Significance
Some biological evolution is slow (millions of years), and some is fast (months to years). The speed at which a protein evolves depends on how stable a protein’s folded structure is, how well it avoids aggregation, and how well-chaperoned it is. What are the mechanisms? We compute fitness landscapes by combining a model of protein-folding equilibria with sequence-change dynamics. We find that adapting to a new environment is fastest for proteins that are least stably folded, because those sit on steep downhill parts of fitness potentials. The modeling shows that cells should adapt to warmer environments faster than to colder ones, explains why increasing a protein’s abundance slows cell evolution, and explains how chaperones accelerate evolution by mitigating this effect.
Keywords: protein evolution, adaptation, substitution rate
Abstract
Proteins evolve at different rates. What drives the speed of protein sequence changes? Two main factors are a protein’s folding stability and aggregation propensity. By combining the hydrophobic–polar (HP) model with the Zwanzig–Szabo–Bagchi rate theory, we find that: (i) Adaptation is strongly accelerated by selection pressure, explaining the broad variation from days to thousands of years over which organisms adapt to new environments. (ii) The proteins that adapt fastest are those that are not very stably folded, because their fitness landscapes are steepest. And because heating destabilizes folded proteins, we predict that cells should adapt faster when put into warmer rather than cooler environments. (iii) Increasing protein abundance slows down evolution (the substitution rate of the sequence) because a typical protein is not perfectly fit, so increasing its number of copies reduces the cell’s fitness. (iv) However, chaperones can mitigate this abundance effect and accelerate evolution (also called evolutionary capacitance) by effectively enhancing protein stability. This model explains key observations about protein evolution rates.
What molecular properties determine the rates of cell evolution? Proteins are known to evolve at different rates, partly based on the functions they perform for the cell, but also depending on their physical properties, such as folding stability and propensity for aggregation (1–8), and also depending on their companion chaperoning (9–16). While some evolution takes place over thousands to millions of years, other evolution can be much faster. Cancer cells evolve over a human lifetime. And pathogenic cells can evolve resistance to drugs in just a few years (17–19) or even faster (18). How do the molecular properties of proteins and chaperones determine the speed of evolution? Here, we develop theory for the rates of protein evolution.
Computing the Evolutionary Equilibria and Dynamics of Protein Sequences
The rate that a protein molecule evolves is given by the dependence on time of the probability that a protein sequence is fixed in a population by the time , through mutation and selection. Before considering the dynamics, we note that the equilibrium distribution of such probabilities will be a Boltzmann-like exponential, as shown (20–23) (and given by an alternative derivation using maximum entropy applied to sequence space in SI Appendix, Eq. S4):
| [1] |
where is the fitness potential, which is related to the fitness landscape (24) by ; is the sequence degeneracy—that is, the number of different sequences of a given fitness; is the selective pressure, proportional to the effective population size, as shown in ref. 20; and is the sum over the statistical weights (relative populations) of the different sequences of the protein. (The fitness landscape is a mathematical surface, often multidimensional, of the cell’s fitness as a function of the different mutations of a given protein.) Eq. 1 gives the equilibrium population of sequences for a given fitness potential. This equilibrium distribution is useful for considering the dynamics below.
A Zwanzig–Szabo–Bagchi-Like Model of Protein Adaptation Rates
We model a protein’s evolutionary kinetics by adapting Zwanzig–Szabo–Bagchi (ZSB) theory applied to the different problem of protein-folding speeds (25, 26). On the one hand, protein-folding dynamics is quite a different process than protein evolution. In folding, a particular protein explores its conformational degrees of freedom, changing its shape, whereas in evolution, a protein undergoes changes of sequence through mutations and selection. However, the dynamics can be modeled by a similar formalism. We define the transition rate from an ancestor sequence to descendant sequence as through a process of mutations and selection steps. Then, the change in population of sequence in a small time interval is given by the master equation expressing the “flow” from different sequences into sequence , minus the flows out from to other sequences,
| [2] |
To solve the dynamics, we need to know the transition rates ; these are dictated by the shape of the fitness potential since the rates are related to the equilibrium probabilities , which is given by Eq. 1. Then, we can solve for two key dynamical quantities: (i) the adaptation time, , or peak time, which is the minimum time required for changes in a sequence , through mutation and selection, to reach the sequence that is optimally adapted to its environment; or (ii) the substitution time, , also called the exit time, which is the average time required for a sequence to change and become any other sequence. The inverse of each of these times is a corresponding rate. The substitution rate is also called the evolution rate. The adaptation rate and substitution rate are measured differently and give different insights. SI Appendix gives the details of the dynamical model; here, we just summarize the main points.
A Protein’s Adaptation Rate Depends Strongly on the Selection Pressure
First, we ask how protein adaptation can sometimes be very fast. For this exploration of principle, it is sufficient to adopt the very simplest model of a fitness landscape that has a single peak. We assume the fitness potential is linear in the number of mutations in a single protein (meaning that the fitness landscape is exponential), with slope and minimum (which is the landscape point of the optimal sequence) (both and are taken to be positive quantities):
| [3] |
The virtue of the linear landscape here is in allowing for a closed-form expression for the adaptation time (SI Appendix, Eqs. S8–S18),
| [4] |
where is the number of possible mutations a residue in the protein can have relative to its starting sequence (), is the total number of residues in the protein, and is the average fixation rate for a single point mutation. (If is large, the adaptation time is independent of the number of mutations; it becomes equally hard to find the peak, no matter what the starting sequence is.)
