Significance
Plant SUVH family H3K9 methyltransferases play a key role in connecting the two epigenetic silencing marks, DNA methylation and H3K9me2. However, the regulation of SUVH protein activities and their precise role in the regulation of DNA methylation remains unclear. In this research, we performed a comprehensive investigation into the structure, biochemistry, and in vivo targeting characteristics of SUVH histone methyltransferases. For binding methylated DNA, we reveal that the SUVH family proteins possess a unique thumb loop-dependent base-flipping mechanism. For methyltransferase function, we reveal that SUVH6 is regulated by a dynamic autoinhibitory domain. Finally, our in vitro DNA-binding assays combined with ChIP-seq data uncover mechanisms to help explain context-biased non-CG DNA methylation in plants.
Keywords: DNA methylation, H3K9me2, SUVH6, plant, histone methyltransferase
Abstract
DNA methylation functions in gene silencing and the maintenance of genome integrity. In plants, non-CG DNA methylation is linked through a self-reinforcing loop with histone 3 lysine 9 dimethylation (H3K9me2). The plant-specific SUPPRESSOR OF VARIEGATION 3–9 HOMOLOG (SUVH) family H3K9 methyltransferases (MTases) bind to DNA methylation marks and catalyze H3K9 methylation. Here, we analyzed the structure and function of Arabidopsis thaliana SUVH6 to understand how this class of enzyme maintains methylation patterns in the genome. We reveal that SUVH6 has a distinct 5-methyl-dC (5mC) base-flipping mechanism involving a thumb loop element. Autoinhibition of H3 substrate entry is regulated by a SET domain loop, and a conformational transition in the post-SET domain upon cofactor binding may control catalysis. In vitro DNA binding and in vivo ChIP-seq data reveal that the different SUVH family H3K9 MTases have distinct DNA binding preferences, targeting H3K9 methylation to sites with different methylated DNA sequences, explaining the context biased non-CG DNA methylation in plants.
DNA methylation is an epigenetic mark that functions in gene silencing, genome imprinting, and suppression of transposable element and repeat sequences (1, 2). In contrast to symmetric CG methylation, which predominates in mammals, plant DNA methylation occurs in all three sequence contexts: CG, CHG (H denotes A, T, or C), and CHH (3–5). All de novo DNA methylation in plants is established by DOMAINS REARRANGED METHYLASE 2 (DRM2), targeted by a plant-specific RNA-directed DNA methylation (RdDM) pathway (2, 6). However, maintenance of plant DNA methylation is carried out in a sequence context-specific manner (2, 7). CG and CHG methylation are mainly maintained by DNA METHYLTRANSFERASE 1 (MET1) and CHROMOMETHYLASE 3 (CMT3), respectively (8–10). CHH methylation is maintained by CMT2 in pericentromeric heterochromatin and DRM2/RdDM at some euchromatic sites (11, 12).
Maintenance of non-CG DNA methylation (CHG and CHH) in plants involves histone 3 lysine 9 dimethylation (H3K9me2), while CG methylation is independent of H3K9me2 (13, 14). CMT3 and CMT2 share a similar regulatory mechanism. Their bromo-adjacent homology (BAH) and chromo domains recognize H3K9me2-containing nucleosomes to target their DNA methyltransferase (MTase) domains to facilitate H3K9me2-directed DNA methylation (11, 15). The de novo RdDM pathway involves two steps: upstream RNA polymerase IV (Pol IV)-dependent siRNA biogenesis, and downstream Pol V-dependent long noncoding scaffold RNA biogenesis and the recruitment of downstream effectors such as DRM2 and chromatin remodelers (16, 17). The Pol IV binding protein SAWADEE HOMEODOMAIN HOMOLOG 1 (SHH1) can recognize H3K9me2 through its SAWADEE domain and further recruit Pol IV to H3K9me2 loci, resulting in the targeting of RdDM by H3K9me2 (18, 19). Altogether, non-CG DNA methylation in plants is targeted to the H3K9me2 mark directly through CMT3 and CMT2 or indirectly through SHH1 (13).
Methylated DNA (mDNA) can target H3K9 methylation via a feedback mechanism involving the plant-specific SUPPRESSOR OF VARIEGATION 3–9 HOMOLOG (SUVH) family H3K9 MTases KRYPTONITE (KYP, also known as SUVH4), SUVH5, and SUVH6 redundantly (20–26). SUVH proteins possess a SET and RING finger-associated (SRA) domain that recognizes mDNA and methylates H3K9 using the pre-SET/SET/post-SET catalytic cassette (20, 25). In this way, non-CG DNA methylation and H3K9 methylation form a self-reinforcing feedback loop that maintains epigenetic silencing in plants (2, 13). In genomewide studies, a kyp suvh5 suvh6 triple mutant, which eliminates H3K9 methylation genomewide, exhibits a global loss of non-CG methylation (14). Reciprocally, a drm1 drm2 cmt3 cmt2 quadruple mutant that eliminates non-CG methylation shows a strong genomewide loss of the H3K9me2 mark (11). CHG methylation is context biased in that CCG methylation levels are significantly lower than CWG (W denotes A or T) methylation levels, which was hypothesized to be attributed to the different mDNA binding abilities of different SUVH proteins (3, 27–30).
Previously, we have analyzed the pathways linking plant DNA methylation and H3K9me2 through structural and functional studies of CMT3, SHH1, and KYP (15, 18, 20). Here, we investigated the structure of the SUVH family H3K9 MTase SUVH6 in complexes with cofactor and mDNA. Comparison of the various complexes revealed that the plant-specific SUVH family MTases exhibit a distinct 5-methyl-dC (5mC) base-flipping mechanism that relies on the thumb loop, in contrast to 5mC base flipping by the mammalian UHRF1 SRA domain, which requires the NKR finger loop. A SUVH6 SET domain protrusion can interact with the two-helix bundle of SUVH6 to inhibit H3 substrate binding, forming part of an autoinhibitory mechanism. The post-SET domain has a disordered conformation in the absence of cofactor, which adopts a fixed conformation upon cofactor binding, a transition that is essential for catalysis. Furthermore, we have shown that different SUVH proteins possess different in vitro mDNA binding and in vivo targeting preferences, revealing a biochemical basis for context-biased non-CG methylation (27).
Results
Structure of SUVH6 in the Absence of DNA.
