu M, Buckley B, Krásný L, Lee JK, et al. 2016. The mechanism of RNA 5′ capping with NAD+, NADH and desphospho-CoA. Nature 535: 444–447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cahová H, Winz ML, Höfer K, Nübel G, Jäschke A. 2015. NAD captureSeq indicates NAD as a bacterial cap for a subset of regulatory RNAs. Nature 519: 374–377. [DOI] [PubMed] [Google Scholar]
- Cantó C, Gerhart-Hines Z, Feige JN, Lagouge M, Noriega L, Milne JC, Elliott PJ, Puigserver P, Auwerx J. 2009. AMPK regulates energy expenditure by modulating NAD+ metabolism and SIRT1 activity. Nature 458: 1056–1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantó C, Jiang LQ, Deshmukh AS, Mataki C, Coste A, Lagouge M, Zierath JR, Auwerx J. 2010. Interdependence of AMPK and SIRT1 for metabolic adaptation to fasting and exercise in skeletal muscle. Cell Metab 11: 213–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantó C, Menzies KJ, Auwerx J. 2015. NAD+ metabolism and the control of energy homeostasis: a balancing act between mitochondria and the nucleus. Cell Metab 22: 31–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen D, Bruno J, Easlon E, Lin SJ, Cheng HL, Alt FW, Guarente L. 2008. Tissue-specific regulation of SIRT1 by calorie restriction. Genes Dev 22: 1753–1757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen YG, Kowtoniuk WE, Agarwal I, Shen Y, Liu DR. 2009. LC/MS analysis of cellular RNA reveals NAD-linked RNA. Nat Chem Biol 5: 879–881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coleman TM, Wang G, Huang F. 2004. Superior 5′ homogeneity of RNA from ATP-initiated transcription under the T7 ɸ2.5 promoter. Nucleic Acids Res 32: e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond DR, Armstrong J, Colman A. 1985. The effect of capping and polyadenylation on the stability, movement and translation of synthetic messenger RNAs in Xenopus oocytes. Nucleic Acids Res 13: 7375–7394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fulco M, Cen Y, Zhao P, Hoffman EP, McBurney MW, Sauve AA, Sartorelli V. 2008. Glucose restriction inhibits skeletal myoblast differentiation by activating SIRT1 through AMPK-mediated regulation of Nampt. Dev Cell 14: 661–673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuichi Y, LaFiandra A, Shatkin AJ. 1977. 5′-Terminal structure and mRNA stability. Nature 266: 235–239. [DOI] [PubMed] [Google Scholar]
- Grudzien-Nogalska E, Kiledjian M. 2017. New insights into decapping enzymes and selective mRNA decay. Wiley Interdiscip Rev RNA 8: e1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall JA, Dominy JE, Lee Y, Puigserver P. 2013. The sirtuin family's role in aging and age-associated pathologies. J Clin Invest 123: 973–979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong SM, Park CW, Kim SW, Nam YJ, Yu JH, Shin JH, Yun CH, Im SH, Kim KT, Sung YC, et al. 2016. NAMPT suppresses glucose deprivation-induced oxidative stress by increasing NADPH levels in breast cancer. Oncogene 35: 3544–3554. [DOI] [PubMed] [Google Scholar]
- Huang F. 2003. Efficient incorporation of CoA, NAD and FAD into RNA by in vitro transcription. Nucleic Acids Res 31: e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jäschke A, Höfer K, Nübel G, Frindert J. 2016. Cap-like structures in bacterial RNA and epitranscriptomic modification. Curr Opin Microbiol 30: 44–49. [DOI] [PubMed] [Google Scholar]
- Jiao X, Xiang S, Oh C, Martin CE, Tong L, Kiledjian M. 2010. Identification of a quality-control mechanism for mRNA 5′-end capping. Nature 467: 608–611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao X, Doamekpor SK, Bird JG, Nickels BE, Tong L, Hart RP, Kiledjian M. 2017. 5′ end nicotinamide adenine dinucleotide cap in human cells promotes RNA decay through DXO-mediated deNADding. Cell 168: 1015–1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Julius C, Yuzenkova Y. 2017. Bacterial RNA polymerase caps RNA with various cofactors and cell wall precursors. Nucleic Acids Res 45: 8282–8290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan JA, Tao X, Tong L. 2006. Molecular basis for the inhibition of human NMPRTase, a novel target for anticancer agents. Nat Struct Mol Biol 13: 582–588. [DOI] [PubMed] [Google Scholar]