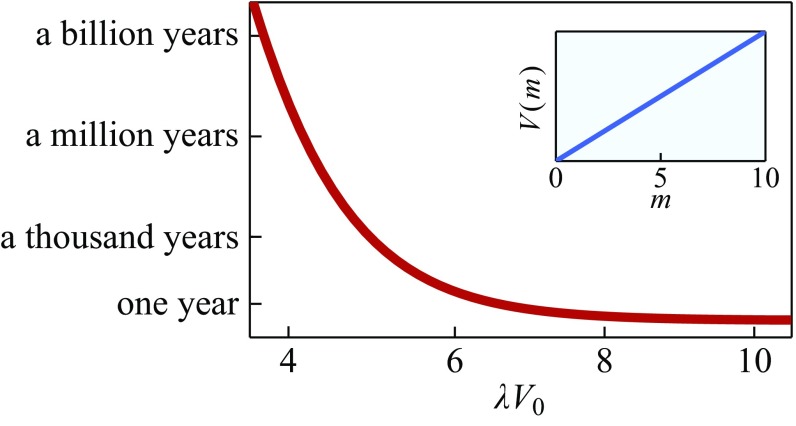
Fig. 1 shows a key conclusion: A protein’s adaptation speed can vary over nine orders of magnitude as a result of only a twofold change in selection pressure . This huge magnification in Eq. 4 is because the adaptation rate is nearly an exponential function of an exponential []. So, even though evolution “would take forever” if fitness landscapes were flat, even a very slight tilt of a fitness landscape gets amplified into a very fast adaptation for protein sequence evolution. [The treatment is valid in the limit of strong selection and weak mutations, for which populations are monomorphic and mutations do not interact with each other. Other contexts require different methods (27).] This general conclusion holds also if instead we had used other hypothetical functional forms of fitness. Here, we have considered just a single isolated protein. Below, we consider situations where mutations happen in multiple proteins.
Fig. 1.
The adaptation time of a protein depends strongly on the selection pressure . The time it takes for a protein to evolve to its optimally adapted sequence, assuming a linear model fitness potential (Inset), if an average random mutation is fixed once every 100 y in the absence of selection pressure is shown (28). We assume that the protein has amino acids and that each residue can be any of the 20 amino acids ().
Proteins Having the Steepest Fitness Landscapes Adapt the Fastest.
Eq. 4 shows another key point, namely, that the adaptation rate increases strongly with the steepness, , of the fitness potential. Metaphorically, a ball rolls faster down a steeper hill than down a shallower hill. (In the limit of a small slope, adaptation will follow a random walk in a large space, requiring an exponentially long time.)
The Least-Stable Proteins Adapt the Fastest Because Their Fitness Landscapes Are the Steepest.
Above, we asked how external pressure affects adaptation speed. Here, we ask how the properties of the protein itself affects its adaptation speed. So, first, we need a model for how fitness depends on protein properties. Ever since the pioneering work of Drummond et al. (1, 3, 6, 21, 29), a major idea has been the misfolding avoidance hypothesis; namely, that a protein’s fitness is substantially due to its folding–unfolding equilibrium. Here, we give a model of the evolution rates. Consider a protein having folding stability, ( for a folded protein) and abundance . Let the number of different types of proteins in the cell be . A well-known result is how the cell’s fitness potential is the following nonlinear function of its folding stability (6):
| [5] |
Eq. 5 simply states that each protein’s fitness potential is proportional to the product of (its abundance, , in the proteome) {its fractional degree of folding, [native/(native + unfolded)]} (the total number of protein types in the cell). [Fitness potential is assumed to be linearly proportional to the number of folded copies of the protein, but only up to the point of overexpression. Folding stability and aggregation are not the only physical contributors to evolution rates; conformational flexibility, which we do not study here, can also affect evolvability, particularly in virus proteins (30–32).]
Here, we model the evolution rates. We combine the fitness potential in Eq. 5 with the principle given by Eq. 4 that proteins undergo the fastest evolutionary adaptation where protein fitness landscapes are steepest.
First, compare two proteins: One protein is more stable than the other. The logic above says that the less-stable protein will accumulate adaptive mutations faster than the more-stable protein. Second, compare a “fit” protein, which is stably folded and well adapted to its environment, to a mutated version of that same protein, which is less stably folded and less fit. The mutant protein will acquire adaptive mutations faster than the well-adapted protein.
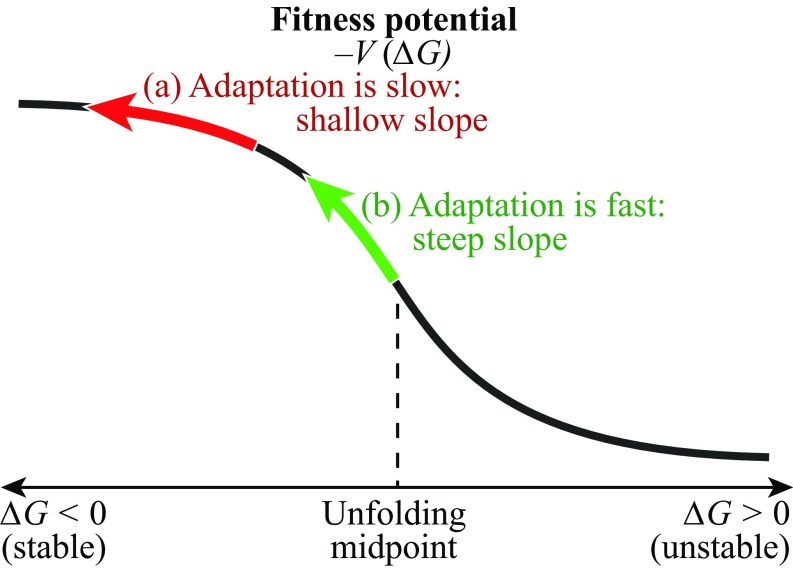
Fig. 2 illustrates that fast adaptation happens where the fitness potential is steep, which is where protein stability is marginal (near , neither stably folded nor substantially unfolded), for a given abundance .