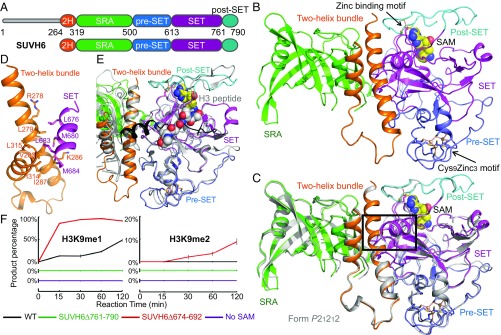
Previously, we have reported the structures of KYP in complex with methylated CHH or CHG (mCHH or mCHG) and the substrate H3 peptide. These results revealed how the binding of mDNA is coupled to deposition of H3K9me2 by the SUVH family histone MTase (20). However, the absence of structures in other functional or regulatory states continues to limit our understanding of the SUVH family histone MTases. Therefore, we chose to study the structure of the SUVH family MTase SUVH6. We used a construct possessing all of the functional domains of SUVH6, including the two-helix bundle, SRA, and the pre-SET/SET/post-SET domains (Fig. 1A). Crystal structures of SUVH6 in complex with the cofactor S-adenosyl methionine (SAM) in two different space groups were determined to 1.9-Å (space group: P21212) and 2.3-Å (space group: P21) resolution, respectively (Fig. 1 B and C and SI Appendix, Table S1). The two crystal forms adopt highly similar structures, with a superimposition root-mean-square deviation (rmsd) of 0.74 Å (Fig. 1C). We will focus on the P21 structure (Fig. 1C), which has fewer disordered regions, in the following discussion of the SUVH6–SAM complex.
Fig. 1.
Overall structure of the DNA-free form of SUVH6. (A) Schematic representation of the domain architecture of SUVH6 (Upper) and the construct used for structural studies in this work (Lower). 2H, two-helix bundle. (B) Overall structure of the SUVH6–SAM complex in form 2 (in space group P21). The two-helix bundle, SRA, pre-SET, SET, and post-SET domains are colored in orange, green, slate, magenta, and cyan, respectively. The SAM is shown in a space-filling representation. The Cys9Zn3 cluster (Lower Right) and the zinc binding motif between SET and post-SET domains (Upper Right) are shown in ball-and-stick representation and are indicated by arrows. (C) Superimposition of SUVH6–SAM complex form 1 (silver, in space group P21212) and form 2 (color scheme as in B, in space group P21) shows almost identical structures, with form 1 being more disordered. Black box highlights the protruding that extends out from the SET domain and interacts with the two-helix bundle. (D) The SET domain protruding loop segment and the two-helix bundle interact through a hydrophobic interface. Interacting residues are represented as stick models. (E) Superimposition of the pre-SET/SET/post-SET domain cassette of SUVH6–SAM complex (color scheme as in B) and KYP–mCHH–SAH-H3 complex (silver). The autoinhibitory SET domain protruding loop is highlighted in black. The H3 peptide is shown in space-filling model. There is significant steric conflict between the SUVH6 autoinhibitory loop and the substrate H3 peptide. (F) Quantified H3K9 methylation assay of SUVH6 (in black), SUVH6 SET domain protrusion deletion construct (SUVH6Δ674–692, in red), and SUVH6 post-SET domain deletion construct (SUVH6Δ761–790, in green), and the condition without cofactor SAM (in magenta). The percentage of the product H3K9me1 (Left) and H3K9me2 (Right) are plotted as means ± SD (n = 3). Reactions were stopped after incubation at the times indicated.
Overall, the structure of SUVH6 resembles those of SUVH9 and KYP (20, 31). The N-terminal two-helix bundle is positioned in the middle of the structure with the SRA domain on one side and the pre-SET/SET/post-SET cassette on the other (Fig. 1B). The SRA domain has a binding cleft that may bind methylated DNA (Fig. 1B). The pre-SET domain has a Cys9Zn3 motif and a SAM molecule is bound between the SET and post-SET domains (Fig. 1B). Three cysteine residues of the post-SET domain and one cysteine residue of the SET domain coordinate a Zn2+ ion and further captures the flexible loop-rich post-SET domain in a fixed conformation (Fig. 1B). The post-SET domain buries the SAM molecule and stabilizes it in the bound state. An extended fragment of the SET domain (residues 668–701, highlighted in Fig. 1C) protrudes from the SET domain. The tip of the protrusion adopts a helical conformation and forms extensive hydrophobic interactions with the two-helix bundle (Fig. 1D), resembling the inactive form of the SUVH9 structure but not the active form KYP–mDNA complex structure (20, 31). The protrusion blocks the histone substrate binding site on SUVH6 (Fig. 1E), therefore representing an autoinhibited form of the enzyme in the substrate-free state. The SET domain protrusion exists in the primary sequences of all other SUVH proteins, except SUVH5 (SI Appendix, Fig. S1), suggesting SUVH5 may have a different regulation mechanism. We performed a mass spectrum-based in vitro histone MTase assay based on a previous protocol (32). The replacement of this autoinhibition loop by a GSGS linker (SUVH6Δ674–692), which is hydrophilic and flexible in nature and can disrupt the loop's interaction with the two-helix bundle mimicking the autoinhibition released conformation, significantly stimulates the activity of SUVH6 (Fig. 1F). We speculate that SUVH6 may exist in a chemical equilibrium between autoinhibited and released conformations with an overall lower MTase activity. Interacting proteins, or other factors, may assist SUVH6 in releasing autoinhibition to modulate activity in vivo. Here we captured the autoinhibited state of SUVH6. In contrast, our previous KYP structures trapped the released conformation, with the autoinhibition loop disordered, allowing us to obtain the substrate H3 peptide-containing complex (20).
Structure of SUVH6 in Complex with a 13-bp mCHG DNA.
