Significance
Circular RNAs (circRNAs) play critical physiologic functions, but it is not known whether human DNA viruses express circRNAs. We surveyed Epstein−Barr virus (EBV) and Kaposi’s sarcoma herpesvirus (KSHV) tumors and cell lines, and found specific circRNAs expressed from both viruses. EBV circular BamHI A rightward transcripts (circBARTs) were expressed in all EBV tumor latency forms, including all EBV-infected posttransplant lymphoproliferative disease tumors tested, whereas EBV circBHLF1 and circLMP2 were more variably expressed. KSHV expressed circvIRF4 constitutively in primary effusion lymphoma cell lines, while the polyadenylated nuclear locus promiscuously generated variable, inducible, and bidirectional circRNAs. Tumor virus circRNAs can be long-lived, unique tumor biomarkers that may also open new research opportunities into understanding how these viruses cause cancer.
Keywords: Epstein−Barr virus, Kaposi sarcoma virus, circular RNA, cancer, herpesvirus
Abstract
Epstein−Barr virus (EBV) and Kaposi’s sarcoma herpesvirus (KSHV) cause ∼2% of all human cancers. RNase R-resistant RNA sequencing revealed that both gammaherpesviruses encode multiple, uniquely stable, circular RNAs (circRNA). EBV abundantly expressed both exon-only and exon−intron circRNAs from the BamHI A rightward transcript (BART) locus (circBARTs) formed from a spliced BART transcript and excluding the EBV miRNA region. The circBARTs were expressed in all verified EBV latency types, including EBV-positive posttransplant lymphoproliferative disease, Burkitt lymphoma, nasopharyngeal carcinoma, and AIDS-associated lymphoma tissues and cell lines. Only cells infected with the B95-8 EBV strain, with a 12-kb BART locus deletion, were negative for EBV circBARTs. Less abundant levels of EBV circRNAs originating from LMP2- and BHLF1-encoding genes were also identified. The circRNA sequencing of KSHV-infected primary effusion lymphoma cells revealed a KSHV-encoded circRNA from the vIRF4 locus (circvIRF4) that was constitutively expressed. In addition, KSHV polyadenylated nuclear (PAN) RNA locus generated a swarm (>100) of multiply backspliced, low-abundance RNase R-resistant circRNAs originating in both sense and antisense directions consistent with a novel hyperbacksplicing mechanism. In EBV and KSHV coinfected cells, exon-only EBV circBARTs were located more in the cytoplasm, whereas the intron-retaining circBARTs were found in the nuclear fraction. KSHV circvIRF4 and circPANs were detected in both nuclear and cytoplasmic fractions. Among viral circRNAs tested, none were found in polysome fractions from KSHV−EBV coinfected BC1 cells, although low-abundance protein translation from viral circRNAs could not be excluded. The circRNAs are a new class of viral transcripts expressed in gammaherpesvirus-related tumors that might contribute to viral oncogenesis.
The oncogenic human herpesviruses, Epstein−Barr virus (human herpesvirus 4, EBV/HHV4) (1) and Kaposi’s sarcoma herpesvirus (human herpesvirus 8, KSHV/HHV8) (2), cause a variety of human malignancies. EBV is associated with a broad spectrum of lymphoid and nonlymphoid malignancies (3, 4). KSHV causes Kaposi’s sarcoma—an endothelial cell tumor—and two hematolymphoid malignancies: primary effusion lymphoma (PEL) and multicentric Castleman’s disease (MCD) (2, 5, 6).
Viral gene expression and viral genome replication are generally tightly limited (latency) in viral tumors. For EBV-associated tumors, at least three latency types (I, II, and III) have been described based on the pattern of EBV proteins expressed (7–9). EBV also expresses small, noncoding RNAs (EBER 1 and 2) and several clusters of miRNA from intronic regions of the BamHI A rightward transcript (BART) (10, 11). In contrast to EBV, KSHV latency is not further subdivided, but differential KSHV gene and protein expression in various tumor types have been described (12–15). Both viruses also cause lymphoproliferative disorders, such as posttransplant lymphoproliferative disease (PTLD) for EBV and MCD for KSHV.
For KSHV, a number of noncoding RNAs (ncRNA) have been found, including 12 miRNAs (16, 17) that are variably detected during latency (18, 19), and the polyadenylated nuclear RNA (PAN) (20) which is expressed abundantly as an early lytic transcript but also at lower levels during latency (21, 22). Additional complementary and anticomplementary transcripts (called K7.3) have been described in the PAN region (23–26).
Covalently closed, nonpolyadenylated circular RNAs (circRNAs) were initially described from plant viroids (27), and, subsequently, the human hepatitis delta virus (28). The circRNAs form through backsplicing of a downstream 5′ splice donor site onto an upstream 3′ splice acceptor site (29) and can act as miRNA sponges (30, 31), as templates for cap-independent translation (32), and as competitive inhibitors to their corresponding linear mRNAs (33). Because they lack free ends, they are particularly resistant to exonuclease activity. Some cellular circRNAs are abundantly expressed in cancer tissues, and may be useful as diagnostic reagents (34).
In this study, we describe the identification and characterization of gammaherpesvirus circRNAs as a new class of viral transcripts. This work was initiated to discover novel human tumor viruses in EBV-negative PTLD by digital transcriptome subtraction (DTS) (35). While Merkel cell polyomavirus was identified by poly(A) DTS (36), no other human cancer viruses since have been found by this approach. We reasoned that circRNA tumor viruses might be missed by poly(A) DTS, and we performed ribosomal RNA-depleted, RNase R-treated RNA sequencing on a small number of EBV-negative and EBV-positive PTLDs. This led to the identification of EBV-encoded novel RNase R-resistant, backspliced junctional RNAs from the BART/RPMS1, BHLF1, and LMP-2 genomic loci. We extended this analysis to KSHV-infected cell lines and identified circRNAs expressed from the KSHV vIRF4 and PAN loci. This study surveys expression of these viral circRNAs in various tumors and describes initial findings on the subcellular localization and polysome association of viral circRNA. We also describe a highly variable, bidirectional backsplicing of the KSHV PAN locus.
Results
EBV circRNA Sequencing.
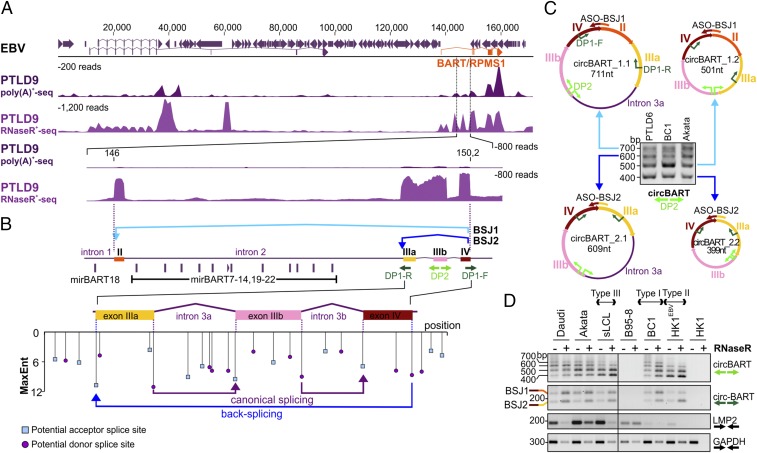
We performed RNA sequencing with two EBV-negative (PTLD4 and PTLD5) and two EBV-positive PTLD (PTLD6 and PTLD9) samples using poly(A)+-selected or RNase R-treated RNA libraries (Fig. 1A, Table 1, and Datasets S1 and S3). RNase R is an exoribonuclease that selectively depletes linear RNAs and enriches circRNAs or lariat RNAs. Backspliced junctions (BSJs) based on EBV genome Mutu sequence (KC207814) were identified using the CIRI2 circRNA prediction algorithm (37, 38). EBV-positive PTLD patient samples each showed two circRNA BSJ candidates from the BART locus: BSJ1 (Mutu: 146,095 to 150,210) and BSJ2 (Mutu: 149,443 to 150,210) (Fig. 1A, Table 1, and Dataset S1).
Fig. 1.