Fig. 2.
The fitness potential for a protein-folding stability sequence space. Having greater folding stability means higher fitness. The green and red arrows indicate that where the slope is steepest on this potential, adaption is fastest. And, it is fastest where proteins are least stable.
The curve in Fig. 2 is general and applicable when both stabilizing and destabilizing directions are accessible to the protein. However, we note that adaptation requires mutations in multiple proteins; therefore, in the next section we make a binary simplification of this landscape, but it does not alter the slope-speed principle.
Cells Should Adapt Faster to a Warmer Environment than to a Colder One
How fast can proteins adapt if cells are put in climates of different temperatures? Some unicellular organisms (mesophiles) live in moderate-temperature environments (C for Escherichia coli), while others (thermophiles) live in hotter environments. Cells grow the fastest at the temperature of their natural environment (33–35), but moved to different environments having different temperatures, they can adapt (36). We compute the speed of adaptation of a cell that is transferred from its normal environment to a new environment having either a higher or lower temperature. We compute rates from SI Appendix, Eq. S16, with a fitness potential given by the thermal folding Eq. 5, and using SI Appendix, Eq. S18 to find its slope along the mutation axis. In this example, we consider a given number of proteins in the cell with either zero or one adaptive mutations to each protein (assuming no epistasis).
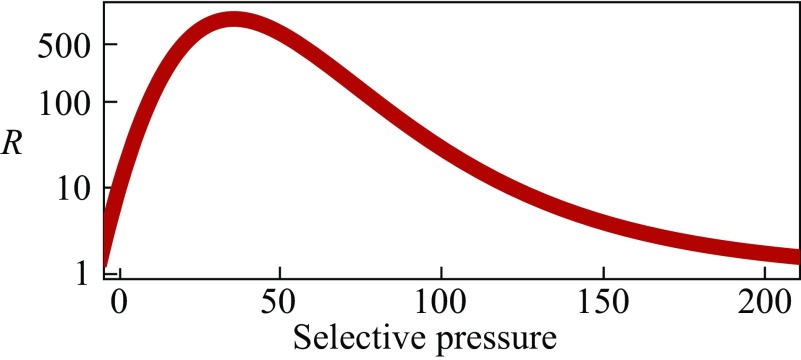
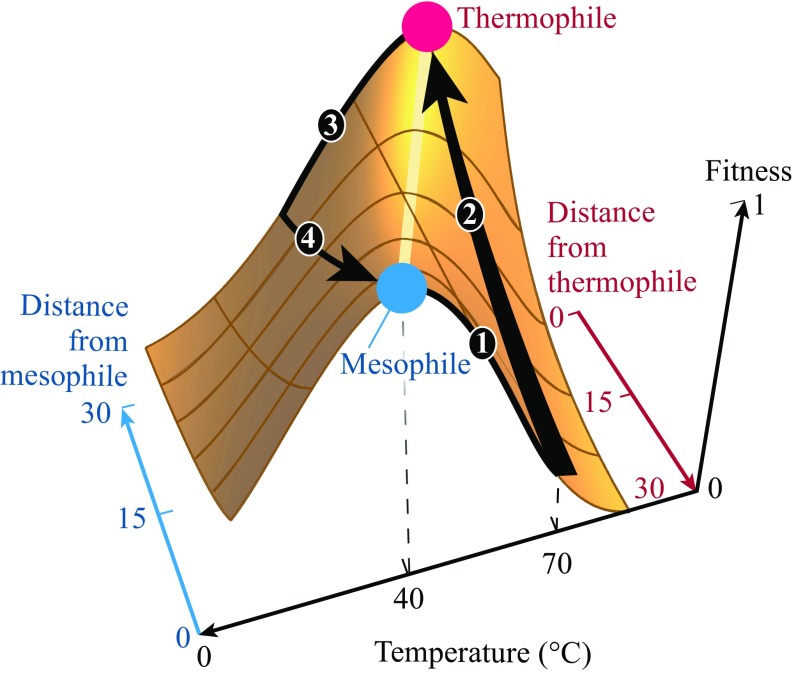
Fig. 3 shows the prediction that cells should be able to adapt much faster to a warmer environment than to a cooler environment. (We are unaware of experiments that bear on this.) Fig. 4 illustrates the reason for this, using a fitness landscape. Start with a healthy mesophilic cell in its normal environment, say, at C, where it is maximally fit. Its proteins are stably folded. Now, upshift its environment to C (path 1) (slowly, in small steps, to avoid killing the cell). Initially, the cell is unfit for its new, warmer environment because its proteins are less stable at this higher temperature, C. Now, mutations accumulate rapidly (30 total, in the model example) because the fitness landscape is steep for proteins that are unstable, leading to fast adaption to the new peak (path 2).
Fig. 3.
Proteins should adapt faster to a warmer than a cooler climate. is the ratio of adaptation rates: (a mesophile adapting to a higher temperature) (a thermophile adapting to a colder temperature). Heat destabilizes folded proteins, putting them onto the steep slopes of fitness landscapes, so cells adapt faster to warmer environments. x axis, the selection pressure per misfolded protein, .