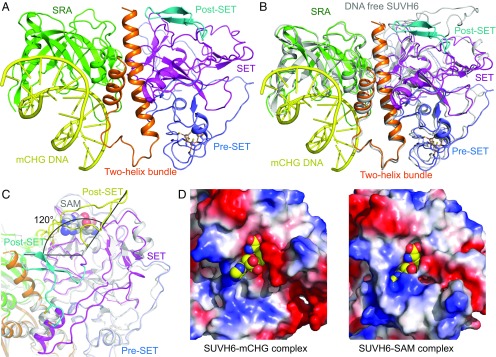
We further obtained the crystal structure of SUVH6 in complex with a 13-bp mCHG DNA with 5′ G/C overhangs (Fig. 2A and SI Appendix, Tables S1 and S2). The DNA duplex is captured by the DNA binding groove of the SRA domain, with additional interactions with a two-helix bundle (Fig. 2A and SI Appendix, Fig. S2A), which is similar to that observed in the KYP–mDNA complex structures (20). As in the KYP complex, we observed a 1:1 complex between SUVH6 and mCHG, in contrast to the 2:1 complexes between the isolated SRA domain of SUVH5 and mDNA (20, 33, 34).
Fig. 2.
Structure of the SUVH6–13-bp mCHG DNA complex. (A) Overall structure of the SUVH6–mCHG DNA complex (ribbon diagram, color code as in Fig. 1B). The mCHG DNA is shown in a yellow ribbon representation. (B) Superimposition of SRA domains of the SUVH6–mCHG DNA complex (color scheme as in A) and SUVH6–SAM complex (silver) indicating an overall similar structure, showing small conformational movements toward the mDNA upon DNA binding. (C) Superimposition of the pre-SET and SET domains of the SUVH6–mCHG DNA complex (color code as in A) and the SUVH6–SAM complex (silver, with the post-SET domain highlighted in yellow). The angle representation shows the post-SET domain undergoes a 120° conformational change. The cysteine residues that coordinate the Zn2+ between SET and post-SET domains are highlighted in stick representation. An enlarged view can be found in SI Appendix, Fig. S2B. (D) Comparison of the cofactor binding pocket of SUVH6 in the mCHG binding state (Left) and SAM state (Right). Left shows a model SAM molecule (in space-filling representation) positioned in an open pocket of the SUVH6–mCHG complex, and Right shows SAM in a closed pocket in the SUVH6–SAM complex, indicating that the cofactor binding pocket is dynamic and undergoes a significant conformational change.
Superimposition of the SRA domains of the DNA-bound and DNA-free forms of SUVH6 revealed that the two-helix bundle and part of the pre-SET/SET/post-SET cassette undergo a small movement toward the bound DNA (Fig. 2B), with DNA binding likely inducing this more compact conformation. We observed a substantial conformational change of the post-SET domain, a rotation of ∼120° with disruption of the zinc coordination between the post-SET and SET domains, compared with the SUVH6–SAM complex structure (Fig. 2C and SI Appendix, Fig. S2B). The closed SAM binding pocket seen in the SUVH6–SAM complex is open in the SAM-free complex (Fig. 2D). The C-terminal fragment of the post-SET domain (residues 774–480) in the SUVH6–mCHG complex contributes to crystal packing. Therefore, we speculate a plausible model in which the post-SET domain moves during cofactor binding and release. In the absence of SAM, the post-SET domain is disordered. The binding of SAM may induce the post-SET domain to both interact with and cover SAM, as well as facilitate formation of the zinc coordination between post-SET and SET domains, which locks the relative positioning between the two domains and stabilizes cofactor binding to promote the MTase reaction. In vitro assays on a post-SET deletion construct of SUVH6Δ761–790 showed a complete loss of activity (Fig. 1F), supporting the idea that the post-SET domain is essential for activity, because it is required for SAM binding.
The Recognition of mCHG DNA by SUVH6.
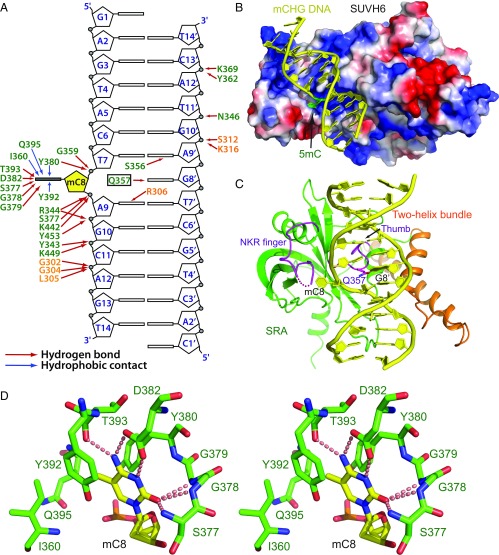
In our SUVH6–mCHG complex (Fig. 2A), mCHG DNA is bound principally by the SRA domain, in a manner similar to previously reported SRA–mDNA complexes (20, 33, 35–37). Overall, the interaction between SUVH6 and mDNA is hydrophilic in nature, with the majority of the specific interactions located near the 5mC flanking region (Fig. 3 A and B). The phosphate groups of mC8, A9, G10, and C11 of the methylated strand form extensive hydrogen bonding interactions with SRA domain residues, while the phosphate group of A12 forms hydrogen bonds with three two-helix bundle residues (Fig. 3A). The backbone phosphates of G10′, T11′, and C13′ of the unmethylated strand make hydrogen bonding interactions with the two-helix segment and SRA domain residues, respectively (Fig. 3A). There are few direct interactions involving in DNA bases. Only the bases of mC8, the orphaned G8′, A9 and A9′ form hydrogen bonds with SUVH6 (Fig. 3A).
Fig. 3.
Molecular basis for the recognition of mCHG DNA by SUVH6. (A) Schematic showing details of the interaction between SUVH6 and mCHG DNA. The residues of the two-helix bundle and the SRA domain are colored in orange and green, respectively. Hydrogen bonds and hydrophobic contacts are shown in red and blue arrows, respectively. (B) Electrostatics surface representation of SUVH6 with the mCHG DNA in ribbon representation showing that the DNA binds in a positively charged surface groove. The mC8 residue is highlighted in green and inserts into a small pocket of the SRA domain. (C) Structural mechanism for the flipping out of 5mC8 by SUVH6. The two-helix bundle, the SRA, and DNA are colored in orange, green, and yellow, respectively. The thumb loop and NKR finger loop are highlighted in magenta. Gln357 of the thumb loop occupies the gap left by the flipped-out 5mC8 and forms hydrogen bonds with the orphaned G8′. The NKR finger loop is partially disordered as shown by a dashed line. (D) Stereoview of the details of the specific recognition of the flipped-out 5mC base. Hydrogen bonds are highlighted by dashed red lines.