Identification and validation of EBV circRNAs. (A) Identification of EBV RNase R-resistant RNAs. Comparison of poly(A)+ RNA (PTLD9polyA+seq) and RNase R-treated RNA (PTLD9RnaseR+seq) from an EBV-positive PTLD sample (PTLD9) reveals RNase R-resistant RNAs that are potential BSJs of circRNAs. CIRI2 analysis using EBV Mutu genome KC207813 identified a minority of these reads to encode actual EBV BSJs (Table 1). An expanded view of the BART (RPMS1) region (146 kb to 150.2 kb) encoding the highest concentration of EBV BSJs revealed low mRNA but high RNase R-resistant RNA abundance. Standardized coverage depths (y axis) are indicated for each alignment. (B) EBV BART BSJ configurations. Shown is a diagram of the EBV BART mRNA coding region from exon II to exon IV. EBV circBART-BSJ1 (light blue) backsplices from exon IV to exon II, while BSJ2 (dark blue) backsplices from exon IV to exon IIIa, to generate circBART RNAs. EBV miRNA (mir)BART 7 to 22 miRNAs encoded by intron 2 are spliced out from the circBARTs. An expanded view of the BSJ2 region shows potential acceptor (blue square) and donor (purple circle) splice sites predicted by MaxEnt, with entropy scores given on the y axis. Divergent primers designed to verify BSJ1 and BSJ2 are shown in dark green (DP1) and light green (DP2), respectively. (C) Characterization of four circBART isoforms. Gel electrophoresis of DP2 RT-PCR products using EBV-positive PTLD6, BC1, and Akata RNAs shows four distinct bands corresponding to circBART_1.1 (711 nt), circBART_2.1 (609 nt), circBART_1.2 (501 nt), and circBART_2.2 (399 nt); circBART_1.1 and circBART_2.1 each contain the 210-nt intron 3a. ASOs targeting unique junction sequences are indicated by colored arrows. (D) The circBARTs are expressed in EBV-infected cell lines having different latency types. RNase R-treated (+) or RNase R-untreated (−) RNAs from cell lines having different latency forms were used for RT-PCR, with the DP2 primer pair (light green arrows) identifying all four circBART forms, or with the DP1 primer pair (dark green arrows) identifying only the two BSJs representing paired circBARTs. Convergent primers (black arrows) were used to measure viral LMP2 and cellular GAPDH mRNA transcripts. RNAs from EBV-uninfected HK1 and the EBV-positive B95-8 cell line, having a deletion of the BART locus, were used as negative controls. bp, base pair; nt, nucleotide.
Table 1.
EBV circRNAs
| Tumor | Proposed circRNA IDs | EBV Mutu (nucleotide no. KC207814), start | EBV Mutu (nucleotide no. KC207814), end | St. | BSJ reads | RPM |
| PTLD6 | circBART_1 | 146,095 | 150,210 | + | 1 | 37.0 |
| circBART_2 | 149,443 | 150,210 | + | 12 | 444.5 | |
| PTLD9 | circBART_1 | 146,095 | 150,210 | + | 9 | 61.3 |
| circBART_2 | 149,443 | 150,210 | + | 33 | 224.7 |
St., strand.
BART-BSJ1 results from the fusion of the 3′ end of exon IV with the 5′ end of exon II (Fig. 1B). BART-BSJ2 is formed by the fusion of the 3′ end of exon IV with the 5′ end of exon IIIa. Potential acceptor and donor splice sites within the BART region were examined using Human Splicing Finder 3.1 (39), which showed high entropy scores for canonical splice sites, including those flanking introns 3a and 3b, as well as for BART-BSJ2 (Fig. 1B), supporting the occurrence of this backsplicing event. BART BSJ1 and BSJ2 junction reads were also sequenced from RNase R-treated RNA of the EBV and KSHV coinfected BC1 cell line with or without sodium butyrate−phorbol ester (NaB/TPA) induction of viral lytic replication (25) (SI Appendix, Fig. S1 and Dataset S1).
Characterization of EBV circBARTs in EBV Cell Lines.
BSJ1 and BSJ2 junction reads of putative circBART_1 and circBART_2 candidates were identified at relatively high levels in both EBV-positive PTLDs [37.0 to 224.7 reads per million (RPM), Table 1] and in latent BC1 cells (between 144 and 320.3 RPM, Dataset S1). Therefore, we designed two junction-spanning divergent primer pairs (DP1 and DP2; Fig. 1B) to further confirm and characterize these circRNAs in different cell lines by reverse transcriptase (RT)-PCR. The DP2 primer pair amplified four bands ranging between 400 bp and 700 bp from PTLD6, BC1, and Akata cell RNAs (Fig. 1C), which were confirmed by cloning and sequencing. The circBART_1.1 (711 nt) and circBART_1.2 (501 nt) contain exons II, IIIa, IIIb, and IV and form the BSJ1 between exons II and IV upon backsplicing; circBART_2.1 (609 nt) and circBART_2.2 (399 nt) lack exon II and form the BSJ1 between exons IIIa and IV. In circBART_1.1 and circBART_2.1, intron 3a between exons IIIa and IIIb is additionally retained (Fig. 1C).
EBV circBART_1 and circBART_2 expression was further examined in RNAs from cell lines having various forms of EBV latency (Fig. 1D). Daudi, Akata, and BC1 have type I EBV latency, whereas PTLD-derived cell lines spontaneously immortalized by EBV (sLCL) express type III latency, and marmoset B95-8 is an EBV producer cell line (40). HK1EBV cells were derived by infecting the EBV-negative HK1 nasopharyngeal carcinoma (NPC) cell line with the EBV Akata strain and have type II latency. We detected three to four bands migrating between 400 bp and 700 bp in all samples except the EBV-uninfected HK1 control cells and the B95-8 cell line, which has a 12-kb deletion within the BART locus (41) from position 139,724 to 151,554 (NC_007605). Junction-spanning DP1 primers amplified bands migrating at 162 bp (BSJ2 of circBART_2.1 and circBART_2.2) and 264 bp (BSJ1 of circBART_1.1 and circBART_1.2) (Fig. 1D). In contrast to circBARTs, linear viral (LMP2) and cellular (GAPDH) transcripts were diminished following RNase R treatment (Fig. 1D).
Characterization of circBARTs in EBV Malignancies.
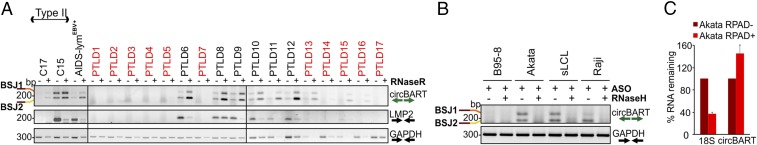
RNA was isolated from 17 PTLDs, including 6 EBV-positive and 11 EBV-negative specimens. EBV status was determined by clinical EBER positivity and RefSeq testing for one sample (PTLD12). All six EBV-positive PTLDs (type III latency) (42) were strongly positive for RNase R-resistant circBART_1 and circBART_2, whereas three of the EBV-negative samples (PTLD13, PTLD15, and PTLD16) were weakly positive (Fig. 2A). Several of these tumors had DNA available for retesting by EBV quantitative (q)PCR, including the three PTLDs clinically reported as EBV-negative by EBER staining but positive for circBART RT-PCR (PTLD13, PTLD15, and PTLD16). PTLD13 and PLTD15 had higher EBV genome copy numbers than EBER-positive PTLD8 and PTLD10 cases, suggesting false negativity for EBER staining. PTLD16 had <0.05 EBV genome copies per cell (see Materials and Methods for details) but retained weak circBART positivity.
Fig. 2.
(A) EBV circBARTs are expressed in EBV-positive tumor samples. RNase R-treated (+) or RNase R-untreated (−) RNAs from 6 EBV-positive and 11 EBV-negative (red font) pathologically diagnosed PTLD, NPC tumor xenoexplants C17 and C15, and an EBV-positive AIDS-associated lymphoma were tested by DP1 RT-PCR for circBART-BSJ1 and circBART-BSJ2 (Top). All six EBV-positive PTLDs were strongly positive, and three of the EBV-negative PTLDs were weakly positive for circBART RNAs. RT-PCR for LMP2 (Middle) and GAPDH (Bottom) mRNAs are shown. RNase R treatment enriched circBART junctions and depleted linear LMP2 and GAPDH products. (B) Confirmation of cyclization for EBV circBARTs by RNase H treatment. In vitro RNase H assays using annealed ASOs targeting BSJ1 and BSJ2 showed depletion of junctional sequences after RNase H treatment as monitored by DP1 PCR for Akata, sLCL, and Raji cell RNAs, but not in B95-8 RNA. GAPDH mRNA amplification was not affected by RNase H treatment. (C) RPAD analysis of EBV circBART. RNase R treatment followed by RPAD diminished linear 18S ribosomal RNA transcripts but increased circBART transcripts. Relative RNA was determined by normalizing the qPCR Ct values RPAD+ RNA to untreated control RNA (RPAD−). The data represent the means ± SD from four replicates.