Fig. 4.
Fitness trajectories for explaining why cells adapt faster to warmer environments than to cooler ones. Paths 1 and 2 show how cell fitness changes upon heating. Path 1: Start with a mesophile preadapted at C, at the peak of its landscape. Increase the temperature to C. The fitness decreases. Path 2: Mutations occur to bring the cell to the peak fitness for C. This is fast because the proteins are destabilized by heating, so the fitness landscape is steep along path 2. Paths 3 and 4 show changes upon cooling. Path 3: Cooling reduces the fitness of a preadapted thermophile. Path 4: The cell now undergoes 30 mutations to bring it to the peak of adaptation for C. However, path 4 is much slower than path 2 because cooling preadapted proteins does not affect their stabilities much. So, adaptation to heat is faster than to cold.
Now, contrast this with cooling. Now, a thermophilic cell starts at C, maximally fit, with its proteins stably folded. Cooling causes this cell to be less fit for its new environment at C (path 3). However, this is not due to protein stability; cooling proteins that are already stable does not change their native populations. Rather, the reduced fitness upon cooling is because of the Arrhenius temperature factor: Cells naturally grow more slowly in colder temperatures (SI Appendix, Eq. S18). Overall, for this cooling situation, the cell’s fitness landscape has a shallow slope (along path 4), and adaptation to the cold through mutations is slow. In summary, cells should adapt to warm climates faster than to colder ones.
The Substitution Rate vs. Adaptation Rate: They Reflect Different Features of Fitness Terrains
For the rest of this work, we now switch attention from the adaptation rate (how fast an arbitrary sequence evolves to become the sequence that has the maximal fitness) to the substitution rate (also called the evolution rate: how fast an arbitrary sequence changes to become fixed as a different arbitrary sequence). This switch allows us to test predictions against experimental data for the properties studied below. Substitution rates are properties of individual proteins, meaning that the accumulation of multiple mutations can take a long time. In contrast, adaptation involves mutations that can occur in parallel throughout the entire proteome, and therefore those changes can happen much faster. For this reason, for the remainder of the work we will be counting the number of mutations in a single protein, as opposed to what was done in the previous section.
More importantly, these two rate properties reflect different features of fitness landscapes. Whereas our model shows that adaptation rates are proportional to the slope of a fitness landscape (see above), substitution rates, instead, are proportional to the average mutational distance of a given protein to its fitness peak (at equilibrium) (see below and SI Appendix, Eq. S27):
| [6] |
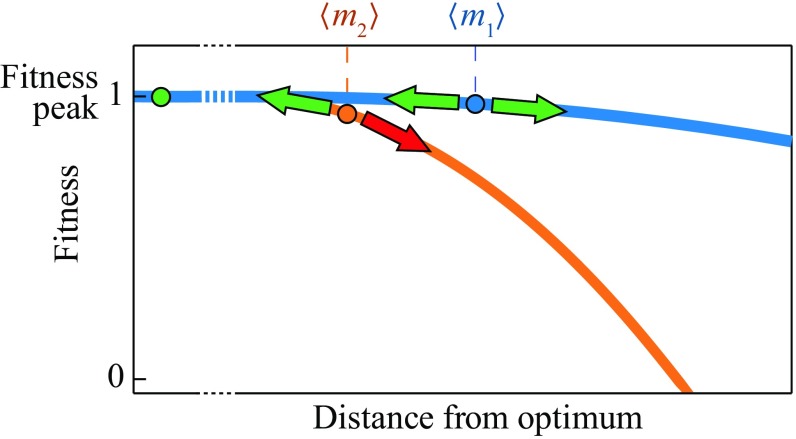
where is a rate quantity used as a fitting parameter and is the average number of sequence mutations from the optimum in a single protein, and hence, the average mutational distance from an hypothetical optimal sequence at equilibrium. Fig. 5 shows the interpretation of . It measures the weight under the curve, so substitution rates are highest on fitness landscape contours that are “high-shouldered”: plateaus of high fitness where slopes are shallow. The bluescape Fig. 5 is high-shouldered, with larger : A mutation in either direction (green arrow) is fit enough, so substitution is fast. The orangescape is not a high plateau or flat. It has smaller : A mutation downhill (red arrow) is too unfit to be fixed. Because of greater access to allowed directions, Eq. 6 says that substitutions happen faster on the bluescape than the orangescape. Moving away from the peak on the bluescape still leads to adaptive mutations, and hence, to substitution; moving away on the orangescape leads to nonadaptive mutations. So, the net substitution speed is greater on the bluescape. This high-shouldering principle is valid beyond the simple model used here to illustrate it.
Fig. 5.
Substitution rates are higher on high-shouldered fitness landscapes. The bluescape has more directions in which mutations are fit and adaptive than the orangescape has, . On the bluescape, mutations can be fixed in either direction (green arrows). On the orangescape, mutations downhill (red arrow) are too unfit to be fixed. Eq. 6 shows that the bluescape has the higher substitution rate than the orangescape.
Substitution rates of amino acids are measurable and have been the basis for the molecular clock idea (37–39) that substitution rates differ among proteins, but are approximately constant for a given protein. Recent work has shown that the average substitution rate is determined not by functional constraints, but by physical ones. Proteins that are more abundant are observed to evolve more slowly than proteins that are less abundant (40). The expression level-rate (E-R) anticorrelation is the observation that increasing expression levels (protein abundances) lead to reduced rates of their evolution. It has been hypothesized that this is a result of either protein misfolding or protein–protein interaction (21, 29, 41).