The 5mC is flipped out from the DNA duplex and inserted into a deep pocket in the SRA domain (Fig. 3B). The thumb loop of the SRA domain inserts into the DNA duplex through the minor groove, with the side chain of Gln357 occupying the gap formed by the flipped-out 5mC, and forming a hydrogen bond with the base of orphaned guanine G8′, which maintains its original B-form conformation (Fig. 3C and SI Appendix, Fig. S2C). By contrast, the mammalian UHRF1 SRA domain uses a thumb loop valine and an NKR finger loop arginine to occupy, from opposite sides, the gap generated by the extruded 5mC, with the NKR loop arginine residue forming hydrogen bonds with the Hoogsteen edge of the orphaned guanine (35–37) (SI Appendix, Fig. S2D). However, in the SUVH6–mCHG complex, only the thumb loop occupies the gap formed by the flipped-out 5mC. The NKR finger loop, which is partially disordered, does not seem to contribute to base flipping (Fig. 3C and SI Appendix, Fig. S2C). In the SUVH5 SRA–mDNA structure, the side chain of thumb loop residue Gln392 occupies the gap left by the flipped out 5mC and is also essential for base flipping (33) (SI Appendix, Fig. S2E). In KYP, although both thumb loop residue Leu176 and NKR finger residue Leu227 insert into the base-flipped gap (SI Appendix, Fig. S2F), our published gel-shift assay showed that only the L176G mutation, and not the Leu227G mutation, disrupted the KYP–mCHH binding, indicating that the thumb loop of KYP is critical for flipping out and stabilizing 5mC (20). Thus, although SRA domains display a generally similar mode of mDNA recognition, the thumb loop of SUVH proteins plays a more important role in flipping out of 5mC, while the NKR finger loop is more important in flipping out of 5mC for UHRF1.
Similar to other SRA–mDNA complexes, the 5mC base is clamped between Tyr392 and Tyr380, involving hydrophobic and stacking interactions (Fig. 3D). The methyl group is accommodated within a small hydrophobic pocket formed by Tyr392, Tyr380, Ile360, and Cβ of Gln395, with the Watson–Crick edge of 5mC fully hydrogen bonded to surrounding residues (Fig. 3D).
SUVH Family H3K9 MTases Have Distinct mDNA Binding Preferences.
There is a clear bias in the sequence context for different non-CG methylation marks in plants, which has been attributed to the methylated DNA binding preferences of the SUVH family proteins (3, 27–29, 38). In Arabidopsis, the CCG methylation level is 20–50% lower than CWG methylation, with kyp mutants showing a much more significant decrease in CWG methylation than CCG (27). It has been suggested that SUVH5 and SUVH6 act redundantly to maintain CCG methylation, while KYP functions principally in binding and maintaining CWG methylation (27). Experiments analyzing binding of the three SUVH proteins to arrays of methylated DNA sequences have either not been quantitatively analyzed or used only the isolated SRA domain (20, 25, 33). To generate a comprehensive comparison of binding activity, we measured the in vitro binding between KYP/SUVH5/6 and different mDNAs using microscale thermophoresis (MST). CHG DNA can be classified into CWG and CCG forms. CCG methylation is both MET1 and CMT3 dependent (27, 38). Following DNA replication, the MET1/DNMT1 family CG maintenance MTase carries out CG methylation immediately, with the fully methylated 5′CCG3′/3′GGC5′ (C means 5mC, hereinafter) yielding two different replication products that can be potentially recognized by SUVH family MTases, CCG/GGC and CCG/GGC (Fig. 4A). Thus, we tested four different DNA sequence contexts: CAT/GTA, CAG/GTC, CCG/GGC, and CCG/GGC, with a DNA containing unmethylated CG, CHG, and CHH sites as control.
Fig. 4.
The three SUVH proteins possess different mDNA binding preferences. (A) Schematic model of CHG methylated DNA during replication. CAG/GTC DNA generates CAG/GTC and CAG/GTC replication products which can be directly recognized by SUVH proteins (Left). CCG/GGC DNA could yield CCG/GGC and CCG/GGC replication products, which are methylated by MET1 to generate CG methylation and then serve as binding targets for the SUVH protein (Right). (B–D) Binding curves between different methylated DNAs and KYP (B), SUVH5 (C), and SUVH6 (D) measured by MST. Data points are shown as means ± SD (n = 3). Curves indicate the calculated fits according to the data points, and the binding affinities (Kd) are listed in the boxes below the binding curves. DNA sequences can be found in SI Appendix, Table S2. C marked in red in the sequence indicates 5mC. NDB, no detectable binding.
KYP exhibits a strong preference for CAG/GTC DNA (Fig. 4B), consistent with the significant decrease of CWG methylation seen in kyp (27). In agreement with previous computational results (39), KYP exhibits relatively lower affinity toward CCG/GGC DNA (Fig. 4B), suggesting that KYP plays a less important role in CCG methylation-mediated H3K9 methylation. KYP recognized CAT/GTA and CCG/GGC sequences with high affinity, although these were marginally weaker than its affinity for CAG/GTC (Fig. 4B). As CCG/GGC can be considered as a combination of a low-affinity fully CG methylation CCG/GGC binding site and a high-affinity hemimethylated CHG CCG/GGC binding site, we propose that KYP does not prefer to bind symmetric fully methylated CG/GC, nor does CG/GC methylation block KYP binding. To validate this idea, we tested KYP binding to a TCG/AGC hemimethylated CG DNA and a TCG/AGC fully methylated CG DNA. Hemimethylated TCG/AGC DNA showed strong binding and the fully methylated TCG/AGC DNA showed a substantially decreased binding affinity (Fig. 4B).