C17 and C15 are two EBV-positive NPC xenografts that retain natively infected latent EBV infection (43, 44); both C17 and C15 were positive for RNase R-resistant circBART PCR products, although the viral gene load for both circBART and LMP2 RNAs was substantially higher in C15 tissue (Fig. 2A). Similarly, an EBV-positive AIDS-associated lymphoma was positive for RNase R-protected circBART products. In contrast, RNase R treatment diminished or eliminated linear viral (LMP2) and cellular (GAPDH) mRNA expression for the tumors.
To further confirm the circularity of circBART_1 and circBART_2, we designed two antisense DNA oligonucleotides (ASO-BSJ1 and ASO-BSJ2; Fig. 1C) targeting the unique junction sites for BSJ1 and BSJ2, respectively. The ASOs were annealed to isolated B95-8 (negative control), Akata, sLCL, and Raji RNAs (Fig. 2B). RNase H, which cleaves DNA:RNA hybrids, abolished DP1 RT-PCR positivity from Akata, sLCL, and Raji RNAs. GAPDH linear amplification products were not affected by RNase H treatment. We also used RNase R treatment followed by polyadenylation and poly(A)+ RNA depletion (RPAD), a method for purifying circRNAs in preference over linear RNAs (45). RPAD treatment of Akata RNA-depleted 18S ribosomal RNA (linear) relative to circBART_2, consistent with BART_2 circularization (Fig. 2C).
Minor EBV-encoded BSJs from BHLF1 and LMP2 were identified from several cell lines and tumors (SI Appendix, Fig. S1 B–D and Dataset S1). Notably, two BSJs from the LMP2 locus (360 nt to 1,682 nt and 1,026 nt to 1,682 nt) were identified by RNase R-protected sequencing of BC1 cells (Dataset S1). On RT-PCR analysis, using DP7 and DP8 primer pairs (SI Appendix, Table S1), multiple BSJs from presumed LMP2-encoded circRNAs were expressed from cell lines (Akata and B95-8) and C15, AIDS-associated lymphoma, and PTLD9 (SI Appendix, Fig. S1 B–D).
KSHV circRNAs Sequencing.
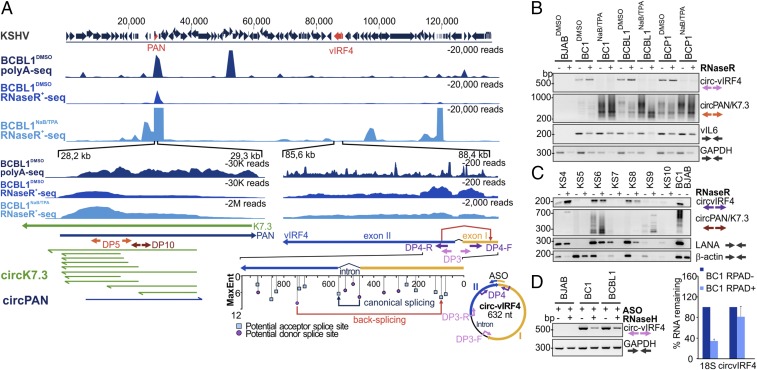
RNAs from DMSO-treated or NaB/TPA-induced KSHV-infected PEL cell lines BCBL1 and BC-1 were treated with RNase R before RNA sequencing to search for KSHV-encoded circRNAs. CIRI2 analysis revealed numerous potential KSHV circRNAs based on backspliced junctional alignments to the BCBL1 KSHV strain (HQ404500) (Table 2 and Dataset S2). Among these, a viral IFN regulatory factor 4 (vIRF4) BSJ read (87,690 nt to 88,321 nt) was detected in untreated cell lines at high levels (220 to 214 RPM). After lytic virus activation, the junction counts were reduced in BC1NaB/TPA from 220 RPM to 13 RPM, and in BCBL1NaB/TPA from 214 RPM to 27 RPM (Dataset S2). Assessment of potential acceptor and donor splice sites in this region showed relatively high entropy scores for the formation of this BSJ, and the complete circvIRF4 was sequenced using the DP9 primer pairs anchored in exon 1 (SI Appendix, Table S1).
Table 2.
KSHV circRNAs
| Cell line | Proposed circRNA IDs | KSHV BCBL1 (nucleotide no. HQ4045004), start | KSHV BCBL1 (nucleotide no. HQ4045004), end | St. | BSJ reads | RPM |
| BCBL1 | circvIRF4 | 87,690 | 88,321 | − | 33 | 214.1 |
| circK7.3_1 | 28,198 | 29,016 | − | 10 | 64.9 | |
| circK7.3_2 | 28,273 | 28,593 | − | 6 | 38.9 | |
| circK7.3_3 | 28,273 | 28,614 | − | 2 | 13.0 | |
| circK7.3_4 | 28,273 | 28,624 | − | 4 | 26.0 | |
| circK7.3_5 | 28,273 | 28,691 | − | 9 | 58.4 | |
| circK7.3_6 | 28,273 | 29,016 | − | 2 | 13.0 | |
| circK7.3_7 | 28,290 | 28,593 | − | 2 | 13.0 | |
| circK7.3_8 | 28,290 | 28,691 | − | 4 | 26.0 | |
| circPAN_1 | 28,406 | 29,044 | + | 7 | 45.4 | |
| circK7.3_9 | 28,519 | 29,016 | − | 2 | 13.0 | |
| circK7.3_10 | 28,692 | 29,016 | − | 6 | 38.9 | |
| BC1 | circvIRF4 | 87,690 | 88,321 | − | 95 | 220.5 |
| circK7.3_7 | 28,273 | 28,593 | − | 2 | 4.6 | |
| circK7.3_8 | 28,290 | 28,691 | − | 2 | 4.6 | |
| circK7.3_10 | 28,519 | 29,016 | − | 5 | 11.6 | |
| circ-miR_1 | 117,854 | 122,054 | − | 36 | 83.5 | |
| circ-miR_2 | 117,854 | 122,169 | − | 206 | 478.1 |
St., strand.
The circvIRF4 maps to the N terminus of its parent transcript: It is a 632-nucleotide intronic−exonic circRNA, with backsplicing flanking the canonical vIRF4 splice donor site (Fig. 3A). The circvIRF4 transcripts detected in latent PEL cells were resistant to RNase R digestion, in contrast to linear KSHV viral interleukin 6 (vIL6) and GAPDH mRNAs (Fig. 3B).
Fig. 3.
Identification and validation of KSHV circRNAs. (A) Identification of KSHV RNase R-resistant RNAs. Comparison of deposited BCBL1 poly(A)+ RNA sequences (SRX2323239, BCBL1polyA+seq) and RNase R-treated RNAs from BCBL1 cells with and without NaB/TPA revealed KSHV RNase R-resistant RNAs with potential BSJs from KSHV circRNAs. Two expanded views (Bottom), spanning the PAN/K7.3 and the vIRF4 regions, have BSJs identified by CIRI2 alignment to the deposited BCBL1 HQ404500 genome (Table 2). For circvIRF4 (nucleotides 85,600 to 88,400), backsplicing from a cryptic donor site in exon 2 to a cryptic acceptor site in exon 1 (red arrow) generates a single 632-bp RNA plasmid. Predicted MaxEnt splice sites with entropy scores are shown, as in Fig. 1A. For circPAN/K7.3 (nucleotides 28,200 to 29,300), multiple cyclized RNAs from both sense and antisense orientations were identified by BSJ analysis (Table 2). Shown here are predicted circRNA sequences in K7.3 sense (green arrows) and PAN sense (dark blue arrows) orientations from untreated BCBL1 cells. A divergent PCR primer pair (DP5) was designed to detect the most common circRNAs from this locus. (B) The circvIRF4 and circPAN/K7.3 are detected in KSHV-positive PELs. RNAs were extracted from DMSO or NaB/TPA-treated KSHV-positive BC1, BCBL1, and BCP1 and KSHV-negative BJAB cell lines and tested with DP3 and DP5 divergent primer RT-PCR (see A for primer location). Nuclease-resistant circvIRF4 was present in all untreated KSHV-positive cell lines but markedly diminished after NaB/TPA induction. In contrast, circPAN/K7.3 products were detected from all KSHV-positive cell lines and markedly increased after NaB/TPA treatment. The circPAN/K7.3 banding patterns varied between cell lines and with virus induction. Viral interleukin-6 (vIL6) and cellular GAPDH mRNA RT-PCR amplification is shown for comparison. (C) KSHV circRNAs in KS patient tissues. LANA mRNAs were detected in six out of seven KS tissues. The circvIRF4 BSJ was found in three KS samples (KS4, KS6, and KS8) which also showed higher levels of LANA mRNA. Various RNase R-resistant circPAN/K7.3 isoforms (∼250 bp to 700 bp) were detected in KS4, KS6, and KS9. BJAB and BC-1 RNAs were used as virus negative and positive controls, respectively. (D) Confirmation of cyclization for circvIRF4. In vitro RNase H assays (Left) using annealed ASO showed depletion of the circvIRF4 junctional sequences after RNase H treatment for BC1 and BCBL RNAs, but not in KSHV-negative BJAB RNAs. RPAD analysis (Right) revealed that ribosomal 18S RNA is selectively diminished compared with circvIRF4 RNA. Data represent the mean ± SD from three replicates.