Abundant Proteins Evolve Slowly
We model the mechanism of the E-R anticorrelation. In a population of cells, many proteins are not peak-fitness sequences. Increasing the abundance of these imperfect proteins reduces the cell’s fitness relative to a perfectly adapted cell. We consider two mechanisms: (i) misfolding, where fitness, , depends on how perfectly a protein sequence folds in its lowest-energy state to maximize hydrophobic–hydrophobic (HH) contacts in the core of its native structure. The deviation from the fitness peak is a count of the number of defects, . (ii) For aggregation and misinteraction, fitness, , depends on how perfectly the protein surface is covered with polar (P) residues, to avoid protein–protein sticking through HH contacts. The deviation from perfect fitness is the number of hydrophobic (H) residues on the surface. Now, to get these fitness landscapes, we use the hydrophobic–polar (HP) lattice model, in which a protein is assumed to have only H or P residues, and different native and mutated protein sequences are enumerated on a 2D square lattice (42). Random mutations over different proteins can reduce either form of “perfectness.” Details are given in SI Appendix, Eqs. S30 and S31. The main distinction between these mechanisms is their dependence on abundance : and . We calculate the substitution rates for these two different mechanisms using Eq. 6.
The E-R Anticorrelation Is Explained by Either Misfolding or Aggregation or Both
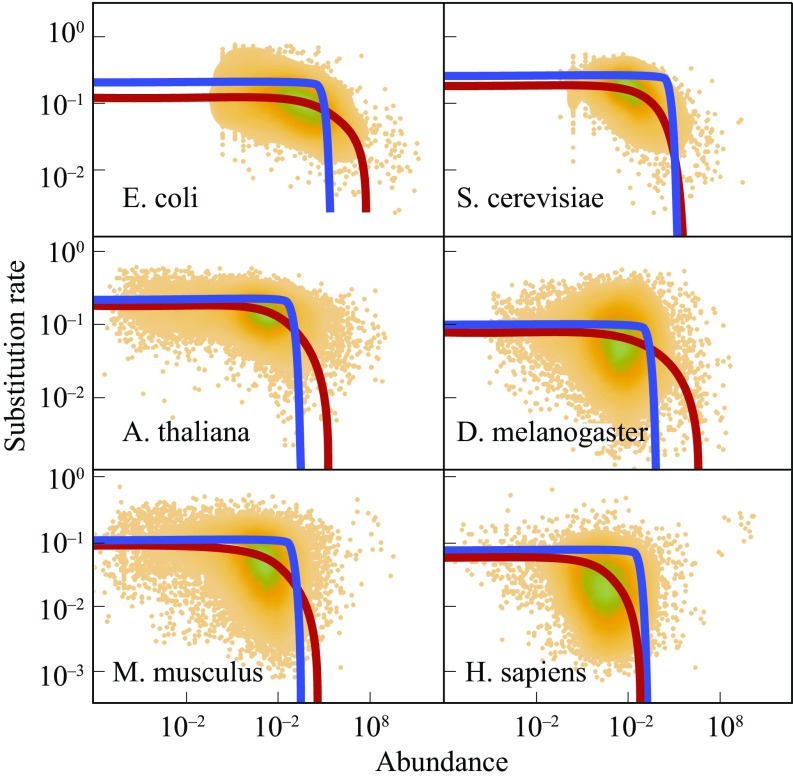
Fig. 6 compares the misfolding and aggregation models to experiments. Both models predict a general E-R anticorrelation. And, both are consistent with the (not very precise) data (29). So, we have no basis for favoring one mechanism over the other. Previous modeling has also observed the E-R anticorrelation, but based on assuming an anticorrelation between and mutational ’s taken from the Protein Data Bank (6, 7). Our more microscopic mechanism here of the full evolutionary landscape allows us also to study aggregation and chaperone effects at a single-protein level.
Fig. 6.
Expression-rate anticorrelation: Abundant proteins evolve more slowly. Experiments (green dots) on different proteins in six organisms, from ref. 29, are shown. Red, misfolding model, . Blue, aggregation model, . The curve parameters are given in SI Appendix, Table S1. Both misfolding and aggregation models are consistent with the data. A. thaliana, Arabidopsis thaliana; D. melanogaster, Drosophila melanogaster; H. sapiens, Homo sapiens; M. musculus, Mus musculus; S. Serevisiae, Saccharomyces cerevisiae.