SUVH5 bound most weakly to CAT/GTA mDNA (Fig. 4C), suggesting that SUVH5 recruitment may only be mildly influenced by mCHH. SUVH5 displayed a preference for CCG mDNA binding, CCG/GGC and CCG/GGC (Fig. 4C). The CAG/GTC site had an intermediate binding affinity (Fig. 4C). We further detected the binding between SUVH5 and the CG methylate sites TCG/AGC and TCG/AGC. Consistently, SUVH5 possessed strong binding to fully methylated CG DNA and slightly weaker binding to hemimethylated CG (Fig. 4C), in agreement with previous isothermal titration calorimetric (ITC)-based binding using the isolated SUVH5 SRA domain (33). These data suggest that the SUVH5 prefers binding to CG methylation-containing CCG mDNA sites, supporting the previous hypothesis that SUVH5, and not KYP, plays a key role in CCG methylation maintenance (27). SUVH6 shows strong and almost identical binding to all tested methylated DNA oligonucleotides (Fig. 4D), suggesting that SUVH6 may function redundantly with both KYP and SUVH5 in all of the sequence contexts and without sequence specificity. KYP, SUVH5, and SUVH6 all showed nondetectable or significantly decreased binding toward unmethylated DNA, consistent with their methylated DNA binding function (Fig. 4 B–D).
H3K9me2 and mC Interdependence of the SUVH MTases in Vivo.
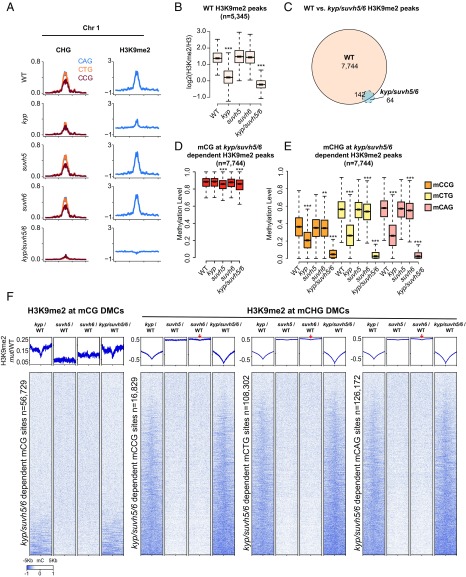
To determine the in vivo relevance of the in vitro binding preferences, we profiled levels of H3K9me2 by chromatin immunoprecipitation coupled to high throughput sequencing (ChIP-seq) in wild type (WT) and kyp, suvh5, suvh6, and kyp/suvh5/6 mutant backgrounds. Parsing the genome into 100-kb bins, we noted that average H3K9me2 levels were reduced and largely eliminated in the kyp and kyp/suvh5/6 mutant backgrounds, respectively (Fig. 5 A and B). In contrast, H3K9me2 levels were largely unaffected in suvh5 and suvh6 single mutants, consistent with the DNA methylation defects observed in these mutants (14, 27), suggesting that suvh5 and suvh6 act redundantly to maintain H3K9me2 levels genomewide (SI Appendix, Fig. S3). To assess whether there are more localized changes in the single mutants, we defined KYP/SUVH5/6-dependent H3K9me2 peaks and plotted levels of DNA methylation in the CG and the three CHG trinucleotide contexts (CCG, CAG, and CTG) (Fig. 5 C–F). In, suvh5, but not kyp or suvh6, we observed small but significant reductions in mCG (Fig. 5D), consistent with the mCG SUVH5 binding preference in vitro (Fig. 4C). In kyp, while there were significant reductions in all CHG contexts, we observed 54% and 55% reductions in median methylation levels for CTC and CAG contexts, respectively, and only a 45% reduction in CCG (Fig. 5E and SI Appendix, Tables S3 and S4). This is consistent with KYP’s lower in vitro binding preference for methylated CCG (Fig. 4B). For suvh6, while there was no effect on CG methylation (Fig. 5D), there was a small but significant reduction in all three CHG contexts (Fig. 5E). Reciprocally, when we mapped kyp/suvh5/6 differentially methylated cytosines (DMCs) and plotted the difference in H3K9me2 levels in mutants vs. wild type, we also observed small reductions in H3K9me2 in suvh6 in all three CHG contexts (Fig. 5F). This is consistent with the ability of SUVH6 to bind methylation equally well in all context (Fig. 4D), while the lack of any effect at CG sites (Fig. 5 D–F) suggests that MET1 and SUVH5 are sufficient to maintain mCG at H3K9me2 sites in vivo (11, 38).
Fig. 5.
Interdependent H3K9me2 and DNA methylation defects in kyp, suvh5, suvh6, and kyp/suvh5/6. (A) Chromosomal distribution of CHG (mCAG, mCGG, and mCTG) methylation and log2 ratio of H3K9me2/H3 in WT, kyp, suvh5, suvh6, and kyp/suvh5/6 in chromosome 1 (bin = 100 kb). (B) Boxplot of log2 ratio of H3K9me2/H3 levels, at WT defined H3K9me2/H3 peaks (using callpeak function in MACS2), in WT, kyp, suvh5, suvh6, and kyp/suvh5/6. ***P value <2.2e−16, **P value <1e−8 with Student’s t test. (C) H3K9me2/H3 peak overlap defined by comparison between WT and kyp/suvh5/6 (using bdgdiff in MACS2). (D) Boxplot for CG methylation levels in WT, kyp, suvh5, suvh6, and kyp/suvh5/6 at kyp/suvh5/6-dependent H3K9me2/H3 peaks [defined by H3K9me2 peaks called in WT but not in kyp/suvh5/6 (n = 7,744), see C]. ***P value <2.2e−16, **P value <1e−8 with Student’s t test. (E) Boxplot for CHG methylation (mCCG, mCTG, and mCAG) levels of kyp/suvh5/6-dependent H3K9me2/H3 peaks in WT, kyp, suvh5, suvh6, and kyp/suvh5/6. ***P value <2.2e−16, **P value <1e−8 with Student’s t test. (F) Heatmap of normalized H3K9me2 levels in kyp/WT, suvh5/WT, suvh6/WT, and kyp/suvh5/6/WT plotted over kyp/suvh5/6-dependent CG and CHG sites (mCCG, mCTG, and mCAG). Red arrows indicate H3K9me2 depletion in suvh6 centered on DMCs.
