Significance
Arp2/3 complex consists of actin-related proteins 2 and 3 with five other subunits. It forms actin filament branches under the regulation of ATP, actin monomers, actin filaments, and a nucleation-promoting factor (NPF). Here we used fluorescence spectroscopy and EM to characterize conformational changes in Arp2/3 complex along the pathway of actin filament branch formation. ATP binding to Arp2/3 complex causes small local changes in Arp2 and Arp3. NPF binding causes a larger conformational change that moves Arp2 closer to Arp3 and favors binding to the side of an actin filament, which allows further rearrangement of the subunits in the complex and growth of the new branch.
Keywords: actin, Arp2/3 complex, WASp, fluorescence, electron microscopy
Abstract
We used fluorescence spectroscopy and EM to determine how binding of ATP, nucleation-promoting factors, actin monomers, and actin filaments changes the conformation of Arp2/3 complex during the process that nucleates an actin filament branch. We mutated subunits of Schizosaccharomyces pombe Arp2/3 complex for labeling with fluorescent dyes at either the C termini of Arp2 and Arp3 or ArpC1 and ArpC3. We measured Förster resonance energy transfer (FRET) efficiency (ETeff) between the dyes in the presence of the various ligands. We also computed class averages from electron micrographs of negatively stained specimens. ATP binding made small conformational changes of the nucleotide-binding cleft of the Arp2 subunit. WASp-VCA, WASp-CA, and WASp-actin-VCA changed the ETeff between the dyes on the Arp2 and Arp3 subunits much more than between dyes on ArpC1 and ArpC3. Ensemble FRET detected an additional structural change that brought ArpC1 and ArpC3 closer together when Arp2/3 complex bound actin filaments. VCA binding to Arp2/3 complex causes a conformational change that favors binding to the side of an actin filament, which allows further changes required to nucleate a daughter filament.
Forces generated by the actin cytoskeleton are crucial to numerous cellular processes including cell migration and endocytosis (1, 2). However, spontaneous nucleation of actin filaments is kinetically unfavorable due to the rate-limiting formation of actin dimers and trimers (3, 4). Cells use proteins such as Arp2/3 complex and formins to overcome the kinetic barrier to nucleation but must regulate these proteins carefully (5).
Arp2/3 complex (6) consists of actin-related proteins Arp2 and Arp3 and five other supporting subunits named ArpC1 (p40), ArpC2 (p34), ArpC3 (p21), ArpC4 (p20), and ArpC5 (p16). Arp2/3 complex is intrinsically inactive, because the supporting subunits hold Arp2 and Arp3 too far apart to nucleate a daughter filament (7). Active Arp2/3 complex binds to the side of a “mother” actin filament, which serves as a base for initiating a new “daughter” filament as a branch at a 70° angle (8). Reconstructions of electron micrographs of branch junctions (9, 10) showed that the center of mass of Arp2 must move ∼30 Å relative to Arp3 to be aligned like subunits along the short-pitch actin filament helix. The crystal structure of inactive Arp2/3 complex suggested that a block of structure including Arp2, ArpC1, ArpC5, and part of ArpC4 might rotate about 30° with respect to the rest of the complex to bring Arp2 and Arp3 together (7, 11). This rotation model was supported by steered molecular dynamics (MD) simulations (12) and 3D reconstruction of electron micrographs of single particles of Arp2/3 complex with nucleation-promoting factors (NPFs) (13). Alternatively, Arp2 might migrate toward Arp3 separately from ArpC1 (11) as assumed during docking of the crystal structure of inactive Arp2/3 complex into the 3D reconstruction of the branch junction (10) and its subsequent refinement by MD simulations (14).
In vitro, three factors cooperate to stimulate the nucleation activity of Arp2/3 complex: actin monomers (15), a mother filament (16), and NPFs such as members of the Wiskott–Aldrich syndrome protein (WASp) family (16–19). These NPFs share a conserved C-terminal VCA motif with one or two verprolin homology (V) sequences that bind actin (20, 21) followed by central (C) and acidic (A) motifs that bind Arp2/3 complex (20, 22). Free VCA is mostly disordered (20) but assumes secondary structure when bound to Arp2/3 complex (22) or actin monomers (21). Arp2/3 complex binds two VCAs (23–25) and GST-VCA dimers are much more active than monomers (23–26). Rho-family GTPases regulate NPFs, thus linking upstream cellular signals to actin polymerization (19, 27, 28). One exceptional NPF, Dip1p from fission yeast, activates Arp2/3 complex to form unbranched filaments without the cooperation of actin filaments (29).
Multiple lines of evidence implicate VCA in the conformational changes that activate Arp2/3 complex. Two-dimensional and 3D models from EM showed differences in the position of Arp2 with and without bound VCA (13, 30). Chemical cross-linking showed that NPFs promote the movement of Arp2 and Arp3 into positions close to the short-pitch actin helix conformation (31). Such cross-linking of Arp2 and Arp3 activates the complex without NPFs (32), and small molecules can prevent both the conformational change required for cross-linking Arp2 and Arp3 and actin filament nucleation (31). ATP, GST-WASp-VCA, and WASp-CA each modestly increased Förster resonance energy transfer (FRET) between YFP fused to the C terminus of ArpC3 and CFP fused to the C terminus of ArpC1 (33). Goley et al. (33) also reported without a figure that actin filaments did not change the energy transfer efficiency (ETeff). FRET was also used to study interactions of VCA with Arp2/3 complex (13, 25). Nevertheless, questions remained about how each ligand contributes to activating Arp2/3 complex.
Here we used single-molecule FRET (smFRET) and bulk FRET measurements and EM to characterize how binding of ATP, WASp-VCA, WASp-CA, and WASp-VCA with bound monomeric actin influences the conformation of Arp2/3 complex. We used pairs of fluorescent dyes conjugated to cysteine residues at the C termini of Arp2 and Arp3 or ArpC1 and ArpC3 for smFRET experiments. smFRET and EM of single particles showed that ATP binding closes the nucleotide-binding cleft of Arp2 without other changes in conformation, while NPF binding favors movement of Arp2 toward Arp3. Ensemble FRET measurements of Arp2/3 complex with a fluorescent dye on a cysteine at the C terminus of Arp3 paired with a dye on a tetracysteine peptide on the C terminus of Arp2 and with a fluorescent dye on a cysteine at the C terminus of ArpC1 paired with a dye on a tetracysteine peptide on the C terminus of ArpC3 showed that binding to an actin filament not only moved Arp2 closer to Arp3 but also rearranged other subunits.
Results
Design of smFRET Pairs for Arp2/3 Complex Conformations.
To investigate the effects of different activating factors on the conformation of Arp2/3 complex, we modified the genome of fission yeast Schizosaccharomyces pombe to add cysteine residues to the C termini of subunits of Arp2/3 complex for labeling with fluorescent dyes (Fig. 1). We started with a strain modified with the mutation ArpC4 C167S that removes the only reactive cysteine of Arp2/3 complex (34). Strain Arp2cys-Arp3cys has single cysteines at the C termini of Arp2 and Arp3, and strain ArpC3cys-ArpC1cys has single cysteines at the C termini of ArpC1 and ArpC3. Both strains grew normally in liquid culture in preparation for biochemical experiments and in spot assays of agar plates (SI Appendix, Fig. S1). Fluorescence microscopy of fixed cells stained with BODIPY-phalloidin showed that both strains have normal-appearing actin patches and contractile rings (SI Appendix, Fig. S2). After purification on three columns, Arp2/3 complex from both strains was >99% pure (Fig. 2A and SI Appendix, Fig. S3). Purified Arp2/3 complex was labeled with Alexa-Fluor-488-C5-maleimide (Alexa 488) and/or Alexa-Fluor-594-C5-maleimide (Alexa 594), using a twofold molar excess of the donor Alexa 488 and a 10-fold molar excess of the acceptor Alexa 594. The Förster distance (R0) for this FRET pair is 54 Å (35). On average each complex had 0.10 donor dye (Alexa 488) and 1.8 acceptor dyes (Alexa 594) (Fig. 2A) distributed among three subpopulations: donor-only, acceptor-only, and double-labeled. This bias in favor of Alexa 594 ensured that most Arp2/3 complexes were labeled either with a donor–acceptor pair or with two acceptor dyes. Acceptor-only complexes are not excited at 488 nm and do not contribute to the fluorescence signal, while donor-only complexes appear as events with an apparent FRET efficiency (ETeff) of 0.
Fig. 1.
Positions of fluorophores on Arp2/3 complex. Front and side views of space-filling models of inactive Arp2/3 complex based on crystal structures [PDB ID codes 1K8K (7) and 4JD2 (43)]. AV calculations of fluorophores (colors) were made with FPS software (36). (A) Fluorophores on the C termini of Arp3 (magenta) and Arp2 (green). (B) Fluorophores on the C termini of ArpC1 (yellow) and ArpC3 (cyan). Interdye distances were calculated using the AV mean fluorophore positions. ETeff values were calculated using Eq. 2.
Fig. 2.