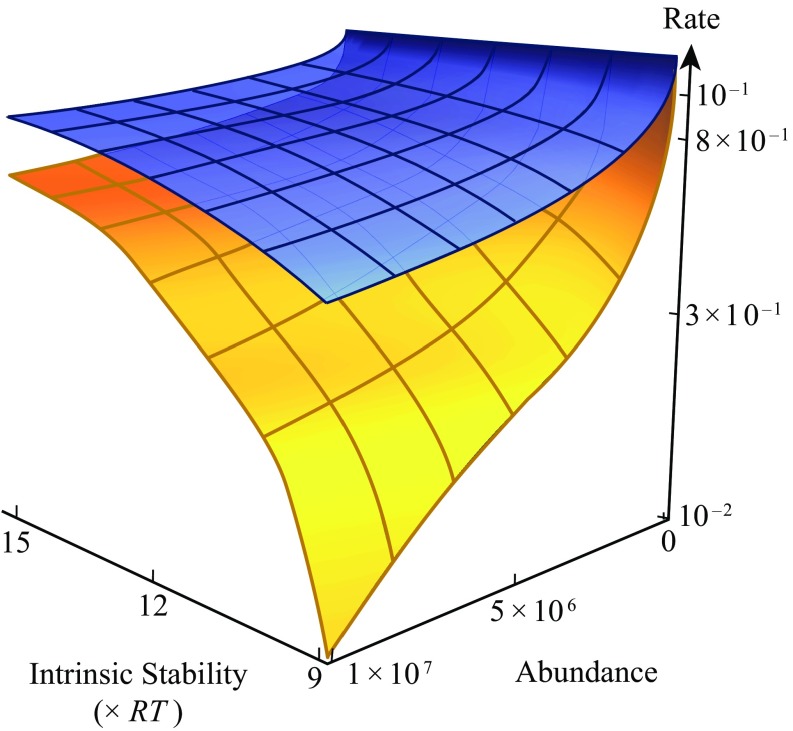
The model explains the E-R anticorrelation as follows. Fig. 7 (yellow surface) shows the substitution rate as functions of both protein stability and abundance. In a very stable protein, a mutation that removes a hydrophobe from the core is usually acceptable (a high-shouldered terrain), so it has a high substitution rate. In contrast, in a weakly stable protein, removing a hydrophobe from the core can unfold the protein, so it is not adaptive (not a high-shouldered terrain), so fewer possible substitutions are acceptable, and the substitution rate is slower. The abundance effect is as follows. This is an integration over all of the proteins in the cell, and most of them are imperfect and not maximally stable. So, increasing the abundance manifests as increasing the concentration of imperfect proteins, which are not high-shouldered, leading to slower average substitution.
Fig. 7.
Yellow, substitution is slower for proteins that are unstable or abundant. Blue, chaperones increase all of the evolution rates. (Larger intrinsic stabilities refer to more negative folding free energies.) Yellow surface, proteins alone. Mutating a stable protein is usually adaptive because the protein can tolerate it. Less-stable proteins are less tolerant of mutations, so their substitution rates are lower. Since most proteins are imperfect, increasing the average abundance decreases cell fitness, leading to a lower substitution rates Blue surface, with chaperones. Chaperones raise the evolution rates of client proteins overall because they raise their average folding stabilities.
Chaperones Are Evolutionary Accelerators
The speed of cell evolution is modulated by chaperones in the cell. Chaperones are biomolecular complexes that help other proteins (their clients) to fold. Experiments show that chaperones are generally evolution accelerators (they have been called evolutionary capacitors). That is, increasing a cell’s chaperone concentrations can speed up the cell’s evolution (9, 10, 12, 14, 43).
What is the mechanism of evolutionary acceleration by chaperones? The blue surface in Fig. 7 shows the effect of adding chaperones within the present model (details of calculations are in SI Appendix, section I). Chaperones are active ATP-driven devices that shift the balance from misfolded and unfolded states to native states of client proteins, resulting in stabilizing the client proteins. And, according to theory above, more-stable proteins have terrains that are more high-shouldered, leading to faster amino acid substitution. Hence, chaperones accelerate evolution. (A subtle point is that while chaperones accelerate substitution, they can slow down adaptation, since chaperones can make “near-perfect” proteins appear perfect to the cell. Such near-perfect proteins have no selective disadvantage in cells with chaperones.)
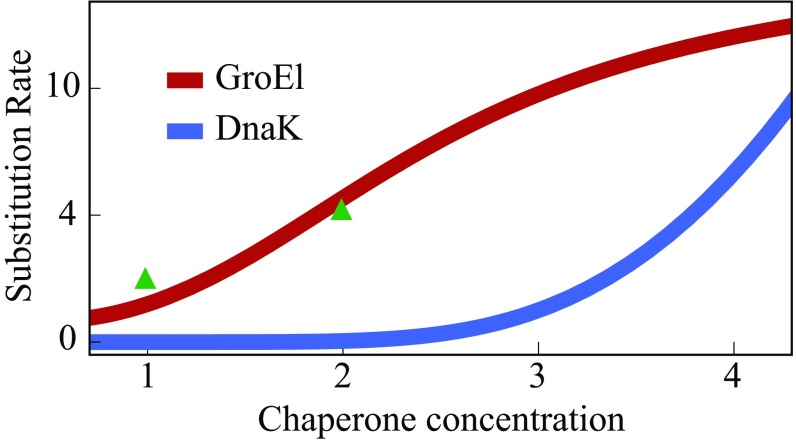
Fig. 8 also shows the prediction that evolutionary acceleration can be different for different types of chaperones. To show this, we use the model of Santra et al. (44) that shows that GroEl binds misfolded protein, sending it either to unfolded (U) or native (N) states, whereas DnaK binds misfolded protein and only sends it to U; SI Appendix, Fig. S2. The model predicts that GroEl is effective at lower concentrations, and on a different set of client proteins, than DnaK. Why the difference in chaperone concentrations? In short, clients of GroEl see only an unstable U and stable N state, so those clients mostly fold. In contrast, clients of DnaK mostly see an M state that is almost as stable as N, so more chaperone is needed to produce more N. That is why GroEl and DnaK should have different values of “evolutionary capacitance” (see SI Appendix, Fig. S5 for a graphical explanation).
Fig. 8.