We also found BSJ reads from the PAN/K7.3 locus (Fig. 3A and Table 2). Specific individual BSJ counts were low; however, the aggregate count of all BSJs from this region was very high. The majority of BSJs were from the complementary strand of the canonical PAN transcript, identified as K7.3 (25), and overlapped within the genome locus spanning 28,198 nt to 29,016 nt (BCBL1, HQ404500) (Fig. 3A and Dataset S2). We found 10 K7.3 and 1 PAN BSJs in latent BC1 and BCBL1 RNAs that would generate predicted circRNAs ∼304 to 819 nucleotides in length (Table 2). BC1 has the lowest number of circRNAs from this region. Following reactivation, the number of circPAN and circK7.3 RNA BSJ reads increased (Dataset S2). In BC1NaB/TPA, a total of 34 circPAN/K7.3 were identified at >500 RPM, 20 of which were also found in BCBL1NaB/TPA at >50 RPM. To validate the circRNA prediction analysis for the PAN/K7.3 region, we designed a divergent primer pair (DP5) that binds to a common region found in the majority of the predicted circPAN and circK7.3 RNAs (Fig. 3A). RT-PCR results using DP5 generated multiple bands ranging between ∼200 bp and 700 bp (Fig. 3B). The number and total intensity of these bands correlated with sequencing read counts, with BC1DMSO showing the least number of PCR products. The majority of circPAN transcripts were resistant to RNase R treatment, and their levels increased, in contrast to circvIRF4, following NaB/TPA treatment (Fig. 3B and Dataset S2). Sequencing analysis of circPAN/K7.3 PCR products cloned from BCBL1 confirmed some of the identified junctions.
Tissues from 10 KS tumors (KS1 to KS3 having degraded RNA, as a result of freeze−thaw during extended storage; KS4 to KS10 were obtained from AIDS and Cancer Specimen Resource) and a KSHV-positive MCD were compared with a PTLD (negative control) and BC1 (positive control) by KSHV circRNA RT-PCR (Fig. 3C and SI Appendix, Fig. S2B). The circvIRF4 was detected in 4 of the 10 KS tumor samples, and RNase R-resistant circPAN/K7.3 isoforms were present in MCD and 6 KS tumors despite diminished RNA integrity for some of the samples (Fig. 3C), as reflected by low β-actin, latency-associated nuclear antigen (LANA), and v-cyclin mRNA levels (SI Appendix, Fig. S2B). To confirm the circularity of circvIRF4, we performed an in vitro RNase H assay with an ASO targeting the unique circvIRF4 junction. This abolished circvIRF4 RT-PCR positivity from BC1 and BCBL1 RNAs, while control cellular GAPDH mRNA was unaffected (Fig. 3D). In addition, RNase R treatment followed by RPAD reduced linear 18S RNA did not significantly reduce circvIRF4 RNA levels.
In addition to circvIRF4 and circPAN/K7.3, a KSHV BSJ from the miRNA locus (Dataset S2) was detected by RT-PCR only in BC1NaB/TPA RNA, but not other cell lines, and was not further explored. In NaB/TPA-treated BC1 and BCBL1 cells, additional candidate BSJ reads were found at low abundance from K4, ORF49, ORF69, K12, ORF71, and ORF72 and from newly described transcripts K1.3, K4.5, K4.7, and K12.5 (Dataset S2).
Subcellular Localization of Viral circRNAs.
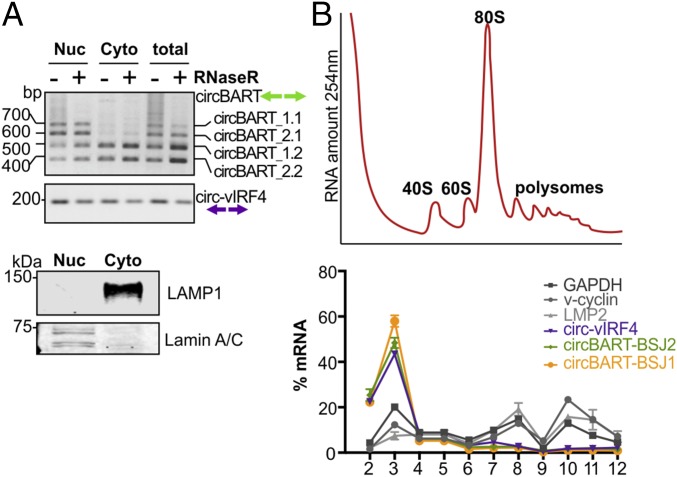
To functionally characterize these viral circRNAs, we isolated nuclear and cytoplasmic fractions of dually infected BC-1 cells (Fig. 4A). EBV circBART_1.1 and circBART_2.1, having a retained intron between exon IIIa and IIIb (Fig. 1B), were detected in the nuclear fraction, whereas entirely exonic circBART_1.2 and circBART_2.2 (Fig. 1B) and circvIRF4 were detected in both nuclear and cytoplasmic fractions (Fig. 4A).
Fig. 4.
Subcellular localization of viral circRNAs. (A) Nuclear vs. cytoplasmic localization of viral circRNAs. RNA extracted from nuclear (Nuc) and cytoplasmic (Cyto) fractions of the KSHV and EBV coinfected BC1 cell line was either treated (+) or untreated (−) with RNase R. BSJs spanning PCR products from intron-retaining circBART_1.1 and circBART_2.1 were detected mainly in the nuclear fraction. Exonic circBART_1.2 and circBART_2.2 were found in both fractions. The circvIRF4 junction-spanning PCR products were detected in both fractions. Protein immunoblotting for lamin A/C (nuclear) and LAMP1 (cytoplasmic) was used to confirm fractionation quality. (B) Polysome profile for viral circRNAs. RNA polysome sucrose gradient profile (254-nm absorbance) of BC1 cell lysates after treatment with CHX (Top). Fraction 1 is at the top of the gradient (free mRNAs) and 12 corresponds to the bottom of the gradient. Polysome RNA fractions are represented by fractions 9 to 12. Quantitative PCR revealed that circvIRF4, circBART_1, and circBART_2 RNAs were not detected in polysomal fractions, whereas mRNAs for translated v-cyclin, LMP2, and GAPDH proteins preferentially fractionated with polysomes (Bottom).
To determine whether the cytoplasmic viral circRNAs were associated with the cellular translation machinery, polysome fractionation was performed. The qRT-PCR analysis of polysome fractions revealed that both KSHV circvIRF4 and EBV circBART-BSJ1 and circBART-BSJ2 partitioned to untranslated fractions (Fig. 4B, fractions 2 to 4), whereas cellular and viral mRNAs were enriched in the polysome fractions (Fig. 4B, fractions 10 to 12).
Cellular circRNAs Identified in EBV/KSHV-Infected Tumors and Cell Lines.
In total, 30,178 human circRNAs were predicted with at least two BSJ reads in all PTLD and lymphoma cell lines sequenced (Dataset S3). Approximately 11% of these (1,385) were shared by all four samples (SI Appendix, Fig. S3B), but, notably, the majority of predicted circRNAs were not overlapping. In part, this may reflect the cellular heterogeneity found within PTLDs (e.g., tumor-infiltrating macrophages and T cells) (SI Appendix, Fig. S3C and Dataset S3). We found 35 and 40 novel human circRNAs that were exclusively detected in EBV-positive and EBV-negative PTLDs, respectively (SI Appendix, Fig. S3C and Dataset S3). We found a total of 22,276 and 13,641 human circRNA BSJs in DMSO- and NaB/TPA-treated BJAB, BC1, and BCBL1 cell lines; 5.3% (1,182) of the human circRNAs from the DMSO-treated samples and 3.7% (505) of the human circRNAs from the NaB/TPA-treated samples were only found in KSHV-infected PELs (SI Appendix, Fig. S4 B and C and Dataset S4). We identified 371 novel cellular circRNAs, in latent and lytic KSHV-positive PELs (SI Appendix, Fig. S4C and Dataset S4).