- Kiledjian M. 2018. Eukaryotic RNA 5′-end NAD+ capping and DeNADding. Trends Cell Biol 28: 454–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Köhrer K, Domdey H. 1991. Preparation of high molecular weight RNA. Methods Enzymol 194: 398–405. [DOI] [PubMed] [Google Scholar]
- Konarska MM, Padgett RA, Sharp PA. 1984. Recognition of cap structure in splicing in vitro of mRNA precursors. Cell 38: 731–736. [DOI] [PubMed] [Google Scholar]
- Kowtoniuk WE, Shen Y, Heemstra JM, Agarwal I, Liu DR. 2009. A chemical screen for biological small molecule-RNA conjugates reveals CoA-linked RNA. Proc Natl Acad Sci 106: 7768–7773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin SJ, Ford E, Haigis M, Liszt G, Guarente L. 2004. Calorie restriction extends yeast life span by lowering the level of NADH. Genes Dev 18: 12–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu SW, Jiao X, Welch S, Kiledjian M. 2008. Analysis of mRNA decapping. Methods Enzymol 448: 3–21. [DOI] [PubMed] [Google Scholar]
- Malygin AG, Shemyakin MF. 1979. Adenosine, NAD and FAD can initiate template-dependent RNA synthesis catalyzed by Escherichia coli RNA polymerase. FEBS Lett 102: 51–54. [DOI] [PubMed] [Google Scholar]
- Muthukrishnan S, Both GW, Furuichi Y, Shatkin AJ. 1975. 5′-Terminal 7-methylguanosine in eukaryotic mRNA is required for translation. Nature 255: 33–37. [DOI] [PubMed] [Google Scholar]
- Neidhardt FC, Ingraham JL, Schaechter M. 1990. Physiology of the bacterial cell: a molecular approach. Sinauer Associates, Sunderland, MA. [Google Scholar]
- Palazzo AF, Lee ES. 2015. Non-coding RNA: what is functional and what is junk? Front Genet 6: 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raynes R, Pombier KM, Nguyen K, Brunquell J, Mendez JE, Westerheide SD. 2013. The SIRT1 modulators AROS and DBC1 regulate HSF1 activity and the heat shock response. PLoS One 8: e54364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwer B, Saha N, Mao X, Chen HW, Shuman S. 2000. Structure-function analysis of yeast mRNA cap methyltransferase and high-copy suppression of conditional mutants by AdoMet synthase and the ubiquitin conjugating enzyme Cdc34p. Genetics 155: 1561–1576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shuman S. 1995. Capping enzyme in eukaryotic mRNA synthesis. Prog Nucleic Acid Res Mol Biol 50: 101–129. [DOI] [PubMed] [Google Scholar]
- Sporty JL, Kabir MM, Turteltaub KW, Ognibene T, Lin SJ, Bench G. 2008. Single sample extraction protocol for the quantification of NAD and NADH redox states in Saccharomyces cerevisiae. J Sep Sci 31: 3202–3211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von der Haar T. 2008. A quantitative estimation of the global translational activity in logarithmically growing yeast cells. BMC Syst Biol 2: 87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vvedenskaya IO, Bird JG, Zhang Y, Zhang Y, Jiao X, Barvik I, Krasny L, Kiledjian M, Taylor DM, Ebright RH, et al. 2018. CapZyme-Seq comprehensively defines promoter-sequence determinants for RNA 5′ capping with NAD. Mol Cell 70: 553–564.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walters RW, Matheny T, Mizoue LS, Rao BS, Muhlrad D, Parker R. 2017. Identification of NAD+ capped mRNAs in Saccharomyces cerevisiae. Proc Natl Acad Sci 114: 480–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T, Zhang X, Bheda P, Revollo JR, Imai S, Wolberger C. 2006. Structure of Nampt/PBEF/visfatin, a mammalian NAD+ biosynthetic enzyme. Nat Struct Mol Biol 13: 661–662. [DOI] [PubMed] [Google Scholar]
- Winz ML, Cahová H, Nübel G, Frindert J, Höfer K, Jäschke A. 2017. Capture and sequencing of NAD-capped RNA sequences with NAD captureSeq. Nat Protoc 12: 122–149. [DOI] [PubMed] [Google Scholar]
- Xiang S, Cooper-Morgan A, Jiao X, Kiledjian M, Manley JL, Tong L. 2009. Structure and function of the 5′→3′ exoribonuclease Rat1 and its activating partner Rai1. Nature 458: 784–788. [DOI] [PMC free article] [PubMed] [Google Scholar]