Increasing chaperone concentration increases a client protein’s evolution rate. Model calculations are given in SI Appendix, based on slightly different mechanisms for GroEl and DnaK (SI Appendix, Fig. S2). The GroEl curve is computed from SI Appendix, Eq. S43, with a 2× overexpression level, compared with experimental data of ref. 10, shown with green triangles. Increases from DnaK are also observed in experiments (15).
Conclusions
We model here how rates of protein evolution depend on folding and aggregation properties. The present model gives a single framework for understanding disparate observations. (i) Adaptation speed depends on the slope of a fitness landscape. This depends strongly on selection pressures. It is fastest for the least-stably folded proteins. Cells that are shifted to warm climates become unstable, so they adapt rapidly. (ii) Substitution speed depends on how high-shouldered the terrain is of a fitness potential. Abundant proteins evolve slowly. This is because most proteins are not perfectly stable, so increasing their abundance shifts the cell to non-high-shouldered terrains of fitness, leading to slow substitution. (iii) This effect is mitigated by chaperones, which increase protein stabilities, increasing their substitution rates. This modeling describes how protein evolution rates depend on their folding and aggregation properties.
Supplementary Material
Acknowledgments
This work was supported by Laufer Center and National Science Foundation Grant MCB1344230.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1810194115/-/DCSupplemental.
References
- 1.Drummond DA, Bloom JD, Adami C, Wilke CO, Arnold FH. Why highly expressed proteins evolve slowly. Proc Natl Acad Sci USA. 2005;102:14338–14343. doi: 10.1073/pnas.0504070102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zeldovich KB, Chen P, Shakhnovich EI. Protein stability imposes limits on organism complexity and speed of molecular evolution. Proc Natl Acad Sci USA. 2007;104:16152–16157. doi: 10.1073/pnas.0705366104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Drummond DA, Wilke CO. Mistranslation-induced protein misfolding as a dominant constraint on coding-sequence evolution. Cell. 2008;134:341–352. doi: 10.1016/j.cell.2008.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yang J-R, Zhuang S-M, Zhang J. Impact of translational error-induced and error-free misfolding on the rate of protein evolution. Mol Syst Biol. 2010;6:421. doi: 10.1038/msb.2010.78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Serohijos AWR, Ryan Lee SY, Shakhnovich EI. Highly abundant proteins favor more stable 3D structures in yeast. Biophys. J. 2013;104:L1–L3. doi: 10.1016/j.bpj.2012.11.3838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Serohijos AWR, Rimas Z, Shakhnovich EI. Protein biophysics explains why highly abundant proteins evolve slowly. Cel Rep. 2012;2:249–256. doi: 10.1016/j.celrep.2012.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yang JR, Liao BY, Zhuang SM, Zhang JZ. Protein misinteraction avoidance causes highly expressed proteins to evolve slowly. Proc Natl Acad Sci USA. 2012;109:E831–E840. doi: 10.1073/pnas.1117408109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Levy ED, De S, Teichmann SA. Cellular crowding imposes global constraints on the chemistry and evolution of proteomes. Proc Natl Acad Sci USA. 2012;109:20461–20466. doi: 10.1073/pnas.1209312109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rutherford SL, Lindquist S. Hsp90 as a capacitor for morphological evolution. Nature. 1998;396:336–342. doi: 10.1038/24550. [DOI] [PubMed] [Google Scholar]
- 10.Tokuriki N, Tawfik DS. Chaperonin overexpression promotes genetic variation and enzyme evolution. Nature. 2009;459:668–673. doi: 10.1038/nature08009. [DOI] [PubMed] [Google Scholar]
- 11.Rutherford SL. Between genotype and phenotype: Protein chaperones and evolvability. Nat Rev Genet. 2003;4:263–274. doi: 10.1038/nrg1041. [DOI] [PubMed] [Google Scholar]
- 12.Sangster TA, Lindquist S, Queitsch C. Under cover: Causes, effects and implications of Hsp90-mediated genetic capacitance. BioEssays. 2004;26:348–362. doi: 10.1002/bies.20020. [DOI] [PubMed] [Google Scholar]
- 13.Gupta AN, et al. Pharmacological chaperone reshapes the energy landscape for folding and aggregation of the prion protein. Nat Commun. 2016;7:12058. doi: 10.1038/ncomms12058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bogumil D, Landan G, Ilhan J, Dagan T. Chaperones divide yeast proteins into classes of expression level and evolutionary rate. Genome Biol Evol. 2012;4:618–625. doi: 10.1093/gbe/evs025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Samer Kadibalban A, Bogumil D, Landan G, Dagan T. DnaK-dependent accelerated evolutionary rate in prokaryotes. Genome Biol Evol. 2016;8:1590–1599. doi: 10.1093/gbe/evw102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Aguilar-Rodríguez J, et al. The molecular chaperone DnaK is a source of mutational robustness. Genome Biol Evol. 2016;8:evw176. doi: 10.1093/gbe/evw176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Martinez JL, Baquero F. Mutation frequencies and antibiotic resistance. Antimicrob Agents Chemother. 2000;44:1771–1777. doi: 10.1128/aac.44.7.1771-1777.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Baym M, et al. Spatiotemporal microbial evolution on antibiotic landscapes. Science. 2016;353:1147–1151. doi: 10.1126/science.aag0822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Greulich P, Waclaw B, Allen R. Mutational pathway determines whether drug gradients accelerate evolution of drug-resistant cells. Phys Rev Lett. 2012;109:1–5. doi: 10.1103/PhysRevLett.109.088101. [DOI] [PubMed] [Google Scholar]
- 20.Sella G, Hirsh AE. The application of statistical physics to evolutionary biology. Proc Natl Acad Sci USA. 2005;102:14475–14475. doi: 10.1073/pnas.0501865102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Echave J, Wilke CO. Biophysical models of protein evolution: Understanding the patterns of evolutionary sequence divergence. Annu Rev Biophys. 2017;46:85–103. doi: 10.1146/annurev-biophys-070816-033819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Barton NH, Coe JB. On the application of statistical physics to evolutionary biology. J Theor Biol. 2009;259:317–324. doi: 10.1016/j.jtbi.2009.03.019. [DOI] [PubMed] [Google Scholar]
- 23.De Vladar HP, Barton NH. The contribution of statistical physics to evolutionary biology. Trends Ecol Evol. 2011;26:424–432. doi: 10.1016/j.tree.2011.04.002. [DOI] [PubMed] [Google Scholar]
- 24.Wright S. 1932. The roles of mutation, inbreeding, crossbreeding, and selection in evolution, Proceedings of the VI International Congress of Genetics (Genetics Society of America, Rockville, MD), Vol 1, 356–366.