Labeled Arp2/3 complexes nucleate and branch actin filaments. (A) Gel electrophoresis of purified and labeled Arp2/3 constructs with data from seven different conditions aligned. Constructs ArpC1cys-ArpC3cys and Arp3cys-Arp3cys were labeled simultaneously with the FRET pair Alexa 488 and Alexa 594. Constructs ArpC1cys-ArpC3tetracys and Arp3cys-Arp2tetracys were labeled sequentially with the FRET pair FlAsH-EDT2 followed by Alexa 568. (Left) Color-coded representation of Arp2/3 complex crystal structure. First gel lane: gel of purified Arp2/3 complex stained with Coomassie blue. Other lanes show the fluorescence of each complex with UV transillumination at 312 nm. EDAS 290 Kodak Molecular Imaging Software was set to SYBR GREEN to image Alexa 488 and FlAsH and to the ethidium bromide channel to image Alexa 568 and Alexa 594. DL, double-labeled; SL, single-labeled. (B) Time course of polymerization of 3 μM actin monomers (10% pyrene-labeled) with 100 nM unlabeled or labeled Arp2/3 complex and 500 nM Wsp1p-VCA at room temperature in KMEI buffer (50 mM KCl, 1 mM MgCl2, 1 mM EGTA, 0.1 mM ATP, 1 mM DTT, and 10 mM imidazole, pH 7.0). (C) TIRF microscopy at intervals of 5 s of 1 μM actin monomers (20% labeled with Alexa-488), 500 nM Wsp1p-VCA and 100 nM of Arp2/3 complex polymerized at room temperature in TIRF buffer [50 mM KCl, 1 mM MgCl2, 1 mM EGTA, 0.1 mM ATP, 1 mM DTT, 10 mM imidazole (pH 7.0), 0.02 mM CaCl2, 15 mM glucose, 0.02 mg/mL catalase, and 0.1 mg/mL glucose oxidase with 0.25% methylcellulose].
Labeling sites were chosen to give FRET signals and to detect relative motions of the subunits. Accessible volume (AV) calculations (36) gave the range of motion of fluorophores conjugated to the C termini of the four Arp2/3 complex subunits in crystal structures (Fig. 1) and estimates of interdye distances in inactive Arp2/3 complex: 62 Å between the dyes on the C termini of Arp2 and Arp3 with a predicted ETeff of 0.46 and 91 Å between dyes on the C termini of ArpC1 and ArpC3 with a predicted ETeff of 0.08 (Fig. 1).
In bulk polymerization assays with pyrene-labeled skeletal muscle actin in the presence of 500 nM fission yeast Wsp1-VCA, Arp2/3 complex with pairs of dyes on two cysteines promoted actin polymerization in a concentration-dependent manner with time courses indistinguishable from unlabeled, wild-type Arp2/3 complex (Fig. 2B). All three samples produced 3.0 nM actin filament ends, similar to 4.4-nm ends with 1,000 nM Wsp1-VCA (37). The lag at the outset of polymerization was longer with Arp2/3 complex with pairs of dyes on a cysteine and a tetracysteine tag, but both produced 2.5-nm actin filament ends. Total internal reflection fluorescence (TIRF) microscopy showed that both labeled complexes formed actin branches with VCA as well as wild-type Arp2/3 complex (Fig. 2C). Thus, neither the addition of C-terminal cysteines nor labeling with Alexa dyes impaired the activity of Arp2/3 complex, although a tetracysteine tag reduced activity slightly.
Energy Transfer Efficiencies Between Labels on Arp2/3 Complex by smFRET.
We measured smFRET of individual double-labeled Arp2/3 complexes at a concentration of ∼75 pM. We fit smFRET histogram peaks with Gaussian distributions and calculated the mean position and width of the nonzero peaks (SI Appendix, Table S1) (see Discussion and SI Appendix for more details and fluorophore controls).
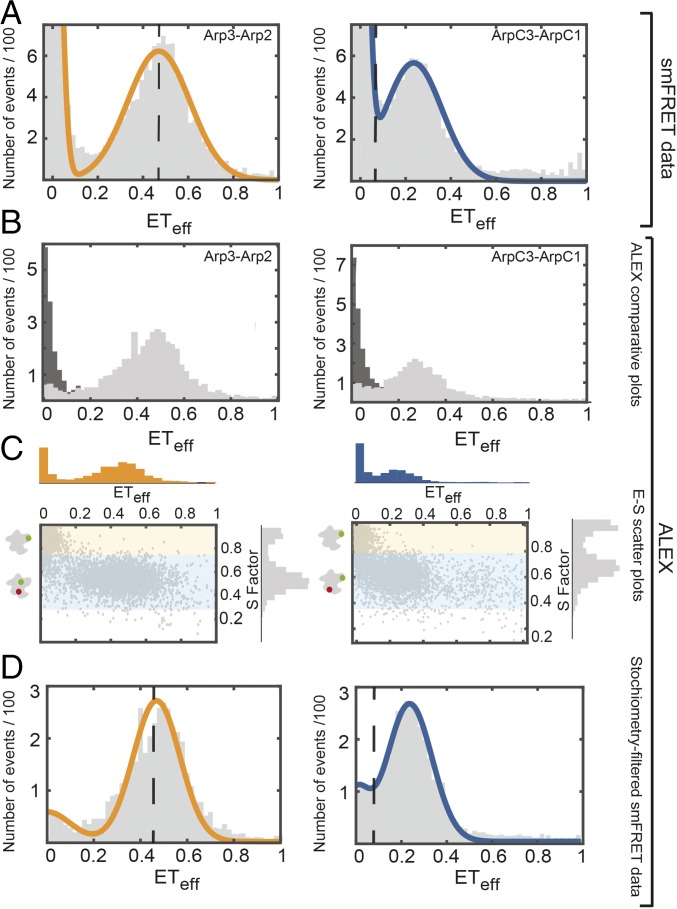
Arp2/3 complex labeled on Arp2 and Arp3 had two ETeff peaks (Fig. 3), one of which was in the donor-only ETeff ≈ 0 position. We used alternative laser excitation (ALEX) to determine if any double-labeled Arp2/3 complex peaks were buried in the ETeff ≈ 0 peak (Fig. 3B). ALEX showed that the first peak comes exclusively from donor-only labeled complexes due to their lack of responsiveness to a 561-nm laser and is thus a measurement artifact (38) (Fig. 3 B and C). The main peak had a mean ETeff of 0.48 ± 0.02 (n: 9) and a width of 0.14 ± 0.03 (n: 9). This agrees with the predicted ETeff of 0.46 for this pair according to the crystal structure. These ETeff values correspond to a distance of 60 ± 10 Å using the Förster equation and assuming κ2 = 2/3 and R0 = 54 Å for the FRET pair Alexa 488/594.
Fig. 3.
smFRET and ALEX measurements show single populations of labeled Arp2/3 complex. (Left) Arp2cys-Arp3cys construct. (Right) ArpC1cys-ArpC3cys construct. Conditions: 75 pM Arp2/3 complex and 40 nM unlabeled Arp2/3 complex in KMET buffer (50 mM KCl, 1 mM MgCl2, 1 mM EGTA, 0.1 mM ATP, 1 mM DTT, and 10 mM Tris⋅HCl, pH 7.0). (A) Distribution of diffusion-based smFRET events with continuous wave, monochromatic excitation. (B) Distributions of donor-only and donor-acceptor molecules sorted by ALEX (38) with donor-only events in dark gray and donor-acceptor events in light gray. (C) Scatter plots of ETeff vs. stoichiometry (E-S) with histograms for E along the x axis and for S along the y axis. Ratio E sorts species according to FRET, and ratio S sorts species according to donor-acceptor stoichiometry. Donor-only species have low E and high S. (D) Stoichiometry-filtered smFRET histograms using ALEX analysis to exclude donor-only species. Orange: fitted curve for the Arp2cys-Arp3cys construct. Blue: fitted curve for the ArpC1cys-ArpC3cys construct. Dashed black lines: locations of ETeff peaks predicted from AV simulations based on the crystal structure of inactive Arp2/3 complex (Fig. 1).
The labeled ArpC3cys-ArpC1cys construct also had two smFRET peaks (Fig. 3A), the zero peak and a peak centered at a mean ETeff of 0.25 ± 0.01 (n: 7) and a mean width of 0.11 ± 0.01 (n: 7). This mean ETeff is higher than the value of 0.08 predicted from the dimensions in the crystal structure and the AV calculations for the dyes on this construct (Fig. 1B), but ALEX confirmed that the low-efficiency peak contained only donor-only labeled complex (Fig. 3B). The observed ETeff value of 0.25 corresponds to an approximate interdye distance of around 70 ± 10 Å, which likely reflects the uncertainties in accurately converting from interdye distance to interresidue distance. For example, fluorophores may interact transiently with the protein, leading to conformational preferences not considered in excluded volume models. Any errors would be systematic, allowing for accurate estimation of relative distance changes between activation states.
Effect of ATP on the Conformation of Arp2/3 Complex.