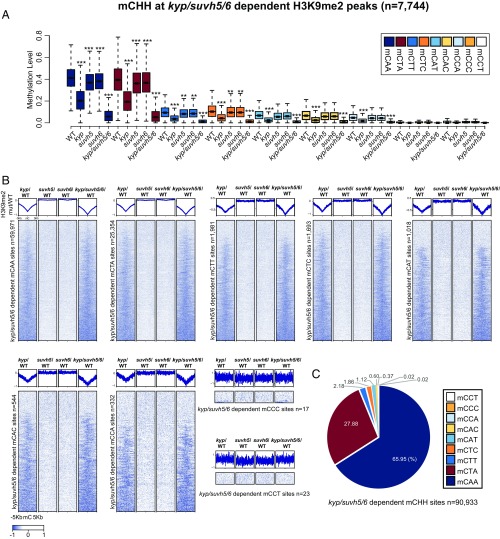
We repeated this analysis for CHH, examining DNA methylation for the nine different CHH trinucletide contexts at KYP/SUVH5/6-dependent H3K9me2 peaks, and reciprocally, H3K9me2 levels at the corresponding DMCs (Fig. 6). We observed reductions in DNA methylation and H3K9me2 primarily in kyp, but also observed small but significant reductions in DNA methylation in both suvh5 and suvh6 in the two most prominent CHH contexts (CAA and CTA), which account for >92% of all kyp/suvh5/6 CHH DMCs. Thus, in contrast to CG and CHG sites, SUVH5 and SUVH6 appear to have similar sequence specificities at CHH sites as KYP, and therefore may serve principally as a redundant back-up system for KYP.
Fig. 6.
Interdependence of H3K9me2 and CHH methylation defects in kyp, suvh5, suvh6, and kyp/suvh5/6. (A) CHH methylation (mCAA, mCTA, mCTT, mCTC, mCAT, mCAC, mCCA, mCCC, and mCCT) levels at kyp/suvh5/6-dependent H3K9me2/H3 peaks in WT, kyp, suvh5, suvh6, and kyp/suvh5/6. ***P value <2.2e−16, **P value <1e−8 with Student’s t test. (B) Heatmap of normalized H3K9me2 levels in kyp/WT, suvh5/WT, suvh6/WT, and kyp/suvh5/6/WT plotted over kyp/suvh5/6-dependent CHH sites (mCAA, mCTA, mCTT, mCTC, mCAT, mCAC, mCCA, mCCC, and mCCT). (C) Proportions of CHH sites (mCAA, mCTA, mCTT, mCTC, mCAT, mCAC, mCCA, mCCC, and mCCT) in kyp/suvh5/6-dependent mCHH sites.
Discussion
CG methylation patterns are replicated in a semiconservative manner and are maintained through binding of the hemimethylated product by UHRF1/VIM1 and the DNA MTase DNMT1/MET1 (40). In contrast, non-CG methylation in plants cannot use the same maintenance strategy. Instead, plants employ the chromatin H3K9me2 as an intermediate in a self-reinforcing loop between H3K9me2 and non-CG methylation for the faithful maintenance of non-CG methylation. The SUVH family H3K9 MTases KYP and SUVH5/6 function in the mDNA-directed H3K9 methylation that forms the self-reinforcing loop, together with H3K9me2-directed DNA methylation by CMT3, CMT2, and RdDM (13).
Our structural data support the idea that SUVH family MTases connect SRA domain binding of mDNA to H3K9 methylation by the pre-SET/SET/post-SET cassette without the involvement of allosteric regulation, as we saw little conformational change upon DNA binding, which was further confirmed by our in vitro activity assay of SUVH6 in both the DNA-free and DNA existence conditions (SI Appendix, Fig. S4). The SET domain histone MTases share a similar reaction mechanism. Although the structures of several H3K9 MTases have been reported, the detailed mechanism of the catalytic process, especially the mechanism of the plant-specific SUVH family H3K9 MTases, is still not fully understood. The cofactor-free form of MTase we observe in our SUVH6–13-bp mCHG complex suggests that release of the cofactor results in disruption of the zinc binding coordination between post-SET and SET domains and that cofactor binding fixes the conformation of post-SET domain (Fig. 2C). This, in turn, explains why SUVH2 and SUVH9 are inactive, as they do not have the post-SET domain and therefore cannot bind cofactor (31, 41). We also observed an autoinhibition loop protruding from the SET domain, which is able to interact with the two-helix bundle through extensive hydrophobic interactions (Fig. 1 C and D). The loop occupies the peptide entrance channel to the catalytic center, suggesting a potential regulatory mechanism for controlling enzymatic activity by regulating the autoinhibitory loop.
CHG DNA methylation shows a sequence context bias in the methylomes of Arabidopsis, rice, and tomato (27) and this has been suggested to be a consequence of the high efficiency with which KYP recognizes CWG and its low efficiency for CCG methylation (27). Here, we provide biochemical support for this hypothesis, and further elaborate upon it. KYP, the dominant H3K9 MTase in Arabidopsis, binds with high affinity to CAG/GTC and CCG/GGC, but with lower affinity to CCG/GGC sites (Fig. 4D), which is half the replication product of CCG/GGC mDNA (Fig. 4A). SUVH5 also binds to CAG/GTC and CCG/GGC, being redundant with KYP, but it can additionally bind with high affinity to CCG/GGC, which KYP cannot (Fig. 4C). Since KYP levels have a greater impact on H3K9 methylation than SUVH5, the complementarily between KYP and SUVH5 for CHG mDNA ensures they target all mCHG subcontexts, while at the same time their redundant binding of CWG methylation sites likely explains the higher genomewide levels of CWG mDNA, compared with CCG mDNA in WT plants.
We observed that the kyp/suvh5/6 triple mutant was clearly stronger than kyp alone, and yet the single suvh5 and suvh6 mutants had only weak individual effects (Figs. 4 and 5). This is consistent with the redundant activities of SUVH5 and SUVH6, but it is not clear whether both are required to maintain the methylation that is kyp independent. It will therefore be important in future work to dissect the pairwise redundancies between KYP, SUVH5, and SUVH6. We have previously shown that many CMT2-dependent CHH sites, characterized by dense H3K9me2, require either CMT3/KYP or MET1 for methylation maintenance (42), implying that SUVH5 and/or SUVH6 may be required to link MET1-dependent CG methylation to H3K9me2. Here, we provide support for this hypothesis, showing that SUVH5 binds strongly to mCG (Fig. 4C) and that suvh5 mutants display mCG methylation defects (Fig. 5D). The in vitro assays revealed that SUVH6 binds the various tested mDNA with almost equal affinity, implying that SUVH6 functions redundantly with KYP and SUVH5 to maintain overall methylation levels, but not in specific sequence contexts, and our in vivo ChIP-seq analysis data from single and triple suvh mutants further confirmed this. However, the mDNA binding preferences represent only one aspect governing SUVH function; other factors, including in vivo expression level, enzymatic activity, binding partners, protein localization, and the likely autoinhibition regulation uncovered here, may also contribute to the regulation of KYP/SUVH5/6 impact in vivo. Indeed, it was reported that the SUVH5 prefers to target transposon sequences, while SUVH6 prefers to target transcribed inverted repeats (21), indicating there are factors in addition to DNA methylation state that likely impact chromatin targeting of SUVH proteins. Although the in vitro mDNA binding affinity of SUVH5 is lower than KYP and SUVH6, for a given chromatin region, SUVH5 will prefer to bind to mCCG over mCWG, while KYP will prefer to bind to mCWG over mCCG. This insight can help explain the overall sequence-biased non-CG methylation patterns observed. Additional studies are required to provide a holistic understanding of the precise mechanisms governing regulation of the different SUVH proteins in different tissues and chromatin contexts.