Discussion
The circRNAs, covalently cyclized single-stranded RNAs, are biologically active molecules. Despite extensive studies to characterize the EBV and KSHV genomes, transcriptomes, and proteomes, viral circRNAs have not been previously described. Our study shows that both EBV and KSHV encode circRNAs that are stably expressed in these viruses’ associated tumors, providing an example of a new form of viral gene expression that may play a role in tumor virus pathogenesis.
PTLDs arise in immunocompromised hosts after transplantation, most often after primary EBV infection. We performed RNase R sequencing on two EBV-negative and two EBV-positive PTLD libraries, which revealed several EBV-derived circRNA, most notably circBARTs that arise from the same gene locus encoding the EBV miRNAs (46). The circBART and BART miRNA are both encoded by the same parental pre-mRNA transcript, so it is not surprising that EBV circBART expression mirrors BART miRNA expression. To generate the circBART RNA, this pre-mRNA undergoes alternative splicing of BART miRNA intron and backsplicing to form the circRNA. Like EBV BART miRNAs, EBV circBARTs are expressed in all forms of EBV tumor latency but are deleted from the genome of the B95-8 EBV strain. EBV genome sequencing revealed the spontaneous occurrence of a BART deletion eliminating miRNA and circBART expression in 1 of 50 lymphoblastoid cell lines (AFB1) (47), suggesting that neither circBARTs nor BART miRNA are obligatory for the maintenance of the EBV genome in cell culture or to achieve in vitro immortalization of B cells. The circBARTs, however, were retained in all EBV-associated tumors we examined, which may indicate that they play important roles in tumor cell reproductive fitness for EBV-positive cancers.
In comparison with LMP2 and BHLF1 loci, the EBV BART locus produces four major circRNA at relative high abundance, which might be a reflection of high transcriptional activity within this region. These consist of two pairs of circRNAs, with each pair sharing the same BSJ, either exon IIIa (BSJ1) or exon II (BSJ2), serving as acceptors to a donor exon IV. Each pair either retains the intron between exons IIIa and IIIb (circBART_1.1 or circBART_2.1) or splices out this intron to become exon-only circRNAs (circBART_1.2 and circBART_2.2). We find that intron-containing circBARTs are retained in the nucleus, whereas exon-only circBARTs and circvIRF4 RNAs are found in both the nucleus and the cytoplasm. Most cellular exonic circRNAs are exported to the cytoplasm either through the nuclear export machinery or released upon nuclear envelope breakdown during mitosis (29, 48). Huang et al. (49) suggest that export of cellular circRNA from the nucleus may be controlled by length measurement mechanisms encoded by RNA helicase proteins UAP56 and URH49. The intron−exon circBART do not reach the length cutoff level found by Huang et al., but circBART nuclear retention may be due to length or presence of an intron (which will also increase circRNA size). In comparison, KSHV circvIRF4, which retains an intron-like feature, is distributed to both nuclear and cytoplasmic fractions. We also surveyed viral circRNA association with BC-1 cell polysomes and did not find evidence for polysome association by qRT-PCR. To fully address whether individual viral circRNAs engage in protein synthesis or polysome targeting, however, requires additional experimentation.
The circRNA sequencing of KSHV-infected PEL cells identified BSJs from the vIRF4 and PAN loci. KSHV vIRF4 resides among three other annotated ORFs: vIRF1, vIRF2, and vIRF3/LANA2 in a ∼10-kb segment of the BC1 genome (83,860 nt to 93,367 nt), which has a complicated transcript organization (15, 50). The KSHV IRFs have sequence and functional similarity to cellular IRFs, but also encode for additional functions (51, 52). The protein-encoding vIRF4 mRNA has only one annotated N-terminally located intron, but neither of these canonical splice donor or acceptor sites are used in the backsplicing of circvIRF4 RNA. Instead, the vIRF4 BSJ uses alternative splice donor and acceptor sequences embedded within exon I and exon II, presumably recognized by the spliceosome, to generate a circRNA.
The KSHV PAN region circRNAs are numerous, and, because they form from both sense (PAN) and antisense (K7.3) transcripts and possess variable BSJs, their analysis is challenging. PAN is a class II transcript (22) that is abundantly expressed during lytic viral replication, accounting for up to 80% of virus-encoded transcripts (26). PAN has not been previously known to be spliced, and we cannot rule out the possibility that promiscuous amplification of circRNA is an artifact of library construction or amplification or may have formed as rare “processing errors” due to the sheer abundance of parental viral pre-mRNAs. If these circRNAs are generated in cells, our findings support the existence of K7.3 transcript. In aggregate, the number of PAN/K7.3 BJSs was abundant, but each individual single BSJ was present at low copy number. We did not find an obvious sequence feature accounting for the hypervariable and bidirectional generation of PAN/K7.3 circRNAs.
Presence of virus-encoded circRNA also raises the possibility for new diagnostic tests to detect these viruses. Five of the six EBV-positive PTLDs (Fig. 2A) were EBER-positive, and the sixth (PTLD12) was EBER-negative but found to have abundant EBV mRNA reads by poly(A) RNA sequencing. Three of the remaining 11 EBER-negative PTLD tissues had weak RT-PCR positivity for circBART RNA. On retesting two of the three “EBV-negative” (PTLD13 and PTLD15) had EBV genome copy numbers comparable to EBV-positive PTLD8 and PTLD10, suggesting false-negative results for the EBER diagnostic test. For the KS tumors tested, several retained KSHV circRNA positivity despite evidence for severe mRNA degradation. While circvIRF4 and circPAN/K7.3 were readily detected in KSHV-infected PEL cell lines, additional studies are needed to determine whether the variable pattern for KS and MCD tumors reflects sample handling or a real difference in tumor gene expression.
The circRNAs are highly resistant to exonuclease degradation and may be retained even in formalin-fixed, paraffin-embedded specimens. Plasma EBV DNA viral loads are highly predictive for NPC (53), and it would be valuable to determine whether circRNA measurements improve prediction for clinical diseases, particularly tumors, over viral DNA determinations. Cellular circRNAs that are induced in various tumor types have shown promise as biomarkers for some cancers (34, 54). We anticipate that viral circRNAs might have even better discriminatory power for detecting EBV- and/or KSHV-infected tumors. We detected KSHV circRNA in PEL cell lines by sequencing a limited number of patient samples including Kaposi sarcoma and MCD by RT-PCR. Detection of these circRNAs in a larger number of various tumor tissues will provide insights to their contributions to KSHV-related malignancies.
We describe here an initial survey of RNA circularization among the human gammaherpesviruses. We do not know what, if any, function they play in virus maintenance, but it will be intriguing to determine whether other nontumorigenic herpesviruses or small DNA viruses also encode viral RNAs. Viral circRNAs may serve as important model systems for RNA biology, since they are expressed from small, well-defined operons, and these circRNA can be manipulated in the context of the viral genome. If these RNAs are found to contribute to viral tumorigenesis, they then represent a possible substrate for targeted therapies that may be more specific and less toxic than current therapeutic approaches.
Materials and Methods
Tumor Samples and Cell Lines.
Seventeen tissue specimens from patients with PTLD, one EBV-positive AIDS-associated lymphoma, three Kaposi’s sarcoma (KS 1 to 3), and MCD were obtained as byproducts of diagnostic or therapeutic procedures performed at Columbia University College of Physicians & Surgeons and at the University of Pittsburgh Medical Center (UPMC) with approval of the Institutional Review Board. These specimens were deidentified before use in our study. Seven pathologically confirmed tissue specimens were obtained from AIDS and Cancer Specimen Resource (ACSR) (KS 4 to 10). Tissues were snap-frozen and stored in liquid nitrogen until use. Assignment of EBV viral status for PTLDs was based on pathology reports and, in one case, based on poly(A) RNA sequencing (PTLD12). Tumor sections from two NPC patient-derived xenograft tumor models, C15 and C17 (43), were kindly provided by Nancy Raab-Traub, University of North Carolina.
EBV-positive Daudi, Raji, and B95-8; KSHV and EBV coinfected BC1; KSHV-positive BCBL1; and EBV/KSHV-negative BJAB cell lines were obtained from the American Type Culture Collection (ATCC). EBV-positive sLCL (55) was a generous gift from Cliona Rooney, Texas Children’s Hospital. Cells were maintained in Roswell Park Memorial Institute (RPMI) 1640 (Cellgro) supplemented with 10% FBS (VWR Seradigm). Recombinant Akata and the HK1 NPC cell line infected with recombinant Akata strain (56) were maintained with 800 µg/mL of neomycin selection in RPMI supplemented with 10% FBS. BC1, BCBL1, Daudi, Raji, and BJAB cell lines were authenticated by the University of Arizona Genetics Core Facility. The Akata and sLCL cell lines showed unique profiles with no matches to any reference in any database and thus were determined not to be contaminated with known cell lines.