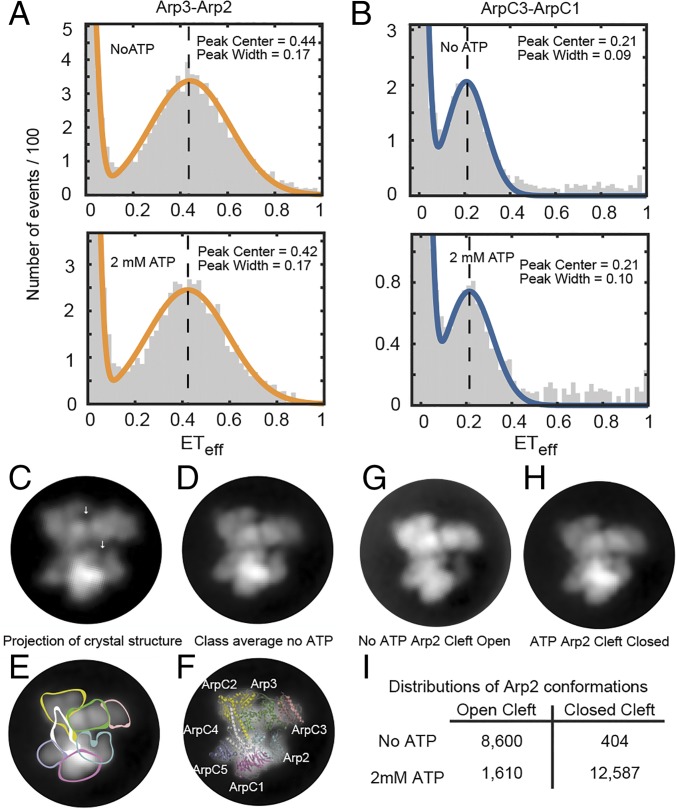
An smFRET experiment showed that a saturating concentration of ATP (2 mM) did not change the smFRET histograms of either the ArpC3cys-ArpC1cys FRET pair (Fig. 4A) or the Arp2cys-Arp3cys FRET pair (Fig. 4B). Neither the mean position nor the width of the ETeff peaks changed for either pair of labeling sites.
Fig. 4.
Effect of ATP on the conformation of Arp2/3 complex. (A and B) smFRET experiments (Upper) without ATP and (Lower) with 2 mM ATP. Conditions: 75 pM Arp2/3 complex and 40 nM unlabeled Arp2/3 complex in KMET buffer (50 mM KCl, 1 mM MgCl2, 1 mM EGTA, 1 mM DTT, and 10 mM Tris⋅HCl, pH 7.0). Vertical dashed lines indicate the mean of the ETeff peaks without ATP as a visual guide. (A) Arp2cys-Arp3cys construct. (B) ArpC1cys-ArpC3cys construct. (C) Low-resolution (20 Å) 2D projection of the crystal structure of bovine Arp2/3 complex with Ca-ATP in the same orientation as the negative stain class averages. The model was made from PDB ID code 4JD2 with mouse GMF deleted. White arrows indicate the nucleotide-binding clefts of Arp2 (lower) and Arp3 (upper). (D–G) Transmission EM of negatively stained Arp2/3 complex without ATP. Virtually all of the particles had the same orientation. After correction for drift, particles were classified and class averages computed. (D) A dominant class average of the projection structure of Arp2/3 complex without ATP computed from 2,066 particles. (E) Subunits are outlined on the class average. (F) A ribbon diagram of the crystal structure of inactive Arp2/3 complex is superimposed on the class average. (G) Dominant class average without ATP (1,636 particles) with the nucleotide-binding cleft of Arp2 open. (H) Dominant class average with ATP with the nucleotide-binding cleft of Arp2 closed. (I) Numbers of particles in two class averages.
EM of negatively stained Arp2/3 complex (Fig. 4 C–G and SI Appendix, Fig. S4) confirmed that binding ATP does not cause large conformational changes. We classified and averaged >10,000 molecules in the presence and absence of ATP. The samples were quite uniform, because the front side of Arp2/3 complex is relatively flat and most particles landed front side down on the grid. The class averages are remarkably similar to a low-resolution, projection image of the crystal structure (Fig. 4C) and clearly resolved Arp2 (including the nucleotide-binding cleft), Arp3 (including the nucleotide-binding cleft), ArpC1, ArpC3, and ArpC5 (Fig. 4D). ArpC2 and ArpC4 appeared as a continuous density as expected in a 2D projection image (Fig. 4C). In the favored orientation Arp3 is viewed from the top, looking down the nucleotide-binding cleft, while the front side of Arp2 is visible. The highest density is in the region where ArpC1, ArpC4, and subdomains 3 and 4 of Arp2 superimpose in the projection, consistent with crystal structures. Our projection view class averages ± ATP are consistent with 2.0-nm resolution 3D reconstructions of negatively stained Arp2/3 complex with ATP (13).
In samples treated with Dowex resin to remove ATP eight of nine class averages (including 96.5% of particles) were very similar with the nucleotide-binding cleft of Arp2 clearly open to various degrees (Fig. 4 D, G, and H and SI Appendix, Fig. S4). Thus, the lack of density for Arp2 subdomains 1 and 2 in maps of most crystal structures (39) arises from a range of hinge angles between the two halves of Arp2. The other class average with 3.5% of particles had no density for Arp2 subdomains 1 and 2 (SI Appendix, Fig. S4A, Lower Right), similar to the “open” class of particles reported by Rodal et al. (30) in their analysis of negatively stained Arp2/3 complex without ATP. The viewing angle is not favorable for detecting differences in the cleft of Arp3.
All nine class averages of Arp2/3 complex in the presence of ATP (14,197 particles; SI Appendix, Fig. S4B) were similar to each other with the Arp2 nucleotide-binding cleft clearly closed and some density bridging the cleft (Fig. 4H). The distance between ArpC1 and ArpC3 was the same in the particles with open (no ATP) and closed (ATP) Arp2 clefts.
VCA/CA Ligands Increase the FRET Efficiency Between Labels on Arp2 and Arp3 More than Between ArpC1 and ArpC3.
We used smFRET to study interactions of Arp2/3 complex with Wsp1p-CA, -VCA, and -VCA cross-linked to an actin monomer (actin-VCA). We made actin-VCA by the method of Boczkowska et al. (40), which was based on careful consideration of the crystal structure of the V motif bound to actin (21). Fluorescence correlation spectroscopy confirmed that VCA binds Arp2/3 complex under our experimental conditions (SI Appendix, Fig. S5). Labeled VCA alone had a diffusion time τ of 0.32 ms. In the presence of 500 nM Arp2/3 complex (τ = 0.82 ms), 58% of labeled VCA diffused more slowly (τ = 0.91 ms), consistent with the higher mass expected from Arp2/3 complex, indicating binding under smFRET experimental conditions.
Fluorescence anisotropy confirmed that Alexa 488-labeled VCA and CA constructs bind to Arp2/3 complex (SI Appendix, Fig. S6 and Tables S3 and S4). Anisotropy values were 0.11 for 50 nM VCA and 0.10 for 50 nM CA. In the presence of 3 μM Arp2/3 complex, the anisotropy values increased to 0.21 for VCA and to 0.22 for CA. These values are similar to the value 0.22 for 100 nM Arp2/3 complex with a single Alexa 488 label. These results and previously reported Kds of less than 2 μM (23) validated the concentrations used to saturate the complex.
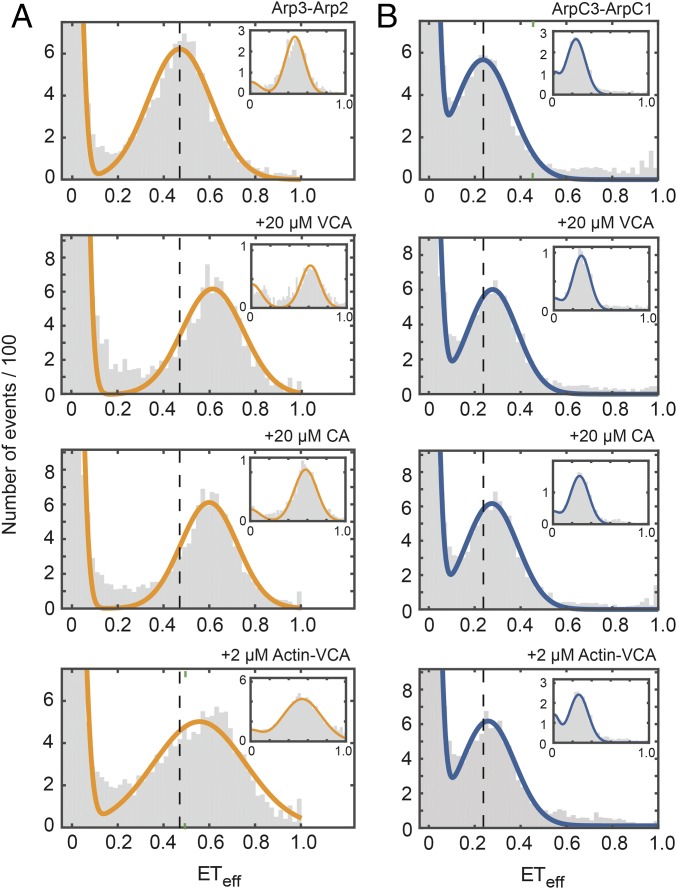
smFRET experiments showed that 20 μM VCA, 20 μM CA, and 2 µM actin-VCA had different effects on the ArpC3cys-ArpC1cys and Arp2cys-Arp3cys constructs (Fig. 5 and SI Appendix, Table S1). All three ligands increased the mean values of the ETeff peak of the Arp2cys-Arp3cys construct, indicating a decrease in the distance between the dyes. VCA increased the ETeff by 0.13, CA by 0.06, and actin-VCA by 0.12. All of the smFRET histograms showed two peaks, but ALEX confirmed that the ETeff ≈ 0 peaks came solely from donor-only labeled complexes (Fig. 5, Insets). The peak widths were generally consistent with shot noise estimates and other potential nonconformational contributions. The broader peak with actin-VCA bound to Arp2cys-Arp3cys may indicate some conformational heterogeneity or slower-timescale dynamics compared with other samples. In contrast, none of these ligands changed the ETeff for the ArpC3cys-ArpC1cys construct substantially. VCA and CA each increased the ETeff by 0.03 and actin-VCA increased ETeff by 0.02.