Materials and Methods
Details are provided in SI Appendix, SI Materials and Methods, including protein expression and purification, crystallization and data collection, structure determination, MST-based binding assay, in vitro histone MTase assay, plant materials, ChIP-seq, bisulfite-seq analysis, and ChIP-seq analysis.
Supplementary Material
Acknowledgments
We thank the staffs from beamlines BL17U1 and BL19U1 of the National Center for Protein Sciences Shanghai at the Shanghai Synchrotron Radiation Facility for their assistance during data collection, and the Proteomic Core Facility of the Shanghai Center for Plant Stress Biology for assistance in the histone methylation assay. This work was supported by the National Key R&D Program of China (Grant 2016YFA0503200 to J.D.), the National Natural Science Foundation of China (Grants 31622032 and 31770782 to J.D.), the Chinese Academy of Sciences (J.D.), the Guangdong Innovation Research Team Fund (Grant 2016ZT06S172 to S.L.), and NIH Grant GM60398 (to S.E.J.). C.J.H. was supported by a European Molecular Biology Organization long-term fellowship (Award ALTF 1138-2014). Z.Z. is supported by a scholarship from the Chinese Scholarship Council.
Footnotes
Conflict of interest statement: S.E.J. and Roger B. Deal were coauthors on a 2016 paper. They did not have any direct research collaboration on this work.
Data deposition: X-ray structures have been deposited in the RCSB Protein Data Bank with the accession codes: 6A5K, 6A5M, and 6A5N. The high throughput sequencing data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE114009).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1809841115/-/DCSupplemental.
References
- 1.Goll MG, Bestor TH. Eukaryotic cytosine methyltransferases. Annu Rev Biochem. 2005;74:481–514. doi: 10.1146/annurev.biochem.74.010904.153721. [DOI] [PubMed] [Google Scholar]
- 2.Law JA, Jacobsen SE. Establishing, maintaining and modifying DNA methylation patterns in plants and animals. Nat Rev Genet. 2010;11:204–220. doi: 10.1038/nrg2719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cokus SJ, et al. Shotgun bisulphite sequencing of the Arabidopsis genome reveals DNA methylation patterning. Nature. 2008;452:215–219. doi: 10.1038/nature06745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lister R, et al. Highly integrated single-base resolution maps of the epigenome in Arabidopsis. Cell. 2008;133:523–536. doi: 10.1016/j.cell.2008.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang X, et al. Genome-wide high-resolution mapping and functional analysis of DNA methylation in Arabidopsis. Cell. 2006;126:1189–1201. doi: 10.1016/j.cell.2006.08.003. [DOI] [PubMed] [Google Scholar]
- 6.Cao X, Jacobsen SE. Role of the Arabidopsis DRM methyltransferases in de novo DNA methylation and gene silencing. Curr Biol. 2002;12:1138–1144. doi: 10.1016/s0960-9822(02)00925-9. [DOI] [PubMed] [Google Scholar]
- 7.Du J. Structure and mechanism of plant DNA methyltransferases. Adv Exp Med Biol. 2016;945:173–192. doi: 10.1007/978-3-319-43624-1_8. [DOI] [PubMed] [Google Scholar]
- 8.Bartee L, Malagnac F, Bender J. Arabidopsis cmt3 chromomethylase mutations block non-CG methylation and silencing of an endogenous gene. Genes Dev. 2001;15:1753–1758. doi: 10.1101/gad.905701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Finnegan EJ, Peacock WJ, Dennis ES. Reduced DNA methylation in Arabidopsis thaliana results in abnormal plant development. Proc Natl Acad Sci USA. 1996;93:8449–8454. doi: 10.1073/pnas.93.16.8449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lindroth AM, et al. Requirement of CHROMOMETHYLASE3 for maintenance of CpXpG methylation. Science. 2001;292:2077–2080. doi: 10.1126/science.1059745. [DOI] [PubMed] [Google Scholar]
- 11.Stroud H, et al. Non-CG methylation patterns shape the epigenetic landscape in Arabidopsis. Nat Struct Mol Biol. 2014;21:64–72. doi: 10.1038/nsmb.2735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zemach A, et al. The Arabidopsis nucleosome remodeler DDM1 allows DNA methyltransferases to access H1-containing heterochromatin. Cell. 2013;153:193–205. doi: 10.1016/j.cell.2013.02.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Du J, Johnson LM, Jacobsen SE, Patel DJ. DNA methylation pathways and their crosstalk with histone methylation. Nat Rev Mol Cell Biol. 2015;16:519–532. doi: 10.1038/nrm4043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stroud H, Greenberg MV, Feng S, Bernatavichute YV, Jacobsen SE. Comprehensive analysis of silencing mutants reveals complex regulation of the Arabidopsis methylome. Cell. 2013;152:352–364. doi: 10.1016/j.cell.2012.10.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Du J, et al. Dual binding of chromomethylase domains to H3K9me2-containing nucleosomes directs DNA methylation in plants. Cell. 2012;151:167–180. doi: 10.1016/j.cell.2012.07.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.He XJ, Ma ZY, Liu ZW. Non-coding RNA transcription and RNA-directed DNA methylation in Arabidopsis. Mol Plant. 2014;7:1406–1414. doi: 10.1093/mp/ssu075. [DOI] [PubMed] [Google Scholar]
- 17.Matzke MA, Mosher RA. RNA-directed DNA methylation: An epigenetic pathway of increasing complexity. Nat Rev Genet. 2014;15:394–408. doi: 10.1038/nrg3683. [DOI] [PubMed] [Google Scholar]