For lytic reactivation, BJAB and KSHV-positive BC1 and BCBL1 cells were incubated with 20 ng/mL of TPA and 3 mM NaB for 48 h; EBV-positive cell lines were incubated with 20 ng/mL of TPA and 5 mM NaB for 48 h. Efficiency of lytic reactivation was measured by qRT-PCR analysis of immediate early (ORF50, ORF39), early (K8, ORF37), and latent (v-cyclin, viral interleukin 6, vIL6) viral transcript expression (SI Appendix, Fig. S2A).
RNA Isolation, poly(A)+ RNA Sequencing, and circRNA Sequencing.
Total RNA was isolated from tumor samples and cell lines using TRIzol (Ambion) followed by treatment with TURBO DNase (Thermo Fisher). RNA quality was confirmed by Agilent TapeStation (Children’s Hospital of Pittsburgh of UPMC, sequencing core facility) and by Agilent 2100 Bioanalyzer (CD Genomics). RNA integrity numbers (RIN) were between 1.9 and 2.1 (A 260/280), and RIN was ≥7.5 for all samples, except BC1NaB/TPA, BCBL1NaB/TPA, and PTLD9 (RIN ≥ 5.7 to 7.3). For poly(A)+ RNA sequencing of PTLD samples, Ion Torrent adapter-ligated libraries were prepared from extracted RNA according to the Ion Total RNA-seq Kit (Life Technologies) following the manufacturer’s instructions and sequenced using Ion PGM sequences at the Children’s Hospital of Pittsburgh of UPMC, sequencing core facility. For circRNA sequencing, ribosome-depleted and RNase R-treated RNA samples were used for library preparation and subsequently sequenced using Illumina HiSeq platform in PE150 sequencing mode (CD Genomics). The accession number for the sequencing data reported in this paper is Gene Expression Omnibus database GSE117798.
Bioinformatic Analysis.
Raw FastQ files were trimmed with Trim Galore, (www.bioinformatics.babraham.ac.uk/projects/trim_galore/) using the following parameters: q = 25, e = 0.1, and length = 50, and the quality control was performed with FastQC tool. CIRI2 algorithm was used for viral and human circRNA prediction (37) (https://sourceforge.net/projects/ciri/files/CIRI2/) with the default settings. In addition to CIRI2, we used CIRCexplorer (57) (https://github.com/YangLab/CIRCexplorer2) algorithm to confirm viral circRNA predictions. RNA-seq reads were aligned to GRCh37 (Hg19; University of California, Santa Cruz Genome Browser), BCBL1 (HQ404500), and Mutu (KC207814) reference genomes using BWA or STAR mappers. Human circRNAs were further analyzed using circBASE (58) to annotate the identified circRNAs in PTLD samples and PEL cell lines.
CLC genomics workbench (Qiagen) was used to align RNA-seq reads to GRCh37 (Hg19), BCBL1 (HQ404500), and Mutu (KC207814) reference genomes and to visualize additional annotation. DMSO-treated poly(A) RNA sequencing data for BCBL1 cell lines (SRX2323239, ref. 59) were obtained from National Center for Biotechnology Information’s Gene Expression Omnibus website.
Potential splice acceptor and donor site analysis was done using Human Splicing Finder (V3.1) (39). Venn diagrams were generated using Biovenn (60) and nVenn (61) programs.
RNase R Treatment and RPAD.
To obtain highly purified circRNAs, 2 µg of RNA was treated with 8 units (U) RNase R (Lucigen) in 1× Rnase R buffer at 37 °C for 30 min. The reaction mixture was heat-inactivated at 65 °C for 20 min or the RNA was precipitated using sodium acetate/ethanol supplemented with 20 µg of glycogen as a carrier. This was followed by polyadenylation (E-PAP, AM1350; Thermo Fisher) with a subsequent poly(A)+ RNA depletion using Poly(A)Purist MAG Kit (AM1922; Thermo Fisher) (RPAD protocol) as described by Panda et al. (45).
cDNA Synthesis, RT-PCR, and qPCR.
One microgram of DNase digested RNA was either treated or untreated with Rnase R and reverse-transcribed using SuperScript IV (Thermo Fisher) with random hexamers in a total volume of 20 µL, according to the manufacturer’s protocol.
All RT-PCRs were performed using 1/40 of the cDNA, Q5 high-fidelity polymerase (NEB) or standard Taq polymerase (NEB). Q5 PCR reactions were performed at the following conditions: initial denaturation at 98 °C for 2 min; followed by 35 cycles of denaturation at 98 °C for 10 s, based on the primer pairs annealing at 65°C to 71 °C for 30 s; extension at 72 °C for 30 s/kb; and a final extension at 72 °C for 5 min. For standard Taq polymerase supplemented with Thermopol buffer (NEB), we performed initial denaturation at 95 °C for 3 min; followed by 25 to 30 cycles of denaturation at 95 °C for 15 s, annealing at 56 °C for 30 s; and extension at 68 °C for 60 s/kb and a final extension at 68 °C for 5 min. As needed, RT-PCR products were gel-extracted and cloned into TOPO-TA vector (Invitrogen) according to the manufacturer’s recommendations.
Synthesized cDNA was analyzed by qPCR using SYBR Green PowerUp Master Mix according to the manufacturer’s instructions (Thermo Fisher). The determined threshold cycle (Ct) values were used to calculate the mRNA fold changes of the NaB/TPA-treated versus DMSO-treated cells using the delta−delta Ct method. The Ct values of GAPDH were used as reference. PCR primers [Integrated DNA Technologies (IDT)] are listed in SI Appendix, Table S1.
EBV DNA copy number was determined by the SYBR green (Thermo Fisher) qPCR absolute quantitation method using a BALF5 plasmid as template for the standard curve. The linear limits of detection were between 4 and 4 × 108 copies per reaction. Reactions were assembled as previously described (62). Input genomic DNA was normalized and compared with a reference cell line (Raji) averaging 50 EBV episomal copies per cell. EBER-positive PTLD8 and PTLD10 measured two and seven copies per cell, respectively. EBER-negative PTLD13, PTLD15, and PTLD16 samples measured 7, 14, and 0.05 copies per cell, respectively. Sample PTLD16 may contain EBV-infected infiltrating B lymphocytes and is more similar in value to the EBER-negative and circRNA-negative PTLD7 measuring 0.001 copy per cell.
Oligonucleotide-Targeted RNase H Cleavage.
ASOs were designed against the unique junction sites for each viral circRNA and contain phosphorothioate linkages for increased stability as well as six nucleotides at each end containing 2′-O-methylated ribose for exo/endonuclease resistance. HPLC-purified (with Na+ salt exchange) ASOs were obtained from IDT. For in vitro RNase H assays, 2 μg of RNA was incubated with 0.4 μg of ASO in 1× RNase H buffer at 37 °C for 20 min. Subsequently, 1 U RNase H (NEB) was added, followed by incubation for an additional 40 min. RNA was purified either using Qiagen RNeasy columns or by sodium acetate/ethanol precipitation with 20 µg of glycogen as carrier. ASO and scrambled controls (IDT) are listed in SI Appendix, Table S1.
Nuclear/Cytoplasmic Fractionation.
Nuclear/cytoplasmic fractionation was performed from 1 × 107 BC1 cells using the NR-PER Nuclear and Cytoplasmic Extraction Reagent (Pierce), according to the manufacturer’s protocol. One microgram of total RNA from each fraction was used for cDNA synthesis, and expression level of the indicated circRNAs in each fraction was analyzed. The quality of the fractionation assay was controlled by immunoblotting for a nuclear marker (Lamin A/C; Cell Signaling) and a cytoplasmic marker (LAMP-1; eBioscience).
Polysome Fractionation.
BC1 cells were incubated with 100 µg/mL of cycloheximide (CHX) for 15 min, harvested, rinsed with ice-cold PBS−CHX, and lysed in 500 µL of polysome lysis buffer (10 mM Hepes pH 7.4, 0.5% Nonidet P-40, 100 nM KCl, 5 nM MgCl2) freshly supplemented with CHX, and protease inhibitor Ribolock RNase Inhibitor (Thermo Fisher). After centrifugation (15 min at 17,000 × g), the cytoplasmic lysates (1 mg of lysate in <400-µL volume) were loaded onto 10 to 50% (wt/vol, 0.9 mL) linear sucrose gradients (10 mM Hepes pH 7.4, 100 mM KCl, 5 mM MgCl2). Gradients were centrifuged for 3 h at 145,000 × g (35,000 rpm in a Sorvall AH-650 rotor), followed by collection of 12 × 0.5 mL fractions. RNA was extracted from the collected fractions as described in RNA Isolation, poly(A)+ RNA Sequencing, and circRNA Sequencing using TRIzol LS reagent (Ambion) and treated with DNase before cDNA synthesis and qRT-PCR. Using the qPCR cycle threshold (Ct) values, the percent distribution for the mRNAs across the gradients was calculated using the delta Ct method (63).