Fig. 5.
VCA/CA ligands increase the FRET efficiency between labels on Arp2 and Arp3 more than between labels on ArpC1 and ArpC3. Distributions of diffusion-based smFRET events for (A) Arp2cys-Arp3cys construct and (B) ArpC1cys-ArpC3cys construct. Conditions: 75 pM labeled Arp2/3 complex in KMET buffer (50 mM KCl, 1 mM MgCl2, 1 mM EGTA, 0.1 mM ATP, 1 mM DTT, and 10 mM Tris⋅HCl, pH 7.0) with top row, 40 nM unlabeled Arp2/3 complex; second row, 20 μM WASP-VCA from fission yeast Wsp1p; third row, 20 μM WASP-CA from fission yeast Wsp1p; and fourth row, 2 μM actin-VCA from fission yeast Wsp1p. (Insets) Representative stoichiometry-filtered ALEX histograms. Gray: representative histograms of smFRET events collected over 60 min. Orange: fitted curves for Arp2cys-Arp3cys construct. Blue: fitted curves for ArpC1cys-ArpC3cys construct. Vertical dashed reference lines indicate the peak center for Arp2/3 complex alone.
EM of Arp2/3 Complex with GST-VCA Shows a Change in Shape.
We analyzed more than 11,000 particles in electron micrographs of negatively stained Arp2/3 complex with ATP and 0.25 µM GST-VCA, a concentration that is not expected to saturate Arp2/3 complex. Visualization of GST tethered by CA to Arp2/3 complex by the flexible V motif of 45 residues depended on the number of class averages calculated (Fig. 6). When divided into 70 class averages of about 170 particles each, an extra mass was associated with the periphery of 34% of the Arp2/3 complexes (Fig. 6A), with 70% of these masses on the “right” side of the complex near Arp2 and Arp3 (Fig. 6B). Rodal et al. (30) also detected a mass next to Arp2 and Arp3 in samples with full-length Las17, while chemical cross-linking revealed sites on both the back and bottom sides of Arp2/3 complex (41). Note that some class averages without an extra mass may actually have GST in different positions and averaged out.
Fig. 6.
Electron micrographs of Arp2/3 complex with GST-VCA. Samples of 50 nM Arp2/3 complex, 0.25 µM GST-VCA, and 0.2 mM ATP were negatively stained, imaged by transmission EM, and sorted into classes. (A and B) Analysis by calculating 70 class averages with the ISAC module in EMAN2/Sparx. (A) Top 24 class averages. (B) Angular distribution of GST around Arp2/3 complex with numbers of class averages on the radial axis. (C–E) Two-dimensional classification of the same dataset with Relion yields 11 well-resolved class averages. (C) One example of six class averages including 7,187 (61%) particles that appear like Arp2/3 complex without GST-VCA. The arrow points at ArpC5. (D) Two examples of class averages of particles with foreshortened ArpC5 (arrow) including 4,596 (39%) particles. (E) Orientations of three line scans used to measure dimensions of Arp2/3 complex class averages: D, diagonal; H, horizontal; V, vertical. (F) Mean dimensions (±1 SD).
When the 11,000 particles were divided into 11 classes, no extra mass appeared in any class, because the randomly positioned GST was averaged out (Fig. 6D and SI Appendix, Fig. S4C). However, the larger numbers of particles in each class produced averages with more details and revealed two large groups that differed in conformation. One conformation with 61% of the particles from seven class averages was indistinguishable from inactive Arp2/3 complex with bound ATP where most subunits are visible (Fig. 6C). The other four class averages with 39% of the particles had a more compact conformation lacking the mass corresponding to ArpC5 projecting from the side of the complex (Fig. 6D). Line scans between ArpC5 and Arp2 (Fig. 6E) confirmed that this dimension was 18% shorter in these group-2 class averages, while two other dimensions were the same (Fig. 6F).
Effect of Actin Filaments on the Conformation of Arp2/3 Complex.
smFRET experiments with labeled Arp2/3 complex bound to actin filaments were not possible due to the slow diffusion of the filaments even when their lengths were restricted with capping protein. Therefore, we used ensemble FRET as an alternative to smFRET to evaluate the effect of actin filaments on the conformation of Arp2/3 complex (Fig. 7 and SI Appendix, Fig. S7).
Fig. 7.
Actin filaments increase the FRET efficiency between dyes on both Arp2/Arp3 and ArpC1/ArpC3. Ensemble ETeff measurements of 20 nM Arp2/3 complex labeled (blue) with FlAsH on ArpC3 and Alexa 568 on ArpC1 or (orange) with FlAsH on Arp2 and Alexa 568 on Arp3. The measurements are differences between the ETeff values of Arp2/3 complex alone and with the each ligand. Conditions: 50 mM KCl, 1 mM MgCl2, 1 mM EGTA, 0.1 mM ATP, 1 mM DTT, and 10 mM imidazole, pH 7.0, unless indicated otherwise. (A) Titration of Arp2/3 complex with Wsp1p-VCA. (B) Titration of Arp2/3 complex with polymerized actin with overnight incubation. (C) Titration of Arp2/3 complex and 10 μM polymerized actin with Wsp1p-VCA. (D) Interleaved scatter plot of differences in mean ETeff values (±1 SD) of labeled Arp2/3 complex alone and in the presence of 2 mM ATP and with 2 mM ATP and various ligands: 20 µM Wsp1p-CA, 20 µM Wsp1p-VCA, 20 µM GST-VCA, 2 µM actin-VCA (actin monomer covalently linked to VCA), 10 µM polymerized actin with 10 µM VCA, or 20 µM polymerized actin. Numbers of repetitions are in parentheses.
Ensemble FRET requires two different types of labeling sites on Arp2/3 complex, so we purified Arp2/3 complex from strains Arp3cys-Arp2tetracys and ArpC1cys-ArpC3tetracys. Strain Arp3cys-Arp2tetracys has a single cysteine at the C terminus of Arp3 and a peptide with four cysteine residues at the C terminus of Arp2, and strain ArpC1cys-ArpC3tetracys has a single cysteine at the C terminus of ArpC1 and the tetracysteine peptide at the C terminus of ArpC3. Purified complexes (Fig. 2A) were labeled in two steps. First, we used a 10-fold molar excess of the donor FlAsH-EDT2 and then a sevenfold molar excess of the acceptor Alexa-Fluor-568-C5-maleimide (Alexa 568). R0 is ∼75 Å for this FRET pair (42). On average, labeling was 50% donor dye (FlAsH-EDT2) and 100% acceptor dye (Alexa 568) (Fig. 2B; see Materials and Methods for details). The C-terminal tetracysteine peptide and labeling with FlAsH dye slowed the initial rate of actin filament nucleation but the orthogonally labeled Arp2/3 complex produced nearly the same numbers of barbed ends as wild-type Arp2/3 complex (Fig. 2B). Ensemble FRET gave results similar to smFRET for comparable samples but the data are less amenable to quantitative interpretation.
Saturating concentrations (2 mM) of ATP did not change the overall ETeff of either construct (Fig. 7D and SI Appendix, Table S2), and the two constructs responded differently to 20 μM CA, 20 μM VCA, 20 μM GST-VCA, and 2 μM actin-VCA (Fig. 7 and SI Appendix, Table S2). All of these ligands increased ETeff of Arp2tetracys-Arp3cys but had no effect or decreased the ETeff for the ArpC1cys-ArpC3tetracys construct. These results are consistent with our smFRET measurements. Titration of Arp2tetracys-Arp3cys with VCA increased ETeff consistent with a Kd of 0.6 μM, while the ETeff of the ArpC1cys-ArpC3tetracys construct did not change (Fig. 7A).
Twenty-micromolar actin filaments increased the ETeff of both ArpC1cys-ArpC3tetracys and Arp3cys-Arp2tetracys (Fig. 7B). The actin filament concentration dependence gave Kds of 6.6 μM for Arp3cys-Arp2tetracys and 4.1 μM for ArpC1cys-ArpC3tetracys, both similar to Kds measured by titrating pyrene–Arp2/3 complex with actin filaments (23).
The double-labeled ArpC1cys-ArpC3tetracys and Arp2tetracys-Arp3cys constructs responded differently to titration with VCA in the presence of 10 µM actin filaments (Fig. 7C). The ETeff of Arp2tetracys-Arp3cys increased up to 0.22 ± 0.03 (n = 3) at 40 μM VCA. A hyperbola fit to this data gave a Kd of 4.4 μM. This Kd was expected to be <0.6 µM based on our previous measurements with Alexa488-labeled Wsp1p-CA (23), so the labels used in the ensemble FRET experiments influenced the reactions, as also noted in polymerization assays (Fig. 2B). However, the ETeff of ArpC1cys-ArpC3tetracys increased only up to 10 μM VCA (Kd of ∼2.0 μM) and declined at higher concentrations of VCA for reasons that we do not understand.