- 18.Law JA, et al. Polymerase IV occupancy at RNA-directed DNA methylation sites requires SHH1. Nature. 2013;498:385–389. doi: 10.1038/nature12178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang H, et al. DTF1 is a core component of RNA-directed DNA methylation and may assist in the recruitment of Pol IV. Proc Natl Acad Sci USA. 2013;110:8290–8295. doi: 10.1073/pnas.1300585110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Du J, et al. Mechanism of DNA methylation-directed histone methylation by KRYPTONITE. Mol Cell. 2014;55:495–504. doi: 10.1016/j.molcel.2014.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ebbs ML, Bender J. Locus-specific control of DNA methylation by the Arabidopsis SUVH5 histone methyltransferase. Plant Cell. 2006;18:1166–1176. doi: 10.1105/tpc.106.041400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ebbs ML, Bartee L, Bender J. H3 lysine 9 methylation is maintained on a transcribed inverted repeat by combined action of SUVH6 and SUVH4 methyltransferases. Mol Cell Biol. 2005;25:10507–10515. doi: 10.1128/MCB.25.23.10507-10515.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jackson JP, Lindroth AM, Cao X, Jacobsen SE. Control of CpNpG DNA methylation by the KRYPTONITE histone H3 methyltransferase. Nature. 2002;416:556–560. doi: 10.1038/nature731. [DOI] [PubMed] [Google Scholar]
- 24.Jackson JP, et al. Dimethylation of histone H3 lysine 9 is a critical mark for DNA methylation and gene silencing in Arabidopsis thaliana. Chromosoma. 2004;112:308–315. doi: 10.1007/s00412-004-0275-7. [DOI] [PubMed] [Google Scholar]
- 25.Johnson LM, et al. The SRA methyl-cytosine-binding domain links DNA and histone methylation. Curr Biol. 2007;17:379–384. doi: 10.1016/j.cub.2007.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Malagnac F, Bartee L, Bender J. An Arabidopsis SET domain protein required for maintenance but not establishment of DNA methylation. EMBO J. 2002;21:6842–6852. doi: 10.1093/emboj/cdf687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gouil Q, Baulcombe DC. DNA methylation signatures of the plant chromomethyltransferases. PLoS Genet. 2016;12:e1006526. doi: 10.1371/journal.pgen.1006526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Song X, Cao X. Context and complexity: Analyzing methylation in trinucleotide sequences. Trends Plant Sci. 2017;22:351–353. doi: 10.1016/j.tplants.2017.03.013. [DOI] [PubMed] [Google Scholar]
- 29.Wendte JM, Schmitz RJ. Specifications of targeting heterochromatin modifications in plants. Mol Plant. 2018;11:381–387. doi: 10.1016/j.molp.2017.10.002. [DOI] [PubMed] [Google Scholar]
- 30.Gruenbaum Y, Naveh-Many T, Cedar H, Razin A. Sequence specificity of methylation in higher plant DNA. Nature. 1981;292:860–862. doi: 10.1038/292860a0. [DOI] [PubMed] [Google Scholar]
- 31.Johnson LM, et al. SRA- and SET-domain-containing proteins link RNA polymerase V occupancy to DNA methylation. Nature. 2014;507:124–128. doi: 10.1038/nature12931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yang Z, et al. Structure of the Arabidopsis JMJ14-H3K4me3 complex provides insight into the substrate specificity of KDM5 subfamily histone demethylases. Plant Cell. 2018;30:167–177. doi: 10.1105/tpc.17.00666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rajakumara E, et al. A dual flip-out mechanism for 5mC recognition by the Arabidopsis SUVH5 SRA domain and its impact on DNA methylation and H3K9 dimethylation in vivo. Genes Dev. 2011;25:137–152. doi: 10.1101/gad.1980311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rajakumara E, Nakarakanti NK, Nivya MA, Satish M. Mechanistic insights into the recognition of 5-methylcytosine oxidation derivatives by the SUVH5 SRA domain. Sci Rep. 2016;6:20161. doi: 10.1038/srep20161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Arita K, Ariyoshi M, Tochio H, Nakamura Y, Shirakawa M. Recognition of hemi-methylated DNA by the SRA protein UHRF1 by a base-flipping mechanism. Nature. 2008;455:818–821. doi: 10.1038/nature07249. [DOI] [PubMed] [Google Scholar]
- 36.Avvakumov GV, et al. Structural basis for recognition of hemi-methylated DNA by the SRA domain of human UHRF1. Nature. 2008;455:822–825. doi: 10.1038/nature07273. [DOI] [PubMed] [Google Scholar]
- 37.Hashimoto H, et al. The SRA domain of UHRF1 flips 5-methylcytosine out of the DNA helix. Nature. 2008;455:826–829. doi: 10.1038/nature07280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yaari R, et al. DNA METHYLTRANSFERASE 1 is involved in (m)CG and (m)CCG DNA methylation and is essential for sporophyte development in Physcomitrella patens. Plant Mol Biol. 2015;88:387–400. doi: 10.1007/s11103-015-0328-8. [DOI] [PubMed] [Google Scholar]
- 39.Zabet NR, Catoni M, Prischi F, Paszkowski J. Cytosine methylation at CpCpG sites triggers accumulation of non-CpG methylation in gene bodies. Nucleic Acids Res. 2017;45:3777–3784. doi: 10.1093/nar/gkw1330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Song J, Teplova M, Ishibe-Murakami S, Patel DJ. Structure-based mechanistic insights into DNMT1-mediated maintenance DNA methylation. Science. 2012;335:709–712. doi: 10.1126/science.1214453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Johnson LM, Law JA, Khattar A, Henderson IR, Jacobsen SE. SRA-domain proteins required for DRM2-mediated de novo DNA methylation. PLoS Genet. 2008;4:e1000280. doi: 10.1371/journal.pgen.1000280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang Y, et al. Large-scale comparative epigenomics reveals hierarchical regulation of non-CG methylation in Arabidopsis. Proc Natl Acad Sci USA. 2018;115:E1069–E1074. doi: 10.1073/pnas.1716300115. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.