Supplementary Material
Acknowledgments
We thank Pierre Busson and Nancy Raab-Traub for providing samples of the C15 and C17 NPC explants, Cliona Rooney for providing the sLCL1 cell line, Robert White for discussion of EBV variant genome sequencing data, Matthias Grabmair for technical assistance in bioinformatics analysis, Huichen Feng for poly(A) RNA sequencing, and Justin Wendzicki for his technical assistance. We were helped in the preparation of this manuscript by Einojuhani Rautavaara and Jóhann Jóhannsson. We thank ACSR, funded by the National Cancer Institute, for providing KS tumor samples. This work used the Bridges system, which is supported by National Science Foundation Award ACI-1445606, at the Pittsburgh Supercomputing Center (64). This study was supported by National Institutes of Health Grants R35 CA197463 (to P.S.M.) and CA170354 (to Y.C.), who are also supported as American Cancer Society Research Professors, National Cancer Institute grant and University of Pittsburgh Skin National Cancer Institute Specialized Programs of Research Excellence (SPORE) Career Enhancement Program funding (to T.T.). This project used the University of Pittsburgh Cancer Center’s shared resources cores supported, in part, by Award P30CA047904.
Footnotes
Conflict of interest: T.T., P.S.M., and Y.C. have made a US patent application based on these findings that has been assigned to the University of Pittsburgh.
Data deposition: The sequences reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, https://www.ncbi.nlm.nih.gov/geo (accession no. GSE117798).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1811728115/-/DCSupplemental.
References
- 1.Epstein MA, Achong BG, Barr YM. Virus particles in cultured lymphoblasts from Burkitt’s lymphoma. Lancet. 1964;1:702–703. doi: 10.1016/s0140-6736(64)91524-7. [DOI] [PubMed] [Google Scholar]
- 2.Chang Y, et al. Identification of herpesvirus-like DNA sequences in AIDS-associated Kaposi’s sarcoma. Science. 1994;266:1865–1869. doi: 10.1126/science.7997879. [DOI] [PubMed] [Google Scholar]
- 3.Young LS, Yap LF, Murray PG. Epstein-Barr virus: More than 50 years old and still providing surprises. Nat Rev Cancer. 2016;16:789–802. doi: 10.1038/nrc.2016.92. [DOI] [PubMed] [Google Scholar]
- 4.Shannon-Lowe C, Rickinson AB, Bell AI. Epstein-Barr virus-associated lymphomas. Philos Trans R Soc Lond B Biol Sci. 2017;372:20160271. doi: 10.1098/rstb.2016.0271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cesarman E, Chang Y, Moore PS, Said JW, Knowles DM. Kaposi’s sarcoma-associated herpesvirus-like DNA sequences in AIDS-related body-cavity-based lymphomas. N Engl J Med. 1995;332:1186–1191. doi: 10.1056/NEJM199505043321802. [DOI] [PubMed] [Google Scholar]
- 6.Soulier J, et al. Kaposi’s sarcoma-associated herpesvirus-like DNA sequences in multicentric Castleman’s disease. Blood. 1995;86:1276–1280. [PubMed] [Google Scholar]
- 7.Young L, et al. Expression of Epstein-Barr virus transformation-associated genes in tissues of patients with EBV lymphoproliferative disease. N Engl J Med. 1989;321:1080–1085. doi: 10.1056/NEJM198910193211604. [DOI] [PubMed] [Google Scholar]
- 8.Young LS, et al. Epstein-Barr virus gene expression in nasopharyngeal carcinoma. J Gen Virol. 1988;69:1051–1065. doi: 10.1099/0022-1317-69-5-1051. [DOI] [PubMed] [Google Scholar]
- 9.Rowe M, et al. Differences in B cell growth phenotype reflect novel patterns of Epstein-Barr virus latent gene expression in Burkitt’s lymphoma cells. EMBO J. 1987;6:2743–2751. doi: 10.1002/j.1460-2075.1987.tb02568.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pfeffer S, et al. Identification of virus-encoded microRNAs. Science. 2004;304:734–736. doi: 10.1126/science.1096781. [DOI] [PubMed] [Google Scholar]
- 11.Edwards RH, Marquitz AR, Raab-Traub N. Epstein-Barr virus BART microRNAs are produced from a large intron prior to splicing. J Virol. 2008;82:9094–9106. doi: 10.1128/JVI.00785-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sarid R, Wiezorek JS, Moore PS, Chang Y. Characterization and cell cycle regulation of the major Kaposi’s sarcoma-associated herpesvirus (human herpesvirus 8) latent genes and their promoter. J Virol. 1999;73:1438–1446. doi: 10.1128/jvi.73.2.1438-1446.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dittmer D, et al. A cluster of latently expressed genes in Kaposi’s sarcoma-associated herpesvirus. J Virol. 1998;72:8309–8315. doi: 10.1128/jvi.72.10.8309-8315.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Parravicini C, et al. Differential viral protein expression in Kaposi’s sarcoma-associated herpesvirus-infected diseases: Kaposi’s sarcoma, primary effusion lymphoma, and multicentric Castleman’s disease. Am J Pathol. 2000;156:743–749. doi: 10.1016/S0002-9440(10)64940-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rivas C, Thlick AE, Parravicini C, Moore PS, Chang Y. Kaposi’s sarcoma-associated herpesvirus LANA2 is a B-cell-specific latent viral protein that inhibits p53. J Virol. 2001;75:429–438. doi: 10.1128/JVI.75.1.429-438.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cai X, et al. Kaposi’s sarcoma-associated herpesvirus expresses an array of viral microRNAs in latently infected cells. Proc Natl Acad Sci USA. 2005;102:5570–5575. doi: 10.1073/pnas.0408192102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Samols MA, Hu J, Skalsky RL, Renne R. Cloning and identification of a microRNA cluster within the latency-associated region of Kaposi’s sarcoma-associated herpesvirus. J Virol. 2005;79:9301–9305. doi: 10.1128/JVI.79.14.9301-9305.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sullivan CS. High conservation of Kaposi sarcoma−associated herpesvirus microRNAs implies important function. J Infect Dis. 2007;195:618–620. doi: 10.1086/511440. [DOI] [PubMed] [Google Scholar]
- 19.Marshall V, et al. Conservation of virally encoded microRNAs in Kaposi sarcoma–associated herpesvirus in primary effusion lymphoma cell lines and in patients with Kaposi sarcoma or multicentric Castleman disease. J Infect Dis. 2007;195:645–659. doi: 10.1086/511434. [DOI] [PubMed] [Google Scholar]
- 20.Sun R, Lin SF, Gradoville L, Miller G. Polyadenylylated nuclear RNA encoded by Kaposi sarcoma-associated herpesvirus. Proc Natl Acad Sci USA. 1996;93:11883–11888. doi: 10.1073/pnas.93.21.11883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Staskus KA, et al. Kaposi’s sarcoma-associated herpesvirus gene expression in endothelial (spindle) tumor cells. J Virol. 1997;71:715–719. doi: 10.1128/jvi.71.1.715-719.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sarid R, Flore O, Bohenzky RA, Chang Y, Moore PS. Transcription mapping of the Kaposi’s sarcoma-associated herpesvirus (human herpesvirus 8) genome in a body cavity-based lymphoma cell line (BC-1) J Virol. 1998;72:1005–1012. doi: 10.1128/jvi.72.2.1005-1012.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Arias C, et al. KSHV 2.0: A comprehensive annotation of the Kaposi’s sarcoma-associated herpesvirus genome using next-generation sequencing reveals novel genomic and functional features. PLoS Pathog. 2014;10:e1003847. doi: 10.1371/journal.ppat.1003847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schifano JM, Corcoran K, Kelkar H, Dittmer DP. Expression of the antisense-to-latency transcript long noncoding RNA in Kaposi’s sarcoma-associated herpesvirus. J Virol. 2017;91:e01698-16. doi: 10.1128/JVI.01698-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dresang LR, et al. Coupled transcriptome and proteome analysis of human lymphotropic tumor viruses: Insights on the detection and discovery of viral genes. BMC Genomics. 2011;12:625. doi: 10.1186/1471-2164-12-625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rossetto CC, Pari GS. PAN’s Labyrinth: Molecular biology of Kaposi’s sarcoma-associated herpesvirus (KSHV) PAN RNA, a multifunctional long noncoding RNA. Viruses. 2014;6:4212–4226. doi: 10.3390/v6114212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sanger HL, Klotz G, Riesner D, Gross HJ, Kleinschmidt AK. Viroids are single-stranded covalently closed circular RNA molecules existing as highly base-paired rod-like structures. Proc Natl Acad Sci USA. 1976;73:3852–3856. doi: 10.1073/pnas.73.11.3852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kos A, Dijkema R, Arnberg AC, van der Meide PH, Schellekens H. The hepatitis delta (delta) virus possesses a circular RNA. Nature. 1986;323:558–560. doi: 10.1038/323558a0. [DOI] [PubMed] [Google Scholar]