Discussion
The crystal structure of inactive Arp2/3 complex suggested that a large conformational change is required to reposition the Arps to initiate the daughter filament (7), so many methods have been used to investigate how nucleotides, NPFs, and mother filaments promote the active conformation. This discussion explains how our work extends the conclusions from previous experiments using X-ray crystallography, Förster energy transfer, chemical cross-linking, radiation footprinting and mass spectrometry, EM of single particles and branch junctions, and MD simulations.
Inactive Arp2/3 Complex Has One Major Conformation.
High-resolution crystal structures provide the most details about the structure of inactive Arp2/3 complex. Crystal packing forces can influence protein conformations, but different crystal forms of bovine Arp2/3 complex alone or associated with nucleotides, VCA, or glial maturation factor (GMF) (7, 23, 39, 43, 44) do not differ substantially. A crystal structure of fission yeast Arp2/3 complex differs only in the absence of Arp2 (45). This conformation of inactive bovine Arp2/3 complex was stable during 10 ns of atomistic, MD simulations except for small fluctuations in local regions, such as the nucleotide-binding clefts of the Arp subunits (12, 46).
Our smFRET data along two dimensions of the complex with small dyes on the C termini of Arp2 and Arp3 or ArpC1 and ArpC3 are consistent with inactive fission yeast Arp2/3 complex having a preferred conformation in solution on the millisecond timescale. Rapid conformational sampling could also give a single “time-averaged” conformation at the millisecond timescale, but large-scale conformational changes are not typical in a multiprotein complex at the 500-µs time limit we tested. Slower conformational changes would result in wide or multimodal smFRET distributions, while the peak widths in our single-molecule ETeff histograms were generally consistent with established noise sources such as Poisson counting error.
Energy transfer between dyes on the C termini of Arp2 and Arp3 match the value predicted from the crystal structure (Fig. 3A). The ETeff between ArpC1 and ArpC3 is higher than expected, so the labeled residues appear to be 20 Å closer together than expected from the crystal structure. However, this is not a concern, because the predicted 91-Å distance between the dyes is outside the range for accurate measurements, and the dyes may have preferred conformations not captured by the AV calculations or the anisotropy/lifetime measurements.
The class averages from our electron micrographs of negatively stained fission yeast apo-Arp2/3 complex showed one major conformation with a small range of hinge angles between the two halves of Arp2. In contrast to the uniformity of these structures, a pioneering EM study of negatively stained budding yeast and bovine Arp2/3 complexes reported the coexistence of an equilibrium mixture three different conformations, one of which was stabilized by coronin (30). All three of their conformations are similar to our class averages for apo-Arp2/3 complex, except that Arp2 appears to be missing or disordered in their open conformation, which was true of only 3.5% of our particles without ATP. Our class averages have more details, because we analyzed more particles and used movies from a direct electron detector camera to correct for drift, methods not available in 2005.
ATP Binding Results in Local Conformational Changes Restricted to Arp2.
Multiple lines of evidence show that nucleotide binding to Arp2/3 complex cause small changes in conformation confined to the Arps. Crystal structures (39, 47), all-atom MD simulations (48), and radiation footprinting with mass spectrometry (49) all showed that central cleft of Arp3 closes around bound ATP or ADP, while ATP bound to subdomains 3 and 4 of Arp2 did not stabilize the disordered subdomains 1 and 2 in the crystals. Our 2D class averages of electron micrographs of fission yeast Arp2/3 complex show that ATP stabilizes subdomains 1 and 2 and closes the cleft of Arp2, in agreement with 3D reconstructions from electron micrographs of frozen-hydrated and negatively stained Arp2/3 complex from budding yeast and Acanthamoeba (13). These samples were homogeneous and indistinguishable from the crystal structures except for the presence of subdomains 1 and 2 of Arp2.
In agreement with this evidence, ATP binding to Arp2/3 complex produced little or no change in FRET efficiency between dye probes on Arp2 and Arp3 or on ArpC1 and ArpC3 in our single-molecule experiments. However, a pioneering study of recombinant human Arp2/3 complex using ensemble FRET measurements of bulk samples with YFP fused to the C terminus of ArpC1 and CFP fused to the C terminus of ArpC3 concluded that “nucleotide binding promotes a substantial conformational change in (Arp2/3) complex” (33). That study reported FRET/CFP ratios, which were determined by dividing the acceptor peak emission by the donor peak emission. The FRET/CFP ratio changed by 0.11 upon binding to ATP. We recalculated ETeff from the data in ref. 27 using Eq. 2 (Materials and Methods) and obtained an ETeff increase of ∼0.02, which is consistent with our data. Although more complicated to prepare, our Arp2/3 complex labeled with small dye molecules was fully active, while Arp2/3 complex fused to fluorescent proteins was 20 times less active than wild-type Arp2/3 complex (33).
ATP binding in the nucleotide cleft of Arp3 can release the C terminus of Arp3 from the barbed end groove (32). This rearrangement might change the ETeff of a dye on the C terminus. However, we did not observe such a change, so the average position of the dye does not change significantly after the release of the C terminus of Arp3. Thus, binding of ATP may trigger local conformational changes that do not cause FRET changes between our specific probes.
Binding of NPFs Reorients Arp2 and Arp3.
The major challenge in the field has been to characterize how NPFs and mother filaments promote the conformational changes that activate Arp2/3 complex to nucleate a daughter filament. There is general agreement that fully active Arp2/3 complex has Arp2 and Arp3 flattened and arranged like successive subunits along the short-pitch helix of an actin filament, as observed at 26-Å resolution in a 3D reconstruction of negatively stained branch junctions (10). Three independent methods show that binding of NPFs to Arp2/3 complex favors a conformation with the Arps moved toward the fully active, short-pitch helix conformation.
Cross-linking.
Chemical cross-linking of Arp2 and Arp3 shows directly that VCA favors a conformation with the Arps near that of a short-pitch actin helix. Rodnick-Smith et al. (32) substituted single cysteines into budding yeast Arp2 and Arp3 that are adjacent when the Arps are in the short-pitch conformation but not inactive Arp2/3 complex. The bifunctional, 8-Å cross-linker bis-maleimidoethane covalently linked the cysteine-substituted Arp2 and Arp3 in minutes and even faster with N-WASP-VCA. Thus, the budding yeast Arp2/3 complex must spontaneously visit the short-pitch conformation, as reflected by its ability to nucleate daughter filaments without NPFs. After cross-linking Arp2/3 complex nucleates actin filaments much better without NPFs, so the cross-link stabilizes an activated state of Arp2/3 complex.
FRET.
Binding of VCA or CA increased ETeff between probes on the C termini of Arp2 and Arp3, so two Arps moved closer together (Fig. 5). However, NPFs made only small changes in FRET between fluorophores on the C termini of ArpC1 and ArpC3 in either smFRET experiments (Fig. 5) or ensemble FRET experiments with dyes (Fig. 7A) or with YFP fused to the C terminus of ArpC3 and CFP fused to the C terminus of ArpC1 (33). We used the reported change in the FRET/CFP ratio of 0.23 (33) and Eq. 2 to calculate an ETeff increase of ∼0.05 upon binding to WASp-CA, in agreement with our experiments. Thus, the distance between the C termini of ArpC1 and ArpC3 changes little when CA or VCA binds.
EM.
EM studies of Acanthamoeba, bovine, and budding yeast Arp2/3 complex reported that NPFs favor a conformation with the Arps closer together than inactive Arp2/3 complex (13, 30). Our class averages of single Arp2/3 complex particles in the presence of GST-VCA fell into two groups: one similar to the ATP-Arp2/3 complex alone (Fig. 6C) and another with foreshortened ArpC5 (Fig. 6D).
Despite this consistent evidence linking NPF binding to rearrangement of Arp2, the nature of the conformational changes is still unclear. One hypothesis is that Arp2 rotates toward Arp3 while associated with ArpC1, ArpC4, and ArpC5 (7). One interpretation of the projection images in Fig. 6D is that rotation of Arp2, ArpC1, ArpC2, and ArpC5 moves ArpC5 behind ArpC1 and moves Arp2 in the Z-direction toward the viewer while remaining in contact with Arp3. An alternative idea is that Arp2 dissociates from these neighbors and “migrates” on its own toward Arp3 with new contacts with ArpC1 and ArpC2 (10, 11). None of the methods has distinguished between these hypotheses. For example, both models predict a change in FRET between the C termini of the Arps coupled with little change between the C termini of ArpC1 and ArpC3 (Fig. 5).
Steered molecular simulations of Arp2/3 complex were used to test these hypotheses. A spring-like force was between Arp2 in the inactive crystal structure and its position in a short-pitch actin helix next to Arp3 (12). During atomistic MD simulations, the force moved Arp2 toward the active target position, along with a block of protein including ArpC1, ArpC5, and part of ArpC4. This block rotated together by twisting a pair of alpha-helices that connect ArpC4 to ArpC2. In 11 experiments steric interference stalled this rotation after about 30°. In one simulation the block of protein with Arp2 moved past this barrier further toward Arp3 in an actin filament-like structure where Arp2 and Arp3 could be cross-linked (32). These results suggest a pathway for the initial part of the conformational change.