- 25.Zwanzig R, Szabo A, Bagchi B. Levinthal’s paradox. Proc Natl Acad Sci USA. 1992;89:20–22. doi: 10.1073/pnas.89.1.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rollins GC, Dill KA. General mechanism of two-state protein folding kinetics. J Am Chem Soc. Aug 2014;136:11420–11427. doi: 10.1021/ja5049434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rouzine IM, Brunet É, Wilke CO. The traveling-wave approach to asexual evolution: Muller’s ratchet and speed of adaptation. Theor Popul Biol. 2008;73:24–46. doi: 10.1016/j.tpb.2007.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee H, Popodi E, Tang H, Foster PL. Rate and molecular spectrum of spontaneous mutations in the bacterium Escherichia coli as determined by whole-genome sequencing. Proc Natl Acad Sci USA. 2012;109:E2774–E2783. doi: 10.1073/pnas.1210309109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang J, Yang J-R. Determinants of the rate of protein sequence evolution. Nat Rev Genet. 2015;16:409–420. doi: 10.1038/nrg3950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tokuriki N, Oldfield CJ, Uversky VN, Berezovsky IN, Tawfik DS. Do viral proteins possess unique biophysical features? Trends Biochem Sci. 2009;34:53–59. doi: 10.1016/j.tibs.2008.10.009. [DOI] [PubMed] [Google Scholar]
- 31.Tokuriki N, Tawfik DS. Protein dynamism and evolvability. Science. 2009;324:203–207. doi: 10.1126/science.1169375. [DOI] [PubMed] [Google Scholar]
- 32.Soskine M, Tawfik DS. Mutational effects and the evolution of new protein functions. Nat Rev Genet. 2010;11:572–582. doi: 10.1038/nrg2808. [DOI] [PubMed] [Google Scholar]
- 33.Sawle L, Ghosh K. How do thermophilic proteins and proteomes withstand high temperature? Biophys J. 2011;101:217–227. doi: 10.1016/j.bpj.2011.05.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dill KA, Ghosh K, Schmit JD. Physical limits of cells and proteomes. Proc Natl Acad Sci USA. 2011;108:17876–17882. doi: 10.1073/pnas.1114477108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ghosh K, De Graff AMR, Sawle L, Dill K. Role of proteome physical chemistry in cell behavior. J Phys Chem B. 2016;120:9549–9563. doi: 10.1021/acs.jpcb.6b04886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Blaby IK, et al. Experimental evolution of a facultative thermophile from a mesophilic ancestor. Appl Environ Microbiol. 2012;78:144–155. doi: 10.1128/AEM.05773-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zuckerkandl E, Pauling L. Horizons in Biochemistry. Academic; New York: 1962. Molecular disease, evolution, and genic heterogeneity; pp. 189–222. [Google Scholar]
- 38.Nei M, Suzuki Y, Nozawa M. The neutral theory of molecular evolution in the genomic era. Annu Rev Genomics Hum Genet. 2010;11:265–289. doi: 10.1146/annurev-genom-082908-150129. [DOI] [PubMed] [Google Scholar]
- 39.Mccandlish DM, Stoltzfus A. Modeling evolution using the probability of fixation: History and implications. Q Rev Biol. 2014;89:225–252. doi: 10.1086/677571. [DOI] [PubMed] [Google Scholar]
- 40.Pal C, Papp B, Hurst LD. Highly expressed genes in yeast evolve slowly. Genetics. 2001;3:125–126. doi: 10.1093/genetics/158.2.927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sikosek T, Chan HS. Biophysics of protein evolution and evolutionary protein biophysics. J R Soc Interface R Soc. 2014;11:20140419. doi: 10.1098/rsif.2014.0419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lau KF, Dill KA. A lattice statistical mechanics model of the conformational and sequence spaces of proteins. Macromolecules. 1989;22:3986–3997. [Google Scholar]
- 43.Wyganowski KT, Kaltenbach M, Tokuriki N. GroEL/ES buffering and compensatory mutations promote protein evolution by stabilizing folding intermediates. J Mol Biol. 2013;425:3403–3414. doi: 10.1016/j.jmb.2013.06.028. [DOI] [PubMed] [Google Scholar]
- 44.Santra M, Farrell DW, Dill KA. Bacterial proteostasis balances energy and chaperone utilization efficiently. Proc Natl Acad Sci USA. 2017;114:E2654–E2661. doi: 10.1073/pnas.1620646114. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.