- 29.Jeck WR, et al. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA. 2013;19:141–157. doi: 10.1261/rna.035667.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hansen TB, et al. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495:384–388. doi: 10.1038/nature11993. [DOI] [PubMed] [Google Scholar]
- 31.Zheng Q, et al. Circular RNA profiling reveals an abundant circHIPK3 that regulates cell growth by sponging multiple miRNAs. Nat Commun. 2016;7:11215. doi: 10.1038/ncomms11215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pamudurti NR, et al. Translation of circRNAs. Mol Cell. 2017;66:9–21.e7. doi: 10.1016/j.molcel.2017.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li Z, et al. Exon-intron circular RNAs regulate transcription in the nucleus. Nat Struct Mol Biol. 2015;22:256–264. doi: 10.1038/nsmb.2959. [DOI] [PubMed] [Google Scholar]
- 34.Zhang Y, et al. Circular RNAs: Emerging cancer biomarkers and targets. J Exp Clin Cancer Res. 2017;36:152. doi: 10.1186/s13046-017-0624-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Feng H, et al. Human transcriptome subtraction by using short sequence tags to search for tumor viruses in conjunctival carcinoma. J Virol. 2007;81:11332–11340. doi: 10.1128/JVI.00875-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Feng H, Shuda M, Chang Y, Moore PS. Clonal integration of a polyomavirus in human Merkel cell carcinoma. Science. 2008;319:1096–1100. doi: 10.1126/science.1152586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gao Y, Zhang J, Zhao F. Circular RNA identification based on multiple seed matching. Brief Bioinform. February 28, 2017 doi: 10.1093/bib/bbx014. [DOI] [PubMed] [Google Scholar]
- 38.Gao Y, Wang J, Zhao F. CIRI: An efficient and unbiased algorithm for de novo circular RNA identification. Genome Biol. 2015;16:4. doi: 10.1186/s13059-014-0571-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Desmet FO, et al. Human Splicing Finder: An online bioinformatics tool to predict splicing signals. Nucleic Acids Res. 2009;37:e67. doi: 10.1093/nar/gkp215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Miller G, Lipman M. Release of infectious Epstein−Barr virus by transformed marmoset leukocytes. Proc Natl Acad Sci USA. 1973;70:190–194. doi: 10.1073/pnas.70.1.190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Raab-Traub N, Dambaugh T, Kieff E. DNA of Epstein-Barr virus VIII: B95-8, the previous prototype, is an unusual deletion derivative. Cell. 1980;22:257–267. doi: 10.1016/0092-8674(80)90173-7. [DOI] [PubMed] [Google Scholar]
- 42.Young LS, Rickinson AB. Epstein-Barr virus: 40 years on. Nat Rev Cancer. 2004;4:757–768. doi: 10.1038/nrc1452. [DOI] [PubMed] [Google Scholar]
- 43.Busson P, et al. Establishment and characterization of three transplantable EBV-containing nasopharyngeal carcinomas. Int J Cancer. 1988;42:599–606. doi: 10.1002/ijc.2910420422. [DOI] [PubMed] [Google Scholar]
- 44.Dittmer DP, et al. Multiple pathways for Epstein-Barr virus episome loss from nasopharyngeal carcinoma. Int J Cancer. 2008;123:2105–2112. doi: 10.1002/ijc.23685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Panda AC, et al. High-purity circular RNA isolation method (RPAD) reveals vast collection of intronic circRNAs. Nucleic Acids Res. 2017;45:e116. doi: 10.1093/nar/gkx297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Qiu J, Smith P, Leahy L, Thorley-Lawson DA. The Epstein-Barr virus encoded BART miRNAs potentiate tumor growth in vivo. PLoS Pathog. 2015;11:e1004561. doi: 10.1371/journal.ppat.1004561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Palser AL, et al. Genome diversity of Epstein-Barr virus from multiple tumor types and normal infection. J Virol. 2015;89:5222–5237. doi: 10.1128/JVI.03614-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Salzman J, Gawad C, Wang PL, Lacayo N, Brown PO. Circular RNAs are the predominant transcript isoform from hundreds of human genes in diverse cell types. PLoS One. 2012;7:e30733. doi: 10.1371/journal.pone.0030733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Huang C, Liang D, Tatomer DC, Wilusz JE. A length-dependent evolutionarily conserved pathway controls nuclear export of circular RNAs. Genes Dev. 2018;32:639–644. doi: 10.1101/gad.314856.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Russo JJ, et al. Nucleotide sequence of the Kaposi sarcoma-associated herpesvirus (HHV8) Proc Natl Acad Sci USA. 1996;93:14862–14867. doi: 10.1073/pnas.93.25.14862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lee HR, et al. Kaposi’s sarcoma-associated herpesvirus viral interferon regulatory factor 4 targets MDM2 to deregulate the p53 tumor suppressor pathway. J Virol. 2009;83:6739–6747. doi: 10.1128/JVI.02353-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lee HR, et al. Kaposi’s sarcoma-associated herpesvirus viral interferon regulatory factor 4 (vIRF4) perturbs the G1-S cell cycle progression via deregulation of the cyclin D1 gene. J Virol. 2015;90:1139–1143. doi: 10.1128/JVI.01897-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chan KCA, et al. Analysis of plasma Epstein-Barr virus DNA to screen for nasopharyngeal cancer. N Engl J Med. 2017;377:513–522. doi: 10.1056/NEJMoa1701717. [DOI] [PubMed] [Google Scholar]
- 54.Barrett SP, Salzman J. Circular RNAs: Analysis, expression and potential functions. Development. 2016;143:1838–1847. doi: 10.1242/dev.128074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gottschalk S, et al. An Epstein-Barr virus deletion mutant associated with fatal lymphoproliferative disease unresponsive to therapy with virus-specific CTLs. Blood. 2001;97:835–843. doi: 10.1182/blood.v97.4.835. [DOI] [PubMed] [Google Scholar]
- 56.Lo AK, et al. Epstein-Barr virus infection alters cellular signal cascades in human nasopharyngeal epithelial cells. Neoplasia. 2006;8:173–180. doi: 10.1593/neo.05625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhang XO, et al. Diverse alternative back-splicing and alternative splicing landscape of circular RNAs. Genome Res. 2016;26:1277–1287. doi: 10.1101/gr.202895.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Glažar P, Papavasileiou P, Rajewsky N. circBase: A database for circular RNAs. RNA. 2014;20:1666–1670. doi: 10.1261/rna.043687.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhou F, et al. Oncolytic reactivation of KSHV as a therapeutic approach for primary effusion lymphoma. Mol Cancer Ther. 2017;16:2627–2638. doi: 10.1158/1535-7163.MCT-17-0041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hulsen T, de Vlieg J, Alkema W. BioVenn–A web application for the comparison and visualization of biological lists using area-proportional Venn diagrams. BMC Genomics. 2008;9:488. doi: 10.1186/1471-2164-9-488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pérez-Silva JG, Araujo-Voces M, Quesada V. nVenn: Generalized, quasi-proportional Venn and Euler diagrams. Bioinformatics. 2018;34:2322–2324. doi: 10.1093/bioinformatics/bty109. [DOI] [PubMed] [Google Scholar]
- 62.Caves EA, et al. Air-liquid interface method to study Epstein-Barr virus pathogenesis in nasopharyngeal epithelial cells. MSphere. 2018;3:e00152-18. doi: 10.1128/mSphere.00152-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Panda AC, Martindale JL, Gorospe M. Polysome fractionation to analyze mRNA distribution profiles. Bio Protoc. 2017;7:e2126. doi: 10.21769/BioProtoc.2126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Nystrom NA, Levine MJ, Roskies RZ, Scott JR. Proceedings of the 2015 XSEDE Conference: Scientific Advancements Enabled by Enhanced Cyberinfrastructure. Assoc Comput Machinery; St. Louis: 2015. Bridges: A uniquely flexible HPC resource for new communities and data analytics; pp. 1–8. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.