Effect of Actin Filament Binding on the Conformation of Arp2/3 Complex.
Full activation of Arp2/3 complex to nucleate a daughter filament requires not only binding a pair of NPFs (23–25) but also binding to the side of an actin filament (16). Assuming that NPF binding is responsible for moving Arp2 part of the way toward Arp3, actin filament binding is the leading candidate to drive the final conformational change toward the fully active structure.
Our ensemble FRET measurements with dye probes on Arp2 and Arp3 or ArpC1 and ArpC3 showed that binding to the side of actin filaments increased the ETeff of both constructs. Thus, the relative positions ArpC1 and ArpC3 change in addition to the change between Arp2 and Arp3 induced by VCA. Two-dimensional (9) and 3D (10) reconstructions of branch junctions were refined by large-scale MD simulations (14) to reveal extensive contacts between Arp2/3 complex and the mother filament. An independent analysis by docking the ArpC2 and ArpC4 heterodimer on the side of an eight-subunit actin filament followed by MD simulations (50) revealed similar interactions. However, high-resolution structures of Arp2/3 complex with bound NPFs and of branch junctions are necessary to clarify the detailed conformational changes associated with NPF and filament binding and to answer questions such as when during activation Arp2 and Arp3 are flattened like subunits in actin filaments (51).
Proposed Conformational Changes.
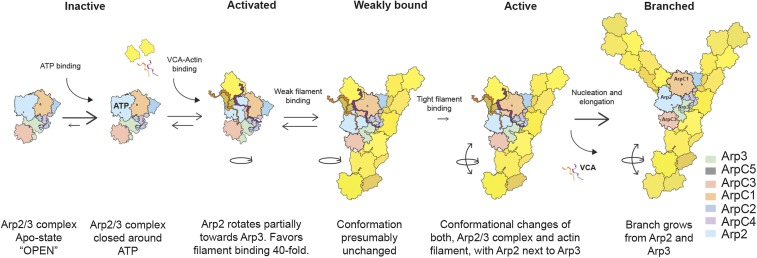
Our results clarify the steps leading to the formation of an actin filament branch (Fig. 8). In solution, Arp2/3 complex has predominantly a single, inactive conformation. ATP binding promotes closure of the nucleotide clefts of both Arps (46–49) and destabilizes the C terminus of Arp3 (32). Interactions with VCA and actin filaments activate Arp2/3 complex to nucleate a daughter filament. Given the concentration of monomers in the cytoplasm and their affinity for the V motif, most VCA will bring along an actin monomer (21, 34).
Fig. 8.
Pathway of actin filament branch formation by Arp2/3 complex. Binding of ATP to inactive Arp2/3 complex closes the nucleotide-binding clefts of Arp2 and Arp3. Binding of VCA or actin-VCA promotes movement of Arp2 to an activated intermediate conformation that favors binding to the side of a mother filament. Weak binding to a mother filament followed a further conformational change that strengthens Arp2/3 complex binding to the filament and favors nucleation of the daughter filament.
VCA binding favors an activated conformation with Arp2 closer to Arp3 but with the distance between ArpC1 and ArpC3 unchanged. A 30° rotation of the block of subunits including Arp2 (7, 12) can explain these changes, the structures observed by EM (Fig. 6), and chemical cross-linking observed between Arp2 and Arp3 (32). However, alternative mechanisms such as migration of Arp2 have not been ruled out. The free energy from VCA binding is used to reach this activated state, but the mechanism is not yet clear. Bound VCA increases the affinity of Arp2/3 complex for the mother filament 20-fold (23), presumably because the conformation of activated complex better matches the side of the filament.
Activated Arp2/3 complex with bound actin-VCA binds rapidly but reversibly to the sides of actin filaments (52). We presume that the interaction is weak, because the conformation of the activated complex is not perfectly matched to the side of the filament. Filaments alone can activate Arp2/3 complex, but the reaction is very slow and unfavorable (8). NTPs favor binding of Arp2/3 complex to the side of a filament by increasing both the rate of the reaction and the affinity of binding. Branching can be very fast when NFPs are concentrated locally such as at sites of endocytosis (40,000 per square micrometer) (53).
Rarely during these brief encounters, activated Arp2/3 complex and the actin filament both undergo the more extensive conformational changes required for tight binding and nucleation of a daughter filament. The interface between Arp2/3 complex and the mother filament buries 9,100 Å2 of surface area (13), which likely provides the free energy change required to reach the fully active state. Given the FRET change we observed between dyes on ArpC1 and ArpC3, interaction with the mother filament is likely to reposition subunits in addition to the Arps. In addition to these conformational changes VCA must also release from filament-bound Arp2/3 complex before growth of the branch (52). Higher-resolution structures of the intermediates and the branch junction should provide more clarity about this pathway.
Materials and Methods
SI Appendix, Supplemental Materials and Methods provides a more detailed account of our methods.
Proteins.
We used standard methods to add a cysteine residue or tetracysteine peptide tag to the C termini of Arp2, Arp3, ArpC1, or ArpC3 (34) and expressed the proteins in fission yeast. We purified Arp2/3 complex from fission yeast (23); actin from skeletal muscle (54); and recombinant Wsp1p-VCA (497Q–574D), Wsp1p-VCAcys (496C–574D), Wsp1-CA, and GST-Wsp1p-VCA from Escherichia coli; and covalently cross-linked an actin monomer to VCA (23). We labeled mutant Arp2/3 complexes with Alexa-Fluor-488-C5-maleimide or/and Alexa-Fluor-594-C5-maleimide. Actin was labeled on Cys374 with pyrene-iodoacetamide (55) or Alexa-Fluor-488-C5-maleimeide (56). Labeling efficiency was determined by absorption (23, 42). We calculated AVs for the conjugated dyes using FPS software (36, 57). We tested labeled Arp2/3 complexes using pyrene actin polymerization assays (58) and TIRF microscopy of single filaments (59).
Ensemble FRET measurements.
Measurements were made in a PTI Alpha-scan fluorimeter (Photon Technology International) (20). FlAsH-EDT2 single-labeled (Arp2FlAsH and ArpC3FlAsH) and FlAsH-EDT2 and Alexa 568 double-labeled complexes (Arp2FlAsH-Arp3Alexa568 and ArpC3FlAsH-ArpC1Alexa568) were diluted to fixed labeled concentrations of 20 nM in KMEI buffer. All experiments had 0.2 mM ATP present, except when testing the effect ATP on constructs where ATP was either at 0 mM or 2 mM. Complexes were incubated for 3 h at room temperature with a range of concentrations of VCA, CA, actin-VCA, GST-VCA, or a fixed concentration of polymerized actin with a titration of VCA. Complexes were incubated overnight (∼12 h) with a range of concentrations of actin filaments. FlAsH-EDT2 was excited at 510 nm and emitted light was collected with a wavelength scan from 520 nm to 650 nm. Changes in donor intensity of double-labeled complexes were calculated by comparing the average fluorescence obtained from 530 nm to 532 nm (emission peak) to single-labeled complexes. SI Appendix gives the equations to calculate ETeff (35).
smFRET measurements.
smFRET measurements were made with an instrument built on an inverted Olympus IX-71 microscope (Olympus) previously described in detail (60). The laser power at 488 nm was set to 25–35 µW before entry into the microscope. Fluorescence emission was collected through the objective and separated from the excitation light by a Z488RDC long-pass dichroic and 500-nm long-pass filter (Chroma). Further separation of donor and acceptor photons was achieved with HQ585LP dichroic, with an ET525/50M band-pass emission filter for donor photons and a 605LP emission filter for acceptor photons. Subsequently, fluorescence emission was focused onto the aperture of an optical fiber with a 100-µm diameter (OzOptics), which was directly coupled to an avalanche photodiode (PerkinElmer) (57). Experiments were done in eight-well chambered cover glasses (Nunc) passivated with polylysine-conjugated polyethylene glycol, which prevents proteins from sticking to chamber surfaces (61). Wells contained 75 pM labeled Arp2/3 complex with an additional 40 nM of unlabeled complex in KMET buffer and ligands described in each experiment. Under these conditions, Arp2/3 complex was stable for the duration of our measurements. Traces were collected in 1-ms time bins for at least 1 h, and with only photon bursts of >30 photons per time bin included in the final histogram.
ETeff values were calculated according to Eq. 1:
| [1] |
where Ia is the acceptor fluorescence intensity, Ib is the donor fluorescence intensity, β is a measurement of donor photon bleed-through into the acceptor channel, and γ accounts for differences in detection efficiency and quantum yield between the fluorophores. The β and γ parameters are instrument-specific and were measured empirically (β = 0.06, γ = 1.3).
ETeff values were compiled into histograms which were fitted with two Gaussian distributions using MATLAB (MathWorks) scripts (57).
We estimated the upper limit for shot noise using the following formula:
| [2] |
where T is the number of photons threshold that defines an smFRET event; we fixed this value at 30 photons. ET is energy transfer obtained in our measurements.
ALEX measurements.
ALEX measurements were made with an instrument based on an Olympus IX-71 inverted microscope (57, 60). Samples were exposed to alternating 100-µs pulses of 488-nm and 561-nm laser light controlled by acousto-optic modulators (Isomet) with 5-µs dark intervals for 60 min. Traditional ETeff values and a stoichiometry (S) ratio were obtained using Eq. 3:
| [3] |
The S ratio characterizes labeling. is the intensity of the acceptor channel during 488-nm excitation, is the intensity of the donor channel during 488-nm excitation, and is the fluorescence intensity of the acceptor channel during 561-nm excitation. Data were collected with LabVIEW software running an in-house script (57).
EM.
Arp2/3 complex was applied to glow-discharged carbon-coated girds (62) and negatively stained with 0.8% (wt/vol) uranyl formate. A Tecnai F20 electron Microscope (FEI Company) operating at 200 kV collected images at a defocus between −0.6 μm and −3.0 μm, at a dose rate of approximately eight counts per pixel per second and a drifting speed <0.2 nm/s on a K2 Summit camera (Gatan Company) in counted mode at a nominal magnification of 29,000×, corresponding to a pixel size of 1.25 Å. Dose-fractionated image stacks were motion-corrected and summed with MotionCorr (63) and binned by a factor of 2. Particles were interactively picked using e2boxer in EMAN2 (64) with a box size of 120 pixels. Particle coordinates were transferred from EMAN2 into RELION2 (65) for 2D analysis. Contrast transfer function (CTF) estimation was performed using CTFFIND4 (66), and particles were CTF-corrected before reference-free 2D classification. Class averages were sorted in RELION2. We made 2D projections the Arp2/3 crystal structure [Protein Data Bank (PDB) ID code 4JD2] (43) after manually removing GMF from the PDB file. The 3D map was converted into a 2D projection with an orientation similar to our EM 2D class averages using the EMAN2 command line programs e2pdb2mrc and e2project3d.
Supplementary Material
Acknowledgments
We thank Shih-Chieh Ti for providing the S. pombe strains used in the ensemble FRET experiments and his advice on the project and three expert reviewers and Roberto Dominquez, who each provided suggestions that substantially improved the paper. This work was supported by National Institute of General Medical Sciences of the National Institutes of Health Grants R01GM026338 (to T.D.P.) and R01NS079955 (to E.R.), an International Fellowship from the American Association of University Women (to S.E.-S.), American Heart Association Fellowship 13PRE16570013 (to L.A.M.), and the Yale Integrated Graduate Program in Physical and Engineering Biology (S.E.-S. and L.A.M.). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1717594115/-/DCSupplemental.
References
- 1.Pollard TD, Borisy GG. Cellular motility driven by assembly and disassembly of actin filaments. Cell. 2003;112:453–465. doi: 10.1016/s0092-8674(03)00120-x. [DOI] [PubMed] [Google Scholar]
- 2.Goley ED, Welch MD. The ARP2/3 complex: An actin nucleator comes of age. Nat Rev Mol Cell Biol. 2006;7:713–726. doi: 10.1038/nrm2026. [DOI] [PubMed] [Google Scholar]
- 3.Pollard TD, Blanchoin L, Mullins RD. Molecular mechanisms controlling actin filament dynamics in nonmuscle cells. Annu Rev Biophys Biomol Struct. 2000;29:545–576. doi: 10.1146/annurev.biophys.29.1.545. [DOI] [PubMed] [Google Scholar]
- 4.Sept D, McCammon JA. Thermodynamics and kinetics of actin filament nucleation. Biophys J. 2001;81:667–674. doi: 10.1016/S0006-3495(01)75731-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chhabra ES, Higgs HN. The many faces of actin: Matching assembly factors with cellular structures. Nat Cell Biol. 2007;9:1110–1121. doi: 10.1038/ncb1007-1110. [DOI] [PubMed] [Google Scholar]
- 6.Machesky LM, Atkinson SJ, Ampe C, Vandekerckhove J, Pollard TD. Purification of a cortical complex containing two unconventional actins from Acanthamoeba by affinity chromatography on profilin-agarose. J Cell Biol. 1994;127:107–115. doi: 10.1083/jcb.127.1.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Robinson RC, et al. Crystal structure of Arp2/3 complex. Science. 2001;294:1679–1684. doi: 10.1126/science.1066333. [DOI] [PubMed] [Google Scholar]
- 8.Mullins RD, Heuser JA, Pollard TD. The interaction of Arp2/3 complex with actin: Nucleation, high affinity pointed end capping, and formation of branching networks of filaments. Proc Natl Acad Sci USA. 1998;95:6181–6186. doi: 10.1073/pnas.95.11.6181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Egile C, et al. Mechanism of filament nucleation and branch stability revealed by the structure of the Arp2/3 complex at actin branch junctions. PLoS Biol. 2005;3:e383. doi: 10.1371/journal.pbio.0030383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rouiller I, et al. The structural basis of actin filament branching by the Arp2/3 complex. J Cell Biol. 2008;180:887–895. doi: 10.1083/jcb.200709092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Aguda AH, Burtnick LD, Robinson RC. The state of the filament. EMBO Rep. 2005;6:220–226. doi: 10.1038/sj.embor.7400363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dalhaimer P, Pollard TD. Molecular dynamics simulations of Arp2/3 complex activation. Biophys J. 2010;99:2568–2576. doi: 10.1016/j.bpj.2010.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xu X-P, et al. Three-dimensional reconstructions of Arp2/3 complex with bound nucleation promoting factors. EMBO J. 2012;31:236–247. doi: 10.1038/emboj.2011.343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pfaendtner J, et al. Key structural features of the actin filament Arp2/3 complex branch junction revealed by molecular simulation. J Mol Biol. 2012;416:148–161. doi: 10.1016/j.jmb.2011.12.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dayel MJ, Mullins RD. Activation of Arp2/3 complex: Addition of the first subunit of the new filament by a WASP protein triggers rapid ATP hydrolysis on Arp2. PLoS Biol. 2004;2:E91. doi: 10.1371/journal.pbio.0020091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Machesky LM, et al. Scar, a WASp-related protein, activates nucleation of actin filaments by the Arp2/3 complex. Proc Natl Acad Sci USA. 1999;96:3739–3744. doi: 10.1073/pnas.96.7.3739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yarar D, To W, Abo A, Welch MD. The Wiskott-Aldrich syndrome protein directs actin-based motility by stimulating actin nucleation with the Arp2/3 complex. Curr Biol. 1999;9:555–558. doi: 10.1016/s0960-9822(99)80243-7. [DOI] [PubMed] [Google Scholar]
- 18.Winter D, Lechler T, Li R. Activation of the yeast Arp2/3 complex by Bee1p, a WASP-family protein. Curr Biol. 1999;9:501–504. doi: 10.1016/s0960-9822(99)80218-8. [DOI] [PubMed] [Google Scholar]
- 19.Rohatgi R, et al. The interaction between N-WASP and the Arp2/3 complex links Cdc42-dependent signals to actin assembly. Cell. 1999;97:221–231. doi: 10.1016/s0092-8674(00)80732-1. [DOI] [PubMed] [Google Scholar]
- 20.Marchand JB, Kaiser DA, Pollard TD, Higgs HN. Interaction of WASP/Scar proteins with actin and vertebrate Arp2/3 complex. Nat Cell Biol. 2001;3:76–82. doi: 10.1038/35050590. [DOI] [PubMed] [Google Scholar]
- 21.Chereau D, et al. Actin-bound structures of Wiskott-Aldrich syndrome protein (WASP)-homology domain 2 and the implications for filament assembly. Proc Natl Acad Sci USA. 2005;102:16644–16649. doi: 10.1073/pnas.0507021102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Panchal SC, Kaiser DA, Torres E, Pollard TD, Rosen MK. A conserved amphipathic helix in WASP/Scar proteins is essential for activation of Arp2/3 complex. Nat Struct Biol. 2003;10:591–598. doi: 10.1038/nsb952. [DOI] [PubMed] [Google Scholar]
- 23.Ti S-C, Jurgenson CT, Nolen BJ, Pollard TD. Structural and biochemical characterization of two binding sites for nucleation-promoting factor WASp-VCA on Arp2/3 complex. Proc Natl Acad Sci USA. 2011;108:E463–E471. doi: 10.1073/pnas.1100125108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Padrick SB, Doolittle LK, Brautigam CA, King DS, Rosen MK. Arp2/3 complex is bound and activated by two WASP proteins. Proc Natl Acad Sci USA. 2011;108:E472–E479. doi: 10.1073/pnas.1100236108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Boczkowska M, Rebowski G, Kast DJ, Dominguez R. Structural analysis of the transitional state of Arp2/3 complex activation by two actin-bound WCAs. Nat Commun. 2014;5:3308. doi: 10.1038/ncomms4308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Higgs HN, Blanchoin L, Pollard TD. Influence of the C terminus of Wiskott-Aldrich syndrome protein (WASp) and the Arp2/3 complex on actin polymerization. Biochemistry. 1999;38:15212–15222. doi: 10.1021/bi991843+. [DOI] [PubMed] [Google Scholar]
- 27.Mullins RD, Pollard TD. Rho-family GTPases require the Arp2/3 complex to stimulate actin polymerization in Acanthamoeba extracts. Curr Biol. 1999;9:405–415. doi: 10.1016/s0960-9822(99)80187-0. [DOI] [PubMed] [Google Scholar]
- 28.Ma L, Rohatgi R, Kirschner MW. The Arp2/3 complex mediates actin polymerization induced by the small GTP-binding protein Cdc42. Proc Natl Acad Sci USA. 1998;95:15362–15367. doi: 10.1073/pnas.95.26.15362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wagner AR, Luan Q, Liu SL, Nolen BJ. Dip1 defines a class of Arp2/3 complex activators that function without preformed actin filaments. Curr Biol. 2013;23:1990–1998. doi: 10.1016/j.cub.2013.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rodal AA, et al. Conformational changes in the Arp2/3 complex leading to actin nucleation. Nat Struct Mol Biol. 2005;12:26–31. doi: 10.1038/nsmb870. [DOI] [PubMed] [Google Scholar]
- 31.Hetrick B, Han MS, Helgeson LA, Nolen BJ. Small molecules CK-666 and CK-869 inhibit actin-related protein 2/3 complex by blocking an activating conformational change. Chem Biol. 2013;20:701–712. doi: 10.1016/j.chembiol.2013.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rodnick-Smith M, Liu SL, Balzer CJ, Luan Q, Nolen BJ. Identification of an ATP-controlled allosteric switch that controls actin filament nucleation by Arp2/3 complex. Nat Commun. 2016;7:12226. doi: 10.1038/ncomms12226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Goley ED, Rodenbusch SE, Martin AC, Welch MD. Critical conformational changes in the Arp2/3 complex are induced by nucleotide and nucleation promoting factor. Mol Cell. 2004;16:269–279. doi: 10.1016/j.molcel.2004.09.018. [DOI] [PubMed] [Google Scholar]
- 34.Beltzner CC, Pollard TD. Pathway of actin filament branch formation by Arp2/3 complex. J Biol Chem. 2008;283:7135–7144. doi: 10.1074/jbc.M705894200. [DOI] [PubMed] [Google Scholar]
- 35.Schuler B, Lipman EA, Eaton WA. Probing the free-energy surface for protein folding with single-molecule fluorescence spectroscopy. Nature. 2002;419:743–747. doi: 10.1038/nature01060. [DOI] [PubMed] [Google Scholar]
- 36.Kalinin S, et al. A toolkit and benchmark study for FRET-restrained high-precision structural modeling. Nat Methods. 2012;9:1218–1225. doi: 10.1038/nmeth.2222. [DOI] [PubMed] [Google Scholar]
- 37.Ti S-C, Pollard TD. Purification of actin from fission yeast Schizosaccharomyces pombe and characterization of functional differences from muscle actin. J Biol Chem. 2011;286:5784–5792. doi: 10.1074/jbc.M110.199794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kapanidis AN, et al. Alternating-laser excitation of single molecules. Acc Chem Res. 2005;38:523–533. doi: 10.1021/ar0401348. [DOI] [PubMed] [Google Scholar]
- 39.Nolen BJ, Pollard TD. Insights into the influence of nucleotides on actin family proteins from seven structures of Arp2/3 complex. Mol Cell. 2007;26:449–457. doi: 10.1016/j.molcel.2007.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Boczkowska M, et al. X-ray scattering study of activated Arp2/3 complex with bound actin-WCA. Structure. 2008;16:695–704. doi: 10.1016/j.str.2008.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Luan Q, Zelter A, MacCoss MJ, Davis TN, Nolen BJ. Identification of Wiskott-Aldrich syndrome protein (WASP) binding sites on the branched actin filament nucleator Arp2/3 complex. Proc Natl Acad Sci USA. 2018;115:E1409–E1418. doi: 10.1073/pnas.1716622115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Granier S, et al. Structure and conformational changes in the C-terminal domain of the beta2-adrenoceptor: Insights from fluorescence resonance energy transfer studies. J Biol Chem. 2007;282:13895–13905. doi: 10.1074/jbc.M611904200. [DOI] [PubMed] [Google Scholar]
- 43.Luan Q, Nolen BJ. Structural basis for regulation of Arp2/3 complex by GMF. Nat Struct Mol Biol. 2013;20:1062–1068. doi: 10.1038/nsmb.2628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jurgenson CT, Pollard TD. Crystals of the Arp2/3 complex in two new space groups with structural information about actin-related protein 2 and potential WASP binding sites. Acta Crystallogr F Struct Biol Commun. 2015;71:1161–1168. doi: 10.1107/S2053230X15013515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nolen B, Pollard TD. Crystal structure and characterization of fission yeast Arp2/3 complex lacking Arp2. J Biol Chem. 2008;283:26490–26498. doi: 10.1074/jbc.M802607200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pfaendtner J, Voth GA. Molecular dynamics simulation and coarse-grained analysis of the Arp2/3 complex. Biophys J. 2008;95:5324–5333. doi: 10.1529/biophysj.108.143313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nolen BJ, Littlefield RS, Pollard TD. Crystal structures of actin-related protein 2/3 complex with bound ATP or ADP. Proc Natl Acad Sci USA. 2004;101:15627–15632. doi: 10.1073/pnas.0407149101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dalhaimer P, Pollard TD, Nolen BJ. Nucleotide-mediated conformational changes of monomeric actin and Arp3 studied by molecular dynamics simulations. J Mol Biol. 2008;376:166–183. doi: 10.1016/j.jmb.2007.11.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kiselar JG, Mahaffy R, Pollard TD, Almo SC, Chance MR. Visualizing Arp2/3 complex activation mediated by binding of ATP and WASp using structural mass spectrometry. Proc Natl Acad Sci USA. 2007;104:1552–1557. doi: 10.1073/pnas.0605380104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Goley ED, et al. An actin-filament-binding interface on the Arp2/3 complex is critical for nucleation and branch stability. Proc Natl Acad Sci USA. 2010;107:8159–8164. doi: 10.1073/pnas.0911668107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Oda T, Iwasa M, Aihara T, Maéda Y, Narita A. The nature of the globular- to fibrous-actin transition. Nature. 2009;457:441–445. doi: 10.1038/nature07685. [DOI] [PubMed] [Google Scholar]
- 52.Smith BA, et al. Three-color single molecule imaging shows WASP detachment from Arp2/3 complex triggers actin filament branch formation. eLife. 2013;2:e01008. doi: 10.7554/eLife.01008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Arasada R, Pollard TD. Distinct roles for F-BAR proteins Cdc15p and Bzz1p in actin polymerization at sites of endocytosis in fission yeast. Curr Biol. 2011;21:1450–1459. doi: 10.1016/j.cub.2011.07.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.MacLean-Fletcher S, Pollard TD. Identification of a factor in conventional muscle actin preparations which inhibits actin filament self-association. Biochem Biophys Res Commun. 1980;96:18–27. doi: 10.1016/0006-291x(80)91175-4. [DOI] [PubMed] [Google Scholar]
- 55.Pollard TD. Polymerization of ADP-actin. J Cell Biol. 1984;99:769–777. doi: 10.1083/jcb.99.3.769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mahaffy RE, Pollard TD. Kinetics of the formation and dissociation of actin filament branches mediated by Arp2/3 complex. Biophys J. 2006;91:3519–3528. doi: 10.1529/biophysj.106.080937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Metskas LA, Rhoades E. Conformation and dynamics of the troponin I C-terminal domain: Combining single-molecule and computational approaches for a disordered protein region. J Am Chem Soc. 2015;137:11962–11969. doi: 10.1021/jacs.5b04471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cooper JA, Walker SB, Pollard TD. Pyrene actin: Documentation of the validity of a sensitive assay for actin polymerization. J Muscle Res Cell Motil. 1983;4:253–262. doi: 10.1007/BF00712034. [DOI] [PubMed] [Google Scholar]
- 59.Kuhn JR, Pollard TD. Real-time measurements of actin filament polymerization by total internal reflection fluorescence microscopy. Biophys J. 2005;88:1387–1402. doi: 10.1529/biophysj.104.047399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Trexler AJ, Rhoades E. Alpha-synuclein binds large unilamellar vesicles as an extended helix. Biochemistry. 2009;48:2304–2306. doi: 10.1021/bi900114z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Middleton ER, Rhoades E. Effects of curvature and composition on α-synuclein binding to lipid vesicles. Biophys J. 2010;99:2279–2288. doi: 10.1016/j.bpj.2010.07.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Aebi U, Pollard TD. A glow discharge unit to render electron microscope grids and other surfaces hydrophilic. J Electron Microsc Tech. 1987;7:29–33. doi: 10.1002/jemt.1060070104. [DOI] [PubMed] [Google Scholar]
- 63.Li X, et al. Electron counting and beam-induced motion correction enable near-atomic-resolution single-particle cryo-EM. Nat Methods. 2013;10:584–590. doi: 10.1038/nmeth.2472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tang G, et al. EMAN2: An extensible image processing suite for electron microscopy. J Struct Biol. 2007;157:38–46. doi: 10.1016/j.jsb.2006.05.009. [DOI] [PubMed] [Google Scholar]
- 65.He S, Scheres SHW. Helical reconstruction in RELION. J Struct Biol. 2017;198:163–176. doi: 10.1016/j.jsb.2017.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Rohou A, Grigorieff N. CTFFIND4: Fast and accurate defocus estimation from electron micrographs. J Struct Biol. 2015;192:216–221. doi: 10.1016/j.jsb.2015.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.